Abstract
Osteoporosis (OP), characterized by reduced bone density and increased fracture risk. Current therapies have limitations due to side effects and variable efficacy, necessitating exploration of novel therapeutic targets and alternatives like Traditional Chinese Medicine (TCM). The molecular mechanisms driving OP remain incompletely understood. This study utilized integrated bioinformatics approaches to identify potentially dysregulated genes in OP and explored in vitro effects of TCM compounds targeting top identified genes. Differentially expressed genes (DEGs) associated with OP were identified by integrating bulk and single-cell RNA sequencing datasets. Potential therapeutic compounds targeting selected DEGs were screened from a TCM database using molecular docking to predict binding affinities. Based on highest binding scores, two compounds were selected for experimental validation. Their effects on chondrogenic differentiation, relevant to the known function of the identified target genes CHAD and COL2A1, were assessed in vitro using ATDC5 cells. Integrated bioinformatics analysis consistently identified CHAD and COL2A1 as significantly downregulated genes in OP datasets, and they were upregulated after teriparatide injection. Molecular docking predicted binding affinities of several TCM compounds to CHAD and COL2A1, with tetrandrine and wogonin showing the highest binding affinities. They were selected for chondrogenesis and qRT-PCR validation, and wogonin was experimentally demonstrated to have better chondrogenic differentiation effects in vitro. This study employed a multi-omics bioinformatics approach to identify CHAD as a potential hub gene of interest associated with osteoporosis. Preliminary in vitro experiments showed that wogonin modulates chondrogenic differentiation, a process related to CHAD’s function, suggesting potential avenues for further investigation into OP therapeutics.
Similar content being viewed by others
Introduction
Osteoporosis (OP) is a systemic skeletal disease that is characterized by deterioration of bone microarchitecture and reduced bone mass, leading to increased bone fragility and fracture risk. OP is highly prevalent in both men and women, especially in postmenopausal women1. In Western populations, up to one in two women and one in three men will experience a fragility fracture during their lifetime2. Recently studies also indicated OP has negative impact on fracture healing, and the underlying mechanism remain unknown3.
Although there are various pharmacological treatments for OP, but it all have series of side effects with long term treatment. For example, bisphosphonates drugs can induce gastric ulcer, creatinine increase, medication-related osteonecrosis of the jaw, and atypical femoral fracture4,5,6. According to the guideline, the preferably treatment for high risk of major osteoporotic fracture is teriparatide (2 years) or romosozumab (1 year)7. Teriparatide also known as parathyroid hormone, but it also has side effects like hypercalcemia and orthostatic hypotension. Therefore, it is crucial to find an alternative treatment for OP due to the side effects of current drugs. Furthermore, the interplay between bone health and cartilage integrity is increasingly recognized; recent genetic studies suggest that osteoporosis itself may causally increase the risk of developing osteoarthritis, a disease primarily affecting cartilage and chondrocytes8. The advantage of Traditional Chinese Medicine (TCM) is composition of natural compound, and has increased in popularity in both Asian and Western countries due to its long-standing use, low cost, and few side effects9. Moreover, there are few TCM compounds had effective outcomes on influence chondrocytes, for example like wogonin, tetrandrine, and curcumin10,11,12. Therefore, it represents promising alternative option for chronic disease like osteoporosis.
Chondroadherin (CHAD) is a cartilage matrix protein thought to mediate adhesion of isolated chondrocytes. Study shown in absence of CHAD leads into distinct skeletal phenotype by impaired hypertrophic differentiation of chondrocytes. It also altered bone turnover in trabecular and cortical bone tissues13. Type II collagen alpha I (COL2A1) also known as a collagen expressed in cartilage, it can interact with CHAD influence collagen fibrillogenesis14. While CHAD and COL2A1 are primarily studied in cartilage, potential links to osteoporosis (OP) pathophysiology are suggested by findings such as decreased COL2A1 in postmenopausal women15. Despite these suggestive observations, the specific role and functional significance of CHAD and its interaction, particularly within the context of osteoporotic bone pathophysiology, remain largely unclear. This gap in understanding limits the exploration of CHAD-related pathways as potential therapeutic avenues for OP. Furthermore, the potential therapeutic relevance of targeting these pathways is suggested by existing research on compounds like wogonin. Wogonin, a flavonoid investigated in this study, has previously demonstrated anti-osteoporosis effects in a zebrafish model16. Wogonin also inhibits osteoclast formation induced by lipopolysaccharide in vitro and in vivo17.
In this study, we leveraged bioinformatics to integrate bulk RNA sequencing and single cell RNA sequencing from different species, to explore potential therapeutic target for OP. Bulk RNA sequencing provides broad view of gene expression but cannot identify the different cell types during analysis. In contrast, single-cell RNA sequencing can capture gene expression information from individual cells, offering insights into gene expression patterns, cell states, and function of specific cell types18. However, this approach produces a vast amount of data. Thus, it is critical to narrow down potential targets across multiple datasets to refine the analysis. This study aims to combine multi-omics and molecular docking to identify therapeutic targets, and it provides new method for osteoporosis treatment discovery.
Materials and methods
Bulk RNA data analysis
Researchers downloaded data from GSE222752 from Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.gov/geo) and PRJCA009927 from BioProject database (BioProject - CNCB-NGDC). GSE222752 and PRJCA009927 datasets provided reliable mRNA sequencing data of 8 weeks ovariectomized mice. Both datasets consisted 3 healthy control bone samples and 3 osteoporosis bone samples, respectively. The GSE222752 data was generated using the GPL21103 (Illumina HiSeq 4000), PRJCA009927 used Illumina HiSeq X Ten platform. PRJCA009927 converted fastq files into mRNA rawcounts after FastQC (fastqc_v0.12.1) screening. Differentially expressed genes (DEGs) from both cortical and trabecular bone groups in the GSE222752 dataset were combined for integrated analysis. However, PRJCA009927 only mentioned bone tissue. Both datasets were annotated with gene symbols, duplicated gene symbols were removed. Researchers used DESeq2 R package to identify Differentially Expressed Genes (DEGs) between osteoporosis and control groups, the criteria for identifying DEGs were set to adjust p value < 0.05 and absolute value of log2 Fold Change > 1. GSE222752 cortical bone and trabecular bone groups DEGs were combined. Find the common DEGs between two datasets for further analyses. DEGs were used for Gene Ontology (GO) and Kyoto Encyclopedia of Gene and Genomes (KEGG) analyses were implemented via g: Profiler19 and DAVID Bioinformatics20,21,22respectively. GO terms were separated into three categories, included Biological Process, Molecular Function, and Cellular Component. Furthermore, those terms were visualized with EnrichmentMap, and interpretated with AutoAnnotate23 in Cytoscape (version 3.10.1). Furthermore, a protein-protein interaction network (PPI) was mapped using Cytoscape based on the STRING database (https://cn.string-db.org/) to study the regulatory relationship between 73 DEGs. Nodes with less than 2 connections were excluded. Hub genes were identified through MCODE in Cytoscape (degree Cutoff = 2, node density = 0.2, K-Core = 2, and Max. Depth = 100). The Cytoscape-MCODE plugin was used to cluster gene to detect densely connected areas in PPI network24. Hub genes expressions were normalized with RPM (Reads per million mapped reads) for statistical analyses.
Single cell RNA-sequencing (sc-RNA Seq) analysis
Researchers obtained sc-RNA Seq datasets GSE169396 and GSE147287 from GEO database. GSE169396 used bone tissue cells extracted from the human femoral head specimens digested with collagenase type II before sequencing. On the other hand, GSE147287 used bone marrow derived mononuclear cells extracted from human femur head, and then purified using CD271+ magnetic microbead for sequencing. Control group consisted 2 samples (GSM5201884 and GSM4423510), and the other 2 samples (GSM5201883 and GSM4423510) assigned in osteoporosis group. The Seurat package (v5.1.0) was employed for quality control and filtering of the feature-barcode matrix. Samples with fewer 200 genes and less than 3 cells were filtered. Afterward, samples were combined for later quality control. Selecting cells that had unique feature counts between 200 and 2500, and mitochondrial gene ratio less than 5%25. First, researchers used original Seurat analysis without integration (resolution = 1). Later, anchor-based CCA integration (resolution = 0.1) was applied to remove batch effects among samples, while PCA was employed for dimensionality reduction, and UMAP was used for visualization. According to the resolution, UMAP been classified into 23 clusters, conservative gene markers of each clusters were identified by FindAllMarkers functions (test.use = “wilcox”, min.pct = 0.5, logfc.threshold = 1.5, Table S2). Later, cell clusters annotation were done by GPTCelltype from GPT4 with conserved gene symbols of each clusters26.
Furthermore, Dotplot was performed with 2 strong gene symbols for each cluster separately by groups. Finally, identifying differences in cell proportions between disease states (healthy vs. osteoporosis), while accounting for the proportions of all of the other cell subsets. The human protein atlas database (HPA, The Human Protein Atlas) was used to search for the tissue and cell distribution of the common DEGs. Tissue sections and cell immunochemistry photograph were downloaded from this database. Furthermore, to understand how stromal cell interacted with others type of cells. The researchers used LIANA-a framework to predict cell-cell communication27. LIANA enabled us to predict the receptors of ligands secreted by stromal cells.
Therapeutic drug prediction
For confirmation of chondrogenic-related genes as therapeutic target from previous data. Researchers collected teriparatide injected versus non-injected sc-RNA seq dataset (GSE169560) for therapeutic validation. The dataset was analyzed using the same Seurat method described in the sc-RNA Seq section above, without dataset integration. The therapeutic drugs targeting common DEGs (CHAD and COL2A1) were mined via Traditional Chinese Medicine Bank (TCMBank)28. The related Chinese Medicine ligands were listed in Table 1. All the ligands 3D structures targeted COL2A1 and CHAD were downloaded through PubChem (PubChem (nih.gov)) for further molecular docking.
For molecular docking analysis, protein structures of CHAD (PDB ID: 1U5M) and COL2A1 (PDB ID: 5LFN) were obtained from the Protein Data Bank (PDB). Dockings were performed using PyRx29which integrates AutoDock 4 and AutoDock Vina as core docking engines. Ligands (wogonin, tetrandrine, curcumin, and isorhapontigenin) structure were imported and preprocessed using Open Babel, which facilitates format conversion and structural optimization from SDF files. For each ligand-protein pair, ten independent docking runs were conducted, each generating nine potential binding poses. The binding pose exhibiting the lowest binding free energy (reported in kcal/mol and presented in heatmap) was selected for subsequent structural analysis, visualization using PyMOL 3.0, and to inform potential mutagenesis design. All docking results were saved in PDBQT format and visualized using PyMOL for structural interpretation.
Cell culture
The ATDC5 mouse chondrocyte cell line was purchased from Jennio Biotech (Guangzhou, China). ATDC5 cell line were maintained in monolayer culture in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS; HyClone, Australia), 100 units/mL penicillin G potassium salt, and 100 µg/mL streptomycin at 37 °C and 5% CO2. The medium was changed to DMEM/HAM’s F-12 (1:1, KeyGEN BioTech, China) after cells seeded on the plates.
Cell toxicity test
ATDC5 cells were seeded in a 96-well plate (10,000 cells/well) and serum-starved overnight. Subsequently, the cells were treated with Wogonin (1 µM–10 µM) or Tetrandrine (100 nM–5 µM) for 12, 24, 36, and 48 h in serum-free medium. Finally, cell viability was assessed using the Cell Counting Kit-8 (CCK8, KeyGEN BioTech, China) according to the manufacturer’s instructions. The absorbance value at 450 nm was detected to analyzed cell viability30.
qRT-PCR
Cells were seeded on 6-wells plate with 80% confluency. The 5 µM wogonin stimulated the cells with 4, 8, 12 h. The total mRNA of each wells were extracted with Takara Minibest Universal RNA extraction Kit (9767, TaKaRa). Afterward, it was reversed-transcribed to DNA template using PrimeScript Fast RT reagent Kit with gDNA Eraser (RR092A, TaKaRa) following to the instructions. qRT-PCR was performed TB Green Premix Ex Taq II Fast qPCR kit (CN830A, TakaRa) with QuantStudio™ 6 Flex Real-Tim PCR system according to the manufacture’s instructions. The relative RNA expression was assessed using the 2−ΔΔCT method (n = 4). The primers used are listed in Table 2.
Alcian blue and alkaline phosphatase staining
For Alcian Blue staining, cells were seeded in 24-well plates (7,600 cells/well) and the medium was supplemented with human insulin (10 µg/mL, Solarbio, China) and ascorbic acid (50 µg/mL, Solarbio, China). The culture medium was replaced every 2 days31. Chondrocytes differentiation period were 10 days, experimental group drugs (Wogonin 5 µM, tetrandrine 1 µM) stimulated for 2 days, control group cultured in DMEM/HAM’s F-12 mediums. After 48 h of drugs stimulations, cells were fixed with methanol for 5 min at room temperature and washed with ddH2O. Subsequently, the fixed cells were stained with Alcian blue solution (Solarbio, China) for 30 min, following by the manufactor’s instructions32.
For Alkaline Phosphatase (ALP) staining, ATDC5 cells were seeded in 24-well plates and cultured in DMEM/HAM’s F-12 medium until they reached 80–90% confluency. Afterward, cells were treated with wogonin (5 µM) or tetrandrine (1 µM) (same concentrations as for chondrogenic stimulation) for 24 h; control cells received standard medium. Cells were then stained for ALP activity using an Alkaline Phosphatase Staining Kit (C3206, Beyotime, China) following the manufacturer’s instructions.
Images of samples stained with Alcian Blue or ALP were captured using an Olympus SZX16 microscope for group comparisons (n = 4, per group for each stain). The stained area within each image was quantified using ImageJ software (National Institutes of Health, USA), applying consistent image analysis parameters (e.g., thresholding) across all images for each respective stain.
Statistical analysis
Data were expressed as mean ± standard error of the mean. Statistical significance was evaluated using simple t-test (GraphPad Prism 10.1.2). Statistical significance was set at P < 0.05 * and P < 0.01 **.
Results
Bulk RNA sequencing analyses
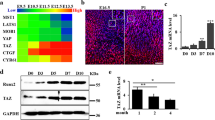
The differentiation expressions genes (DEGs) of GSE222752 and PRJCA009927 datasets were expressed by volcano maps (Fig. 1A and B). The common DEGs of both datasets were analysis with Venn Diagram (Fig. 1C), the results showed 73 common DEGs (Table S3). G-profiler analysis and Cytoscape clustering with biological process, molecular function, and cellular components. The results indicated those genes were associated with following: development muscle cell, contraction muscle process, actin filament binding, complex myosin supramolecular, and synapse extracellular matrix (Figure S1). The PPI network was visualized with Cytoscape, and hub genes were identified with MCODE function. MCODE identified three modules, the hub genes including MYBPC1, DNTT, and CHAD (Fig. 1D). After RPM normalization, CHAD and MYBPC1 showed significantly lower in the OVX group. However, DNTT showed significantly upregulated (Fig. 1E).
Bulk RNA Sequence Analysis. GSE222752 dataset cortical and trabecular bones Control VS OVX volcano maps (A). PRJCA009927 dataset Control VS OVX volcano map (B). Venn diagram of two datasets (C). Protein-Protein Interaction map of 73 common genes, identified 3 modules from MCODE hub-gene analysis (D). Hub genes expressions after RPM normalization, it includes CHAD, MYBPC1, and DNTT (E).
Single cell RNA sequencing analyses
The combination of 4 datasets were 20,554 cells (control) and 15,059 cells (osteoporosis) with UMAP shown in. After CCA integration were completed, 8304 cells (control) and 3970 cells (osteoporosis) met the criteria (Fig. 2A). The cell type annotation identifications were narrow down in 14 major cell types with GPTCelltype, including erythroid cells, erythrocytes, erythroblasts, adipocytes, M2 macrophages, stromal cells, B cells, Dendritic cells, T cells, granulocytes, plasma cells, neutrophils, macrophages, pDCs shown in Fig. 2B for later analysis. Moreover, two conservative gene markers of each clusters were shown with dot plot, and separately by groups (Fig. 2C). Cell type composition analysis demonstrated erythrocyte and adipocytes counts upregulated the most in osteoporosis group; macrophages and neutrophils counts were more in control group. Interestingly, the stromal cells were more in the osteoporosis group (Fig. 2D).
Single Cell RNA Sequence Analysis. Umap visualization separate by normal and osteoporosis groups before and after CCA integration (A). Umap visualization with cell clusters annotation and dotplot (B,C). Cell type composition graph showed erythrocytes and adipocytes upregulated in osteoporosis groups. However, macrophage and neutrophil downregulated (D). Differential gene expressions analysis in stromal cell showed 5 common genes compared with previous Bulk RNA Sequence results (E). Gene expressions of 5 common genes, including CHAD, ACAN, COL2A1, HAPLN1, FKBP5 (F).
Stromal cells contain varies types of cells, including osteoblast, osteocytes, and chondrocytes. For identification of DEGs in stromal cell cluster, researchers subset stromal cells data, and used FindMarkers function (logfc.threshold = 1.5, min.pct = 0.1). This analysis identified 1594 DEGs in stromal cells. Notably, five of these DEGs overlapped with those identified in the bulk RNA-seq datasets (GSE222752 and PRJCA009927, Fig. 2E). These overlapping genes were COL2A1, FKBP5, CHAD, ACAN, and HAPLN1. Furthermore, most of these overlapping genes (specifically, COL2A1, CHAD, ACAN, HAPLN1) belonged to the significant interaction modules previously identified by MCODE analysis of the bulk RNA-seq data, modules in which CHAD was found to be a key hub gene. Within the stromal cell data, these four genes (COL2A1, CHAD, ACAN, HAPLN1) also showed significantly higher expression in the control group compared to the OP group, with COL2A1 exhibiting a particularly strong difference (Fig. 2F). Consistent with their known roles, the Human Protein Atlas (HPA) database confirms that both ACAN and COL2A1 proteins are highly expressed in normal human cartilage (Figure S2). Our finding of lower expression of these cartilage-related genes in the OP group’s stromal cells therefore suggests potential alterations involving chondrocytes or cartilage matrix production within the stromal compartment during osteoporosis. Finally, LIANA analysis showed no direct known ligand or receptor interacts with CHAD.
Therapeutic drug prediction
For therapeutic target prediction, researchers included parathyroid hormone treated and non-treated Single cell RNA sequencing dataset (Fig. 3A,B). The cell type compositions showed chondrocytes increased significantly after treated with parathyroid hormone, and it also influenced COL2A1 and CHAD expressions (Fig. 3C,D). Ingredients associated with COL2A1 and CHAD were wogonin, Isorhapontigenin, tetrandrine, and curcumin. However, wogonin and tetrandrine showed highest binding affinity through Autodock Vina. Tetrandrine binding affinity was − 6.6 kcal/mol in COL2A1, and − 6.9 kcal/mol in CHAD; wogonin binding affinity was − 6.4 kcal/mol in COL2A1, and − 6.4 kcal/mol in CHAD (Fig. 3E). Therefore, these two ingredients were used for later in vitro experiments. However, results show no significant hydrogen bond with both tetrandrine and curcumin in CHAD (Fig. 3F).
Therapeutic target predictions. Umap visualizations of teriparatide injected VS non-injected separated by groups and cell types (A,B). COL2A1 and CHAD expressions were higher in teriparatide injected group (C,D). Heatmap of compounds binding affinities showed wogonin and tetrandrine were highest (E). Pymol visualizations of each compounds h-bond with CHAD and COL2A1 (F).
In vitro experiment
In vitro experiment demonstrated ATDC5 cell were not toxic under tetrandrine 1µM and wogonin 5µM (Fig. 4A). Thus, these two concentrations were used for later chondrocyte stimulation. According to qRT-PCR results, wogonin can influence COL2A1 and CHAD expressions (Fig. 4B). Alcian Blue staining was performed using 7,600 cells/well, and treatment with ascorbic acid and insulin for 10 days induced significant chondrogenic differentiation. Notably, wogonin further enhanced this effect, whereas tetrandrine did not produce a similar enhancement (Fig. 4C). To assess potential osteogenic effects, an ALP assay was performed on ATDC5 cells treated with wogonin and tetrandrine. No significant changes in ALP activity were observed compared to control cells under the tested conditions (Figure S8).
ATDC5 cell experiments. CCK-8 experiment showed cell were not toxic under tetrandrine 1 µM and wogonin 5 µM, n = 4 (A). qRT-PCR results showed wogonin significantly upregulated COL2A1 and CHAD expressions, n = 4 (B). Alcian Blue staining showed 5µM wogonin has chondrogenic effect, but it not under tetrandrine stimulation, n = 4 (C).
Discussion
Bulk RNA sequencing identified three distinct gene modules based on differential gene expression. While COL2A1 emerged as the most significantly differentially expressed gene in single cell RNA sequencing, CHAD was identified as a hub gene within one of these modules. CHAD is a cell binding, leucine-rich repeat protein found in the territorial matrix of articular cartilage, and it was shown to bind two sites on collagen type II14. CHAD deficiency mice revealed lower cortical thickness and mechanical strength. It also leads to a distinct characterization by widening of the epiphyseal growth plate with possible impaired of hypertrophic differentiation of chondrocytes13.
Cell composition provide macrophages, neutrophils, adipocytes, and erythrocytes relationship in osteoporosis. Recent studies showed immune cells were highly associate with bone remodeling. Both macrophages and neutrophils can differentiate into osteoclast through inflammatory cytokines stimulation33. Osteoclast are well known factor of osteoporosis, it can be slow down by inhibition of osteoclast drug like bisphosphonates34. However, in our results showed the biomarker of osteoclast matrix metalloproteinase 9 (MMP9) expression were no differences (Figure S3). Moreover, skeletal lineage cells will differentiate into adipocytes with aging leading to osteoporosis35. Interesting, there are not much studies showed erythrocytes connection with osteoporosis, further research was needed.
Stromal cell consisted osteocytes, osteoblast, chondrocytes, stem cells etc. In our study, chondrocytes higher in control group. On contrary, osteoblasts markers like COL1A1 and ALPL were higher in the osteoporosis group (Figure S7). Given that chondrocytes and osteoblasts originate from a common mesenchymal progenitor, it was important to assess whether wogonin might also induce osteogenic differentiation in this cell model. Therefore, we performed an ALP assay. The results (Figure S8) indicated that neither wogonin nor tetrandrine significantly altered ALP activity in ATDC5 cells, suggesting that the observed pro-chondrogenic effects were not accompanied by a shift towards an osteoblastic lineage under these experimental conditions. This reinforces the interpretation that wogonin primarily promoted chondrogenesis in this context. Therefore, Chondrocytes might act as important therapeutic target in osteoporosis, and its being validated through teriparatide treated datasets. Furthermore, osteoporosis can impair and slow down bone healing process36. Type II collagen progenitor cells has been identified as a crucial factor during bone healing, with COL2-positive lineage cells decline both with age and during the fracture healing. Notably, 95.5% of CD31-positive blood vessels in long bone are associated with COL2-positive linage cells, highlighting their significant role in vascular support during bone regeneration37. It is also important to identify the potential receptors for COL2A1, and interactions between cells. Therefore, the researchers used LIANA to identify the interaction between cells (Figure S4). Cell types interactions showed control group stromal cells were highly interacted with macrophages, T cells, and M2 macrophages. On the other hand, osteoporosis group showed stromal cells were highly interacted with macrophages, stromal cells, and T cells. The ligand-receptors interactions showed control group COL2A1 sent by stromal cells can bind to CD44 on M2 macrophages, macrophages, and T cells, which indicated COL2A1-CD44 interaction might play a crucial role in OP.
CD44, which mediates hyaluronan, also can interact with type II collagen38. CD44 on macrophage serves as a primary receptor for type II collagen, and it expressed at higher level in control group (Figure S5). Downregulating CD44 can inhibited human amniotic mesenchymal stem cells differentiated into chondrocytes39. Interestingly, CD44 is also widely expressed on surfaces of osteoclast precursors, where it is strongly associate with MMP939. Previous study suggests that MMP9 should expression should be elevated in osteoporosis group40. However, our analysis showed no significantly differences in MMP9 between both groups. This finding suggests that osteoclast activation, typically MMP9, may not be the key factor for osteoporosis progression.
Through molecular docking identified highest binding affinity to CHAD and COL2A1 was tetrandrine not wogonin, but there was no significant chondrogenic induction. This might due to the molecular weight of tetrandrine is larger than wogonin, and molecular docking binding affinity result will be better if the compound weight is higher41. Nevertheless, wogonin showed better results in alcian blue staining. Also, the molecular weight of wogonin is 284.26 g/mol, which is similar to bisphosphonate, makes it more appropriate than tetrandrine. Moreover, the qRT-PCR results also confirm our staining results. It upregulates chondrogenic genes like COL2A1 and CHAD. This suggests that wogonin may act through a specific pathway that enhance cartilage formation, making it promising candidate for osteoporosis treatment. A potential anti-inflammatory mechanism for wogonin may involve enhancing M2 macrophages within the immune system11. The relevance of these cells is highlighted by findings that M2 macrophages are downregulated in ovariectomized (OVX) osteoporotic mice, but their levels are restored by 17β-estradiol pretreatment42. Furthermore, M2 polarization can also be induced by factors such as type II collagen, which increases the expression of M2-related genes and pro-chondrogenic cytokines like TGF-β and IGF43. Besides wogonin, curcumin with lower binding affinity also show anti-osteoporosis44.
Despite the promising results, several limitations need to be addressed. For instance, while the molecular docking simulations identified wogonin as most promising therapeutic agents, the in vivo and human efficacy of wogonin in osteoporosis models remains to be validated. Future studies should focus on the mechanistic understanding of how COL2A1 and CHAD influenced on macrophage M1/M2 differentiation. On the other hand, the COL2A1 could be biomarkers for osteoporosis development due to its downregulation in menopause women15.
Conclusion
This study utilized multi-omics approaches and cross-species analysis to identified key therapeutic target CHAD specifically for OP. Additionally, we employed numerous TCM bank compounds for molecular docking to identified wogonin as best compound for targeting CHAD and COL2A1. Wogonin emerged as a promising compound with high potential for treating osteoporosis through chondrogenic induction, supported by experimental validation (Fig. 5).
Schematic overview of the study design for identifying potential osteoporosis therapeutic targets and compounds. Integrated bioinformatics analysis of human single-cell and murine bulk RNA sequencing datasets was employed to identify osteoporosis-associated genes, highlighting chondrogenesis-related genes such as CHAD and COL2A1. These targets were used for screening a Traditional Chinese Medicine (TCM) database, followed by molecular docking to prioritize candidate compounds. Subsequent in vitro validation of a top candidate, wogonin, demonstrated its effects on chondrogenic differentiation, suggesting its potential for further investigation.
Data availability
Bulk mRNA Sequence data researchers downloaded data from GSE222752 and PRJCA009927 of GEO database (http://www.ncbi.nlm.gov/geo) and BioProject database (https://ngdc.cncb.ac.cn/bioproject/). Osteoporosis single Cell RNA Sequence datasets GSE169396 and GSE147287 from GEO database, including control group consisted 2 samples (GSM5201884 and GSM4423510), and the other 2 samples (GSM5201883 and GSM4423510) assigned in osteoporosis group. Moreover, researchers collected teriparatide injected versus non-injected from GSE169560.
Abbreviations
- OP:
-
Osteoporosis
- TCM:
-
Traditional Chinese Medicine
- CHAD:
-
Chondroadherin
- COL2A1:
-
Type II collagen alpha I
- RNA:
-
Ribonucleic acid
- GEO:
-
Gene expression omnibus
- DEGs:
-
Differentially expressed genes
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of gene and genomes
- PPI:
-
Protein-protein interactions
- RPM:
-
Reads per million mapped reads
- sc-RNA Seq:
-
Single Cell RNA-sequencing
- PCA:
-
Principal component analysis
- HPA:
-
Human protein atlas
- qRT-PCR:
-
Quantitative reverse transcription polymerase chain reaction
- MCODE:
-
Molecular complex detection
- MYBPC1:
-
Myosin binding protein C1
- DNTT:
-
DNA nucleotidylexotransferase
- OVX:
-
Ovariectomy
- pDCs:
-
Plasmacytoid dendritic cells
- FKBP5:
-
FKBP prolyl isomerase 5
- ACAN:
-
Aggrecan
- HAPLN1:
-
Hyaluronan and proteoglycan link protein 1
- ALP:
-
Alkaline Phosphatase
References
Zengin, A., Prentice, A. & Ward, K. A. Ethnic differences in bone health. Front. Endocrinol. 6, 24. https://doi.org/10.3389/fendo.2015.00024 (2015).
Kanis, J. A. et al. A systematic review of hip fracture incidence and probability of fracture worldwide. Osteoporos. Int.. 23, 2239–2256. https://doi.org/10.1007/s00198-012-1964-3 (2012).
Gorter, E. A., Reinders, C. R., Krijnen, P., Appelman-Dijkstra, N. M. & Schipper, I. B. The effect of osteoporosis and its treatment on fracture healing a systematic review of animal and clinical studies. Bone Rep. 15, 101117. https://doi.org/10.1016/j.bonr.2021.101117 (2021).
Harris, S. T. et al. Effects of risedronate treatment on vertebral and nonvertebral fractures in women with postmenopausal osteoporosis: a randomized controlled trial. Vertebral efficacy with risedronate therapy (VERT) study group. Jama 282, 1344–1352. https://doi.org/10.1001/jama.282.14.1344 (1999).
Lyles, K. W. et al. Zoledronic acid and clinical fractures and mortality after hip fracture. N. Engl. J. Med. 357, 1799–1809. https://doi.org/10.1056/NEJMoa074941 (2007).
McClung, M. R., Balske, A., Burgio, D. E., Wenderoth, D. & Recker, R. R. Treatment of postmenopausal osteoporosis with delayed-release risedronate 35 mg weekly for 2 years. Osteoporos. Int. 24, 301–310. https://doi.org/10.1007/s00198-012-2175-7 (2013).
Foessl, I., Dimai, H. P. & Obermayer-Pietsch, B. Long-term and sequential treatment for osteoporosis. Nat. Rev. Endocrinol. 19, 520–533. https://doi.org/10.1038/s41574-023-00866-9 (2023).
Qu, Y. et al. Osteoporosis and osteoarthritis: a bi-directional Mendelian randomization study. Arthritis Res. Therapy. 25, 242. https://doi.org/10.1186/s13075-023-03213-5 (2023).
Li, K. et al. Traditional Chinese medicine in osteoporosis intervention and the related regulatory mechanism of gut Microbiome. Am. J. Chin. Med. 51, 1957–1981. https://doi.org/10.1142/s0192415x23500866 (2023).
Shi, F., Ni, L. & Gao, Y. M. Tetrandrine attenuates cartilage degeneration, osteoclast proliferation, and macrophage transformation through inhibiting P65 phosphorylation in Ovariectomy-induced osteoporosis. Immunol. Investig. 51, 465–479. https://doi.org/10.1080/08820139.2020.1837864 (2022).
Wang, D. et al. Construction of Wogonin Nanoparticle-Containing Strontium-Doped nanoporous structure on titanium surface to promote osteoporosis fracture repair. Adv. Healthc. Mater. 11, e2201405. https://doi.org/10.1002/adhm.202201405 (2022).
Li, Z. et al. Cell death regulation: A new way for natural products to treat osteoporosis. Pharmacol. Res. 187, 106635. https://doi.org/10.1016/j.phrs.2022.106635 (2023).
Hessle, L. et al. The skeletal phenotype of Chondroadherin deficient mice. PloS One. 8, e63080. https://doi.org/10.1371/journal.pone.0063080 (2014).
Mansson, B., Wenglén, C., Mörgelin, M., Saxne, T. & Heinegård, D. Association of Chondroadherin with collagen type II. J. Biol. Chem. 276, 32883–32888. https://doi.org/10.1074/jbc.M101680200 (2001).
Kojima, T., Kojima, M., Noda, K., Ishiguro, N. & Poole, A. R. Influences of menopause, aging, and gender on the cleavage of type II collagen in cartilage in relationship to bone turnover. Menopause (New York N Y). 15, 133–137 (2008).
Yang, Y., Shen, L., Wang, P. & Tao, Y. Anti-osteoporosis bioactivity evaluation in zebrafish model of Raw and salt-processed Achyranthes bidentata followed by liquid chromatography-mass spectrometry analysis and correlation analysis. Biomed. Chromatog. BMC. 37, e5742. https://doi.org/10.1002/bmc.5742 (2023).
Jang, S. et al. Wogonin inhibits osteoclast formation induced by lipopolysaccharide. Phytother. Res. 24, 964–968. https://doi.org/10.1002/ptr.3013 (2010).
Chen, G. et al. Front. Genet. 10, 317, https://doi.org/10.3389/fgene.2019.00317 (2019).
Kolberg, L. et al. g:Profiler-interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 51, W207–W212. https://doi.org/10.1093/nar/gkad347 (2023).
Sherman, B. T. et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216–W221. https://doi.org/10.1093/nar/gkac194 (2022).
Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M. KEGG: biological systems database as a model of the real world. Nucleic Acids Res. 53, D672–d677. https://doi.org/10.1093/nar/gkae909 (2025).
Kanehisa, M. Toward Understanding the origin and evolution of cellular organisms. Protein Science: Publication Protein Soc. 28, 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Reimand, J. et al. Pathway enrichment analysis and visualization of omics data using g:profiler, GSEA, cytoscape and enrichmentmap. Nat. Protoc. 14, 482–517. https://doi.org/10.1038/s41596-018-0103-9 (2019).
Bader, G. D. & Hogue, C. W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 4, 2. https://doi.org/10.1186/1471-2105-4-2 (2003).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304. https://doi.org/10.1038/s41587-023-01767-y (2024).
Hou, W. & Ji, Z. Assessing GPT-4 for cell type annotation in single-cell RNA-seq analysis. Nat. Methods. 21, 1462–1465. https://doi.org/10.1038/s41592-024-02235-4 (2024).
Dimitrov, D. et al. Comparison of methods and resources for cell-cell communication inference from single-cell RNA-Seq data. Nat. Commun. 13, 3224. https://doi.org/10.1038/s41467-022-30755-0 (2022).
Lv, Q. et al. TCMBank: bridges between the largest herbal medicines, chemical ingredients, target proteins, and associated diseases with intelligence text mining. Chem. Sci. 14, 10684–10701. https://doi.org/10.1039/d3sc02139d (2023).
Dallakyan, S. & Olson, A. J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 1263, 243–250. https://doi.org/10.1007/978-1-4939-2269-7_19 (2015)
Li, M. et al. Extracellular vesicles from apoptotic BMSCs ameliorate osteoporosis via transporting regenerative signals. Theranostics 14, 3583–3602. https://doi.org/10.7150/thno.96174 (2024).
Okita, K. et al. Ascorbic acid enhances chondrocyte differentiation of ATDC5 by accelerating insulin receptor signaling. Cell. Biol. Int. 47, 1737–1748. https://doi.org/10.1002/cbin.12067 (2023).
Meng, Q. et al. Digoxin protects against intervertebral disc degeneration via TNF/NF-κB and LRP4 signaling. Front. Immunol. 14, 1251517. https://doi.org/10.3389/fimmu.2023.1251517 (2023).
Fischer, V. & Haffner-Luntzer, M. Interaction between bone and immune cells: implications for postmenopausal osteoporosis. Semin. Cell Dev. Biol. 123, 14–21. https://doi.org/10.1016/j.semcdb.2021.05.014 (2022).
Rachner, T. D., Khosla, S. & Hofbauer, L. C. Osteoporosis: now and the future. Lancet (London England). 377, 1276–1287. https://doi.org/10.1016/s0140-6736(10)62349-5 (2011).
Wu, S., Ohba, S., Matsushita, Y. & Single-Cell, R. N. A. S. Reveals the skeletal cellular dynamics in bone repair and osteoporosis. Int. J. Mol. Sci. 24 https://doi.org/10.3390/ijms24129814 (2023).
Chandran, M. et al. Impact of osteoporosis and osteoporosis medications on fracture healing: a narrative review. Osteoporos. Int. 35, 1337–1358. https://doi.org/10.1007/s00198-024-07059-8 (2024).
Li, X. et al. Type II collagen-positive progenitors are important stem cells in controlling skeletal development and vascular formation. Bone Res. 10, 46. https://doi.org/10.1038/s41413-022-00214-z (2022).
Xu, Y., Wang, A. T. & Xiao, J. H. CD44 mediates hyaluronan to promote the differentiation of human amniotic mesenchymal stem cells into chondrocytes. Biotechnol. Lett. 45, 411–422. https://doi.org/10.1007/s10529-022-03322-2 (2023).
Samanna, V., Ma, T., Mak, T. W., Rogers, M. & Chellaiah, M. A. Actin polymerization modulates CD44 surface expression, MMP-9 activation, and osteoclast function. J. Cell. Physiol. 213, 710–720. https://doi.org/10.1002/jcp.21137 (2007).
Sabry, M. et al. Matrix metalloproteinase 9 a potential major player connecting atherosclerosis and osteoporosis in high fat diet fed rats. PloS One. 16, e0244650. https://doi.org/10.1371/journal.pone.0244650 (2021).
Xu, M., Shen, C., Yang, J., Wang, Q. & Huang, N. Systematic investigation of Docking failures in Large-Scale Structure-Based virtual screening. ACS Omega. 7, 39417–39428. https://doi.org/10.1021/acsomega.2c05826 (2022).
Dou, C. et al. Estrogen Deficiency-Mediated M2 macrophage osteoclastogenesis contributes to M1/M2 ratio alteration in ovariectomized osteoporotic mice. J. Bone Mineral. Res. 33, 899–908. https://doi.org/10.1002/jbmr.3364 (2018).
Dai, M., Sui, B., Xue, Y., Liu, X. & Sun, J. Cartilage repair in degenerative osteoarthritis mediated by squid type II collagen via Immunomodulating activation of M2 macrophages, inhibiting apoptosis and hypertrophy of chondrocytes. Biomaterials 180, 91–103. https://doi.org/10.1016/j.biomaterials.2018.07.011 (2018).
Wang, K. The potential therapeutic role of Curcumin in osteoporosis treatment: based on multiple signaling pathways. Front. Pharmacol. 15, 1446536. https://doi.org/10.3389/fphar.2024.1446536 (2024).
Author information
Authors and Affiliations
Contributions
KL: Writing – original draft, Methodology, Formal analysis, Data curation. AM: Writing – review & editing, Investigation, Formal analysis, Funding acquisition. CW: Writing- original draft, Validation, Methodology, Data curation. WS: Validation, Methodology, Data curation. YZ: Writing – review & editing, Investigation, Formal analysis. YW: Writing – original draft, Investigation, Formal analysis. SY: Methodology, Data curation. DH: Visualization, Methodology. YL: Writing – review & editing, Investigation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lan, KF., Su, WH., Zhong, Y. et al. Integrated bioinformatics analysis identifies CHAD association with osteoporosis and in vitro chondrogenic effects of Wogonin. Sci Rep 15, 24262 (2025). https://doi.org/10.1038/s41598-025-05861-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-05861-w