Abstract
Accurate segmentation of colorectal polyps in colonoscopy images represents a critical prerequisite for early cancer detection and prevention. However, existing segmentation approaches struggle with the inherent diversity of polyp presentations, variations in size, morphology, and texture, while maintaining the computational efficiency required for clinical deployment. To address these challenges, we propose a novel dual-stream architecture, Bilateral Convolutional Multi-Attention Network (BiCoMA). The proposed network integrates both global contextual information and local spatial details through parallel processing streams that leverage the complementary strengths of convolutional neural networks and vision transformers. The architecture employs a hybrid backbone where the convolutional stream utilizes ConvNeXt V2 Large to extract high-resolution spatial features, while the transformer stream employs Pyramid Vision Transformer to model global dependencies and long-range contextual relationships. Our model employs Spatial Refinement (SR) modules to process high-resolution convolutional features \({\bf{C}}_1\) and \({\bf{C}}_2\), thereby preserving critical boundary information through asymmetric convolutional processing. Channel Refinement (CR) and Non-Local Attention (NLA) mechanisms are integrated into transformer features \({\bf{P}}_3\) and \({\bf{P}}_4\) to enhance discriminative capacity and capture global contextual relationships. These refined multi-scale representations from both streams are progressively integrated through a hierarchical decoder incorporating a Pyramidal Attention Block (PAB) for multi-scale feature processing and Convolutional Block Attention Modules (CBAM) for enhanced feature discrimination. The fusion strategy employs systematic upsampling with lateral connections, enabling adequate information flow from semantic understanding to fine-grained spatial localization. Channel alignment operations through \(1 \times 1\) and \(3 \times 3\) convolutions ensure computational efficiency while preserving essential semantic information. The proposed BiCoMA architecture achieves state-of-the-art performance across five benchmark datasets (Endoscene, ClinicDB, ColonDB, ETIS, and Kvasir-SEG), demonstrating superior generalization capabilities and practical computational requirements for real-time clinical applications.
Similar content being viewed by others
Introduction
Visual intelligence-assisted detection systems have revolutionized medical imaging through the sophisticated application of deep learning architectures that can analyze intricate visual patterns and subtle morphological variations in endoscopic imagery1. This technological breakthrough holds particular significance for colorectal cancer (CRC), which constitutes the third most frequently diagnosed cancer globally and remains the second leading cause of cancer-related mortality worldwide2. The vast majority of CRC cases develop from precancerous lesions known as colorectal polyps—abnormal tissue growths that proliferate from the mucosal epithelium of the colonic wall. These polyps, especially adenomatous variants, exhibit substantial malignant transformation potential, rendering their early identification and prophylactic removal a cornerstone of cancer prevention strategies. Colonoscopic examination serves as the definitive gold standard for polyp detection and therapeutic intervention, with early-stage diagnosis significantly improving patient outcomes and elevating five-year survival rates to exceed 90%3.
However, polyp detection and segmentation during colonoscopy remain highly operator-dependent tasks characterized by substantial inter-observer variability. Manual delineation of polyp boundaries is inherently subjective and prone to errors due to variations in clinician expertise, procedural fatigue, and the intrinsic difficulty of interpreting ambiguous visual cues within complex mucosal environments. These limitations manifest as polyp miss rates ranging between 6% and 27%, particularly for small, flat, or morphologically diverse lesions that present subtle visual characteristics4. Consequently, there is an urgent clinical need for automated and accurate polyp segmentation systems to augment diagnostic decision-making and reduce interpretive variability5.
Automated polyp segmentation presents multifaceted challenges due to the heterogeneous nature of polyp presentations across diverse endoscopic conditions. Polyps exhibit remarkable morphological diversity, ranging from minute sub-millimeter nodules to large irregular masses, with varying architectural patterns including sessile, pedunculated, and flat configurations6. Recent deep learning approaches have addressed some of these challenges through specialized attention mechanisms. BMANet7 introduced boundary-guided multi-level attention to enhance boundary delineation by integrating low-level spatial features with high-level semantic information. LACINet8 focused on lesion-aware contextual interactions to mitigate background interference while maintaining computational efficiency through asymmetric multi-branch strategies. NA-SegFormer9 leveraged neighborhood attention mechanisms within Transformer architectures to capture long-range dependencies essential for accurate polyp localization. Additionally, FAENet10 exploited frequency-domain processing to separate high and low-frequency components, enabling precise boundary delineation through cross-component attention mechanisms.
Despite these advances in encoder-decoder frameworks and attention-based architectures11,12, several fundamental limitations persist that hinder clinical deployment. Current methods often struggle with inadequate multi-scale feature integration, leading to suboptimal performance on polyps with diverse size distributions. The challenge of preserving fine-grained boundary details while maintaining global contextual understanding remains unresolved, particularly when dealing with polyps obscured within surrounding mucosa. Furthermore, existing approaches frequently exhibit computational inefficiencies that limit real-time clinical applicability. At the same time, their reliance on single-stream processing architectures constrains their ability to simultaneously capture both local spatial details and global semantic relationships essential for robust polyp segmentation.
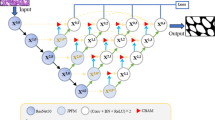
Beyond architectural limitations, several practical challenges hinder clinical deployment of state-of-the-art models. Many high-performing methods exhibit poor generalization across different colonoscopy datasets due to variations in equipment, imaging protocols, and patient populations13. Additionally, transformer-based models, although capable of modeling long-range dependencies14, often suffer from high computational complexity and latency, limiting their feasibility in real-time applications15,16. To address these limitations, we propose a novel Bilateral Collaborative Streams with Multi-Modal Attention Network (BiCoMA) that effectively balances spatial precision, semantic depth, and computational efficiency. Our approach integrates convolutional inductive biases with transformer-based global modeling while preserving scale-specific information through a refined attention and fusion strategy. We further enhance learning via multi-scale supervision, ensuring robust performance across varying lesion types and imaging conditions as shown in Fig. 1.
Contributions
The main contributions of our work are summarized as follows:
-
Dual-stream hybrid architecture: We introduce Bilateral Convolutional Multi-Attention Network (BiCoMA). This innovative dual-stream network combines the strong local feature extraction of ConvNeXt V2 Large with the global contextual modeling abilities of Pyramid Vision Transformer. This dual-stream backbone allows our network to process complementary visual information through parallel pathways, where the convolutional stream captures detailed spatial and boundary features, while the transformer stream models long-range dependencies and semantic relationships. This effectively addresses the multi-scale and complex appearance variations of polyps in challenging endoscopic environments.
-
Specialized multi-level attention mechanisms: We present a comprehensive attention strategy that employs specialized mechanisms tailored for different feature types and processing stages within the BiCoMA network hierarchy. For high-resolution convolutional features, Spatial Refinement modules utilize asymmetric convolutions to boost boundary sensitivity and maintain vital spatial details. For transformer features, Channel Refinement and Non-Local Attention mechanisms enhance discriminative power and gather global contextual information. Furthermore, Pyramidal Attention Blocks facilitate multi-scale feature processing via dual DASPP blocks and Multi-Head Self-Attention, while Convolutional Block Attention Modules offer sequential channel and spatial attention refinement across the hierarchical decoder.
-
Progressive fusion with attention-guided integration: We design an advanced hierarchical decoder that uses progressive fusion with systematic upsampling and lateral connections to effectively combine features from both streams. The decoder features Pyramidal Attention Blocks for handling multi-scale features and Convolutional Block Attention Modules for improved feature discrimination at each fusion point. This attention-guided progressive fusion effectively combines spatial details from the convolutional stream with contextual information from the transformer stream. This ensures accurate segmentation of different polyp shapes and precise boundary outlining.
-
Multi-scale supervision with hierarchical optimization: We use a detailed four-level supervision approach that integrates weighted Dice and binary cross-entropy losses at different decoder stages within the BiCoMA pathway. This multi-scale supervision ensures balanced optimization across various feature resolutions, addressing challenges in endoscopic image segmentation and maintaining consistency between broad semantic understanding and detailed spatial data from both streams. The hierarchical supervision enables the network to learn representations at multiple scales, enhancing our dual-stream model’s robustness against the variability in polyp appearances and the challenging imaging conditions often encountered in clinical environments.
-
State-of-the-art performance across diverse clinical scenarios: Through extensive testing on five diverse and challenging polyp segmentation benchmarks, Endoscene, ClinicDB, ColonDB, ETIS, and Kvasir-SEG, where our proposed BiCoMA consistently outperforms existing methods. It proves exceptionally effective in accurately detecting polyps under complex visual conditions such as varying light, reflections, partial occlusion, and diverse morphologies. Quantitative results reveal marked improvements in Dice score, IoU, F-measure, MAE, S-measure, and E-measure across all datasets, establishing a new standard for automated polyp segmentation in real-world, complex endoscopic imaging scenarios with our innovative dual-stream architecture and focused attention mechanisms.
Related work
Our research builds upon advances in three related areas: segmentation backbone architectures, polyp-specific modeling strategies, and adaptive multi-scale integration mechanisms.
Advances in segmentation architectures
Medical image segmentation has undergone significant evolution, propelled mainly by advancements in deep learning, particularly convolutional neural networks (CNNs). A foundational milestone was established by Long et al.17, who introduced fully convolutional networks (FCNs) for dense, pixel-wise predictions. This innovation shifted segmentation from patch-based methods to end-to-end trainable architectures. Expanding upon this idea, Ronneberger et al.18 developed the widely recognized U-Net, which adopted an encoder-decoder structure with skip connections. These connections allowed for the effective integration of spatial details from the encoding path into the decoding process, significantly improving segmentation quality. U-Net rapidly became a cornerstone model in medical imaging applications. To further reduce the semantic gap between encoder and decoder outputs, Zhou et al.19 proposed U-Net++, which introduced nested and densely connected skip pathways. This refined feature aggregation and enhanced performance across diverse segmentation tasks. Attention to boundary precision—a critical aspect in clinical diagnostics—led to targeted innovations. Fang et al.20 incorporated explicit area and boundary constraints to enhance edge localization, while Hatamizadeh et al.21 proposed customized loss functions designed to optimize boundary accuracy during training. The integration of attention mechanisms marked another pivotal advancement. By using spatial and channel attention modules, networks could selectively emphasize meaningful anatomical structures while down-weighting irrelevant background features. These modules improved model focus and segmentation performance, especially in complex or low-contrast medical images. In recent developments, hybrid architectures that combine CNNs with transformer-based components have gained momentum. Models such as TransUNet22 and MedT23 leverage self-attention to capture long-range dependencies, while convolutional layers preserve local spatial coherence. Concurrently, new CNN variants like ConvNeXt24 demonstrate that with architectural refinements, pure convolutional networks can match or even exceed transformer-based models in both accuracy and efficiency. Additionally, the Pyramid Vision Transformer (PVT)25 employs hierarchical attention to effectively model multi-scale contextual information, making it particularly well-suited for dense prediction challenges in medical segmentation.
Polyp segmentation strategies
Polyp segmentation extends general medical imaging methods but faces distinct challenges because of the significant variability in polyp size, texture, shape, and imaging conditions. U-Net variants remain dominant in this domain, with specialized adaptations addressing polyp-specific requirements. MSNet26 uses dense multi-scale connections to handle scale variations, while PraNet27 enhances focus on difficult boundaries through innovative reverse attention mechanisms that specifically target challenging polyp edges. Multi-scale representation has proven crucial in polyp segmentation due to the diverse morphologies encountered in clinical practice. ResUNet++28 fuses multi-level features across decoder stages to capture both fine-grained details and global context, while PPFormer29 blends CNNs and transformers for improved scale robustness. Recent transformer-based approaches like Polyp-PVT30 have successfully adapted Pyramid Vision Transformer for polyp segmentation, demonstrating the potential of hierarchical attention mechanisms for this domain. Contemporary methods have introduced specialized attention mechanisms tailored for polyp characteristics. BMANet7 proposed boundary-guided multi-level attention to enhance boundary delineation by integrating low-level spatial features with high-level semantic information. LACINet8 focused on lesion-aware contextual interactions to mitigate background interference while maintaining computational efficiency through asymmetric multi-branch strategies. FAENet10 exploited frequency-domain processing to separate high and low-frequency components, enabling precise boundary delineation through cross-component attention mechanisms. For boundary refinement, techniques have evolved from probability map corrections such as SANet31 with shallow attention mechanisms to sophisticated multi-task learning approaches that combine edge and region supervision14,15. While frequency-aware processing remains underexplored in this domain, recent advances demonstrate its potential for separating fine-grained detail from broader contextual cues. Lightweight networks such as those in13,32 reduce parameters but often sacrifice boundary accuracy, illustrating the ongoing challenge of balancing efficiency with precision in clinical deployment scenarios.
Adaptive multi-scale feature fusion
Given the significant scale variability of polyps ranging from small sessile lesions to large pedunculated masses, multi-scale fusion plays a pivotal role in achieving accurate segmentation across diverse clinical scenarios. Vision models have introduced sophisticated methods like adaptive kernel convolutions33 and progressive fusion modules34, which have inspired successful adaptations in medical imaging applications. Feature pyramid networks and their variants have established the foundation for multi-scale processing in medical segmentation. Fang et al.35 applied pyramid-based strategies to address scale discrepancies in medical imaging, while He et al.36 proposed dynamic adaptive mechanisms for intelligent scale handling. These approaches demonstrate the importance of learnable fusion strategies that can adapt to varying imaging conditions and pathological presentations. Attention-guided fusion mechanisms have emerged as particularly effective for polyp segmentation. In the domain of saliency detection, Khan et al.37 and Sinha et al.38 showcased the benefits of multi-level semantic integration through complex attention mechanisms. Progressive fusion strategies have shown particular promise for medical applications, where maintaining both global context and local details is crucial. However, existing methods often employ either fixed fusion schemes that lack adaptability or overly complex attention mechanisms that compromise computational efficiency.
Proposed methodology
Network overview
The BiCoMA architecture implements a dual-stream framework that combines ConvNeXt V2 Large for spatial detail extraction and Pyramid Vision Transformer for global contextual modeling, as illustrated in Fig. 2. This design addresses the limitations of single-stream approaches by simultaneously capturing fine-grained spatial information and long-range dependencies essential for robust polyp segmentation. The network processes input images through parallel pathways: the convolutional stream generates high-resolution features \({\bf{C}}_1\) and \({\bf{C}}_2\) that undergo Spatial Refinement (SR), while the transformer stream produces features \({\bf{P}}_3\) and \({\bf{P}}_4\) refined through Channel Refinement (CR) and Non-Local Attention (NLA) modules. Before feature integration, channel alignment is performed through convolutional operations where \(1 \times 1\) convolutions standardize channel dimensions while \(3 \times 3\) convolutions provide feature refinement, reducing high-dimensional features to 1024 channels before PAB processing for computational efficiency. Progressive fusion operates through a hierarchical decoder that systematically integrates features from both streams. The Pyramidal Attention Block serves as the central integration hub with dual DASPP blocks and Multi-Head Self-Attention, while CBAM modules at each decoder level provide channel and spatial attention refinement. The systematic upsampling pathway, equipped with lateral connections, guarantees that semantic information impacts all processing stages. It ends with a \(1 \times 1\) convolution and sigmoid activation to produce the binary segmentation output. The dual-stream architecture allows for parallel processing, while attention mechanisms offer focused enhancement without adding significant overhead, ensuring the model remains suitable for real-time clinical use.
Hybrid dual-stream backbone
Polyp segmentation is inherently challenging due to the need for precise boundary localization alongside a comprehensive understanding of the surrounding anatomical context. Traditional single-stream networks often struggle to balance local detail preservation with global semantic comprehension. While convolutional neural networks (CNNs) are adept at extracting spatial features, they typically fall short in modeling long-range dependencies. Conversely, transformers can capture global relationships effectively but may lose fine spatial granularity. To overcome these limitations, we employ a dual-stream framework that leverages the strengths of both approaches in parallel.
Our method integrates ConvNeXt V2 Large for detailed spatial feature learning with Pyramid Vision Transformer (PVT) for capturing contextual information. This architecture facilitates concurrent processing of complementary features, allowing the model to maintain a high level of spatial fidelity while also understanding the broader scene context. Such a configuration enhances feature diversity without incurring significant computational overhead.
Convolutional stream (ConvNeXt V2 large): this stream exploits the hierarchical structure of ConvNeXt V2 Large to extract spatially rich features. Incorporating modern components such as depthwise separable convolutions and layer normalization, the architecture is optimized for boundary-sensitive tasks. The input images, resized to 224\(\times\)224 pixels, are passed through ConvNeXt V2 stages, producing the following intermediate representations:
-
\({\bf{C}}_1 \in \mathbb {R}^{B \times 192 \times 56 \times 56}\): Encodes fine spatial textures and edge details, which are particularly valuable for detecting small or ambiguous polyps and ensuring accurate boundary delineation;
-
\({\bf{C}}_2 \in \mathbb {R}^{B \times 384 \times 28 \times 28}\): Represents mid-level patterns, capturing geometric and structural features that help in identifying medium-scale polyps while retaining sufficient spatial context.
The increase in channel dimensions reflects deeper semantic abstraction as the feature maps progress through the convolutional hierarchy.
Transformer stream (PVT): operating in parallel, this stream utilizes the Pyramid Vision Transformer to enhance contextual reasoning. PVT is designed with a multi-stage architecture that gradually reduces spatial dimensions while expanding feature richness. The inclusion of self-attention mechanisms allows the model to establish long-range interactions, crucial for distinguishing polyps from visually similar regions such as mucosal folds or noise artifacts.
This pathway generates the following feature representations:
-
\({\bf{P}}_3 \in \mathbb {R}^{B \times 320 \times 56 \times 56}\): retains spatial resolution while embedding global dependencies, facilitating context-aware discrimination of polyps from background clutter;
-
\({\bf{P}}_4 \in \mathbb {R}^{B \times 512 \times 7 \times 7}\): captures abstract semantic features that describe the shape, location tendencies, and morphological patterns of polyps across varied imaging scenarios.
By combining the convolutional stream’s spatial acuity with the transformer’s global interpretability, this dual-pathway architecture establishes a robust and comprehensive feature foundation. It serves as an effective precursor to later-stage attention modules and fusion operations, ultimately enhancing the segmentation performance of the entire framework.
Spatial refinement module
The convolutional feature maps \({\bf{C}}_1 \in \mathbb {R}^{B \times 192 \times 56 \times 56}\) and \({\bf{C}}_2 \in \mathbb {R}^{B \times 384 \times 28 \times 28}\) retain valuable spatial information that is essential for detecting subtle polyp boundaries. However, without targeted refinement, these high-resolution features may not fully exploit their potential for precise edge delineation. To address this, we utilized Spatial Refinement (SR) modules from the study37 that enhance spatial discrimination while maintaining efficiency, as illustrated in Fig. 3.
Each SR module employs a dual-branch architecture based on asymmetric convolutions to emphasize directional features. Specifically, for a given convolutional output \({\bf{C}}_i\) where \(i \in \{1, 2\}\), two parallel convolutional branches are applied:
These intermediate representations are then passed through a second round of cross-branch convolution to capture broader spatial interactions:
To synthesize the refined spatial information, the outputs of both branches are combined using element-wise addition, followed by a sigmoid activation to generate a spatial attention map:
The resulting attention weights are applied to the original feature map using element-wise multiplication. A residual connection is also introduced to retain the initial information:
Here, \(\odot\) denotes element-wise multiplication. This mechanism allows the model to selectively amplify informative regions while preserving the base structure of the original feature map.
Channel refinement module
Transformer features \({\bf{P}}_3 \in \mathbb {R}^{B \times 320 \times 56 \times 56}\) and \({\bf{P}}_4 \in \mathbb {R}^{B \times 512 \times 7 \times 7}\) contain rich channel information that requires selective enhancement to improve polyp detection accuracy. We apply Channel Refinement (CR) module of the study37 to these features, as shown in Fig. 4, using a squeeze-and-excitation approach that recalibrates channels based on their importance for polyp segmentation. The CR module processes each transformer feature \({\bf{P}}_i\) (where \(i \in \{3, 4\}\)) through several steps. First, adaptive average pooling creates global channel descriptors:
These descriptors pass through a bottleneck structure with two fully connected layers:
Here, \(\text {FC}_1\) reduces channels by factor \(r=16\) while \(\text {FC}_2\) restores the original dimension. Channel attention weights are obtained via sigmoid activation:
The attention weights modulate the original features with a residual connection:
This design offers four main advantages: enhanced discrimination of polyp-relevant channels, reduced background interference, context-adaptive recalibration, and better representation of diverse polyp morphologies. The resulting features \(\tilde{{\bf{P}}}_3\) and \(\tilde{{\bf{P}}}_4\) retain spatial dimensions while gaining improved channel selectivity for polyp segmentation tasks.
Non-local attention module
Long-range spatial dependencies are critical for polyp segmentation, as understanding anatomical context helps distinguish polyps from similar structures. We apply Non-Local Attention (NLA) modules to the channel-refined features \(\tilde{{\bf{P}}}_3\) and \(\tilde{{\bf{P}}}_4\), following the approach in12. The NLA mechanism allows each spatial location to gather information from all other positions, creating comprehensive contextual representations.
For each feature \(\tilde{{\bf{P}}}_i\) where \(i \in \{3, 4\}\), we first generate query, key, and value representations:
Attention weights are calculated using scaled dot-product attention with relative position encodings:
Here, \(d_k\) represents key dimensionality, while \(\text {rel}_i\) and \(\text {rel}_w\) encode relative spatial relationships. The softmax normalization ensures proper attention distribution across spatial locations.
The final NLA output combines attention results with input features via a residual connection:
where \({\bf{W}}\) is the output projection matrix, C provides normalization, and \(\theta\), \(\phi\), \(\psi\) represent feature transformations for attention computation.
The resulting features \(\hat{{\bf{P}}}_3\) and \(\hat{{\bf{P}}}_4\) offer four key benefits: improved global context understanding for polyp-structure differentiation, long-range dependency modeling for occlusion handling, enhanced spatial relationship awareness across the entire image, and increased robustness to imaging variations. This global attention mechanism ensures comprehensive spatial interaction modeling, essential for accurate polyp detection.
Pyramidal attention block (PAB)
To enhance multi-scale feature representation and contextual understanding, we integrate a Pyramidal Attention Block (PAB) adapted from a video summarization study39. The PAB module combines the attention-enhanced features from both convolutional and transformer streams, producing comprehensive multi-scale representations that capture polyp characteristics across different spatial resolutions. The module addresses the challenge of integrating features with varying spatial dimensions and semantic levels while preserving critical information from both pathways. The PAB architecture consists of three key components working in parallel to process the concatenated features from both streams. The input to PAB comprises the spatially-refined convolutional features \(\tilde{{\bf{C}}}_1\), \(\tilde{{\bf{C}}}_2\), and the attention-enhanced transformer features \(\hat{{\bf{P}}}_3\), \(\hat{{\bf{P}}}_4\), which are first aligned to a common spatial resolution through interpolation operations.
Dual DASPP blocks: two dense atrous spatial pyramid pooling (DASPP) blocks process the input features at multiple scales using dilated convolutions with different dilation rates \(\{1, 6, 12, 18\}\). The DASPP blocks capture polyp features at various receptive field sizes while preserving spatial resolution:
where \(\text {Conv}_{d_i}\) represents convolution with dilation rate \(d_i\), enabling the network to capture polyp features ranging from fine-grained details to broader contextual patterns.
Multi-head self-attention block: a transformer-based MHSA block models long-range dependencies and contextual relationships within the multi-scale features40. The MHSA operation enhances the network’s ability to distinguish polyp regions from complex backgrounds by establishing relationships between distant spatial locations:
where LN denotes layer normalization and the residual connection preserves original feature information.
Feature integration: the outputs from both DASPP blocks and the MHSA block are concatenated and processed through a \(1 \times 1\) convolution to generate the final PAB output:
The PAB module offers enhanced multi-scale representations that integrate spatial details from the convolutional stream with contextual information from the transformer stream, yielding robust feature representations essential for precise polyp segmentation across diverse morphologies and imaging conditions.
Feature flow inside the CA and optimized SA modules in the CBAM module41. Input features undergo parallel max and average pooling, followed by MLP-based interaction and skip connection. The outputs are merged and refined to enhance spatial and channel-wise representations.
Convolutional block attention module (CBAM)
To enhance feature relevance and minimize background interference, we incorporate the Convolutional Block Attention Module (CBAM) at each decoder fusion stage41. CBAM applies a lightweight yet effective attention mechanism in two consecutive phases—channel attention followed by spatial attention as shown in Fig. 5. This sequential strategy enables the model to selectively amplify polyp-relevant information while suppressing less useful features.
For a given decoder-level feature map \({\bf{F}}\), the CBAM framework performs the following operations:
Channel attention sub-module: this component is designed to model inter-channel dependencies by leveraging global context. Two types of pooling operations—average pooling and max pooling—are applied independently across the spatial dimensions to generate channel-wise descriptors. These descriptors are then passed through a shared multi-layer perceptron (MLP) to produce attention weights:
Here, \(\sigma\) denotes the sigmoid activation function. The MLP comprises two fully connected layers with a bottleneck design (i.e., reduced dimensionality). The output attention map \({\bf{M}}_c\) is then used to rescale the original feature map:
Spatial attention sub-module: after enhancing the channel-wise importance, the spatial attention module focuses on identifying significant spatial locations. This is achieved by applying average and max pooling along the channel axis, followed by concatenation and a convolution operation with a large receptive field:
This spatial attention map \({\bf{M}}_s\) highlights informative regions within the feature map. It is element-wise multiplied with the channel-attended feature \({\bf{F}}'\) to yield the final refined output:
By sequentially applying both channel-wise and spatial attention, CBAM enhances the network’s ability to isolate features related to polyps while filtering out irrelevant background artifacts. This results in improved segmentation accuracy, especially in challenging endoscopic environments where contrast and texture variations are subtle.
Progressive feature fusion and hierarchical decoding
The BiCoMA decoder systematically merges features from both convolutional and transformer streams using a multi-stage fusion process. This method combines spatial details from ConvNeXt V2 with contextual information from PVT while preserving resolution and semantic content during decoding.
Feature alignment: the fusion begins with refined features from both pathways: spatial features \(\tilde{{\bf{C}}}_1 \in \mathbb {R}^{B \times 192 \times 56 \times 56}\) and \(\tilde{{\bf{C}}}_2 \in \mathbb {R}^{B \times 384 \times 28 \times 28}\) from SR modules, plus context features \(\hat{{\bf{P}}}_3 \in \mathbb {R}^{B \times 320 \times 56 \times 56}\) and \(\hat{{\bf{P}}}_4 \in \mathbb {R}^{B \times 512 \times 7 \times 7}\) from NLA modules. We use \(1 \times 1\) convolutions to align channel dimensions across scales.
High-resolution fusion: same-resolution features \(\tilde{{\bf{C}}}_1\) and \(\hat{{\bf{P}}}_3\) (both \(56 \times 56\)) are combined through addition and refined with CBAM:
This preserves spatial details while adding global context for boundary detection.
Mid-level integration: mid-resolution features combine \(\tilde{{\bf{C}}}_2\) with upsampled deep features through PAB processing:
PAB’s dual DASPP blocks and MHSA handle multi-scale processing efficiently.
Deep feature processing: the deepest transformer features undergo PAB enhancement:
Hierarchical decoding: top-down processing with lateral connections progressively combines multi-scale features:
CBAM refinement at each stage maintains feature quality and suppresses irrelevant information.
Output generation: final upsampling and \(1 \times 1\) convolution produce the segmentation mask:
where \(\bf{Y}_{\text {final}} \in \mathbb {R}^{B \times 1 \times 224 \times 224}\) represents the binary polyp segmentation output. This progressive approach effectively combines multi-scale spatial information with global context for accurate polyp detection.
Experimental evaluation protocol
To assess the effectiveness of our proposed BiCoMA framework, we conduct extensive experiments across five publicly available benchmark datasets. All datasets undergo consistent preprocessing to ensure uniformity during the training and testing phases. Our evaluation protocol includes detailed configuration of computational resources and careful tuning of hyperparameters, which provides fair and reproducible comparisons with existing state-of-the-art methods. For performance evaluation, we employ six widely accepted quantitative metrics: Mean Absolute Error (MAE), weighted F-measure (\(\hbox {F}^{w}_{\beta }\)), Structure-measure (S\(\alpha\)), Mean Enhanced-alignment Measure (mE\(\xi\)), mean Dice coefficient, and mean Intersection over Union (IoU). These metrics collectively provide a comprehensive perspective on detection accuracy, structural similarity, and segmentation quality. In addition to performance benchmarking, we perform ablation studies to investigate the contribution of each significant component within our architecture. This is complemented by both numerical and visual comparisons against recent leading methods in the field. Overall, the results demonstrate that BiCoMA consistently delivers superior performance across all datasets and evaluation metrics, reinforcing its robustness and generalization capability in polyp segmentation tasks.
Technical configuration and training procedure
All experiments were performed on a high-performance workstation equipped with an NVIDIA GeForce RTX 4090 GPU (24GB VRAM) to manage computationally demanding tasks. Considering the considerable variation in polyp sizes within the dataset, a multi-scale training approach was adopted to improve the model’s capacity to generalize across diverse polyp dimensions. Input images were resized to 224 \(\times\) 224 pixels. Optimal performance was achieved after 100 training epochs with a batch size of 16, providing an effective balance between computational efficiency and model accuracy. Following systematic experimentation and a review of relevant domain literature, we adopted the AdamW optimizer with a learning rate of 0.0005 and a weight decay coefficient of 0.1.
Benchmark datasets and comparative baselines
We evaluate our method using five publicly available and challenging benchmark datasets, following the established evaluation protocols of PraNet27: KvasirSEG42, ClinicDB43, ColonDB44, Endoscene45, and ETIS46. The KvasirSEG dataset contains high-resolution polyp images captured during various endoscopic procedures, while ClinicDB includes images from clinical endoscopic sessions. The ColonDB, Endoscene, and ETIS datasets offer complementary resources for cross-dataset validation. Our training protocol utilized a combined dataset of 1,450 samples, consisting of 900 images from KvasirSEG and 550 from ClinicDB. For validation, we used 162 images (100 from KvasirSEG and 62 from ClinicDB), alongside the entire ColonDB dataset and selected subsets from EndoScene and ETIS. For comparison, we benchmarked our model against several leading polyp segmentation approaches, including U-Net18, UNet++47, PraNet27, ACSNet48, UACANet49, Polyp-PVT30, BDG-Net50, SSFormer51, PVT-CASCADE52, MEGANet53, and EMCAD54.
Quantitative evaluation criteria
We employ six well-established metrics to provide a comprehensive performance characterization. These metrics are:
-
1.
Mean absolute error (MAE): this metric quantifies the pixel-wise divergence between the predicted segmentation and the ground truth:
$$\begin{aligned} \text {MAE} = \frac{1}{H \times W}\sum _{i=1}^{H}\sum _{j=1}^{W}|P(i,j) - G(i,j)| \end{aligned}$$where P represents the prediction matrix and G denotes the ground truth matrix, both with dimensions \(H \times W\).
-
2.
Weighted F-measure (\(\hbox {F}^{w}_{\beta }\)): this metric incorporates spatial context through weighted precision and recall, with \(\beta ^2 = 0.3\) to prioritize precision, which is particularly relevant for medical image analysis:
$$\begin{aligned} F^{w}_{\beta } = \frac{(1+\beta ^2)\cdot \text {Precision}^w\cdot \text {Recall}^w}{\beta ^2\cdot \text {Precision}^w + \text {Recall}^w} \end{aligned}$$The weighted F-measure offers enhanced sensitivity to boundary accuracy.
-
3.
Structure-measure (S\(\alpha\)): this metric evaluates structural similarity through a combination of region-aware (\(S_r\)) and object-aware (\(S_o\)) components:
$$\begin{aligned} S_{\alpha } = \alpha \cdot S_r + (1-\alpha )\cdot S_o,\quad \alpha =0.5 \end{aligned}$$It assesses the preservation of structural integrity in segmented regions.
-
4.
Mean enhanced-alignment measure (mE\(\xi\)): this metric integrates local and global information using adaptive thresholding:
$$\begin{aligned} E_{\xi } = \frac{1}{H\times W}\sum _{i=1}^{H}\sum _{j=1}^{W}\phi (P(i,j),G(i,j)) \end{aligned}$$where \(\phi\) represents the enhanced alignment function, providing a comprehensive assessment of alignment quality.
-
5.
Mean dice coefficient: this metric evaluates region overlap as the harmonic mean of precision and recall:
$$\begin{aligned} \text {Dice} = \frac{2|P \cap G|}{|P| + |G|} \end{aligned}$$where \(|P \cap G|\) denotes true positive pixels, and |P| and |G| represent the total pixels in the prediction and ground truth masks, respectively. This metric is especially valuable for evaluating segmentation in imbalanced datasets.
-
6.
Mean intersection over union (IoU): this metric measures segmentation precision as the ratio of intersection to union:
$$\begin{aligned} \text {IoU} = \frac{|P \cap G|}{|P \cup G|} \end{aligned}$$where \(|P \cap G|\) represents true positive pixels, and \(|P \cup G|\) encompasses all pixels in either the prediction or ground truth. IoU provides a more stringent evaluation of segmentation quality than the Dice coefficient.
These six metrics are calculated at the dataset level to ensure robust comparative analysis. Higher values indicate superior performance for all metrics except MAE, where lower values are preferable. This multi-metric approach enables a comprehensive evaluation across complementary dimensions: pixel accuracy (MAE), boundary precision (\(\hbox {F}^{w}_{\beta }\)), structural coherence (S\(\alpha\), mE\(\xi\)), and region overlap (Dice/IoU).
Ablation studies
To comprehensively evaluate the contribution of each architectural component and design choice, we conducted extensive ablation experiments across five benchmark datasets: Endoscene, ClinicDB, ColonDB, ETIS, and Kvasir-SEG. Our ablation studies are organized into four key aspects: (1) backbone feature analysis, (2) progressive module integration, (3) attention mechanism comparison, and (4) fusion strategy evaluation. The ablation studies demonstrate that our BiCoMA architecture achieves optimal performance through the synergistic combination of dual-stream processing, specialized attention mechanisms, progressive fusion, and multi-scale supervision. Each component contributes meaningfully to the overall performance, with particularly significant gains on challenging datasets like ETIS, where comprehensive feature integration is crucial for accurate polyp segmentation.
Backbone feature analysis
We first analyze the contribution of individual feature streams and their combinations to understand the effectiveness of our dual-stream architecture. Table 1 presents the performance of different feature configurations. The analysis reveals that PVT features P3 and P4 consistently outperform ConvNeXt V2 features C1 and C2 across all datasets, with an average improvement of +1.6% mDice, demonstrating the effectiveness of transformer-based global context modeling. However, the ConvNeXt V2 features show superior performance on boundary-critical tasks, particularly evident in the qualitative results. The combination of the same-resolution features C1+P3 achieves better performance than cross-resolution pairing C2+P4, indicating the importance of spatial alignment in feature fusion. Most significantly, the complete dual-stream configuration with all four features C1+C2+P3+P4 achieves optimal performance with +2.9% average improvement over the best single stream, validating our hypothesis that complementary feature representations from both convolutional and transformer paradigms are essential for comprehensive polyp segmentation.
Progressive module integration analysis
We systematically evaluate the contribution of each attention module by progressively integrating components into our dual-stream backbone. Table 2 presents the performance evolution as modules are added sequentially. The Spatial Refinement modules applied to convolutional features C1 and C2 provide consistent improvements across all datasets with an average gain of +1.3% mDice, demonstrating the effectiveness of asymmetric convolution processing for boundary enhancement. Adding Channel Refinement modules to transformer features P3 and P4 yields additional improvements of +1.6% average, highlighting the importance of channel-wise feature recalibration for semantic understanding. The Non-Local Attention modules contribute significantly with +1.5% average improvement, particularly excelling on challenging datasets like ETIS with +1.8% gain, validating the importance of global contextual modeling. The Pyramidal Attention Block integration provides substantial improvements, especially on ETIS (+1.6%) and ColonDB (+1.2%), demonstrating its effectiveness in multi-scale feature processing. Finally, the combination of CBAM and multi-scale supervision achieves the complete model performance with total improvements ranging from +4.5% on ClinicDB to +9.2% on ETIS, confirming the synergistic effect of our comprehensive attention mechanism design.
Attention mechanism comparison
We compare different attention strategies to validate our design choices. Table 3 presents the performance of various attention mechanisms applied to our dual-stream backbone. Individual attention mechanisms show modest improvements over the baseline, with self-attention achieving the best single-type performance (+1.9% average) due to its global modeling capability. Combined attention strategies demonstrate superior performance, with spatial and self-attention combination achieving +3.9% average improvement, indicating the complementary nature of different attention types. However, our specialized design with SR, CR, and NLA modules tailored explicitly for convolutional and transformer features achieves +4.3% average improvement over the baseline, outperforming generic attention combinations. The complete model with PAB and CBAM integration further enhances performance to +6.5% average improvement, demonstrating that specialized attention mechanisms designed for specific feature types and processing stages significantly outperform conventional attention approaches in polyp segmentation tasks.
Fusion strategy evaluation
We analyze the impact of different fusion strategies in our hierarchical decoder. Table 4 compares various approaches to combining dual-stream features. Simple fusion methods like concatenation and element-wise addition achieve modest improvements of +2.0% and +2.6% respectively over baseline, demonstrating the basic benefit of feature combination. The Feature Pyramid Network approach shows better performance with +3.6% improvement, indicating the value of hierarchical processing. Progressive fusion without attention mechanisms achieves +4.4% improvement, confirming the effectiveness of our systematic upsampling and lateral connection strategy. The integration of individual attention modules PAB and CBAM in progressive fusion yields +5.4% and +4.8% improvements respectively, highlighting their complementary roles in multi-scale processing and feature refinement. The complete progressive fusion with both PAB and CBAM achieves optimal performance with +6.5% improvement, demonstrating that the combination of specialized multi-scale processing through PAB and sequential attention refinement through CBAM provides the most effective strategy for integrating complementary features from our dual-stream architecture.
Multi-scale supervision impact
Finally, we evaluate the contribution of multi-scale supervision at different decoder levels. Table 5 shows the progressive performance enhancement. Starting from single-level supervision at the final output, each additional supervision level contributes incremental improvements, with two-level supervision providing +0.4% average gain and three-level supervision adding another +0.3% improvement. The progression continues with four-level supervision, achieving optimal performance with +1.1% total average improvement over single-level supervision. The most significant benefits are observed on challenging datasets, particularly ETIS, with +2.5% improvement, where multi-scale supervision helps the model learn hierarchical feature representations essential for detecting subtle polyps against complex backgrounds. The consistent but moderate improvements across supervision levels demonstrate that our multi-scale strategy effectively enhances gradient flow and feature refinement throughout the hierarchical decoder without causing optimization conflicts, validating the importance of comprehensive supervision for training deep segmentation networks on medical imaging tasks.
Comparison with state-of-the-art methods
In this section, we compare the performance of our proposed network against various SOTA approaches, highlighting both quantitative and qualitative analyses.
Quantitative analysis
We conducted a comprehensive quantitative evaluation of our proposed BiCoMA method against state-of-the-art approaches across five challenging benchmark datasets using six complementary evaluation metrics. Tables 6 and 7 present detailed performance comparisons spanning traditional CNN architectures, modern Transformer-based methods, and hybrid frameworks published between 2015 and 2024. Our BiCoMA model demonstrates consistent superiority across all evaluation metrics and datasets, achieving the highest performance on all 5 datasets for both mDice and mIoU. On the most challenging ETIS dataset, our method achieves 0.810 mDice, representing substantial improvements of +0.2% over PVT-CASCADE, +2.1% over MEGANet-ResNet, and +1.6% over the recent PVT-EMCAD-B2. Similarly, on Endoscene, we achieve 0.908 mDice, outperforming the strongest competitor UACANet-S by +0.6%. The results reveal clear performance trends across different architectural paradigms, where traditional CNN methods show significant limitations, particularly on complex datasets like ETIS, with UNet achieving 0.398 mDice versus BiCoMA achieving 0.810, while our dual-stream hybrid approach consistently outperforms pure Transformer methods, validating our design philosophy of combining ConvNeXt V2 Large spatial precision with PVT global understanding through specialized attention mechanisms.
The comprehensive evaluation in Table 7 demonstrates our BiCoMA model’s superior performance across all complementary metrics. Our method achieves the highest \(\hbox {F}^{w}_{\beta }\) scores on Endoscene with 0.892, ColonDB with 0.829, and ETIS with 0.788 while maintaining the lowest MAE across all datasets, ranging from 0.005 to 0.019, indicating optimal precision-recall balance and minimal segmentation errors. Our model’s exceptional ability to preserve object structure is evidenced by achieving the highest S\(\alpha\) scores on four datasets: Endoscene with 0.946, ClinicDB with 0.949, ColonDB with 0.878, and ETIS with 0.889, with consistently high mE\(\xi\) scores ranging from 0.922 to 0.985, validating accurate spatial correspondence crucial for clinical decision-making. When compared to the most recent methods, our BiCoMA approach consistently outperforms state-of-the-art techniques, achieving improvements of +2.1% on Endoscene, +1.1% on ClinicDB, +4.0% on ColonDB, +2.1% on ETIS, and +1.8% on Kvasir-SEG in mDice scores against MEGANet-ResNet. The systematic integration of convolutional spatial details with transformer global context through our hierarchical attention mechanisms ensures optimal performance across diverse evaluation criteria, demonstrating that our proposed BiCoMA architecture with dual-stream processing, specialized attention mechanisms including SR, CR, and NLA, and progressive fusion modules including PAB and CBAM establishes a new benchmark for automated polyp segmentation across diverse clinical scenarios.
Qualitative evaluation
To evaluate the visual quality of segmentation outputs, we provide qualitative comparisons in Figs. 6 and 7. Figure 6 presents a side-by-side comparison of our BiCoMA framework with six well-established baseline models—UNet, UNet++, SEA, PolyPVT, MegaNet RNet, and UACANet-L—across six diverse and challenging test cases. In the first scenario, which involves a small-sized polyp, BiCoMA precisely delineates the complete region with smooth and well-defined boundaries. By contrast, UNet and UNet++ fail to detect the lesion, and SEA yields fragmented segmentation. For the second case featuring a large, oval-shaped polyp, our method maintains high shape fidelity and clear edge representation. Other models either miss parts of the region or exhibit distorted boundary contours. In the third example involving multiple small polyps, BiCoMA accurately captures all instances, whereas competing methods tend to overlook certain areas or produce broken outputs. Cases four to six further demonstrate BiCoMA’s robustness across varied imaging challenges, such as lighting inconsistencies, variable polyp scales, and background noise. In these instances, BiCoMA consistently generates smooth, complete, and coherent segmentations, while alternative methods show common issues such as jagged contours, partial coverage, or missed detections.
Figure 7 extends this evaluation to five benchmark datasets: Endoscene, ClinicDB, ColonDB, ETIS, and Kvasir-SEG. Each dataset is represented with two sample cases to illustrate performance across different clinical conditions. On the Endoscene dataset, which includes challenging lighting conditions, our method succeeds in detecting small and faint polyps. In ClinicDB, BiCoMA accurately segments large polyps with boundaries that closely align with ground truth masks. For ColonDB, where polyps often blend into the background due to low contrast, BiCoMA leverages its dual-stream design to effectively resolve subtle boundaries. ETIS contains some of the most challenging cases, with complex textures and low visibility. Even in these situations, our model identifies polyps that are otherwise hard to discern. Finally, on Kvasir-SEG, which features a wide range of polyp shapes and appearances, BiCoMA demonstrates high adaptability, accurately segmenting both rounded and irregular structures. Overall, these qualitative results highlight BiCoMA’s strong generalization ability and visual consistency. This performance is attributed to the synergy between its dual-stream architecture, attention modules (SR, CR, NLA), and progressive fusion blocks (PAB, CBAM), which together enhance feature discrimination, boundary accuracy, and robustness against visual variability found in real-world endoscopic imagery.
Conclusion
We propose the Bilateral Convolutional Multi-Attention network (BiCoMA) for precise colorectal polyp segmentation in colonoscopy images. By combining ConvNeXt V2 Large and Pyramid Vision Transformer in a dual-stream architecture, BiCoMA effectively merges local spatial details with global contextual understanding to address diverse polyp variations. The convolutional stream captures high-resolution spatial features, while the transformer stream models long-range dependencies. Specialized attention mechanisms, including Spatial Refinement for boundary enhancement, Channel Refinement and Non-Local Attention for semantic understanding, Pyramidal Attention Block for multi-scale processing, and Convolutional Block Attention Modules for progressive refinement, optimize feature representations at multiple levels. Extensive experiments on five benchmark datasets show that BiCoMA achieves state-of-the-art performance across all evaluation metrics, with particularly notable improvements on challenging datasets like ETIS. The consistent excellence across various clinical scenarios establishes BiCoMA as an effective solution for automated polyp segmentation. Future work will focus on real-time video analysis, uncertainty quantification, and adaptation to broader endoscopic imaging applications.
Data availability
All five datasets used in this study are publicly available benchmark datasets for polyp segmentation research. All datasets were used in accordance with their respective licensing agreements and ethical guidelines for medical image research. No additional permissions were required for the use of these publicly available benchmark datasets. The datasets are commonly used in the computer vision and medical imaging research community for polyp segmentation algorithm development and evaluation. The specific datasets and their access information are as follows: Kvasir-SEG Dataset: This dataset is freely available for research purposes and can be accessed through the official Kvasir dataset repository at https://datasets.simula.no/kvasir-seg/. The dataset contains 1,000 polyp images with corresponding ground truth segmentation masks. CVC-ClinicDB Dataset: This dataset is publicly available for academic research and can be obtained from the Computer Vision Center (CVC) at https://polyp.grand-challenge.org/CVCClinicDB/. The dataset comprises 612 images extracted from colonoscopy videos with manually annotated polyp segmentation masks. CVC-ColonDB Dataset: This dataset is available for research purposes and can be accessed through the link https: //www.kaggle.com/datasets/longvil/cvc-colondb. The dataset contains 380 images with corresponding ground truth annotations for polyp segmentation evaluation. CVC-EndoScene Dataset: This dataset is publicly available for academic research and can be downloaded from the repository https://service.tib.eu/ldmservice/dataset/endoscene. The dataset includes 912 images with pixel-level annotations for comprehensive polyp segmentation evaluation. ETIS-Larib Dataset: This dataset is freely available for research purposes and can be accessed through the repository https://www.kaggle.com/datasets/nguyenvoquocduong/etis-laribpolypdb. The dataset contains 196 polyp images with corresponding segmentation ground truth masks.
References
Ying, Z. et al. A denoising unet model with convnext block for MRI reconstruction. In 2024 International Annual Conference on Complex Systems and Intelligent Science (CSIS-IAC). 682–687 (IEEE, 2024).
Hossain, M. S. et al. Colorectal cancer: A review of carcinogenesis, global epidemiology, current challenges, risk factors, preventive and treatment strategies. Cancers 14, 1732 (2022).
Kim, N. H. et al. Miss rate of colorectal neoplastic polyps and risk factors for missed polyps in consecutive colonoscopies. Intest. Res. 15, 411 (2017).
Misawa, M. et al. Artificial intelligence-assisted polyp detection for colonoscopy: Initial experience. Gastroenterology 154, 2027–2029 (2018).
Khan, H. et al. Visionary vigilance: Optimized yolov8 for fallen person detection with large-scale benchmark dataset. Image Vis. Comput. 149, 105195 (2024).
Zhao, X. et al. \(m^{2}\)snet: Multi-scale in multi-scale subtraction network for medical image segmentation. arXiv preprint arXiv:2303.10894 (2023).
Wu, Z. et al. Bmanet: Boundary-guided multi-level attention network for polyp segmentation in colonoscopy images. Biomed. Signal Process. Control 105, 107524 (2025).
Li, W., Lu, W., Chu, J. & Fan, F. Lacinet: A lesion-aware contextual interaction network for polyp segmentation. IEEE Trans. Instrum. Meas. 72, 1–12 (2023).
Liu, D., Lu, C., Sun, H. & Gao, S. Na-segformer: A multi-level transformer model based on neighborhood attention for colonoscopic polyp segmentation. Sci. Rep. 14, 22527 (2024).
Tang, R. et al. A frequency attention-embedded network for polyp segmentation. Sci. Rep. 15, 4961 (2025).
Wang, K.-N. et al. Dlgnet: A dual-branch lesion-aware network with the supervised gaussian mixture model for colon lesions classification in colonoscopy images. Medical Image Anal. 87, 102832 (2023).
Khan, H., Usman, M. T. & Koo, J. Bilateral feature fusion with hexagonal attention for robust saliency detection under uncertain environments. Inf. Fusion 103165 (2025).
Shah, S. et al. Effect of computer-aided colonoscopy on adenoma miss rates and polyp detection: A systematic review and meta-analysis. J. Gastroenterol. Hepatol. 38, 162–176 (2023).
Yang, H. et al. Boosting medical image segmentation via conditional-synergistic convolution and lesion decoupling. Comput. Med. Imaging Graph. 101, 102110 (2022).
Murugesan, B. et al. Psi-net: Shape and boundary aware joint multi-task deep network for medical image segmentation. In 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). 7223–7226 (IEEE, 2019).
Usman, M. T., Khan, H., Singh, S. K., Lee, M. Y. & Koo, J. Efficient deepfake detection via layer-frozen assisted dual attention network for consumer imaging devices. In IEEE Transactions on Consumer Electronics (2024).
Long, J., Shelhamer, E. & Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 3431–3440 (2015).
Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5–9, 2015, Proceedings, Part III 18. 234–241 (Springer, 2015).
Zhou, Z., Siddiquee, M. M. R., Tajbakhsh, N. & Liang, J. Unet++: Redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans. Med. Imaging 39, 1856–1867 (2019).
Fang, Y., Chen, C., Yuan, Y. & Tong, K.-Y. Selective feature aggregation network with area-boundary constraints for polyp segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, October 13–17, 2019, Proceedings, Part I 22. 302–310 (Springer, 2019).
Hatamizadeh, A., Terzopoulos, D. & Myronenko, A. End-to-end boundary aware networks for medical image segmentation. In International Workshop on Machine Learning in Medical Imaging. 187–194 (Springer, 2019).
Chen, J. et al. Transunet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306 (2021).
Valanarasu, J. M. J., Oza, P., Hacihaliloglu, I. & Patel, V. M. Medical transformer: Gated axial-attention for medical image segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part I 24. 36–46 (Springer, 2021).
Woo, S. et al. Convnext v2: Co-designing and scaling convnets with masked autoencoders. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 16133–16142 (2023).
Wang, W. et al. Pyramid vision transformer: A versatile backbone for dense prediction without convolutions. In Proceedings of the IEEE/CVF International Conference on Computer Vision. 568–578 (2021).
Zhao, X., Zhang, L. & Lu, H. Automatic polyp segmentation via multi-scale subtraction network. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part I 24. 120–130 (Springer, 2021).
Fan, D.-P. et al. Pranet: Parallel reverse attention network for polyp segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention. 263–273 (Springer, 2020).
Jha, D. et al. Resunet++: An advanced architecture for medical image segmentation. In 2019 IEEE International Symposium on Multimedia (ISM). 225–2255 (IEEE, 2019).
Cai, L. et al. Using guided self-attention with local information for polyp segmentation. In International Conference on Medical Image Computing and Computer-assisted Intervention. 629–638 (Springer, 2022).
Dong, B. et al. Polyp-pvt: Polyp segmentation with pyramid vision transformers. arXiv preprint arXiv:2108.06932 (2021).
Wei, J. et al. Shallow attention network for polyp segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part I 24. 699–708 (Springer, 2021).
Usman, M. T., Khan, H., Rida, I. & Koo, J. Lightweight transformer-driven multi-scale trapezoidal attention network for saliency detection. Eng. Appl. Artif. Intell. (2025).
Li, T. et al. Microstructure and mechanical properties of particulate reinforced nbmocrtial high entropy based composite. Entropy 20, 517 (2018).
Jiang, K. et al. Multi-scale progressive fusion network for single image deraining. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 8346–8355 (2020).
Fang, X. & Yan, P. Multi-organ segmentation over partially labeled datasets with multi-scale feature abstraction. IEEE Trans. Med. Imaging 39, 3619–3629 (2020).
He, J., Deng, Z. & Qiao, Y. Dynamic multi-scale filters for semantic segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision. 3562–3572 (2019).
Khan, H., Usman, M. T., Rida, I. & Koo, J. Attention enhanced machine instinctive vision with human-inspired saliency detection. Image Vis. Comput. 152, 105308 (2024).
Sinha, A. & Dolz, J. Multi-scale self-guided attention for medical image segmentation. IEEE J. Biomed. Health Inform. 25, 121–130 (2020).
Khan, H., Hussain, T., Khan, S. U., Khan, Z. A. & Baik, S. W. Deep multi-scale pyramidal features network for supervised video summarization. Expert Syst. Appl. 237, 121288 (2024).
Vaswani, A. et al. Attention is all you need. Adv. Neural Inf. Process. Syst. 30 (2017).
Woo, S., Park, J., Lee, J.-Y. & Kweon, I. S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV). 3–19 (2018).
Jha, D. et al. Kvasir-seg: A segmented polyp dataset. In MultiMedia Modeling: 26th International Conference, MMM 2020, Daejeon, South Korea, January 5–8, 2020, Proceedings, Part II 26. 451–462 (Springer, 2020).
Bernal, J. et al. WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Comput. Med. Imaging Graph. 43, 99–111 (2015).
Tajbakhsh, N., Gurudu, S. R. & Liang, J. Automated polyp detection in colonoscopy videos using shape and context information. IEEE Trans. Med. Imaging 35, 630–644 (2015).
Vázquez, D. et al. A benchmark for endoluminal scene segmentation of colonoscopy images. J. Healthc. Eng. 2017, 4037190 (2017).
Silva, J., Histace, A., Romain, O., Dray, X. & Granado, B. Toward embedded detection of polyps in WCE images for early diagnosis of colorectal cancer. Int. J. Comput. Assist. Radiol. Surg. 9, 283–293 (2014).
Zhou, Z., Rahman Siddiquee, M. M., Tajbakhsh, N. & Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 20, 2018, Proceedings 4. 3–11 (Springer, 2018).
Zhang, R. et al. Adaptive context selection for polyp segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, October 4–8, 2020, Proceedings, Part VI 23. 253–262 (Springer, 2020).
Kim, T., Lee, H. & Kim, D. Uacanet: Uncertainty augmented context attention for polyp segmentation. In Proceedings of the 29th ACM International Conference on Multimedia. 2167–2175 (2021).
Qiu, Z. et al. Bdg-net: Boundary distribution guided network for accurate polyp segmentation. In Medical Imaging 2022: Image Processing. Vol. 12032. 792–799 (SPIE, 2022).
Wang, J. et al. Stepwise feature fusion: Local guides global. In International Conference on Medical Image Computing and Computer-Assisted Intervention. 110–120 (Springer, 2022).
Rahman, M. M. & Marculescu, R. Medical image segmentation via cascaded attention decoding. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision. 6222–6231 (2023).
Bui, N.-T., Hoang, D.-H., Nguyen, Q.-T., Tran, M.-T. & Le, N. Meganet: Multi-scale edge-guided attention network for weak boundary polyp segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision. 7985–7994 (2024).
Rahman, M. M., Munir, M. & Marculescu, R. Emcad: Efficient multi-scale convolutional attention decoding for medical image segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 11769–11779 (2024).
Ma, J. et al. Segment anything in medical images. Nat. Commun. 15, 654 (2024).
Xiong, X. et al. Sam2-unet: Segment anything 2 makes strong encoder for natural and medical image segmentation. arXiv preprint arXiv:2408.08870 (2024).
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (RS-2025-00518960) and also supported by Princess Nourah Bint Abdulrahman University Researchers Supporting Project number (PNURSP2025R733).
Author information
Authors and Affiliations
Contributions
R.K. conceived the original idea, designed the BiCoMA architecture, implemented the bilateral collaborative streams framework, conducted the comprehensive experiments, analyzed the results, and wrote the original manuscript. N.A. contributed to the multi-modal attention mechanism design, assisted in dataset preparation and preprocessing, performed ablation studies, and contributed to the quantitative analysis. Y.I.D. developed the intelligent feature fusion decoder modules, implemented the multi-scale progressive supervision strategy, conducted comparative analysis with state-of-the-art methods, and contributed to the qualitative evaluation. M.Y.L. supervised the overall research direction, provided critical insights for the architectural design, contributed to the experimental validation methodology, and reviewed and edited the manuscript. I.U. coordinated the research project, provided expertise in medical imaging applications, contributed to the clinical interpretation of results, and assisted in manuscript preparation and revision. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Khan, R., Alzaben, N., Daradkeh, Y.I. et al. Bilateral collaborative streams with multi-modal attention network for accurate polyp segmentation. Sci Rep 15, 34182 (2025). https://doi.org/10.1038/s41598-025-15401-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-15401-1