Abstract
GRWD1, a novel WD40-repeat-containing protein designated glutamate-rich WD repeat, is highly expressed in CRC and participates in a series of oncogenic activities. However, the cause of GRWD1 overexpression and its oncogenic mechanism in CRC remains elusive. This study revealed that GRWD1 was correlated with inflammation and was progressively upregulated during the progression of Azoxymethane/Dextran sodium sulfate (AOM/DSS) in a mouse model. Moreover, GRWD1 was activated by the IL-6/STAT3 signal pathway in CRC cells. Besides, it promoted the degradation of p53 and further induced GLUT1 to facilitate aerobic glycolysis in CRC. Taken together, GRWD1 played an oncogenic role in tumorigenesis of CRC and represented a promising therapeutic target.
Similar content being viewed by others
Introduction
According to the latest global cancer statistics, Colorectal Carcinoma (CRC) is the third most prevalent cancer and one of the leading causes of cancer-related mortality1. Besides, its incidence and mortality rates have steadily risen in recent decades in numerous countries, including China1,2. Notably, the incidence of early-onset colon carcinoma is increasing by 1–4% per year, signifying that it poses a growing threat to young adults1,3. Although early detection of colon cancer and surgery for primary diseases are beneficial to patient survival, the high rate of metastasis and acquired drug resistance have significantly decreased 5-year survival rates4,5. Therefore, there is an urgent need to elucidate the molecular mechanism underlying the pathogenesis of colon carcinoma.
CRC progression involves numerous factors, some of which include an imbalance in microbiomes in the colon, tobacco, and alcohol consumption, and inflammation6,7, while others are genetic changes including chromosomal instability (CIN), microsatellite instability (MSI) and DNA methylation8. It is well-established that patients with inflammatory bowel disease (IBD) have higher odds of developing colon cancer9. IL-6 is a critical cytokine participating in the development of IBD and colon cancer10. Indeed, several studies have described that IL-6 levels were elevated in tumor tissue and the serum of CRC patients11.
Genetic changes lead to the tumors’ progression from benign to malignant stages which involve many abnormal stimulating signaling pathways, such as the RAS pathway, the p53 pathway and the inflammatory pathway12. In the classic IL-6 signaling pathway, IL-6 binds to its receptor IL-6R, thus inducing the formation of a heterohexameric complex comprising two molecules each of IL-6, IL-6R, and the IL-6 receptor subunit-β (gp130). The formation of this complex drives the activation of the JAK/STAT3 signaling pathway. Notably, phosphorylated STAT3 is a transcription factor stimulating a series of target genes13,14. The JAK–STAT3 signaling pathway has recently been shown to play a central role in inflammation-mediated cancer, metabolism, cell proliferation, and invasion11.
GRWD1 is a novel WD40-repeat-containing protein designated glutamate-rich WD repeat that is likely to be involved in ribosome biosynthesis15. Recent studies documented that GRWD1 could negatively regulate the expression of p53 via direct interaction with the p53 DNA-binding domain. In addition, it also abrogated RPL11‐mediated p53 stabilization and may consequently be a potential oncogene16,17. Moreover, GRWD1 was highly expressed in non-small cell lung cancer (NSCLC) and related to poor differentiation and prognosis18. Our previous studies determined that GRWD1 was not only highly expressed in colon carcinoma tissues and several human colon cancer cell lines but also correlated with proliferation, cell cycle arrest, apoptosis, migration, and invasion19. However, the underlying molecular mechanisms of GRWD1 remain enigmatic.
Our results revealed that IL-6/STAT3 could induce GRWD1 expression in CRC cells, which in turn negatively regulated p53 protein stability, thereby promoting GLUT1 expression and facilitating glycolysis in colon carcinoma.
Materials and methods
Antibodies and reagents
The primary antibodies used in this study are listed below: GRWD1 (Sigma, #HPA042643), GLUT1 (CST, #12939), p53 (CST, #9282), p-STAT3 (CST, #9145), STAT3 (CST, #9139), α-actinin (Proteintech, #11313-2-AP).
As for other reagents, Pifithrin-β (#HY-16702), Stattic (#HY-13818) and Cycloheximide (#66-81-9) were purchased from MCE. Recombinant Human IL-6 (#200-06) was purchased from Peprotech. Azoxymethane (#A5486) was obtained from Sigma. Dextran sodium sulfate (#0216011080) was purchased from MP Biomedicals.
Data acquisition
To examine the protein expression levels of GRWD1 in normal and tumor samples across diverse cancer types, we employed data from the Clinical Proteomic Tumor Analysis Consortium (CPTAC). Furthermore, to evaluate the differential mRNA expression of GRWD1 in colorectal cancer, we utilized the BEST online analysis platform (https://rookieutopia.com/app_direct/BEST/) for processing and visualizing the pertinent clinical data.
Spatial transcriptomics analysis
In spatial transcriptomics analysis, cell types within spatial transcriptomics slices were characterized by the predominant cell type present in each microregion. Specifically, if malignant cells constituted the majority within a microregion, that region was designated as “malignant cells.” Conversely, if endothelial cells were most prevalent, the region was labeled as “endothelial cells.” The cell type exhibiting the highest proportion in each microregion was determined, and the spatial distribution of microregion information across each slice was visualized utilizing the SpatialPlot function from the Seurat package. The expression profile of GRWD1 across various microregions was visualized using the SpatialFeaturePlot function from the Seurat package.
Additionally, to assess the expression levels of GRWD1 in spatial transcriptomics slices of gastrointestinal tumors, we calculated the average expression of GRWD1 for each cell type within each slice. The data underwent z-score standardization using the scale function, and the outcomes were visualized employing the pheatmap package. To further validate the expression of GRWD1 in colorectal cancer (CRC) using spatial transcriptomics slices, tissue regions were classified based on the proportion of malignant cells within microregions. Specifically, a microregion was designated as “malignant” if the deconvolution analysis indicated a proportion of 1 for malignant cells, as “normal” if the proportion was 0, and as a “mixed malignant region” for any other proportion. Differences in expression levels between these groups were assessed using the Wilcoxon test, and the mean expression levels were illustrated through bar charts.
Patient and tissue specimens
A total of 28 paired tissue samples, including CRC tissues, adjacent paranormal tissues, and normal tissues were collected at The First Affiliated Hospital of Xiamen University. Tissue samples were used for qPCR, Western blot and IHC analyses. This study was approved by the Ethics Committee of The First Affiliated Hospital of Xiamen University (No. XMYY-2022KYSB097) and was conducted following the ethical guidelines of the Declaration of Helsinki. Informed consent was obtained from all subjects or their legal guardian(s).
Gene function enrichment
To further investigate the association between abnormal expression of the GRWD1 gene and the biological functions of CRC, we collected 14 different functional states of tumor cells from the CancerSEA database. Using the z-score algorithm, we calculated the activity of the given pathways based on the gene sets for these 14 functional states, obtaining the combined z-score. We further standardized the scores using the scale function, defining them as gene set scores and calculated the Pearson correlation between GRWD1 expression and each of the gene set scores. Additionally, in the TCGA-COADREAD cohort, tumor samples were extracted. Using a cutoff value of 0.3, the top 30% of samples with the highest GRWD1 expression were defined as the high-expression group, and the bottom 30% with the lowest GRWD1 expression were defined as the low-expression group. Differential analysis was performed using the limma package to obtain the log2 fold change (log2FC) for each gene. All genes were sorted according to their log2FC, and GSEA enrichment analysis was performed (c2.all.v7.5.1.symbols.gmt). Gene signals or pathways with a p-value less than 0.05 were considered significantly enriched.
Transcriptional regulator analysis
To investigate the transcriptional regulatory pattern of GRWD1, we integrated four tools—hTFtarget, ENCODE, GTRD, and Chip_Atlas—to explore the potential transcriptional regulatory mechanisms of GRWD1 by analyzing the overlapping transcription factors across these four datasets.
Survival analysis
To assess the potential relationship between GRWD1 and SLC2A1, we first divided the TCGA-COAD READ cohort into four groups based on a binary classification: (GRWD1 high, SLC2A1 high; GRWD1 high, SLC2A1 low; GRWD1 low, SLC2A1 high; GRWD1 low, SLC2A1 low). The survival curve differences between the groups were tested using the log-rank test.
Cell culture and treatment
All cell lines were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Human colon carcinoma cell lines RKO and SW480 were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin sulfate. Cells were incubated in a humidified atmosphere containing 5% CO2 at a temperature of 37 °C.
For experimental treatments, cells were seeded in appropriate plates. To assess protein degradation, cells were treated with the protein synthesis inhibitor Cycloheximide (CHX) at a concentration of 50 μg/ml for the indicated times (e.g. 0, 10, 20, 30, 60 and 120 min). To inhibit STAT3 signaling, cells were treated with stattic, a specific STAT3 inhibitor that blocks STAT3 activation by inhibiting its phosphorylation and dimerization20, at a concentration of 1 μmol/L for 4 h. Stattic was dissolved in DMSO and diluted in culture medium (final DMSO concentration < 0.1%). Control cells received an equivalent volume of DMSO. Following treatments, cells were harvested for follow-up analysis.
Plasmids and cell transfection
The specific shRNA targeting GRWD1 was constructed by the Public Protein/Plasmid Library and cloned into the plko-puro vector, shRNA sequences were shown in Supplementary Table 1. Full-length GRWD1 was synthesized by the Public Protein/Plasmid Library and cloned into the pLVX-puro vector to generate GRWD1 overexpression.
HEK293T cells were transfected by the resulting lentiviral vectors and packing plasmids using Lipofectamine 2000 (Invitrogen, #11668-019). The medium was replaced after 8 h. After 48 h, supernatants were collected and filtered. RKO and SW480 cells were infected by viral supernatant and selected using puromycin.
Western blot analysis
Cells and tissue samples were lysed in RIPA lysis buffer supplemented with a protease inhibitor (Roche, #04693116001), phosphatase inhibitor cocktail (Roche, #04906845001) and PMSF. The lysates were centrifuged at 12,000 rpm, and the supernatant was collected. Proteins were quantified using a Bicinchoninic Acid (BCA) Assay (Pierce BCA Protein Assay Kit, Thermo Fisher Scientific, #23227). Cell lysates (10 µg) were separated by 10% SDS-PAGE and transferred to PVDF membranes. After blocking in 5% nonfat milk at room temperature for 1 h, the membranes were incubated with primary antibodies at 4 °C overnight. Membranes were sequentially washed 3 times with 1X PBS containing 0.1% Tween20 (v/v), incubated with horseradish peroxidase-conjugated secondary antibodies at room temperature for 1 h, and washed to eliminate unbound antibodies. Bound antibody complexes were visualized using the enhanced chemiluminescence detection system (ECL) and imaged using the Bio-Rad ChemiDoc XRS System. Notably, we used different membranes with parallel loading to detect p-STAT3 and STAT3 respectively. All samples were from the same lysates and processed under the same conditions of electrophoresis and transmembrane.
Hematoxylin and Eosin (H&E) and Immunohistochemistry (IHC) analysis
Patients’ tissues and the distal colon tissues of CAC mice were fixed in formalin and embedded in paraffin. To investigate the different stages of colitis and cancer, 5 μm-thick sections were used for staining. Hematoxylin and eosin staining were performed for H&E analysis. For IHC, the samples were deparaffined, endogenous peroxidase activity was quenched, and antigen retrieval was conducted. After blocking, slices were incubated with anti-GRWD1 antibody at 4 ℃ overnight. Positive signals were visualized using the DAB kit.
Real-time quantitative PCR
According to the manufacturer’s instructions, total RNA was extracted from cells and tissues using the RNAsimple Total RNA Kit (TIANGEN, #DP419). The mRNA was reverse-transcribed using the FastQuant RT Kit (with gDNase) (TIANGEN, #KR116-02). Quantitative real-time RT-PCR was conducted on a 7500 Real-Time PCR system using SuperReal Premix Plus (SYBR Green) (TIANGEN, #FP205-02). All primers used in this study were shown in Supplementary Table 2.
Mouse model for colitis-associated cancer (CAC)
Azoxymethane (AOM) and Dextran sodium sulfate (DSS) were used to induce colitis-associated cancer in mice21. 8-week-old male mice were purchased from the SLAC (Shanghai, China) and were randomly assigned to 4 groups (n = 5): normal group, inflammation group, precancer group and tumor group. Experimental mice were intraperitoneally (IP) injected with 10 mg/kg AOM, and one week later, they were supplied with distilled water containing 2% DSS for 7 days. Then, they were treated with standard drinking water for 2 weeks. The cycle was repeated three times. The mice were euthanized by exposure to the gradually increasing concentrations of carbon dioxide at different time points according to the progression of CAC. Experiments were repeated independently three times. The animal experiments were approved by the Institutional Animal Care and Use Ethics Committee of Xiamen University (XMULAC20180077). All methods were reported in accordance with ARRIVE guidelines (https://arriveguidelines.org) for the reporting of animal experiments.
Glucose uptake and lactate release analysis
The Glucose Uptake Colorimetric Assay Kit (Biovision, #K676) was used according to the manufacturer’s instructions to detect glycolysis in colon carcinoma cells. Briefly, cells were quantified and plated in a 96-well plate. After 24 h, cells were starved in a serum-free medium overnight to enhance glucose uptake. Next, glucose-starved cells were washed with PBS and treated with 2-deoxyglucose (2-DG) for 20 min at 37 °C. Then the accumulated 2-DG6P was oxidized to generate NADPH, amplified with an enzymatic recycling reaction. The glucose concentration was analyzed at an absorbance of 412 nm under a microplate reader.
Lactate release analysis was used with the L-lactate Assay Kit (Biovision, #K607). Cells were plated in a 96-well plate. The medium was collected and added to the reaction mix. Samples were incubated for 30 min at room temperature and the absorbance values were measured at 570 nm using a microplate reader.
Extracellular acidification rate (ECAR) and oxygen consumption rate (OCR)
The Seahorse XF Cell Mito Stress Test Kit (Agilent, #103020) and Glycolysis Stress Test Kit (Agilent, #103015) were used to examine the cellular mitochondrial function and glycolytic capacity. Following the manufacturer’s instructions, cells were seeded onto 96-well plates at a density of 2 × 104 and incubated overnight. Then, they were washed with Seahorse buffer, which consists of DMEM, phenol red, 25 mM glucose, 2 mM sodium pyruvate, and 2 mM glutamine. 10 mM glucose, 1 μM oligomycin, and 100 mM 2-deoxy-glucose (2-DG) were added to detect extracellular acidification rate (ECAR). 1 μM oligomycin, 1 μM FCCP, and 1 μM rotenone were introduced to determine the oxygen consumption rate (OCR). The ECAR and OCR were calculated after normalization to the cell number. All values were expressed as the mean ± SD.
Statistical analysis
Statistical analyses were conducted using GraphPad Prism software. Comparisons between groups were analyzed using a Student’s t-test. Data were presented as mean ± SD. A p-value of < 0.05 was considered statistically significant. All methods were carried out in accordance with the relevant guidelines and regulations.
Results
GRWD1 is overexpressed in the clinical colon carcinoma (CRC) tissues
To detect the association between the expression of GRWD1 and the status of patients with CRC, mRNA expression levels of the GRWD1 gene in normal versus tumor tissues derived from colorectal cancer (CRC) within the GSE71187 and GSE87211 datasets were analyzed. The findings revealed a statistically significant elevation in GRWD1 mRNA expression in tumor tissues compared to normal tissues (Fig. 1A). Subsequently, we conducted a comparative assessment of GRWD1 expression at the protein level across various cohorts, distinguishing between tumor and normal tissues. The results indicated that, in CRC, GRWD1 protein expression was elevated in tumor tissues, corroborating the transcriptomic analysis (Fig. 1B).
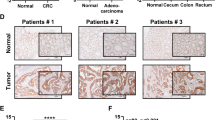
Expression patterns of GRWD1 in colon cancer tissues. (A) Analysis of GRWD1 mRNA expression in normal and CRC tissues using GSE71187 and GSE87211 datasets. (B) Examination of GRWD1 protein levels across different cancer types, including COAD, utilizing the CPTAC database (**, p < 0.01; ***, p < 0.001). (C) Assessment of GRWD1 expression in the malignant cellular microregions of CRC tissues. (D) Investigation of GRWD1 expression pattern in representative CRC sections and statistical analysis of GRWD1 in malignant regions. (E) mRNA levels of GRWD1 in colon cancer tissues (T), para-cancer tissues (P), and paired normal tissues (N) of 16 patients. (F) Western blot analysis of GRWD1 protein expression in clinical patient tissue samples (N, normal tissue; P, para-cancer tissue; T, tumor tissue). The statistical grey value of GRWD1 protein expression is displayed in the lower right corner. (G) IHC analysis of normal and tumor tissues (T) of colon cancer. Red boxes represent enlarged photos of yellow boxes. The area within the red dotted line represents para-cancer tissue (P).
In spatially analyzed tumor tissue sections of gastrointestinal malignancies, we identified a preferential expression of GRWD1 within the malignant cellular microregions of CRC tissues (Fig. 1C). In representative CRC sections, the cell-type feature maps revealed the predominant cellular characteristics of distinct microregions, thereby elucidating the composition of the tumor microenvironment at a sub-resolution scale. Furthermore, the expression pattern of GRWD1 closely paralleled that observed in tumor cells, implying that GRWD1 may predominantly be expressed within the tumor cells of CRC. Additionally, statistical analysis utilizing tissue region segmentation revealed that GRWD1 expression was significantly elevated in malignant regions (Fig. 1D). The original HE image from the spatial transcriptomics analysis was presented in Supplementary Figure S1.
Consistent with these findings, qPCR analysis exposed that GRWD1 was highly expressed in tumor tissues of CRC patients (Fig. 1E). Similarly, the protein expression level of GRWD1 in 28 pairs of CRC and adjacent normal and para-carcinoma tissues was detected using western blot analysis. The analysis determined that GRWD1 protein expression was significantly upregulated in CRC tissues compared to both normal and para-carcinoma tissues (Fig. 1F). Besides, tissue samples of CRC patients were examined using IHC, the positive signal of GRWD1 expression showed brown color, and the nuclei of the cells were stained blue by hematoxylin. In normal colon tissues, GRWD1 was hardly expressed, and in tissues from CRC patients, GRWD1 was slightly expressed in the area of para-cancer tissues, and its expression was significantly elevated in tumor tissues (Fig. 1G). Taken together, these results indicated that GRWD1 overexpression may be an essential indicator of colon carcinoma.
GRWD1 is induced in CAC mouse model and correlates with activated STAT3
To investigate the connection between GRWD1 and the malignant features of CRC, we quantified the abilities of tumors by the z-score algorithm. As indicated, GRWD1 expression was significantly positively correlated with the scores of malignant features such as cell cycle, proliferation, and metastasis in CRC which were consistent with our previous results19. Notably, GRWD1 also correlated with inflammation (Fig. 2A).
Expression of activated STAT3 and GRWD1 in the CAC mouse model and CRC tissues. (A) The correlation analysis between GRWD1 expression and several tumor malignant features. (B) The experimental scheme of the AOM/DSS mouse model. (C) Appearance of longitudinally opened colons of different groups (N, normal group; I, inflammation group; P, precancer group; T, tumor group). (D) HE and IHC analysis of tissues of the AOM/DSS mouse model. The blue arrow indicated lymphocytic infiltration. The green arrow indicated dysplasia. The red arrow indicated adenomas with hyperchromatic nuclei. The area of red boxes in photos of 100 × was enlarged to 400 × . (E) qPCR analysis of GRWD1 mRNA expression at different stage groups of mice (n = 5). (F) The identification of potential TFs of GRWD1. (G) The correlation between STAT3 and GRWD1 in TCGA-COAD databases. (H) GRWD1, phosphorylated-STAT3 (p-STAT3), and STAT3 expression in distal colon tissues were analyzed by Western blot. The statistical grey value of protein expression was depicted on the bottom. (I) The correlation of the statistical grey value between GRWD1 and p-STAT3/STAT3 in clinical patient tissue samples (n = 28) was calculated.
As is well documented, chronic inflammation drives tumorigenesis22. For instance, patients with chronic inflammatory bowel diseases are at an increased risk of developing colitis-associated cancer (CAC)23. Therefore, an AOM/DSS mouse model of CAC was constructed to investigate the role of GRWD1 during CAC progression. A single injection of AOM was administered to induce mutagenesis, followed by repetitive treatment with DSS, which could generate pronounced inflammation in the colon. Throughout AOM/DSS treatment, the continuous process in the colon presented “normal-inflammation-dysplasia-carcinoma” as the development of CRC in humans, inflammation in the colonic mucosa was observed in week 2, atypical hyperplasia appeared at the 6th week, and at the end of the 18th week, mice developed CRC24. Therefore, we divided mice into four groups according to their CAC progression stages, namely “normal,” “inflammation,” “precancer,” and “tumor” groups. Mice of the inflammation group were sacrificed on day 17th, and the precancer group was sacrificed on day 49th. In addition, at the end of the modeling on day 100th, the tumor group was sacrificed (Fig. 2B). Longitudinally opened colons of different stages were observed. The appearance of the intestines in the normal and inflammation groups was similar. The distal intestines of mice in the precancer group were congested, presenting as red and having raised polyps in the distal part of the intestines. AOM-induced tumors occur most frequently in the left-sided colon and rectum, which is consistent with the predominant localization of CRC in humans25. As anticipated, all mice of the tumor group developed CRC, and tumors were found at the distal colon and rectum in the tumor group (Fig. 2C). HE staining displayed the pathological features of each group. Although there was no significant difference in the appearance of the intestines of mice in the normal and inflammation groups, HE staining indicated that compared with the normal group, inflammatory infiltrates especially lymphocytes were visualized in the inflammation group. In the precancer group, dysplasia could be observed in representative images. Besides, features of neoplasia, such as hyperchromatic nuclei and increased nuclei/cytoplasmic ratios, were identified in the tumor group (Fig. 2D). Furthermore, the expression of GRWD1 of the distal colon in CAC progression was examined. The mRNA expression level of GRWD1 was slightly stimulated in the inflammation and precancer groups and markedly higher in the tumor group (Fig. 2E). On the other hand, IHC analysis indicated that GRWD1 expression was barely detectable in the normal group and slightly expressed in the inflammation and precancer groups. In the tumor group, GRWD1 was highly expressed (Fig. 2D).
In order to figure out the reason why GRWD1 was gradually upregulated in the CAC mouse model, we analyzed the putative transcription factors of GRWD1 using datasets including hTFtarget, ENCODE, GTRD, and Chip_Atlas. The findings revealed the identification of 159, 119, 489, and 535 putative transcription factor (TF) genes by each respective tool, as detailed in Supplementary File 1. However, the intersection of these tools resulted in the identification of only 66 target genes including STAT3 (Fig. 2F). In addition, the TCGA database indicated GRWD1 had an obvious correlation with STAT3 in COAD (Fig. 2G). Previous reports demonstrated that IL-6 was essential to tumor development of CAC, of which transcription factor STAT3 was the major protumorigenic of IL-626. Therefore we detected the phosphorylated STAT3, which was also induced in the progression of the AOM/DSS mouse model27. As expected, p-STAT3 was highly stimulated in experimental groups. In addition, the protein levels of GRWD1 were also gradually induced during the progression of CAC (Fig. 2H). To further verify the relationship between the IL-6/STAT3 pathway with GRWD1 expression in CRC, we detected p-STAT3, STAT3 and GRWD1 levels in the tissues of CRC patients. The grey value of GRWD1 protein expression had a positive relationship with p-STAT3/STAT3 expression that indicated the level of activated STAT3 (Supplementary Fig S2, Fig. 2I).
Overall, our results signaled that GRWD1 may play a role in the pathogenesis of CAC and highly correlate with the activated STAT3.
GRWD1 is up-regulated through the IL-6/STAT3 signaling pathway
To determine whether GRWD1 overexpression was upregulated with IL-6/STAT3 stimulation in colon cancer, CRC cells SW480 and RKO were exposed to the inflammatory cytokine IL-6. Firstly, GRWD1 mRNA level was assessed following IL-6 treatment. In SW480 cells, GRWD1 expression was induced after IL-6 therapy for 4 h (Fig. 3A). Likewise, GRWD1 expression was up-regulated following IL-6 treatment for 2 h in RKO cells (Fig. 3B). Moreover, changes in protein expression levels were also evaluated after IL-6 stimulation. After IL-6 treatment, p-STAT3 expression was up-regulated, whilst GRWD1 was marginally upregulated in both colon cancer cells (Fig. 3C,D). Previous studies demonstrated that the IL-6/STAT3 pathway is aberrantly hyperactivated in several types of cancer13. Therefore, IL-6 and the STAT3 inhibitor stattic were used to determine the relationship between the GRWD1 and the IL-6/STAT3 pathway. The results unveiled that GRWD1 expression was induced following IL-6 treatment and inhibited following exposure to stattic, indicating that IL-6 may regulate GRWD1 expression via STAT3 phosphorylation (Fig. 3E).
GRWD1 expression following IL-6 treatment. qPCR analysis of GRWD1 mRNA expression following IL-6 (30 μg/L) treatment at pre-defined time points in SW480 cells (A) and RKO cells (B). Western blot analysis of p-STAT3, STAT3, and GRWD1 protein levels after IL-6 treatment at indicated time points in SW480 cells (C) and RKO cells (D). The statistical grey value of relative protein expression was calculated by comparing the IL-6 group with the vehicle group, as shown on the right. (E) Western blot analysis of p-STAT3, STAT3, and GRWD1 protein expression after treatment with IL-6 (30 μg/L)and the STAT3 inhibitor stattic (1 μmol/L) for 6 h in SW480 cells. The statistical grey value of protein expression is delineated on the right.
GRWD1 knockdown Inhibits aerobic glycolysis of CRC cells
To further elucidate the potential role of GRWD1 in CRC, we performed gene set enrichment analysis (GSEA) with the high GRWD1 expression group. The results indicated biological processes such as “DNA synthesis,” “cell cycle,” “glucose metabolism,” “aerobic glycolysis,” and “glycolysis and gluconeogenesis” were significantly enriched in the GRWD1 high expression group (Fig. 4A). As previously reported, tumor cells predominantly rely on aerobic glycolysis for energy production, even in the presence of oxygen, with increased glycolysis correlating with aggressive tumor progression, treatment resistance, and unfavorable prognosis in various cancers13,28. We hypothesized that GRWD1 affects the aerobic glycolysis of CRC cells and further accelerates the progression of CRC. To test this hypothesis, GRWD1 knockdown and overexpression cell lines of RKO and SW480 were established (Fig. 4B,C). Firstly, a Seahorse XFe96 analyzer was employed to determine the effect of GRWD1 on ECAR and OCR. The results uncovered that GRWD1 knockdown reduced the overall glycolytic flux, and the glycolysis, glycolytic capacity, and glycolytic reserve were also significantly inhibited with GRWD1 knockdown (Fig. 4D). However, in the Cell Mito Stress Test, OCR and several stages including basal respiration, ATP production, and maximum respiration were higher following GRWD1 knockdown (Fig. 4E). Furthermore the impact of GRWD1 on glucose uptake and lactate release was investigated. The results showed that glucose uptake was inhibited in GRWD1-knockdown CRC cells; conversely, GRWD1 overexpression promoted glucose uptake in CRC cells (Fig. 4F). Concerning lactate release, GRWD1 knockdown repressed lactate release, whereas overexpression of GRWD1 exerted a positive effect on lactate release (Fig. 4G). These data collectively indicated that GRWD1 promoted glycolytic flux and limited mitochondrial respiratory capacity, signifying that GRWD1 expression activated glycolysis in CRC cells.
The effect of GRWD1 on glycolysis in CRC cells. (A) The GSEA enrichment analysis of GRWD1 high expression with biological processes. GRWD1 knockdown (B) and overexpression (C) in SW480 and RKO cells. (D) The values of ECAR, glycolysis, glycolytic capacity and glycolytic reverse in GRWD1 knockdown cells were represented. (E) The relative OCR, basal respiration, ATP production, and maximum respiration were shown in GRWD1 knockdown cells. (F) The effect of GRWD1 on glucose uptake. (G) The effect of GRWD1 on lactate release.
GRWD1 upregulates GLUT1 expression in CRC
To discover the underlying relationship between GRWD1 and glycolysis, the TCGA databases were employed to analyze the correlation between GRWD1 and the glycolysis-related genes GLUT1 (SLC2A1), PGK1, and G6PD in COAD and READ patients. The results showed GRWD1 expression significantly with G6PD (R = 0.493, p < 0.001), SLC2A1 (R = 0.273, p < 0.001) and PGK1 (R = 0.236, p < 0.001) (Fig. 5A). Moreover, the correlation between GRWD1 and glycolysis-related gene expression was further explored in CRC cells. Based on our results, GRWD1 knockdown inhibited the expression of all three genes, while overexpression of GRWD1 exclusively upregulated the mRNA expression of GLUT1 (Fig. 5B,C). Transmembrane transporter-GLUT is essential for glucose transport, which is significantly upregulated in tumor cells to increase glucose uptake. GLUT1 (known as SLC2A1) is highly expressed in many types of cancers29. Previous studies showed chronic inflammation led to the reprogramming of the glucose metabolism of cancer cells, which promoted tumorigenesis30. Combined with our previous study, IL-6 could induce GRWD1 expression in CRC. Therefore, we hypothesized that IL-6 might affect GRWD1 upregulation and further promote aerobic glycolysis through GLUT1 in CRC cells. To verify our hypothesis, IL-6 was introduced to stimulate GRWD1-knockdown and -overexpressed cells and detect the mRNA expression of GLUT1. Interestingly, GLUT1 expression had been affected. Compared with the control group, GRWD1 knockdown inhibited GLUT1 mRNA. Following IL-6 treatment, the expression level of GLUT1 was increased in the control group, whereas GRWD1 knockdown inhibited GLUT1 upregulation even though under IL-6 treatment (Fig. 5D). Besides, overexpression of GRWD1 further promoted the mRNA expression of GLUT1 following IL-6 stimulation (Fig. 5E). Overall, the data indicated that IL-6 influenced GLUT1 through GRWD1.
The effect of GRWD1 on GLUT1 expression. (A) The correlation between GRWD1 and SLC2A1, PGK1, and G6PD mRNA expression was analyzed using the TCGA databases. qPCR analysis of GRWD1 knockdown (B) and overexpression (C) on GLUT1, PGK1 and G6PD mRNA level. qPCR analysis of GLUT1 mRNA expression following IL-6 (30 μg/L) treatment for 4 h in GRWD1 knockdown (D) and overexpression (E) cells. (F) The four groups of GRWD1 and GLUT1 expression in the TCGA-COADREAD cohort were represented on the left. The Kaplan–Meier survival curves showed the PFS (on the middle) and DFS (on the right) by the expression levels of GRWD1 and GLUT1.
Additionally, the Kaplan–Meier survival curve showed the PFS (Progression-Free Survival) and DFS (Disease-Free Survival) of patients stratified by the expression levels of GRWD1 and SLC2A1. Patients with high expression of both GRWD1 and SLC2A1 (in red) had significantly poorer survival compared to those with low expression of both markers (in blue) (Fig. 5F). Besides, we also analyzed all four GRWD1/SLC2A1 subgroups in both PFS and DFS, in both analyses, the group of GRWD1 + /SLC2A1 + patients had the worst prognosis. In addition, the GRWD1 + /SLC2A1 − group showed no significant survival difference versus the GRWD1-/SLC2A1 −. These results reinforced the oncogenic function of GRWD1 required GLUT1-mediated metabolism of CRC (Supplementary Fig S3). Taken together, GRWD1 promoted glycolysis of CRC cells, which may correlate with GLUT1.
GRWD1 promotes glycolysis in a p53/GLUT1-dependent manner
We also figured out the reason why GRWD1 had an effect on GLUT1. Previous studies demonstrated that GRWD1 negatively regulated p5317. It was also confirmed that p53 downregulated GLUT1 gene expression31. Therefore, changes in the protein and mRNA levels of p53 expression were analyzed. Based on our results, GRWD1 knockdown promoted the protein expression level of p53. However, p53 expression was lower while GRWD1 was overexpressed (Fig. 6A). qPCR analysis determined that GRWD1 had no effect on p53 mRNA level (Fig. 6B), indicating that GRWD1 chiefly influenced the protein level of p53, consistent with the results of a previous study17. Furthermore, the impact of GRWD1 on the protein stability of p53 was examined. Cycloheximide (CHX) is a small molecule that could restrict the translation elongation of eukaryotic protein synthesis. Once protein synthesis is inhibited with CHX, the expression of proteins is decreased by degradation32. Following CHX treatment, GRWD1 knockdown decreased the degradation velocity of p53 (Fig. 6C).
The effect of GRWD1 on the p53/GLUT1 signaling pathway. (A) Western blot analysis of p53 protein level following GRWD1 knockdown and overexpression. (B) qPCR analysis of p53 mRNA expression following GRWD1 knockdown and overexpression. (C) Western blot of p53 degradation after treatment with CHX (50 μg/mL) following GRWD1 knockdown for the indicated times (0, 10, 20, 30, 60 and 120 min). (D) Western blot analysis of p53 and GLUT1 protein levels following PFT-β (40 μM) treatment for 4 h and 6 h. The statistical value of the representative expression of the protein is shown on the right. (E) qPCR analysis of GLUT1 mRNA expression following treatment with the p53 inhibitor PFT-β (40 μM) for 6 h. (F) p53 and GLUT1 protein expression levels were detected following GRWD1 knockdown and PFT-β treatment. Grey values were analyzed to present a representative expression of the protein. (G) GRWD1, p53, and GLUT1 expression were assessed in tumors of previous nude mouse models, which were subcutaneously injected with CRC cells. The statistical grey value of protein expression was shown on the right. (H) Glucose uptake of GRWD1-knockdown cells following PFT-β treatment. (I) Lactate release of GRWD1-knockdown cells following PFT-β treatment.
To determine the influence of GRWD1 on the glycolysis of CRC cells through GLUT1, the relationship between p53 and GLUT1 was initially examined. Herein, a specific inhibitor of p53 termed PFT-β was used33. Western blot analysis showed that following PFT-β treatment, the expression level of GLUT1 steadily increased (Fig. 6D). As reported, PFT-β could also block p53-dependent transcriptional activity34. qPCR analysis indicated that PFT-β promoted an increase in GLUT1 mRNA level (Fig. 6E). Moreover, GRWD1 knockdown upregulated the expression of p53 and further inhibited that of GLUT1. Of note, the result was reversed after combination with PFT-β. Following GRWD1 knockdown, GLUT1 expression was induced after PFT-β treatment, demonstrating that GRWD1 may regulate GLUT1 through p53 (Fig. 6F).
Our previous study established a subcutaneous tumor mouse model, indicating that GRWD1 knockdown inhibited CRC proliferation in vivo19. Therefore, the expression of p53 and GLUT1 was examined in tumors, and the results revealed that p53 was upregulated in the GRWD1 knockdown group. Meanwhile, GLUT1 expression was lower (Fig. 6G). Besides, the impact of GRWD1 on p53/GLUT1-dependent glycolysis was examined. The results showed that PFT-β facilitated glucose uptake and lactate release despite GRWD1 knockdown (Fig. 6H,I). In summary, GRWD1 facilitated glycolysis by promoting the degradation of p53 and further upregulating the expression of GLUT1.
IL6/STAT3-activated GRWD1 further affects the p53/GLUT1 axis
Finally, the correlation between IL-6/STAT3 with GRWD1 and p53/GLUT1 was analyzed by treating CRC cells with IL-6 and stattic. The findings demonstrated that p-STAT3 expression was significantly induced in response to IL-6 treatment. Meanwhile, the expression of GRWD1 and GLUT1 was upregulated, while that of p53 was inhibited. Indeed, stattic repressed the positive effect of IL-6, which resulted in the down-regulation in the level of GRWD1 and GLUT1, whereas p53 expression was increased compared with the control group. In conclusion, the IL-6/STAT3 signaling pathway positively affected GRWD1 expression, which repressed p53 protein levels and further induced GLUT1 expression (Fig. 7A).
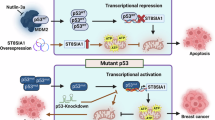
The effect of IL-6/STAT3 on GRWD1/p53/GLUT1 axis. (A) RKO cells were treated with IL-6 (30 μg/L) and stattic (1 μmol/L) for 6 h, and the expression levels of multiple proteins were detected using Western blot. The statistical grey value of protein expression is shown on the right. (B) The working model for IL-6-stimulated GRWD1 mediated aerobic glycolysis through p53-GLUT1 of CRC, created by Figdraw.
Briefly, our research demonstrated that GRWD1 was progressively activated in the inflammatory-associated pathogenesis of CAC, particularly in the IL-6/STAT3 signal pathway. GRWD1 overexpression mediated aerobic glycolysis by promoting the degradation of p53 and subsequently upregulating GLUT1, which accelerated various malignant phenotypes in CRC (Fig. 7B).
Discussion
Our earlier study concluded that GRWD1 was highly expressed in CRC tissues and played an essential oncogenic role in CRC19. Previous investigations also focused on GRWD1 as an oncogene with high expression that promoted malignant phenotypes in a variety of cancers. In gastric cancer (GC), GRWD1 upregulated ADAM metallopeptidase domain 17 (ADAM17) expression and released Notch1 intracellular domain (NICD) which further facilitated GC cell proliferation and metastasis35. In non-small cell lung cancer and triple negative breast cancer, GRWD1 activated the Notch pathway that further promoted cell proliferation and migration18,36. However, the reasons why GRWD1 is highly expressed in cancers are rarely elucidated. Therefore, our study aimed to identify the mechanism by which GRWD1 is overexpressed in CRC.
We analyzed the correlation between GRWD1 and the malignant features of CRC, interestingly we found GRWD1 had a positive relationship with inflammation. Chronic inflammation has been established as a risk factor for tumorigenesis. Similarly, infectious diseases, dietary factors, and inflammatory bowel disease (IBD) can cause CRC22. The AOM/DSS mouse model is a widely used model for the study of inflammation-associated colon carcinogenesis37. In our constructed AOM/DSS mouse model, GRWD1 expression progressively increased with disease progression, indicating that the inflammatory environment may play an essential role in GRWD1 expression. Besides, IL-6 treatment also induced GRWD1 expression in CRC cells. We noticed the difference in the temporal kinetics of the p-STAT3 and GRWD1 protein levels after IL-6 treatment. Although both SW480 and RKO cells are CRC cell lines, they originate from different patients with unique genetic backgrounds and epigenetics38. Transcriptional activation of target genes downstream of STAT3 and subsequent rates of protein expression may be influenced by a variety of cellular intrinsic factors. For example, the decrease in GRWD1 observed in RKO cells at 6 h (although p-STAT3 remained high) may suggest the feedback regulation. In contrast, in SW480 cells, this negative feedback may be absent, resulting in the continued accumulation of GRWD1. It was also possible that in SW480 cells, it may take longer to accumulate from STAT3 phosphorylation to transcription of the GRWD1 gene. In contrast, in RKO cells, this signaling may be more efficient, resulting in a faster response. This also inspired us that the expression of GRWD1, which was a downstream effector of p-STAT3, may also be regulated by other mechanisms in the different cellular microenvironments.
According to an earlier study, IL-6 is frequently expressed and associated with STAT3 in CRC39. Our data demonstrated that inhibition of STAT3 phosphorylation using stattic potently suppressed GRWD1 expression, confirming the GRWD1 upregulation dependent on active STAT3 signaling downstream of IL-6. However, our current experiments have not yet identified specific regulatory sites where STAT3 directly bound to the GRWD1 promoter region. We speculate that several possible mechanisms may also exist. For example, STAT3 may regulate the expression of other transcription factors, which in turn directly bind and activate the GRWD1 promoter. Besides, STAT3 signaling might induce epigenetic modifications of the GRWD1 genomic region, for example, histone methylation, making it more accessible to other transcription factors, without phosphorylated STAT3 directly binding. Previous studies have shown that inhibition of STAT3 phosphorylation can reduce pentraxin-3 (PTX3) enhancer methylation and its expression activity in CRC cells40. Furthermore, the IL-6/STAT3 pathway frequently interacts with other signaling cascades including Ras, PI3K/AKT, etc41. STAT3 activation might affect the activity of other signaling pathways that directly regulate GRWD1 transcription. This hypothesis needs to be further verified in future studies.
Malignant cells typically require more energy, thus accelerating malignant phenotypes, including cell proliferation, migration, and apoptosis. Irrespective of the oxygenation status, tumor cells present high glycolytic activity28. Glycolysis is faster and more efficient in generating ATP, which can rapidly provide energy to tumor cells. Metastasis is a leading cause of CRC-related death. Several studies have evinced that colon cancer metastasis is closely associated with glycolytic metabolism28,42. Our previous study found that GRWD1 promotes the proliferation and metastasis of CRC cells. GSEA further indicated high expression of GRWD1 was associated with aerobic glycolysis. Therefore, we hypothesized that GRWD1 may affect glycolysis in CRC. As expected, our results displayed that GRWD1 promoted glucose uptake and lactate release in CRC cells. Additionally, the results of the ECAR and OCR analyses also pointed out that GRWD1 increased extracellular acidification and inhibited the oxygen consumption rate. The Warburg effect increases the levels of GLUTs, including GLUT1, which in turn increases the glucose uptake of tumor cells43. PGK1 is a major glycolytic enzyme that promotes the proliferation and invasion of cancer cells44. G6PD, as a limiting-rate enzyme, influences PPP flux, a primary metabolic pathway of glucose45,46. The correlation between GRWD1 and these three genes was analyzed using the TCGA and GETx database, and qPCR analysis demonstrated that GRWD1 principally promoted GLUT1 expression in CRC. Furthermore, the survival analysis of GRWD1 + /SLC2A1 + patients had the worst prognosis which demonstrated the clinical relevance of GRWD1 affecting the glycolytic pathway through GLUT1 and its synergistic effect as a poor prognostic factor. In addition, the GRWD1 + /SLC2A1 − group showed no significant survival difference versus GRWD1 −/SLC2A1 −, which was consistent with our experimental data that GRWD1 relied on GLUT1 to enhance glycolysis and promoted CRC progression. In the absence of GLUT1, high GRWD1 expression failed to drive tumor progression, explaining why GRWD1 alone would not confer a poor prognosis. This result indicated that GRWD1 potentially contributed to the glycolysis of CRC cells through GLUT1.
In addition, the mechanism underlying the regulatory relationship between GRWD1 and GLUT1 was evaluated. Previous reports evinced that GRWD1 negatively regulated p53 expression and inhibited the transcriptional activity of p5317. Besides, p53 can directly suppress the transcription of GLUT1 and glucose transport31. Therefore, we speculated that GRWD1 modulated the levels of GLUT1 through p53. This hypothesis was validated by treating GRWD1 knockdown cells with the p53 inhibitor PFT-β. Meanwhile, PFT-β treatment reversed the glycolysis of GRWD1 knockdown cells, which meant GRWD1 regulated the glucose metabolism that relied on p53 transcriptional activity. Interestingly, the IL-6/STAT3 signaling pathway stimulated GRWD1 expression, further altering the protein expression levels of the downstream proteins p53 and GLUT1.
In CRC, TP53 is one of the most important driver genes47. As reported, 43.28% of CRC patients have TP53 variation48. Most TP53 variations encoded missense mutation proteins with loss-of-function (LOF), dominant negative effects (DNE) and gain-of-function (GOF)49. The p53 protein in SW480 cells has two point missense mutations R273H and P309S, and RKO cells express wild-type p5350,51. The results of our investigation demonstrated that IL-6 could induce GRWD1 expression in both SW480 and RKO cells. Additionally, GRWD1 facilitated glycolysis in these two cell lines, indicating that GRWD1 had a significant impact on CRC cells regardless of the p53 expression pattern. We confirmed that IL-6-upregulated GRWD1 may change the level of p53 and, in turn, affect the expression of GLUT1 in RKO cells. However, it is not clear if this mechanism is present in mutant p53 colon cancer cells. Indeed, the functions of mutant p53 are involved in various signal pathways, and even different mutations of p53 have distinct roles, which is a complicated mechanism47,52. As reported, the two most common hotspots TP53 mutations, p53R273H and p53R175H, had different effects on the development of CRC. Compared with p53R175H and p53KO cells, p53R273H upregulated a unique gene signature comprising 140 genes in CRC cells53. Therefore, in p53 mutation CRC cells, there may exist other novel pathways that promote aerobic glycolysis via GRWD1, which require further investigations.
Our findings emphasized that GRWD1 was an essential oncogene and potential diagnostic target in CRC, which also provided new strategies for treatment. Our previous study indicated high expression of GRWD1 led to drug resistance in CRC cells. Therefore, novel small molecular inhibitors targeting GRWD1 could be identified. Moreover, targeting STAT3 activation that inhibits GRWD1 expression may also contribute to CRC treatment54. Meanwhile, molecules that disrupt the interaction of wild-type p53 with its E3 ubiquitin ligase may be employed to treat CRC cells with IL6-stimulated GRWD1 high expression in the future52. Combining conventional therapies with these novel target inhibitors may improve drug resistance in GRWD1 high-expression CRC cells.
In summary, GRWD1 was overexpressed in CRC and served as a tumor oncogene. Mechanically, IL-6/STAT3 promoted GRWD1 expression that further facilitated the degradation of p53, in turn upregulating GLUT1, which mediated aerobic glycolysis in CRC cells. Targeting GRWD1 and its associated factors may be promising for CRC treatment.
Data availability
The data supporting this study’s findings are available from the corresponding author upon reasonable request.
Abbreviations
- GRWD1:
-
Glutamate-rich WD40 repeat-containing 1
- CRC:
-
Colorectal carcinoma
- AOM/DSS:
-
Azoxymethane/dextran sodium sulfate
- CAC:
-
Colitis-associated cancer
- GLUT1:
-
Glucose transporter type 1
- IL-6:
-
Interleukin-6
- ECAR:
-
Extracellular acidification rate
- OCR:
-
Oxygen consumption rate
- CHX:
-
Cycloheximide
- PFT-β:
-
Pifithrin-β
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249. https://doi.org/10.3322/caac.21660 (2021).
Xia, C. et al. Cancer statistics in China and United States, 2022: Profiles, trends, and determinants. Chin. Med. J. 135, 584–590. https://doi.org/10.1097/CM9.0000000000002108 (2022).
Levine, O. & Zbuk, K. Colorectal cancer in adolescents and young adults: Defining a growing threat. Pediatr. Blood Cancer 66, e27941. https://doi.org/10.1002/pbc.27941 (2019).
Martini, G. et al. Present and future of metastatic colorectal cancer treatment: A review of new candidate targets. World J. Gastroenterol. 23, 4675–4688. https://doi.org/10.3748/wjg.v23.i26.4675 (2017).
Shin, A. E., Giancotti, F. G. & Rustgi, A. K. Metastatic colorectal cancer: Mechanisms and emerging therapeutics. Trends Pharmacol. Sci. 44, 222–236. https://doi.org/10.1016/j.tips.2023.01.003 (2023).
Kastrinos, F., Kupfer, S. S. & Gupta, S. Colorectal cancer risk assessment and precision approaches to screening: Brave new world or worlds apart?. Gastroenterology 164, 812–827. https://doi.org/10.1053/j.gastro.2023.02.021 (2023).
Krzystek-Korpacka, M., Diakowska, D., Kapturkiewicz, B., Bebenek, M. & Gamian, A. Profiles of circulating inflammatory cytokines in colorectal cancer (CRC), high cancer risk conditions, and health are distinct. Possible implications for CRC screening and surveillance. Cancer Lett. 337, 107–114. https://doi.org/10.1016/j.canlet.2013.05.033 (2013).
Lin, J. K., Chang, S. C., Yang, Y. C. & Li, A. F. Loss of heterozygosity and DNA aneuploidy in colorectal adenocarcinoma. Ann. Surg. Oncol. 10, 1086–1094. https://doi.org/10.1245/aso.2003.12.014 (2003).
Shah, S. C. & Itzkowitz, S. H. Colorectal cancer in inflammatory bowel disease: Mechanisms and management. Gastroenterology 162, 715–730. https://doi.org/10.1053/j.gastro.2021.10.035 (2022).
Waldner, M. J. & Neurath, M. F. Master regulator of intestinal disease: IL-6 in chronic inflammation and cancer development. Semin. Immunol. 26, 75–79. https://doi.org/10.1016/j.smim.2013.12.003 (2014).
Wang, S. W. & Sun, Y. M. The IL-6/JAK/STAT3 pathway: Potential therapeutic strategies in treating colorectal cancer (Review). Int. J. Oncol. 44, 1032–1040. https://doi.org/10.3892/ijo.2014.2259 (2014).
Colussi, D., Brandi, G., Bazzoli, F. & Ricciardiello, L. Molecular pathways involved in colorectal cancer: Implications for disease behavior and prevention. Int. J. Mol. Sci. 14, 16365–16385. https://doi.org/10.3390/ijms140816365 (2013).
Johnson, D. E., O’Keefe, R. A. & Grandis, J. R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 15, 234–248. https://doi.org/10.1038/nrclinonc.2018.8 (2018).
Garbers, C., Aparicio-Siegmund, S. & Rose-John, S. The IL-6/gp130/STAT3 signaling axis: Recent advances towards specific inhibition. Curr. Opin. Immunol. 34, 75–82. https://doi.org/10.1016/j.coi.2015.02.008 (2015).
Takafuji, T., Kayama, K., Sugimoto, N. & Fujita, M. GRWD1, a new player among oncogenesis-related ribosomal/nucleolar proteins. Cell Cycle 16, 1397–1403. https://doi.org/10.1080/15384101.2017.1338987 (2017).
Fujiyama, H. et al. GRWD1 directly interacts with p53 and negatively regulates p53 transcriptional activity. J. Biochem. 167, 15–24. https://doi.org/10.1093/jb/mvz075 (2020).
Kayama, K. et al. GRWD1 negatively regulates p53 via the RPL11-MDM2 pathway and promotes tumorigenesis. EMBO Rep. 18, 123–1376. https://doi.org/10.15252/embr.201642444 (2017).
Wang, Q. et al. GRWD1 promotes cell proliferation and migration in non-small cell lung cancer by activating the Notch pathway. Exp Cell Res 387, 111806. https://doi.org/10.1016/j.yexcr.2019.111806 (2020).
Zhou, X. et al. Clinical significance and oncogenic activity of GRWD1 overexpression in the development of colon carcinoma. Onco Targets Ther. 14, 1565–1580. https://doi.org/10.2147/OTT.S290475 (2021).
Schust, J., Sperl, B., Hollis, A., Mayer, T. U. & Berg, T. Stattic: A small-molecule inhibitor of STAT3 activation and dimerization. Chem. Biol. 13, 1235–1242. https://doi.org/10.1016/j.chembiol.2006.09.018 (2006).
Snider, A. J. et al. Murine model for colitis-associated cancer of the colon. Methods Mol. Biol. 1438, 245–254. https://doi.org/10.1007/978-1-4939-3661-8_14 (2016).
Schmitt, M. & Greten, F. R. The inflammatory pathogenesis of colorectal cancer. Nat. Rev. Immunol. 21, 653–667. https://doi.org/10.1038/s41577-021-00534-x (2021).
Hirano, T. et al. Immunological mechanisms in inflammation-associated colon carcinogenesis. Int. J. Mol. Sci. https://doi.org/10.3390/ijms21093062 (2020).
Wu, S., Luo, W., Wu, X., Shen, Z. & Wang, X. Functional phenotypes of peritoneal macrophages upon AMD3100 treatment during colitis-associated tumorigenesis. Front. Med. 9, 840704. https://doi.org/10.3389/fmed.2022.840704 (2022).
Neufert, C. et al. Inducible mouse models of colon cancer for the analysis of sporadic and inflammation-driven tumor progression and lymph node metastasis. Nat. Protoc. 16, 61–85. https://doi.org/10.1038/s41596-020-00412-1 (2021).
Grivennikov, S. et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 15, 103–113. https://doi.org/10.1016/j.ccr.2009.01.001 (2009).
Xin, J. Critical signaling pathways governing colitis-associated colorectal cancer: Signaling, therapeutic implications, and challenges. Dig. Liver Dis. 55, 169–177. https://doi.org/10.1016/j.dld.2022.08.012 (2023).
Zhou, D. et al. The significance of glycolysis in tumor progression and its relationship with the tumor microenvironment. Front. Pharmacol. 13, 1091779. https://doi.org/10.3389/fphar.2022.1091779 (2022).
Meng, Y. et al. The progress and development of GLUT1 inhibitors targeting cancer energy metabolism. Future Med. Chem. 11, 2333–2352. https://doi.org/10.4155/fmc-2019-0052 (2019).
Vaughan, R. A. et al. Tumor necrosis factor alpha induces Warburg-like metabolism and is reversed by anti-inflammatory curcumin in breast epithelial cells. Int. J. Cancer 133, 2504–2510. https://doi.org/10.1002/ijc.28264 (2013).
Schwartzenberg-Bar-Yoseph, F., Armoni, M. & Karnieli, E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 64, 2627–2633. https://doi.org/10.1158/0008-5472.can-03-0846 (2004).
Miao, Y. et al. Cycloheximide (CHX) chase assay to examine protein half-life. Bio Protoc. 13, e4690. https://doi.org/10.21769/BioProtoc.4690 (2023).
Fernandez-Cruz, M. L. et al. Biological and chemical studies on aryl hydrocarbon receptor induction by the p53 inhibitor pifithrin-alpha and its condensation product pifithrin-beta. Life Sci. 88, 774–783. https://doi.org/10.1016/j.lfs.2011.02.019 (2011).
Da Pozzo, E. et al. p53 functional inhibitors behaving like pifithrin-beta counteract the Alzheimer peptide non-beta-amyloid component effects in human SH-SY5Y cells. ACS Chem. Neurosci. 5, 390–399. https://doi.org/10.1021/cn4002208 (2014).
Ding, H., Feng, Z. & Hu, K. GRWD1 over-expression promotes gastric cancer progression by activating notch signaling pathway via up-regulation of ADAM17. Dig. Dis. Sci. 69, 821–834. https://doi.org/10.1007/s10620-023-08208-5 (2024).
Yao, L. & Tian, F. GRWD1 affects the proliferation, apoptosis, invasion and migration of triple negative breast cancer through the Notch signaling pathway. Exp. Ther. Med. 24, 473. https://doi.org/10.3892/etm.2022.11400 (2022).
De Robertis, M. et al. The AOM/DSS murine model for the study of colon carcinogenesis: From pathways to diagnosis and therapy studies. J. Carcinog. 10, 9. https://doi.org/10.4103/1477-3163.78279 (2011).
Ahmed, D. et al. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2, e71. https://doi.org/10.1038/oncsis.2013.35 (2013).
Waldner, M. J., Foersch, S. & Neurath, M. F. Interleukin-6–a key regulator of colorectal cancer development. Int. J. Biol. Sci. 8, 1248–1253. https://doi.org/10.7150/ijbs.4614 (2012).
Rubino, M. et al. Epigenetic regulation of the extrinsic oncosuppressor PTX3 gene in inflammation and cancer. Oncoimmunology 6, e1333215. https://doi.org/10.1080/2162402X.2017.1333215 (2017).
Xu, J., Lin, H., Wu, G., Zhu, M. & Li, M. IL-6/STAT3 Is a promising therapeutic target for hepatocellular carcinoma. Front. Oncol. 11, 760971. https://doi.org/10.3389/fonc.2021.760971 (2021).
Chen, Y. J. et al. Interaction between glycolysis-cholesterol synthesis axis and tumor microenvironment reveal that gamma-glutamyl hydrolase suppresses glycolysis in colon cancer. Front. Immunol. https://doi.org/10.3389/fimmu.2022.979521 (2022).
Chen, X., Qian, Y. & Wu, S. The Warburg effect: Evolving interpretations of an established concept. Free Radic. Biol. Med. 79, 253–263. https://doi.org/10.1016/j.freeradbiomed.2014.08.027 (2015).
Zhang, K., Sun, L. & Kang, Y. Regulation of phosphoglycerate kinase 1 and its critical role in cancer. Cell. Commun. Signal 21, 240. https://doi.org/10.1186/s12964-023-01256-4 (2023).
Song, J., Sun, H., Zhang, S. & Shan, C. The multiple roles of glucose-6-phosphate dehydrogenase in tumorigenesis and cancer chemoresistance. Life https://doi.org/10.3390/life12020271 (2022).
Liu, B. et al. Pan-cancer analysis of G6PD carcinogenesis in human tumors. Carcinogenesis 44, 525–534. https://doi.org/10.1093/carcin/bgad043 (2023).
Nakayama, M. & Oshima, M. Mutant p53 in colon cancer. J. Mol. Cell Biol. 11, 267–276. https://doi.org/10.1093/jmcb/mjy075 (2019).
Kim, K. M. et al. Clinical significance of p53 protein expression and TP53 variation status in colorectal cancer. BMC Cancer 22, 940. https://doi.org/10.1186/s12885-022-10039-y (2022).
Tang, J. et al. Trp53 null and R270H mutant alleles have comparable effects in regulating invasion, metastasis, and gene expression in mouse colon tumorigenesis. Lab. Invest. 99, 1454–1469. https://doi.org/10.1038/s41374-019-0269-y (2019).
Rochette, P. J., Bastien, N., Lavoie, J., Guerin, S. L. & Drouin, R. SW480, a p53 double-mutant cell line retains proficiency for some p53 functions. J. Mol. Biol. 352, 44–57. https://doi.org/10.1016/j.jmb.2005.06.033 (2005).
Solomon, H. et al. Mutant p53 gain of function underlies high expression levels of colorectal cancer stem cells markers. Oncogene 37, 1669–1684. https://doi.org/10.1038/s41388-017-0060-8 (2018).
Liebl, M. C. & Hofmann, T. G. The role of p53 signaling in colorectal cancer. Cancers https://doi.org/10.3390/cancers13092125 (2021).
Hassin, O. et al. Different hotspot p53 mutants exert distinct phenotypes and predict outcome of colorectal cancer patients. Nat. Commun. 13, 2800. https://doi.org/10.1038/s41467-022-30481-7 (2022).
Gargalionis, A. N., Papavassiliou, K. A. & Papavassiliou, A. G. Targeting STAT3 signaling pathway in colorectal cancer. Biomedicines https://doi.org/10.3390/biomedicines9081016 (2021).
Funding
This research was financially supported by the Natural Science Foundation of Fujian Province, China (Grant No. 2022J011362), the National Natural Science Foundation of China (Grant No. 82270664), the Project of Xiamen Cell Therapy Research Center, Xiamen, China (Grant No. 3502Z20214001).
Author information
Authors and Affiliations
Contributions
Jin Shang: Conceptualization; Data curation; Formal analysis; Investigation; Validation; Visualization; Writing—original draft. Hong-Yue Ren: Data curation; Formal analysis; Investigation; Methodology; Validation; Visualization. Fa-Hui Liu: Data curation; Formal analysis; Methodology; Validation; Visualization. Dong-Hua Liu and Shi-Hang Tang: Data curation; Formal analysis; Investigation; Methodology; Validation. Guo-Xian Guan: Conceptualization; Resources; Project administration; Writing—review and editing. Guo-Qiang Su: Conceptualization; Validation; Visualization; Resources; Project administration. Dong-Yan Shen: Conceptualization; Formal analysis; Visualization; Funding acquisition; Project administration; Writing—original draft. Xin Zhou: Conceptualization; Data curation; Formal analysis; Investigation; Methodology; Funding acquisition; Project administration ; Resources; Writing—review and editing. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The Ethics Committee of The First Affiliated Hospital of Xiamen University (Xiamen, China) approved this study. The feasibility of tissue sampling in the experiment is strictly adhered to and standardized ethically according to the Helsinki Declaration. Each patient signed a document requesting their informed consent, ensuring their understanding and agreement. Animal experiments were approved by and performed following the Institutional Animal Care and Use Ethics Committee of Xiamen University. All animal experimental methods were carried out in accordance with the ARRIVE principles. All methods were carried out in accordance with relevant guidelines and regulations.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shang, J., Ren, HY., Liu, FH. et al. IL6/STAT3 induced GRWD1 mediates aerobic glycolysis via P53/GLUT1 signal axis in colon carcinoma. Sci Rep 15, 33534 (2025). https://doi.org/10.1038/s41598-025-18114-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-18114-7