Abstract
Community-acquired pneumonia (CAP) remains one of the leading causes of death among pediatric patients under the age of 5, making timely and accurate diagnosis crucial for subsequent treatment. While numerous artificial intelligence diagnostic methods for pneumonia have shown reliable results, most of them still exhibit significant limitations: (a) They only rely on single-modality data, which have limitations in capturing comprehensive clinical information; (b) They do not consider differences in postures among pediatric patients, resulting in constrained diagnostic performance and generalizability. To address these challenges, we construct a real-world pediatric CAP dataset from tertiary hospital records, and develop a multimodal framework for the precise diagnosis of pediatric CAP. In order to simulate clinical diagnostic workflows, the developed model not only integrates frontal chest X-ray images but also considers laboratory test results and clinical texts, enabling comprehensive diagnosis of diverse symptoms, enhancing diagnostic accuracy and generalizability. Experimental results based on the constructed dataset demonstrate that our multimodal approach achieves an impressive diagnostic result, with an accuracy of 94.2%, showing significant improvement over single-modality baselines and validating the potential as an auxiliary diagnostic tool to enhance the clinical practice for pediatric CAP screening.
Similar content being viewed by others
Introduction
Pneumonia, as a severe lower respiratory tract infection, is one of the leading causes of death among children under five globally, with most cases being community-acquired pneumonia (CAP)1. Studies have indicated that severe pneumonia can lead to complications, including respiratory failure, cardiac failure, and toxic encephalopathy2,3. Furthermore, CAP may potentially deter the development of the respiratory and immune systems, which is related to long-term pulmonary function decline and an increased risk of asthma4. Notably, CAP in children exhibits a high incidence during epidemics, causing substantial healthcare pressure. This phenomenon is particularly exacerbated in regions with limited medical infrastructure. As a result, there is an urgent need to develop rapid, accurate, and automated diagnostic tools to relieve physician workload while enhancing early diagnostic capabilities for CAP in children in underserved areas.
The standard clinical diagnostic procedure for CAP in children involves medical case information collection, laboratory tests, and chest radiography examination. For radiographic examinations, chest X-ray (CXR) has significant advantages, such as cost-effectiveness, rapid implementation, and low radiation exposure, serving as a primary screening tool for CAP detection. However, even for experienced physicians, detecting pneumonia through CXR imaging only is still challenging and time-consuming5. With the rapid development of artificial intelligence (AI), innovative solutions are being proposed to face these challenges. For example, most researchers have focused on medical imaging by providing large-scale datasets containing pneumonia radiographic images6,7. While multiple AI diagnostic models based on CXR images have achieved impressive performance8,9,10, most of them rely on single radiographic modalities only, which means ignoring other clinical information for consideration, unable to conduct a comprehensive case analysis. This limitation is magnified among children, who post significant physical variations during growth11. Consequently, it may reduce the diagnostic accuracy when using CXR images alone for pediatric patients, increasing the risk of misdiagnosis.
The clinical appearances of CAP typically include common symptoms such as fever and pulmonary wet rales12,13. Physicians record examination results in clinical texts or Electronic Medical Records (EMRs) in clinical practice, where symptoms are documented as crucial indicators for CAP diagnosis. Furthermore, CAP can trigger systemic inflammatory responses that lead to abnormal results in laboratory tests, such as higher C-reactive protein levels14. Therefore, medical textual data and laboratory tests contain critical features for clinical CAP diagnosis, prompting some researchers to conduct preliminary investigations on diagnostic approaches using laboratory data and medical record information.
However, several diseases may exhibit similar laboratory abnormalities or clinical symptoms which resemble CAP15,16. Consequently, relying on these modalities only or traditional screening criteria for CAP diagnosis may reduce diagnostic specificity. Multimodal approaches could face these challenges by implementing a comprehensive analysis by integrating CXR imaging, clinical texts, and laboratory tests. This has the potential to receive more generalizable and more accurate diagnostic results, thereby offering significant insights for enhancing CAP diagnostic accuracy.
To address these issues, this study built a multimodal CAP database with comprehensive clinical information for patients aged 0–13 years, which contains frontal CXR imaging, laboratory testing data, and medical records to simulate the clinical decision-making workflow. Then, we developed a multimodal auxiliary diagnostic model for precise analysis of pediatric CAP and assist physicians’ decision-making process, which is trained and evaluated on our constructed multimodal database. The main contributions of this work are:
-
We construct a genuine pediatric CAP multimodal dataset. Samples within the dataset contain frontal CXR images, laboratory tests, and clinical texts. The following data preprocessing and augmentations provide rich multimodal information that enables multimodal methods to detect posture differences effectively.
-
We develop a multimodal diagnostic model that simulates the clinical diagnostic procedure. The model incorporates three primary medical data sources in regular clinical processes: CXR imaging features captured through ResNet-50, laboratory test features extracted by a deep neural network (DNN), and contextual semantic relationships in clinical documentation analyzed using bidirectional gated recurrent units (Bi-GRU). Then, these modality features will be element-wise concatenated, enabling the classifier to generate precise predictions with enhanced generalizability compared to single-modal approaches.
-
We provide explanations on each modality to visualize the decision-making processes of the developed multimodal model that provides potential diagnostic information to physicians.
-
The designed method has been evaluated in our collected dataset and achieved an impressive diagnostic accuracy of 94.2%, demonstrating the potential assistance to physicians for pediatric diagnostic tasks.
Related work
This section will review the relevant work, including traditional CAP scoring systems, CXR images, medical data (including medical texts and laboratory test indicators), and multimodal diagnostic approaches.
Traditional CAP scoring systems
To facilitate the rapid screening of CAP, several scoring systems have been proposed with validations and support from authoritative institutions such as the Infectious Diseases Society of America. These include the Pneumonia Severity Index (PSI), which contains multidimensional indicators such as demographics and comorbidities findings to assess the risk and severity of CAP. Then, simpler criteria like CURB-65 that rely on basic clinical signs are designed, incorporating confusion, respiratory rate, age, and blood pressure information. However, these criteria are designed for adults and fail to account for specific indicator variations in age among children, such as the heart rate. Therefore, several assessment models exclusive to pediatric pneumonia are proposed and further validated in clinical practice17,18. Despite their validity having been initially evaluated, these models still rely on patients’ characterization performance while ignoring key diagnostic factors such as radiological imaging19. Moreover, existing research has demonstrated superior performance compared to these criteria using machine learning methods20. Therefore, these models are suitable for screening in emergency situations rather than the precise diagnosis of CAP in normal clinical practice.
CXR pneumonia diagnosis
Due to the economic efficiency and rapid availability, CXR has become a primary screening tool in most hospitals. In CXR diagnosis, pneumonia can result in distinct imaging variations, with relatively significant features including parenchymal infiltration, pulmonary consolidation, and obscuration of silhouette signs, which AI systems can capture effectively. Hence, with the development of AI and medical technology, high-quality CXR medical imaging datasets, such as MIMIC-CXR and ChestX-ray are emerging rapidly6,7. Based on these datasets, some researchers have proposed AI models, including convolutional neural networks (CNNs), Transformers, and generative adversarial networks (GANs), to achieve automated pneumonia diagnosis based on CXR8,10,21. To further enhance the diagnostic capabilities of models, several improvements based on existing architectures have been proposed. For instance, Zhang et al. developed a pneumonia multi-classification diagnostic model by adopting an Encoder-Decoder-Encoder structure derived from U-Net22. This approach leverages the Encoder-Decoder framework to extract pixel-level interpretations of pathological features from radiographic images, followed by an additional Encoder that performs secondary feature extraction and diagnosis based on these pixel-wise explanations. This dual-stage processing not only enhances model interpretability but also significantly improves diagnostic performance23. Moreover, some studies proposed using image segmentation as a preprocessing step for CXR images to prevent misdiagnosis caused by erroneous pulmonary region identification. Lee et al., for example, implemented UNet++ to segment CXR images, eliminating irrelevant information outside the pulmonary area, thereby enabling models to better capture clinically significant regions of interest (ROIs). Then, a Transformer-based Position Aware Feature Encoding (PAFE) module is implemented to achieve practical severity prediction of pneumonia in different lung regions24.
However, existing datasets do not consider pediatric CXR situations. Specifically, the physical variations at different ages are significantly distinct due to their growth and development stages11. Hence, relying solely on these datasets for training diagnostic models may lead to insufficient capture of physical variations, which increases the risk of misdiagnosis. This limited the clinical applicability in real-world CAP in children’s diagnostic scenarios.
Medical record feature extraction
Clinical documentation, which includes medical texts and laboratory tests, also contributes to one of the critical diagnostic factors for pneumonia in clinical settings. However, these data always exist as unstructured text, which cannot be analyzed directly. Hence, Natural Language Processing (NLP) techniques are needed to extract diagnostic information and features25. Recently, researchers have employed methodologies including Named Entity Recognition (NER)26,27, dictionary mapping27, and regular expressions for feature extraction28. Then, the extracted features will be analyzed using models like Long-Short Term Memory (LSTM)29, Gated Recurrent Units (GRU)30, and Bidirectional Transformer Encoders (BERT)31. These approaches could effectively capture relationships among texts through the model’s integrated memory or attention mechanisms. Moreover, these methods have shown specific adaptive abilities to tackle challenges such as terminological variations. However, the symptoms recorded in clinical documents of other illnesses may resemble the CAP. For example, systemic inflammatory response syndrome can also cause increased C-reactive protein levels15, while interstitial lung disease may similarly manifest pulmonary wet rales16.
Multimodal pneumonia diagnosis
In clinical practice, taking the diagnosis of CAP in children as an example, the accurate diagnosis requires a comprehensive analysis of multiple factors, including age, gender, clinical symptoms, physical examination findings, laboratory results, and imaging data. Generally, existing multimodal diagnostic methods have improved diagnostic performance compared to single modality diagnosis in various disease detection. Most of these approaches integrate diverse medical data sources, which have been mentioned above. For instance, Q. Wang et al. developed a multimodal that used CT imaging, age, gender, and patient-reported symptoms for pneumonia analysis and achieved 0.946 average diagnostic accuracy on the private dataset32. A.P. Narmadha and N. Gobalakrishnan then expanded the modalities by integrating several laboratory indicators. They proposed a Dual Way Hybrid Efficient Transformer architecture to diagnose multiple pulmonary diseases accurately33. Moreover, H. Ren et al. introduced a training strategy based on age, observing the performance in pneumonia diagnosis between young and elderly groups, and emphasised the importance of splitting age groups during model training34. J. Shao et al. constructed a relatively comprehensive multimodal dataset by integrating clinical reports, laboratory tests, CT imaging, and demographic data35. They also implemented a mid-fusion multimodal model consisting of 2D-3D SwinTransformer and Multimodal Cross-Attention Module (MCAM). Then, by validating the multimodal approach on the proposed datasets, an impressive performance was achieved in identifying pneumonia, tuberculosis, and other pulmonary infections. Moreover, they figured out that several indicators play important roles in predicting severe infection outcomes and present a further analysis and discussion.
However, the physical differences during the growth of children suggest that applying existing models may raise the risk of misdiagnosis. Consequently, some scholars have utilised pediatric pneumonia multimodal datasets collected from medical institutions to train and validate diagnostic models, attempting to achieve a precise diagnosis36. However, these studies have not fully analyzed the medical data generated during clinical diagnosis. Therefore, improving the utilisation of medical data in the diagnostic workflow remained a critical issue that urgently needs to be addressed in the multimodal diagnosis of pediatric pneumonia, including CAP.
Data collection
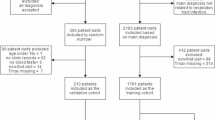
We collected 1,790 hospitalised cases at Fujian Provincial Children’s Hospital from July 2023 to December 2024. Then, followed by the strict exclusion criteria, we finally picked up 1,590 cases to construct the multimodal dataset. Figure 1 illustrates the specific inclusion and exclusion criteria for the sample selection, which can be divided into four phases:
-
Collected Modalities. We collected the comprehensive medical data for each patient, including clinical texts, CXR, and laboratory tests. For clinical texts, it contains demographic information (gender and age, precise to the month), symptoms (fever, cough, and other discovery description will be included. For instance,“fever for 5 days, cough for 3 days, severe convulsion”), physical examination records (containing pulmonary auscultation and any possible findings. For example,“coarse breath sounds in both lungs with moist rales”). Then, laboratory test results such as blood routine tests (e.g., C-reactive protein, monocyte count, neutrophil levels) are given. Finally, the frontal CXR imaging data before hospitalization are captured as the CXR modality.
-
Case Exclusion. We reviewed all collected patients’ data and excluded samples with missing CXR examinations before admission, unclear CXR images, incomplete medical records, or infected with other diseases (e.g., pulmonary tuberculosis).
-
Experts Review. To ensure the accurate diagnosis of CAP, we conducted a re-diagnosis by two respiratory specialists (each with over 10 years of clinical experience) independently. A third chief respiratory expert physician will determine the final diagnosis if the two reviewers’ results are different. Based on the final diagnoses, the dataset was divided into CAP and non-CAP groups to train and validate the developed multimodal diagnostic model.
-
Data Anonymization and Duplication Check. We removed personal information from each patient’s data, which is stored in a separate file. Then, an additional check is performed to remove duplicate medical records. Specifically, each patient is assigned a unique ID in the database. After the collection process, we identify and remove potential repeated acquisition cases by checking the case ID.
Methodology
Architecture overview
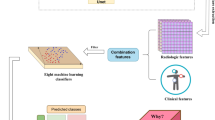
To maximise the utilisation of collected patient medical data and simulate clinical reasoning processes for diagnosis, we develop a multimodal model targeted for CAP in children’s diagnosis. Figure 2 illustrates the architecture of our multimodal model. The model uses frontal CXR images, laboratory test results, and clinical texts. These inputs are jointly processed through the multimodal model to generate a probability distribution indicating the patient’s likelihood of CAP. For CXR images, we chose ResNet-50 as the backbone. Its composition of convolution layers can extract extensive diagnostic information. Image augmentations could further adapt to pediatric CXR images with significant posture variations, enabling accurate identification of lesion areas. Text information is extracted using Bidirectional GRU (Bi-GRU). By analyzing words in two independent Gated Recurrent Unit (GRU) modules that process vectors in reversed directions, it effectively extracts rich diagnostic features among contextual semantic dependencies using the memory and forgetting mechanism. We adapted a deep neural network (DNN) for the laboratory indicators and a multimodal classifier for its ability to perform non-linear and comprehensive analyses of multidimensional data, which commonly occur in laboratory indicators and features produced by heterogeneous models. Therefore, the multimodal model could effectively utilize multimodal data using different strategies, producing accurate diagnoses in our constructed dataset.
Architecture of the multimodal auxiliary diagnosis model for pediatric CAP. The model takes laboratory test indicators, medical text, and CXR images as inputs and outputs the non-CAP or CAP probability distribution. During the feature extraction phase, ResNet-50 is used to extract features from CXR images, while laboratory test results and medical texts are processed using a Deep Neural Network (DNN) and a bidirectional GRU (Bi-GRU), respectively. Then, in the multimodal diagnosis phase, the model concatenates the extracted feature vectors and feeds the fused feature into a DNN classifier to obtain the probability distribution of the diagnostic results. Moreover, explanations on each modality would be provided.
CXR image augmentation
A dataset with sufficient cases is one of the key factors in enhancing the diagnosis model. However, variations such as physical development, posture variations during imaging, and environment/equipment differences would result in significant divergence of feature distributions among CXR images. Hence, we deployed a series of image preprocessing in the training set during dataset preparation, which aims to simulate physical variability and diverse imaging conditions produced from different diagnostic mechanisms, thereby enhancing the generalization and diagnostic capability of the image feature extraction model.
Random scaling and padding
Firstly, the input CXR image will be compressed:
Where \(ar_{new}\) represents the ratio of width and height to the compressed image, w and h represent the original width and height of the input image, respectively. \(jitter = 0.3\) controls the degree of distortion, while \(rand(\cdot )\) indicates the random number in a specific range.
Then, a scaling factor is sampled:
Finally, adjust the size of the compressed image based on the aspect ratio:
Where \(h_{new}\) and \(w_{new}\) represent the height and width of the new image, respectively. Finally, images are resized to the dimensions \(h_{new} \times w_{new}\).
Random rotation
To enhance the generalization of the multimodal model, CXR images have a 25% probability of being rotated within the range of \([-10^{ \circ },10^{ \circ }]\) using bicubic interpolation.
Color augmentation
Color enhancement was applied to emphasise lung regions in order to improve diagnostic performance. In this experiment, we randomly adjusted the hue, saturation, and value channels to CXR images:
Where \(r_h\), \(r_v\) and \(r_v\) denote the modification factors for hue, saturation, and value, computed using maximum offsets \(h=0.1\), \(s=0.7\) and \(v=0.3\), respectively, scaled by random variables \(u_h\), \(u_s\), \(u_v\) uniformly sampled from \([-1,1]\).
CXR feature extraction
The preprocessed CXR image will be processed through the ResNet-50, which is modified to extract deep image features for further multimodal diagnosis. It contains two parts, as shown in Figure 3.
Feature extraction
In this part, the input preprocessed CXR image represents \(x \in \mathbb {R} ^ {B \times C \times H \times W}\), where B denotes the batch size, \(C=3\) represents the number of channels and \(H=W=512\) demonstrate the height and width of the input CXR images, respectively. Firstly, the image will be processed by an extraction module (Including a convolutional layer with \(7\times 7\) kernel, Batch Normalization, ReLU activation and Max Pooling transformation), followed by several residual processing: \(y = F(x) + x\), where x, y denotes the input image and output feature respectively, and F(x) represents the transformations inside the residual block. Finally, the feature will be extracted by average pooling, producing the tensor \(x \in \mathbb {R} ^ {B \times 2048 \times 1 \times 1}\).
Feature decoder
In this part, feature vectors will be further decoded utilising a fully connected (FC) layer, followed by Leaky ReLU activation and Batch Normalisation. The FC layer could effectively interpret and compress the feature tensor to the vector \(x_{c} \in \mathbb {R} ^ {B \times 128}\), while maintaining diagnostic information. Then, the combined operation of the activation function and normalisation applied to the vector \(x_{c}\) helps prevent gradient vanishing and accelerates model convergence. Finally, the processed vector will be further concatenated with vectors from other modalities in the multimodal diagnosis phase.
Laboratory test features analysis
Laboratory data mainly consists of specific blood test indicators for patients. Due to the difference in patients’ symptom appearances, their examination indicators differ from one patient to another. If a patient lacks a particular indicator, the data for that will be imputed using the median method. Specifically, we will calculate and fill the average value of the corresponding indicator for cases in the same age group. Then, the processed data will be normalized for convergence acceleration. The preprocessed laboratory data are analyzed using the DNN that is composed of five layers of FC layers. For each layer:
Where x represents the input features, y indicates the features after processing of the FC layer, W is the weights of an individual FC layer, b for bias, and f(x) denotes the nonlinear activation function LeakyReLU and Batch Normalization. For the first FC layer, the feature will be expanded from \(x \in \mathbb {R} ^ {B \times N}\) to \(x \in \mathbb {R} ^ {B \times 8N}\) to analysis with sufficient information, where \(N=8\) represents the number of laboratory indicators. The following FC layers would extract and compress the feature, which its length is halved (\(kN \rightarrow \frac{1}{2}kN\)) for each layer, in order to have a comprehensive indicators reviewing, until producing the diagnostic vector \(x\in \mathbb {R}^{B \times N}\). Finally, a convolution layer which projects the laboratory feature to \(x_{l}\in \mathbb {R} ^ {B \times 32}\), followed by a batch normalization for the convergence acceleration of the model.
Clinical text feature extraction
Patient data includes gender, age, clinical symptoms (e.g. pulmonary auscultation), and physical examination results (e.g. fever), which also play an important role in diagnosing CAP. These data are compacted yet composed of unstructured texts that cannot be directly analyzed. To utilize these textual information, we constructed an extractor based on bidirectional GRU (Bi-GRU), with a vocabulary mapping table, shown in Figure 4.
Unformatted words mapping
It is necessary to format the unstructured texts before analysis. A practical way to transform is by creating a word dictionary. First, the collected text data were segmented into words, which was done by a Chinese language processing tool, jieba, in Python. Then, duplicate words extracted were removed. Finally, each unique word was encoded and stored in a word dictionary. We collected 642 unique Chinese words in total, containing useful diagnostic information describing symptoms like fever and auscultation. After preparing the dictionary, word mapping transforms unformatted texts into tensors, representing useful medical information. Moreover, because the model only accepts fixed-length tensors, we filled 0, which represents empty words, in the remaining, to form the word vector limited in 27 words \(x \in \mathbb {R} ^ {B \times 27}\). Finally, the mapped feature will be embedded for better feature extraction.
Sequence feature extraction
To effectively extract diagnostic knowledge from word vectors, we employed a 2-layer Bi-GRU to analyze the word index vectors. Then, feature normalization is conducted for convergence acceleration, then fed into a FC layer with Leaky ReLU activation and Batch Normalization, producing the normalized output \(x_p \in \mathbb {R} ^ {B \times 32}\).
Multimodal feature classification
Before diagnosing the CAP, it is necessary to connect the CXR image features, laboratory data features, and patient data features extracted from various modalities to ensure that the classifier can more effectively capture key information and improve diagnostic performance:
Where \(x_{fused}\) represents the concated features, and \(x_{c} \in \mathbb {R} ^ {B \times 128}\) denotes the extracted CXR image features, \(x_{l} \in \mathbb {R} ^ {B \times 32}\) is the laboratory data features, and \(x_{p} \in \mathbb {R} ^ {B \times 32}\) indicates the patient data features. For diagnosing CAP in children based on fused features, a DNN classifier with two FC layers is proposed. It takes the fused features \(x_{fused} \in \mathbb {R} ^ {B \times 178}\) as input, then extracted into the shape of \(x \in \mathbb {R} ^ {B \times 32}\) by the first FC layer. Finally outputs the CAP diagnosis result with precise probability distributions \(y \in \mathbb {R} ^ {B \times 2}\) produced by second FC layer with softmax activation.
Experiment
Datasets
This study collected 1,590 patients at Fujian Provincial Children’s Hospital, including 797 non-CAP patients and 793 CAP cases. Each case includes pre-admission CXR imaging data, corresponding laboratory test results, and clinical symptom text records. In the constructed dataset, most non-CAP patients are aged from 5 to 8 years, while CAP patients are mainly from 3 to 7 years old, as Figure 5 illustrates. For CXR images, the original resolution was 1024\(\times\)1024 pixels. However, images were compressed to 512\(\times\)512 pixels for model input to balance model computational load and accuracy.
In the laboratory test data, the indicators are different due to different symptoms. Hence, we selected eight indicators performed in over 95% of patients, which were relatively significant for CAP diagnosis (Pearson correlation coefficient \(|r| \ge 0.25\)) as the indicators fed into the model, shown in Figure 6 and Figure 7. Moreover, the figure also shows the distribution of these indicators in non-CAP and CAP patients. Notably, most of these data exhibited significant differences in distribution between negative and positive groups, such as C-reactive protein (CRP) and monocyte count (MO#), demonstrating the great potential of laboratory test indicators in CAP diagnosis. As for the clinical text records, important diagnostic factors, such as patients’ physical examination information, lung auscultation findings, fever status, duration, and so on, are included.
Model training setup and evaluation metrics
To validate the diagnostic performance and the generalization of the multimodal model, we conducted a 5-fold cross-validation experiment: all unique data were evenly divided at the patient level into five subsets according to the case ID to prevent data duplication. Four subsets were used for training, while the remaining one was reserved for validation. This training and validation process will be repeated five times. Moreover, the average performance will be recognised as the final experimental result, which is calculated from 5-fold cross-validation. In this study, we use cross-entropy to calculate the loss between the predictions and the actual results. Then, the AdamW optimizer was employed with \(\beta _{1}=0.9\), \(\beta _{2}=0.999\), and \(10^{-4}\) for the learning rate37. During training, we set 100 epochs, which provides sufficient time for the multimodal model to converge. In this experiment, we present the results that perform the lowest loss in validation sets for further analysis. Finally, to comprehensively validate the model’s performance, we use the following metrics:
-
Accuracy. The ratio of correctly diagnosed samples to the total number of cases. This would assess the model’s overall diagnostic performance.
$$\begin{aligned} \text {Accuracy} = \frac{TP + TN}{TP + TN + FP + FN} \end{aligned}$$(7) -
Recall. The ratio of correctly diagnosed in CAP patients. This would evaluate the model’s ability to distinguish CAP patients to check for missed diagnoses.
$$\begin{aligned} \text {Recall} = \frac{TP}{TP + FN} \end{aligned}$$(8) -
Precision. The ratio of predicted as CAP that are actually CAP, assessing the error rate of wrong diagnose.
$$\begin{aligned} \text {Precision} = \frac{TP}{TP + FP} \end{aligned}$$(9) -
F1-Score. The harmonic mean of the precision and recall.
$$\begin{aligned} \text {F1} = 2 \times \frac{\text {Precision} \times \text {Recall}}{\text {Precision} + \text {Recall}} \end{aligned}$$(10) -
Receiver Operating Characteristic Curve (ROC Curve). The curve utilising the recall and \(1 - Specificity\) as coordinates to decribe the classification performance at different probability threshold.
-
Area Under Curve (AUC). The area under the ROC curve. Larger AUC represents the better diagnostic performance.
Experiment setup
In this experiment, we build the diagnostic model using PyTorch, a Python deep learning framework. The hardware platform of this study are (CPU) Intel Core i9-13900K with (GPU) Nvidia RTX 4090, and the operating system is Windows 11.
Accurate CAP diagnosis
We first compare the performance in CXR image diagnosis among multiple models, shown in Table 1. It shows the validity of diagnosing CAP utilising ResNet-50, which reached the highest performance in comparison. Then, the multimodal model integrating our modified ResNet-50 achieved the highest results in diagnosing CAP in children, with AUC, accuracy, recall, precision, and F1-Score of 0.979, 0.942, 0.953, 0.932, and 0.943, respectively, shown in Table 2. Then, Table 3 displays the diagnostic performance for children of different ages, validating the precise detection of the physical variations and performance in diagnosing according to the results. Notably, while results aged from 0 to 11 achieved considerable outcomes, performance that was aged above 11 further even achieved 100% performance. This could occur due to the relatively insignificant physical development of these children. Therefore, the model could effectively capture all CAP-related features from the multimodal data. Furthermore, the number of samples collected in this range is small, which only takes 4.65% of the total cases. Hence, the identification performance of these cases could be coincidental and statistical artifacts, and caution should be exercised against overinterpreting these results.
Ablation studies
To prove the efficiency and performance of the utilised CXR recognition model, we firstly compare the results among other popular backbones for comparison, including DenseNet-121 (Dense-121)38, MobileNetV3-Large (MNv3-L)39, EfficientNetv2_Medium (Effv2_m)40, Vision Transformer (ViT)41, Swin Transformer v2 (STv2_s)42, and Multi-Axis Vision Transformer (MaxViT_t)43. Then, a t-test on accuracy is conducted compared to the performance of ResNet-50. Results from Table 1 denote that the modified ResNet-50, which is integrated into the multimodal model, achieved the reasonable performance, further validating the robustness of our multimodal model within the constructed dataset. It is noteworthy that the EfficientNetv2_Medium achieved the highest diagnostic accuracy. However, its improvement is not significant according to the t-test result, while the computational requirements are larger than ResNet-50; in addition, its performance on F1-Score, which is very close to ResNet-50, further supports these findings.
Finally, to ensure contributions of each modality could result in performance improvement, we conducted ablation experiments on the model, which involved each modality with their combinations: diagnose with CXR data, clinical text data, laboratory test data only, then diagnosing using CXR data with clinical text data, CXR data with laboratory test data, and clinical text data with laboratory test data. The results are shown in Table 2 and Figure 8. Moreover, to demonstrate the necessity of CXR image augmentation, we compared the diagnostic performance on CXR images with aggressive (scale factors are between 0.5 and 2.0) and without the image augmentations, denoted as“Aggressive augmentations”and“CXR w/o augmentations”, respectively. The experiments showed that analysis only relying on CXR, laboratory tests, or clinical text had already achieved a pretty high level of performance, with accuracy rates all above 83%, preliminarily demonstrating the diagnostic significance of modal data. With appropriate image augmentations, the ResNet-50 could more effectively capture CAP-related information, resulting in a more robust performance. Moreover, we evaluated the diagnostic performance of multimodal models combining two modalities. As a result, all of their performance improved significantly compared to single-modality models, with all of their accuracy being above 86%. Finally, we compared them with our multimodal model, which includes all three modalities mentioned in the constructed dataset. This demonstrates that our developed model achieved further improvement in diagnostic performance compared to the multimodal models combining two modalities. The corresponding T-test can be found on Table 4.
Interpretability
To enhance the transparency and assist physicians’ diagnostic processes of our multimodal diagnostic model, we analyzed our model using a series of interpretability tools. Specifically, we deployed Gradient-weighted Class Activation Mapping (GradCAM)44 to our ResNet-50 model for CXR images explanation, then implemented SHapley Additive exPlanations (SHAP) to analyze the indicators that contributed to the DNN’s decision for lab test indicators. Finally, we presented the saliency map to inspect tokens’ contributions to the decision of Bi-GRU.
CXR interpretability
The interpretations of CXR are shown in Figure 9. In this figure, we demonstrate GradCAM activation maps that show that the model made correct and incorrect decisions. Specifically, the color’s depth represents the significance of the contribution in the corresponding region. It can be observed that:
-
For the correctly classified examples, the model successfully captured uniform high-density opacity with relatively ill-defined margins, which is a typical situation for CAP. Then, normal patients’ activations are focused on lung-irrelevant regions of the CXR images, avoiding providing misleading information during multimodal fusion.
-
For the incorrectly classified examples, although a part of the model’s activation has captured the lung region, its main focus is on regions outside the lungs. Similar to the normal cases, the model mistakenly recognized the ambiguous shadows as suspicious lesions. These misinterpretations could interrupt the analysis process of the classifier, producing incorrect results.
Laboratory test indicators interpretability
The SHAP visualisations of lab test indicators are shown in Figure 10. In this figure, each data point has its unique SHAP and feature value. The SHAP value measures the weights contributed to the final decision. If the specific indicator would increase the CAP diagnostic result, its SHAP value is significant. The feature value represents the original value of the feature itself. It can be observed that:
-
Data points with high feature values for CRP, MO%, and NE% are clustered in the region of high, positive SHAP values, demonstrating their positive impact on the model’s CAP decisions. Conversely, lower values of BA%, and EO% are associated with CAP outcomes in the model’s decision-making process. These findings are consistent with the Pearson correlation heatmap presented in Figure 7.
-
The remaining indicators, absolute counts NE# corresponding to NE% and LY%, demonstrated a relatively insignificant distribution in the figure. Consequently, these indicators could be analyzed using the result of relative value among other indicators in the DNN model.
-
It is worth noting that several data points exhibit trends that diverge from the overall SHAP distributions, which are caused by the unique characteristics of specific samples or incorrect predictions by the model. Therefore, these highlight that the reconfirmation is required for suspect cases in real practice.
Clinical texts interpretability
To inspect mechanisms of the Bi-GRU model in processing clinical text, we employed a saliency map-based interpretability framework. In Figure 11, four representative samples are given, including correctly and incorrectly diagnosed cases. In the saliency map, the significance of the token is colored. The deeper background color represents the corresponding token that puts more significance on the model’s decision. Through observation:
-
For the sample correctly diagnosed as CAP, Bi-GRU successfully captured the important tokens. For instance, ’Fever’, ’cough’, ’coarse breathing’, and ’wet rales’ are key diagnostic factors for CAP screening.
-
For the sample incorrectly diagnosed as normal, although Bi-GRU captured critical tokens, ’cough’ and ’Difficulty breathing’, the absence of a specific auscultation description resulted in misdiagnosis.
-
For the sample correctly diagnosed as normal, although the Bi-GRU captured key words, it successfully captured contextual information, such as ’no rales’, and finally made a non-CAP prediction.
-
For samples that were misdiagnosed as CAP, the model detected typical CAP-related tokens such as ’fever’ and ’convulsion’. However, the model assigns higher weights to these CAP-like symptoms, while overlooking the descriptions of lung auscultation information, which could rule out the possibility of CAP, leading to false positive diagnoses. Therefore, a re-check is required for cases irrelevant to CAP but similar in appearance in clinical practice.
Therefore, the proposed Bi-GRU successfully analyzed deterministic descriptions related to CAP, while external noises will lead to potential misdiagnoses. This could be addressed by expanding our dataset’s scale and diversity, which will be discussed in the following section.
Ethical declarations
This study was reviewed and approved by the ethics committee at Fujian Children’s Hospital (2024ETKLRK12007). All procedures performed in this study involving human participants were following the ethical standards of the ethics committee at Fujian Children’s Hospital and with the 1964 Declaration of Helsinki and its later amendments. All data are de-identified, the ethics committee of Fujian Children’s Hospital approved this study as a retrospective review with a waiver for patient informed consent.
Discussions and limitations
Community-acquired pneumonia (CAP) is a prevalent disease that strains hospital resources, especially during peak periods. Moreover, CAP triggers a series of health risks, particularly in children with weak immune systems4. To address this issue, we collect a multimodal dataset and develop the corresponding multimodal diagnostic model incorporating modified ResNet-50 for CXR image analysis, Bi-GRU for symptom text feature extraction, and a DNN model for lab test indicators recognition with a multimodal feature classifier for accurate CAP analysis.
Regarding the dataset, we collected real-world clinical data from Fujian Provincial Children’s Hospital (a tertiary hospital). Then, the extracted data are further reviewed by medical experts in the field. Therefore, from a medical diagnostic perspective, our multimodal model contains an“local expert-level knowledge database”for diagnosing CAP in children, which is crucial for developing a highly accurate diagnostic model.
As for the results of comparison studies, it is observed that the performance of ResNet-50 is almost the best among the criteria, while its computational requirements of ResNet-50 are reduced compared to the others. Therefore, selecting ResNet-50 for incorporation into the multimodal model represents an efficient solution. In addition, we evaluated several Vision Transformer variations. However, their performance did not surpass ResNet-50, potentially due to insufficient training data for the Transformer-based model’s attention mechanisms, which require large-scale datasets. Although CXR and lab tests diagnosis model successfully captured valid information related to CAP, such as physical variations, their performance is still significantly lower than the text modality. This may arise due to the imbalance of age distributions in the dataset, potentially resulting in slightly biased analysis during feature extraction, while the format of text descriptions are relatively consistent. Then, results from ablation studies demonstrated that each modality has successfully contributed to the precise analysis, as a result of increased performance by adding modalities.
However, limitations were revealed in our designed method. From the dataset perspective, although we have constructed a high-quality multimodal CAP dataset, these data demonstrated potential regional limitations due to the source of a single medical institution (single-center study). Therefore, the overall performance and generalizability require further validation for external applications, such as different races, regions, and medical systems, since the influences of local demographics are introduced. Moreover, analytical results from age-specific validation indicate the imbalanced distribution of cases. As mentioned in this paper, significant physical heterogeneity exists across children’s age groups, which could constrain the performance. Therefore, promising results for cases older than 11 should be considered cautiously. Then, from the model perspective, it demonstrates high performance according to experiment results. Nevertheless, our model was primarily designed to systematically validate the feasibility of a multimodal approach for diagnosis using our constructed dataset. To further enhance both the generalizability and diagnostic performance of the model, more sophisticated designs such as intermediate multimodal fusion strategies that facilitate cross-modal feature interaction and online large language models (LLM) empowered solutions could be explored. Additionally, although the ResNet-50 is used in our CXR image modality analysis, which is relatively lightweight, potential remains to reduce computational complexity. Therefore, a model compression strategy, such as Knowledge Distillation, could be further implemented.
Based on the analysis, our future work focuses on expanding our dataset and constructing a more reasonable and lightweight multimodal diagnostic system. Specifically, we will continue to collect cases on a broader time range and seek cooperation with other medical institutions to construct a balanced and diverse pediatric CAP dataset. Followed by this, constructing a robust and lightweight system using reasonable techniques should be conducted for further evaluation in order to fully demonstrate the potential of the multimodal diagnostic model.
Conclusion
In conclusion, we constructed a pediatric CAP dataset from a tertiary hospital, and trained data on the developed multimodal auxiliary diagnosis model, aiming to validate the feasibility of developing a multimodal auxiliary tool that could potentially assist the diagnosis process for pediatric CAP. The designed approach could simulate clinical diagnosis procedure by integrating the chest X-ray image, laboratory indicators, and clinical texts. After evaluation, the developed model has shown impressive results in our constructed dataset. Therefore, this method has the potential for improvement in clinical practice for pediatric CAP diagnosis. Nevertheless, several improvements are required, such as expanding the dataset and constructing a more advanced model. That said, our multimodal model demonstrated potential in assisting pediatric CAP diagnostic tasks.
Data availability
The data that support the findings of this study are available from Fujian Children’s Hospital, but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the author Yanying Rao (email: raoyanying@fjmu.edu.cn) upon reasonable request and with permission of Fujian Children’s Hospital.
References
Chee, E., Huang, K., Haggie, S. & Britton, P. N. Systematic review of clinical practice guidelines on the management of community acquired pneumonia in children. Paediatr. Respir. Rev. 42, 59–68 (2022).
Corrales-Medina, V. F. et al. Cardiac complications in patients with community-acquired pneumonia: incidence, timing, risk factors, and association with short-term mortality. Circulation 125, 773–781 (2012).
Reyes, L. F. et al. Severe pneumococcal pneumonia causes acute cardiac toxicity and subsequent cardiac remodeling. Am. J. Respir. Critical Care Medicine 196, 609–620 (2017).
van Meel, E. R. et al. Early-life respiratory tract infections and the risk of school-age lower lung function and asthma: a meta-analysis of 150 000 european children. Eur. respiratory journal 60 (2022).
Ayan, E. & Ünver, H. M. Diagnosis of pneumonia from chest x-ray images using deep learning. In 2019 Scientific meeting on electrical-electronics & biomedical engineering and computer science (EBBT), 1–5 (Ieee, 2019).
Wang, X. et al. Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the IEEE conference on computer vision and pattern recognition, 2097–2106 (2017).
Cid, Y. D. et al. Development and validation of open-source deep neural networks for comprehensive chest x-ray reading: a retrospective, multicentre study. The Lancet Digit. Heal. 6, e44–e57 (2024).
Feng, Y. et al. Pcxrnet: Pneumonia diagnosis from chest x-ray images using condense attention block and multiconvolution attention block. IEEE J. Biomed. Heal. Informatics 26, 1484–1495 (2022).
Fu, Y., Xue, P., Zhang, Z. & Dong, E. Pka 2-net: Prior knowledge-based active attention network for accurate pneumonia diagnosis on chest x-ray images. IEEE J. Biomed. Heal. Informatics 27, 3513–3524 (2023).
Chen, S., Ren, S., Wang, G., Huang, M. & Xue, C. Interpretable cnn-multilevel attention transformer for rapid recognition of pneumonia from chest x-ray images. IEEE J. Biomed. Heal. Informatics 28, 753–764 (2023).
Lafond, D., Descarreaux, M., Normand, M. C. & Harrison, D. E. Postural development in school children: a cross-sectional study. Chiropr. & osteopathy 15, 1–7 (2007).
Karetzky, M., Cunha, B. A. & Brandstetter, R. D. The pneumonias (Springer, 1993).
Florin, T. A. et al. Reliability of examination findings in suspected community-acquired pneumonia. Pediatrics 140 (2017).
Almirall, J. et al. Contribution of c-reactive protein to the diagnosis and assessment of severity of community-acquired pneumonia. Chest 125, 1335–1342 (2004).
Sierra, R. et al. C-reactive protein used as an early indicator of infection in patients with systemic inflammatory response syndrome. Intensive care medicine 30, 2038–2045 (2004).
Epler, G. R., Carrington, C. B. & Gaensler, E. A. Crackles (rales) in the interstitial pulmonary diseases. Chest 73, 333–339 (1978).
Williams, D. J. et al. Predicting severe pneumonia outcomes in children. Pediatrics 138, e20161019 (2016).
Florin, T. A. et al. Predicting paediatric pneumonia severity in the emergency department: a multinational prospective cohort study of the pediatric emergency research network. The Lancet Child & Adolesc. Heal. 9, 383–392 (2025).
Ilg, A. et al. Performance of the curb-65 score in predicting critical care interventions in patients admitted with community-acquired pneumonia. Annals of emergency medicine 74, 60–68 (2019).
Wang, D., Willis, D. R. & Yih, Y. The pneumonia severity index: Assessment and comparison to popular machine learning classifiers. Int. J. Med. Informatics 163, (2022).
Sanchez, K. et al. Cx-dagan: Domain adaptation for pneumonia diagnosis on a small chest x-ray dataset. IEEE Transactions on Med. Imaging 41, 3278–3288 (2022).
Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical image computing and computer-assisted intervention–MICCAI 2015: 18th international conference, Munich, Germany, October 5-9, 2015, proceedings, part III 18, 234–241 (Springer, 2015).
Zhang, X. et al. Cxr-net: a multitask deep learning network for explainable and accurate diagnosis of covid-19 pneumonia from chest x-ray images. IEEE journal of biomedical and health informatics 27, 980–991 (2022).
Lee, J. B., Kim, J. S. & Lee, H. G. Covid19 to pneumonia: Multi region lung severity classification using cnn transformer position-aware feature encoding network. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 472–481 (Springer, 2024).
Landolsi, M. Y., Hlaoua, L. & Ben Romdhane, L. Information extraction from electronic medical documents. state of the art and future research directions. Knowl. Inf. Syst. 65, 463–516 (2023).
Lample, G., Ballesteros, M., Subramanian, S., Kawakami, K. & Dyer, C. Neural architectures for named entity recognition. arXiv preprint (2016).
Sun, Q. & Bhatia, P. Neural entity recognition with gazetteer based fusion. arXiv preprint arXiv:2105.13225 (2021).
Murtaugh, M. A., Gibson, B. S., Redd, D. & Zeng-Treitler, Q. Regular expression-based learning to extract bodyweight values from clinical notes. J. biomedical informatics 54, 186–190 (2015).
Liu, Z. et al. Entity recognition from clinical texts via recurrent neural network. BMC medical informatics and decision making 17, 53–61 (2017).
Jagannatha, A. N. & Yu, H. Bidirectional rnn for medical event detection in electronic health records. In Proceedings of the conference. Association for Computational Linguistics. North American Chapter. Meeting, vol. 2016, 473 (2016).
Roy, A. & Pan, S. Incorporating medical knowledge in bert for clinical relation extraction. In Proceedings of the 2021 conference on empirical methods in natural language processing, 5357–5366 (2021).
Wang, Q., Yang, D., Li, Z., Zhang, X. & Liu, C. Deep regression via multi-channel multi-modal learning for pneumonia screening. IEEE Access 8, 78530–78541 (2020).
Narmadha, A. & Gobalakrishnan, N. Het-rl: Multiple pulmonary disease diagnosis via hybrid efficient transformers based representation learning model using multi-modality data. Biomed. Signal Process. Control. 100, (2025).
Ren, H. et al. Chexmed: A multimodal learning algorithm for pneumonia detection in the elderly. Inf. Sci. 654, (2024).
Shao, J. et al. A multimodal integration pipeline for accurate diagnosis, pathogen identification, and prognosis prediction of pulmonary infections. The Innov. 5 (2024).
Li, J. et al. Assessing severity of pediatric pneumonia using multimodal transformers with multi-task learning. Digital Health 10, 20552076241305170 (2024).
Loshchilov, I. & Hutter, F. Decoupled weight decay regularization. arXiv preprint arXiv:1711.05101 (2017).
Huang, G., Liu, Z., Van Der Maaten, L. & Weinberger, K. Q. Densely connected convolutional networks. In 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2261–2269 (2017).
Howard, A. et al. Searching for mobilenetv3. CoRR (2019).
Tan, M. & Le, Q. Efficientnetv2: Smaller models and faster training. ICML 5 (2021).
Dosovitskiy, A. et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929 (2020).
Liu, Z. et al. Swin transformer v2: Scaling up capacity and resolution. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, 12009–12019 (2022).
Tu, Z. et al. Maxvit: Multi-axis vision transformer. In European conference on computer vision, 459–479 (Springer, 2022).
Selvaraju, R. R. et al. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE international conference on computer vision, 618–626 (2017).
Funding
This work was sponsored by Fujian provincial health technology project (Grant number: 2024GGA063). Startup Fund for scientific research, Fujian Medical University (Grant number: 2024QH1247).
Author information
Authors and Affiliations
Contributions
Wang Y. conducted the data analysis, ablation studies, comparison studies, and paper writing. Rao Y. collected the multimodal datasets and conducted the initial experiment. Zhu Y. designed and constructed the multimodal model. Tang Q. and Xu Z. reviewed the manuscript, provided technology support, and communicated with Fujian Children’s Hospital. Wu J., Qiao B., and Wu X. finished the data analysis and comparison studies.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, Y., Rao, Y., Zhu, Y. et al. Multimodal-based auxiliary diagnosis for pediatric community acquired pneumonia. Sci Rep 15, 39152 (2025). https://doi.org/10.1038/s41598-025-23006-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-23006-x