Abstract
Accurate and efficient disease diagnosis remains a critical challenge in the healthcare sector. With the growing availability of biomedical data, machine learning techniques have become invaluable tools for developing intelligent disease detection systems. Researchers have applied various algorithms, including artificial neural networks (ANNs), to improve classification accuracy. To further improve ANN performance, various optimization methods are applied to enhance learning and avoid the local minima problem, as each model demonstrates distinct performance characteristics. Therefore, this paper presents a hybrid Bio inspired Ropalidia Marginata Optimization-based hybrid neural network (RMO-NN) aimed at improving medical data classification. The proposed RMO-NN incorporates biologically inspired task allocation and dominance hierarchy mechanisms from RMO to optimize neural network learning performance effectively and reducing classification errors. To validate its effectiveness, the RMO-NN is tested on three large-scale medical datasets such as breast cancer, diabetes, and blood transfusion datasets and three medical images datasets. The performance of the proposed model is compared against two established metaheuristic neural models: Cuckoo Search Neural Network (CSNN) and Artificial Bee Colony Neural Network (ABCNN). The proposed RMO-NN model outperforms CSNN and ABCNN in terms of accuracy, MSE, SD, and convergence speed. And for medical images datasets the proposed is further validated with various start of art deep learning models. The results highlight the proposed model perform better on biomedical data classification tasks. The Proposed method significantly outperforms baseline approaches, achieving substantial accuracy, while introducing a novel RMO algorithm.
Similar content being viewed by others
Introduction
The healthcare sector continues to generate massive volumes of complex data, offering new opportunities for improved diagnostics, disease prediction, and optimized treatment strategies. Modern machine learning (ML) techniques are central to harnessing this data, offering advanced predictive capabilities, better pattern recognition, and enhanced support for clinical decision-making1,2. However, its performance can be constrained by challenges such as high dimensionality, redundant features, and the tendency to converge on local optima3,4. These methods help lower the cost and save time in medical testing. By analyzing large amounts of medical and imaging data, machine learning models support doctors in making quick and accurate decisions5,6. In recent years, various approaches have been developed to enhance the diagnosis and treatment of numerous diseases, including cancer4. Multiple types of cancer have been documented in the literature and are typically classified based on the type of affected cells for example, breast cancer7,8. However, mortality rates have remained relatively stable, largely due to early detection, which plays a crucial role in improving patient outcomes9. To enhance early detection, computerized analysis tools known as computer-aided detection/diagnostic (CAD) systems are increasingly used to support clinical decision-making7. The development of CAD systems for breast cancer typically involves key modules such as feature extraction, feature selection, and classification, which are active areas of research10. Among these, the classifier is the most critical component, as it directly influences diagnostic accuracy. Various classification algorithms have been employed for the automated detection of breast cancer, including K-nearest neighbor (K-NN)11, artificial neural networks (ANN)12, and support vector machines (SVM)13,14.
In recent years, ANN has seen notable performance improvements using randomly initialized parameters, which enhance both learning speed and generalization15. ANNs enhance the accuracy of mammography and ultrasound interpretation, aiding in the identification of suspicious lesions16,17. Selecting optimal parameters for ANNs primarily involves determining the best network topology, which includes choosing appropriate initial weights, the number and configuration of hidden layers and nodes, an effective training algorithm, and a suitable learning rate. The existing research methods are used to identify ideal ANN parameters to enhance the accuracy of classification models. Commonly adopted techniques include trial-and-error, network constructive and destructive methods, and brute-force approaches. However, these traditional methods often result in inefficient training processes18. The performance of ANN classifiers greatly depends on selecting the right input feature subsets, along with other design parameters19. Due to the limitations of conventional techniques in managing large parameter spaces, only a few studies have explored genetic algorithms20,21 for simultaneous optimization of multiple parameters, particularly in breast cancer classification tasks. The use of swarm intelligence based methods, such as artificial bee colony algorithms22, which have shown improvements in parameter selection and classification accuracy. Additionally, it examines the application of other evolutionary techniques like particle swarm optimization23 and genetic algorithms21 for fine-tuning ANN parameters. These optimized approaches have been successfully applied in various breast cancer diagnostic systems to assess disease severity. Building on these optimization strategies, it is crucial to address the remaining challenges in ANN-based systems, which are further outlined in the following problem formulation.
Problem formulation
Despite advancements, current research still struggles with achieving consistent performance across multiple datasets and balancing computational cost with model accuracy. Many hybrid ML systems lack adaptability, require extensive manual tuning, or fail to generalize across different biomedical domains. Lately, ANN have achieved significant performance gains by employing randomly initialized parameters, which boost both the speed of learning and the ability to generalize. The training and hyperparameter optimization of ANNs models represent some of the most complex challenges in machine learning. Traditionally, many studies have relied on gradient-based backpropagation techniques for this task24. However, these methods face significant limitations, including susceptibility to local minima in multi-objective cost functions, high computational cost due to numerous gradient calculations25, and a requirement for continuous cost functions. Given that training ANNs model is an NP-hard problem, there has been a growing interest in using metaheuristic (MH) algorithms for optimizing their structure and parameters. MH algorithms offer a robust approach to estimating optimal configurations, including hyperparameters, weights, layer counts, neuron numbers, and learning rate26. The integration of ANNs with metaheuristic algorithms gained traction for improving training and optimization efficiency. However, major issues include high computational complexity27, overfitting remains a concern, reporting reduced generalization in medical diagnosis tasks28. Scalability and training time increase with larger ANN models, as found by29. Additionally, hyperparameter sensitivity and lack of theoretical convergence, present further limitations in hybrid ANN-metaheuristic approaches. To solve these limitations this research develops a novel hybrid model that combines bio-inspired RMO inspired by wasp dominance hierarchies, hybridized neural networks to improve classification accuracy and efficiency on large medical datasets by incorporating intelligent weights and bias values to ANN model. To address these limitations and further enhance the effectiveness of ANN-based diagnostic systems, the following research motivation outlines the foundation behind developing a robust hybrid model.
Research motivation
ML techniques, especially artificial neural networks (ANNs), have advanced disease prediction by detecting complex patterns in biomedical data. However, challenges like high dimensionality, redundant features, and local optima limit their effectiveness. Early cancer detection, particularly breast cancer, benefits from ML-driven computer-aided detection systems, where classifier accuracy is crucial. Traditional ANN training using gradient-based methods faces issues such as local minimum and high computational cost. Metaheuristic (MH) algorithms have emerged to optimize ANN training but suffer from overfitting, scalability problems, and hyperparameter sensitivity. There remains a need for hybrid models that balance accuracy and efficiency across diverse datasets. This research proposes a novel hybrid model combining bio-inspired Ropalidia Marginata Optimization (RMO) with neural networks. This approach aims to enhance classification accuracy and computational efficiency for large-scale medical datasets, addressing current ML limitations in healthcare analytics. Based on these motivations, the research aims to address the following key questions to evaluate the effectiveness and applicability of the proposed hybrid RMO-ANN model. The main differences of RMO with other established bio-inspired algorithms such as PSO, GWO, and ACO in terms of their mechanisms, benefits, and behaviors is given as:
-
Mechanism: Unlike PSO, which models particle movement based on velocity and global/local best positions, RMO simulates the decentralized leadership and task allocation of Ropalidia marginata wasps, where any individual can temporarily assume leadership without centralized control. This differs from GWO, which uses strict hierarchical leadership (alpha, beta, delta, and omega) to guide hunting, and ACO, which relies on pheromone trails and probabilistic path selection.
-
Benefits: RMO’s flexible leader transition mechanism offers higher adaptability to dynamic optimization environments and reduces the risk of premature convergence that can occur in PSO or GWO when leader solutions stagnate. Its decentralized information sharing also enhances robustness compared to the more rigid structures in GWO and pheromone-based reinforcement in ACO.
-
Behavior: In exploration exploitation balance, RMO dynamically shifts roles and influence levels among agents, promoting diversity in search patterns, whereas PSO often favors exploitation once good solutions are found, and ACO can overly reinforce early paths. RMO’s stochastic leader reassignment encourages continual exploration while maintaining search efficiency.
Research questions
-
How can bio-inspired Ropalidia Marginata Optimization (RMO) be effectively hybridized with artificial neural networks to improve classification accuracy in medical datasets?
-
To what extent the RMO-enhanced neural networks by selecting their weights and bias values, and how does this impact model efficiency?
-
How does the proposed RMO-based hybrid model compare to traditional gradient-based and other methods in terms of training time, accuracy, and MSE?
-
Can the RMO-hybrid neural network model consistently achieve robust performance across multiple benchmark biomedical datasets, including breast cancer detection task. To address these research questions, this paper proposes a novel approach detailed in the following main contribution.
Paper contribution
This paper introduces a novel Ropalidia Marginata Optimization-based Neural Network (RMO-NN) for biomedical data classification. The proposed RMO is inspired by the dominance hierarchy and task allocation behavior of Ropalidia Marginata wasps. Ropalidia marginata colonies consist of a single egg-laying queen and numerous non-reproductive workers. Queens and workers are morphologically identical, and individuals can change between roles30,31. Unlike most primitively eusocial species, the queens of RM are remarkably docile, non-aggressive, and minimally interactive32. Males are distinguished by their weaker mandibles and lack of stingers. Female workers and queens are morphologically similar but differ in behavior30,33. Females generally serve as workers, performing tasks like foraging, nest building, and larval care34. The proposed bio-inspired algorithm is hybridized with neural networks to enhance learning performance, reduce classification error, and eliminate local minima problem. In this study, the RMO algorithm is employed exclusively for optimizing the weights and bias values of the artificial neural network, rather than for feature selection. The role of RMO in the proposed RMO-NN framework is to enhance the learning process by efficiently navigating the high-dimensional weight space, avoiding local minima, and improving convergence speed. All datasets are used in their complete feature form after preprocessing, ensuring that performance improvements are attributable to the optimized network parameters rather than to changes in the input feature set. The RMO-NN model is validated on three large-scale medical datasets breast cancer, diabetes, and blood transfusion and outperforms established metaheuristic models like Cuckoo Search Neural Network (CSNN) and Artificial Bee Colony Neural Network (ABCNN) in terms of accuracy, mean squared error (MSE), standard deviation (SD), and convergence speed. This research contributes a robust, scalable, and efficient hybrid classification framework for disease diagnosis, demonstrating the value of incorporating bio-inspired optimization techniques in machine learning for healthcare applications. The main contribution of this paper is given as below:
-
Proposed a novel Ropalidia Marginata Optimization-based Neural Network (RMO-NN) that integrates the dominance hierarchy behavior of Ropalidia Marginata wasps into neural network learning. This biologically inspired hybrid approach enhances the model’s ability to avoid local minimum and improves classification performance across various medical datasets.
-
Validated RMO-NN on three benchmark classification datasets Breast Cancer, Pima Indian Diabetes, and Blood Transfusion and three more medical images datasets. Where it consistently outperformed other neural and metaheuristic models (CSNN, ABCNN, LM, ERN) in terms of accuracy, Mean Squared Error (MSE), and Standard Deviation (SD).
-
RMONN demonstrated lower standard deviation across all datasets, indicating more robust and stable classification performance. It maintained high generalization ability while reducing overfitting, a common limitation in traditional hybrid ANN-metaheuristic models.
-
Despite being a hybrid model, RMONN achieved faster or comparable convergence in terms of MSE and required moderate CPU time, outperforming methods like LM and ERN in execution time while maintaining higher accuracy showcasing its practical utility for real-time medical diagnostic applications.
Peper organization
The paper is organized as follows: Sect. “Related work” reviews related work on neural networks and metaheuristic optimization in medical classification. Sect. “Methods and materials” details the methods and materials. Sect. “Termination” analyzes the performance results. Finally, Sect. “Conclusions and Future Work” concludes the study and outlines future research directions.
Related work
In recent years, machine learning (ML) has been widely applied to biomedical data, aiming to improve diagnosis, prognosis, and treatment planning. Studies such as35 have demonstrated that deep learning models can extract complex features from medical images and clinical data, outperforming traditional statistical methods. Similarly36, reviewed various feature selection (FS) methods, emphasizing their critical role in improving classifier performance by reducing dimensionality and eliminating irrelevant attributes. Traditional classifiers such as Support Vector Machines (SVM) and Random Forests (RF) have also been widely used for disease classification37. Similarly, in38 this article addresses the challenge of multi-class imbalance in medical datasets by proposing a rebalancing framework using SCUT (SMOTE and Cluster-based Under sampling Technique), SHAP-RFE for feature selection, and DES-MI for dynamic ensemble classification. The framework introduces enhancements to SCUT and classifier selection, achieving superior performance across eight imbalanced datasets with notable improvements in accuracy, G-mean, and AUC. Where else in39 this study tackles early liver disease detection using single and ensemble machine learning models on the Indian Liver Patient Dataset (ILPD). Various preprocessing and feature selection techniques were applied to improve model accuracy. A two-level ensemble stacking model outperformed others, achieving up to 94.12% accuracy with feature selection. The proposed approach demonstrated strong predictive performance, indicating its effectiveness in liver disease detection. Further in40 this research proposes an improved breast cancer diagnosis method using Association Classification (AC) enhanced with ensemble filter and wrapper-based feature selection techniques. A new bootstrapping strategy and the WARF method are used to select optimal discriminative features. Two models WARF-PCBA and hybrid PSO-WARF-PCBA are developed and evaluated on UCI breast cancer datasets. Results show these models consistently outperform existing AC algorithms in prediction accuracy and efficiency. Another paper in41 introduces a Cuckoo Search Back-Propagation (CSBP) algorithm, inspired by cuckoo bird behavior, to improve BP training by enhancing convergence speed and avoiding local minima. CSBP is evaluated using OR and XOR datasets and compared with other hybrid methods, including artificial bee colony-based BP. Results show that CSBP significantly improves the computational efficiency of the BP training process. Further in42 this paper presents an in-depth analysis of the Firefly Algorithm (FA), a popular nature-inspired optimization method known for its effectiveness across various domains. FA has shown strong performance in solving complex problems in biomedical engineering and healthcare. The study reviews its variants, applications, and enhancements, aiming to inspire further research and innovation using FA in these critical fields. Although in43 this paper proposes a machine learning model for accurate recognition of Parkinson’s disease, addressing limitations of existing methods. It employs a hybrid feature selection algorithm combining Relief and ant-colony optimization, with a SVM trained on the selected features. Using K-fold cross-validation, the model achieved 99.50% accuracy on a real-world dataset, outperforming existing approaches and proving highly effective for Parkinson’s disease detection. Further in18 this paper reviews the use of swarm intelligence-based metaheuristic optimization algorithms for enhancing ANN models in breast cancer diagnosis. It emphasizes the importance of feature selection and hyperparameter optimization in improving classification accuracy and reducing computation time. The review compares various ANN-based approaches, highlighting their strengths, limitations, and evaluation metrics. It also outlines future research directions in optimized ANN models for more effective breast cancer detection. Another paper in44 presents a novel approach to improving cardiovascular disease (CVD) prediction by integrating Gray Wolf Optimization (GWO) with machine learning models. Using a patient dataset, the study applies GWO to optimize hyperparameters and feature selection for models like SVM, Decision Tree, and K-NN. The results show that the GWO-enhanced model achieves higher prediction accuracy 87% compared to traditional models like SVM (78%), Random Forest (76%), and K-NN (56%). This demonstrates GWO’s effectiveness in enhancing CVD prediction. Although in45this article addresses the challenge of high-dimensional gene data in bioinformatics for medical diagnosis, where identifying relevant genes is difficult due to redundancy and irrelevance. It proposes a Multi-Objective Binary Cuckoo Search Algorithm (MOBCSA) for gene selection, which optimizes both classification accuracy and the number of selected genes. MOBCSA extends the standard cuckoo search algorithm by using an S-shaped transfer function for binary search and includes an external archive and adaptive crowding distance to maintain solution diversity. Tested on six biomedical datasets with three classifiers, MOBCSA outperformed four state-of-the-art multi-objective feature selection methods, achieving classification accuracy between 92.79% and 98.42% while selecting fewer genes (15.67 to 27.88 on average). Another paper in46 proposes a Graylag Goose Optimization (GGO) algorithm for feature selection to improve heart disease classification accuracy, with LSTM identified as the best-performing classifier. The GGO-LSTM achieved 99.58% accuracy, outperforming six alternative optimizers and validated through statistical tests and visual analysis. Where else in47 this study introduces Cardiovascular disease snake optimization (CVD-SO), a framework combining snake optimization and machine learning for efficient feature selection and classification of cardiovascular disease data. The proposed model achieved 99.9% accuracy, demonstrating its potential to enhance early diagnosis and reduce CVD-related mortality. Similarly in48 proposes the Modified Al-Biruni Earth Radius (MBER) algorithm for feature selection to enhance eye state (open/closed) classification from EEG data. Optimized with KNN as the fitness function, MBER outperformed five competing algorithms, achieving 96.12% accuracy and demonstrating superior robustness through statistical validation. Further another paper in49 proposes a multilayer multitask LSTM model with PSO-based feature selection to predict mechanical ventilation need, mortality, and ventilation duration in ICU patients using MIMIC-III data. Using the first 24 h of data yielded the best results, with the model achieving high accuracy (0.944 and 0.971) and strong performance across precision, recall, F-score, and AUC. Additionally this study in50 applies four binary optimization algorithms for feature selection in orthopedic disease diagnosis, with BBFS reducing average error by 47.29% compared to others. The optimized RF model (BFS-RF) achieved the best performance, reaching 99.41% accuracy, significantly improving upon the original classifier. Recently the study in51 addresses breast cancer’s global health impact and proposes a deep learning framework combining transfer learning and Grey Wolf Optimization (GWO) for improved diagnosis. Using CNN architectures such as ResNet and Inception on mammographic images, along with WBCD-based feature selection via GWO, the method enhances classification accuracy, robustness, and generalization. Experimental results show high performance (precision 0.942, sensitivity of 0.982, accuracy of 0.965, AUC of 0.971), indicating its potential to improve early detection, reduce diagnostic errors, and strengthen healthcare outcomes. This research introduces Ropalidia Marginata Optimization (RMO), a wasp-inspired algorithm that improves exploration–exploitation balance and outperforms classic metaheuristics. However, its application in hybrid neural networks for biomedical classification remains limited and untested against other bio-inspired models on large clinical datasets, highlighting a key area for future study.
Methods and materials
In this article, a new Ropalidia Marginata Optimization-Neural Network (RMO-NN) method is formulated for medical data classification. At the preliminary stage, the presented RMO-NN method initially pre-processes the input data using null value removal, normalization, and transformation techniques. Moreover, the presented RMO-NN technique applies the RMO model for selecting the most effective subset of features, using the dominance hierarchy of the wasp-inspired approach to balance exploration and exploitation. Finally, the reduced feature set is passed into a Multi-Layer Perceptron (MLP) or Feedforward Neural Network (FNN), where RMO is again employed to optimize the weights and biases for enhanced learning performance.
Ropalidia Marginata optimization (RMO) algorithm
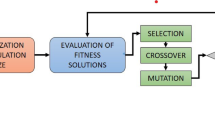
Ropalidia marginata (RM) are dark reddish wasps with yellow markings, including spots on joints and a yellow ring on the lower abdomen52. Ropalidia marginata colonies consist of a single egg-laying queen and numerous non-reproductive workers. Queens and workers are morphologically identical, and individuals can change between roles30,31. Unlike most primitively eusocial species, the queens of RM are remarkably docile, non-aggressive, and minimally interactive32. Males are distinguished by their weaker mandibles and lack of stingers. Female workers and queens are morphologically similar but differ in behavior30,33. Females generally serve as workers, performing tasks like foraging, nest building, and larval care34. Workers self-regulate foraging through aggression and can mate with males, remaining inseminated even if they do not reproduce53. Where the queen is physically dominant and aggressive, the RM queen is remarkably meek and docile in behavior54. Despite being docile, she can maintain a reproductive monopoly and is the only egg layer in the colony. Upon removal of the queen, one of the workers becomes extremely aggressive but immediately drops her aggression if the queen is returned55. If the queen is not returned, this hyperaggressive individual will develop her ovaries, lose almost all her aggression, and become the next queen of the colony. Males, produced in smaller numbers, stay in the nest briefly before leaving to mate with females from other colonies. They do not contribute to colony maintenance and depend on workers for food52. Occasionally, they cannibalize larvae but are unsuited for tasks like foraging or defense due to their physical limitations56. This research proposed the nature-inspired behavior of Ropalidia marginata (RM), a species of social wasp as an optimization technique. These wasps exhibit decentralized and efficient task allocation, cooperation, and resource utilization in their colonies. The RMO algorithm leverages these behaviors to solve optimization problems effectively. Ropalidia marginata colonies demonstrate unique hierarchical behaviors: Fig. 1 graphicly represent RMO Cooperation Communication, and Task Switching behavior. Figure 2 give proposed RMO algorithm Flowchart.
-
1.
Decentralized leadership: Task distribution is achieved without a central leader, allowing dynamic adaptability.
-
2.
Task switching: Wasps switch tasks based on the colony’s immediate needs.
-
3.
Cooperation and communication: Efficient sharing of resources and information ensures colony survival and productivity.
These behaviors inspire the core mechanisms of the RMO algorithm, such as adaptive exploration, cooperation, and exploitation. Below is an explanation of how the algorithm works. The Key Steps in the RMO Algorithm 1 as given as: Here is a mathematical formulation of the Ropalidia Marginata Optimization (RMO) algorithm:
Notation.
\(N\) : Number of agents (wasps).
\(X_{i}\) : Position of the \(i\). agent in the search space \(\left( {i = 1,2, \ldots \ldots \ldots .N} \right).\)
\(f\left( X \right)\) : Objective function to minimize or maxize.
\(Role_{i}\) : Role of the \(i\) agent (queen\worker).
\(X_{best}\) : Position of the current best solution.
\(\alpha ,\beta\) : Control parameters for exploration and exploitation.
Initialization
Randomly initialize the positions of agents(wasps) to create an initial population representing potential solutions (wasps). Assign each agent a random position within the search space and evaluate their fitness using the problem’s objective function as defined in Eq. (1).
where \(\left( {X_{min} ,X_{max} } \right)\) are the lower and upper bounds of the search space.
-
i.
Compute the fitness for each agent:
$$f\left( {X_{i} } \right) \forall i \in \left\{ {1,2, \ldots ,N} \right\}$$(2) -
ii.
Assign initial roles based on fitness:
-
a.
Queen: Agents with the best fitness values.
-
b.
worker: Remaining agents.
-
a.
Position update rules
-
i.
Exploiter update (local search)
\(If Rolei = queen\) The agent refines its position:
$$X_{i}^{t + 1} = X_{i}^{t} + \alpha \cdot \left( {X_{best} - X_{i}^{t} } \right) + \varepsilon$$(3)where: where\(t\) is the current iteration, \(\alpha\) controls the step size toward the best solution,
-
\(\varepsilon \sim Uniform\left( { - \delta ,\delta } \right)\) is a small random perturbation for local search.
-
-
ii.
Explorer Update (Global Search)
\(If Rolei = worker\) The agent explores the global space:
$$X_{i}^{t + 1} = X_{i}^{t} + \beta \cdot rand\left( { - 1,1} \right) \cdot \left( {X_{max} - X_{min} } \right)$$(4)where: i. \(\beta\) controls the exploration step size, ii. \(rand\left( { - 1,1} \right)\) is a random value in [− 1,1].
Fitness evaluation and role switching
-
i.
Evaluate the fitness of all agents:
$$f\left( {X_{i}^{t + 1} } \right)\;\forall i \in \left\{ {1,2, \ldots ,N} \right\}$$(5) -
ii.
Update the best solution:
$$X_{best} = argmin f\left( {X_{i}^{t + 1} } \right)$$(6) -
iii.
Dynamically reassign roles:
-
a.
Agents with fitness close to \(f\left({X}_{best}\right)\) become queen.
-
b.
Remaining agents become worker.
-
a.
Termination
The algorithm continues until a termination criterion is met, such as:
-
i.
Stop
$$if:t \ge T$$(7)
Proposed hybrid RMO-NN algorithm
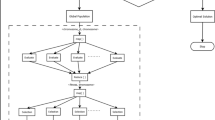
The Proposed Hybrid RMO-NN algorithm represents a synergistic integration of the Ropalidia Marginata Optimization (RMO) algorithm with a neural network (NN) to improve the learning efficiency and predictive accuracy of the NN. In this hybrid model, the RMO algorithm is utilized to optimize the weights and biases of the neural network, addressing common challenges such as local minima and slow convergence associated with traditional gradient-based training methods. By leveraging the adaptive role-switching behavior inspired by Ropalidia Marginata wasps, the RMO algorithm enhances the global search capability of the training process. This leads to more robust optimization, ultimately resulting in improved generalization performance of the neural network across various complex tasks. RMO is used to search for optimal NN parameters (weights, biases) in the solution space. The neural network computes outputs using forward propagation with standard NN equations:
Hybrid RMO-NN algorithm: mathematical model
-
1.
ANN as objective function
The neural network forward pass is computed as:
Where \(X\) is the input nodes, \(W,{ }b\) is the weight bias value and \({\varvec{\sigma}}\) is the activation function to the ANN model. The output of the network is:
The fitness function (loss to minimize) is:
where:
\(f\left( \theta \right):\) Mean squared error (MSE).
where \(\theta\) is optimized by RMO as per (13)-(18).
Randomly initialize the positions of agents (wasps) to create an initial population representing potential solutions (wasps). Assign each agent a random position within the search space and evaluate their fitness using the problem’s objective function as defined in Eq. (1). Ans compute the fitness for each agent:
Exploiter update (local search)
\(If Rolei = queen\) The agent refines its position:
where:
-
\(t\) is the current iteration,
-
\(\alpha\) controls the step size toward the best solution,
-
\(\epsilon \sim Uniform(-\delta ,\delta )\) is a small random perturbation for local search.
Explorer update (global search)
\(If Rolei = worker\) The agent explores the global space:
where: \(\beta\) controls the exploration step size, and \(rand\left( { - 1,1} \right)\) is a random value in [− 1,1].
Evaluate the fitness of all agents:
Update the best solution:
Dynamically reassigned roles: Agents with fitness close to \(f\left({X}_{best}\right)\) become queen. And the remaining agents become worker. The algorithm continues until a termination criterion is met, such as:
The Key Steps in the RMO-NN is shown in Algorithm 2. Figure 3. Give the proposed RMO-NN algorithm Flowchart.
Computational complexity
Let \({\varvec{N}}\) denote the population size (number of agents), \({\varvec{T}}\) the maximum number of iterations, \({\varvec{D}}\) the total number of trainable parameters in the neural network (weights + biases),\(n\) the number of training samples, and \(L\) the number of layers in the network. The dominant cost of the Hybrid RMO-NN algorithm arises during the optimization process. In the initialization phase, the population is generated in \(O(N \cdot D\)) time, and the initial fitness of all agents is computed via forward propagation at a cost of \(O\left( {N \cdot n \cdot \cos t_{NN} } \right),\) where \(\cos t_{NN} \approx \mathop \sum \limits_{l = 1}^{L} \left( {m_{l.1} .m_{l} } \right)\) for fully connected layers. During each iteration, the role-based position updates (queen and worker) require \(O\left( {N \cdot D} \right)\) time, followed by a forward pass for each agent, costing \(O\left( {N \cdot n \cdot \cos t_{NN} } \right).\) Additional operations such as finding the best agent or reassigning roles \(O\left( {N\log N} \right)\) are negligible compared to the forward pass cost. Overall, the time complexity of the Hybrid RMO-NN is:
The space complexity is \(O\left( {N \cdot D} \right)\) due to the storage of all candidate solutions in the population. This linear dependence on \(N,\) \(T,\) and \(n\) implies that scalability is primarily constrained by the network size and dataset volume, making parallelization strategies beneficial for large-scale applications.
Data collection
Data collection plays a crucial role in this research, ensuring the accuracy, reliability, and relevance of the information aligned with the study’s objectives. Effective data collection methods enhance the validity of the findings by enabling well-informed and meaningful conclusions. In this study, three benchmark classification datasets are used for simulation purposes. These datasets, are obtained from the UCI Machine Learning Repository, include Breast Cancer57, Pima Indian Diabetes58 and Blood Transfusion dataset59. The Breast Cancer dataset was created by William H. Wolberg from the microscopic examination of breast tissue samples to diagnose breast cancer57. This dataset aims to classify tumors as either benign or malignant based on continuous clinical attributes. It contains 699 instances, each with 9 attributes and a binary output class (benign or malignant). The Pima Indian diabetes dataset aims to predict the onset of diabetes in Pima Indian women based on diagnostic measurements. This dataset includes 768 instances with 8 input attributes and 2 output classes. The blood transfusion dataset goal is to predict whether a blood donor will donate blood in March 2007 based on their past donation history or not. It includes 748 instances with 4 input features and 2 output classes. Table 1. Give the used datasets descriptions in this paper.
Data preprocessing
To further validated the performance the proposed model three more images datasets such as oral cancer and two breast cancer images datasets are proposed in this paper. Preprocessing was applied to all input images to enhance the consistency of feature extraction and improve classification performance. The original dataset images, with dimensions of 1100 × 825, were resized to 256 × 256 to match the input layer requirements of the CNN used for feature extraction. Given the limited size of the dataset, data augmentation was employed to expand the number of training samples. Various transformation operations were applied to generate new images and enrich the dataset. For experimentation, a publicly available dataset from the Kaggle repository “Available: https://www.kaggle.com/code/ shivam17299/oral-cancer-lips-and-tongue-images-dataset/data”60 was used to evaluate the proposed model for oral cancer classification. This dataset consists of 131 images of the mouth and tongue, categorized into 87 cancerous and 44 non-cancerous samples. Figure 4 shows representative examples from this dataset. After applying data augmentation techniques61,62 the dataset size increased to 1,310 images. The images were divided into training and testing sets: with 90:10. And for the Breast Histopathology Images in this study, we employed two publicly available datasets from the Kaggle repository to evaluate the proposed model. This dataset contains 277,524 image patches derived from 162 whole-slide breast cancer images at 40 × magnification. Out of these, 198,738 patches are IDC-negative and 78,786 patches are IDC-positive used in 70:30% for training and testing shown in Fig. 5. Each patch is labeled with a patient ID, origin coordinates, and class designation. This work utilizes the DDSM Mammography dataset, which is publicly available on Kaggle at https://www.kaggle.com/datasets/skooch/ddsm-mammography. The images from the DDSM database were preprocessed and then supplied to different classifiers for breast cancer recognition. The DDSM dataset contains 1,950 images in total, distributed equally across three categories: 650 normal, 650 benign, and 650 malignant. Preprocessing was applied to eliminate background noise and enhance the contrast between cancer cells and surrounding tissue, which helps in localizing the region of interest (ROI). Figure 6 shows representative examples from this dataset.
Feature extraction
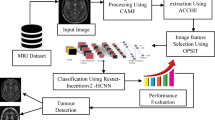
To extract the most relevant features from the input images, a convolutional neural network (CNN) model was employed60,63. In CNN architecture, the block serves as the fundamental unit, and multiple blocks are combined to form a complete cellular structure. The network design is typically organized by factorizing it into cells, which are further divided into blocks, thus defining the search space. There is no universal standard for the size or composition of these cells and blocks; instead, they are tailored to the specific characteristics of the dataset. A block may include operations such as convolution, separable convolution, max pooling, average pooling, and identity mapping. In essence, the block transforms a pair of inputs into a feature map through element-wise operations. For example, if a block of size \(H \times W\) is processed by a cell with a stride of 1, the resulting feature map will maintain the same dimensions \((H \times W).\) However, when the stride is increased to 2, the spatial dimensions of the output are reduced by half. In this study, the CNN architecture was selected based on the proposed optimization algorithm (described in the following section). The general CNN structure used for feature extraction is illustrated in Fig. 7.
Convolutional neural network (CNN) Architecture used in feature extraction63.
In proposed methodology in this work, medical image datasets were collected from the Kaggle repository, including oral cancer images of the mouth and tongue, as well as breast cancer mammography scans. The datasets contained both cancerous and non-cancerous samples, which served as the input for the proposed system. Before classification, preprocessing was applied to the images to prepare them for analysis. This involved resizing all images to a fixed dimension (256 × 256) so that they could fit the input layer of the neural network, as well as applying data augmentation techniques such as rotation, flipping, and scaling to increase the number of samples and improve the model’s robustness. The preprocessed images were then passed through a convolutional neural network (CNN), which automatically extracted meaningful features such as textures, shapes, and patterns that distinguish cancerous from non-cancerous samples. These extracted features were subsequently fed into an artificial neural network (ANN) for final classification, with the ANN’s parameters optimized using the RMO algorithm to enhance accuracy and generalization. Finally, the performance of the system was evaluated using several metrics, including accuracy, precision, recall, and F-score, ensuring a comprehensive assessment of the model’s ability to detect and classify cancer. Figure 8 illustrates the architecture of the proposed methodology used in the paper for medical images dataset.
Performance parameters
To evaluate the performance and accuracy of the proposed model, accuracy, Mean Squared Error (MSE) and Standard Deviation (SD) are employed to compare and validate the results. The generalization accuracy of each simulation run across all algorithms is calculated and expressed as a percentage relative to the defined range limits. The formulas for MSE, SD, and accuracy are provided below.
Results and simulation
This section presents the evaluation of the proposed algorithms RMONN, against the CSNN, LM, ERN and ABCNN. The performance of these algorithms is assessed in terms of Mean Squared Error (MSE), standard deviation (SD), and average accuracy across. The simulation results and comparative analyses of the proposed algorithms are presented in the subsequent section. The hyperparameter settings used for all algorithms in this research are given in Table 2.
Performance analysis of WBC classification problem
The data set used in this study contains information for classifying tumors as either benign or malignant, based on selected continuous variables. Seven input features and two output classes were chosen, with a total of 699 cases analyzed. The input attributes for each case include single epithelial cell size, uniformity of cell size, frequency of bare nuclei, clump thickness, cell shape, bland chromatin, number of marginal adhesions, normal nuclei, and mitosis. For this classification problem, the neural network architecture employed consists of 9 input nodes, 5 hidden nodes, and 2 output nodes, with target error of 0.00001 and total of 1000 number of iterations was set for the model.
Table 3 presents a comparative evaluation of the proposed models used in this paper with various machine learning algorithms applied to breast cancer classification, using three key metrics: accuracy, MSE, and SD. These metrics reflect not only the correctness of predictions but also the reliability and stability of the models across multiple runs. Among the earlier models in the literature, the Deep Neural Network with Restricted Boltzmann Machine (DNN-RBM) achieved a high accuracy of 98.24%, although the MSE and SD values were not reported. This performance is comparable to that of the Elam recurrent Neural (ERN), which attained 98.00% accuracy with a low MSE of 0.0140 and SD of 0.0130, indicating both high precision and consistency. The Levenberg–Marquardt (LM) model also demonstrated strong results with 95.20% accuracy and a relatively low error rate MSE of 0.0280, though its SD of 0.0142 suggests slightly more variation. Other models such as CSNN achievd an accuracy of 91.61%, with MSE of 0.0626, and ABCNN reached accuracy upto 85.31%, with MSE of 0.1080 showed moderate performance, whereas models like ABC-BP had lower consistency, evident from its high SD (0.459) despite a decent accuracy of 92.02%. The ABC-LM model, in contrast, provided a solid balance between accuracy (93.83%) and extremely low MSE (0.0139) and SD (0.0010), making it more stable and dependable.
An exceptional model in terms of precision was CSBPERN, which reported 97.37% accuracy with an incredibly low MSE (0.00072) and SD (0.0004), indicating almost perfect prediction accuracy with minimal variability. On the other hand, models like PSO-MLP, GSA-MLP, and ICA-MLP consistently underperformed, each recording only 80% accuracy and relatively high MSE values around 0.179–0.190, rendering them less suitable for critical diagnostic tasks. Among the models evaluated, bSCWDTO-KNN achieved the highest classification accuracy of 97.64%, accompanied by relatively low error values of MSE 0.369, SD of 0.2763. This indicates that the hybrid approach leveraging the Weighted Differential Tuned Optimization (WDTO) framework is particularly effective in enhancing KNN’s decision boundaries. Similarly, bGA-KNN and bSBO-KNN also demonstrated strong performance with accuracies of 96.12% and 95.43%, respectively, suggesting that evolutionary strategies like Genetic Algorithm (GA) and SBO are capable of significantly improving classification outcomes.
On the other hand, the bGWDTO-KNN model shows two contrasting outcomes. In the first case, it reports an accuracy of 95.23% with minimal error values of MSE 0.245, SD of 0.1365, reflecting stable convergence and effective error minimization. However, in another reported scenario, the same method yields a substantially lower accuracy of 71.64%, accompanied by much higher error levels MSE of 0.5811, SD of 0.40078. The proposed RMO-based models such as RMONN, RMOBPERN, RMOLMBP, and RMOLM surpassed most existing approaches. RMONN achieved the highest accuracy of 98.60%, with low MSE of 0.0184 and SD of 0.0022, demonstrating both excellent predictive power and consistency. Similarly, RMOBPERN matched this accuracy, though with a slightly higher MSE of 0.042, but showed outstanding stability with a minimal SD 0.0001. RMOLMBP and RMOLM also exhibited high accuracy of 97.20% and 96.50%, respectively, along with low MSE and SD values, underscoring the robustness of the RMO-based architectures. In conclusion, the analysis clearly highlights the superiority of RMO-enhanced neural networks for breast cancer risk detection, combining high accuracy, low error rates, and strong consistency qualities essential for dependable medical diagnostic systems. Figure 9 shows the graphically representation of accuracy, MSE and SD convergence performance on breast cancer classification Problem.
Performance analysis of Pima Indian diabetes classification problem
Performance analysis on diabetes classification problem
The PIMA dataset, formally known as the Pima Indians Diabetes Database (PIDD), is a benchmark medical dataset widely used in the field of machine learning for predicting the onset of diabetes. It originates from a study conducted by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and focuses specifically on female patients of Pima Indian heritage aged 21 and older. The dataset contains 768 records, each described by eight clinical attributes that are risk factors for diabetes, including the number of pregnancies, plasma glucose concentration, blood pressure, skinfold thickness, serum insulin levels, body mass index (BMI), diabetes pedigree function, and age. The target variable indicates whether or not a patient was diagnosed with diabetes (1 = diabetic, 0 = non-diabetic). Due to its well-structured format and medical relevance, the PIMA dataset has become a standard benchmark for evaluating the performance of various classification algorithms, particularly in diabetes prediction and health analytics research. Its relatively small size and moderate complexity make it especially suitable for testing both traditional machine learning models and advanced optimization-based approaches.
Table 4 presents a comparative evaluation of the proposed RMO-based neural network models against a wide range of existing neural network architectures, machine learning models, and bio-inspired optimization-based KNN classifiers across different studies. In the earlier studies65, baseline neural architectures such as ABCNN, CSNN, ERN, and LM demonstrated moderate performance, with accuracies ranging from 65.09% to 73.87%. Among these, CSNN achieved the highest accuracy of 73.87% and relatively low MSE of 0.1505), indicating stronger predictive ability compared to ABCNN and ERN. The hybrid ABC-LM achieved only 65.09% accuracy, though it recorded one of the lowest MSE values of 0.14, suggesting some stability despite limited correctness. Hybrid feature selection and ensemble approaches from69 further improved results, with BMNABC + ODF achieving the best performance at 77.21% accuracy, followed closely by BMNABC + NB of 76.43% and BMNABC + C4.5 of 76.17%. These models demonstrate that integrating feature selection with conventional classifiers provides tangible performance gains. Similarly, the use of PCA + Naïve Bayes70 reached 79.13% accuracy, showcasing the effectiveness of dimensionality reduction in strengthening traditional classifiers. More advanced temporal learning approaches, such as Mean Imputation + LSTM69, achieved 85.00% accuracy, highlighting the advantages of recurrent neural networks in handling missing values and sequential patterns. By contrast, Mean Imputation + RB-Bayes71 and Mean Imputation + NB72 reported lower accuracies of 72.90% and 76.30%, respectively. Metaheuristic-optimized neural networks19 also provided notable improvements over conventional baselines. CAPSO-MLP achieved 74.68% accuracy with relatively low error MSE of 0.204, outperforming PSO-MLP 74.03% and ICA-MLP 66.23%. However, GSA-MLP underperformed with 56.49% accuracy, reflecting weak generalization and poor optimization capacity. Further evaluation of bio-inspired KNN models66 revealed modest accuracies 60–66%, with bGWO_GA-KNN achieving the highest at 65.74%, alongside an MSE of 3.426 and SD of 0.2655. Other methods such as bSCWDTO-KNN achieved accuracy of 65.00% and bGWO-KNN of 65.09% also performed competitively, though optimization methods like bMVO-KNN of 60.03% and bWOA-KNN of 61.67% lagged behind, indicating reduced adaptability. A more significant advancement was observed in67, where bGWDTO-KNN achieved 87.23% accuracy with a notably low error MSE of 0.256, and SD of 0.1475. However, the same model performed less consistently in68, where accuracy dropped to 75.64% with higher error margins MSE of 0.5825, and SD of 0.4407, pointing to dataset sensitivity and variability in optimization efficiency. In contrast, the proposed RMO-based models demonstrated superior performance across all evaluation criteria. RMONN achieved an impressive accuracy of 88.38%, with a low MSE of 0.1235 and SD of 0.0313, reflecting both predictive strength and stability. RMOBPERN also provided strong results at 70.31% accuracy, though with slightly higher error values MSE of 0.3073. Most notably, RMOLMBP delivered the highest accuracy overall at 97.20%, with extremely low error metrics MSE of 0.049, and SD of 0.0011, confirming its robustness and reliability in classification tasks. Even RMOLM, despite achieving a moderate accuracy of 71.88%, demonstrated improved error minimization compared to several traditional baselines. In summary, while prior models including hybrid metaheuristic-based KNN classifiers, optimized MLPs, and ensemble learning approaches provided incremental improvements over conventional classifiers, the RMONN architecture clearly outperformed all existing benchmarks. The results establish that the integration of robust optimization with neural network models significantly enhances classification accuracy, minimizes error, and ensures stable convergence. Thus, the proposed frameworks represent a state-of-the-art advancement for medical data classification, offering reliable and interpretable solutions for real-world clinical applications. Figure 10 graphically represent the accuracy, MSE and SD convergence performance on Diabetes classification problem.
Performance analysis of the blood transfusion dataset classification problem
The Blood Transfusion dataset, sourced from the UCI Machine Learning Repository, comprises data aimed at predicting whether a blood donor will donate blood in March 2007 based on their historical donation behavior. This dataset includes 748 instances, each characterized by four input features and classified into two output classes. The input attributes are: Recency (number of months since the last donation), Frequency (total number of donations made), Monetary (total volume of blood donated in cubic centimeters), and Time (number of months since the first donation). For the classification task, neural network architecture is commonly employed, typically consisting of four input nodes corresponding to the features, a variable number of hidden nodes (usually ranging from 5 to 10 depending on the specific algorithm or tuning strategy), and two output nodes representing the binary classification outcome. During training, a target error threshold is defined to guide the convergence of the model, ensuring that learning continues until the desired accuracy or minimum error is achieved. This setup supports effective modeling of donor behavior and helps in developing predictive systems for blood donation programs.
Table 5 presents a systematic performance analysis of various machine learning models applied to the Blood Transfusion Dataset classification problem, using three key metrics: accuracy, MSE, and SD. These metrics collectively evaluate each model’s predictive power, error rate, and stability across multiple runs. Among the earlier models, ABCNN delivered the highest accuracy at 70.54%, with a low MSE of 0.1758 and a moderate SD of 0.0244, indicating a balanced trade-off between performance and consistency. The CSNN model closely followed, achieving 76.47% accuracy with a comparable MSE of 0.1760 and a slightly higher SD of 0.0425, suggesting marginally greater variability in predictions. In contrast, the LM and ERN models showed relatively lower performance, with accuracies of 74.87% and 74.33% respectively. Additionally, both had higher MSE values 0.2513 for LM and 0.2567 for ERN indicating greater prediction errors, despite their low standard deviations (0.0169 for LM and 0.0125 for ERN), which reflect stable but less accurate outputs. The proposed RMONN significantly outperformed the traditional approaches, achieving a much higher accuracy of 76.98%, a lower MSE of 0.15, and a comparatively higher SD of 0.1144. This result demonstrates superior predictive capability and reduced error, though with a slightly increased variability, possibly due to the model’s sensitivity to complex data patterns. In conclusion, RMONN proves to be the most effective model for the blood transfusion classification task, combining high accuracy and low error, making it a promising candidate for real-world donor prediction systems, despite a modest increase in standard deviation compared to traditional models. Figure 11 described the accuracy, MSE, SD convergence performance on Blood Transfusion classification dataset.
Statistical significance analysis
To verify the performance improvements of the proposed algorithms both t-test and the Wilcoxon signed-rank test at a significance level of α = 0.05 are applied. As summarized in Table 6, the results demonstrate that all proposed models achieved statistically significant improvements over the baselines CSNN and ABCNN, with both tests yielding extremely small p-values (p < 0.001). This confirms that the observed performance gap is highly robust and unlikely to be due to chance. Against stronger baselines such as CSBPERN and DNN-RBM, the proposed models also achieved competitive performance, with RMONN and RMOBPERN consistently showing statistically significant improvements (p < 0.01 across both tests). In contrast, RMOLMBP and RMOLM achieved strong gains over CSNN and ABCNN but showed only marginal significance in some comparisons with CSBPERN and DNN-RBM (e.g., RMOLMBP vs. CSBPERN, Wilcoxon p = 0.050; RMOLM vs. DNN-RBM, Wilcoxon p = 0.0078). These findings highlight that RMONN and RMOBPERN are the most robust performers, while RMOLMBP and RMOLM remain competitive but less consistently superior to the strongest baselines. The combination of parametric (t-test) and non-parametric (Wilcoxon) analyses strengthens the validity of these results by accounting for both normality and non-normality in the data distribution, ensuring that the observed improvements are both reliable and statistically sound.
The statistical analysis in Table 7 confirms the superiority of the proposed models over the strongest baselines. Both parametric (t-test) and non-parametric (Wilcoxon signed-rank test) analyses at α = 0.05 demonstrate that RMONN (88.38%) and RMOLMBP (97.20%) achieved highly significant improvements across all baseline comparisons (p < 0.001), indicating that their performance gains are robust and unlikely to be due to chance. In particular, RMOLMBP provided the highest and most consistent improvements, with extremely small p-values across tests, establishing it as the most reliable proposed model. In contrast, RMOBPERN (70.31%) and RMOLM (71.88%) underperformed compared to the baselines, with statistical tests confirming significant disadvantages (p < 0.05). These results highlight that while RMONN and RMOLMBP offer statistically validated performance advantages, RMOBPERN and RMOLM remain less competitive. The combined use of t-test and Wilcoxon test strengthens the validity of these conclusions by ensuring robustness to different data distribution assumptions.
Table 8 presents the computational time (in seconds) required by different classification algorithms when applied to three benchmark datasets: Breast Cancer, Diabetes, and Blood Transfusion. Computational time is a critical evaluation metric in machine learning as it reflects the efficiency and scalability of an algorithm in real-world applications. The computational time analysis across three datasets highlights significant variations among the proposed algorithms. Lightweight models such as LM, RM0NN, and RNN consistently achieve the shortest execution times, making them more suitable for real-time or resource-limited applications. In contrast, computationally heavy models like RMOBPERN and RMOBPLM require considerably longer processing time, which may limit their practicality despite potential accuracy benefits. Algorithms such as ABCNN, CSNN, and RMOLM fall in the moderate range, balancing efficiency with complexity. Overall, LM and RM0NN emerge as the most computationally efficient, while the heavier models demand careful consideration depending on the application context.
Table 9 provides a comparative analysis of four proposed algorithms RMONN, RMOLM, RMOLMBP, and RMOBPERN based on their average classification accuracy and MSE under a tenfold cross-validation scheme across three datasets such as Breast Cancer, Diabetes, and Blood Transfusion. Accuracy quantifies predictive performance, while MSE evaluates error magnitude, where lower values indicate higher reliability. For the Breast Cancer dataset, all algorithms demonstrate high performance, with RMOBPERN achieving 97.72% highest accuracy and RMOLMBP showing 0.000013 the lowest MSE, confirming strong generalization. On the Diabetes dataset, accuracies are relatively moderate, where RMOLM attains 76.82% the highest accuracy, while RMOLMBP provides the 0.121630 lowest error rate, indicating efficient error minimization. For the Blood Transfusion dataset, RMOBPERN delivers 79.81% accuracy, whereas RMOLMBP again secures 0.136699 the lowest MSE, highlighting its stability. In summary, the results from tenfold cross-validation suggest a performance trade-off: RMOBPERN consistently outperforms in terms of classification accuracy, whereas RMOLMBP excels in minimizing errors through lower MSE values. This indicates that model selection should depend on whether the application prioritizes predictive accuracy or error minimization for optimal decision-making.
Performance analysis on oral cancer classification problem
Table 10 presents a comprehensive performance comparison between the proposed RMONN model and several established deep learning and machine learning approaches for oral cancer classification. The evaluation metrics considered include Accuracy, Sensitivity, Specificity, Precision, and F-score, which together provide a holistic assessment of model robustness. From the deep learning models, AlexNet achieved accuracy of 0.945, F-score of 0.957, ResNet-50 reached accuracy of 0.944, F-score of 0.957, and VGGNet have accuracy of 0.940, F-score 0.957 demonstrate relatively strong predictive performance, with particularly high sensitivity of 0.984, indicating their effectiveness in correctly identifying positive oral cancer cases. However, these models exhibit comparatively lower specificity values ranging from 0.845 to 0.879, suggesting some limitations in accurately distinguishing non-cancerous cases. The DBN model achieves a balanced performance with Accuracy of 0.935 and an F-score of 0.953, benefiting from both relatively high sensitivity of 0.943, and specificity of 0.917. Among traditional machine learning methods, SVM-Linear achieved accuracy of 0.912, F-score of 0.924 and K-NN has accuracy of 0.888, F-score of 0.880 show moderate classification capability, while LD reached accuracy of 0.874, F-score of 0.841 and DT attained accuracy of 0.860, F-score of 0.786 lag behind in predictive performance, primarily due to reduced sensitivity. When integrated with the proposed optimization techniques (PSOBER), machine learning models show mixed outcomes. For example, PSOBER-SVM and PSOBER-K-NN yield accuracy values of 0.9315 and 0.9298, respectively, with improved specificity of 0.94), but they suffer from substantially reduced sensitivity less than 0.67, resulting in low F-scores of 0.4444 and 0.6000. This trade-off indicates that while the PSOBER optimization enhances the models’ ability to reject false positives, it compromises their ability to detect true positive cancer cases. In contrast, the proposed RMONN model outperforms all competing approaches, achieving the highest overall accuracy of 0.9653, alongside strong sensitivity of 0.871 and specificity of 0.976. Notably, RMONN also demonstrates superior precision of 0.962 and an F-score of 0.9511, indicating a more balanced trade-off between detecting true positives and minimizing false positives. This demonstrates the robustness of the proposed method in both early detection of oral cancer and reduction of misclassifications, establishing it as a superior alternative to existing deep learning and machine learning classifiers. Figure 12. illustrating the confusion matrix classification performance of the proposed RMONN model on the oral cancer test dataset. Figure 13.
Performance analysis histopathology images testing data classification problem
Table 11 presents a comparative evaluation of different deep learning and machine learning models for medical image classification. The baseline models, such as VGGNet, LeNet, and CNN, demonstrate moderate performance, with VGGNet achieving the highest accuracy among them the 0.8598 and a relatively low loss of 0.3531. However, LeNet and CNN show lower accuracy values of 0.7545 and 0.7232, respectively, accompanied by higher loss values, indicating weaker generalization ability. More advanced architectures reported in the literature, including GRU, VGG16, and ResNet50, show notable improvements, achieving accuracy values above 0.91. These models also exhibit strong sensitivity and specificity, reflecting balanced performance in identifying both positive and negative cases. In contrast, the proposed RMONN model outperforms all compared approaches across nearly all evaluation metrics. It achieves the highest accuracy of 0.9734, with superior precision of 0.9511, recall of 0.9325, and F-score of 0.9722, alongside robust sensitivity of 0.9601 and specificity of 0.9561. Additionally, RMONN reports the lowest loss of 0.0234, highlighting its effectiveness and stability during training. In conclusion, while existing deep learning architectures provide competitive results, the proposed RMONN demonstrates a substantial performance gain, establishing it as a more reliable and accurate model for medical image classification tasks. Figure 14. illustrating the confusion matrix classification performance of the proposed RMONN model on Breast Histopathology Images Testing Data. Similarly Figs. 15 and 16 show the accuracy and loss comparison on Breast Histopathology Images on testing Data.
Performance analysis of DDSM mammography breast cancer classification problem
Table 12 compares the performance of various algorithms across learning rates LR of 0.01 and optimization methods such as Adam, RMSprop, and SGDM. Performance is assessed using accuracy and loss values. For Google Net, the Adam optimizer achieves the best accuracy of 0.9009 with a relatively low loss of 0.2195. However, its performance deteriorates under RMSprop accuracy is 0.7541, and loss of 0.8687. And further declines with SGDM accuracy of 0.7219, and loss of 1.7342, indicating optimizer sensitivity. Similarly, AlexNet exhibits strong performance across optimizers. With Adam, it achieves an accuracy of 0.9106, while RMSprop further improves accuracy to 0.9294, although with higher loss of 1.3740. Under SGDM, accuracy falls to 0.7549, with significantly increased loss of 1.7325. This demonstrates that AlexNet benefits most from RMSprop in terms of accuracy, albeit at the expense of stability in loss. The PSO-MLP model achieves accuracy of 0.9021 with Adam and the lowest loss of 0.1972 among all compared methods, suggesting efficient learning with this optimizer. Similarly, ACO-MLP reaches accuracy of 0.8614 and loss of 0.2234 using Adam, though no results are reported for RMSprop or SGDM. In contrast, the proposed RMONN model consistently delivers superior performance across all optimization strategies. With Adam, it achieves the highest accuracy 0.9215 and the lowest loss 0.1135 across all models and methods. RMSprop also provides strong results accuracy of 0.9124, and loss of 0.2113, while SGDM yields slightly lower performance accuracy of 0.8614, and loss of 0.2135 but still outperforms Google Net and AlexNet under the same conditions. Figure 17. illustrating the confusion matrix classification performance of the proposed RMONN on DDSM Mammography breast cancer classification testing data. Similarly Figs. 18 and 19 give accuracy and Loss Comparison on DDSM Mammography breast cancer Testing Data.
Conclusions and future work
Despite advancements, achieving consistent performance across datasets while balancing accuracy and computational cost remains challenging. Hybrid ML systems often lack adaptability, require extensive tuning, and struggle to generalize in biomedical domains. ANN optimization covering topology, weights, and hyperparameters remains complex, with traditional trial-and-error methods proving inefficient. Recent work explores evolutionary swarm intelligence, and optimization methods, to enhance classification accuracy. However, persistent challenges such as high-dimensional feature spaces, premature convergence to local optima, and increased computational demands continue to hinder the full potential of ML applications in clinical settings. To address these limitations, this study introduces a novel model Ropalidia Marginata Optimization-based Neural Network (RMO-NN) inspired by the hierarchical dominance and decentralized task management observed in Ropalidia marginata wasps. This biologically inspired hybrid approach integrates a swarm intelligence optimization algorithm with neural network learning, aiming to improve classification performance by accelerating convergence, minimizing errors, and eliminating irrelevant or redundant features. The RMO-NN model was rigorously tested on three standard medical datasets: breast cancer, diabetes, and blood transfusion. And further its performance is validated on three more medical images datasets. Its performance was benchmarked against well-established metaheuristic models, including CSNN and ABCNN and some state-of-the-art deep learning models in literature. Results revealed that RMO-NN consistently outperformed its counterparts across all datasets. Specifically, it achieved an outstanding 98.60% accuracy with MSE of 0.0184 for breast cancer classification, 78.38% accuracy MSE of 0.1535 for diabetes prediction, and 77.54% accuracy with MSE of 0.15 for the blood transfusion dataset. Additionally, the RMO-NN demonstrated improved model stability through lower standard deviation (SD) values and faster convergence rates, underscoring its reliability and generalization capability in medical diagnostics. Although the proposed RMO-NN framework achieves competitive performance, including a 98.60% accuracy for breast cancer classification. Several limitations should be acknowledged. First, the evaluation was conducted on relatively small, structured benchmark datasets, and the generalization ability to large-scale, high-dimensional, or unstructured data (e.g., medical imaging, wearable sensor data) remains to be verified. Second, while the RMO algorithm effectively tunes neural network weights and biases, it can be computationally more demanding than gradient-based optimizers, particularly for deep architecture or real-time applications. Third, the approach’s performance is sensitive to RMO hyperparameter settings, yet no systematic sensitivity analysis was performed in this study. Furthermore, the current model does not incorporate explainability or domain-specific interpretability, which is essential for decision-making in high-stakes fields such as healthcare. Finally, external validation on independent datasets and evaluation in real-world clinical environments are necessary to fully establish robustness and practical applicability.
Future work will focus on extending the RMO-NN framework to larger and more complex datasets, optimizing computational efficiency, integrating explainable AI (XAI) methods, and conducting prospective clinical studies to assess real-world performance. Additionally, adaptive hyperparameter tuning and hybrid optimization strategies will be explored to enhance both training speed and generalization capability. To extend the impact and applicability of RMO-NN, the following research work are proposed:
-
Extension to deep learning architectures: applying the RMO optimization mechanism to more complex neural frameworks such as deep neural networks (DNNs) and convolutional neural networks (CNNs) could enhance performance in medical imaging tasks like tumor detection in MRI or X-rays.
-
Multi-modal and heterogeneous data integration:expanding the model to handle a variety of data types including genetic, imaging, and electronic health records to enable a more holistic and personalized diagnostic approach.
-
Application to time-series medical data: extending RMO-NN to handle sequential and temporal patterns in data such as ECG, EEG, and continuous vital sign monitoring.
-
Real-time diagnostic integration: embedding RMO-NN into real-time clinical decision support systems, with emphasis on optimizing inference speed and system reliability in live hospital environments.
-
Comparative studies on imbalanced data: conducting rigorous benchmarking against competing algorithms on datasets with varying imbalance ratios, including rare disease detection scenarios, to evaluate robustness and bias mitigation capabilities.
-
To further extend our study to include real-world datasets such as hospital records, datasets covering multi-class conditions, and those with imbalanced class distributions, to better assess the model’s performance under the complexities and challenges typical of real-world clinical scenarios.
Data availability
In this study, three benchmark classification datasets are used for simulation purposes. These datasets, are obtained from the UCI Machine Learning Repository. https://archive.ics.uci.edu/
References
Alzubaidi, L. et al. Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. J. Big Data 8, 1–74 (2021).
Esteva, A. et al. A guide to deep learning in healthcare. Nat. Med. 25(1), 24–29 (2019).
Yu, Y., Li, M., Liu, L., Li, Y. & Wang, J. Clinical big data and deep learning: Applications, challenges, and future outlooks. Big Data Mining Anal 2(4), 288–305 (2019).
Atteia, G. et al. Adaptive dynamic dipper throated optimization for feature selection in medical data. Comput. Mater. Continua. 75(1), 1883–1900 (2023).
M. H. Mohamed et al., Towards an Accurate Liver Disease Prediction Based on Two-level Ensemble Stacking Model, 2024.
Avanzo, M. et al. Artificial intelligence applications in medical imaging: A review of the medical physics research in Italy. Physica Med. 83, 221–241 (2021).
Bacha, S. & Taouali, O. A novel machine learning approach for breast cancer diagnosis. Measurement 187, 110233 (2022).
R. A. Smith et al., Cancer screening in the United States, 2017: a review of current American Cancer Society guidelines and current issues in cancer screening, vol. 67, no. 2, pp. 100–121, 2017.
Zheng, B., Yoon, S. W. & Lam, S. S. Breast cancer diagnosis based on feature extraction using a hybrid of K-means and support vector machine algorithms. Expert Syst. Appl. 41(4), 1476–1482 (2014).
Wan K, Zunaidi I., An efficient data mining approaches for breast cancer detection and segmentation in Mammogram, 2017.
Nandagopal, V., Geeitha, S., Kumar, K. V. & Anbarasi, J. Feasible analysis of gene expression–A computational based classification for breast cancer. Measurement 140, 120–125 (2019).
Thawkar, S. & Ingolikar, R. Classification of masses in digital mammograms using firefly based optimization. Int. J. Image Gr. Signal Process. 14(2), 25 (2018).
Vijayarajeswari, R., Parthasarathy, P., Vivekanandan, S. & Basha, A. A. Classification of mammogram for early detection of breast cancer using SVM classifier and Hough transform. Measurement 1(146), 800–805 (2019).
Abdar, M. & Makarenkov, V. CWV-BANN-SVM ensemble learning classifier for an accurate diagnosis of breast cancer. Measurement 1(146), 557–570 (2019).
Mishra, K. N., Mishra, A., Ray, S., Kumari, A. & Waris, S. M. Enhancing cancer detection and prevention mechanisms using advanced machine learning approaches. Inform. Med. Unlocked. 1(50), 101579 (2024).
Bharati S, Podder P, Mondal M., Artificial neural network based breast cancer screening: A comprehensive review, 2020.
Kokabi, M., Tahir, M. N., Singh, D. & Javanmard, M. Advancing healthcare: Synergizing biosensors and machine learning for early cancer diagnosis. Biosensors 13(9), 884 (2023).
Veeranjaneyulu, K., Lakshmi, M. & Janakiraman, S. Swarm intelligent metaheuristic optimization algorithms-based artificial neural network models for breast cancer diagnosis: emerging trends, challenges and future research directions. Arch. Comput. Methods Eng. 32(1), 381–398 (2025).
Beheshti, Z., Shamsuddin, S. M., Beheshti, E. & Yuhaniz, S. S. Enhancement of artificial neural network learning using centripetal accelerated particle swarm optimization for medical diseases diagnosis. Soft Comput. 18(11), 2253–2270 (2014).
Ahmad, F., Mat Isa, N. A., Hussain, Z. & Sulaiman, S. N. A genetic algorithm-based multi-objective optimization of an artificial neural network classifier for breast cancer diagnosis. Neural Comput. Appl. 23(5), 1427–1435 (2013).
Ahmad, F., Mat Isa, N. A., Hussain, Z., Osman, M. K. & Sulaiman, S. N. A GA-based feature selection and parameter optimization of an ANN in diagnosing breast cancer. Pattern Anal. Appl. 18(4), 861–870 (2015).
Stephan, P., Stephan, T., Kannan, R. & Abraham, A. A hybrid artificial bee colony with whale optimization algorithm for improved breast cancer diagnosis. Neural Comput. Appl. 33(20), 13667–13691 (2021).
Menghour, K. & Souici-Meslati, L. Hybrid ACO-PSO based approaches for feature selection. Int. J. Intell. Eng. Syst. 9(3), 65–79 (2016).
A. Zaras, N. Passalis, and A. Tefas, Neural networks and backpropagation, in Deep learning for robot perception and cognition: Elsevier, 2022, pp. 17–34.
Momeni, A., Rahmani, B., Malléjac, M., Del Hougne, P. & Fleury, R. Backpropagation-free training of deep physical neural networks. Science 382(6676), 1297–1303 (2023).
Kaveh, M. & Mesgari, M. S. Application of meta-heuristic algorithms for training neural networks and deep learning architectures: A comprehensive review. Neural Process Lett. 55(4), 4519–4622 (2023).
Kowalski, P. A., Kucharczyk, S. & Mańdziuk, J. Constrained hybrid metaheuristic algorithm for probabilistic neural networks learning. Inf. Sci. 713, 122185 (2025).
Mjahed, O., Hadaj, S. E., Guarmah, E. M. & Mjahed, S. Bio-Inspired hybridization of artificial neural networks for various classification tasks. Stud. Inform. Control. 31(3), 21–30 (2022).
Mohammed, S. J. et al. Application of metaheuristic algorithms and ANN model for univariate water level forecasting. Adv. Civil Eng. 2023(1), 9947603 (2023).
Bhadra, A. et al. Regulation of reproduction in the primitively eusocial wasp Ropalidia marginata: On the trail of the queen pheromone. J. Chem. Ecol. 36, 424–431 (2010).
Gadagkar, R., Sharma, N. & Pinter-Wollman, N. Queen succession in the Indian paper wasp Ropalidia marginata: On the trail of the potential queen. J. Biosci. 47(2), 18 (2022).
Gadagkar, R. The Social Biology of Ropalidia marginata: Toward Understanding the Evolution of Eusociality (Harvard University Press, 2001).
Gadagkar R, Gadgil M, Joshi NV, Mahabal AS., Observations on the natural history and population ecology of the social wasp Ropalidia marginata (Lep.) from peninsular India (Hymenoptera: Vespidae), vol. 91, pp. 539–552, 1982.
R. Gadagkar, Science as a hobby: How and why I came to study the social life of an Indian primitively eusocial wasp, pp. 845–858, 2011.
Almbaidin, H. et al. Application of artificial intelligence in the diagnosis of haematological disorders. Biomed. Res. Therapy 11(12), 6989–7002 (2024).
Cheng, X. A comprehensive study of feature selection techniques in machine learning models. Insights Comput. Signals Syst. 1(1), 10.70088 (2024).
Rajalakshmi, R., Sivakumar, P., Kumari, L. K. & Selvi, M. C. A novel deep learning model for diabetes melliuts prediction in IoT-based healthcare environment with effective feature selection mechanism. J. Supercomput. 80(1), 271–291 (2024).
Edward, J., Rosli, M. M. & Seman, A. A new multi-class rebalancing framework for imbalance medical data. IEEE Access 11, 92857–92874 (2023).
M. H. Mohamed et al., Towards an accurate liver disease prediction based on two-level ensemble stacking model, IEEE Access, 2024.
Sowan, B., Eshtay, M., Dahal, K., Qattous, H. & Zhang, L. Hybrid PSO feature selection-based association classification approach for breast cancer detection. Neural Comput. Appl. 35(7), 5291–5317 (2023).
N. M. Nawi, A. Khan, and M. Z. Rehman, A new back-propagation neural network optimized with cuckoo search algorithm, In Computational Science and Its Applications–ICCSA 2013: 13th International Conference, Ho Chi Minh City, Vietnam, June 24–27, 2013, Proceedings, Part I 13, 2013, pp. 413–426: Springer.
Nayak, J., Naik, B., Dinesh, P., Vakula, K. & Dash, P. B. Firefly algorithm in biomedical and health care: Advances, issues and challenges. SN Comput. Sci. 1(6), 311 (2020).
Ul Haq, A. et al. Recognition of the Parkinson’s disease using a hybrid feature selection approach. J. Intell. Fuzzy Syst. 39(1), 1319–1339 (2020).
N. K. Chauhan and N. Kaur, Cardiovascular disease prediction utilizing machine learning algorithm using grey wolf optimization method, In AIP Conference Proceedings, 2024, vol. 3209, no. 1: AIP Publishing.
Abdulwahab, H. M., Ajitha, S., Saif, M. A. N., Murshed, B. A. H. & Ghanem, F. A. MOBCSA: Multi-objective binary cuckoo search algorithm for features selection in bioinformatics. IEEE Access 12, 21840–21867 (2024).
Elshewey, A. M. et al. Enhancing heart disease classification based on greylag goose optimization algorithm and long short-term memory. Sci. Rep. 15(1), 1277 (2025).
Tarek, Z., Alhussan, A. A., Khafaga, D. S., El-Kenawy, E. S. & Elshewey, A. M. A snake optimization algorithm-based feature selection framework for rapid detection of cardiovascular disease in its early stages. Biomed. Signal Process. Control. 102, 107417 (2025).
Elshewey, A. M., Alhussan, A. A., Khafaga, D. S., Elkenawy, E. S. & Tarek, Z. EEG-based optimization of eye state classification using modified-BER metaheuristic algorithm. Sci. Rep. 14(1), 24489 (2024).
El-Rashidy, N., Tarek, Z., Elshewey, A. M. & Shams, M. Y. Multitask multilayer-prediction model for predicting mechanical ventilation and the associated mortality rate. Neural Comput. Appl. 37(3), 1321–1343 (2025).
Elshewey, A. M. & Osman, A. M. J. S. R. Orthopedic disease classification based on breadth-first search algorithm. Sci. Rep. 14(1), 23368 (2024).
Hassan, E., Saber, A., El-Sappagh, S. & El-Rashidy, N. Optimized ensemble deep learning approach for accurate breast cancer diagnosis using transfer learning and grey wolf optimization. Evol. Syst. 16(2), 59 (2025).
Sen, R. & Gadagkar, R. Males of the social wasp Ropalidia marginata can feed larvae, given an opportunity. Anim. Behav. 71(2), 345–350 (2006).
R. Gadagkar, K. Chandrashekara, S. Chandran, and S. Bhagavan, Serial polygyny in the primitively eusocial wasp Ropalidia marginata: implications for the evolution of sociality, in Queen number and sociality in insects: Oxford University, 1993, pp. 189–214.
Gadagkar, R. Evolution of social behaviour in the primitively eusocial wasp Ropalidia marginata: Do we need to look beyond kin selection?. Philos. Transac. Royal Soc. B Biol. Sci. 371(1687), 20150094 (2016).
Lamba, S. et al. A possible novel function of dominance behaviour in queen-less colonies of the primitively eusocial wasp Ropalidia marginata. Behav. Process. 74(3), 351–356 (2007).
Sen R, Gadagkar R. Natural history and behaviour of the primitively eusocial wasp Ropalidia marginata (Hymenoptera: Vespidae): A comparison of the two sexes. vol. 44, no. 15–16, pp. 959–968, 2010.
Wolberg WH, Mangasarian OL., Multisurface method of pattern separation for medical diagnosis applied to breast cytology, vol. 87, no. 23, pp. 9193–9196, 1990.
Chang, V., Bailey, J., Xu, Q. A. & Sun, Z. Pima Indians diabetes mellitus classification based on machine learning (ML) algorithms. Neural Comput. Appl. 35(22), 16157–16173 (2023).
Huang, Z. et al. Analysis of a large data set to identify predictors of blood transfusion in primary total hip and knee arthroplasty. Transfusion 58(8), 1855–1862 (2018).
Öksüz, C., Urhan, O. & Güllü, M. K. Brain tumor classification using the fused features extracted from expanded tumor region. Biomed. Signal Process. Control. 72, 103356 (2022).
Saber, A., Sakr, M., Abo-Seida, O. M., Keshk, A. & Chen, H. A novel deep-learning model for automatic detection and classification of breast cancer using the transfer-learning technique. IEEe Access 9, 71194–71209 (2021).
Rashid, J. et al. Skin cancer disease detection using transfer learning technique. Appl. Sci. 12(11), 5714 (2022).
Myriam, H. et al. Advanced meta-heuristic algorithm based on Particle Swarm and Al-biruni Earth Radius optimization methods for oral cancer detection. IEEE Access. 11, 23681–23700 (2023).
Khan, A. et al. Optimizing connection weights of functional link neural network using APSO algorithm for medical data classification. J. King Saud Univ. Comput. Inf. Sci. 34(6), 2551–2561 (2022).
Nawi, N. M., Khan, A., Rehman, M. Z., Chiroma, H. & Herawan, T. Weight optimization in recurrent neural networks with hybrid metaheuristic Cuckoo search techniques for data classification. Math. Probl. Eng. 2015(1), 868375 (2015).
Abdelhamid, A. A. et al. Innovative feature selection method based on hybrid sine cosine and dipper throated optimization algorithms. IEEE access 11, 79750–79776 (2023).
R. Alkanhel et al., Network intrusion detection based on feature selection and hybrid metaheuristic optimization, vol. 74, no. 2, 2023.
Atteia, G. et al. Adaptive dynamic dipper throated optimization for feature selection in medical data. Comput. Mater. Continua 75, 1883–1900 (2023).
Mousa, A., Mustafa, W. & Marqas, R. B. A comparative study of diabetes detection using the Pima Indian diabetes database. Methods 26(2), 277–288 (2023).
Rajni, R. & Amandeep, A. Pima Indians diabetes mellitus classification based on machine learning (ML) algorithms. Int. J. Electr. Comput. Eng 35(22), 16157–16173 (2023).
Rajni, R. & Amandeep, A. RB-Bayes algorithm for the prediction of diabetic in Pima Indian dataset. Int. J. Electr. Comput. Eng. 9(6), 4866–4872 (2019).
Sisodia, D. & Sisodia, D. S. Prediction of diabetes using classification algorithms. Procedia Comput. Sci. 132, 1578–1585 (2018).
Khan, A. et al. A computational classification method of breast cancer images using the VGGNet model. Front. Comput. Neurosci. 16, 1001803 (2022).
Kaddes, M., Ayid, Y. M., Elshewey, A. M. & Fouad, Y. J. S. R. Breast cancer classification based on hybrid CNN with LSTM model. Sci. Rep. 15(1), 4409 (2025).
R. Rajakumari, L. J. I. A. Kalaivani, and S. Computing, Breast cancer detection and classification using deep CNN techniques, vol. 32, no. 2, 2022.
Acknowledgements
The authors extend their appreciation to the University of Agriculture Peshawar and Universiti Sains Malaysia for the support of this research.
Funding
The authors extend their appreciation to the Universiti Sains Malaysia for the support of this research under the Ministry of Higher Education Malaysia for Fundamental Research’s Grant Scheme, project code FRGS/1/2024/ICT03/USM/02/1.
Author information
Authors and Affiliations
Contributions
Maria Ali contributed to the original draft preparation and methodology. Abdullah Khan was responsible for supervision and conceptualization. Muhammad Imran conducted validation, while Dzati Athiar Ramli handled the investigation. Javed Iqbal Bangash and Arshad Khan reviewed and edited the manuscript. All authors have read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ali, M., Khan, A., Ramli, D.A. et al. A hybrid bio inspired neural model based on Ropalidia Marginata behavior for multi disease classification. Sci Rep 15, 42004 (2025). https://doi.org/10.1038/s41598-025-26030-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-26030-z