Abstract
Head and neck squamous cell carcinoma (HNSCC) is a highly prevalent malignancy with poor prognosis, largely driven by lymph node metastasis (LNM). Despite its clinical significance, the underlying mechanisms of LNM remain elusive. In this study, we used single-cell transcriptomic data to dissect the cellular and molecular interactions within metastatic lymph nodes (MET). Specifically, we analyzed Single-cell RNA sequencing (scRNA-seq) data from GSE195655, GSE140042, GSE227156, and GSE159929 (n = 41) to delineate cellular heterogeneity, intercellular communication networks, and functional enrichment in primary tumors (PT), MET, and non-metastatic lymph nodes (LN). Our analysis identified a subpopulation of tumor-associated macrophages (TAMs) distinctly enriched in MET, characterized by high expression of SPP1. Functional analysis revealed that this TAM subpopulation promotes angiogenesis through specific ligand-receptor interactions with endothelial cells (ECs), involving the SPP1-ITGa9b1 and FN1-ITGa2b1 signaling axes. Furthermore, we leveraged bulk RNA-seq for prognostic research. Immunohistochemistry (IHC) confirmed the increased density and number of blood and lymphatic vessels in MET. Spatial analysis via multiplex immunohistochemistry (mIHC) confirmed the preferential localization of SPP1+ TAMs near ECs in MET. These results suggest a potential communication between TAMs and ECs that contributes to LNM in HNSCC, providing critical insights for HNSCC prognosis and precision treatment.
Similar content being viewed by others
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the most common head and neck malignancy tumor1. In HNSCC, lymph node metastasis (LNM) rate is approximately 29–36% with early stage of primary tumor site and significantly affect the prognosis of patients2. The presence of metastatic lymph nodes (MET) has been established as a predictor of poor patient outcomes3,4. LNM is thought to involve sequential stages, comprising: (1) intravasation from the primary site into the circulation, (2) survival in the blood circulation, (3) extravasation into distal tissues, and (4) colonization. Recent studies have emphasized the role of metastatic niche formation in LNM as a process characterized by the formation of a specialized lymph node microenvironment, marked by features such as lymphangiogenesis, angiogenesis, immune suppression, and remodeling of the extracellular matrix4,5,6. As both blood and lymphatic vessels serve as crucial conduits for tumor cell dissemination, high levels of lymphangiogenic growth factors and high lymphatic vessel density in sentinel lymph nodes indicate that LNM are more likely to occur in HNSCC4. Nevertheless, a comprehensive understanding of the mechanisms that regulate the vascular and lymphatic vessel systems to promote metastatic activity in HNSCC lymph nodes remains elusive.
Single-cell RNA sequencing (scRNA-seq) has emerged as a powerful tool for dissecting the cellular heterogeneity and molecular mechanisms in tumor microenvironment. Of particular interest are tumor-associated macrophages (TAMs), which exhibit remarkable functional plasticity in the metastatic niche formation. Emerging evidence suggests that TAMs exhibit functional heterogeneity, with subsets involved in tumor cell homing and outgrowth7, immune regulation8,9,10, angiogenesis11,12, ECM remodeling13,14, and inflammation15. Among these, the interaction between TAMs and endothelial cells (ECs) is particularly intriguing. Macrophage-EC interactions have been shown to regulate vascular remodeling in various pathological contexts, and specifically in cervical cancer primary sites, TAM-lymphatic endothelial cell (LEC) interactions enhance lymphangiogenesis, thereby facilitating metastatic dissemination16,17. However, to uncover the cell-cell interaction within the metastatic sites and the heterogeneity among patients of HNSCC needs further investigation. Could TAM subsets orchestrate metastatic niches through EC communication, thus generating a pro-vascular microenvironment?
Through this systematic approach, we identified a uniquely enriched subpopulation of SPP1+ TAMs in MET that exhibited robust spatial co-localization and functional interplay. Mechanistically, our analyses revealed that these SPP1+ TAMs might promote both angiogenesis and lymphangiogenesis through the signaling axis SPP1-ITGa9b1 and FN1-ITGa2b1. Moreover, the clinical relevance of these findings was reinforced by the observation that an SPP1+ TAM-specific gene signature was significantly associated with poor prognosis and increased LNM risk. Furthermore, we experimentally validated the presence and spatial distribution of this SPP1+ TAM phenotype and associated vascular and lymphatic system alterations in clinical specimens from HNSCC patients with LNM.
Methods
Data acquisition
The Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) was utilized to retrieve scRNA-seq. Specifically, scRNA-seq data from HNSCC from 41 samples were sourced from GSE19565518, GSE140042, GSE227156, and GSE15992919. Patient metadata and bulk RNA sequencing data of the TCGA-HNSC and GEO cohort (GSE65858)20 were downloaded using TCGAbiolink and GEOquery21 (https://tcga-data.nci.nih.gov/tcga/, https://www.ncbi.nlm.nih.gov/geo/).
Patient samples and ethical statement
Paraffin-embedded MET and paired LN tissue were obtained from 19 HNSCC cases at the Hospital of Stomatology, Sun Yat-sen University (Guangzhou, Guangdong, China). Written informed consent was obtained from each patient prior to this study, and this study was approved by the medical ethics committee of the Hospital of Stomatology, Sun Yat-sen University (approval no. ERC-[2017]−26). All patient studies were conducted in accordance with the Declaration of Helsinki and local legislation.
ScRNA-seq data pre-processing, dimension reduction, and clustering
The quality control, statistical analysis, and exploration of the scRNA-seq data are all completed by Seurat (v4.3.0, https://satijalab.org/seurat/) package in R software (v4.4.2, https://www.r-project.org/), and 104,794 cells are included in the analysis based on the following quality control standards: (1) cells with mitochondrial genome < 10% and nFeature_RNA > 500 were included, nUMI > 500, (2) genes that expressed at least in 3 cells were included. Doublets were identified and removed by DoubletFinder package (v2.0.3).
The filtered gene expression matrix was log-normalized by the NormalizeData function in Seurat; FindVariableFeatures to find 2000 highly variable genes. We used the ScaleData function to remove gene expression fluctuations caused by the proportion of mitochondrial and ribosomal gene expression, and the different cycles of cells. Then we integrated the data, and Harmony (https://github.com/immunogenomics/harmony) was used to remove the batch effect. The local inverse Simpson’s Index (LISI) (https://github.com/immunogenomics/LISI) was used to evaluate the batch effect. RunPCA performs linear dimensionality reduction on the expression matrix and selects the principal components for subsequent analysis; Cells were clustered using FindNeighbors and FindClusters function. RunUMAP unified manifold approximation and projection (UMAP) is used for nonlinear dimensionality reduction. FindAllMarkers function was used to identify marker genes for the individual clusters. By querying the CellMarker databases (http://117.50.127.228/CellMarker/) and literature as the main method, and SingleR software (https://github.com/dviraran/SingleR) and Garnett (https://cole-trapnell-lab.github.io/garnett/) automatic annotation as the auxiliary method, we find the cell types and corresponding marker genes in the corresponding tissues for cell annotation.
Tissue preference scoring
To evaluate the tissue preference of each cell cluster, we calculated the ratio of observed to expected cell numbers (Ro/e score) using the STARTRAC-dist index22. The expected number of cells was calculated based on the assumption of random distribution across tissues. Ro/e scores were categorized into five levels to indicate tissue enrichment: “+++” (Ro/e > 1), “++” (0.8 < Ro/e ≤ 1), “+” (0.2 ≤ Ro/e ≤ 0.8), “+/-” (0 < Ro/e < 0.2), and “-” (Ro/e = 0). Higher Ro/e scores indicate a stronger tissue preference of the corresponding cell cluster.
Functional enrichment analysis
For comprehensive pathway enrichment analysis, we employed the Gene Set Variation Analysis (GSVA) approach utilizing the Molecular Signatures Database (MSigDB) (https://www.gsea-msigdb.org/gsea/msigdb) HALLMARK, GO, KEGG collection (v2024.1). Gene expression data were filtered to include genes with mean expression > 0.5, present in at least one-third of samples, and showing variance > 0.1 across samples. GSVA was implemented using the ‘gsva’ R package (v2.0.7, https://www.bioconductor.org/packages/release/bioc/html/GSVA.html) with Gaussian kernel transformation. To identify differentially enriched pathways between different groups, we applied linear modeling using the ‘limma’ package (v3.62.2, https://www.bioconductor.org/packages/release/bioc/html/limma.html). Statistical analysis was performed using empirical Bayes moderation, with significant pathways identified based on adjusted P-value < 0.05.
Pseudo-sequential analysis
To investigate the developmental trajectory of macrophage subpopulations, we performed pseudotime analysis using Monocle2 (v2.26, https://cole-trapnell-lab.github.io/monocle-release/), Monocle3(v1.4.26, https://cole-trapnell-lab.github.io/monocle3/), CytoTRACE (v0.3.1, https://cytotrace.stanford.edu/) and ClusterGVis (v0.1.4, https://github.com/junjunlab/ClusterGVis) algorithms. Cell subsets for pseudotime analysis were chosen as indicated.
For specific analysis of TAMs populations, gene expression values were scaled log10(normalized gene expression) within each subgroup, and expression dynamics were visualized along pseudotime.
Cell–cell communication analysis
CellphoneDB (v5.0) was utilized with default parameters to quantify cellular communication probabilities. In this study, cell cross-talk interaction was performed using CellPhoneDB according to the manufacturer’s manual (https://www.cellphonedb.org/).
TCGA analysis and gene-set scoring
Patient metadata and bulk RNA sequencing data of the TCGA-HNSC cohort were downloaded from the TCGA Data Portal (https://tcga-data.nci.nih.gov/tcga/). And the NCBI GEO database (GSE65858) (https://www.ncbi.nlm.nih.gov/geo/). Samples of GSE65858 were 270 HNSCC patients with clinical information. Gene-set scoring analysis was evaluated using single-sample gene set enrichment analysis (ssGSEA) based on top100 genes differentially expressed in the active cluster in scRNA-seq in R package gsva (v2.0.7). The optimal cutpoint for patient stratification and active cluster gene set scores was determined using the surv_cutpoint function in the survminer R package (v0.5.0, https://github.com/kassambara/survminer). Kaplan-Meier survival plots were generated using the survfit function in the survival R package (v3.7-0, https://www.rdocumentation.org/packages/survival) and visualized using the ggsurvplot function in the survminer R package (v0.5.0). Statistical significance was assessed using the log-rank test.
Histologic analyses
Immunohistochemistry (IHC) staining: HNSCC MET tissue sections were deparaffinized, and antigen retrieval was performed using EDTA antigen retrieval buffer (pH 9.0) in a microwave oven. The tissue sections were then stained following the manufacturer’s protocol of DAB Horseradish Peroxidase Color Development Kit. Primary antibodies anti-CD34 (ab81289, Abcam, 1:2000 dilution), anti-LYVE1 (ab219556, Abcam, 1:2000 dilution) and anti-CD163(16646-1-AP, Proteintech, 1:2000 dilution) were used for staining.
Multiplex Immunohistochemistry (mIHC) staining: For mIHC analysis, 4 μm-thick formalin-fixed paraffin-embedded (FFPE) sections of HNSCC MET tissues were processed using the Triple Staining Kit (Melabio, Nanjing, China) based on tyramide signal amplification (TSA) technology. The following primary antibodies were applied: anti-SPP1 (22952-1-AP, Proteintech, 1:300 dilution), anti-CD163 (16646-1-AP, Proteintech, 1:2000 dilution), and anti-CD31 (ER31219, HuaBio, 1:1000 dilution). Nuclear counterstaining was performed using DAPI. Images were captured using TissueFAXS Spectra S system, and 1–3 representative fields from whole-slide scans were selected for quantitative analysis using StrataQuest analysis software. The mIHC staining results and analyses were independently validated by two certified pathologists.
Statistics analysis
All statistical analyses were performed using R language (v4.4.2), Python language (v3.9), and SPSS (v27, https://www.ibm.com/products/spss), and p < 0.05 was considered statistically significant.
Results
Landscape of HNSCC primary tumor and lymph node metastasis microenvironment
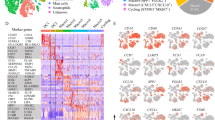
The overall workflow of this study is represented in Fig. 1A. To comprehensively characterize the cellular heterogeneity and microenvironmental dynamics in HNSCC, an integrated analysis of scRNA-seq data derived from multiple anatomical sites was performed: 18 metastatic lymph nodes (MET), 6 non-metastatic lymph nodes (LN), and 17 human HNSCC primary tumors (PT) (dataset: GSE195655, GSE140042, GSE227156, GSE159929). After rigorous quality control and batch-effect correction, a total of 104,794 high-quality cells were retained for downstream analysis, comprising 50,365 cells from MET, 15,961 cells from LN, and 38,468 cells from PT. (Fig. 1B) Batch effects were effectively mitigated by Harmony, as demonstrated by a local inverse Simpson’s Index (LISI) exceeding 1 for all cells (Supfig. 1 A).
The single-cell landscape of HNSCC patients with lymph node metastases. (A) the pipeline of this research, (B) UMAP plots displays the 9 major cell types by canonical markers, colored by different cell type(left) and sampling sources(right), (C) Plots displaying the cellular frequencies between PT, LN and MET, (D) Tissue preference of each cluster measured by Ro/e. (E) The dotplot displays the expression of conanical marker genes (columns), with rows representing cell subpopulations. Dot size: percent of cells expressing gene and dot color: scaled average expression.
Dimensionality reduction and unified manifold approximation and projection (UMAP)-based clustering revealed nine distinct cell populations: T cells (CD3D, CD3E), B cells (MS4A1, CD79A), plasma cells (JCHAIN, TNFRSF17), mast cells (TPSAB1, TPSB2), tumor cells (S100A2, KRT17), macrophages (CD163, C1QC), fibroblasts (COL1A1, ACTA2), dendritic cells (DCs) (CD83, CCR7), and endothelial cells (ECs) (PECAM1, VWF) (Fig. 1B and E). As shown in the dotplot, the marker genes for each cell type confirmed the accuracy of the cell type annotation (Fig. 1E). Comparative analysis of cellular compositions across tissue sites revealed distinct organizational patterns. The primary tumor sites exhibited a relatively heterogeneous distribution of cell types, while T cells and B cells were presented as the predominant cell populations in both LN and MET (Fig. 1C). Quantitative assessment of cellular frequencies demonstrated significant enrichment of macrophages (9.1%), plasma cells (7.1%), fibroblasts (5.2%), DCs (5.3%) and ECs (1.6%) in MET compared to LN (Fig. 1C). To validate these findings, we employed the Ratio of observed to expected (Ro/e) score, the ratio of observed to randomly expected cell numbers, to assess tissue-specific preferences of each cell type22. Except for the predominant T and B cells, DCs, ECs, macrophages, and mast cells were significantly enriched in MET (Fig. 1D), underscoring their potential functional significance in the establishment of the metastatic niche.
The accumulation of SPP1+ TAM in HNSCC lymph node with metastasis
Macrophages represent a crucial component of the lymph node immune landscape, orchestrating both immune regulation and microenvironmental remodeling, but their phenotypic diversity and functional roles in HNSCC metastasis remain incompletely understood. Our analysis revealed that macrophages constitute the predominant non-lymphoid population within MET (9.1%) (Fig. 1C) Furthermore, comparative GSVA enrichment analysis of TAMs from different anatomical sites demonstrated that MET-derived TAMs exhibit molecular signatures more closely aligned with PT TAMs than those from LN (Fig. 2D). Specifically, pathways including “ANGIOGENESIS”, “IL6_JAK_STAT3_SIGNALING”, “TNFA_SIGNALING_VIA_NFKB”, “EPITHELIAL_MESENCHYMAL_TRANSITION”, “HYPOXIA”, showed remarkable enrichment (Fig. 2D). All of which prompted a detailed investigation of their role in metastatic niche formation.
SPP1 + TAMs are preferentially enriched in MET. (A) UMAP visualization of TAM subclusters colored by cell types (left) and sampling sources (right). (B) Bar plot showing the relative frequencies of TAM subclusters across different tissues (PT, LN, and MET). (C) Tissue preference analysis of TAM subclusters using Ro/e score. (D) Radar plot comparing GSVA enrichment scores of key pathways among TAM populations from different tissues. (E) Dot plot showing the expression patterns of top 5 marker genes (columns) across TAM subclusters (rows). (F) Heatmap depicting the scaled mean expression of TAM function-associated genes across different subclusters. (G) Heatmap showing significantly enriched signaling pathways (rows) across TAM subclusters (columns) identified by GSVA analysis. (H) Kaplan-Meier analysis of 5-year overall survival (OS) in TCGA-HNSC cohort (upper) and GEO cohort (GSE65868, lower) grouped by the expression levels of TAM1 gene set signature. Statistical significance was determined by two-sided log-rank test.
We identified 8,640 macrophages (LISI > 1, Supfig. 1B), which clustered into six distinct subpopulations (Fig. 2A). While TAM2, TAM3, exhibited no strong tissue preference, TAM5 was enriched in lymph node tissue, TAM1 was specifically enriched in MET, and TAM4 and TAM6 were prominent subpopulations within PT (Fig. 2B and C). This finding suggests a functional interplay between TAM subpopulations and the tissue microenvironment, with TAM1 potentially playing a more prominent role in the establishment of MET. Moreover, differential gene expression analysis revealed distinct transcriptomic profiles among the TAM clusters (Fig. 2E and F). TAM1 exhibited genes including FN1, SPP1, MMP7, and CHI3L1, which were previously proven to be closely related to metastasis niche formation by regulating vascular, immune, and ECM microenvironment23,24,25,26 (Fig. 2E and F). TAM2 displayed enhanced expression of antigen presentation-related genes, including C1QA, C1QB, HLA-DQB1, and HLA-DQA1 (Fig. 2F). TAM3 and TAM4 both expressed VEGFA, EREG, and CLEC5A, which are related to angiogenesis11(Fig. 2E and F). TAM3 also highly expressed AREG, as a significant M1 marker27 (Fig. 2E). In addition, TAM4 showed elevated expression of chemotactic factors including CXCL1, CXCL5, CXCL8, IL6 suggest its pro-inflammatory function (Fig. 2E). TAM5 was characterized by elevated expressed of FOLR2, MAF, CETP which presented as canonical marker of resident-tissue macrophages(RTM)-TAMs24 (Fig. 2E). TAM6 uniquely expressed ISG15, IFI6 and IFIT1, all of which are canonical IFN-γ TAM-related gene24 (Fig. 2F). Additionally, we use the AddModuleScore function in Seurat confirmed these trends, TAM1, TAM3, TAM4 score highly in angiogenesis (Supfig. 2). TAM1 also exhibited relatively high expression of genes related to cells adhesion to ECs (Supfig. 2).
To further investigate functional differences, we performed gene set variation analysis (GSVA) for each TAM cluster. A set of distinct pathway enrichment characteristics was identified (Fig. 2G). “VEGF SIGNALING PATHWAY”, “LYMPHANGIOGENESIS PATHWAY”, “LYMPHATIC ENDOTHELIAL CELL DIFFERENTIATION”, “BLOOD VESSEL ENDOTHELIAL DIFFERENTIATION”, were enriched in TAM1_SPP1 and TAM4_CXCL5, suggesting a strong angiogenesis ability. And they are both enriched in pathways like “METASTASIS EMT UP”, “ECM RECEPTOR INTERACTION”, “ENDOTHELIAL CELL MATRIX ADHESION”. While exhibiting similar pro-angiogenesis, pro-metastatic, and pro-adhesive functions, these TAM subpopulations may be functionally specialized based on location. In contrast, TAM2_C1QA exhibited enrichment in the antigen presentation-related pathway, TAM3_AREG and TAM5_CETP enriched in immune activation and anti-tumor activity. TAM4_CXCL5, TAM5_CETP, and TAM6_ISG15 were enriched in inflammatory regulation-related pathways, including “CYTOKINE CYTOKINE RECEPTOR INTERACTION”, highlighting their immune regulation function. These results align with prior studies implicating TAM5_CETP as an RTM-TAMs subset and TAM6_ISG15 as IFN-γ TAMs, both of which play roles in immune modulation and inflammatory responses24. Together, these findings support the presence of multiple polarization states in TAMs.
Previous studies have demonstrated that SPP1+ TAMs are crucial determinants of tumor progression and clinical outcomes24. Our analysis revealed a distinct TAM1_SPP1 subpopulation that was uniquely enriched in MET compared to LN and PT. This subpopulation exhibited a complex molecular signature, including not only angiogenesis-related genes (SPP1, FN1, MMP7) but also markers associated with poor prognosis (CHI3L)15. To evaluate the clinical relevance of the TAM1_SPP1 signature, we derived a top 100 gene set and applied ssgsea function. In the TCGA-HNSC dataset, patients with high TAM1_SPP1 gene signature scores exhibited significantly poorer overall survival compared to those with low scores (Fig. 2H). Consistent with this finding, a higher gene signature score was also associated with a significantly increased risk of LNM (Supfig. 3). Furthermore, we validated these results using the independent GSE65858 cohort (Fig. 2I). Given its specific enrichment in metastatic sites and correlation with poor prognosis, we focused our subsequent analyses on understanding how TAM1_SPP1 contributes to metastatic niche formation in lymph nodes.
Development trajectory of macrophages
To investigate the differentiation dynamics and lineage progression of TAMs within lymph nodes, we separated TAMs from METs, and employed Monocle2 and Monocle3 for trajectory inference, followed by CytoTRACE to assess cellular differentiation states. This pseudotime analysis showed that the TAM3_AREG were mainly at the beginning of the projected timeline trajectory, whereas TAM1_SPP1, TAM4_CXCL5, TAM6_ISG15 occupied the opposite end with a notably higher CytoTRACE score (Fig. 3A and B, Supfig. 4). Along the inferred progression path, cells transition through intermediate populations TAM2_C1QA (Fig. 3A, Supfig. 4).
SPP1+ TAMs are derived from AREG+ TAMs. (A) Trajectory analysis of TAM populations from MET. Two-dimensional visualization of pseudotime ordering generated by Monocle3. (B) Cells colored by CytoTRACE scores indicating differentiation potential. (C) Left panel: Genes are clustered based on their temporal expression patterns. Middle panel: Heatmap showing dynamic gene expression patterns along pseudotime progression. Right panel: GOBP (green) and KEGG (yellow) pathway enrichment analysis with pseudotime phase. (D) Temporal expression dynamics of key transcription factors, immune-related genes, and angiogenesis-related genes during TAM transition. Red lines represent TAM1_SPP1 signature genes; blue lines represent TAM3_AREG signature genes.
We next investigated the transcriptional changes associated with transitional states. TAM3_AREG were predominantly at initial phase, characterized by upregulated expression of AREG, FCN1, EREG, VCAN (Fig. 3C). The subsequent phase mainly characterized by upregulation of antigen presentation gene like HLA-DQA1 and HLA-DQB1, canonical RTM-TAMs gene FOLR2, and CETP representing TAM5_CETP (Fig. 3C). Pathway enrichment analysis revealed significant involvement in processes such as “T cell proliferation”, “Antigen presentation” and “Defense response to tumor cell” indicating immune activation. The final phase exhibited high expression of SPP1, FN1 representing TAM1_SPP1, as well as CXCL8, CXCL5, IL6, ISG15, and IFIT1, characteristic of TAM4_CXCL5 and TAM6_ISG15. This phase showed enrichment in pathways including “Lymphangiogenesis”, “Response to hypoxia”, “Leukocyte transendothelial migration” and “Focal adhesion”, suggesting a role for these TAMs in migrating towards ECs and promoting angiogenesis. Additionally, transcription factors associated with macrophage hypoxia and pro-angiogenesis, including HIF1A, NFKB1, and TGFB1, were upregulated in this final phase.
To further analyze gene expression changes along the pseudotime trajectory in specific clusters, we compared gene expression profiles of TAM3_AREG and TAM1_SPP1, representing the extremes of macrophage polarization. Unsurprisingly, this analysis revealed significant differences in the expression of the angiogenesis marker SPP1 between TAM1_SPP1 and TAM3_AREG, with SPP1 serving as a terminal marker for TAM1_SPP1 (Fig. 3D). The upregulation of transcription factors HIF1A, STAT3, and TGFB1 in the final phase of TAM1_SPP1 development, correlating with SPP1 expression and M2 polarization as previously reported, highlights their potential role in promoting metastasis28,29.
Changes in endothelial cell subset in METs
A total of 1,692 ECs were identified (LISI > 1, Supfig. 1 C). Comparative analysis between ECs from LN and MET revealed significant enrichment pathways including “ANGIOGENESIS”, “EPITHELIAL MESENCHYMAL TRANSITION”, “TGF BETA SIGNALING”, all of which are closely associated with tumor metastasis (Fig. 4C). Given that tumor cells in HNSCC can disseminate to lymph nodes via both blood vascular and lymphatic vessels, we further categorized ECs into two distinct subpopulations: blood endothelial cells (BECs) and lymphatic endothelial cells (LECs), to clarify their respective contributions to metastasis (Fig. 4A). Cell composition analysis revealed a marked increase in LECs within MET, whereas BECs predominated in the primary tumor site (Supfig. 5). The dotplot of canonical marker genes for BECs and LECs confirmed the accuracy of this classification (Fig. 4B).
SPP1+ TAMs exhibit strong interaction with endothelial cells in metastatic lymph nodes. (A) UMAP visualization of ECs populations. Left: Cells colored by subtype classification. Right: Cells colored by tissue origin. (B) Dot plot showing expression patterns of canonical EC marker genes across identified subpopulations. (C) Bar plot comparing GSVA enrichment scores between LN and MET ECs. Statistical analysis performed using limma. (D) Dot plot visualization of predicted ligand-receptor interactions between TAM1_SPP1 and BECs or LECs.
Interaction relationship of TAMs with endothelial cells
To further elucidate the mechanistic basis of TAM–EC crosstalk, we analyzed cell–cell communication between TAM1_SPP1 and other cell clusters using CellPhoneDB. Remarkably, ligand–receptor analysis revealed consistent interactions between TAM1 and both BECs and LECs mediated by SPP1-ITGa9b1, FN1-ITGa2b1, VEGFA-KDR, and VEGFA-NRP2 signaling axes. In addition, CXCL8-ACKR1 interactions were enriched in BECs, and CCL4-ACKR2 in LECs (Fig. 4D). BECs and LECs demonstrate overlapping functions but also exhibit specialized roles in facilitating lymph node metastasis. Critically, our integrated analysis suggests that TAM1_SPP1 contributes to the formation of a metastatic niche within lymph nodes through the expression of chemotactic and adhesion-related genes (CCL4, FN1) and pro-angiogenic factors (SPP1, CXCL8, VEGFA). This crosstalk with ECs potentially facilitates HNSCC metastasis and provides valuable insights into the cellular mechanisms underlying lymph node metastasis in HNSCC.
SPP1+ TAM increases in MET and is spatially correlated with endothelial cells
Given the critical role of ECs in mediating tumor cell extravasation within lymph nodes, we observed a significant enrichment of ECs in MET (Fig. 1C), accompanied by a markedly increased vessel network density (Fig. 5A and B). But there is no significant difference in the number of CD163+ cells between MET and LN (Fig. 5B), nor a significant relationship between the vessel network density and the number of CD163+ TAMs (Supfig. 6).
Spatial characterization of vascular networks and SPP1+ TAM distribution in lymph node metastases. (A) Representative immunohistochemistry (IHC) images showing CD34+ blood vessels, LYVE1+ lymphatic vessels and the number of CD163+ cells in LN and MET. The dashed line represents border of the tumor tissue. (B) Quantitative comparison between LN and MET samples. Upper: CD34+ blood vessel area (n = 19). Middle: Percentage of LYVE1+ lymphatic vessel coverage (n = 19). Lower: The number of CD163+ cells (n = 19). (median, IQR) (C) Representative mIHC images showing co-localization of SPP1 and CD163 in TAMs from LN and MET specimens. (D) Spatial analysis using StrataQuest software to visualize TAM-EC interactions. White lines connect SPP1+ TAMs and CD31+ ECs located within 20 μm of CD31+ ECs in LN (upper) and MET (lower). (E) Boxplots (median, IQR) compare CD163+ cells numbers at different distances from endothelial cells in LN (upper)(n = 19) and MET (lower) (n = 19). (F) Scatter plots showing the distribution of SPP1+ TAMs at distinct distances from endothelial cells (0–20 μm, 20–50 μm, 50–100 μm) in LN (upper) and MET (lower). (G) Box plots (median, IQR) comparing SPP1+ TAM numbers at different distances from endothelial cells in LN (upper)(n = 19) and MET (lower)(n = 19). (H) Box plots (median, IQR) comparing the number of SPP1+ TAMs within 0-20 μm (upper) and 20-50 μm (lower) of ECs between LN and MET (n = 19). Statistical significance determined by Wilcoxon test (*p < 0.05, **p < 0.01, ***p < 0.001).
To further investigate the spatial relationship between SPP1+ TAMs and the metastatic niche environment, we performed mIHC analysis (Fig. 5C). The SPP1+ TAMs and those within 20 μm from ECs were connected using white lines by TissueGnostics panoramic tissue cell quantitative analysis system software and StrataQuest software (Fig. 5D). First, we quantified the number of CD163+ TAMs at varying distances from ECs. Our analysis revealed a significant difference in CD163+ TAMs abundance between the 0–20 μm and 50–100 μm proximity zones to ECs in metastatic samples. Notably, we observed a gradient pattern, with CD163+ TAMs density progressively decreasing as the distance from ECs increased across the measured intervals (Fig. 5E). In contrast, LN showed no significant variation in CD163+ TAMs distribution within 100 μm of ECs (Fig. 5E). Further analysis of SPP1+ TAMs also revealed a distance-dependent distribution pattern in metastatic tissues, with significantly higher numbers within 20 μm of ECs and a gradual decrease at greater distances (Fig. 5F and G). This spatial pattern was absent in LN (Fig. 5F and G). Direct comparison between MET and LN showed significant enrichment of SPP1+ TAMs within 20 μm of ECs in metastatic samples, while no significant difference was observed at 20–50 μm (Fig. 5H). Together, these findings demonstrate a clear spatial association between SPP1+ TAMs and ECs, further supporting the role of this interaction in promoting the formation of the metastatic niche.
Discussion
Though LNM represents a critical prognostic factor in HNSCC. While most studies have focused on the tumor microenvironment in primary tumors, the mechanisms underlying metastasis site remain poorly understood. This analysis revealed a distinct population of SPP1+ TAMs that was specifically enriched at metastatic sites. This subset exhibits a pronounced pro-angiogenic signature, demonstrates spatial proximity to ECs, and maintains intimate function interactions with ECs, suggesting its crucial role in metastatic niche formation. The clinical significance of this subpopulation was further validated by its strong correlation with poor patient outcomes.
TAMs were considered to have critical regulatory functions in both tissue homeostasis and tumor microenvironment. They are highly heterogeneous and associated with immune, vascular, and tissue remodeling functions. In this study, we identified and characterized a distinct SPP1+ TAMs population specifically enriched in MET. Previous research has established various mechanisms by which TAMs promote metastasis through vascular system regulation24,30. These include collaboration with ECs to enhance lymphangiogenesis, tumor adhesions11,31, and vascular permeabilization23. However, prior studies have predominantly focused on primary tumor sites rather than metastatic sites. Our study novelly focused on its pro-angiogenesis and lymphangiogenesis role in lymph node in HNSCC, which accounts for an important reason in metastasis environment establishment. The SPP1+ TAMs are enriched in angiogenesis-related pathways, ECs regulation, and cellular adhesion/extravasation functions. The enrichment of “METASTASIS EMT UP” pathway further supports their role in metastatic niche formation. Importantly, analysis of TCGA and GEO cohort data reveals that high gene signature score of SPP1+ TAMs correlates with poor patient survival and higher LNM risk, underscoring their clinical significance in driving lymph node metastasis. While previous studies have documented perivascular TAMs in primary tumors and distant metastatic sites32, we quantitatively characterized their unique spatial distribution within metastatic lymph nodes, demonstrating significant enrichment within 0–20 μm of vascular structures. This intimate spatial relationship provides compelling visual evidence for their functional interactions in orchestrating the metastatic niche formation.
Beyond SPP1+ TAMs, we also identified other TAM subclusters, including antigen-presentation-related TAM2_C1QA, angiogenesis-related TAM3_AREG, pro-inflammatory TAM4_CXCL5, TAM5_CETP24,33,34 as one subcluster of RTM, and IFN-γ-related TAM6_ISG1535,36. Notably, both TAM3_AREG and TAM4_CXCL5 displayed pro-angiogenic functions while showing distinct tissue preferences: TAM3 in normal lymph nodes and TAM4 in primary tumor sites. This observation might be due to the heterogeneity of macrophage subsets in physiological angiogenesis regulation, where different macrophage populations exhibit tissue-specific functions in vascular remodeling37. Moreover, our pseudotime analysis revealed a potential developmental trajectory in which TAM1_SPP1 may evolve from TAM3_AREG through an intermediate phagocytic state characterized by C1QA+ TAMs. These findings suggest that TAMs populations in HNSCC exist along a phenotypic continuum, with distinct subsets executing diverse functions during tumor progression. Some findings indicate that tumor cells can secrete extracellular vesicles that induce a pro-angiogenic phenotype in TAMs38,39,40, while neurotransmitters might modulate TAM function to promote tumor proliferation and neovascularization41.
To understand the interactions between TAMs and ECs in lymph node metastasis, we analyzed their communication patterns through ligand-receptor analysis. While both BECs and LECs are crucial stromal components controlling vessel permeability and lymph flow, which play important parts in LNM. While most prior studies focused primarily on LECs16,42,43, our analysis revealed that TAM1_SPP1 interacts with both cell types through SPP1-ITGa9b1 and FN1-ITGa2b1 pathways. SPP1, a secreted non-collagenous glycoprotein that is highly expressed in tumor and stromal cells, recent studies that have revealed that TAM-derived SPP1 plays multiple roles in tumor development, including promoting growth, inflammation, blood vessel formation, and metastatic dissemination44,45. FN1, as a secretive protein closely related to ECM remodeling, also plays an important role in immune cell recruitment and angiogenesis regulation23,24,46. In a single cell research in pancreatic cancer, FN1+ SPP1+ TAMs was also found to communicate with ECs via FN1/SPP1-ITGAV/ITGB1 to promote metastasis46. These molecular interactions suggest that SPP1+ TAMs orchestrate vascular development and remodeling, potentially facilitating lymph node metastasis through multiple coordinated pathways. Importantly, experimental studies targeting ITGa9 and ITGa2 have demonstrated that inhibition of these receptors can effectively reduce tumor metastasis47,48, highlighting promising therapeutic opportunities. These findings not only deepen our understanding of the mechanisms underlying metastasis but also point to potential therapeutic strategies targeting these specific pathways.
LNM serves as a crucial prognostic indicator in cancer, with clinical evidence showing approximately 40% incidence, occult metastasis rates ranging from 15–34%49, and the substantial neck recurrence rate of 17–20% within 5 years post-treatment, which underscores the urgent need for improved therapeutic strategies. In light of these challenges, targeted therapy against the lymphatic microenvironment could serve as a preventive strategy to reduce neck recurrence in both cN0 patients who opted out of neck dissection and pN+ patients who underwent neck dissection. Furthermore, in cN+ patients, combining this targeted approach with neoadjuvant immunochemotherapy could potentially control or even downstage nodal status preoperatively, thereby improving overall prognosis. Our findings advance understanding of pro-metastatic niche formation in lymph nodes, potentially revealing novel therapeutic targets within specific TAM and EC subclusters. Notably, while nanoparticle-based lymph node targeting has shown promise in enhancing antigen presentation50, this delivery strategy might be able to extend beyond immunostimulation to target pro-metastatic niche inhibition.
However, several critical questions remain to be addressed in future research. Firstly, while our findings suggest important roles for SPP1+ TAMs, their precise biological functions in lymphangiogenesis and angiogenesis require further in vivo and in vitro experimental validation. Spatial transcriptomic analysis could provide valuable insights into the relationship between these cells and their microenvironmental niches. Furthermore, the developmental origin of SPP1+ TAMs, whether from tissue-resident macrophages or circulating monocytes, and the factors driving their formation remain important questions for investigation. We will do these works in our ongoing long-term investigations.
Conclusions
Our analysis has revealed distinct TAMs heterogeneity in HNSCC, with particular emphasis on the SPP1+ TAM population enriched in MET. These SPP1+ TAMs emerge as critical mediators of HNSCC metastasis and serve as valuable prognostic biomarkers. Mechanistically, we reveal a potential mechanism whereby SPP1+ TAMs orchestrate metastatic niche formation in lymph nodes through the coordinated regulation of angiogenesis and lymphangiogenesis, facilitated by their spatial proximity to ECs. These findings not only advance our understanding of HNSCC metastasis but also identify SPP1+ TAMs as promising therapeutic targets for preventing metastatic progression.
Data availability
The datasets generated and/or analysed during the current study are available in the GEO (GEO, [https://www.ncbi.nlm.nih.gov/geo/](https:/www.ncbi.nlm.nih.gov/geo)) and TCGA(([https://tcga-data.nci.nih.gov/tcga/](https:/tcga-data.nci.nih.gov/tcga)) repository. ScRNA-seq data including GSE195655(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE195655), GSE140042(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE140042), GSE227156(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE227156) and GSE159929([https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE159929](https:/www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE159929)). BulkRNA-seq data including GSE6585820(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE65858) and TCGA-HNSC(https://portal.gdc.cancer.gov/).
Abbreviations
- BEC:
-
Blood endothelial cell
- DC:
-
Dendritic cell
- EC:
-
Endothelial cell
- ECM:
-
Extracellular Matrix
- EMT:
-
Epithelial-to-mesenchymal transition
- GEO:
-
Gene Expression Omnibus
- GSVA:
-
Gene set variation analysis
- HNSCC:
-
Head and neck squamous cell carcinoma
- IHC:
-
Immunohistochemistry
- LEC:
-
Lymphatic endothelial cell
- LISI:
-
Local inverse Simpson’s Index
- LN:
-
Lymph nodes
- LNM:
-
Lymph node metastasis
- MET:
-
Metastatic lymph nodes
- mIHC:
-
Multiplex immunohistochemistry
- PCA:
-
Principal components analysis
- PT:
-
Primary tumor
- RTM:
-
Resident-tissue macrophage
- ssGSEA:
-
Single-sample gene set enrichment analysis
- TAM:
-
Tumor-associated macrophage
- TSA:
-
Tyramide signal amplification
- UMAP:
-
Uniform manifold approximation and projection
References
Johnson, D. E. et al. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers. 6, 92 (2020).
He, Y. P., Zhou, P., Guan, L. M. & Wu, S. G. Clinical and dosimetric feasibility of sparing submandibular gland in patients with oral cavity squamous cell carcinoma. Ann. Med. 57, 2445186 (2025).
Xing, Y. et al. Relationship between level of lymph node metastasis and survival in locally advanced head and neck squamous cell carcinoma. Cancer 122, 534–545 (2016).
Hu, C., Huang, Q. & Sun, Q. The regulation of lymph node Pre-Metastatic niche formation in head and neck squamous cell carcinoma. Front. Oncol. 12, 852611 (2022).
Celià-Terrassa, T. & Kang, Y. Metastatic niche functions and therapeutic opportunities. Nat. Cell. Biol. 20, 868–877 (2018).
Gillot, L., Baudin, L., Rouaud, L., Kridelka, F. & Noël, A. The Pre-Metastatic niche in lymph nodes: formation and characteristics. Cell. Mol. Life Sci. 78, 5987–6002 (2021).
Lu, X. & Kang, Y. Chemokine (C-C motif) ligand 2 engages CCR2 + stromal cells of monocytic origin to promote breast cancer metastasis to lung and bone. J. Biol. Chem. 284, 29087–29096 (2009).
Maus, R. L. G. et al. Identification of novel, immune-mediating extracellular vesicles in human lymphatic effluent draining primary cutaneous melanoma. OncoImmunology 8, e1667742 (2019).
Dieterich, L. C. Mechanisms of extracellular vesicle-mediated immune evasion in melanoma. Front. Immunol. 13, 1002551 (2022).
Morrissey, S. M. et al. Tumor-derived exosomes drive immunosuppressive macrophages in a pre-metastatic niche through glycolytic dominant metabolic reprogramming. Cell. Metab. 33, 2040–2058e10 (2021).
Hongu, T. et al. Perivascular Tenascin C triggers sequential activation of macrophages and endothelial cells to generate a pro-metastatic vascular niche in the lungs. Nat. Cancer. 3, 486–504 (2022).
You, H., Baluszek, S. & Kaminska, B. Supportive roles of brain macrophages in CNS metastases and assessment of new approaches targeting their functions. Theranostics 10, 2949–2964 (2020).
Shu, C. et al. C3a-C3aR signaling promotes breast cancer lung metastasis via modulating carcinoma associated fibroblasts. J. Exp. Clin. Cancer Res. 39, 11 (2020).
Fujita, S., Sumi, M., Tatsukawa, E., Nagano, K. & Katase, N. Expressions of extracellular matrix-remodeling factors in lymph nodes from oral cancer patients. Oral Dis. 26, 1424–1431 (2020).
Dong, L. et al. SPP1 + TAM regulates the metastatic colonization of CXCR4 + Metastasis-Associated tumor cells by remodeling the lymph node microenvironment. Adv. Sci. (Weinh). 11, 2400524 (2024).
Jalkanen, S. & Salmi, M. Lymphatic endothelial cells of the lymph node. Nat. Rev. Immunol. 20, 566–578 (2020).
Chen, X. J. et al. A novel lymphatic pattern promotes metastasis of cervical cancer in a hypoxic tumour-associated macrophage-dependent manner. Angiogenesis 24, 549–565 (2021).
Horny, K. et al. Mesenchymal-Epithelial transition in lymph node metastases of oral squamous cell carcinoma is accompanied by ZEB1 expression. J. Transl Med. 21, 267 (2023).
He, S. et al. Single-cell transcriptome profiling of an adult human cell atlas of 15 major organs. Genome Biol. 21, 294 (2020).
Wichmann, G. et al. The role of HPV RNA transcription, immune response-related gene expression and disruptive TP53 mutations in diagnostic and prognostic profiling of head and neck cancer. Int. J. Cancer. 137, 2846–2857 (2015).
Colaprico, A. et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 44, e71 (2016).
Zhang, L. et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 564, 268–272 (2018).
Patras, L., Shaashua, L., Matei, I. & Lyden, D. Immune determinants of the pre-metastatic niche. Cancer Cell. 41, 546–572 (2023).
Ma, R. Y., Black, A. & Qian, B. Z. Macrophage diversity in cancer revisited in the era of Single-Cell omics. Trends Immunol. 43, 546–563 (2022).
Roles of matrix. metalloproteinase-7 (MMP-7) in cancer. Clin. Biochem. 92, 9–18 (2021).
Ma, B. et al. CHI3L1 enhances melanoma lung metastasis via regulation of T cell co-stimulators and CTLA-4/B7 axis. Front. Immunol. 13, 1056397 (2022).
Yuan, G. et al. LPS-induced extracellular AREG triggers macrophage pyroptosis through the EGFR/TLR4 signaling pathway. Front Immunol 16, (2025).
Colegio, O. R. et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 513, 559–563 (2014).
He, H. et al. Multi-dimensional single-cell characterization revealed suppressive immune microenvironment in AFP-positive hepatocellular carcinoma. Cell. Discov. 9, 60 (2023).
Bied, M., Ho, W. W., Ginhoux, F. & Blériot, C. Roles of macrophages in tumor development: A Spatiotemporal perspective. Cell. Mol. Immunol. 20, 983–992 (2023).
García-Silva, S. et al. Melanoma-Derived small extracellular vesicles induce lymphangiogenesis and metastasis through an NGFR-dependent mechanism. Nat. Cancer. 2, 1387–1405 (2021).
Huang, J. et al. LncRNA-MALAT1 promotes angiogenesis of thyroid cancer by modulating Tumor-Associated macrophage FGF2 protein secretion. J. Cell. Biochem. 118, 4821–4830 (2017).
Bugatti, M. et al. A population of TIM4 + FOLR2 + Macrophages localized in tertiary lymphoid structures correlates to an active immune infiltrate across several cancer types. Cancer Immunol. Res. 10, 1340–1353 (2022).
Nalio Ramos, R. et al. Tissue-resident FOLR2 + macrophages associate with CD8 + T cell infiltration in human breast cancer. Cell 185, 1189–1207e25 (2022).
Li, J. et al. cGAS-ISG15-RAGE axis reprogram necroptotic microenvironment and promote lymphatic metastasis in head and neck cancer. Exp. Hematol. Oncol. 13, 63 (2024).
Wei, J. et al. Cohort-based pan-cancer analysis and experimental studies reveal ISG15 gene as a novel biomarker for prognosis and immunotherapy efficacy prediction. Cancer Immunol. Immunother. 74, 168 (2025).
Martin, P. & Gurevich, D. B. Macrophage regulation of angiogenesis in health and disease. Semin Cell. Dev. Biol. 119, 101–110 (2021).
Ludwig, N. et al. TGFβ + small extracellular vesicles from head and neck squamous cell carcinoma cells reprogram macrophages towards a pro-angiogenic phenotype. J. Extracell. Vesicles. 11, e12294 (2022).
Huber, D. et al. The role of plasma-derived small extracellular vesicles in pre-metastatic niche formation through modulation of macrophages in head and neck squamous cell carcinoma. Br. J. Cancer. 133, 121–130 (2025).
You, Y. et al. M1-like tumor-associated macrophages cascade a mesenchymal/stem-like phenotype of oral squamous cell carcinoma via the IL6/Stat3/THBS1 feedback loop. J. Exp. Clin. Cancer Res. 41, 10 (2022).
Dong, Y. et al. Tumor-derived GABA promotes lung cancer progression by influencing TAMs polarization and neovascularization. Int. Immunopharmacol. 126, 111217 (2024).
Qiu, C. et al. RGS5 + lymphatic endothelial cells facilitate metastasis and acquired drug resistance of breast cancer through oxidative stress-sensing mechanism. Drug Resist. Updat. 77, 101149 (2024).
Wei, J. C. et al. Tumor-associated lymphatic endothelial cells promote lymphatic metastasis by highly expressing and secreting SEMA4C. Clin. Cancer Res. 23, 214–224 (2017).
Wei, J. et al. Osteopontin mediates glioblastoma-associated macrophage infiltration and is a potential therapeutic target. J. Clin. Invest. 129, 137–149 (2019).
Nallasamy, P. et al. Pancreatic tumor microenvironment factor promotes cancer stemness via SPP1-CD44 axis. Gastroenterology, 161, 1998–2013 (2021).
Ainiwaer, A., Qian, Z., Wang, J., Zhao, Q. & Lu, Y. Single-cell analysis uncovers liver susceptibility to pancreatic cancer metastasis via myeloid cell characterization. Discov Oncol. 15, 696 (2024).
Guo, P., Moses-Gardner, A., Huang, J., Smith, E. R. & Moses, M. A. ITGA2 as a potential nanotherapeutic target for glioblastoma. Sci. Rep. 9, 6195 (2019).
Navarro, N. et al. Integrin alpha9 emerges as a key therapeutic target to reduce metastasis in rhabdomyosarcoma and neuroblastoma. Cell. Mol. Life Sci. 79, 546 (2022).
Yu, Y. F. et al. Frequency of lymph node metastases at different neck levels in patients with oral squamous cell carcinoma: a systematic review and meta-analysis. Int. J. Surg. 111, 1285–1300 (2025).
Li, Z. Z. et al. Nanoparticles targeting lymph nodes for cancer immunotherapy: strategies and influencing factors. Small 20, e2308731 (2024).
Acknowledgements
Not applicable.
Funding
This work was financially supported by the National Natural Science Foundation of China grants (No.82103558).
Author information
Authors and Affiliations
Contributions
L.Y., G.L., R.T. and Z.L. were responsible for the experimental design. Z.L. and R.T. evaluated mIHC and IHC experiments of HNSCC tissues. M.M. and X. X. performed the statistical analyses. Z.L. wrote the manuscript with input from all authors. L.Y., G.L., S.Z, J.M., D.W. contributed to the manuscript editing, response of reviewers’ comments, collection and analysis of clinical cases. All authors have approved the content of the manuscript and agreed to its publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Clinical samples and data were obtained with written consent from each patient and used in accordance with protocols approved by the Subcommittee on Research and Animal Care of the Hospital of Stomatology, Sun Yat-sen University. (approval no. ERC-[2017]-26). All studies were conducted in accordance with the Declaration of Helsinki and local legislation.
Consent for publication
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lao, Z., Tan, R., Maitikuerban, M. et al. Single-cell identification of an endothelial cell proximal SPP1+ macrophage population defines the metastatic vascular niche in lymph nodes. Sci Rep 15, 43896 (2025). https://doi.org/10.1038/s41598-025-27796-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-27796-y