Abstract
Understanding Alzheimer’s disease (AD) from its earliest stages is essential for uncovering initial mechanisms of pathology and developing interventions. Here, we use the humanized AppNL-G-F mouse model, which develops early amyloid pathology along a predictable timeline, to characterize epigenetic changes in the brain and blood at early pre-symptomatic, as well as later, stages of disease progression. We identified alterations in chromatin accessibility, gene expression, and DNA methylation before and after amyloidosis, in the absence of advanced age. Despite broadly stable hippocampal cell composition, AppNL-G-F mice exhibit major gene expression differences preceding amyloid plaque deposition, particularly in pathways related to mitochondrial function and protein biosynthesis. In later stages of pathology, immune pathways were upregulated, consistent with established neuroinflammatory processes in AD. Extensive DNA methylation changes were also detected in both blood and hippocampus at early and late pathology stages. Many blood methylation differences at early stages overlapped brain cis-regulatory elements and mapped near differentially expressed hippocampal genes, with enrichment in neuronal development and synaptic pathways, underscoring a potential link between blood methylation and brain physiology and supporting the potential of blood DNA methylation as an early biomarker of amyloidosis. Notably, five genes, including Rbfox1 and Camta1, showed coordinated epigenetic dysregulation in both brain and blood prior to amyloidosis, highlighting them as potential early blood-based biomarkers.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) is a complex progressive neurodegenerative disorder and the most common cause of dementia, affecting millions of people worldwide1. AD is characterized by hallmark pathological features, including extracellular amyloid-β (Aβ) plaques, intracellular tau neurofibrillary tangles, neuroinflammation, and widespread synaptic and neuronal loss2. Despite decades of intensive research, the precise mechanisms driving the onset and progression of AD remain elusive, including whether intracellular or extracellular Aβ causally contributes to the disease beyond being a hallmark3, and effective therapeutic options are limited. One of the greatest challenges in AD research lies in uncovering the molecular and cellular changes that arise during the preclinical phase, a prolonged asymptomatic stage during which pathological features, such as Aβ deposition, develop4. This phase, which can span several decades, represents a critical window for therapeutic intervention aimed at preventing or delaying the onset of clinical symptoms. However, studying preclinical AD, presents several challenges, particularly when working with human subjects, including difficulties in identifying at-risk individuals, limited access to relevant tissues, the presence of confounding factors (e.g., aging and lifestyle), variability in disease progression, and the subtlety of molecular changes at early stages of pathology. Overcoming these barriers is critical for understanding the early mechanisms driving AD and enabling the development of effective strategies to combat later symptoms.
To address these challenges, a range of genetically modified animal models, particularly transgenic mice, have been developed5. While no single model fully replicates the complexity of AD, each successfully mimics key features of the condition making their detailed characterization crucial for understanding AD mechanisms. A prominent model is the AppNL-G-F mouse6 which carries a knock-in of the human amyloid precursor protein (APP) with three specific mutations: the Swedish mutation (KM670/671NL), which increases the overall production of Aβ40 and Aβ42; the Arctic mutation (E693G), which promotes the formation of insoluble Aβ fibrils; and the Beyreuther/Iberian (I716F) mutation, which raises the ratio of Aβ42 to Aβ40. The AppNL-G-F model exhibits early-onset amyloid pathology starting at 2 months (~ 8 weeks) of age, with neuropathology progressing to saturation by 7 months (~ 28 weeks). Cognitive impairments typically manifest by 6 months (~ 24 weeks)7,8. This predictable disease timeline makes the AppNL-G-F model useful for studying molecular changes across all stages of pathology, particularly the preclinical stages. Importantly, due to the early onset of disease in this model the confounding effects of advanced age are minimized, offering a robust platform for investigating early molecular events leading to amyloidosis and for identifying blood-based biomarkers of brain dysregulation before the onset of clinical symptoms. However, despite its promise, the AppNL-G-F model has not yet been used to examine epigenetic remodeling before and after amyloid onset. This gap limits our understanding of both the model itself and the epigenetic mechanisms driving and contributing to AD pathology, ultimately hindering efforts to identify early epigenetic biomarkers and therapeutic targets.
In this study, we leveraged a combination of single-nucleus and bulk epigenetic and transcriptomic data from a cohort of AppNL-G-F mice and their wild-type littermates to characterize molecular differences across three ages, ranging from postnatal week 3 (an early pre-symptomatic stage prior to detectable Aβ pathology6) to postnatal week 24 (representing advanced pathology with near-saturation of Aβ and the onset of cognitive decline)6. Our study specifically focuses on the hippocampus, a brain region critical for memory, learning, and spatial navigation9 which is known to exhibit early and pronounced vulnerability in AD, with significant tissue loss and connectivity disruptions10. Our findings reveal substantial early epigenetic and gene expression dysregulation in the hippocampus of AppNL-G-F mice prior to the onset of severe amyloidosis. Importantly, we also identified early DNA methylation signals in peripheral blood of the AppNL-G-F mice that warrant further study as potential blood-based biomarkers for preclinical AD, paving the way for development of blood-based diagnostic tools in preclinical AD.
Results
Hippocampal cell composition remains largely stable during progression of amyloid pathology
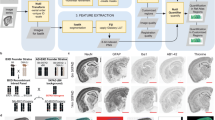
To investigate epigenetic and transcriptomic changes preceding and following onset of severe amyloidosis in the brain and blood of the AppNL-G-F mouse model, we collected hippocampus and blood from 30 AppNL-G-F and 30 wild-type littermates. The cohort included equal numbers of male and female, sampled at postnatal week 3 (weaning), 8, and 24 (n = 5 per sex/genotype/age combination; Fig. 1A). All assays conducted in this study (Fig. 1B) were performed on tissue from the same 60 individual animals. We selected postnatal week 3 (W3) to represent an early pre-symptomatic stage, prior to detectable Aβ pathology6, enabling investigation of initial molecular changes preceding severe amyloidosis in the hippocampus and blood. Postnatal week 8 (W8) represents an intermediate stage associated with early progression of Aβ pathology in the AppNL-G-F model, while postnatal week 24 (W24) represents advanced pathology, approaching Aβ saturation and coinciding with onset of cognitive decline6 (Fig. 1A).
Changes in brain cell composition, including neuron loss and gliosis, have been reported in AD11. To investigate changes in brain cell-composition and chromatin accessibility in the AppNL-G-F mice before and during progression of Aβ pathology, we used hippocampus single-cell ATAC-seq (snATAC-seq) to study AppNL-G-F (n = 5 per sex) and wild-type littermates (n = 5 per sex) at W3, W8 and W24. To improve cell yields in our snATAC-seq assay, nuclei from biological replicates were pooled to create one pool per sex/genotype/age combination yielding a total of 12 pools, comprising 14,907 cells. Given the lack of reported sex differences in amyloidosis patterns in the AppNL-G-F model6 and to improve statistical robustness, male and female pools were treated as replicates within each age and genotype combination. To identify and characterize cell types within our snATAC-seq dataset, we conducted unsupervised clustering and identified 21 cell clusters (Fig. 2A). Cross-reference of cluster-specific accessible chromatin regions with known marker genes (Fig. 2B; Supplementary Fig. 1)11,12 identified the following cell-types: microglia (n = 615 cells), oligodendrocyte (n = 1,729), precursor/immature oligodendrocyte (n = 349), endothelial (n = 127), astrocyte (n = 771), Cajal-Retzius cells (n = 63), excitatory neurons (n = 8,070) and inhibitory neurons (n = 1,362) (Fig. 2B; Supplementary Table 1). Although most clusters were successfully annotated, four (C8, C9, C10 and C19) could not be confidently classified due to ambiguous or inconsistent chromatin accessibility patterns (Fig. 2B; Methods). These clusters, which comprise a total of 471 cells, likely represent rare or transitional cell types (C8 and C10), as well as technical noise (C9 and C19) and were therefore excluded from downstream analysis.
snATAC-seq reveal cell-specific changes in chromatin accessibility in early vs. late Aβ pathology. (A) Uniform manifold approximation and projection (UMAP) dimensionality reduction after iterative LSI of snATAC–seq data from 12 sample pools. Each dot represents a single nucleus (n = 14,907), colored by its corresponding cluster (left) or cell type (right). Bar plot shows the number of cells per cluster, with corresponding cluster colors and assigned cell type (B) Genomic tracks display chromatin accessibility at a subset of marker genes used to annotate cell types in this study. (C) Estimated average hippocampus cell composition is shown for each time point and genotype. The n refers to the number of nuclei recovered after quality control filtering. Cell composition for WT at W8 is not presented due to low nuclei count (n = 88) (D) MA plots of differential snATAC-seq peaks reveal chromatin accessibility differences in excitatory neurons during early pathology (W3), while in late pathology (W24), these differences are exclusive to inhibitory neurons. (E) Significant gene ontology (GO) terms associated with differentially accessible regions in inhibitory neurons at W24.
We used cell distribution across clusters to estimate cellular composition of the hippocampus across ages and genotypes (Fig. 2C; Supplementary Table 1). Across all samples, excitatory neurons were the most prevalent cell type (61.0% ± 0.44; mean ± SEM), followed by inhibitory neurons in most cases (12.6% ± 0.73%; mean ± SEM). Due to low cell recovery from wild-type mice at W8 (< 100 cells), this time point was excluded from downstream snATAC-seq analyses, limiting our ability to draw conclusions about chromatin accessibility patterns at this stage (Fig. 2C; Supplementary Table 1). We noted a mild trend towards increase in microglia and oligodendrocyte abundance in AppNL-G-F mice with increased age (Fig. 2C), consistent with progressive amyloid pathology and expected neuroinflammatory changes. However, statistical analysis of cell type proportions revealed no significant differences in any cell types between genotypes or across time points (adjusted p > 0.05; Supplementary Table 1).
Chromatin accessibility changes are detected in inhibitory neurons later in amyloidosis
To identify condition-specific changes in chromatin accessibility, we first performed marker peak analysis and identified snATAC-seq peaks unique to each age/genotype/cell-type condition (q < 0.1). At W3, significant marker peaks were identified exclusively in excitatory neurons (n = 6), while at W24, most marker peaks were found in inhibitory neurons (n = 37; Supplementary Table 1). The scarcity of marker peaks in other cell types is likely due to the low and inconsistent number of cells representing those cell types in our dataset (Supplementary Table 1). Marker peaks corresponding to W3 excitatory neurons in AppNL-G-F mice overlapped with several genes relevant to AD pathology and neurodegeneration, such as the mammalian target of rapamycin (mTOR) and Smpd313,14. Considering that marker peaks were predominantly detected in excitatory and inhibitory neurons, and that other cell types had insufficient counts, we focused subsequent differential snATAC-seq analysis on neurons at W3 and W24 (Supplementary Table 1). Consistent with our previous findings, in early stages of pathology (W3), we only detected three significantly different AppNL-G-F vs. WT chromatin accessibility peaks, all of which were identified in excitatory neurons (FDR < 0.1) and exhibited reduced accessibility in AppNL-G-F mice compared to WT (Fig. 2D). One of these differential peaks was found at the promoter of the Kdm6a gene, which codes for an X-linked H3K27 histone demethylase whose dysregulation disrupts neurodevelopment (Supplementary Fig. 2; Supplementary Table 1)15,16.
In contrast to W3, differential accessibility peaks at W24 were more numerous and confined to inhibitory neurons (n = 542; Fig. 2D; Supplementary Table 1). These peaks were roughly evenly split between regions of increased (~ 69%) and decreased accessibility (~ 41%) in AppNL-G-F. GO term analysis of these regions revealed enrichment of several pathways related to neuron projection, neurogenesis, and synaptic signaling (Fig. 2E; Supplementary Table 2), indicating major changes in function of inhibitory neurons during later amyloidosis. Several of the differential accessibility regions at W24 overlapped directly with AD-relevant genes (Supplementary Table 1). For example, inhibitory neurons of AppNL-G-F mice exhibited higher accessibility at a putative enhancer located in the intron of Optn (Optineurin) gene (log2fold change = 4.34, FDR = 0.07; Supplementary Fig. 2), a receptor that increases expression of autophagic genes to reduce neurotoxicity and inflammation in AD17. Moreover, AppNL-G-F mice showed reduced accessibility at a putative intronic enhancer at the Ptk2b gene, also known as Pyk2 or Protein tyrosine kinase 2 beta (log2fold change = − 2.48, FDR = 0.02; Supplementary Fig. 2), which is a susceptibility gene for late-onset Alzheimer’s disease. PTK2B is implicated in AD through its involvement in synaptic plasticity, calcium signaling, and neuroinflammatory pathways, all of which are disrupted in AD18,19,20. There were no overlap between differential peaks identified at W3 and W24, indicating a shift in epigenetic dysregulation AD pathology progresses, with differential accessibility peaks increasingly identified in inhibitory neurons in association with genes involved in neurogenesis, synaptic signaling, and neuroinflammation.
Significant gene expression changes precede neuropathology in the hippocampus of App NL-G-F mice
The overall stability we observed in hippocampal cell composition across genotypes and ages alleviates concerns that differential gene expression in bulk tissue from our model would be drastically skewed by cell composition differences that have been previously reported in AD11,21. Thus, to investigate gene expression changes in the hippocampus before and during AD pathology, we performed RNA-seq data on bulk hippocampus tissue of the same 60 AppNL-G-F and wild-type mice used in our snATAC-seq assay (n = 5 per sex, genotype and age group; Supplementary Table 3). Our initial assessment of overall effect of sex revealed only 27 genes with significant differential expression between males and females (adjusted p ≤ 0.05), suggesting minimal sex-related transcriptional differences (Supplementary Table 3). Consequently, we conducted differential expression analyses using both sexes as biological replicates, while including sex as a covariate in the model. We found that the number of differentially expressed genes (DEGs) between AppNL-G-F and WT mice varied drastically across the three timepoints.
The highest number of DEGs were identified at W3 (n = 2,628; adjusted p ≤ 0.05), despite lack of detectable amyloidosis at this age6, suggesting complex gene regulatory changes occur even before severe amyloid accumulation and neuropathology. DEGs at W3 were evenly split between being over-expressed (49.7%; n = 1,306) and under-expressed (50.3%; n = 1,322) in AppNL-G-F mice (Fig. 3A; Supplementary Table 3). The numbers of DEGs were much lower at W8 (n = 4) and W24 (n = 156). The four DEGs identified at W8 were all under-expressed in AppNL-G-F hippocampus compared to controls, while at W24 nearly all DEGs (94%; n = 147) were over-expressed (Fig. 3A). Across disease stages, only 17 DEGs were shared across timepoints, all between W3 and W24 (Fig. 3A), and several (8 out of 17) displayed opposing expression changes between these ages, suggesting dynamic regulation across early and late amyloid pathology. Among these, Cx3cr1 was most notable, showing significant downregulation at both W3 and W24. This chemokine receptor, primarily expressed in microglia, plays a key role in modulating microglial activation and amyloid-β (Aβ) clearance22. Prior studies in AD mouse models have shown that CX3CR1 deficiency exacerbates Aβ pathology, tau hyperphosphorylation, neuritic dystrophy, synaptic loss, neurodegeneration, and cognitive impairment22,23.
Gene expression changes in AppNL-G-F hippocampus precede severe Aβ pathology and highlight brain dynamics during disease progression. (A) A table summarizes differentially expressed genes (DEGs) across timepoints of pathology and a Venn diagram illustrates DEGs shared across timepoints (B) Selected significantly enriched gene ontology (GO) terms for DEGs at each timepoint highlight key pathway shifts during disease progression. (C) Clustering of genes with significant genotype-by-age interactions reveals major trajectories of gene expression changes throughout disease progression.
We cross-referenced our DEGs against those reported in a recent study by Rico et al. 202424, which profiled gene expression in the hippocampus of AppNL-G-F males at 6 weeks and 7 months of age. We found several shared DEGs, specifically, 4 of their 36 DEGs at 6 weeks and 32 of their 292 DEGs at 7 months were also differentially expressed in our dataset at one or more time points, with the majority overlapping at W3 (Supplementary Table 3). Of note, Fabp7 was a DEG in our study at W3 and was also differentially expressed at both 6 weeks and 7 months in Rico et al. 2024. FABP7 (Fatty Acid Binding Protein 7) is consistently upregulated in AD mouse models and in brains of human AD patients, especially near amyloid plaques25,26. This protein has been implicated in neuroinflammation and altered lipid metabolism, processes that may drive early hippocampal dysfunction25,26. We also compared our findings with a recent meta-analysis based on bulk gene expression data from frontal and temporal lobes of human AD brains27 and observed that 96 of the 760 DEGs reported in AD patient brains were also identified in our RNA-seq analysis (Supplementary Table 3). Among these, a notable example is Aqp4, which has been implicated in tau pathology as well as amyloid-β deposition and clearance in mouse models28,29,30, and is also considered a potential therapeutic target for AD in humans31.
We next investigated the correspondence between our bulk RNA-seq and snATAC-seq results (Supplementary Table 4). Of the DEGs identified, 41 also overlapped a significant differential chromatin accessibility region in excitatory neurons at W3 or inhibitory neurons at W24. Three of these genes, Kdm6a, Lars2, and Hvcn1, showed both transcriptional and chromatin changes at the same age: Kdm6a at W3, and Lars2 and Hvcn1 at W24. All three of these genes have been implicated in AD or neurodegeneration. Notably, elevated Kdm6a expression is associated with resilience to age-related cognitive decline in male mice32 and AD-related toxicity in mouse models, particularly in females, as the gene escapes X-chromosome inactivation33. In humans, genetic variation in KDM6A is linked to higher brain expression and correlates with reduced cognitive decline in aging and preclinical AD in both sexes33; Lars2, encoding mitochondrial leucyl-tRNA synthetase, is critical for mitochondrial protein synthesis and its reduced expression is associated with mitochondrial dysfunction and cognitive decline in AD mouse models, while its overexpression has been proposed as a potential therapeutic strategy for AD patients34; and increased Hvcn1 expression is linked to neuroinflammation and microglial-mediated neurodegeneration in neurodegenerative and ischemic mouse models35,36. The remaining 38 DEGs associated with differential accessibility, were differentially expressed in bulk hippocampus at W3, but exhibited chromatin changes in inhibitory neurons at W24, reflecting either shifts in cellular vulnerability during disease progression or dataset limitations (e.g., low, and variable cell counts in our snATAC-seq assay). Several of the genes in this group, such as Rbfox137 and Auts238, are implicated in neurodevelopmental disease, suggesting that key AD and neurodevelopmental genes are altered at the epigenetic and transcriptional levels, spanning early stages before amyloid pathology (W3) through later stages of disease progression (W24). Altogether, the large number of DEGs identified at W3, along with the variable patterns of gene expression observed from W3 to W24, suggest that widespread gene dysregulation precedes severe neuropathology and that disruption of various genes contributes to AD as the brain advances through stages of pathology.
Gene expression profiles reveal distinct pathways involved in pre- and post-amyloidosis
To shed light on the physiological states of the AppNL-G-F brain as it transitions from exhibiting no symptoms to severe amyloid pathology6, we set to identify over-represented biological pathways among differentially expressed genes (DEGs; q ≤ 0.05) at each time point using EnrichR39 (Supplementary Table 5). DEGs identified at W3, which comprised of both upregulated and downregulated genes, were enriched in several biological pathways, even though Aβ pathology is lowest at this stage. Significantly enriched biological pathways (q < 0.01) included major pathways of protein biosynthesis and catabolism including “gene expression”, “mRNA splicing”, “translation”, and “proteasomal protein catabolic process”, likely reflecting alteration in gene expression paradigms before or in response to the earliest traces of amyloidosis. The rest of significant pathways were mostly relevant to “mitochondria function and organization” (Fig. 3B; Supplementary Table 5), consistent with previous reports in AD patients suggesting mitochondria and metabolic dysregulation as one of the earliest mechanisms leading to pathology40,41. Many of the DEGs corresponding to these pathways have been previously implicated in cellular respiration (e.g., components of the electron transport chain, such as Ndufa8 and Cyc1), or mitochondrial organization (e.g., translocase of inner mitochondrial membrane genes, such as Timm23 and Timm44), indicative of extensive disruption of mitochondrial function in our model preceding amyloid neuropathology. At W8, the number of DEGs (n = 4) was too small to provide reliable gene ontology enrichment. However, DEGs at W24, which were mostly upregulated in AppNL-G-F, revealed significant enrichment of several biological pathways, predominantly associated with immune and neuroinflammatory response (Fig. 3B; Supplementary Table 5). Notable genes included those implicated in microglia activation (e.g., Trem2), phagocytosis (e.g. Tlr2) and cytokine signaling (e.g. Csf1r). Additionally, DEGs at W24 were significantly associated with synapse pruning, aligning with the cognitive decline observed in the AppNL-G-F model around this age40,41.
We also identified 209 genes with significant “age-by-genotype interaction” (q < 0.05), representing genes whose expression is affected differently during aging in AppNL-G-F versus WT mice, and clustered them based on their expression trajectories (Fig. 3C; Supplementary Table 6). Most of these genes were also identified as a DEG in at least one time point (Supplementary Table 6), showing a dynamic relationship with age in AppNL-G-F mice. Specifically, 121 of the genes with significant interaction were differentially expressed at W3, one at W8, 72 at W24, and 12 at both W3 and W24. Among the gene clusters with significant age-by-genotype interaction, two clusters (clusters 1 and 2) contained genes with highly divergent AppNL-G-F expression at W3, exhibiting opposite age-dependent changes in expression between genotypes (Fig. 3C; Supplementary Table 6). Given the early and distinct expression divergence of these genes, they may be involved in the onset and progression of neuropathy. While we did not detect significant enrichment of biological pathways among the genes in these clusters (q < 0.05), they did include several genes implicated in neurodegeneration, such as Crebbp, Atxn2, and Mt342,43,44. Four other clusters (clusters 6, 7, 8, and 9) represented genes whose expression trajectory culminated in highly divergent expression at W24 (Fig. 3C; Supplementary Table 6). These genes were enriched in several immune response pathways, including B-cell proliferation, dendritic cell antigen processing/presentation, and phagocytosis (Supplementary Table 6). This suggests that the immune response follows a markedly different trajectory and reaches a highly divergent state in brains predisposed to Aβ pathology.
DNA methylation patterns in brain and blood of App NL-G-F mice differ from wild-types and are linked to hippocampal gene regulation
We used whole genome bisulfite sequencing (WGBS; Supplementary Table 7), on the same individuals used for snATAC-seq and RNA-seq, to investigate DNA methylation changes in the hippocampus and blood of AppNL-G-F mice throughout disease progression, and to determine if methylation changes in the blood reflect gene dysregulation in the brain. To explore the role of DNA methylation in disease progression of the AppNL-G-F model, we first identified significant differentially methylated regions (DMRs; difference in methylation ≥ 10% and q ≤ 0.05) in hippocampus and blood of AppNL-G-F mice relative to WT, while including sex as a covariate (Supplementary Table 8). To better infer the functional roles of DMRs, each was assigned to the gene with the nearest transcription start site, assuming it to be the most likely regulatory target. In the hippocampus, we identified 537 significant DMRs between AppNL-G-F and wild-type mice at W3, 276 at W8 and 510 at W24. In the blood, there were 664 DMRs at W3, 3,476 at W8 and 262 at W24 (Fig. 4A; Supplementary Table 8). Approximately 5% of genes associated with hippocampal DMRs in the brain, and 4% of DMR-associated genes in the blood were shared across two or all three time points in the same tissue, suggesting some persistence of DNA methylation dysregulation throughout disease progression (Fig. 4A). In the hippocampus, over half of the DMRs were hypermethylated in each time point (Fig. 4B). Based on visual inspection of the data, the overlap of hippocampal DMRs with genomic features showed broadly similar patterns across all three time points, with roughly equal numbers of DMRs falling into genic vs. intergenic regions. Among the DMRs overlapping genes (i.e., genic), most overlapped intronic regions. The majority of intergenic DMRs were distal to genes, suggesting they may overlap distal gene regulatory elements (Fig. 4C). In the blood, DMRs at W3 and W24 showed broadly similar methylation changes and genomic feature overlaps as DMRs in the hippocampus. However, blood DMRs at W8 were more numerous, mostly hypomethylated and predominantly genic, with nearly half overlapping exons (Fig. 4A and C).
AppNL-G-F mice show dynamic DNA methylation changes in hippocampus and blood. (A) Venn diagram shows DMR-containing genes shared across different timepoints in blood and hippocampus. (B) Bar heights shows number of DMRs in each timepoint and tissue. Shade within each bar indicates proportion of DMRs that are hypermethylated (darker shade) vs. hypomethylated (white) in AppNL-G-F mice, with exact values also shown. (C) Pie charts show disruption of DMR overlapping various gene features. Intergenic DMRs located > 3 kb from the start or end of a gene were classified as “distal intergenic”. Intergenic DMRs located < 3 kb from a gene start or end were categorized as being “upstream” or “downstream”, respectively. (D) Upset plot shows number of gene associated with at least one DMR across three timepoints and two tissues, illustrating the extent of overlap between these groups. (E) Dot plots display the statistical significance of the top five most enriched biological Gene Ontology (GO) pathways associated with DMRs in the hippocampus and blood.
We next evaluated the correspondence between DNA methylation in the hippocampus and the blood. Overall, we found average gene promoter methylation in blood and brain of individuals to be significantly correlated regardless of age and genotype (Supplementary Fig. 3). We identified several genes that were differentially methylated in both hippocampus and blood (although their DMRs were not necessarily located at the same exact genomic position). Most of these genes showed differential methylation in the blood at W8 (Fig. 4D), likely due to the high number of blood DMRs at this age. Notably, W3 showed the second-highest number of DMR-associated genes shared between the brain and blood (Fig. 4D), indicating the potential to detect early blood-based epigenetic signals that reflect brain epigenetic state during the initial stages of disease. When we restricted the analysis to DMRs overlapping the exact same genomic regions, rather than those associated with the same genes, we identified only seven shared DMRs between brain and blood, with two detected at the same age (Supplementary Table 8; Supplementary Fig. 4). One of these, is a 1 kb non-coding DMR located approximately 15 kb upstream of the Tmprss15 gene, displaying 32% hypomethylation in the brain and 37% hypomethylation in the blood of AppNL-G-F mice at W3. This hypomethylation remains significant in the blood by W8. (Supplementary Table 8; Supplementary Fig. 4). Tmprss15 is thought to play a role in neurogenesis and/or APP metabolism and has been found duplicated in some early-onset AD cases associated with Down syndrome45. Despite containing DMRs, Tmprss15 does not exhibit differential expression or chromatin accessibility based on our RNA-seq or snATAC-seq data. Another DMR shared between blood and brain was observed at W8 overlapping the Eif4A3 gene (Supplementary Fig. 4). This region is 11% hypomethylated in blood, but 11% hypermethylated in brain. Eif4A3 is an RNA-binding protein implicated in RNA metabolism, splicing, and nonsense-mediated decay46, as well as axon development47, processes that may contribute to broader mechanisms of neurodegeneration. While Eif4A3 does not show differential chromatin accessibility, it is significantly downregulated in the hippocampus at W3 in our RNA-seq data (Supplementary Table 4). The rest of the DMRs shared between brain and blood were associated with Mettl27, Tns1, Srp54b, Gm32357 and Rai2 genes (Supplementary Table 8; Supplementary Fig. 4). These genes do not exhibit differential expression or accessibility in our datasets (Supplementary Table 4) and, to our knowledge, lack strong ties to neurodevelopment or neurodegenerative disease.
Building on our RNA-seq findings, we examined the relationship between hippocampal gene expression and DNA methylation across all DMRs. We identified 104 DEG that were associated with at least one DMR, though not necessarily at the same age. Of these, 43 genes exhibited concurrent changes in DNA methylation and gene expression within the hippocampus at the same age: 39 genes at W3, 3 at W24, and 1 at both W8 and W24 (Supplementary Table 4). Of note, several of the differentially methylated and expressed genes at W3 have been previously implicated in AD, including Atxn1, Rbfox1 and Ep300. Loss of Atxn1 in mouse models increases BACE1 expression and promotes amyloidogenic processing of APP, exacerbating Aβ pathology in AD-vulnerable brain regions48. Similarly, loss or reduced expression of Rbfox1 is linked to increased amyloid burden and widespread dysregulation of neuronal splicing programs that affect synaptic genes49,50, while the histone acetyltransferase Ep300 (p300) modulates chromatin states and cognitive-related and amyloid-reducing pathways51,52. Beyond these individual genes, we observed a broader pattern across all three time points and in both blood and hippocampus, in which promoter DNA methylation levels were significantly negatively correlated with hippocampal gene expression (Supplementary Fig. 5). These patterns suggest that DNA methylation in both the blood and brain is broadly, and at times locally, associated with levels of gene expression in the hippocampus. Based on a published single-cell atlas of adult mouse cerebrum cis-regulatory elements (i.e. CREs)53, we found that 42% of our hippocampus DMRs at W3, 44.2% of DMRs at W8 and 38.2% of W24 DMRs overlap with at least one CRE identified in one or more mouse brain cell types. Similarly, in the blood, 33.6% of our DMRs at W3, 75.5% of DMRs at W8, and 28.2% at W24 overlapped with brain CREs53, indicating that methylation changes detected in blood and hippocampus of AppNL-G-F may impact function of brain gene regulatory elements. Additionally, the distance between differentially expressed genes in the hippocampus of AppNL-G-F mice and the nearest DMRs in either blood or brain was significantly shorter than expected by chance (Wilcoxon signed-rank test, p < 0.05). All in all, these observations suggest that some DNA methylation differences in AppNL-G-F hippocampus and blood are tied to genotype-dependent alterations in hippocampal gene regulation and expression. Gene ontology analysis of hippocampal DMRs revealed notable enrichment in pathways related to neurodevelopment, synapse organization and cell projection at both early (W3) and late (W24) disease stages (Benjamini–Hochberg adjusted p < 0.05; Fig. 4E). In line with the limited number of DEGs at W8, hippocampus of AppNL-G-F mice also exhibited the fewest DMRs at W8, with no significant gene ontology pathways identified at this stage. Of note, blood DMRs at early stages of neuropathology (W3 and W8) also showed significant enrichment in pathways related to neurogenesis and cell projection (Benjamini–Hochberg adjusted p < 0.05), but no significant pathways were recovered for blood DMRs at W24 (Supplementary Table 9; Fig. 4E). Altogether, these observations suggest that epigenetic dysregulation associated with neuronal function arises early and persists through stages of neuropathology and that early epigenetic alterations in the blood could reflect neuronal dysfunction in AppNL-G-F mice, highlighting the potential utility of blood methylation as biomarkers for detecting early amyloidosis.
Integration of multi-omics data identifies potential early blood-based epigenetic biomarkers for Aβ pathology
To identify genes exhibiting epigenetic and transcriptional dysregulation in AppNL-G-F mice, we integrated our multi-omics datasets by linking each differentially expressed genes to its closest differential snATAC-seq peaks and differentially methylated regions in both the hippocampus and blood across all time points (Supplementary Table 4). To pinpoint the subset of genes with potential for early detection of Aβ pathology in the blood, we focused specifically on the W3 time point, aiming to capture molecular signatures prior to the emergence of major histopathological or cognitive symptoms6. We established stringent criteria to maximize reliability and generalizability of our candidate biomarkers: First, the candidate gene must contain a DMR in the blood of AppNL-G-F mice at W3, allowing for early epigenetic detection in the blood. Second, the candidate genes should contain at least one DMR in the hippocampus at W3, though exact sequence overlap between blood and hippocampus DMRs is not necessary. Third, the candidate gene linked to DMRs should be differentially expressed in the hippocampus of AppNL-G-F mice at W3.
Through the integration of our datasets, we identified five genes —Rbfox1, Camta1, Diaph2, Mkl2 and Manea— that exhibit significant epigenetic changes in the blood, as well as significant dysregulation in the brain at W3, making them potential candidates for development of early blood-based biomarkers (Fig. 5A; Supplementary Table 10). Of note, two of these genes (Rbfox1 and Camta1) are also implicated in neurodevelopment and/or neuropathology, with Rbfox1 particularly standing out due to its documented role in early and preclinical AD50. Blood of AppNL-G-F mice showed a 1 kb region of significant hypomethylation in an intron of Rbfox1, overlapping a putative mouse brain CRE (Fig. 5B;53). Average methylation at this DMR did not correlate with expression of Rbfox1 in the hippocampus across individuals (Fig. 5B). However, AppNL-G-F mice showed significant, though mild, heightened expression of Rbfox1 in the hippocampus at W3 (log2 fold change = 0.15, FDR = 0.05; Supplementary Table 4) along with an intronic hypomethylated DMR, distinct from the blood DMR (Fig. 5B). Camta1, a calcium-responsive transcriptional regulator highly expressed in the brain, showed hypomethylated intronic DMRs in both hippocampus and blood at W3 (Fig. 5C), with mild expression increase in the hippocampus (log2 fold change = 0.15, FDR = 0.02; Supplementary Table 4;54). Of note, average methylation at the Camta1 W3 blood DMR was significantly correlated with hippocampal Camta1 expression across all individuals (r = 0.35, p = 0.04; Fig. 5C). While no clear link exists between Diaph2 and AD, methylation at the W3 Diaph2 blood DMR was positively associated with hippocampal expression across all individuals (r = 0.42, p = 0.027; Fig. 5D), indicating a potential functional link between this DMR and hippocampal gene expression. All five genes identified as potential biomarkers display additional hyper- and/or hypomethylated DMRs at W8 and/or W24, hinting at prolonged and cross-tissue dysregulation of these genes during amyloidosis progression (Supplementary Table 4).
Identification of candidate early blood-based biomarker genes through integration of gene expression and DNA methylation data. (A) Venn diagram displays the number of genes meeting the criteria for candidate early blood-based biomarkers for amyloidosis. (B) Left: UCSC genome browser screenshots provide a zoomed-out and zoomed-in view of the candidate biomarker DMR associated with Rbfox1, alongside additional brain or blood DMRs identified at other time points. The CREs track displays mouse brain cis-regulatory elements from Li et al. 2021. Right: A scatterplot shows the average methylation at the focal DMR versus the normalized expression of the corresponding gene in the hippocampus. (C, D) Similar UCSC genome browser views and methylation versus expression plots are shown for the candidate biomarkers associated with Camta1 (C) and Diaph2 (D).
It should be noted that our data integration also identified many additional candidate genes that met only a subset of our stringent biomarker criteria. Several of these genes were highly relevant to AD and/or neurodevelopment (Supplementary Table 10). For instance, among the 48 genes that contained DMRs in both the blood and brain of AppNL-G-F mice at W3 but were not DEGs, at least six have been widely implicated in neurodevelopment and neurological diseases: Abca13, Adgrl3, Cntnap2, Dab1, Dpp6 and Lingo255,56,57,58,59,60. Additionally, of the 48 candidate marker genes differentially methylated in the blood (but not hippocampus) and differentially expressed in the hippocampus at W3, several are linked to neurodevelopment and neuropathology, including Anks1b, Atxn2, Auts2, Dcc, Gabbr2, Hdac, Nfia, and Slc16a238,44,61,62,63,64,65,66. Overall, our findings highlight five promising candidate genes, as well as many additional genes of interest, that merit further investigation as potential early biomarkers or contributors to AD pathology.
Discussion
Understanding Alzheimer’s disease (AD) during its preclinical stages is critical for uncovering the early mechanisms of neuropathology and identifying biomarkers for early diagnosis and intervention. However, studying these early stages in humans poses major challenges, including limited access to relevant tissues, such as the brain, and the inability to predict when disease onset will occur. These barriers underscore the need for robust, well-characterized animal models. The AppNL-G-F knock-in mouse model presents a valuable system in this regard, as it develops early-onset amyloid pathology along a predictable timeline without overexpression of APP beyond physiological levels and in the absence of endogenous murine APP (Fig. 1A). This model provides a controlled system to investigate the mechanisms preceding severe AD neuropathology, independent of advanced aging. While this model has been extensively characterized with respect to various phenotypes including cognition67 behavior68, synaptic function68, and even the microbiome8, its epigenetic landscape before and after amyloidosis onset remains largely unexplored, limiting our understanding of the epigenetic basis of AD and the development of early blood-based epigenetic biomarkers. In this study, we apply a multiomic approach to examine epigenetic and gene expression changes in the hippocampus and peripheral blood of epigenetics of AD pathology and development of blood-based early epigenetic biomarkers. All assays were performed on the same set of 60 animals (5 biological replicates per condition), enabling direct comparisons across data types.
Despite the progressively worsening amyloid deposition reported previously in the brain of AppNL-G-F mice6,7,69, we observed no significant differences in hippocampal cell composition between genotypes or across the time points examined. While subtle compositional changes cannot be ruled out and might be detected in larger datasets, our findings suggest they are unlikely to be substantial, supporting the use of bulk tissue analyses for gene expression and DNA methylation without major confounding from cell type shifts. Despite the overall stability in cell composition, the hippocampus of AppNL-G-F mice showed changes in chromatin accessibility with age in neuronal cells. Other cell types, particularly glial cells, such as microglia and astrocytes, likely also undergo significant changes during disease progression in our model, as observed in human AD studies70. However, we were unable to test them due to the low and variable non-neuronal cell counts in our snATAC-seq assay. In early pathology stages (W3), we noted a few chromatin accessibility changes in excitatory neurons, suggesting excitatory neurons to be susceptible to early Aβ deposition, aligning with previous findings in a tau pathology mouse model71, reports of selective neuronal vulnerability in neurodegenerative diseases72, as well as a higher hippocampal activity and pathological neuronal hyperexcitability in earlier stages of AD73,74. Later in pathology (W24), changes in chromatin accessibility were only detected in inhibitory neurons. These changes may reflect the increasing cellular stress due to Aβ burden and worsening neural network dysfunction, which likely contributes to the cognitive impairment observed in the AppNL-G-F model around this age6. Epigenetic modifications in inhibitory neurons may represent compensatory mechanisms aimed at maintaining inhibitory control and mitigating excitatory neuron dysregulation or excitotoxicity75,76, as Aβ pathology is known to disrupt the delicate balance between excitatory and inhibitory neuronal activity in AD75,76. These findings signify the importance of understanding temporal and cell-type-specific mechanisms in AD for developing effective and targeted therapeutic strategies. Our findings suggest that early-stage therapies targeting excitatory neurons may help preserve function in early stages of the disease, while later interventions supporting inhibitory neurons could stabilize neural networks and slow disease progression.
Surprisingly, the most extensive gene expression differences we found was in the hippocampus of three-week old AppNL-G-F mice, despite the lack of cortical amyloid plaques at this age6. This extends recent findings from Rico et al. 2024, which reported hippocampal transcriptional changes beginning as early as 6 weeks in AppNL-G-F males24. Although the overlap between DEGs from this study and those reported in previous analyses of AppNL-G-F mice and AD patients was modest (likely in part reflecting differences in experimental design and analysis) our results highlight regulatory disruptions in many AD-relevant genes and identify alterations in metabolism and protein biosynthesis pathways as among the earliest transcriptional changes in the hippocampus, consistent with patterns reported in both mouse models24 and AD patients77,78. The pronounced gene expression differences at W3 might reflect genotype-by-age differences in expression trajectories, developmental transitions unique to this age, and greater variability or non-linear regulatory dynamics during disease progression. Nevertheless, our findings show that major molecular dysregulation precedes the onset of neuropathological symptoms, underscoring the importance and promise of studying early stages of disease. The small number of significant DEGs at W8 may be attributable to technical limitations in our study. However, if validated by further research, this finding could indicate that W8 represents a transitional stage in disease progression in this model, during which compensatory mechanisms begin to fail, giving way to neuroinflammation and synaptic dysfunction. Further studies focused on this timepoint could help clarify the molecular events happening during this period. Consistently, at W24, when amyloid deposition is the most pronounced, the changes in expression profile mostly showed enrichment of immune and neuroinflammatory pathways. The upregulation of genes associated with microglial activation, phagocytosis, and cytokine signaling at this later stage aligns with the high level of Aβ plaques reported at this age and the well-documented role of neuroinflammation in later stages of AD pathology79. The observed changes in synapse pruning pathways at W24 further align with the cognitive deficits reported in the AppNL-G-F model at this age6, suggesting that the immune response may play a role in synaptic loss and dysfunction during AD progression. Overall, the variations in gene expression patterns in early versus late amyloidosis suggest that the brain undergoes drastically different physiological phases as symptoms emerge and worsen.
Our whole-genome bisulfite sequencing revealed that significant DNA methylation differences also exist between AppNL-G-F and wild-type mice, spanning all stages of disease in both the hippocampus and blood. In the hippocampus, differentially methylated regions in early (W3) and late (W24) stages showed enrichment of neurodevelopment and neuron connectivity pathways. These findings suggest persistent epigenetic dysregulation linked to hippocampal neuronal function throughout disease progression. Interestingly, hippocampus of W8 AppNL-G-F mice exhibited the fewest DMRs and no significant pathway enrichment, supporting the idea that this stage may represent a transitional phase, a hypothesis warranting further investigation. In the blood we observed dynamic methylation changes across timepoints, with a striking increase in DMRs at W8. Although direct overlaps between blood and brain DMRs were rare, the broad associations between blood methylation and hippocampal gene regulation, overlap of blood DMRs with brain cis-regulatory elements, and the proximity of DMRs to hippocampal DEGs, suggest that blood methylation patterns broadly reflect gene dysregulation in the brain. Furthermore, both blood and brain DMRs were enriched in pathways related to neuronal development in the earliest stage of amyloidosis, underscoring a link between peripheral epigenetic signals in the blood and early brain dysregulation. These findings highlight the potential utility of blood DNA methylation as a biomarker for detecting early stages of amyloidosis and related neuropathological processes.
By integrating our multi-omics data, we identified DMRs at Rbfox1, Camta1, Diaph2, Mkl2 and Manea as the most promising biomarker candidate genes exhibiting epigenetic dysregulation in both brain and blood before onset of severe amyloidosis. Notably, the current mouse and human literature clearly implicates Rbfox1 and Camta1 in neurodevelopment and/or neuropathology80,81,82,83,84,85,86, with Rbfox1 (RNA Binding Fox-1 Homolog 1) being particularly significant50. Rbfox1 encodes a neuronal RNA-binding protein, and recent human studies have identified it as a novel locus associated with AD during preclinical stages50. In AD patients, RBFOX1 is notably localized around amyloid-β plaques and reduced RBFOX1 expression correlates with a higher amyloid-β burden50. In DNA methylation human studies, RBFOX1 has been shown to exhibit differential DNA methylation in the blood of individuals with AD and mild cognitive impairment80, as well as in cortical brain tissue of AD patients81. Camta1 (calmodulin-binding transcription activator 1) is a transcription factor regulating gene expression in response to calcium signaling. Dysregulated calcium signaling is associated with several hallmarks of AD in both patients and mouse models, including amyloid-β deposition, tau hyperphosphorylation, synaptic dysfunction, and apoptosis, suggesting that CAMTA1 may contribute to AD progression82. CAMTA1 has been implicated in episodic memory83 an immediate recall, as well as various neurological and neurodegenerative diseases, including intellectual disability, ataxia, and amyotrophic lateral sclerosis84,85,86. Additionally, in humans CAMTA1 is thought to regulate neuroprotective genes under stress, and significant differences in CAMTA1 DNA methylation have been found in the peripheral blood of stroke patients87, in the blood of individuals with mild-cognitive impairment80 and in the blood and brain cortex (including prefrontal cortex) of AD patients80,81,88, highlighting its potential relevance as a candidate for further investigation in AD and amyloidosis. Moving forward, validating these candidates in humans, and elucidating their functional roles in early disease progression will be critical. Nevertheless, our findings underscore the potential of DNA methylation at these loci as early indicators of amyloidosis and provide a foundation for further research into the mechanisms driving their epigenetic dysregulation.
Despite the intriguing findings of our study, several limitations warrant consideration. First, the AppNL-G-F mouse model used in this study mirrors familial autosomal dominant AD, which accounts for only 1–5% of all AD cases, raising questions about the generalizability of our findings to sporadic and late-onset AD. Moreover, this model does not develop tau pathology, a hallmark of AD, potentially overlooking critical amyloid-tau interactions. Furthermore, our analysis was limited to three timepoints and one brain region, which may miss key transitions in gene expression and epigenetic regulation occurring at other intervals or in other tissues. Second, the brain’s heterogeneous cellular composition and the differential vulnerability of cell types to AD pathology89 highlight the need for single nucleus approaches with improved resolution and sample sizes, as well as reduced technical variability. Such methods would provide more precise insights into cell-type-specific dynamics, addressing the limitations of bulk tissue analysis. Also, for our snATAC-seq assay we pooled hippocampal nuclei from mice of the same genotype and sex to obtain sufficient input material. Although we aimed to contribute equal material from each animal, pooling can introduce uneven representation across individuals, and we recovered variable cell numbers across conditions. This limitation constrained the resolution of our snATAC-seq analysis, and our chromatin accessibility findings should therefore be interpreted with caution and validated in future studies. Finally, we used both male and female samples as replicates to enhance statistical power, and because we did not detect major sex differences in gene expression patterns. While this strategy may obscure potential sex-specific epigenetic differences, it does not compromise our goal of identifying universal mechanisms of early pathology that are relevant to both sexes. Nonetheless, given the well-established sex differences in AD risk and progression90, future research should employ larger sample sizes to enable the investigation of whether distinct molecular and epigenetic mechanisms drive pathology development in males versus female AppNL-G-F mice.
While subject to some limitations, our findings nevertheless illustrate the unique strength of a multi-omics approach for uncovering epigenetic and transcriptional alterations that arise before overt neuropathology in an amyloidosis mouse model. Many of the genes we found affected early in disease were previously implicated in AD or neurodevelopment in experimental mouse models and patients. Moreover, our results point to the potential of blood-based epigenetic biomarkers as minimally invasive tools for early diagnosis and intervention in Alzheimer’s disease. Future research should further validate these findings in human cohorts and elucidate the functional consequences of these epigenetic and gene expression changes in both brain and peripheral tissues, ultimately advancing our understanding of Alzheimer’s disease and informing strategies for intervention before irreversible damage occurs.
Methods
Animal use approval statement
All procedures involving live animals complied with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and with IACUC approval at Oregon Health & Sciences University.
ARRIVE accordance statement
All methods are reported in accordance with ARRIVE guidelines.
Mouse breeding paradigm and tissue collection
The mice were maintained on a 12/12 h light/dark schedule (lights on at 06:00). Laboratory chow (PicoLab Rodent diet 20, #5053; PMI Nutrition International, St. Louis, MO, USA) and water were provided ad libitum. We bred hAPP knock-in mice containing the Swedish, Iberian and Arctic mutation (i.e. AppNL-G-F) on a C57BL/6 J background, generated by Dr. Saito6,91 and shared with us with C57BL/6 J wild-type (WT) mice from Jax. Heterozygous breeding pairs were set up to generate homozygous AppNL-G-F and WT littermates. We genotyped all offspring and only used homozygous AppNL-G-F and WT individuals for downstream analysis. The genotyping protocols are available on the Riken Institute web site6,91. Briefly, we used primers reported in Table 1 to amplify part of exon 17 containing the Artic mutation. The PCR products were digested with the MboII restriction enzyme and genotype was determined based on digestion fragments (WT genotype is cut into 171 bp and 67 bp fragments, while presence of the Arctic mutation prevents digestion). For genotyping of the Iberian mutations, we used PCR to amplify part of exon 17 containing the Iberian mutation (Table 1). The PCR products were digested with the BsaBI restriction enzyme (WT amplicon is cut into 171 bp and 67 bp fragments, while presence of the Iberian mutation prevents digestion). Due to COVID19-related modified operations, we shipped some biopsy samples for genotyping to Transnetyx, Cordova, TN.
At postnatal week 3, 8, or 24 (W3, W8 or W24), we collected n = 5 females and n = 5 males per genotype (homozygous AppNL-G-F vs. WT), for a total of 60 individuals. The mice were euthanized by cervical dislocation. Trunk blood was collected into EDTA-treated tubes. Blood was centrifuged at 5,500 g for 10 min and the supernatant was transferred to a new tube and stored at − 80 °C until assay. For each animal, the hippocampus from both hemispheres was dissected, finely chopped and combined into a single sample. The chopped hippocampus tissue from each individual was then split into three portions before freezing and storage at − 80 °C. Each portion was used for one of the three assays (i.e., snATAC-seq, RNA-seq, or WGBS) ensuring that all assays were performed on the exact same individuals.
Single-nucleus assay for transposase-accessible chromatin sequencing (snATAC-seq) library preparation and analysis
Fresh frozen hippocampus aliquots of the same 60 subject described above were used for snATAC-seq analysis. Briefly, to obtain sufficient nuclei, we pooled hippocampus from replicates within each sex/age/genotype combination, resulting in 12 final pools (one male and one female pool per age and genotype condition). Tissue pools were homogenized separately in 2 mL chilled NIB (10 mM HEPES, pH 7.2, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL [v/v; Sigma-Aldrich, Cat#I8896], 0.1% Tween-20, and 1 × protease inhibitor [Roche, Cat#11873580001]) and 10 mM D( +)-Glucosamine hydrochloride [Sigma Aldrich G1414] in a 7 mL dounce-homogenizer on ice for 10 min. The homogenate was then strained through a 35 µm strainer and counted using a hemacytometer and trypan blue. The samples were centrifuged at 500 × g for 10 min at 4 °C. Samples were aspirated, resuspended in ice-cold PBS-BSA buffer (0.5%. Bovine serum albumin (BSA), in 1 × PBS, and 30 mM D( +)-Glucosamine hydrochloride) to obtain 50,000 nuclei/5 ul of PBS-BSA-glucosamine. We tagmented 50,000 nuclei for each condition by adding 5 ml ETB3 and 5 ml of an individual Tn5 (Table 2) at 37 °C for 1 h (f/c of D-glucosamine was 10 mM) and iced 5 min. Nuclei were spun down (3 min, 500xg, 4 °C) and washed in 1.5 mL TMG buffer (36% TAPS premix (4X TAPS-TD buffer (132 mM TAPS (N-[Tris(hydroxymethyl)methyl]-3-aminopropanesulfonic acid, Sigma-Aldrich, Cat#T0647-100G) pH = 8.5, 264 mM potassium acetate, 40 mM magnesium acetate), 64% glycerol)/SCALE wash buffer twice. Nuclei were counted with a hemacytometer and trypan blue. Samples were multiplexed and processed as written in Step 2 (Gem Generation and Barcoding) of the 10 × Genomics Single Cell ATAC v2 Kit protocol. 15 ml of each multiplexed pool of nuclei was added to 60 ml of the 10 × Master Mix. We proceeded with the 10 × protocol. For Step 4.1.c (Sample Index PCR), we substituted Sample Index N, Set A Reagent – with a ScaleBio S700 index primer compatible with the ScaleBio tagmentation (Table 2). Libraries were processed as described in the 10 × Genomics Single Cell ATAC v2 Kit. They were quantified via the Qubit dsDNA High Sensitivity assay (Thermo Fisher Q32851) and via the Agilent Tapestation 4150 D500 tape (Agilent 5067–5592). Libraries were sequenced on the Illumina NextSeq2000 for 650 pM with a P2-200 flow cell (Illumina Inc., 20,046,812). ScaleBio tagmented libraries were sequenced paired-end with 85 cycles for read 1, 125 cycles for read 2, 8 cycles for index 1, and 16 cycles for index 2.
Raw snATAC-seq data was processed using the previously described scitools package92. Briefly, the sequencing reads were demultiplexed, and the read names were replaced with the cell barcode and a unique identifier. Reads were then mapped to the mouse genome (mm10) using BWA-MEM93. The resulting BAM files were filtered to remove low quality and duplicated reads. Additionally, we removed all barcodes with less than a specified number of reads. We used the ArchR (v 1.01) package94 to filter cells with TSS enrichment < 2, unique fragments < 1000 and doublets. Using ArchR, iterative LSI dimension reduction and clustering was performed using the 500 bp tile matrix, producing 21 clusters that were visualized via UMAP. Gene activity scores were determined and used to find marker genes for each cluster. Cell clusters were manually annotated via manual curation using top marker genes and publicly available data12,53. Out of the 21 clusters, four (clusters 8, 9, 10, and 19) could not be confidently classified and were broadly called “unclassified”. Clusters 9 and 19 were presumed to represent technical noise due to their UMAP distribution and lack of distinct marker genes (Supplementary Table 1). Clusters 8 and 10 were labeled as Trpm3-defined and Grm4-defined, respectively, based on their top marker genes (Supplementary Table 1). Sample pools with fewer that 100 cells after filtering were excluded from downstream analysis. This resulted in exclusion of the wild-type pool at W8 (n = 88 cells). Cell composition of each sample pool was determined as percentage of total cells in the pool belonging to any cluster. Cell type proportion analysis was performed using the propeller method from the speckle v1.6.0 R library95 and calculated per sample, followed by arcsine square-root transformation to stabilize variance. A linear model was fit using limma v3.62.196, with experimental group (time point + genotype) and sex included in the model. Differential proportion testing was performed using contrasts between conditions of interest, followed by empirical Bayes moderation. Statistical significance was assessed using moderated t-tests, and results were extracted with the topTable function.
Next, peaks were called at the pseudo-bulk level using all cells grouped by age/genotype combination, and cell-type. This was accomplished by using the addGroupCoverages function with parameters minCells = 30, maxCells = 100, minReplicates = 2, maxReplicates = 3, sampleRatio = 0.8 along with the addReproduciblePeakSet functions with parameters peaksPerCell = 500 and minCells = 20. Marker peaks/genes for each cell type and age/genotype combination were determined. Next, we focused only on excitatory and inhibitory neurons (granule neurons were included as excitatory neurons). Peaks were called once again at the pseudo-bulk level grouped by age/genotype and cell type. Parameters for the addGroupCoverages function were minCells = 20, maxCells = 100, minReplicates = 4, maxReplicates = 8, sampleRatio = 0.8, and parameters for addReproduciblePeakSet were peaksPerCell = 500 and minCells = 20. Pairwise comparisons were performed using ArchR’s getMarkerFeatures to identify regions with significantly different chromatin accessibility between WT and AppNL-G-F at each age, within excitatory neurons and inhibitory neurons.
Bulk RNA-seq library preparation and analysis
Total RNA was extracted from each of the 60 fresh frozen hippocampus aliquots without pooling as described above, using the NEB Monarch Total RNA Miniprep Kit (Cat T2010S, New England Biolabs). Briefly, samples were weighed and pulverized in a bead basher with DNA/RNA protection buffer. Pulverized samples were incubated with Proteinase K at 55 °C for 5 min, followed by a 3 min centrifugation at 16,000 g. Supernatants were mixed with lysis buffer and centrifuged in gDNA removal columns. The flow through was mixed with 100% Ethanol and centrifuged in RNA columns at maximum speed. The columns were incubated with DNAse I/DNAse I buffer for 15 min at room temperature, followed by addition of priming buffer and centrifugation. Finally, the column was washed twice with a wash buffer and the RNA was eluted in water. RNA concentrations were measured with Nanodrop (Thermofisher) and integrity was confirmed (RIN > 7) with the Bioanalyzer RNA kit (Agilent). Samples were stored at − 80 °C until further use. RNA-seq libraries were generated blindly from 1ug of total RNA per animal, beginning with an rRNA depletion of samples using the NEBNext rRNA Depletion Kit v2 for human/mouse/rat (New England Biolabs) according to manufacturer’s protocol. Samples were then purified with NEBNext RNA Sample Purification Beads (New England Biolabs). The RNA was then fragmented, primed, and reverse transcribed using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs). Libraries were prepared using the NEBNext Ultra II (New England Biolabs), with size selection to 300 bp using Ampure beads (Beckman Coulter). The final libraries were quantified using Qubit (Thermofisher) and 150 bp paired-end sequenced (30–80 million reads per sample, Supplementary Table 3) on the Illumina Nova-Seq 6000 Platform at the Massively Parallel Sequencing Shared Resource (MPSSR) at Oregon Health and Science University (OHSU).
Raw reads were QC’d using FastQC97 and aligned to the mm10 genome using STAR98 (–runThreadN 8, –readFilesCommand zcat –outSAMstrandField intronMotif –outFilterIntronMotifs RemoveNoncanonicalUnannotated –quantMode GeneCounts –twopassMode Basic). The raw gene counts table was generated using custom bash scripts and filtered by the filterByExpr function from edgeR99 with default parameters and defining group membership as the combination of age and genotype. DESeq2100 was used to perform differential analysis. Pairwise comparisons were performed to compare gene expression between wild-type and AppNL-G-F at each age separately with sex as a covariate. The LRT test from DESeq2 was used to find genes with significant ageXgenotype interaction, followed by clustering according to the expression profile pattern using the degPatterns function from DEGreport101.
Whole genome bisulfite sequencing (WGBS) library preparation and analysis
DNA from each of the 60 hippocampus samples was extracted using the NEB Monarch Genomic DNA Purification Kit (Cat T3010, New England Biolabs) according to manufacturer’s protocol and without pooling. Extracted gDNA was quantified with Qubit (Thermofisher) and Nanodrop (Thermofisher) and stored at − 80 °C. DNA from 60 blood samples was extracted using the Purgene Genomic DNA Purification from Blood kit (Cat D-5500, Qiagen) based on a modified version of the Puregene DNA purification from blood protocol (Gentra Systems). Briefly, cell lysis solution was added directly to the frozen blood at a 5:1 ratio and incubated at 55 °C overnight, followed by 40 min or 1,000 rpm agitation. The samples were cooled on ice and protein precipitation solution was added (1:3 ratio) followed by 1,000 rpm agitation and 16,000 g centrifugation for 1 min. The lysate was moved to a 1:1 ratio of isopropyl alcohol and centrifuged to pellet the DNA. Pellets were washed with 80% ethanol, air dried, and resuspended at 65 °C with 1,000 rpm agitation. The gDNA samples were then quantified with Qubit (Thermofisher) and Nanodrop (Thermofisher) and stored at − 80 °C. To generate whole genome bisulfite sequencing (WGBS) libraries for both hippocampus and blood,100 ng of gDNA per animal was randomly sheared with a Bioruptor Pico Sonicator (Diagenode) at 30:30 on/off for 15 cycles. Libraries were blindly prepared with the NEBNext Ultra II Modules (New England Biolabs) and the NEBNext Methylated Adaptor (New England Biolabs), with size-selection to 200 bp using Ampure beads (Beckman Coulter). Bisulfite conversion and subsequent cleanup was performed with the EZ DNA Methylation-Gold Kit (Zymo Research). The converted libraries were PCR amplified using NEBNext Q5U polymerase and the NEBNext Multiplex Oligos for Illumina (New England Biolabs) for unique library barcoding. The final libraries were quantified using Qubit (Thermofisher) and then normalized and multiplexed for 100 bp paired-end sequencing on the Illumina Nova-Seq 6000 Platform (Supplementary Table 7).
The blood and the hippocampus WGBS datasets were analyzed separately, but similarly. Raw sequencing reads were trimmed with TrimGalore102, and then aligned to the mm10 reference genome with Bismark103 using default parameters. Deduplication was performed with deduplicate_bismark, followed by bismark_methylation_extractor with parameters –ignore 2, –ignore_r2 2, and –ignore_3prime_r2 2. Analysis for identifying Differentially Methylated Regions (DMRs) was performed with methylKit104 with the coverage files from Bismark methylation extractor data serving as input, and applying a Chi-square test with overdispersion correction to obtain a balance between sensitivity and specificity105. The genome was tiled into non-overlapping 1 kb tiles, that were merged based on the following parameters: CpG coverage threshold ≥ 1X, covered CpGs in the tile ≥ 0, and tile present in ≥ 3 samples per group, resulting in 2,364,188 1 kb tiles in the blood samples and 2,363,219 tiles in the hippocampus samples (corresponding to ~ 87% of the mouse genome). Sex was included as a covariate.
Overlap with gene features was determined using genomation106, with default settings. Intersection with the mouse cerebrum atlas of gene regulatory elements53 was done using BEDtools107.
Gene Ontology term analysis
Gene ontology (GO) term analysis of DEGs from RNA-seq data was conducted using EnrichR39,108,109 with default settings, with all annotated genes in the GO Biological Process 2024 as background. A subset of significant pathways (q < 0.01) was visualized across time points in dot plots using the ggplot2 library110 in R. To assess and visualize the enrichment of biological functions associated with differential snATAC-seq peaks and WGBS DMRs, which include both coding and non-coding regions, we utilized gProfiler111. The gProfler analysis were performed using the whole mm10 genome as background, with an ordered query (sorted by adjusted p-values from most to least significant), using a 0.05 significance threshold based on the Benjamini–Hochberg false discovery rate correction. Dot plots of a subset of significant biological pathways were generated using ggplot2110 in R.
Multi-omics data integration
To integrate findings across our snATAC-seq, RNA-seq and WGBS datasets and prioritize candidate biomarker genes based on concordant transcriptional and epigenetic changes, we linked DEGs from bulk RNA-seq to nearby differentially accessible regions (from snATAC-seq) and DMRs across both brain and blood. For each DEG, we used the BEDtools107 closest command with the -d option to identify the nearest differential accessibility or DMR and compute the absolute distance between features. Separate analyses were conducted for each omics modality (snATAC-seq, brain WGBS, blood WGBS) and added to one comprehensive table. Only regions identified as significantly differential were considered for integration. These integrated results are provided in Supplementary Table 4, enabling downstream filtering or prioritization based on co-occurrence or proximity of regulatory signals.
To investigate the distance between DMRs and DEGs, we used the closest-features program (–closest –delim ‘\t’ –dist) from the BEDOPS toolkit112 to calculate distance of each DEG from the closest DMR. These observed distances were then compared using the Wilcoxon signed-rank test to distances obtained against a randomly shuffled set of DMRs, generated by BEDtools shuffle (-chrom -noOverlapping)107.
To assess overlap of DMRs with putative gene regulatory elements, we utilized the published single nucleus atlas of adult mouse cerebrum cis-regulatory elements (CREs)53, and used bedtools intersect107 was used to identify DMRs with ≥ 1 bp overlap in enhancer regions annotated in at least one cell type.
Data availability
Raw and processed RNA-seq, snATAC-seq and WGBS data generated as part of this study are available on the NCBI Gene Expression Omnibus (GEO; [https://www.ncbi.nlm.nih.gov/geo/](https://www.ncbi.nlm.nih.gov/geo)) under accession numbers GSE290305, [GSE290154](https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE290154) and GSE290397, respectively.
References
Knopman, D. S. et al. Alzheimer disease. Nat. Rev. Dis. Primers 7, 33. https://doi.org/10.1038/s41572-021-00269-y (2021).
Vitek, M. P. Increasing amyloid peptide precursor production and its impact on Alzheimer’s disease. Neurobiol Aging 10, 471–473. https://doi.org/10.1016/0197-4580(89)90102-4 (1989).
Small, S. An Open Letter to Dennis J. Selkoe, M.D., <https://www.alzforum.org/news/community-news/open-letter-dennis-j-selkoe-md.> (2024).
Gale, S. A. Language and meaning: asymptomatic alzheimer’s disease in the clinic and society. J. Alzheimers Dis. 99, 489–492. https://doi.org/10.3233/JAD-240195 (2024).
Bryan, K. J., Lee, H. G., Perry, G., Smith, M. A. & Casadesus, G. in Methods of Behavior Analysis in Neuroscience Frontiers in Neuroscience (ed J. J. Buccafusco) (2009).
Saito, T. et al. Single app knock-in mouse models of alzheimer’s disease. Nat. Neurosci. 17, 661–663. https://doi.org/10.1038/nn.3697 (2014).
Holden, S. et al. Apolipoprotein E isoform-dependent effects on human amyloid precursor protein/abeta-induced behavioral alterations and cognitive impairments and insoluble cortical abeta42 levels. Front. Aging Neurosci. 14, 767558. https://doi.org/10.3389/fnagi.2022.767558 (2022).
Kundu, P. et al. Integrated analysis of behavioral, epigenetic, and gut microbiome analyses in App(NL-G-F), App(NL-F), and wild type mice. Sci. Rep. 11, 4678. https://doi.org/10.1038/s41598-021-83851-4 (2021).
Maguire, E. A., Burgess, N. & O’Keefe, J. Human spatial navigation: Cognitive maps, sexual dimorphism, and neural substrates. Curr. Opin. Neurobiol. 9, 171–177. https://doi.org/10.1016/s0959-4388(99)80023-3 (1999).
Samuel, W., Masliah, E., Hill, L. R., Butters, N. & Terry, R. Hippocampal connectivity and Alzheimer’s dementia: Effects of synapse loss and tangle frequency in a two-component model. Neurology 44, 2081–2088. https://doi.org/10.1212/wnl.44.11.2081 (1994).
Johnson, T. S. et al. Combinatorial analyses reveal cellular composition changes have different impacts on transcriptomic changes of cell type specific genes in Alzheimer’s Disease. Sci. Rep. 11, 353. https://doi.org/10.1038/s41598-020-79740-x (2021).
O’Connell, B. L. et al. Atlas-scale single-cell chromatin accessibility using nanowell-based combinatorial indexing. Genome Res. 33, 208–217. https://doi.org/10.1101/gr.276655.122 (2023).
Baloni, P. et al. Multi-Omic analyses characterize the ceramide/sphingomyelin pathway as a therapeutic target in Alzheimer’s disease. Commun. Biol. 5, 1074. https://doi.org/10.1038/s42003-022-04011-6 (2022).
Stoffel, W. et al. SMPD3 deficiency perturbs neuronal proteostasis and causes progressive cognitive impairment. Cell Death Dis. 9, 507. https://doi.org/10.1038/s41419-018-0560-7 (2018).
Lindgren, A. M. et al. Haploinsufficiency of KDM6A is associated with severe psychomotor retardation, global growth restriction, seizures and cleft palate. Hum Genet 132, 537–552. https://doi.org/10.1007/s00439-013-1263-x (2013).
Van Laarhoven, P. M. et al. Kabuki syndrome genes KMT2D and KDM6A: Functional analyses demonstrate critical roles in craniofacial, heart and brain development. Hum. Mol. Genet. 24, 4443–4453. https://doi.org/10.1093/hmg/ddv180 (2015).
Xu, Y. et al. OPTN attenuates the neurotoxicity of abnormal Tau protein by restoring autophagy. Transl. Psychiatry 12, 230. https://doi.org/10.1038/s41398-022-02004-x (2022).
Chami, M. Calcium Signalling in Alzheimer’s Disease: From pathophysiological regulation to therapeutic approaches. Cells https://doi.org/10.3390/cells10010140 (2021).
Heneka, M. T. et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 14, 388–405. https://doi.org/10.1016/S1474-4422(15)70016-5 (2015).
Skaper, S. D., Facci, L., Zusso, M. & Giusti, P. Synaptic plasticity, dementia and Alzheimer disease. CNS Neurol. Disord. Drug Targets 16, 220–233. https://doi.org/10.2174/1871527316666170113120853 (2017).
Yap, C. X. et al. Brain cell-type shifts in Alzheimer’s disease, autism, and schizophrenia interrogated using methylomics and genetics. Sci. Adv. 10, eadn7655. https://doi.org/10.1126/sciadv.adn7655 (2024).
Liu, Z., Condello, C., Schain, A., Harb, R. & Grutzendler, J. CX3CR1 in microglia regulates brain amyloid deposition through selective protofibrillar amyloid-beta phagocytosis. J. Neurosci. 30, 17091–17101. https://doi.org/10.1523/JNEUROSCI.4403-10.2010 (2010).
Puntambekar, S. S. et al. CX3CR1 deficiency aggravates amyloid driven neuronal pathology and cognitive decline in Alzheimer’s disease. Mol. Neurodegener 17, 47. https://doi.org/10.1186/s13024-022-00545-9 (2022).
Gutierrez Rico, E., Joseph, P., Noutsos, C. & Poon, K. Hypothalamic and hippocampal transcriptome changes in App(NL-G-F) mice as a function of metabolic and inflammatory dysfunction. Neuroscience 554, 107–117. https://doi.org/10.1016/j.neuroscience.2024.07.007 (2024).
Hamilton, H. L. et al. FABP7 drives an inflammatory response in human astrocytes and is upregulated in Alzheimer’s disease. Geroscience 46, 1607–1625. https://doi.org/10.1007/s11357-023-00916-0 (2024).
Moreno-Rodriguez, M., Perez, S. E., Malek-Ahmadi, M. & Mufson, E. J. APOEepsilon4 alters ApoE and Fabp7 in frontal cortex white matter in prodromal Alzheimer’s disease. J Neuroinflammation 22, 25. https://doi.org/10.1186/s12974-025-03349-y (2025).
Aguzzoli Heberle, B. et al. Systematic review and meta-analysis of bulk RNAseq studies in human Alzheimer’s disease brain tissue. Alzheimers Dement 21, e70025. https://doi.org/10.1002/alz.70025 (2025).
Pedersen, T. J., Keil, S. A., Han, W., Wang, M. X. & Iliff, J. J. The effect of aquaporin-4 mis-localization on Abeta deposition in mice. Neurobiol. Dis. 181, 106100. https://doi.org/10.1016/j.nbd.2023.106100 (2023).
Iliff, J. J. et al. Impairment of glymphatic pathway function promotes tau pathology after traumatic brain injury. J. Neurosci. 34, 16180–16193. https://doi.org/10.1523/JNEUROSCI.3020-14.2014 (2014).
Ishida, K. et al. Glymphatic system clears extracellular tau and protects from tau aggregation and neurodegeneration. J. Exp. Med. https://doi.org/10.1084/jem.20211275 (2022).
Lan, Y. L. et al. Aquaporin 4 in astrocytes is a target for therapy in Alzheimer’s disease. Curr. Pharm. Des. 23, 4948–4957. https://doi.org/10.2174/1381612823666170714144844 (2017).
Shaw, C. K. et al. X chromosome factor Kdm6a enhances cognition independent of its demethylase function in the aging XY male brain. J. Gerontol. A Biol. Sci. Med. Sci. 78, 938–943. https://doi.org/10.1093/gerona/glad007 (2023).
Davis, E. J. et al. A second X chromosome contributes to resilience in a mouse model of Alzheimer’s disease. Sci. Transl. Med. https://doi.org/10.1126/scitranslmed.aaz5677 (2020).
Qian, W. et al. Regulating Lars2 in mitochondria: a potential Alzheimer’s therapy by inhibiting tau phosphorylation. Neurotherapeutics 21, e00353. https://doi.org/10.1016/j.neurot.2024.e00353 (2024).
Wu, L. J. et al. The voltage-gated proton channel Hv1 enhances brain damage from ischemic stroke. Nat Neurosci 15, 565–573. https://doi.org/10.1038/nn.3059 (2012).
Liu, J. et al. Microglial Hv1 proton channel promotes cuprizone-induced demyelination through oxidative damage. J Neurochem 135, 347–356. https://doi.org/10.1111/jnc.13242 (2015).
O’Leary, A. et al. Behavioural and functional evidence revealing the role of RBFOX1 variation in multiple psychiatric disorders and traits. Mol. Psychiatry 27, 4464–4473. https://doi.org/10.1038/s41380-022-01722-4 (2022).
Oksenberg, N. & Ahituv, N. The role of AUTS2 in neurodevelopment and human evolution. Trends Genet 29, 600–608. https://doi.org/10.1016/j.tig.2013.08.001 (2013).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 14, 128. https://doi.org/10.1186/1471-2105-14-128 (2013).
Wang, W., Zhao, F., Ma, X., Perry, G. & Zhu, X. Mitochondria dysfunction in the pathogenesis of Alzheimer’s disease: Recent advances. Mol. Neurodegener 15, 30. https://doi.org/10.1186/s13024-020-00376-6 (2020).
Moreira, P. I., Carvalho, C., Zhu, X., Smith, M. A. & Perry, G. Mitochondrial dysfunction is a trigger of Alzheimer’s disease pathophysiology, Biochimica et Biophysica Acta BBA Mol. Basis Disease 1802(1), 2–10 (2010).
Rouaux, C., Loeffler, J. P. & Boutillier, A. L. Targeting CREB-binding protein (CBP) loss of function as a therapeutic strategy in neurological disorders. Biochem.Pharmacol. 68(6), 1157–1164 (2004).
Koh, J. Y. & Lee, S. J. Metallothionein-3 as a multifunctional player in the control of cellular processes and diseases. Mol. Brain 13, 116. https://doi.org/10.1186/s13041-020-00654-w (2020).
Laffita-Mesa, J. M., Paucar, M. & Svenningsson, P. Ataxin-2 gene: A powerful modulator of neurological disorders. Curr. Opin. Neurol. 34, 578–588. https://doi.org/10.1097/WCO.0000000000000959 (2021).
Wiseman, F. K. et al. A genetic cause of Alzheimer disease: Mechanistic insights from Down syndrome. Nat. Rev. Neurosci. 16, 564–574. https://doi.org/10.1038/nrn3983 (2015).
Ye, J. et al. Eukaryotic initiation factor 4A–3: A review of its physiological role and involvement in oncogenesis. Front Oncol 11, 712045. https://doi.org/10.3389/fonc.2021.712045 (2021).
Alsina, F. C. et al. The RNA-binding protein EIF4A3 promotes axon development by direct control of the cytoskeleton. Cell Rep 43, 114666. https://doi.org/10.1016/j.celrep.2024.114666 (2024).
Suh, J. et al. Loss of Ataxin-1 Potentiates Alzheimer’s Pathogenesis by Elevating Cerebral BACE1 Transcription. Cell 178, 1159-1175 e1117. https://doi.org/10.1016/j.cell.2019.07.043 (2019).
Fogel, B. L. et al. RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum. Mol. Genet. 21, 4171–4186. https://doi.org/10.1093/hmg/dds240 (2012).
Raghavan, N. S. et al. Association between common variants in RBFOX1, an RNA-binding protein, and brain amyloidosis in early and preclinical Alzheimer disease. JAMA Neurol. 77, 1288–1298. https://doi.org/10.1001/jamaneurol.2020.1760 (2020).
Xu, D. C. et al. Histone acetylation in an Alzheimer’s disease cell model promotes homeostatic amyloid-reducing pathways. Acta Neuropathol. Commun. 12, 3. https://doi.org/10.1186/s40478-023-01696-6 (2024).
Cho, S. B. Comorbidity genes of Alzheimer’s disease and type 2 diabetes associated with memory and cognitive function. Int. J. Mol. Sci. https://doi.org/10.3390/ijms25042211 (2024).
Li, Y. E. et al. An atlas of gene regulatory elements in adult mouse cerebrum. Nature 598, 129–136. https://doi.org/10.1038/s41586-021-03604-1 (2021).
Bas-Orth, C., Tan, Y. W., Oliveira, A. M., Bengtson, C. P. & Bading, H. The calmodulin-binding transcription activator CAMTA1 is required for long-term memory formation in mice. Learn Mem. 23, 313–321. https://doi.org/10.1101/lm.041111.115 (2016).
Knight, H. M. et al. A cytogenetic abnormality and rare coding variants identify ABCA13 as a candidate gene in schizophrenia, bipolar disorder, and depression. Am. J. Hum. Genet. 85, 833–846. https://doi.org/10.1016/j.ajhg.2009.11.003 (2009).
Maussion, G. et al. Implication of LRRC4C and DPP6 in neurodevelopmental disorders. Am. J. Med. Genet. A 173, 395–406. https://doi.org/10.1002/ajmg.a.38021 (2017).
Moreno-Alcazar, A. et al. Brain structural and functional substrates of ADGRL3 (latrophilin 3) haplotype in attention-deficit/hyperactivity disorder. Sci. Rep. 11, 2373. https://doi.org/10.1038/s41598-021-81915-z (2021).
Nawa, Y. et al. Rare single-nucleotide DAB1 variants and their contribution to Schizophrenia and autism spectrum disorder susceptibility. Hum. Genome. Var. 7, 37. https://doi.org/10.1038/s41439-020-00125-7 (2020).
Poot, M. Connecting the CNTNAP2 Networks with Neurodevelopmental Disorders. Mol. Syndromol. 6, 7–22. https://doi.org/10.1159/000371594 (2015).
Vilarino-Guell, C. et al. LINGO1 and LINGO2 variants are associated with essential tremor and Parkinson disease. Neurogenetics 11, 401–408. https://doi.org/10.1007/s10048-010-0241-x (2010).
Carbonell, A. U. et al. Haploinsufficiency in the ANKS1B gene encoding AIDA-1 leads to a neurodevelopmental syndrome. Nat. Commun. 10, 3529. https://doi.org/10.1038/s41467-019-11437-w (2019).
Dini, G. et al. NFIA haploinsufficiency: case series and literature review. Front. Pediatr. 11, 1292654. https://doi.org/10.3389/fped.2023.1292654 (2023).
Li, Y. et al. Association between DCC polymorphisms and susceptibility to autism spectrum disorder. J. Autism. Dev. Disord. 50, 3800–3809. https://doi.org/10.1007/s10803-020-04417-3 (2020).
Singh, R. et al. HAT and HDAC: enzyme with contradictory action in neurodegenerative diseases. Mol. Neurobiol. 61, 9110–9124. https://doi.org/10.1007/s12035-024-04115-6 (2024).
Yamamoto, T. et al. SLC16A2 mutations in two Japanese patients with Allan-Herndon-Dudley syndrome. Hum. Genome Var. 1, 14010. https://doi.org/10.1038/hgv.2014.10 (2014).
Yoo, Y. et al. GABBR2 mutations determine phenotype in rett syndrome and epileptic encephalopathy. Ann. Neurol. 82, 466–478. https://doi.org/10.1002/ana.25032 (2017).
Jacob, S. et al. Neural oscillations during cognitive processes in an App knock-in mouse model of Alzheimer’s disease pathology. Sci. Rep. 9, 16363. https://doi.org/10.1038/s41598-019-51928-w (2019).
Boxall, A. G., MA. The AppNLGF mouse model of amyloid pathology: age-related behavioural characterisation. Alzheimer’s & Dementia (2023).
Masuda, A. et al. Cognitive deficits in single App knock-in mouse models. Neurobiol. Learn. Mem. 135, 73–82. https://doi.org/10.1016/j.nlm.2016.07.001 (2016).
Smith, A. M. et al. Diverse human astrocyte and microglial transcriptional responses to Alzheimer’s pathology. Acta Neuropathol. 143, 75–91. https://doi.org/10.1007/s00401-021-02372-6 (2022).
Fu, H. et al. A tau homeostasis signature is linked with the cellular and regional vulnerability of excitatory neurons to tau pathology. Nat. Neurosci. 22, 47–56. https://doi.org/10.1038/s41593-018-0298-7 (2019).
Morrison, B. M., Hof, P. R. & Morrison, J. H. Determinants of neuronal vulnerability in neurodegenerative diseases. Ann. Neurol. 44, S32-44. https://doi.org/10.1002/ana.410440706 (1998).
Kamondi, A., Grigg-Damberger, M., Loscher, W., Tanila, H. & Horvath, A. A. Epilepsy and epileptiform activity in late-onset Alzheimer disease: clinical and pathophysiological advances, gaps and conundrums. Nat. Rev. Neurol. 20, 162–182. https://doi.org/10.1038/s41582-024-00932-4 (2024).
Putcha, D. et al. Hippocampal hyperactivation associated with cortical thinning in Alzheimer’s disease signature regions in non-demented elderly adults. J. Neurosci. 31, 17680–17688. https://doi.org/10.1523/JNEUROSCI.4740-11.2011 (2011).
Harris, S. S., Wolf, F., De Strooper, B. & Busche, M. A. Tipping the scales: peptide-dependent dysregulation of neural circuit dynamics in Alzheimer’s disease. Neuron 107, 417–435. https://doi.org/10.1016/j.neuron.2020.06.005 (2020).
Palop, J. J., Chin, J. & Mucke, L. A network dysfunction perspective on neurodegenerative diseases. Nature 443, 768–773. https://doi.org/10.1038/nature05289 (2006).
Olagunju, A. S. et al. Mitochondrial dysfunction: a notable contributor to the progression of Alzheimer’s and Parkinson’s disease. Heliyon 9, e14387. https://doi.org/10.1016/j.heliyon.2023.e14387 (2023).
Swerdlow, R. H. Mitochondria and mitochondrial cascades in Alzheimer’s disease. J Alzheimers Dis 62, 1403–1416. https://doi.org/10.3233/JAD-170585 (2018).
McGeer, E. G. & McGeer, P. L. Neuroinflammation in Alzheimer’s disease and mild cognitive impairment: a field in its infancy. J Alzheimers Dis 19, 355–361. https://doi.org/10.3233/JAD-2010-1219 (2010).
Madrid, A. et al. Whole genome methylation sequencing in blood from persons with mild cognitive impairment and dementia due to Alzheimer’s disease identifies cognitive status. Alzheimers Dement 21, e14474. https://doi.org/10.1002/alz.14474 (2025).
Shireby, G. et al. DNA methylation signatures of Alzheimer’s disease neuropathology in the cortex are primarily driven by variation in non-neuronal cell-types. Nat. Commun. 13, 5620. https://doi.org/10.1038/s41467-022-33394-7 (2022).
Ge, M. et al. Role of calcium homeostasis in Alzheimer’s disease. Neuropsychiatr. Dis. Treat 18, 487–498. https://doi.org/10.2147/NDT.S350939 (2022).
Huentelman, M. J. et al. Calmodulin-binding transcription activator 1 (CAMTA1) alleles predispose human episodic memory performance. Hum. Mol. Genet. 16, 1469–1477. https://doi.org/10.1093/hmg/ddm097 (2007).
Alves, G., Ornellas, M. H. & Liehr, T. The role of calmodulin binding transcription activator 1 (CAMTA1) gene and its putative genetic partners in the human nervous system. Psychogeriatrics 22, 869–878. https://doi.org/10.1111/psyg.12881 (2022).
Long, C. et al. Ataxia and Purkinje cell degeneration in mice lacking the CAMTA1 transcription factor. Proc. Natl. Acad. Sci. U S A 111, 11521–11526. https://doi.org/10.1073/pnas.1411251111 (2014).
Wijnen, I. G. M. et al. De novo variants in CAMTA1 cause a syndrome variably associated with spasticity, ataxia, and intellectual disability. Eur. J. Hum. Genet. 28, 763–769. https://doi.org/10.1038/s41431-020-0600-5 (2020).
Liu, Y. et al. CAMTA1 gene affects the ischemia-reperfusion injury by regulating CCND1. Front. Cell. Neurosci. 16, 868291. https://doi.org/10.3389/fncel.2022.868291 (2022).
Zhang, L. et al. Epigenome-wide meta-analysis of DNA methylation differences in prefrontal cortex implicates the immune processes in Alzheimer’s disease. Nat Commun 11, 6114. https://doi.org/10.1038/s41467-020-19791-w (2020).
Saura, C. D., A; Capilla-Lopez MD; Parra-Damas, A. Revealing cell vulnerability in Alzheimer’s disease by single-cell transcriptomics. Seminars Cell Development. Biol. 139, 73–83 (2023).
Farrer, L. A. et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. J. Am. Med. Assoc. 278, 1349–1356 (1997).
Saito, T., Matsuba, Y., Yamazaki, N., Hashimoto, S. & Saido, T. C. Calpain activation in Alzheimer’s model mice is an artifact of app and presenilin overexpression. J. Neurosci. 36, 9933–9936. https://doi.org/10.1523/JNEUROSCI.1907-16.2016 (2016).
Sinnamon, J. R. et al. The accessible chromatin landscape of the murine hippocampus at single-cell resolution. Genome Res. 29, 857–869. https://doi.org/10.1101/gr.243725.118 (2019).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. https://doi.org/10.1093/bioinformatics/btp324 (2009).
Granja, J. M. et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet. 53, 403–411. https://doi.org/10.1038/s41588-021-00790-6 (2021).
Phipson, B. et al. propeller: Testing for differences in cell type proportions in single cell data. Bioinformatics 38, 4720–4726. https://doi.org/10.1093/bioinformatics/btac582 (2022).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. https://doi.org/10.1093/nar/gkv007 (2015).
Andrews, G. P. et al. The development of an inline Raman spectroscopic analysis method as a quality control tool for hot melt extruded ramipril fixed-dose combination products. Int. J. Pharm. 566, 476–487. https://doi.org/10.1016/j.ijpharm.2019.05.029 (2019).
Dobin, A. et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. https://doi.org/10.1093/bioinformatics/bts635 (2013).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. https://doi.org/10.1093/bioinformatics/btp616 (2010).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome. Biol. 15, 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
DEGreport: Report of DEG analysis. R package version 1.42.0 (2024).
TrimGalore.
Krueger, F. & Andrews, S. R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572. https://doi.org/10.1093/bioinformatics/btr167 (2011).
Akalin, A. et al. methylKit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome. Biol. 13, R87. https://doi.org/10.1186/gb-2012-13-10-r87 (2012).
Wreczycka, K. et al. Strategies for analyzing bisulfite sequencing data. J. Biotechnol. 261, 105–115. https://doi.org/10.1016/j.jbiotec.2017.08.007 (2017).
Akalin, A., Franke, V., Vlahovicek, K., Mason, C. E. & Schubeler, D. Genomation: A toolkit to summarize, annotate and visualize genomic intervals. Bioinformatics 31, 1127–1129. https://doi.org/10.1093/bioinformatics/btu775 (2015).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. https://doi.org/10.1093/bioinformatics/btq033 (2010).
Kuleshov, M. V. et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90-97. https://doi.org/10.1093/nar/gkw377 (2016).
Xie, Z. et al. Gene set knowledge discovery with enrichr. Curr. Protoc. 1, e90. https://doi.org/10.1002/cpz1.90 (2021).
ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag New York, 2016).
Kolberg, L., Raudvere, U., Kuzmin, I., Vilo, J. & Peterson, H. gprofiler2 -- an R package for gene list functional enrichment analysis and namespace conversion toolset g: Profiler. F1000Res https://doi.org/10.12688/f1000research.24956.2 (2020).
Neph, S. et al. BEDOPS: High-performance genomic feature operations. Bioinformatics 28, 1919–1920. https://doi.org/10.1093/bioinformatics/bts277 (2012).
Acknowledgements
We thank Dr. Saido at the Riken Institute for sharing the App mouse model. Libraries were generated by the KCVI Epigenetics Consortium at OHSU and sequenced at the OHSU Massively Parallel Sequencing Shared Resource and at Novogene Corporation. Data analysis carried out in this study used computational infrastructure supported by the Office of Research Infrastructure Programs, Office of the Director, of the National Institutes of Health under Award Number S10OD034224. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was supported by the NIA award 1R21AG065681-01A1 awarded to LC and partially supported by R21 AG079158-01A1 and DOD HT94252410812 awarded to JR.
Author information
Authors and Affiliations
Contributions
LC, MO and JR conceived and designed the study. CEL, AP, AON, SH, KK, SNA, KJW, KAN, JH, and SW performed the experiments. BAD, AA, and MO analyzed the data. KVC and MO prepared the figures. MO and LC wrote the manuscript, with contribution and feedback from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Okhovat, M., Layman, C.E., Davis, B.A. et al. Epigenetic and transcriptomic alterations precede amyloidosis in the Alzheimer’s disease AppNL-G-F knock-in mouse model. Sci Rep 15, 44934 (2025). https://doi.org/10.1038/s41598-025-28244-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-28244-7