Abstract
The influence of deep learning in the medical and molecular biology sectors is swiftly growing and holds the potential to improve numerous crucial domains. Osteoporosis is a significant global health issue, and the current treatment options are highly restricted. Transplanting genetically engineered MSCs has been acknowledged as a highly promising therapy for osteoporosis. We utilized a random walk-based technique to discern genes associated with ossification. The osteogenic value of these genes was assessed on the basis of information found in published scientific literature. GO enrichment analysis of these genes was performed to determine if they were enriched in any certain function. Immunohistochemical and western blot techniques were used to identify and measure protein expression. The expression of genes involved in osteogenic differentiation was examined via qRT‒PCR. Lentiviral transfection was utilized to suppress the expression of the FOXA1 gene in hBMSCs. An in vivo mouse model of ovariectomy was created, and radiographic examination was conducted to confirm the impact of FOXA1 knockdown on osteoporosis. The osteogenic score of each gene was calculated by assessing its similarity to osteo-specific genes. The majority of the genes with the highest rankings were linked with osteogenic differentiation, indicating that our approach is useful for identifying genes associated with ossification. GO enrichment analysis revealed that these pathways are enriched primarily in bone-related processes. FOXA1 is a crucial transcription factor that controls the process of osteogenic differentiation, as indicated by similarity analysis. FOXA1 was significantly increased in those with osteoporosis. Downregulation of FOXA1 markedly augmented the expression of osteoblast-specific genes and proteins, activated the ERK1/2 signaling pathway, intensified ALP activity, and promoted mineral deposition. In addition, excessive expression of FOXA1 significantly reduced ALP activity and mineral deposits. Using a mouse model in which the ovaries were surgically removed, researchers reported that suppressing the FOXA1 gene in bone marrow stem cells (BMSCs) prevented the loss of bone density caused by ovariectomy. This finding was confirmed by analyzing the bone structure via micro-CT. Furthermore, our approach can distinguish genes that exhibit osteogenic differentiation characteristics. This ability can aid in the identification of novel genes associated with osteogenic differentiation, which can be utilized in the treatment of osteoporosis. Computational and laboratory evidence indicates that reducing the expression of FOXA1 enhances the process of bone formation in bone marrow-derived mesenchymal stem cells (BMSCs) and may serve as a promising approach to prevent osteoporosis.
Similar content being viewed by others
Introduction
Machine learning is a fundamental division of artificial intelligence that focuses on creating algorithms and methods capable of autonomously acquiring knowledge and enhancing performance through data analysis1. Machine learning technologies surpass standard rule-based computer programs by effectively extracting concealed patterns and regularities from vast quantities of data. These methods autonomously generate mathematical models to facilitate predictions and judgments2 Recently, owing to substantial advancements in computing power and the widespread acquisition of extensive data, machine learning approaches have made considerable strides in areas such as computer vision, natural language processing, and bioinformatics2,3,4.
Osteoporosis is a prevalent and long-lasting metabolic condition that places significant social and individual burdens, including the expenses and health issues associated with fractures. The International Osteoporosis Foundation’s current estimations indicate that osteoporosis is responsible for causing 2 million fractures in the United States5 and 3.5 million in the European Union annually6. Bisphosphonate (BP) therapy, which specifically targets and inhibits the action of osteoclasts to prevent bone loss, is the primary treatment for osteoporosis. Consequently, prolonged utilization of BP may inhibit bone remodeling, leading to increased susceptibility to fractures, and carry a certain risk of severe adverse effects7. There is ample evidence suggesting that the loss of the osteogenic differentiation ability of bone marrow-derived mesenchymal stem cells (BMSCs) is a significant factor contributing to osteoporosis8,9. Bone marrow-derived mesenchymal stem cells (BMSCs) have the capacity for self-renewal and the ability to develop into many cell types, including osteoblasts, adipocytes, and chondrocytes10,11, which play critical roles in bone formation and are regulated by genetic factors12. Gene-modified BMSCs have the potential to be a promising method for preventing osteoporosis by improving the bone-forming abilities of BMSCs8,13,14,15.
Nevertheless, conducting tests to screen genes that control the osteogenic differentiation of BMSCs is both time-consuming and expensive. As a result, there is a pressing need to create a dependable approach for quickly identifying genes associated with osteogenic differentiation.
An increasing number of studies have focused on identifying genes that are involved in osteogenic differentiation. However, the majority of these studies have focused primarily on human bone marrow-derived mesenchymal stem cells (hBMSCs) and samples that have been induced to undergo osteogenic differentiation. These studies have typically utilized traditional bioinformatics tools and methods, such as differential expression analysis and weighted gene coexpression analysis16,17, resulting in a uniform pattern in the investigation of osteogenic differentiation. Therefore, devising a new methodology for the identification and interpretation of these genes associated with ossification is imperative.
Transcription factor networks are activated during development to create transcriptional programmes that are specific to different cell types. Forkhead box protein A1 (FOXA1) is a transcription factor belonging to the forkhead/winged-helix family (which includes FOXA1, FOXA2, and FOXA3). It is highly conserved and plays a significant role in controlling cell development, differentiation, and organogenesis18. FOXA1 is characterized as a pioneer factor that attaches to compacted chromatin, facilitating the recruitment of other transcription factors to the DNA19,20.
FOXA1 mutations frequently occur in hormone receptor-driven cancers that impact the bladder and prostate, breast20 and salivary glands21. These mutations play crucial roles in facilitating the development of prostate cancer by modulating androgen receptor-driven oncogenesis and promoting the beginning and spread of metastatic prostate malignancies22.
This work utilized a random walk-based method to evaluate genes associated with the process of osteogenic differentiation in human bone marrow-derived mesenchymal stem cells (hBMSCs). Every gene is assessed and prioritized according to its resemblance to osteo-specific genes. Our analysis revealed that the majority of these highly ranked genes have been linked to osteogenic differentiation in previously published scientific literature. This finding indicates that the algorithm used is dependable in identifying genes connected to ossification. FOXA1 is a candidate gene that plays crucial roles in osteogenic differentiation. Previous research has confirmed the critical role of FOXA1 in the osteogenic differentiation of stem cells through various analytical methods23,24. Immunohistochemistry (IHC) suggested a strong association between FOAX1 and osteoporosis. Experimental evidence confirms that reducing the expression of FOXA1 enhances the process of bone formation in bone marrow-derived mesenchymal stem cells (BMSCs) and protects against bone loss caused by ovariectomy.
Results
The random walk-based algorithm prioritizes osteogenic differentiation-related genes and identifies FOXA1 as an osteogenic differentiation-related transcription factor
This study used a random walk-based algorithm to identify genes associated with osteogenic differentiation in human bone marrow-derived mesenchymal stem cells (hBMSCs). The approach involved two main steps: first, an affinity matrix was constructed via the human protein‒protein interaction (PPI) network, and then, the osteo-scores for genes were calculated on the basis of mutual information in the PPMI matrix. The initial stage involves downloading the human protein-protein interaction (PPI) network (Fig. 1A) from the STRING database25. The Random Walk with Restart (RWR) technique is subsequently employed to recover the topological structure of the human Protein-Protein Interaction (PPI) network and deduce an affinity matrix of genes(Fig. 1A). This affinity matrix is then utilised to compute the Positive Pointwise Mutual Information (PPMI) matrix (Fig. 1B). Eleven osteo-specific genes—COL1A1, RUNX2, SP7, SPP1, BGLAP, ALPL, ALPP, BMP2, IBSP, DLX5, and CTNNB1—were used as reference genes for osteo-scoring (Fig. 1B). These genes, which were previously linked to osteogenic differentiation, helped prioritize other candidate genes with similar topological structures in the PPI network. The PPMI matrix, reflecting the co-occurrence probability between genes, was used to rank genes on the basis of their potential for osteogenesis. Genes such as SP7, BGLAP, IBSP, and DLX5 presented greater similarity to osteo-specific genes, indicating their strong association with osteogenesis. Among the top 2000 genes analyzed, RUNX2, COL1A1, and BMP2 presented greater similarity to osteo-specific genes, suggesting their crucial role in osteogenic differentiation (Fig. 1B).
(A) A diagram illustrating the prioritization of osteogenic differentiation-related genes. Human PPI network used in this study to identify osteogenic differentiation-related genes. Different color indicates different clusters. Size of the node is proportional with the degree.A diagram illustrating the RWR algorithm. It initializes from a start point, and then walk to its neighbor with probability or return to the start point with probability. Red node is the start point. Directed edge is the path of random walk. Affinity matrix produced by RWR algorithm. (B) Osteo-specific scoring of genes based on PPMI matrix of these 11 osteo-specific genes. Top 100 genes ranked by osteo-scoring that is calculated based on PPMI with 11 osteo-specific genes. Boxplot showing the distribution of PPMI of top 2000 osteogenic differentiation-related genes. (C) Top 10 genes identified based on RWR algorithm. Heatmap shows the PPMI matrix of the top 10 genes that are ranked based on osteo-specific scoring. The 11 osteo-specific genes are annotated as column names.
To validate the reliability of this approach, the identified candidate genes were cross-referenced with published literature. A majority of the top 100 genes were associated with osteogenic differentiation, including SOST, MGP, COL10A1, DSPP, and FGF18 (Fig. 1C). The gene SOST, identified as the important osteogenic gene in our analysis, is involved in bone production and is linked to diseases such as sclerosteosis and osteoporosis. The MGP is crucial for regulating calcification, and its downregulation has been associated with osteoporotic fractures (Fig. 1C). Genes such as COL10A1 and DSPP also play significant roles in bone formation and are linked to bone-related disorders.
Gene Ontology (GO) enrichment analysis of the top 30 genes identified by our algorithm revealed that they are enriched in biological pathways related to bone development, including osteoblast differentiation, bone mineralization, collagen binding, and endochondral ossification (Fig. 2A). These findings confirm that our approach is effective in identifying genes with osteogenic potential. Furthermore, differential expression analysis of the top 30, 50, 70, and 100 ranked genes revealed that higher-ranked genes presented greater variability in log-fold change (logFC) and were more likely to exhibit statistically significant differential expression during osteogenic differentiation (Fig. 2B). These genes, particularly those with higher osteo-scores, are more prone to activation or downregulation during the osteogenic process.
(A) GO functional enrichment analysis of the top 30 osteo genes. (B) Comparison between osteo genes and other genes during osteogenic differentiation. Two indexes logFC and p-value are used to measure the extend of differential expression between hBMSCs and osteogenic induced samples. (A-D) Density curve shows the distribution of logFC between top 30, 50, 70, 100 osteo genes and other genes. (E-H) Density curve shows the distribution of p-value between top 30, 50, 70, 100 osteo genes and other genes. (C) FOXA1 is an osteogenic differentiation-related transcription factor. Twenty-six genes of the top 500 osteo genes are found to be transcription factor. Regulation network of transcription factor FOXA1. PPI network of FOXA1. PPMI of FOXA1 with 11 osteo-specific genes.
Most of the top-ranked candidate genes with an osteo-specific score higher than 6, as listed in the Supplementary Material Table, are confirmed to be involved in osteogenic differentiation, except for six osteo-specific scoring examples, including transcription factors FOXA1. Our analysis revealed that FOXA1 interacts with osteo-specific genes such as ALPL, BMP2, COL1A1, RUNX2, DLX5, and SPP1, indicating its role in osteogenesis (Fig. 2C). FOXA1 was found to directly regulate 20 genes, including BCL2, MYC, and CFTR (Fig. 2C), further supporting its potential function as an osteogenic transcription factor. Our findings suggest that FOXA1 could play a significant role in regulating osteogenesis, warranting further investigation into its relationship with bone formation and osteoporosis.
FOXA1 is significantly increased in osteoporosis model mice
Immunohistochemistry (IHC) confirmed a significant increase in FOXA1 expression in osteoporotic mice compared with that in sham animals, suggesting a strong association between FOXA1 expression and bone metabolism in osteoporosis (Fig. 3A-E).
The Immunohistochemistry staining of FOXA1 expression in osteoporosis mice. (A–B) FOXA1 expression significantly increased in osteoporosis mice compared to sham animals. (C) The level of relative integral optical density(IOD) statistical analysis. (D–E) Microcomputed tomography analysis for bone mass in sham and OVX mice. The OVX mice bone mass significantly decreased compared to sham mice. (F) The level of BMD statistical analysis. All data are means ± SDs (n = 3). * p < 0.05, **p < 0.01, and ***p < 0.001 versus the control group.
Knockdown of FOXA1 in hBMSCs
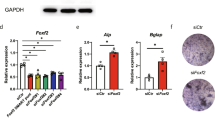
To elucidate the function of FOXA1 in osteogenic development, a lentiviral vector was used to effectively suppress the expression of FOXA1 in human bone marrow-derived mesenchymal stem cells (hBMSCs), in accordance with our previous studies23. The level of FOXA1 expression was measured via quantitative reverse transcription polymerase chain reaction (qRT‒PCR) upon transinfection. The mRNA expression of FOXA1 was markedly lower in the KD-FOXA1 group (FOXA1 knockdown) than in the NC (negative control) group, as shown in Fig. 4A.
FOXA1 knockdown enhanced the osteo-specific gene. (A–F) The mRNA expression levels (normalised to that of 18 S) of FOXA1, COL1A1, RUNX2, SP7, OPN in FOXA1-NC and FOXA1-KD groups after 4 days of osteogenesis. All data are means ± SDs (n = 3). * p < 0.05, **p < 0.01, and ***p < 0.001 versus the control group.
Knockdown of FOXA1 increased the levels of osteo-specific genes and proteins and activated the ERK1/2 signaling pathway
To examine the impact of reducing FOXA1 expression on the process of bone formation, we measured the expression levels of specific genes associated with bone development, such as RUNX2, COL1A1, OPN, OCN, and SP7, via quantitative real-time PCR (qRT‒PCR). On the fourth day, the FOXA1-knockdown group presented significantly elevated mRNA levels of SP7, OCN, OPN, RUNX2, and COL1A1 than did the FOXA1-knockdown group (P < 0.05, Fig. 4B–F). These findings suggest that a reduction in FOXA1 expression promotes the transcription of genes critical for osteogenic differentiation. In addition, the levels of proteins associated with osteogenesis, including COL1A1, RUNX2, SP7, and p-ERK1/2, were greater in the FOXA1-KD group than in the KD-NC group on day 7 (Fig. 5A–C). The increased levels of COL1A1 and RUNX2, which are major components of the bone matrix and transcriptional regulators of osteogenesis, further support the notion that FOXA1 depletion enhances osteogenic differentiation. SP7, a transcription factor essential for osteoblast differentiation, was also upregulated, suggesting that FOXA1 knockdown promotes the progression of osteogenic differentiation. Moreover, the activation of ERK1/2, a key signaling pathway involved in cellular proliferation and differentiation, was observed, as indicated by increased levels of phosphorylated ERK1/2 (p-ERK1/2). The upregulation of p-ERK1/2 in the FOXA1-KD group suggested that FOXA1 knockdown might trigger the ERK1/2 signaling pathway, which is known to play a pivotal role in osteoblast differentiation and bone formation.
FOXA1 knockdown enhanced the osteo-specific protein levels, activated the ERK1/2 signalling pathway. (A) The protein expression levels (normalised to that of GAPDH) of FOXA1, COL1A1, RUNX2 and SP7 in FOXA1-NC and FOXA1-KD groups after 4 days of osteogenesis. (B) The level of p-ERK1/2 (normalised to that of ERK1/2) increased after 4 days of osteogenesis in FOXA1-KD group. (C) The level of protein statistical analysis. All data are means ± SDs (n = 3). * p < 0.05, **p < 0.01, and ***p < 0.001 versus the control group.
Knockdown or overexpression of FOXA1 did not affect hBMSC proliferation
The proliferation of human bone marrow-derived mesenchymal stem cells (hBMSCs) was assessed on days three, five, and seven following lentiviral particle infection via Cell Counting Kit-8 (CCK-8) staining. There was no notable difference in the rates of cell proliferation between the FOXA1 KD-NC and FOXA1-KD groups, as shown in Fig. 6A. Similarly, the FOXA1 OE-NC and FOXA1-OE groups presented no significant variation in proliferation rates, as depicted in Fig. 6B. This observation indicated that modulation of FOXA1 expression does not significantly affect hBMSC proliferation.
The proliferation by FOXA1 knockdown and overexpression of hBMSCs. (A–B) The CCK-8 assay revealed that hBMSCs proliferation was not affected by FOXA1 knockdown (FOXA1-KD) and overexpression (FOXA1-OE). All data are means ± SDs (n = 3). * p < 0.05, **p < 0.01, and ***p < 0.001 versus the control group.
Knockdown of FOXA1 enhanced alkaline phosphatase (ALP) activity and promoted calcium deposit formation
ALP activity, which serves as an indicator of early bone formation, was assessed on the seventh day. Compared with the control group, the group in which FOXA1 was knocked down presented considerably greater ALP activity (p < 0.05), as confirmed by ALP staining (Fig. 7A and B).
Stain effects of FOXA1 knockdown and overexpression on osteogenic differentiation of hBMSCs. (A-B) Knockdown of FOXA1 significantly enhanced hBMSC ALP activity (after 7 days of osteogenesis) and calcium deposits. (after 14 days of osteogenesis) in ALP and ARS staining. whereas overexpression of FOXA1 significantly reduced hBMSC ALP activity and calcium deposits in ALP and ARS staining. All data are means ± SDs (n = 3). * p < 0.05, **p < 0.01, and ***p < 0.001 versus the control group.
Calcium deposits were observed via alizarin red (ARS) staining on day 14. The group in which FOXA1 was knocked down presented a greater abundance of calcium deposits than did the control group. This observation was further corroborated by quantitative analysis, as shown in Fig. 7A and B.
Transplantation of MSCs with FOXA1 knockdown prevents bone loss in OVX mice
A total of forty female C57BL/6 mice, aged 12 weeks, were categorized into four groups according to the following procedure: sham + PBS, OVX + PBS, OVX + KD-NC, and OVX + KD. Among the four study groups, the measurement of bone mass via microcomputed tomography revealed that, compared with the sham + PBS group, the OVX + PBS group presented a decrease in bone density 8 weeks after OVX, as anticipated. Compared with OVX + PBS, the introduction of MSCs with FOXA1 knockdown (OVX + KD) effectively reduced bone loss caused by ovariectomy (OVX) after 8 weeks. At 8 weeks, the OVX + KD group, which underwent MSC transplantation, presented significantly greater bone mass than the OVX + KD-NC group did (P < 0.05). Surprisingly, compared with the OVX + PBS group, the OVX + KD-NC group also exhibited no bone loss. HE staining also revealed similar results. These findings suggest that MSCs themselves may play a role in the development of osteoporosis induced by OVX in mice, as shown in Fig. 8A–C.
Transplantation of MSCs knockdown FOXA1 prevents bone loss of OVX mice. (A–B) Microcomputed tomography analysis for bone mass in OVX mice. The transplantation of MSCs with FOXA1 knockdown increased the BV/TV and Tb.Th decreased the BS/BV and Tb.Sp (n = 10). (C) The transplantation of MSCs with FOXA1 knockdown increased the trabecular number. All data are means ± SDs. * p < 0.05, **p < 0.01, and ***p < 0.001 versus the control group.
Discussion
Presently, an increasing number of studies have focused on identifying genes that are associated with osteogenic differentiation. However, the majority of these studies focused on analyzing the expression profiles of bone marrow-derived mesenchymal stem cells (BMSCs) and samples that have been induced to undergo osteogenesis16,17. This study focuses primarily on the protein‒protein interaction (PPI) network and categorizes genes that exhibit topological similarity with osteo-specific genes as ossification-related genes26,27. This approach utilizes the topological structure of the PPI network to identify genes, providing a novel avenue for gene classification. In addition, other data, such as the protein’s geometric structure, DNA sequence, and miRNA regulation network, which have distinct gene identification, can also be used to identify genes associated with osteogenesis28. This finding offers a new approach for identifying genes related to bone formation in the future.
Our random walk-based method uniquely integrates protein-protein interaction (PPI) networks with gene expression data, capturing the topological relationships between genes. This approach is crucial for identifying genes involved in osteogenesis, especially those interacting with other regulatory genes. In contrast, ARACNE and GENIE3 primarily use gene expression data. ARACNE uses mutual information to detect direct gene-gene interactions, but it can be sensitive to noise, potentially missing subtle relationships. GENIE3, based on random forest algorithms, excels at handling large, high-dimensional datasets but also focuses only on gene expression, overlooking network interactions29,30. Our method’s focus on PPI network topology allows it to uncover indirect gene interactions and novel osteogenesis-related genes, providing insights that expression-based methods may miss. Our approach’s integration of network structure offers a distinct advantage in identifying less obvious gene relationships in osteogenesis.
Our investigation highlights a set of 11 genes specifically linked to osteo-related functions. The key indicators of osteogenic differentiation that have been clearly identified include RUNX2, COL1A1, SP7, SPP1, BGLAP, ALPL, ALPP, IBSP, BMP2, DLX5, and CTNNB1, all of which are strongly associated with osteogenesis31,32,33,34.
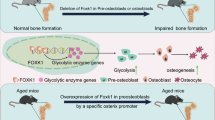
The genes listed in the Supplementary Material Table, which have been validated, are predominantly transcription factors closely associated with osteogenic differentiation. Our investigation identified FOXA1 as a significant and innovative indicator of osteogenic differentiation among the top transcription factors, showing similarity to six osteo-specific genes and aligning with their functional profiles. Additionally, immunohistochemical labeling validated the substantial elevation of FOXA1 in osteoporosis, suggesting a strong correlation between FOXA1 expression and bone metabolism in this condition. Foxa1 is an evolutionarily conserved winged helix transcription factor that plays regulatory roles in development, differentiation, and bone metabolism35,36,37. FOXA1 positively regulates the majority of genes that are controlled by FOXA1, including important mediators of ERK signaling, such as GRB2 and BRAF. GRB2, which is responsible for facilitating Ras activation, and BRAF, a downstream target of Ras, are both essential for complete ERK activation38,39. PAK1, a protein kinase, is directly targeted by FOXA1, and its activity is regulated by FOXA1. PAK1 binds to the GRB2 adaptor protein, and this joint control of GRB2 and PAK1 has a substantial effect on the subsequent ERK signaling cascade40. FOXA1 transcriptionally suppresses IL-8, the expression of which would otherwise stimulate the MAPK/ERK pathway. The FOXA1, IL-8 and MAPK/ERK pathways may be promising targets for therapeutic intervention in related diseases, such as prostate cancer progression to NEPC41. Moreover, FOXA1 is essential for the activation of ERK phosphorylation and the preservation of cell viability in MDA-MB-453 cells40. The MEK-ERK MAPK signaling pathway plays a crucial role in regulating the functions of osteoblasts and osteoclasts42,43,44. FOXA1 enhances cell proliferation by interacting with the androgen receptor (AR) and stimulating the Notch pathway by modulating AR expression. Both FOXA1 and AR have the ability to attach directly to the promoter and enhancer regions located before MYC35. Through the androgen receptor, androgens stimulate the growth of cells similar to osteoblasts in a controlled environment and trigger the development of osteoblasts while inhibiting the function, activity, and lifespan of osteoclasts45,46. Myc, a well-established Wnt target gene, has been shown to enhance the process of osteogenic differentiation in skeletal development and control the accumulation of bone mass after birth47,48,49,50. The combination of MYC and BMP2 has a synergistic effect on promoting osteoblast development and matrix mineralization in human bone marrow-derived mesenchymal stem cells (hBMSCs)48. Overexpressing Foxa1 led to an increase in the expression of HSP72, whereas knocking down Foxa1 via antisense oligonucleotides resulted in a decrease in HSP72 expression in MCF-7 cells. HSP72, also known as HSPA1A, is a member of the HSP70 family. It promotes the process of osteogenic differentiation in bone marrow mesenchymal stem cells by activating the Wnt/β-catenin signaling pathway51. Our work revealed that reducing the presence of FOXA1 greatly increased the expression of crucial genes that control the initiation of bone formation. This leads to increased ALP activity and the formation of mineral deposits, providing unequivocal evidence that FOXA1 plays a vital role in encouraging the differentiation of bone cells. When introduced into the bodies of mice with ovariectomy-induced osteoporosis, FOXA1-silenced BMSCs effectively prevent bone loss. In the ovariectomy (OVX)-induced mouse model, FOXA1-KD BMSCs were transplanted in vivo and successfully induced more bone loss than FOXA1-NC BMSCs did. These findings demonstrate that FOXA1 KD can counteract osteoporosis by increasing osteogenic activity, with particularly pronounced effects under pathological conditions of estrogen deficiency. The in vitro and in vivo experimental evidence supporting the correlation between the FOXA1 gene and osteogenic differentiation confirms the accuracy and validity of the prediction method. These findings not only validate the molecular role of FOXA1 KD but also provide critical insights for potential future clinical applications.
Bisphosphonates (BP) are presently the primary treatment for osteoporosis, as they decrease osteoclast activity, leading to an antiresorptive effect52. Nevertheless, solely inhibiting the development of osteoclasts might impede the process of bone turnover, hence increasing the risk of fractures and potentially causing severe complications such as bisphosphonate-related osteonecrosis of the jaw or atypical femur fractures53,54,55. From this viewpoint, mesenchymal stem cells (MSCs) have great potential for treating osteoporosis, as they can stimulate anabolic processes following MSC transplantation. The osteogenic development of bone marrow mesenchymal stem cells (BMSCs) is strongly linked to osteoporosis, and these cells have been demonstrated to be exceptional cell sources for therapeutic purposes52,56. An improved prediction method for osteogenic differentiation-associated genes in BMSCs will allow researchers to identify new and appropriate targets for more efficient induction of bone tissue regeneration and treatment of related disorders.
Multiple studies have demonstrated that the systemic transplantation of mesenchymal stem cells (MSCs) in mice with osteoporosis effectively repairs impaired MSCs in the recipient’s bone marrow and enhances bone density in osteoporotic animals57,58,59,60. Reducing the expression of FOXA1 in MSCs is anticipated to improve the effectiveness of this method, as FOXA1-silenced BMSCs have a greater ability to differentiate into bone cells.
We used new and more advanced machine learning methods to further explore osteogenic differentiation genes and found that FOXA1 is extremely important in osteogenic differentiation, which is the same as FXOA1 reported in the original article23, but the methods used were completely different. This similarity in the results further demonstrates the reliability of our study, meaning that both studies provide significant and reliable results for osteogenic differentiation-related biomarkers. These candidate biomarkers are worthy of future investigation. Moreover, we further conducted different animal experiments and used the OVX model to further verify osteogenic differentiation. The results demonstrated that FOXA1 expression was significantly increased in osteoporotic mice compared with sham-operated animals, indicating a strong association between FOXA1 expression and bone metabolism in osteoporosis. This may be attributed to the impaired osteogenic differentiation and increased senescence of stem cells observed in osteoporosis, which was accompanied by a notable elevation in FOXA1 levels. Furthermore, our study revealed that FOXA1 knockdown promoted osteogenesis in BMSCs and mitigated bone loss induced by ovariectomy. These findings hold great importance and are expected to inspire biologists, clinicians, and researchers to further advance the study of osteogenic differentiation in the future.
Nevertheless, additional research is needed to gain a deeper understanding of the regulatory mechanisms and various areas involved. Furthermore, additional research is needed to study the impact of reducing FOXA1 in MSCs on the process of osteoblast development.
Method
Data collection
The PPI network of humans, consisting of 19,344 genes, was obtained from the STRING database (version 11.0). It is subsequently simplified through the removal of loops and many edges. In our investigation, we collected 11 osteo-specific genes to capture and extract their osteogenic characteristics. The genes encoding the following genes were identified: RUNX2, COL1A1, SP7, SPP1, BGLAP, ALPL, ALPP, BMP2, IBSP, DLX5, and CTNNB1. The osteo-specific genes are utilized for assessing similarity to prioritize and identify genes relevant to osteogenesis.
The expression profiles of human bone marrow-derived mesenchymal stem cells (hBMSCs) and samples stimulated to undergo osteogenesis were obtained from the Gene Expression Omnibus database (GSE80614)61 for the analysis of differential expression. The sample consisted of 6 human bone marrow-derived mesenchymal stem cells (hBMSCs) and 30 samples that were stimulated to differentiate into osteogenic cells. To determine the nature of a gene as a transcription factor, the roster of transcription factors was acquired from the TRRUST database62. This study also includes a collection of networks that represent the interactions between a transcription factor and its target.
Random walk-based algorithm identifies osteogenic differentiation-related genes
Features are defined as the topological structures of genes included in the human protein‒protein interaction (PPI) network. To precisely capture and extract the topological structures of genes, a method based on random walks is employed to encode genes into a representation consisting of vectors with a high number of dimensions63. Upon receiving a network, the vertices are initially arranged in a random order. The transition matrix B, which quantifies the likelihood of transitioning from node i to node j, can be derived from the adjacency matrix A and is defined as follows:
At each iteration, a predetermined restart probability is assigned to each initialized node. This probability considers both the global and local topological structure information of genes in the human PPI network to use the inherent indirect and direct interactions between genes. Therefore, after each iteration, a vertex moves to the next vertex with a certain probability and returns to the starting vertex with another probability. This can be mathematically expressed as:
where \(\:{p}_{k}\) is a vector representing the probability of reaching other vertices after k steps of iterations and where \(\:{p}_{0}\) is the initialized vector with the starting point being 1. Vector \(\:{p}_{k}\) can be updated iteratively to capture the underlying associations between genes. The local and global topological structure of the network can be captured on the basis of the parameter \(\:{\upalpha\:}\), where a large value strongly emphasizes the local structure. Finally, it outputs an affinity matrix, indicating the co-occurrence probability between genes. RWR is implemented via the dnet R package64.
After the co-occurrence matrix between genes is obtained, the PPMI matrix is computed for similarity representation, which can be deemed a matrix factorization method65. The mathematical description can be defined as follows:
where \(\:\text{M}\) is the co-occurrence matrix. \(\:{N}_{i}\) and \(\:{N}_{j}\) are the i-th and j-th column vectors in the matrix. The value in the PPMI matrix indicates the similarity between different genes in the PPI topological structure. To properly prioritize and identify osteogenic genes, we marked genes with different scores on the basis of their similarity with osteo-specific genes. We then set a similarity threshold of 1 to identify whether a gene is similar to an osteospecific gene. The number of similar osteo-specific genes was counted for each gene to evaluate the significance of osteogenic potential, which is used for gene ranking.
Functional enrichment analysis of these newly discovered osteogenic genes
To determine whether these potential genes are concentrated in particular functions and states, we conducted a Gene Ontology (GO) enrichment analysis on the top 30 genes identified by the RWR algorithm. We established a threshold of 0.01 as the level of statistical significance for the Benjamini‒Hochberg adjusted p value. Pathways with a Benjamini‒Hochberg adjusted p value of less than 0.01 were considered statistically significant. GO enrichment analysis is conducted by utilizing many categories, such as cellular component, molecular function, and biological process. Gene Ontology (GO) enrichment analysis was performed via clueGO software66.
Differential expression analysis between hBMSCs and osteogenic-induced samples
To determine whether these putative genes are more inclined to be upregulated or downregulated during the process of osteogenic differentiation, we conducted a differential expression study comparing hBMSCs with osteogenically produced samples. The limma software was used for differential expression analysis67. During the osteogenic differentiation of hBMSCs, two indicators, the logFC and p value, were chosen to assess the degree of differential expression of each gene.
Cell culture, reagents, and antibodies
HBMSCs and MBMSCs were obtained from Cyagen (Guangzhou, China). These cells have the ability to develop into osteoblasts, adipocytes, and chondrocytes when exposed to particular inducing conditions. Cultured bone marrow stromal cells (BMSCs) were placed in growth media (Cyagen Biosciences, Guangzhou, China) in culture flasks and kept in an environment with 5% CO2 at 37 °C. The cells were then transferred to new flasks after they reached approximately 80–90% confluence. Cells from passages two to six were utilized in future research.
Specific antibodies against Osterix (SP7, 28694-1-AP, 1:1000) were purchased from PROTEINTECH (Chicago, USA). Runt-related transcription factor 2 (RUNX2, 12556, 1:1000), extracellular signal-regulated kinase 1/2 (ERK1/2, 9102, 1:1000), and phospho-ERK1/2 (p-ERK1/2, 9101, 1:1000) were obtained from Cell Signaling Technology (Danvers, MA, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH, ab8245, 1:10000), collagen type I alpha 1 (COL1A1, ab138492, 1:1000) and FOXA1 (ab170933, 1:1000) were purchased from Abcam (Cambridge, UK).
Osteogenic differentiation protocol
Human bone marrow mesenchymal stem cells (hBMSCs) were grown in either six- or twelve-well cell culture plates at a density of 3E4 cells per square centimeter. The cells were then incubated for 48 h in growth medium at 37 °C with a 5% concentration of carbon dioxide. Afterwards, the cells were placed in a culture medium that promoted the development of bone cells (DMEM with 10% FBS, 100 IU/ml penicillin/streptomycin, 100 nM dexamethasone, 0.2 mM ascorbic acid, and 10 mM β-glycerophosphate). The cells were passaged by periodically replacing the osteogenic induction media with fresh media every 2 days.
Lentiviral packaging and cell infection
hBMSCs were transfected with lentiviral particles obtained from OBiO Technology (Shanghai, China) for the purpose of overexpressing or knocking down FOXA1. The lentiviral package included overexpression control particles (FOXA1-overexpressing negative control group, OE-NC), lentiviral particles for overexpressing FOXA1 (FOXA1-overexpressing group, OE), knockdown control particles (FOXA1-knockdown negative control group, KD-NC), and lentiviral particles for knocking down FOXA1 (FOXA1-knockdown group, KD). Upon reaching 50% confluence, the growth media of the hBMSCs was supplemented with lentiviral particles containing 5 µg/ml polybrene, following the guidelines provided by the manufacturer. Following a three-day period, the cells were subjected to screening with puromycin (2 g/mL) and subsequently passaged for additional investigations. FOXA1 expression was quantified via quantitative real-time PCR (qRT‒PCR).
The lentiviral package for silencing FOXA1 in mouse BMSCs (mBMSCs) was obtained from GeneChem (Shanghai, China). The mBMSCs were infected by culturing them in growth media containing lentiviral particles and polybrene (5 g/mL) following the instructions provided by the manufacturer. After three days, the cells were subjected to puromycin (5 g/mL) screening and then transferred for further experiments. GFP fluorescence was used to measure the transduction efficiency.
Cell counting Kit-8
The cells were placed in 96-well plates with growth media (Cyagen) at a concentration of 5,000 cells per well. A solution containing 10% CCK-8 (Beyotime) was subsequently added to the wells, and the cells were subjected to a 2-hour incubation at 37 °C. The assessment of cell proliferation was conducted via a microplate reader (ELX808; BioTek, USA) at a wavelength of 450 nm.
RNA extraction and quantitative RT‒PCR
The quantification of gene expression related to the development of osteoblasts was conducted via quantitative reverse transcription polymerase chain reaction (qRT‒PCR). The RNAiso reagent from Takara Bio in Kusatsu, Japan, was utilized to extract and measure the total cellular RNA, together with the NanoDrop 2000 instrument. The absorbance of each sample at 260 nm was measured according to the instructions provided by the manufacturer (Thermo Fisher Scientific, MA, USA). A total of 20 µl of reaction mixture was used to reverse transcribe total RNA into complementary DNA (cDNA). The template for the qRT‒PCR was 1 µl of cDNA, which was mixed with Power SYBR® Green PCR Master Mix from Takara. The qRT‒PCR was performed in triplicate via the ABI 7500 System from Thermo Fisher Scientific. The housekeeping genes utilized in this experiment were 18 S or GAPDH. The primers used in this study were synthesized by Sangon Biotech (Shanghai, China). The primer sequences are provided in Table 1. The qRT‒PCR was conducted at 95 °C for 30 s, followed by 45 cycles of 95 °C for 5 s and 60 °C for 30 s. The 2-▵▵Ct method was used to assess the expression levels of all the genes.
Western blotting analysis
The cells were broken down at a low temperature for 30 min using a RIPA buffer that included substances to prevent the activity of proteases and phosphatases (Boster, Wuhan, China). Centrifugation was performed at a speed of 14,000 revolutions per minute for 10 min at 4 °C to separate the lysates and obtain the supernatants. The proteins were separated via 10% sodium dodecyl sulfate‒polyacrylamide gel electrophoresis and subsequently deposited onto a polyvinylidene fluoride membrane (Millipore, Shanghai, China). The membranes were subsequently blocked in Tris-buffered saline containing 5% nonfat milk and 0.1% Tween for 2 h at room temperature. The membranes were subsequently incubated with primary antibodies overnight at 4 °C. After being washed three times with a solution containing 0.1% Tween in Tris-buffered saline, the samples were treated with horseradish peroxidase-conjugated secondary antibodies (anti-rabbit; Boster) at room temperature for one hour. The proteins were then detected via an enhanced chemiluminescent detection reagent (Millipore, Sigma) and an XRS chemiluminescence detection system (Bio-Rad Laboratories, Hercules, CA, USA).
ALP staining
For ALP staining, the cells were cultured in osteogenic induction media for 7 days in 24-well plates. Afterwards, the cells were immobilized with 4% paraformaldehyde (Beyotime, Shanghai, China) for 30 min at room temperature and washed three times with phosphate-buffered saline. The cells were stained via a BCIP/NBT ALP color development kit from Beyotime, Shanghai, China. An ALP activity assay (Beyotime) was used to assess ALP activity, following the instructions provided by the manufacturer.
ARS staining
Human bone marrow mesenchymal stem cells (HBMSCs) were transferred to 24-well plates and cultured in medium that promotes the formation of bone tissue for a period of 14 days. The cells were treated with 4% paraformaldehyde (Beyotime, Shanghai, China) for 20–30 min at room temperature after being rinsed three times with PBS. Alizarin Red S solution (Beyotime) was subsequently added, and the mixture was allowed to incubate for 15 min at room temperature. Next, the solution was rinsed three times with PBS. The ARS stain was subsequently exposed to a 10% solution of cetylpyridinium chloride for 1 h at ambient temperature. The resulting solutions were then collected, distributed onto a 96-well plate, and assessed at a wavelength of 560 nm via a microplate reader (ELX808; BioTek). The data were standardized to the control group.
Immunohistochemistry (IHC)
The spine slices (10 μm) were treated with xylene to remove the paraffin and then soaked in a series of ethanol solutions (100%, 95%, 80%, and 75%) for 5 min each to restore moisture. This was followed by antigen recovery. After being immersed in a 3% H2O2 solution at 37 °C for 15 min, the sections were subsequently washed with PBS. Next, the sections were incubated with goat serum for a period of 30 min at 37 °C. Finally, the sections were incubated overnight at 4 °C with a primary rabbit anti-FOXA1 antibody (diluted at a ratio of 1:100; PROTEINTECH, USA). The following day, the sections were treated with a secondary goat anti-rabbit antibody (Solarbio, Beijing, China) at 37 °C for 45 min. This was followed by treatment with an ABC working solution (Solarbio) at 37 °C for 25 min. Finally, the sections were incubated with 3,3’-diaminobenzidine (3,3’-diaminobenzidine; Solarbio). The visual fields were chosen at random from each sample via a Leica DM600B automated microscope (Leica, Wetzlar, Germany). The expression levels of FOXA1 were measured via Image-Pro Plus 6.0 (Media Cybernetics, USA). The mean density value, which is obtained by dividing the integrated optical density by the relevant area, was computed for each visual field.
In vivo evaluation in animals
The Zhejiang Centre of Laboratory Animals provided a total of 40 C57/B16 mice, all of which were female and 12 weeks old. The Institutional Animal Care and Use Committee of the Zhejiang Centre of Laboratory Animals (ZJCLA-IACUC-20030053) approved all the animal research.
A total of 40 female C57/B16 mice were utilized to produce a mouse ovariectomized model. Ten female mice in the sham group were subjected to anesthesia, incisions, bilateral ovary exposure, and incision closure without the bilateral ovaries being removed. The mice were randomly and equally divided into four groups: (1) the sham + PBS group (injected with PBS, n = 10), (2) the OVX + PBS group (injected with PBS, n = 10), (3) the OVX + KD-NC group (transplanted with mBMSCs with FOXA1 KD-NC, n = 10), and (4) the OVX + KD group (transplanted with mBMSCs with FOXA1-KD, n = 10). The mouse ovariectomized model was created following previously documented procedures74-79, and the surgical procedures were carried out by two skilled orthopedic surgeons. In summary, the mice were rendered unconscious by injecting 0.3% pentobarbital sodium (30 mg/kg body weight) into their peritoneal cavity. The control group of mice experienced anesthesia, incisions, bilateral ovary exposure, and incision closure without undergoing bilateral ovariectomy. Thirty female mice in the ovariectomized model group were exposed by making two small incisions in the dorsal skin and muscle layers near the midline. The bilateral ovaries of the mice were surgically removed following closure of the uterine horn. Next, the muscle incisions were stitched together, and the skin was sutured. On study day 0, a total of 6–7 × 105 NC or FOXA1-KD mMSCs in 100 µl of PBS were administered intravenously into the lateral tail veins of the mice in the KD-NC or KD OVX groups over a period of 10–12 s. The PBS OVX group mice and the sham group mice received an injection of 100 µl of PBS. The injection was administered on a weekly basis for a total of eight consecutive weeks. The mice were euthanized in a CO2 chamber, and samples from the experimental subjects were collected for further investigations.
Radiographic analysis
After harvest, a total of 40 four-week samples (10 from each group) were sent for examination via microcomputed tomography (µCT) for a duration. The femurs were scanned via a U-CT system (MILabs U-CT, Netherlands). The datasets were reconstructed via MILabs Rec 10.16 software. Trabecular BMD, BS/BV, BV/TV, Tb.Th, and Tb.Sp were calculated from the region of interest (ROI). The µCT data were evaluated, and three-dimensional bone structure image slices were reconstructed via IMALYTICS Preclinical 2.1 software.
Histological evaluation
Following collection, the samples (n = 10 per group) were fixed in 4% paraformaldehyde (Beyotime) at 4 °C for 36 h. Decalcification was performed via the use of ethylenediaminetetraacetic acid (EDTA) decalcifying solution (E1171, Solarbio, Beijing, China) for one week at room temperature. The samples were then processed for paraffin embedding following standard protocols. Serial sections were cut at a thickness of 4 μm, mounted onto polylysine-coated glass slides, and deparaffinized. Hematoxylin and eosin (HE) staining was subsequently performed on consecutive tissue sections as described in previous studies68.
Statistical analysis
GraphPad Prism v.7.0 was used for statistical analysis (GraphPad Software, San Diego, CA, USA). The experiments were performed at least in triplicate. The data are presented as the means ± SDs. Statistical significance was determined via a 2-tailed Student’s t test when 2 groups were compared, and 1-way ANOVA followed by Tukey’s post hoc test was used when more than 2 groups were compared. A value of P < 0.05 was considered to indicate significance.
Data availability
Data is provided within the manuscript or supplementary information files.
Abbreviations
- hBMSCs:
-
Human bone mesenchymal stem cells
- mBMSCs:
-
Mouse bone mesenchymal stem cells
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- Osx/SP7:
-
Osterix
- OCN:
-
Osteocalcin
- ALP:
-
Alkaline phosphatase
- ARS:
-
Alizarin red staining
- OPN:
-
Osteopontin
- RUNX2:
-
Runt-related transcription factor 2
- COL1A1:
-
collagen type I alpha 1
- ERK:
-
extracellular signal-regulated kinase MAPK
- p-ERK:
-
phospho-ERK
- GEO:
-
Gene Expression Omnibus
- qRT‒PCR:
-
Quantitative real-time PCR
References
Xu, Y. et al. Artificial intelligence: A powerful paradigm for scientific research. Innovation 2, 100179 (2021).
Taye, M. Understanding of machine learning with deep learning: Architectures, workflow, applications and future directions. Computers 12, 91 (2023).
Hu, J. et al. Voronoi-based multi-robot autonomous exploration in unknown environments via deep reinforcement learning. IEEE Trans. Veh. Technol. 69, 14413–14423 (2020).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Gómez-Barrena, E. et al. Bone fracture healing: Cell therapy in delayed unions and nonunions. Bone 70, 93–101 (2015).
Svedbom, A. et al. Osteoporosis in the European union: A compendium of country-specific reports. Arch. Osteoporos. 8, 137 (2013).
Prasarn, M. L. et al. Bisphosphonate-associated femur fractures have high complication rates with operative fixation. Clin. Orthop. Relat. Res. 470, 2295–2301 (2012).
Qadir, A. et al. Senile osteoporosis: The involvement of differentiation and senescence of bone marrow stromal cells. LID – 10.3390/ijms21010349 [doi] LID – 349.
Kiernan, J., Davies, J. E. & Stanford, W. L. Concise review: Musculoskeletal stem cells to treat age-related osteoporosis. Stem Cells Transl Med. 6, 1930–1939 (2017).
Bianco, P. et al. Bone marrow stromal stem cells: Nature, biology, and potential applications. Stem Cells. 19, 180–192 (2001).
Liao, J. et al. The fabrication of biomimetic biphasic CAN-PAC hydrogel with a seamless interfacial layer applied in osteochondral defect repair. Bone Res. 5, 17018 (2017).
Zhang, W. et al. IGFBP7 regulates the osteogenic differentiation of bone marrow-derived mesenchymal stem cells via Wnt/beta-catenin signaling pathway. FASEB J. 32, 2280–2291 (2018).
Cho, S. W. et al. Transplantation of mesenchymal stem cells overexpressing RANK-Fc or CXCR4 prevents bone loss in ovariectomized mice. Mol. Ther. J. Am. Soc. Gene Ther. 17, 1979–1987 (2009).
Li, F. et al. Effect of stem cell therapy on bone mineral density: A meta-analysis of preclinical studies in animal models of osteoporosis. PLoS One. 11, e0149400 (2016).
Jiang, Y. et al. Advances in mesenchymal stem cell transplantation for the treatment of osteoporosis. Cell. Prolif. 54, e12956 (2021).
Fan, T. et al. Bioinformatics analysis of the biological changes involved in the osteogenic differentiation of human mesenchymal stem cells. J. Cell. Mol. Med. 24, 7968–7978 (2020).
Yang, W. et al. Co-expression network analysis identified key genes in association with mesenchymal stem cell osteogenic differentiation. Cell. Tissue Res. 378, 513–529 (2019).
Hannenhalli, S. & Kaestner, K. H. The evolution of Fox genes and their role in development and disease. Nat. Rev. Genet. 10, 233–240 (2009).
Iwafuchi-Doi, M. et al. The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol. Cell. 62, 79–91 (2016).
Arruabarrena-Aristorena, A. et al. FOXA1 mutations reveal distinct chromatin profiles and influence therapeutic response in breast cancer. Cancer Cell. 38, 534–550 (2020). e9.
Parolia, A. et al. Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature 571, 413–418 (2019).
Adams, E. J. et al. FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes. Nature 571, 408–412 (2019).
Li, L. et al. Knockdown of FOXA1 enhances the osteogenic differentiation of human bone marrow mesenchymal stem cells partly via activation of the ERK1/2 signalling pathway. Stem Cell. Res. Ther. 13, 456 (2022).
Sun, L. et al. Similarity-based metric analysis approach for predicting osteogenic differentiation correlation coefficients and discovering the novel osteogenic-related gene FOXA1 in BMSCs. Peer J. 12, e18068 (2024).
Szklarczyk, D. et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucl. Acids Res. 45, D362–D368 (2017).
Wang, T., Zhang, X. & Bikle, D. D. Osteogenic differentiation of periosteal cells during fracture healing. J. Cell. Physiol. 232, 913–921 (2017).
He, Y. D. et al. Site-specific function and regulation of Osterix in tooth root formation. Int. Endod J. 49, 1124–1131 (2016).
Balachander, G. M. et al. Unravelling microRNA regulation and miRNA-mRNA regulatory networks in osteogenesis driven by 3D nanotopographical cues. Biomater. Sci. 12, 978–989 (2024).
Margolin, A. A. et al. ARACNE: An algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinform. 7 (Suppl 1), S7 (2006).
Huynh-Thu, V. A. & Geurts, P. dynGENIE3: Dynamical GENIE3 for the inference of gene networks from time series expression data. Sci. Rep. 8, 3384 (2018).
Abdallah, B. M. et al. Skeletal (stromal) stem cells: An update on intracellular signaling pathways controlling osteoblast differentiation. Bone 70, 28–36 (2015).
Chan, W. C. W. et al. Regulation and role of transcription factors in osteogenesis. Int. J. Mol. Sci. ;22. (2021).
Lowery, J. W. & Rosen, V. The BMP pathway and its inhibitors in the skeleton. Physiol. Rev. 98, 2431–2452 (2018).
Ryoo, H. M., Lee, M. H. & Kim, Y. J. Critical molecular switches involved in BMP-2-induced osteogenic differentiation of mesenchymal cells. Gene 366, 51–57 (2006).
Qiu, M. et al. FOXA1 promotes tumor cell proliferation through AR involving the notch pathway in endometrial cancer. BMC Cancer. 14, 78 (2014).
Friedman, J. R. & Kaestner, K. H. The Foxa family of transcription factors in development and metabolism. Cell. Mol. Life Sci. 63, 2317–2328 (2006).
Xu, J. et al. The role of Forkhead box family in bone metabolism and diseases. Front. Pharmacol. 12, 772237 (2021).
Xu, T. R. et al. Inferring signaling pathway topologies from multiple perturbation measurements of specific biochemical species. Sci. Signal. 3, ra20 (2010).
Nagata, Y., Honda, Y. & Matsuda, R. FGF2 induces ERK phosphorylation through Grb2 and PKC during quiescent myogenic cell activation. Cell. Struct. Funct. 35, 63–71 (2010).
Naderi, A., Meyer, M. & Dowhan, D. H. Cross-regulation between FOXA1 and ErbB2 signaling in estrogen receptor-negative breast cancer. Neoplasia 14, 283–296 (2012).
Kim, J. et al. FOXA1 inhibits prostate cancer neuroendocrine differentiation. Oncogene 36, 4072–4080 (2017).
De Simone, A. et al. Control of osteoblast regeneration by a train of Erk activity waves. Nature 590, 129–133 (2021).
Lu, N. & Malemud, C. J. Extracellular signal-regulated kinase: A regulator of cell growth, inflammation, chondrocyte and bone cell receptor-mediated gene expression. Int. J. Mol. Sci. ;20. (2019).
Kim, J. M. et al. The ERK MAPK pathway is essential for skeletal development and homeostasis. Int. J. Mol. Sci. ;20. (2019).
Vanderschueren, D. et al. Androgens and bone. Endocr. Rev. 25, 389–425 (2004).
Kung, A. W. Androgen and bone mass in men. Asian J. Androl. 5, 148–154 (2003).
Day, T. F. et al. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev. Cell. 8, 739–750 (2005).
Piek, E. et al. Osteo-transcriptomics of human mesenchymal stem cells: Accelerated gene expression and osteoblast differentiation induced by vitamin D reveals c-MYC as an enhancer of BMP2-induced osteogenesis. Bone 46, 613–627 (2010).
Zhu, E. D., Demay, M. B. & Gori, F. Wdr5 is essential for osteoblast differentiation. J. Biol. Chem. 283, 7361–7367 (2008).
Zhou, Z. Q. et al. Sequential and coordinated actions of c-Myc and N-Myc control appendicular skeletal development. PLoS One. 6, e18795 (2011).
Hang, K. et al. Role of the heat shock protein family in bone metabolism. Cell. Stress Chaperones. 23, 1153–1164 (2018).
Drake, M. T., Clarke, B. L. & Khosla, S. Bisphosphonates: Mechanism of action and role in clinical practice. Mayo Clin. Proc. ;83:1032-45. (2008).
Saita, Y., Ishijima, M. & Kaneko, K. Atypical femoral fractures and bisphosphonate use: Current evidence and clinical implications. Ther. Adv. Chronic Dis. 6, 185–193 (2015).
Xu, H. et al. Targeting strategies for bone diseases: Signaling pathways and clinical studies. Signal. Trans. Target. Ther. 8, 202 (2023).
Noble, J. A., McKenna, M. J. & Crowley, R. K. Should denosumab treatment for osteoporosis be continued indefinitely? Ther. Adv. Endocrinol. Metab. 12, 20420188211010052 (2021).
Phetfong, J. et al. Osteoporosis: The current status of mesenchymal stem cell-based therapy. Cell. Mol. Biol. Lett. 21, 12 (2016).
Shen, M. et al. Injection of synthetic mesenchymal stem cell mitigates osteoporosis in rats after ovariectomy. J. Cell. Mol. Med. 22, 3751–3757 (2018).
Antebi, B., Pelled, G. & Gazit, D. Stem cell therapy for osteoporosis. Curr. Osteoporos. Rep. 12, 41–47 (2014).
Agata, H. et al. Intra-bone marrow administration of mesenchymal stem/stromal cells is a promising approach for treating osteoporosis. Stem Cells Int. 2019, 4214281 (2019).
Cho, S. W. et al. Transplantation of mesenchymal stem cells overexpressing RANK-Fc or CXCR4 prevents bone loss in ovariectomized mice. Mol. Ther. 17, 1979–1987 (2009).
van de Peppel, J. et al. Identification of three early phases of cell-fate determination during osteogenic and adipogenic differentiation by transcription factor dynamics. Stem Cell. Rep. 8, 947–960 (2017).
Han, H. et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucl. Acids Res. 46, D380–D386 (2018).
Valdeolivas, A. et al. Random walk with restart on multiplex and heterogeneous biological networks. Bioinformatics 35, 497–505 (2019).
Fang, H. & Gough, J. The ‘dnet’ approach promotes emerging research on cancer patient survival. Genome Med. 6, 64 (2014).
Bullinaria, J. A. & Levy, J. P. Extracting semantic representations from word co-occurrence statistics: A computational study. Behav. Res. Methods. 39, 510–526 (2007).
Bindea, G. et al. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 (2009).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucl. Acids Res. 43, e47 (2015).
Chen, E. E. M. et al. Knockdown of SIRT7 enhances the osteogenic differentiation of human bone marrow mesenchymal stem cells partly via activation of the Wnt/β-catenin signaling pathway. Cell. Death Dis. 8, e3042 (2017).
Acknowledgements
We appreciate all the clients from the Clinical Research Center of the Second Affiliated Hospital, Zhejiang University School of Medicine.
Funding
The Medical Health Science and Technology Project of Zhejiang Provincial Health Commission (Grant No. 2021KY939) and National Natural Science Foundation of China (Grant No. 82402761) funds were received in support of this work. The authors declare that they have no relevant financial activities outside the submitted work.
Author information
Authors and Affiliations
Contributions
JH, LDL and ZJP designed the research; LJL, RJL and YX performed the in vitro experiments; LLJ, RJL and YX performed the in vivo experiments; JH and YX analyzed the data; LJL and RJL wrote the paper; JH, LDL and ZJP revised the paper. All authors have read and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest related to this work.
Ethics approval and consent to participate
All animal experiments were performed in accordance with the Zhejiang Center of Laboratory Animals. All experimental procedures were approved by the Institutional Animal Care and Use Committee of Zhejiang Center of Laboratory Animals (ZJCLA-IACUC-20030053). All applicable institutional and/or national guidelines for the care and use of animals were followed. Additionally, all methods were reported in accordance with the ARRIVE guidelines for animal experiments.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, L., Lin, R., Xu, Y. et al. FoxA1 knockdown promotes BMSC osteogenesis in part by activating the ERK1/2 signaling pathway and preventing ovariectomy-induced bone loss. Sci Rep 15, 4594 (2025). https://doi.org/10.1038/s41598-025-88658-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-88658-1