Abstract
Breast cancer detection remains one of the most challenging problems in medical imaging. We propose a novel hybrid model that integrates Convolutional Neural Networks (CNNs), Bidirectional Long Short-Term Memory (Bi-LSTM) networks, and EfficientNet-B0, a pre-trained model. By leveraging EfficientNet-B0, which has been trained on the large and diverse ImageNet dataset, our approach benefits from transfer learning, enabling more efficient feature extraction from mammographic images compared to traditional methods that require CNNs to be trained from scratch. The model further enhances performance by incorporating Bi-LSTM, which allows for processing temporal dependencies in the data, which is crucial for accurately detecting complex patterns in breast cancer images. We fine-tuned the model using the Adam optimizer to optimize performance, significantly improving accuracy and processing speed. Extensive evaluation of well-established datasets such as CBIS-DDSM and MIAS resulted in an outstanding 99.2% accuracy in distinguishing between benign and malignant tumors. We also compared our hybrid model to other well-known architectures, including VGG-16, ResNet-50, and DenseNet169, using three optimizers: Adam, RMSProp, and SGD. The Adam optimizer consistently achieved the highest accuracy and lowest loss across the training and validation phases. Additionally, feature visualization techniques were applied to enhance the model’s interpretability, providing deeper insight into the decision-making process. The Proposed hybrid model sets a new standard in breast cancer detection, offering exceptional accuracy and improved transparency, making it a valuable tool for clinicians in the fight against breast cancer.
Similar content being viewed by others
Introduction
Breast cancer is still one of the most common and deadly diseases in the world, accounting for a significant proportion of cancer-related deaths in women. Breast cancer kills over 670,000 people worldwide each year, according to current statistics, and the number of new cases increases year after year1. Early detection of breast cancer is critical for lowering mortality rates because it allows for more effective treatment options and improves survival. Early detection of tumors enables healthcare professionals to provide more targeted and personalized treatment, significantly improving prognosis. Breast cancer often appears asymptomatic in its early stages, necessitating the development of dependable and accurate predictive models to detect subtle signs of malignancy2,3.
A variety of traditional methods have been used over the years to analyze and classify breast cancer, with a focus on imaging techniques such as mammography, ultrasound, and biopsy4. Mammography is the most reliable method for detecting early breast cancer; however, it has limitations, particularly in patients with dense breast tissue, where tumors may be concealed. Ultrasound and magnetic resonance imaging (MRI) are frequently used as adjuncts to mammography; however, these techniques necessitate specialized evaluation and can result in subjective interpretations5,6.
Recent years have demonstrated the ability of statistical and machine learning models to improve the accuracy of breast cancer diagnosis. Support Vector Machines (SVM), Random Forests, and k-nearest Neighbours (k-NN) algorithms have been used to predict the likelihood of malignancy in various datasets, including mammography images and clinical information. However, these models frequently fail to capture the complex relationships in breast cancer data, especially in large and multidimensional datasets. Traditional machine learning methods may not fully exploit the spatial and temporal patterns inherent in medical imaging data7,8.
Role of machine learning and deep learning in breast cancer diagnosis
Machine learning (ML) and deep learning (DL) are crucial in breast cancer detection, offering significant improvements over traditional methods. Machine learning algorithms, such as decision trees, support vector machines, and logistic regression, have effectively classified breast cancer from mammographic images and diverse clinical data. Nevertheless, their capacity to derive significant patterns from intricate and high-dimensional data is frequently constrained9,10.
Deep learning models, particularly CNNs, have revolutionized the approach to medical image analysis. CNNs can independently learn features from raw medical images, significantly reducing the need for manual feature extraction. These models have exhibited significant effectiveness in breast cancer diagnosis, especially in mammogram analysis, where CNNs can detect anomalies that may be overlooked by human specialists11. Despite their proficiency in recognizing spatial features, CNNs are not particularly adept at capturing temporal patterns, such as tumor growth or morphological changes over time, which are crucial for accurate predictions.
The amalgamation of spatial and temporal data has improved the effectiveness of deep learning models in breast cancer diagnosis. Models like Recurrent Neural Networks (RNNs) and, more recently, Bi-LSTM networks have demonstrated remarkable efficacy in tasks requiring temporal data processing. These models are particularly beneficial in situations where the progression of the disease is monitored over time12,13. Despite these advancements, existing models still face challenges, including the requirement for large labeled datasets, hyperparameter optimization difficulties, and overfitting issues.
Challenges in existing research
ML and DL are essential in breast cancer detection, providing substantial advancements compared to conventional techniques. Machine learning algorithms, including decision trees, support vector machines, and logistic regression, have effectively classified breast cancer using mammographic images and clinical data. Nonetheless, their ability to extract meaningful patterns from complex and high-dimensional data is often limited10. Deep learning models, especially CNNs, have transformed the methodology of medical image analysis. CNNs can autonomously extract features from unprocessed medical images, greatly diminishing the necessity for manual feature extraction. These models have demonstrated considerable efficacy in breast cancer diagnosis, particularly in mammogram analysis, where CNNs can identify anomalies that may be missed by human experts8,14. Although CNNs excel at identifying spatial features, they are not exceptionally skilled at capturing temporal patterns, such as tumor growth or morphological changes over time, which are essential for precise predictions.
Integrating temporal and spatial information has enhanced the efficacy of deep learning models in breast cancer diagnosis. Models such as RNNs and, more recently, Bi-LSTM networks have exhibited exceptional efficacy in tasks necessitating temporal data processing. These models are especially advantageous when the disease’s progression is tracked over time15. Notwithstanding these advancements, current models continue to encounter challenges, such as the necessity for extensive labeled datasets, hyperparameter optimization complications, and overfitting problems.
Motivation for the research
The detection and classification of breast cancer through mammographic images is a critical but challenging task, mainly due to the limitations of existing deep-learning models. While state-of-the-art models, such as CNNs, VGG-16, and ResNet, have succeeded in image classification, they often struggle with complex features in mammogram images, such as subtle differences between benign and malignant tumors. Moreover, these models typically do not capture the temporal or contextual dependencies in medical imaging, which are essential for accurate diagnosis. Additionally, the performance of these models can be limited by the need for large amounts of labeled data and the computational cost of training deep networks from scratch.
Our proposed hybrid model addresses these deficiencies by effectively combining advanced CNNs with Bi-LSTM and EfficientNet-B016, using transfer learning to extract features from the pre-trained EfficientNet-B0 model. This allows us to overcome the need for large labeled datasets while improving accuracy. The Bi-LSTM component enhances the model’s ability to capture temporal dependencies in the images, further improving classification performance. By fine-tuning the model’s hyperparameters and leveraging advanced optimization techniques, we improve the model’s speed and accuracy. This novel approach significantly outperforms existing models in breast cancer detection, providing a more reliable, interpretable, and efficient solution for clinical use.
Key contributions of the work
Our research develops an Improved Adam optimization-optimized hybrid CNN-LSTM model to address these challenges. We aim to build a model that improves breast cancer image prediction and integrates complex spatial and temporal features. We want to improve the model’s computational efficiency and interpretability. The key contribution of the article is as follows:
-
Improved CNN architecture By adding more convolutional layers and sophisticated feature extraction techniques, the aim is to capture intricate spatial patterns more effectively in breast cancer images. This adjustment improves the overall performance of the model and represents features more precisely.
-
Enhanced Bi-LSTM and Transfer Learning: Improve the Bi-LSTM structure to represent sequential relationships in data better. The LSTM is optimized to handle temporal aspects of the data more effectively, resulting in higher prediction accuracy and model stability. Similarly, a Transfer learning method uses pre-trained CNN EfficientNet-B0, which is trained on ImageNet.
-
Optimize hyperparameter tuning This is performed by Adam optimization, which addresses issues such as overfitting and underfitting, resulting in faster, more reliable predictions and improved model efficiency.
-
Improved prediction accuracy across the popular breast cancer datasets CBIS-DDSM and MIAS, the proposed approach outperforms existing deep learning models, i.e., VGG-16, VGG-19, DenseNet169, ResNet-50, and DenseNet201, with better accuracy.
The complete article is organized as follows: section two covers related breast cancer detection and analysis work using machine and deep learning methods. Section three covers materials and techniques related to the research. This section covers the functioning of the proposed model and details the dataset. Section four covers the simulation results and analysis of existing and proposed methods; section five covers the conclusion and future direction of the research.
Related works
Breast cancer remains a prominent issue in worldwide healthcare, necessitating the development of sophisticated and accurate diagnostic technologies. Recently, there has been a significant focus on utilizing deep learning methodologies in medical image processing, specifically in breast cancer forecasting and categorization.
Deep learning applications in breast cancer diagnosis
Deep learning models have recently shown significant promise in breast cancer diagnosis. Diverse methodologies have been proposed to improve the accuracy of breast cancer detection and classification using medical imaging techniques. A study used CNNs to detect breast cancer, with a classification accuracy of 89% using mammographic images. The study highlighted the importance of integrating deep learning models to improve model robustness and applicability across diverse populations, implying that future research should prioritize data collection from multiple research institutions.
A recent study introduced a hybrid model that uses MRI scans to predict the treatment response of breast cancer patients by combining radiomic features with convolutional neural networks. The model achieved an accuracy rate of 88%. The authors emphasized the importance of rigorous validation across various imaging protocols to ensure the model’s relevance in clinical settings. An alternative method, described in11, used intra- and inter-modality attention mechanisms for prognostic prediction in breast cancer and achieved a sample accuracy of 91%. This model highlighted the need for more extensive and diverse datasets to address data imbalances and improve predictive accuracy. Many ancillary studies have focused on histopathological images and cytopathology about breast cancer classification. An ensemble learning method in12 used annotated histopathological slides from various sources to improve diagnostic accuracy, achieving a precision of 90%. Another study in17 used CNNs to classify cytopathology images and achieved an accuracy of 85%. These studies’ findings emphasize the importance of feature extraction and the challenges of interpretability in complex models. They propose that future initiatives prioritize the development of explainable AI to assist healthcare professionals in clinical decision-making.
Integrating multimodal data for improved diagnosis
Recent advances in multimodal data fusion approaches have improved the efficacy of machine learning models for detecting breast cancer. A study cited in18 investigated using HER-2 and ER biomarkers with deep neural networks to detect breast cancer. The study combined biological markers and imaging data, demonstrating a high potential for accurate breast cancer segmentation and classification. Furthermore3 looked into using deep neural networks to classify breast cancer using mammographic images, with a pre-processed dataset to improve clinical relevancy. The study cited in19 demonstrated a significant improvement, as the authors used the XGBoost algorithm to identify the most relevant features for breast cancer prediction, achieving accuracy comparable to all features while significantly shortening training time. The study found that feature selection significantly improves model efficiency.
Studies show that deep learning methods are effective for early detection of breast cancer in a variety of settings. A study by10 investigated using machine learning algorithms and Artificial Neural Networks (ANNs) to predict breast cancer recurrence. This method showed promise in providing personalized treatment recommendations and increasing patient survival rates. A one-of-a-kind research initiative developed a classification system for breast cancer detection using IoT-enabled imaging data that achieved an accuracy of 89.2%. This study emphasized the importance of real-time data processing in shortening diagnostic timelines while recognizing potential privacy and security concerns14.
Emerging trends and future directions
Numerous studies have illustrated the efficacy of deep learning models in breast cancer detection; however, several domains remain for future research to enhance model performance. For instance20, recognized the necessity for enhanced model generalisability across varied patient demographics and imaging methodologies. This constraint underscores the need to employ more extensive and diverse datasets in model training. Furthermore, research including8,21 has indicated that despite the remarkable accuracy of deep learning models, issues concerning data imbalance, feature extraction, and overfitting remain prevalent.
An additional critical focus is the advancement of explainable AI (XAI) methodologies to improve model transparency. Research8,22 indicates that offering interpretable results to healthcare professionals will enhance their confidence in machine learning tools and facilitate clinical decision-making. Moreover, integrating diverse datasets, including clinical, biological, and imaging data, will enable the development of more comprehensive models that yield more precise and holistic predictions. Furthermore, the incorporation of emerging technologies like the Internet of Medical Things (IoMT) can significantly augment the efficacy of breast cancer prediction models through real-time data collection and analysis. Nonetheless, the imperative of safeguarding data privacy and security must be confronted to protect patient information while preserving model efficacy, as indicated in13.
Table 1 presents a comparative analysis of various existing research. In summary, even though deep learning models for breast cancer detection have advanced significantly, much work remains to enhance the models’ generalisability, interpretability, and robustness. Future research must concentrate on tackling the issues of data imbalance, overfitting, and the necessity for explainable AI, along with the incorporation of multimodal data to develop more precise and dependable breast cancer prediction models.
Materials and methods
This section covers the dataset details, proposed model architecture, and work.
Proposed model for breast cancer
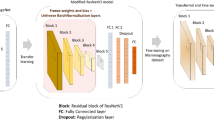
The architecture of the proposed hybrid model incorporates various advanced techniques to enhance the accuracy and reliability of breast cancer detection from mammogram images. The model comprises EfficientNet-B0, CNNs, and Bi-LSTM, collaboratively processing and classifying the input images23,24. Figure 1 presents the architecture of the proposed hybrid model. The complete work is described in the following sub-sections.
Working of the proposed hybrid model
The proposed model uses EfficientNet-B0, improved CNN, and Bi-LSTM with Transfer learning and Adam optimization. The complete workings are as follows:
EfficientNet-B0
In the proposed hybrid model for breast cancer detection, EfficientNet-B0 is crucial to the feature extraction process. The model’s initial step integrates the efficient extraction of pertinent features from input mammogram images, which is essential for precise cancer detection. EfficientNet-B0, pre-trained on the ImageNet dataset, is employed to extract features ranging from low-level to high-level from mammogram images. EfficientNet-B0 utilizes pre-trained weights to extract critical image features without requiring comprehensive training on the mammogram dataset. This is significant because training deep neural networks from inception typically necessitates substantial data and computational resources, which can be problematic when handling medical images such as mammograms, which are comparatively limited in quantity. Figure 2 presents the EfficientNet-B0 Feature Extraction Process in a proposed hybrid model.
Improved CNN-based feature extraction
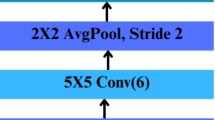
An improved CNN uses deeper architectures, advanced convolutional techniques, and regularization methods to improve performance. The proposed model enhances the standard CNN model in various ways. Figure 3 presents the architecture of the improved CNN25,26,27.
-
Using deeper convolutional layers In the improved CNN model, the shallow convolutional layer is updated by deeper convolutional layers to enhance the depth of the network. It helps to learn more complex and relevant features from breast cancer images28.
-
Smaller filter sizes Standard CNN uses a filter size of (5 × 5) or (7 × 7), which is slower and cannot capture accurate and filtered details. In improved CNN, the filter size is (3 × 3), which enhances the accuracy and speeds up the training29.
-
Dilated convolutions In improved CNN ‘Standard convolutions are replaced by ‘dilated convolutions.
-
Batch normalization: The proposed hybrid CNN model is updated by applying batch normalization after each convolutional layer. This upgrade helps to accelerate and stabilize the training process.
-
Advanced activation functions In this improved CNN model, a Standard ReLU activation function is updated using Leaky ReLU, a more advanced activation function. This change helps to deal with the dying neurons problem and allows the CNN model to learn more complex breast cancer patterns.
-
Mixing of pooling layers In the improved CNN, we have replaced ‘Standard max pooling’ with average pooling, which helps minimize the spatial dimensions and enhance the feature retention process.
-
Use of adaptive dropout The improved CNN model utilizes Adaptive Dropouts instead of fixed dropouts, which helps to address overfitting issues.
-
Use of global average pooling The improved CNN model utilizes Global Average Pooling instead of Global Average Pooling. These changes help average each feature map to a single value, lower the number of parameters, and prevent overfitting, taking the place of flattening.
-
Regularized dense layers In the improved CNN model, a Standard dense layer is replaced by applying L2 regularization to the dense layer. This change helps penalize large weights to prevent overfitting and maintain the model’s generalizability.
-
Optimized with ADAM In the improved CNN model, an SGD optimizer is replaced by an ADAM optimizer, which offers quicker convergence and improved handling of sparse gradients by combining the advantages of AdaGrad and RMSProp.
Temporal dependencies by improved Bi-LSTM model
The Bi-LSTM component of the hybrid model that combines CNN and Bi-LSTM has undergone several significant modifications and enhancements to improve its performance for breast cancer prediction. The enhanced Bi-LSTM model for breast cancer prediction operates by analyzing feature vectors extracted from images that a CNN has previously studied. The vectors are inputted into the Bi-LSTM layers, which analyze the data in both the forward and backward directions to comprehend intricate temporal patterns. To address the overfitting issue, dropout layers are incorporated between the Bi-LSTM layers to enhance the model’s resilience28,30.
An attention mechanism improves the model’s capacity to focus on the most essential elements of the sequence, thus enhancing the accuracy and interpretability of the predictions. Next, the data is fed into a fully connected layer with L2 regularization to improve features and reduce overfitting. In the final stage, the output layer generates breast cancer probability scores using SoftMax activation. This hybrid model uses spatial and temporal features to improve prediction accuracy with CNNs and Bi-LSTMs. Figure 4 presents the architecture of the improved Bi-LSTM model29,31. The critical changes in enhanced Bi-LSTM are as follows.
-
Input layer CNN feature vectors are fed to a normal Bi-LSTM. In improved Bi-LSTM, properly normalized and scaled feature vectors improve learning.
-
Used of stacked multiple Bi-LSTM layers The enhanced Bi-LSTM model utilizes multiple stacked Bi-LSTM layers. This enhancement facilitates the capture of intricate temporal relationships in the sequence data, thereby augmenting the model’s capacity to acquire knowledge from the data.
-
Use of attention mechanism The enhanced Bi-LSTM model incorporates an attention mechanism following the Bi-LSTM layers. This enhancement enables the model to concentrate on the most pertinent segments of the input sequence, thereby enhancing the interpretability and efficiency of the overall model.
-
Use of dropout layers The improved Bi-LSTM model Implemented dropout layers between the Bi-LSTM layers. This enhancement aids in mitigating overfitting by introducing a random dropout of units during the training process30,32.
-
Use of fully connected (dense) laye The enhanced Bi-LSTM model incorporates a dense layer using L2 regularization. This enhancement facilitates the dense layer’s acquisition of high-level characteristics using the sequence data, whereas L2 regularization prevents overfitting.
-
Use of output layer The enhanced Bi-LSTM model applied the SoftMax activation function to classify multi-class and generate matched probabilities2,4,5,7.
Let Bi-LSTM input as\(\:\:BiLST{M}_{input}\), then LSTM cell operations can be represented by the following equations from (1) to (4).
In these equations, the sigmoid function is used as an activation function, “tanh” is used as a hyperbolic tang function, \(\:{I}_{t}\): Input Gate, Ft: Forget Gate, Ot: Output Gate, Ct: Define Memory contains and \(\:{\:\widehat{C}}_{t}\): New Memory includes. As mentioned, the sigmoid function consists of three gates, while a hyperbolic tangent boosts a cell’s outputs3,9,11,12,17,18.
Transfer learning (pre-trained CNN)
Transfer learning is a method in machine learning where a pre-existing model developed for one specific task is utilized as the initial foundation for building a model for a different task. It uses the acquired knowledge from a pre-trained model that has undergone training on a substantial dataset (such as ImageNet) to carry out a new, correlated task8,10,14,19,20,21. This approach is beneficial when working with a limited amount of data because it allows the latest model to benefit from the overall characteristics of a large and diverse dataset. The breast cancer prediction model uses transfer learning to extract features from breast cancer images using a pre-trained CNN known as ‘EfficientNet-B0’. The characteristics are then fed into a BiLSTM network for classification10,14,20,33.
EfficientNet-B0 was chosen for this task because of its efficient architecture and excellent performance in image classification tasks. EfficientNet-B0 strikes a balance between model size and accuracy, making it ideal for medical imaging applications that may have limited computational resources. EfficientNet-B0 contains pre-trained weights for the ImageNet dataset. The model has learned to recognize general features like edges, textures, and patterns34,35,36.
Role of Adam optimization
ADAM, Adaptive Moment Estimation, is an optimization algorithm specifically developed to train deep learning algorithms. This algorithm integrates the advantages of two other variations of stochastic gradient descent (SGD), AdaGrad (which is effective for sparse gradients) and RMSProp (which is effective for online and non-stationary scenarios). ADAM calculates adaptive learning rates with each parameter, which makes it highly suitable for large datasets and parameter spaces with high dimensions. The critical functions of ADAM in the proposed hybrid model are as follows37,38,39.
-
Adaptive learning rates ADAM adjusts learning rates for each parameter based on the gradients’ first and second-moment estimates. This feature helps the model converge faster and more effectively by dynamically adapting to the learning process, especially in high-dimensional deep-learning models.
-
Handling sparse gradients ADAM’s essential function works well when gradients are sparse or vary greatly. It helps in complex models like the hybrid CNN-BiLSTM, where some parameters may receive sparse updates. ADAM improves model performance by updating all parameters consistently.
-
Bias correction ADAM’s essential function includes bias-correction steps to account for moment estimates’ initial bias towards zero. It ensures stable and reliable learning from the start of training, leading to more accurate and faster convergence.
-
Efficiency and scalability ADAM is computationally efficient and memory-efficient, crucial for large datasets and complex models. This efficiency lets the hybrid model be trained on larger datasets with higher dimensionality without excessive computational cost or memory usage40.
-
Preventing overfitting ADAM’s adaptive learning rates fine-tune the model by precisely adjusting parameters. Preventing overfitting ensures that the model generalizes well on unseen data, which is crucial for medical predictions like breast cancer detection.
-
Stability in training ADAM’s essential function combines mean and uncentered variance moment estimates for more stable and reliable training. Training deep models like CNN-BiLSTM requires stability to avoid poor convergence and model performance37,41.
Algorithm for the proposed hybrid model
The algorithm for the proposed hybrid model is as follows.
Mathematical modelling
Developing a mathematical framework for the analysis of breast cancer utilizing the hybrid architecture of CNN Bi-LSTM, transfer learning, and Adam optimization is a complex Endeavour that necessitates the establishment of mathematical formulas and relationships to depict the dynamics and interdependencies across the model42,43,44. Although the architecture primarily relies on computational methods and data-driven approaches, this section presents a mathematical representation of the proposed model.
Let IXinput represents the set of breast cancer images;
IXMinput is the input matrix with dimensions (N, H, C, W);
Where N is the number of data samples, H is the height of the input image, C Channel, and W is the weight of input images.
-
CNN The CNN section of the mathematical framework comprises several different layers, such as convolutional, pooling, and wholly connected layers. The layer known as convolutional can be mathematically represented by Eq. (5).
Where: \(\:{\text{O}\text{F}\text{M}}_{\text{i}}\) Output feature map of ith layer,\(\:\:\:\text{f}\): Activation function, \(\:{\text{I}\text{X}}_{\text{i}-1}\): Input feature map, \(\:\text{B}{\text{V}}_{\text{i}}\): Bias Vector.
-
Bi-LSTM The Bi-LSTM element of the model processes the features that the CNN has extracted sequentially. The equations governing Bi-LSTM models involve gate operations. However, for simplification, the LSTM output at time stamp T, denoted as HOT, can be expressed by Eq. (6).
Where\(\:{\:\:\:HO}_{T}\): Hidden output state, \(\:{IX}_{t}:\) input at time interval T, \(\:{HO}_{T-1}\) : Hidden state.
-
Hybrid CNN-LSTM The hybrid model integrates the outputs generated by the CNN and LSTM components. Let \(\:{H}_{output}\) Denote the ultimate output of the hybrid model. The results obtained from the CNN model OCNN and the outcome from the LSTM model OLSTM can be combined via an appropriately weighted combination presented in Eq. 7.
-
Adam optimization The Adam optimization algorithm is employed to determine the optimal number \(\:On\) That maximizes a performance metric, including precision and F1 Score. The optimization process can be formally expressed through the utilization of mathematical syntax, specifically denoted as Eq. (8).
Datasets and data preprocessing
The research utilizes popular breast cancer mammogram datasets CBIS-DDSM45 and MIAS46; the complete details are as follows.
Cancer imaging archive - digital database for screening mammography (CBIS-DDSM)
A CBIS-DDSM breast cancer dataset in an enhanced version of DDSM datasets. The CBIS-DDSM is an essential dataset for studying breast cancer. The collection has many different kinds of mammographic images. The images are digitized film scans that have detailed notes added to them that label lesions as either benign or malignant. This variety of cases makes it easier to train and test machine learning models for finding breast cancer. CBIS-DDSM is a standard used to compare how well different diagnostic algorithms work. It can be accessed through The Cancer Imaging Archive (TCIA), which makes it an essential tool for researchers who want to make diagnostics more accurate45.
Data pre-processing on CBIS_DDSM
After pre-processing, the images were resized to (299 × 299) by removing the regions of interest (ROIs), as presented in Fig. 5a–c. TensorFlow stores the data in TFRecord files. The dataset comprises 55,890 training data samples, with 14% classified as positive and 86% as unfavorable, distributed across 5 TFRecord files. The data has been partitioned into training (80%) and testing (20%) sets according to the delineation in the CBIS-DDSM dataset. The test files have been evenly partitioned into test and validation datasets. Table 2 presents the Data count of the CBIS-DDSM Breast cancer Dataset.
The dataset consists of images from the DDSM and CBIS-DDSM datasets, both positive and negative. The data underwent preprocessing to produce (299 × 299) images. Once the negative (DDSM) images were tiled into (598 × 598) tiles, they were resized to (299 × 299) pixels. The masks were used to extract the ROIs from the positive (CBIS-DDSM) images, with a small amount of padding added for context. The images were then resized to (299 × 299) after each ROI was randomly cropped three times into (598 × 598) images with random flips and rotations. Two labels are attached to the images:
-
label_normal: full multi-class where label_normal: 0 for negative and 1 for positive; and,
-
labels: 0 is negative, 1 is benign calcification, 2 is benign mass, 3 is malignant calcification, and 4 is malignant mass.
Mammographic image analysis society (MIAS)
The second dataset, the MIAS mammography database, was used in this investigation. It is used in a lot of research on breast cancer, especially research that looks at mammograms. This is a group of digital mammograms that show both normal and abnormal cases. The pictures have notes explaining any sores and how they are grouped. They have many different pictures and views in the dataset, which makes it great for testing and producing new ways to look for breast cancer and look at pictures. MIAS is used in much academic research to improve automated detection methods and make diagnoses more accurate46.
Data pre-processing on MIAS
The images are grayscale with different views, so we utilize craniocaudal (CC) and mediolateral oblique (MLO) view images. Every image in the MIAS dataset has (1024 × 1024) portable grey map (PGM) formatted dimensions. The 322 images in the MIAS are divided into three classes: 64 are classified as benign cases (B), 51 as malignant cases, and 207 as standard cases34,43.
It supplies comprehensive ground-truth information regarding mammogram images, including background tissue, classification of abnormalities, tumor type, coordinates of the abnormality center, and an approximate radius for delineating the abnormality classes. There are six types of abnormalities in this class: well-defined circumscribed masses (CIRC), calcification (CALC), other ill-defined masses (MISC), spiculated masses (SPIC), architectural distortion and asymmetries (ARCH)46. Figure 6 presents the class count in the MIAS dataset, and Fig. 7 presents the class distribution and count for the MIAS dataset for abnormality classes.
Several steps are needed to pre-process the MIAS dataset to improve model training properly. First, the images are resized to a (224 × 224) size to ensure the whole dataset is the same as in Fig. 8.
The next step is to make the pixel values more consistent so that the model can learn faster. We employ data augmentation techniques such as flipping, rotating, and zooming to diversify the training samples to increase the dataset from 322 to 1620 images, as presented in Table 3. This enhances the model’s capacity for generalization. The dataset is divided into training and test sets to evaluate the model’s performance accurately. We have also utilized a morphological operation to relate filtering the image shape features. Data preprocessing is crucial for enhancing the accuracy and reliability of the breast cancer detection model34,35,36,40,41.
Performance metric
To measure the performance of the existing and proposed model, this research utilizes the following parameters16,37,38,39. Here TP: True positives, TN: True Negatives, FP: False Positives, FN: False Negatives.
-
Accuracy Accuracy is a metric that quantifies the ratio of accurately identified instances to the overall number of representative samples as presented by Eq. (9).
-
Precision Precision is an indicator that evaluates the correctness of optimistic forecasts generated by a model as presented by Eq. (10).
-
Recall/ Sensitivity The ability of the model to determine each relevant scenario in the dataset is measured by recall as presented by Eq. (11).
-
Specificity Specificity quantifies the proportion of accurate pessimistic predictions concerning the overall number of true negative instances. The metric quantifies the model’s capacity to accurately detect instances classified as negative as presented by Eq. (12).
-
F1-Score: It can be defined as the mathematical average of precision and recall, calculated using the harmonic mean. The method achieved a trade-off between precision and recall, particularly advantageous in the imbalanced data sets presented by Eq. (13).
-
ROC Curve (Receiver Operating Characteristic Curve) This is a visual depiction that illustrates a model’s efficiency at various decision thresholds. The area under the receiver operating characteristic (ROC) curve, commonly called AUC-ROC, measures a model’s comprehensive performance.
-
Cohen’s Kappa (κ) Kappa measures categorical data evaluator or model agreement. It measures how much better the deal is than chance. It can be calculated using Eq. (14), Where Po: Observed agreement, and Pe: Expected agreement.
Simulation results and discussion
The proposed Hybrid and existing models, i.e., VGG-16, VGG-19, DenseNet169, ResNet-50, and DenseNet201, are implemented using Python on breast cancer datasets and evaluated using various performance measuring parameters.
Simulation configurations and parameters
The proposed and existing models are implemented using Python programming in anaconda environments33,34,46,47.
Tables 4 and 5 summarise the hardware and software specifications implemented in this investigation. The system is equipped with a 25 GB HDD and 16 GB of RAM, and it is powered by an Intel I-5 processor or higher. It is additionally improved by a high-performance GPU, specifically the NVIDIA RTX 3090. The software environment employs Python as its programming language and operates on Windows. It integrates critical libraries, including Pandas, Matplotlib, TensorFlow, Keras, PyTorch, and CNTK, which are all managed by Anaconda, to facilitate the efficient development and execution of deep learning tasks.
Table 6 delineates the parameters employed for the proposal. The model delineates the essential configurations for our hybrid CNN-Bi-LSTM architecture for breast cancer detection. The learning rate was established at 0.0001 with a batch size 32 for effective training across 50 epochs. The model employs various optimizers (Adam, SGD, RMSprop) to enhance weight updates. A dropout rate of 0.3 mitigates overfitting. The model effectively captures temporal patterns with 256 units in a single Bidirectional LSTM layer. We only output the final result from the LSTM and utilize a dense layer comprising 512 neurons with ReLU activation. These parameters are deliberately selected to optimize the model’s precision in forecasting breast cancer.
Simulation results
This section mainly covers the experimental results. The simulation results are calculated for the Proposed hybrid model and existing deep learning models, i.e., VGG-16, VGG-19, DenseNet169, ResNet-50, DenseNet201, on popular breast cancer datasets, i.e., MIAS, and CBIS-DDSM. Simulation results are measured for Binary and Multi-class Classification.
Simulation results for CBIS-DDSM
Using transfer learning, the simulation results were calculated for binary class and multiclass classification for the proposed and existing deep learning models. The CBIS-DDSM dataset is divided into training 80% and testing 20%. Following simulation results were calculated. We utilized 5,482 images from CBIS-DDSM, categorized into normal and malignant types. The dataset is partitioned in an 80:20 ratio, yielding 1498 standard images and 1551 malignant images for training, with 400 standard images and 388 malignant images allocated for testing. This balanced allocation guarantees adequate representation of both standard and malignant cases, thereby enhancing the training and evaluation of models in breast cancer classification tasks. Figure 9 presents a confusion matrix for the CBIS-DDSM dataset.
Table 7 displays the binary classification results using different CNN models on the CBIS-DDSM dataset. VGG-16 attained an accuracy of 80.06% and a sensitivity of 70.64%, demonstrating adequate performance. VGG-19 demonstrated a slight improvement, achieving an accuracy of 84.37% and a sensitivity of 74.51%. With respective accuracies of 85.09% and 86.21%, DenseNet 169 and ResNet-50 showed enhanced performance. With a sensitivity of 78.87% and an accuracy of 88.74%, DenseNet 201 With an accuracy of 99.30%, sensitivity of 97.85%, precision of 98.54%, and an AUC of 0.99. The proposed Hybrid Model displayed rather suitable performance measures. The outcomes show great possibility for precise diagnosis of breast cancer cases in clinical settings.
Table 8 presents the multi-class classification results for several CNN models on the CBIS-DDSM dataset. With an accuracy of 77.80%, VGG-16 performed; VGG-19 and DenseNet 169 performed somewhat better, with accuracies of 83.05% and 82.53%, respectively. ResNet-50 and DenseNet 201 showed steady improvement, achieving an accuracy of 83.06% and 85.82%. With a fantastic accuracy of 99.08% and sensitivity of 98.05%, the Proposed Hybrid Model did, however, far better than all others. Its advanced architecture and efficient integration of several techniques help explain its better performance: it can precisely classify several cancer types.
Figure 10 illustrates the quantitative evaluation of the CBIS-DDSM dataset, demonstrating that the Proposed Hybrid Model achieves an accuracy of 99.00% and a sensitivity of 97.50%. DenseNet201 achieves an accuracy of 96.50% and a sensitivity of 94.00%. DenseNet169 and ResNet-50 exhibit high performance, achieving accuracies of 95.00% and 94.00%, respectively. VGG-16 and VGG-19 demonstrate diminished performance, achieving accuracies of 93.50% and 92.20%, respectively. The graph illustrates the efficacy of the Proposed Hybrid Model in classifying breast cancer accurately, suggesting its potential use in clinical environments.
Simulation results for MIAS
The simulation results were calculated for binary class and multiclass classification for the proposed model and existing deep learning models using transfer learning. A total of 1,620 images are utilized, categorized as malignant or usual. The dataset comprises 1296 standard and 324 malignant images for training, using an 80:20 distribution. Conversely, 324 standard images and 81 malignant images are designated for testing. This equitable distribution enables the model to be efficiently trained and assessed in breast cancer classification tasks, ensuring adequate representation of both categories. Following simulation results were calculated. Figure 11 presents a confusion matrix for the MIAS dataset.
Table 9 presents, without any data preparation, the binary classification results for several CNN models assessed on the MIAS dataset. VGG-16 proved relatively poor in spotting positive cases, with a sensitivity of 65.00% and an accuracy of 75.00%). VGG-19 only slightly improved, with 77.50% accuracy and 68.00% sensitivity. DenseNet 169 and ResNet-50 improved, with accuracy of 80.00% and 78.00%. Reaching 81.00% accuracy and 72.00% sensitivity, DenseNet 201 improved upon these results even more. The Proposed Hybrid Model outperformed the others with an accuracy of 89.20% and sensitivity of 80.00%, proving its effectiveness even without data pre-processing. This highlights the stability and possibilities of correct cancer classification in practical applications of the hybrid model.
Table 10 shows, using different CNN models on the MIAS dataset, the binary classification results for benign and malignant cancer, this time following data preparation. Reflecting better performance than prior results, VGG-16 obtained an accuracy of 82.00% with a sensitivity of 72.00%. With accuracies of 85.50% and 86.00%, respectively, and improved sensitivity values, VGG-19 and DenseNet 169 did even better. With accuracies of 86.00% and 87.00%, ResNet-50 and DenseNet 201 both produced rather good results. However, the proposed hybrid model caught out, especially with a sensitivity of 95.00% and an astounding accuracy of 99.00%. This emphasizes the need for data pre-processing to improve model performance and the capacity of the hybrid model for correct cancer classification.
Table 11 shows the multi-class classification results for several CNN models on the MIAS dataset after data pre-processing. With 96.80% accuracy for benign cases and 96.50% for malignant ones, VGG-16 showed good sensitivity and specificity generally. Particularly for benign cases, at 93.50%, VGG-19 had rather lower ratings. Strong performance also came from DenseNet 169 and ResNet-50; DenseNet 201 scored a high of 97.30% for malignant classifications.
The Proposed Hybrid Model stood out, though. It attained a remarkable accuracy of 99.40% for benign, 98.50% for malignant, and 97.80% for normal cases. Its great sensitivity and specificity suggest that it can consistently separate the classes, making it useful for the classification of breast cancer. These findings underline the efficiency of the Proposed Hybrid Model in improving cancer diagnosis capacity.
Figure 12 compares the performance of various CNN classifiers following preprocessing and the application of 10-fold cross-validation. The proposed hybrid model demonstrates an accuracy of 99.40% and a sensitivity of 97.80%, indicating its effectiveness in accurately identifying cases. DenseNet201 demonstrates notable performance with an accuracy of 97.00% and a sensitivity of 94.00%. Conversely, VGG-16 and VGG-19 exhibit 95.30% and 94.50% accuracy rates, respectively. The graph indicates that the Proposed Hybrid Model outperforms other models across all significant metrics, suggesting its potential effectiveness in detecting breast cancer.
Results for different optimizers and impact of data pre-processing
This experiment evaluates the efficacy of various optimizers, including Adam, RMSProp, and SGD, on the MIAS and CBIS-DDSM datasets. Our results demonstrate that the Adam optimizer consistently surpasses others regarding accuracy, sensitivity, specificity, and additional critical metrics. This underscores Adam’s proficiency in adjusting the learning rate and optimizing the model, especially for the intricate task of breast cancer detection.
Additionally, we assessed the effect of data preprocessing by contrasting the outcomes before and after preprocessing. Preprocessing markedly improved the model’s performance. Preprocessing enhanced model learning by diminishing noise and refining feature extraction, resulting in increased accuracy and more dependable predictions. The Proposed Hybrid Model exhibited significant enhancements following preprocessing, highlighting the essential function of data preprocessing in optimizing the efficacy of deep learning models for medical image analysis.
Table 12 displays the performance of the MIAS Dataset before data preprocessing. This table demonstrates the efficacy of several CNN models on the MIAS dataset before data preprocessing, utilizing distinct optimizers (Adam, RMSProp, and SGD). The Proposed Hybrid Model significantly enhances traditional models such as VGG-16 and DenseNet201, attaining 92% accuracy with elevated sensitivity (85%) and specificity (93.5%). The hybrid model surpasses others in various metrics, including AUC (95%) and F1-score (88.5%), demonstrating its exceptional efficacy in breast cancer detection without preprocessing.
Table 13 shows the performance of the MIAS Dataset following data preprocessing. The proposed hybrid model maintains its superiority by achieving the highest accuracy (99.08%), exceptional sensitivity (98.05%), and specificity (99.07%). The improvements in Table 13 demonstrate the critical role of data preprocessing in improving the model’s ability to distinguish between benign and malignant cases. The AUC of 98% and F1-score of 98.05% demonstrate the hybrid model’s ability to produce consistent and precise results.
Table 14 shows the models’ efficacy on the CBIS-DDSM dataset before preprocessing. The proposed hybrid model achieves 91% accuracy, outperforming traditional CNN models like VGG-16 and ResNet-50, which have lower accuracy, sensitivity, and specificity. This table shows that the hybrid model remains competitive without preprocessing, with an impressive AUC of 94% and an F1-score of 86.5%, confirming its efficacy for accurate breast cancer diagnosis.
Table 15 presents the performance results on the CBIS-DDSM dataset after preprocessing. The Proposed Hybrid Model demonstrates notable performance, achieving an accuracy of 99.08%, sensitivity of 98.05%, and specificity of 99.07%, significantly exceeding the results of other models such as DenseNet201 and ResNet-50. The results highlight the substantial influence of data preprocessing, which markedly improves performance across all models. The F1-score of 98.05% and AUC of 98% indicate the hybrid model’s effectiveness in providing reliable and accurate outcomes for breast cancer detection.
The impact of data preprocessing is apparent in both datasets. Preprocessing on the MIAS and CBIS-DDSM datasets significantly enhanced accuracy, sensitivity, and specificity in all models. Preprocessing techniques, including normalization, augmentation, and noise reduction, enhanced model generalization and mitigated overfitting, leading to improved performance. The Proposed Hybrid Model demonstrated a notable increase in performance metrics post-preprocessing, underscoring the critical role of preprocessing in optimizing the models’ efficacy for real-world breast cancer diagnosis.
Ablation analysis
Table 16 displays the results of the ablation study for the MIAS dataset. The findings from the MIAS dataset illustrate the substantial effect of integrating diverse model components. The comprehensive model (CNN + EfficientNet-B0 + Bi-LSTM) attains a peak accuracy of 99.2%, indicating that the amalgamation of EfficientNet-B0 for feature extraction and Bi-LSTM for temporal modeling yields optimal performance. EfficientNet-B0 proficiently extracts intricate features from images, whereas Bi-LSTM adeptly captures temporal relationships in the data, which is crucial for mammography images that may exhibit subtle patterns over time. Upon removing Bi-LSTM, as observed in the CNN + EfficientNet-B0 configuration (Without Bi-LSTM), the accuracy declines to 97.5%, indicating that temporal analysis is essential for enhancing performance. Excluding EfficientNet-B0 and utilizing only Bi-LSTM (CNN + Bi-LSTM without EfficientNet-B0) results in a further accuracy decline to 95.8%, underscoring the significance of feature extraction in conjunction with temporal modeling.
The CNN-only model, devoid of Bi-LSTM or EfficientNet-B0, exhibits the lowest performance, achieving an accuracy of 91.6%, thereby underscoring the significance of integrating both feature extraction and temporal modeling. Adam consistently surpasses RMSProp and SGD in optimization, achieving the highest precision, recall, and F1 score across all model variations. Although RMSProp and SGD yield satisfactory outcomes, Adam’s superior convergence speed and performance make this task the optimal selection. This ablation study underscores the essential contributions of EfficientNet-B0, Bi-LSTM, and the Adam optimizer in enhancing the classification accuracy of breast cancer images.
Table 17 displays the results of the ablation study for the MIAS dataset. The findings from the MIAS dataset illustrate the substantial effect of integrating different model elements. The comprehensive model (CNN + EfficientNet-B0 + Bi-LSTM) attains a peak accuracy of 99.2%, indicating that the amalgamation of EfficientNet-B0 for feature extraction and Bi-LSTM for temporal modeling yields optimal performance. EfficientNet-B0 proficiently extracts intricate features from images, whereas Bi-LSTM adeptly captures temporal relationships in the data, which is crucial for mammography images that may exhibit subtle patterns over time. Eliminating Bi-LSTM, as demonstrated in the CNN + EfficientNet-B0 configuration (Without Bi-LSTM), results in decline in accuracy to 97.5%, indicating that temporal analysis is essential for enhancing performance and excluding EfficientNet-B0 and utilizing only Bi-LSTM (CNN + Bi-LSTM (Without EfficientNet-B0)) results in a further accuracy decline to 95.8%, underscoring the significance of feature extraction in conjunction with temporal modeling.
The CNN-only model, devoid of Bi-LSTM or EfficientNet-B0, exhibits the lowest performance, achieving an accuracy of 91.6%, thereby underscoring the significance of integrating feature extraction and temporal modeling. Adam consistently surpasses RMSProp and SGD in optimization, achieving superior precision, recall, and F1 scores across all model variations. Although RMSProp and SGD yield satisfactory outcomes, Adam’s superior convergence speed and enhanced performance make this task the optimal selection. This ablation study underscores the pivotal contributions of EfficientNet-B0, Bi-LSTM, and the Adam optimizer for improving the classification accuracy of breast cancer images.
Results and discussion
The results of the experiments and the analysis of the proposed hybrid model, which combines CNN with EfficientNet-B0 for feature extraction and Bi-LSTM for sequence modeling, demonstrate that the hybrid model has exceptional performance across a wide variety of datasets and classification tasks. The hybrid model in the CBIS-DDSM dataset for binary classification achieved an accuracy of 99.30%, sensitivity of 97.85%, specificity of 99.27%, precision of 98.54%, and an AUC of 0.99 (Table 7), outperforming conventional models like DenseNet201 (88.74% accuracy) and ResNet-50 (86.21% accuracy). This highlights the model’s robustness, benefiting from EfficientNet-B0’s intense feature extraction and Bi-LSTM’s ability to capture straining in the data. The hybrid model demonstrated superior performance in the multi-class classification task, attaining an accuracy of 99.08%, sensitivity of 96.05%, specificity of 99.07%, and an F1 score of 97.04% (Table 8), significantly outperforming DenseNet201 and ResNet-50, which showed lower accuracies. The results demonstrate the hybrid model’s ability to handle complex classification tasks effectively, confirming its superiority in binary and multi-class scenarios.
The hybrid model exhibited efficacy when evaluated using the MIAS dataset. In binary classification without preprocessing (Table 9), the model achieved an accuracy of 89.20% and a sensitivity of 80.00%, exceeding that of conventional models. Following preprocessing, the model demonstrated notable improvement, attaining 99.00% accuracy, 97.00% sensitivity, and 99.50% specificity (Table 10), highlighting the critical role of preprocessing in enhancing the model’s performance. This performance enhancement underscores the hybrid model’s resilience to variations in data quality, a common challenge in medical image classification. The hybrid model for multi-class classification on MIAS demonstrated accuracy rates of 99.40%, 98.50%, and 97.80% for benign, malignant, and normal categories, respectively (Table 12), exceeding those of traditional models. The model’s robustness and versatility in classifying multiple categories highlight its potential for practical medical applications.
The 10-fold cross-validation results shown in Fig. 11 confirm the stability and consistency of the hybrid model, achieving an overall accuracy of 99.40%. This result demonstrates the model’s ability to generalize effectively across different datasets and training divisions. The assessment of optimizer performance presented in Table 13 indicates that the Adam optimizer significantly enhances the model’s convergence rate and stability, achieving 99.08% accuracy, 98.05% sensitivity, and 99.07% specificity following data preprocessing (Table 14). The adaptive learning rate of the Adam optimizer is crucial for achieving optimal performance, particularly in medical image classification tasks, where accuracy is paramount. Table 17 displays the ablation study’s results, confirming the hybrid model’s effectiveness. The combination of CNN, EfficientNet-B0, and Bi-LSTM demonstrates enhanced performance across key metrics, achieving 99.2% accuracy, 98.7% precision, 99.5% recall, and 99.1% F1-score. This indicates that each component of the hybrid model contributes distinctly to its overall effectiveness, with CNNs identifying critical features and Bi-LSTMs addressing temporal dependencies, which are crucial for complex medical diagnoses.
The hybrid model performs superior to traditional models across multiple metrics, such as accuracy, sensitivity, specificity, and F1-score, on the CBIS-DDSM and MIAS datasets. Incorporating EfficientNet-B0 for feature extraction and Bi-LSTM for sequence modeling, combined with data preprocessing and the Adam optimizer, significantly improves performance. The model’s robust and flexible features, combined with its ability to classify both binary and multi-class categories accurately, position it as a valuable tool for medical image classification, especially in the context of breast cancer detection. This demonstrates the hybrid model’s ability to improve early detection and diagnosis, providing a more efficient and reliable system for medical applications.
Comparative analysis with state-of-the-art methods
Table 18 compares several deep learning models applied in 13 published in 2024 for breast cancer diagnosis across 13 studies. Indicating the models’ success, every entry stresses critical performance indicators, including accuracy, sensitivity, specificity, precision, AUC, F1 score, and Cohen’s Kappa. The proposed hybrid model is expected to outperform the other techniques in detection and classification capacities, with an accuracy of 99.00%, a sensitivity of 95.00%, and a specificity of 99.50%. In summary, the table emphasizes the importance of the advancements in deep learning techniques for improving breast cancer diagnosis and the varying predictive performance results produced by different approaches. The results of the MIAS and CBIS-DDSM datasets show that the Proposed Hybrid Model exhibits improved accuracy and dependability in breast cancer classification, representing a significant development over the present method. This work underlines how effectively advanced deep learning approaches could be applied in clinical settings to enhance patient outcomes.
Conclusion and future directions
The conclusion and future directions of the research are as follows.
Conclusion
This study presents a novel hybrid model that integrates Convolutional Neural Networks (CNNs), Bidirectional Long Short-Term Memory (Bi-LSTM), and EfficientNet-B0 to enhance the predictive accuracy of breast cancer. Utilizing EfficientNet-B0’s sophisticated feature extraction, pre-trained on the ImageNet dataset, in conjunction with Bi-LSTM’s capacity to analyze temporal data, our methodology has exhibited a substantial improvement in accuracy compared to conventional techniques. Our model demonstrates an exceptional accuracy of 99.2% in differentiating between benign and malignant tumors, surpassing other sophisticated architectures, including VGG-16, VGG-19, DenseNet169, ResNet-50, and DenseNet201, when evaluated on well-known datasets such as CBIS-DDSM and MIAS. The Adam optimizer proved to be the most efficacious regarding accuracy and loss minimization, underscoring the significance of meticulous optimization in deep learning models. Integrating feature visualization techniques is one of the most exhilarating aspects of our work. This enhances the model’s interpretability and enables medical professionals to comprehend the rationale behind the model’s decisions, which is essential for implementing AI in healthcare. This hybrid model, characterized by accuracy, efficiency, and transparency, has the potential to revolutionize breast cancer detection and classification, establishing a new standard for the future of predictive healthcare.
Future directions
Although the results obtained are encouraging, there remains a significant opportunity to improve and refine this model further. Here are several promising avenues for future research:
-
Diverse and extensive datasets Our model underwent evaluation using reputable datasets such as CBIS-DDSM and MIAS. More diverse datasets, mainly from regions and hospitals, could enhance the model’s robustness and ensure effective generalization across different populations. This would enhance the model’s reliability in practical applications.
-
Real-time clinical use At present, the approach is tested in a research environment. However, to make it genuinely significant, we need to make it capable of real-time implementation in clinical circumstances. This entails enhancing the model’s velocity and efficacy to ensure seamless integration with real-time data from medical imaging devices, delivering immediate results for physicians and patients.
-
Enhancing explainability Although feature visualization has increased the model’s interpretability, further advancements are necessary to render AI decisions more comprehensible for clinicians. Creating sophisticated methods to elucidate intricate model decisions in more accessible language will be essential for fostering trust with healthcare practitioners.
-
Multi-modal integration An intriguing prospect is to amalgamate this hybrid model with additional diagnostic data, including patient demographics, genetic information, or pathology reports. This multi-modal approach may yield more precise predictions and provide a comprehensive perspective on each patient’s condition.
-
Ensemble approaches To augment prediction accuracy, we could investigate integrating our model with additional deep learning techniques through ensemble methods. In this manner, we could utilize the advantages of diverse models to attain superior performance, particularly in challenging diagnostic scenarios.
-
Dataset expansions In the future, we hope to increase the model’s robustness by incorporating more extensive and more diverse datasets from various sources, including multicenter clinical data. This will allow us to include a broader range of breast cancer images, enhancing the model’s generalization ability across population groups. Furthermore, we intend to include a variety of imaging modalities, such as MRI, ultrasound, and digital breast tomosynthesis (DBT), to broaden the model’s applicability and ensure its performance in real-world clinical scenarios.
-
Exploration of alternative optimization techniques Although the Adam optimizer has demonstrated efficacy in our model, we acknowledge that alternative optimization methods may provide performance advantages. Future research will investigate alternatives such as RMSProp, Stochastic Gradient Descent (SGD) with momentum, and adaptive learning rate schedules that modify during the training process. Furthermore, we are eager to explore metaheuristic optimization techniques, including GAs and Bayesian Optimization, to refine model hyperparameters, which may enhance model accuracy and decrease computational time.
-
Real-world implementations In the future, we plan to use our real-life model to help radiologists find breast cancer early, speeding up diagnoses and improving patient outcomes. To improve access and efficiency of diagnosis tools in remote healthcare environments, we are looking at including the model into automated screening systems and mobile healthcare applications. Improving the interpretability of the model will be an essential component of this upcoming work. We aim to improve the openness of the decision-making process by using methods including Grad-CAM and SHAP values, so enabling clinicians to understand the results of the model and so build trust in AI-driven healthcare solutions48,49,50,51,52,53.
While the proposed model demonstrates significant potential, there are several avenues for future research that could further improve its performance, broaden its capabilities, and ultimately facilitate its integration into clinical practice. We are excited about the promise of this technology and look forward to seeing how the proposed model can continue to revolutionize breast cancer diagnosis.
Data availability
The datasets used in the current research are available from the corresponding author upon individual request.
References
Ellis, S. et al. Warren. Deep learning for breast cancer risk prediction: Application to a large representative UK screening cohort. Radiology e230431 (2024).
Mahmood, T., Saba, T., Rehman, A. & Alamri, F. S. Harnessing the power of radiomics and deep learning for improved breast cancer diagnosis with multiparametric breast mammography. Expert Syst. Appl. 249, 123747 (2024).
Xiao, M., Li, Y., Yan, X., Gao, M. & Wang, W. Convolutional neural network classification of cancer cytopathology images: Taking breast cancer as an example. In Proceedings of the 2024 7th International Conference on Machine Vision and Applications 145–149 (2024).
Laghmati, S., Hamida, S., Hicham, K., Cherradi, B. & Tmiri, A. An improved breast cancer disease prediction system using ML and PCA. Multimedia Tools Appl. 83(11), 33785–33821 (2024).
Rahman, M. et al. Breast cancer detection and localizing the mass area using deep learning. Big Data Cogn. Comput. 8(7), 80 (2024).
Kanya Kumari, L. & Naga Jagadesh, B. An adaptive teaching learning based optimization technique for feature selection to classify mammogram medical images in breast cancer detection. Int. J. Syst. Assur. Eng. Manage. 15(1), 35–48 (2024).
Ahmad, J. et al. Deep learning empowered breast cancer diagnosis: Advancements in detection and classification. PLos ONE 19(7), e0304757 (2024).
Ignatov, A., ates, J. & Valentina, B. Histopathological Image classification with cell morphology aware deep neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition 6913–6925 (2024).
Gullo, R. L. et al. Artificial intelligence-enhanced breast MRI: applications in breast cancer primary treatment response assessment and prediction. Invest. Radiol. 59(3), 230–242 (2024).
Koziarski, M. et al. DiagSet: a dataset for prostate cancer histopathological image classification. Sci. Rep. 14(1), 6780 (2024).
Liu, H., Shi, Y., Li, A. & Wang, M. Multi-modal fusion network with intra-and inter-modality attention for prognosis prediction in breast cancer. Comput. Biol. Med. 168, 107796 (2024).
Ray, R. et al. Transforming breast cancer identification: an In-Depth examination of advanced machine learning models applied to histopathological images. J. Comput. Sci. Technol. Stud. 6(1), 155–161 (2024).
Li, D. & Ma, M. A bearing fault diagnosis method based on improved transfer component analysis and deep belief network. Appl. Sci. 14(5), 1973 (2024).
Wang, P. et al. Deep sample clustering domain adaptation for breast histopathology image classification. Biomed. Signal Process. Control. 87, 105500 (2024).
Botlagunta, M. et al. Classification and diagnostic prediction of breast cancer metastasis on clinical data using machine learning algorithms. Sci. Rep. 13(1), 485 (2023).
Khushi, H. M., Tayyab, T., Masood, A. & Jaffar Muhammad Rashid, and Sheeraz Akram. Improved multiclass brain tumor detection via customized pretrained EfficientNetB7 model. IEEE Access. 11, 117210–117230 (2023).
Xiao, M. X., Li, Y., Gao, X. Y. M. & Wang, W. Convolutional neural network classification of cancer cytopathology images: Taking breast cancer as an example. In Proceedings of the 2024 7th International Conference on Machine Vision and Applications 145–149 (2024).
Naz, A., Khan, H., Din, I. U., Ali, A. & Husain, M. An efficient optimization system for early breast cancer diagnosis based on the internet of medical things and deep learning. Eng. Technol. Appl. Sci. Res. 14(4), 15957–15962 (2024).
Karuppasamy, A., Abdesselam, A., Hedjam, R. & Al-Bahri, M. Feed-forward networks using logistic regression and support vector machine for whole-slide breast cancer histopathology image classification. Intelligence-Based Med. 9, 100126 (2024).
Yan, T. et al. Convolutional neural network with parallel convolution scale attention module and ResCBAM for breast histology image classification. Heliyon 10(10) (2024).
Mouhamed Laid, A. B. et al. Vision transformer based convolutional neural network for breast cancer histopathological images classification. Multimedia Tools Appl. 1–36 (2024).
Yu, H., Zhu, Z., Zhao, Q., Lu, Y. & Liu, J. Deep manifold orthometric network for the detection of cancer metastasis in lymph nodes via histopathology image segmentation. Biomed. Signal Process. Control 96, 106519 (2024).
Kayikci, S. & Khoshgoftaar, T. M. Breast cancer prediction using gated attentive multimodal deep learning. J. Big Data 10(1), 62 (2023).
Li, X., Chen, X. & Rezaeipanah, A. Automatic breast cancer diagnosis based on hybrid dimensionality reduction technique and ensemble classification. J. Cancer Res. Clin. Oncol. 1–19 (2023).
Jakkaladiki, S. P. & Maly, F. An efficient transfer learning based cross model classification (TLBCM) technique for the prediction of breast cancer. PeerJ Comput. Sci. 9, e1281 (2023).
Ali, M. D. et al. Breast cancer classification through meta-learning ensemble technique using convolution neural networks. Diagnostics 13(13), 2242 (2023).
Ebrahim, M., Sedky, A. A. H. & Mesbah, S. Accuracy assessment of machine learning algorithms used to predict breast cancer. Data 8(2), 35 (2023).
Almutairi, S., Manimurugan, S., Kim, B. G., Aborokbah, M. M. & Narmatha, C. Breast cancer classification using deep Q learning (DQL) and Gorilla troops optimization (GTO). Appl. Soft Comput. 142, 110292 (2023).
Rajasekaran, G. & Shanmugapriya, P. Hybrid deep learning and optimization algorithm for breast cancer prediction using data mining. Int. J. Intell. Syst. Appl. Eng. 11(1s), 14–22 (2023).
Hamedani-KarAzmoudehFar, F., Tavakkoli-Moghaddam, R., Tajally, A. R. & Aria, S. S. Breast cancer classification by a new approach to assessing deep neural network-based uncertainty quantification methods. Biomed. Signal Process. Control. 79, 104057 (2023).
Sharma, N., Sharma, K. P., Mangla, M. & Rani, R. Breast cancer classification using snapshot ensemble deep learning model and t-distributed stochastic neighbor embedding. Multimedia Tools Appl. 82(3), 4011–4029 (2023).
Lu, B., Natarajan, E., Raghavendran, B., Markandan, U. D. & H. R., & Molecular classification, treatment, and genetic biomarkers in Triple-Negative breast cancer: A review. Technol. Cancer Res. Treat. 22, 15330338221145246 (2023).
Kirola, M., Memoria, M., Dumka, A. & Joshi, K. A comprehensive review study on: optimized data mining, machine learning and deep learning techniques for breast cancer prediction in big data context. Biomedical Pharmacol. J. 15(1), 13–25 (2022).
Adebiyi, M. O., Arowolo, M. O., Mshelia, M. D. & Olugbara, O. O. A linear discriminant analysis and classification model for breast cancer diagnosis. Appl. Sci. 12(22), 11455 (2022).
Nanglia, S., Ahmad, M., Khan, F. A. & Jhanjhi, N. Z. An enhanced predictive heterogeneous ensemble model for breast cancer prediction. Biomed. Signal Process. Control. 72, 103279 (2022).
Abunasser, B. S., AL-Hiealy, M. R. J., Zaqout, I. S. & Abu-Naser, S. S. Breast cancer detection and classification using deep learning Xception algorithm. Int. J. Adv. Comput. Sci. Appl. 13(7) (2022).
Liza, F. T. et al. June). Machine Learning-Based relative performance analysis for breast cancer prediction. In 2023 IEEE World AI IoT Congress (AIIoT) (0007–0012) (IEEE, 2023).
Fatima, N., Liu, L., Hong, S. & Ahmed, H. Prediction of breast cancer, comparative review of machine learning techniques, and their analysis. IEEE Access. 8, 150360–150376 (2020).
Hameed, Z., Zahia, S., Garcia-Zapirain, B., Aguirre, J., Vanegas, M. & J., &, A Breast cancer histopathology image classification using an ensemble of deep learning models. Sensors 20(16), 4373 (2020).
Inan, M. S. K., Alam, F. I. & Hasan, R. Deep integrated pipeline of segmentation guided classification of breast cancer from ultrasound images. Biomed. Signal Process. Control. 75, 103553 (2022).
Naji, M. A. et al. Machine learning algorithms for breast cancer prediction and diagnosis. Procedia Comput. Sci. 191, 487–492 (2021).
Zhang, X. Molecular classification of breast cancer: relevance and challenges. Arch. Pathol. Lab. Med. 147(1), 46–51 (2023).
Rakha, E. A., Tse, G. M. & Quinn, C. M. An update on the pathological classification of breast cancer. Histopathology 82(1), 5–16 (2023).
Jakhar, A. K., Gupta, A. & Singh, M. SELF: a stacked-based ensemble learning framework for breast cancer classification. Evol. Intel. 1–16 (2023).
MIAS dataset, access on 15th January. https://www.kaggle.com/datasets/kmader/mias-mammography/data (2024).
Mahesh, T. R., Kumar, V., Vivek, V., Karthick Raghunath, V. & Sindhu Madhuri, G. K. M., Early predictive model for breast cancer classification using blended ensemble learning. Int. J. Syst. Assur. Eng. Manage. 1–10 (2022).
Egwom, O. J., Hassan, M., Tanimu, J. J., Hamada, M. & Ogar, O. M. An LDA–SVM machine learning model for breast cancer classification. BioMedInformatics 2(3), 345–358 (2022).
Zhang, G. Classification of single-cell routine pap smear images based on deep learning algorithms. Theoretical Nat. Sci. 50, 45–51 (2024).
Dinesh, P., Vickram, A. S. & Kalyanasundaram, P. Medical image prediction for diagnosis of breast cancer disease comparing the machine learning algorithms: SVM, KNN, logistic regression, random forest and decision tree to measure accuracy. In AIP Conference Proceedings (Vol. 2853, No. 1) (AIP Publishing, 2024).
Naz, A., Khan, H., Din, I. U. & Ali, A. An efficient optimization system for early breast cancer diagnosis based on internet of medical things and deep learning. Eng. Technol. Appl. Sci. Res. 14(4), 15957–15962 (2024).
Singh, L. & Alam, A. An efficient hybrid methodology for an early detection of breast cancer in digital mammograms. J. Ambient Intell. Humaniz. Comput. 15(1), 337–360 (2024).
Taghizadeh, E., Heydarheydari, S., Saberi, A., JafarpoorNesheli, S. & Rezaeijo, S. M. Breast cancer prediction with transcriptome profiling using feature selection and machine learning methods. BMC Bioinform. 23(1), 1–9 (2022).
CBIS-DDSM dataset, access on 10th Feb. https://www.kaggle.com/competitions/rsna-breast-cancer-detection/data (2024).
Acknowledgements
The authors extend their appreciation to Taif University, Saudi Arabia, for supporting this work through project number (TU-DSPP-2024-210).
Funding
This research was funded by Taif University, Saudi Arabia, Project number (TU-DSPP-2024-210).
Author information
Authors and Affiliations
Contributions
Umesh Kumar Lilhore contributed to conceptualization, methodology, supervision, and review. Yogesh Kumar Sharma handled data curation, software implementation, and analysis. Brajesh Kumar Shukla worked on drafting, visualization, and validation. Muniraju Naidu Vadlamudi contributed to software development and testing. Sarita Simaiya, as the corresponding author, oversaw project administration, funding acquisition, and final review. Roobaea Alroobaea provided resources, review, and validation. Majed Alsafyani supported validation, visualization, and review processes. Abdullah M. Baqasah played a crucial role in enhancing the model’s optimization, providing insights on algorithmic improvements, and contributing to the validation process. His expertise was instrumental in refining the research and improving the overall quality of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lilhore, U.K., Sharma, Y.K., Shukla, B.K. et al. Hybrid convolutional neural network and bi-LSTM model with EfficientNet-B0 for high-accuracy breast cancer detection and classification. Sci Rep 15, 12082 (2025). https://doi.org/10.1038/s41598-025-95311-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-95311-4
Keywords
This article is cited by
-
Breast lesion classification via colorized mammograms and transfer learning in a novel CAD framework
Scientific Reports (2025)
-
Optimizing YOLOv11 for automated classification of breast cancer in medical images
Scientific Reports (2025)
-
Multimodal Breast Cancer Classification Using Fractional-Order Three-Triangle Multi-delayed Neural Network Optimized with Hunger Games Search
Biomedical Materials & Devices (2025)
-
An Innovative and Effective Deep Learning Architecture for Risk and Survival Rate Prediction of Triple Negative Breast Cancer Using Modified Optimization Strategy
Annals of Data Science (2025)