Abstract
N6-methyladenosine (m6A) has been established as a critical regulator in various human cancers. However, the role of m6A modification in renal cell carcinoma (RCC) and its interaction with long non-coding RNA LHX1-DT (LHX1-DT) remains unclear. Differentially expressed lncRNAs and m6A levels were identified through microarray analysis. The interaction between IGF2BP2 and LHX1-DT was examined via RNA immunoprecipitation and luciferase reporter assays. LHX1-DT expression was found to be downregulated in RCC tissues, and reduced expression of LHX1-DT was associated with poor overall survival in RCC patients. Functional assays demonstrated that overexpression of LHX1-DT significantly inhibited RCC cell proliferation and invasion. The m6A reader protein IGF2BP2, mediated by METTL14, recognized the m6A modification site on LHX1-DT and promoted its stability. Additionally, LHX1-DT acted as a competing endogenous RNA (ceRNA) by sponging miR-590-5p, which in turn downregulated PDCD4, thereby inhibiting RCC cell proliferation and invasion. LHX1-DT serves as an independent prognostic biomarker for RCC, and the IGF2BP2/LHX1-DT/miR-590-5p/PDCD4 axis represents a novel therapeutic target for RCC progression.
Similar content being viewed by others
Introduction
Renal cell carcinoma (RCC) is a highly prevalent malignancy, with a global incidence of 434,419 and a mortality rate of 155,702 in 20221. It is projected that China will see 48,894 new cases and 16,179 due to RCC in 20242. In the early stages, most patients are asymptomatic, and by the time of diagnosis, the disease has typically progressed to an advanced stage. Surgical resection remains one of the most effective treatment options for patients in the early stage. However, for those in advanced stages, survival can only be extended through targeted therapy or immunotherapy3, unfortunately, the prognosis is generally poor. Therefore, further investigation into the pathogenesis of RCC is essential.
Long non-coding RNAs (lncRNAs) are a class of RNA molecules longer than 200 nucleotides that lack protein-coding potential4. In addition to the well-established competing endogenous RNA (ceRNA) mechanism5, lncRNAs can also regulate gene transcription by interacting with transcription factors6,7, RNA polymerases8, or directly with DNA9, thereby modulating transcriptional activity and influencing gene expression10,11. Moreover, Some lncRNAs modulate chromatin structure and gene accessibility through their interaction with chromatin modification-related proteins12. For instance, Wang et al. 13 demonstrated that lncHand2 recruits the chromatin remodeling complex Ino80 to activate the expression of Nkx1-2, subsequently affecting the c-Met signaling pathway. Additionally, lncRNAs can influence the expression of adjacent genes by acting on enhancers or silencers14. Finally, lncRNAs may function as molecular scaffolds, interacting with proteins and other RNAs to regulate their stability and activity15,16.
MicroRNAs (miRNAs) are small ncRNAs that bind to complementary sequences on target messenger RNAs (mRNAs), leading to their degradation or translational repression17. In cancer, miRNAs can function as oncogenes or tumor suppressors, regulating critical cellular processes such as cell proliferation, apoptosis, migration, and differentiation18,19,20,21,22. Furthermore, miRNAs can fine-tune multiple signaling pathways by simultaneously targeting several genes including those in the PI3K/AKT, Ras/MAPK, and p53 pathways23,24,25. This ability to regulate gene networks positions miRNAs as key modulators in cancer biology and as potential therapeutic targets.
N6-methyladenosine (m6A) is one of the most prevalent internal RNA modifications and is widely found in eukaryotic mRNAs, lncRNAs, and small RNAs. It regulates various biological processes by influencing RNA stability, splicing, transport, and mRNA translation. The m6A modification is mediated by a specific group of enzymes, often referred to as “writers” (methyltransferase), “erasers” (demethylase), and “readers”. The m6A methyltransferases include METTL3, METTL14, WTAP, VIRMA, and RBM15/RBM15B, while the demethylases include FTO and ALKBH5. The m6A readers consist of the YTHDF family, YTHDC family, HNRNP family, and IGF2BP family. Notably, the IGF2BP family, which comprises IGF2BP1, IGF2BP2, and IGF2BP3, binds to m6A and promotes mRNA stability and translation26,27,28.
Recently, the role of m6A methylation in the progression of RCC has gained increasing attention. Some investigators have revealed the effects of m6A-regulated lncRNAs on the malignant behavior of RCC. For example, Qiu et al. 29 constructed a prognostic risk model consisting of seven m6A-related lncRNAs, which could be helpful in managing RCC patients and guiding individualized immunotherapy. Similarly, Lin et al. 30 reported that two m6A-related lncRNAs, selected through weighted gene co-expression network analysis from The Cancer Genome Atlas (TCGA), were associated with the overall survival (OS) in RCC patients. However, most of these studies were based on data analysis by TCGA or Gene Expression Omnibus (GEO), which lacked the support of functional cell experiments. In addition, almost all studies on m6A modification in RCC has focused on protein-coding genes, with limited Studies addressing RCC-related m6A methylation and lncRNAs.
Results
The m6A levels are downregulated in RCC tissues and cell lines
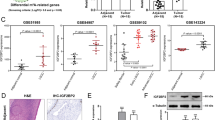
Recent advancements have revealed that m6A methylation was involved in the onset and progression of various human cancers31,32,33,34,35. To further investigate this, we assessed the m6A levels in RCC tissues and cell lines. The colorimetry assay was applied to detect the overall m6A levels in RCC tissues, adjacent normal renal tissues, normal renal tubular epithelial cells (HK-2), 293 T cells, and RCC cell lines (Caki-1, 786-O, and ACHN). The results demonstrated that the overall m6A levels in RCC tissues and cell lines were significantly lower than those in adjacent normal renal tissues, HK-2 cells and 293 T cells (Fig. 1A, B). Next, we examined the mRNA and protein expressions of METTL3, METTL14, WTAP, FTO, ALKBH5, and other key m6A regulatory factors in RCC tissues and adjacent normal renal tissues. The results showed that the mRNA and protein expression of METTL14 in RCC tissues were significantly lower than those in adjacent normal renal tissues (Fig. 1C, D). Subsequently, we evaluated the expression of METTL14 in HK-2 cells, 293 T cells, and RCC cell lines. The results indicated that METTL14 mRNA expression in RCC cell lines was significantly lower than in HK-2 cells and 293 T cells (Fig. 1E). Similar results were observed in both RCC tissues and cell lines by western blot (Fig. 1F, G). Concurrently, METTL14 overexpressed plasmid was transfected into Caki-1 cells (Fig. 1H). The results suggested that the m6A levels were significantly increased following METTL14 overexpression in Caki-1 cells (Fig. 1I).
A The m6A level was identified in RCC tissues and adjacent normal renal tissues by qRT-PCR. B The m6A level was identified in normal renal tubular epithelial cells, 293 T cells, and RCC cell lines by qRT-PCR. C, D The qRT-PCR (C) and western blot (D) analysis of the relative expression of 10 m6A regulatory factors in RCC tissues and adjacent normal renal tissues. E The qRT-PCR analysis of the relative expression of METTL14 in HK-2 cells, 293 T cells, and RCC cell lines. F The protein expression levels of METTL14 in RCC tissues and adjacent normal renal tissues. G The protein expression levels of METTL14 in HK-2 cells, 293 T cells, and RCC cell lines. H Verification of METTL14 overexpression by western blot assay. I The m6A level was detected after METTL14 overexpression by qRT-PCR.
These findings collectively demonstrated that the downregulation of m6A levels in RCC tissues and cell lines may play a crucial role in regulating the onset and progression of RCC.
LHX1-DT is a key target gene regulated by m6A modification and related to poor prognosis in RCC patients
Recent studies have revealed that m6A modification was closely linked to lncRNA expression. Several research groups have found that m6A modification can regulate lncRNA expression by affecting its stability36,37,38,39. In light of this, we performed m6A-mRNA and lncRNA epitranscriptomic microarray analysis on 3 paired RCC tissues and adjacent normal renal tissues. The results showed that the m6A levels were up-regulated in 2290 lncRNAs and down-regulated in 1660 lncRNA. The top 10 up-regulated and 15 down-regulated lncRNAs based on m6A levels are listed in Fig. 2A, with LHX1-DT showing the most obviously downregulation in m6A levels. The differentially expressed genes (DEGs) of the two groups were displayed in the volcano plots (Fig. 2B). Meanwhile, another microarray analysis was performed to compare lncRNA expression profiles between the aforementioned 3 pairs of samples. The results revealed 1688 up-regulated and 2931 down-regulated lncRNAs in the two groups, respectively. The top 15 up-regulated and 15 down-regulated lncRNAs were listed in Fig. 2C. Notably, the expression of LHX1-DT in RCC tissues was significantly lower than that in paired adjacent normal renal tissues. The DEGs between the two groups were displayed in the volcano plots (Fig. 2D). Further experiments revealed that LHX1-DT expression remained largely unchanged after treatment with 5-aza-dC, a DNA methyltransferase inhibitor (Supplementary Fig. 1A), implying that DNA methylation doesn’t participate in LHX1-DT modulation. Similarly, broad-spectrum histone deacetylase (HDAC) inhibitors (SAHA and NaB), did not affect LHX1-DT expression (Supplementary Fig. 1B), revealing that histone acetylation doesn’t involve in LHX1-DT regulation in RCC cells. These findings suggest that LHX1-DT may be a target gene regulated by m6A modification in RCC.
A The heatmap showing the top 10 upregulated and 15 down-regulated m6A levels of lncRNAs in RCC tissues compared with adjacent normal renal tissues analyzed by RNA sequencing. B The volcano plot showing the m6A expression profile of lncRNAs in RCC samples. C The heatmap showing the top 15 up-regulated and 15 down-regulated lncRNAs in RCC tissues compared with adjacent normal renal tissues analyzed by RNA sequencing. D The volcano plot showing the expression profle of lncRNAs in RCC samples. E, F The m6A methylation level of LHX1-DT in RCC tissues and adjacent normal renal tissues (E), HK-2 cells and RCC cell lines (F) were determined by MeRIP-qPCR assays. G, H Relative expression of LHX1-DT in 52 paired RCC tissues (G) and three cell lines (H) compared with adjacent normal renal tissues and HK2 cells respectively by qRT-PCR. I Kaplan–Meier analysis of the correlation between LHX1-DT expression and overall survival in a cohort of 52 RCC patients.
However, the biological and clinical function of LHX1-DT in RCC progression remain unclear. To further explore this, we assessed the m6A level of LHX1-DT in RCC tissues and cell lines. The m6A level of LHX1-DT was measured by meRIP using m6A antibody to pull down the m6A modified RNA from RCC tissues, adjacent normal renal tissues, HK-2 cells and RCC cell lines (Caki-1, 786-O, and ACHN). The results demonstrated that the m6A level of LHX1-DT in RCC tissues and cell lines were significantly lower than those in the paired adjacent normal renal tissues and HK-2 cells (Fig. 2E, F). As expected, the expression of LHX1-DT in RCC tissues (52 paired samples) and cell lines were significantly lower than those in the paired adjacent normal renal tissues and HK-2 cells (Fig. 2G, H). Low level of LHX1-DT was found to be positively correlated with advanced Fuhrman grade (Table 1). Kaplan-Meier (KM) analysis further revealed that high LHX1-DT expression was associated with better OS in RCC patients (Fig. 2I).
These findings suggest that LHX1-DT was an independent prognostic factor regulated by m6A modification, and its expression is correlated with a favorable prognosis in RCC patients.
LHX1-DT inhibits RCC cell proliferation and invasion in vitro and in vivo
To investigate the biological function of LHX1-DT in RCC, we performed cell proliferation and invasion assays. First, 786-O and Caki-1 cells were transfected with functional LHX1-DT-cDNA and LHX1-DT-shRNA. The qRT-PCR results confirmed that LHX1-DT expression was effectively regulated (Fig. 3A, B). The cell proliferation assay results that overexpression of LHX1-DT dramatically inhibited cell proliferation, while LHX1-DT knockdown significantly promoted cell proliferation in 786-O and Caki-1 cells (Fig. 3C, D). Similarly, transwell invasion assays in 786-O and Caki-1 cells yielded comparable results (Fig. 3E, F). These data indicate that LHX1-DT inhibits RCC cell proliferation and invasion in vitro.
A, B Verification of LHX1-DT overexpression (A) and knockdown (B) by qRT-PCR. C, D Cell proliferation assay of 786-O and Caki-1 cells transfected with functional LHX1-DT-cDNA (C) and LHX1-DT-shRNA (D). E, F Cell invasion assay of 786-O and Caki-1 cells transfected with functional LHX1-DT-cDNA (E) and LHX1-DT-shRNA (F). G Tumors collected from nude mice are exhibited. H, I Tumor volume (H) and weight (I) were dramatically decreased in LHX1-DT overexpression group. J Representative images of IHC staining of Ki-67 from the tumor sections. K LHX1-DT expression in tumors detected by qRT-PCR.
To further explore the effect of LHX1-DT in vivo, orthotopic xenograft mouse models were established to verify the anti-tumor role of LHX1-DT. Caki-1 cells were labeled with luciferase and transfected with functional LHX1-DT-cDNA. These cells were then injected into the renal capsule of nude mice. Tumor size was monitored using an In Vivo Imaging System. After 6 weeks, the results showed a significant reduction in tumor growth, volume, and weight in the LHX1-DT overexpression group compared to the control group (Fig. 3G–I). Additionally, Ki67 staining showed decreased in the LHX1-DT overexpression group compared to the control group (Fig. 3J). qRT-PCR analysis confirmed that LHX1-DT expression was elevated in the overexpression group (Fig. 3K). In summary, the above data indicated that LHX1-DT inhibits RCC cells proliferation and invasion both in vitro and in vivo.
LHX1-DT acts as a ceRNA to absorb miR-590-5p
To explore how LHX1-DT exerts its function, we predicted its subcellular localization using the online tool lncATLAS (http://lncatlas.crg.eu/). The results suggested that LHX1-DT was primarily localized in the cytoplasm (Fig. 4A). This was confirmed by a nuclear-cytoplasmic fractionation assay (Fig. 4B), which was further validated by a FISH assay (Fig. 4C). Recent studies have reported that cytoplasmic lncRNA can function either as tumor suppressor or an oncogene in human cancers, often acting as miRNA sponges40,41,42. Therefore, we hypothesized that LHX1-DT might inhibit RCC cell proliferation and invasion by sponging miRNAs. To test this, We conducted a RIP assay using a specific anti-AGO2 antibody, a key component of the miRNA-mediated mRNA translation repression and destabilization complex (Fig. 4D). The results showed that endogenous LHX1-DT was more enriched in AGO2 compared to the control (Fig. 4E). These data demonstrated that LHX1-DT may act as a ceRNA to inhibit RCC cell development.
A LHX1-DT was predicted to be located mainly in the cytoplasm using the bioinformatics tools in lncATLAS. B The qRT-PCR analysis of subcellular LHX1-DT expression in the nucleus and cytoplasm of RCC cells. C Subcellular localization of LHX1-DT in RCC cells detected by RNA-FISH. LHX1-DT was stained red (Cy3), and nuclei were stained blue (DAPI). D Schematic diagram of the Ago2-RIP process. E Fold enrichment of LHX1-DT in 786-O and Caki-1 cells. F Venn diagram showing the predicted target of LHX1-DT. G Relative expression of miR-590-5p in 52 paired RCC tissues compared with adjacent normal renal tissues by qRT-PCR. H RIP assay was performed using AGO2 antibody in 786-O and Caki-1 cells, then the enrichment of miR-590-5p was detected. I RIP assay was performed using AGO2 antibody in 786-O and Caki-1 cells transfected with miR-590-5p mimics or mimics NC, then the enrichment of LHX1-DT was detected. J The correlation between miR-590-5p and LHX1-DT was evaluated, and regression analysis was applied (r = −0.2984, P = 0.0317). K Expression level of miR-590-5p in 786-O and Caki-1 cells after transfection with overexpression plasmids of LHX1-DT. L Expression level of miR-590-5p in 786-O and Caki-1 cells treated with siRNAs of LHX1-DT. M The binding sites of LHX1-DT and miR-590-5p. N, O Luciferase reporter activity of wild-type (WT) or mutated (MUT) LHX1-DT co-transfected with miR-590-5p mimics in 786-O (N) and Caki-1 (O) cells. P LHX1-DT was pulled down with biotinylated miR-590-5p, and miR-590-5p was also pulled down with biotinylated LHX1-DT in 786-O and Caki-1 cells. Q, R The addition of functional FOXQ1-cDNA partially rescued the growth (Q) and invasion (R) of 786-O and Caki-1 cells transfected with miR-590-5p mimics. S, T The addition of LHX1-DT-shRNA partially rescued the growth (S) and invasion (T) of 786-O and Caki-1 cells transfected with an miR-590-5p inhibitor.
Then, we searched for the potential miRNAs that could bind to LHX1-DT. Using DIANA Tools, 51 miRNAs were predicted to interact with LHX1-DT. Among these, forty miRNAs were differentially expressed in RCC tissues compared to adjacent normal renal tissues as analyzed using the GEO database (GSE12105). miR-590-5p and miR-545-5p were identified as common miRNAs in both datasets (Fig. 4F). The qRT-PCR analysis revealed that miR-590-5p was significantly upregulated in 52 paired RCC tissues compared to adjacent normal renal tissues, while miR-545-5p showed no significant change (Fig. 4G and Supplementary Fig. 2A). A RIP assay using AGO2 antibody and IgG as a negative control confirmed that AGO2 can interact with miR-590-5p (Fig. 4H). Hence, miR-590-5p was selected for further investigation. To explore whether LHX1-DT is part of the AGO2-miR-590-5p complex, we conducted an AGO2 assay. The results confirmed that LHX1-DT was more enriched in the miR-590-5p mimics group (Fig. 4I). Next, we found that the level of miR-590-5p was negatively correlated with LHX1-DT expression in both RCC tissues and adjacent normal renal tissues (Fig. 4J and Supplementary Fig. 2B). Overexpression of LHX1-DT led to a marked downregulation of miR-590-5p, while silencing of LHX1-DT resulted in an upregulation of miR-590-5p (Fig. 4K, L). A luciferase reporter assay was designed with wild-type (LHX1-DT-WT) and mutated miR-590-5p binding sites (LHX1-DT-MUT) (Fig. 4M). The results indicated that the luciferase activity of the wild-type group was significantly decreased upon miR-590-5p mimic treatment, whereas the luciferase activity of the mutant group remained unchanged, suggesting that LHX1-DT targets miR-590-5p in a sequence-specific manner (Fig. 4N, O). Moreover, biotin-coupled miR-590-5p successfully pulled down the binding of LHX1-DT, while biotin-coupled LHX1-DT successfully pulled down miR-590-5p in 786-O and Caki-1 cells (Fig. 4P). These findings confirm that LHX1-DT acts as a ceRNA for miR-590-5p in RCC.
Subsequently, a rescue assay was performed to investigate the role of miR-590-5p in RCC cell proliferation and invasion. The expression of miR-590-5p was upregulated and downregulated by miR-590-5p mimics or inhibitors in 786-O and Caki-1 cells, respectively (Supplementary Fig. 2C, D). Overexpression of miR-590-5p promoted cell proliferation and invasion, whereas upregulation of LHX1-DT partially reversed the promotive effect of miR-590-5p on RCC cell proliferation and invasion (Fig. 4Q, R). Conversely, silencing miR-590-5p inhibited cell proliferation and invasion, and LHX1-DT silencing partially reversed the inhibitory effect of miR-590-5p on cell proliferation and invasion in both 786-O and Caki-1 cells (Fig. 4S, T). The above data collectively demonstrated that LHX1-DT acts as a miRNA sponge for miR-590-5p, thereby modulating RCC progression.
IGF2BP2 binds to LHX1-DT and regulates its expression in a m6A-dependent manner
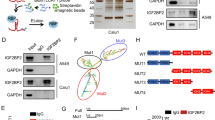
We found that lncRNAs may be targets regulated through an m6A modification mechanism, although the exact underlying mechanism remains complex36,38. Building upon this, we investigated the m6A modification of LHX1-DT. As previously mentioned, METTL14 expression is reduced in RCC. Therefore, we explored the effect of METTL14 on LHX1-DT expression. The qRT-PCR results indicated that overexpression of METTL14 significantly increased both m6A levels and LHX1-DT expression in 786-O and Caki-1 cells (Fig. 5A, B). A positive correlation between METTL14 and LHX1-DT levels was also observed in RCC tissues (Fig. 5C). These findings demonstrate that m6A modification plays a role in the downregulation of LHX1-DT.
A, B The m6A (A) and expression of LHX1-DT (B) were detected after METTL14 overexpression by qRT-PCR. C The correlation between METTL14 and LHX1-DT was evaluated, and regression analysis was applied (r = 0.09209, P = 0.0304). D, E Reduction of LHX1-DT RNA stability in METTL14 knockdown 786-O (D) and Caki-1 (E) cells as compared to control. Cells were treated with 5 μg/mL actinomycin-D and RNA was isolated at 0 h, 2 h and 4 h. F Relative expression of LHX1-DT was detected by RIP assay. G RIP analysis showing the enrichment of LHX1-DT on IGF2BP2 in the indicated cells. H, I Reduction of LHX1-DT RNA stability in IGF2BP2 knockdown 786-O (H) and Caki-1 (I) cells as compared to control. Cells were treated with 5 μg/mL actinomycin-D and RNA was isolated at 0 h, 2 h and 4 h. J Relative expression of IGF2BP2 in 523 RCC tissues compared with 100 adjacent normal renal tissues in the GEPIA2 dataset. K The putative wild-type m6A sites and designed mutant m6A sites in LHX1-DT. L The RIP analysis shows the enrichment of LHX1-DT on IgG and IGF2BP2 in the LHX1-DT-Wt or LHX1-DT-Mut RCC cells. M The qRT-PCR analysis of LHX1-DT expression in the LHX1-DT-Wt or LHX1-DT-Mut RCC cells with or without METTL14 or IGF2BP2 knockdown. N Schematic representation of mutated LHX1-DT of pmirGLO vector to investigate the m6A roles on LHX1-DT expression. O The luciferase activities of different mutated LHX1-DT reporters in the indicated groups.
Next, we examined the stability of LHX1-DT following METTL14 manipulation. The results showed that METTL14 knockdown dramatically decreased the half-life of LHX1-DT in 786-O and Caki-1 cells treated with actinomycin-D (Fig. 5D, E), indicating that METTL14 regulates LHX1-DT expression by modulating LHX1-DT its stability. Therefore, we investigated potential m6A readers that could recognize methylated LHX1-DT and regulate its stability. Recent studies have shown that IGF2BP family can recognize m6A modifications and increase RNA stability39,43. RIP assays revealed that LHX1-DT was dramatically enriched in IGF2BP2 instead of IGF2BP1 and IGF2BP3 (Fig. 5F). The deletion of METTL14 decreased the enrichment of LHX1-DT in IGF2BP2 (Fig. 5G), suggesting that METTL14-induced m6A modification regulates the recognition of methylated LHX1-DT by IGF2BP2. Meanwhile, inhibition of IGF2BP2 reduced the half-life of LHX1-DT in 786-O and Caki-1 cells (Fig. 5H, I), a result similar to METTL14 knockdown. These data indicated that IGF2BP2 functions as an m6A reader for LHX1-DT. Next, IGF2BP2 expression was found to be downregulated in RCC according to the GEPIA2 dataset (http://gepia2.cancer-pku.cn/#index) (Fig. 5J). In addition, to investigate whether the potential m6A modification sites predicted by SRAMP were associated with the interaction between LHX1-DT and IGF2BP2, we mutated all predicted m6A modification sites (Fig. 5K). The results of RIP assay indicated that the direct binding between LHX1-DT and IGF2BP2 was disrupted by mutations of the m6A modification sites (Fig. 5L). METTL14 or IGF2BP2-mediated modulation of LHX1-DT was impaired after mutation of the m6A modification sites (Fig. 5M). To identify which specific m6A modification site was responsible for stabilizing LHX1-DT, three LHX1-DT mutants were designed (Fig. 5N). Luciferase reporter assay showed that site 1 (‘UUGGACU’) plays a critical role in regulating LHX1-DT stability (Fig. 5O).
In all, the above data provide strong evidence that IGF2BP2 binds to LHX1-DT and enhances its stability in a m6A dependent manner.
LHX1-DT inhibits RCC progression by targeting miR-590-5p/PDCD4 axis
It is well established that miRNAs perform a variety of biological functions through targeting mRNAs44,45. To investigate the mechanism underlying the LHX1-DT/miR-590-5p axis in RCC progression, we employed three online analysis tools and identified 11 mRNAs that could potentially be targets of miR-590-5p, including PDCD4 (Fig. 6A), a known tumor suppressor in RCC46,47,48. The qRT-PCR results revealed that the expression levels of PDCD4 and BTG2 were lower in RCC tissues compared to adjacent normal renal tissues, with PDCD4 showing a more pronounced decrease (Fig. 6B). Using 52 paired RCC samples, we confirmed that PDCD4 expression was significantly decreased in RCC tissues compared to adjacent normal renal tissues (Fig. 6C). The result of KM analysis showed that low PDCD4 level indicated poorer OS according to the GEPIA2 dataset (http://gepia2.cancer-pku.cn/#index) (Fig. 6D). Moreover, miR-590-5p levels were found to be negatively correlated with PDCD4 expression in RCC tissues (Fig. 6E). We also observed that both the mRNA and protein levels of PDCD4 were downregulated or upregulated by the introduction of miR-590-5p mimics or inhibitors in 786-O and Caki-1 cells, respectively (Fig. 6F, G). To further validate the predicted binding sites of miR-590-5p in the 3’-untranslated region (3’-UTR) of PDCD4, we conducted a luciferase reporter assay (Fig. 6H). The results revealed that the luciferase activitity of PDCD4-WT construct was significantly reduced upon treatment with miR-590-5p mimics, while the activity of the PDCD4-MUT construct remained unchanged (Fig. 6I, J). Overexpression of PDCD4 partially reversed the promotive effect of miR-590-5p mimics on cell proliferation and invasion (Fig. 6K, L), while knockdown of PDCD4 partially reversed the inhibitory effect of miR-590-5p inhibitors on cell proliferation and invasion in 786-O and Caki-1 cells (Fig. 6M, N). These findings confirm that PDCD4 is a direct target of miR-590-5p.
A Venn diagram showing the predicted target genes of miR-590-5p. B The qRT-PCR analysis of the relative expression of 11 possible targets of miR-590-5p in RCC tissues and adjacent normal renal tissues. C Relative expression of PDCD4 in 52 paired RCC tissues compared with adjacent normal renal tissues by qRT-PCR. D Kaplan–Meier analysis of the correlation between PDCD4 expression and OS in RCC patients according to the GEPIA2 dataset. E The correlation between PDCD4 and miR-590-5p was evaluated, and regression analysis was applied (r = −0.2868, P = 0.0393). F The expression of PDCD4 was detected after silencing or overexpression of miR-590-5p by qRT-PCR. G Western blot assay showing protein levels of PDCD4 after silencing or overexpression of miR-590-5p. H The binding sites of PDCD4 and miR-590-5p. I, J Luciferase reporter activity of wild-type (WT) or mutated (MUT) PDCD4 co-transfected with miR-590-5p mimics in 786-O (I) and Caki-1 (J) cells. K, L The addition of functional PDCD4-cDNA partially rescued the growth (K) and invasion (L) of 786-O and Caki-1 cells transfected with miR-590-5p mimics. M, N The addition of PDCD4-shRNA partially rescued the growth (M) and invasion (N) of 786-O and Caki-1 cells transfected with an miR-590-5p inhibitor. O The correlation between LHX1-DT and PDCD4 was evaluated, and regression analysis was applied (r = 0.8933, P < 0.0001). P Western blot assay showing protein levels of PDCD4 after overexpression or silencing of LHX1-DT. Q The expression of PDCD4 was detected after overexpression or silencing of LHX1-DT by qRT-PCR. R Protein level of PDCD4 was determined by western blot assay after co-transfection with functional LHX1-DT-cDNA and miR-590-5p mimics. S Protein level of PDCD4 was determined by western blot assay after co-transfection with LHX1-DT-shRNA and an miR-590-5p inhibitor. T The expression of PDCD4 was detected after co-transfection with functional LHX1-DT-cDNA and miR-590-5p mimics. U The expression of PDCD4 was detected after co-transfection with LHX1-DT-shRNA and an miR-590-5p inhibitor.
We further found a positive correlation between PDCD4 and LHX1-DT expression in RCC tissues (Fig. 6O). The results indicated that both the protein and mRNA levels of PDCD4 were upregulated or downregulated by transfection of functional LHX1-DT-cDNA or LHX1-DT-shRNA in 786-O and Caki-1 cells, respectively (Fig. 6P, Q). Overexpression of LHX1-DT partially reversed the inhibitory effect of miR-590-5p mimics on PDCD4 expression (Fig. 6R, S), while knockdown of LHX1-DT partially reversed the promotive effect of miR-590-5p inhibitors on PDCD4 expression in both protein and mRNA levels (Fig. 6T, U). In conclusion, these data suggested that the inhibitory effect of LHX1-DT on RCC progression was primarily mediated through the miR-590-5p/PDCD4 axis.
PDCD4 is responsible for LHX1-DT-mediated inhibition of cell proliferation and invasion
To investigate whether LHX1-DT inhibits RCC progression in a PDCD4-dependent manner, we transfected functional LHX1-DT-cDNA and PDCD4-knockdown vector in 786-O and Caki-1 cells. The results revealed that downregulation of PDCD4 partially reversed the inhibitory effect of LHX1-DT overexpression on RCC cell proliferation (Fig. 7A). Similar results were obtained using a transwell invasion assay in both 786-O and Caki-1 cells (Fig. 7B). Conversely, upregulation of PDCD4 partially reversed the promotive effect of LHX1-DT knockdown on RCC cell proliferation and invasion (Fig. 7C, D). These findings demonstrated that LHX1-DT functions as a tumor suppressor lncRNA that inhibits RCC cell proliferation and invasion through the miR-590-5p/PDCD4 signaling pathway (Fig. 8A).
A, B The addition of PDCD4-shRNA partially rescued the growth (A) and invasion (B) of 786-O and Caki-1 cells transfected with functional LHX1-DT-cDNA. C, D The addition of functional PDCD4-cDNA partially rescued the growth (C) and invasion (D) of 786-O and Caki-1 cells transfected with LHX1-DT-shRNA.
The expression of MELLT14, a key regulatory factor of m6A, is downregulated, and the level of m6A is also downregulated. The m6A reader protein IGF2BP2, mediated by METTL14, recognized the m6A modification site on LHX1-DT and promoted its stability and expression. Additionally, LHX1-DT acted as a ceRNA by sponging miR-590-5p, which in turn downregulated PDCD4, thereby inhibiting RCC cell proliferation and metastasis.
Discussion
Recently, m6A modification of RNAs has been recognized as a novel layer of epigenetic regulation. This physiological process plays crucial roles in modulating cell differentiation, proliferation, and self-renewal by controlling RNA stability, translation, and splicing49,50,51. While previous studies have primarily focused on the involvement of m6A in the modification of mRNAs, recent studies indicate that m6A also plays a role in the modification of ncRNAs52,53,54,55. However, the potential role of m6A modification and lncRNAs in RCC remains unclear.
To explore the role of m6A modification in RCC, we examined the m6A levels in RCC tissues and cell lines. The results showed that the overall m6A levels were significantly lower in RCC tissues and cell lines compared to normal renal tissues and cell lines. Subsequently, we excavated the role of m6A modification and lncRNAs in RCC. Through microarray analysis, we identified thousands of abnormally expressed lncRNAs. Considering that the m6A level of LHX1-DT was the most obviously downregulated one, suggesting that LHX1-DT may regulate the biological behaviors of RCC through m6A modification.
LHX1-DT is transcribed from the bidirectional promoter of the LIM Homeobox 1 gene. Studies have reported that during cardiomyocyte differentiation, LHX1-DT interacts with PHF6 to facilitate the replacement of histone H2A with H2A.Z, thereby promoting LHX1 transcription56. Moreover, LHX1-DT has been found to play a significant role in the progression of thyroid cancer, breast cancer (BC), and colorectal cancer57,58,59. In clinical analysis, we found that LHX1-DT expression was downregulated in RCC. Low level of LHX1-DT was related to advanced Fuhrman grade and a poor OS, which could be used as an independent prognostic factor for RCC patients. In the functional assays, we found that overexpression of LHX1-DT inhibited RCC cell proliferation and invasion both in vitro and in vivo.
Another valuable discovery was that m6A modified LHX1-DT expression was mediated by the m6A reader IGF2BP2. Firstly, using METTL14 overexpression transfection, we validated that m6A modification was involved in the downregulation of LHX1-DT. Secondly, RNA stability assay and qRT-PCR demonstrated that suppression of IGF2BP2 reduced the stability of LHX1-DT and decreased its expression. Thirdly, RIP assay showed that IGF2BP2 significantly interacted with LHX1-DT, highlighting a molecular interaction betweeen IGF2BP2 and LHX1-DT. Finally, luciferase reporter assay showed that an m6A modification site participatded in regulating LHX1-DT stability.
Next, we sought to explore the potential downstream target of LHX1-DT. Based on previous functional analysis and bioinformatics predictions of lncRNAs, we speculated that LHX1-DT might act as a sponge for miRNAs to inhibit the proliferation and invasion of RCC cells. After screening, we discovered that miR-590-5p might be regulated by LHX1-DT. The qRT-PCR results showed that miR-590-5p was significantly increased in RCC tissues compared to adjacent normal renal tissues, and it exhibited a negative correlation with LHX1-DT. Moreover, luciferase reporter assay and RNA pull down assay revealed direct binding between LHX1-DT and miR-590-5p. Finally, we demonstrated that LHX1-DT actes as a miRNA sponge for miR-590-5p, regulating RCC progression through rescue assay.
Using bioinformatics prediction and qRT-PCR detection, we identified that PDCD4 as a potential target mRNA of miR-590-5p. PDCD4 is a tumor suppressor gene involved in the regulation of tumor cell proliferation, invasion, and apoptosis. For instance, in BC, miR-421 targets PDCD4 to regulate cell proliferation60. Additionally, SKP2 gene promotes proliferation and inhibits apoptosis of BC cells by facilitating the ubiquitination and degradation of PDCD461. Furthermore, a study by Zennami et al. 62 revealed that in castration-resistant prostate cancer (PCa), AR promotes the expression of miR-21, which then targets and suppresses PDCD4, ultimately inhibiting PCa cell apoptosis. Western blot assay confirmed that PDCD4 can be regulated by miR-590-5p in RCC. Luciferase reporter assay showed the direct binding site of miR-590-5p and PDCD4 3’-UTR, which further confirmed that PDCD4 was a target of miR-590-5p. Moreover, we also verified that LHX1-DT exerts its tumor suppressor role by inhibiting RCC cell proliferation and invasion through miR-590-5p/PDCD4 axis.
LncRNAs, as key transcriptional regulators, play pivotal roles in the initiation, development, and drug resistance of tumors, and have become promising targets for clinical diagnosis and therapeutic intervention. There is growing evidence that specific lncRNAs exhibit significant expression alterations at the early stages of tumorigenesis, enabling them to be used for early cancer screening, pathologic classification, and prognostic assessment63,64,65. In the therapeutic domain, lncRNA-targeted approaches, such as antisense oligonucleotides (ASOs), small interfering RNAs (siRNAs), and CRISPR/Cas-based systems, have shown powerful anti-tumor efficacy in various models. For example, ASO-mediated targeting of mitochondria-associated lncRNAs, such as ASncmtRNA, has led to complete tumor regression in mouse RCC models and has entered Phase I clinical trials in solid tumors, including RCC, with preliminary data showing favorable tolerance66. Moreover, the combination of nanodelivery systems further improved the tissue specificity and delivery efficiency of lncRNA-directed therapy. The representative platform uses self-assembled nanoparticles composed of PEI-PDLLA and DSPE-mPEG amphiphilic copolymers that can co-deliver siRNA targeting lncRNA CCAT1 and curcumin. By enhancing apoptosis and inhibiting cell proliferation, a synergistic effect against colorectal cancer is generated, while also providing excellent biocompatibility and imaging capabilities67. Although large-scale clinical studies are still needed, current findings lay the foundation for the application of lncRNA-based precision therapy in RCC and highlight its broad potential in the era of personalized medicine.
In conclusion, our study elucidated the critical role of m6A modification and lncRNA in the regulation of RCC proliferation and invasion. We demonstrated that the m6A reader IGF2BP2 could increase the stability of LHX1-DT via recognition of a specific m6A modification site, thereby suppressing RCC progression through the miR-590-5p/PDCD4 axis. These findings highlight the functional significance of m6A-modified LHX1-DT and provide novel insights into the epigenetic regulation of RCC by m6A methylation.
Methods
Clinical samples
All RCC samples and matched adjacent normal renal tissues were acquired from patients who underwent radical nephrectomy at the Department of Urology, Shengjing Hospital of China Medical University from April 2020 to January 2025. All specimens were immediately deposited at −80 °C in the Department of Biobank, Shengjing Hospital of China Medical University after surgical excision. None of the patients was received chemotherapy or radiation before surgery. The pathological diagnoses were obtained from two pathologists. The use of all specimens were authorized by the Ethics Committee of Medical Research and New Technology of Shengjing Hospital of China Medical University (Ref#2020PS280K) and Declaration of Helsinki, and ethic informed consent was signed by all patients. All data and information collected during the study were kept strictly confidential to minimize potential risks to patients. All patients agreed that the data from their samples could be used for experimental studies and paper presentations.
Cell lines and cell culture
Human RCC cells (Caki-1, 786-O, and ACHN), human renal tubular epithelial cells (HK-2), and 293 T cells were purchased from American Type Culture Collection (ATCC, Manassas, USA). Cells were all maintained in DMEM medium (Invitrogen, Grand Island, USA), supplemented with 10% fetal bovine serum (FBS; Clark Bioscience, Richmond, USA) and 1% penicillin/streptomycin (Invitrogen, Grand Island, USA). Cells were cultivated in a humidified atmosphere at 37 °C with 5% CO2. Cells were routinely identified for bacteria and mycoplasma infection under ATCC’s instructions.
Antibodies
The antibodies used in our manuscript are listed below: METTL3 (1:500, proteintech, cat#15073-1-AP), METTL14 (1:500, proteintech, cat#26158-1-AP), WTAP (1:500, proteintech, cat#60188-1-PBS), FTO (1:500, proteintech, cat#27226-1-AP), ALKBH5 (1:500, proteintech, cat#82083-1-RR), YTHDF1 (1:500, proteintech, cat#17479-1-AP), YTHDF2(1:500, proteintech, cat#24744-1-AP), YTGDF3 (1:500, proteintech, cat#25537-1-AP), YTHDC1 (1:500, proteintech, cat#14392-1-AP), YTHDC2 (1:500, proteintech, cat#83970-1-RR), AGO2 (proteintech, cat#67934-1-Ig), GAPDH (1:500, CST, cat#2118S) and PDCD4 (1:500, abcam, cat#80590).
Plasmid and cell transfection
Human lncRNA coding sequence (NR_135671.1), METTL14, and PDCD4 were subcloned into the lentiviral vector pHBLV-CMV-MCS-EF1-puro designed and constructed by Hanhan. Hairpin RNA (shRNA) of METTL14, LHX1-DT, IGF2BP2, and PDCD4 were synthesized and cloned into the pHBLV-U6-MCS-PGK-PURO vector (Hanheng). sh-METTL14: Sense: 5’-GATCCGGATGAACTAGAAATGCAACACTCGA GTGTTGCATTTCTAGTTCATCCTTTTTG-3’; Antisense: 5’-AATTCAAAAAGGATGAA CTAGAAATGCAACACTCGAGTGTTGCATTTCTAGTTCATCCG-3’. sh-LHX1-DT: Sense: 5’-GATCCGAGACCGCGTTTAGAGGATCACTCGAGTGATCCTCTAAACGC GGTCTCTTTTTG-3’; Antisense: 5’-AATTCAAAAAGAGACCGCGTTTAGAGGATCA CTCGAGTGATCCTCTAAACGCGGTCTCG-3’. sh-IGF2BP2: Sense: 5’-GATCCGCA TGATTCTTGAAATCATGCCTCGAGGCATGATTTCAAGAATCATGCTTTTTG-3’. Antisense: 5’-AATTCAAAAAGCATGATTCTTGAAATCATGCCTCGAGGCATGATTTC AAGAATCATGCG-3’. sh-PDCD4: Sense: 5’-GATCCGGAGGTGGATGTGAAAGATC CCTCGAGGGATCTTTCACATCCACCTCCTTTTTG-3’; Antisense: 5’-AATTCAAAAA GGAGGTGGATGTGAAAGATCCCTCGAGGGATCTTTCACATCCACCTCCG-3’. miR-590-5p inhibitor: 5’-CUGCACUUUUAUGAAUAAGCUC-3’; miR-590-5p mimics: 5’-GAGCUUAUUCAUAAAAGUGCAG-3’.
Empty lentiviral vector was used as a negative control. 48 h after viral transduction, cells were selected with 2 μg/mL puromycin. Human RCC cells were transduced with above lentivirus according to the manufacturer’s instructions. Stably transfected cell lines were obtained and confirmed by quantitative real-time polymerase chain reaction (qRT-PCR) and western blotting for subsequent experiments.
RNA extraction and qRT-PCR
Total RNAs were extracted from tissues and cell lines by Trizol (Invitrogen, Grand Island, USA) according to the manufacturer’s instructions. Then, RNAs were reversed to cDNA by SuperscriptTM III Reverse Transcriptase (Invitrogen, Grand Island, USA). QRT-PCR was conducted by a Bio-Rad CFX96 system (Bio-Rad, Hercules, USA) with SYBR green to measure the mRNA expression. GAPDH was used as a internal control. MiRNAs were extracted by PureLink™ miRNA Isolation Kit (Invitrogen, Grand Island, USA) according to the manufacturer’s instructions. U6 and/or RPL32 were used as internal controls. Relative quantification of lncRNA, miRNA, and mRNA expression was analyzed by the 2−△△CT method. The sequences of primers were listed as follows: LHX1-DT: Forward: 5’-CTCCTGCTCCGTTGTATCGT-3’; Reverse: 5’-TGT ATCTCTCCGGGTCTCTGT-3’. IGF2BP2: Forward: 5’-AGTGGAATTGCATGGGAAAA TCA-3’; Reverse: 5’-CAACGGCGGTTTCTGTGTC-3’. PDCD4: Forward: 5’-ACAGGT GTATGATGTGGAGGA-3’; Reverse: 5’-TTCTCAAATGCCCTTTCATCCAA-3’. METTL3: Forward: 5’-ACACTGCTTGGTTGGTGTCA-3’; Reverse: 5’-GCGAGTGCC AGGAGATAGTC-3’. METTL14: Forward: 5’-AGATTGCAGCACCTCGATCA-3’; Reverse: 5’-GAAGTCCCCGTCTGTGCTAC-3’. WTAP: Forward: 5’-GTAATGGTAGC TCCTCCCGC-3’; Reverse: 5’-ACCCCGCACTGAGTTGATTT-3’. FTO: Forward: 5’-A CTTGGCTCCCTTATCTGACC-3’; Reverse: 5’-TGTGCAGTGTGAGAAAGGCTT-3’. ALKBH5: Forward: 5’-GGACAACTATAAGGCGGGCA-3’; Reverse: 5’-GGACACAGG GTAAGGTTCGG-3’. YTHDF1: Forward: 5’-TGGATGATGCTGCTGGTTGT-3’; Reverse: 5’-CAACGGTCCCTCATTCCACA-3’. YTHDF2: Forward: 5’-AGTAGGGCAA CAGACACAGC-3’; Reverse: 5’-TTATGACCGAACCCACTGCC-3’. YTHDF3: Forward: 5’-AACAAGGACCTCAGCCACAG-3’; Reverse: 5’-TGT TCTGGTTGAAGCCTGCT-3’. YTHDC1: Forward: 5’-CCATGCTCCACCTCCTCAAG-3’; Reverse: 5’-GCCACTGAC AACAGCTTGTG-3’. YTHDC2: Forward: 5’-ACGAGGTGGTGGTGACATTC-3’; Reverse: 5’-TCTCCTTTGGCCCTGTCAAC-3’.
Stem-loop transcription was used to detect miR-590-5p. Primer sequences for miR-590-5p was: stem-loop reverse transcription primers: 5’-GTCGTATCCAGTGCA GGGTCCGAGGTATCGCACTGGATACGACCTGCAC-3’; Forward: 5’-CGCGGAGC TTATTCATAAAA-3’; Reverse: 5’-ATCCAGTGCAGGGTCCGAGG-3’.
Western blot assay
RCC cells were collected and extracted by RIPA lysis buffer. Then, the protein was quantified by BCA assay. After transferred onto a PVDF membrane, total protein was incubated with specific antibodies at 4 °C overnight. The second day, the membranes were incubated with anti-rabbit or anti-mouse IgG secondary antibodies for 1 h. The bands were detected by a chemiluminescent imaging system (Thermo Fisher Scientific, Shanghai, China).
RNA microarray analysis
Human m6A-mRNA&lncRNA epitranscriptomic microarray analysis was applied by Aksomics. Total RNA from each sample was quantified using the NanoDrop ND-1000. The sample preparation and microarray hybridization were performed based on the Arraystar’s standard protocols. Results were provided in Sample QC report. After washing, slides were scanned with Agilent Scanner G2505C. Differentially m6A-methylated mRNAs and lncRNAs based on “m6A methylation level” and “m6A quantity” passing fold change (FC) and statistical significance cutoffs were identified and compiled. The default thresholds were |FC | ≥ 1.5 and p-values ≤ 0.05.
Cell proliferation assay
The transfected RCC cells were incubated in a 24-well plate (1800 cells/well). MTT reagents were dropped into the plates at day 1, 2, 3, and 4. The liquid was discarded after 2 h incubation. The absorbance was detected at 570 nm.
RNA immunoprecipitation (RIP)
RIP was carried out with a Magna RIP Kit (Millipore, Billerica, MA) according to the manufacturer’s protocol. RCC cells were lysed with RIP lysis buffer and incubated with conjugated beads for 4 h at 4 °C. The protein was washed off with proteinase K. Finally, RNA was purifed and subjected to qRT-PCR for next analysis.
Methylated RIP (meRIP)
M6A antibody was used to pull down m6A modified RNA. The rest experiment was carried out according to the above RIP procedure.
Luciferase reporter assay
Bioinformatics methods were used to analyze and predict the possible binding sites of transcription factors in promoter regions. Primers were designed to clone the desired target promoter fragment from genomic DNA by qRT-PCR, and this fragment was inserted into pGL3-basic vector (Promaga, Madison, WI). Mutation plasmid was also constructed by QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies, Palo Alto, CA). RCC cells were incubated in 24-well plates and the cDNA were transfected with lipofectamine 3000 transfection reagent (Invitrogen, Carsbad, CA). Luciferase activity was detected 36 h after transfection by Dual-Luciferase Assay (Promaga, Madison, WI).
Cell invasion assay
RCC cells were seeded in a 6-well plate and incubated for 72 h. The upper chamber (8.0 µm pore size, BD Corning, Corning, NY) were coated with matrigel (1:20, BD Biosciences, Franklin Lakes, NJ) 2 h before plating the cells. Then the cells were collected and transferred into the upper chamber. 750 µL of 10% FBS medium was dropped into the lower chamber and incubated for 12 h. The invaded cells were permeabilized by methanol and stained with crystal violet. Then the invaded cells were counted and photographed.
Animal studies
The animal studies were carried out according to the institutional ethics guidelines, which were approved by the Animal Care Committee of China Medical University (Ref#2020PS328K). Eight female BALB/c nude mice (4 weeks old) were purchased from Shanghai Laboratory Animal Center Co. Ltd. (Shanghai, China). The mice were first weighed and numbered in order of weight. Then, the mice were randomly divided into treatment group and control group for experimental observation. A total of 1 × 106 Caki-1 cells were injected into subrenal capsule. After 6 weeks, the mice were harvested and measured for further analysis. Euthanasia methods was inhaled drugs. The mice were placed in a closed container where they could be clearly seen and euthanized with an overdose of isoflurane anesthesia, ensuring that unnecessary suffering was not caused. Tumor volume was measured with the formula: volume = (lengt × width2) × 0.52.
Immunohistochemical (IHC) staining
Tumor sections were cut into a 5 μM thin slides, deparaffinized and rehydrated for antigen retrieval. Then, the tissue slides were added with primary antibodies. Secondary antibodies were used after washing with wash buffer. Next, diaminobenzidine (Zymed, South San Francisco, CA) was added into tumor slides, which were counter-stained with hematoxylin for further calculated.
Data availability
The data of human m6A-mRNA and lncRNA epitranscriptomic microarray analysis in our manuscript were deposited in the GEO datasets with the accession numbers GSE289147 and GSE289145, respectively.
Code availability
Not applicable.
References
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74, 229–263 (2024).
Wu, Y. et al. Comparative analysis of cancer statistics in China and the United States in 2024. Chin. Med. J. 137, 3093–3100 (2024).
Pontes, O., Oliveira-Pinto, S., Baltazar, F. & Costa, M. Renal cell carcinoma therapy: Current and new drug candidates. Drug Discov. Today 27, 304–314 (2022).
Cabili, M. N. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927 (2011).
Nejadi Orang, F. & Abdoli Shadbad, M. CircRNA and lncRNA-associated competing endogenous RNA networks in medulloblastoma: a scoping review. Cancer Cell Int. 24, 248 (2024).
He, H. et al. LncRNA ZNF503-AS1 acts as a tumor suppressor in bladder cancer by up-regulating Ca2+ concentration via transcription factor GATA6. Cell. Oncol.44, 219–233 (2021).
Yu, Z. et al. Exosomal LOC85009 inhibits docetaxel resistance in lung adenocarcinoma through regulating ATG5-induced autophagy. Drug Resist. Updat. 67, 100915 (2023).
Niu, Y. et al. HIF2-Induced Long Noncoding RNA RAB11B-AS1 Promotes Hypoxia-Mediated Angiogenesis and Breast Cancer Metastasis. Cancer Res. 80, 964–975 (2020).
Mas, A. M. & Huarte, M. lncRNA-DNA hybrids regulate distant genes. EMBO Rep. 21, e50107 (2020).
Long, Y., Wang, X., Youmans, D. T. & Cech, T. R. How do lncRNAs regulate transcription?. Sci. Adv. 3, eaao2110 (2017).
Tang, Y. et al. Dissection of FOXO1-Induced LYPLAL1-DT Impeding Triple-Negative Breast Cancer Progression via Mediating hnRNPK/β-Catenin Complex. Res.6, 0289 (2023).
Hu, X. et al. METTL3-stabilized super enhancers-lncRNA SUCLG2-AS1 mediates the formation of a long-range chromatin loop between enhancers and promoters of SOX2 in metastasis and radiosensitivity of nasopharyngeal carcinoma. Clin. Transl. Med 13, e1361 (2023).
Wang, Y. et al. Long noncoding RNA lncHand2 promotes liver repopulation via c-Met signaling. J. Hepatol. 69, 861–872 (2018).
Yan, P. et al. LncRNA Platr22 promotes super-enhancer activity and stem cell pluripotency. J. Mol. Cell. Biol. 13, 295–313 (2021).
Luo, X.-J. et al. M6A-modified lncRNA FAM83H-AS1 promotes colorectal cancer progression through PTBP1. Cancer Lett. 598, 217085 (2024).
Shi, Z. et al. Loss of LncRNA DUXAP8 synergistically enhanced sorafenib induced ferroptosis in hepatocellular carcinoma via SLC7A11 de-palmitoylation. Clin. Transl. Med. 13, e1300 (2023).
Tang, G. siRNA and miRNA: an insight into RISCs. Trends Biochem. Sci. 30, 106–114 (2005).
Lee, Y. S. & Dutta, A. MicroRNAs in cancer. Annu. Rev. Pathol. 4, 199–227 (2009).
Bica-Pop, C. et al. Overview upon miR-21 in lung cancer: focus on NSCLC. Cell. Mol. Life Sci.: CMLS 75, 3539–3551 (2018).
Naghizadeh, S. et al. The role of miR-34 in cancer drug resistance. J. Cell. Physiol. 235, 6424–6440 (2020).
Xiong, S., Hu, M., Li, C., Zhou, X. & Chen, H. Role of miR‑34 in gastric cancer: From bench to bedside (Review). Oncol. Rep. 42, 1635–1646 (2019).
Zhang, L., Liao, Y. & Tang, L. MicroRNA-34 family: a potential tumor suppressor and therapeutic candidate in cancer. J. Exp. Clin. Cancer Res.: CR 38, 53 (2019).
Akbarzadeh, M., Mihanfar, A., Akbarzadeh, S., Yousefi, B. & Majidinia, M. Crosstalk between miRNA and PI3K/AKT/mTOR signaling pathway in cancer. Life Sci. 285, 119984 (2021).
Soleimani, A. et al. The potential role of regulatory microRNAs of RAS/MAPK signaling pathway in the pathogenesis of colorectal cancer. J. Cell. Biochem. 120, 19245–19253 (2019).
La, T. et al. A p53-Responsive miRNA Network Promotes Cancer Cell Quiescence. Cancer Res. 78, 6666–6679 (2018).
Fang, Z. et al. Role of m6A writers, erasers and readers in cancer. Exp. Hematol. Oncol. 11, 45 (2022).
Wang, Y. et al. Epigenetic modification of m6A regulator proteins in cancer. Mol. Cancer 22, 102 (2023).
Li, R. et al. The relationship between the network of non-coding RNAs-molecular targets and N6-methyladenosine modification in tumors of urinary system. Cell Death Dis. 15, 275 (2024).
Qiu, Y. et al. Integrated analysis on the N6-methyladenosine-related long noncoding RNAs prognostic signature, immune checkpoints, and immune cell infiltration in clear cell renal cell carcinoma. Immun. Inflamm. Dis. 9, 1596–1612 (2021).
Lin, G., Wang, H., Wu, Y., Wang, K. & Li, G. Hub Long Noncoding RNAs with m6A Modification for Signatures and Prognostic Values in Kidney Renal Clear Cell Carcinoma. Front Mol. Biosci. 8, 682471 (2021).
An, Y. & Duan, H. The role of m6A RNA methylation in cancer metabolism. Mol. Cancer 21, 14 (2022).
Han, J. et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer 18, 110 (2019).
Chang, G. et al. YTHDF3 Induces the Translation of m6A-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell 38, https://doi.org/10.1016/j.ccell.2020.10.004 (2020).
Wang, X., Liu, Y., Zhou, M., Yu, L. & Si, Z. m6A modified BACE1-AS contributes to liver metastasis and stemness-like properties in colorectal cancer through TUFT1 dependent activation of Wnt signaling. J. Exp. Clin. Cancer Res.: CR 42, 306 (2023).
Lin, Y. et al. Pan-cancer analysis reveals m6A variation and cell-specific regulatory network in different cancer types. Genomics Proteomics Bioinformatics 22, qzae052 (2024).
Wu, Y. et al. m6A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol. Cancer 18, 87 (2019).
Zheng, Z.-Q. et al. Long noncoding RNA FAM225A promotes nasopharyngeal carcinoma tumorigenesis and metastasis by acting as ceRNA to Sponge miR-590-3p/miR-1275 and Upregulate ITGB3. Cancer Res. 79, 4612–4626 (2019).
Ni, W. et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m6A reader YTHDF3. Mol. Cancer 18, 143 (2019).
Hu, X. et al. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 27, 1782–1794 (2020).
Mahmoud, M. M., Sanad, E. F., Elshimy, R. A. A. & Hamdy, N. M. Competitive endogenous role of the LINC00511/miR-185-3p axis and miR-301a-3p from liquid biopsy as molecular markers for breast cancer diagnosis. Front. Oncol. 11, 749753 (2021).
Brex, D. et al. LINC00483 has a potential tumor-suppressor role in colorectal cancer through multiple molecular axes. Front. Oncol. 10, 614455 (2020).
Wang, K., Jin, W., Song, Y. & Fei, X. LncRNA RP11-436H11.5, functioning as a competitive endogenous RNA, upregulates BCL-W expression by sponging miR-335-5p and promotes proliferation and invasion in renal cell carcinoma. Mol. Cancer 16, 166 (2017).
Lang, C. et al. m6 A modification of lncRNA PCAT6 promotes bone metastasis in prostate cancer through IGF2BP2-mediated IGF1R mRNA stabilization. Clin. Transl. Med. 11, e426 (2021).
Wang, Y., Chen, J., Wang, X. & Wang, K. miR-140-3p inhibits bladder cancer cell proliferation and invasion by targeting FOXQ1. Aging 12, 20366–20379 (2020).
Mohamed, A. A. et al. MiR-155 and MiR-665 role as potential non-invasive biomarkers for hepatocellular carcinoma in Egyptian patients with chronic hepatitis C virus infection. J. Transl. Int. Med. 8, 32–40 (2020).
Li, X. et al. Down-regulation of PDCD4 expression is an independent predictor of poor prognosis in human renal cell carcinoma patients. J. Cancer Res. Clin. Oncol. 138, 529–535 (2012).
Li, X. et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor PDCD4 and promotes cell transformation, proliferation, and metastasis in renal cell carcinoma. Cell. Physiol. Biochem. 33, 1631–1642 (2014).
Gao, P., Huang, Y., Hou, Y., Li, Q. & Wang, H. Circular RNA ITCH is a tumor suppressor in clear cell renal cell carcinoma metastasis through miR-106b-5p/PDCD4 Axis. J. Immunol. Res. 2021, 5524344 (2021).
Cai, T. et al. The N6-methyladenosine modification and its role in mrna metabolism and gastrointestinal tract disease. Front. Surg. 9, 819335 (2022).
Wang, Y. et al. N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat. Neurosci. 21, 195–206 (2018).
Xu, Y. et al. Regulation of N6-methyladenosine in the differentiation of cancer stem cells and their fate. Front. Cell Dev. Biol. 8, 561703 (2020).
Fan, H.-N. et al. METTL14-mediated m6A modification of circORC5 suppresses gastric cancer progression by regulating miR-30c-2-3p/AKT1S1 axis. Mol. Cancer 21, 51 (2022).
Shi, W., Tang, Y., Lu, J., Zhuang, Y. & Wang, J. MIR210HG promotes breast cancer progression by IGF2BP1 mediated m6A modification. Cell Biosci. 12, 38 (2022).
Liu, N. et al. LncRNA CARMN m6A demethylation by ALKBH5 inhibits mutant p53-driven tumour progression through miR-5683/FGF2. Clin. Transl. Med. 14, e1777 (2024).
Liu, J. et al. N6 -methyladenosine-modified lncRNA ARHGAP5-AS1 stabilises CSDE1 and coordinates oncogenic RNA regulons in hepatocellular carcinoma. Clin. Transl. Med. 12, e1107 (2022).
Yu, Q. et al. Long non-coding RNA LHX1-DT regulates cardiomyocyte differentiation through H2A.Z-mediated LHX1 transcriptional activation. iScience 26, 108051 (2023).
Zhang, Y. et al. Identification of long non-coding RNA expression profiles and co-expression genes in thyroid carcinoma based on The Cancer Genome Atlas (TCGA) database. Med. Sci. Monit. 25, 9752–9769 (2019).
Yang, X. et al. Individualized prediction of survival by a 10-long non-coding RNA-based prognostic model for patients with breast cancer. Front. Oncol. 10, 515421 (2020).
Sun, Y., Peng, P., He, L. & Gao, X. Identification of lnc RNAs related to prognosis of patients with colorectal cancer. Technol. Cancer Res. Treat. 19, 1533033820962120 (2020).
Wang, Y., Liu, Z. & Shen, J. MicroRNA-421-targeted PDCD4 regulates breast cancer cell proliferation. Int. J. Mol. Med. 43, 267–275 (2019).
Li, C. et al. SKP2 promotes breast cancer tumorigenesis and radiation tolerance through PDCD4 ubiquitination. J. Exp. Clin. Cancer Res.38, 76 (2019).
Zennami, K. et al. PDCD4 is an androgen-repressed tumor suppressor that regulates prostate cancer growth and castration resistance. Mol. Cancer Res.17, 618–627 (2019).
Slavik, H. et al. Transcriptomic profiling revealed Lnc-GOLGA6A-1 as a novel prognostic biomarker of meningioma recurrence. Neurosurgery 91, 360–369 (2022).
Guo, X. et al. A liquid biopsy signature for the early detection of gastric cancer in patients. Gastroenterology 165, 402–413.e13 (2023).
Ghasemian, A. et al. Long non-coding RNAs and JAK/STAT signaling pathway regulation in colorectal cancer development. Front. Genet. 14, 1297093 (2023).
Chen, Y., Li, Z., Chen, X. & Zhang, S. Long non-coding RNAs: from disease code to drug role. Acta Pharm. Sin. B 11, 340–354 (2021).
Jia, F. et al. Self-assembled fluorescent hybrid nanoparticles-mediated collaborative lncRNA CCAT1 silencing and curcumin delivery for synchronous colorectal cancer theranostics. J. Nanobiotechnol. 19, 238 (2021).
Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant No. 82072835) to K.W., 345 Talent Project of Shengjing Hospital of China Medical University (Grant No. M0366) to K.W., Outstanding Scientific Fund of Shengjing Hospital (Grant No. 202205) to K.W., Natural Science Foundation of Liaoning Province (Grant No. 2024-MSLH-562) to K.W., and Department of Science and Technology of Liaoning Province (2022-BS-121) to D.D.
Author information
Authors and Affiliations
Contributions
K.W. and X.C. conceived the experiments; D.D. and J.W. supervised the experiments; C.Z. and R.L. conducted the experiments; X.Y. and C.Z. analyzed the data and figures. J.X. supports software and validation. K.W. and C.Z. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhu, C., Li, R., You, X. et al. m6A reader IGF2BP2-stabilized lncRNA LHX1-DT inhibits renal cell carcinoma (RCC) cell proliferation and invasion by sponging miR-590-5p. npj Precis. Onc. 9, 193 (2025). https://doi.org/10.1038/s41698-025-00958-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41698-025-00958-x