Abstract
This study investigates the role of Gαi3 in bladder cancer, focusing on its expression, functions, and mechanisms. Bioinformatics reveal that Gαi3 is overexpressed in bladder cancer, linked to poorer survival and higher tumor grades. Single-cell sequencing data confirm Gαi3 overexpression in bladder cancer epithelial cells. Additionally, Gαi3 is upregulated in tissues from locally treated patients and various bladder cancer cells. Silencing Gαi3 with targeted shRNA inhibited cell proliferation, migration, and invasion, activating caspase-mediated apoptosis. CRISPR/Cas9-mediated knockout of Gαi3 also exhibited significant anti-cancer effects, while overexpression enhanced cell proliferation and mobility. Gαi3 activates the Akt-mTOR pathway; silencing or knockout reduced phosphorylation of Akt and S6K, while overexpression increased it. The anti-cancer effects of Gαi3 shRNA were lessened by reactivating Akt-mTOR with a constitutively active Akt1 mutant. We observed a stronger binding affinity between the Gαi3 promoter and the transcription factor GATA4 in bladder cancer tissues. GATA4 silencing lowered Gαi3 expression, while GATA4 overexpression increased it. In vivo, Gαi3 silencing significantly hindered the growth of bladder cancer xenografts in nude mice, along with inactivation of the Akt-mTOR pathway and increased apoptosis. These results highlight Gαi3’s functional role in bladder cancer progression.
Similar content being viewed by others
Introduction
Bladder cancer is a common malignancy1,2, with long-term survival rates for patients with advanced or metastatic disease remaining low3,4. Treatment for non-muscle-invasive bladder cancer typically involves complete tumor resection followed by intravesical immunotherapy, chemotherapy, and maintenance therapy5,6,7. For muscle-invasive cases, radical cystectomy and neoadjuvant chemotherapy are the primary approaches5,6,7, supplemented by additional therapies. Recently, immunotherapy has emerged as a promising option, particularly for patients resistant to first-line chemotherapy5,6,7. The genetic diversity and complex tissue microenvironment of bladder cancer highlight the disease’s intricacies6,8,9,10. A thorough understanding of the underlying molecular biology is essential for developing new, targeted therapeutic strategies6,8,9,10.
Current targeted therapies for bladder cancer focus on disrupting specific molecular pathways and genetic alterations intrinsic to the disease. Fibroblast growth factor receptor (FGFR) inhibitors, including erdafitinib, target FGFR mutations prevalent in bladder cancer11. Immune checkpoint inhibitors, including pembrolizumab and atezolizumab, inhibit PD-1/PD-L1 pathways, thereby enhancing the immune system’s ability to combat cancer cells12. HER2 inhibitors like trastuzumab are employed for tumors with HER2 overexpression, while vascular endothelial growth factor (VEGF) inhibitors, such as bevacizumab, aim to obstruct tumor angiogenesis. mTOR inhibitors, including everolimus, target critical pathways for cancer cell proliferation and survival13. Current targeted therapies for bladder cancer face limitations due to the genetic heterogeneity of the disease, leading to variable patient responses and the development of resistance, which diminishes long-term efficacy. It is therefore important to identify other key targets.
The inhibitory heterotrimeric G protein (Gαi) family has three distinct subunits: Gαi1, Gαi2, and Gαi314,15. Gαi proteins are known to associate with G protein-coupled receptors (GPCRs) and play a significant role in downregulating adenylate cyclase (AC) activity, which in turn lowers intracellular levels of cyclic AMP (cAMP) [1]. Recent research has highlighted that Gαi1 and Gαi3 are pivotal for signal transduction from various receptor tyrosine kinases (RTKs), such as the epidermal growth factor receptor (EGFR), fibroblast growth factor receptor (FGFR), keratinocyte growth factor receptor (KGFR), stem cell factor (SCF) receptor c-kit, brain-derived neurotrophic factor (BDNF) receptor TrkB, and vascular endothelial growth factor receptor 2 (VEGFR2)16,17,18,19,20,21. Gαi proteins are integral to the mediation of oncogenic signaling cascades, including the Akt-mTOR pathway16,17,18,19,20,21. This discovery underscores their critical role in the transmission of signals that promote tumorigenesis and cancer progression.
Beyond their association with receptor tyrosine kinases (RTKs)16,17,18,19,20,21, Gαi1 and Gαi3 have been found to engage with a variety of non-RTK receptors. These include the leucine-rich repeat-containing G-protein coupled receptor 4 (LGR4) for R-spondin 3 (RSPO3), CD146 for Netrin-1, the interleukin-4 (IL-4) receptor, and the Toll-Like Receptor 4 (TLR4) for lipopolysaccharide (LPS)22,23,24,25. Through these interactions, Gαi proteins are instrumental in the activation of the Akt-mTOR signaling pathway, highlighting their critical role as oncogenic drivers and their potential as therapeutic targets in cancer treatment. Recent studies have demonstrated that Gαi proteins are often overexpressed in a variety of human cancers, underscoring their essential functions in tumor initiation and progression26,27,28. This research is dedicated to exploring the expression patterns and functional importance of Gαi3 in bladder cancer, aiming to provide comprehensive insights into its potential as a biomarker and a target for therapeutic strategies.
Results
Gαi3 overexpression correlates with key clinical parameters of bladder cancer
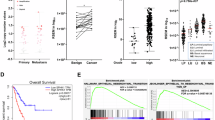
We initially queried The Cancer Genome Atlas bladder cancer (TCGA-BLCA) database to retrieve Gαi3 (GNAI3) expression data. The analysis revealed that the number of Gαi3 mRNA transcripts in bladder cancer tissues (“Tumor”) was significantly higher than in normal bladder tissues (“Normal”) (Fig. 1A). Furthermore, paired data corroborated this finding, showing higher Gαi3 expression in bladder cancer tissues (“Tumor”) compared to their paired normal counterparts (Fig. 1B). Gαi3 expression was higher in high-grade bladder cancer tissues than that in the low-grade ones (Fig. 1C). We also utilized publicly available Gene Expression Omnibus (GEO) datasets for validation. Using data from GSE13507, we confirmed increased Gαi3 expression in high-grade bladder cancer tissues (Fig. 1D). Additionally, Gαi3 expression in bladder cancer tissues was significantly higher in cases of disease progression (PD) but relatively lower in cases of complete response (CR) (Fig. 1E).
TCGA-BLCA cohort analysis demonstrates Gαi3 mRNA transcript levels in bladder cancer tissues (“Tumor”) and normal bladder tissues (“Normal”) across specific patients (A–C, E), and the GSE13507 cohort analysis demonstrating Gαi3 mRNA transcripts in low- and high-grade bladder cancer tissues (D). Kaplan-Meier survival curves of overall survival (OS, F), disease-specific survival (DSS, G) and progression-free interval (PFI, H) for bladder cancer patients from TCGA-BLCA cohort with low Gαi3 expression (blue) and high Gαi3 expression (red) are shown. Subgroup Kaplan–Meier survival analyses were conducted for papillary (I) and non-papillary (J) bladder cancer patients, male (K) and female (L) patients, and for patients ≤ 70 (M) and >70 (N) years age. Kaplan-Meier survival curves of overall survival (OS) for bladder cancer patients from GSE13507 and GSE169455 cohorts with low Gαi3 expression (blue) and high Gαi3 expression (red) are shown (O, P). “TPM” stands for transcripts per million. “CI” stands for confidence interval. HR hazard rate, PD progressive disease, SD stable disease, PR partial response, CR complete response. *P < 0.05. **P < 0. 01. ***P < 0.001.
The Kaplan Meier Survival analyses of the TCGA-BLCA cohort showed that Gαi3 overexpression in bladder cancer tissues was correlated with poorer overall survival of patients (Fig. 1F), reduced disease-specific survival (DSS) (Fig. 1G), and shorter progression-free interval (PFI) (Fig. 1H). The stratification by Gαi3 expression for the main survival plot was determined using the surv_cutpoint function (survminer), which identifies a threshold for maximal survival curve separation.
Furthermore, elevated Gαi3 expression consistently predicted poorer overall survival across various bladder cancer patient subgroups. This association was observed in both papillary and non-papillary bladder cancers (Fig. 1I, J), in both female and male patients (Fig. 1K, L), and was also evident when stratifying patients by age (≤70 and >70 years) (Fig. 1M, N). Furthermore, survival analysis using data from both GSE13507 and GSE169455 cohorts demonstrated that high Gαi3 expression was associated with poorer overall survival (Fig. 1O, P), corroborating our findings from the TCGA cohort. These findings suggest that elevated Gαi3 levels are associated with poorer clinical outcomes in bladder cancer patients.
Single cell sequencing shows Gαi3 overexpression in epithelial cells of bladder cancer
Further bioinformatics analysis was conducted on the GEO single cell dataset GSE222315, which includes bladder cancer (BLCA) and control normal bladder samples (“NORM”). The dimensionality reduction graph illustrated cell annotations (Fig. 2A), revealing that Gαi3 is expressed across various cell populations in both bladder cancer and control samples (Fig. 2B). We performed manual cell type annotation based on the expression of classic cell type-specific marker genes. Figure 2B demonstrates a dot plot visualizing the expression levels of these key markers across the identified cell clusters. The specific markers used for annotation include: Epithelial cells (EPCAM, KRT18, KRT5, KRT17, KRT13, KRT20, UPK1A, UPK1B, UPK3B, UPK3A, UPK2, VIM), Endothelial cells (PECAM1), Fibroblasts (COL3A1, COL1A1, PDGFRA, RGS5, ACTA2), B lymphocytes (CD79A), Myeloid cells (LYZ), T lymphocytes (CD3D), and Mast cells (TPSAB1) (Fig. 2B).
The UMAP (Uniform Manifold Approximation and Projection) plot of cell annotations derived from single-cell data of bladder cancer (GSE222315, A) illustrates the expression distribution of Gαi3. A dot plot showed the expression levels of cell type-specific marker genes across the identified cell clusters (B). The distribution is further visualized in UMAP plot (C), Dot plot (D) for normal (“NORM”) and bladder cancer (“BLCA”) samples. A violin plot highlights the expression levels of Gαi3 in normal (“NORM”) and bladder cancer (“BLCA”) epithelial cell subsets (E), with the Wilcoxon rank-sum test was performed. Volcano plot showing genes differentially expressed between Gαi3+ and Gαi3- epithelial cells (F). Red points are upregulated, blue are downregulated, grey are not significant (F). Top enriched GO Biological Process terms for the DEGs, sorted by adjusted P value (G). Top enriched KEGG pathways for the DEGs, sorted by adjusted P value (H).
The dot plot analysis demonstrated that Gαi3 is highly expressed in epithelial cell populations and myeloid immune cell populations (Fig. 2C). In comparison to the NORM group, Gαi3 expression was significantly elevated in the epithelial cell population of the BLCA group (Fig. 2D, E).
Epithelial cells were further extracted and separated into Gαi3 positive (Gαi3+) and Gαi3 negative (Gαi3-) cell populations based on the presence or absence of Gαi3 expression (Fig. 2F). Differential analysis was conducted on these Gαi3+ and Gαi3- populations, identifying differentially expressed genes (DEGs) with thresholds of adjusted P value < 0.05 and log2FC > 1 (Fig. 2F). We then performed enrichment analysis on the differentially expressed genes to identify associated biological processes and signaling pathways. Gene Ontology (GO) analysis of biological processes (Fig. 2G) revealed significant enrichment of terms including tyrosine phosphorylation, T-helper 17 immune response regulation, and epithelial cell migration (Fig. 2G). Key enriched KEGG pathways included Cytokine-cytokine receptor interaction, JAK-STAT signaling, Akt-mTOR signaling, and TGF-beta signaling pathways (Fig. 2H).
Gαi3 is overexpressed in both human bladder cancer tissues and cells
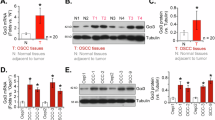
We also assessed the expression of Gαi3 in human bladder cancer tissues of locally-treated patients. Twelve patients undergoing tumor resection were included in the study. Both cancerous tissues (“T”) and adjacent normal bladder tissues (“N”), as reported previously29, were separated. Part of the tissue samples were homogenized using a tissue lysis buffer. The results showed that Gαi3 mRNA levels in cancerous tissues were over three times higher than those in normal tissues (Fig. 3A). Furthermore, increased Gαi3 protein levels were observed in cancerous tissues from four representative patients (“T1/T2/T3/T4”, Fig. 3B), whereas normal tissues from the same patients showed relatively low expression (“N1/N2/N3/N4”, Fig. 3B). When aggregating the blotting data from all twelve tissue groups, a significant upregulation of Gαi3 protein was evident in the bladder cancer tissues (Fig. 3C).
Gαi3 mRNA (A) and protein (B, C) expression were analyzed in human bladder cancer tissues (“T”) and adjacent normal bladder tissues (“N”) of twelve (12) primary patients undergoing tumor resection. Gαi3 mRNA and protein expression were also measured in primary (priBlCa-1, priBlCa-2, priBlCa-3) and immortalized (T24) bladder cancer cells, as well as in the bladder epithelial cells (priBEC-1 and priBEC-2) (D, E). Data were presented as mean ± standard deviation (SD). A–C each group consisted of 12 patient tissue sets (n = 12). D, E n = 5 represents five biological replicates. *P < 0.05 compared to “N” tissues/priBEC-1 cells.
The expression of Gαi3 in human bladder cancer cells was also investigated. The data indicated a significant increase in Gαi3 mRNA expression in both primary (priBlCa-1/priBlCa-2/priBlCa-3, see our previous study29) and T24 immortalized cancer cells (Fig. 3D), compared to the low levels found in priBEC-1 and priBEC-2 bladder epithelial cells (Fig. 3D). Additionally, Gαi3 protein upregulation was detected in both primary and immortalized bladder cancer cells (Fig. 3E), whereas priBEC-1 and priBEC-2 epithelial cells exhibited low expression levels (Fig. 3E). Hence, Gαi3 is overexpressed in both human bladder cancer tissues and cells.
Gαi3 shRNA induces significant antitumor effects in bladder cancer cells
To investigate the functional effect of Gαi3 in bladder cancer cells, we utilized a shRNA-mediated technique to silence Gαi3 expression. Human primary bladder cancer cells, priBlCa-129, were transfected with three distinct lentiviral shRNAs targeting Gαi3: shGαi3-S1, shGαi3-S2, and shGαi3-S327,30. Post-puromycin selection, stable cells were generated and cultured. qPCR assays indicated a substantial reduction, exceeding 80%, in Gαi3 mRNA levels in priBlCa-1 cells expressing shGαi3-S1, shGαi3-S2, or shGαi3-S3 compared to controls transfected with scramble control shRNA (shC) (Fig. 4A). The mRNA levels of Gαi1 and Gαi2 remained unchanged (Fig. 4A). Western blot analysis confirmed the effective silencing of Gαi3 protein in stable priBlCa-1 cells (Fig. 4B), while the protein levels of Gαi1 and Gαi2 were unaffected (Fig. 4B).
Primary (priBlCa-1, priBlCa-2, priBlCa-3) and established (T24) human bladder cancer cells, as well as bladder epithelial cells (priBEC-1 and priBEC-2), were subjected to stable transduction with Gαi3-specific lentiviral shRNAs (shGαi3-S1, shGαi3-S2, or shGαi3-S3, representing different sequences) or a scramble control shRNA (“shC”). The expression levels of targeted mRNAs and proteins were subsequently measured using qPCR (A, G, J) and western blotting (B), respectively. Following a specified culture period, cell viability (C, L), cell proliferation (assessed by the ratio of EdU-positive nuclei; D, H, K), cell migration, and invasion (“Transwell” assays; E, F, I) were evaluated. Parental control cells were denoted as “Pare.” Data were presented as mean ± standard deviation (SD). n = 5 stands for five biological repeats. Statistical significance was indicated by *P < 0.05 compared to the “shC” group, while “N.S.” denoted non-significant differences (P > 0.05). Each experiment was repeated five times, yielding consistent results. Scale bar = 100 μm.
The viability of priBlCa-1 cells, assessed via CCK-8 assays, was significantly reduced following Gαi3 knockdown (Fig. 4C). Additionally, a marked decrease in cell proliferation was observed in priBlCa-1 cells expressing shGαi3, as evidenced by a reduced ratio of EdU-stained nuclei (Fig. 4D). Furthermore, “Transwell” assays revealed that Gαi3 shRNA hindered in vitro migration (Fig. 4E) and invasion (Fig. 4F) of priBlCa-1 cells. As anticipated, shC did not significantly alter the expression of Gαi1, Gαi2, or Gαi3 (Fig. 4A, B) or the functional properties of the bladder cancer cells (Fig. 4C–F).
Subsequent experiments aimed to determine if Gαi3 silencing would elicit comparable effects in other bladder cancer cell lines. Primary human bladder cancer cells derived from two additional patients, designated as priBlCa-2 and priBlCa-329, as well as the immortalized bladder cancer cell line T24, were subjected to stable transduction with lentiviral Gαi3 shRNA (by “shGαi3-S3”). The results indicated robust downregulation of Gαi3 mRNA in both primary and immortalized bladder cancer cells (Fig. 4G), without affecting Gαi1 and Gαi2 mRNA levels (Fig. 4G). This knockdown of Gαi3 led to a pronounced reduction in cell proliferation, as demonstrated by a decreased ratio of EdU-positive nuclei (Fig. 4H), and inhibited cell migration, as evidenced by “Transwell” assays (Fig. 4I), in both primary bladder cancer cells and the T24 cell line.
Additionally, primary bladder epithelial cells, priBEC-1 and priBEC-2 (as reported previously29), were transduced with shGαi3-S3. This resulted in a significant reduction of Gαi3 mRNA expression (Fig. 4J), without affecting Gαi1 and Gαi2 mRNA levels (Fig. 4J). Unlike in bladder cancer cells, Gαi3 silencing in these epithelial cells by shGαi3-S3 did not inhibit cell proliferation, as assessed by EdU incorporation (Fig. 4K), nor did it affect cell viability, as measured by CCK-8 assays (Fig. 4L). Together these results showed that Gαi3 shRNA induced significant antitumor effects in bladder cancer cells.
Gαi3 shRNA induces apoptosis activation in bladder cancer cells
In Fig. 3 we showed that Gαi3 silencing inhibited bladder cancer cell viability and proliferation, outcomes frequently associated with the activation of programmed cell death (apoptosis). Furthermore, early studies have shown that Gαi3 knockdown or knockout can induce apoptosis in other cancer cells26,27. Experiments were therefore conducted to evaluate whether Gαi3 shRNA elicits apoptosis in bladder cancer cells. Remarkably, there was a significant elevation in caspase-3 activity within stable priBlCa-1 cells expressing shGαi3-S1, shGαi3-S2, and shGαi3-S3 (Fig. 5A). Further Western blot analysis revealed that Gαi3 shRNA induced the cleavage of both caspase-3 and poly (ADP-ribose) polymerase 1 (PARP-1) in priBlCa-1 cells (Fig. 5B), suggesting apoptosis initiation. Supporting this observation, we detected a marked increase in histone-bound DNA content (Fig. 5C) and cytosolic cytochrome C levels (Fig. 5D) in Gαi3-silenced priBlCa-1 cells. Additionally, the accumulation of JC-1 green monomers supported mitochondrial depolarization in Gαi3-depleted priBlCa-1 cells (Fig. 5E), further corroborating the induction of apoptosis. This was further confirmed by a rise in TUNEL-positive nuclei in priBlCa-1 cells treated with shGαi3-S1, shGαi3-S2, and shGαi3-S3 (Fig. 5F). Moreover, FACS analysis demonstrated an increased proportion of Annexin V-positive priBlCa-1 cells following Gαi3 knockdown (Fig. 5G). In contrast, shC did not trigger apoptosis in priBlCa-1 cells (Fig. 5A–G).
Primary (priBlCa-1, priBlCa-2, priBlCa-3) and established (T24) human bladder cancer cells, as well as the bladder epithelial cells (priBEC-1 and priBEC-2), were subjected to stable transduction with Gαi3-specific lentiviral shRNAs (shGαi3-S1, shGαi3-S2, or shGαi3-S3, representing different sequences) or a scramble control shRNA (“shC”), following 72 h, the activity of caspase-3 (A, H), the expression levels of apoptosis-related proteins (B), the content of histone-bound DNA (measured by ELISA assay, C), cytosolic cytochrome C levels (measured by ELISA assay, D), and mitochondrial depolarization (assessed by JC-1 staining assay, E) were evaluated. Cell apoptosis was quantified using nuclear TUNEL staining (F, I, J) and Annexin V FACS assays (G). Parental control cells were denoted as “Pare.” Data were presented as mean ± standard deviation (SD). n = 5 stands for five biological repeats. Statistical significance was indicated by *P < 0.05 compared to the “shC” group, while “N.S.” denoted non-significant differences (P > 0.05). Each experiment was repeated five times, yielding consistent results. Scale bar = 100 μm.
Similar outcomes were observed in other primary bladder cancer cells, priBlCa-2 and priBlCa-3, as well as the immortalized T24 cell line. Silencing of Gαi3 using shGαi3-S3 (see Fig. 4) resulted in a significant increase in caspase-3 activity (Fig. 5H). Additionally, significant apoptosis activation was evident in these bladder cancer cells expressing shGαi3-S3, as indicated by a substantial rise in TUNEL-positive nuclei staining (Fig. 5I). In contrast, Gαi3 silencing in bladder epithelial cells, priBEC-1 and priBEC-2, by shGαi3-S3 did not lead to apoptosis activation, as evidenced by the minimal change in the ratio of TUNEL-positive nuclei (Fig. 5J). These results showed that Gαi3 silencing induced apoptosis activation in bladder cancer cells.
Gαi3 knockout hinders the progression of bladder cancer cells
To further substantiate the critical function of Gαi3 in bladder cancer cells, we utilized the CRISPR/Cas9 gene-editing technique to achieve complete knockout (KO) of Gαi3. Following the introduction of the CRISPR/Cas9-Gαi3-KO-puro construct27,30 into Cas9-expressing priBlCa-1 primary cells, we successfully established two single stable clones, designated as “koGαi3-C1” and “koGαi3-C2”, through screening for Gαi3 KO. As illustrated, Gαi3 protein expression was completely depleted in koGαi3-C1 and koGαi3-C2 priBlCa-1 cells (Fig. 6A), while the expression levels of Gαi1 and Gαi2 proteins remained unaffected (Fig. 6A). The proliferation in priBlCa-1 cells was inhibited following the CRISPR/Cas9-mediated Gαi3 KO, as indicated by a marked decrease in nuclear EdU staining (Fig. 6B). Additionally, results from the “Transwell” assays revealed a significant impairment in the in vitro migration (Fig. 6C) and invasion (Fig. 6D) capabilities of priBlCa-1 cells after Gαi3 KO. The observed accumulation of JC-1 green monomers further supported mitochondrial depolarization in the koGαi3-C1 and koGαi3-C2 priBlCa-1 cells (Fig. 6E). Moreover, apoptosis activation was confirmed in ko-Gαi3 priBlCa-1 cells, as evidenced by a significantly elevated ratio of TUNEL-positive nuclei (Fig. 6F).
Cas9-expressing primary bladder cells, priBlCa-1, were used to establish two stable colonies named “koGαi3-C1” and “koGαi3-C2.” These colonies harbored the CRISPR/Cas9-Gαi3-KO-puro construct, with a control group transfected with the CRISPR/Cas9 empty vector, referred to as “koC”. The expression levels of Gαi1, Gαi2, and Gαi3 proteins were assessed using western blotting assays (A). Following this, the cells were cultured for specified periods, and various assays were performed to evaluate cell proliferation (measured by the ratio of EdU-positive nuclei, B), cell migration and invasion (measured via “Transwell” assays, C, D), mitochondrial depolarization (analyzed using the JC-1 staining assay, E), and apoptosis (determined by the ratio of TUNEL-positive nuclei, F). Additionally, Cas9-expressing bladder epithelial cells (priBEC-1 and priBEC-2) containing either the CRISPR/Cas9-Gαi3-KO-puro construct (“koGαi3”) or the control CRISPR/Cas9 empty vector (“koC”) were established. The expression levels of Gαi1 and Gαi3 proteins were also evaluated using western blotting assays (G). Subsequently, these cells were cultured for specified durations, and their cell viability (H) and proliferation (I) were assessed. Data were presented as mean ± standard deviation (SD). n = 5 stands for five biological repeats. Statistical significance was indicated by *P < 0.05 compared to the “koC” group, while “N.S.” denoted non-significant differences (P > 0.05). Each experiment was repeated five times, yielding consistent results. Scale bar = 100 μm.
In bladder epithelial cells, priBEC-1 and priBEC-2, the identical CRISPR/Cas9-Gαi3-KO-puro construct was employed to generate Gαi3 KO epithelial cells, designated as “koGαi3”. Compared to the control epithelial cells, referred to as koC, the expression of Gαi3 protein was completely absent in koGαi3 priBEC-1 and priBEC-2 cells (Fig. 6G). The expression of Gαi1 protein remained virtually unchanged (Fig. 6G). Unlike the results observed in cancerous cells, the KO of Gαi3 did not reduce the viability (measured by CCK-8 OD, Fig. 6H) nor did it affect the proliferation (assessed by nuclear EdU incorporation, Fig. 6I) of the epithelial cells. Collectively, these findings underscore the indispensable role of Gαi3 in the progression of bladder cancer cells, highlighting its potential as a therapeutic target.
Gαi3 overexpression enhances oncogenic behaviors in bladder cancer cells
Given the potent anti-bladder cancer effects observed upon silencing or KO of Gαi3, we hypothesize that overexpression of Gαi3 may exert pro-tumorigenic properties. To investigate this, a lentivirus-mediated Gαi3-overexpressing construct (termed “oeGαi3”) was introduced into priBlCa-1 primary bladder cancer cells. Post-puromycin selection, two stable cell selection, “oeGαi3-stb slc1” and “oeGαi3-stb slc2”, were established. qPCR (Fig. 7A) and Western Blotting (Fig. 7B) assays confirmed a significant upregulation of Gαi3 mRNA and protein levels in these oeGαi3-expressing priBlCa-1 cells, in comparison to cells transduced with a lentiviral empty vector (“Vec”). The expression levels of Gαi1 and Gαi2 remained unaltered (Fig. 7A, B).
The priBlCa-1 primary bladder cancer cells were transduced with either a lentiviral Gαi3-overexpressing construct (“oeGαi3-stb slc1” and “oeGαi3-stb slc2”, representing two stable cell selections) or an empty vector (“Vec”). The mRNA and protein expression levels of Gαi1, Gαi2, and Gαi3 were subsequently assessed (A, B). Following designated culture periods, cell proliferation was evaluated using EdU staining assays (C), while in vitro cell migration (D) and invasion (E) were also measured. Similarly, primary bladder cancer cells (priBlCa-2 and priBlCa-3) and the established T24 human bladder cancer cell line were transduced with either the lentiviral Gαi3-overexpressing construct (“oeGαi3”) or the empty vector (“Vec”). The mRNA expression levels of Gαi1 and Gαi3 were analyzed (F, G). After a specified culture period, cell proliferation (H) and in vitro cell migration (I) were assessed similarly. Data were presented as mean ± standard deviation (SD). n = 5 stands for five biological repeats. Statistical significance was indicated by *P < 0.05 compared to the “Vec” group, while “N.S.” denoted non-significant differences (P > 0.05). Each experiment was repeated five times, yielding consistent results. Scale bar = 100 μm.
The overexpression of Gαi3 in priBlCa-1 cells resulted in enhanced cellular proliferation, as evidenced by increased nuclear EdU incorporation (Fig. 7C). Additionally, “Transwell” assays demonstrated that both cell migration (Fig. 7D) and invasion (Fig. 7E) were accelerated in oeGαi3-expressing priBlCa-1 cells. These pro-tumorigenic effects were consistently observed in other primary bladder cancer cells, priBlCa-2 and priBlCa-3, as well as in the immortalized T24 bladder cancer cell line. Overexpression of Gαi3 via the Gαi3-overexpressing construct (“oeGαi3”) led to a significant increase in Gαi3 mRNA levels (Fig. 7F), without affecting Gαi1 mRNA expression (Fig. 7G). This upregulation of Gαi3 was associated with augmented cellular proliferation (Fig. 7H) and migration (Fig. 7I), underscoring the role of Gαi3 in promoting oncogenic behaviors in bladder cancer cells.
Gαi3 is important for Akt-mTOR activation in in bladder cancer cells
Gαi3 has been identified to interact with both RTKs and non-RTK receptors, facilitating the activation of Akt-mTOR signaling pathway. The above single cell sequencing data showed that DEGs of Gαi3+ and Gαi3- bladder cancer epithelial cells were enriched in Akt-mTOR signaling pathway. Given the known involvement of Akt-mTOR hyperactivation in the initiation and progression of bladder cancer31, we investigated the potential influence of Gαi3 on Akt-mTOR activation within bladder cancer cells. In priBlCa-1 cells, silencing Gαi3 via shGαi3-S1, shGαi3-S2, or shGαi3-S3 led to a reduction in the phosphorylation of Akt at Ser-473 and S6K at Thr-389, while the total levels of Akt1 and S6K proteins remained unchanged (Fig. 8A). Additionally, CRISPR-sgRNA-mediated KO of Gαi3 markedly diminished Akt-mTOR activation, evidenced by decreased Akt-S6K phosphorylation in koGαi3-C1 and koGαi3-C2 priBlCa-1 cells (Fig. 8B), without altering the total expression of Akt1 and S6K (Fig. 8B). Conversely, overexpression of Gαi3 in priBlCa-1 cells, labeled as “oeGαi3-stb slc1” and “oeGαi3-stb slc2,” resulted in enhanced Akt-S6K phosphorylation (Fig. 8C). These findings underscore the critical role of Gαi3 in modulating Akt-mTOR signaling in bladder cancer cells.
The priBlCa-1 primary bladder cancer cells with Gαi3-specific lentiviral shRNA (shGαi3-S1, shGαi3-S2, or shGαi3-S3, representing different sequences), a scramble control shRNA (“shC”), the Cas9-expressing construct plus the CRISPR/Cas9-Gαi3-KO-puro construct (“koGαi3-C1” and “koGαi3-C2”, two stable colonies), the CRISPR/Cas9 empty vector (“koC”), a lentiviral Gαi3-overexpressing construct (“oeGαi3-stb slc1” and “oeGαi3-stb slc2,” representing two stable cell lines) or an empty vector (“Vec”) were cultured and expression of listed proteins was shown (A–C). shGαi3-S3-expressing priBlCa-1 cells were stably transduced with or without a constitutively-active S473D mutant Akt1 (caAkt1). The expression levels of the specified proteins were shown (D). These transduced cells were then cultured for the indicated durations, and assays were conducted to evaluate cell proliferation using EdU-nuclei staining (E), migration using the “Transwell” (F), and apoptosis using TUNEL-nuclei staining (G). Data were presented as mean ± standard deviation (SD). n = 5 stands for five biological repeats. Statistical significance was indicated by *P < 0.05 compared to the “shC”/“koC”/“Vec” cells. #P < 0.05. Each experiment was repeated five times, yielding consistent results. Scale bar = 100 μm.
To elucidate the connection between Gαi3-induced bladder cancer cell growth and the activation of the Akt-mTOR pathway, we introduced a constitutively active mutant of Akt1 (S473D, referred to as caAkt129) into Gαi3-silenced priBlCa-1 cells using stable transduction (“shGαi3-S3). Our findings revealed that caAkt1 fully reinstated Akt-S6K phosphorylation levels without impacting the expression of Gαi3 protein in priBlCa-1 cells (Fig. 8D). Furthermore, caAkt1 significantly mitigated the effects of Gαi3 silencing, almost reversing the observed decreases in cell proliferation (Fig. 8E), migration (Fig. 8F), and reducing apoptosis (Fig. 8G). These observations suggest that the activation of the Akt-mTOR signaling pathway is a crucial mechanism by which Gαi3 promotes the growth and survival of bladder cancer cells.
The dominant negative Gαi3 mutation inhibits the Akt-mTOR pathway, reduces cell proliferation and migration, and promotes apoptosis in bladder cancer cells
The dominant negative (dn) Gαi3 mutant alters the conserved Glycine (G) residue to a Threonine (T) in the G3 box, disrupting the interaction between Gαi1/3 and its associated proteins16,32. This modification led to the disruption of Gαi3’s interaction with various receptor tyrosine kinases (RTKs), ultimately blocking the Akt-mTOR signaling pathway19,21,23,30. To explore these effects, priBlCa-1 primary bladder cancer cells were transduced with a lentivirus encoding dnGαi3, followed by puromycin selection, resulting in two stable cell selections, “dnGαi3-Slc1” and “dnGαi3-Slc2”, both of which expressed dnGαi3 (Fig. 9A). In these dnGαi3-expressing cells, decreased levels of phosphorylated Akt and S6K confirmed the inhibition of the Akt-mTOR pathway (Fig. 9B). Expression of Gαi2 protein as well as total Akt1 and S6K1 was unchanged (Fig. 9B). Functional assays further revealed that dnGαi3 expression significantly reduced cell proliferation (measured by nuclear EdU incorporation, Fig. 9C) and migration (Fig. 9D). Increased nuclear TUNEL staining also indicated apoptosis induction in dnGαi3-expressing priBlCa-1 cells (Fig. 9E). These findings demonstrate that the dominant negative Gαi3 mutation effectively suppressed Akt-mTOR signaling, reduced cell proliferation and migration, and promoted apoptosis in bladder cancer cells.
The priBlCa-1 primary bladder cancer cells were transduced with either a lentiviral dominant negative Gαi3 construct (“dnGαi3-Slc1” and “ dnGαi3-Slc1”, representing two stable selection) or an empty vector (“Vec”). The expression of listed proteins was shown (A, B). Following designated culture periods, cell proliferation was evaluated using nuclear EdU staining assays (C); In vitro cell migration (“Transwell” assays, D) and apoptosis (nuclear TUNEL staining assays, E) were also measured. Data were presented as mean ± standard deviation (SD). n = 5 stands for five biological repeats. Statistical significance was indicated by *P < 0.05 compared to the “Vec” group. Each experiment was repeated five times, yielding consistent results. Scale bar = 100 μm.
Increased binding between GATA4 and the Gαi3 promoter in bladder cancer tissues and cells
We next explored the possible transcriptional mechanisms driving increased Gαi3 overexpression observed in bladder cancer tissues and cells. Previous studies have suggested that GATA4, a transcription factor, may play a crucial role for Gαi3 expression27,33. Thus, priBlCa-1 primary bladder cancer cells were infected with lentivirus carrying GATA4-specific shRNA (“shGATA4-S1” or “shGATA4-S2”, two different sequences) or a non-targeting shRNA control (“shC”), and stable cells were selected using puromycin. Both GATA4 mRNA (Fig. 10A) and protein (Fig. 10B) levels were significantly diminished in cells expressing shGATA4-S1/S2 when compared to the control group. This reduction was paralleled by a significant decrease in Gαi3 mRNA (Fig. 10A) and protein (Fig. 10B) levels. These findings strongly indicate that GATA4 is a critical transcriptional regulator of Gαi3 expression in bladder cancer cells.
The priBlCa-1 primary bladder cancer cells were subjected to stable transduction with GATA4-specific lentiviral shRNAs (shGATA4-S1, shGATA4-S2, representing different sequences), a scramble control shRNA (“shC”), a lentiviral GATA4-overexpressing construct (“oeGATA4”) or an empty vector (“Vec”). The expression levels of targeted mRNAs and proteins were subsequently measured (A–D). Chromatin immunoprecipitation (ChIP) assays presented the relative levels of GATA4-bound Gαi3 promoter in primary (priBlCa-1, priBlCa-2, priBlCa-3) and immortalized (T24) bladder cancer cells, or in the bladder epithelial cells (priBEC-1 and priBEC-2) (E) as well as in the designated bladder cancer tumor tissues (“T1/T2/T3”) and matched adjacent normal bladder epithelial tissues (“N1/N2/N3”) (F), with results quantified. Parental control cells were denoted as “Pare”. Data were presented as mean ± standard deviation (SD). n = 5 stands for five biological repeats. Statistical significance was indicated by *P < 0.05 compared to the “shC”, “Vec”, “priBEC-1” or “N” group. Each experiment was repeated five times, yielding consistent results.
To further validate the hypothesis, priBlCa-1 cells were transduced with a lentivirus encoding GATA4, and after puromycin selection, stable “oeGATA4” cells showed a marked increase in both GATA4 and Gαi3 expression at the mRNA (Fig. 10C) and protein (Fig. 10D) levels. Importantly, a chromatin immunoprecipitation (ChIP) assay results revealed a substantial rise in GATA4 protein binding to the Gαi3 promoter region in various primary and established bladder cancer cells (Fig. 10E). Furthermore, examination of bladder cancer tissue samples from three patients (“T1/T2/T3”) showed significantly greater GATA4 binding to the Gαi3 promoter compared to nearby normal pancreatic tissues (“N1/N2/N3”) (Fig. 10F). These findings collectively suggest that the enhanced interaction between GATA4 and the Gαi3 promoter may play a pivotal role in driving Gαi3 overexpression in bladder cancer.
Gαi3 silencing impedes primary bladder cancer xenograft growth in nude mice
To underscore the pivotal role of Gαi3 in in vivo growth of bladder cancer cells, we injected five million priBlCa-1 cells expressing either shGαi3-S3 or shC subcutaneously into the flanks of nude mice. Observations commenced ten days following cell inoculation (“Day-10”). The resulting tumor growth curve, recording tumor volumes every six days (“Day-10” to “Day-52”), demonstrated a significant reduction in the growth of shGαi3-S3-expressing priBlCa-1 xenografts compared to those expressing shC (Fig. 11A). Quantifying the daily growth rate further confirmed the substantial suppression of tumor progression in the shGαi3-S3-expressing priBlCa-1 xenografts (Fig. 11B). At Day-52, all tumors were separated and weighted. the weights of the shGαi3-S3 priBlCa-1 xenografts were significantly lower than those of the Cas9-C priBlCa-1 xenografts (Fig. 11C), while the overall body weights of the mice were comparable between the two groups (Fig. 11D). A significant decrease in Gαi3 mRNA and protein expression was observed in the shGαi3-S3 priBlCa-1 xenografts, with Gαi1 expression levels remaining unchanged (Fig. 11E, F).
The stable priBlCa-1 cells expressing Gαi3 shRNA (“shGαi3-S3”) or control shRNA (“shC”) were injected subcutaneously into the flanks of nude mice, with each mouse receiving five million cells. Each experimental group comprised ten mice (n = 10). Observations began ten days after the initial cell inoculation (Day-10). The volumes of the priBlCa-1 xenografts (A) and the body weights of the mice (D) were measured and recorded at six-day intervals, spanning from Day-10 to Day-52. The daily growth rate of the priBlCa-1 xenografts was assessed (B). On Day-52, the xenografts were surgically removed, and their weights were measured (C). For further analysis, two xenografts from each group (designated as “1#” and “2#”) were selected, and protein and mRNA expression analyses were conducted on the tissue lysates from these harvested xenograft tissues (E–G, J). Additionally, sections of the xenografts were subjected to immunohistochemistry (IHC) staining to detect p-Akt (Ser-473, H) or nuclear Ki-67 (I). TUNEL/DAPI fluorescence staining was also performed (K). Data were presented as the mean ± standard deviation (SD). Statistical significance was indicated by *P < 0.05 when compared to the “shC” group, whereas “N. S.” signified non-statistically significant differences (P > 0.05). A–D The sample size was ten mice per group. E–K each xenograft was sectioned into five pieces and analyzed individually. Scale bar = 100 μm.
Further examination unveiled a pronounced decline in the phosphorylation levels of Akt and S6K1 in the shGαi3-S3 priBlCa-1 xenograft tissues (Fig. 11G). Moreover, IHC images confirmed reduction of the phosphorylation level of Akt in shGαi3-S3 priBlCa-1 xenograft sections (Fig. 11H). The nuclear Ki-67 staining within the Gαi3-silenced priBlCa-1 xenograft tissues was significantly decreased, indicating proliferation inhibition (Fig. 11I). In contrast, there was a significant elevation in the levels of cleaved-Caspase-3 and cleaved-PARP-1 in the shGαi3-S3-expressing priBlCa-1 xenograft tissues (Fig. 11J). To further corroborate the induction of apoptosis, tissue fluorescence staining assays revealed a higher proportion of TUNEL-positive nuclei in the shGαi3-S3 priBlCa-1 xenograft tissue sections (Fig. 11K). Collectively, these findings highlight that silencing Gαi3 markedly impeded the growth of priBlCa-1 xenografts in nude mice, inactivated the Akt-mTOR signaling pathway and induced apoptosis.
Discussion
The current landscape of targeted therapy for bladder cancer features significant advancements and have transformed treatment by focusing on specific molecular alterations, leading to improved outcomes5,6,8,12,34. Their importance lies in providing personalized treatment options for patients with specific genetic mutations or biomarker expressions. However, limitations include resistance development, variability in patient responses, and a limited number of approved targets5,6,8,12,34. Therefore, identifying novel therapeutic targets is of paramount urgency.
Recent studies have underscored the critical function of Gαi proteins, particularly Gαi1 and Gαi3, in the signal transduction processes linked to various RTKs16,17,18,19,20 and non-RTK receptors22,23,24,25. These proteins play a crucial role in driving oncogenic pathways activated by cell surface receptors16,17,18,19,20,22,23,24,25. They are notably overexpressed in several types of cancers, including glioma, osteosarcoma, cervical cancer, and nasopharyngeal carcinoma, where their elevated levels are associated with a grim prognosis17,26,27,28,30. Research has demonstrated that silencing or knocking out Gαi3 hinders the growth of osteosarcoma, cervical cancer, and glioma cells, both in vitro and in vivo. Conversely, overexpression of Gαi3 is correlated with increased cancer cell growth, further emphasizing its role in tumor progression17,26,27,28,30. These findings highlight the potential of targeting Gαi3 for therapeutic interventions in human cancer.
The current research identifies Gαi3 as a novel biomarker and critical therapeutic target for bladder cancer. Bioinformatics analyses reveal that Gαi3 is significantly overexpressed in bladder cancer, correlating with worse survival rates, higher tumor grades, and other key clinical parameters. Further examination of single-cell sequencing data shows elevated Gαi3 expression in epithelial cells of bladder cancer. Additionally, Gαi3 is upregulated in bladder cancer tissues from locally-treated patients and various bladder cancer cells. Targeted shRNA silencing of Gαi3 or CRISPR/Cas9-mediated KO of Gαi3 inhibited cell proliferation, migration, and invasion, while inducing caspase-mediated apoptosis in bladder cancer cells. Conversely, Gαi3 overexpression enhanced primary bladder cancer cell proliferation, migration, and invasion. In vivo, Gαi3 silencing markedly reduced the growth of subcutaneous xenografts of primary bladder cancer cells in nude mice. These findings offer fresh insights into the role of Gαi3 in bladder cancer progression.
The Akt-mTOR signaling cascade plays a pivotal role in the progression of bladder cancer through a cascade of molecular events initiated by the overexpression/sustained activation of growth factor receptors, including EGFR and FGFR331,35. Upon ligand binding, PI3K is activated, leading to the production of PIP3 and subsequent recruitment and phosphorylation of Akt31,35. Activated Akt inhibits TSC2, resulting in the activation of mTORC1, which promotes cell growth, proliferation, and survival by stimulating protein synthesis and inhibiting apoptosis31,35. This pathway also reprograms cellular metabolism to support rapid tumor growth and enhances angiogenesis via upregulation of VEGF31,35. Furthermore, the Akt-mTOR pathway contributes to therapeutic resistance, highlighting its importance as a target for therapeutic strategies in bladder cancer treatment31,35.
Early investigations have thoroughly shown the interaction of Gαi3 with various RTKs (including FGFR, EGFR and VEGFR)16,18,19,20,24,28 and non-RTK receptors22,23,24, highlighting its critical involvement in the activation of the Akt-mTOR signaling pathway. In this research, we have shown that Gαi3 is essential for the activation of the Akt-mTOR signaling pathway in bladder cancer cells. DEGs of Gαi3+ and Gαi3- bladder cancer epithelial cells were enriched in Akt-mTOR signaling pathway. Silencing or KO of Gαi3 led to a marked reduction in the phosphorylation levels of both Akt and S6K, which are pivotal components of this pathway. Ectopic overexpression of Gαi3, on the other hand, resulted in elevated phosphorylation levels. Furthermore, re-activation of the Akt-mTOR pathway through the introduction of caAkt1 was able to counteract the anti-bladder cancer effects induced by Gαi3 shRNA. The dominant-negative mutant of Gαi3 has been found to interfere with its interaction with various RTKs19,21,23,24,30, resulting in decreased activation of downstream signaling pathways. In this study, we also demonstrated that dnGαi3 specifically inhibited Akt-mTOR signaling, effectively reducing bladder cancer cell growth and proliferation. Additionally, we observed a significant reduction in Akt and S6K phosphorylation in bladder cancer xenografts where Gαi3 had been silenced. This emphasizes the critical role of Gαi3 in sustaining Akt-mTOR pathway activity in bladder cancer.
Early studies have proposed GATA4 as a key transcription factor of Gαi3. Guo et al. demonstrated that GATA4 promotes Gαi3 transcription, playing a vital role in tooth development33. Similarly, Yao et al. discovered that GATA4, in association with phosphoenolpyruvate carboxykinase 1 (PCK1), enhances Gαi3 transcription in endothelial cells36. Our research indicates that GATA4-mediated transcriptional activation of Gαi3 could be a key driver of Gαi3 overexpression in bladder cancer. In primary human bladder cancer cells, silencing GATA4 led to a marked reduction in both Gαi3 mRNA and protein levels, while overexpression of GATA4 resulted in elevated Gαi3 expression. Furthermore, ChIP assays revealed an increased binding of GATA4 to the Gαi3 promoter in bladder cancer tissues and different bladder cells. This enhanced interaction appears to be a crucial mechanism contributing to Gαi3 upregulation in bladder cancer.
In summary, the study identifies Gαi3 as a crucial therapeutic target for bladder cancer, showing that its overexpression correlates with poor prognosis, while its silencing or KO inhibited bladder cancer cell growth both in vitro and in vivo.
Methods
Reagents and chemicals
The Cell Counting Kit-8 (CCK-8) was obtained from Dojindo Co. (Kumamoto, Japan). Puromycin, polybrene, medium, antibiotics, serum, and RNA reagents were sourced from Sigma-Aldrich (St. Louis, MO). EdU, DAPI, TUNEL, JC-1, and Annexin V/PI fluorescence dyes were purchased from Invitrogen Thermo-Fisher (Shanghai, China). All antibodies, viral constructs and mRNA primers were provided by Dr. Cao27,30.
Single cell RNA sequencing (scRNA-seq)
The raw scRNA-seq data were obtained from GEO (GSE222315). We used the Seurat R package for downstream analysis. Following initial quality control, the gene-cell expression matrix was normalized using NormalizeData with the LogNormalize method (scale factor 10,000). Highly variable features were identified using FindVariableFeatures, and data were scaled using ScaleData. Dimensionality reduction involved principal component analysis (PCA) [top 30 principal components (PC)] and Uniform Manifold Approximation and Projection (UMAP) for visualization. Cells were clustered using FindNeighbors and FindClusters (resolution 0.5). Cell types were manually annotated by comparing cluster-specific genes to known markers. Within the epithelial cell population, differential expression analysis between Gαi3-positive and negative cells was performed using FindMarkers (P < 0.05, log2FC > 1), and resulting upregulated genes were used for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis via Enrichr (https://maayanlab.cloud/Enrichr/).
Human tissues
As reported29, 12 patients (65–76 years old) with muscle-invasive bladder cancer (T2-T4a, N0, M0) undergoing tumor resection surgery were included. Cancer and adjacent normal bladder tissues were carefully isolated under microscopy. Tissue lysates were subjected to Western Blotting and qPCR assays. All patients were treated at Changzhou No.2 People’s Hospital of Nanjing Medical University, and informed-consent was obtained. The study protocols were approved by the Ethical Review Board of Changzhou No.2 People’s Hospital of Nanjing Medical University (No. JCY-2021-032), in line with the Declaration of Helsinki.
Cells
T24 immortalized bladder cancer cells were obtained from the Shanghai Institutes for Biological Sciences and were cultured in DMEM with 8% FBS, gentamicin, penicillin/streptomycin, L-glutamine, and HEPES, in a humidified incubator at 37 °C and 5% CO2. For primary human cells, primary cancer tissues and adjacent normal epithelial tissues were isolated, rinsed with PBS, minced, and digested using collagenase-I and DNase. The resulting single-cell suspension was filtered through a 70 µm strainer, collected, washed, and resuspended in culture medium. Primary bladder cancer cells were derived from three patients (priBlCa-1, priBlCa-2, priBlCa-3), and primary epithelial cells from two patients (priBEC-1, priBEC-2), as reported previously29. Cell phenotype validation included tests for mycoplasma, microbial contamination, STR profiling, population doubling time, and morphology assessment every 8 weeks. Protocols were approved by the Ethical Review Board of Changzhou No. 2 People’s Hospital of Nanjing Medical University, adhering to the Declaration of Helsinki.
Western blotting
As reported previously29,37, protein lysates (30 µg) from cells and tissues were separated on SDS-PAGE gels and transferred to PVDF blots. Membranes were blocked, incubated with designated primary antibodies overnight at 4 °C, followed by secondary antibodies at room temperature for 45 min. Protein bands were visualized using an enhanced chemiluminescence (ECL) system and quantified with ImageJ software. Figure S1 shows uncropped blotting images.
qPCR
As reported previously29,37, RNA was extracted from cells and tissues using TRIzol, converted to cDNA with a Takara kit, and analyzed by qPCR using SYBR Green PCR Master Mixes on the ABI-7900 system. GAPDH served as a reference gene, and the 2−ΔΔCt method determined relative gene expression. Primers were supplied by Dr. Cao27,30. Following mRNA primers were utilized, Gαi1, Forward, 5′-TTAGGGCTATGGGGAGGTTGA-3′, Reverse, 5′-GGTACTCTCGGGATCTGTTGAAA-3′; Gαi2, Forward, 5′-CAGCAAGATGTGCTACGGACC3′, Reverse, 5′-CACCAAGTCATAGGCGCTCA3′ and Gαi3, Forward, 5′-CACTTCACCTGTGCCACAGACA-3′, Reverse, 5′-GTCTGGTCTCAACACTCCACAC-3′.
Gαi3 silencing via shRNA
Three distinct shRNAs targeting human Gαi3 (shGαi3-S1, targeting: CAGCTCAATGATTCTGCTTCATATT; shGαi3-S2, targeting: AACTTCAATCATTCTCTTCCTTAAC; and shGαi3-S3, targeting TAGCAGGAGTGATTAAACGGTTATG) were provided by Dr. Cao from Soochow University27,30, using the GV369 construct. Bladder cancer and epithelial cells were cultured in a medium with FBS and polybrene to enhance transduction. Cells were infected with the lentivirus at an MOI of 10 for 40 h. After infection, cells were kept in a puromycin-containing medium for ten days to select stable colonies. Knockdown efficiency of Gαi3 was confirmed using qPCR and Western Blotting.
Gαi3 overexpression
To overexpress Gαi3 [both wild-type or dominant negative (“dn”) mutant], we used a lentiviral vector containing the wild-type or dn mutant Gαi3 gene, provided again by Dr. Cao27,30. Bladder cancer cells were grown in medium with FBS and polybrene to boost viral transduction. These cells were infected with the Gαi3-expressing lentivirus at an MOI of 10 for 40 h. After infection, cells were cultured in puromycin-containing medium for 7-8 days to select for stable Gαi3-overexpressing cells. We confirmed Gαi3 expression in these cells using qPCR and/or Western Blotting assays.
Gαi3 knockout
Bladder cancer cells at 50–60% confluence were grown in complete medium with polybrene and infected with Cas9-expressing lentivirus to create stable cells. These cells were then transduced with a CRISPR/Cas9-Gαi3-KO lentivirus containing sgRNA targeting Gαi327,30. After puromycin selection, cells were isolated into 96-well plates at the single-cell level. Sequencing around the target site and Western Blotting confirmed Gαi3 KO. Thereafter, two stable single-cell-derived Gαi3 knockout colonies, namely koGαi3-C1 and koGαi3-C12, were established, both showing complete Gαi3 protein depletion. Control cells were infected with a CRISPR/Cas9-empty vector lentivirus (“koC”), as previously described27,30. Gαi3 KO in the epithelial cells was through the same procedure, resulting in one stable single cell clone.
GATA4 silencing or overexpression
Two distinct shRNAs targeting human GATA4 (shGATA4-S1 and shGATA4-S2) were obtained from Dr. Cao at Soochow University36, using the GV369 construct. Bladder cancer and epithelial cells were cultured in a medium supplemented with FBS and polybrene for enhanced transduction. Cells were infected with the lentivirus at an MOI of 10 for 40 h. Subsequently, cells were maintained in a puromycin-containing medium for ten days to select stable cell colonies. For GATA4 overexpression, the lentivirus with GATA4-expressing construct (provided from Dr. Cao36) was added to cultured primary human bladder cancer cells, and stable cells were formed using the same protocol. The efficiency of GATA4 knockdown or overexpression was confirmed by qPCR and Western blotting.
CCK-8 viability assay
Cells (at 3 × 103 cells/well) were plated in 96-well plates and cultured for designated durations. CCK-8 reagent (15 µL/well) was added for 2 h, and absorbance at 450 nm was measured using a microplate reader.
Nuclear TUNEL/EdU staining
Treated cancer or epithelial cells were seeded on coverslips in 24-well plates and cultured for specified durations. Cells were fixed with 4% formaldehyde-PBS for 12 min at room temperature and permeabilized with 0.1% Triton X-100 in PBS for 6 min. Nuclei were stained with TUNEL, EdU, or DAPI dyes. Fluorescent images were taken with a fluorescence microscope.
Transwell assays
Transwell chambers (Corning, NY, US) were used, with 150 μL of cell suspension (10,000 cells) added to each chamber. The lower chamber contained 600 μL of medium with 10% FBS. After 24 h of incubation, non-migrated cells were removed, and migrated cells were stained with 0.25% crystal violet for 10 min, washed with PBS, and photographed. For in vitro cell invasion assays, Transwell inserts were coated with Matrigel (100 µg/cm², Sigma).
Caspase-3 activity assay
As reported previously29,37, total cell or tissue lysates were analyzed using Caspase fluorescent assay kits (BD Bioscience, Suzhou, China). Caspase-3 activity was measured fluorometrically with the substrate Ac-DEVD-AMC.
Flow cytometry
As reported previously29,37, bladder cancer cells or epithelial cells were centrifuged, resuspended, and stained with Annexin V-APC (5 µL) and/or propidium iodide (PI, 5 µL) (Sigma). Apoptosis was evaluated using a CytoFLEX flow cytometer (Beckman, Shanghai, China).
Cytochrome C detection
The Cytochrome C ELISA assay was conducted with a commercial kit (Biyuntian, Wuxi, China). Cytosol lysates were added to a 96-well plate, and non-specific binding sites were blocked. The plate was then washed, and a primary antibody specific for cytochrome C was added, followed by a HRP-conjugated secondary antibody. A TMB substrate solution initiated the colorimetric reaction. Absorbance was measured at 450 nm using a microplate reader.
Mitochondrial depolarization detection (ΔΨm)
As reported previously29,37, the JC-1 dye assay (Invitrogen) was used to assess mitochondrial membrane potential (MMP) reduction. After treatment, cells were stained with JC-1 dye (4.0 μg/mL) and washed to remove excess dye. The green fluorescence intensity of JC-1, indicating decreased MMP (ΔΨm), was measured at 490 nm using a fluorescence microscope.
Constitutively active mutant Akt1 (caAkt1)
As reported previously29,37, the bladder cancer cells were infected with a lentivirus carrying the constitutively active S473D mutant of Akt1 (“caAkt1”) or an empty vector. Stable caAkt1-expressing cells were selected using puromycin.
Chromatin immunoprecipitation (ChIP)
ChIP was employed to investigate the interaction between (GATA binding protein 4) and the Gαi3 (GNAI3) promoter. The experimental protocol adhered to established methodologies36,38. Briefly, lysates derived from both cells and tissues were homogenized, followed by the fragmentation of genomic DNA39. These lysates were subsequently diluted and subjected to immunoprecipitation using an anti-GATA4 antibody, enabling the enrichment of GATA4-bound DNA fragments. Quantitative PCR (qPCR) was then utilized to assess the association between GATA4 and the putative Gαi3 promoter DNA sequence (TGCTATC)33, with results normalized to control values.
Xenograft studies
Five million priBlCa-1 cells were subcutaneously injected into the flanks of BALB/c nude mice (5–6 weeks old, 18–19 grams, evenly divided by sex) obtained from the Shanghai Institutes for Biological Sciences. Tumors developed within 10 days post-injection. Tumor volumes were recorded every six days. For IHC staining, tumors were fixed, embedded in paraffin, and sectioned at 4 μm. Sections were deparaffinized, rehydrated, and subjected to antigen retrieval. After blocking, sections were incubated with a primary antibody, followed by a secondary antibody, and detection was performed using DAB. The IHC protocols for human cancer tissues were the same. Alternatively, sections were stained with TUNEL (Biyuntian, Wuxi, China), washed, and counterstained with DAPI to visualize nuclei. The sections were mounted with an anti-fade medium and examined under a fluorescence microscope. To prepare animals for the procedure, a freshly mixed anesthetic cocktail of 100 mg/kg Ketamine and 10 mg/kg Xylazine was administered intraperitoneally (IP). Maintaining animal well-being was a priority: a warming pad set to 37 °C was used during the procedure and recovery to ensure normothermia, and ophthalmic ointment was applied to prevent corneal drying. All animal experiments were approved by the IACUC and the Ethics Review Board of Nanjing Medical University (No. JCDW-2021-019).
Statistical analysis
Investigators performed blinded group allocations for all in vitro experiments, which were replicated five times. Data following a normal distribution were presented as mean ± SD (standard deviation). Statistical analyses were conducted using SPSS version 23.0 (SPSS Co., Chicago, IL). An unpaired Student’s t-test compared two groups, while one-way ANOVA with Scheffe’ and Tukey tests compared multiple groups. Statistical significance was defined as P < 0.05.
Data availability
No datasets were generated or analysed during the current study.
References
Kamat, A. M. et al. Bladder cancer. Lancet 388, 2796–2810 (2016).
Sharma, A., Sharma, K. L., Gupta, A., Yadav, A. & Kumar, A. Gallbladder cancer epidemiology, pathogenesis and molecular genetics: recent update. World J. Gastroenterol. 23, 3978–3998 (2017).
Siegel, R. L., Wagle, N. S., Cercek, A., Smith, R. A. & Jemal, A. Colorectal cancer statistics, 2023. CA Cancer J. Clin. 73, 233–254 (2023).
Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 73, 17–48 (2023).
Tran, L., Xiao, J. F., Agarwal, N., Duex, J. E. & Theodorescu, D. Advances in bladder cancer biology and therapy. Nat. Rev. Cancer 21, 104–121 (2021).
Lenis, A. T., Lec, P. M., Chamie, K. & Mshs, M. D. Bladder cancer: a review. JAMA 324, 1980–1991 (2020).
Sanli, O. et al. Bladder cancer. Nat. Rev. Dis. Prim. 3, 17022 (2017).
Patel, V. G., Oh, W. K. & Galsky, M. D. Treatment of muscle-invasive and advanced bladder cancer in 2020. CA Cancer J. Clin.70, 404–423 (2020).
Cormio, A. et al. Mitochondrial dysfunctions in bladder cancer: Exploring their role as disease markers and potential therapeutic targets. Crit. Rev. Oncol./Hematol. 117, 67–72 (2017).
Caldow Pilgrim, C. H., Groeschl, R. T., Quebbeman, E. J. & Gamblin, T. C. Recent advances in systemic therapies and radiotherapy for gallbladder cancer. Surg. Oncol. 22, 61–67 (2013).
Casadei, C. et al. Targeted therapies for advanced bladder cancer: new strategies with FGFR inhibitors. Ther. Adv. Med. Oncol. 11, 1758835919890285 (2019).
Lopez-Beltran, A. et al. Immune checkpoint inhibitors for the treatment of bladder cancer. Cancers 13, 131 (2021).
Zangouei, A. S., Barjasteh, A. H., Rahimi, H. R., Mojarrad, M. & Moghbeli, M. Role of tyrosine kinases in bladder cancer progression: an overview. Cell Commun. Signal.18, 127 (2020).
Fan, H. et al. Heterotrimeric Galpha(i) proteins are regulated by lipopolysaccharide and are anti-inflammatory in endotoxemia and polymicrobial sepsis. Biochim. Biophys. Acta 1813, 466–472 (2011).
Tontson, L., Babina, A., Vosumaa, T., Kopanchuk, S. & Rinken, A. Characterization of heterotrimeric nucleotide-depleted Galpha(i)-proteins by Bodipy-FL-GTPgammaS fluorescence anisotropy. Arch. Biochem. Biophys. 524, 93–98 (2012).
Cao, C. et al. Galpha(i1) and Galpha(i3) are required for epidermal growth factor-mediated activation of the Akt-mTORC1 pathway. Sci. Signal. 2, ra17 (2009).
Liu, Y. Y. et al. microRNA-200a downregulation in human glioma leads to Galphai1 over-expression, Akt activation, and cell proliferation. Oncogene 37, 2890–2902 (2018).
Marshall, J. et al. Antidepression action of BDNF requires and is mimicked by Galphai1/3 expression in the hippocampus. Proc. Natl. Acad. Sci. USA 115, E3549–E3558 (2018).
Sun, J. et al. Galphai1 and Galphai3mediate VEGF-induced VEGFR2 endocytosis, signaling and angiogenesis. Theranostics 8, 4695–4709 (2018).
Zhang, Y. M. et al. Requirement of Gαi1/3-Gab1 signaling complex for keratinocyte growth factor-induced PI3K-AKT-mTORC1 activation. J. Invest. Dermatol. 135, 181–191 (2015).
Shan, H. J. et al. SCF/c-Kit-activated signaling and angiogenesis require Galphai1 and Galphai3. Int. J. Biol. Sci. 19, 1910–1924 (2023).
Xu, G. et al. Galphai1/3 mediation of Akt-mTOR activation is important for RSPO3-induced angiogenesis. Protein cell 14, 217–222 (2023).
Li, Y. et al. Galphai1/3 mediate Netrin-1-CD146-activated signaling and angiogenesis. Theranostics 13, 2319–2336 (2023).
Bai, J. Y. et al. Requirement of Galphai1 and Galphai3 in interleukin-4-induced signaling, macrophage M2 polarization and allergic asthma response. Theranostics 11, 4894–4909 (2021).
Li, X. et al. Galphai1 and Galphai3 regulate macrophage polarization by forming a complex containing CD14 and Gab1. Proc. Natl. Acad. Sci. USA 112, 4731–4736 (2015).
Yin, D. P. et al. Overexpressed Galphai1 exerts pro-tumorigenic activity in nasopharyngeal carcinoma. Cell Death Dis. 14, 792 (2023).
Zhang, J. et al. Identification of Galphai3 as a novel molecular therapeutic target of cervical cancer. Int. J. Biol. Sci. 18, 5667–5680 (2022).
Wang, Y. et al. Neuronal-driven glioma growth requires Galphai1 and Galphai3. Theranostics 11, 8535–8549 (2021).
Zhang, L. et al. A first-in-class TIMM44 blocker inhibits bladder cancer cell growth. Cell Death Dis. 15, 204 (2024).
Bian, Z. J. et al. Identification of Galphai3 as a promising target for osteosarcoma treatment. Int. J. Biol. Sci. 18, 1508–1520 (2022).
Sathe, A. & Nawroth, R. Targeting the PI3K/AKT/mTOR pathway in bladder cancer. Methods Mol. Biol. 1655, 335–350 (2018).
Zhang, Y. M. et al. Requirement of Galphai1/3-Gab1 signaling complex for keratinocyte growth factor-induced PI3K-AKT-mTORC1 activation. J. Investig. Dermatol. 135, 181–191 (2015).
Guo, S. et al. Role of GATA binding protein 4 (GATA4) in the regulation of tooth development via GNAI3. Sci. Rep. 7, 1534 (2017).
Comperat, E. et al. Current best practice for bladder cancer: a narrative review of diagnostics and treatments. Lancet 400, 1712–1721 (2022).
Ching, C. B. & Hansel, D. E. Expanding therapeutic targets in bladder cancer: the PI3K/Akt/mTOR pathway. Lab. Investig. 90, 1406–1414 (2010).
Yao, J. et al. The requirement of phosphoenolpyruvate carboxykinase 1 for angiogenesis in vitro and in vivo. Sci. Adv. 8, eabn6928 (2022).
Mi, Y. Y. et al. A first-in-class HBO1 inhibitor WM-3835 inhibits castration-resistant prostate cancer cell growth in vitro and in vivo. Cell Death Dis. 14, 67 (2023).
Wang, Y. et al. G protein inhibitory alpha subunit 2 is a molecular oncotarget of human glioma. Int. J. Biol. Sci. 19, 865–879 (2023).
Chen, M. et al. Identification of G protein subunit alpha i2 as a promising therapeutic target of hepatocellular carcinoma. Cell Death Dis. 14, 143 (2023).
Acknowledgements
We thank Dr. Cao at Soochow University for reagents and other suggestions of this study. This study was supported by Qinghai Province basic research project (Grant number: 2024-ZJ-743); National Natural Science Foundation (No. 81902565), Jiangsu Province 333 Project (No. RC202202), the Basic research project of Changzhou Medical Center of Nanjing Medical University (CMCB202313 and CMCB202319), Qinghai Province health system suitable technology promotion project (2023-wjtg-07), Top Talent of Changzhou “The 14th Five-Year Plan” High-Level Health Talents Training Project (2022CZBJ058 and 2022CZBJ057), Changzhou Health Commission Young Talent Project (No. CZQM2020065), Jiangsu Province Postdoctoral Research Foundation (2021K588C), Changzhou Sci&Tech Program (CJ20220146), the Innovation team funding of Changzhou (XK201803) and Science and Technology Project of Changzhou Health Commission (ZD 202440).
Author information
Authors and Affiliations
Contributions
All listed authors contributed to designing the study and conducting all experiments. All authors drafted the article, critically reviewed it for significant intellectual contributions, and provided final approval for the version submitted to the journal.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, L., Zhang, L., Gao, S. et al. Gαi3: a crucial biomarker and therapeutic target in bladder cancer. npj Precis. Onc. 9, 181 (2025). https://doi.org/10.1038/s41698-025-00989-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41698-025-00989-4