Abstract
Ewing sarcoma is an aggressive bone tumor of adolescence characterized by a hallmark EWSR1::ETS fusion oncogene. The resulting chimeric oncoprotein drives tumorigenesis by reshaping transcriptional and epigenetic landscapes. However, how it is transcriptionally regulated and whether additional master transcription factors (MTFs) form a core regulatory circuit (CRC) in Ewing sarcoma remain unclear. Using an extensive panel of Ewing sarcoma cell lines and primary tumors, we mapped super-enhancers and identified enrichment of GGAA microsatellites, confirming their specificity to Ewing sarcoma as compared to other pediatric cancers and normal tissues. Integrating transcriptomic, epigenetic, 3D chromatin conformation, and dependency data, we predicted a set of MTFs potentially forming a CRC. However, functional validation demonstrated that these MTFs neither establish auto-regulatory loops nor confer robust proliferative dependencies typical of CRCs in other pediatric tumors. Instead, EWSR1::FLI1 emerged as an “hegemonic” oncoprotein, regulating expression of these MTFs without reciprocal regulation. Knockdown of EWSR1::FLI1 strongly shifted H3K27ac profiles toward mesenchymal states, whereas silencing individual or combined MTFs did not alter cell growth or EWSR1::FLI1 expression. These findings highlight the absence of a classical CRC in Ewing sarcoma and emphasize EWSR1::FLI1 as the dominant oncoprotein and a major vulnerability in this disease.
Similar content being viewed by others
Introduction
Ewing sarcoma is a highly aggressive bone sarcoma that most frequently occurs during adolescence. Genetically, it is characterized by a translocation involving FET (FUS, EWSR1, TAF15) and ETS (E-twenty-six) family members, with EWSR1::FLI1 present in 85% of cases1,2. Consequently, a chimeric oncoprotein combining the EWSR1 transcriptional activation and disordered domains with an ETS DNA-binding domain is expressed in all Ewing sarcoma cells. These cells exhibit plastic behaviors, primarily regulated by EWSR1::FLI1 direct and indirect mechanisms3. Ewing sarcoma cells proliferate optimally at specific levels or activity of EWSR1::FLI1 but acquire mesenchymal features and eventually die when this oncogene is either silenced (EWSR1::FLI1 KD) or overexpressed (EWSR1::FLI1 OE)3,4,5,6,7. Fluctuations in EWSR1::FLI1 expression enhance migration and metastasis in preclinical models, although this has yet to be demonstrated in patients. Mechanistically, post-transcriptional/translational modifications, competitive DNA binding, and 3D chromatin alterations can affect EWSR1::FLI1 level or activity5,6,8,9,10. However, the transcriptional regulation of EWSR1::FLI1 remains poorly understood. Beyond its pathognomonic translocation, Ewing sarcoma exhibits few recurrent genetic alterations, primarily involving STAG2, CDKN2A, TP53, and chromosomes 1q, 8, 12, and 16q11,12,13. The pioneer transcription factor (TF) activity of EWSR1::FLI1, which generates neo-enhancers by binding GGAA microsatellites (mSats), plays a pivotal role in tumorigenesis by altering the epigenetic and 3D chromatin landscape8,14,15,16,17,18. The high variability in mSat composition among tumors, however, leads to strong intertumoral transcriptional heterogeneity19,20 as demonstrated at mSats that regulate EGR2, MYBL2, SOX6, and PRC121,22,23,24. Despite this variability, Ewing sarcoma shows distinct methylation and transcriptomic profiles relative to other sarcomas or pediatric cancers18,25,26.
In normal tissues and various cancers, epigenetic profiling has proven effective in defining cell identity through core regulatory circuitry (CRC) analyses. There, a restricted set of master transcription factors (MTFs), typically co-enriched at super-enhancers (SEs), dominate the control of the gene expression program27,28,29. These MTFs collectively regulate their own expression, forming interconnected auto-regulatory loops. Altering CRCs, for example, through MTFs abrogation, typically alters cellular identity or viability29. In several cancers, including pediatric ones, CRCs have been characterized and functionally validated. The depletion of their respective MTFs typically leads to dependency phenotypes. Transcriptional auto-regulatory loops and dependencies are observed as early as 24 and 48 h, respectively, following MTF silencing, and these effects persist over extended periods of time30,31,32,33. For example, in alveolar rhabdomyosarcoma (aRMS), the fusion oncogene PAX3::FOXO1 (or PAX7::FOXO1) binds SEs together with other MTFs, including MYOD, MYOG, and MYCN, to sustain proliferation, and their depletion leads to altered proliferation and cell death33,34. In neuroblastoma (NB), subgroup-specific CRCs have been identified. For instance, the noradrenergic (NorNB) CRC encompasses PHOX2A/B, HAND1/2, and GATA2/3 MTFs, while the mesenchymal (MesNB) CRC is driven, among others, by factors of the AP1 family30,35. Single-cell data in these cancers support these findings and have also identified intermediate/transitioning cells, highlighting the strong plasticity of these cancer cell populations36,37,38. In Ewing sarcoma, a CRC including KLF15, TCF4, and NKX2-2 MTFs that cooperate with EWSR1::FLI1 was reported in A-673 and EW-8 cells39. Short-term siRNA silencing of individual MTFs decreased cell viability, which was, however, not observed in long-term CRISPR knockout experiments40. Given the transcriptional heterogeneity of Ewing sarcoma, studying multiple models is crucial for identifying CRCs19,41. Here, we investigated a Ewing sarcoma CRC using a large set of Ewing sarcoma cell lines and tumors, compared to other normal and pediatric cancer samples. Through integrative analyses involving transcriptomics, epigenetics, 3D chromatin conformation, and silencing approaches, we show that the predicted CRC does not point towards Ewing sarcoma vulnerabilities. This contrasts with findings in NB and RMS, where CRC analyses uncovered functional dependencies. Here, we use the term hegemonic (from the Greek hēgemonikos, meaning ruling, dominant, or supreme) to describe the overarching influence of the EWSR1::ETS oncoprotein, which overrules the CRC principles in Ewing sarcoma. Altogether, the hegemonic EWSR1::ETS fusion oncoprotein operates upstream of the predicted CRC, represents a key vulnerability, and drives Ewing sarcoma identity.
Results
The epigenetic landscape of Ewing sarcoma
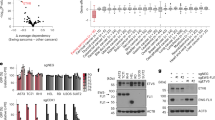
To identify key drivers of Ewing sarcoma identity, we investigated SEs based on H3K27ac ChIP-Seq profiles performed in a panel of 20 Ewing sarcoma cell lines, six primary Ewing sarcoma tumors, two Ewing sarcoma models derived from MSCs (MSC-EWSR1::FLI1), and six EWSR1::FLI1-silenced Ewing sarcoma cell lines (EWSR1::FLI1-KD)8,19,20,42,43. Since MSCs have been shown to be one of the putative cells of origin for Ewing sarcoma43, SEs from four MSC samples were also included in this analysis. As a comparison cohort, SEs identified in 56 normal tissues/cells and other pediatric cancers, including 25 NB and 20 RMS, were used (Supp. Data 1). Principal component analysis (PCA) of normalized SE intensity distinguished clusters corresponding to normal or tumor cell identities (Fig. 1A, B). PC1 primarily separated MSC and EWSR1::FLI1-KD samples from the other entities, while PC2 segregated Ewing sarcoma samples. Gene enrichment analyses of SE-associated genes contributing most to PC1 and PC2 confirmed their mesenchymal and Ewing sarcoma signatures, which correspond to these two axes, respectively (Supp. Data 2). PC1, 2, and 4 also isolated RMS and NorNB samples into distinct groups, while MesNB samples clustered closer to MSC samples (Supp. Fig. 1A, B). Ewing sarcoma cell lines, tumors, and MSC-EWSR1::FLI1 models formed a separate cluster with broad dispersion, reflecting high intertumoral heterogeneity. Silencing EWSR1::FLI1 strongly reshaped the H3K27ac-based SE landscape (Fig. 1A and Supp. Fig. 1C). In line with previous transcriptomic studies41,43, EWSR1::FLI1-KD cells formed an intermediate population, bridging Ewing sarcoma cell lines and MSCs. These results highlight the critical role of the fusion oncogene in defining the epigenetic landscape of Ewing sarcoma (Fig. 1A). Through a comprehensive and agnostic analysis of mSats (all possible repeats of [4-mers]4x and [3-mers]5x) in SEs, we confirmed that only GGAA mSats were strongly enriched in Ewing sarcoma. No mSat enrichment was detected in non-Ewing sarcoma samples (Supp. Fig. 2). These findings further support previous data highlighting the unique specificity of GGAA mSats for Ewing sarcoma3. The frequency of this mSat was significantly higher in SEs than in enhancers even after size normalization (Fig. 1C). These findings are consistent with previous studies on Ewing sarcoma cell lines, which did not perform size normalization44,45. In Ewing sarcoma cell lines, 34% of SEs contained mSats, while mSats were identified in only 3% of enhancers. Furthermore, FLI1- or ERG-ChIP-Seq data from 20 Ewing sarcoma cell lines (Supp. Data 3) showed a significant enrichment of fusion oncoprotein binding at mSats in SEs compared to enhancers (Fig. 1D).
A, B PCA of normalized intensity in Super-Enhancers for Ewing sarcoma (WT in blue and EWSR1::FLI1-KD in red), NB (green), MSC (orange), RMS (purple) and public control data (gray). C Boxplot showing the mean of the normalized number of mSats per region per sample (20 Ewing sarcoma cell lines). Only H3K27ac peaks in super-enhancer or enhancer regions were considered. Outliers are not shown. D Boxplot of normalized number of mSats ETS peaks per region per sample (20 Ewing sarcoma cell lines). Only H3K27ac and ERG/FLI1 peaks in Super-Enhancer or Enhancer regions were considered. Outliers are not shown. A paired Wilcoxon test was used to compare super-enhancers and enhancers in Ewing sarcoma (C, D). E Example of a super-enhancer specific to Ewing sarcoma with high density of GGAA mSats (VRK1 locus). IGV was used to represent the track for FLI1 ChIP-Seq (blue), H3K27ac ChIP-Seq (green) and SEs (green bars) for the control (no Dox) and EWSR1::FLI1-KD (+Dox) condition in A-673/TR/shEF1 cell line. The black barplot represents the number of consecutive GGAA in the human genome. F–H Super-enhancer ranked by mean intensity in Ewing cell lines (F), norNB cell lines (G), and aRMS cell lines and tumors (H). Super-enhancers associated with transcription factors are shown in red. The size of the points corresponds to the adjusted p value adjusted of genes with Ewing sarcoma (F), NB (G), and RMS (H) dependency based on DepMap 23Q4 data. Only genes with p adjust <1e-5 for Ewing sarcoma data and RMS, and p adjust <1e-20 for NB are displayed.
Top-ranked SEs in Ewing sarcoma emphasized previously reported highly expressed or direct EWSR1::ETS targets such as BCL11B46,47, CAV148, CCND149, JAK150, NKX2-251,52, PRKCB53, or VRK18 (Fig. 1E). Additional top hits included HMCN1, DLG2, and IGF2BP1, whose functions in Ewing sarcoma remain unknown or poorly characterized (Fig. 1F and Supp. Data 4). As expected, SE-associated genes in Ewing sarcoma cell lines and in EWSR1::FLI1-KD cells were respectively enriched in Ewing sarcoma/proliferation and migratory/mesenchymal signatures (Supp. Fig. 3A, B). To explore the relationship between SEs and their associated gene dependencies, we used the Dependency Map (DepMap) CRISPR/Cas9-KO data (21 days) and selected genes with a negative dependency (Tstat <0) in Ewing sarcoma40,54. Among these 291 identified dependency genes, 23 were associated with SEs (8%). This proportion is similar to that reported in aRMS (113 dependent genes, 16 associated with SEs, 14%) or NorNB (756 dependent genes, 69 associated with SEs, 9%) (Supp. Data 5). However, in notable contrast with NorNB and aRMS, where SE-associated TFs were readily identified among their respective top ten DepMap dependencies (PHOX2A/B, HAND2, GATA3 for NorNB and MYOD1, MYOG for aRMS) (Fig. 1G, H), in Ewing sarcoma, none of the top ten dependencies were SE-associated TFs, despite a similar representation of TFs across all three entities (Supp. Fig. 3C–E). It should be noted that SEs are typically absent at the EWSR1, FLI1, and ERG loci in Ewing sarcoma.
Absence of a classical CRC in Ewing sarcoma
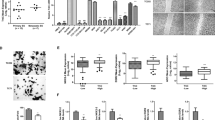
A CRC comprises MTFs that typically self-regulate and interact with each other to define cell identity and fate. To identify an Ewing sarcoma-specific CRC, we applied CRC mapper28 to our data. We selected MTF candidates identified in at least 50% of Ewing sarcoma cell lines but absent in more than 70% of controls (Supp. Data 6, 7). Twelve MTF candidate genes were identified for the Ewing sarcoma CRC (Fig. 2A). Most of these genes were also identified in Ewing sarcoma tumors and one of the two MSC-EWSR1::FLI1 CRCs, but were not predicted in MSC, NB, or RMS CRCs. Based on the expression of these 12 MTFs, no sub-clustering within primary tumors could be identified (Fig. 2B). Interestingly, silencing EWSR1::ETS in seven Ewing sarcoma cell lines led to a reduction in the expression of nine of these 12 MTFs (Fig. 2C), suggesting their direct regulation by EWSR1::FLI1. To evaluate this, we examined EWSR1::FLI1/ERG-ChIP-Seq and H3K27ac-HiChIP data9,19 (Supp. Fig. 4). EWSR1::FLI1/ERG mSat peaks were present in nine SEs associated with these 12 MTFs (Fig. 2D). Cis-regulatory promoter-enhancer (P-E) chains (composed of sequential promoter-enhancer and enhancer-enhancer interactions) incorporating at least one EWSR1::FLI1 mSat-bound element within the first five interactions, were readily identified for ten MTFs in A-673 and/or TC-71 H3K27ac-HiChIP data9 (Fig. 2E). This confirms the direct interaction of EWSR1::FLI1 with the promoters of these MTF genes. Some TFs, such as POU3F2, did not display EWSR1::FLI1 binding in their closest associated SE. Instead, additional EWSR1::FLI1-bound mSats were connected to these TF promoters through long-range P-E chains (Fig. 2D, E and Supp. Fig. 4). By integrating TFs that1 contain mSat peaks within their SEs (Fig. 2D)2, exhibit expression specific to Ewing sarcoma across multiple datasets (Supp. Fig. 5A), and3 are identified as dependencies in Ewing sarcoma by DepMap (Supp. Data 5), we selected six MTFs (TCF12, SOX6, POU3F1, POU3F2, NKX2-2, and IKZF2) as putative members of the Ewing sarcoma CRC, based on fulfilling at least two out of these three criteria (Fig. 2F).
A Heatmap showing the 12 genes predicted to be specific to Ewing sarcoma CRC (≥50% of the Ewing sarcoma cell lines) compared to control data (<30%). Each row represents a gene, and each column a sample. Black and white boxes represent respectively the presence and the absence of the gene in the CRC. The code color at the top is the same as in Fig. 1A. B Spearman correlation plot of expression of 132 Ewing sarcoma tumors for the 12 candidate TFs. C Boxplot of log2 fold change for the 12 previous candidate genes in each of the seven Ewing cell lines expressing EWSR1::ETS compared to EWSR1::ETS-KD oncogene (RNA-Seq data). D Heatmap of the number of FLI1/ERG mSat peaks in super-enhancer (SE) associated with the 12 candidate TF. NA values correspond to TFs without an associated SE in the given cell line. E Heatmap of position of FLI1 mSat peaks in enhancer-promoter chains in A-673 and TC-71 for the 12 candidate TFs (H3K27ac-HiChIP)9. F Venn diagram showing the selection of CRC candidates, corresponding to six MTFs that meet at least two of the three criteria (gray area).
To determine if candidate MTFs colocalize with EWSR1::FLI1-bound and SE regions, we performed ChIP-Seq for SOX6, POU3F2, IKZF2, and EWSR1::FLI in EW-1 cell line (Supp. Fig. 5B). Co-localization of MTFs and EWSR1::FLI1 peaks was observed within the SEs associated with these MTFs (Fig. 3A and Supp. Fig. 5C). The three MTFs (POU3F2 & SOX6 & IKZF2) colocalised in 50% of top 100 Ewing sarcoma SEs (Fig. 3B). Using quantitative real-time PCR, we confirmed that silencing of EWSR1::FLI1 reduced the expression of all, except TCF12, MTFs in EW-1 and TC-71 cell lines (Fig. 3C, D), as observed in RNA-Seq data (Fig. 2C). We then individually silenced these six MTFs in EW-1 and TC-71. However, none of the silencing of candidate MTFs led to a significant decreased expression of the others in both cell lines, hence challenging the interconnected regulatory concept of CRC (Fig. 3E, F). Interestingly, silencing IKZF2 led to increased expression of the other MTFs, the reverse of positive gene regulation that is expected in CRC models. Moreover, the expression of EWSR1::FLI1 was not modulated upon silencing of these genes (Fig. 3E, F and Supp. Data 8). Despite evidence of co-localization, the lack of co-transcriptional regulation highlights their absence of auto-regulatory function typically expected for MTFs. To evaluate the dependency of EW-1 and TC-71 cell lines on these candidate MTFs, we silenced them and monitored cell proliferation using real-time live imaging microscopy (IncuCyte), but no significant differences were observed compared to the control conditions (Fig. 3G, H). Short-term endpoint cell count experiments confirmed these results (Fig. 3I, J). Silencing of EWSR1::FLI1 reduced cell proliferation in EW-1 and TC-71 cells, in agreement with previous findings in other Ewing sarcoma cell lines3,55,56. Altogether, our data show that, despite partial co-localization, these candidate MTFs do not appear as co-regulatory elements nor acute dependencies in Ewing sarcoma. Nevertheless, these results are restricted to individual MTF considerations. We, therefore, asked if combinatorial silencing of these MTFs would be necessary to impact Ewing sarcoma CRC. Among them, POU3F1 and POU3F2 share 95% homology in their DNA-binding domains. We hypothesized that they could complement each other’s functions. We performed individual and combined silencing of POU3F1 and POU3F2 in the EW-1 and TC-71 cell lines and confirmed their specificity and efficiency by RT-qPCR and western blot experiments (Fig. 4A–D). However, combined silencing of POU3F1 and POU3F2 did not alter the expression of the other putative MTFs (Fig. 4C, D). Similarly, dual silencing of POU3F1 and POU3F2 did not alter proliferation in the EW-1 cell line and had a slight decrease in TC-71 (Fig. 4E, F). Ultimately, we asked whether simultaneous silencing of all MTF candidates could impact EWSR1::FLI1 level and cell proliferation. A siRNA pool targeting IKZF2, NKX2-2, POU3F1, POU3F2, SOX6, and TCF12 was transfected into EW-1 and TC-71 cell lines. Of note, EWSR1::FLI1 level remained unchanged in the siRNA pool condition (Fig. 5A, B), highlighting the absence of early transcriptional regulatory circuits of these MTFs on the fusion oncogene. Similarly, no impact on proliferation rate was observed under these conditions (Fig. 5C–F). Since a modest but significant increase in the expression of some MTF candidates was observed upon IKZF2 inhibition in TC-71 (Fig. 3F), we sought to rule out any compensatory mechanism from this gene. To this end, we transfected EW-1 and TC-71 cells with a siRNA pool excluding siIKZF2 and confirmed consistent silencing of the remaining MTFs (Supp. Fig. 6A, B). No or minimal impact on proliferation was observed under these conditions in both cell lines. (Supp. Fig. 6C–F). Altogether, these results show that the predicted CRC does not control the proliferation of Ewing sarcoma cells, at least in the short term.
A Heatmap of normalized ChIP-Seq intensity in EW-1 cells for the indicated TFs. 2065 peaks identified in super-enhancers for the three MTF candidates (POU3F2, SOX6, IKZF2) are displayed. The peaks are ranked by their intensity (order: FLI1, mean (POU3F2, SOX6, IKZF2)). Read density is displayed within a 5 kb window around the peak center, and color scale intensities are shown in normalized coverage. The black bar indicates peaks with mSat. Regions with MYC amplification are removed. B Barplot showing the distribution of co-bound TF within the top 100 SEs (EW-1 cell line): 1 (only POU3F2, SOX6 or IKZF2), 2 (POU3F2 & SOX6, POU3F2 & IKZF2 or SOX6 & IKZF2), 3 (POU3F2 & SOX6 & IKZF2). Knockdown of EWSR1::FLI1 leads to a decrease in CRC gene expression in EW-1 (C) and TC-71 (D) Ewing cells. Gene expression analysis by RT-qPCR was performed after transfection with siEF1 and compared to siRNA Control (siCT), and results were normalized using RPLP0 (48 h post-transfection for EW-1, n = 3 and 72 h post-transfection for TC-71, n = 6). E, F Heatmap of RT-qPCR analysis in EW-1 and TC-71, respectively, after transfection with siRNAs and comparing them to siRNA Control (siCT) and normalizing the results using RPLP0 (48 h post-transfection, n = 3). G, H show proliferation rate using Incucyte technology in EW-1 and TC-71, respectively, after transfecting cells with siRNAs (n = 3). I, J Cell count analysis after siRNAs transfection (3 days post-transfection for EW-1 and 2 days post-transfection for TC-71, n = 3). The data were shown as the mean ± SD, *P < 0.05; **P < 0.01; ***P < 0.001 : two-tailed Student’s t-test. ns non-significant.
A, B Western blot analysis for POU3F1 and POU3F2 following siRNA transfection (48 h) in EW-1 and TC-71 cell lines (siF1 = siPOU3F1 and siF2 = siPOU3F2; # indicates different siRNAs). C, D RT-qPCR-based heatmap in EW-1 and TC-71 cells upon combined POU3F1/2 silencing compared to siCT conditions (48 h post-transfection, n = 2 or 3). E, F Cell count in EW-1 and TC-71 cells upon single or combined POU3F1/2 silencing compared to siCT conditions (48 h post-transfection, n = 2, 3, or 4).
A, B RT-qPCR of MTFs and EWSR1::FLI1 gene expression in EW-1 (A) and TC-71 (B) upon pooled silencing of all MTFs (siPool) compared to siRNA control conditions (siCT) (n = 3). C, D Proliferation rates in EW-1 (C) and TC-71 (D) cells measured using Incucyte technology after siPool compared to siCT (n = 3). EW-1 (E) and TC-71 (F) cell counts upon siPool compared to siCT (n = 3). Data were shown as mean ± SD; *P < 0.05; **P < 0.01; ***P < 0.001 (two-tailed Student’s t-test); ns not significant. EW-1 data shown 72 h post-transfection, and TC-71 data shown 48 h post-transfection.
Predicted CRC and hegemonic oncogene in Ewing sarcoma
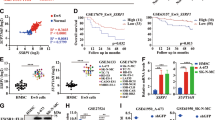
To investigate whether long-term inhibition of TFs could affect Ewing sarcoma cells, we explored the DepMap database40,54. Among our 12 MTF candidates (Fig. 2A), only POU3F1 and POU3F2 were reported as dependencies in Ewing sarcoma (among 28 TFs, 291 genes, Supp. Fig. 7A). Notably, these two genes exhibit a moderate effect size as compared to EWSR1::FLI1, which ranks as the top Ewing sarcoma dependency in DepMap (Fig. 6A). However, the fusion oncogene is not part of our Ewing sarcoma CRC since SEs were mostly absent around EWSR1 and FLI1 loci (Figs. 1F, 6A). The above results are in striking contrast with aRMS and norNB, where MTFs from their CRCs rank among each cancer’s top ten dependencies (Fig. 6B, C) and have been thoroughly validated30,31,32,33,34,35,37,40. Given the well-known heterogeneity of Ewing sarcoma, we investigated whether each tumor might harbor a distinct, individualized CRC. For each cell line, we (1) listed TFs involved in any of the Top 10 predicted CRC and (2) retrieved their TF dependency (gene effect) scores from the DepMap database. However, comparing gene effect scores of CRC TFs with those of non-CRC TFs at the individual cell level revealed no significant enrichment (Supp. Fig. 7B).
A–C Bar plots of the top 20 genes with dependencies enriched in Ewing sarcoma (A), NB (B), and RMS (C), using the CRISPR (DepMap 23Q4 public + score, chronos) dataset. The y-axis represents the log10 of the adjusted p value from DepMap (dependency in the lineage compared to all other models). CRC genes are shown in brown, TFs in orange, and other genes in gray. D Heatmap of TFs involved in different differentiation processes, with only those TFs that have motifs in JASPAR reported. Each column represents a sample, and each row represents a gene. Black boxes indicate TFs that are members of the CRC in the sample. E Alternative CRC regulation in Ewing sarcoma.
Lastly, we investigated if TFs involved in developmental/differentiation pathways of MSCs, the putative cell of origin of Ewing sarcoma, could be enriched in our CRC data. However, TFs controlling adipogenic, osteogenic, or chondrogenic pathways were not enriched in Ewing sarcoma compared to the NB or RMS (Fig. 6D). Altogether, using a broad panel of Ewing sarcoma models, we demonstrate here that the predicted CRC identifies MTFs with predominantly inactive acute transcriptional auto-regulatory and proliferative roles, despite all being direct targets of EWSR1::ETS (Fig. 6E). Instead, we highlight here that the EWSR1::ETS fusion oncoprotein acts as the overarching and unique TF (hegemonic oncoprotein) that regulates a broad panel of heterogeneous target genes and represents the major vulnerability in Ewing sarcoma.
Discussion
Our findings reveal that the concept of CRC is challenged in Ewing sarcoma and diverges from those observed in other pediatric cancers, such as NB and aRMS. Several distinctions emerge from our analyses that may account for these discrepancies:
Unlike NB or aRMS, Ewing sarcoma harbors SEs that are highly enriched in GGAA microsatellites. We also demonstrated that these mSat-SEs are more enriched in EWSR1::ETS fusion protein binding compared to typical enhancers. These non-canonical elements diverge from evolutionarily conserved regulatory sequences found in developmental gene networks and contribute to a more “neomorphic” mode of oncogenic transcriptional control. By systematically scanning for entity-specific microsatellites in SEs, we confirmed the uniqueness of this feature in Ewing sarcoma. Additionally, some genes associated with SEs appear specific to Ewing sarcoma, as they are defined as Ewing sarcoma dependencies. While genes such as BCL11B and VRK1 are well characterized, others, such as IGF2BP1 and LSM6, remain poorly studied, yet they may offer valuable insights into the biology and vulnerabilities of Ewing sarcoma.
In pediatric tumors, MTFs typically highlight lineage-driven programs. For example, in aRMS, myogenic regulators such as PAX3, MYOD1, and MYOG are co-opted32, while in NB, TFs such as HAND2, PHOX2B, and GATA3 govern noradrenergic differentiation and form part of the norNB CRC30,35. Notably, these MTFs consistently rank among the top dependencies in their respective cancers, demonstrating strong lineage addiction mechanisms57. Interestingly, this observation also emerges in analyses of pediatric circulating tumor DNA (ctDNA). In NB and RMS, fragmentomic studies readily detect lineage-specific MTF footprints58. By contrast, previously reported MTFs in Ewing sarcoma39 have not proven discriminatory in ctDNA, whereas a broader GGAA-mSat-based enhancer signature robustly distinguishes Ewing sarcoma from other entities58,59.
In Ewing sarcoma, the precise cell of origin remains debated. Although bone marrow–derived MSCs can undergo transformation, and EWSR1::FLI1 silencing reverts 3D chromatin conformation toward an MSC-like architecture43,51,60, Ewing sarcoma does not appear to co-opt canonical MSC lineage TFs as principal dependencies. The non-evolutionarily conserved nature of GGAA mSats may explain this divergence, as these elements do not align with standard developmental gene regulatory modules. In contrast, other sarcomas, including RMS, exhibit strong skeletal muscle lineage associations based on phylogeny and gene expression data61. Although EWSR1::FLI1 silencing enables osteogenic differentiation in vitro41, raising the possibility of a cryptic MSC differentiation program, our SE analyses (including in EWSR1::FLI1-KD cells) did not detect typical MSC lineage-associated MTFs with major functional roles in Ewing sarcoma. These data further support that the fusion oncoprotein’s neomorphic transcriptional activity, rather than a lineage-based CRC, dominates Ewing sarcoma biology.
The expression and enhancer usage of candidate MTFs vary considerably among Ewing sarcoma cell lines, reflecting the extensive intertumoral heterogeneity reported in transcriptomic and methylation studies. Clustering based on MTF or SE occupancy did not reveal biologically meaningful subtypes, underscoring the heterogeneity of Ewing sarcoma epigenomes and highlighting the challenge of defining a universal CRC in this disease. Variability in mSat lengths among Ewing sarcoma cell lines may partly explain this heterogeneity19,21. We also hypothesized that cell line-specific vulnerabilities and CRCs might exist in Ewing sarcoma. However, leveraging DepMap data did not reveal such CRCs, although further validation in individual models is needed to fully exclude this hypothesis. Interestingly, the most critical dependencies reported in Ewing sarcoma, aside from the EWSR1::ETS fusion itself, include its E3 ligase TRIM8 and the competing transcription factor ETV6, reinforcing the pivotal role of this fusion oncoprotein5,6,7,40. In NB and aRMS, CRCs have been robustly identified30,32,35 and their MTFs are consistently ranked among the strongest dependencies and can be observed as early as 48 h post-silencing in aRMS. However, in Ewing sarcoma, more heterogeneous results have been reported. For instance, silencing of individual MTFs can inhibit proliferation in a subset of Ewing sarcoma lines. Specifically, SOX6 knockdown impedes proliferation in SOX6high lines (RD-ES, TC-32) but not in SOX6low lines (A-673), with effects observed 96 hours post-silencing and confirmed in long-term in vivo experiments23. Similarly, SOX2 expression is highly heterogenous in Ewing sarcoma tumors, and only 20% express the protein at high levels62. SOX2 silencing reduced proliferation within 48 hours in A-673 and RD-ES but showed little effect in other models40,54,63. These results are consistent with a mechanism in which phenotypes correlate with expression levels across diverse cancers rather than being specific to Ewing sarcoma. For example, SOX2 expression strongly correlates with its knockout dependency across all cancer cell types (Pearson correlation between SOX2 log₂(TPM + 1) and SOX2 gene effect (Chronos): −0.506, p = 8.1 × 10−73 64. While this does not rule out functional roles for such TFs in a subset of Ewing sarcoma, it aligns with our findings and the general concept of CRC in defining specific (cancer) entities and vulnerabilities.
A CRC comprising TCF4, NKX2-2, and KLF15 was previously reported in Ewing sarcoma. This CRC was inferred from A-673 ChIP-Seq data, refined using CCLE expression databases, and validated in A-673 and EW-8 cells39. While we also identified these three MTFs in our putative Ewing sarcoma CRC, silencing NKX2-2 in TC-71 or EW-1 cells had little to no effect on short-term proliferation and failed to co-regulate the other MTFs. Differences in methods, timing, or cell lines may explain these discrepancies. However, consistent with our findings, long-term knockout of these three MTFs does not impair proliferation in Ewing sarcoma cell lines40,54. Our results also agree with previous studies showing that acute NKX2-2 silencing does not impair proliferation, although clonogenic growth assays and xenograft experiments reveal a strong phenotype52. Our findings should, however, not be interpreted to suggest that MTFs have no functional role in Ewing sarcoma. Long-term or context-specific assays, such as clonogenic growth, in vivo tumor formation, or studies under stress conditions, may reveal additional dependencies. Future efforts to map Ewing sarcoma dependencies should consider single-cell or spatially resolved technologies, as our bulk analyses may overlook important nuances. It also remains possible that under specific microenvironmental conditions, during metastasis, or within other cancer hallmarks, certain MTFs become more critical. For instance, POU3F2 may play a role in metastasis, particularly in the context of STAG2 loss65. Beyond NKX2-2 and SOX6, whose functions in Ewing sarcoma have already been described and discussed above, the roles of IKZF2, TCF12, and POU3F1 remain to be explored in this cancer.
Our study demonstrates that Ewing sarcoma stands as an exception to the “lineage addiction” paradigm. Rather than engaging with a developmental TF network, the EWSR1::ETS fusion reshapes chromatin by mobilizing a set of GGAA mSats, creating neo-enhancers that drive a neomorphic transcriptional program. Classic auto-regulatory loops involving multiple MTFs do not appear central, as their short-term silencing showed limited impact on Ewing sarcoma cells. Overall, our results suggest that Ewing sarcoma lacks the conventional CRC architecture observed in other pediatric cancers and is instead dominated by a “hegemonic” driver oncoprotein. EWSR1::ETS enforces tumor identity through non-canonical, microsatellite-rich SEs, with minimal reciprocal regulation by MTFs. Despite being still a challenge, our results combined with previous reports support the notion that targeting these dependencies or the microsatellite-based enhancers offer promising therapeutic strategy in Ewing sarcoma.
Methods
Cell culture
The Ewing sarcoma TC-71 cell line was obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ). The EW-1 cell line was obtained from the International Agency for Research on Cancer (IARC). TC-71 and EW-1 were cultured at 37 °C in 5% CO2 using RPMI (Merk-Sigma, R8758) supplemented with 10% fetal bovine serum (Merk-Sigma, F7524). Cells were routinely tested negative for mycoplasma contamination by qPCR (conducted by Eurofins Genomics). For information on other cell lines, refer to the papers by refs. 9,19.
siRNA
The siRNA experiment was performed as described in our previous paper9. Cells were transfected using lipofectamine RNAiMAX (Invitrogen, 13778030) with siCT ON-TARGET plus Non targeting control (Dharmacon, D-001810-01-20) or All-stars negative Control siRNA, (Qiagen, 1027281), siIKZF2 (SMARTpool Dharmacon, L-006946-00-0005), siNKX2-2 (SMARTpool Dharmacon, L-011341-00-0005), siPOU3F1#3 (Qiagen, SI00690221), siPOU3F1#4 (Qiagen, SI00690228), siPOU3F1#5 (Qiagen, SI04166673), siPOU3F1#6 (Qiagen, si04321653), siPOU3F2 (SMARTpool Dharmacon, L-020029-00-0005), siSOX6 (SMARTpool Dharmacon, L-015101-01-0005), siTCF12 (SMARTpool Dharmacon, L-006356-00-0005), siEWSR1-FLI1 fusion type 1 (7/6) for TC-71 or type 2 (7/5) for EW-1 (Qiagen, custom design, see ref. 9). For a 24-well plate, siRNA transfection mix was prepared by adding 1 µL of Lipofectamine RNAiMAX to 50 µL of Opti-Mem (Thermo Fisher, ref 31985062) and combining it for 20 min with 50 µl of Opti-Mem and siRNA mix. This mix was then added to 400 µL of the respective cell media. The final concentration of siRNA was 50 nM in the single knockdown experiments and 25 nM of each siRNA in the dual knockdown experiments. For the siRNA pools targeting five or six genes, 25 nM of each siRNA was used, for total concentrations of 125 and 150 nM, respectively. The siRNA controls were transfected at the same total concentrations (125 or 150 nM, respectively). All experiments were conducted without antibiotics and scaled up when necessary. The cells were harvested after 48 or 72 h post-transfection.
Cell proliferation
EW-1 and TC-71 cell growth was measured using an IncuCyte Zoom (Essen Bioscience). Cells were plated at 25,000 cells/well in a 24-well plate or 50,000 cells/well in a 12-well plate. The cells were imaged every hour using a 4X objective for 2 or 3 days. Cell proliferation curves were generated using the IncuCyte software for confluence analysis (percentage). Cell counts were also performed at the end of the experiment (48 or 72 h) using a Vi-Cell counter (Beckman Coulter) based on cell viability assessed with trypan blue.
RT-QPCR
RT-qPCR was performed following RNA extraction using RNeasy Plus Mini Kit (Qiagen, ref 74134) and reverse transcription was carried out using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, ref 4368814), according to the supplier’s recommendations. Power SYBR Green PCR Master Mix (Applied Biosystems, ref 4367659) was used for RT-qPCRs, and Oligonucleotides were purchased from MWG Eurofins Genomics (Supp. Data 9). Reactions were run on a CFX384 Touch Real-Time PCR Detection System (Bio-Rad) and analyzed using the Bio-Rad CFX Manager 3.1 software.
Western blotting
Cells were trypsinized, counted, washed with ice-cold PBS and lysed in Laemmli buffer (50 mM Tris-HCL, 2.5 mM EDTA, 2.55 mM EGTA, 2% SDS, 5% Glycerol, 1% Bromophenol blue, protease inhibitor cocktail tablets and 2 mM DL-Dithiothreitol solution) at 10 million cells/ml as described in our previous paper9. Nitrocellulose membranes (BIO-RAD, 1704159) were incubated overnight at 4 °C with rabbit anti-POU3F1 (Abcam, ab259952), rabbit anti-POU3F2 (Cell signaling, 12137), and rabbit anti-vinculin (Abcam, ab129002). Membranes were then incubated for 1 h at room temperature with respective anti-rabbit immunoglobulin G horseradish peroxidase (HRP)-coupled secondary antibody (1:3000, NA934, Cytiva). Proteins were visualized using SuperSignal West Pico Plus (Thermo Scientific, 34580) and a ChemiDoc Imaging System (Bio-Rad).
ChIP-Seq
Chromatin Immunoprecipitation (ChIP) experiments were performed using the iDeal ChIP-Seq kit for transcription factors and for histones (Diagenode), following the manufacturer’s instructions and as described in our previous study9. Briefly, Ewing cell lines were fixed for 10 min with 1% methanol-free formaldehyde (28908, Thermo Scientific). Chromatin was sonicated (Bioruptor, Diagenode) for 20 cycles (30 s on, 30 s off) at the “high” setting to generate DNA fragments with an average size of 150–300 bp. The antibodies used for ChIP were Rabbit polyclonal anti-FLI1 (ab15289, Abcam), rabbit polyclonal anti-IKZF2 Helios (A303-618A, Thermo Fisher), rabbit monoclonal anti-POU3F2/BRN2 (12137, Cell Signaling), rabbit polyclonal anti-SOX6 (ab30455, Abcam), and rabbit polyclonal anti-H3K27ac (ab4729, Abcam). For ChIP sequencing, libraries were generated using the TruSeq ChIP library preparation kit (Illumina) and sequenced on an Illumina HiSeq 2500 or NovaSeq 6000 (single-end, 100 bp).
ChIP-seq processing
Sequencing data were processed using the nextflow ChIP-Seq pipeline66. Briefly, reads were mapped with bwa (version 2.19.0)67 to the human genome (version hg19). Duplicates or low-quality reads were removed. Peaks were called with MACS2 (version 2.2.6)68, 2008) using the “narrow” option for transcription factors and the “broad” option for the H3K27ac histone mark. The fragment size was estimated from the data using MACS for transcription factors, while a fixed fragment size of 200 bp was used for histones. ChIP-Seq data were normalized according to their respective input DNA samples. The ChIP-Seq signal tracks were generated by macs2 using the bdgcmp option (and –m FE to compute fold enrichment between the ChIP and the control). Files were converted to binary format (BigWig) using bedGraphToBigWig. ETS (FLI1 and ERG) ChIP-Seq peaks were considered as mSat peaks if they contained at least four consecutive GGAA. Super-enhancers were identified by applying the ROSE algorithm to H3K27ac peaks29. Core regulatory circuits (CRC) were detected using CRCmapper28 with an updated DNA motif database (Jaspar version 2022)69. The top 10 CRCs were considered to avoid overly stringent results. Super-enhancers were annotated according to CRCmapper (closest active genes) and compared using BEDTools70. Super-enhancers overlapping by at least 50% were merged. For the PCA and SE curve, we used the ROSE signal normalized by the maximum value in each sample. For each group, we averaged the normalized signal and ranked the regions to generate the SE curve. We consider only the top 500 SEs in the paper. For IGV visualization, the tracks were normalized relative to the input. For the heatmap, the coverage was normalized by the total number of mapped reads. Transcription factors were annotated using HumanTFDB (version 3.0)71. Dependencies for Neuroblastoma, Rhabdomyosarcoma, and Ewing sarcoma were extracted from DepMap (DepMap, Broad (2023). DepMap 23Q4 Public. Figshare + . Dataset. https://doi.org/10.25452/figshare.plus.24667905.v2). ChIP-Seq tracks were displayed using IGV72. For the differentiation factors (Fig. 6D), transcription factors with known motifs were extracted from various papers34,73,74,75,76,77,78. For each TF, we checked whether the predicted de novo motif (best hit) was similar to the expected motif from the JASPAR database69. For the motif predictions, we used meme-chip79 on the top 5000 peaks (±100 bp around summit positions).
Expression data analysis
For RNA-Seq from Ewing cell lines, reads were aligned with STAR 2.5.380 to the human genome (GRCh37/hg19). The count matrix generated by STAR used the human gene annotation v19 of GENCODE. Genes with fewer than ten reads were excluded. Data were normalized, and differential analysis was performed using the Wald test with DESEQ2 1.20.081. For the published Affymetrix data of inducible models19, the GSE176190 dataset was analyzed using GEO2R. Expression analyses from Ewing tumors, HPA, GTEx and TCGA were processed as described by ref. 18.
Gene dependency analysis
The dataset “CRISPR (DepMap Public 23Q4+Score Chronos)“ was used for gene dependency analyses (DepMap, Broad (2023). DepMap 23Q4 Public. Figshare + . Dataset. https://doi.org/10.25452/figshare.plus.24667905.v2)40,54). We downloaded dependencies enriched in Ewing Sarcoma, Neuroblastoma and Rhabdomyosarcoma. In the paper, we considered only genes with a negative T-statistic (dependency higher in the considered tumor type compared to the control dataset).
Data availability
The H3K27ac ChIP-seq public data used in the paper are indicated in the Supp. Data 1 (extracted from GSE106925, GSE116344, GSE129155, GSE150783, GSE176400, GSE61953, GSE83728, GSE90683, and Encode data). Concerning RNA-seq, public data for A-673/TR/shEF1 (WT vs EWSR1::FLI1-KD, PRJNA521683), A-673 (WT vs EWSR1::FLI1-KD, GSE133228), and MSC vs MSC-EWSR1::FLI1 (GSE150783) were used. The additional ChIP-seq data (H3K27ac, FLI1, POU3F2, SOX6, and IKZF2) and RNA-seq data generated for this study were available on GSE293632.
References
Delattre, O. et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 359, 162–165 (1992).
Grünewald, T. G. P. et al. Ewing sarcoma. Nat. Rev. Dis. Prim. 4, 5 (2018).
Apfelbaum, A. A., Wrenn, E. D. & Lawlor, E. R. The importance of fusion protein activity in Ewing sarcoma and the cell intrinsic and extrinsic factors that regulate it: a review. Front. Oncol. 12, 1044707 (2022).
Franzetti, G. A. et al. Cell-to-cell heterogeneity of EWSR1-FLI1 activity determines proliferation/migration choices in Ewing sarcoma cells. Oncogene 36, 3505–3514 (2017).
Gao, Y. et al. ETV6 dependency in Ewing sarcoma by antagonism of EWS-FLI1-mediated enhancer activation. Nat. Cell Biol. 25, 298–308 (2023).
Lu, D. Y. et al. The ETS transcription factor ETV6 constrains the transcriptional activity of EWS-FLI to promote Ewing sarcoma. Nat. Cell Biol. 25, 285–297 (2023).
Seong, B. K. A. et al. TRIM8 modulates the EWS/FLI oncoprotein to promote survival in Ewing sarcoma. Cancer Cell 39, 1262–1278.e7 (2021).
Riggi, N. et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell 26, 668–681 (2014).
Surdez, D. et al. STAG2 mutations alter CTCF-anchored loop extrusion, reduce cis-regulatory interactions and EWSR1-FLI1 activity in Ewing sarcoma. Cancer Cell 39, 810–826.e9 (2021).
Yu, L., Davis, I. J. & Liu, P. Regulation of EWSR1-FLI1 function by post-transcriptional and post-translational modifications. Cancers 15, 382 (2023).
Brohl, A. S. et al. The genomic landscape of the Ewing sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet. 10, e1004475 (2014).
Crompton, B. D. et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 4, 1326–1341 (2014).
Tirode, F. et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 4, 1342–1353 (2014).
Boulay, G. et al. Cancer-specific retargeting of BAF complexes by a prion-like domain. Cell 171, 163–178.e19 (2017).
Gangwal, K. & Lessnick, S. L. Microsatellites are EWS/FLI response elements: genomic “junk” is EWS/FLI’s treasure. Cell Cycle 7, 3127–3132 (2008).
Guillon, N. et al. The oncogenic EWS-FLI1 protein binds in vivo GGAA microsatellite sequences with potential transcriptional activation function. PLoS ONE 4, e4932 (2009).
Tomazou, E. M. et al. Epigenome mapping reveals distinct modes of gene regulation and widespread enhancer reprogramming by the oncogenic fusion protein EWS-FLI1. Cell Rep. 10, 1082–1095 (2015).
Vibert, J. et al. Oncogenic chimeric transcription factors drive tumor-specific transcription, processing, and translation of silent genomic regions. Mol. Cell. 82, 2458–2471.e9 (2022).
Orth, M. F. et al. Systematic multi-omics cell line profiling uncovers principles of Ewing sarcoma fusion oncogene-mediated gene regulation. Cell Rep. 41, 111761 (2022).
Sheffield, N. C. et al. DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma. Nat. Med. 23, 386–395 (2017).
Grünewald, T. G. P. et al. Chimeric EWSR1-FLI1 regulates the Ewing sarcoma susceptibility gene EGR2 via a GGAA microsatellite. Nat. Genet. 47, 1073–1078 (2015).
Li, J. et al. Therapeutic targeting of the PLK1-PRC1-axis triggers cell death in genomically silent childhood cancer. Nat. Commun. 12, 5356 (2021).
Marchetto, A. et al. Oncogenic hijacking of a developmental transcription factor evokes vulnerability toward oxidative stress in Ewing sarcoma. Nat. Commun. 11, 2423 (2020).
Musa, J. et al. Cooperation of cancer drivers with regulatory germline variants shapes clinical outcomes. Nat. Commun. 10, 4128 (2019).
Koelsche, C. et al. Sarcoma classification by DNA methylation profiling. Nat. Commun. 12, 498 (2021).
Watson, S. et al. Transcriptomic definition of molecular subgroups of small round cell sarcomas. J. Pathol. 245, 29–40 (2018).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Saint-André, V. et al. Models of human core transcriptional regulatory circuitries. Genome Res. 26, 385–396 (2016).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Boeva, V. et al. Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nat. Genet. 49, 1408–1413 (2017).
Durbin, A. D. et al. Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nat. Genet. 50, 1240 (2018).
Gryder, B. E. et al. Histone hyperacetylation disrupts core gene regulatory architecture in rhabdomyosarcoma. Nat. Genet. 51, 1714–1722 (2019).
Gryder, B. E. et al. PAX3-FOXO1 establishes myogenic super enhancers and confers BET bromodomain vulnerability. Cancer Discov. 7, 884–899 (2017).
Pomella, S. et al. Genomic and epigenetic changes drive aberrant skeletal muscle differentiation in rhabdomyosarcoma. Cancers 15, 2823 (2023).
van Groningen, T. et al. Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat. Genet. 49, 1261–1266 (2017).
Danielli, S. G. et al. Single-cell profiling of alveolar rhabdomyosarcoma reveals RAS pathway inhibitors as cell-fate hijackers with therapeutic relevance. Sci. Adv. 9, eade9238 (2023).
Thirant, C. et al. Reversible transitions between noradrenergic and mesenchymal tumor identities define cell plasticity in neuroblastoma. Nat. Commun. 14, 2575 (2023).
Yuan, X. et al. Single-cell profiling of peripheral neuroblastic tumors identifies an aggressive transitional state that bridges an adrenergic-mesenchymal trajectory. Cell Rep. 41, 111455 (2022).
Shi, X. et al. EWS-FLI1 regulates and cooperates with core regulatory circuitry in Ewing sarcoma. Nucleic Acids Res. 48, 11434–11451 (2020).
Dharia, N. V. et al. A first-generation pediatric cancer dependency map. Nat. Genet. 53, 529–538 (2021).
Tirode, F. et al. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 11, 421–429 (2007).
Boulay, G. et al. Epigenome editing of microsatellite repeats defines tumor-specific enhancer functions and dependencies. Genes Dev. 32, 1008–1019 (2018).
Sole, A. et al. Unraveling Ewing sarcoma tumorigenesis originating from patient-derived mesenchymal stem cells. Cancer Res. 81, 4994–5006 (2021).
Kennedy, A. L. et al. Functional, chemical genomic, and super-enhancer screening identify sensitivity to cyclin D1/CDK4 pathway inhibition in Ewing sarcoma. Oncotarget 6, 30178–30193 (2015).
Lin, L. et al. Super-enhancer-associated MEIS1 promotes transcriptional dysregulation in Ewing sarcoma in co-operation with EWS-FLI1. Nucleic Acids Res. 47, 1255–1267 (2019).
Baldauf, M. C. et al. Robust diagnosis of Ewing sarcoma by immunohistochemical detection of super-enhancer-driven EWSR1-ETS targets. Oncotarget 9, 1587–1601 (2018).
Wiles, E. T., Lui-Sargent, B., Bell, R. & Lessnick, S. L. BCL11B is up-regulated by EWS/FLI and contributes to the transformed phenotype in Ewing sarcoma. PLoS ONE 8, e59369 (2013).
Tirado, O. M. et al. Caveolin-1 (CAV1) is a target of EWS/FLI-1 and a key determinant of the oncogenic phenotype and tumorigenicity of Ewing’s sarcoma cells. Cancer Res. 66, 9937–9947 (2006).
Sanchez, G. et al. Alteration of cyclin D1 transcript elongation by a mutated transcription factor up-regulates the oncogenic D1b splice isoform in cancer. Proc. Natl. Acad. Sci. USA 105, 6004–6009 (2008).
Sancéau, J., Hiscott, J., Delattre, O. & Wietzerbin, J. IFN-beta induces serine phosphorylation of Stat-1 in Ewing’s sarcoma cells and mediates apoptosis via induction of IRF-1 and activation of caspase-7. Oncogene 19, 3372–3383 (2000).
Showpnil, I. A. et al. EWS/FLI mediated reprogramming of 3D chromatin promotes an altered transcriptional state in Ewing sarcoma. Nucleic Acids Res. 50, 9814–9837 (2022).
Smith, R. et al. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing’s sarcoma. Cancer Cell 9, 405–416 (2006).
Surdez, D. et al. Targeting the EWSR1-FLI1 oncogene-induced protein kinase PKC-β abolishes ewing sarcoma growth. Cancer Res 72, 4494–4503 (2012).
Tsherniak, A. et al. Defining a cancer dependency map. Cell 170, 564–576.e16 (2017).
Kovar, H. et al. EWS/FLI-1 antagonists induce growth inhibition of Ewing tumor cells in vitro. Cell Growth Differ. 7, 429–437 (1996).
Stoll, G. et al. Systems biology of Ewing sarcoma: a network model of EWS-FLI1 effect on proliferation and apoptosis. Nucleic Acids Res. 41, 8853–8871 (2013).
Garraway, L. A. & Sellers, W. R. Lineage dependency and lineage-survival oncogenes in human cancer. Nat. Rev. Cancer 6, 593–602 (2006).
George, S. L. et al. Stratified medicine pediatrics: cell-free DNA and serial tumor sequencing identifies subtype-specific cancer evolution and epigenetic states. Cancer Discov. 15, 717–732 (2025).
Peneder, P. et al. Multimodal analysis of cell-free DNA whole-genome sequencing for pediatric cancers with low mutational burden. Nat. Commun. 12, 3230 (2021).
Sanalkumar, R. et al. Highly connected 3D chromatin networks established by an oncogenic fusion protein shape tumor cell identity. Sci. Adv. 9, eabo3789 (2023).
Taylor, B. S. et al. Advances in sarcoma genomics and new therapeutic targets. Nat. Rev. Cancer 11, 541–557 (2011).
Sannino, G. et al. Gene expression and immunohistochemical analyses identify SOX2 as major risk factor for overall survival and relapse in Ewing sarcoma patients. EBioMedicine 47, 156–162 (2019).
Ren, C. et al. Inhibition of SOX2 induces cell apoptosis and G1/S arrest in Ewing’s sarcoma through the PI3K/Akt pathway. J. Exp. Clin. Cancer Res. 35, 44 (2016).
DepMap 23Q4 public. figshare https://plus.figshare.com/articles/dataset/DepMap_23Q4_Public/24667905/2 (2023).
Adane, B. et al. STAG2 loss rewires oncogenic and developmental programs to promote metastasis in Ewing sarcoma. Cancer Cell 39, 827–844.e10 (2021).
Servant, N. et al. bioinfo-pf-curie/ChIP-seq: v1.0.4. Zenodo https://zenodo.org/records/10357860 (2023).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Zhang, Y. et al. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 (2008).
Castro-Mondragon, J. A. et al. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 50, D165–D173 (2022).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Hu, H. et al. AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 47, D33–D38 (2019).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Almalki, S. G. & Agrawal, D. K. Key transcription factors in the differentiation of mesenchymal stem cells. Differentiation 92, 41–51 (2016).
Chan, W. C. W., Tan, Z., To, M. K. T. & Chan, D. Regulation and role of transcription factors in osteogenesis. Int. J. Mol. Sci. 22, 5445 (2021).
Goridis, C. & Rohrer, H. Specification of catecholaminergic and serotonergic neurons. Nat. Rev. Neurosci. 3, 531–541 (2002).
Green, J. D. et al. Multifaceted signaling regulators of chondrogenesis: Implications in cartilage regeneration and tissue engineering. Genes Dis. 2, 307–327 (2015).
Rohrer, H. Transcriptional control of differentiation and neurogenesis in autonomic ganglia. Eur. J. Neurosci. 34, 1563–1573 (2011).
van de Peppel, J. et al. Identification of three early phases of cell-fate determination during osteogenic and adipogenic differentiation by transcription factor dynamics. Stem Cell Rep. 8, 947–960 (2017).
Machanick, P. & Bailey, T. L. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697 (2011).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Acknowledgements
We thank H. Kovar and the Childhood Cancer Repository for providing EwS cell lines and G. Boulay and N. Riggi for sending us their data. We thank N. Gruel, C. Thirant, NEH Djerir, C. Pierre-Eugene and all members of the Diversity and Plasticity of Pediatric Tumors and Integrative Functional Genomics of Cancer laboratories for helpful discussions. With regard to funding, this work was supported by Institut Curie, Institut National de la Santé et de la Recherche Médicale (INSERM), Ligue Nationale Contre le Cancer (Equipe labellisée and program Ligue EAC 2020), Institut National du Cancer (PLBIO19-192), Agence Nationale de la Recherche (ANR-10-EQPX-03), Institut Curie Génomique d’Excellence (ICGex), and Société française de lutte contre les leucémies et cancers de l’enfant et de l’adolescent. This project also received support from European funding as follows: ERA-NET TRANSCAN JTC-2011 (01KT1310), TRANSCAN JTC-2014 (TRAN201501238), TRANSCAN JTC-2017 (TRANS201801292), EEC (HEALTH-F2-2013-602856), H2020-lMI2-JTl-201 5-07 (116064—ITCC P4), and H2020-SC1-DTH-2018-1 (SEP-210506374—iPC). We are indebted to the following associations for providing essential support: L’Etoile de Martin, la Course de l’Espoir, M la vie avec Lisa, ADAM, Couleur Jade, Dans les pas du Géant, Courir pour Mathieu, Marabout de Ficelle, Olivier Chape, Les Bagouz A Manon, l’association Enfance & Cancer, Fédération Enfants Cancers Santé, Les Amis de Claire, Un Elan pour Lucas, and Amarape. The laboratory of D.S. acknowledges funding through the Swiss National Science Foundation (10000473) and the Swiss Cancer League (KFS-5422-08-2021-R) The laboratory of T.G.P.G. acknowledges funding through the Barbara and Wilfried Mohr foundation, the PROTECT team supported by the Cancer Grand Challenges partnership funded by Cancer Research UK, the National Cancer Institute, and the European Research Council (ERC CoG 2023 #101122595).
Author information
Authors and Affiliations
Contributions
O.D. and D.S. conceived the study. S.G. performed all bioinformatic analyses. S.Z. performed most of the experiments. M.F.O., K.L., C.L.B., A.S.F., V.P., M.M.A., and D.S. contributed to experiments. V.R. and S.B. were involved in the sequencing part. S.M., M.W., I.J.L., E.B., and T.G.P.G. played an important role in interpreting the results and helped with the discussion. M.F.O., F.C.-A., and T.G.P.G. provided cell line models. S.G., S.Z., O.D., and D.S. wrote the paper and drafted the figures and tables. O.D. and D.S. supervised the study and data analysis. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Grossetete, S., Zaidi, S., Orth, M.F. et al. The hegemonic EWSR1::ETS oncoprotein overrules core regulatory circuitry principles in Ewing sarcoma. npj Precis. Onc. 10, 36 (2026). https://doi.org/10.1038/s41698-025-01226-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41698-025-01226-8