Abstract
The efficacy of immunotherapy in bladder cancer (BCa) is highly influenced by tumor heterogeneity, underscoring the urgent need for mechanistic insights and precise molecular subtyping. Adenosine-to-inosine RNA editing (AIRE), a critical post-transcriptional mechanism driving protein diversity, offers a promising lens for decoding the immune microenvironment in BCa. In this study, we established an integrative framework combining molecular, imaging, and pathological data to identify and validate a novel immune-related BCa subtype. Based on multi-center transcriptomic datasets, we employed NMF, WGCNA, and Lasso algorithms to classify tumors into two distinct immune subtypes characterized by different levels of immune cell infiltration and divergent responses to immunotherapy. Mechanistic validation revealed that ANXA1 and RASAL2 may contribute to immunosuppressive niche formation via the TNF-NFκB-CXCL8 axis, potentially acting as molecular triggers for subtype differentiation. Furthermore, radiomic and histopathological analyses at the imaging and tissue levels supported the molecular findings, providing orthogonal validation of subtype-specific characteristics. Collectively, this study introduces an AIRE-based immune molecular subtype of BCa and systematically characterizes it across multiple dimensions. By achieving a closed-loop from identification to validation and clinical translation, our findings offer a new perspective for patients most likely to benefit from immunotherapy and advancing precision treatment strategies in bladder cancer.

Similar content being viewed by others
Introduction
Worldwide, bladder cancer (BCa) is the most common malignancy carcinoma in the urinary tract. The majority of patients had non-muscle invasive bladder cancer (NMIBC), while others were muscle invasive bladder cancer (MIBC)1. Compared with NMIBC, MIBC has worse clinical outcomes and higher rate of tumor progression2,3. Consequently, it is imperative to find molecular mechanisms and therapeutic targets for BCa’s malignant biology and aggressive progression.
In recent years, immunotherapy—particularly immune checkpoint inhibitors (ICIs)—has emerged as a key strategy for treating advanced bladder cancer, especially in patients who are unresponsive to conventional chemotherapy such as platinum-based regimens4. Agents targeting PD-1/PD-L1 and CTLA-4 have demonstrated durable responses in a subset of patients; however, the overall response rate remains suboptimal5. Therefore, a deeper understanding of the mechanisms underlying ICI efficacy is critical. Several mechanisms have been proposed to explain the limited benefit of ICIs: Insufficient tumor immunogenicity6. Defects in antigen processing and presentation pathways7, and an immunosuppressive tumor microenvironment (TME)8. Among these, the immunosuppressive nature and high degree of heterogeneity within the TME have recently gained increasing attention as key determinants of resistance to immunotherapy. Deciphering the complexity and functional states of the TME is thus essential for improving predictive biomarkers and guiding personalized therapeutic strategies.
TME, a cellular environment that contains tumor cells, is mainly composed of blood vessels, extracellular matrix (ECM), lymphatic vessels, stromal cells etc.9,10. Tumor growth and metastasis are influenced by TME11. Different carcinoma types and locations often produce different TMEs which lead to highly aggressive and motile tumors9,12. Precision medicine in BCa is hindered by high TME heterogeneity13,14. Therefore, an improved theoretical basis for precision medicine can be achieved by clarifying the heterogeneity and characteristics of TME.
RNA editing for specific nucleotides occurs post-transcriptionally without affecting the DNA template15. An RNA editing type of adenosine to inosine (A-to-I) accounts for more than 50% of human RNA editing, in which translation recognizes I as G16. Among metazoans, the Adenosine deaminase (ADAR) enzymes are highly conserved in the A-to-I conversion of RNA17. These enzymes include ADAR1, ADAR2, and ADAR318. Human diseases such as cancer are caused by deregulated RNA editing19,20,21. Thus, the heterogeneity of TME in BCa can be elaborated from the perspective of AIRE.
The development of molecular subtypes will explain the TME heterogeneity and further define precision medicine22. At present, molecular classifications such as TCGA, CIT, Lund, Baylor, and UNC types are well established and well proven23. However, it still faces several problems when it comes to clinical applications like complex detection methods, high costs, and inevitable individual differences24. As a result, more sensitive and specific molecular classifications as well as more efficient and rapid methods are required. In recent years, radiomics has experienced rapid development in the area of medical and has been applied to determine the biological behavior of many types of tumors25,26,27. A promising approach would be to optimize radiological assessment to identify suitable algorithm for oncogenic expression and other prognostic factors. In contrast to the subjective evaluation of imaging characteristics, radiomics provides better objective information and can extract the high-dimensional radiomics features (RFs) of images that cannot be seen with the naked eye. Additionally, it is a non-invasive and preoperative method for assessing tumor heterogeneity. There have been previous studies that used CT/MRI to construct radiomic signatures for biologically relevant behaviors, including lymph node metastasis, muscle-invasive status, prognosis, and tumor stages28. These new RFs have the potential function to in identifying biomarker, malignant progression as well as evaluate molecular subtypes of BCa patients. T2-weighted (T2WI) MRI is applied to detect the location and size of tumor, morphology, and growth pattern in BCa29. Then the dynamic contrast-enhancement (DCE) is also applied to detect the change of NMIBC into the MIBC.
The clinical diagnosis of BCa currently relies heavily on histopathological examination, where pathologists analyze tissue samples using medical microscopes and direct visual assessment. However, certain histopathological patterns may exhibit a degree of ambiguity, potentially leading to misinterpretation30. Traditional immunochemical techniques sometimes prove insufficient in complex differential diagnoses. To address this, automated pathology imaging systems present a practical and effective alternative. By utilizing high-throughput medical image processing, these systems extract valuable high-dimensional data, playing a critical role in advancing precision medicine31,32. Automated pathology imaging systems are increasingly being applied to analyze medical images for various cancers, such as breast cancer33, lung adenocarcinoma34, and skin cancer35. Chen et al.’s study further validates the effectiveness of machine learning-derived pathomic features in diagnosing BCa and predicting patient survival36.
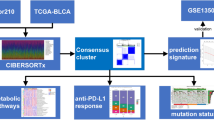
In this study, based on BCa patient cohorts from Shanghai Tenth People’s Hospital (STPH) and multiple public datasets, we identified two novel immune-related molecular subtypes of bladder cancer from the perspective of RNA editing: the “immune-dysfunctional” subtype and the “immune desert” subtype. We validated the differences between these subtypes in terms of prognosis, metastasis risk, chemotherapy response, drug sensitivity, and immunotherapy efficacy. Furthermore, we explored the underlying molecular mechanisms of the “immune-dysfunctional” subtype, focusing on the key genes ANXA1 and RASAL2, which mediate an immunosuppressive tumor microenvironment. Finally, using MRI and H&E-stained pathological images from our in-house BCa cohort, we investigated the imaging and pathological characteristics of the two subtypes and constructed an integrated radiopathomic model to predict immune molecular subtypes, aiming to support precision diagnosis and treatment.
Results
Developing and exploring the characteristics of RE subtypes
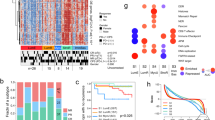
At first, nine prognosis-related AIRE sites were screened by Unicox analysis as shown in the Fig. 1A. Chromosome 1 has the largest number of significant prognostic related ATIRE sites which were shown in the Fig. 1B, C. After the LASSO, the nine ATIRE sites were finally selected for identifying RE subtypes (Fig. 1D and Supplementary Fig. S2A). Then based on the consensus clustering results, the At an optimal grouping, TCGA-BCa patients could be categorized into two molecular subgroups (Cluster A and Cluster B) (k = 2) (Fig. 1E and Supplementary Fig. S2B) Based on the consensus clustering results, TCGA-BCa patients could be optimally grouped into two molecular subgroups (Cluster A and Cluster B) (k = 2) (Fig. 1E and Supplementary Fig. S2B). Kaplan–Meier curve for the TCGA-BCa cohort (samples with missing clinical parameters were removed) showed significant differences between clusters, and patients in subtype B have worse prognostic outcomes (Fig. 1F) (P < 0.001). To explore the characteristics of the two subtypes, we used GSVA to probe the possible molecular pathways involved. The heatmap of immune clusters revealed that Cluster B might involve T cell, B cell receptor signaling and ECM receptor interaction (Fig. 1G). This finding suggests that Cluster B may be associated with immune infiltration and TME, therefore we further verified the difference between the two subtypes in immune cell infiltration. The outcome of the CIBERSORT algorithm suggest that subtype B is significantly higher than subtype A in tumor-infiltrating immune cells (TIICs) (including CD4+, CD8+ T cells, Monocytic cells etc.), while the results of the MCP algorithm are similar to the former (Fig. 1H, I). To investigate the correlation between RE subtypes and classical molecular subtypes, the results of the comparison show that patients in RE subtypes intersected with Basal and Luminal subtypes in the UNC subtyping system (Supplementary Fig. S2C). Basal correlation scores (Ba/Sq-Score) were significantly higher in samples from ClusterB (P < 0.01), while Luminal subtype correlation scores (LumNS-Score, LumU-Score and LumP-Score) were significantly higher in samples from ClusterA (Supplementary Fig. S2D, P < 0.01).
A Univariate Cox regression identified nine prognostic ATIRE sites. B, C These sites were primarily located on Chromosome 1. D LASSO regression was used for feature selection. E Consensus clustering based on the nine ATIRE sites revealed two molecular subtypes (Cluster A and B) in the TCGA-BCa cohort. F Kaplan–Meier analysis showed worse prognosis in Cluster B (P < 0.001). G GSVA analysis highlighted immune-related and ECM signaling pathways enriched in Cluster B. H, I Immune infiltration analysis (CIBERSORT, MCP-counter) showed increased CD4+, CD8+ T cells and monocytes in Cluster B. J, K Venn analysis and WGCNA identified four gene modules; the turquoise module was most correlated with ImmuneScore, subtypes, and T stage. L The turquoise module negatively correlated with immune and clinical features. M, N NMF clustering based on 29 turquoise module genes further identified two distinct clusters (optimal k = 2). Results are presented as mean ± SD. ns indicates P > 0.05; * indicates P < 0.05; ** indicates P < 0.01; *** indicates P < 0.001.
Developing and exploring the RNA editing-related subtypes
RE subtypes were correlated with classical subtypes and immune infiltration. To further establish more precise molecular subtypes, we performed the differential analysis of ClusterA and ClusterB samples. We intersected the differential genes with MIBC and NMIBC differential genes from our center (Supplementary Fig. S2E). And the Venn diagram demonstrated 659 intersected genes (Fig. 1J). Subsequently, we used WGCNA based on intersecting genes to screen the RE-related mRNAs associated with ImmuneScore, the classical subtype and malignant progression. A total of 397 BCa samples were identified for subsequent analysis (Supplementary Fig. S2F). During the downscaling analysis, β = 5 was deemed to be the optimal soft threshold, the gene clustering results suggested that 639 genes were classified into 4 modules (Fig. 1K), with the turquoise module having the strongest correlation with ImmuneScore (R = −0.47 P < 0.001), Basal/Luminal subtypes (R = -0.7, P < 0.001) and T stage (R = −0.22, P < 0.001) (Fig. 1L). Subsequently, based on the 29 genes in the turquoise module, we used NMF clustering to re-cluster the TCGA-BCa cohort (k = 2 is the optimal number of groups) (Fig. 1M, N). The newly identified RE-related subtypes were classified as Cluster1 (C1) and Cluster2 (C2). And the Kaplan–Meier curve showed a worse clinical prognostic outcome for C2 compared to C1. (P < 0.001) (Fig. 2A). Then, we compared the levels of TIICs in the C1 and C2 sets based on three algorithms (ssGSEA, MCPcounter, and CIBERSORT). And the results suggested that the C2 set had higher levels of TIICs (including CD4+, CD8+ T cells, Macrophages cells) with immune function activation levels (Fig. 2B, C and Supplementary Fig. S2I). The subsequent GSVA results provided a reasonable explanation for the high immune system activation status of C2 (Supplementary Fig. S2G). To further investigate the differences in immunotherapy sensitivity between C1 and C2, we compared the difference in the expression of PD-L1 between RE-related subtypes, and the results suggested that PD-L1 were significantly more expressed in C2 samples (P < 0.001) (Supplementary Fig. S2H). Furthermore, in C2 samples, characteristic genes of Basal subtype (CD44, CDH3, STAT3, KRT5 and KRT14) were significantly overexpressed while that of Luminal subtype (KRT20, FGFR3, CD24, PPARG and ERBB2) were significantly downexpressed (Supplementary Fig. S2J).
A Kaplan–Meier survival analysis revealed significantly worse prognosis in Cluster C2 compared to C1 (P < 0.001). B, C Immune cell infiltration analysis using ssGSEA, MCPcounter, and CIBERSORT showed higher levels of TIICs (e.g., CD4+, CD8+ T cells, macrophages) and immune activation in C2. D PCA-based RE score model divided TCGA-BCa patients into high and low RE score groups (n = 197 and n = 198, respectively); heatmap illustrates associations with clinical features. E GO analysis of low RE score signature genes revealed enrichment in T cell activation and intercellular adhesion; KEGG analysis showed ECM receptor interaction, TGF-β, and T cell receptor pathways. F Validation in GSE31684, GSE32894, GSE48075, and STPH datasets showed that high RE score samples had better OS (P < 0.05). G FPS validation in an independent STPH cohort confirmed better prognosis in high RE score samples. H Immunotherapy cohort analysis (IMvigor210, GSE78220, GSE176307) revealed lower OS and response rates in high RE score samples; combined low PD-L1 and high RE score predicted poor outcomes. I In the TCGA-PRAD cohort, patients with low TMB and low RE score had worse OS. Results are presented as mean ± SD. ns indicates P > 0.05;* indicates P < 0.05; ** indicates P < 0.01; *** indicates P < 0.001.
Validating the RE score’s role in immune infiltration and clinical significance
The final TCGA-BCa patients were classified into a low RE score group (n = 198) and a high RE score group (n = 197) by performing PCA on RE-related subtypes and calculating RE scores. Additionally, the heatmap illustrates the correlation between RE clusters and clinicopathological features (Fig. 2D). Moreover, the subtype A (High score group) principal component genes were also identified such as ATAT1, MZF1, NSUN5P1, PLA2G6, TMC7, and ZNF296, and the principal component genes of subtype B (low RE scoring group) included AGER, ASIC1, ANXA1, and RASAL2. The outcome of Unicox analysis shows that the grouping of RE scores, age, T stage, and N stage significantly predicted the survival outcomes of the BCa patients. The RE score was able to quantify the RE clusters as expected. Based on the results of both algorithms, a negative correlation was found between RE scores and TIICs, including CD4+, CD8+ T cells, Treg cells, NK cells and macrophages as well. GO analysis of the principal component genes in the low-scoring group revealed that functions of PCA genes were mainly contain the activation and proliferation of T cells. And they were significantly associated with intercellular adhesion (Fig. 2E). KEGG analysis suggested that the low-scoring group might involve ECM receptor interaction, TGF-β pathway and T cell receptor signaling pathway, which might explain the molecular mechanisms of the above findings (Supplementary Fig. S3A). In addition, RE score was also negatively correlated with bladder cancer malignant progression (T stage and N stage), which was consistent with our previous WGCNA screening results (Supplementary Fig. S3B).
RE scores are a crucial indicator of BCa patient prognosis and immunotherapy effectiveness
GSE31684, GSE32894, GSE48075 and STPH’s data set were used for analysis of the prognosis of BCa samples. From the results, the overall survival (OS) of high RE score samples were better than low RE Score samples (P < 0.05) which suggested that RE score is a key predictor of the prognosis in BCa patients (Fig. 2F). And we also validate the FPS of different RE scores samples in our center’s BCa cohort (Fig. 2G). Subsequently, we performed an immune cell infiltration analysis using our STPH’s data, and the results showed that RE scores were negatively associated with TIICs (including CD8+, CD4+ T cells, and B lymphocytes) (Supplementary Fig. S3C). To investigate the predictive effectiveness of RE score on immunotherapy outcome, we validated it in the BCa immunotherapy cohort (IMvigor210, GSE78220, GSE176307), and the results revealed that the OS and response rate of immunotherapy was lower in the high RE score samples compared with low RE score samples. The OS of the PD-L1 high expression and the low score samples were better than that of the PD-L1 low expression and RE high score samples (Fig. 2H). Based on the TCGA-PRAD cohort, the study found that patients with low PD-L1 and low RE Score had lower overall survival compared to those with high PD-L1 and high RE Score (Supplementary Fig. S3D). Additionally, patients with low tumor mutation load and low RE Score had worse overall survival compared to those with high tumor mutation load and high RE Score (Supplementary Fig. S4A, B). The correlation analysis showed a significant relationship between PD-L1 and RE Score (Supplementary Fig. S4C). This evidence confirms that RE score is an important indicator of response to immunotherapy in BCa patients.
Validation of the principal component genes within subtypes at the cellular level and prediction of RE subtype-sensitive drugs
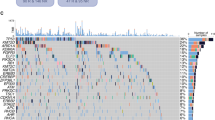
The clinical characteristics of RE subtypes were further verified at the cellular level from TCGA-BCa-cohort. The result suggested that RASAL2, ANXA1 and ASIC1 were highly expressed in tumor tissues compared with normal tissues (Fig. 3A). In the data obtained from our research center, it was observed that the expression levels of RASAL2, ANXA1, and ASIC1 were significantly increased in MIBC samples, while the expression of AGER was significantly decreased in MIBC compared to NMIBC (Fig. 3B). In addition, RT-qPCR verified that RASAL2 and ANXA1 were significantly overexpressed in bladder cancer cell lines (T24, 5637, RT4, U3) (Fig. 3C). The findings from immunofluorescence analysis indicate that RASAL2 and ANXA1 are predominantly localized in the cytoplasmic region (Supplementary Fig. S5A, B). In order to explore the role of RASAL2, ANXA1 in the oncogenic progression of bladder cancer, we utilized siRNA to suppress the expression of mRNAs and assessed the efficacy of knockdown (Supplementary Fig. S5C, D). The result of IHC in bladder cancer patients indicated a significant upregulation of RASAL2, ANXA1 in high-grade bladder cancer in comparison to low-grade bladder cancer (Fig. 3D). The findings of the EdU and colony formation assay indicated that the proliferation of bladder cancer cell lines (T24 and 5637) was suppressed upon knockdown of RASAL2, ANXA1 (Fig. 3E, F). Additionally, the results of the Transwell, 3D-Transwell and Wound healing assays demonstrated that the invasion and migration of bladder cancer cell lines (T24 and 5637) were impeded by the knockdown of RASAL2, ANXA1 (Fig. 3G, H and Supplementary Fig. S5E). These observations provide further evidence for the involvement of RASAL2 and ANXA1 in facilitating the progression of bladder cancer. Correlation analysis demonstrates the correlation between RE Score and A to I RNA editing sites (Supplementary Fig. S6A). The prediction results for RE subtype sensitivity suggested that high RE scores samples was more sensitive to Dasatinib, PLX-4720, AZD8005 drugs whereas low RE scores samples was sensitive to Navitoclax, Wee1-inhibitor, and Vorinostat drugs (Supplementary Fig. S6B). We then analyzed the commonly used drugs for bladder cancer using different algorithms, and the results suggested that low RE scores samples was more sensitive to gemcitabine, cisplatin, docetaxel, and paclitaxel drugs (Supplementary Fig. S6C). All the experiments were statistically significant with P < 0.05.
A, B RASAL2, ANXA1, and ASIC1 were upregulated in tumor tissues and MIBC samples; AGER was downregulated in MIBC. C RT-qPCR confirmed overexpression of RASAL2 and ANXA1 in bladder cancer cell lines. D IHC showed higher RASAL2 and ANXA1 expression in high-grade vs. low-grade tumors. E, F EdU and colony formation assays showed reduced proliferation after RASAL2 and ANXA1 knockdown. G, H Transwell and 3D-Transwell assays revealed impaired invasion and migration upon gene knockdown. Results are presented as mean ± SD. ns indicates P > 0.05; * indicates P < 0.05; ** indicates P < 0.01; *** indicates P < 0.001.
The principal component genes RASAL2 and ANXA1 potentially promote immunosuppression in bladder cancer via the TNF-NF-κB-CXCL8 signaling axis
To investigate the effects of principal component genes on CD8+ T cells, experiments were conducted in vitro. BCa cells (T24 and 5637) with RASAL2 and ANXA1 knocked down were co-cultured with CD8+ T cells. The proportion of remaining tumor cells after co-culturing was detected by flow cytometry. The results showed that fewer BCa cells survived after co-culture when RASAL2 and ANXA1 were knocked down (Fig. 4A, B). Meanwhile, the number of CD8+ T cells migrating to the lower chamber increased (Fig. 4C). To explore the pathways through which principal component genes affect the function of CD8+ T cells, GSEA hallmark analysis was performed on these genes. The analysis indicated that both RASAL2 and ANXA1 were enriched in the TNF signaling pathway (Fig. 4D). To further elucidate the mechanisms by which RASAL2 and ANXA1 regulate the tumor immune microenvironment, a screening of chemokines associated with these genes was performed using publicly available datasets. Among the candidates, CXCL2, CXCL11, and CXCL8 were found to be significantly and positively correlated with both RASAL2 and ANXA1 expression (Fig. 4E). To validate these associations, the expression levels of the identified chemokines were examined following knockdown of RASAL2 and ANXA1 in bladder cancer cell lines. Quantitative PCR analysis revealed that CXCL8 expression was significantly decreased in both T24 and 5637 cells after gene silencing (Fig. 4F). Consistent with these results, enzyme-linked immunosorbent assay (ELISA) confirmed a marked reduction in CXCL8 protein secretion upon knockdown of RASAL2 and ANXA1. To determine whether RASAL2 and ANXA1 modulate CXCL8 expression via the TNF-NF-κB signaling axis, rescue experiments were conducted. Western blot analysis demonstrated that TNF stimulation restored the nuclear translocation of p65 and p50, which was impaired following knockdown of RASAL2 and ANXA1. Furthermore, ELISA results indicated that TNF treatment could reverse the downregulation of CXCL8 induced by the knockdown of these genes (Fig. 4G, H). Taken together, these findings support the hypothesis that RASAL2 and ANXA1 positively regulate CXCL8 expression through the TNF–NF-κB pathway, thereby contributing to an immune-dysfunctional tumor microenvironment by inhibiting CD8+ T cells migration and cytotoxic function in bladder cancer (Fig. 4I).
A, B Flow cytometry showed enhanced CD8⁺ T cell-mediated killing of BCa cells (T24, 5637) after RASAL2 and ANXA1 knockdown. C Transwell assays revealed increased CD8⁺ T cell migration toward knockdown BCa cells. D GSEA indicated enrichment of RASAL2 and ANXA1 in the TNF signaling pathway. E Correlation analysis identified CXCL8, CXCL2, CXCL11, and CXCL8 as chemokines positively associated with RASAL2 and ANXA1. F qPCR and ELISA confirmed reduced CXCL8 expression and secretion upon knockdown in T24. G, H TNF stimulation restored NF-κB nuclear translocation and rescued CXCL8 expression in knockdown T24 cells. I The schematic summary of the proposed mechanism is drawn by Figdraw: RASAL2 and ANXA1 promote CXCL8 expression via TNF–NF-κB signaling, impairing CD8⁺ T cell function in the tumor microenvironment. Results are presented as mean ± SD. ns indicates P > 0.05; * indicates P < 0.05; ** indicates P < 0.01; *** indicates P < 0.001.
Predicting RE-related subtypes by constructing a radiomics model
A total of 3562 radiomic features were extracted from T2WI and axial DCE sequences, with 1781 features from each sequence. The intra- and inter-observer ICCs ranged from 0.762 to 0.908, indicating good reproducibility. The top 40 features was selected by the mRMR algorithm (Fig. 5A). Subsequently, the least absolute LASSO regression was applied, and 9 features with non-zero coefficients were identified for the construction of the LASSO signature and correlation plots show the correlation of radiomic features with RE scores (Fig. 5B and Supplementary Fig. S7). The LASSO model showed favorable predictive performance, with an AUC of 0.79 in the training set and 0.75 in the validation set (Fig. 5C). It also demonstrated high sensitivity and specificity in identifying RE-related subtypes, supporting its utility as a reliable predictive model (Fig. 5D). Further validation was performed using confusion matrices based on our institutional patient cohort, indicating good classification accuracy in both training and validation sets (Fig. 5E, F). To further investigate imaging differences between subtypes, tumor morphology was manually delineated. Results revealed that tumors in the high RE score subgroup exhibited an exophytic growth pattern, resembling narrow-based papillary tumors, whereas tumors in the low RE score subgroup showed an endophytic pattern, similar to wide-based flat tumors (Fig. 5G, H).
A The top 40 radiomic features were selected using the mRMR algorithm from a total of 3562 features extracted from T2WI and axial DCE sequences. B LASSO regression identified 9 features with non-zero coefficients for signature construction. C The LASSO model demonstrated favorable predictive performance with an AUC of 0.79 in the training set and 0.75 in the validation set. D The radiomic model showed high sensitivity and specificity in distinguishing RE-related subtypes. E, F Confusion matrices confirmed accurate classification in both training and validation cohorts. G, H Tumor morphological differences between subtypes were observed: high RE score tumors exhibited exophytic, papillary-like patterns, while low RE score tumors displayed endophytic, flat-like patterns.
Pathomics combined with radiomics for the joint prediction of immunotherapy efficacy in RE subtypes
To gain deeper insights into the pathological characteristics and to predict different RE subtypes in BCa patients, we developed an integrated pathomics model based on H&E-stained tumor sections from our institutional cohort. The RetCCL model was employed to extract deep learning features from the WSIs of each sample (Fig. 6A). After performing PCA dimensionality reduction on these features, we found that an SVM model based on six PCA components showed competitive performance in predicting the RE subtypes, with an accuracy of 0.788 and 0.784 in the training and validation datasets, respectively (Fig. 6B, C). The AUC values for the training and validation datasets were 0.87 and 0.81, respectively (Fig. 6D). Waterfall plots and confusion matrices further demonstrated the performance of this pathomics-based SVM model in predicting RE subtypes (Fig. 6E, F). Histopathological examination revealed distinct morphological differences between the two subtypes: patients with high RE scores exhibited densely clustered tumor cells, while those with low RE scores had a greater presence of peritumoral stromal components, including fibrous connective tissue and immune cell infiltration (Fig. 6G).
A H&E-stained WSIs were analyzed using the RetCCL model to extract deep learning features from the WSIs. B, C The pathomics-based SVM model achieved high accuracy, sensitivity, specificity, PPV, and NPV in both training and validation cohorts. D ROC curves showed AUCs of 0.87 and 0.81 for the training and validation sets, respectively. E, F Confusion matrices demonstrated reliable classification of high and low RE score subtypes. G Representative histopathological images revealed distinct morphology: high RE score tumors showed dense tumor cell clusters, while low RE score tumors exhibited more stromal infiltration. H Schematic overview of the integrated pathomics–radiomics model incorporating treatment response–related parameters. I, J The integrated model showed improved predictive performance (AUC = 0.89 in training and 0.85 in validation) with enhanced diagnostic metrics compared to the pathomics model alone.
To further improve the accuracy of predicting RE subtypes, we combined the pathomics-based SVM model with a radiomics-based LASSO model to construct a combined pathomics–radiomics model (Fig. 6H). This integrated model demonstrated superior predictive performance in both the training and validation cohorts, showing consistently high accuracy, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and AUC (Fig. 6I–K). The integrated model provided enhanced predictive capability for RE subtype classification, offering valuable guidance for subsequent precision therapy
Constructing a nomogram to predict prognosis of ICI treatment for bladder cancer
In order to translate the RE subtypes clinically, we constructed nomogram based on TCGA-BCa set with IMvigor210 respectively. Multivariate and univariate Cox regression were used to screen for clinical characteristics (Supplementary Fig. S8A, B), and the nomogram created based on the grouping of RE scores, age, N stage, and T stage predicted at 1-year versus 3-year OS. The obtained actual C-index was 0.83 ± 0.13, indicating that the predicted OS matched the actual. Finally, the calibration curve with the DCA curve showed the good utility of Nomogram in predicting OS in BCa patients (Supplementary Fig. S8E). Similarly, we produced a Nomogram consistent with predicting free PFS for BCa patients after immunotherapy. After univariate and multifactorial Cox regression screening, we included subgroups of RE scores, the presence or absence of cisplatin treatment, and the Baseline-ECOG-Score in the model (Supplementary Fig. S8C, D). The test results showed a Nomograms C-index of 0.815, indicating that the predicted PFS matched the actual. The calibration chart and DCA curve support the above conclusion (Supplementary Fig. S8F).
Disscussion
Despite advances in BCa treatment, effective options for advanced disease remain limited beyond radical cystectomy. Immunotherapy has shown promise, but significant individual variability hampers its widespread application37. Tumor heterogeneity, particularly within the TME, is a major cause of differential responses38,39,40. A-to-I RNA editing, a key mechanism driving protein diversity, may shape the TME and contribute to such heterogeneity41. Thus, characterizing TME features from the perspective of RNA editing offers novel insights into BCa biology and supports the development of precision immunotherapeutic strategies.
TME heterogeneity is well reflected by existing molecular subtyping42,43. There are, however, several issues that may limit its clinical application, including the following, such as complex detection methods, high costs, and inevitable individual differences. We developed a new molecular subtyping system based on RNA editing was first established that accurately predicts BCa immunophenotypes, treatment possibilities, and clinical outcomes. In addition, we established a RE score to identify RE-related subtypes in patients and modeled the prediction of this score by RFs derived from MRI, leading to clinical translation. The RE score stratified the basal and luminal subtypes to some extend corresponding to their clinical features. The validation confirmed the feasibility of the novel subtypes. It is also important to note that the RE score predicts the clinical response of BCa patients to different treatment regimens, such as chemotherapy, targeted therapy, and immunotherapy. The Basal subtype has low urogenital differentiation, as reflected by low RE scores. RB1 mutations are highly common in the group with low RE scores, suggesting that neoadjuvant chemotherapy might benefit them. The results confirmed that this subtype is sensitive to chemotherapeutic agents commonly used in BCa. Interestingly, several immune activation pathways were enriched in the low RE subgroup (Cluster2), which may explain the inflammatory phenomenon of TME in this subtype. Furthermore, we found the low RE score group had an inflammatory phenotype, which may lead to increased sensitivity to immunotherapy that was later validated in the subsequent immunotherapy set. Overall, the use of RE scores to distinguish specific molecular subtypes can guide a precision therapy.
The RE editing process has been shown to activate an anti-tumor immune response, resulting in an inflammatory phenotype in the TME of patient44,45,46. In the present study, RE-related subtypes and RE scores reflect many of the hallmarks of immunity in BCa. The pathway enrichment results suggested that RE-related subtype (ClusterB) were significantly enriched in TGF-β, activation of T lymphocytes, B lymphocytes and EMT-related pathways. The main component gene function of the low RE score group was mainly enriched in the proliferative regulation and intercellular adhesion of T cells. The TGF-β pathway has shown to regulate the TME and alter ICB sensitivity47,48, revealing that the RNA editing results in the low RE score group could affect the proliferation and activation of T and B lymphocytes. Consistent with this, the assessment of immune cell infiltration and activation suggested that RE scores were negatively correlated with CD4+, CD8+ T cells, NK cells, macrophage infiltration and immune-related responses, such as promoting inflammation, T cell activation, immune checkpoint activation, and immune response. Therefore, we were able to consider a low RE score (Cluster2) to indicate an inflammatory phenotype. The inflammatory phenotype is more sensitive to immunotherapy49,50. Similarly, we found that RE scores were negatively correlated with immune checkpoints. Furthermore, we confirmed the role of RE scores in several ICB treatment cohorts such as IMvigor210, where, as expected, the lower the RE score, the more sensitive the ICB treatment. Therefore, RE score may be a potentially important predictor of ICB treatment in BCa patients.
Interestingly, although the RE-low subtype displayed a higher degree of immune cell infiltration, its clinical prognosis remained poor. This paradox can be explained by the presence of an “exhausted” immune phenotype, in which infiltrating CD8⁺ T cells are functionally impaired, characterized by diminished cytotoxicity and sustained expression of immune checkpoints such as PD-1, TIM-3, and LAG-351,52. To elucidate the molecular basis of this phenotype in the RE-low subtype, we identified ANXA1 and RASAL2 as key genes significantly upregulated in bladder cancer tissues and cell lines. Both genes have been reported to promote tumor progression and immune evasion: ANXA1 upregulates PD-L1 to facilitate immune escape in breast cancer, lung cancer, and melanoma53, and shapes an immunosuppressive microenvironment via Treg recruitment in pancreatic cancer54. RASAL2 is strongly linked to poor prognosis and drug resistance in gastric and colorectal cancers55,56. Pathway analysis further suggested that ANXA1 and RASAL2 may cooperatively activate the TNF-NF-κB pathway, a well-known driver of immunosuppressive TME. NF-κB activation facilitates the recruitment of MDSCs and Tregs through chemokine induction, promotes chronic inflammation, and impairs effector T cell responses, ultimately enabling immune escape57,58. Moreover, chemokine correlation analysis revealed a strong positive association between ANXA1/RASAL2 and CXCL8. CXCL8 plays a dual role in immune regulation. In the early stages of acute inflammation or infection, CXCL8 clears pathogens and repairs damaged tissues by activating various pathways through its receptors CXCR1 and CXCR259,60. For instance, it activates the NF-κB pathway to recruit immune cells such as neutrophils and monocytes to the infection site. Additionally, it activates the MAPK pathway, further enhancing the inflammatory response and promoting immune cell migration. The PI3K/Akt pathway is also activated, supporting immune cell survival and function, thereby boosting their pathogen-fighting abilities and enhancing local immune responses. In the tumor microenvironment, CXCL8 recruits immunosuppressive cells such as MDSCs and Treg through CXCR1 and CXCR2. This occurs while activating the TNF-NF-κB, JAK-STAT, and PI3K-AKT pathways, promoting the accumulation of immune-suppressive cells, inhibiting CD8+ T cell proliferation and cytotoxicity, and facilitating tumor immune evasion59,61. However, in the bladder cancer RE-low subtype examined in this study, CXCL8 is upregulated via the TNF–NF-κB pathway through ANXA1/RASAL2. This upregulation subsequently recruits MDSCs and Tregs, induces CD8+ T cell exhaustion, and exhibits clear immunosuppressive functions. NF-κB has been demonstrated to directly regulate CXCL8 transcription62,63. Our subsequent western blot and ELISA analyses confirmed that ANXA1 and RASAL2 enhance CXCL8 expression through TNF-NF-κB signaling, suggesting that both factors promote immunosuppressive cell recruitment and CD8⁺ T cell exhaustion, ultimately facilitating tumor immune evasion.
Radiomics is a novel field that acquired imaging features from medical imaging to predict cancer biology or essential markers25,26,64.It has been reported that RFs can be applied to assess the expression level of essential markers in breast cancer65, lung cancer66,and hepatocellular carcinoma67. Radiomics has been proven useful in prediction of biological behaviors in BCa according to MRI. To date, the MRI based radiomics signature has been developed for the first time to predict BCa molecular subtypes. To evaluate a tumor comprehensively, in comparison to CT scans, MRIs provide different functional parameters, orientations, and angles68. It has been found that T2WI is more accurate at evaluating biological form of the tumor. The previous study shows that T2WI-based radiomic method can use to predict Ki67 expression in thyroid cancer69 and bladder cancer70. Therefore, previous results provide a fundamental theoretical basis for our prediction of RE score based on T2WI imaging histological features. Based on the LASSO model with nine features in our study, four T2WI-based features and five DCE-based features were used in this study respectively. Furthermore, we demonstrated how wavelet filters can reveal multiple levels of tumor biology in the LASSO model with five of nine RFs wavelet-filtered71. In comparison to radiologists’ subjective evaluations of tumor behavior and low-dimensional radiomics, wavelet-filtered features have more information about tumor heterogeneity and biological behavior72,73. The validation results showed that the LASSO model not only performed well in predicting REscore, but also became an important tool for predicting the survival outcomes of BCa patients when combined with Nomogram. In conclusion, the use of radiomic features capable of predicting RE scores may help in treatment decisions in clinical practice and achieve better clinical translation.
In recent years, pathomics has made significant advances in extracting image features related to BCa and applying them to predict patient recurrence and survival outcomes. Chen et al. employed pathomics to enhance the diagnostic accuracy of BCa and utilized the neutrophil-to-lymphocyte ratio to predict the survival of BCa patients36,74. Wang et al. established a deep learning-based pathomics model to predict early recurrence of NMIBC75. In the current study, we utilized pathomics to extract features from grouped patients, achieving an AUC value of 0.81 (95% CI: 0.67–0.96). To better predict the efficacy of immunotherapy for patients, we integrated radiomics with the results of tumor tissue feature extraction from pathomics. Radiomics analysis of tumor morphology revealed that in high-risk individuals, tumors predominantly exhibited an exophytic solid mass growing into the bladder lumen, characterized as “cold tumors” in immunotherapy. In contrast, low-risk individuals had tumors with a predominantly flat morphology, classified as “hot tumors” in immunotherapy. Pathomics results showed that the histological sections of high-risk individuals were almost entirely composed of tumor cells, while those of low-risk individuals were rich in immune cells.
Of course, this study had several drawbacks. First, the sample size of our center cohort is limited. Second, the clinical information of our center cohort is not perfect and lacks information on specific treatments received by patients, and further follow-up is needed to improve the information of this cohort. Third, the specific changes in transcriptomics due to RNA editing were not validated in vivo and need further exploration in the future.
In conclusion, novel molecular subtypes of BCa based on adenosine to inosine RNA editing can reflect the biology of TME in BCa and provide a new perspective for precision medicine. The RE score can quantify RNA editing-related subtypes and stratify the survival outcomes and ICB treatment sensitivity of BCa patients. The radiomic features based on T2WI sources can predict the RE score and help to achieve clinical application.
Methods
Patient cohorts
This retrospective cohort study integrated transcriptomic, radiomic, and pathologic analyses. A total of 111 patients diagnosed with BCa by board-certified pathologists at Shanghai Tenth People’s Hospital (STPH) between November 2019 and July 2021 were included. The inclusion criteria were as follows: (1) patients who underwent radical transurethral resection or cystectomy; (2) histologically and immunohistochemically confirmed diagnosis of BCa; (3) preoperative multiparametric MRI performed within 20 days prior to surgery; (4) availability of complete clinical and pathological information; and (5) high-quality H&E-stained whole-slide images (WSIs), with some patients potentially contributing multiple WSIs. Exclusion criteria included: (1) poor-quality MRI images; (2) patients who received radiotherapy or chemotherapy before MRI examination; (3) unclear tumor boundaries; (4) WSIs without identifiable BCa lesions; and (5) WSIs with low scanning quality or substantial missing values in extracted features.
Transcriptomic data acquisition
Formalin-fixed, paraffin-embedded (FFPE) tissue samples were obtained BCa cases at STPH. Total RNA was extracted using the RNeasy FFPE Kit (Qiagen), followed by ethanol elution. Paired-end libraries were constructed from 100 ng/mL of total RNA using the ABclonal RNA-seq Library Preparation Kit and sequenced on the Illumina NovaSeq 6000 platform.
Radiomic data acquisition
All MRI scans were obtained in Digital Imaging and Communications in Medicine (DICOM) format for further analysis
Pathologic data acquisition
All slides were scanned at 40× magnification using an automatic digital slide scanner (KF-PRO-120/005, KFBIO Co., Ltd.) and stored in svs format.
Public datasets and data integration
The overall workflow of this study is illustrated in Supplementary Fig S1. RNA editing data for TCGA-BCa were downloaded from Synapse. Transcriptomic datasets were collected from The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov), comprising 411 tumor and 19 normal urothelial carcinoma samples, as well as from GEO datasets GSE32894, GSE31684, GSE48075, GSE176307, and GSE78220 (https://www.ncbi.nlm.nih.gov/geo). Clinical data and tumor mutational burden (TMB) information were obtained from the UCSC Xena platform (http://genome.ucsc.edu). The IMvigor210 cohort, which includes transcriptomic data from 348 urothelial carcinoma patients treated with immune checkpoint blockade (ICB) therapy, was obtained from http://research.pub.Gene.com/IMvigor210orebiologies/. Immunohistochemistry and immunofluorescence data were also collected from the Human Protein Atlas (HPA; https://www.proteinatlas.org). Incomplete data entries were excluded from the analysis. This study was approved by the Ethics Committees of STPH (Approval No. 24KT68).Written informed consent was obtained from all participants.
MpMRI-based radiomics features extraction and selection
The segmented region of interest (ROI) was read by experienced radiologists. The ITK-SNAP software (The number of version: 3.6.1) was applied to determine the boundaries of the tumor on the DCE images and T2WI images. In the evaluation, the areas of necrosis and vessels were excluded. The largest BCa tumor size was selected for feature extraction for patients with multiple BCa tumors76. In order to establish the volume of interest (VOI), ROIs were stacked up from each patient. To confirm feature extraction consistency, the radiologists repeated screening and segmentation of 40 randomized BCa patients. For data homogeneity, all DICOMs were normalized and resampled to the same resolution before radiomic features extraction. PyRadiomics (http://PyRadiomics.readthedocs.io/en) is used to extract four types of RFs (including shape, wavelet, and textural). As a final step, the delay phase of DCE and axial T2WI images were used to extract 3562 radiomic features.
All eligible cases were randomly assigned to a training set and a validation set in a 7:3 ratio. Before the construction of the signature, these RFs extracted above were standardized by Z-score([(x – μ)/σ], x represents the rRF value, μ represents the mean of RF values, and σ represents the corresponding standard deviation estimated from the training cohort only. The same μ and σ were then applied to the validation set. Inter- and intraclass correlation coefficients were calculated on the training set to evaluate the consistency of feature extraction among intra- and inter-observers. The minimum redundancy maximum relevance (mRMR) algorithm was used on the training set to obtain the relevant and non-redundant RFs.
Deep learning-based pathological features extraction
In terms of pathology, the patch extraction, color normalization, and feature extraction can be referenced from the research team’s previous literature77.In this study, valid patches were resized to 224 × 224 pixels and normalized using Reinhard color normalization before being input into the RetCCL model, a pre-trained ResNet-50 model78. The fully connected layer of the ResNet-50 was replaced with an identity layer to extract 2048-dimensional patch-level feature vectors. At the WSI level, the patch-level feature vectors were aggregated to form WSI-level features. For each of the 2048 features, seven statistical measures were computed: mean, median, interquartile range (IQR), standard deviation, range, first quartile (Q1), and third quartile (Q3), resulting in a total of 2048 × 7 = 14,336 features for each WSI, which were used for downstream analysis.
Construction and evaluation of the pathology-based model and integrated model
The deep learning-based pathological features were standardized. Principal component analysis (PCA) was used to reduce the feature dimensions, and support vector machine (SVM) models were optimized through a grid search. The grid search spanned a range of PCA components (5–20), SVM kernels (linear, rbf, sigmoid), regularization parameters (C = 0.1, 1, 10), and kernel-specific parameters (gamma = scale, auto; coef0 = 0.0, 0.1, 0.5, 1.0). Each combination was evaluated using area under the receiver operating characteristic curve (AUC) and accuracy on the validation set. The model with the highest validation AUC, and accuracy in case of ties, was selected as the best model. All analyses were performed using Python (version 3.1) with scikit-learn (version 0.24). Radiomics and pathological risk scores were combined using logistic regression to predict RE subtype. The logistic regression model was developed using the training set and evaluated on both the training and validation sets.
Evaluation of the level of immune infiltration
Basing on the TCGA-BCa cohorts, the estimated score and immune score were constructed by “ESTIMATE” and “ssGSEA” (R packet). Furthermore, MCPcounter was also applied to evaluate the immune cell infiltrate for validation. CIBERSORT deconvolution was used to determine the immune response activation, infiltration of immunocyte subtypes, and immunocyte subtype composition of BCa samples. PD-L1 and Microsatellite Instability (MSI)-related genes expression were analyzed in different subtypes to verify the effectiveness of immunotherapy.
Consensus cluster
On the basis of mRNA expression levels in BCa patients from TCGA-BCa, an unsupervised consistent clustering analysis was conducted. Clusters were obtained by the R package “consensusClusterPlus” and tested with 100 replicates with pltem = 0.8. A nonsupervised NMF clustering method was performed on the metadata set using the “NMF” package. The best cluster type was determined by the coexistence correlation coefficient K = 2. Principal component analysis (PCA) was applied to evaluate molecular clusters’ ability to discriminate different subtypes and construct RNA editing Score. The formula was shown as follow:
S represent the expression level of RE-related subtypes principal component genes.
Identifying the classical molecular subtypes of BCa and screening for subtype-specific sensitive drugs. This study applied the Consensus MIBC and BCa subtyping R packages to subtype BCa. In addition, we extracted and predicted BCa-related drugs through the DrugBank database and the “Oncopredict” R packages to further analyze the effect of RE subtypes and RE Score in predicting treatment opportunities.
Cell culture
BCa (T24, 5637, J82, UMUC3, RT4) cell lines and normal human bladder epithelial (SV-HUC-1) were obtained from the Chinese Academy of Sciences. RPMI-1640 (Gibco, USA) mixed with 10% fetal bovine serum and 1% penicillin/streptomycin was used to culture BCa cell lines. The SV-HUC-1 cell line was cultured in a defined Kerationocyte-SFM (Invitrogen, USA) containing 1% keratinocyte growth supplement. All cells were cultured at 37 °C and 5% CO₂.
Isolation and culture of CD8+ T cells
Peripheral blood mononuclear cells (PBMCs), isolated from healthy donor whole blood by Ficoll density-gradient centrifugation, were activated for 48 h in 24-well plates pre-coated with 1 μg/ml CD3/CD28 antibodies. Activated T cells were subsequently cultured in X-VIVO medium supplemented with 3% fetal bovine serum, 1% penicillin–streptomycin (Beyotime, Shanghai), 10 ng/ml recombinant human IL-7, and 5 ng/ml recombinant human IL-15. CD8+ T cells were stained with anti-CD8 and anti-CD3 antibodies and purified by FACS Aria III; the sorted cells were maintained in the same X-VIVO complete medium.
Flow cytometry for assessing T cell-mediated tumor cell killing
Tumor cells (1 × 10⁵ per well) were seeded into 12-well plates with 1.5 mL of medium and allowed to adhere overnight. T cells were added at a 1:1 effector-to-target (E:T) ratio, and the volume was adjusted to 2.5 mL. After 24 h of co-culture, suspended T cells were collected by PBS washing and centrifugation at 1800 rpm for 5 min. Adherent tumor cells were harvested by centrifugation at 800 rpm for 5 min. All collected cells were resuspended in 50 μL PBS. Surface staining for CD3 was performed using an APC-conjugated antibody (50 μL) to identify T cells. The GFP signal was used to assess the remaining tumor cells. After 30 min of staining at 4 °C, cells were washed, fixed with 1% paraformaldehyde, and analyzed by flow cytometry.
Quantification of CXCL8 via ELISA
CXCL8 concentrations were measured using a commercial ELISA kit (HUABIO, EH0014, China) according to the manufacturer’s instructions. Samples and standards were loaded into 96-well plates pre-coated with anti-human CXCL8 monoclonal antibodies and incubated at room temperature for 2 h. After washing, biotin-labeled detection antibodies were added, followed by a 1-h incubation. Streptavidin-HRP conjugate was then applied for 30 min. Subsequent addition of TMB substrate enabled color development in the dark for 15 min. The reaction was stopped using 2N sulfuric acid, and absorbance was recorded at 450 nm using a microplate reader. A standard curve was generated from known recombinant CXCL8 concentrations, and sample values were interpolated accordingly. Each sample was tested in duplicate, and average values were reported.
CD8⁺ T cell chemotaxis via transwell migration assay
Transwell migration assays were conducted using 24-well low-attachment plates and 5-μm-pore inserts. Inserts were pre-equilibrated with 200 μL and 600 μL of serum-free medium in the upper and lower chambers, respectively, and incubated at 37 °C for 1–2 h. CD8⁺ T cells were prepared, counted, and resuspended to a final density of 5 × 10⁷ cells/mL. After removing the pre-equilibration medium, 200 μL of the T cell suspension was placed in the upper chamber, while tumor cell supernatants were added to the lower chamber as chemoattractants. After 2 h of incubation, cells that migrated to the lower chamber were collected, centrifuged at 1800 rpm for 5 min, and stained with 20 μL of crystal violet. Following a 30-min staining period, 5 μL of the suspension was mounted on slides, covered with coverslips, and imaged under a microscope.
Data processing software
The Flow chart shown in the Supplementary Fig 1. R version 4.2.0 (Action of the Toes) with Strawberry Perl version-5.30.0 (64-bit) (URL: https://www.perl.org/) was used in this study. All R packages were sourced from public websites and downloaded from Bioconductor (URL: http://www.bioconductor.org/). All statistical and data processing software is open source and can be downloaded at the URL provided above.
Identification of mRNAs with differential expression
EDGER R package (http://bioconductor.org/package/EDGER/) wasapplied to screen the differential expressed genes (DEGs) between normal and tumor samples in the TCGA-BCa as well as between MIBC and NMIBC samples from our center. After normalization of the raw data, the differences of genes expression between two types of samples were analyzed with false discovery rate (FDR) < 0.01 and log2 |fold change|>1. Prognosis-related genes were selected according to tumor invasiveness (NMIBC vs. MIBC) and RE-related genes via Venn diagram.
Weighted gene co-expression network approach
Based on the “WGCNA” R package v1.67, gene co-expression networks were constructed based on the Basal/Luminal grouping, Immune score, and T-stage. First, To calculate Pearson correlation coefficients, all paired genes with similar co-expression were compared on the basis of their co-expression importance. The appropriate β value(0.9) was chosen to establish the scale-free network. After this, a topological overlap matrix (TOM) was generated, which measures how well networks are connected. An estimated gene dendrogram was created, similar modules with with correlations >0.7 were ombined. As a last step, the TOM’s difference measure was performed to group genes that have similar expression profiles into modules. Module eigengenes (ME) constitute the expression pattern of all genes. The module with the highest association with three indicators (P < 0.05) was selected for further analysis.
Enrichment analysis
Biological pathways between clusters were explored by gene set variation analysis (GSVA). GO (Gene Ontology) describes biological processes(BP), molecular functions(MF), and cellular components(CC) of genes The Kyoto Encyclopedia of Genes and Genomes (KEGG) were used to annotate involved pathways. Furthermore, the FDR threshold (0.05) was applied to c5.cp.kegg.v5.2.symbols.gmt, which is the reference gene set.
Construction of nomogram
To predict BCa patients’ prognoses based on RE score and other clinical factors, univariate and multivariate Cox regression analyses were performed. Calibration plots and consistency index (C-index) were used to validate the nomogram.
Transwell assay
Polyester membrane cell embedding dishes with 24-well plates (Insert: 8.0 um; diameter: 6.5 mm, JET, China) were used to test cells’ invasion and migration ability. 6 × 104 cells were inoculated into the upper chamber covered with 200 mg/mL Matrigel (1:8, Yepsen, China) and resuspended using a serum-free DMEM medium. Basolateral chambers were then filled with DMEM medium containing 10% FBS. Matrigel-invaded cells were fixed with 90% ethanol for 10 min for 15 min and stained with 0.5% crystal violet after 16 h of incubation. Then five randomly selected fields of view were selected and counted, and this experiment was repeated three times.
Real time quantitative PCR
In accordance with the manufacturer’s instructions, total RNA of cells was extracted with TRIzol® reagent (Thermo Fisher Scientific, Inc, USA). RT-qPCR was conducted using SYBR®Green PCR Master Mix Reagents (Applied Thermo Scientific) in the 7900HT Fast Real-Time PCR System following the recommended protocols. GAPDH was applied as a control for calculating the normalization of relative mRNA expressions. The 2−ΔΔCt method was used to analyze the results. The list of prime was shown in Supplementary Table S1
Western blot
Total protein was extracted using SDS lysis buffer (Beyotime, Nantong, China), and protein concentration was quantified using the BCA Protein Assay Kit (Beyotime, Nantong, China). Equal amounts of protein (40 μg per lane) were separated on 12% SDS-PAGE gels and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Burlington, MA, USA). Membranes were blocked in 5% non-fat milk at room temperature to reduce non-specific binding. Subsequently, membranes were incubated with primary antibodies overnight at 4 °C. The following day, membranes were washed three times with PBST (10 min per wash) and then incubated with corresponding HRP-conjugated secondary antibodies for 1 h at room temperature. After a second round of washing (three times, 10 min each), protein bands were visualized using a silver stain detection system (Beyotime, Nantong, China). Details of the antibodies used for this assay are provided in Supplementary Table S2.
Statistical analysis
The heatmaps were drawn using Cran (https://cran.r-project.org/web/packages). Disease progression was used as an endpoint with clinical data obtained from the TCGA database. Kaplan–Meier (K-M) analysis was conducted using the “survival” and “survminer” packages. The specificity and sensitivity of ROC and DCA curves were used to assess the reliability of nomogram as the prognostic factors. The area under the ROC and DCA curves were calculated using “timeROC” and “ggDCA” packages. Independent t-tests were applied to determine the differences among clinical parameters. P value less than 0.05 was considered to be statistically significant.
Data availability
All public database sources can be found in Methods. The radiomic data from STPH can be found in Supplementary Material.
Code availability
The data and code used in this project are still being utilized in ongoing studies; therefore, we are unable to make them publicly available at this time. If access is needed, please contact the corresponding author directly to discuss. The underlying code used in this study is not publicly available, but can be obtained from the corresponding author upon reasonable request by qualified researchers.
References
Zhu, C. Z., Ting, H. N., Ng, K. H. & Ong, T. A. A review on the accuracy of bladder cancer detection methods. J. Cancer 10, 4038–4044 (2019).
Stenzl, A. et al. The updated EAU guidelines on muscle-invasive and metastatic bladder cancer. Eur. Urol. 55, 815–825 (2009).
Choi, W. et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 25, 152–165 (2014).
Maiorano, B. A. et al. Immune-Checkpoint Inhibitors in Advanced Bladder Cancer: Seize the Day. Biomedicines 10, https://doi.org/10.3390/biomedicines10020411 (2022).
Gómez de Liaño Lista, A. et al. Clinical outcome after progressing to frontline and second-line anti-PD-1/PD-L1 in advanced urothelial cancer. Eur. Urol. 77, 269–276 (2020).
Alsaafeen, B. H., Ali, B. R. & Elkord, E. Resistance mechanisms to immune checkpoint inhibitors: updated insights. Mol. Cancer 24, 20 (2025).
Gettinger, S. et al. Impaired HLA class I antigen processing and presentation as a mechanism of acquired resistance to immune checkpoint inhibitors in lung cancer. Cancer Discov. 7, 1420–1435 (2017).
Ma, K. et al. Turning cold into hot: emerging strategies to fire up the tumor microenvironment. Trends Cancer 11, 117–134 (2025).
Zhang, Y. et al. Targeting the tumour immune microenvironment for cancer therapy in human gastrointestinal malignancies. Cancer Lett. 458, 123–135 (2019).
Tan, H. Y. et al. Targeting tumour microenvironment by tyrosine kinase inhibitor. Mol. Cancer 17, 43 (2018).
Guo, S. & Deng, C. X. Effect of Stromal Cells in Tumor Microenvironment on Metastasis Initiation. Int. J. Biol. Sci. 14, 2083–2093 (2018).
Cheng, Y. Q. et al. Modifying the tumour microenvironment and reverting tumour cells: new strategies for treating malignant tumours. Cell Prolif. 53, e12865 (2020).
Liu, Z. et al. A novel TGF-β risk score predicts the clinical outcomes and tumour microenvironment phenotypes in bladder cancer. Front. Immunol. 12, 791924 (2021).
Upadhrasta, S. & Zheng, L. Strategies in developing immunotherapy for pancreatic cancer: recognizing and correcting multiple immune “defects” in the tumor microenvironment. J. Clin. Med. 8, https://doi.org/10.3390/jcm8091472 (2019).
Kurkowiak, M. et al. The effects of RNA editing in cancer tissue at different stages in carcinogenesis. RNA Biol. 18, 1524–1539 (2021).
Hosaka, T., Tsuji, H. & Kwak, S. RNA editing: a new therapeutic target in amyotrophic lateral sclerosis and other neurological diseases. Int. J. Mol. Sci. 22, https://doi.org/10.3390/ijms222010958 (2021).
Yang, Y., Okada, S. & Sakurai, M. Adenosine-to-inosine RNA editing in neurological development and disease. RNA Biol. 18, 999–1013 (2021).
Nishikura, K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79, 321–349 (2010).
Chen, Y., Wang, H., Lin, W. & Shuai, P. ADAR1 overexpression is associated with cervical cancer progression and angiogenesis. Diagn. Pathol. 12, 12 (2017).
Chen, L. et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 19, 209–216 (2013).
Han, L. et al. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell 28, 515–528 (2015).
Deepak, K. G. K. et al. Tumor microenvironment: challenges and opportunities in targeting metastasis of triple negative breast cancer. Pharm. Res. 153, 104683 (2020).
Kamoun, A. et al. A consensus molecular classification of muscle-invasive bladder cancer. Eur. Urol. 77, 420–433 (2020).
Satyal, U., Sikder, R. K., McConkey, D., Plimack, E. R. & Abbosh, P. H. Clinical implications of molecular subtyping in bladder cancer. Curr. Opin. Urol. 29, 350–356 (2019).
Zhang, Q. W. et al. Personalized CT-based radiomics nomogram preoperative predicting Ki-67 expression in gastrointestinal stromal tumors: a multicenter development and validation cohort. Clin. Transl. Med. 9, 12 (2020).
Sun, R. et al. Radiomics to predict outcomes and abscopal response of patients with cancer treated with immunotherapy combined with radiotherapy using a validated signature of CD8 cells. J. Immunother. Cancer 8, https://doi.org/10.1136/jitc-2020-001429 (2020).
Chen, K., Li, J. & Li, L. Artificial intelligence for disease X: progress and challenges. J. Transl. Int. Med. 12, 534–536 (2024).
Aerts, H. J. et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 5, 4006 (2014).
Lee, M. et al. Non-contrast magnetic resonance imaging for bladder cancer: fused high b value diffusion-weighted imaging and T2-weighted imaging helps evaluate depth of invasion. Eur. Radio. 27, 3752–3758 (2017).
Humphrey, P. A., Moch, H., Cubilla, A. L., Ulbright, T. M. & Reuter, V. E. The 2016 WHO classification of tumours of the urinary system and male genital organs-part B: prostate and bladder tumours. Eur. Urol. 70, 106–119 (2016).
Lambin, P. et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 48, 441–446 (2012).
Gillies, R. J., Kinahan, P. E. & Hricak, H. Radiomics: images are more than pictures, they are data. Radiology 278, 563–577 (2016).
Ehteshami Bejnordi, B. et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318, 2199–2210 (2017).
Yu, K. H. et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat. Commun. 7, 12474 (2016).
Brinker, T. J. et al. Deep neural networks are superior to dermatologists in melanoma image classification. Eur. J. Cancer 119, 11–17 (2019).
Chen, S. et al. Clinical use of machine learning-based pathomics signature for diagnosis and survival prediction of bladder cancer. Cancer Sci. 112, 2905–2914 (2021).
Lopez-Beltran, A., Cookson, M. S., Guercio, B. J. & Cheng, L. Advances in diagnosis and treatment of bladder cancer. BMJ 384, e076743 (2024).
Lee, Y. C. et al. The dynamic roles of the bladder tumour microenvironment. Nat. Rev. Urol. 19, 515–533 (2022).
Song, Q., Zhou, R., Shu, F. & Fu, W. Cuproptosis scoring system to predict the clinical outcome and immune response in bladder cancer. Front. Immunol. 13, 958368 (2022).
Lin, T. et al. Synergy of tumor microenvironment remodeling and autophagy inhibition to sensitize radiation for bladder cancer treatment. Theranostics 10, 7683–7696 (2020).
Takeda, S. et al. Activation of AZIN1 RNA editing is a novel mechanism that promotes invasive potential of cancer-associated fibroblasts in colorectal cancer. Cancer Lett. 444, 127–135 (2019).
da Costa, J. B. et al. Molecular tumor heterogeneity in muscle invasive bladder cancer: Biomarkers, subtypes, and implications for therapy. Urol. Oncol. 40, 287–294 (2022).
Kang, H. W., Kim, W. J., Choi, W. & Yun, S. J. Tumor heterogeneity in muscle-invasive bladder cancer. Transl. Androl. Urol. 9, 2866–2880 (2020).
Xu, X., Wang, Y. & Liang, H. The role of A-to-I RNA editing in cancer development. Curr. Opin. Genet Dev. 48, 51–56 (2018).
Zhang, M. et al. RNA editing derived epitopes function as cancer antigens to elicit immune responses. Nat. Commun. 9, 3919 (2018).
Peng, X. et al. A-to-I RNA editing contributes to proteomic diversity in cancer. Cancer Cell 33, 817–828.e817 (2018).
Xia, J. et al. TGFβ signaling activation correlates with immune-inflamed tumor microenvironment across human cancers and predicts response to immunotherapy. Cell Cycle, 22, 57–72 (2022).
Chen, X. et al. Secretion of bispecific protein of anti-PD-1 fused with TGF-β trap enhances antitumor efficacy of CAR-T cell therapy. Mol. Ther. Oncolytics 21, 144–157 (2021).
Gaudreau, P. O. et al. WISP1 is associated to advanced disease, EMT and an inflamed tumor microenvironment in multiple solid tumors. Oncoimmunology 8, e1581545 (2019).
Tilsed, C. M. et al. Retinoic acid induces an IFN-driven inflammatory tumour microenvironment, sensitizing to immune checkpoint therapy. Front. Oncol. 12, 849793 (2022).
Thommen, D. S. & Schumacher, T. N. T cell dysfunction in cancer. Cancer Cell 33, 547–562 (2018).
Fourcade, J. et al. Upregulation of Tim-3 and PD-1 expression is associated with tumor antigen-specific CD8+ T cell dysfunction in melanoma patients. J. Exp. Med. 207, 2175–2186 (2010).
Xiao, D. et al. ANXA1 promotes tumor immune evasion by binding PARP1 and upregulating Stat3-induced expression of PD-L1 in multiple cancers. Cancer Immunol. Res. 11, 1367–1383 (2023).
Zheng, Y. et al. Glioma-derived ANXA1 suppresses the immune response to TLR3 ligands by promoting an anti-inflammatory tumor microenvironment. Cell Mol. Immunol. 21, 47–59 (2024).
Cao, L. et al. Helicobacter pylori-induced RASAL2 through activation of nuclear factor-κB promotes gastric tumorigenesis via β-catenin signaling axis. Gastroenterology 162, 1716–1731.e1717 (2022).
Pan, Y. et al. RASAL2 promotes tumor progression through LATS2/YAP1 axis of hippo signaling pathway in colorectal cancer. Mol. Cancer 17, 102 (2018).
Xia, Y., Shen, S. & Verma, I. M. NF-κB, an active player in human cancers. Cancer Immunol. Res. 2, 823–830 (2014).
Ebrahimi, N. et al. Targeting the NF-κB pathway as a potential regulator of immune checkpoints in cancer immunotherapy. Cell Mol. Life Sci. 81, 106 (2024).
Cambier, S., Gouwy, M. & Proost, P. The chemokines CXCL8 and CXCL12: molecular and functional properties, role in disease and efforts towards pharmacological intervention. Cell Mol. Immunol. 20, 217–251 (2023).
Yang, Q. et al. The CXCL8/MAPK/hnRNP-K axis enables susceptibility to infection by EV-D68, rhinovirus, and influenza virus in vitro. Nat. Commun. 16, 1715 (2025).
Han, Z. J. et al. Roles of the CXCL8-CXCR1/2 axis in the tumor microenvironment and immunotherapy. Molecules 27, https://doi.org/10.3390/molecules27010137 (2021).
Collins, T. S., Lee, L. F. & Ting, J. P. Paclitaxel up-regulates interleukin-8 synthesis in human lung carcinoma through an NF-kappaB- and AP-1-dependent mechanism. Cancer Immunol. Immunother. 49, 78–84 (2000).
Yi, T. et al. Activation of lncRNA lnc-SLC4A1-1 induced by H3K27 acetylation promotes the development of breast cancer via activating CXCL8 and NF-kB pathway. Artif. Cells Nanomed. Biotechnol. 47, 3765–3773 (2019).
Liu, S. et al. Evaluation of Vesical Imaging-Reporting and Data System (VI-RADS) scoring system in predicting muscle invasion of bladder cancer. Transl. Androl. Urol. 9, 445–451 (2020).
Fan, M. et al. Joint prediction of breast cancer histological grade and Ki-67 expression level based on DCE-MRI and DWI radiomics. IEEE J. Biomed. Health Inf. 24, 1632–1642 (2020).
Gu, Q. et al. Machine learning-based radiomics strategy for prediction of cell proliferation in non-small cell lung cancer. Eur. J. Radio. 118, 32–37 (2019).
Hu, X. X. et al. Whole-tumor MRI histogram analyses of hepatocellular carcinoma: Correlations with Ki-67 labeling index. J. Magn. Reson. Imaging 46, 383–392 (2017).
Kijima, S. et al. Preoperative evaluation of colorectal cancer using CT colonography, MRI, and PET/CT. World J. Gastroenterol. 20, 16964–16975 (2014).
Meyer, H. J., Schob, S., Höhn, A. K. & Surov, A. MRI texture analysis reflects histopathology parameters in thyroid cancer - a first preliminary study. Transl. Oncol. 10, 911–916 (2017).
Zheng, Z. et al. Magnetic resonance imaging-based radiomics signature for preoperative prediction of Ki67 expression in bladder cancer. Cancer Imaging 21, 65 (2021).
van Griethuysen, J. J. M. et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 77, e104–e107 (2017).
Kim, T. Y., Cho, N. H., Jeong, G. B., Bengtsson, E. & Choi, H. K. 3D texture analysis in renal cell carcinoma tissue image grading. Comput. Math. Methods Med. 2014, 536217 (2014).
Bhattacharjee, S. et al. Multi-Features Classification of Prostate Carcinoma Observed in Histological Sections: Analysis of Wavelet-Based Texture and Colour Features. Cancers 11, https://doi.org/10.3390/cancers11121937 (2019).
Chen, S. et al. A novel nomogram based on machine learning-pathomics signature and neutrophil to lymphocyte ratio for survival prediction of bladder cancer patients. Front. Oncol. 11, 703033 (2021).
Wang, G. Y. et al. Prediction of non-muscle invasive bladder cancer recurrence using deep learning of pathology image. Sci. Rep. 14, 18931 (2024).
Wang, H. et al. Elaboration of a multisequence MRI-based radiomics signature for the preoperative prediction of the muscle-invasive status of bladder cancer: a double-center study. Eur. Radio. 30, 4816–4827 (2020).
Zheng, Z. et al. Pathology-based deep learning features for predicting basal and luminal subtypes in bladder cancer. BMC Cancer 25, 310 (2025).
Wang, X. et al. RetCCL: clustering-guided contrastive learning for whole-slide image retrieval. Med. Image Anal. 83, 102645 (2023).
Acknowledgements
For their contributions to the preliminary planning of this study, we would like to thank Wenchao Ma and Haotian Chen. This work was supported in part by a grant from the National Key Research and Development Program of China (#2022YFB3804504); National Natural Science Foundation of China, Youth Project (#82302304, #82101838, #82203367, #82203403); Experimental Animal Fund of Shanghai Science and Technology Commission (#22140903800); M.J.R. Young Scientist Research Fund Grant (grant numbers: MJR202310015); Doctoral Workstation Foundation of Guangdong Second Provincial General Hospital, China (#2022BSGZ011); Science and Technology Program of Guangzhou, China (#2024A04J4159); Tongji University Medicine-X Interdisciplinary Research Initiative (2025-0553-YB-02)
Author information
Authors and Affiliations
Contributions
(I) Conception and design: J.L.; (II) administrative support: X.D.Y. and S.B.; (III) provision of study materials or patients: S.W.S., Z.R.G., Y.Y. and Y.D.G.; (IV) collection and assembly of data: Z.T.Z. and W.Y. (V) data analysis and interpretation: H.W. and S.Y.M.; (VI), manuscript writing: All authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, J., Sun, S., Guo, Y. et al. Integrative molecular–radiopathomic characterization of AIRE-driven immune subtypes in bladder cancer. npj Digit. Med. 8, 766 (2025). https://doi.org/10.1038/s41746-025-02091-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41746-025-02091-2