Abstract
Accurate delineation of pulmonary nodules in chest computed tomography (CT) is essential for early lung cancer diagnosis and treatment planning. However, voxel-wise segmentation methods often produce fragmented masks and inconsistent topology due to low contrast, anatomical variability, and imaging noise. We propose CoreFormer, a segmentation framework that models nodules through structural core anchoring and geodesic shape decoding. CoreFormer identifies the intrinsic topological core of each nodule and generates continuous boundaries guided by anatomy-aware geodesic paths. It is built upon a Swin Transformer backbone and a dual-branch decoder consisting of a Structural Core Predictor and a Context-Aware Shape Decoder, enhanced by Feature Manifold Regularization for discriminative feature learning. Extensive experiments on four public datasets-LIDC-IDRI, LNDb, Tianchi-Lung (MosMedData), and NSCLC-Radiomics-demonstrate that CoreFormer achieves state-of-the-art boundary accuracy and topological fidelity, offering robust and high-fidelity pulmonary nodule segmentation.
Similar content being viewed by others
Introduction
Lung cancer remains the leading cause of cancer-related mortality worldwide, and a patient’s prognosis is heavily dependent on the stage at which the disease is detected1. In this context, pulmonary nodules-small, localized opacities in the lung tissue-have emerged as critical early indicators of potential malignancy. The accurate and reproducible delineation of these nodules in computed tomography (CT) scans is therefore a cornerstone of modern thoracic oncology. It provides essential information for differential diagnosis, guides clinical decisions regarding biopsies or follow-up imaging, and is indispensable for treatment planning, such as defining surgical margins or radiation targets2,3. However, manual segmentation by radiologists is a labor-intensive and time-consuming process that is prone to significant inter- and intra-observer variability, making it a bottleneck in high-throughput screening programs and longitudinal studies. This has created an urgent need for automated, reliable, and efficient segmentation algorithms.
The advent of deep learning, particularly convolutional neural networks (CNNs), has catalyzed a paradigm shift in medical image analysis, leading to significant advances in automated pulmonary nodule segmentation4,5. The predominant approach has been to frame the task as a dense, voxel-wise classification problem, typically employing encoder-decoder architectures like the U-Net6 and its 3D variants7. While these models have achieved considerable success, their performance is often hampered by their reliance on local convolutional operations, which can struggle to capture global shape context. This fundamental limitation leads to several persistent challenges: (1) the generation of fragmented or noisy masks, particularly for nodules with low contrast or ambiguous borders with adjacent structures like blood vessels or the pleural wall; (2) a failure to preserve topological consistency, resulting in anatomically implausible shapes for small or irregularly formed nodules; and (3) a heavy reliance on large datasets with exhaustive, voxel-level annotations, which are exceptionally expensive and difficult to acquire in the medical domain.
To overcome the limitations of discrete, grid-based representations, a new class of methods based on continuous or implicit representations has recently gained prominence in computer vision and medical imaging8,9,10,11. These techniques model object boundaries as the zero-level set of a continuous function, such as a Signed Distance Function (SDF), which inherently ensures smooth and topologically coherent surfaces. However, existing implicit models are often suboptimal for medical applications. Their typical reliance on a global coordinate system and a simple Euclidean distance metric fails to account for the fact that lesion morphology is intimately intertwined with the local anatomical context. The “distance" from a nodule’s center to its boundary is rarely a straight line but rather a path that must navigate complex tissue structures.
In this work, we propose CoreFormer, a novel segmentation framework that addresses the limitations of both voxel-wise and conventional implicit methods by introducing the principles of structural core anchoring and geodesic shape decoding. CoreFormer revolutionizes the implicit representation paradigm by modeling each nodule not from an abstract single point, but from its intrinsic structural core-a more descriptive and topologically stable representation like its skeleton or centerline. Our key insight is that a nodule’s complex topology is better captured by this rich structural anchor, and its boundary is more faithfully defined by a continuous field that evolves along geodesically optimal paths. These paths are learned to respect the underlying anatomy, effectively navigating through and around different tissue types. This formulation fundamentally enhances the model’s ability to segment nodules with irregular, complex, or attached morphologies where traditional methods often fail.
The CoreFormer architecture is built upon a powerful Swin Transformer backbone, which excels at capturing both local details and long-range dependencies in a hierarchical manner. This is followed by a sophisticated dual-branch decoder designed for synergistic operation. The first branch, the Structural Core Predictor, leverages high-level semantic features to identify the nodule’s topological skeleton. The second branch, the Context-Aware Shape Decoder, then uses this predicted core as an anchor, synthesizing it with multi-scale features to compute a continuous SDF. This decoding is achieved using learned, anatomy-aware geodesic distances and attention mechanisms. To further enhance robustness, we introduce a novel training scheme, Feature Manifold Regularization, which explicitly structures the latent feature space to ensure high separability between nodular and non-nodular tissue features.
We conducted a comprehensive evaluation of CoreFormer on four public benchmark datasets: LIDC-IDRI2, LNDb12, Tianchi-Lung, and NSCLC-Radiomics. The results demonstrate that our approach consistently and significantly outperforms a wide range of strong baselines across multiple metrics, including Dice, Hausdorff distance, and boundary precision. Our work underscores the superiority of a paradigm that combines strong topological priors with anatomy-aware shape decoding for achieving high-fidelity medical image segmentation.
Our key contributions are summarized as follows: We introduce CoreFormer, a novel segmentation model that moves beyond point-based representations by utilizing a structural core anchor and a learned, geodesic shape decoding process. This unique combination achieves state-of-the-art accuracy and produces topologically consistent segmentations for pulmonary nodules. We design a sophisticated and synergistic architecture featuring a dual-branch decoder for explicit core prediction and context-aware shape generation. The model is further enhanced by a novel Feature Manifold Regularization scheme that promotes a more discriminative and robust feature space, improving overall performance. We provide extensive empirical evidence on multiple real-world CT datasets, demonstrating that CoreFormer not only sets a new standard in segmentation accuracy but also excels in boundary fidelity and topological integrity, highlighting its robustness and clinical potential.
Pulmonary Nodule Segmentation: Segmentation of pulmonary nodules in chest CT scans is a long-standing problem in medical imaging. Early approaches relied on classical image processing techniques, such as thresholding, region growing, or level sets13,14, which are sensitive to noise and lack generalization to nodules with diverse shapes and intensities. With the emergence of large-scale public datasets such as LIDC-IDRI2 and LUNA164, deep learning-based methods have become dominant.
CNNs, especially 2D and 3D U-Net architectures6,7, have demonstrated strong performance in volumetric medical segmentation tasks. Extensions such as V-Net15, Dense V-Net16, and Attention U-Net17 further enhance representation learning and spatial awareness. However, these voxel-wise segmentation models often produce spatially inconsistent masks for small or irregular nodules, due to their reliance on local receptive fields and discrete prediction structures.
Continuous and Implicit Representations: To overcome the limitations of discrete voxel grids, recent research has explored continuous and implicit representations for medical image segmentation. Inspired by neural implicit functions8,9, methods such as Neural Implicit Segmentation10 and SIREN-based organ modeling18 represent anatomical structures as continuous fields, enabling smoother boundary reconstructions. In19, authors proposed STUNet for shape-aware implicit nodule segmentation, combining CNNs with signed distance fields. These methods offer superior geometric flexibility but often operate on global coordinate systems or rely on a simple Euclidean distance metric. Our work significantly advances this direction by proposing a locally-anchored implicit model where the distance metric is not Euclidean but a learned, anatomy-aware geodesic distance, and the anchor is a structured, topological prior (the core) rather than a simple coordinate or latent code.
Center-Guided and Shape-Aware Models: Center-guided representations have recently emerged as a promising paradigm, particularly in instance segmentation and keypoint localization20,21. In the medical domain, models such as CENet22 predict lesion centers to guide segmentation boundaries, improving robustness to surrounding noise and occlusion. Similarly, radial or polar representations have been applied to organs with centralized topology (e.g., the liver or eyes)23,24. While effective, these methods typically rely on a single point (a zero-dimensional prior), which is insufficient for capturing the morphology of elongated, branching, or highly irregular lesions. For instance, in the case of a complex, bi-lobed or “dumbbell-shaped" lesion, a single predicted center point could erroneously fall into the non-lesion space between the lobes, providing a poor and misleading anchor for shape generation and failing to capture the object’s complete topology.
In contrast, our method generalizes this concept by replacing the single-point center with a one-dimensional structural core, providing a much richer and more stable topological prior. A predicted core in the dumbbell example would naturally form a line spanning both lobes, perfectly capturing the underlying structure. Consequently, our shape decoding process, which follows learned geodesic paths from this core, is fundamentally more powerful and flexible than simpler radial or polar evolutions.
Label Efficiency and Feature Learning: Label efficiency is crucial in medical image analysis, where expert annotations are expensive. Semi-supervised approaches aim to leverage unlabeled data through consistency regularization25, self-training26, or pseudo-labeling27. More recently, shape-constrained semi-supervised frameworks28,29 have shown that integrating strong priors can lead to improved generalization. Our work contributes to this area from a different perspective. Instead of focusing on semi-supervised algorithms, we enhance label efficiency by building powerful inductive biases directly into our architecture via the structural core and geodesic path priors. Furthermore, our proposed Feature Manifold Regularization acts as a strong form of supervision on the feature space itself, forcing the model to learn more discriminative and better-organized representations. This structured feature space leads to improved generalization and more robust performance, especially in data-scarce regimes.
Results
Experimental setup
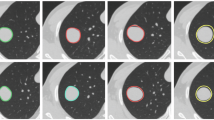
Datasets: To rigorously evaluate the generalization capability of our model, we selected four public datasets with significant heterogeneity, as illustrated in Fig. 1. These datasets present a notable domain gap: LIDC-IDRI2 and LNDb12 primarily contain nodules from lung cancer screening programs, which vary in size and texture. In contrast, NSCLC-Radiomics30 consists of large, irregularly shaped tumors from diagnosed non-small cell lung cancer patients, while MosMedData31 features diffuse, amorphous ground-glass opacities characteristic of COVID-19 lesions. This diversity in pathology, lesion morphology, and imaging protocols creates a challenging testbed for assessing the model’s robustness and its ability to generalize to unseen data distributions, which is a critical requirement for clinical applicability.
LIDC-IDRI includes 1,018 thoracic CT scans with pulmonary nodules annotated independently by four radiologists. Following prior works4, we extract nodules ≥ 3 mm and retain voxels with three or more annotators’ consensus. Scans are resampled to an isotropic spacing of 1 × 1 × 1 mm.
LNDb contains 294 chest CT scans acquired during a lung cancer screening program in Portugal. It provides expert-annotated masks and diagnosis metadata. Nodules ≥ 3 mm are extracted and resampled to a uniform voxel size. Due to limited volume, we use 5-fold cross-validation.
NSCLC-Radiomics30 consists of 422 CT scans of non-small cell lung cancer (NSCLC) patients, with radiologist-delineated tumor contours provided in RTSTRUCT format. We convert annotations into binary masks and extract lesion-centered patches. The diverse scanner vendors and clinical variability make it ideal for testing cross-domain robustness.
MosMedData31 offers 1,110 chest CT studies including COVID-19 patients and healthy controls. A labeled subset contains infection region annotations, which we use to assess generalization to non-nodular, irregular lesions. Although this task differs from nodule segmentation, it stresses boundary coherence and shape awareness under pathological changes.
*Only a subset of 50 studies contain expert-annotated segmentation masks. A detailed summary of the datasets used in our experiments is provided in Table 1.
Preprocessing and Data Augmentation: A standardized preprocessing pipeline was applied to all CT volumes to ensure consistency and optimize them for the neural network. Initially, the raw Hounsfield Unit values were clipped to a range of [−1000, 400] to isolate the lung parenchyma and other relevant soft tissues while discarding extreme values corresponding to air or bone. The clipped volumes were then normalized to a floating-point range of [0, 1] to stabilize the training process. For computational efficiency and to maintain a consistent input size, we extracted 3D patches of 64 × 64 × 64 voxels, each centered on a region of interest (ROI). To enhance the model’s robustness and prevent overfitting, especially given the variability in medical imaging, we employed a comprehensive online data augmentation strategy. This included random affine transformations, such as rotations (up to ±10∘) and scaling (up to ±20%), to simulate variations in patient positioning and scanner geometry. Additionally, we applied intensity jittering to account for differences in contrast and brightness, and elastic deformations to model non-rigid anatomical variations.
Ground-Truth and Implementation Details: To ensure reproducibility, we provide key implementation details here. The ground-truth skeletons required for supervising the Structural Core Predictor were generated using the skeletonize_3d function from the Python scikit-image library, a standard morphological thinning algorithm. For the Feature Manifold Regularization described in section “Feature Manifold Regularization”, dynamic sub-manifold discovery was performed using the K-Means clustering algorithm, with the number of clusters (k = 3) chosen via cross-validation.
Training Protocol: All models were implemented within the PyTorch deep learning framework, utilizing the MONAI library for its specialized tools in medical image analysis. Our proposed CoreFormer model was trained end-to-end using the Adam optimizer, which is well-suited for high-dimensional parameter spaces. The initial learning rate was set to 1 × 10−4 with a weight decay of 1 × 10−5 for regularization, and a batch size of 4 was used. To facilitate stable convergence, a learning rate scheduler was employed to automatically reduce the learning rate whenever the validation loss reached a plateau. Each model was trained for a maximum of 300 epochs, with performance on a validation set monitored every 10 epochs. An early stopping mechanism, triggered by a lack of improvement in the validation Dice score, was used to select the best-performing model checkpoint and prevent overfitting. For a rigorous and fair comparison, all baseline methods, including 3D U-Net, V-Net, nnU-Net, and STUNet, were trained under the exact same data splits, augmentation schemes, and optimization settings.
Evaluation Metrics: To conduct a thorough and multifaceted assessment of segmentation quality, we employed four widely-accepted metrics that collectively evaluate volumetric overlap, boundary accuracy, and contour fidelity. For volumetric accuracy, we used the Dice Similarity Coefficient (DSC), which measures the spatial overlap between the predicted mask (P) and the ground-truth mask (G). It is defined as:
To specifically assess the precision of the predicted boundaries, a critical factor for clinical applications, we utilized three complementary metrics. The Hausdorff Distance (HD) measures the maximum surface distance between the predicted and ground-truth boundaries, thus quantifying the worst-case localization error. The Average Symmetric Surface Distance (ASSD) computes the average of the bidirectional boundary distances, offering a more global measure of contour alignment. Finally, the Boundary F1 Score (BF1) calculates the precision-recall F1 score for boundary voxels within a 2-voxel tolerance, providing a direct evaluation of the contour’s correctness.
Hardware and Computational Cost: All experiments were conducted on a workstation with dual NVIDIA RTX 3090 GPUs. While the full training of CoreFormer on LIDC-IDRI took ~6 h, the inference performance is critical for clinical deployment. For a 64 × 64 × 64 input patch, the average inference time of CoreFormer is ~720 ms, broken down as follows: Swin Transformer Backbone (350 ms), Structural Core Predictor (60 ms), Cost Map Generation & FMM (210 ms), and the Context-Aware Shape Decoder (100 ms). For comparison, a baseline 3D U-Net takes ~410 ms. While CoreFormer is more computationally intensive, we argue this modest increase is a justifiable trade-off for the significant gains in accuracy and reliability, with the total time remaining well within practical limits for clinical workflows.
Comparison with state-of-the-art methods
To situate CoreFormer within the current landscape of medical image segmentation, we conducted a rigorous comparison against a comprehensive suite of state-of-the-art (SOTA) methods. These baselines span classic convolutional architectures (3D U-Net, V-Net), attention-augmented models (Attention U-Net), highly optimized frameworks (nnU-Net), advanced implicit representation techniques (STUNet), and leading Transformer-based approaches (TransUNet, UNETR). Our analysis is presented in three parts: performance on established nodule segmentation benchmarks, evaluation of cross-domain generalization, and a statistical analysis of the results.
Performance on Nodule Segmentation Benchmarks: As detailed in Table 2, CoreFormer establishes a new state-of-the-art on both the LIDC-IDRI and LNDb datasets, demonstrating superior performance across all evaluation metrics.
On the large-scale LIDC-IDRI dataset, CoreFormer achieves a Dice score of 88.7%, surpassing the next-best method, STUNet, by a significant margin of 1.6%. More importantly, the improvements in boundary-specific metrics are particularly pronounced. CoreFormer achieves the lowest Hausdorff Distance (2.54 mm) and Average Symmetric Surface Distance (0.93 mm), and the highest Boundary F1 Score (87.6%). This substantial enhancement in boundary fidelity directly validates our core hypothesis: the proposed geodesic shape decoding, anchored to a structural core, generates smoother, more anatomically plausible contours than methods relying on voxel-wise classification or simpler implicit representations.
This superior performance is consistently maintained on the LNDb dataset, which features different acquisition protocols and higher image noise. CoreFormer again leads in all metrics, achieving a Dice score of 86.4% and a Boundary F1 Score of 86.1%. The consistent gains across both datasets underscore the robustness of our framework. While Transformer-based models like UNETR show competitive results, they are outperformed by CoreFormer, suggesting that their powerful feature extraction capabilities, while beneficial, lack the explicit geometric and topological priors needed for high-fidelity segmentation. Similarly, while STUNet confirms the value of continuous representations, CoreFormer’s use of a structural skeleton as an anchor-rather than a single point-provides a more stable and descriptive prior for complex nodule morphologies, leading to more accurate and reliable shape generation.
Cross-Dataset Generalization: To assess the real-world utility and robustness of our model, we performed a challenging cross-dataset generalization experiment. Models were trained exclusively on LIDC-IDRI and then directly evaluated on two unseen target datasets-NSCLC-Radiomics and MosMedData-without any fine-tuning. As shown in Table 3, CoreFormer demonstrates exceptional generalization capabilities.
On NSCLC-Radiomics, which contains large, non-spherical lung tumors, CoreFormer achieves the best performance across all metrics, with a Dice score of 83.2% and a Boundary F1 score of 80.3%. This result is particularly significant, as it shows that the inductive biases of our model, designed for nodules, successfully transfer to more complex and varied tumor morphologies. The emphasis on smooth geometric evolution from a core structure allows it to handle these challenging shapes better than conventional voxel-based methods, which often produce fragmented boundaries.
Similarly, on the MosMedData dataset, which features highly irregular and multi-focal infectious lesions from COVID-19 patients, CoreFormer again delivers the top results (78.8% Dice, 77.0% BF1). This task is challenging for models that lack strong shape priors. Our method’s core-conditioned geodesic modeling acts as a powerful form of regularization, suppressing noisy predictions and preserving shape coherence even for amorphous structures. These results confirm that CoreFormer’s architectural principles provide a versatile and robust foundation that generalizes far beyond its initial training domain.
Statistical Significance and Prediction Stability: To verify the reliability of our findings, we conducted a statistical analysis on the LIDC-IDRI test set using paired t-tests, with the results presented in Table 4. The analysis reveals that CoreFormer not only achieves the highest mean performance but also exhibits the greatest prediction stability. With a mean Dice score of 88.7% and the lowest standard deviation (2.6), our model demonstrates consistently high accuracy across diverse nodule appearances, unlike competing models which show higher variance.
Crucially, the performance improvements are statistically significant. The computed p values for both Dice and HD comparisons against all baseline methods are well below the 0.05 threshold, with most being less than 0.01. For instance, the improvement over a strong baseline like STUNet is significant for both Dice (p = 0.021) and HD (p = 0.034). This statistical validation confirms that the performance gains achieved by CoreFormer are not attributable to random chance or dataset bias. The combination of superior mean accuracy, lower prediction variance, and statistically significant improvements provides compelling evidence that our proposed method introduces a meaningful and robust advancement over existing segmentation models.
Ablation studies
To rigorously validate the architectural design of CoreFormer and quantify the contribution of its individual components, we conducted a series of ablation studies on the LIDC-IDRI dataset. These experiments systematically deconstruct the framework to isolate the impact of the Structural Core Predictor, the Context-Aware Shape Decoder (CAS-Decoder), and the Feature Manifold Regularization scheme.
Dissection of CoreFormer’s Architectural Components: Our analysis, summarized in Table 5 and Fig. 2, demonstrates that each component of the CoreFormer architecture plays a critical and synergistic role in achieving state-of-the-art performance. The full, unmodified model serves as the benchmark, achieving a Dice score of 88.7% and a Boundary F1 score of 87.6%, with the lowest boundary errors (2.54 mm HD and 0.93 mm ASSD).
Removing the Structural Core Predictor and relying on a geometric center forces the model to anchor its shape generation process without anatomical guidance. This results in a notable performance drop (Dice to 86.9%, BF1 to 84.9%) and a significant increase in boundary error (HD to 3.12 mm). The qualitative results in Fig. 3 (column “w/o SCP”) visually confirm this, showing segmentation masks that are noticeably offset and asymmetric, underscoring the importance of learning an intrinsic, anatomically-aware core.
This figure visually contrasts the segmentation masks produced by the full CoreFormer model with those from ablated variants against the ground truth (GT). The variants shown are: a model without the Context-Aware Shape Decoder ('w/o CAS-Decoder') and a model without the Structural Core Predictor ('w/o SCP'). This comparison illustrates the improvements in boundary adherence and topological consistency achieved by the full model.
Ablating the Context-Aware Shape Decoder (CAS-Decoder) leads to a further decline across all metrics (Dice to 86.1%, BF1 to 83.7%). As seen in Fig. 3 (column “w/o Decoder”), this variant produces irregular and fragmented masks. This highlights the decoder’s crucial function in evolving a smooth and continuous boundary along learned geodesic paths, a capability that a conventional decoder lacks.
The removal of the Feature Manifold Regularization during training results in the poorest performance among the module removal variants (Dice of 85.4%). Although this module is not active during inference, its absence during training leads to a less discriminative feature space, which consequently hampers the downstream prediction and decoding tasks. This confirms that explicitly structuring the latent space is vital for robust feature learning.
Finally, replacing the entire dual-branch decoder with a standard Voxel-wise Classifier, which mimics a conventional U-Net-like architecture, causes the most substantial performance degradation (Dice to 85.0%, BF1 to 81.9%). This variant struggles to maintain spatial continuity and fails to leverage any geometric priors, resulting in the jagged, fragmented, and anatomically inconsistent masks shown in Fig. 3. The sharp decline in boundary metrics (HD of 3.58 mm, ASSD of 1.41 mm) unequivocally demonstrates the limitations of purely local, voxel-level predictions and highlights the superiority of our global, shape-aware generation process.
Superiority of the Dual-Branch Geodesic Decoder: To further isolate and emphasize the contribution of our novel decoder architecture, we conducted a direct comparison between our dual-branch decoder and a conventional voxel-wise decoder across all four evaluation datasets. The results, presented in Table 6, show a consistent and significant performance advantage for our proposed design.
On the LIDC-IDRI dataset, our decoder architecture boosts the Dice score by 3.7 points and the Boundary F1 score by a remarkable 5.7 points, while reducing the Hausdorff Distance by over 1 mm. This trend is robustly maintained across the other datasets. On LNDb, the improvements (+3.1% Dice, +5.7% BF1) demonstrate strong generalization to different scanner protocols. The benefits are even more pronounced on the cross-domain datasets. For the large, complex tumors in NSCLC-Radiomics, our decoder yields a 3.6-point gain in Dice and a 5.3-point gain in BF1. For the amorphous COVID-19 lesions in MosMedData, it achieves a 2.4-point increase in Dice and a 4.5-point increase in BF1.
These results validate the core intuition behind our method. Voxel-wise decoders make independent, local decisions, which often leads to noisy and topologically inconsistent predictions. In contrast, our continuous, geodesic formulation regularizes the entire segmentation process, evolving the boundary smoothly from a learned anatomical anchor. The consistently larger gains in boundary-centric metrics (HD and BF1) compared to the volumetric Dice score highlight that our method specifically remedies the primary structural weaknesses of conventional segmentation approaches, resulting in more robust, accurate, and anatomically plausible results.
Boundary and topology consistency analysis
Beyond volumetric accuracy, a critical measure of a segmentation model’s clinical utility is its ability to preserve structural and topological fidelity. To investigate this, we performed a detailed analysis using both boundary-aware and topology-sensitive metrics on the LIDC-IDRI dataset.
Quantitative Analysis: The quantitative results, presented in Table 7, show that CoreFormer consistently and significantly outperforms all baseline methods in preserving structural integrity. Our model achieves a Boundary F1 Score of 87.6%, a substantial improvement over strong baselines like STUNet (78.2%) and nnU-Net (76.7%). This indicates that the boundaries generated by CoreFormer are more accurately aligned with the ground truth, a crucial requirement for applications like treatment planning where precise margins are essential. Furthermore, CoreFormer records the lowest boundary errors, with a Hausdorff Distance of 2.54 mm and an Average Symmetric Surface Distance of 0.93 mm. These superior results reflect a reduction in both maximum and average boundary deviations, which can be directly attributed to our model’s geodesic decoder that generates smooth, anatomically-guided contours.
A key differentiator of our method is its ability to maintain topological correctness. The Euler Similarity Index (ESI), which measures the similarity of topological features like connected components and holes, is highest for CoreFormer at 0.94. In contrast, conventional voxel-based methods like 3D U-Net and V-Net score much lower (0.81 and 0.83, respectively), as their local, pixel-wise decision process often leads to spurious fragments or the incorrect merging of structures. This demonstrates the efficacy of our center-driven, continuous representation in producing outputs that are not only accurate but also structurally and topologically faithful.
Qualitative Analysis: The quantitative improvements are strongly supported by qualitative evidence, as shown in Fig. 4. Across several challenging cases, CoreFormer consistently produces segmentation masks that are visually superior to those of other state-of-the-art methods like UNETR and STUNet. The boundaries generated by our model are smoother and more tightly aligned with the true anatomical contours of the nodules, even in regions with low contrast or complex shapes. In contrast, the baseline methods, while generally accurate, often exhibit minor deviations, jagged edges, or less precise delineation, particularly for smaller or irregularly shaped nodules.
For instance, in the challenging case involving a nodule with an internal void, our model correctly preserves this topological feature, whereas conventional voxel-based methods often fail, either filling the hole or fragmenting the mask. This visual evidence reinforces our quantitative findings, demonstrating that CoreFormer’s geodesic shape evolution, guided by a structural core prior, naturally enforces a level of global coherence and smoothness that is absent in methods relying on local information alone. This results in segmentations that are not only more accurate by the numbers but are also more anatomically plausible and visually reliable, which is a critical requirement for deployment in clinical decision-making pipelines.
Model interpretability via attention visualization
To better understand the model’s decision-making process and verify that it learns clinically relevant features, we visualized its internal attention mechanisms. As shown in the heatmaps in Fig. 5, the model demonstrates a strong ability to focus its computational resources precisely on the regions of interest. The high-activation areas (shown in red and yellow) closely align with the ground-truth nodule locations across various cases, including those with irregular shapes and subtle appearances. Notably, the model not only attends to the core of the nodule but also accurately highlights its boundaries and peripheral features, such as spiculation. This indicates that our model has learned to identify key diagnostic characteristics, providing a more interpretable and trustworthy segmentation result.
For each case (row), we show the original CT image, the ground-truth mask (GT Mask), and the corresponding attention heatmap. The heatmaps demonstrate that the model accurately focuses on the pulmonary nodule area, with heightened attention on the prominent features along the nodule’s boundary, validating its ability to capture clinically relevant information for segmentation.
Semi-supervised segmentation performance
A key challenge in medical imaging is the high cost of acquiring expert annotations, making it crucial for models to perform well with limited labeled data. To evaluate CoreFormer’s label efficiency, we conducted a semi-supervised segmentation experiment on the LIDC-IDRI dataset, progressively reducing the proportion of labeled training data from 100% down to just 5%. We compared our method against several strong baselines: a standard fully-supervised Voxel Baseline (U-Net), Mean Teacher, which leverages consistency between a student model and an exponential moving average of its weights (the teacher), FixMatch, which combines pseudo-labeling with consistency regularization, and ST++, another advanced semi-supervised framework.
The results, presented in Table 8 and visualized in Fig. 6, demonstrate the decisive superiority of CoreFormer across all levels of supervision. In the extremely data-scarce regime with only 5% of labels, CoreFormer achieves a Dice score of 68.5%, outperforming the standard U-Net baseline by a remarkable 10.4 points and surpassing even advanced methods like ST++ by 5.5 points. This substantial performance gap is maintained as more labels are introduced, with CoreFormer leading all competitors at the 10% and 25% levels. This indicates that our model is significantly more effective at leveraging unlabeled data.
This strong performance in low-data regimes stems directly from the powerful inductive biases inherent in our model’s architecture. The structural core anchoring and geodesic shape decoding act as a potent shape regularizer. While conventional consistency-based methods can propagate errors through noisy pseudo-labels at the pixel level, CoreFormer’s structural prior constrains the shape generation process to be anatomically plausible. By forcing the segmentation to evolve coherently from a predicted core, our method effectively suppresses noise and preserves semantic boundaries, enabling a more stable and effective learning signal from unlabeled data.
Furthermore, the scalability of CoreFormer is evident as the amount of labeled data increases. The performance gap between our method and the baselines persists even with full supervision, where CoreFormer achieves the highest Dice score of 88.8%. This confirms that the architectural benefits are not merely a low-data crutch but provide a fundamental advantage across the entire supervision spectrum. These results underscore the robustness and label efficiency of our framework, positioning it as a highly practical solution for clinical scenarios where annotated data is often a limited and valuable resource.
Discussion
In this paper, we introduced CoreFormer, a novel framework that reconceptualizes pulmonary nodule segmentation through the principles of structural core anchoring and geodesic shape decoding. By first identifying a nodule’s intrinsic topological skeleton and subsequently evolving its boundary along learned, anatomy-aware geodesic paths, our method directly addresses the critical limitations of conventional voxel-wise approaches, namely their tendency to produce fragmented boundaries and topological inconsistencies, especially for complex lesions. This is realized through an architecture that integrates a Swin Transformer backbone with a sophisticated dual-branch decoder and a novel Feature Manifold Regularization scheme, ensuring high structural fidelity and precise boundary delineation even under challenging imaging conditions.
Through extensive experiments on multiple public datasets, CoreFormer was shown to consistently outperform a wide range of strong segmentation baselines across both volumetric and boundary-specific metrics, empirically validating the efficacy of our architectural design. This superior performance stems from the powerful inductive biases embedded within our framework. The explicit modeling of a structural core and geodesic paths, coupled with a highly structured feature space, endows the model with exceptional generalization capabilities and significantly improved label efficiency. This makes our framework particularly well-suited for the medical domain, where the scarcity of comprehensive annotations remains a major bottleneck.
In advancing the state-of-the-art, CoreFormer demonstrates the profound impact of integrating deep topological priors with anatomy-aware implicit modeling. Future work will focus on extending this powerful paradigm to more complex, multi-class segmentation of thoracic structures and to the dynamic tracking of lesions over time for monitoring lung cancer progression. Furthermore, integrating our framework with shape-aware uncertainty modeling could further enhance its reliability for real-world clinical deployment. Ultimately, CoreFormer offers a robust and powerful solution that paves the way for more reliable automated analysis in clinical practice.
Methods
Overall architecture
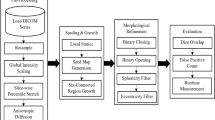
As shown in Fig. 7, the overall framework of CoreFormer comprises a hierarchical Swin Transformer backbone, a dual-branch decoder, and a feature manifold regularization module. Our proposed framework introduces a novel paradigm for pulmonary nodule segmentation by shifting from traditional voxel-wise classification to a structured, shape-aware generative process. The core of our method lies in a cascaded architecture that first identifies the intrinsic topological core of a nodule and then grows the segmentation boundary outward in a context-aware manner. The entire architecture, as illustrated in the main framework diagram, consists of three integral components: a hierarchical Swin Transformer backbone for feature extraction, a dual-branch decoder comprising a Structural Core Predictor and a Context-Aware Shape Decoder (CAS-Decoder), and a training-specific Feature Manifold Regularization module designed to learn a highly discriminative latent space. The final segmentation is supervised by a loss \({{\mathcal{L}}}_{Seg}\), while the training process is augmented by a core prediction loss and the regularization loss \({{\mathcal{L}}}_{Reg}\).
The input 3D CT volume is processed by a hierarchical Swin Transformer backbone, yielding multi-scale feature maps (FLow, FMid, FHigh). These features feed into a dual-branch decoder. The Structural Core Predictor uses high-level features FHigh to predict a core probability map PCore, from which a structural Core Anchor S is derived. The Context-Aware Shape Decoder (CAS-Decoder) takes the anchor S and all feature maps as input, utilizing a pipeline of a lightweight CNN, Fast Marching Method (FMM), an Attention Block, and an MLP to generate the final predicted mask. During training, the Feature Manifold Regularization module provides an additional supervisory signal, \({{\mathcal{L}}}_{Reg}\), to structure the feature space.
Hierarchical Swin Transformer backbone
The foundation of our model is a hierarchical Swin Transformer backbone, engineered to efficiently extract both fine-grained local details and long-range global semantic context from the input 3D CT volume. The input volume is initially divided into non-overlapping 3D patches, which are then projected into a high-dimensional embedding space via a linear embedding layer. These tokenized embeddings are processed through a series of Swin Transformer Encoder Layers. As detailed in the framework diagram, each encoder layer is built upon a standard Multi-head Self-Attention (MSA) module and a Shifted-Window based Multi-head Self-Attention (SW-MSA) module. This windowing mechanism restricts self-attention computation to local windows, significantly improving computational efficiency, while the shifting strategy facilitates cross-window connections, enabling the model to learn global interactions. To produce a hierarchical representation, patch merging layers are inserted between stages, which reduce the spatial resolution of the feature maps while increasing their channel depth. This process yields a set of multi-scale feature maps, denoted as FLow, FMid, and FHigh. FLow retains high-resolution spatial information crucial for precise boundary delineation, whereas FHigh captures abstract, high-level semantic information essential for understanding the nodule’s overall structure and location.
Dual-branch core and shape decoder
Our dual-branch decoder is the centerpiece of the framework, responsible for translating hierarchical features into a precise segmentation mask. It operates in two sequential stages: core prediction and shape decoding.
The first stage, the Structural Core Predictor, aims to identify a descriptive structural “skeleton" of the nodule, providing a robust anchor for subsequent steps. Taking the highest-level semantic features FHigh as input, this predictor employs a lightweight fully convolutional decoder to generate a dense core probability map, \({P}_{Core}\in {[0,1]}^{H{\prime} \times W{\prime} \times D{\prime} }\). Each voxel in PCore represents the probability of that location being part of the nodule’s structural centerline. At inference time, the Core Anchor S is derived from this probability map through a fully automated, two-step online process integrated into the pipeline. First, the map is binarized using a confidence threshold of 0.5. Second, we apply the same 3D morphological thinning algorithm used for generating the ground-truth skeletons to the binarized map. This ensures consistency between training and inference and yields a sparse set of connected voxels representing the final anchor. This module is supervised by a dedicated loss, \({{\mathcal{L}}}_{Core}\), computed between the predicted map PCore and a ground-truth skeleton.
The second stage, the Context-Aware Shape Decoder (CAS-Decoder), executes the final shape generation using the Core Anchor S as a geometric prior. This module first concatenates the multi-scale features (FLow, FMid, FHigh) and processes them with a lightweight CNN to generate a voxel-wise traversal cost map, with a Softplus activation ensuring all costs are positive. Subsequently, the Fast Marching Method (FMM) algorithm is employed, using S as the source to efficiently compute the shortest geodesic distance from the core to every other voxel across this learned cost map. An Attention Block then aggregates features along these geodesic paths to form a contextually rich representation for each voxel. Finally, a concluding MLP takes this context-aware feature and the geodesic distance as input to regress a final continuous signed distance field, which is thresholded to produce the final predicted mask.
Feature manifold regularization
To learn a more discriminative and semantically structured feature space, we introduce a Feature Manifold Regularization module, which is exclusively active during the training phase. The overall workflow of the feature manifold regularization process is illustrated in Fig. 8, which shows how features are sampled, clustered, and organized into within- and between-manifold graphs. This module explicitly sculpts the latent space of the high-level features FHigh by enforcing intra-class compactness and inter-class separability, as detailed in the regularization diagram. The process unfolds in four steps. First, for each training batch, a Semantics-Aware Sampling strategy is employed. Leveraging the ground truth masks, we sample a representative set of feature vectors from FHigh that belong to distinct, pre-defined semantic classes, primarily ‘Nodular` and ‘Non-nodular` tissues. Second, these sampled features undergo Dynamic Sub-manifold Discovery. For features within a major class (e.g., all ‘Nodular` samples), we apply an unsupervised clustering algorithm to identify finer-grained sub-groups, which may correspond to different nodule characteristics. This allows the model to adaptively recognize the inherent heterogeneity within a single class. Third, based on this clustering, we perform Multi-Level Adjacency Graph Construction. A within-manifold graph (Gw) is built by connecting feature vectors within each discovered sub-cluster, defining pairs that should be pulled together. A between-manifold graph (Gb) is built by connecting feature vectors from different major classes (e.g., ‘Nodular` vs. ‘Non-nodular`), defining pairs that should be pushed apart. Finally, a composite regularization loss \({{\mathcal{L}}}_{Reg}\) is formulated from these graphs. The compactness loss, \({{\mathcal{L}}}_{Comp}\), penalizes large distances between connected nodes in Gw, thereby enforcing tightness within each sub-manifold (Equation 1):
Simultaneously, the separability loss, \({{\mathcal{L}}}_{Sep}\), uses a margin-based formulation to penalize small distances between nodes in Gb, forcing a clear separation between the feature representations of different classes (Equation 2):
where m is a predefined margin. The total regularization loss is the sum \({{\mathcal{L}}}_{Reg}={{\mathcal{L}}}_{Comp}+{{\mathcal{L}}}_{Sep}\), guiding the backbone to learn features that are not only effective for downstream tasks but are also inherently more robust and separable.
a A set of high-dimensional feature vectors are sampled from the backbone’s output FHigh using ground truth masks for guidance. b These features are dynamically clustered, identifying not only primary classes (`Nodular̀, `Non-nodular̀) but also finer-grained sub-manifolds within them. c Two graphs are constructed: a within-manifold graph (Gw, blue arrows) connecting samples in the same sub-cluster, and a between-manifold graph (Gb, cyan arrows) connecting samples from different primary classes. d The regularization loss function then optimizes the feature space to enforce within-manifold compactness (minimizing \({{\mathcal{L}}}_{Comp}\)) and between-manifold separability (maximizing inter-cluster distance, related to \({{\mathcal{L}}}_{Sep}\)).
Training objective
The overall model is trained end-to-end by optimizing a composite loss function that combines the objectives from the primary segmentation task, the auxiliary core prediction task, and the feature regularization module. The total loss \({{\mathcal{L}}}_{total}\) is a weighted sum of these individual components (Equation 3):
Here, \({{\mathcal{L}}}_{Seg}\) is the primary segmentation loss (e.g., a combination of Dice and cross-entropy loss) computed on the final predicted mask. \({{\mathcal{L}}}_{Core}\) is the loss for the Structural Core Predictor, ensuring the accurate localization of the nodule’s skeleton. \({{\mathcal{L}}}_{Reg}\) is the feature manifold regularization loss described previously. The terms λcore and λreg are scalar hyperparameters that balance the contribution of the auxiliary tasks. Based on empirical validation on a held-out set to optimally balance the learning objectives, these weights were set to λcore = 1.0 and λreg = 0.1 in our experiments. This ensures that all components of our architecture are synergistically optimized to produce accurate, robust, and topologically sound segmentations.
Ethics approval and consent to participate
This study uses publicly available, de-identified CT imaging datasets (LIDC-IDRI and LNDb), which do not require ethical approval or informed consent under current data-sharing regulations.
Data availability
All datasets used in this study are publicly accessible from the following sources:- LIDC-IDRI: https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI- LNDb: https://lndb.grand-challenge.org/- Tianchi-Lung: https://tianchi.aliyun.com/competition/entrance/231601/ information- NSCLC-Radiomics: https://wiki.cancerimagingarchive.net/display/Public/ NSCLC-Radiomics Code for the CoreFormer framework, training scripts, and evaluation protocols will be made available upon reasonable request. A public GitHub repository will be released after publication to promote reproducibility.
Code availability
Code for the CoreFormer framework, training scripts, and evaluation protocols will be made available upon reasonable request. A public GitHub repository will be released after publication to promote reproducibility.
References
Henschke, C. I. & McCauley, D. I. et al. Early lung cancer action project: overall design and findings from baseline screening. Lancet 354, 99–105 (1999).
Armato, S. G., McLennan, G. & Bidaut, L. et al. The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans. Med. Phys. 38, 915–931 (2011).
Gierada, D. S. et al. Pulmonary nodule detection and volumetry: influence of reconstruction techniques on nodule volume accuracy. Radiology 267, 326–333 (2013).
Setio, A. A. A., Traverso, A. & de Bel, T. et al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: the luna16 challenge. Med. image Anal. 42, 1–13 (2017).
Liu, C., Zhang, Y. & Zhang, Y. et al. Deep learning in medical ultrasound analysis: A review. Engineering 6, 261–275 (2020).
Ronneberger, O., Fischer. P. & Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, et al. editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Cham: Springer International Publishing; 2015. p. 234–41.
Çiçek, Ö., Abdulkadir, A., Lienkamp, S. S., Brox, T. & Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In: Ourselin, S. et al. editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016. p. 424–32 (Cham: Springer International Publishing,2016).
Park, J. J., Florence, P. R., Straub, J., Newcombe, R. A., Lovegrove, S., DeepSDF: Learning Continuous Signed Distance Functions for Shape Representation. In 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). 165–174 (2019).
Mescheder, L., Oechsle, M., Niemeyer, M., Nowozin S., Geiger, A., Occupancy Networks: Learning 3D Reconstruction in Function Space, In 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 4455–4465 (Long Beach, CA, USA, 2019).
Ma, J. et al. Neural implicit segmentation of 3d medical volumes with structured shape prior. Med. Image Anal. 71, 102049 (2021).
Hao, J. et al. Plus: Plug-and-play enhanced liver lesion diagnosis model on non-contrast ct scans. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2025, 461–471 (Springer Nature Switzerland, 2026).
Pedrosa, J. et al. Lndb: a lung nodule database on computed tomography. arXiv preprint arXiv:1911.13750 https://doi.org/10.48550/arXiv.1911.08434 (2019).
Ahmed, S., Subhan, F., Su’ud, M. M., Alam, M. M. & Waheed, A. A. Survey of lung nodules detection and classification from CT scan images. CSSE. 48:1483–1511 (2024).
Farag, A. A. et al. A novel approach for lung nodule segmentation using level sets. IEEE Trans. Image Process. 22, 5202–5213 (2013).
Milletari, F., Navab, N. & Ahmadi, S.-A. V-net: fully convolutional neural networks for volumetric medical image segmentation. In International Conference on 3D Vision (3DV) 565–571 (IEEE, 2016).
Gibson, E., Li, W. & Sudre, C. et al. Automatic multi-organ segmentation on abdominal CT with dense V-networks. IEEE Trans. Med. imaging 37, 1822–1834 (2018).
Oktay, O. et al. Attention U-Net: learning where to look for the pancreas. arXiv preprint https://doi.org/10.48550/arXiv.1804.03999 (2018).
Sitzmann, V. et al. Implicit neural representations with periodic activation functions. NeurIPS 33, 7462–7473 (2020).
Zhang, Q. et al. Stunet: Shape-aware implicit segmentation for pulmonary nodules. Med. Image Anal. 82, 102635 (2023).
Tian, Z., Shen, C., Chen H. & He, T. FCOS: Fully Convolutional One-Stage Object Detection, In 2019 IEEE/CVF International Conference on Computer Vision (ICCV), 9626–9635 (Seoul, Korea (South), 2019.
Zhou, X., Wang, D. & Krähenbühl, P. Objects as points. In arXiv preprint https://doi.org/10.48550/arXiv.1904.07850 (2019).
Li, Y. et al. Cenet: Center-enhanced convolutional network for lesion segmentation. IEEE Trans. Med. Imaging 41, 250–262 (2022).
Xie, E. et al. PolarMask: Single Shot Instance Segmentation With Polar Representation, In 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 12190−12199 (Seattle, WA, USA, 2020).
Chen, X. et al. Radial U-net for anatomical-aware retinal layer segmentation. Biomed. Opt. Express 13, 2140–2154 (2022).
Bai, W. et al. Semi-supervised learning for Network-based cardiac MR image segmentation. In medical image computing and Computer-assisted intervention − MICCAI 2017 (eds Descoteaux M. et al.) 253–260 (Cham, Springer International Publishing, 2017).
Li, X. et al. Shape-aware semi-supervised 3d segmentation for medical images. Med. Image Anal. 64, 101747 (2020).
Zhang, S., Yue, J., Wang, C., Liu X. & Wang, G. Box2Pseudo: A Semi-Supervised Learning Framework for Pulmonary Nodule Segmentation with Box-Prompt Pseudo Supervision, In 2023 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 1696–1703, (Istanbul, Turkiye, 2023).
Liu, X. et al. Semi-supervised segmentation with shape-aware consistency learning. Med. Image Anal. 78, 102411 (2022).
Tang, Y. et al. Rethinking semi-supervised medical image segmentation: a deep generative approach. Med. Image Anal. 78, 102383 (2022).
Aerts, H. J. et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 5, 4006 (2014).
Morozov, S. P. et al. Mosmeddata: Chest CT scans with COVID-19 related findings dataset. arXiv preprint https://doi.org/10.48550/arXiv.2005.06465 (2020).
Acknowledgements
This work was supported by National Key R&D Program of China (2023YFC2508604), Ningbo Natural Science Foundation (2022J256) and National Natural Science Foundation of China (82360460).
Author information
Authors and Affiliations
Contributions
Y.X. and M.Y. conceptualized the study, designed the methodology, and participated in securing research funding (Conceptualization, Methodology, Funding acquisition). C.X., C.Y. and Y.H. carried out data acquisition, curation, and investigation (Investigation, Data curation) and provided key resources, instruments, and technical support (Resources, Software). F.Y., L.J. and J.Z. drafted the initial manuscript and generated visualizations (Writing—Original Draft, Visualization). X.L. and B.Y. supervised the project, coordinated collaborations, and ensured administrative support (Supervision, Project administration). All authors contributed to reviewing and revising the manuscript critically for important intellectual content (Writing—Review and Editing) and approved the final version for submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xi, Y., Xu, C., Ye, F. et al. CoreFormer high fidelity pulmonary nodule segmentation with structural core priors and geodesic implicit fields. npj Digit. Med. 9, 48 (2026). https://doi.org/10.1038/s41746-025-02221-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41746-025-02221-w