Abstract
Idiopathic pulmonary fibrosis (IPF) is an irreversible lung condition that progresses over time, which ultimately results in respiratory failure and mortality. In this study, we found that PLAC8 was downregulated in the lungs of IPF patients based on GEO data, in bleomycin (BLM)-induced lungs of mice, and in primary murine alveolar epithelial type II (pmATII) cells and human lung epithelial cell A549 cells. Overexpression of PLAC8 facilitated autophagy and inhibited apoptosis of pmATII cells and A549 cells in vitro. Moreover, inhibition of autophagy or overexpression of p53 partially abolished the effects of PLAC8 on cell apoptosis. ATII cell-specific overexpression of PLAC8 alleviated BLM-induced pulmonary fibrosis in mice. Mechanistically, PLAC8 interacts with VCP-UFD1-NPLOC4 complex to promote p53 degradation and facilitate autophagy, resulting in inhibiting apoptosis of alveolar epithelial cells and attenuating pulmonary fibrosis. In summary, these findings indicate that PLAC8 may be a key target for therapeutic interventions in pulmonary fibrosis.
Similar content being viewed by others
Introduction
The most prevalent type of idiopathic interstitial pneumonia is idiopathic pulmonary fibrosis (IPF), a chronic, progressive, and irreversible lung disease characterized by progressive lung scarring in the interstitium of the lungs1. Extensive extracellular matrix and abnormal alveolar architecture lead to a decline in lung function and ultimately respiratory failure and death2. The pathogenetic mechanism of IPF is still unknown with few treatment options3. Now researchers have started to assume that the sustained or repetitive injury of epithelial cells including alveolar epithelial cells is one of the ways to trigger the initiation of IPF. The injured epithelial cells interact with multiple cell types through many signaling pathways, and fibroblasts and myofibroblast differentiation eventually are activated4,5. Alveolar epithelial type II (ATII) cells are epithelial stem cells that have a regenerative role in the alveolus to maintain lung homeostasis. ATII cell loss and aberrant activation will lead to dysregulated repair and pathogenic fibroblast activation6. Evidence supports that p53 is a critical factor in promoting apoptosis and senescence of ATII cells, which leads to pulmonary fibrosis7,8.
Autophagy is a common cellular homeostatic process that destroys the cellular substrates via autophagosome capture and fusion to degradative lysosomes. Autophagy functions as a pro-survival mechanism in the face of cellular stress, such as ER stress, starvation, and hypoxia9,10,11. Impaired autophagy has been seen as determined by p62 accumulation in lung tissues of IPF patients, which suggests that autophagy might be a pathogenic characteristic of IPF12. In bleomycin (BLM)-induced mouse fibrotic lung tissue, BLM activates autophagy determined by decreased p62 and increased LC3. More importantly, Atg4b deficiency impairs autophagy, which leads to exacerbating bleomycin-induced lung fibrosis13. In addition, in mouse type II alveolar epithelial line MLE-12 cells and human lung epithelial line A549 cells, BLM upregulates LC3II and downregulates p62 protein expression in vitro, and transcription factor EB (TFEB) activation by Torin 1 increases autophagic flux and improves BLM-induced pulmonary fibrosis in mice14. Autophagy may be dysregulated in aberrant epithelial cells and affect the apoptosis of ATII cells in IPF15. Meng et al also claimed that LC3B is mainly localized in Surfactant protein C (SPC)-positive cells (ATII cell marker), and inhibition of autophagy by 3-MA aggravates pulmonary fibrosis induced by BLM in mice16.
In this study, we found that placenta associated 8 (PLAC8) is markedly decreased in the lung tissues of IPF patients using the microarray dataset GSE110147 from Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/). Moreover, we established a pulmonary fibrosis murine model using BLM and collected the lung tissues for mRNA-seq analysis. The results showed that PLAC8 was also significantly decreased in BLM-treated mice. PLAC8 has a pro-survival function by promoting autophagic activity. For instance, in human macrophage THP-1 cells, forced expression of PLAC8 inhibits LPS-stimulated IL-1β and IL-18 expression, and inhibition of autophagy using 3-MA reduces the effects of PLAC8 in THP-1 cells17. PLAC8 promotes autophagy in human trophoblast cells by regulating p53 degradation, which results in increasing cell proliferation of trophoblast cells18. PLAC8 is a highly conserved protein between humans and murine and is expressed in lung tissue19. Therefore, we hypothesized that the downregulated PLAC8 in fibrotic lungs might be involved in impaired autophagy activity in IPF and induce apoptosis in alveolar epithelial cells.
In this study, we verified that overexpression of PLAC8 promotes autophagy and inhibits the apoptosis of primary mouse alveolar type 2 epithelial cells (pmATII) and A549 cells. Inhibition of autophagy or overexpression of p53 partially abolished the effects of PLAC8 on apoptosis. ATII cell-specific overexpression of PLAC8 using adenovirus alleviated BLM-induced pulmonary fibrosis in mice. Coimmunoprecipitation/mass spectrum (COIP/MS) and COIP analysis identified PLAC8 can interact with protein p53, VCP, UFD1 and NPLOC4 in lung epithelial cell A549 cells. We demonstrated that PLAC8 may engage in joint VCP-UFD1-NPLOC4 complex to promote p53 degradation and facilitate autophagy, resulting in inhibiting apoptosis of alveolar epithelial cells and attenuating pulmonary fibrosis.
Results
PLAC8 is decreased in lung tissues of IPF patients and BLM-induced fibrotic lungs in mice
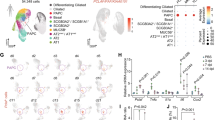
To investigate which factor plays important roles in the initiation of fibrosis in BLM-induced pulmonary fibrosis in mice, we examined differentially expressed genes (DEGs) in control and BML-treated groups by RNA sequencing (Fig. 1a). Volcano plots and heatmaps showed the DEGs in Fig. 1b, c. The DEGs were presented in Supplementary data 1. We analyzed a public microarray dataset GSE110147 and obtained the DEGs between lung tissues of IPF patients and control donors (Fig. 1d). By crossing the DEGs in our result and GSE110147, we found that PLAC8 was decreased both in lung tissues from BLM-treated mice and IPF patients (Fig. 1b, d–f). The distribution of DEGs following BLM treatment in different mouse chromosomes was displayed and PLAC8 is located on chromosome 5 (Fig. 1g). The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of common DEGs demonstrated that the DEGs were mainly enriched in the regulation of immune responses, cell adhesion, and extracellular matrix, all of which are pathways related to pulmonary fibrosis (Fig. 1h, i).
a Schematic diagram of the RNA sequencing of lung tissues from BLM-treated mice. The lung tissues were collected on day 3 after BLM treatment. (The schematic pictures were downloaded from sciDraw, mouse: https://doi.org/10.5281/zenodo.3925901; syringe: https://doi.org/10.5281/zenodo.4152947). b The volcano plot of mRNA profiling between the lung tissues of BLM-treated and control mice (n = 6 for the control group, n = 5 for the BLM group). The blue and red dots represent downregulated and upregulated DEGs, respectively. c The heatmap of the expression patterns of DEGs. d Microarray dataset GSE110147 from the Gene Expression Omnibus (GEO) database contained lung tissues of IPF patients and control lung tissues (n = 11 for the control group, n = 22 for the IPF group). The volcano plot of mRNA profiling between the lung tissues of IPF patients and control donors. The blue and red dots represent downregulated and upregulated differentially expressed genes (DEGs), respectively. e The expression of PLAC8 in the GSE110147 dataset. f Venn diagram exhibited the common gene between the GSE110147 dataset and mRNA-seq from BLM-treated mice. g DEG distribution in mouse chromosomes following BLM treatment. The red and blue bars represent upregulated and downregulated DEGs, respectively. h, i KEGG enrichment analysis and GO enrichment analysis (BP biological processes, CC cellular component, MF molecular function).
To confirm the altered expression of PLAC8 in lung tissues of BLM-treated mice, we established a BLM-induced fibrosis model and examined the expression of PLAC8 in the 3, 7, and 14 days after BLM treatment. Hematoxylin and eosin (HE) and Masson staining on day 14 demonstrated that BLM induced fibrosis in mouse lungs (Fig. 2a, b). In addition, the apoptosis of ATII cells was observably increased (Fig. 2c, d). The ATII marker SPC expression slightly decreased on day 3, but markedly on day 7 and day 14 after BLM treatment. Importantly, the expression of PLAC8 was decreased starting from day 3 and had a time-dependent decline from day 3 to day 14 (Fig. 2e). Moreover, apoptosis and autophagy were induced by BLM in fibrotic lungs. The expression change of pro-apoptotic protein Cleaved caspase-3 was observed on day 3 and the autophagy marker LC3II was slightly increased when p62 was decreased starting from day 3 (Fig. 2f). It’s indicated that BLM-induced initiation of apoptosis and autophagy in lung tissues at the early stage of lung fibrosis. Next, we demonstrated that PLAC8 was downregulated in alveolar epithelial cells (E-cadherin, epithelial cell marker) (Fig. 2g, h). The results suggested that BLM decreased PLAC8 expression in alveolar epithelial cells and PLAC8 likely plays an important role in pulmonary fibrosis.
a, b Hematoxylin and eosin (HE) and Masson’s trichrome staining of lung tissues on day 14 post-BLM administration. c Immunofluorescence analysis of the Surfactant protein C (SPC, green) and TUNEL (red) in lung tissues. The white arrows were pointing to the double positive cells. d The percentages of double TUNEL + SPC+ positive cells in the SPC+ positive cells were determined. e, f Immunoblotting and semi-quantitative analysis of SPC, PLAC8, cleaved caspase-3, LC3, and p62 protein levels in lungs on indicated days after BLM treatment. g Immunofluorescence analysis of the PLAC8 (green) and E-cadherin (red) in lung tissues on day 3 after BLM treatment. h The percentages of double PLAC8 + E-cadherin + positive cells in the E-cadherin+ positive cells were determined. Data represent the mean ± SD (6 mice per group; representative of three independent experiments). Unpaired t-test applied for (d and h), and Tukey’s multiple comparison test after the one-way ANOVA was conducted for (e, f). *P < 0.05. NS non-significant.
Overexpression of PLAC8 inhibits BLM-induced apoptosis of pmATII cells and A549 cells in vitro
In vitro assay, BLM significantly decreased PLAC8 in pmATII cells and A549 cells determined by RT-qPCR and western blot (Fig. 3a–d). Therefore, we tried to uncover the role of PLAC8 in BLM-induced cell damage. PLAC8 was overexpressed in pmATII cells and A549 cells by adenovirus and overexpressed plasmid, respectively (Fig. 3e–h). BLM induced the apoptosis of pmATII cells and A549 cells and forced expression of PLAC8 suppressed the apoptosis after BLM treatment (Fig. 3i–l). The results indicated that PLAC8 can inhibit BLM-induced apoptosis of pmATII cells and A549 cells in vitro.
a–d The pmATII cells and A549 cells were treated with 50 μM BLM for 24 h. RT-qPCR analysis and immunoblotting analysis of PLAC8 levels. e, f The mRNA and protein levels of PLAC8 in pmATII cells after infection of adenovirus containing the mouse PLAC8 overexpression vector (oe-PLAC8) or negative control (oe-NC). g, h The mRNA and protein levels of PLAC8 in A549 cells after transfection of PLAC8 overexpression plasmid (OE-PLAC8) or negative control (OE-NC). Immunoblotting analysis of cleaved caspase-3 protein levels in PLAC8-overexpressing pmATII cells (i) and A549 cells (j) after BLM treatment. Cell apoptosis was detected by TUNEL staining of pmATII cells (k) and A549 cells (l). Data represent the mean ± SD (n = 3 per group; representative of three independent experiments. Unpaired t-test applied for (a and c), and Tukey’s multiple comparison test after the one-way ANOVA was conducted for (e–j). *P < 0.05.
Overexpression of PLAC8 facilitates autophagy in BLM-treated pmATII cells and A549 cells
Induction of autophagy is a survival-promoting pathway when the cell is damaged. PLAC8 has been shown to inhibit apoptosis by promoting autophagy. Therefore, we presumed that PLAC8 can facilitate autophagy in BLM-treated ATII cells. To verify our hypothesis, we detected the autophagic activity by monitoring the autolysosomes using transmission electron microscopy (TEM). Autolysosome was found in BLM-treated pmATII cells and A549 cells, and more autolysosomes were found in PLAC8-overexpressing cells (Fig. 4a, b). The mCherry-GFP-LC3B reporter was used as a probe to estimate the populations of autophagosomes (yellow fluorescence) and autolysosomes (red fluorescence) in A549 cells. BLM treatment boosted LC3II expression and LC3B puncta formation, and PLAC8 overexpression accelerated autophagic flux as evidenced by reduced yellow autophagosome fluorescence and increased red autolysosome fluorescence compared to the NC group under BLM administration (Fig. 4c). Moreover, overexpression of PLAC8 reduced LC3II and p62 levels in pmATII and A549 cells (Fig. 4d, e). Moreover, inhibition of autophagy by CQ reversed the PLAC8-prevented cell apoptosis of pmATII cells and A549 cells, as evaluated by cleaved caspase-3 expression and TUNEL staining (Fig. 4f–i). These data indicate that forced expression of PLAC8 inhibits cell apoptosis by facilitating autophagy in BLM-treated pmATII cells and A549 cells.
a, b Transmission electron microscopy (TEM) images of PLAC8-overexpressing pmATII cells and A549 cells following BLM treatment. c A549 cells were transfected with OE-PLAC8 or OE-NC and infected with adenovirus-containing a mCherry- EGFP-LC3B reporter, and cells then were treated with BLM for 24 h. Autophagic flux was measured by counting the LC3 puncta including yellow fluorescence dots (autophagosomes) and red fluorescence dots (autolysosomes) under a microscope. Immunoblotting analysis of LC3 and p62 protein levels in pmATII cells (d) and A549 cells (e). The PLAC8-overexpressed pmATII cells (f) and A549 cells (h) were treated with BLM and autophagy inhibitor CQ (20 μM) for 24 h, and cell apoptosis was detected by TUNEL staining. g, i Immunoblotting analysis of cleaved caspase-3 protein levels after BLM and CQ treatment. Data represent the mean ± SD (n = 3 per group; representative of three independent experiments. Tukey’s multiple comparison test after the one-way ANOVA was conducted for (c–i). *P < 0.05. NS non-significant.
PLAC8 inhibits cell apoptosis by decreasing p53 expression in BLM-treated pmATII cells and A549 cells
In this study, we attempted to determine whether PLAC8 affects apoptosis and autophagy by regulating p53 expression. Our results showed that a time-dependent upregulation of the p53 expression in lung tissues was observed in BLM-treated mice (Fig. 5a). In pmATII cells and A549 cells, BLM also induced the p53 expression and overexpression of PLAC8 downregulated p53 (Fig. 5b, c). Forced expression of p53 increased apoptosis and aggravated the autophagy dysregulation in PLAC8-overexpressing pmATII cells (Fig. 5d–f) and A549 cells (Fig. 5g–i). These findings indicated that PLAC8 inhibits cell apoptosis by suppressing p53 expression in BLM-treated pmATII cells and A549 cells.
a Immunoblotting analysis of p53 protein levels in lungs on days 0, 3, 7, and 14 after BLM treatment. The PLAC8-overexpressed pmATII cells (b) and A549 cells (c) were treated with BLM for 24 h, and p53 protein levels were detected by immunoblotting analysis. The pmATII cells were infected with oe-PLAC8 with or without oe-TP53 before being treated with BLM. Immunoblotting analysis of cleaved caspase-3 (d), LC3, and p62 (e) protein levels were detected. f Cell apoptosis was detected by TUNEL staining. A549 cells were transfected with OE-PLAC8 with or without OE-TP53 before being treated with BLM. Immunoblotting analysis of cleaved caspase-3 (g), LC3, and p62 (h) levels was detected. i Cell apoptosis was detected by TUNEL staining. Data represent the mean ± SD (n = 3 per group; representative of three independent experiments. Tukey’s multiple comparison test after the one-way ANOVA was conducted for (a–h). *P < 0.05. NS non-significant.
Overexpression of PLAC8 mitigates BLM-induced pulmonary fibrosis in mice
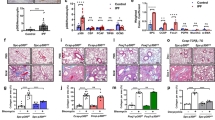
To evaluate the therapeutic potential of PLAC8 for pulmonary fibrosis, mice were given ATII cell-specific overexpression of PLAC8 plasmid driven by adenovirus contain Surfactant protein B (SPB) promoter (Ad-SPB-oePLAC8) or negative control Ad-SPB-oeNC one day before BLM treatment to specifically overexpress PLAC8 in mouse lung ATII cells. Subsequently, mice received BLM by intratracheal instillation (I.T.) on the next day, and the pulmonary fibrosis was assessed on day 14 after BLM treatment (Fig. 6a). We found that overexpression of PLAC8 in ATII cells inhibited pulmonary fibrosis, evaluated by HE and Masson’s trichrome staining (Fig. 6b–e). When compared to the Ad-SPB-oeNC-treated group, the lung/body coefficient was decreased in the Ad-SPB-oePLAC8 group (Fig. 6f). Collagen content as determined by hydroxyproline content and it was also decreased by PLAC8 overexpression (Fig. 6g). We confirmed that PLAC8 was overexpressed by Ad-SPB-oePLAC8 in lung tissues by western blot. Moreover, the protein levels of fibrosis markers α-SMA, collagen I and fibronectin were decreased (Fig. 6h, i). PLAC8 inhibited the p53 expression and further induced autophagy under BLM treatment (Fig. 6j, k). These results indicated that ATII-specific overexpression of PLAC8 could mitigate BLM-induced pulmonary fibrosis in mice.
a Schematic diagram of the construction of alveolar epithelial type II (ATII)-specific overexpression of PLAC8 under the Surfactant protein B (SPB) promoter and BLM-induced mouse lung fibrosis in mice. (The schematic pictures were downloaded from sciDraw, mouse: doi.org/10.5281/zenodo.3925901; syringe: 10.5281/zenodo.4152947). b, c Hematoxylin and eosin (HE) staining of lung tissues on day 14 post-BLM administration and quantification of lung fibrosis by the Ashcroft score. d, e Masson’s trichrome staining of lung tissues and quantification of lung fibrosis. f Lung/body coefficient was calculated. g Collagen measurement by the hydroxyproline assay in lungs. Immunoblotting analysis of PLAC8 and α-SMA (h), collagen I and fibronectin (i), p53 (j), LC3, and p62 (k) in lung tissues of mice. Data represent the mean ± SD (6 mice per group; representative of three independent experiments). Tukey’s multiple comparison test after the one-way ANOVA was conducted for (c–k). *P < 0.05.
Bioinformatic analysis of PLAC8-interacting proteins in A549 cells
In this study, to investigate the regulatory mechanism of PLAC8 affecting p53 expression in ATII cells, we performed a COIP/MS assay in PLAC8-overexpressed A549 cell lysate with an anti-PLAC8 antibody, and a total of 2161 PLAC8-specific binding proteins were identified using MS analysis (Fig. 7a, b and Supplementary data 2). Bioinformatic analysis of PLAC8-interacting proteins by the GO and KEGG enrichment analyses was carried out. PLAC8-interacting proteins are significantly enriched in the lysosome, autophagy, ubiquitin-mediated proteolysis, and apoptosis in KEGG analysis (Fig. 7c, d). Additionally, PLAC8-interacting proteins in GO analysis are significantly mainly enriched in p53 binding, protein acetylation, protein ubiquitination, DNA-binding transcription factor binding, lysosome organization, and regulation of autophagy (Fig. 7e). The data indicated that PLAC8-interacting proteins may participate in cell apoptosis, autophagy, acetylation- and ubiquitination-regulated protein activity or degradation, and transcription factor activity. However, p53 was not found in PLAC8-interacting proteins in A549 cells from the MS analysis. This may be because the expression of p53 is very low in A549 cells in the absence of BLM treatment (Fig. 5c), and overexpression of PLAC8 further downregulated p53 expression, thus MS analysis did not detect p53 fragment signals. Since the expression of p53 is mainly mediated by acetylation and ubiquitination of p53, we analyzed the common proteins in GO terms of biological process (BP) including autophagy-related pathways, protein ubiquitination, and protein acetylation. Many proteins are co-enriched in at least two GO terms of BP including autophagy, autophagosome maturation, lysosome organization, and proteasome-mediated ubiquitin-dependent protein catabolic process (Fig. 7f). However, PLAC8-interacting proteins, which can influence protein acetylation, are quite different from autophagy-related proteins. Next, we analyzed the proteins in GO terms of molecular function (MF) from Fig. 7e including p53 binding, DNA-binding transcription factor binding, ubiquitin-dependent protein binding, and ubiquitination-like modification-dependent protein binding. TP53BP1, SMARCB1, HDAC1, TP53BP2, GSK3B, DNAJB2, VCP, IDE, and USP15 are the common proteins in these MF pathways (Fig. 7g), and protein-protein interaction (PPI) network analysis by STRING database (https://cn.string-db.org/) indicated that these proteins, except DNAJB2, might interact with p53 and influence p53 expression or activity (Fig. 7h).
a Schematic diagram of the COIP/MS analysis of PLAC8-interacting proteins in A549 cells. b Percentages of specific PLAC8-interacting proteins in A549 cells. c KEGG enrichment analysis of PLAC8-interacting proteins. d The proteins enriched in Lysosome, Apoptosis, and Autophagy pathways in KGEE from (c). e GO enrichment analysis of PLAC8-interacting proteins. f The proteins enriched in Autophagy, autophagosome maturation, lysosome organization, and proteasome-mediated ubiquitin-dependent protein catabolic process pathways in GO analysis of BP from (e). g The proteins enriched in p53 binding, DNA-binding transcription factor binding, Ubiquitination-like modification-dependent protein binding, and Ubiquitination-dependent-protein binding in GO analysis of MF from (e). h Protein-protein interaction (PPI) network analysis of protein from (g) from the STRING database (https://cn.string-db.org/).
PLAC8 inhibits p53 expression through VCP-medicated ubiquitin-mediated protein degradation
Based on the literature search, we found that the interaction between VCP and PLAC8 has not yet been revealed. We selected VCP as the target protein of PLAC8 for further study. First, we identified that PLAC8 can interact with VCP in A549 cells. It is well known that the VCP-UFD1-NPLOC4 complex facilitates protein degradation by the ubiquitin-proteasome system and it has been reported that VCP can interact and reduce p53 expression20,21. Therefore, we searched the PLAC8-interacting protein list and found that UFD1 and NPLOC4 also bind with PLAC8 in A549 cells. We hypothesized that PLAC8 can interact with the VCP-UFD1-NPLOC4 complex and facilitate p53 degradation exposed to BLM. To verify our hypothesis, using an anti-PLAC8 antibody to precipitate the proteins in BLM-treated A549 cells, we found that VCP, UFD1, NPLOC4, and p53 could be coimmunoprecipitated and BLM reduced the interaction among PLAC8 and p53, VCP, UFD1 and NPLOC4 (Fig. 8a, b). It also suggested that p53 was not found in the MS analysis data because of the low expression of p53 in A549 cells without BLM treatment. We demonstrated that overexpression of PLAC8 reduced the accumulation of ubiquitinated p53 and increased degradation of p53 (Fig. 8c, d), whereas knockdown of VCP or UFD1 resulted in the accumulation of ubiquitinated p53 and inhibited the p53 degradation (Fig. 8e–g). Next, we demonstrated that the knockdown of VCP weakened the effects of PLAC8 on A549 cells by impairing autophagy and inducing apoptosis of BLM-treated cells (Fig. 8h, i). In summary, the results indicated that PLAC8 inhibits p53 expression through binding with VCP-UFD1-NPLOC4 complex and facilitating p53 degradation.
a Coimmunoprecipitation (COIP) verified the interaction among PLAC8, p53, VCP, UFD1, and NPLOC4 in BLM-treated cells. b Protein-protein interaction (PPI) network analysis of PLAC8, p53, VCP, UFD1, and NPLOC4 in ATII cells based on STRING database, COIP/MS analysis, and COIP experimentally determined. c A549 cells were transduced with OE-PLAC8 and incubated with MG132 and BLM, the ubiquitinated p53 was detected. d The p53 degradation was measured in PLAC8 overexpressing cells after incubation with CHX and BLM. e Cells were transfected with OE-PLAC8 and siRNA against VCP (siVCP) or UFD1 (siUFD1) and then treated with MG132 and BLM. The accumulation of ubiquitinated p53 was detected. f, g Immunoblotting analysis of the p53 expression and degradation in OE-PLAC8 and siVCP transfected cells. h Immunoblotting analysis of LC3 and p62 in A549 cells. i Cell apoptosis was detected by TUNEL staining. Data represent the mean ± SD (n = 3 per group; representative of three independent experiments). Tukey’s multiple comparison test after the one-way ANOVA was conducted for (f and h), Sidak’s test multiple comparison test two-way ANOVA was conducted for (d and g). *P < 0.05. NS non-significant.
Discussion
Injury of alveolar epithelial cells is a critical step for the initiation of IPF. Alveolar epithelial type I (ATI) cells are responsible for maintaining the structural integrity of the epithelial surface area and gas exchange. ATII cells are the primary progenitors for ATI cells5,22. ATII cells are more likely to suffer from apoptosis, senescence, and abnormal differentiation, which will impair the renewal capacity of ATII cells in IPF lungs. Growing evidence supports that the reduction of the apoptosis of ATII cells relieves pulmonary fibrosis. Chen et al found that Testis-specific serine kinase 4 (TSSK4) is selectively upregulated in ATII cells in fibrotic lungs of mice and knockdown of TSSK4 ameliorates bleomycin (BLM)-induced lung fibrosis through inhibiting the apoptosis of ATII cells23. ATII-specific knockout of endoplasmic reticulum (ER) chaperone Grp78 accelerates tamoxifen-induced lung fibrosis in mice by inducing ER stress, apoptosis, and senescence in ATII cells24. In addition, thyroid hormone improves ATII cell mitochondrial function and inhibits mitochondria-regulated apoptosis in BLM-treated mice25. Our results showed that PLAC8 was decreased in fibrotic lung both in IPF patients and BLM-treated mice. Overexpression of PLAC8 promoted autophagy and reduced BLM-induced apoptosis in pmATII cells and A549 cells. Moreover, specific overexpression of PLAC8 in ATII cells in mice relieved the BLM-induced pulmonary fibrosis.
Growing literature demonstrates that PLAC8 can inhibit cell apoptosis. Overexpression of PLAC8 reduces cell apoptosis in breast cancer cells, and knockout of PLAC8 enhances radio-sensitivity of nasopharyngeal carcinoma cells by promoting apoptosis26,27. In addition, the downregulation of PLAC8 reduced the cell viability of rat insulinoma cell lines INS1 cells and RINm5F cells28. Autophagy functions as a pro-survival role and anti-inflammatory effect on injured cells. PLAC8 reduced the inflammatory factors IL-1b and IL-18 production by promoting autophagy activation17. Overexpression of PLAC8 promotes autophagic activity and increases the cell viability of trophoblast cells by aggravating p53 degradation18. Knockdown of PLAC8 increases LC3II accumulation, and p62 expression and inhibits autophagosome-lysosome fusion in pancreatic cancer cells29. In CNE-2 cells, however, knockout of PLAC8 induced autophagy, increased LC3II expression, and decreased p62 expression30, probably due to proteins having different behavior and functions depending on the different cellular contexts. Our results showed that PLAC8 inhibited apoptosis and improved autophagy dysregulation in ATII cells, an autophagy inhibitor CQ partially abolished the roles of PLAC8 in apoptosis and autophagy. It’s suggested that PLAC8 inhibits apoptosis by promoting autophagy in BLM-induced ATII cells.
The expression of p53 is increased in lung tissues and isolated alveolar epithelial cells from IPF patients8,31. It has been reported that p53 is upregulated in lung tissues in silica-induced and BLM-induced pulmonary fibrosis, and downregulation of p53 could alleviate pulmonary fibrosis in both pulmonary fibrosis models32,33. In damaged ATII cells, p53 can induce cell apoptosis, senescence, and epithelial-mesenchymal transition (EMT)8,34,35, which will contribute to the initiation of pulmonary fibrosis. Caveolin‑1‑derived 7‑mer peptide (CSP7) improves pulmonary fibrosis by inhibiting autophagy dysregulation in alveolar epithelial cells. CSP7 decreases p53, LC3II, and p62 expression, and the knockout of p53 in alveolar epithelial cells abrogates the effects of CSP7 on autophagy36. Consistent with their report, our results showed that p53 protein expression was increased in mouse primary ATII cells and human A549 cells. Moreover, the forced expression of p53 partially reversed the effects of PLAC8 on BLM-induced cell apoptosis and autophagy, determined by increased cleaved caspase-3, LC3II accumulation, and p62 expression.
COIP/MS analysis was used to mine potential regulators in alveolar epithelial cells that may be involved in the regulation of p53 by PLAC8. The data showed that PLAC8-interacting proteins are enriched pathways including apoptosis, autophagy, lysosome organization, autophagosome maturation, proteasome assembly and ubiquitin-mediated proteolysis, and p53 binding. The protein p53 degradation can be mediated by the ubiquitin-proteasome system (UPS) and autophagy-lysosomal (AL) pathway37,38. Ubiquitin is a unifying factor between the UPS and selective autophagy. Numerous ubiquitylated proteins are known to be targets of both the degradative systems, which could be degraded by the UPS and AL pathways39. Therefore, in ATII cells, overexpression of PLAC8 may decrease p53 expression by promoting protein degradation. We identified some common PLAC8-interacting proteins, enriched in molecular function pathways, that might interact with p53 and regulate its expression and activity. In this study, we verified the VCP and its cofactors UFD1 and NPLOC4 can bind with PLAC8 and p53 in BLM-treated A549 cells. VCP forms a complex with UFD1 and NPLOC4 to promote the degradation of ubiquitinated proteins through delivery to the proteasome20,40,41. VCP can interact with p53, and the knockdown of VCP upregulates the p53 protein level21. We found that the knockdown of VCP and UFD1 inhibited PLAC8-increased p53 degradation and autophagy. Moreover, siVCP promoted cell apoptosis in PLAC8 overexpressing cells. It indicated that PLAC8 mediated p53 expression, at least partially, through interacting with the VCP-UFD1-NPLOC4 complex.
This study is limited by several factors. A549 cell line is not ATII cell line, but it is served as a model for type II alveolar epithelial cells in vitro assay. There is an inherent experimental constraint because of its cancerous character. Thus, the pmATII cells were isolated from mice to verify the role of PLAC8 in ATII cells along with A549 cells. Additionally, our results cannot strictly distinguish between alveolar epithelial type I cells (ATI cells) and alveolar epithelial type II cells (ATII cells) in terms of PLAC8 function. We mainly focused the role of PLAC8 on apoptosis and autophagy in ATII cells as these cells are stem cells that can differentiate into ATI cells. Therefore, we isolated pmATII cells for in vitro validation tests.
In conclusion, PLAC8 is decreased in in the IPF patients’ lungs and BLM-treated lungs. Overexpression of PLAC8 inhibited cell apoptosis of pmATII cells and A549 cells in vitro and reduced BLM-induced lung fibrosis in vivo. Mechanistic studies indicated that PLAC8 inhibits apoptosis of ATII cells and promotes autophagy through inhibiting p53 expression. In addition, we identified PLAC8-interacting proteins in A549 cells and provided more potential targets of PLAC8 in IPF. Our findings are anticipated to be of important value for better understanding the pathogenesis of IPF and the potential clinical use of IPF given the significance of PLAC8 downregulation in alveolar epithelial cell for cell apoptosis.
Methods
Cell isolation and culture
The male 7-week-old C57BL/6 mice were used to isolate pmATII following prior descriptions42. Briefly, the lungs were broken down and minced in dispase (MX1006, MKBio, China), and the cell suspension was then passed through nylon meshes that ranged in size from 100-μm, 20-μm to 10-μm. Cell pellets were resuspended after being centrifuged at 200 × g for 10 min with the filtered cell suspension. Macrophages and white blood cells were eliminated using the CD45 MicroBead (130-052-301, Miltenyi Biotec GmbH, Germany), and endothelial cells were eliminated using the CD31 MicroBead (130-097-418, Miltenyi Biotec GmbH) following the manufacturer’s recommendations. The purified pmATII cells were identified by expressing epithelial marker prosurfactant protein C (pro-SPC) instead of the CD45 and CD31 using immunofluorescence staining assay (Supplementary Fig. 1). The purified cells were cultured in the DMEM complete medium supplied with 10% FBS. A549 cells (iCell Bioscience, Shanghai, China) were cultured in F-12K medium with 10% FBS.
Construction of adenovirus, recombinant plasmids, and siRNA
Herein, an adenovirus vector pDC315 (VT1500, YouBio, Changsha, China) was used to construct the Ad-SPB-oePLAC8, which will effectively and specifically overexpress PLAC8 in mouse lung ATII cells in vivo. The Ad-SPB-oePLAC8 and negative control Ad-SPB-oeNC were constructed by General Biol (Anhui, China). Briefly, the MCMV promoter in pDC315 had been removed by digestion with XbaI and EcoRI, and the synthesized SPB promoter fragment was digested with XbaI and EcoRI and inserted into pDC315 (Ad-SPB-pDC315). The CDS of mouse Plac8 was ligated with Ad-SPB-pDC315 by digestion with NheI and BamHI. The resulting plasmid, Ad-SPB-oePLAC8 was constructed and the Ad-SPB-pDC315 was considered as negative control and termed Ad-SPB-oeNC. In vitro assay, adenovirus vectors containing the CDS of mouse Plac8 and Tp53 were constructed to overexpress PLAC8 (Ad-oe-PLAC8) or p53 (Ad-oe-TP53) in pmATII cells. Adenoviruses were subject to be packaged, multiplied, purified, and gotten ready to use. Recombinant plasmids overexpressed PLAC8 (OE-PLAC8) and TP53 (OE-TP53) were constructed to overexpress human PLAC8 and TP53 in A549 cells. The empty plasmid was considered to be a negative control (OE-NC). PmATII cells were infected with the appropriate adenovirus, and A549 cells were transfected with indicated overexpressing plasmids for 24 h. The mRNA and protein levels of target genes were detected using real-time reverse transcriptase-polymerase chain reaction (RT-qPCR) and western blot, respectively. The siRNA against VCP (siVCP, GGGCACAUGUGAUUGUUAUTT) and UFD1 (siUFD1, GGAGAGCGUCAACCUUCAATT) and negative control sequence (siNC) were synthesized by JTS Scientific (Wuhan, China).
Animal experiments
Male C57BL/6 mice that were 8 weeks old and pathogen-free were purchased from Liaoning Changsheng Biotechnology (Benxi, China). The mice were kept in an environment with a 12-h light/dark cycle and temperature was maintained at 22 ± 1 °C, and unfettered access to food and water. Mice were randomly divided into control mice and model mice that had received BLM treatment. Mice from the BLM group received 3 U/kg BLM (B802467, MACKLIN, China) in 50 μL by intratracheal instillation (I.T.) to induce lung fibrosis. The procedure was carried out on day 0. An identical quantity of saline solution was given to the control mice. The mice were sacrificed at 3, 7, and 14 days after being exposed to bleomycin or saline.
Mice were randomly assigned to four groups to study the effects of PLAC8 on lung fibrosis: Control, BLM, BLM + Ad-SPB-oeNC, and BLM + Ad-SPB-oePLAC8 Adenovirus (5 × 109 pfu) carrying the Plac8 overexpression vector driven by SPB promoter (Ad-SPB-oePLAC8) or control vector (Ad-SPB-oeNC) was given to mice from BLM + Ad-SPB-oePLAC8 or BLM + Ad-SPB-oeNC group by I.T. on day −1. The BLM or saline was administered to mice on day 0. Prior to sacrifice, mice were weighed (body weight) on day 14. Subsequently, the isolated lungs were weighed (lung weight). The lung/body coefficient of mice was calculated using the following formula = lung weight/body weight × 100%. After that, lung tissues were used for further analyses.
Cell treatments
The pmATII cells and A549 cells were received 50 μM BLM for 24 h and then subjected to mRNA and protein detection using RT-qPCR and western blot. A549 cells were transfected for 24 h with the overexpressing vectors (OE-PLAC8, OE-TP53) or siRNAs (siVCP, siUFD1), and pmATII cells were infected with the adenovirus (Ad-oe-PLAC8, Ad-oe-TP53) before being treated with BLM for 24 h. To detect the effect of autophagy on apoptosis, A549 cells were transfected with OE-PLAC8 for 24 h, and then treated with BLM with or without autophagy inhibitor chloroquine (CQ, 20 μM) for 24 h. Subsequently, the cells were gathered for TUNEL staining and cleaved caspase 3 detection. To detect cellular autophagic flux, A549 cells were transfected with the OE-PLAC8 and then infected with an adenovirus-containing mCherry-GFP-LC3B reporter (WZ Bioscience, Jinan, Shandong) for 24 h. After treatment with BLM, the cells were fixed and the numbers of autophagosomes (yellow dots) and autolysosomes (red dots) in A549 cells were counted using a fluorescence microscope. In addition, the evaluation of autophagy in situ in the PLAC8 overexpressing pmATII cells and A549 cells was also carried out using a transmission Electron Microscopy (H-7650, Hitachi, Japan). To demonstrate the effect of PLAC8 and the VCP-UFD1-NPLOC4 complex on p53 ubiquitination degradation, A549 cells were transfected with OE-PLAC8, siVCP, and siUFD1, and treated with a proteasomal inhibitor MG132 (10 μM) and BLM for 24 h. The ubiquitination of p53 was detected using a COIP assay. In other experiments, A549 cells were added with 20 μg/mL cycloheximide (CHX, protein synthesis inhibitor) and BLM, and cultured at different times. Subsequently, the protein levels p53 were measured and normalized to the protein levels at 0 h.
Hematoxylin and eosin (HE) and Masson’s trichrome staining
The lung tissues from the mice were separated, instantly fixed, and then embedded in paraffin for histological investigation. To evaluate the overall lung architecture, the lung tissues were cut into 5 μm sections and stained with hematoxylin and eosin Y. Ashcroft score was used to evaluate lung fibrosis43. In Masson’s trichrome staining for evaluating tissue fibrosis, the sections were stained with hematoxylin, ponceau-acid fuchsin solution, phosphomolybdic acid, and aniline blue solution in order. The results were observed under the microscope and the images were photographed.
Hydroxyproline measurement
A hydroxyproline assay kit was purchased from Nanjing Jiancheng Bioengineering Institute (A030, Nanjing, China) and was used to measure the amount of hydroxyproline in the lungs of mice.
Immunofluorescence and TUNEL staining
Lung sections were blocked for 15 or 30 min at room temperature with 1% BSA (A602440-0050, sangon) and co-incubated with the primary mouse anti-E-cadherin (60335-1-Ig, Proteintech, Wuhan, China) and rabbit anti-PLAC8 antibody (orb462681, Biorbyt, Cambridge, UK) at 4 °C overnight. Subsequently, the sections were treated with s either FITC- or Cy3-conjugated second antibodies. For TUNEL staining in lung tissues, sections were stained with TUNEL solution and rabbit anti-SPC primary antibody (10774-1-AP, Proteintech) in order. After washing, the sections were stained second antibody to capture the anti-SPC staining. The cell apoptosis of ATII cells in vitro was detected using a TUNEL staining assay as well. For nuclear staining, the sections and cell slides were stained with DAPI (D106471, Aladdin, China).
RT-qPCR
Total RNA was extracted from lung tissues using TRIpure reagent (RP1001, BioTeke, China) and cells, and BeyoRTTM II M-MLV reverse transcriptase (D7160L, Beyotime) was used following the manufacturer’s instructions to perform reverse-transcriptase reactions. The PCR Master Mix (PC1150, Solarbio), primers, cDNA, and SYBR Green were mixed and the RT-qPCR was performed. The RT-qPCR primers are as follows:
Mus Plac8: Forward, 5′- GGCACCAACAGTTATCG -3′,
Reverse 5′- AAAGGTCCCACAGAGGC -3′,
Homo PLAC8: Forward, 5′- GGCATCCCTGGATCTATT -3′,
Reverse 5′- GAAAGTACGCATGGCTCT -3′.
The relative level of GAPDH expression was used to normalize the target gene’s relative expression.
Western blot
RIPA lysis buffer was used to extract the total protein from lung tissues and cells, and separated by SDS-PAGE. The protein gels were transferred to PVDF membranes (IPVH00010, Millipore). The membranes were incubated with Western Blocking Buffer (SW3010, Solarbio) for 1 h followed by incubation with primary antibodies: anti-SPC (10774-1-AP), anti-PLAC8 (12284-1-AP, Proteintech), anti-Cleaved Caspase-3 (AF7022, Affinity), anti-LC3B (A19665, Abclonal, Wuhan, China), anti-p62 (A19700, ABclonal), anti-p53 (AF0865, Affinity), anti-α-SMA (AF1032, Affinity), anti-Collagen I (A1352, ABclonal), and anti-Fibronectin (A16678, ABclonal), GAPDH (60004-1-Ig, Proteintech) were incubated overnight at 4 °C. The secondary antibodies were separately incubated with membranes for 1 h. The proteins were observed with ECL reagent (PE0010, Solarbio) and gel imaging system.
GEO data, mRNA-seq, and COIP/MS analysis and preprocessing
Microarray dataset GSE110147 from GEO contained twenty-two samples of IPF patients and eleven normal lung tissues44. DEGs in lung tissues from the IPF group and control group were identified with |log2FC| ≥ 1 and p-value < 0.05 using GEO2R. The lung tissues of control mice and BLM-treated mice from day 3 were subjected to mRNA sequencing by Lianchuan Biological Technology (Hangzhou, China). Using |log2FC| ≥ 1 and p-value < 0.05 as the threshold, DEGs were screened out. After identifying the common DEGs from GSE110147 and mRNA-seq of lung tissues from BLM-treated and Control mice, the GO and KEGG pathway enrichment of DEGs were conducted by “R package”.
To analyze the PLAC8 specifically interacting proteins, A549 cells were transfected with OE-PLAC8 and COIP assay was performed. Subsequently, IgG and PLAC8 antibody (#13885, Cell Signaling Technology, MA, USA) precipitated proteins were analyzed using MS by Qinglianbio (Beijing, China). PLAC8-specific binding proteins were identified by the exclusion of IgG-bound proteins in A549 cells and annotated using GO and KEGG enrichment analyses. The interaction among PLAC8, p53, VCP, UFD1, and NPLOC4 in A549 cells exposed to BLM was detected using an anti-PLAC8 antibody. Additionally, a COIP assay using an anti-p53 antibody was performed to detect the ubiquitination of p53 in indicated experiments and the precipitates were immunoblotted with an anti-Ubi antibody (10201-2-AP, Proteintech).
Statistics and reproducibility
GraphPad Prism 8 (San Diego, USA) was used for data analysis. To assess the difference between the two groups, unpaired, two-tailed Student’s t-tests were used. To examine the differences between more than two groups, a one-way ANOVA analysis using the Tukey multiple comparison test was applied. Two-way ANOVA followed by Sidak’s test were used to evaluate significance for two groups with two variables. N = 3 biological repetitions per group for in vitro assay and N = 6 mice per group for in vivo assay. For the in vitro assay, each group consists of three biological repeats, and for the in vivo assay, each group consists of six mice. The means ± standard deviations (SD) represent the results. The threshold for statistical significance was set at P < 0.05.
Data availability
Microarray dataset GSE110147 contained twenty-two samples of IPF patients and eleven normal lung tissues were downloaded from Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/). The high-throughput data has been deposited in the GEO database (GSE282477) and are available in Supplementary Data 1 and 2. The source data for the graphs analyzed in this study can be found in Supplementary Data 3. The uncropped blot images are provided in Supplementary Fig. 2 in Supplementary Information. The other data will be made available from the corresponding author upon reasonable request.
References
Martinez, F. J. et al. Idiopathic pulmonary fibrosis. Nat. Rev. Dis. Prim. 3, 17074 (2017).
Richeldi, L., Collard, H. R. & Jones, M. G. Idiopathic pulmonary fibrosis. Lancet 389, 1941–1952 (2017).
King, T. E. Jr., Pardo, A. & Selman, M. Idiopathic pulmonary fibrosis. Lancet 378, 1949–1961 (2011).
Mei, Q., Liu, Z., Zuo, H., Yang, Z. & Qu, J. Idiopathic pulmonary fibrosis: an update on pathogenesis. Front. Pharmacol. 12, 797292 (2021).
Moss, B. J., Ryter, S. W. & Rosas, I. O. Pathogenic mechanisms underlying idiopathic pulmonary fibrosis. Annu. Rev. Pathol. 17, 515–546 (2022).
Parimon, T., Yao, C., Stripp, B. R., Noble, P. W. & Chen, P. Alveolar epithelial type II cells as drivers of lung fibrosis in idiopathic pulmonary fibrosis. Int. J. Mol. Sci. 21, 2269 (2020).
Jiang, C. et al. Serpine 1 induces alveolar type II cell senescence through activating p53-p21-Rb pathway in fibrotic lung disease. Aging Cell 16, 1114–1124 (2017).
Shetty, S. K. et al. p53 and miR-34a feedback promotes lung epithelial injury and pulmonary fibrosis. Am. J. Pathol. 187, 1016–1034 (2017).
Guo, J. et al. miR-346 functions as a pro-survival factor under ER stress by activating mitophagy. Cancer Lett. 413, 69–81 (2018).
Klarer, A. C. et al. Inhibition of 6-phosphofructo-2-kinase (PFKFB3) induces autophagy as a survival mechanism. Cancer Metab. 2, 2 (2014).
Liu, H. et al. Long non-coding RNA MALAT1 mediates hypoxia-induced pro-survival autophagy of endometrial stromal cells in endometriosis. J. Cell Mol. Med. 23, 439–452 (2019).
Hill, C. et al. Autophagy inhibition-mediated epithelial-mesenchymal transition augments local myofibroblast differentiation in pulmonary fibrosis. Cell Death Dis. 10, 591 (2019).
Cabrera, S. et al. Essential role for the ATG4B protease and autophagy in bleomycin-induced pulmonary fibrosis. Autophagy 11, 670–684 (2015).
Wang, K. et al. Identification of ANXA2 (annexin A2) as a specific bleomycin target to induce pulmonary fibrosis by impeding TFEB-mediated autophagic flux. Autophagy 14, 269–282 (2018).
Zhao, H., Wang, Y., Qiu, T., Liu, W. & Yao, P. Autophagy, an important therapeutic target for pulmonary fibrosis diseases. Clin. Chim. Acta 502, 139–147 (2020).
Meng, Y. et al. Autophagy attenuates angiotensin II-induced pulmonary fibrosis by inhibiting redox imbalance-mediated NOD-like receptor family pyrin domain containing 3 inflammasome activation. Antioxid. Redox Signal. 30, 520–541 (2019).
Segawa, S. et al. Placenta specific 8 suppresses il-18 production through regulation of autophagy and is associated with adult still disease. J. Immunol. 201, 3534–3545 (2018).
Feng, X. et al. PLAC8 promotes the autophagic activity and improves the growth priority of human trophoblast cells. Faseb J 35, e21351 (2021).
Rogulski, K. et al. Onzin, a c-Myc-repressed target, promotes survival and transformation by modulating the Akt-Mdm2-p53 pathway. Oncogene 24, 7524–7541 (2005).
Meyer, H., Bug, M. & Bremer, S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 14, 117–123 (2012).
Valle, C. W. et al. Critical role of VCP/p97 in the pathogenesis and progression of non-small cell lung carcinoma. PLoS ONE 6, e29073 (2011).
Barkauskas, C. E. et al. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 123, 3025–3036 (2013).
Chen, H. et al. TSSK4 upregulation in alveolar epithelial type-II cells facilitates pulmonary fibrosis through HSP90-AKT signaling restriction and AT-II apoptosis. Cell Death Dis 12, 938 (2021).
Borok, Z. et al. Grp78 loss in epithelial progenitors reveals an age-linked role for endoplasmic reticulum stress in pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 201, 198–211 (2020).
Yu, G. et al. Thyroid hormone inhibits lung fibrosis in mice by improving epithelial mitochondrial function. Nat. Med. 24, 39–49 (2018).
Mao, M. et al. PLCA8 suppresses breast cancer apoptosis by activating the PI3k/AKT/NF-κB pathway. J. Cell Mol. Med. 23, 6930–6941 (2019).
Shen, L. J. et al. PLAC8 gene knockout increases the radio-sensitivity of xenograft tumors in nude mice with nasopharyngeal carcinoma by promoting apoptosis. Am. J. Transl. Res. 13, 5985–6000 (2021).
Tatura, M. et al. Placenta-specific 8 is overexpressed and regulates cell proliferation in low-grade human pancreatic neuroendocrine tumors. Neuroendocrinology 110, 23–34 (2020).
Kinsey, C. et al. Plac8 links oncogenic mutations to regulation of autophagy and is critical to pancreatic cancer progression. Cell Rep. 7, 1143–1155 (2014).
Huang, M. L. et al. Plac8-mediated autophagy regulates nasopharyngeal carcinoma cell function via AKT/mTOR pathway. J. Cell Mol. Med. 24, 7778–7788 (2020).
Nakashima, N. et al. The p53-Mdm2 association in epithelial cells in idiopathic pulmonary fibrosis and non-specific interstitial pneumonia. J. Clin. Pathol. 58, 583–589 (2005).
Bhandary, Y. P. et al. Regulation of alveolar epithelial cell apoptosis and pulmonary fibrosis by coordinate expression of components of the fibrinolytic system. Am. J. Physiol. Lung Cell. Mol. Physiol. 302, L463–473 (2012).
Geng, F. et al. Quercetin alleviates pulmonary fibrosis in mice exposed to silica by inhibiting macrophage senescence. Front. Pharmacol. 13, 912029 (2022).
Yao, C. et al. Senescence of alveolar type 2 cells drives progressive pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 203, 707–717 (2021).
Shaikh, S. B., Prabhu, A. & Bhandary, Y. P. Curcumin targets p53-fibrinolytic system in TGF-β1 mediated alveolar epithelial mesenchymal transition in alveolar epithelial cells. Endocr. Metab. Immune Disord. Drug Targets 21, 1441–1452 (2021).
Venkatesan, S., Fan, L., Tang, H., Konduru, N. V. & Shetty, S. Caveolin-1 scaffolding domain peptide abrogates autophagy dysregulation in pulmonary fibrosis. Sci. Rep. 12, 11086 (2022).
Abbas, R. & Larisch, S. Killing by degradation: regulation of apoptosis by the ubiquitin-proteasome-system. Cells 10, 3465 (2021).
Luo, P. et al. HMGB1 represses the anti-cancer activity of sunitinib by governing TP53 autophagic degradation via its nucleus-to-cytoplasm transport. Autophagy 14, 2155–2170 (2018).
Korolchuk, V. I., Menzies, F. M. & Rubinsztein, D. C. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 584, 1393–1398 (2010).
Mirsanaye, A. S. et al. VCF1 is a p97/VCP cofactor promoting recognition of ubiquitylated p97-UFD1-NPL4 substrates. Nat. Commun. 15, 2459 (2024).
Riemer, A. et al. The p97-Ufd1-Npl4 ATPase complex ensures robustness of the G2/M checkpoint by facilitating CDC25A degradation. Cell Cycle 13, 919–927 (2014).
Lehmann, M. et al. Chronic WNT/β-catenin signaling induces cellular senescence in lung epithelial cells. Cell Signal 70, 109588 (2020).
Ashcroft, T., Simpson, J. M. & Timbrell, V. Simple method of estimating severity of pulmonary fibrosis on a numerical scale. J. Clin. Pathol. 41, 467–470 (1988).
Cecchini, M. J., Hosein, K., Howlett, C. J., Joseph, M. & Mura, M. Comprehensive gene expression profiling identifies distinct and overlapping transcriptional profiles in non-specific interstitial pneumonia and idiopathic pulmonary fibrosis. Respir. Res. 19, 153 (2018).
Acknowledgements
This work was supported by the Science and Technology Project of Liaoning Province [2024-MS-022, 2023-MS-01]. We thank scidraw.io for the schematic pictures of mouse and syringe.
Author information
Authors and Affiliations
Contributions
W.Sun and Y.C. designed research; W.Sun, B.Z., Z.H., and W.Song performed experiments and collected samples; W.Sun, B.Z., and L.C. analyzed data and wrote the original paper. Y.C. supervised the project and revised the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
All animal experiments were conducted under the Guide for the Care and Use of Laboratory Animals and this study was approved by the Ethics Committee of Shengjing Hospital of China Medical University (2023PS005K).
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Elah Pick and Mengtan Xing.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sun, W., Zhao, B., He, Z. et al. PLAC8 attenuates pulmonary fibrosis and inhibits apoptosis of alveolar epithelial cells via facilitating autophagy. Commun Biol 8, 48 (2025). https://doi.org/10.1038/s42003-024-07334-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-07334-8
This article is cited by
-
Emodin: A Promising Natural Compound for Combating Fibrotic Diseases
Current Medical Science (2026)
-
How cells die determines the consequences of tissue repair: roles of programmed cell death in lung injury on the progression of pulmonary fibrosis
Molecular and Cellular Biochemistry (2025)