Abstract
Intestinal farnesoid X receptor (FXR) signaling plays a critical role in maintaining intestinal microbiota stability. In this study, we investigated the probiotic Lactobacillus rhamnosus GG (LGG) and its ability to promote intestinal adaptation and stabilize the gut microbiota by activating intestinal FXR signaling in short bowel syndrome (SBS). In patients with type I SBS, fecal microbial α-diversity was decreased, Proteobacteria abundance was increased, and Firmicutes, Actinobacteria, and Bacteroidetes abundance levels were decreased. In vitro, LGG supernatant (LGGs) upregulated FXR expression in Caco-2 cells and ileum organoids. In vivo, LGG supplementation significantly improved intestinal morphology in wild-type (WT) SBS mice, including increased villus height, crypt depth and goblet cell numbers. Serum fibroblast growth factor 15 (FGF15) levels increased and fecal Proteobacteria abundance decreased, while secondary bile acids rose and primary bile acids declined in WT SBS mice after LGG supplementation. In addition, LGG supplementation also increased occludin and FXR expression in WT SBS mice, but not in intestinal FXR knockout (FXRInt-KO) SBS animals. SBS disrupts FXR signaling and gut microbiota equilibrium. LGG counteracts these effects by activating intestinal FXR, which stabilizes microbiota composition, protects the mucosal barrier, and promotes intestinal adaptation both in vitro and in vivo.

Similar content being viewed by others
Introduction
Short bowel syndrome (SBS) is a malabsorptive condition resulting from extensive bowel resection or functional exclusion, in which the residual intestine is unable to maintain adequate nutrition or meet the growth demands of children. Common symptoms include chronic diarrhea, acid-base imbalances, fluid/electrolyte disturbances, and impaired absorption/metabolism of nutrients1,2. In pediatric SBS, the residual intestinal length is typically less than 25% of the predicted gestational age-adjusted norm, or patients often require prolonged parenteral nutrition (PN) (>42 days) following intestinal resection or due to intestinal failure3. Major etiologies of SBS include neonatal necrotizing enterocolitis, intestinal atresia, gastroschisis, and midgut volvulus.
Due to altered intestinal structure and function, impaired peristalsis and compensatory expansion of intestine which do not effectively for bacterial elimination and lead to intestinal microbiota dysbiosis in SBS1,4. When compared with healthy populations, intestinal microbiota diversity is reduced in SBS patients, and harmful bacteria such as Proteobacteria are dominant. The abundance of other opportunistic bacteria in the Enterobacteriaceae family, such as Escherichia coli, Shigella, Klebsiella, and Proteobacteria, is significantly increased, whereas the abundance of beneficial bacteria in Firmicutes, such as Clostridium is considerably decreased5. This disbiosis drives small bowel bacterial overgrowth6, and further causes intestinal inflammation, hematogenous infection, PN associated liver disease and multiple metabolic disorders7,8.
Patients with SBS exhibit abnormal bile acid metabolism, characterized by elevated fecal primary bile acid and depleted secondary bile acid9. Metagenomics analyses reveal a marked reduction in commensal genera with bile acid-modifying function, including decreased abundance of Bacteroides, Eubacterium, and Clostridium species abundance. These taxa possess bile salt hydrolase (BSH) activity and/or 7α-dehydroxylation capacity, which are critical for converting primary bile acids into secondary bile acids7. Gut dysbiosis disrupts BSH-mediated deconjugation and 7α-dehydroxylation, blocking secondary bile acid synthesis. As a result, reduced levels of lithocholic acid (LCA) and deoxycholic acid (DCA) lead to diminished farnesoid X receptor (FXR) activity and decreased serum fibroblast growth factor 19 (or FGF15 in mice) levels. FGF15/19, a FXR-dependent mediator, suppresses intrahepatic bile acid synthesis. Impaired feedback regulation of the FXR–FGF15/19 axis elevates bile acid production, exacerbates hepatic cholestasis, and promotes intestinal failure-related liver damage10,11. Therefore, stabilizing gut microbiota and enhancing intestinal FXR–FGF15/19 signaling in SBS may ameliorate dysregulated bile acid metabolism and strengthen the intestinal mucosal barrier.
As a widely used clinical probiotic, Lactobacillus rhamnosus GG (LGG) mitigates gut microbiota dysbiosis and preserves colonic barrier integrity in inflammatory bowel disease and diarrhea disorders12,13,14. Furthermore, LGG reduces hepatic bile acid accumulation via FXR signaling, a mechanism that prevents liver injury and fibrosis15. However, its therapeutic role in SBS remains unclear. In this study, we investigated the dysregulation of the gut microbiota-bile acid-FXR axis in SBS and hypothesized that LGG exerts its effects through two distinct pathways: 1. Direct FXR stimulation: LGG components activate FXR in Caco-2 cells and intestinal organoids in vitro. 2. Indirect FXR modulation: LGG restores microbiota-bile acid equilibrium in vivo by enriching beneficial taxa (e.g., Firmicutes) and suppressing pathogens (e.g., Proteobacteria). This study aimed to elucidate the molecular mechanisms by which LGG maintains intestinal homeostasis through the FXR pathway in SBS.
Results
Biochemical abnormalities and impaired intestinal FXR signaling in SBS patients
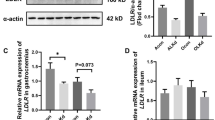
Complete data from 53 children with enterostomy were collected, including 11 in the SBS group (type I SBS) and 42 in the control group. No significant differences were observed between groups in gender, corrected gestational age, primary disease, and enterostomy methods. However, gestational week, birth weight, and weight-for-age percentages before stoma closure were significantly lower in the SBS group compared to the control group (Table S1). Analysis of biochemical indicators before stoma closure showed that retinol binding protein (P = 0.021, t = 2.384) and high density lipoprotein cholesterol (HDL-C) (P < 0.001, t = 4.170) were significantly lower in the SBS group compared to the control group. In contrast, total bilirubin (P < 0.001, t = 4.589), direct bilirubin (P < 0.001, t = 4.532), alanine aminotransferase (ALT) (P = 0.002, t = 3.304), aspartate aminotransferase (AST) (P < 0.001, t = 3.759), gamma-glutamyltransferase (GGT) (P = 0.017, t = 2.480), and alkaline phosphatase (ALP) (P = 0.010, t = 2.691) were higher in the SBS group (Fig. 1a). RT-PCR analysis of FXR signaling components revealed that FXR expression (P = 0.004, t = 3.045) and its downstream genes organic solute transporter α (OST-α) (P = 0.003, t = 3.124), organic solute transporter β (OST-β) (P = 0.002, t = 3.393), and intestinal bile acid-binding protein transport (I-BABP) (P = 0.015, t = 2.543) were lower in the SBS group compared to the control group (Fig. 1b).
Intestinal microbiome dysbiosis in SBS feces
At the phylum level, Proteobacteria, Firmicutes, Actinobacteriota, and Bacteroidota (Fig. 2a) were the main bacteria in enterostomal fecal samples. At the genus level, Klebsiella, Escherichia-Shigella, Enterobacter, Enterococcus, Streptococcus, Morganella, Clostridium, and Lactobacillus predominated (Fig. 2b). Significant differences in species richness existed between SBS and control groups. Both the Chao1 and observed_species indices were significantly decreased in the SBS group relative to the control group (P < 0.01). The Shannon and Simpson indices showed no significant abundance or evenness differences between groups (Fig. 2c). According to the Principal Coordinate Analysis (PCoA) profile of β-diversity based on Jaccard and permutational multivariate analysis (PERMANOVA), a significant difference in the microbiome was observed between the control and SBS groups (Fig. 2d). Specifically, gut microbiota diversity was significantly lower, and the microbiota profiles were distinct in SBS patients compared to controls.
a At the phylum level, the predominant bacterial colonies are Proteobacteria, Firmicutes, Actinobacteriota, and Bacteroidota. b At the genus level, the main bacterial colonies are Klebsiella, Escherichia-Shigella, Enterobacter, Enterococcus, Streptococcus, Morganella, Clostridium, and Lactobacillus. c α-diversity analysis: The Chao1 and Observed_species indices indicate that the species richness in the SBS group is lower than that in the control group, while the Shannon and Simpson indices reveal no significant differences in species diversity and evenness between the two groups. d β-diversity analysis: PCoA and PERMANOVA showed the significant difference among the control and SBS groups, P = 0.004. e Phylum with significant differences in the relative abundances between groups: Phylum with significant differential relative abundances are Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidota. f LDA scores for genera differentiating SBS (purple) and controls (green). Threshold: LDA > 4.0. g Taxonomic cladogram form LEfSe analysis: Green nodes indicate control-enriched taxa, purple nodes represent SBS-enriched taxa. *P < 0.05, **P < 0.01.
At the phylum level, the relative abundance of Proteobacteria was signigicantly higher in the SBS group compared to the control group. In contrast, the abundances of Firmicutes, Actinobacteriota, and Bacteroidota were significantly lower in the SBS group (Fig. 2e). A linear discriminant analysis (LDA) threshold of 4.0 was set to identify significant differences attributable to influential species. The LDA scores, ranked from high to low, were Gammaproteobacteria, Proteobacteria, Enterobacteria, Morganella, and Pseudomonadale in the SBS group. Firmicutes, Clostridia, and Clostridiales (Fig. 2f, g) were more abundant in the control group.
LGGs improves Caco-2 viability and tight junction protein expression
Caco-2 cells were stimulated with lipopolysaccharides (LPS) at concentrations of 50, 100, and 150 μg/mL, which reduced the expression of tight junction protein occludin and claudin-1 (P < 0.05). Specifically, 100 μg/mL LPS significantly reduced the expression of occludin (P = 0.008) and claudin-1 (P = 0.001) (Fig. S1). We observed that Lactobacillus acidophilus supernatant (LAs), LGGs, and Bifidobacterium adolescentis supernatant (BAs) at concentrations of 2%, 4%, 6%, and 8% improved Caco-2 cell proliferation (CCK-8 assays). Cell viability was enhanced with increased probiotic concentrations (supernatants), which efficiently antagonized injury caused by 100 μg/mL LPS in Caco-2 cells (Fig. S1). Furthermore, 4% LGGs enhanced tight junction protein expression in Caco-2 cells, which was better than 4% LAs and 4% BAs (Fig. S1).
LGG reduces apoptosis and activates FXR signaling in Caco-2 cell
As shown in Fig. S2, Caco-2 cells were stimulated with 100 μg/mL LPS or 4% LGGs. Compared to the untreated groups, LPS promoted apoptosis (P < 0.01), whereas the LGG-supplemented group reduced apoptosis (P < 0.01). Obeticholic acid (OCA, 20 μM) was included as a positive control to comparing the effects of OCA and LGGs effects toward FXR signaling in Caco-2 cells. We observed that FXR, SHP, and OST-α mRNA expression levels in the LGGs group were significantly higher than levels in the untreated group. Additonally, FGF19 mRNA levels increased in the OCA and LGGs combination group (Fig. S2).
LGGs upregulates FXR expression and its downstream genes and maintains structural integrity in mouse ileal organoids
Ileum tissues from C57BL/6 mice and intestinal crypts were embedded in 3D Matrigel culture, and organoids were matured after 5 days in culture. LGGs supplemented at different concentrations (2%, 4%, and 8%) did not cause significant morphological disruption but significantly increased organoid size. However, when the FXR inhibitor beta-muricholic acid (β-MCA) concentration reached 0.8 mM, organoids were damaged, exhibiting irregular margins, obscured villus boundaries, and decreased organoid size (Fig. 3a, b). FXR gene expression levels and its downstream genes (OST-α, OST-β, and I-BABP) were activated by 2% LGGs. However, gene expression and downstream signaling pathways were significantly decreased at 0.4 mM β-MCA (Fig. 3c, d). Additonally, FXR protein expression was reduced (P = 0.003) while ASBT protein expression was increased (P = 0.006) in ileum organoids supplemented with 0.4 mM β-MCA, as visualized by immunofluorescence. No significant differences were observed between the 2% LGGs+0.4 mM β-MCA group and the NC group (P = 0.935, P = 0.438, Fig. 3e, g). Additonally, 100 μg/mL LPS significantly reduced protein expression of villin (P = 0.011) and occludin (P = 0.001) in mouse ileum organoids, and 2% LGGs increased the expression of villin (P = 0.024) and the protein level of occludin had no difference between 2% LGGs + 100 μg/mL LPS group and NC group (P = 0.127, Fig. 3f, h).
a Morphological changes of ileum organoids treated with LGGs (2%, 4%, 8%) or β-MCA (0.1, 0.2, 0.4, 0.8 mM) for 24 h. LGGs preserved organoid structure at all concentrations, whereas 0.8 mM β-MCA induced epithelial damage (arrows: disrupted edges). Scale bar: 200 μm. b Size of organoids cultured with LGG and β-MCA. Data expressed as mean ± SEM. c qPCR analysis of FXR and its downstream targets (OST-α, OST-β, I-BABP) in organoids treated with 2% LGGs for 24 h. d Suppression of FXR signaling pathway mRNA levels by β-MCA (0.4 mM, 24 h). FXR, OST-α, OST-β, and I-BABP expression decreased. e Immunofluorescence staining of FXR (red) and ASBT (green) in organoids treated with LGGs (2%) or β-MCA (0.4 mM). Nuclei counterstained with DAPI (blue). Scale bar: 100 μm. f Occludin (green) and Villin (red) expression in organoids exposed to LGGs (2%) or LPS (100 μg/mL). Scale bar: 100 μm. g, h Quantification of fluorescence intensity for FXR, ASBT, Occludin, and Villin. *P < 0.05, **P < 0.01 vs. untreated control.
LGG effects on ileum morphology and intestinal FXR signaling in SBS
The experimental design for WT mice is shown in Fig. 4a. Compared to the NC group, ileum samples exhibited lymphocytic infiltration, villi edema, and disordered arrangement (visualized by H&E staining) at 2 weeks after successful modeling in the WT SBS group. After LGG supplementation, the villi structures became neatly arranged, and edema was lessened (Fig. 4b, f). Statistical analyses revealed that both ileum villus length and crypt depth were increased in the SBS group compared to the NC group: 757.3 ± 73.53 μm vs. 546.8 ± 59.65 μm and 179.7 ± 18.96 μm vs. 91.1 ± 8.67 μm, respectively (P < 0.01). Villus height and crypt depth levels were greater in the SBS + LGG group compared to the SBS group: 918.4 ± 86.73 μm vs. 757.3 ± 73.53 μm and 222.0 ± 28.90 μm vs. 179.7 ± 18.96 μm, respectively (P < 0.01). PAS staining showed that goblet cell numbers decreased in the SBS group, but improved after LGG supplementation (Fig. 4c, e). Residual intestine lengths were measured after 2 weeks of modeling. Residual jejunum and ileum lengths in the SBS + LGG group were longer than those in the SBS group (8.7 ± 0.75 cm vs. 6.1 ± 0.77 cm, 8.4 ± 0.34 cm vs. 7.1 ± 1.33 cm, respectively, P < 0.01). However, no significant differences were observed in colon length (5.4 ± 0.51 cm vs. 5.7 ± 1.03 cm, P > 0.05) (Fig. 4d). Western blotting demonstrated that occludin protein expression in ileum samples increased in the SBS group, while levels increased when SBS mice were supplemented with LGG (Fig. 4g, h). Immunofluorescence analysis showed that FXR protein levels in SBS ileum samples were decreased, but increased after LGG supplementation (Fig. 4i, j). Serum FGF15 levels in the SBS + LGG group were higher than those in the SBS group (269.2 ± 58.37 ng/mL vs. 89.29 ± 23.82 ng/mL, P < 0.01), as measured by ELISA (Fig. 4k).
a Experimental design: WT mice were divided into four groups (NC normal control, LGG LGG gavage [109 CFU/day], SBS short bowel resection (SBR), SBS + LGG SBR with LGG treatment). Interventions lasted 14 days post-surgery (n = 6/group). b–f Histological analysis of jejunum, ileum, and colon on postoperative day (POD) 14. b, f H&E staining: Villus height (↑) and crypt depth (↑) of ileum in SBS + LGG vs. SBS (P < 0.01). Scale bar: 200 μm. c, e PAS staining: Goblet cell density (↑) of ileum in SBS + LGG vs. SBS. Scale bar: 100 μm d Residual jejunum, ileum, and colon length (cm): SBS + LGG > SBS (P < 0.05). g, h Western blot analysis of intestinal proteins: Occludin (tight junction protein) expression (↑) in SBS + LGG vs. SBS (P < 0.01). i, j Immunofluorescence of FXR (red) in intestinal epithelium: Nuclear FXR intensity (↑) in SBS + LGG vs. SBS (P < 0.01). Nuclei counterstained with DAPI (blue). Scale bar: 100 μm. k ELISA analysis of plasma FGF15 levels: SBS + LGG > SBS (P < 0.01). * indicated comparison with NC group and # indicates comparison with SBS group. * and # indicated P < 0.05, ** and ## indicated P < 0.01.
LGG modulates intestinal microbiota disorder in SBS
At the phylum level, the main bacterial communities in WT fecal samples were Bacteroidota, Firmicutes, Proteobacteria, Desulfobacterota, and Campilobacterota (Fig. 5a). At the genus level, the predominant taxa were Muribaculaceae, Bacteroides, Escherichia-Shigella, Helicobacter and Lachnospiraceae-NK4A136-Group (Fig. 5b). For α-diversity analyses, the Chao1, Observed_species, Shannon, and Simpson indices were used. All indices in the NC and LGG groups were significantly higher than those in the SBS group (P < 0.01). The Shannon and Simpson indices showed that microbial diversity in the SBS + LGG group was increased compared to the SBS group (Fig. 5c). For β-diversity analyses, PCoA and PERMANOVA were performed among the WT, LGG, SBS, SBS + LGG groups, which showed statistically significant differences in microbial communities between groups (P < 0.001), indicating distinct microbiota compositions across groups. (Fig. 5d).
a At the phylum level, the predominant bacterial colonies are Bacteroidota, Firmicutes, Proteobacteria, Desulfobacterota, and Campylobacterota. b At the genus level, the main bacterial colonies include Muribaculaceae, Bacteroides, Escherichia-Shigella, Helicobacter. c α diversity analysis: The SBS group exhibited lower Chao1, Observed_species, Shannon, and Simpson indices compared to the NC group. After supplementing SBS mice with LGG, the Shannon and Simpson indices increased. d β-diversity analysis based on PCoA and PERMANOVA among the WT, LGG, SBS, SBS + LGG groups, P = 0.001. e Phylum with significant differences in the relative abundances between groups are Bacteroidota, Proteobacteria, Desulfobacterota, and Campylobacterota. * indicated comparison with the WT normal group. # indicated comparison with the WT LGG group. *, #P < 0.05. **, ##P < 0.01. f, g Analysis the influence of species abundance by LEfSe test. f Histigram of LDA scores for differentially abundant genera between groups. Threshold: LDA > 4.0. g Cladogram for taxonomic representation of significant differences between groups. Green, purple, orange and yellow represent core micrbes in NC, LGG, SBS and SBS + LGG groups, respectively. Olive green indicates microbes shared between groups.
At the phylum level, when compared with NC, LGG, and SBS + LGG groups, the SBS group exhibited elevated abundance of Proteobacteria and Campilobacterota, while the relative abundance of Bacteroidota and Desulfobacterota was decreased (Fig. 5e). LEfSe was used to identify differentially abundant bacterial taxa, with a LDA threshold of 4.0 to evaluate the influence of significantly different species. The LDA scores from high to low were as follows: Enterobacterales, Enterbacteriaceae, and Escherichia_Shigella in SBS group. In the SBS + LGG group, they were Lachnospirales, Lachnospiraceae, Bacteroides, and Bacteroidaceae (Fig. 5f, g).
Bile acid metabolism alterations are mediated by LGG supplementation in WT mice
The gut microbiota plays an important role in intestinal bile acid metabolism. In this study, we investigated the effects of LGG on fecal bile acids in SBS mice. Using targeted metabolomics analysis, changes in a total of 15 bile acids were detected. Principal Component Analysis (PCA) score plots revealed clear clustering of bile acids in each group, with distinct separation between the NC, LGG, SBS, and SBS + LGG groups (Fig. 6a). Orthogonal Partial Least Squares-Discriminant Analysis (OPLS-DA) score plots demonstrated clear distinctions between the NC vs. LGG, NC vs. SBS, and SBS vs. SBS + LGG groups, despite small differences within groups (Fig. 6a). Furthermore, the levels of alpha-muricholic acid (α-MCA), tauro-apha-muricholic acid (T-α-MCA), LCA, DCA, Ursodeoxycholic acid (UDCA), Hyodeoxycholic acid (HDCA), isoLCA, and 7-ketoLCA were significantly decreased in the SBS group compared to the NC group (P < 0.05) (Fig. 6b).
a PCA and OPLS-DA score chart of cecum bile acids. The PCA plot showed that each group was clustered, but there was still partial overlap. OPLS-DA score chart showed that NC vs. LGG, NC vs. SBS, SBS vs. SBS + LGG groups were significantly different. b Determination of ileocecal stool bile acids in groups. The level of α-MCA, T-α-MCA, LCA, DCA, UDCA, HDCA, isoLCA, 7-ketoLCA and NorDCA were decreased in SBS group when compared with NC group. The level of α-MCA, CDCA, UDCA, 7-ketoLCA were decreased in SBS + LGG group when compared with SBS group. *P < 0.05, **P < 0.01, ***P < 0.001.
The composition of principal bile acids is shown in Fig. 7a. In SBS animals, the proportion of secondary bile acids was decreased, including a two-fold decrease in the proportion of DCA, a 3.5-fold decrease in the abundance of 12-ketoLCA. Conversely the proportion of primary bile acids in SBS animals was increased compared to NC animals, including a 1.7-fold increase in the proportion of β-MCA and a 7.5-fold increase in the proportion of cholic acid (CA). In SBS + LGG animals the proportion of secondary bile acids was increased, including a 1.5-fold increase in the proportion of DCA, 3.7-fold increase in the abundance of 12-ketoLCA. Conversely the proportion of primary bile acids in SBS + LGG animals was decreased compared to SBS animals, including a 1.2-fold decrease in the proportion of β-MCA and a 1.7-fold decrease in the proportion of CA.
a Bile acid composition in each group. The proportion of secondary bile acids were decrease and primary bile acids were increased in SBS animals. LGG reversed the changes of bile acids in SBS mice. b Heatmap of spearman correlation association between bile acids and gut microbiota. NC vs. LGG vs. SBS vs. SBS + LGG, Muribaculaceae was positively correlated with NorDCA, HDCA, isoLCA, DCA, T-β-MCA, and α-MCA levels. NC vs. LGG, the levels of Prevotellaceae_UCG-001 and Helicobacter were positively correlated with LCA, isoLCA, DCA, HDCA, 7-ketolLCA, UDCA, and 12-ketolLCA. Bacteroides was negatively correlated with β-MCA and CA levels. SBS vs. SBS + LGG showed that Escherichia-Shigella were positively correlated with α-MCA levels. Muribaculaceare and Alistipes were negatively correlated with the level of α-MCA. *P < 0.05, **P < 0.01, ***P < 0.001.
Association analysis between fecal bile acids and intestinal bacteria
Spearman’s rank correlations analysis was performed to explore the potential functional relationships between the 7 distinctive microbial species and the top 15 differential metabolites. Additionally, hierarchical clustering heatmap results were used to investigate the relationship between gut microbiota and bile acids in ileocecal fecal samples of WT mice at the genus level.
As shown in Fig. 7b, correlation analysis between intestinal microbiota and bile acid levels in fecal samples from NC group, LGG group, SBS group, and SBS + LGG group was performed at the genus level. Muribaculaceae showed a positive correlated with the levels of NorDCA, HDCA, isoLCA, DCA, T-β-MCA and α-MCA (P < 0.05). In the correlation analysis of fecal samples from the NC group and LGG group, the levels of Prevotellaceae_UCG-001 and Helicobacter were positively correlated with LCA, isoLCA, DCA, HDCA, 7-ketolLCA, UDCA, and 12-ketolLCA (P < 0.05). In contrast, Bacteroides was negatively correlated with the levels of β-MCA and CA. Furthermore, correlation analysis of fecal samples from the SBS group and SBS + LGG group revealed that Escherichia-Shigella levels were positively correlated with α-MCA levels (P < 0.01), while Muribaculaceare and Alistipes were negatively correlated with α-MCA levels (P < 0.05).
The protective effect of LGG on mouse intestines was attenuated in FXRInt-KO mice
Intestinal villin and PCNA protein levels were effectively reduced in FXRInt-KO mice compared to the WT group (P = 0.016 and P = 0.015, respectively), while occludin was undetected in both groups (Fig. 8a, c, d). In FXRInt-KO mice, the mRNA levels of FXR downstream genes (OST-α, OST-β, and I-BABP) were downregulated while ASBT levels were upregulated (Fig. 8b). LGG supplementation increased the expression of FXR (P = 0.005), OST-α (P = 0.001), and OST-β (P = 0.036) in the WT SBS + LGG group compared to the WT SBS group. However, no significant differences were observed in FXRInt-KO mice among the K, KL, KS, and KS + L groups (Fig. 8e, f). (* compared with the WT group, # compared with the WT SBS group; *, # P < 0.05, **, # P < 0.01). Immunofluorescence analysis revealed that LGG supplementation significantly increased villin (red fluorescence, P = 0.001) and occludin (green fluorescence, P = 0.040) expression in WT SBS mice, while no increase in occludin expression was observed in FXRInt-KO SBS mice (Fig. 8g, h).
a Animal experiment program illustrating the ileum FXR pathway and protein expression between WT and FXRInt-KO mice. b Significant downregulation of FXR and its downstream targets (OST-α/β, I-BABP) and upregulation of ASBT in FXRInt-KO mice (P < 0.01). c, d Reduced levels of villin and PCNA in FXRInt-KO mice, *P < 0.05. e Experimental design: FXRInt-KO mice were divided into four groups (K knockout, KL knockout with LGG gavage [109 CFU/day], KS knockout with SBR, KS + L knockout with SBR and LGG treatment. Interventions lasted 14 days post-surgery (n = 6/group). f No improvement in FXR pathway genes by LGG gavege following FXRInt-KO SBS modeling. * indicated comparison with the WT normal group and # indicates comparison with WT SBS group. g, h Immunofluorescence of villin (red) and occludin (green) in intestinal epithelium: Villin and occludin intensity (↑) in SBS + LGG vs. KS or KS + L (P < 0.01). Nuclei counterstained with DAPI (blue). Scale bar: 50 μm. * indicated comparison with the WT SBS group and # indicates comparison with WT SBS + LGG group. *, #P < 0.05, **, ##P < 0.01.
Gut microbiota alterations are mediated by LGG supplementation in FXRInt-KO mice. At the phylum level, the the predominant microbial communities in fecal samples from FXRInt-KO mice were Bacteroidota, Firmicutes, Desulfobacterota, Proteobacteria, and Campilobacterota (Fig. 9a). At the genus level, Muribaculaceae, Lachnospiraceae-NK4A136-group, Escherichia-Shigella, and Alistipes were the dominant taxa (Fig. 9b). α-diversity was assessed using the Chao 1, Observed_species, Shannon, and Simpson indices. The Chao1 and Observed_species indices indicated that microbial abundance in the KS group was significantly lower compared to the K, KL, and KS + L groups (P < 0.05). However, abundance in the KS + L group showed a slightly increase following LGG supplementation when compared with the KS group (P < 0.05). The Shannon and Simpson indices showed no significant differences in bacterial diversity among the KS, KS + L, and NC groups (P > 0.05) (Fig. 9c). β-diversity was evaluated using PCoA, which demonstrated significant differences in microbial composition and abundance between the K and KL, KS, and KS + L groups (P < 0.001, PERMANOVA) (Fig. 9d).
a At the phylum level, the predominant bacterial colonies are Bacteroidota, Firmicutes, Desulfobacterota, Proteobacteria, and Campylobacterota. b At the genus level, the main bacterial colonies include Muribaculaceae, Lachnospiraceae, Escherichia-Shigella, Alistipes, Parabacteroides, Bacteroides. c α-diversity analysis:The Chao1 and Observed_species indices indicate that the species richness in the KS group is reduced, and supplementation with LGG can mitigate the decrease in species richness between groups. The Shannon and Simpson indices show no significant differences in the diversity and evenness of microbial communities between the KS group and the KS + L group. d β-diversity analysis: PCoA and PERMANOVA showed the significant difference among the groups, P = 0.001. e Phylum with significant differences in the relative abundances between groups are Desulfobacterota, Proteobacteria, and Verrucomicrobia. f, g Analysis the influence of species abundance by LEfSe test. f Histigram of LDA scores for differentially abundant genera between groups. Threshold: LDA > 4.0. g Cladogram for taxonomic representation of significant differences between groups. Green, purple, orange, and yellow represent core micrbes in K, KL, KS and KS + L groups, respectively. Olive green indicates microbes shared between groups. * indicated comparison with the K group. *P < 0.05, **P < 0.01.
At the phylum level, the abundance of Proteobacteria in the KS group was significantly higher compared to the K, KL, and KS + L groups, while Desulfbacterota abundance was highest in the SBS + LGG group. Additionally, Verrucomicrobiota was highly expressed in the K group compared to the others (Fig. 9e). A LDA threshold of 4.0 was used to evaluate the influence of significantly different species. In the KS group, the scores from high to low were as follows: Proteobacteria, Gammaproteobacteria, Enterobacterales, Enterobacteriaceae, and Escherichia-Shigella. In the KS + L group, the dominant taxa were Desulfovibrionaceae, Desulfobacterota, Desulfovibrionia, and Desulfovibrionales. The K group was predominatly by Muribaculaceae from the Bacteroidetes phylum (Fig. 9f, g).
Discussion
SBS is caused by congenital or acquired intestinal deficiencies and a reduction in effective intestinal absorptive areas, leading to nutrient absorption disorders, bloodstream infections, metabolic disturbances, and intestinal function-associated liver damage3.The ultimate goal of multidisciplinary SBS treatments is to accelerate intestinal adaptation, increase the proportion of enteral nutrition, reduce related complications, and reintroduce intestinal autonomy16.
Intestinal microbiota plays crucial roles in intestinal immunity, as well as energy, lipid, and glucose metabolism. They are a key mediator in intestinal homeostasis as their composition is an essential factor affecting clinical outcomes in SBS5,17. When beneficial bacteria decrease, pathogens bacteria increase, releasing LPS into the intestines. This can lead to damaged intestinal epithelial cells, increased intestinal permeability, and an imbalance in the intestinal microbiota, allowing LPS to enter the circulation. Consequently, this results in intestinal nutrient malabsorption, liver damage, bloodstream infections, and prolonged PN dependence times18,19. Piper et al. reported that microbial diversity in the feces from children with SBS was reduced, with a predominance of Proteobacteria, and a marked increase in opportunistic pathogens, such as Escherichia, Shigella, Klebsiella and Proteobacteria. In contrast, Clostridium from the Firmicutes phylum was significantly reduced5,20. The microbial density of the small intestinal (SI) microbiota follows a gradient, increasing from going from the duodenum (~103 cells/g) to the terminal ileum (~108 cells/g)12. At the phylum level, the SI microbiota is predominantly composed of Firmicutes and Actinobacteria13. Key genera include Streptococcus, Actinomyces, Gemella, and Lactobacillus13. In our study, fecal samples were collected from children with enterostomy, and alterations in intestinal microbiota between SBS and non-SBS patients with small intestinal stoma were compared. Proteobacteria was the dominant phylum in the SBS group, whereas Firmicutes, Actinobacteria, and Bacteroidetes were dominant in the non-SBS group. At the genus level, Enterobacteriaceae was predominant in the SBS group. These findings on gut microbiome composition in SBS are consistent with earlier studies5,20. Small intestinal microbiota have important roles in SBS prognoses. Bacterial overgrowth can increase the risk of bacterial translocation and bloodstream infections14,21, impair digestion and absorption functions, and delay or prevent weaning from PN22.
Due to the significant loss of intestinal tissue, the number of FXR+ intestinal epithelial cells decreased, and the expression of FGF19 in plasma was reduced. Concurrently, the abundance of harmful bacteria, such as Proteobacteria, which produce endotoxins, increased in the intestine. This led to inflammatory injured in the residual intestine, a reduced utilization rate of bile acid transport-related proteins, disordered bile acid metabolism, and persistent diarrhea9,23. We confirmed that FXR gene levels and its downstream genes (I-BABP and OST-α/β) were suppressed in the type I SBS group with proximal enterostomy. Pereira-Fantini et al. found that the primary bile acid chenodeoxycholic acid (CDCA) was abundant in the feces of SBS patients, while the proportion of the secondary bile acid DCA decreased significantly, both acting as FXR agonists24. Similarly, in our WT SBS mice, we observed an abundance of the primary bile acid CA and β-MCA, along with a decrease in the secondary bile acid DCA. Intestinal FXR was not activated due to the altered bile acid composition9. Although CDCA can alleviate intestinal barrier damage and reduce intestinal inflammation, high physiological concentration of bile acids has huge adverse effects including DNA damage, apoptosis, and oxidative/nitrative stress21,25,26. Therefore, in the context of SBS, it may be necessary to explore and incorporate additional FXR agonists beyond bile acids.
Probiotics are live microorganisms that provide health benefits when administered in adequate amounts. They help maintain intestinal barrier integrity, inhibit pathogenic bacterial growth, promote beneficial microbial colonization, and regulate metabolic and immune functions27. However, their efficacy in SBS remains controversial. Some studies report no significant infection-prevention effects of probiotics in SBS patients, and their use may increase the risk of complications such as D-lactic acidosis28,29. In contrast, other studies have demonstrated that probiotics and their metabolites produce short-chain fatty acids (SCFAs), which reduce intestinal bacterial translocation, maintain barrier integrity, and promote adaptive remodeling in SBS30,31,32.
Previous study has demonstrated that pre-treatment with LGG reduces apoptosis in intestinal crypts and preserves intestinal epithelial stem cells following irradiation33. Additionally, LGG enriches gut microbiota taxa with bile salt hydrolase (BSH) activity, thereby enhancing bile acid metabolism and excretion15. In our study, LGGs proved more effective than BAs and LAs in stabilizing tight junctions (e.g., occludin, claudin-1) in intestinal epithelial cells. Furthermore, our SBS mouse model demonstrated that LGG supplementation conferred therapeutic benefits through improving disordered intestinal microbiota, upregulating intestinal tight junction protein, and enhancing gut barrier integrity. These findings suggest that LGG may offer clinical advantages for SBS patients, including suppression of pathogenic overgrowth and protection against dysbiosis-driven complications. The precise mechanism by which LGG enhances the small intestinal barrier in SBS remains incompletely understood. Intriguingly, we found that LGG directly activates FXR and its downstream signaling pathways in both Caco-2 cells and mouse ileal organoids. This activation was independent of bile acid-mediated FXR modulation and instead involved direct antagonism of β-MCA, a known FXR inhibitor. In WT SBS mice, LGG supplementation significantly upregulated FXR downstream genes (OST-α, OST-β, I-BABP) and elevated serum FGF15 levels, and enhanced intestinal barrier integrity via increased occludin expression and FXR signaling activation. In FXRInt-KO SBS mice, LGG failed to increase occludin levels, activate FXR-related genes, or restore gut microbiota abundance. LGG’s ability to stabilize gut microbiota and protect the intestinal mucosal barrier depends on functional FXR signaling. FXR activation by LGG likely maintains epithelial cell stability and microbiota equilibrium, mitigating SBS-associated dysbiosis.
The interaction between the gut microbiome and bile acids modulates the balance of FXR agonists and antagonists, enabling the gut microbiome to influence host metabolism34. For instance, Lactobacillus strains (e.g., L. reuteri and L. plantarum) reduce hepatic bile acid synthesis and enhance bile acid transport by downregulating FXR antagonists (T-α/β-MCA) and activating ileal/hepatic FXR signaling. These strains also alleviate dysbiosis, with L. reuteri enriching Muribaculaceae and L. plantarum boosting Akkermansia abundance35. In SBS, impaired conversion of primary bile acids (e.g., CA) to secondary bile acids—such as LCA and DCA, which are potent FXR agonists—compromises intestinal FXR activation7. Our findings demonstrate that LGG intervention in SBS mice that alters fecal bile acid profiles, increasing secondary bile acids (DCA, keto-LCA) and reducing primary bile acids, activates FXR signaling via enhanced secondary bile acid production, and modulates microbiota composition: Secondary bile acids (LCA, DCA, keto-LCA) positively correlate with Prevotellaceae_UCG-001 (beneficial taxa). The opportunistic pathogen Escherichia-Shigella associates with primary bile acid α-MCA (an FXR antagonist).
The mechanisms by which LGG regulates intestinal microbiota involve both adhesion to intestinal epithelial cells and the production of bioactive metabolites. While LGG is known to colonize the intestinal mucosa and modulate the gut microbiota36, our study highlights the role activating FXR and promoting intestinal epithelial cell homeostasis. This suggests that LGG’s beneficial effects may be mediated by both colonization and soluble metabolites. Future studies should investigate the extent of LGG colonization in our model to further elucidate its mechanisms of action. Although specific activated FXR components due to LGG have not been identified, in in vitro and in vivo studies, we confirmed that LGG could not only activate FXR signaling alone, but also activate FXR by altering gut microbiota to affect the proportion of bile acids. Our next step is to analyze LGG components that activate FXR signaling pathways in the intestine.
In summary, this study revealed that LGG maintained intestinal epithelial cell integrity and improved disordered intestinal microbiota in SBS, potentially through the activation of intestinal FXR signaling. Our findings provide new insights into the mechanisms of LGG in the small intestine.
Methods
Statistics and Reproducibility
Clinical data
Stool, blood, and intestinal tissue samples were collected from enterostomy patients at Nanjing Children’s Hospital, an affiliated hospital of Nanjing Medical University, between June 2021 and October 2022. The SBS group (type I SBS group) was dependent on PN for >42 days or had a residual bowel length (length of small intestine expected for gestational age) of <25%. The remaining enterostomy patients were assigned to the control group. To estimate the normal small intestine length based on age, the following formula was used: Length (cm) = 418.183 − (9575.69.2/weeks)18.
Clinical data included the following parameters: gender, age, birth weight, corrected gestational age, medical history, and biochemical parameters. The research protocol was approved by the local medical ethics committee of Nanjing Children’s Hospital (Ethics No. 202205051-1). Clinical data were collected only after obtaining informed consent from patients or their guardians, confirmed by the signing of an informed consent form.
Inclusion criteria
Patients who underwent enterostomy after birth due to neonatal necrotizing enterocolitis, intestinal atresia, Hirschsprung’s disease, meconium ileus, and congenital intestinal malrotation. Exclusion criteria: Patients with enterostomy and concurrent conditions such as biliary atresia, severe congenital heart disease, or congenital inherited metabolic disease. Fecal samples were collected prior to enterostomy closure. None of the patients had used antibiotics within 6 weeks before sample collection. Intestinal tissues were collected intraoperatively from proximal enterostomy sites and stored at −80 °C.
Caco-2 cell culture study
Caco-2 cells (ATCC HTB-37) were obtained from the Key Laboratory of the Affiliated Children’s Hospital of Nanjing Medical University. The frozen cells were thawed in a 37 °C water bath, seeded into culture flasks, and cultured in Dulbecco’s Modified Eagle Medium (DMEM, Thermo Fisher, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, USA) and 1% penicillin-streptomycin (Beyotime, China). When cells reached 70% confluency, they were stimulated with different concentrations of lipopolysaccharide (LPS, Beyotime, China) (2, 10, 50, 100, and 150 μg/mL) for 24 h. Tight junction protein expression (occludin, Abclonal, China; claudin-1, Santa Cruz, USA) was analyzed by western blotting. Based on the effects of different LPS concentrations on tight junction proteins in Caco-2 cells, 100 μg/mL LPS was selected for subsequent experiments.
Probiotic selection
LA, LGG and BA (ATCC 4356, ATCC 53103, ATCC 15704, USA) were provided as gifts by the State Key Laboratory of Innovative Drug Technology, China National Institute of Pharmaceutical Industry, Shanghai, China. The strains were cultured at 37 °C in MRS medium (Solarbio, China) for 48–72 h under anaerobic conditions. Bacterial density was measured using a microplate reader (OD620 nm). The cultures were terminated during the exponential growth phase, and the culture supernatants and bacterial pellets were separated by centrifugation at 5000 rpm for 5 min at 4 °C. The pH value of the supernatant was measured and adjusted to 7.0 using 3% NaOH. The supernatants were filtered through a 0.22 μm membrane and stored at −20 °C for future use. The bacterial pellets were frozen at −20 °C. Caco-2 cells were seeded into 96-well plates at a density of 2 × 10⁵ cells/mL and incubated in 5% CO₂ at 37 °C. Probiotic supernatants from LAs, LGGs, and BAs were collected during the exponential growth phase and added to Caco-2 cells at various concentrations (2%, 4%, 6%, and 8%) for 24 h. Cell viability was measured using a Cell Counting Kit-8 (CCK-8, Beyotime, China) assay. Based on the results, 4% probiotic supernatants were selected for further studies. Occludin and claudin-1 (Santa Cruz, USA) protein expression levels were compared among Caco-2 cells from different groups: Normal, LPS, LAs, LAs+LPS, BAs, BAs+LPS, LGGs, and LGGs+LPS.Caco-2 cells were obtained from the Key Laboratory of Affiliated Children’s Hospital of Nanjing Medical University. The frozen cells were resuscitated in a 37 °C water bath, seeded into culture flasks, and cultured in Dulbecco’s Modified Eagle Medium (Thermo Fisher, USA), supplemented with 10% fetal bovine serum (Gibco, USA) and 1% penicillin-streptomycin (Beyotime, China). When cells reached 70% confluency, they were stimulated with different concentration s of LPS (Beyotime, China) (2, 10, 50, 100, 150 μg/mL) for 24 h. Tight junction protein expression (occludin, Abclonal, China; claudin-1, Santa Cruz, USA) was analyzed by western blotting. Based on the effects of different LPS concentration on tigh junction proteins in Caco-2 cells, 100 μg/mL LPS was selected for subsequent experiments. Cells were seeded into 96-well plates at a density of 2 × 105 cells/mL and incubated in 5% CO2 at 37 °C. Probiotic supernatants from LAs, LGGs, and BAs were collected during the exponentially growing phase and added to Caco-2 cells at various concentrations (2%, 4%, 6%, and 8%) for 24 h. Cell viability was measured using a Cell Counting Kit-8 (CCK-8, Beyotime, China) assay. Based on the results, 4% probiotic supernatants were selected for further studies. Occludin and claduin-1 (Santa Cruz, USA) protein expression levels were compared among Caco-2 cells from different groups: Normal, LPS, LAs, LAs+LPS, BAs, BAs+LPS, LGGs, and LGGs+LPS.
Cell viability assay
A Caco-2 cell suspension (100 μL) was added to each well of a 96-well plate (6 wells per group) at a density of 2 × 10⁴ cells/mL. The cells were cultured at 37 °C in a CO₂ incubator for 24 h to allow adherence and growth. When cell density reached ~70%, the medium was replaced with DMEM containing 1% fetal bovine serum. Then, 100 μg/mL LPS (Beyotime, China) and various concentrations of LAs, LGGs, and BAs (2%, 4%, 6%, and 8%) were added, respectively, and incubation continued for 24 h. Subsequently, 10 μL of CCK-8 reagent (Beyotime, China) was added to each well and incubated for 2 h at 37 °C. Optical density was measured at 450 nm using a microplate reader.
Cell apoptosis experiment
Caco-2 cells were fixed with 4% paraformaldehyde (Servicebio, China) for 30 min, washed twice with phosphate-buffered saline (PBS), and then treated with immunostaining permeabilization solution (Beyotime, China) for 5 min at room temperature. After washing twice with PBS, 50 μL of TUNEL assay solution (Beyotime, China) was added, and the cells were incubated at 37 °C for 60 min in the dark. Following another PBS wash, the slides were mounted with anti-fluorescence quenching mounting medium (Beyotime, China) and observed under a fluorescence microscope.
Extraction and culture of mouse ileum organoids
The terminal ileum tissue (10 cm) was extracted and placed in sterile phosphate-buffered saline (PBS). The mesenteric membrane was removed using microscissors, and the intestinal contents were flushed repeatedly with sterile PBS using a 1 mL pipette. The intestinal tube was cut open longitudinally and rinsed repeatedly until the liquid became clear. The tissue was then cut into 0.3–0.5 cm fragments and transferred to a 50 mL centrifuge tube. The fragments were washed 10–15 times with 10 mL of sterile PBS. After each wash, the suspension was allowed to settle for 30 s, and the supernatant was completely aspirated and discarded.
Next, 10 mL of cell dissociation solution (Stemcell, Canada) was added to the tube, and the fragments were gently mixed for 20 min at room temperature (~25 °C) to form a suspension. The suspension was filtered through a 70 μm nylon mesh, and the filtrate was collected. This filtration step was repeated 4 times to maximize the yield of crypt fractions. The crypts were resuspended in Matrigel (Cellada, China), and 50 μL of the suspension (containing ~50–100 crypts) was seeded into each well of a 24-well plate. After the Matrigel solidified, 500 μL of IntestiCult™ Organoid Growth Medium (Stemcell, Canada) was added to each well for culture.
In vitro organoid studies
Ileal crypts extracted from WT mouse were cultured and passaged. LGGs in exponential growth phase (2%, 4%, and 8%) was added to organoid culture wells after organoid maturation. Additionally, different concentrations of the FXR inhibitor β-MCA (0.1, 0.2, 0.4, and 0.8 mM, MCE, USA) were used to stimulate organoids. After 24 h, mRNA was extracted from organoid tissues to assess the effects of LGG and the FXR inhibitor on FXR signaling. Ileum organoids were then treated with LGGs (2%) and β-MCA (0.4 mM) and divided into the following groups: normal control (NC), LGGs, β-MCA, and LGGs+β-MCA. After 24 h of incubation, FXR and ASBT expression was detected by immunofluorescence. Organoids were also treated with LGGs (2%) or LPS (100 mg/mL) for 24 h and divided into NC, LGGs, LPS, and LGGs+LPS groups. Occludin and villin (Santa Cruz, USA) expression were detected by immunofluorescence.
Establishment of mouse SBS model
Male mice weighing 25 to 30 g were fasted for 8 h before surgery. Mice were anesthetized with 2% isoflurane (RWD, China), and a 1 cm longitudinal abdominal incision was made. The entire small intestine was gently pulled out of the abdominal cavity using a cotton swab to clamp the intestinal tube, avoiding the use of forceps to prevent damage to the intestinal wall. The small intestine was externalized, and a 75% resection was performed, starting 2 cm distal to the ligament of Treitz and ending 5–6 cm proximal to the ileocecal junction. The first-level mesenteric vascular arch was ligated using 9–0 monofilament sutures, and the third-level mesenteric microvessels were cauterized with an electric cautery to minimize intraoperative bleeding. The intestinal tube was cut with micro-scissors, and a 20 cm segment was removed. If residual intestinal contents were present in the proximal and distal enteric cavities, they were flushed with 0.9% sodium chloride using a 5 mL syringe to prevent postoperative intestinal obstruction. Intestinal continuity was restored through an end-to-end, single-layer, interrupted anastomosis using 10–0 monofilament sutures. The abdominal wall muscle layer and skin were sutured with 4–0 silk sutures. Immediately after surgery, each mouse received 5 mg/kg carprofen subcutaneously for pain management, followed by 1 mL of glucose-saline solution (5% glucose and 0.9% sodium chloride) administered subcutaneously. Starting on postoperative day 2, mice were injected with 1 mL of glucose-saline solution subcutaneously for 6 consecutive days.
Contruction of FXR knockout model
According to the design scheme, gRNA was designed, constructed, and transcribed in vitro. Simultaneously, a homologous recombination vector (Donor vector) was constructed, and the correctness of the vector sequence was verified by sequencing. Female C57BL/6J mice (donors) were injected with PMSG and hCG and then housed with male C57BL/6J mice. Concurrently, female C57BL/6 mice without hormone injection were housed with vasectomized male mice to obtain pseudopregnant female mice (recipients). The superovulated donor female mice were sacrificed, and fertilized eggs were collected. The Cas/gRNA/loxp-donor samples (designed and constructed by Jiangsu Jichi Pharmaceutical Kang Co., Ltd.) were prepared and tested in vitro. These samples were microinjected into the pronuclei of fertilized eggs from C57BL/6Gpt background mice using a microinjection instrument. The pseudopregnant female mice were anesthetized with isoflurane, and the fertilized eggs were transplanted into their oviducts. Three weeks later, the recipient female mice gave birth to F0 generation mice. At 5–7 days old, tail and toe samples were collected from the F0 mice, and genomic DNA was extracted for PCR and sequencing to confirm the genotype. After reaching sexual maturity, F0 generation mice were mated with wild-type C57BL/6J mice to obtain stable F1 generation heterozygous mice. Fxr-flox heterozygous mice were screened using PCR technology. Intestine-specific Cre recombinase-expressing mice (Intestine Cre) were crossed with FXR-flox mice. The genotype of the offspring was identified by PCR, and intestine-specific FXR knockout mice (Intestine-Fxr fl/fl Cre-positive) were selected.
Animal studies
Twenty-four 10-week-old male WT and twenty-four FXRInt-KO C57BL/6 mice were used in the study. WT mice were divided into four groups: NC, LGG-supplemented (LGG), 75% small bowel resection (SBR), and 75% SBR with LGG supplementation (SBS + LGG). FXRInt-KO mice were divided into four groups: knockout control (K), knockout with LGG supplementation (KL), knockout with 75% SBR (KS), and knockout with 75% SBR and LGG supplementation (KS + L). SBS was induced by 75% small bowel resection. The small intestine was removed from 2–3 cm proximal to the Treitz ligament to 5–6 cm distal of the ileocecal junction, and end-to-end intestinal anastomosis was performed using 10–0 sutures. Starting from day 2 after surgery, live LGG (in physiological saline) was administered to mice via oral garage at a dose of 109 CFU/day for 2 weeks. Mice in the NC, SBS, K, and KS groups were gavaged with an equivalent volume of physiological saline. After 2 weeks, mice were humanely euthanized by exsanguination under anesthesia, and plasma, intestinal tissue, and ileocecal feces were collected and stored at −80 °C.
Cell viability assay
A Caco-2 cell suspension (100 μL) was added to per well of a 96-well plate (6 well per group) at density of 2 × 104 cells/mL. The cells were cultured at 37 °C in CO2 incubator for 24 h and adhered to growth. When cell density was about 70%, the medium was changed with DMEM containing 1% fetal bovine serum. Then 100 μg/mL LPS (Beyotime, China) and various concentrations of LAs, LGGs, and BAs (2%, 4%, 6%, 8%) were added, respectively, and incubation continued for 24 h. 10 μL CCK8 reagent (Beyotime, China) was added to each well and incubated for 2 h at 37 °C. Optical density was measured at 450 nm by microplate reader.
Assess FGF15 in plasma
Plasma samples were collected from WT mice and the expression levels of FGF15 in plasma was detected using an ELISA kit (Mlbio, China). The procedure was performed according to the manufacturer’s instructions.
Intestinal morphology examination
In the WT NC, LGG, SBS, and SBS + LGG groups, villus height and crypt depth in the jejunum and ileum were examined using hematoxylin & eosin (H&E) staining, while goblet cell numbers were calculated using Periodic acid-Schiff (PAS) staining. Tissues from the proximal jejunum, distal ileum, and proximal colon were fixed with 4% paraformaldehyde, dehydrated, embedded in paraffin, and sectioned. Intestinal structure was evaluated by H&E staining and the number of goblet cells was assessed by PAS staining. Images were captured using an OLYMPUS BX51 and ZEN laser confocal microscope. Villus height and crypt depth in the jejunum and ileum were measured using Image J software, and goblet cell numbers were counted.
PCR
Intestinal tissue from the proximal enterostomy of humans and the terminal ileum of mice was collected. Total RNA were extracted using TRIzol@Reagent (Life technologies, USA) after homogenizing the tissue samples, and RNA concentration was quantified using a NanoDrop 1000 spectrophotometer. Reverse transcription was performed using Vazyme reverse transcription kit (Vazyme, China) and quantitative PCR (qPCR) was conducted using AceQ@qPCR SYBR Green Master Mix (Vazyme, China). Primers were synthesized by Tsingke (Beijing), and the primer sequences are listed in Tables S2 and S3.
Immunofluorescence detection
Immunofluorescence analysis was performed to assess proteins expression in ileum organoids and intestinal tissues. The slides were blocked in 3% bovine serum albumin and 0.03% Triton-X100 for 30 min at room temperature, followed by incubation with primary antibodies: Occludin (1:500 dilution; Abclonal, China) and Villin (1:500 dilution; Santa Cruz, USA), FXR (1:500 dilution; Santa Cruz, USA) and ASBT (1:500 dilution; Abclonal, China) at 4 °C overnight. Secondary antibodies (anti-mouse or anti-rabbit, 1:500 dilution; Beyotime, China) were incubated for 1 h at room temperature, and nuclei were stained with DAPI. To explore the role of LGGs in FXR signaling, organoids were pretreated with 0.4 mM β-MCA, and FXR and ASBT expression was determined by immunofluorescence staining. The effect of 2% LGG on Occludin and Villin in the ileum organoids stimulated with 100 μg/mL LPS was examined. Additonally, the expression of the surface proteins Occludin and Villin in intestinal epithelial cells was evaluated in 75% SBR model on postoperative day 14(POD14).
Western blot
Cells and tissues were lysed in RIPA buffer (Beyotime, China). After centrifugation at 4 °C for 30 min, the supernatant was collected by centrifuging the mixture for 15 min at 4 °C. The protein concentration was determined by BCA Protein Assay kit (Yeasen, China), and SDS-sample loading buffer (Beyotime, China) was added and the samples were boiled at 100 °C for 5 min. Samples were electrophoresed on 8%-10% SDS-PAGE gel (YEASEN, China) for 30 min at 80 V and then for 45 min-1 h at 120 V. The proteins were transferred to PVDF membrane at 300 mA for 1.5 h. The membrane was blocked with 5% nonfat dry milk at room temperature for 2 h, and then primary antibodies Occludin (dilution, 1:1000; Santa Cruz, USA), Claudin-1 (dilution, 1:1000; Santa Cruz, USA), Villin (dilution, 1:1000; Santa Cruz, USA) and PCNA (dilution, 1:1000; Santa Cruz, USA) were incubated at 4 °C overnight. After three washes with TBST for 10 min, secondary antibodies (dilution, 1:2000; Beyotime, China) were added and incubated at room temperature for 1 h. After TBST wash (3 × 10 min), hypersensitive ECL luminescent solution (NCM, China) was added for developing imaging.
Gut microbiota detection
Fecal samples from wild-type mice in each group were collected in sterile tubes, with 6 samples per group. Gene sequencing (16S rDNA) was performed by OE Biotech Co., Ltd (China). Fecal DNA was extracted using the MagPure Soil DNA LQ Kit, and the V3-V4 regions of the 16S rRNA gene were amplified by PCR. QIIME2 (version 2020.11) was used for microbiome bioinformatics analysis. After NovaSeq sequencing to generate raw sequence data, primers were trimmed using Cutadapt software. High-quality paired-end reads were further processed using DADA2 within QIIME2 for quality control, including quality filtering, denoising, merging, and chimera removal. The relative abundance of all species in each sample was normalized to 1 (i.e., 100%). A community structure distribution map was generated by plotting bar graphs based on the proportion of each species in the sample. At the phylum level, the top 10 most abundant species were selected to create bar charts. QIIME2 software was used to calculate four alpha diversity indices for each sample: Chao1, Observed_species, Simpson, and Shannon. Boxplots were generated to compare species richness and evenness among samples. Higher Chao1 and Observed_species values indicate greater species richness, while higher Simpson and Shannon values reflect higher diversity and evenness of community distribution. Beta diversity was assessed using the Bray-Curtis distance metric in R and QIIME2 software. Principal coordinate analysis (PCoA) was used to visualize differences in microbial community structure among samples. LEfSe was used to identify differentially abundant taxa between two or more groups. A linear discriminant analysis (LDA) score greater than the set threshold was defined as a differential species, representing statistically significant biomarkers. The bar graph length indicates the effect size of significantly different species. Circles radiating from the center represent taxonomic levels from phylum to genus (or species). Each small circle at a taxonomic level represents a classification, with its diameter proportional to the relative abundance of the taxon.
Metabolomics analysis of fecal bile acids
Fecal samples were collected, with 6 samples per group. Shanghai Bipu Biotechnology Co., Ltd. was commissioned to perform bile acid metabolomics analysis. Bile acid standards were weighed, and a standard stock solution was prepared using methanol (final concentration: 1 mg/mL). The stock solution was then serially diluted to create ten standard curve points. A 50 mg fecal sample was mixed with 400 μL of methanol pre-cooled at −20 °C. The mixture was centrifuged at 12,000 rpm for 10 min at 4 °C. Then, 600 μL of water was added to 300 μL of the supernatant, and the mixture was vortexed for 30 s. An appropriate amount of the supernatant was diluted 5-fold with 30% methanol, filtered through a 0.22 μm membrane, and the filtrate was collected for analysis. LC-MS was used to detect 28 fecal bile acids, including:
Primary bile acids: Cholic acid (CA), Chenodeoxycholic acid (CDCA), Allocholic acid (ACA), Alpha-muricholic acid (α-MCA), Beta-muricholic acid (β-MCA), Taurocholic acid (TCA), Taurochenodeoxycholic acid (TCDCA), Tauro-alpha-muricholic acid (T-α-MCA), Tauro-beta-muricholic acid (T-β-MCA).
Secondary bile acids
Lithocholic acid (LCA), Deoxycholic acid (DCA), Ursodeoxycholic acid (UDCA), Hyodeoxycholic acid (HDCA), Glycolithocholic acid (GLCA), Glycodeoxycholic acid (GDCA), Glycoursodeoxycholic acid (GUDCA), Taurolithocholic acid (TLCA), Taurodeoxycholic acid (TDCA), Taurohyodeoxycholic acid (THDCA), Tauroursodeoxycholic acid (TUDCA).
Other bile acids
Allolithocholic acid (alloLCA), Isolithocholic acid (isoLCA), Nordeoxycholic acid (NorDCA), 7-ketolithocholic acid (7-ketoLCA), 12-ketolithocholic acid (12-ketoLCA), 6-ketolithocholic acid (6-ketoLCA), Beta-ursodeoxycholic acid (β-UDCA), Norcholic acid (NorCA), Dehydrocholic acid (DHCA), 7,12-diketolithocholic acid (7,12-diketoLCA), 6,7-diketolithocholic acid (6,7-diketoLCA), Ursocholic acid (UCA), Glycocholic acid (GCA), Taurocholate-3-sulfate disodium salt (LCA-3S).
Statistical analysis
Statistical analysis was performed in GraphPad Prism 6 software (CA, USA). Counting card information using the chi-square test. Independent sample t tests were used to compare continuous variables between two groups. One-way analysis of variance was performed for multiple variables and followed by Tukey’s post hoc tests for multiple comparisons. The non-normal distributiong data were conducted with Mann-Whitney U test. Correlation between the level of fecal microbial species and bile acids were evaluated by Spearman’s rank correlation analysis. P < 0.05 were statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The source data underlying Figs. 1–9 is provided as a Source Data file. The data used in this study have been deposited in the NCBI database with the following accession numbers: Raw sequencing data: SRA accession SRR 28257074-SRR 28257095 (22), SRR 28276859-SRR 28276882 (24), SRR 28262099-28262122 (24). Project metadata: BioProject PRJNA1085009, PRJNA1085271, PRJNA1085787.
References
Carter, J. et al. Gut microbiota modulation of short bowel syndrome and the gut-brain axis. Nutrients 15, 2581 (2023).
Vanuytsel, T. Disease modifying therapy in short bowel syndrome: finally coming of age?. Lancet Gastroenterol. Hepatol. 4, 328–329 (2019).
Caporilli, C., Giannì, G., Grassi, F. & Esposito, S. An overview of short-bowel syndrome in pediatric patients: focus on clinical management and prevention of complications. Nutrients 15, 2341 (2023).
Neelis, E. et al. The gut microbiome in patients with intestinal failure: current evidence and implications for clinical practice. Jpen. J. Parenter. Enter. Nutr. 43, 194–205 (2019).
Piper, H. G. et al. Severe gut microbiota dysbiosis is associated with poor growth in patients with short bowel syndrome. J. Parenter. Enter. Nutr. 41, 1202–1212 (2017).
Efremova, I. et al. Epidemiology of small intestinal bacterial overgrowth. World J. Gastroenterol. 29, 3400–3421 (2023).
Huang, Y., Jiao, J., Yao, D., Guo, F. & Li, Y. Altered fecal microbiome and metabolome profiles in rat models of short bowel syndrome. Front. Microbiol. 14, 1185463 (2023).
Thänert, R. et al. Antibiotic-driven intestinal dysbiosis in pediatric short bowel syndrome is associated with persistently altered microbiome functions and gut-derived bloodstream infections. Gut Microbes 13, 1940792 (2021).
Pereira-Fantini, P. M. et al. Altered FXR signalling is associated with bile acid dysmetabolism in short bowel syndrome-associated liver disease. J. Hepatol. 61, 1115–1125 (2014).
Mutanen, A., Lohi, J., Heikkilä, P., Jalanko, H. & Pakarinen, M. P. Loss of ileum decreases serum fibroblast growth factor 19 in relation to liver inflammation and fibrosis in pediatric onset intestinal failure. J. Hepatol. 62, 1391–1397 (2015).
Bettag, J. et al. Novel therapeutic approaches for mitigating complications in short bowel syndrome. Nutrients 14, 4660 (2022).
Ruigrok, R., Weersma, R. K. & Vich Vila, A. The emerging role of the small intestinal microbiota in human health and disease. Gut Microbes 15, 2201155 (2023).
Villmones, H. C. et al. Species level description of the human ileal bacterial microbiota. Sci. Rep. 8, 4736 (2018).
Cole, C. R. et al. The rate of bloodstream infection is high in infants with short bowel syndrome: relationship with small bowel bacterial overgrowth, enteral feeding, and inflammatory and immune responses. J. Pediatr. 156, 941–947.e941 (2010).
Liu, Y. et al. Probiotic Lactobacillus rhamnosus GG prevents liver fibrosis through inhibiting hepatic bile acid synthesis and enhancing bile acid excretion in mice. Hepatology 71, 2050–2066 (2020).
Le Beyec, J., Billiauws, L., Bado, A., Joly, F. & Le Gall, M. Short bowel syndrome: a paradigm for intestinal adaptation to nutrition?. Annu. Rev. Nutr. 40, 299–321 (2020).
Marchix, J., Goddard, G. & Helmrath, M. A. Host-gut microbiota crosstalk in intestinal adaptation. Cell. Mol. Gastroenterol. Hepatol. 6, 149–162 (2018).
Struijs, M. C., Diamond, I. R., de Silva, N. & Wales, P. W. Establishing norms for intestinal length in children. J. Pediatr. Surg. 44, 933–938 (2009).
Hajam, I. A. et al. Anti-inflammatory properties of plasma from children with short bowel syndrome. Pathogens 10, 1021 (2021).
Engstrand Lilja, H., Wefer, H., Nyström, N., Finkel, Y. & Engstrand, L. Intestinal dysbiosis in children with short bowel syndrome is associated with impaired outcome. Microbiome 3, 18 (2015).
Payne, C. M., Bernstein, C., Dvorak, K. & Bernstein, H. Hydrophobic bile acids, genomic instability, Darwinian selection, and colon carcinogenesis. Clin. Exp. Gastroenterol. 1, 19–47 (2008).
Kaufman, S. S. et al. Influence of bacterial overgrowth and intestinal inflammation on duration of parenteral nutrition in children with short bowel syndrome. J. Pediatr. 131, 356–361 (1997).
Hvistendahl, M. K. et al. Bile acid-farnesoid X receptor-fibroblast growth factor 19 axis in patients with short bowel syndrome: the randomized, glepaglutide phase 2 trial. J. Parenter. Enter. Nutr. 46, 923–935 (2022).
Pereira-Fantini, P. M. et al. Farnesoid X receptor agonist treatment alters bile acid metabolism but exacerbates liver damage in a piglet model of short-bowel syndrome. Cell. Mol. Gastroenterol. Hepatol. 4, 65–74 (2017).
Han, B. et al. Gut microbiota-related bile acid metabolism-FXR/TGR5 axis impacts the response to anti-α4β7-integrin therapy in humanized mice with colitis. Gut microbes 15, 2232143 (2023).
Ajouz, H., Mukherji, D. & Shamseddine, A. Secondary bile acids: an underrecognized cause of colon cancer. World J. Surg. Oncol. 12, 164 (2014).
Gorreja, F. & Walker, W. A. The potential role of adherence factors in probiotic function in the gastrointestinal tract of adults and pediatrics: a narrative review of experimental and human studies. Gut Microbes 14, 2149214 (2022).
Piper, H. G. et al. The impact of lactobacillus probiotics on the gut microbiota in children with short bowel syndrome. J. Surg. Res 251, 112–118 (2020).
Merras-Salmio, L. & Pakarinen, M. P. Infection prevention and management in pediatric short bowel syndrome. Front. Pediatr. 10, 864397 (2022).
Wu, J. et al. Soluble mediators from lactobacillus rhamnosus gorbach-goldin support intestinal barrier function in rats after massive small-bowel resection. J. Parenter. Enter. Nutr. 42, 1026–1034 (2018).
Reddy, V. S., Patole, S. K. & Rao, S. Role of probiotics in short bowel syndrome in infants and children-a systematic review. Nutrients 5, 679–699 (2013).
Bartholome, A. L., Albin, D. M., Baker, D. H., Holst, J. J. & Tappenden, K. A. Supplementation of total parenteral nutrition with butyrate acutely increases structural aspects of intestinal adaptation after an 80% jejunoileal resection in neonatal piglets. J. Parenter. Enter. Nutr. 28, 210–222 (2004).
Riehl, T. E. et al. Lactobacillus rhamnosus GG protects the intestinal epithelium from radiation injury through release of lipoteichoic acid, macrophage activation and the migration of mesenchymal stem cells. Gut 68, 1003–1013 (2019).
Fogelson, K. A., Dorrestein, P. C., Zarrinpar, A. & Knight, R. The gut microbial bile acid modulation and its relevance to digestive health and diseases. Gastroenterology 164, 1069–1085 (2023).
Ye, X. et al. FXR signaling-mediated bile acid metabolism is critical for alleviation of cholesterol gallstones by lactobacillus strains. Microbiol. Spectr. 10, e0051822 (2022).
Capurso, L. Thirty years of lactobacillus rhamnosus GG: a review. J. Clin. Gastroenterol. 53 (Suppl 1), S1–s41 (2019).
Acknowledgements
This work was supported by the Natural Science Foundation of Hubei Province under award number 2023AFB908, China; the Wuhan Knowledge Innovation Project of Hubei province under award number 2023020201010199, China; the General project of Jiangsu Provincial Health Commission under award number M2022037, China; the Top Talents of Jiangsu Provincial Health Committee’s Six One Project under award number LGY2020019, China.
Author information
Authors and Affiliations
Contributions
L.L.G. was involved with conceptualization, methodology, data curation, and writing—original draft preparation. X.F.D. was involved with the conceptualization, project administration. H.F.W. was involved with investigation, data curation, and writing-review. R.Y.Z. and H.X. were performed the data analyses and generated the tables and figures. W.W.J. and W.B.T. were involved with conceptualization, funding acquisition, writing-review and editing, and supervision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Laura Grasa and the other, anonymous, reviewer for their contribution to the peer review of this work. Primary Handling Editor: Tobias Goris. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gui, L., Duan, X., Wang, H. et al. Lactobacillus rhamnosus GG maintains gut microbiota stability and promotes intestinal adaptation via activated intestinal farnesoid X receptor signaling in short bowel syndrome. Commun Biol 8, 816 (2025). https://doi.org/10.1038/s42003-025-08254-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08254-x