Abstract
Shrimp aquaculture is critically important for global food security, but viral diseases like white spot syndrome virus (WSSV) cause devastating economic losses, highlighting the urgent need for effective disease control strategies. While trained immunity has been observed in invertebrates like shrimp after exposure to pathogens, the underlying molecular mechanisms remain elusive. Here we reveal that lysine acetyltransferase KAT8-mediated histone H3K27ac is critical for antiviral defense in shrimp Marsupenaeus japonicus. We demonstrate that ultraviolet-inactivated WSSV (UV-WSSV) induces antiviral trained immunity in the shrimp via KAT8-dependent H3K27ac. UV-WSSV training enhances glycolysis and the tricarboxylic acid (TCA) cycle, increasing acetyl-CoA production. This acetyl-CoA fuels KAT8 activity, depositing H3K27ac marks at the promoter of the NF-κB-like transcription factor Dorsal. This epigenetic modification upregulates Dorsal expression, leading to the enhanced production of the antiviral cytokine Vago5 and antimicrobial peptides (AMPs) upon subsequent WSSV challenge. Furthermore, H3K27ac directly activates key glycolytic genes (Hk2, Pk, Ldh), creating a feedforward loop that sustains metabolic reprogramming. Our work reveals a conserved KAT8-H3K27ac axis driving trained immunity in invertebrates through integrated metabolic-epigenetic crosstalk, analogous to mammalian systems. These findings provide a crucial theoretical foundation for developing antiviral vaccines and sustainable immunostimulants to control disease in shrimp aquaculture.

Similar content being viewed by others
Introduction
The vertebrate immune system involves both innate and adaptive immune responses. Invertebrates typically possess only innate immunity against invasive pathogens and lack immunological memory. However, research over the past two decades has demonstrated that innate immunity can exhibit memory, a phenomenon called innate immune memory, or trained innate immunity1. Trained immunity is characterized by faster and more effective immune responses upon restimulation and provides long-term protection against foreign invaders2,3. The mechanisms of trained immunity are well known in the innate immune cells of mammals. Trained immunity is driven by the integration of signaling, metabolic, epigenetic and founctional rewiring: Immunological stimuli are initially recognized by different pattern recognition receptors (PRRs). These receptors trigger specific trained immunity-inducing signaling pathways that are crucial to trained phenotypes and subsequently mediate metabolic reprogramming (including glycolysis, the tricarboxylic acid (TCA) cycle, and fatty acid metabolism) and epigenetic modifications [including DNA methylation, histone methylation (such as H3K4me3), and acetylation (such as H3K27ac)]. This leads to a rapid and augmented innate immune response upon reinfection2,4. Trained immunity has also been found in invertebrates, including insects and shrimp5,6. For example, multiple lines of evidence establish that Marsupenaeus japonicus develops significantly enhanced resistance to WSSV upon secondary challenge when primed through oral delivery or immunization with formalin- or UV-inactivated WSSV7,8,9. The innate immune memory appeared to persist over a longer time span, or sometimes across generations10. Several possible mechanisms have been proposed to explain the induction of innate immune memory in invertebrates. These include the upregulation of immune-related pathways (such as Toll, Imd, JNK and JAK/STAT pathways) and phenotypic changes in immune cell populations (such as increased abundance of granulocytes)11,12,13, however, it is unclear whether epigenetic modifications and metabolic reprogramming are involved in the innate immune memory of invertebrates. On the other hand, although the study of trained immunity has taken many advances over the past decade, there are many key knowledge gaps that hamper our ability to use trained immunity to combat pathogens14.
Lysine acetylation is an evolutionarily conserved post-translational modification (PTM) that occurs in both prokaryotes and eukaryotes. It is jointly regulated by lysine acetyltransferases (KATs) and lysine deacetylases (KDACs)15,16. Lysine acetyltransferase 8 (KAT8), also known as MYST1 or MOF (males absent on the first), is important in catalyzing the acetylation of histone H4 at K16 (H4K16) in mammalian cells17,18,19. It is also responsible for the acetylation of nonhistone substrates such as p5320 and interferon regulatory factor 3 (IRF3)21. KAT8 is involved in diverse biological processes, ranging from DNA damage repair, DNA transcription, and embryonic development to apoptosis22,23. However, the function of KAT8 in trained innate immunity is unclear.
Penaeid shrimp species are valuable for marine aquaculture. However, diseases caused by different pathogens, especially white spot disease (WSD), cause high mortality rates and significant economic losses24,25. White spot syndrome virus (WSSV) is an infectious pathogen of shrimp and other crustaceans. Approximately 80% of cultured shrimp losses are due to WSSV26,27. There are currently no effective control strategies for WSSV in shrimp aquaculture. The study of trained immunity in shrimp will facilitate the development of more reliable, cost-effective, and sustainable disease control strategies (such as the application of antiviral vaccines) in shrimp aquaculture. In the present study, we established a trained immunity model using ultraviolet light-inactivated WSSV (UV-WSSV) in the shrimp Marsupenaeus japonicus. We analyzed the profiles of H3K27ac modifications and identified that KAT8 is involved in the antiviral trained immunity of shrimp. The roles and underlying mechanisms of KAT8 in UV-WSSV-induced antiviral trained immunity were analyzed.
Results
H3K27ac and H4K16ac modifications mediated by KAT8 are enhanced in WSSV infected shrimp
Alterations in epigenetic modifications and reprogramming of cellular metabolism are two pillars of trained immunity. In our previous study, we reported that inactivated WSSV induced trained immunity in shrimp9. To reveal the mechanisms of trained immunity induced by inactivated WSSV, we first detected changes in histone acetylation, including histone pan-acetylation (pan-Hac), H3K27ac, H4K16ac, and H3K9ac, in the hemocytes of shrimp challenged with WSSV via the use of corresponding antibodies. The results revealed that the abundance of all of the above modifications was increased in the hemocytes of WSSV-infected shrimp, and H3K27ac and H4K16ac abundance were particularly high at 12 h post-WSSV infection (Fig. 1a, a’). Therefore, we focused on H3K27ac and H4K16ac in the following study.
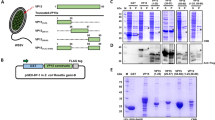
a Epigenetic modification patterns of pan-Hac, H3K27ac, H4K16ac, and H3K9ac in hemocytes of shrimp challenged with WSSV were analyzed by western blotting. a’ Statistical analysis of panel a based on three replicates. b–d RNA interference efficiency against Kat8 (b), Tip60 (c), and Kat2a (d) in shrimp hemocytes determined by qPCR. The qPCR data were normalized using Ef-1α as the endogenous control gene. e Abundances of H3K27ac and H3K9ac marks were measured in the hemocytes of shrimp after Kat8, Tip60, and Kat2a knockdown following WSSV infection via western blotting. dsGfp was used as the control. Bottom panel: Statistical analysis of the data in panel e based on three replicates. f Kat8-RNAi efficiency in hemocytes and intestinal tissues analyzed using qPCR; dsGfp injection was used as the control. g Modification levels of H3K27ac and H4K16ac were measured in the hemocytes and intestinal tissues of shrimp after Kat8 knockdown following WSSV infection, as determined by western blotting. dsGfp was used as a control. The samples were extracted at 24 hpi from Kat8 knockdown shrimp following WSSV infection. g’ The bands of three replicates of (g) were digitalized using ImageJ software and analyzed. The data are presented as the mean value ± SD (n = 3). P ≤ 0.05 (Student’s t-test).
To identify the lysine acetyltransferases (KATs) critical for histone H3K27ac epigenetic modification in shrimp, RNA interference (RNAi) screening was performed. Nine KATs were identified in our shrimp transcriptome sequencing data and online sources of the genome sequences of M. japonicus (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (Supplementary Fig. 1). We chose three KATs (KAT8, Tip60, and KAT2a) that were potentially involved in this modification for further study. After the expression of Kat8, Tip60, and Kat2a was knocked down in shrimp following WSSV infection (Fig. 1b–d), the H3K27ac level in shrimp hemocytes was significantly lower in dsKat8 injection group compared with that in the dsGfp group, no obvious change in H3K27ac was detected after RNAi of the other KATs (Fig. 1e).
Next, we detected H4K16ac in addition to H3K27ac after the knockdown of Kat8 (Fig. 1f), and the results revealed that H4K16ac and H3K27ac modifications were significantly decreased in shrimp hemocytes and the intestinal tissues at 24 h after WSSV infection (Fig. 1g, g’). Hence, KAT8 is also involved in H4K16ac modification in shrimp. The 6-alkylsalicylate MG149, an inhibitor of KAT828,29 was used to confirm these results after optimizing the injection dosage of the inhibitor (Supplementary Fig. 2a). Treatment with MG149 significantly inhibited H3K27ac and H4K16ac modifications in a dose-dependent manner in shrimp hemocytes (Supplementary Fig. 2b, b’) and the intestinal tissues (Supplementary Fig. 2c, c’). These results suggest that histone H3K27 and H4K16 acetylation mediated by KAT8 were increased in WSSV infected shrimp.
KAT8 is significantly upregulated and inhibits WSSV replication in shrimp
Shrimp KAT8 is a relatively conserved protein with 65% similarity to the amino acid sequence of Homo sapiens, and contains the CHROMO (Chromatin organization modifier) domain and histone acetyltransferase activity domain (Supplementary Fig. 3a, b). Phylogenetic analysis revealed that M. japonicus KAT8 clustered with human KAT8 and was closely related to Litopenaeus vannamei KAT8 (Supplementary Fig. 3c).
To characterize KAT8 tissue distribution, expression dynamics, and subcellular localization, we first generated specific polyclonal antibodies against the recombinantly expressed the protein (Supplementary Fig. 4a). Subsequent analyses revealed widespread distribution of both Kat8 mRNA and KAT8 protein across hemocytes and multiple tissues, including the heart, gills, stomach, and intestine (Supplementary Fig. 4b). Following WSSV infection, Kat8 transcript levels increased from 6 hpi, peaked at 24 hpi, and subsequently declined (Supplementary Fig. 4c). Consistent with transcriptional upregulation, KAT8 protein levels also increased from 6 hpi, and reaching maximal expression at 24 hpi (Supplementary Fig. 4d–f). The subcellular distribution of KAT8 was analyzed by immunocytochemical and western blot assays. As shown in Supplementary Fig. 4g, g’, KAT8 localized to both the cytoplasm and nucleus of normal shrimp hemocytes. However, 24 h post WSSV infection, its localization shifted predominantly to the nucleus. KAT8 was also detected in the cytosol and nucleus of intestinal cells by western blotting, and similar results were obtained (Supplementary Fig. 4h, h’). Our previous study revealed that KAT8 expression is regulated by the transcription factor Forkhead box O (FOXO) in insects30. The FOXO-binding element (FOXOBE) was also identified in the promoter of the shrimp Kat8 gene via the JASPAR website (http://jaspar.genereg.net/) (Supplementary Fig. 4i). To determine whether WSSV promotes KAT8 transcription through FOXO, we knocked down FoxO using RNAi and examined Kat8 expression. The results showed that FoxO knockdown markedly reduced Kat8 expression in shrimp hemocytes and the intestinal tissues at 24 h post-WSSV infection (Supplementary Fig. 4j). These results demonstrate that KAT8 expression is regulated by FOXO in response to WSSV infection and function in the nucleus for H3K27ac modification.
To explore the functions of KAT8 in shrimp infected with WSSV, RNAi of Kat8 was performed, followed by WSSV replication analysis via the expression of Vp28 (envelope protein-encoding gene of WSSV) and Ie1 (immediate early gene 1 of WSSV) as indicators. The results revealed that the expression of Vp28 (Fig. 2a) and Ie1 (Fig. 2b) increased significantly in shrimp hemocytes and the intestinal tissues at 24 hpi. Similar results were obtained for the expression of the VP28 protein (Fig. 2c). The number of WSSV copies was significantly increased in the intestinal tissues of Kat8-knockdown shrimp challenged with WSSV (Fig. 2d). Compared with that of the control shrimp, the survival of the Kat8-knockdown shrimp significantly decreased (Fig. 2e). These results suggest that KAT8 has antiviral functions mediated through H3K27ac modification.
a, b The expression of WSSV Vp28 (a) and Ie1 (b) was detected in Kat8-knockdown shrimp at 24 hpi via qPCR. Ef-1α was used as the endogenous control gene. c The expression level of the VP28 protein was detected in shrimp challenged with WSSV after Kat8 knockdown using western blotting with anti-VP28 Abs. The samples were extracted from Kat8 knockdown shrimp 24 h after WSSV infection. Bottom panel: Statistical analysis of the data in panel c based on three replicates. The data are presented as the mean ± SD (n = 3). d The copy number of WSSV in the intestinal tissues of Kat8 knockdown and dsGfp-injected shrimp was analyzed using qPCR. e Survival rate of the Kat8-RNAi shrimp infected with WSSV and injected with dsGfp, which was used as the control. f, g The expression of Vp28 (f) and Ie1 (g) in the hemocytes and intestinal tissues of MG149-treated shrimp was detected at 24 hpi via qPCR. Ef-1α was used as the endogenous control gene. h The expression level of the VP28 protein was detected after treatment with MG149 challenged with WSSV using western blotting with anti-VP28 Abs. Bottom panel: Statistical analysis of the data in panel h based on three replicates. The data are presented as the mean ± SD (n = 3). i Survival rate of MG149-treated shrimp challenged with WSSV (n = 30) and the control group injected with PBS (n = 30). The data are presented as the mean ± SD (n = 3) and significant differences were analyzed by Student’s t test. Significant differences of survival rate were analyzed using the log-rank test with GraphPad Prism 8.0.2 software.
To verify the role of KAT8 in shrimp infected with WSSV, we treated shrimp with the KAT8 inhibitor MG149 followed by analysis of WSSV replication. The results showed that WSSV replication increased significantly in the hemocytes and intestinal tissues of MG149-treated shrimp infected with WSSV (Fig. 2f, g). The VP28 protein in hemocytes and intestinal tissues was also increased in the MG149 injection group, followed by WSSV infection (Fig. 2h). The survival rate of MG149-treated shrimp infected with WSSV was significantly lower than that of control shrimp (Fig. 2i). These results suggested that KAT8 upregulated in WSSV-infected shrimp, and significantly reduced WSSV infection and increased the shrimp survival rate.
Resistance to WSSV infection is increased in UV-WSSV-trained shrimp through H3K27ac mediated by KAT8
To decipher the underlying mechanisms of trained immunity in the shrimp M. japonicus, we established a model of trained immunity using WSSV inactivated by ultraviolet light (UV-WSSV) as an inducer9 (Fig. 3a, b). Compared with that of the control shrimp, the survival rate of UV-WSSV-trained shrimp significantly increased after infection with WSSV (Fig. 3c). These results suggest that UV-WSSV induced trained immunity in shrimp.
a Diagram showing the model of trained immunity in M. japonicus induced by UV-WSSV. The sample collection times after initial training are shown. b Survival rates of shrimp injected with UV-WSSV and PBS-injected shrimp used as the controls (n = 40). c Survival rates of the shrimp in the UV-WSSV training model (UV-WSSV + WSSV) and the control (PBS + WSSV) (n = 30/group). d Abundance of H3K27ac, H4K16, and H3K9ac modifications in the hemocytes of shrimp in the UV-WSSV training model (UV-WSSV + WSSV) and the control (PBS + WSSV), as measured by western blotting. d’ Western blot bands of H3K27ac modification shown in d were digitalized on the basis of three independent data points obtained by scanning. e Expression of Kat8 at the mRNA level was detected in shrimp hemocytes in the UV-WSSV-trained immunity model via qPCR with Ef-1α as the internal control gene. f The H3K27ac mark was detected in the hemocytes of Kat8-knockdown shrimp in the trained immunity model, as determined by western blotting. Bottom panel: Statistical analysis of the data in (f), which was based on three replicates. g The expression of Kat8 and Vp28 at the mRNA level was measured by qPCR in shrimp hemocytes at 12 hpi after KAT8 knockdown, followed by UV-WSSV training. Ef-1α was used as the endogenous control gene. h Expression of VP28 at the protein level was measured by western blotting in shrimp hemocytes 12 hpi after knockdown of Kat8, followed by UV-WSSV training. Bottom panel: Statistical analysis of the data presented in (h) on the basis of three replicates after digitalization using ImageJ software. The data are presented as the mean ± SD (n = 3). Significant differences were determined by Student’s t test for pairwise comparisons and ANOVA for multiple comparisons. i Survival rates of the Kat8-knockdown shrimp in the UV-WSSV trained group (dsKat8 + UV-WSSV + WSSV) using the UV-WSSV trained group as the control (UV-WSSV + WSSV) (n = 30/group). Kat8 was firstly knocked down in shrimp using RNAi, followed by UV-WSSV training and WSSV infection. Shrimp survival rates were then determined.
To explore whether epigenetic modification was involved in UV-WSSV-induced trained immunity in shrimp, we detected variations in H3K27ac, H4K16ac, and H3K9ac marks in the hemocytes of UV-WSSV-trained shrimp using the training model and observed that only the H3K27ac level in hemocytes increased significantly 12 h after UV-WSSV training and it was restored to the basal level on the 5th d (120 h after UV-WSSV injection); after infection with WSSV, the H3K27ac level in the UV-WSSV group was significantly greater than that in the PBS control group at 3 h postinfection. This finding suggested a faster and stronger response of H3K27ac to the second stimulus with live WSSV (Fig. 3d, d’). However, there was no significant change in H4K16ac and H3K9ac in the UV-WSSV-induced training model. These results suggest that H3K27ac modification, but not H4K16ac or H3K9ac, is involved in UV-WSSV-induced trained immunity, which enhances shrimp resistance to WSSV infection.
The expression patterns of KAT8 were subsequently analyzed in UV-WSSV-trained shrimp, and the analysis revealed that Kat8 was upregulated at 12 hpi and then declined to basal levels at 120 h after training. Kat8 was then upregulated from 3 to 24 h after WSSV infection (Fig. 3e), suggesting a faster and stronger response after infection with WSSV, which is similar to the increase in H3K27ac in the training model. These results indicate that KAT8 might be involved in trained immunity.
To determine whether KAT8 contributes to antiviral trained immunity, we knocked down Kat8 expression via RNAi, and assessed H3K27ac modification and WSSV replication. RNAi-mediated Kat8 knockdown abrogated UV-WSSV-induced increase in H3K27ac levels in shrimp hemocytes (Fig. 3f). Compared to UV-WSSV-trained controls, Kat8-knockdown shrimp exhibited significantly elevated Vp28 and Ie1 mRNA expression (Fig. 3g), increased VP28 protein levels (Fig. 3h), and markedly reduced survival rates (Fig. 3i). These findings were corroborated using the KAT8 inhibitor MG149 as an independent loss-of-function approach (Supplementary Fig. 5a–c). Collectively, these results indicate that KAT8 mediated H3K27ac modification is required for maintaining antiviral trained immunity in UV-WSSV-primed shrimp.
The glycolysis is promoted in shrimp trained with UV-WSSV
Metabolic reprogramming and epigenetic modifications are the two pillars of trained immunity4. To determine whether metabolic rewiring (such as glycolysis) is involved in UV-WSSV-induced trained immunity, we first detected the consumption of glucose and the production of lactic acid (LA) in the hemocytes of UV-WSSV-trained shrimp. The results showed that the contents of glucose decreased and LA increased significantly at 12 h and 24 h post UV-WSSV training (Fig. 4a, b). Next, we analyzed the expression patterns of key genes involved in glycolysis [hexokinase2 (HK2), pyruvate kinase (PK), L-lactate dehydrogenase (LDH) and pyruvate dehydrogenase (PDH)], and the results showed that Hk2, Pk, Ldh and Pdh were upregulated in UV-WSSV-trained shrimp (Fig. 4c). We then detected the contents of glycolytic metabolite pyruvate (PA) in the hemocytes of UV-WSSV-trained shrimp. The results indicated that the contents of PA increased at 12 h post UV-WSSV training (the first stage of the training model) and increased significantly at 3 h or 6 h post WSSV infection (the 2nd stage of the training model) (Fig. 4d). To further confirm the involvement of glycolysis in the trained immunity, the UV-WSSV-trained shrimp were treated with 2-deoxy-D-glucose (2-DG), a glucose analog that competitively inhibits hexokinase in the glycolysis pathway31, the expression of Pk, Ldh and Pdh (Fig. 4e), and the contents of PA (Fig. 4f), LA (Fig. 4g) and acetyl-CoA (Fig. 4h), and H3K27ac modification were significantly decreased (Fig. 4i) in the shrimp. The results indicated that glycolysis is involved in H3K27ac modification in UV-WSSV-induced trained immunity.
a, b Contents of glucose (a) and LA (b) in hemocytes of UV-WSSV trained shrimp collected at 3 h, 6 h, 12 h and 24 h after training. c Expression of Hk2, Pk, Ldh and Pdh at the mRNA level was measured in hemocytes of shrimp at 12 h after UV-WSSV training via qPCR with Ef-1α as the endogenous control gene. d Contents of PA in hemocytes collected at 6 h and 12 h after UV-WSSV training and at 3 h and 6 h after WSSV infection in UV-WSSV-trained shrimp. e Expression of Pk, Ldh and Pdh at the mRNA level was measured via qPCR in hemocytes of 2-DG-pretreated shrimp followed by UV-WSSV training, with Ef-1α as the endogenous control gene. f–h Contents of PA (f), LA (g) and acetyl-coenzyme A (Ac-CoA) (h) in hemocytes of 2-DG-pretreated shrimp followed by UV-WSSV training. i Influence of 2-DG on H3K27ac in shrimp. UV-WSSV-trained shrimp were pretreated with 2-DG. PBS was used as a control. At 12 h after training, the enrichment of H3K27ac in hemocytes of the shrimp was examined using western blotting. Bottom panel: Statistical analysis of the data in panel i based on three replicates. j Hif-1α and Hk2 expression was detected in Hif-1α-knockdown shrimp 12 h after UV-WSSV training via qPCR. k The abundance of H3K27ac was detected in Hif-1α-knockdown shrimp 12 h after UV-WSSV training using western blotting. Bottom panel: Statistical analysis of the data in panel k based on three replicates. The data are presented as the mean ± SD (n = 3) and significant differences were analyzed by Student’s t test.
In our previous study, we reported that Hypoxia-inducible factor 1 alpha (HIF-1α) interacts with the binding site of the HK promoter and initiates the transcription of HK32. To investigate how glycolysis is upregulated in shrimp trained with UV-WSSV, we detected the expression of Hk after Hif-1α was knocked down and found a significant decrease in the Hk mRNA level in hemocytes (Fig. 4j). H3K27ac modification was reduced after Hif-1α was knocked down (Fig. 4k), indicating that HK expression was regulated by Hif-1α, a key transcription factor of the mTOR signaling pathway, thereby activating glycolysis in UV-WSSV-trained shrimp. These data demonstrated that glycolysis is involved in the trained immunity induced by UV-WSSV training in shrimp.
The TCA cycle is involved in trained immunity in shrimp induced with UV-WSSV
To determine whether the TCA cycle is involved in UV-WSSV-induced trained immunity, we first analyzed the expression patterns of key genes involved in the TCA cycle [citrate synthase (CS), isocitrate dehydrogenase (IDH), 2-oxoglutarate dehydrogenase (OGDH) and ATP-citrate synthase (ACLY)], and the results showed that Cs, Idh, Ogdh and Acly were upregulated in UV-WSSV-trained shrimp (Fig. 5a). We also detected the contents of key metabolites of TCA cycle, citric acid (CA), and acetyl-coenzyme A (Ac-CoA) in the hemocytes of UV-WSSV-trained shrimp. The results revealed that the contents of CA (Fig. 5b) and Ac-CoA (Fig. 5c) increased significantly at 6 h or 12 h post UV-WSSV training and at 3 h or 6 h post WSSV infection. To further confirm the involvement of the TCA cycle in the H3K27ac modification induced by UV-WSSV training, shrimp were pretreated with SB204990 (SB), a specific inhibitor of ACLY enzyme33. We found that the contents of Ac-CoA (Fig. 5d) and the level of H3K27ac (Fig. 5e, e’) were declined in UV-WSSV trained shrimp. Next, we knocked down the TCA cycle key enzyme Cs through RNAi and found that the expression of Ogdh and Acly (Fig. 5f), the contents of CA (Fig. 5g) and Ac-CoA (Fig. 5h) were decreased and the H3K27ac modification (Fig. 5i, i’) were significantly inhibited after Cs was successfully knocked down in UV-WSSV trained shrimp. Overall, these data indicate that TCA cycle plays important roles in the UV-WSSV-induced trained immunity by promoting H3K27ac modification via supply of the cofactor (Ac-CoA) of KAT8.
a Expression of Cs, Idh, Ogdh and Acly at the mRNA level was measured in hemocytes of shrimp at 12 h after UV-WSSV training via qPCR with Ef-1α as the endogenous control gene. b, c Contents of CA (b) and Ac-CoA (c) in hemocytes collected at 6 h and 12 h after UV-WSSV training and at 3 h and 6 h after WSSV infection in UV-WSSV-trained shrimp. d Contents of Ac-CoA in hemocytes of SB-pretreated shrimp followed by UV-WSSV training. e The abundance of H3K27ac was detected in SB-pretreated shrimp 12 h after UV-WSSV training using western blotting. e’ Statistical analysis of the data in panel e based on three replicates. f Expression of Cs, Idh, Ogdh and Acly at the mRNA level was measured in hemocytes of Cs-RNAi shrimp followed by UV-WSSV training via qPCR, with Ef-1α as the endogenous control gene. g, h Contents of CA (g) and Ac-CoA (h) in hemocytes of Cs-RNAi shrimp followed by UV-WSSV training. i H3K27ac modification in hemocytes were detected via western blotting in Cs-RNAi shrimp. i’ Statistical analysis of the data panel i based on three replicates. The data are presented as the mean ± SD (n = 3) and significant differences were analyzed using Student’s t test.
Identification of target genes regulated by H3K27ac modification
To reveal the underlying mechanisms mediated by the site-specific histone acetylation, the target genes directly regulated by H3K27ac modification were characterized through a chromatin immunoprecipitation sequencing (ChIP-seq) assay. The hemocytes from UV-WSSV- and PBS-trained shrimp were subjected to genome-wide ChIP-seq analysis. The results revealed that the H3K27ac signal intensity was greater in the transcription start site (TSS)-related region than in the control region (Fig. 6a). H3K27ac modification in the promoter region of genes increased after UV-WSSV training; most of the DNAs bound to H3K27ac were in intergenic regions, and 36.93% of the DNAs bound to H3K27ac were located within the promoter sequences in UV-WSSV-trained shrimp, while 29.96% was in the control (Fig. 6b). The promoter of 2361 genes bound to H3K27ac (Fig. 6c), indicating that the expression of these target genes was regulated by site-specific histone acetylation (H3K27ac). Gene Ontology (GO) functional analysis revealed that the enriched target genes were related to cellular components (nucleus, cytosol, cytoplasm and plasma membrane), cell migration and RNA transcription (Fig. 6d). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis indicated that the target genes regulated by H3K27ac were enriched in cellular signaling pathways and metabolic pathways (Supplementary Fig. 6a), suggesting the regulatory roles of histone acetylation in trained immunity. A comparison of the gene expression of UV-WSSV trained- and control shrimp revealed that 2305 genes were upregulated in UV-WSSV-trained shrimp. To confirm the genes regulated by H3K27ac modification in UV-WSSV trained shrimp, integrative Genomics Viewer (IGV) tracks and ChIP-qPCR analysis were performed. The results revealed significant enrichment of H3K27ac signal peaks around the transcription start sites (TSSs) of casein kinase I (Ckl), eukaryotic translation initiation factor 4E (Eif4E), Ras-related protein 21 (Rab21), NF-κB signaling pathway related genes [ubiquitin-conjugating enzyme E2 A (UbE2A), inhibitor of nuclear factor kappa-B kinase (IKKβ) and transcription factor p65 (Dorsal)] in UV-WSSV-trained shrimp, and a significant difference in the H3K27ac peak was detected between the UV-WSSV-trained and PBS groups in IGV track analysis (Supplementary Fig. 6b–g). Same results were obtained in ChIP-qPCR assay (Fig. 6e–j). The results suggest that the above genes are the target genes of H3K27ac modification.
a Heatmap of genome-wide enrichment analysis of DNAs bound to H3K27ac in PBS- and UV-WSSV-trained shrimp. TSS, transcription start site; TTS, transcription end site. b Genome-wide distribution of the DNAs bound to H3K27ac in UV-WSSV-trained shrimp (with PBS training as the control). c Venn diagrams showing the number of target genes whose promotors were bound to H3k27ac. d GO analysis of the target genes regulated by H3K27ac. e–j Effects of UV-WSSV training on the expression of Ckl (e), Eif4E (f), Rab21 (g), UbE2A (h), Ikkβ (i) and Dorsal (j) analyzed by ChIP-qPCR. All bar chart data are shown as mean ± SD of three replicates (n = 3). p-value was determined by Student’s t test.
KAT8-mediated H3K27ac suppresses WSSV replication by enhancing expression of NF-κB-like transcription factor Dorsal in UV-WSSV trained shrimp
Humoral immunity is important for the resistance of invertebrates to pathogen infection and is achieved by a range of antiviral genes and antimicrobial peptides (AMPs) which mainly regulated by NF-κB signaling34,35,36,37. To investigate whether genes related to the NF-κB signaling pathway are regulated by the H3K27ac modification mediated by KAT8, we performed ChIP-qPCR assays using H3K27ac antibody after knocked down Kat8 via RNAi, and found that the enrichments of H3K27ac in the promoter region of UbE2A, Ikkβ and Dorsal were significantly reduced in hemocytes of UV-WSSV trained shrimp (Fig. 7a–c).
a–c ChIP‒qPCR was used to detect enrichment of the DNA fragments in the promoter regions of UbE2A (a), Ikkβ (b) and Dorsal (c) using the DNA templates obtained by ChIP with the H3K27ac antibody after the knockdown of Kat8. IgG was used as a control. d Expression of UbE2A, Ikkβ, Dorsal, Relish, Vago5, Ficolin and Amps mRNAs in hemocytes of Kat8-RNAi shrimp infected with WSSV analyzed by qPCR at 24 hpi, with Ef-1α as the endogenous control gene. e–h The expression profiles of Dorsal (e), Vago5 (f), Ficolin (g) and CrusI-5 (h) in the hemocytes of UV-WSSV-trained shrimp were analyzed via qPCR with Ef-1α as the endogenous control gene. i Expression of Dorsal, Vago5, Ficolin and Amps at the mRNA level was measured in hemocytes of MG149-pretreated shrimp followed by UV-WSSV training via qPCR, with Ef-1α as the endogenous control gene. The assays were repeated three times, and the data are expressed as the mean value ± SD (n = 3). Significant differences were analyzed using Student’s t test.
To further validate the above findings, we examined the expression of NF-κB-like signaling components (UbE2A and Ikkβ), transcription factors (Dorsal and Relish), and Dorsal/Relish-regulated effector genes (Vago5, Ficolin, Amps) in Kat8-silenced shrimp. We found that the expression levels of UbE2A, Ikkβ, Dorsal, Vago5, Ficolin, and Amps, including Anti-lipopolysaccharide factor B1 (Alf-B1), Alf-C2, Alf-D2, Alf-E2, CrusI-1, CrusI-3, and CrusI-5, decreased significantly in the Kat8-silenced shrimp compared with those in the control group. However, there was no difference in the expression of Relish or Alf-C1 (Fig. 7d). Thus, Dorsal, the NF-κB-like transcription factor, was further studied. After knockdown of Dorsal by RNAi in shrimp, the expression of Vago5 and the effectors Ficolin, Amps including Alf-C1, Alf-D2, Alf-E2, CrusI-1, CrusI-3, and CrusI-5, was significantly reduced (Supplementary Fig. 7a, b). To verify whether Dorsal as well as Vago5, Ficolin and AMPs are involved in shrimp trained immunity, we detected the expression patterns of Dorsal, Vago5, Ficolin and Amps in the UV-WSSV trained model and found that Dorsal, Vago5, Ficolin, CrusI-5 (Fig. 7e–h), and Alf-D2, Alf-E2, CrusI-1, and CrusI-3 (Supplementary Fig. 7c–f) exhibited faster and stronger immune responses in the trained immunity of shrimp. After pretreatment with the MG149 inhibitor, the increased expression of Dorsal, Vago5, Ficolin and Amps was inhibited in the training model (Fig. 7i). These results suggest that KAT8-mediated H3K27ac participates antiviral trained immunity by regulating the Dorsal–Vago5–Ficolin and Dorsal–AMPs axes against WSSV in UV-WSSV trained shrimp.
KAT8-mediated H3K27ac promotes the expression of key enzymes Hk2, Pk and Ldh in glycolysis
In the ChIP-seq analysis, significant enrichment of H3K27ac signal peaks was detected around the TSSs of Hk2, Pk, Ldh, Pdh, Cs, Idh and Ogdh, and the peaks of Hk2, Pk and Ldh were significantly increased in UV-WSSV-trained shrimp (Fig. 8a–c). To confirm ChIP-seq results, a ChIP-qPCR assay was performed using H3K27ac antibody after Kat8-RNAi. The results indicated that the expression of Hk2, Pk and Ldh was upregulated in UV-WSSV-trained shrimp (Fig. 8d–f), but decreased following KAT8 knockdown (Fig. 8g–i). Thus, glycolysis key genes, Hk2, Pk and Ldh are also target genes of H3K27ac, suggesting a positive regulatory loop for the expression of the key enzyme-encoding genes in metabolic pathways in trained immunity. The expression of key enzymes involved in glycolysis was positively regulated in a feedforward manner in shrimp trained immunity.
a–c Integrative Genomics Viewer (IGV) tracks showing the H3K27ac modification peak in the TSSs of Hk2 (a), Pk (b) and Ldh (c) in UV-WSSV-trained shrimp (with PBS as the control). The arrows indicate the peak regions of H3K27ac around the TSS and the enrichment at the target gene promoters. The number represents the peak height. d–f Effects of UV-WSSV training on the expression of Hk2 (d), Pk (e) and Ldh (f) analyzed by ChIP-qPCR. g–i Effects of Kat8-RNAi on the expression of Hk2 (g), Pk (h) and Ldh (i) analyzed by ChIP-qPCR. The data are presented as the mean ± SD (n = 3). Statistical significance was analyzed by Student’s t test for pairwise comparisons, and p < 0.05 was considered a significant difference.
Taken together, in UV-WSSV-induced trained immunity, glycolysis and the TCA cycle are activated and provide Ac-CoA to increase KAT8 mediated H3K27ac modification, which is deposited at the promoter regions of Dorsal and glycolysis enzymes to enhance gene transcription and metabolic pathways. As a transcription factor, Dorsal promotes the expression of Vago5 and Amps and inhibits the replication of WSSV in shrimp (Fig. 9).
(Left) In UV-WSSV-trained shrimp (the 1st stage of trained immunity), the activation of the glycolysis and TCA pathways provides energy and a cofactor for H3K27ac modification of KAT8, and epigenetic modifications are deposited at the Dorsal promoter region to increase Dorsal transcription and promote the expression of Vago5 and AMPs in shrimp. H3K27ac is also deposited at the Hk2, Pk, and Ldh promoter regions to increase glycolysis in a feedforward manner. (Right) At the 2nd stage of the trained immunity, H3K27ac mediated by KAT8 enhances in shrimp infected with WSSV, and promotes transcription of Dorsal to positively regulate expression of antiviral effectors against WSSV proliferation. HK Hexokinase, PK Pyruvate kinase, LDH L-lactate dehydrogenase, PDH Pyruvate dehydrogenase, ACLY ATP-citrate synthase.
Discussion
In the present study, using a model of trained immunity in shrimp induced by UV-WSSV, we demonstrated that UV-WSSV-induced trained immunity in shrimp resulted in a faster and stronger immune response against WSSV infection. UV-WSSV training in shrimp enhanced the glycolysis and TCA cycle metabolic pathways and increased Ac-CoA concentrations. Acting as a cofactor of the enzyme, Ac-CoA promoted the abundance of KAT8 mediated H3K27ac, which was deposited in the promoter region of Dorsal and increased the transcription of Dorsal to further upregulate the expression of the interferon-like cytokine Vago5 and AMP against WSSV infection in shrimp. Moreover, the key enzymes of glycolysis HK2, PK and LDH were also positively regulated by the H3K27ac modification, and further promoted the metabolic pathway in a feedforward manner. This is the first systematic study on the mechanism of invertebrate trained immunity.
The acetylation of histones (such as H3K27ac, H3K9ac, and H4K16ac) is associated with active transcriptional activity38,39,40,41,42. KATs (also known as histone acetyltransferases, HATs) and histone deacetylases (HDACs) dynamically regulate these modifications18. H3K27ac modification is associated with enhanced immune responses in β-glucan-trained murine macrophages43, but which KAT responsible for the modification is unclear. We performed RNAi to identify which the KATs and found that H3K27ac and H4K16ac modifications were significantly reduced after the knockdown of Kat8 and MG149 treatment. KAT8 (MOF or MYST1) is a member of the multiprotein complex primarily responsible for H4K16 acetylation, and it also acetylates nonhistone substrates such as p53 and IRF320,21. A recombinant human MOF also has histone acetyltransferase activity directed toward histones H2A, H3, and H444, but the specific acetylation sites of histones H2A and H3 are unknown. In the present study, we found that Kat8 expression was regulated by FOXO in shrimp infected with WSSV. KAT8 was distributed in the cytosol and nucleus and significantly increased in the nuclei of hemocytes and intestinal cells of shrimp challenged with WSSV. Knockdown of the KAT8 gene by RNAi reduced H3K27ac and H4K16ac modifications and promoted WSSV replication in the hemocytes and intestinal tissues of shrimp challenged with WSSV. KAT8 inhibitor MG149 injection produced the same results. However, in our trained immunity model, although the transcription level of Kat8 in hemocytes was significantly increased and the survival rate of UV-WSSV-trained shrimp was increased, the levels of H3K9ac and H4K16ac were not increased. These results suggest that UV-WSSV-induced trained immunity enhances H3K27ac modification and results in resistance to WSSV infection in shrimp. These results indicate that H3K27ac, mediated by KAT8, plays an important role in maintaining antiviral trained immunity in shrimp. Here, we also revealed that KAT8 is responsible for specific site modifications and that H3K27 acetylation and a feedforward loop for metabolic pathways exist in trained immunity.
KAT8 is a catalytic subunit of two independent protein complexes: MSL (male-specific lethal) and NSL (non-specific lethal), and plays a critical role in epigenetic regulation through the two complexes. It is generally believed that both MSL and NSL exert their activity via H4K16ac modification22. In our study, we found although KAT8 catalyzes H4K16ac and H3K27ac modifications, only the latter plays an important role in the trained immunity induced by UV-WSSV. Whether the modification is the complex-dependent need further study. To elucidate the antiviral mechanism and target genes of H3K27ac modification in UV-WSSV-trained shrimp, a genome-wide ChIP‑seq method was used. The results revealed that the H3K27ac peaks in the promoter regions of 2305 genes significantly increased, while 3170 genes significantly decreased in shrimp trained with UV-WSSV. Surprisingly, we found that an NF-κB signaling pathway related genes UbE2A, Ikkβ and Dorsal were the target genes of the H3K27ac modification. Kat8-RNAi and ChIP‒qPCR with H3K27ac antibody confirmed these results. Humoral immunity is important in invertebrate resistance to pathogen infection and is achieved by a series of AMPs and antiviral genes34,45,46. Three main families of AMPs, i.e., penaeidins, crustins, and ALFs, have been identified in shrimp. These AMPs provide resistance to bacteria, fungi, and viruses37,47. AMP expression is regulated mainly by the NF-κB (Toll/Drosal and IMD/Relish) signaling pathways in Drosophila48. In this study, we detected the expression of NF-κB-like transcription factors, including Dorsal and Relish, and related interferon-like Vago5, as well as the downstream antiviral factor Ficolin, and the expression of related Amps in UV-WSSV-trained shrimp. The results revealed that UV-WSSV training promoted the expression of Dorsal and downstream Vago5, Ficolin, and associated Amps, including Alf-D2, Alf-E2, CrusI-1, CrusI-3, and CrusI-5, in the hemocytes of shrimp. Thus, KAT8 directly activates Dorsal expression by modifying the level of H3K27ac in the Dorsal promoter region, which subsequently promotes the expression of downstream Vago5, Ficolin and Amps to exert antiviral effects. Vago5, an interferon-like cytokine identified in shrimp, activates the JAK/STAT signaling pathway and induces the effector Ficolin through integrin to resist WSSV replication34. The faster and stronger expression of Dorsal associated with Vago5 and Amps in the trained immunity of shrimp is responsible for the significantly higher survival rate of UV-WSSV-trained shrimp than of PBS-trained shrimp.
Metabolic reprogramming is also a key step in regulating trained immunity49. To confirm the results, the metabolites and related enzymes in metabolic pathways were analyzed. We found that after training with UV-WSSV, the consumption of glucose and the production of LA significantly increased, indicating an enhanced glycolysis in shrimp. Subsequently, we detected the expression patterns of key genes involved in glycolysis, including Hk2, Pk, Ldh and Pdh, and found that the expression of the key genes of glycolysis was upregulated in UV-WSSV-trained shrimp. We subsequently observed that the key transcription factor Hif-1α of the mTOR signaling pathway enhances glycolysis by regulating the expression of Hk2 in UV-WSSV-trained shrimp. We also found that expression of genes encoding key enzymes in the TCA cycle were significantly increased after UV-WSSV training, and knocking down the TCA cycle key enzyme gene Cs significantly reduced the content of key enzymes Ogdh and Acly, and CA and Ac-CoA production as well as H3K27ac modification. Therefore, the TCA cycle is involved in UV-WSSV induced trained immunity. In the ChIP-seq analysis, genes encoding key enzymes involved in glycolysis (HK2, PK, LDH and PDH) and the TCA cycle (CS, IDH and OGDH) were identified as target genes of H3K27ac. In addition, Hk2, Pk and Ldh showed significant enrichment of H3K27ac signal peaks after UV-WSSV training. All in all, the enhancement of the glycolysis and TCA cycle metabolic pathways provides the acetyl group for H3K27ac modification and the energy for antiviral trained immunity.
Trained immunity mechanisms appear to have been conserved throughout the evolutionary process50. Upon stimulation, the activation of gene transcription is accompanied by several metabolic pathways and epigenetic changes in histone epigenetic modifications at the promoters and enhancers of proinflammatory genes14. However, different stimuli can activate different trained immunity programs. Here, we found that UV-WSSV training promoted the glycolysis and TCA pathways and H3K27ac modification, ultimately promoted the expression of antiviral effectors, which is somewhat similar to β-glucan-induced trained immunity in monocytes/macrophages of mammals. It is reported that the PRRs involved in the initiation of trained immunity in mammals include dectin-1 for β-glucan, nucleotide-binding oligomerization domain 2 (NOD2) for Bacille Calmette-Guerin (BCG) and TLR4 for LPS51. However, the corresponding molecules of dectin-1 and NOD2 were not identified in shrimp. There might be different PRRs in invertebrates that induce trained immunity.
Previous research demonstrated that M. japonicus primed with formalin-inactivated WSSV exhibited significantly enhanced resistance when challenged at 10 days post-vaccination, though this protection was lost by 30 days post-priming8. In contrast, shrimp trained with low-dose live WSSV and challenged with a higher viral dose from 1 week to 3 months post-exposure showed significantly elevated survival rates during re-challenges conducted between 3 weeks and 2 months post-training. However, this protective effect dissipated by 3 months52. A comprehensive review analyzing immunological memory in cultured decapods following administration of live, inactivated, and subunit vaccines revealed that protective effects persist for 7–30 days in shrimp and crayfish5. Collectively, these studies document immunological memory phenotypes, whereas our findings establish the mechanism of trained immunity in invertebrates.
In conclusion, metabolic rewiring (activation of glycolysis and the TCA cycle) and epigenetic reprogramming (H3K27ac modification) are involved in UV-WSSV-induced antiviral trained immunity in shrimp (Fig. 9). This study provides evidence for the conservation of mechanisms of trained immunity in mammals and invertebrates. The results of this study reveal the mechanism of training immunity in shrimp and provide a basis for the development of antiviral vaccines for disease prevention and control in shrimp aquaculture.
Limitations of the study
In the process of trained immunity, inducers (such as pathogens and pathogen-associated molecular patterns) are generally recognized by PRRs and initiate a series of intracellular cascades that trigger the upregulation of metabolic pathways. The metabolites obtained from these pathways play crucial roles in the modulation of enzymes involved in epigenetic rewiring, which leads to an increase in transcriptional activation-associated marks (such as H3K27ac and H3K4me3), causing the transcription of proinflammatory genes against pathogens. In this study, we revealed that metabolic pathways and epigenetic modifications (H3K27ac) are involved in trained immunity and induce the transcription of NF-κB-like transcription factor to regulate antiviral effectors expression, but the PRRs of inactivated WSSV in trained immunity need to be identified.
Methods
Animals and virus
Healthy shrimp (M. japonicus, approximately 10 g each) obtained from the Aquatic Product Market of Qingdao, Shandong Province, China, were used in this study. The shrimp were raised in seawater. Prior to experimentation, M. japonicus were acclimated for more than 1 d in a temperature-controlled recirculating aquaculture system (RAS) under the following conditions: water temperature 20–22 °C, salinity 35‰ (Natural seawater), dissolved oxygen ≥5 mg/L (maintained via air stones). Shrimp were fed twice daily with commercial feed at 3%–5% of body weight. WSSV suspensions were prepared in sterile phosphate-buffered saline (PBS). Shrimp were artificially infected via intramuscular injection of purified WSSV35.
Immune challenge and sample collection
Healthy shrimp were intramuscularly injected at the third abdominal segment, with approximately 1.0 × 108 copies of WSSV. PBS-injected shrimp were used as controls. Shrimp hemolymph was collected from the ventral sinus into cold anticoagulant (0.45 M NaCl, 10 mM KCl, 10 mM EDTA, and 10 mM HEPES [pH 7.45]) with a syringe, followed by centrifugation at 800 × g for 8 min at 4 °C to obtain the hemocyte precipitate. Other shrimp tissues (heart, hepatopancreas, gills, stomach, and intestine) were collected simultaneously for the following experiments. The collected tissues were either used immediately or snap-frozen in liquid nitrogen and stored in an ultra-low temperature freezer (−80 °C) for further study.
Establishment of the trained immunity model
WSSV was inactivated by exposure to ultraviolet light for 30 min, and the shrimp trained immunity model was established9. The shrimp were divided into two groups. Each shrimp in the experimental group was injected with 50 μl of UV-WSSV (approximately 1.0 × 105 copies), and the shrimp in the control group were injected with 50 µL of PBS. Both groups of shrimp were injected with WSSV (1.0 × 107 copies in 50 µL PBS) 5 days after the first injection. Then, shrimp hemocytes and other tissues were collected at different time points for analysis.
RNA extraction and cDNA synthesis
Total RNA was extracted from hemocytes or ∼100 mg of tissue using TRIzol Reagent (Bioteke, Beijing, China), and the first strand of cDNA was synthesized via a cDNA Synthesis Kit (5x All-in-One RT MasterMix; Applied Biological Materials-abm, Vancouver, Canada) with 2 μg of total RNA according to the manufacturer’s instructions. Prior to reverse transcription, RNA purity was assessed by UV Spectrophotometry (A260/A280 ratio) and integrity was verified via agarose gel electrophoresis, and genomic DNA contamination was eliminated using DNase I.
Identification and analysis of lysine acetyltransferases (KATs) in shrimp
Shrimp KAT sequences in M. japonicus were obtained from both our transcriptome data and publicly available genome data. The transcriptome data were generated by MeRIP-seq using the following procedure: Gills were collected from the shrimp at 24 h post-injection with PBS or UV-WSSV. The samples were flash-frozen in liquid nitrogen and sent to Oebiotech (Shanghai, China) for sequencing. Total RNA was extracted using Trizol™ (Invitrogen, USA). Polyadenylated mRNA was then enriched from the total RNA using the Ambion™ Dynabeads™ mRNA Purification Kit. The enriched mRNA was fragmented and used as a template for cDNA synthesis. Sequencing was performed according to the manufacturer’s protocol on the Illumina NovaSeq™ 6000 platform (Lc-Bio Technologies Co., Ltd., Hangzhou, China). The Input control generated through MeRIP-seq for transcriptome dataset; which has been deposited in NCBI under accession number PRJNA1294137). Human KAT sequences (KAT2a, KAT7, KAT8, CBP/P300, MYST3, Tip60, ATAT, ESCO2 and HAT1) obtained from NCBI were used as query sequences for BLAST analyses. These analyses were performed against our local transcriptome dataset using local BLAST, and against the publicly available Penaeus japonicus genome sequence (https://blast.ncbi.nlm.nih.gov/Blast.cgi) using BLAST. All identified M. japonicus KAT sequences are provided in Supplementary Data 3. Sequence alignments and phylogenetic trees of KATs from different species were constructed using MEGA 7.0 software (http://www.megasoftware.net/).
The sequence of open reading frame (ORF) of Kat8 was amplified via a pair of primers (Kat8-EX-F and Kat8-EX-R; Supplementary Table 1) for sequence confirmation. The corresponding cDNA was subsequently translated to obtain the protein sequence using the ExPASy translation tool (https://web.expasy.org/translate/). The molecular mass and isoelectric point of the peptide were determined using the online tool ExPASy (http://web.expasy.org/compute_pi/). The domain architecture prediction was performed with SMART.
Tissue distribution and expression profile analysis
Semiquantitative RT‒PCR was performed to study the tissue distribution of Kat8 mRNAs via the specific primers listed in Supplementary Table 1 (Kat8-RT-F and Kat8-RT-R). β-actin was used as the control with primers (β-actin-RT-F and β-actin-RT-R) (Supplementary Table 1). RT-PCR amplifications were performed in 50 μL reaction volumes using TransStart FastPfu DNA Polymerase (TransGen Biotech, Beijing, China) Each reaction contained: 1 μL cDNA template (synthesized from 200 ng RNA), 10 μL of 5 × TransStart FastPfu Buffer, 4 μL of 2.5 mM dNTPs (final concentration 0.2 mM), 1 μL of each 10 μM primer (final concentration 0.2 μM each), 1 μL of FastPfu DNA Polymerase (2.5 units), and nuclease-free water to a final volume of 50 μL. Thermal cycling conditions were as follows: 94 °C for 3 min; 35 cycles of 94 °C for 30 s, 54 °C for 30 s, and 72 °C for 30 s; final extension at 72 °C for 10 min; followed by a hold at 4 °C. PCR products were analyzed using 1.2% agarose gel electrophoresis.
Quantitative real-time RT‒PCR (qRT‒PCR, qPCR) analysis was performed in triplicate using SYBR Green PCR Master Mix (TaKaRa, Dalian, China) on a fluorescence quantitative PCR instrument (Analytik Jena, Germany) with gene-specific primers listed in Supplementary Table 1. qPCR amplifications were carried out in 10 μL reactions containing 1 μL cDNA template (synthesized from 100 ng RNA), 3.6 μL RNase-Free Water, 0.2 μL of each primer, and 5 μL qPCR SuperMix. Thermal cycling conditions were as follows: 95 °C for 10 min; 40 cycles of 95 °C for 10 s, 60 °C for 50 s, and 75 °C for 2 s, followed by a melting curve analysis from 68 °C to 95 °C with continuous fluorescence monitoring. PCR data were analyzed using the 2−ΔΔCt method, and Ef-1α mRNA expression levels were used as internal references. Each experiment was performed in triplicate and analyzed using Student’s t test or one-way ANOVA.
RNA interference assay
To analyze KAT8 function, we performed RNAi assays. The Kat8 fragments were amplified with primers (Kat8-RI-F and Kat8-RI-R) linked to the T7 promoter (Supplementary Table 1). The double-stranded RNA fragments were synthesized using an in vitro T7 Transcription Kit (EP0111, Thermo Fisher Scientific, Waltham, MA, USA) with the DNA template. The dsRNA was synthesized in a 50 μL reaction mixture containing: 1 μg DNA template, 2.5 μL of each 25 mM NTP, 4 μL T7 RNA polymerase (50 U/μL), 2 μL RNase inhibitor (20 U/μL), 20 μL 5 × Transcription Buffer, and DEPC-treated water to a final volume of 50 μL. The reaction mixture was incubated at 37 °C for 5 h or overnight. The dsRNA products were purified by phenol-chloroform extraction prior to shrimp injection. Shrimp were then administered 50 μg of dsRNA via microsyringe injection into the abdominal hemocoel, followed by an identical second injection 24 h later. The control group was injected with an equal amount of control dsRNA. The RNA interference efficiency was examined via qPCR or western blotting at 24 h after the second injection. To test the function of KAT8 in virus infection, WSSV infection (1.0 × 108 copies per shrimp) was performed at 24 h after the second dsRNA injection. The same method was used for RNAi of Foxo, Dorsal, Hif-1α and Cs. The expression levels of associated genes were then analyzed via qPCR with the primers listed in Supplementary Table 1. The qPCR conditions for Foxo, Dorsal, Hif-1α, and Cs were identical to those detailed in “Tissue distribution and expression profile analysis” section.
Recombinant expression and antibody preparation
For the recombinant expression of KAT8, a pair of primers (KAT8-EX-F and KAT8-EX-R) (Supplementary Table 1) was designed to amplify a fragment from the open reading frame (ORF) of KAT8. The PCR products were purified using a quick DNA purification kit (Sangon Biotech, Shanghai, China). The purified fragments were ligated into the pET-30a vector via a ClonExpress II One Step Cloning Kit (C112-01, Vazyme, Nanjing, China). The recombinant plasmids were subsequently transformed into E. coli Rosetta (DE3) cells. The expression of recombinant KAT8 was induced using 0.5 mM isopropyl-β-D-thiogalactopyranoside at 28 °C for 8 h. The recombinant protein was then purified53 and used for antiserum preparation54.
Protein extraction, SDS‒PAGE, and western blotting
Proteins from hemocytes and other organs were extracted with radioimmunoprecipitation assay (RIPA) lysis buffer (R0020, Solarbio, Beijing, China) supplemented with 20 nM phenylmethylsul fonylfluoride (PMSF) and a protease inhibitor cocktail (RM02916, ABclonal, Wuhan, China). The histones were extracted from hemocytes and the intestine according to the manufacturer’s protocol (BB31171, BestBio, Shanghai, China)55.
The homogenized samples were centrifuged at 12,000 × g for 10 min at 4 °C, and the supernatants were obtained. Protein extracts were mixed with protein loading buffer (5X) for sodium dodecyl phosphate polyacrylamide gel electrophoresis (SDS‒PAGE) and heated at 100 °C for 10 min. Then, 30 μg of protein sample was separated by SDS‒PAGE. The proteins in the SDS‒PAGE gels were transferred to a nitrocellulose (NC) membrane using transfer buffer (25 mM Tris–HCl, 20 mM glycine, 0.037% mM SDS, and 20% ethanol) for 1 h. The NC membranes were incubated in 5% skim milk in TBST (150 mM NaCl, 10 mM Tris-HCl, pH 8.0) for 2 h. Then, they were incubated with the indicated primary antiserum: Anti-KAT8 with 1:100 dilution, anti-ACTB with 1:250 dilution, and anti-VP28 with 1:250 dilution, prepared in our laboratory; anti-histone-H3 polyclonal antibody (17168-1-AP, proteintech, Wuhan, China, 1:5000), anti-acetyl lysine antibody (ICP0380, Immunechem, 1:500), anti-acetyl-histone H3-K27 rabbit pAb (A7253, ABclonal, Wuhan, China, 1:5000), anti-acetyl-histone H3-K9 rabbit pAb (A7255, ABclonal, Wuhan, China, 1:5000), and anti-acetyl-histone H4-K16 rabbit pAb (PTM-122, PTM BIO, Hangzhou, China, 1:1000) and shaken gently overnight at 4 °C. After they were washed three times with TBST, the membranes were incubated with the secondary Ab (ZB2308 ZSGB-Bio, Beijing, China, 1:5,000) for 1 h at room temperature. The protein bands were visualized with enhanced chemiluminescence (ECL) (Tanon, Shanghai, China), using a chemiluminescence instrument (Tanon, Shanghai, China).
Inhibitor assay
The KAT8 inhibitor MG149 (HY-15887) was purchased from MedChemExpress (Monmouth Junction, NJ, USA) and was dissolved in PBS. MG149 was injected in vivo at different concentrations in a volume of 50 μl (0, 1, 5, and 10 μM). The same volume of PBS was used as the control. Shrimp were first treated with MG149 for 6 h. Subsequently, the shrimp were stimulated with 1.0 × 108 copies WSSV for 24 h. Hemocytes and intestinal samples were collected and detected via qPCR and western blotting.
To suppress glycolysis, the shrimp were intramuscularly injected with 100 μl of 36 mM 2-deoxy-D-glucose (2-DG) (M5140, AbMole, Houston, USA) for 24 h; to inhibit ACLY and thus Ac-CoA production and H3K27 acetylation, we treated shrimp with SB (TargetMol, Boston, USA) at a concentration of 100 M for 48 h, and the hemocytes of the shrimp were collected 12 h after UV-WSSV training for detection. PBS was used as a control.
Immunocytochemistry assay
To examine the subcellular localization of KAT8 in the hemocytes of shrimp after WSSV infection, we performed immunocytochemical assays34. Shrimp were injected with WSSV (1.0 × 108 copies), with PBS injected as the control. At 24 h after injection, the hemocytes were collected and analyzed.
Chromatin immunoprecipitation (ChIP)-seq
ChIP assays were performed by Shandong Xiuyue Biotechnology Co., Ltd., according to the standard crosslinking ChIP protocol with modifications56. Briefly, hemocytes from UV-WSSV-trained shrimp were harvested and crosslinked with 1% formaldehyde for 10 min at room temperature, and PBS-trained shrimp were used as controls. After sonication, immunoprecipitation was performed with an anti-H3K27ac antibody (8173, Cell Signaling Technology, USA). The immunoprecipitated complex was washed, and the DNA was extracted and purified with a MinElute PCR Purification Kit (QIAGEN, #28006). The ChIP-Seq library was prepared using the TruePrep DNA Library Prep Kit V2 for Illumina (Vazyme, #TD501) according to the manufacturer’s instructions. For ChIP-seq, extracted DNA was ligated to specific adaptors, followed by deep sequencing on the Illumina NovaSeq 6000 platform via 150 bp paired ends.
ChIP-seq data analysis
Raw data (raw reads) in fastq format were first processed through in-house Perl scripts. In this step, clean reads were obtained by removing reads containing adapters, reads containing poly-N sequences and low-quality reads from the raw data. Moreover, the Q20, Q30 and GC contents of the clean data were calculated. All of the downstream analyses were based on high-quality, clean data. Clean reads were mapped to the reference genome using Bowtie2 software. Macs2 was used to call peaks with P < 0.05.
We analyzed the differentially accessible peaks in 3 steps. First, the peak files of each sample were merged via bedtools software. Second, the counts of the reads across the bed were determined for each sample using bedtools multicov. Finally, differentially accessible peaks were assessed via DESeq2. Regions were considered differentially accessible at P < 0.05.
The HOMER findMotifsGenome.pl tool was used for motif analysis. The input files were the peak file and the genome fasta file. The DNA sequence was extracted according to the peak file, and the sequence was compared with the Motif database to obtain the motifs. Peaks were annotated via the ChIPseeker package.
GO and KEGG enrichment analysis
Gene Ontology (GO) analysis was performed to elucidate the biological implications of unique genes in the significant or representative profiles of the genes in the experiment57. We downloaded the GO annotations from NCBI, UniProt (http://www.uniprot.org/) and Gene Ontology (http://www.geneontology.org/). Fisher’s exact test was applied to identify the significant GO categories, and FDR was used to correct the p-values.
Pathway analysis was used to determine the significant pathways associated with the genes according to the KEGG database. We used Fisher’s exact test to select the significant pathway, and the threshold of significance was defined by the p-value and FDR58.
ChIP‒qPCR assay
A ChIP assay was performed using a ChIP Assay Kit (Beyotime Biotechnology, Wuhan, China) according to the manufacturer’s instructions. Shrimp were trained with UV-WSSV (1.0 × 105 copies), and PBS was used as the control. The hemocytes were collected 12 h post-UV-WSSV training and pooled for ChIP analysis. qPCR assays were used to detect enrichment of the DNA fragments in the Dorsal promoter sequence using DNA obtained from ChIP with the anti-H3K27ac antibody via Dorsal promoter region primers (Supplementary Table 1). IgG antibody was used as a control. The ChIP-qPCR assays for Ckl, Eif4E, Rab21, UbE2A, Iκκβ, Hk2, Pk and Ldh were performed with same method.
Survival rate analysis
The survival rates of Kat8-RNAi- and MG149-treated shrimp challenged with WSSV were determined. Shrimp were randomly divided into two groups (30 shrimp/group) with three replicates: the experimental group, which was injected with dsKat8/MG149, and the control group, which was injected with dsGfp/PBS. At 24 h after the injection of dsKat8/dsGfp or at 6 h after the injection of MG149/PBS, the shrimp were injected with WSSV (1.0 × 107) for virus infection. The number of dead shrimp was counted every 12 h for 5 d or 6 d. Survival rates were analyzed by Kaplan‒Meier methodology using GraphPad Prism 8.0 software (San Diego, CA, USA).
Measurement of glucose, PA, LA, CA, and acetyl-CoA contents
To detect the contents of metabolites involved in glycolysis and the TCA cycle in UV-WSSV-trained shrimp, hemocytes were collected at different time points. The intracellular contents of glucose, PA, LA, CA, and acetyl-CoA were quantified via the Glucose Content Assay Kit (BC2500, Solarbio), PA Content Assay Kit (BC2200, Solarbio), LA Content Assay Kit (BC2230, Solarbio), CA Content Assay Kit (BC2155, Solarbio), and A-CoA (Acetyl Coenzyme A) ELISA Kit (E-EL-0125, Elabscience, Wuhan, China), respectively, according to the manufacturer’s instructions, with a microplate spectrophotometer at 505, 520, 575, 545, and 450 nm, respectively.
Statistics and reproducibility
Statistical analyses were performed using GraphPad Prism 8.0 software. Statistical significance was analyzed by Student’s t test for pairwise comparisons or one-way ANOVA for multiple comparisons. The results are presented as the mean ± standard deviation (SD) from three replicates, and a p-value less than 0.05 was considered statistically significant (ns, not significant). The western blotting bands were digitalized on the basis of three independent replicates with ImageJ software (National Institutes of Health, https://imagej.nih.gov/ij/download.html). All in vivo experiments included three independent biological replicates (n = 3 shrimp per replicate group) and technical replicates. For the survival assay, the survival curves are shown as Kaplan‒Meier plots with the log-rank test.
Ethics statement
All animal experiments were performed in accordance with protocols approved by the Institutional Animal Care & Welfare Committee at the School of Life Sciences of Shandong University (SYDWLL-2021-53).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All of the data generated or analyzed during this study are included in this published article and its supplementary information files. The numerical source data for graphs and charts are available in Supplementary Data 1. ChIP-seq data presented in the study were deposited in NCBI GEO under the accession number GSE273689. The transcriptome data were deposited in NCBI SRA with accession number PRJNA1294137. All ChIP-Seq datasets generated for this study have been processed in detail and are provided as Supplementary Data 2. Uncropped and unedited blot/gel images (Supplementary Figs. 8–29) are provided in the Supplementary Information. Nine KAT sequences are provided as Supplementary Data 3. All other data are available from the corresponding author on reasonable request.
References
Netea, M. G., Quintin, J. & van der Meer, J. W. Trained immunity: a memory for innate host defense. Cell. Host. Microbe 9, 355–361 (2011).
Netea, M. G. et al. Trained immunity: A program of innate immune memory in health and disease. Science 352, aaf1098 (2016).
Dominguez-Andres, J., Joosten, L. A. & Netea, M. G. Induction of innate immune memory: the role of cellular metabolism. Curr. Opin. Immunol. 56, 10–16 (2019).
Netea, M. G. et al. Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 20, 375–388 (2020).
Chang, Y. H., Kumar, R., Ng, T. H. & Wang, H. C. What vaccination studies tell us about immunological memory within the innate immune system of cultured shrimp and crayfish. Dev. Comp. Immunol. 80, 53–66 (2018).
Pham, L. N., Dionne, M. S., Shirasu-Hiza, M. & Schneider, D. S. A specific primed immune response in Drosophila is dependent on phagocytes. PLos. Pathog. 3, 8059–8072 (2007).
Sudheer, N. S. et al. Expression profile of bio-defense genes in Penaeus monodon gills in response to formalin inactivated white spot syndrome virus vaccine. Antivir. Res. 117, 60–68 (2015).
Namikoshi, A. et al. Vaccination trials with Penaeus japonicus to induce resistance to white spot syndrome virus. Aquaculture 229, 25–35 (2004).
Zang, S. et al. Metabolomic investigation of ultraviolet ray-inactivated white spot syndrome virus-induced trained immunity in Marsupenaeus japonicus. Front. Immunol. 13, 885782 (2022).
Little, T. J., Hultmark, D. & Read, A. F. Invertebrate immunity and the limits of mechanistic immunology. Nat. Immunol. 6, 651–654 (2005).
Rodrigues, J., Brayner, F. A., Alves, L. C., Dixit, R. & Barillas-Mury, C. Hemocyte differentiation mediates innate immune memory in Anopheles gambiae mosquitoes. Science 329, 1353–1355 (2010).
Boutros, M., Agaisse, H. & Perrimon, N. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev. Cell. 3, 711–722 (2002).
Steiner, H. Peptidoglycan recognition proteins: on and off switches for innate immunity. Immunol. Rev. 198, 83–96 (2004).
Vuscan, P., Kischkel, B., Joosten, L. A. B. & Netea, M. G. Trained immunity: General and emerging concepts. Immunol. Rev. 323, 164–185 (2024).
Narita, T., Weinert, B. T. & Choudhary, C. Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell. Biol. 20, 156–174 (2019).
Allis, C. D. et al. New nomenclature for chromatin-modifying enzymes. Cell 131, 633–636 (2007).
Li, X. et al. MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Mol. Cell. Biol. 30, 5335–5347 (2010).
Su, J., Wang, F., Cai, Y. & Jin, J. The functional analysis of histone acetyltransferase MOF in tumorigenesis. Int. J. Mol. Sci. 17, 99 (2016).
Dou, Y. et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 121, 873–885 (2005).
Sykes, S. M. et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell. 24, 841–851 (2006).
Huai, W. et al. KAT8 selectively inhibits antiviral immunity by acetylating IRF3. J. Exp. Med. 216, 772–785 (2019).
Radzisheuskaya, A. et al. Complex-dependent histone acetyltransferase activity of KAT8 determines its role in transcription and cellular homeostasis. Mol. Cell. 81, 1749–1765 (2021).
Yoo, L., Mendoza, D., Richard, A. J. & Stephens, J. M. KAT8 beyond acetylation: A survey of its epigenetic regulation, genetic variability, and implications for human health. Genes-Basel 15, 639 (2024).
Li, W. et al. Virus replication cycle of white spot syndrome virus in secondary cell cultures from the lymphoid organ of Litopenaeus vannamei. J. Gen. Virol. 96, 2844–2854 (2015).
Tassanakajon, A., Somboonwiwat, K., Supungul, P. & Tang, S. Discovery of immune molecules and their crucial functions in shrimp immunity. Fish. Shellfish. Immunol. 34, 954–967 (2013).
Dey, B. K., Dugassa, G. H., Hinzano, S. M. & Bossier, P. Causative agent, diagnosis and management of white spot disease in shrimp: A review. Rev. Aquacult. 12, 822–865 (2019).
Joseph, T. C., James, R., Rajan, L. A., Surendran, P. K. & Lalitha, K. V. White spot syndrome virus infection: Threat to crustacean biodiversity in Vembanad Lake, India. Biotechnol. Rep. (Amst.) 7, 51–54 (2015).
van den Bosch, T. et al. A 6-alkylsalicylate histone acetyltransferase inhibitor inhibits histone acetylation and pro-inflammatory gene expression in murine precision-cut lung slices. Pulm. Pharmacol. Ther. 44, 88–95 (2017).
Liu, L. et al. A crosstalk between BRG1 and MOF contributes to ROS production in a mouse model of non-alcoholic steatohepatitis. Antioxid. Redox, Signal. 30, 1539–1552 (2019).
Liu, T.-W., Zhao, Y.-M., Jin, K.-Y., Wang, J.-X. & Zhao, X.-F. KAT8 is upregulated and recruited to the promoter of Atg8 by FOXO to induce H4 acetylation for autophagy under 20-hydroxyecdysone regulation. J. Biol. Chem. 300, 105704 (2024).
Zhu, Z. J., Jiang, W. P., McGinley, J. N. & Thompson, H. J. 2-Deoxyglucose as an energy restriction mimetic agent: effects on mammary carcinogenesis and on mammary tumor cell growth in vitro. Cancer Res 65, 7023–7030 (2005).
Zhang, P. et al. WSSV exploits AMPK to activate mTORC2 signaling for proliferation by enhancing aerobic glycolysis. Commun. Biol. 6, 361 (2023).
Shares, B. H., Busch, M., White, N., Shum, L. & Eliseev, R. A. Active mitochondria support osteogenic differentiation by stimulating β-catenin acetylation. J. Biol. Chem. 293, 16019–16027 (2018).
Gao, J. et al. Interferon functional analog activates antiviral Jak/Stat signaling through integrin in an arthropod. Cell. Rep. 36, 109761 (2021).
Gao, J., Wang, J. X. & Wang, X. W. MD-2 homologue recognizes the white spot syndrome virus lipid component and induces antiviral molecule expression in shrimp. J. Immunol. 203, 1131–1141 (2019).
Wang, H. Y., Xiao, B., Chen, S. H., He, J. G. & Li, C. Z. Identification of an ortholog of MALT1 from shrimp that induces NF-κB-mediated antiviral immunity. Viruses-Basel 15, 2361 (2023).
Jiang, H.-S., Lv, L.-X. & Wang, J.-X. Anti-lipopolysaccharide factor D from kuruma shrimp exhibits antiviral activity. Mar. Life. Sci. Tech. 4, 52–61 (2021).
Li, B., Carey, M. & Workman, J. L. The role of chromatin during transcription. Cell 128, 707–719 (2007).
Novakovic, B. et al. beta-Glucan reverses the epigenetic state of LPS-induced immunological tolerance. Cell 167, 1354–1368.e1314 (2016).
Dion, M. F., Altschuler, S. J., Wu, L. F. & Rando, O. J. Genomic characterization reveals a simple histone H4 acetylation code. P Natl Acad. Sci. USA 102, 5501–5506 (2005).
Corona, D. F., Clapier, C. R., Becker, P. B. & Tamkun, J. W. Modulation of ISWI function by site-specific histone acetylation. Embo. Rep. 3, 242–247 (2002).
Smith, E. R., Allis, C. D. & Lucchesi, J. C. Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J. Biol. Chem. 276, 31483–31486 (2001).
Benjaskulluecha, S. et al. Screening of compounds to identify novel epigenetic regulatory factors that affect innate immune memory in macrophages. Sci. Rep.-uk. 12, 1912 (2022).
Neal, K. C., Pannuti, A., Smith, E. R. & Lucchesi, J. C. A new human member of the MYST family of histone acetyl transferases with high sequence similarity to Drosophila MOF. Biochim. Biophys. Acta 1490, 170–174 (2000).
Irazoqui, J. E., Urbach, J. M. & Ausubel, F. M. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat. Rev. Immunol. 10, 47–58 (2010).
Li, C., Weng, S. & He, J. WSSV–host interaction: Host response and immune evasion. Fish. Shellfish. Immunol. 84, 558–571 (2019).
Tassanakajon, A., Amparyup, P., Somboonwiwat, K. & Supungul, P. Cationic antimicrobial peptides in penaeid shrimp. Mar. Biotechnol. 13, 639–657 (2011).
Lemaitre, B. & Hoffmann, J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 (2007).
Taks, E. J. M., Moorlag, S. J. C. F. M., Netea, M. G. & Van der Meer, J. W. M. Shifting the immune memory paradigm: Trained immunity in viral infections. Annu Rev. Virol. 9, 469–489 (2022).
Netea, M. G., Schlitzer, A., Placek, K., Joosten, L. A. B. & Schultze, J. L. Innate and adaptive immune memory: an evolutionary continuum in the host’s response to pathogens. Cell. Host. Microbe 25, 13–26 (2019).
Mcbride, M. A. et al. Bacteria- and fungus-derived PAMPs induce innate immune memory via similar functional, metabolic, and transcriptional adaptations. J. Leukoc. Biol. 115, 358–373 (2024).
Wu, J. L., Nishioka, T., Mori, K., Nishizawa, T. & Muroga, K. A time-course study on the resistance of Penaeus japonicus induced by artificial infection with white spot syndrome virus. Fish. Shellfish Immun. 13, 391–403 (2002).
Du, X. J. et al. Identification and molecular characterization of a peritrophin-like protein from fleshy prawn (Fenneropenaeus chinensis). Mol. Immunol. 43, 1633–1644 (2006).
Du, X. J., Zhao, X. F. & Wang, J. X. Molecular cloning and characterization of a lipopolysaccharide and beta-1,3-glucan binding protein from fleshy prawn (Fenneropenaeus chinensis). Mol. Immunol. 44, 1085–1094 (2007).
Wei, F. et al. Molecular cloning and subcellular localization of six HDACs and their roles in response to salt and drought stress in kenaf (Hibiscus cannabinus L.). Biol. Res. 52, 20 (2019).
Lee, T. I., Johnstone, S. E. & Young, R. A. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 1, 729–748 (2006).
Ashburner, M. et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Draghici, S. et al. A systems biology approach for pathway level analysis. Genome Res. 17, 1537–1545 (2007).
Acknowledgements
We thank Dr. Xiang-Mei Ren of the Core Facilities for Life and Environmental Sciences, State Key Laboratory of Microbial Technology of Shandong University for help with the microplate spectrophotometer technique, and Dr. Yiteng Xu from the Core Facility and Service Platform at School of Life Sciences, Shandong University for help in confocal microscopy imaging. This work was supported by grants from the National Natural Science Foundation of China (31930112) and Provincial Natural Science Foundation of Shandong (ZR2022QC232).
Author information
Authors and Affiliations
Contributions
L.X.L., J.X.W. and X.F.Z. designed the research. J.X.W., X.F.Z. and P.Z. supervised all aspects of the research. L.X.L. performed most of the experiments. Y.M., Y.S. and H.W. helped to analyze data. P.Z. and Y.M. performed Western blot, and qPCR. L.X.L. and J.X.W. wrote and finalized the manuscript. All persons have made contributions to this work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Eirini Trompouki and David Favero. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lv, LX., Zhang, P., Ma, Y. et al. Histone H3K27 acetylation mediated by KAT8 maintains antiviral trained immunity in shrimp induced by inactivated white spot syndrome virus. Commun Biol 8, 1314 (2025). https://doi.org/10.1038/s42003-025-08767-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08767-5