Abstract
Severe COVID-19 associates with humoral immune response dysregulation. While antibodies confer protection against SARS-CoV-2, evidence also support their putative contribution to disease severity. Our study demonstrates that higher levels of S2-IgG, and S2-, RBD-, and Nucleocapsid-IgA differentiate severe and non-severe cases. However, no major antibody functional differences are found between both COVID-19 manifestations. Enhanced Fc-dependent functions in severe cases are primarily driven by increased antibody titers. No differences in antibody avidity are found between severe and non-severe cases, but a gradation in binding strength across specificities suggests that early anti-RBD, -S2, and -Nucleocapsid antibodies may originate from different pathways. In golden Syrian hamsters, S2 immunization or transfer of RBD-depleted antibodies isolated from severe and non-severe cases promote a faster clinical recovery after SARS-CoV-2 challenge, despite a transient initial weight loss. These findings indicate that antibodies are not major determinants of COVID-19 severity and suggest additional factors influencing disease outcomes.
Similar content being viewed by others
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) rapidly induces a humoral immune response upon infection. SARS-CoV-2-specific antibodies can be detected one week after symptom onset, reaching peak levels within the first month after infection1,2,3,4,5,6. Thereafter, antibody titers progressively decrease with different kinetics that depend on isotype, specificity, and disease severity, being IgG the most stable over time5,6,7,8. In contrast to other infections, IgM does not predominate during the initial phase of the humoral response, and both IgA and IgG can be detected early after infection3,8,9. In addition, neutralizing humoral responses are rapidly elicited, remain stable over time1,4,10, and correlate with the levels of binding antibodies, particularly anti-RBD IgG.
Coronavirus disease 2019 (COVID-19) severity has been associated with sex, age, and differences in magnitude, kinetics, and antibody-dependent effector functions11,12,13,14. Individuals with severe COVID-19 exhibit greater titers of anti-SARS-CoV-2 antibodies, including neutralizing antibodies, compared to those with milder disease, suggesting the contribution of humoral responses to COVID-19 pathogenesis14,15,16,17,18,19,20,21. Severe COVID-19 cases have also been associated with deregulated complement activation and deposition22,23,24, macrophage/monocyte activation25,26, impaired NK function27, neutrophilia and NETosis20,28,29, and high levels of hyperinflammatory cytokines and chemokines15,19,30. Importantly, antibodies can directly or indirectly participate in all these processes by interacting with complement and Fc receptors expressed on the surface of immune cells. Accordingly, Liu et al. showed that passive administration of anti-SARS-CoV Spike IgGs to SARS-CoV-infected macaques modified the alveolar macrophage response and promoted acute lung injury via Fcγ receptors (FcγR)31. Similarly, severe COVID-19 is also characterized by elevated levels of afucosylated anti-SARS-CoV-2 IgG32,33,34. These afucosylated IgG, when part of immunocomplexes and combined with TLR-3 agonists, stimulate macrophages to secrete high levels of proinflammatory cytokines (e.g. IL6, TNF, or IL1β), a hallmark of COVID-19 severity32. This process is strongly dependent on the engagement of immunocomplexes with FcγRIIIa and FcγRIIa on the macrophage cell surface32,33,34. Besides the putative role of anti-SARS-CoV-2 IgG in COVID-19 pathogenesis, IgA has also emerged as a contributing factor to severe COVID-1935. IgA antibodies are detected promptly after infection3,36 and serum IgA may activate neutrophils to release neutrophil extracellular traps (NETs)37, a process that has also been associated with severe COVID-1920. Despite all evidence, most studies support the protective role of antibodies in preventing SARS-CoV-2 infection or severe disease development through mechanisms such as neutralization, antibody-dependent cellular phagocytosis (ADCP) or cytotoxicity (ADCC)38,39,40. Therefore, the contribution of the humoral response to COVID-19 severity is not clear, probably due to the interplay of multiple dysregulated immune pathways involving both the innate and the adaptative arms of the immune system.
To further investigate the relative contribution of SARS-CoV-2 antibodies to COVID-19 severity, we designed a multifaceted study where we analyzed the levels of antibodies binding to different SARS-CoV-2 antigens in individuals with mild or severe COVID-19 and assessed their ability to mediate Fc-dependent effector functions (i.e., ADCP and ADCC). Additionally, we conducted two in vivo experiments using golden Syrian hamsters (GSHs) to evaluate the impact of active immunization or the use of passive transfer of human immunoglobulin on the development of severe COVID-19-like disease upon SARS-CoV-2 infection.
Results
Cohort description
A total of 112 SARS-CoV-2-infected patients were initially included in the current study and classified into two groups according to the NIH criteria for COVID-19 severity. Group S consisted of patients with severe COVID-19, all of whom had oxygen saturation below 94%, while Group M included those with mild COVID-19, characterized by the absence of shortness of breath, dyspnea, or abnormal chest imaging. Samples were collected up to 31 days post-diagnosis, which was confirmed by PCR, antigen test, or antibody detection. To better pinpoint the mechanisms responsible for the development of severe COVID-19, 13 patients were excluded due to prior vaccination, active cancer, or primary immunodeficiencies. Thus, Group S (n = 74) comprised 74.3% men with an average age of 54 years (range 29–80), while Group M (n = 25) consisted of 44% men with an average age of 47 years (range 25–92). Most participants (64% in Group S and 56% in Group M) exhibited at least one risk factor associated with severe COVID-19, such as diabetes or obesity, whereas around 30% of patients from each group had multiple risk factors. Individuals with comorbidities were receiving the appropriate care for these diseases at the time of sample collection. While 69% of Group S were administered at least two drugs to treat COVID-19 symptomatology, only 12% of Group M received similar treatment. Common treatments in both groups included corticosteroids and antivirals (e.g. Remdesivir). Additionally, all individuals from Group S required some form of assisted ventilation (e.g. nasal cannula, Venturi mask, invasive or non-invasive mechanical ventilation). Table 1 summarizes the main characteristics of the study groups.
Severe COVID-19 is associated with greater levels of SARS-CoV-2-specific antibodies compared to mild disease
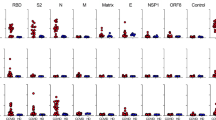
To characterize SARS-CoV-2-specific humoral responses elicited in Groups S and M, we first quantified plasma levels of IgG, IgA, and IgM that targeted SARS-CoV-2 Spike S2 and RBD subunits as well as Nucleocapsid and Envelope proteins (Fig. 1A–D). While we did not detect significant differences in IgM responses between the two groups, we observed higher levels of anti-S2 IgG antibodies (p = 0.0049) in Group S compared to those in Group M. Additionally, Group S also exhibited elevated levels of S2-, RBD-, and Nucleocapsid-specific IgA (p = 0.0011, p = 0.0311, and p = 0.0407, respectively). We did not observe any isotype differences against Envelope. Besides measuring antigen-specific antibody levels, we assessed plasma neutralization activity of Group S and M against the ancestral strain of SARS-CoV-2 (WH1). Our analysis revealed no significant differences in the neutralizing antibody capacity between groups (Fig. 1E).
Comparison of IgG, IgA, and IgM binding antibody levels (from left to right) in unvaccinated individuals with severe (Group S, blue squares) and mild COVID-19 (Group M, red circles) targeting (A) S2, (B) RBD, (C) Nucleocapsid, and (D) Envelope. Binding antibody levels in plasma samples are expressed as arbitrary units/ml. E Plasma neutralizing activity was evaluated against SARS-CoV-2 WH-1. Differences between both groups were analyzed using Peto&Peto censored samples test. Geometric mean with its geometric standard deviation is shown. The number of individuals with detectable antibodies out of the group total is indicated below each group. Dotted lines indicate the upper and lower limit of quantification. Undetectable samples are represented as half this threshold, and samples above the upper limit are represented as twice this threshold.
Thus, although we did not find differences in plasma neutralizing activity, individuals with severe COVID-19 exhibited greater humoral responses, characterized by significantly elevated IgA levels targeting Spike and Nucleoprotein as well as increased S2-specific IgG titers compared to those with mild disease.
The avidity of the anti-SARS-CoV-2 humoral response varies depending on the antigen, and is not associated with disease severity
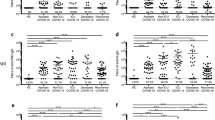
Next, we evaluated the antibody avidity of samples from Group S and M with detectable plasma antibodies against RBD, S2, or Nucleocapsid (Fig. 2). No notable differences in IgG or IgA binding strength to any of these proteins were observed between groups (Fig. 2A–C). However, we noted significant differences in antibody avidity among antigens within each group (Fig. 2D, E). Anti-RBD IgG and IgA antibodies exhibited the lowest avidity among the tested SARS-CoV-2 antigens in both study groups. In Group S, a gradation in avidity was detected for antigen-specific IgG responses, with anti-S2 IgG exhibiting intermediate avidity, and anti-Nucleocapsid IgG displaying the highest binding strength.
Antigen-specific IgG and IgA avidity (left to right) was evaluated in individuals with detectable antibodies specific against (A) RBD, (B) S2, and (C) Nucleocapsid. The number of patients with detectable antibodies in each category is indicated below each group. Comparison of (D) IgG or (E) IgA antibody avidity among antigens within each group (Group M on the left, Group S on the right). Differences between Group S and M were analyzed using Mann-Whitney U test and among antigens with Kruskal Wallis test corrected for false discovery rate (FDR). Mean with standard deviation is shown.
Enhanced Fc-dependent activity of RBD- and S2-specific antibodies in individuals with severe COVID-19 may result from elevated plasma antibody levels
To determine functional differences of SARS-CoV-2-specific antibodies elicited in patients with severe or non-severe COVID-19, we measured their capability to mediate ADCP and ADCC. First, we defined the overall phagocytic activity of plasma antibodies specific to RBD, S2, and Nucleocapsid in Group S and M using the human promyeloblast HL60 cell line, since these cells express both FcγR and FcαR (Supplementary Fig, 1). We observed that RBD- and S2-specific antibodies showed enhanced ADCP in Group S compared to Group M (p = 0.0167 and p = 0.0096, respectively, Fig. 3A). To discriminate the contribution of each isotype to ADCP activity and if these differences were related to intrinsic antibody traits, we corrected antigen-specific phagocytic activity using the levels of antigen-specific IgG and IgA antibodies. We observed a slight increase in ADCP activity in Group S compared to Group M after correcting with anti-RBD binding IgG antibodies (p = 0.0563, Fig. 3B). No additional differences were detected after normalizing with IgA or against any of the other proteins (Fig. 3C). Next, we measured overall plasma ADCC against RBD, S2, and Nucleocapsid from Groups S and M (Fig. 3D) using CD16 + KHYG-1 cell line as effector cells41. We confirmed our ADCC data using primary NK cells isolated from blood donors and a subset of samples that were randomly selected. Both assays showed a strong correlation (Supplementary Fig. 2A, B). The results of these assays showed that RBD-specific antibodies from Group S induced slightly greater, although not significant, NK cell degranulation (CD107+) than those from Group M (Fig. 3D). After adjusting ADCC activity with RBD-specific IgG levels (Fig. 3E), Group S and M displayed similar ADCC activity. These results indicate that functional differences in ADCP and ADCC activity described above might be related to antibodies titers rather than to intrinsic antibody features.
A Total ADCP was evaluated in plasma samples against RBD, S2, and Nucleocapsid (from left to right) using the HL60 cell line. ADCP was normalized using antigen-specific (B) IgG, or (C) IgA antibody titers. D Total ADCC was evaluated in plasma samples against RBD, and S2 (left to right) using primary NK cells. E ADCC was normalized using antigen-specific IgG antibody titers. Only plasma from patients with detectable antigen-specific antibodies were assayed, the number of individuals is indicated below each group. Differences between groups were analyzed using Mann–Whitney U test. Mean with standard deviation is shown.
We also assessed whether purified IgG and IgA from a randomly selected subset of severe or mild COVID-19 individuals could enhance the production of pro-inflammatory cytokines (i.e., IP-10, IL-6, and IL12/IL23 p40) in peripheral blood monocyte-derived macrophages. Macrophages were stimulated for 24 h with purified antibodies, either in the presence or absence of infectious SARS-CoV-2. No significant differences between Groups S and M were observed in cytokine production in cell cultures stimulated with immunoglobulins, irrespective of the presence of virus (Fig. 4). These findings suggest that both IgG and IgA, purified from COVID-19 individuals, exhibited a similar capacity to stimulate macrophages to produce proinflammatory cytokines, regardless of their severity status.
Quantification of IL-12/IL-23 p40, IL-6, and IP-10 by Luminex from peripheral blood monocyte- derived macrophages cultures for 24 h in the presence of infectious SARS-CoV-2. Purified immunoglobulins were assessed at 10 µg/mL. Each dot represents one patient. Stimulation with LPS or no stimulation (mock) were included as controls. Data were analyzed using the Kruskal Wallis test corrected for FDR. Mean with standard deviation is shown. Data are representative of three independent experiments.
Infusion of purified RBD-depleted human IgG and IgA from COVID-19 patients induce an early transient weight loss in SARS-CoV-2-challenged golden Syrian hamsters followed by a faster disease recovery
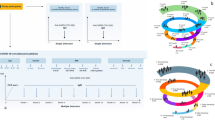
To determine whether antibodies could promote COVID-19 severity in vivo, we purified IgG and IgA antibodies from severe or mild COVID-19 and infused them into GSHs 24 h before SARS-CoV-2 challenge. To minimize the activity of neutralizing antibodies, we removed RBD-specific antibodies from all four antibody fractions by cross-adsorption to RBD-coated Sepharose. As expected, anti-S2 antibody levels remained consistent, while anti-RBD antibodies were effectively depleted, as they were undetectable by ELISA in the cross-adsorbed samples (Supplementary Fig. 3). Sixty-six GSHs were used in this study. Sixty GSHs were divided into five groups (n = 12/group, 1:1 female-to-male ratio). Groups 1 and 2 were treated with purified anti-RBD-depleted human IgG and IgA from Group S, respectively, while Groups 3 and 4 received the same fractions obtained from Group M (Fig. 5A). Group 5 served as positive controls of infection and received 1x PBS. Six animals remained uninfected throughout the entire study and were used as negative controls (Group 6). All animals, except those in Group 6, were intranasally inoculated with SARS-CoV-2 D614G (Cat01 isolate, 104 TCID50/animal) 24 h after the antibody administration, and were followed up for 14 days post-challenge. After inoculation, human IgG levels (Fig. 5B) slowly decreased over time. At day 4 post-challenge, animals from Groups 1 and 3 had between 314 and 452 µg/mL of human IgG, except for one animal in Group 1, which had undetectable levels. Animals that were euthanized at day 14 post-infection presented between 74 to 215 µg/ml of human IgG in circulation. Conversely, a rapid decline of human IgA was observed in vivo. Specifically, Groups 2 and 4 had over 10-fold less human IgA in circulation by day 4 compared to IgG-inoculated GSHs, with levels ranging from 7.5 to 36 µg/ml. By day 7 and thereafter, human IgA was undetectable by ELISA.
A Overview of infection timeline. Blood drops indicate collection of biological samples, and needles represent the administration of either human immunoglobulin or infectious virus. Created in BioRender. Carrillo, J. (2025) https://BioRender.com/2sf3jvn. B Human IgG and IgA levels in sera from GSHs after SARS-CoV-2 infection over time. Antibody levels are represented as µg/ml. Dotted lines indicate the limit of quantification. Undetectable samples are represented as half this threshold. Mean with standard deviation is shown. C Weight variation follow up of SARS-CoV-2-infected GSHs after the infusion of purified human antibodies. D Comparison of GSH weight changes among groups on days 1 to 6. Differences among groups were analyzed using Kruskal–Wallis test corrected for FDR. Mean with standard deviation is shown.
Although all GSHs challenged with SARS-CoV-2 progressively lost weight after challenge, those GSHs treated with IgG, regardless of their origin, recovered weight slightly faster than the positive control group (Group 5) that had not received any human antibody prior to infection (Fig. 5C). On day 1 after challenge, we observed significant weight reduction across all antibody-treated GSHs compared to infected controls (G1 vs G5: p = 0.046; G2 vs G5: p = 0.029; G3 vs G5: p = 0.006; G4 vs G5: p = 0.0001). By day 2, GSHs in Groups 2, 3, and 4 continued to exhibit significant weight loss compared to both control groups (Fig. 5D). Group 1, however, only showed weight loss compared to the uninfected control group (G1 vs G6: p = 0.0069). Notably, all treated male GSHs -whether receiving IgG or IgA- showed reduced weight compared to controls (Supplementary Fig. 4A, C). Interestingly, these weight differences were not observed in female GSHs on day 2 post-challenge, suggesting that these early weight changes may be sex-dependent (Supplementary Fig. 4B, D). To investigate whether weight variation might be a consequence of antibody injection, we performed a second experiment in which IgG and IgA isolated from a pool of pre-pandemic plasma samples (n = 15) were administered to 12 GSHs (IgG n = 6; IgA n = 6, 50% each sex). No weight reduction was observed in any animal, with the exception of one male GSH treated with IgA. This animal showed a rapid and sustained weight reduction compared with day 0, reaching a reduction peak of 7% by day 7 post-antibody administration. This weight reduction was not attributed to antibody preparation, since the dynamic was different to the one previously observed (Fig. 5 and Supplementary Fig. 4), and could be due to unrelated factors such as hierarchy in the animal litter, littermate aggressions, or other unidentified factors. The overall trend of all groups was to rise weight, reaching statistical significance by day 10 post antibody administration (Supplementary Fig. 5).
To comprehensively assess the potential impact of RBD-depleted human antibodies in vivo, we monitored viremia by RT-qPCR in three relevant samples: oropharyngeal swabs, nasal turbinates, and lungs on days 4, 7, and 14 after viral challenge (Fig. 6A). To increase statistical power, male and female GSHs were analyzed together, since sex-segregated analysis did not show statistically significant differences (Supplementary Fig. 6). Our analysis revealed that viremia kinetics in all three tissues significantly declined over time, with no major differences among groups. Interestingly, GSHs treated with IgG or IgA from Group M (Groups 3 and 4, respectively) had significantly greater viral loads on day 4 post-infection in the lungs compared to SARS-CoV-2-challenged controls (Group 5, p = 0.026 and p = 0.009, respectively, Fig. 6A, right panel), suggesting that these antibody preparations might contribute to enhance early viral replication or accumulation in the lungs. Conversely, animals treated with IgG from Group S (Group 1) had significantly lower viremia in nasal turbinate on day 14 (p = 0.0034, Fig. 6A, middle panel), indicating that IgG from patients with severe COVID-19 may offer partial protection. To determine the presence of infectious viral particles, we performed viral titration on Vero cells. Infectious particles were rarely detected in oropharyngeal swabs at any time point and were only occasionally found in nasal turbinate and lung samples on days 7 and 14 post-challenge (Fig. 6B). However, they were consistently detected in nasal turbinates and lungs on day 4, with no significant differences observed among the SARS-CoV-2 challenged groups.
A Viral load was analyzed in oropharyngeal swab, nasal turbinate, and lungs (left to right). Levels of SARS-CoV-2 gRNA are expressed as log10 viral RNA copies/ml. Differences among groups were analyzed using Kruskal Wallis and Conover’s post-hoc tests corrected for FDR. Mean with standard deviation is shown. B Infectious virus titers were determined in the same tissues as in (A) at days 4, 7, and 14 post-infection. Data is shown as log10 median tissue culture infectious dose per milliliter (TCID50/ml). Dotted line indicates limit of quantification. Differences between groups were analyzed with the Kruskal–Wallis test followed by Conover’s post-hoc analysis adjusted by FDR. C Hematoxylin and eosin staining was used to perform a histopathologic analysis of nasal turbinate and lungs (left to right). Lesion score: 0 indicates absence of damage, while 1, 2, and 3 indicate mild, moderate, and severe lesions, respectively. D Nucleocapsid detection in nasal turbinate and lungs (left to right) by immunohistochemistry. Staining score: 0 indicates the absence of Nucleocapsid, while 1, 2, and 3 indicate low, moderate, and high levels of this viral antigen, respectively. Histopathologic and immunohistochemical differences among groups were analyzed using asymptotic generalized Pearson chi-squared test corrected for FDR. Mean with standard deviation is shown.
We also conducted histopathology and immunohistochemical analysis on the collected tissue samples to evaluate the extent and severity of tissue damage. Sex-disaggregated graphs are shown in Supplementary Fig. 7. We observed a significant progressive reduction in tissue damage over time in nasal turbinates across all groups. However, this reduction was delayed in the lungs and was only observed on day 14 post-challenge (Fig. 6C). No significant differences were observed among groups at any time point. Active viral replication was evaluated by detecting the presence of Nucleocapsid by immunohistochemistry in nasal turbinate and lungs (Fig. 6D). In both tissues, we detected similar kinetics for the immunohistochemical score. Throughout the experiment, the presence of Nucleocapsid progressively decreased across all groups, with no significant differences observed among them.
Overall, our results showed that antibodies isolated from severe COVID-19 individuals did not enhance disease severity in GSHs. Although antibody-treated GSHs showed a more rapid and significant weight loss after SARS-CoV-2 challenge, compared to untreated but infected GSHs, this effect was equally observed in groups receiving antibody preparations from either severe or mild COVID-19 individuals. This weight reduction was transient and antibody-treated animals showed slightly faster recovery compared to SARS-CoV-2-challenged controls; particularly, those animals infused with IgG preparations. Thus, RBD-depleted IgG antibodies isolated from COVID-19 individuals with either mild or severe disease partially protected GSHs from disease development upon SARS-CoV-2 challenge.
S2 vaccination induced an early and transient weight loss in male GSHs after SARS-CoV-2 challenge but protected animals against disease
Since SARS-CoV-2 infection induced higher levels of anti-S2 antibodies in individuals with severe COVID-19 compared to those with milder symptoms, we investigated whether immunization-induced S2-specific antibodies could exacerbate disease severity after SARS-CoV-2 infection. To address this point, 54 GSHs (1:1 sex ratio) were divided into four groups (Fig. 7A). Group 1 (n = 18) was vaccinated with a recombinant S2 protein mixed with aluminium hydroxide as an adjuvant and received a booster dose 20 days apart. Group 2 (n = 18) received two doses of the adjuvant 20 days apart. Groups 3 (n = 12) and 4 (n = 6) served as non-immunized controls. Groups 1–3 were intranasally inoculated with SARS-CoV-2 D614G (Cat01 isolate, 104 TCID50/animal) six days after the vaccine booster, and they were followed up for 14 days post-challenge. Anti-S2 and anti-RBD antibodies were quantified by ELISA (Fig. 7B). As expected, anti-S2 antibodies were detected exclusively in S2-immunized animals (Group 1) prior to SARS-CoV-2 challenge. In Groups 2 and 3, these antibodies were detected by day 4 post-infection, with levels progressively increasing through day 14 (Fig. 7B). Interestingly, while S2-specific antibodies in Group 1 did not continue to increase after infection, they remained significantly higher than in Group 2 on days 4 and 7 post-infection. Anti-RBD antibodies were not present in any GSH before SARS-CoV-2 challenge but appeared by day 7 post-challenge, indicating that S2 vaccination did not hamper the elicitation of RBD antibodies upon infection (Fig. 7B). Only one GSH from the S2-immunized Group 1 had detectable neutralizing activity, although at very low titers, prior to intranasal challenge with SARS-CoV-2, indicating that monomeric S2 immunization induced mainly non-neutralizing antibodies. All animals from Groups 1, 2, and 3 developed similar neutralizing antibody titers after infection, reaching maximum levels on day 7 post-infection (Fig. 7C).
A Overview of vaccine strategy and infection timeline. Blood drops indicate collection of biological samples, and needles represent immunization or infectious virus administration. Created in BioRender. Carrillo, J. (2025) https://BioRender.com/w7tt4aa. B Levels of vaccine-induced S2- and RBD-specific IgG antibodies (left to right) of all groups after vaccination and challenge. C Neutralizing antibody titers in GSHs pre- and post-infection. Dotted lines indicate the upper and lower limit of quantification. Undetectable samples are represented as half this threshold, and samples above the upper limit are represented as twice this threshold. Differences among groups were analyzed using Peto&Peto censored samples test with pairwise multiple comparisons, and longitudinal analysis were performed with Prentice-Wilcoxon and Conover’s post-hoc tests. Mean with standard deviation is shown.
To investigate the effect of S2 immunization in SARS-CoV-2-induced disease progression, we monitored GSH weight until day 14 after challenge (Fig. 8). Overall, Group 1 showed a faster recovery rate compared to Groups 2 and 3. S2-immunized animals started gaining weight on days 3-4 post-infection and continued until the end of the experiment (day 14). In contrast, the control groups (Groups 2 and 3) started their recovery on days 6-7 after challenge (Fig. 8A, B). These results indicate that S2 immunization protected SARS-CoV-2-infected GSHs, accelerating their recovery. However, we observed a greater weight loss in S2-immunized Group 1 males compared to the control groups (Groups 3 and 4) on days 1 to 3 post-challenge (day 1: p = 0.0003, and p = 0.0194; day 2: p = 0.0032, and p = 0.0017; day 3: p = 0.0174 and p = 0.0018, respectively, Fig. 8C). Interestingly, these differences were sex-dependent and temporary, since they were not observed in females (Fig. 8D), and S2-immunized animals (both male and female) recovered weight after day 4 post-challenge (Fig. 8A–D).
Overall weight variation percentage in SARS-CoV-2-infected GSH (A) males and (B) females until day 14 post-challenge. Comparison of weight changes among groups in days 1 to 4 in GSH (C) males and (D) females. Differences among groups were analyzed using Kruskal Wallis test corrected for FDR. Mean with standard deviation is shown.
In addition to monitoring the clinical outcomes, we quantified viral load in oropharyngeal swabs, nasal turbinate, and lungs upon euthanasia on days 4, 7, and 14 after viral challenge (Fig. 9). To increase statistical power, we grouped males and females from each study group. Sex-disaggregated graphs are shown in Supplementary Figs. 8, 9. Our analysis revealed a significant decline in viral load over time in oropharyngeal swab, nasal turbinate, and lungs in all infected groups (Fig. 9A). Additionally, while we did not observe differences among groups on days 4 or 14 after infection in any tissue, Group 1 had lower viral load on day 7 post-infection in lungs compared to Group 2 (p = 0.034) (Fig. 9A). These findings are consistent with previous results, as the animals in this group exhibited weight recovery on day 4 post-challenge, earlier than both control groups (Fig. 8).
A Viral load was analyzed from oropharyngeal swab, nasal turbinate, and lungs (left to right). Levels of SARS-CoV-2 gRNA are expressed as log10 viral RNA copies/ml. Differences among groups were analyzed using Kruskal Wallis and Conover’s post-hoc tests corrected for FDR. Mean with standard deviation is shown. B Titers of infectious virus were evaluated in the same three tissues indicated in (A) at days 4, 7, and 14 post-infection. Data is shown as log10 median TCID50/ml. Dotted line indicates limit of quantification. Differences between groups were analyzed using the Kruskal–Wallis test followed by Conover’s post-hoc analysis adjusted by FDR. C Hematoxylin and eosin staining was used to perform a histopathologic analysis of nasal turbinate and lungs (left to right). Lesion score: 0 indicates absence of damage, while 1, 2, and 3 indicate mild, moderate, and severe lesions, respectively. D Nucleocapsid detection in nasal turbinate and lungs (left to right) by immunohistochemistry. Staining score: 0 indicates the absence of Nucleocapsid, while 1, 2, and 3 indicate low, moderate, and high levels of this viral antigen, respectively. Histopathologic and immunohistochemical differences among groups were analyzed using asymptotic generalized Pearson chi-squared test corrected for FDR. Mean with standard deviation is shown.
Infectious viral particles were detected in nasal turbinate and lung samples on day 4 after challenge, and to a lesser extent in oropharyngeal swabs (Fig. 9B). By days 7 and 14, infectious particles were rarely observed, indicating viral clearance over time. On day 4, higher titers of infectious viruses were found in nasal turbinate and lungs compared to oropharyngeal swab. Notably, reduced levels of infectious particles were observed in nasal turbinate and lungs of S2-immunized SARS-CoV-2-challenged GSHs compared to unimmunized controls (p = 0.066) and adjuvant-only controls (p = 0.035), respectively (Fig. 9B). Next, we evaluated inflammation and severity of the lesions on each sample using a semi-quantitative histopathological score (Fig. 9C and Supplementary Fig. 9A, B). In nasal turbinate, the most severe lesions were observed on day 4, whereas in lungs peaked on day 7. Tissue lesions were significantly reduced over time in all infected groups, with no major differences observed among groups at any time point. Additionally, as proxy of local viral replication, we quantified the magnitude and distribution of Nucleocapsid using immunohistochemistry in both nasal turbinate and lungs (Fig. 9D and Supplementary Fig. 9C, D). An overall greater immunohistochemical score was identified in nasal turbinate compared to lungs, consistent with the results of infectious viral particles analysis. However, Nucleocapsid staining declined over time in all infected groups in both tissues. According to viral load data, S2-immunized animals showed reduced Nucleocapsid staining on both nasal turbinate and lungs compared to those GSHs that only received aluminium hydroxide (Fig. 9D).
Overall, our results showed that S2 immunization did not enhance disease severity in GSHs upon SARS-CoV-2 challenge. Although S2-immunized male GSHs underwent a more pronounced initial weight loss following viral challenge, this weight reduction was transient, and S2-immunized GSHs exhibited a more rapid weight recovery and faster viral load clearance compared to control animals, indicating that they were protected from severe disease development.
Discussion
While most SARS-CoV-2 infections result in no clinical signs or mild COVID-19, a smaller yet significant number of SARS-CoV-2-infected individuals develop severe disease, which can eventually lead to death42,43,44. Although the mechanisms that promote severe COVID-19 are not fully understood, they are associated with dysregulated immune responses that impact both innate and adaptive immunity15,17. High levels of proinflammatory cytokines and chemokines, along with dysfunctional cell activation, homing, and distribution, have been observed in SARS-CoV-2-infected individuals who progress to severe COVID-1916,21,44,45. A dysregulated humoral response has also been associated with severe disease, although its exact role is not completely understood. Early studies reported the presence of higher titers of anti-SARS-CoV-2 antibodies in individuals with severe disease compared to those with mild COVID-191,7,10,13,14,46,47. Here, we confirm this statement as individuals with severe COVID-19 exhibited greater levels of anti-Spike S2 IgGs, and IgA responses targeting the Spike and Nucleocapsid. We did not identify differences in IgM responses or in IgG levels targeting other SARS-CoV-2 antigens (i.e., Nucleoprotein and Envelope) or RBD, which can explain the similarity in neutralizing antibody titers. Whereas Zevou and colleagues linked anti-Nucleoprotein IgA to severe COVID-1935, we also observed greater IgA levels against RBD and S2 in severe cases compared to subjects with mild COVID-19. Despite this association, and the fact that anti-SARS-CoV-2 IgA responses are detected early after infection3,36, it is not clear whether IgA responses might enhance COVID-19 severity. Serum IgA antibodies can activate neutrophils to release neutrophil extracellular traps (NETs)37, a process associated with COVID-19 severity20. However, early neutralizing responses are dominated by IgA36, particularly in dimeric form, which is more effective in neutralization activity and prevalent in nasopharinx48. This fact may explain the correlation between nasal anti-RBD IgA responses and symptom resolution in mild COVID-1949. Additionally, broadly neutralizing IgA antibodies have been isolated from convalescent COVID-19 patients48,50, indicating a potential protective role.
Beside specificity, isotype, and neutralizing activity, antibodies can influence COVID-19 progression through interactions with Fc receptors. Severe COVID-19 cases have been associated with high levels of proinflammatory afucosylated anti-SARS-CoV-2 IgG, which exhibit enhanced binding to FcγR32,33,34, potentially promoting systemic inflammation through FcγR-mediated uptake of the virus by monocytes and macrophages51. However, Zohar and colleagues14 proposed an alternative scenario, reporting that non-survivors of severe COVID-19 had reduced IgG responses and impaired Fc-dependent activity. Here, we have assessed the capacity of antibodies to mediate Fc-dependent functions (i.e., ADCP and ADCC) and found that plasma from individuals with severe COVID-19 showed higher ADCP and ADCC activities compared to those with mild disease. Importantly, these differences disappeared when ADCP and ADCC values were corrected for antigen-specific IgG or IgA levels, indicating that antibody titers, rather than intrinsic antibody properties, drive the increased Fc-dependent activities observed in Group S. Accordingly, we did not observe significant differences in cytokine production by PBMC-derived macrophages stimulated with IgG or IgA purified from individuals with severe and mild COVID-19, even in the presence of SARS-CoV-2 viral particles.
Kaneko and coworkers described a lack of germinal centers in individuals with COVID-1952, which could partly explain the impaired humoral response observed in deceased patients with severe COVID-1914. We analyzed the avidity of anti-S2, -RBD and -Nucleocapsid IgG and IgA as a surrogate marker of humoral response maturation. No significant differences in antibody avidity were found between both groups, indicating that the greater levels of S2-specifc IgG and RBD-, S2-, and Nucleocapsid-specific IgA antibodies observed in severe cases may not be attributed to stronger antibody binding. However, significant variation in binding strength was noted among the different antigen-specific antibody responses. The lower avidity of RBD responses aligns with previous findings showing that neutralizing responses in individuals with COVID-19 are often dominated by germline-coded antibodies53, and that extrafollicular humoral responses correlate with neutralizing activity54. However, whether these differences might be the result of a differential contribution of the germinal center to the antibody response against different viral antigens, would need further investigation.
To investigate whether anti-SARS-CoV-2 antibodies can enhance the severity of SARS-CoV-2 infection, we designed an in vivo experiment using GSHs since these animals develop moderate disease upon SARS-CoV-2 challenge. Polyclonal IgG and IgA were isolated from pooled plasma of individuals with severe and mild COVID-19 and administered to GSHs 24 h before infection. Given the protective role of neutralizing antibodies40,55 and the dominance of anti-RBD antibodies in this response8,38,54, we depleted anti-RBD antibodies before infusion into GSHs. Overall, antibody-treated animals recovered weight at a similar or slightly faster rate than the controls. Interestingly, these GSHs experienced sharper weight reduction during the first days after SARS-CoV-2 challenge regardless of the antibody origin. However, no enhanced tissue damage was observed in nasal turbinate or lung tissues on days 4, 7, or 14 post-challenge. Early anti-SARS-CoV-2 antibodies might modulate initial antiviral responses, transiently affecting animal physiology (e.g appetite and food intake56) and promoting disease recovery. Intriguingly, passive prophylactic intranasal administration of neutralizing dimeric IgA (B8dIgA clone) to GSHs reduced lung viral load but increased tissue damage and the presence of infectious viral particle in nasal turbinate, likely through a trans-infection process involving both dendritic cells and nasal turbinate epithelial cells57. Additionally, passive administration of anti-SARS-CoV Spike IgGs promoted acute lung injury in SARS-CoV-infected non-human primates31. However, our data do not fully support antibody-mediated enhancement of disease in GSHs, as we did not observe an increase in tissue damage, or other evidence beyond the early and transient weight loss. Moreover, antibody-treated animals recovered from the disease faster than controls. Nevertheless, we cannot completely exclude the involvement of antibodies, particularly IgA, given the poor pharmacokinetics of human IgA in GSHs, which may limit its long-term effect.
Our study has proved that individuals with severe COVID-19 develop greater titers of anti-S2 IgG and IgA antibodies than those who develop mild COVID-19. S2 region is the most conserved region of the Spike protein and elicits the most potent cross-reactive humoral responses across all beta coronaviruses. Although some S2-specific neutralizing antibodies have been described58, the majority of anti-S2 antibodies are non-neutralizing8. To assess whether anti-S2 immune responses might contribute to disease enhancement, we immunized GSHs with the S2 subunit and subsequently challenged them with SARS-CoV-2. We used a recombinant monomeric S2 glycoprotein produced in house alongside with aluminum hydroxide, a Th2 adjuvant widely used in human vaccines59 and previously associated with disease enhancement60. We observed that S2 immunization partially protected GSHs from SARS-CoV-2-induced disease. S2-immunized GSHs did not develop potent neutralizing humoral responses. However, these animals exhibited faster weight recovery compared to controls, particularly females. In addition, S2-immunized animals showed reduced viral load and less tissue damage in nasal turbinates and lungs on day 7 post-challenge. Despite these findings, we observed a pronounced weight reduction in males during the first three days after challenge. Accordingly, Ebening and colleagues described that GSHs immunized with non-stabilized Spike protein and aluminum as adjuvant developed vaccine-associated enhanced respiratory pathology within the first four days after SARS-CoV-2 challenge61. This pathology was mediated by a Th2-immune response, characterized by a significant eosinophil accumulation in the lungs. A direct comparison between the results published by Ebening et al. and ours is not possible since we did not analyze viral load and tissue damage before day 4 post-challenge, and Ebening’s study did not provide data beyond day 4. Additionally, Ebening and colleagues did not present sex-disaggregated data. Nevertheless, it is unlikely that the early and transient weight loss described here results from an antibody-mediated disease enhancement process, as S2-immunized GSHs exhibited faster disease recovery and less tissue damage on day 7 post-SARS-CoV-2 challenge. Thus, S2 immunization might enhance early antiviral immune responses associated with a transient weight reduction, ultimately leading to faster recovery. Interestingly, this effect appears to be more pronounced in males than in females, probably due to sex differences in food intake upon viral infection. In line with this, male mice showed a more drastic reduction in food consumption after endotoxin-exposure related to higher levels of IL-6 and TNF62.
Our study shows some limitations, such as the lack of GSH sampling during the first days after challenge and the use of young animals. Our data do not fully support the role of humoral responses in disease enhancement. However, since samples were only collected on days 4, 7, and 14 after infection, we cannot completely rule out the possibility of enhanced tissue damage occurring at early time points. Additionally, the use of young animals may have limited the clinical effects observed in antibody-treated and S2-immunized animals. Like humans, older GSHs develop more severe disease following SARS-CoV-2 challenge, and the efficacy of SARS-CoV-2 vaccine is reduced in aged individuals63. Therefore, the disease enhancement effect of humoral responses may be more pronounced in older populations compared to younger ones. In addition, participants were not screened for prior SARS-CoV-2 infection, which could be a confounding factor in our analysis. The capacity of the present study to reach some statistically significant differences during in vitro antibody characterization might be limited since no formal sample size calculation was performed and both study groups (S and M) were unbalanced regarding the number of included participants.
In summary, we demonstrated that patients with severe COVID-19 display high levels of S2-specific IgG, and IgA responses targeting the Spike (RBD and S2 domains) as well as the Nucleocapsid, which appears to be responsible for the increased Fc-dependent activity reported in these individuals. However, no major differences were observed between antibodies from individuals with severe and mild COVID 19, beyond their levels, suggesting that these antibodies are not major determinants of disease severity. Differences in the avidity among anti-RBD, -S2 and -N antibody responses suggest the coexistence of several SARS-CoV-2 B cell activation pathways that might depend on the target viral antigens. Finally, preexisting or early anti-S2 non-neutralizing antibodies may contribute to disease protection in GSHs, with factors such as sex and age significantly modulating the final disease outcome. Further research is needed to uncover the factors that regulate the balance between antibody-dependent disease prevention and progression, particularly during the early steps of the immune response.
Materials and methods
Study design and participants
This work includes plasma samples from participants of two different cohorts: the KING cohort extension and the AbSeverCOVID cohort, which were conducted at the Hospital Universitari Germans Trias i Pujol (Badalona, Spain) with previous Institutional Review Board approval (PI-20-217 and PI-21–130, respectively). All ethical regulations relevant to human research participants were followed. In this observational study, we initially included 112 patients (age > 18 years) that had been infected with SARS-CoV-2. Patients with the following conditions were excluded: previous SARS-CoV-2 vaccination, primary immunodeficiencies, or an active form of cancer. Participants were not screened for prior SARS-CoV-2 infection. All participants provided written informed consent and had a documented positive RT-qPCR result from nasopharyngeal swab, positive antigen, and/or antibody detection. No formal sample size calculation was performed. Samples were collected within 31 days post-onset of symptoms between March 13th 2020, and August 31st 2021. Additional details of all patients are described in Table 1.
Antigen-specific human IgG, IgA, and IgM ELISA
Plates were coated with mouse anti-histidine tag monoclonal antibody (ThermoFisher MA1-21315) at 2 μg/ml and incubated at 4 °C overnight. The next day, plates were washed and 1% bovine serum albumin (BSA) in 1x PBS (blocking buffer) was added for 2 h at room temperature. The following SARS-CoV-2 proteins from SinoBiological were diluted to 1 μg/ml: N (40588-V08B), and S2 (40590-V08B) and RBD subunits (40592-V08H). Envelope protein was purchased from Novus Biologicals (NBP2-90986) and was also diluted to 1 μg/ml. While the corresponding protein was added to half a plate and incubated overnight at 4 °C, the other half received blocking buffer to establish absorbance background. Diluted plasmas, serially diluted standards (plasma samples from convalescent SARS-CoV-2 individuals with high anti-SARS-CoV-2 antibodies titers), and positive (plasma from a SARS-CoV-2 infected individual different to the one used for standard) and negative (pool of ten pre-pandemic plasmas) controls were then added and incubated for 1 h at room temperature. After washing, HRP goat anti-human IgG, IgA, or IgM antibodies (Jackson ImmunoResearch 109-036-098, 109-035-011, and 109-036-129, respectively) were added to adequate wells. Plates were incubated for 30 min at room temperature and washed again. Lastly, plates were developed with o-phenylenediamine dihydrochloride (OPD, Sigma Aldrich P8412-50TAB). The reaction was stopped with 2N H2SO4 and all plates were read at 429 nm with noise correction at 620 nm using an EnSight Multimode Plate Reader and the Kaleido Data Acquisition and Analysis Software (PerkinElmer).
Antigen-specific human IgG and IgA Avidity ELISA
For antibody avidity determination we used the guanidine HCl method64 with modifications. Plates were coated with S2, RBD, or Nucleocapsid at 1 μg/mL (SinoBiological 40590-V08B, 40592-V08H, and 40588-V08B, respectively) and incubated overnight at 4 °C. The following day, blocking buffer was added to all wells for 2 h at room temperature. Samples that had detectable SARS-CoV-2-specific IgG or IgA antibodies were diluted to 0.5 arbitrary units/ml and incubated for 1 h. Each sample was assessed in quadruplicate. Of the four wells, two were treated with 2 M guanidine HCl for 15 min, while the other two received 1x PBS. After washing, plates were incubated with HRP goat anti-human IgG or IgA (Jackson ImmunoResearch 109-036-098 and 109-035-011, respectively). Bound antibodies were detected using OPD. The reaction was stopped with 2 N H2SO4 and read at 429 nm and 620 nm, as described above. Avidity index was calculated as the ratio between mean signal obtained with and without guanidine HCl treatment.
Pseudovirus-based neutralization assay
Plasma neutralizing antibody titers against SARS-CoV-2 WH1 strain were determined using an HIV-based luciferase reporter pseudovirus generated as previously described1. Serial dilutions of heat-inactivated plasma samples were preincubated with 200 TCID50 of pseudovirus supernatant for 30 min at 37 °C before adding this mixture onto human ACE2-overexpressing HEK293T cells. After 48 h, cells were lysed with BriteLite plus luciferase reagent (PerkinElmer 6066761) and luminescence was measured using an EnSight plate reader. Plasma neutralization capacity was calculated and expressed as inhibitory dilution 50 (ID50) using Prism 9.0.2 (GraphPad Software, USA).
Antibody-dependent cellular phagocytosis
The ability of plasma antibodies to mediate phagocytosis was determined using SARS-CoV-2-coated fluorescent beads (Luminex MC10026-01) and HL-60 cells. Fluorescent beads were coated with RBD, S2, or Nucleocapsid (SinoBiological 40590-V08B, 40592-V08H, and 40588-V08B, respectively) as indicated by the manufacturer. Next, 2 × 104 antigen-coated fluorescent beads were incubated with diluted plasma (1: 1000) for 30 min at room temperature. HL-60 cells were then added onto each well and incubated for 4 h at 37 °C. Antigen-free beads were used to establish background. Cells were then fixed with 1% formaldehyde and acquired using a LSRII flow cytometer (BD Biosciences). Phagocytosis was calculated based on bead-uptake percentage of cells. Normalization with IgG or IgA values was performed by dividing ADCP values by the levels of IgG or IgA and multiplied by 100.
Antibody-dependent NK cell activation
To detect antigen-specific NK cell activation, we combined diluted plasma (1:100) with 105 magnetic beads (ThermoFisher 14305D) coated with S2, RBD, or Nucleocapsid (SinoBiological 40590-V08B, 40592-V08H, and 40588-V08B, respectively) for 30 min at room temperature, while shaking. Antigen-free beads were used to establish background. Next, antibody-bead immunocomplexes were washed, and incubated with the human CD16 + KHYG-1 cell line41 (50,000 cells per well), CD107a PE (BD Biosciences 555801, clone H4A3), and GolgiStop (BD Biosciences 554724) for 4 h at 37 °C. This experiment was also performed using a subset of plasma and untouched primary NK cells isolated from blood donors using the RossetteSep human NK cell enrichment cocktail kit (StemCell 15025). Lastly, cells were washed with 1x PBS and fixed with 1% formaldehyde before acquisition on LSRII flow cytometer (BD Biosciences). When using purified primary cells, we conducted a 20-min staining prior to cell fixation with the following antibodies: CD14 PerCPCy5.5 (BD Biosciences 550787, clone M5E2), CD3 BV510 (BD Biosciences 566779, clone OKT3), CD19 BV510 (Biolegend 363020, clone SJ25C1), and CD56 APC (BD Biosciences 555518, clone B159). Antibody-dependent NK degranulation percentage was defined as CD107-expressing CD16 + KHYG-1 cells or CD3-CD19-CD14-CD56+ cells. Normalization with IgG was performed by dividing ADCC values by the levels of IgG multiplied by 100.
Macrophage cytokine and chemokine measurement
Monocyte-derived macrophages were isolated from blood donors with magnetic beads by negative selection (Miltenyi 130-117-337) from three donors (one donor per experiment) and cultured in 24-well plates (250.000 cells/well in 500 µl) in presence of 100 ng/ml of recombinant human M-CSF (R&D systems 216-MC-025) in RPMI with 10% heat inactivated FBS at 37°C and 5% CO2. Medium feeding and M-CSF replacing was performed on days 3 and 6 of macrophage differentiation. On day 7, SARS-CoV-2 clinical isolate Delta grown in Vero E6 cells (GISAID EPI_ISL_3342900; at a MOI of 2) or supplemented media (no virus) was incubated for 1 h at 37°C and 5% CO2 with purified IgG or IgA (at 10 µg/ml) from five individuals from either Group S or M. LPS (10 ng/ml, Sigma-Aldrich L4391) was used as a positive control. After this time, the mix of samples with virus or media were transferred to the macrophage’s wells and incubated for 24 h at 37°C with 5% CO2. Supernatants were subsequently collected at 24 h post-infection to perform cytokines analysis by Luminex (Life Technologies ProcartaPlex 14 plex).
Animal studies
All animal studies were performed under the approval of the ethical committee on animal experimentation of Generalitat de Catalunya (code 11433), and the authorization of IRTA-CReSA’s biosecurity committee (CBS-128). We have complied with all relevant ethical regulations for animal use. A formal protocol describing the research question, key design features, and analysis plan was not prepared in advance. However, the project, including the described information, was presented in advance to “La Maraton de TV3” (Barcelona, Spain) for funding. All procedures were performed in an animal BSL-3 facility, and 8-week-old GSHs (males and females) (HsdHan®:AURA) were obtained from Envigo and kept in a 12-h light-dark cycle, at 22 °C and 70% relative humidity. Animals were housed in social groups with environmental enrichment, and they were fed ad libitum. The order of treatment administration and weigh determination in each study group was ramdonly assigned. Animal treatment and monitoring were not blinded. However, all in vitro determinations (i.e viral load, ELISA and histopathological analysis) were performed in a blinded fashion manner. The primary outcome was the percentage of weight reduction on day 7 post-SARS-CoV-2 challenge. Sample size calculation was performed using the epitool software (https://epitools.ausvet.com.au/samplesize) assuming equal variance among groups, a power of 0.8, a risk alpha of 0.05, a variance of 5 and a 10% reduction of body weight compared with uninfected animals. A 20% weight reduction, severe respiratory distress, lack of response to stimuli or blood in faeces were established as humane endpoints. None animal reached humane endpoint.
S2 immunization
After acclimatation (one week), a total of 54 GSHs (1:1 sex ratio) were ramdomly distributed into four groups. Group 1 (n = 18) was vaccinated twice (three weeks apart) with S2 protein mixed with aluminium hydroxide gel (AlOH, InvivoGen vac-alu-50). Group 2 (n = 18) received sterile 1x PBS mixed with AlOH at the same time points indicated for Group 1. Group 3 (n = 12) and Group 4 (n = 6) were not immunized and were used as positive and negative control groups, respectively. Twenty-seven days after the first dose, all animals (except Group 4) were intranasally inoculated with 104 TCID50/animal of a B.1 SARS-CoV-2 isolate (Cat01). The SARS-CoV-2 Cat01 isolate (GISAID ID EPI_ISL_510689) used in all GSH experiments was isolated from the nasopharyngeal aspirate specimen of a patient with laboratory-confirmed COVID-19 in Barcelona (Spain). Virus was amplified in Vero E6 cells. Viral titres were determined by the median TCID assay. Animals were monitored for weight and clinical signs for 14 days. Animals were euthanised on days 4, 7, and 14 by pentobarbital administration (200 mg/Kg) under isofluorane sedation, and blood was collected for immunological analyses. Oropharyngeal swabs, nasal turbinate, and lungs were collected for virological and histological analyses.
S2 production and purification
S2 subunit with a 8xhistidine tail was cloned into a pcDNA3.4 backbone and transfected following the Expi293 transfection manufacturer’s protocol. Five days after, this histidine-tag protein was purified using Ni Sepharose Excel resin (Cytiva 17371201).
Human antibody administration
After acclimatation (one week), forty-eight GSHs (1:1 sex ratio) were ramdomly distributed among four groups (n = 12/group). Each group received purified RBD-depleted IgG or IgA antibodies isolated from pooled plasma of individuals from either Group S or M. Twelve additional GSHs were administered with purified IgG (n = 6, 1:1 sex ratio) or IgA (n = 6 1:1 sex ratio) isolated from pre-pandemic plasmas. Antibodies were administered 24 h prior to viral challenge at 30 mg/kg. The positive and negative control groups used in this study were the same ones described in S2 immunization section. Doses were adjusted according to antibody concentration and weight of each animal. Animals were euthanised as indicated above.
Sequential human Ig purification and RBD-specific Ig depletion
Two pools of plasmas, one per study group, containing five plasma samples from different individuals (10 mL from each one) with high levels of anti-S2 antibodies, were prepared for sequential IgG and IgA purification. An additional pool of 15 pre-pandemic plasmas (2 mL each one) was prepared for control IgG and IgA preparations. Purifications were performed using protein G (Cytiva GE17-0618-01) and anti-IgA affinity matrix (ThermoFisher 2943972005), respectively. Once we obtained purified pooled IgG and IgA from individuals with severe and mild COVID-19, we depleted these pools from RBD-specific immunoglobulins using NHS-activated Sepharose (Cytiva 17090601) that had been previously coated with RBD. Purifications were performed according to the manufacturer’s protocol.
Antigen-specific GSH IgG ELISA
Half of the plates were coated with S2 (SinoBiological 40590-V08B) or RBD subunits (SinoBiological 40592-V08H) at 1 μg/ml, while the other half received 1x PBS to establish absorbance background. Plates were incubated at 4 °C overnight. The following day, plates were washed and blocked with blocking buffer for 2 h at room temperature. Next, diluted plasmas, serially diluted standards, and controls were then added and incubated at 4 °C overnight. After washing, a HRP goat anti-hamster antibody (Jackson ImmunoResearch 107-035-142) was added to all wells for 1 h at room temperature. Plates were washed again and developed with OPD (Sigma Aldrich P8412-50TAB). The reaction was stopped with 2 N H2SO4 and all plates were read at 429 nm with noise correction at 620 nm.
Quantification of human IgG- or IgA-specific antibodies in GSH
Half of the plates were coated with purified anti-human IgG (Jackson ImmunoResearch 109-006-098) or IgA antibodies (Jackson ImmunoResearch 109-005-011) at 1 μg/ml. The other half plate covered with 1x PBS. Plates were incubated at 4 °C overnight. The following day, plates were washed and blocked with blocking buffer for 2 h at room temperature. Next, diluted plasmas, serially diluted standards, and controls were then added and incubated at 4 °C overnight. After washing, the corresponding HRP goat anti-human IgG or IgA (Jackson ImmunoResearch 109-036-098 or 109-035-011, respectively) was added to all wells for 1 h at room temperature. Plates were washed again and developed with OPD (Sigma Aldrich P8412-50TAB). The reaction was stopped with 2 N H2SO4 and all plates were read at 429 nm with noise correction at 620 nm.
Measurement of viral RNA load (RT-qPCR)
Automated RNA extraction of each extracted tissue (oropharyngeal swab, nasal turbinate, and lungs) was performed using the IndiMag pathogen kit (Indical) and Biosprint 96 workstation, according to the manufacturer’s instructions. gRNA RT-qPCR was based on Corman et al.65, adapted to the AgPath-ID One-Step RT-PCR Kit (Life Technologies 4387424). The prime and probe were designed to match the envelope gene in the SARS-CoV-2 genome and were as follows: Primer 1 5’ ACAGGTACGTTAATAGTTAATAGCGT 3’; Primer 2 5’ ATATTGCAGCAGTACGCACACA 3’; Probe 5’ FAM-ACACTAGCCATCCTTACTGCGCTTCG-TAMRA 3’. Ten-fold serial dilutions of a SARS-CoV-2 plasmid starting at 2 × 105 copies/μL was used as standard (Integrated DNA Technologies, 10006896). Alternatively, in the absence of the commercial plasmid, the SARS-CoV-2 Cat02 isolate (GISAID ID EPI_ISL_47147) was utilized, which had been previously quantified with the same protocol.
Pathology and immunohistochemistry
Formalin-fixed tissue samples (lungs and nasal turbinates) were processed for histopathology, and haematoxylin and eosin stained slides were examined under an optical microscope. A semi-quantitative score based on the amount of inflammation and the severity of the observed lesions was assigned to each sample (0 = none, 1 = mild, 2 = moderate, 3 = severe)66. Immunohistochemistry was used to detect SARS-CoV-2 Nucleoprotein using the rabbit monoclonal antibody (SinoBiological 40143-R019) at 1: 15,000, followed by an Envision + /HRP-conjugated anti-rabbit secondary antibody (Agilent-Dako K4065). A semi-quantitative score was assigned to each tissue sample, according to the amount and distribution of the Nucleoprotein-specific staining (0 = absence of viral antigen; 1 = low amount, multifocal localization; 2 = moderate amount, multifocal localization; 3 = high amount, multifocal to diffuse localization)66.
Statistics and reproducibility
Binding antibody titers between Groups S and M were analyzed using the Peto-Peto test for censored samples. Avidity, ADCP, and ADCC activity differences between groups were assessed with the Mann–Whitney U test, while avidity differences among antigens were evaluated using the Kruskal-Wallis test with false discovery rate (FDR) correction. Dose-response neutralization curves were fitted to a logistic equation via non-linear regression. A significance threshold of 0.05 was applied for all statistical tests, and all p-values were reported as two-tailed. Statistical analyses were conducted using the R software environment and GraphPad Prism v8.0.
In our animal studies, neutralizing and binding antibody titers in GSHs were compared across groups using the Peto-Peto test with pairwise multiple comparisons, and longitudinal data were analyzed using the Prentice-Wilcoxon test with Conover’s post-hoc correction. Weight differences among groups were examined using the Kruskal Wallis test with FDR correction or the Friedman test with FDR correction (control group treated with antibodies from pre-pandemic samples). Viral loads and TCID50 values were analyzed with the Kruskal Wallis test followed by Conover’s post-hoc analysis, both FDR-adjusted. Histopathology and immunohistochemistry results were evaluated using the asymptotic generalized Pearson chi-squared test, also corrected for FDR. No data were excluded from the analysises.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data are included in the published version of the article, and its supplementary information files. All data can be found in Supplementary Data. .1
References
Pradenas, E. et al. Stable neutralizing antibody levels 6 months after mild and severe COVID-19 episodes. Med. 2, 313–320.e4 (2021).
Long, Q.-X. et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 26, 845–848 (2020).
Ma, H. et al. Serum IgA, IgM, and IgG responses in COVID-19. Cell Mol. Immunol. 17, 773–775 (2020).
Suthar, M. S. et al. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Rep. Med. 1, 100040 (2020).
Amellal, H. et al. Kinetics of specific anti-SARS-CoV-2 IgM, IgA, and IgG responses during the first 12 months after SARS-CoV-2 infection: a prospective longitudinal study. PLoS ONE 18 (2023).
Isho, B. et al. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci. Immunol. 5 (2020).
Röltgen, K. et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci. Immunol. 5, 1–20 (2020).
Qi, H., Liu, B., Wang, X. & Zhang, L. The humoral response and antibodies against SARS-CoV-2 infection. Nat. Immunol. 23, 1008–1020 (2022).
Sun, B. et al. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg. Microbes Infect. 9, 940–948 (2020).
Trinité, B. et al. SARS-CoV-2 infection elicits a rapid neutralizing antibody response that correlates with disease severity. Sci. Rep. 11, 2608 (2021).
Williamson, E. J. et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature 584, 430–436 (2020).
Richardson, S. et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA 323, 2052–2059 (2020).
Atyeo, C. et al. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity 53, 524–532.e4 (2020).
Zohar, T. et al. Compromised humoral functional evolution tracks with SARS-CoV-2 mortality. Cell 183, 1508–1519.e12 (2020).
Blanco-Melo, D. et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181, 1036–1045.e9 (2020).
Schulte-Schrepping, J. et al. Severe COVID-19 is marked by a dysregulated myeloid. Cell Compartment. Cell 182, 1419–1440.e23 (2020).
Rydyznski Moderbacher, C. et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell 183, 996–1012.e19 (2020).
Lucas, C. et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 584, 463–469 (2020).
Lu, Q. et al. SARS-CoV-2 exacerbates proinflammatory responses in myeloid cells through C-type lectin receptors and Tweety family member 2. Immunity 54, 1304–1319.e9 (2021).
Veras, F. P. et al. SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology. J. Exp. Med. 217 (2020).
Krämer, B. et al. Early IFN-α signatures and persistent dysfunction are distinguishing features of NK cells in severe COVID-19. Immunity 54, 2650–2669.e14 (2021).
Gralinski, L. E. et al. Complement activation contributes to severe acute respiratory syndrome coronavirus pathogenesis. mBio. 9, 1–15 (2018).
Holter, J. C. et al. Systemic complement activation is associated with respiratory failure in COVID-19 hospitalized patients. Proc. Natl Acad. Sci. USA 117, 25018–25025 (2020).
Carvelli, J. et al. Association of COVID-19 inflammation with activation of the C5a–C5aR1 axis. Nature 588, 146–150 (2020).
Chen, S. T. et al. A shift in lung macrophage composition is associated with COVID-19 severity and recovery. Sci. Transl. Med. 14 (2022).
Sefik, E. et al. Inflammasome activation in infected macrophages drives COVID-19 pathology. Nature 606, 585–593 (2022).
Lee, M. J. & Blish, C. A. Defining the role of natural killer cells in COVID-19. Nat. Immunol. 24, 1628–1638 (2023).
Zuo, Y. et al. Neutrophil extracellular traps in COVID-19. JCI Insight 5 (2020).
LaSalle, T. J. et al. Longitudinal characterization of circulating neutrophils uncovers phenotypes associated with severity in hospitalized COVID-19 patients. Cell Rep. Med. 3 (2022).
Yang, L. et al. The signal pathways and treatment of cytokine storm in COVID-19. Signal Transduct. Target Ther. 6, 255 (2021).
Liu, L. et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight 4 (2019).
Larsen, M. D. et al. Afucosylated IgG characterizes enveloped viral responses and correlates with COVID-19 severity. Science 371 (2021).
Hoepel, W. et al. High titers and low fucosylation of early human anti-SARS-CoV-2 IgG promote inflammation by alveolar macrophages. Sci. Transl. Med. 13, 1–17 (2021).
Chakraborty, S. et al. Proinflammatory IgG Fc structures in patients with severe COVID-19. Nat. Immunol. 22, 67–73 (2021).
Zervou, F. N. et al. SARS-CoV-2 antibodies: IgA correlates with severity of disease in early COVID-19 infection. J. Med Virol. 93, 5409–5415 (2021).
Sterlin, D. et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 13, 2223 (2021).
Stacey, H. D. et al. IgA potentiates NETosis in response to viral infection. Proc. Natl Acad. Sci. USA 118 (2021).
Tortorici, M. A. et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science 370, 950–957 (2020).
Zohar, T. & Alter, G. Dissecting antibody-mediated protection against SARS-CoV-2. Nat. Rev. Immunol. 20, 392–394 (2020).
Khoury, D. S. et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 27, 1205–1211 (2021).
Alpert, M. D. et al. A novel assay for antibody-dependent cell-mediated cytotoxicity against HIV-1- or SIV-infected cells reveals incomplete overlap with antibodies measured by neutralization and binding assays. J. Virol. 86, 12039–12052 (2012).
Berlin, D. A., Gulick, R. M. & Martinez, F. J. Severe Covid-19. N. Engl. J. Med. 383, 2451–2460 (2020).
Huang, C. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020).
Chen, G. et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 130, 2620–2629 (2020).
Kuri-Cervantes, L. et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 5 (2020).
Qu, J. et al. Profile of immunoglobulin G and IgM antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 71, 2255–2258 (2020).
Pradenas, E. et al. Clinical course impacts early kinetics,magnitude, and amplitude of SARS-CoV-2 neutralizing antibodies beyond 1 year after infection. Cell Rep. Med. 3, 100523 (2022).
Wang, Z. et al. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci. Transl. Med. 13, 1555 (2021).
Fröberg, J. et al. SARS-CoV-2 mucosal antibody development and persistence and their relation to viral load and COVID-19 symptoms. Nat. Commun. 12, (2021).
Planchais, C. et al. Potent human broadly SARS-CoV-2–neutralizing IgA and IgG antibodies effective against Omicron BA.1 and BA.2. J. Exp. Med. 219 (2022).
Junqueira, C. et al. FcγR-mediated SARS-CoV-2 infection of monocytes activates inflammation. Nature 606, 576–584 (2022).
Kaneko, N. et al. Loss of Bcl-6-expressing T follicular helper. Cells Germinal Cent. COVID-19. Cell 183, 143–157.e13 (2020).
Andreano, E. & Rappuoli, R. Immunodominant antibody germlines in COVID-19. J. Exp. Med. 218 (2021).
Woodruff, M. C. et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat. Immunol. 21, 1506–1516 (2020).
Regev-Yochay, G. et al. Correlates of protection against COVID-19 infection and intensity of symptomatic disease in vaccinated individuals exposed to SARS-CoV-2 in households in Israel (ICoFS): a prospective cohort study. Lancet Microbe 4, e309–e318 (2023).
Krapić, M., Kavazović, I. & Wensveen, F. M. Immunological mechanisms of sickness behavior in viral infection. Viruses 13, 2245 (2021).
Zhou, B. et al. SARS-CoV-2 hijacks neutralizing dimeric IgA for nasal infection and injury in Syrian hamsters1. Emerg. Microbes Infect. 12 (2023).
Li, C.-J. & Chang, S.-C. SARS-CoV-2 spike S2-specific neutralizing antibodies. Emerg. Microbes Infect. 12, 2220582 (2023).
Guerrero Manriquez, G. G. & Tuero, I. Adjuvants: friends in vaccine formulations against infectious diseases. Hum. Vaccin Immunother. 17, 3539–3550 (2021).
Dillard, J. A. et al. Adjuvant-dependent impact of inactivated SARS-CoV-2 vaccines during heterologous infection by a SARS-related coronavirus. Nat. Commun. 15 (2024).
Ebenig, A. et al. Vaccine-associated enhanced respiratory pathology in COVID-19 hamsters after TH2-biased immunization. Cell Rep. 40 (2022).
Kuo, S. M. Gender difference in bacteria endotoxin-induced inflammatory and anorexic responses. PLoS ONE 11 (2016).
Usai, C. et al. Immunisation efficacy of a stabilised SARS-CoV-2 spike glycoprotein in two geriatric animal models. NPJ Vaccines 9, 48 (2024).
Dauner, J. G. et al. Development and application of a GuHCl-modified ELISA to measure the avidity of anti-HPV L1 VLP antibodies in vaccinated individuals. Mol. Cell Probes 26, 73–80 (2012).
Corman, V. M. et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 25, 2000045 (2020).
Brustolin, M. et al. Protection against reinfection with D614- or G614-SARS-CoV-2 isolates in golden Syrian hamster. Emerg. Microbes Infect. 12, 1–36 (2021).
Acknowledgements
The authors would like to thank all the patients who participated in this study, the scientific and technical services at IrsiCaixa, and the people who are supporting our research through the MaratoTV3 (618/C/2021), CERCA Programme/Generalitat de Catalunya, Direcció General de Recerca i Innovació en Salut (SLD0015 and SLD0016), Centro de Investigación Biomédica en Red de Enfermedades Infecciosas, CIBERINFEC, Ministerio de Ciencia e Innovación and Unión Europea—NextGenerationEU, and the following crowdfunding projects: “YomeCorono”, BonPreu/Esclat, and Correos. In addition, this research was supported partially by the AGAUR (Generalitat de Catalunya), 2021 SGR 00452 (GRC. GRUP DE RECERCA BÀSICA I CLINICA A LES MALALTIES INFECCIOSES. CAN and EP were supported by Generalitat de Catalunya and Fons Social Europeu (2020 FI_B_0742) and National Agency for Research and Development of Chile (ANID: 72180406), respectively. NPL was funded by Juan de la Cierva postdoctoral fellowship (FJC2021-047205-I). MM was granted with RYC2020-028934-I/AEI/10.13039/501100011033 from Spanish Ministry of Science and Innovation and State Research Agency and the European Social Fund “investing in your future”. Drawing in Figs. 5 and 7 were created with BioRender.com.
Author information
Authors and Affiliations
Contributions
Study conception, design, and funding: J.C., B.C., J.S., J.V., N.I.U.; Manuscript draft preparation: N.P.L., J.C.; Data acquisition, analysis, and interpretation: N.P.L., F.T.F., C.U., D.R.R., S.B., E.A.E., J.C., E.P., C.A.N., N.R., G.C., M.P., M.L.R., M.Mat., B.R., AH., J.A.C., B.M., J.M., L.M., M.Mas., N.I.U., J.B., J.S., J.V., J.C.
Corresponding authors
Ethics declarations
Competing interests
N.I.U. is supported by institutional funding from Pharma Mar, HIPRA, Amassence, and Palobiofarma. The authors declare no other competing interests.
Peer review
Peer review information
Communications Biology thanks Marco Polo Peralta and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Isabela Pedroza-Pacheco and Dario Ummarino. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Pedreño-López, N., Tarrés-Freixas, F., Usai, C. et al. Anti-SARS-CoV-2 antibodies from severe COVID-19 individuals or S2 immunizations do not worsen disease in hamsters. Commun Biol 8, 1643 (2025). https://doi.org/10.1038/s42003-025-09034-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-09034-3