Abstract
Prokaryotic Argonaute proteins (pAgos) are programmable nucleases with potential for the development of novel nucleic acid manipulation tools. Previous studies have shown that several pAgos exhibit strong preferences for either DNA or RNA targets, and effective cleavage of plasmid or linear double-stranded DNA (dsDNA) is typically limited to regions with low GC content. Here, we characterize a pAgo, GgeAgo from the thermophilic organism Geobacillus genomosp. 3. GgeAgo utilizes 5’ phosphorylated DNA guides to cleave both DNA and RNA with high efficiency and specificity under a wide range of conditions. Notably, GgeAgo efficiently cleaves both plasmid and linear dsDNA with GC content up to 53% at elevated temperatures, and retains detectable activity even at 64% GC. Furthermore, GgeAgo demonstrates robust activity in practical applications, including DNA cloning, nucleic acid detection, and enrichment of rare variants, with high specificity and sensitivity. These results suggest that GgeAgo holds promise as an effective DNA and RNA manipulation tool for biotechnology.

Similar content being viewed by others
Introduction
Argonaute (Ago) and CRISPR-Cas proteins are two major programmable nucleases that use DNA or RNA guides to recognize and cleave their targets, playing a central role in protecting host cells from invading nucleic acids1,2,3,4. Cas proteins utilize RNA guides transcribed from the CRISPR arrays to destroy invading foreign DNA or RNA5,6. Eukaryotic Argonaute proteins (eAgos) are core components of the RNA-induced silencing complex (RISC), and bind small RNAs processed from viral RNA or transcribed from genomic loci as guides to silence RNA targets7,8,9. In contrast, most studied prokaryotic Argonautes (pAgos) use small DNA guides (gDNAs) generated from invader elements or genomic sequences to act on DNA targets10,11,12,13,14,15,16,17,18,19,20. Some pAgos have also been found to use RNA guides (gRNAs)2,21,22,23, while a smaller subset employs gDNAs with a preference for RNA targets3,24,25. Previous genomic studies have shown that the diversity of pAgos is far greater than that of eAgos, with genes encoding pAgo proteins present in 9% of bacterial and 32% of archaeal genomes26. As programmable nucleases with diverse binding and cleavage activities, several pAgos have successfully been used in various molecular applications, including nucleic acid detection, molecular cloning, and DNA assembly27,28,29,30,31,32,33,34,35. Notably, compared to the Cas nucleases that use longer RNA guides, pAgos employ shorter DNA oligonucleotides as guides, which are more stable, cost-effective, and easier to synthesize16,28,29. Additionally, pAgos do not require specific motifs in the target sequence for guide binding or target recognition. This simplicity, combined with their programmability and specificity, positions pAgos as promising candidates for developing novel nucleic acid manipulation tools.
Phylogenetic analyses have revealed that pAgos can be classified into three major clades, including long-A (active), long-B (inactive) and short pAgos4,8,26. The long pAgo proteins contain six main domains, the N-terminal, L1 (linker 1), PAZ (Piwi-Argonaute-Zwille), MID (Middle) and PIWI (P-element induced wimpy testis), while short pAgos lack the PAZ and N domains but include the MID and PIWI domains1,26. The MID and PAZ domains are responsible for binding the 5’ and 3’ ends of the guide molecule, respectively. Long-A pAgos possess a conserved catalytic tetrad DEDX (X is D, H, or K) in the PIWI domain, which coordinates two essential divalent metal ions necessary for catalytic activity. When programmed with small nucleic acid guides, active pAgos can cleave complementary targets between the 10th and 11th positions of the guide strand in most cases1,4. However, since pAgos lack helicase function and contain only a single nuclease domain, they require two complementary guides to simultaneously nick the upper and lower strands of the target DNA sequence to generate double-strand breaks (DSBs)16,35.
Many studies have attempted to harness and develop pAgos as DNA-guided DNA nuclease tools27,29. Initial efforts primarily focused on pAgos from (hyper)thermophiles, such as PfAgo (Pyrococcus furiosus) and TtAgo (Thermus thermophilus), for targeting and cleaving plasmid DNA in vitro, leveraging high-temperature conditions for DNA denaturation12,30,31,34. Interestingly, several pAgos from mesophilic organisms, such as CbAgo (Clostridium butyricum) and KmAgo (Kurthia massiliensis), have demonstrated the ability to cleave supercoiled DNA substrates with low GC content16,17,18. In addition, PfAgo can cleave linear double-stranded DNA (dsDNA) by unwinding it at 95 °C30. TtAgo and CbAgo have shown cleavage activity on linear dsDNA but require the addition of specific single-strand binding proteins ET-SSB or the RecBC helicase, respectively, to facilitate the process35,36. Despite the characterization of over two dozen pAgos to date, many still face limitations in efficiently cleaving dsDNA targets with high GC content. While some pAgos, such as TtAgo and KmAgo, can also cleave RNA, their activity rates are generally lower compared to DNA substrates. In particular, a small subset of pAgos exhibits a preference for RNA cleavage at mesophilic temperatures in vitro. This group includes MbpAgo (Mucilaginibacter paludis), PliAgo (Pseudooceanicola lipolyticus) and PnyAgo (Pedobacter nyackensis), which hold potential as DNA-guided RNA nucleases3,24. These pAgos can cleave RNA substrates at mesophilic temperatures in vitro, with their activity influenced by the secondary structure of the RNA target, particularly in regions with partially double-stranded elements.
In this study, we identified a novel pAgo, GgeAgo from the thermophilic organism Geobacillus genomosp. 3. Biochemical characterization showed that GgeAgo uses gDNAs to cleave both single-stranded DNA (ssDNA) and RNA with high activity and specificity under a broad temperature range. Remarkably, GgeAgo mediates precise, guide-dependent cleavage of both plasmid and linear dsDNA with GC content up to 53% at elevated temperatures, while retaining detectable activity even at 64% GC. We further demonstrate its utility in DNA cloning and nucleic acid detection, highlighting GgeAgo as a promising tool for DNA and RNA manipulation in biotechnology.
Results
GgeAgo uses DNA guides to process ssDNA and ssRNA targets
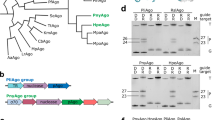
To identify highly active pAgos as programmable nucleases for nucleic acid manipulation, we chose the protein sequence of TtAgo (WP_011229221.1) as the query, given its demonstrated DNA and RNA cleavage activity with single-nucleotide precision. We used the web interface of the BLASTp program to search for thermophilic pAgos. GgeAgo from thermophilic bacteria, Geobacillus genomosp. 3, was chosen as the candidate for it is closely related to the omnipotent KmAgo by phylogenetic analysis (Supplementary Fig. 1a). Multiple sequence alignments showed that GgeAgo contains the conserved catalytic residues (D502, E537, D571, D686) in the PIWI domain (Supplementary Fig. 1b). We cloned the gene encoding GgeAgo into the pET-28a plasmid, expressed and purified the protein from E. coli. The purified GgeAgo showed high purity, consistent with the predicted molecular weight 82.3 kDa (Supplementary Fig. 1c, d). The catalytically inactive variant (GgeAgo_DM) was obtained by substituting the first and third residues of the DEDD catalytic tetrad residues (D502A/D571A) (Supplementary Fig. 1b).
To determine the endonuclease activity of GgeAgo, we performed in vitro cleavage assays using a set of synthetic guide and target oligonucleotides selected from the previous studies of the pAgos12,18 (Fig. 1a, b). GgeAgo was loaded with 5’ phosphorylated (5’ P) or 5’ hydroxylated (5’ OH) gDNAs or gRNAs (18 nt in length) at 50 °C for 10 min, followed by the addition of complementary 5’-end FAM-labeled 45-nt ssDNA or RNA targets. After incubation for 30 min at 50 °C, GgeAgo was shown to act as a programmable nuclease, using gDNAs to process almost all DNA and RNA targets (Fig.1c, d). In contrast, no cleavage products were observed with GgeAgo_DM, or in the absence of GgeAgo protein or guides (Fig. 1c–e). In addition, GgeAgo showed a strong preference for 5’P-guides, similarly to most thermophilic pAgos12,13,20,37. Cleavage occurred precisely at the expected site between positions 10 and 11 relative to the 5’ end of the guide, which is the canonical cleavage site observed in other characterized Ago proteins (Supplementary Fig. 1e, f). Therefore, GgeAgo can potentially be exploited for DNA and RNA manipulation with 5’P-gDNAs.
a Schematic diagram of the workflow during in vitro cleavage assay. GgeAgo cleaves a 45 nt substrate with 18 nt guides to generate 34 nt product. b Scheme of the guide and target nucleic acids. 5’-phosphorylated DNA guides were used in most experiments; the black triangle indicates the cleavage site. M1 and M2 correspond to the cleavage products of FAM-labeled DNA and RNA targets. c GgeAgo exhibits DNA-guided DNA endonuclease activity. d GgeAgo exhibits DNA-guided RNA endonuclease activity. e Catalytic dead mutant GgeAgo_DM with substitutions of two out of four catalytic tetrad residues (D502A/D571A) causes the loss of activity. Positions of the cleavage products and target are indicated on the left of the gels. GgeAgo or GgeAgo_DM, guide and target were mixed in a 4:2:1 molar ratio (800 nM GgeAgo preloaded with 400 nM guide in 5 mM Mn2+ for 10 min at 50 °C, plus 200 nM target) and incubated for 30 min at 50 °C. Lanes M1 and M2 contain chemically synthesized 34 nt DNA and RNA corresponding to the cleavage products of the DNA and the RNA target, respectively.
GgeAgo binds guides and functions efficiently under a wide range of reaction conditions
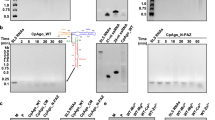
To determine the optimal reaction conditions for GgeAgo mediated target cleavage, the effects of temperature and the guide length on the cleavage efficiency were tested. Analysis of the temperature-dependent cleavage revealed that GgeAgo loaded with 5’P-gDNA was most active at 50-75 °C (Fig. 2a, b). DNA cleavage activity increased from 30 to 85 °C but was lost at 90 °C (Fig. 2a and Supplementary Fig. 2a). Moreover, GgeAgo was able to cleave DNA efficiently at physiological temperatures for 15 min, indicating that the ssDNA cleavage activity of GgeAgo is not strictly dependent on high-temperature conditions. For RNA substrates, GgeAgo displayed efficient cleavage above 50 °C, but the RNA target could be degraded under high temperature, especially over 75 °C (Fig. 2b and Supplementary Fig. 2b). Therefore, most experiments were performed at 50 °C.
Effects of temperature on DNA (a) and RNA (b) cleavage by GgeAgo. c Effects of the 5’P-gDNA length on nucleic acid cleavage by GgeAgo. Target cleavage was performed at 50 °C with guides of various lengths as indicated. The assays in a–c were performed for 15 min (DNA Target) or 20 min (RNA Target) at indicated temperatures. Data are represented as the means ± SD from three independent experiments. d Co-purified nucleic acids from GgeAgo expressed in E. coli treated with enzymes as indicated. M, ssDNA marker; -, untreated; R, RNase A; D, DNase I; DR, both nucleases. The protein expression was induced for 16 h at 18 °C or 6 h at 37 °C.
We further studied the effect of 5’P-gDNA length on target cleavage by GgeAgo. In contrast to previously reported eAgos and pAgos that function efficiently across a broad range of gDNA lengths (15–30 nt), GgeAgo exhibited maximum cleavage activity with 18 or 19 nt guides, with reduced efficiency observed for both shorter and longer guides (Fig. 2c and Supplementary Fig. 2c, d). The cleavage position was not shifted if shorter or longer gDNA were used, and efficient cleavage was only observed with guides of 16–21 nt (Supplementary Fig. 2c, d). To explore this preference for guide length exhibited by GgeAgo, the three-dimensional (3D) models built using AlphaFold338 revealed that the GgeAgo PAZ domain contains an extra loop compared to PfAgo13, which is able to adopt long guides for cleavage activity (Supplementary Fig. 3a). We further compared the structure models of GgeAgo binary complex with 16, 18 and 21 nt gDNA. The extra stick-out loop in GgeAgo likely narrowed the channel to bind the 3’-end of the guides, which may explain the preference for GgeAgo to use gDNA of this specific length (Supplementary Fig. 3b).
Previous studies have shown that pAgo proteins can bind small guide nucleic acids when expressed in either their native hosts or a heterologous E. coli system12,15,17,20,39,40. To investigate whether GgeAgo also associated guide nucleic acids when expressed in E. coli, we measured the A260/A280 ratio of the Ni-NTA purified GgeAgo expressed at 18 °C using the Thermo Scientific NanoDrop 8000 Spectrophotometer. The measured ratio was 0.55. Given that GgeAgo functions optimally at high temperatures, we also expressed GgeAgo at 37 °C, and the ratio of A260/280 of the purified GgeAgo was 0.58. Next, we extracted the co-purified nucleic acids and analyzed equivalent amounts of nucleic acids with DNase I or RNase A treatment (Fig. 2d). Small DNAs with a length around 18 nt in all situations were consistent with in vitro experiments, but RNAs with undefined length were observed in GgeAgo expressed at 37 °C, possibly due to the low RNA targeting activity of GgeAgo at 18 °C. Therefore, future work will involve cloning and sequencing the co-purified nucleic acids to determine their precise lengths and sequences, which will provide deeper insights into GgeAgo’s guide selection and cleavage mechanisms.
As divalent metal ions are crucial co-factors for Ago protein activity, we next investigated the performance of GgeAgo with various cations. GgeAgo exhibited DNA cleavage activity in the presence of Mn2+, Co2+, and Mg2+, while RNA cleavage was supported by Mn2+ and Mg2+, with Mn2+ showing the highest activity (Fig. 3a and Supplementary Fig. 4a). Titration of Mn2+ and Mg2+ showed that GgeAgo was active at Mn2+ concentrations ≥0.1 mM and displayed increased cleavage activity at Mn2+ concentrations >2.5 mM (Supplementary Fig. 4b). However, GgeAgo was still unable to efficiently cleave DNA or RNA target at Mg2+ concentrations up to 10 mM (Supplementary Fig. 4c). Analysis of target cleavage under various ionic strength conditions revealed that GgeAgo was active at 25–250 mM NaCl concentrations for DNA cleavage with the highest activity observed at 100 mM NaCl, while it retained comparable activity at 25–250 mM NaCl for RNA cleavage (Supplementary Fig. 4d). Finally, analysis of target cleavage at various pH conditions revealed that pH did not noticeably influence the efficiency of DNA cleavage within the tested range (6.5–9.0), but the highest activity of RNA cleavage was observed at pH 7.5, with degradation occurring at pH above 8.0 (Supplementary Fig. 4e). Therefore, the optimal conditions (10 mM HEPES-NaOH, 100 mM NaCl, 5 mM MnCl2, pH 7.5) were determined for the cleavage kinetic assays with 5’P or 5’OH guides.
a DNA-guided nucleic acid cleavage by GgeAgo with various divalent cation. The reactions were performed for 15 min (DNA Target) or 20 min (RNA Target) at 50 °C. b Kinetics of nucleic acid cleavage using 5’P-gDNA by GgeAgo measured with FAM-labeled target ssDNA or RNA at 50 °C. The data were fitted to a single-exponential equation, and the resulting kobs value is shown. c Binding of GgeAgo to 18 nt gDNAs by fluorescence polarization assay. GgeAgo binds 5’P-gDNA and 5’OH-gDNA with average KD values of 8.23 ± 1.47 and 11.14 ± 1.88 nM, respectively. Binding of GgeAgo-gDNA complex to DNA target (d) and RNA Target (e) by fluorescence polarization assay. GgeAgo_DM preloaded 5’P-gDNA or 5’OH-gDNA binds target DNA with average KD values of 8.69 ± 1.28 and 19.98 ± 2.29 nM, and RNA with 10.65 ± 1.11 and 32.21 ± 4.90 nM, respectively. Results are from three independent experiments. Error bars represent the SD.
Under single-turnover conditions with 5’P-gDNA, the observed rates (kobs) of DNA cleavage (0.234 ± 0.041 min–1) were almost three times faster than RNA cleavage (0.076 ± 0.011 min–1) at 50 °C (Fig. 3b and Supplementary Fig. 5a). However, only weak cleavage activity was detected when using 5’OH-gDNA, with significantly lower rates compared to 5’P-gDNA (Supplementary Fig. 5b). To explore the role of 5’ phosphate group in gDNA binding by GgeAgo, the equilibrium dissociation constants (Kd) were measured using fluorescence polarization assays (Fig. 3c). The results showed that the Kd value of GgeAgo associated with 5’P-gDNA (Kd: 8.23 ± 1.47 nM) was slightly lower than that for 5’OH-gDNA (Kd: 11.14 ± 1.88 nM) (Supplementary Fig. 5c). Furthermore, we measured the Kd value for target binding by the GgeAgo-gDNA complex (Fig. 3d, e). The Kd values of GgeAgo with 5’P-gDNA binding DNA and RNA targets were 8.69 ± 1.28 nM and 10.65 ± 1.11 respectively, notably lower than those of GgeAgo loaded with 5’OH-gDNA (19.98 ± 2.29 nM and 32.21 ± 4.90 nM, respectively) (Supplementary Fig. 5d). The higher affinity of GgeAgo for targets when using 5’P-gDNA likely explains its preference for 5’ phosphorylated guides. In addition, the real Kd values for guide binding by GgeAgo and target binding by the GgeAgo–gDNA complex are probably lower, owing to the detection limits of measurements.
Effects of the 5’-nucleotide of the guide and guide-target mismatches on target cleavage
Previous studies have demonstrated that some pAgos have a certain bias for the 5’-nucleotide of the guide1,12,19,41,42. To explore whether GgeAgo has a preference for the 5’-nucleotide of gDNAs, four gDNA variants with 5’-A, 5’-T, 5’-G, or 5’-C but otherwise identical sequences were used (Supplementary Table S1). Under single-turnover conditions with 5’P-gDNA, GgeAgo was able to cleave almost all complementary targets (DNA or RNA) mediated by all four gDNAs, though with the kobs values: 5’T > 5’A > 5’G > 5’C (Fig. 4a, b and Supplementary Fig. 6a–d).
Preferences for the 5’ nucleotide of the guide on DNA (a) and RNA (b) cleavage. The kobs values were determined from the single-exponential fits of the data. Effects of mismatches on DNA (c) and RNA (d) cleavage by GgeAgo. Data are the means ± SD from three independent measurements. The reactions were performed for 15 min (DNA Target) or 20 min (RNA Target) at 50 °C. *P < 0.05, ***P < 0.001 and ****P < 0.0001, compared to (c), the control reactions with guide containing no mismatches, using Student’s t test.
Precise recognition and cleavage of target nucleic acids are essential for the programmable nucleases used in DNA or RNA manipulation. To assess the specificity of GgeAgo, the effects of the guide-target mismatches on its cleavage activity were analyzed. In the effector complex of Agos, the guide molecule can be divided into five functionally distinct regions: the 5’ anchor (position 1), the seed region (positions 2–8), the central region (positions 9–12), the 3’ supplementary region (positions 13–15), and 3’ tail region (positions 16–18)16,17. Firstly, we performed the DNA and RNA cleavage at 50 °C for a fixed time with a set of gDNAs, each introduced a single-nucleotide mismatch at a certain position (Supplementary Table S1). Mismatches in the partial central and 3’ supplementary regions affected the cleavage efficiency (Fig. 4c, d and Supplementary Fig. 7a, b). The higher efficiency of RNA target cleavage was observed when the mismatches in positions 2–10, 13 and 16–17, possibly because such mismatches affect target positioning and/or induce conformational changes in the active site of GgeAgo during RNA catalysis16. Surprisingly, a dramatic decrease in cleavage efficiency was observed at position 11 for both DNA and RNA targets, and at positions 12 and 15 for RNA targets. To further explore these effects, the cleavage kinetics were performed with gDNAs bearing single-nucleotide mismatch at these critical positions (Supplementary Fig. 8a, b). The results revealed that the cleavage rates were markedly reduced, particularly for mismatches at position 12, which had the greatest impact on RNA cleavage efficiency. Moreover, Vladimir Panteleev et al. demonstrated that elevated temperatures can enhance the fidelity of target recognition by TceAgo during cleavage43. Thus, we examined the cleavage kinetics with the identical gDNAs bearing mismatch at elevated temperatures. Interestingly, while higher temperatures increased the fidelity of RNA target recognition, they reduced the fidelity for DNA targets (Supplementary Fig. 8c, d). In addition, we introduced dinucleotide mismatches to the gDNA at position 8-15, and dinucleotide mismatches at position 11–13 completely abolished DNA cleavage (Supplementary Fig. 8e). Overall, GgeAgo can serve as a tool to allow specific detection and cleavage of DNA and RNA targets.
GgeAgo mediates double-strand DNA cleavage and manipulation
Thermophilic or mesophilic pAgos have been successfully used to generate breaks in plasmid DNA at specific sites defined by paired gDNAs, with their cleavage efficiency being influenced by the GC content of the target dsDNA12,13,15,18,30,35,36. To determine the ability of GgeAgo to process double-stranded DNA, we first performed the cleavage reactions with GgeAgo loaded with a pair of gDNAs targeting the low GC region (29%) of the supercoiled plasmid. In the presence of gDNAs, GgeAgo completely linearized the plasmid within the 29% GC region (Supplementary Fig. 9a, lanes 4–5). In the absence of gDNAs, the substrate plasmid was converted from a supercoiled to relaxed (open circle, OC) state, accompanied by degradation—indicative of the guide-independent “chopping” activity observed in apo-pAgo proteins (without loading guides) (Supplementary Fig. 9a, lane 3)39. Previous studies have shown that pAgo loaded with guides could be more thermostable than their apo forms18,24. To investigate the thermostability of apo-GgeAgo versus GgeAgo-gDNA complex, we treated them at 50–90 °C for 30 min prior to performing DNA target cleavage. GgeAgo without gDNA lost its cleavage activity at temperatures above 65 °C (Supplementary Fig. 9b, upper panel). In contrast, GgeAgo preloaded with gDNA retained efficient DNA cleavage up to 75 °C (Supplementary Fig. 9b, lower panel). Moreover, no plasmid processing was observed in the absence of gDNAs after pre-incubation at 70 °C for 30 min, while complete plasmid cleavage occurred when gDNAs were present (Supplementary Fig. 9a, lanes 6–8). Thus, apo-GgeAgo can be inactivated by pre-treatment at 70 °C for 30 min, thereby eliminating the guide-independent ‘chopping’ activity. In the next set of experiments, we sought to determine the reaction temperature at which GgeAgo can use gDNAs to cleave plasmids with specific and efficient activity. GgeAgo exhibited specific and efficient plasmid cleavage between 37 and 70 °C, with activity increasing at higher temperatures. However, plasmid degradation was observed at temperatures above 70 °C (Supplementary Fig. 9c). Thus, we performed plasmid cleavage by GgeAgo-gDNA complexes at 70 °C after pre-treating the complexes at 70 °C for 30 min.
We further analyzed the dependence of plasmid cleavage on the GC-content of the target sites. Five sets of guides were designed to target regions with varying GC content in the commonly used plasmid pUC19 (Fig. 5a and Supplementary Table S2). After the incubation with two GgeAgo-gDNA complexes, the product was digested with a restriction enzyme (Hind III or Sca I) (Fig. 5b). Surprisingly, GgeAgo could cleave almost all plasmid DNA in regions with a GC-content below 45%, and the cleavage efficiency in the regions with 53% GC-content exceeded 50%. The cleavage site of the linear plasmid product was confirmed by DNA Sanger sequencing, which showed that GgeAgo cleaves at the expected position with high precision (Supplementary Fig. 10a). However, cleavage efficiency dropped significantly in regions with 64% GC content. Previous studies have shown that TtAgo exhibits a strong preference for plasmid substrates with lower GC-content and even for ssDNA substrates36. We therefore analyzed the effect of GC-content on the cleavage of ssDNA substrates by GgeAgo. These ssDNA targets (55 nt) are complementary to the gDNAs targeting DNA sites with different GC content in plasmid pUC19. We found that GC content did not significantly affect ssDNA cleavage efficiency (Supplementary Fig. 11a). Furthermore, we analyzed the effect of GC-content on the cleavage of linear dsDNA substrates by GgeAgo. Efficient cleavage of linear dsDNA substrates with GC-content below 53% was observed (Fig. 5c and Supplementary Fig. 11b, c). Hence, the reason GgeAgo cannot act on dsDNA substrates with GC-content above 64% may be that the dsDNA with high GC-content cannot be efficiently unwound at 75 °C, leading to the target inaccessibility to GgeAgo.
a Schematic overview of the pUC19 target plasmid. The target sites are indicated by blue polylines, while percentages indicate the GC content of the 80-bp segments where target sites are located (Lower). The target sites (indicated in blue) and complementary DNA guides used in this experiment (indicated in red), and black triangles indicate the cleavage sites (Upper). b Plasmid cleavage at DNA sites with different GC content by GgeAgo-gDNA complexes at 70 °C for 30 min. Cleavage products were digested with Hind III or Sca I and analyzed by agarose gel electrophoresis. The product sizes are indicated below; positions of specific cleavage products are indicated with asterisks; M, molecular weight marker; Lin, linearized plasmid. c Linear dsDNA with different GC content cleavage by GgeAgo-gDNA complexes at 70 °C for 30 min.
Based on the high activity and precision of GgeAgo for plasmid cleavage, we next investigated whether GgeAgo can be used for the insertion of DNA fragments at desired sites in the plasmid. For this experiment, we selected a GFP expression cassette (~1 kb in size) as the insert and the pUC19 plasmid as the target. First, the plasmid was linearized by GgeAgo with designed gDNAs at different locations with varied GC-content. The resulting digested products were used directly for DNA fragment insertion without additional purification steps. Second, two homologous regions of 20 bp, compatible with the ends of the digested plasmid, were introduced into the insert via PCR. The PCR products were then mixed with the digested plasmid and treated with 0.5 U T5 exonuclease in an ice-water bath for 7 min, following our previously established TLCL method44. Third, the mixture was transformed into E. coli to generate recombinant plasmids (Fig. 6a). The green positive clones expressing GFP proteins were counted to evaluate the efficiency of the cloning process (Fig. 6b). When plasmid DNA regions with a GC-content below 53% as target sites, the cloning accuracy exceeded 50% (Fig. 6c). Sanger sequencing of plasmids from positive clones revealed correct and seamless cloning (Supplementary Fig. 10b), demonstrating the potential of GgeAgo for precise and efficient plasmid manipulation.
a Schematic overview of site-specific insertion of DNA fragments into target plasmid using GgeAgo. First, the plasmid was digested by GgeAgo with a pair designed gDNAs at the desired location. Second, two homologous regions of 20 bp compatible with the ends of the digested plasmid were introduced into the destination DNA (the insert) via PCR. The PCR products were then mixed with the digested plasmid and treated with T5 exonuclease, following transformation into E. coli as our previously established TLCL method44. b The representative transformation plates showing cloning results with 10-fold dilution. GFP expression cassette as the insert fragment cloned into the different site of the pUC19 plasmid, digested by the GgeAgo-gDNA complex. The positive clones are green. c The cloning efficiency of the transformation. Colonies are defined as the total colony-forming unit (CFU). Cloning efficiency is the percentage of the number of positive clones (green) to the total number of clones. Data are the means ± SD from two experimental replicates.
In summary, GgeAgo can cleave dsDNA with the precision similar to restriction endonucleases or the Cas9 nuclease, but without strict sequence requirements (although the cleavage efficiency depends on the GC-content of the cleavage site).
GgeAgo-mediated virus RNA detection and variant allele enrichment
pAgos have been developed as molecular diagnostic tools based on their stepwise endonuclease activity, in which the products of the input guide-directed cleavage can serve as secondary guides, enabling free pAgos to cleave reporter molecules31,45,46,47. To evaluate the stepwise endonuclease activity of GgeAgo, the partially synthesized RNA fragment of the S (Spike) gene of SARS-Cov2 was chosen as the target to develop a GgeAgo Improved Nucleic acid detection system-GAIN (Fig. 7a and the synthesized sequence shown in Supplementary Fig. 12a). In this design, the target RNA is amplified via RT-PCR, then the antisense strand of the S gene amplicon is cleaved by GgeAgo with a designed gDNA to generate the secondary gDNA. This process generates secondary gDNA that directs GgeAgo to cleave a fluorescent reporter. First, we explored the effect of temperature on stepwise endonuclease activity, with the highest signals obtained at 75 °C (Supplementary Fig. 13a). Next, we determined the limit of detection (LoD) of the GAIN system using serial dilutions of the target RNA. The LoD was found to be 5 copies per reaction, with positive detection in 4 out of 9 replicates (Fig. 7c).
a Schematic illustration of the virus RNA detection workflow using GgeAgo. The target RNA is pre-amplified by RT-PCR, followed by GgeAgo cleavage. The antisense strand of amplicon is cleaved by GgeAgo with a designed gDNA (black line), generating secondary gDNAs (gray background) that direct GgeAgo to cleave a fluorescent reporter. b Scheme illustration of virus RNA variant alleles enrichment using GgeAgo. Before RT-PCR amplification, the wild-type (WT) RNA is selectively removed by GgeAgo with a gDNA (Blue line) targeting WT RNA. The remaining steps are the same as in (a). c Detection sensitivity of the strategy in a using diluted synthetic mock SARS-Cov2 S gene WT RNA. d Detection of the D614G mutant RNA with different mutation frequencies by the strategy in (b). Data are the means ± SD from three biological replicates. *P < 0.05, **P < 0.01 and ****P < 0.0001, compared to the control, using Student’s t test.
Based on the high specificity of GgeAgo for target RNA recognition, we developed a mutant enrichment version of GAIN (enGAIN) to discriminate between wild-type (WT) and the single-base mutant (Fig. 7b and Supplementary Fig. 12b). Using the SARS-Cov2 S gene-D614G mutant as an example, we designed a gDNA-WT to guide GgeAgo to specficly cut WT RNA before the GAIN reaction. According to the base-mismatch experiments, we first performed the cleavage reaction at 60 °C for 5 min, and WT and D614G mutant RNA can be distinguished by GgeAgo with gDNA-WT (Supplementary Fig. 13b). However, a small amount of WT RNA residue remained detectable. Subsequently, enrichment reactions were optimized at 70 °C. After 5 min of reaction, no detectable WT RNA was observed, yet there was no significant difference in fluorescent signals after the GAIN reaction (Supplementary Fig. 13c, d). This may be that there was still residual WT RNA, which cannot be detected by the SYBR-Gold dye. Unexpectedly, the maximum signal differentiation was achieved when the enrichment reactions were performed for 25 min. To validate whether enGAIN can be applied to detect a mutation carried at different frequencies, we used mixed templates of WT and the D614 mutant to obtain mutant frequencies of 0%, 0.01%, 0.1%, 1%, 10%, 25%, and 100% (Fig. 7d). The results demonstrated that enGAIN could reliably detect the D614G mutation at a frequency as low as 0.1%, and even 0.01%, with positive detection in 2 out of 9 replicates. This finding suggests that enGAIN, which can distinguish mutants with low mutation frequencies, is a powerful tool for the investigation of infectious viral variants.
Discussion
In this study, we identified a novel thermophilic GgeAgo and demonstrated that it can exclusively use 5’P-gDNA to efficiently cleave both DNA and RNA target with high precision and specificity. Previously, most studied thermophilic pAgos exhibited a strong preference for DNA targets, although AaAgo, MpAgo, TtAgo and TpsAgo can also cleave RNA with lower activity in vitro21,42,48,49. GgeAgo is able to cleave almost all DNA and RNA target with comparable activity at 60 °C (Supplementary Fig. 8c, d). Furthermore, the biochemical characterization revealed that GgeAgo can target single-stranded DNA and RNA under a wide range of reaction conditions, with cleavage efficiency and precision modulated by temperature, divalent ions, the length and phosphorylation of the gDNA and its complementarity to the target.
Although derived from thermophilic organisms, GgeAgo can function efficiently over a wide temperature range for DNA target (30–85 °C), whereas higher temperatures were required for efficient RNA cleavage (above 50 °C). GgeAgo cleaved DNA target greater than RNA target below 50 °C, while it cleaved RNA target greater than DNA above 55 °C. Besides, GgeAgo cleaved DNA three times faster than RNA at 50 °C, but can cleave DNA and RNA with comparable velocity at 60 °C (Fig. 3b and Supplementary Fig. 8c, d). Previously, eAgo (KpAgo, Kluyveromyces polysporus) and pAgos (such as KmAgo, MbpAgo) were used for the cleavage of highly structured RNA17,18,24,50. However, the cleavage efficiency of these proteins was found to be dependent on the secondary RNA structure at mesophilic temperatures in vitro. In contrast, GgeAgo demonstrates efficient RNA cleavage at elevated temperatures that promote the unwinding of secondary structures, thereby enabling unbiased RNA processing. Therefore, GgeAgo could cleave complex RNA targets, independent of secondary structure formation. GgeAgo is most active with 16–21 nt gDNA for both DNA and RNA target, with cleavage efficiency dropping dramatically for shorter guides, consistent with other studied pAgos3,37. We also showed that GgeAgo associates with small DNAs of a length around 18 nt when the protein expressed in bacterial cells at 18 °C or 37 °C, and DNAs longer than 45 nt were also observed in association with GgeAgo. Accordingly, RNAs of undefined length are only detected in GgeAgo samples purified from bacterial cells induced at 37 °C. These results suggest that GgeAgo may function as a DNA-guided nuclease in vivo like other pAgos14,17,39, obtaining short DNA fragments as guides from invading genetic elements such as plasmids or viruses and then cleaving the complementary DNA/RNA targets as a prokaryotic immune defense system.
GgeAgo preferentially utilizes 16–21 nt long 5’-phosphorylated guide DNAs and exhibits optimal activity in the presence of Mn2+, similarly to previously studied pAgos1,13. GgeAgo can also use 5’OH guides, albeit at extremely low efficiencies. Interestingly, although GgeAgo shows only slightly higher binding affinity for 5’P-gDNA than for 5’OH-gDNA, the presence of the 5’-phosphate significantly enhances target binding. This may be the reason why GgeAgo prefers 5’P-gDNA and suggest that GgeAgo using 5’P-gDNA to bind target could form a more stable ternary complex. Further structural and biochemical studies are needed to elucidate the molecular basis of this preference. Furthermore, GgeAgo can function efficiently at various ionic strength and pH conditions, indicating that its performance is less affected by environmental conditions.
Unlike other thermophilic pAgos such as TtAgo, MjAgo, and TpsAgo, which show strong sequence preferences for the 5’-nucleotide of the guide1,12,19,42, GgeAgo has no strong preference for the 5’-guide nucleotide. This is more similar to mesophilic pAgos like CbAgo and KmAgo16,17,18,51, allowing for greater flexibility in targeting any desired sequence. Importantly, single mismatches in the central region of the guide greatly affect the nuclease activity of GgeAgo, whereas mismatches in the seed and 3’-supplementary regions have little or no effect on cleavage efficiency. Interestingly, high temperatures not only enhance the catalytic activity of GgeAgo for both DNA and RNA targets but also improve its ability to discriminate against mismatched RNA targets, although the same effect is not observed for DNA targets. In addition, dinucleotide mismatches at position 11–13 can completely abolish DNA cleavage. Given the strong effects of mismatches on GgeAgo dependent cleavage, accurate design of guide oligonucleotides could enable discrimination of closely related target sequences, making GgeAgo a valuable tool for enriching rare DNA and RNA variants.
Some pAgos can interact with double-stranded DNA only if its melting is facilitated by various factors, including low GC-content, increased temperature, or supercoiling12,15,16,17,18,30,39,42. However, GgeAgo can efficiently cut both plasmid and linear dsDNA with a GC content up to 53% and retain a detectable activity at 64% GC when programmed with corresponding 5’P-gDNA. Thus, GgeAgo can be used for site-specific double-stranded DNA cleavage under these conditions. Furthermore, GgeAgo can efficiently cut single-strand DNA with a GC content up to 64%. This suggested that the reduced activity on dsDNA substrates with GC content above 64% may be due to inefficient unwinding of high-GC regions at 75 °C, limiting target accessibility. Therefore, rational design to improve the thermal stability of GgeAgo would potentially overcome the limitation.
Finally, we have demonstrated that GgeAgo can serve as a versatile tool for DNA-guided DNA and RNA cleavage in applications such as DNA cloning and nucleic acid detection. A GgeAgo-based DNA cloning method was developed to easily achieve efficient insertion of DNA fragments into the desired site on a plasmid when the GC content of the target site is below 53%. Moreover, we established a GgeAgo-based system for RNA detection and rare mutant enrichment with high sensitivity and specificity, highlighting its potential for future practical applications. In conclusion, GgeAgo is a unique programmable nuclease with high activity and high specificity for DNA and RNA targets, offering significant promise for various DNA and RNA manipulation applications.
Methods
Protein expression and purification
The gene encoding GgeAgo (WP_023817613.1) from Geobacillus genomosp. 3 was codon-optimized for expression in E. coli. The optimized genes were synthesized by Wuhan Genecreate Biotechnology Co., Ltd, and cloned into pET28a expression vectors in frame with the N-terminal 6×His tag. GgeAgo double mutant (GgeAgo_DM: D502A, D571A) gene was obtained by PCR mediated site-directed mutagenesis and cloned in the same way, and verified by DNA sequencing.
For protein expression, the strain E. coli Rosetta (DE3) (Novagen) was utilized to express GgeAgo or GgeAgo_DM proteins. Cultures were grown in the Luria-Bertani (LB) broth containing 50 μg/mL kanamycin at 37 °C until the OD600 reached 0.7–0.8. Induction was achieved by adding 0.5 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG), followed by incubation at 18 °C for 16 h with continuous shaking to facilitate expression, and subsequently harvested via centrifugation. The collected cells were stored at −80 °C for subsequent protein purification. The cell pellet was resuspended in Buffer A (20 mM Tris–HCl pH 7.0, 500 mM NaCl, 20 mM imidazole), supplemented with 10% glycerol and 1 mM PMSF, then subjected to sonication (SCIENTZ-IID: 400 W, 2 s on/4 s off for 30 min) for disruption. The resulting lysate was centrifuged at 16,000 rpm for 60 min, and the supernatant was loaded onto Ni-NTA agarose resin with rotation for 1 h. The beads were washed with Buffer A containing 50 mM imidazole, and the protein was eluted with Buffer A containing 200 mM imidazole. Fractions containing GgeAgo were concentrated using an Amicon 50 K filter unit (Millipore) and diluted in 20 mM Tris-HCl, pH 7.0, to reduce salt concentration to 150 mM NaCl. The diluted protein was loaded onto a Heparin column (HiTrap Heparin HP, Cytiva) equilibrated with Buffer B1 (20 mM Tris-HCl pH 7.0, 150 mM NaCl). The column was washed with 10 column volumes of Buffer B1 and eluted with a linear NaCl gradient (0.15–1.0 M) through mixing Buffer B1 and Buffer B2 (20 mM Tris-HCl pH 7.0, 1 M NaCl). Fractions containing GgeAgo were concentrated using an Amicon 50 K filter unit (Millipore). Finally, the protein preservation solution was exchanged with Buffer C (20 mM Tris-HCl pH 7.0, 500 mM NaCl), adjusted to 1.5 mg/mL, aliquoted and flash-frozen in liquid nitrogen. The purification protocol for GgeAgo_DM was identical.
Analysis of single-stranded nucleic acid cleavage by GgeAgo
The sequences of all guides and targets utilized in the assays were synthesized by Sangon and Genscript (refer to Table S1). When required, guides were 5’-phosphorylated with ATP using T4 polynucleotide kinase (New England Biolabs). The cleavage assays were performed as described previously with some modification24. Unless otherwise indicated, 800 nM GgeAgo was pre-incubated with 400 nM gDNA or gRNA for 10 min at 50 °C to assemble GgeAgo-guide complexes in reaction buffer RB (10 mM HEPES-NaOH pH 7.5, 100 mM NaCl, 5 mM MnCl2, 5% glycerol). The cleavage reactions were initiated by adding 200 nM target DNA or RNA at 50 °C. For analysis of the temperature dependence of target cleavage, the samples were incubated at different temperatures using a PCR thermocycler (T100, Bio-Rad). Kinetic analyses of target cleavage were performed in single-turnover reaction conditions, and the data were fitted to the single-exponential equation: Y = Cmax × [1 – exp(–kobs × t)], where Y is the cleavage efficiency at a given time point, Cmax is the maximum cleavage, and kobs is the observed rate constant. To investigate the effect of various divalent cations, 5 mM Mg2+, Ni2+, Co2+, Cu2+, Fe2+, Ca2+, or Zn2+ were added to the reaction buffer instead of Mn2+. To determine the effect of mismatches on the target cleavage, a set of DNA guides was employed, each containing a single or double mismatched nucleotide at a certain position. All reactions were carried out at 50 °C if not indicated, and the sample were quenched after the indicated time intervals by the addition of equal volumes of 2× RNA loading dye (95% formamide, 18 mM EDTA, and 0.025% sodium dodecyl sulfate, and 0.025% bromophenol blue), followed by heating for 5 min at 95 °C. The cleavage products were resolved by 20% denaturing PAGE, pre-stained with SYBR Gold (Invitrogen) for unlabeled target or visualized with GelDoc Go (Bio-Rad). The gels were analyzed by the NIH program ImageJ and Prism 9 (GraphPad).
Co-purification nucleic acids with GgeAgo
Co-purification nucleic acids were extracted as described previously, with slight modifications15,17. Briefly, following the first Ni-NTA purification step in Buffer C, 5 mg GgeAgo was supplemented with CaCl2 and proteinase K (Zomanbio) to final concentrations of 5 mM CaCl2 and 0.5 mg/ml proteinase K. The sample was then incubated for 60 min at 55 °C. The nucleic acids present in the top layer were isolated from the organic fraction by adding Roti-phenol/chloroform/isoamyl alcohol (pH 7.5–8.0) at a 25:24:1 ratio, followed by centrifugation at 12,000 rpm for 15 min. The nucleic acids were subsequently precipitated using ethanol precipitation with 99% ethanol added at a 1:2 ratio along with 0.5% linear polymerized acrylamide as a co-precipitating agent. The mixture was then incubated overnight at −20 °C, and centrifuged at 12,000 rpm for 30 min. The resulting nucleic acid pellet was washed twice with 500 μl of 70% ethanol and solved in 25 μl nuclease-free water. The concentration of the co-purified nucleic acids was determined by Nanodrop 8000. For subsequent analysis, 50 ng nucleic acids were treated with either 100 μg/ml RNase A (Thermo Fisher Scientific), 2 units DNase I (NEB), or both for 1 h at 37 °C, followed by resolution on 20% denaturing PAGE and staining with SYBR Gold. To enhance the binding of nucleic acids by GgeAgo, protein expression was induced for 6 h at 37 °C.
Guide and target binding assays by fluorescence polarization
Equilibrium binding of guide and target by GgeAgo was determined by measuring the changes in fluorescence polarization using SPARK microplate reader (TECAN), equipped with excitation filter of 485/20 nm and emission filter of 535/20. For guide binding assays, WT GgeAgo and the guide were incubated in buffer RB with 5 mM Mn2+ for 30 min at 37 °C in black 96-well half-area plates (Corning). The concentration of the internally FAM-labeled fluorescent guide was fixed at a final 2.5 nM in 100 μL reaction system, whereas the concentration of GgeAgo varied. Following incubation, the samples were read at 535 nm at an excitation of 485 nm. For target binding assays, inactive GgeAgo_DM and label-free guide (1:1) with varying concentrations were incubated for 10 min at 50 °C. Subsequently, 2.5 nM 5’-FAM-labeled target was added to the reaction for 1 h at 37 °C in buffer RB with 5 mM Mn2+. To determine the apparent dissociation constants (Kd) for guide binding by GgeAgo and target binding by the GgeAgo–gDNA complex, the data were normalized by substraction of the initial polarization of free fluorescein-labeled guide or target and fitted using the one-site special binding equation, utilizing Prism 9 (GraphPad).
Analysis of double-stranded DNA cleavage by GgeAgo
The gDNAs used in plasmid or linear double-stranded DNA cleavage were listed in Table S2. The linear double-stranded DNA targets were generated by PCR with primers in Table S3, and purified using Gel Extraction kit (Omega). For the cleavage reaction, 8 pmol GgeAgo was loaded with 4 pmol forward or reverse DNA guide in RB buffer at 50 °C for 10 min. Then, the two separate reactions were mixed, followed by addition of 200 ng target plasmid or linear double-stranded DNA, and incubated for 30 min at indicated temperatures. For plasmid cleavage, the resultant products were subsequently digested with Hind III or Sca I (NEB) for 1 h at 37 °C. The cleaved products were mixed with 6×DNA loading dye (NEB), followed by 1.0% or 1.5% agarose gel electrophoresis and EB staining.
DNA fragment cloning using GgeAgo
200 ng plasmid pUC19 was linearized, as described above, by GgeAgo-gDNA complex in regions with different GC-content for 30 min at 70 °C, and used directly as vector backbones for DNA fragment cloning without purification. The insert GFP expression cassette (Table S4) used in all performed cloning was amplified via PCR to introduce 20 bp homologous ends corresponding to the digested backbones. Then, the PCR products were treated with Dpn I (NEB) to remove the plasmid template. The DNA fragment cloning was performed by our previous method-TLCL44. Briefly, the amplified DNA fragments (1 μL) and linear plasmid (3 μL) with the same homologous end were incubated in reaction buffer containing 0.5 U T5 exonuclease (NEB) for 7 min on ice-water bath, followed by the addition of 50 μL of E. coli DH5α competent cells for transformation. The cloning efficiency was assessed by calculating the total colonies. The cloning accuracy was assessed by determining the percentage of positive colonies against the total colonies.
RNA detection and variant allele enrichment using GgeAgo
To investigate nucleic acid detection using GgeAgo, a partially synthesized RNA fragment of SARS-Cov2 S-gene as the mock RNA was pre-amplified using the One Step RT-PCR kit (Yeasen) according to the manufacturer’s protocol. In brief, 5 μL of the viral RNA, 10 μL RT-PCR mix, primer pairs for S gene (400 nM for each primer, Table S5) were mixed. 20 μL reaction was performed as follows: 1 cycle of reverse transcription at 55 °C for 15 min; 95 °C for 5 min, 45 thermo-cycles of denaturing at 95 °C for 15 s, extending at 60 °C for 2 s. Subsequently, a mixture comprising 60 pmol purified GgeAgo pre-loading 3 pmol 5’ phosphorylated input gDNA, 10 pmol fluorescent reporter and 2.5 μL of 10 ×RB buffer was added to the PCR products to a final volume of 25 μL. The reaction was carried out at 75 °C for 15 min, followed by the fluorescence intensity detection of each sample with Real-time fluorescence quantitative PCR instrument (Bio-Rad). For variant alleles enrichment, the mixture of WT and D614G mutant RNA was cleaved by GgeAgo pre-loading WT gDNA before RT-PCR amplification (Table S6). The reference control without target was measured to determine the background fluorescence values. The actual measured fluorescence values were normalized by subtracting the background fluorescence values.
Statistical information
Statistical analyses were performed using GraphPad Prism 9 and Microsoft Excel. The two-tailed Student’s t test was utilized to compare differences between two groups, with significance set at a p < 0.05. All data are shown as means ± standard deviation from three independent replicates.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Change history
13 January 2026
In this article the author name Yibin Cheng was incorrectly written as Yibin Chen. Also, the affiliation “Key Laboratory of Biocatalysis and Enzyme Engineering” was incorrect; it should have been “State Key Laboratory of Biocatalysis and Enzyme Engineering”. The original article has been corrected.
References
Lisitskaya, L., Aravin, A. A. & Kulbachinskiy, A. DNA interference and beyond: structure and functions of prokaryotic Argonaute proteins. Nat. Commun. 9, 5165 (2018).
Agapov, A. et al. Prokaryotic Argonaute nuclease cooperates with co-encoded RNase to acquire guide RNAs and target invader DNA. Nucleic Acids Res. 52, 5895–5911 (2024).
Lisitskaya, L. et al. Programmable RNA targeting by bacterial Argonaute nucleases with unconventional guide binding and cleavage specificity. Nat. Commun. 13, 4624 (2022).
Swarts, D. C. et al. The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol. 21, 743–753 (2014).
Wang, J. Y., Pausch, P. & Doudna, J. A. Structural biology of CRISPR-Cas immunity and genome editing enzymes. Nat. Rev. Microbiol. 20, 641–656 (2022).
Makarova, K. S. et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 18, 67–83 (2020).
Olina, A. V., Kulbachinskiy, A. V., Aravin, A. A. & Esyunina, D. M. Argonaute proteins and mechanisms of RNA interference in Eukaryotes and Prokaryotes. Biochemistry 83, 483–497 (2018).
Bobadilla Ugarte, P., Barendse, P. & Swarts, D. C. Argonaute proteins confer immunity in all domains of life. Curr. Opin. Microbiol. 74, 102313 (2023).
Ghildiyal, M. & Zamore, P. D. Small silencing RNAs: an expanding universe. Nat. Rev. Genet 10, 94–108 (2009).
Koonin, E. V., Makarova, K. S. & Wolf, Y. I. Evolutionary genomics of defense systems in archaea and bacteria. Annu Rev. Microbiol. 71, 233–261 (2017).
Hegge, J. W., Swarts, D. C. & van der Oost, J. Prokaryotic Argonaute proteins: novel genome-editing tools? Nat. Rev. Microbiol. 16, 5–11 (2018).
Swarts, D. C. et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature 507, 258–261 (2014).
Swarts, D. C. et al. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Res. 43, 5120–5129 (2015).
Kuzmenko, A. et al. DNA targeting and interference by a bacterial Argonaute nuclease. Nature 587, 632–637 (2020).
Hegge, J. W. et al. DNA-guided DNA cleavage at moderate temperatures by Clostridium butyricum Argonaute. Nucleic Acids Res. 47, 5809–5821 (2019).
Kuzmenko, A., Yudin, D., Ryazansky, S., Kulbachinskiy, A. & Aravin, A. A. Programmable DNA cleavage by Ago nucleases from mesophilic bacteria Clostridium butyricum and Limnothrix rosea. Nucleic Acids Res. 47, 5822–5836 (2019).
Kropocheva, E., Kuzmenko, A., Aravin, A. A., Esyunina, D. & Kulbachinskiy, A. A programmable pAgo nuclease with universal guide and target specificity from the mesophilic bacterium Kurthia massiliensis. Nucleic Acids Res. 49, 4054–4065 (2021).
Liu, Y. et al. A programmable omnipotent Argonaute nuclease from mesophilic bacteria Kurthia massiliensis. Nucleic Acids Res. 49, 1597–1608 (2021).
Willkomm, S. et al. Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein. Nat. Microbiol. 2, 17035 (2017).
Zander, A. et al. Guide-independent DNA cleavage by archaeal Argonaute from Methanocaldococcus jannaschii. Nat. Microbiol. 2, 17034 (2017).
Kaya, E. et al. A bacterial Argonaute with noncanonical guide RNA specificity. Proc. Natl. Acad. Sci. USA 113, 4057–4062 (2016).
Lapinaite, A., Doudna, J. A. & Cate, J. H. D. Programmable RNA recognition using a CRISPR-associated Argonaute. Proc. Natl. Acad. Sci. USA 115, 3368–3373 (2018).
Wang, L. et al. A bacterial Argonaute with efficient DNA and RNA cleavage activity guided by small DNA and RNA. Cell Rep. 41, 111533 (2022).
Li, W. et al. A programmable pAgo nuclease with RNA target preference from the psychrotolerant bacterium Mucilaginibacter paludis. Nucleic Acids Res. 50, 5226–5238 (2022).
Liu, Q. et al. A programmable pAgo nuclease with RNA target-cleavage specificity from the mesophilic bacterium Verrucomicrobia. Acta Biochim. Biophys. Sin. 55, 1204–1212 (2023).
Ryazansky S., Kulbachinskiy A. & Aravin A. A. The expanded universe of prokaryotic argonaute proteins. mBio 9, 10−1128 (2018).
Kropocheva, E. V. et al. Prokaryotic Argonaute Proteins as a Tool for Biotechnology. Mol. Biol. 56, 854–873 (2022).
Li, Y. et al. Comparison of CRISPR/Cas and Argonaute for nucleic acid tests. Trends Biotechnol. 41, 595–599 (2023).
Graver, B. A., Chakravarty, N. & Solomon, K. V. Prokaryotic Argonautes for in vivo biotechnology and molecular diagnostics. Trends Biotechnol. 42, 61–73 (2024).
Enghiad, B. & Zhao, H. Programmable DNA-guided artificial restriction enzymes. ACS Synth. Biol. 6, 752–757 (2017).
He, R. et al. Pyrococcus furiosus Argonaute-mediated nucleic acid detection. Chem. Commun. 55, 13219–13222 (2019).
Enghiad, B. et al. PlasmidMaker is a versatile, automated, and high throughput end-to-end platform for plasmid construction. Nat. Commun. 13, 2697 (2022).
Liu, Q. et al. Argonaute integrated single-tube PCR system enables supersensitive detection of rare mutations. Nucleic Acids Res. 49, e75 (2021).
Song, J. et al. Highly specific enrichment of rare nucleic acid fractions using Thermus thermophilus argonaute with applications in cancer diagnostics. Nucleic Acids Res. 48, e19 (2020).
Vaiskunaite, R., Vainauskas, J., Morris, J. J. L., Potapov, V. & Bitinaite, J. Programmable cleavage of linear double-stranded DNA by combined action of Argonaute CbAgo from Clostridium butyricum and nuclease deficient RecBC helicase from E. coli. Nucleic Acids Res. 50, 4616–4629 (2022).
Hunt, E. A., Evans, T. C. Jr. & Tanner, N. A. Single-stranded binding proteins and helicase enhance the activity of prokaryotic argonautes in vitro. PLoS ONE 13, e0203073 (2018).
Guo, X. et al. A hyperthermophilic argonaute from ferroglobus placidus with specificity on guide binding pattern. Front. Microbiol. 12, 654345 (2021).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Swarts, D. C. et al. Autonomous generation and loading of DNA guides by bacterial argonaute. Mol. Cell 65, 985–998.e986 (2017).
Jolly, S. M. et al. Thermus thermophilus Argonaute Functions in the completion of DNA replication. Cell 182, 1545–1559.e1518 (2020).
Olovnikov, I., Chan, K., Sachidanandam, R., Newman, D. K. & Aravin, A. A. Bacterial argonaute samples the transcriptome to identify foreign DNA. Mol. Cell 51, 594–605 (2013).
Sun, Y. et al. An Argonaute from Thermus parvatiensis exhibits endonuclease activity mediated by 5’ chemically modified DNA guides. Acta Biochim. Biophys. Sin. 54, 686–695 (2022).
Panteleev, V., Kropocheva, E., Esyunina, D. & Kulbachinskiy, A. Strong temperature effects on the fidelity of target DNA recognition by a thermophilic pAgo nuclease. Biochimie 209, 142–149 (2023).
Yu, F. et al. TLTC, a T5 exonuclease-mediated low-temperature DNA cloning method. Front. Bioeng. Biotechnol. 11, 1167534 (2023).
Xun, G. et al. Argonaute with stepwise endonuclease activity promotes specific and multiplex nucleic acid detection. Bioresour. Bioprocess 8, 46 (2021).
Jang, H. et al. ANCA: artificial nucleic acid circuit with argonaute protein for one-step isothermal detection of antibiotic-resistant bacteria. Nat. Commun. 14, 8033 (2023).
Fang, M. et al. Characterization and application of a thermophilic Argonaute from archaeon Thermococcus thioreducens. Biotechnol. Bioeng. 119, 2388–2398 (2022).
Yuan, Y. R. et al. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell 19, 405–419 (2005).
Wang, Y. et al. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 456, 921–926 (2008).
Dayeh, D. M., Cantara, W. A., Kitzrow, J. P., Musier-Forsyth, K. & Nakanishi, K. Argonaute-based programmable RNase as a tool for cleavage of highly-structured RNA. Nucleic Acids Res. 46, e98 (2018).
Lisitskaya, L. et al. Bacterial Argonaute nucleases reveal different modes of DNA targeting in vitro and in vivo. Nucleic Acids Res. 51, 5106–5124 (2023).
Acknowledgements
This work was supported by China National Key Research and Development (R&D) Program (2023YFA0913700); Project of Technological Innovation Plan in Hubei Province (2024BCA001); the Natural Science Foundation of Wuhan City (2024040701010046); the China Postdoctoral Science Foundation (2023M741100); and Hubei Provincial Science Fund for Distinguished Young Scholars (2024AFA101); Major Special Project for the Development of the Agricultural Microbiology Industry in Hubei Province (NYWSWZX2025-2027-10); open project funding of the State Key Laboratory of Biocatalysis and Enzyme Engineering (SKLBEE2021016).
Author information
Authors and Affiliations
Contributions
W.Q.L. conceived the project and supervised it with L.X.M. and F. W. W.Q.L., J.H., Y.Y.H., and Y.W. designed and performed biochemical experiments, analyzed the data, and wrote the manuscript. H.R.J., M.C., J.J.Z., F.J.L., X.L., Y.B.C., T.L., and G.L.Y. provided materials and assistance. All authors reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
L.X.M., W.Q.L., J.H., and F.W. applied for a patent based on the use of GgeAgo for DNA and RNA manipulation. The remaining authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Huan Bao and Mengtan Xing.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hu, J., Hu, Y., Wang, Y. et al. A programmable pAgo nuclease with high activity and specificity for efficient DNA and RNA manipulation. Commun Biol 8, 1685 (2025). https://doi.org/10.1038/s42003-025-09082-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-09082-9