Abstract
Glioblastoma (GBM) is a highly aggressive primary brain tumor with a dismal prognosis, particularly in its mesenchymal (MES) subtype, which correlates strongly with poor survival. Despite this, the mechanisms preserving MES identity remain poorly understood. Here, we show that alpha-actinin 1 (ACTN1) is upregulated in MES GBM and drives proneural-to-mesenchymal transition (PMT). Using patient samples and multiple GBM cell lines, we find that ACTN1 overexpression promotes proliferation, invasion, and tumorigenesis, while its silencing diminishes these malignant traits and shifts gene expression away from MES markers. Mechanistically, we identify ubiquitin-specific peptidase 14 (USP14) as a pivotal deubiquitinase (DUB) that stabilizes ACTN1 by removing its ubiquitin chains. Pharmacological inhibition of USP14 with IU1 reduces ACTN1 protein levels, impairs MES-associated phenotypes, and suppresses tumor progression in vitro and in intracranial xenograft models. Clinically, elevated USP14 and ACTN1 expression correlates with poorer survival in GBM patients, highlighting the USP14–ACTN1 axis as a key driver of PMT and a promising therapeutic target for this devastating disease.
Similar content being viewed by others
Introduction
Glioblastoma (GBM) is the most aggressive primary brain tumor in adults, with a median survival of only 15 to 20 months1. Standard treatment consists of maximal surgical resection, followed by high-dose radiation therapy and concurrent or adjuvant temozolomide chemotherapy. Despite these intensive interventions, GBM prognosis remains poor, with a 5-year survival rate under 5%2,3. Gene expression profiling has categorized GBM into three subtypes: classical, mesenchymal (MES), and proneural (PN)4,5,6. Of these, the PN subtype is linked with relatively better outcomes, while MES represents the most malignant form, closely associated with worse survival rates7. Similar to the epithelial-mesenchymal transition (EMT) observed in other cancers, GBM’s PN subtype can transition to the MES subtype8,9. This proneural-to-mesenchymal transition (PMT) is a pivotal factor driving glioblastoma invasion, progression, and poor prognosis10. However, the specific mechanisms underlying PMT in GBM are largely unexplored and warrant further investigation.
Alpha-actinins (ACTNs) are cytoskeletal proteins involved in various non-muscle functions, including cytokinesis, cell adhesion, and migration, in addition to their established role in sarcomere structure11. Four isoforms of α-actinin exist: ACTN1, ACTN2, ACTN3, and ACTN412. Among these, ACTN1 plays a critical role in controlling cellular processes, such as cell division, adhesion, and migration13. Elevated ACTN1 levels are associated with poor prognosis in cancers such as oral squamous cell carcinoma and acute myeloid leukemia14,15. Recent studies reveal ACTN1’s involvement in the progression of cancers like head and neck squamous cell carcinoma13, breast cancer16, hepatocellular carcinoma17, and gastric cancer18, as well as its prognostic significance in malignant gliomas19. However, the exact mechanisms by which ACTN1 drives these malignant behaviors, particularly in supporting the mesenchymal identity of GBM, are not fully understood. ACTN1 is known to be regulated by post-translational modifications (PTMs), including the ubiquitin-proteasome pathway mediated by E3-ubiquitin ligase Cullin320. The stability of ACTN1 in GBM cells and its impact on PMT, however, require further investigation.
Deubiquitinases (DUBs) are essential enzymes that reverse ubiquitination, a process critical for regulating the ubiquitin signaling pathway involved in cancer initiation and progression21. Human DUBs are divided into six families: ubiquitin-specific proteases (USPs), ubiquitin carboxyterminal hydrolases (UCHs), ovarian tumor proteases (OTUs), Machado-Joseph disease proteases (MJDs), JAB1/MPN/Mov34 metalloenzymes (JAMMs), and the MINDY family22,23. Among these, ubiquitin-specific peptidase 14 (USP14), a notable member of the USP family, plays a significant role in modulating several oncogenic processes, such as cell invasion, proliferation, drug resistance, immune evasion, apoptosis, and autophagy24,25,26,27. Recent studies highlight USP14’s ability to deubiquitinate NAP1L1, which promotes glioma cell proliferation28. In glioblastoma stem cells, USP14 enhances stem-like properties, tumorigenicity, and resistance to radiotherapy by stabilizing MST4-phosphorylated ALKBH529. However, whether USP14 regulates ACTN1, a potential driver of the mesenchymal identity in glioblastoma, remains unknown. Exploring USP14’s role in ACTN1 regulation could provide valuable insights into tumor progression and therapeutic approaches.
In this study, we observed that ACTN1 is upregulated in GBM, correlating positively with MES subtype markers and inversely with PN markers, indicating its role in the MES phenotype. Elevated ACTN1 levels suggest that it may promote malignant GBM progression. We also identified USP14 as a critical post-translational regulator of ACTN1 stability in GBM cells. Mechanistically, USP14 binds to and stabilizes ACTN1, thereby supporting GBM’s mesenchymal identity. Furthermore, we demonstrated that IU1, a small-molecule inhibitor targeting USP14’s catalytic activity24,30, promotes ACTN1 degradation and suppresses the mesenchymal phenotype of GBM in both in vitro and in vivo models. Our findings suggest that IU1 holds promise as a lead compound for developing anti-cancer drugs targeting glioblastoma with high ACTN1 expression. Collectively, this study positions the USP14/ACTN1 axis as a potential therapeutic target to counter mesenchymal identity and malignant progression in GBM.
Results
High ACTN1 expression in the Mesenchymal (MES) subtype GBM reflects MES characteristics
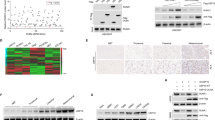
To investigate ACTN1’s function in glioblastoma (GBM), we analyzed its protein levels across 10 normal brain tissues (NBTs) and 64 GBM patient samples, including three subtypes: 25 mesenchymal (MES), 20 classical (CL), and 19 proneural (PN) samples, by IB analysis. Results indicated that ACTN1 expression was significantly reduced in NBTs compared to GBM subtypes, with distinct variations observed among GBM subtypes. The MES subtype displayed the highest levels of ACTN1, while PN had the lowest (Fig. 1A; Supplementary Fig. 1A). This pattern suggests a specific upregulation of ACTN1 in the MES subtype, possibly contributing to its aggressive features.
A Immunoblot (IB) analysis of ACTN1 protein in normal brain tissues (NBTs) and glioblastoma (GBM) subtypes, with β-actin as a loading control. B IB of ACTN1 levels in normal human astrocytes (NHAs), four GBM cell lines, and two primary GBM samples, with β-actin as a loading control. n = 3. C IB of ACTN1 and MES (YKL-40, MET, COL5A1) and PN (Olig2, PDGFRα) markers in selected MES (LN229, GBM2) and PN (U251, GBM1) GBM cells, with β-actin as control. n = 3. D IB of ACTN1 and MES/PN markers in LN229 and GBM2 cells following transduction with ACTN1-targeting shRNAs (shACTN1#1 and shACTN1#2), with β-actin control. n = 3. E, F Transwell assays of invasion and migration abilities in LN229 and GBM2 cells with ACTN1 knockdown or control, scale bar = 100 μm, with quantifications on the right. n = 6. G CCK8 assay measuring the proliferation of specified cells. n = 3. H Representative bioluminescent images of intracranial GBM xenografts from modified LN229 and GBM2 cells, with scale bars for photon emissions. Each group included 10 mice. I Quantification of bioluminescence in LN229- and GBM2-derived tumors across groups. J Kaplan-Meier survival curves for mice with indicated modifications. **p < 0.01; ***p < 0.001.
To deepen our understanding of ACTN1 expression, we assessed it in normal human astrocytes (NHAs) alongside multiple GBM cell lines. Here, U251, T98G, and primary GBM1 cells represented PN subtypes, while U87MG, LN229, and primary GBM2 cells were classified as MES subtypes31. Notably, ACTN1 expression was significantly higher in GBM cells compared to NHAs, with particularly high levels in MES subtype cells (U87MG, LN229, and GBM2) (Fig. 1B). This observation suggests a link between ACTN1 expression and MES subtype-specific markers, potentially indicating a role in promoting aggressive traits in MES GBM cells. To explore the association between ACTN1 and subtype-specific markers, we compared ACTN1 levels in MES (LN229, GBM2) and PN (U251, GBM1) cells. MES cells (LN229, GBM2) exhibited higher ACTN1 and MES markers (YKL-40, MET, COL5A1), whereas PN cells (U251, GBM1) displayed reduced ACTN1 levels and increased PN marker expression (Olig2, PDGFRα) (Fig. 1C). These results suggest ACTN1 may have a functional role in mesenchymal features in GBM.
ACTN1 enhances invasion, migration, proliferation, and tumorigenesis in GBM cells
Given that the proneural-to-mesenchymal transition (PMT) contributes to the invasiveness and growth of glioblastoma, we explored ACTN1’s role in this process. Depleting ACTN1 by two non-overlapping lentiviral shRNAs in LN229 and GBM2 cells led to decreased expression of MES markers (YKL-40, MET, COL5A1) while increasing PN markers (Olig2, PDGFRα) (Fig. 1D). In contrast, ACTN1 overexpression in U251 and GBM1 cells increased MES markers while reducing PN markers (Supplementary Fig. 1B). Transwell assays further demonstrated that ACTN1 silencing reduced invasion and migration in LN229 and GBM2 cells (Fig. 1E, F), whereas its overexpression enhanced these capabilities in U251 and GBM1 cells (Supplementary Fig. 1C, D). These findings indicate that ACTN1 positively influences GBM cell invasiveness and migration, aligning with MES subtype characteristics. Additionally, ACTN1 knockdown decreased proliferation in LN229 and GBM2 cells, while its overexpression significantly boosted proliferation in U251 and GBM1 cells (Fig. 1G; Supplementary Fig. 1E), suggesting a role in promoting tumorigenic features. To examine tumorigenic potential in vivo, equal numbers of luciferase-labeled LN229 and GBM2 cells, transduced with either shCtrl or shACTN1, were injected into mouse brains. Xenografts derived from ACTN1-depleted cells showed notably slower tumor growth (Fig. 1H, I), and mice with these xenografts had significantly improved survival rates compared to controls (Fig. 1J). These results support that ACTN1 contributes to tumor growth and aggressiveness in GBM, specifically in the MES subtype.
Identification of USP14 as a potential DUB of ACTN1
To investigate whether ACTN1 stability is regulated by the ubiquitin-proteasome system (UPS), we performed a cycloheximide (CHX) chase assay in U251 and GBM1 cells. ACTN1 protein levels declined rapidly post-CHX treatment, becoming nearly undetectable within 8 hours. However, treatment with the UPS inhibitor MG132 effectively prevented this degradation, indicating that ACTN1 stability is regulated by proteasomal pathways (Supplementary Fig. 2A). To identify deubiquitinases (DUBs) that may interact with ACTN1, we utilized the BioGRID database, identifying USP2, USP14, BAP1, and ZRANB1 as potential candidates. Of these, only USP14 was observed to interact with ACTN1 in HEK293T cells, suggesting it as the primary DUB associated with ACTN1 (Fig. 2A). Analysis of USP14 expression across NBTs and GBM subtypes showed its highest expression in MES GBM (Fig. 2B; Supplementary Fig. 2B), further indicating its relevance in aggressive GBM subtypes. Moreover, correlation analysis of clinical specimens encompassing NBTs and GBM subtypes revealed a significant positive correlation between ACTN1 (Fig. 1A) and USP14 (Fig. 2B) protein levels (Supplementary Fig. 2C).
A Immunoprecipitation (IP) and IB analysis from HEK293T cells transfected with Flag-tagged DUBs (USP2, USP14, BAP1, ZRANB1) and probed with ACTN1 and Flag antibodies. B IB analysis of USP14 protein levels in NBTs and GBM subtypes, with β-actin control. C HEK293T cells were transfected with varying amounts of Flag-tagged USP14 (WT or C114A) for IB analysis. n = 3. D Quantitative analysis of USP14 and ACTN1 mRNA in LN229 and GBM2 cells with USP14 knockdown. n = 3. E IB showing ACTN1 protein reduction following USP14 knockdown in LN229 and GBM2 cells, with MG132 treatment reversing this effect. n = 3. F IB analysis of USP14 and ACTN1 in cells transfected with USP14-WT or C114A after USP14 knockdown. n = 3. G IB showing accelerated ACTN1 degradation following USP14 knockdown in LN229 and GBM2. n = 3. H Quantification of ACTN1 normalized to β-actin. I Overexpression of USP14-WT, but not USP14-C114A, stabilized ACTN1 in GBM1 cells, with quantification on the right. n = 3. **p < 0.01; ***p < 0.001; n.s., not significant.
Given USP14’s known deubiquitinating activity, we hypothesized it might stabilize ACTN1 by removing ubiquitin chains. In HEK293T cells, overexpression of wild-type USP14 (USP14-WT), but not the inactive mutant C114A, led to a dose-dependent increase in ACTN1 levels (Fig. 2C). Further experiments revealed that silencing USP14 in LN229 and GBM2 cells significantly decreased ACTN1 protein levels without affecting mRNA levels, implying that USP14 stabilizes ACTN1 post-translationally (Fig. 2D, E). Treatment with MG132 restored ACTN1 levels, reinforcing that USP14 stabilizes ACTN1 by preventing proteasomal degradation (Fig. 2E). Furthermore, compared to control LN229 and GBM2 cells, USP14-deficient cells exhibited significantly reduced levels of ACTN1, which could be effectively restored by USP14-WT overexpression, but not USP14-C114A (Fig. 2F). CHX chase assays revealed that USP14 depletion accelerated ACTN1 degradation in MES GBM (Fig. 2G, H; Supplementary Fig. 2D, E), whereas ectopic expression of USP14-WT, but not USP14-C114A, markedly increased ACTN1 stability in both HEK293T and PN GBM cells (Fig. 2I; Supplementary Fig. 2F, G). These findings establish USP14 as a stabilizing factor for ACTN1 in GBM.
USP14 interacts with ACTN1
To confirm a direct interaction between USP14 and ACTN1, double immunofluorescence staining was performed, showing co-localization in the nucleus through confocal microscopy (Fig. 3A). Co-immunoprecipitation (Co-IP) assays demonstrated that His-tagged ACTN1 could be detected in Flag-USP14 WT or Flag-USP14 C114A precipitates in HEK293T cells, suggesting the interaction does not require USP14’s deubiquitinase (DUB) activity (Fig. 3B). Additionally, we examined endogenous USP14 and ACTN1 in LN229 and GBM2 cell lines, which showed a consistent association (Fig. 3C). Next, GST pull-down assay demonstrated that GST-ACTN1 can be directly bound to USP14-WT or USP14-C114A, but not to GST alone (Fig. 3D), indicating that DUB activity of USP14 is not required for its interaction with ACTN1. To verify the binding domains, we generated truncation mutants of Flag-USP14 and His-ACTN1 to narrow down their interaction sites (Fig. 3E, G). Results showed that the M4 region of USP14 (amino acids 352–428) and the M1 segment of ACTN1 (amino acids 31–245) were both necessary and sufficient for this interaction (Fig. 3F, H), confirming a robust direct association.
A Immunofluorescence (IF) images of USP14 and ACTN1 in LN229 and GBM2 cells, with nuclei labeled by DAPI (blue), scale bar = 10 μm. B IP and IB of HEK293T cells transfected with His-ACTN1 alone or with Flag-USP14 WT or C114A to detect interaction. n = 3. C Co-immunoprecipitation showing USP14-ACTN1 interaction in LN229 and GBM2 cells. n = 3. D GST pull-down using purified GST-ACTN1 or GST control incubated with Flag-USP14 (WT or C114A). IB: anti-Flag detects USP14 bound to GST-ACTN1 but not GST. Coomassie staining shows input and integrity of GST fusion proteins. n = 3. E, G Diagram of full-length (FL) Flag-tagged USP14, His-tagged ACTN1, and deletion mutants. F, H Co-IP analysis identifying USP and CH sequences in USP14 and ACTN1 as essential for interaction. n = 3.
USP14 deubiquitinates ACTN1 and specifically cleaves Lys27-, Lys48-, and Lys63-linked polyubiquitin chains
Considering USP14’s function as a deubiquitinase (DUB), we next examined its effect on ACTN1 ubiquitination. Loss of USP14 resulted in a significant increase in ACTN1 ubiquitination in LN229 and GBM2 cells (Fig. 4A). To further investigate, we co-expressed His-ACTN1 and HA-ubiquitin with either USP14-WT or USP14-C114A in U251 and GBM1 cells. Wild-type USP14 reduced ACTN1 ubiquitination, whereas the C114A mutant had no effect (Fig. 4B). In addition, we assessed endogenous ACTN1 ubiquitination in GBM cells: USP14 knockdown increased endogenous ACTN1 ubiquitination (Supplementary Fig. 3A), while overexpression of USP14-WT markedly decreased endogenous ACTN1 ubiquitination, with no change upon overexpression of USP14-C114A (Supplementary Fig. 3B). Previous studies suggest that ACTN1 can be ubiquitylated and targeted for degradation by the E3 ligase Cullin3. To determine if USP14 counteracts this activity, we overexpressed USP14, which largely reversed the ubiquitination induced by Cullin3 (Fig. 4C).
A IP and IB of LN229 and GBM2 cells transfected with shRNAs and HA-Ub to detect ACTN1 ubiquitination; MG132 was used pre-harvest. n = 3. B IB showing USP14-mediated deubiquitination of ACTN1 in U251 and GBM1 cells transfected with His-ACTN1, HA-Ub, and either Flag-USP14 WT or Flag-USP14 C114A. n = 3. C ACTN1 ubiquitination analysis in LN229 and GBM2 cells with E3 Cullin3 and USP14. n = 3. D GST fusion assay to detect deubiquitination of ACTN1 by USP14-WT or USP14-C114A. n = 3. E IB showing specific ubiquitin linkages cleaved by USP14 in LN229 cells transfected with His-ACTN1, Flag-USP14, and various HA-Ub mutants. n = 3. F, G Analysis of polyubiquitin linkages in ACTN1 with USP14 and Ub mutants (WT, 27R, 48R, 63R, or 3KR). n = 3. H, I In vitro deubiquitination assay of ACTN1 by USP14, assessing Lys27 and Lys48. n = 3. J IB of ACTN1 in cells transfected with Ub-WT or Ub-Lys27/48/63R under control or shUSP14 conditions. n = 4.
In vitro ubiquitination assays confirmed that only wild-type USP14, and not the C114A variant, effectively removed ubiquitin from ACTN1 (Fig. 4D). We further assessed USP14’s ability to cleave different ubiquitin linkages—seven distinct types formed via specific lysine residues32 (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, Lys63). USP14 selectively cleaved Lys27-, Lys48-, and Lys63-linked polyubiquitin chains on ACTN1 (Fig. 4E), suggesting that USP14 targets these specific linkages in ACTN1 ubiquitination.
To further verify, we used ubiquitin mutants with single and combined lysine mutations (K27R, K48R, K63R, and a triple-mutant 3KR, which lacks all three Lys residues). While each single mutation (27R, 48R, 63R) reduced deubiquitination by USP14, the 3KR mutant completely abolished it (Fig. 4F, G). In an in vitro deubiquitination assay, USP14-WT efficiently removed the Lys27-, Lys48-, and Lys63-linked chains on ACTN1 (Fig. 4H, I; Supplementary Fig. 4A). Expressing of lysine-resistant ubiquitin variants (Lys27R, Lys48R, Lys63R) in USP14-depleted LN229 and GBM2 cells partially mitigated the USP14 loss–induced downregulation of ACTN1 (Supplementary Fig. 4C–H), whereas the triple lysine-resistant ubiquitin mutant (Lys27/48/63R) completely reversed the decrease in ACTN1 protein upon USP14 knockdown (Fig. 4J; Supplementary Fig. 4B). These findings indicate that USP14 targets the Lys27-, Lys48-, and Lys63-linked ubiquitin chains on ACTN1, and that these three linkages are critical for USP14-mediated stabilization of ACTN1. Collectively, our findings demonstrate that USP14 is essential for ACTN1 deubiquitination, selectively cleaving key ubiquitin linkages to stabilize the protein.
ACTN1 Mediates the Impact of USP14 on PMT in GBM
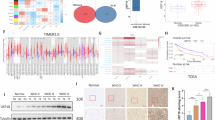
To further understand USP14’s role in glioblastoma, we examined its effects on mesenchymal (MES) subtype cells. MES cells (LN229, GBM2) showed higher USP14 expression compared to proneural (PN) cells (U251, GBM1) (Supplementary Fig. 5A). To explore if ACTN1 could counteract USP14 loss in MES cells, we performed knockdown experiments. Reduced USP14 expression led to decreased MES markers (YKL-40, MET, COL5A1) and an increase in PN markers (Olig2, PDGFRα), changes that were reversed by ACTN1 overexpression (Fig. 5A). Conversely, USP14 upregulation increased MES markers and reduced PN markers, which was counteracted by ACTN1 knockdown (Fig. 5B).
A IB analysis of USP14, ACTN1, and MES/PN markers in LN229 and GBM2 cells transduced with shUSP14 and vector controls or ACTN1. n = 3. B IB of the same markers in U251 and GBM1 cells with USP14 overexpression, showing reversal with ACTN1 knockdown. n = 3. C Quantification of invasion and migration via transwell assay in cells with vector or USP14, with or without ACTN1 knockdown. n = 6. D, E Transwell assay results showing invasion and migration with shUSP14, with vector controls or ACTN1 in LN229 and GBM2 cells, scale bar = 100 μm. n = 6. F CCK8 proliferation assay measuring the proliferation of specified cells. n = 3. G Bioluminescent imaging of intracranial xenografts from LN229 cells with shUSP14 and vector controls or ACTN1, scale bar for photon emission. Each group included 10 mice. H Quantification of bioluminescence in LN229- and GBM2-derived xenografts across groups. I Kaplan-Meier survival analysis for indicated groups of mice. *p < 0.05; **p < 0.01; ***p < 0.001.
Our findings led us to hypothesize that ACTN1 mediates USP14’s effects on PMT. Transwell and CCK8 assays demonstrated that USP14 overexpression enhanced invasiveness, migration, and proliferation in U251 and GBM1 cells, effects that were nullified by ACTN1 knockdown (Fig. 5C; Supplementary Fig. 5B-D). Conversely, USP14 inhibition reduced the invasiveness, migration, and proliferation of LN229 and GBM2 cells, which was restored by ACTN1 overexpression (Fig. 5D–F). To verify this effect in vivo, we used an orthotopic xenograft model. Bioluminescence imaging revealed that tumors from USP14-depleted LN229 and GBM2 cells showed significantly slower growth compared to controls, but ACTN1 overexpression reversed this effect (Fig. 5G, H; Supplementary Fig. 5E). Kaplan-Meier survival analysis showed that mice injected with USP14-depleted cells had prolonged survival compared to controls; however, survival decreased significantly when ACTN1 was overexpressed in these cells (Fig. 5I). These results underscore ACTN1 as a mediator of USP14’s role in promoting PMT in GBM cells.
Pharmacological Inhibition of USP14 by IU1 Enhances ACTN1 Ubiquitination and Suppresses PMT
To further validate our findings on USP14, we used IU1, a small-molecule inhibitor of USP14’s catalytic activity, to assess its impact on ACTN1 stabilization. First, we confirmed IU1’s effectiveness in inhibiting USP14’s deubiquitinating activity. As shown in Fig. 6A, IU1 treatment markedly reduced USP14’s ability to remove ubiquitin from ACTN1. Accordingly, ACTN1 protein levels decreased in LN229 and GBM2 cells following IU1 treatment, with partial restoration by co-treatment with MG132 (Fig. 6B). Additionally, IU1 and CHX co-treatment significantly shortened ACTN1’s half-life (Fig. 6C), indicating that IU1, similar to USP14 depletion, promotes ACTN1 ubiquitination and proteasomal degradation.
A IB showing USP14-mediated deubiquitination of ACTN1 is inhibited by IU1. n = 3. B IB analysis of ACTN1 in LN229 and GBM2 cells treated with IU1 in the presence or absence of MG132. n = 3. C IB showing IU1 effect on ACTN1 stability, with quantification normalized to β-actin. n = 3. D, E Transwell assay of invasion and migration in LN229 and GBM2 cells with vehicle or IU1, reconstituted with control or ACTN1, scale bar = 100 μm. n = 6. F CCK-8 assay of cell proliferation in LN229 and GBM2 cells with vehicle or IU1, reconstituted with control or ACTN1. n = 3. G IB showing expression of ACTN1, MES, and PN markers in LN229 and GBM2 cells with vehicle or IU1, reconstituted with control or ACTN1. n = 3. H Bioluminescent imaging of intracranial xenografts treated with IU1, with control or ACTN1 reconstitution, scale bars for photon emission. Each group included 10 mice. I Kaplan-Meier survival curves for mice treated with vehicle or IU1, reconstituted with control or ACTN1. **p < 0.01; ***p < 0.001.
IU1 treatment also impaired the invasive, migratory, and proliferative abilities of LN229 and GBM2 cells, effects that were counteracted by ACTN1 overexpression (Fig. 6D–F). Furthermore, IU1 treatment reduced ACTN1 levels and MES markers while increasing PN markers, with ACTN1 overexpression reversing these effects (Fig. 6G). In an in vivo model, mice with intracranial tumors derived from luciferase-labeled LN229 and GBM2 cells treated with 40 mg/kg IU1 exhibited reduced tumor growth compared to vehicle-treated controls; ACTN1 overexpression reversed this effect (Fig. 6H; Supplementary Fig. 6A, B). Kaplan-Meier survival curves demonstrated that IU1 treatment significantly extended survival compared to vehicle-treated mice, with survival benefits negated by ACTN1 overexpression (Fig. 6I). We also assessed whether combining IU1 treatment with ACTN1 knockdown confers additional benefit in vivo. In intracranial models using LN229 and GBM2 cells, IU1 treatment produced similar intracranial tumor progression dynamics and survival regardless of ACTN1 knockdown status (Supplementary Fig. 6C–F), likely because inhibition of USP14 activity by IU1 maintained ACTN1 at persistently low levels in tumor cells. Taken together, these results indicate that IU1-induced ACTN1 ubiquitination disrupts PMT and thereby limits GBM progression.
USP14 correlates positively with ACTN1 levels and links to poor prognosis in GBM patients
To assess the clinical significance of USP14 in relation to ACTN1, we evaluated USP14 and ACTN1 expression in PN and MES subtype GBM tissues. MES samples showed significantly higher USP14 and ACTN1 levels compared to primary PN tissues (Fig. 7A). Using IHC staining across 19 PN and 25 MES subtype GBM specimens, we confirmed that MES tumors predominantly expressed high levels of both USP14 and ACTN1 (Fig. 7B, C), with a strong positive correlation in expression levels (Fig. 7D–F). Kaplan-Meier survival analysis of 64 GBM samples showed that patients with elevated USP14 expression had significantly shorter overall and progression-free survival compared to those with low USP14 levels (Fig. 7G). Likewise, Kaplan-Meier analysis stratified by ACTN1 expression demonstrated that high ACTN1 expression corresponded with reduced overall and progression-free survival, indicating poorer outcomes for patients with elevated ACTN1 levels (Fig. 7H). Together, these findings underscore the importance of USP14 as a key deubiquitinating enzyme stabilizing ACTN1, which promotes PMT and negatively impacts prognosis in GBM through the USP14/ACTN1 axis.
A IHC staining for USP14 and ACTN1 in PN and MES GBM specimens. B, C Relative USP14 and ACTN1 protein levels in PN and MES GBM specimens. D, E Relative USP14 and ACTN1 levels in GBM specimens with high and low expression. F Correlation analysis between USP14 and ACTN1 in GBM tissues. G Kaplan-Meier progression-free and overall survival plots showing shorter survival in USP14 high-expressing GBM patients. H Survival analysis in GBM patients based on ACTN1 expression. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. High/Low definition: for D, E, G, and H, High and Low groups were defined by median dichotomization of the per-sample IHC intensity score within the cohort (>,median = High; ≤,median = Low).
Discussion
Intratumor heterogeneity, characterized by diverse genotypes and cell subtypes within a single tumor, is influenced by intrinsic cellular factors, inflammatory cytokines, and therapeutic interventions such as radiotherapy, chemotherapy, and anti-angiogenesis therapy33,34. This heterogeneity remains a significant barrier in developing effective treatment plans for glioblastoma (GBM). PMT, a process in GBM that mimics the epithelial-mesenchymal transition (EMT) seen in other cancers, endows GBM cells with enhanced aggressive traits linked to increased invasion, proliferation, recurrence, therapy resistance, and poor prognosis10,35. Thus, deciphering the molecular mechanisms that drive PMT is crucial for identifying therapeutic targets and designing new GBM treatment strategies.
Previous studies have shown that ACTN1 expression in grade II/III gliomas correlates negatively with patient survival, indicating that targeting ACTN1 could potentially improve outcomes in these patients19. However, ACTN1’s specific expression patterns and functions in GBM remain unclear. Our results reveal that ACTN1 plays a pivotal role in driving the transition from PN to MES differentiation, which is associated with aggressive malignancy in GBM. We observed that ACTN1 expression is notably higher in MES cells (LN229, GBM2) than in PN cells (U251, GBM1), with MES cells expressing MES markers and PN cells expressing PN markers. Notably, ectopic expression of ACTN1 in PN cells induced MES phenotypes, while ACTN1 knockdown in MES cells led to a reduction in MES markers, an increase in PN markers, and significant inhibition of tumorigenic properties in both in vitro and in vivo models, underscoring ACTN1’s role in driving PMT. Collectively, our data implicate ACTN1 as a key contributor to the mesenchymal phenotype in GBM, although the downstream circuitry remains to be fully defined. As an actin-filament crosslinker, ACTN1 likely promotes mesenchymal traits by modulating cell morphology, motility, and focal-adhesion–associated signaling. Consistent with this, ACTN1 depletion reduced MES markers and increased PN markers, indicating that ACTN1 helps sustain a MES gene-expression program—likely as a downstream consequence of these signaling changes.
ACTN1 stability is known to be regulated by post-translational modifications (PTMs) like ubiquitination, with the E3-ubiquitin ligase Cullin3 mediating this process in conditions like nemaline myopathy20. However, how ACTN1 stability is maintained in GBM remains unclear. The counter-process of ubiquitination, or deubiquitination, has yet to be fully explored for ACTN1 regulation. Through BioGRID database analysis36, we identified USP14 as a novel deubiquitinase that stabilizes ACTN1. USP14 has emerged as a broadly oncogenic deubiquitinase across multiple tumor contexts. In gastric cancer, USP14 stabilizes the m6A reader IMP2, boosting CXCL2 secretion to drive resistance to anti–PD-1 therapy37. In hepatocellular carcinoma, disrupting the AKT–USP14–TUBA1A axis promotes TUBA1A degradation and suppresses liver tumorigenesis in vivo38. In triple-negative breast cancer, lovastatin disrupts the USP14–Survivin complex, increasing Survivin polyubiquitination; silencing or inhibiting USP14 with IU1 synergizes with lovastatin to curb tumor growth39. In head and neck squamous cell carcinoma, USP14 bound to both RELA and IκBα and reduced IκBα K48-ubiquitination, leading to the degradation of IκBα, thereby activating NF-κB signaling40. These studies reinforce a convergent theme: USP14 stabilizes pro-tumor substrates (e.g., IMP2, TUBA1A, Survivin) and pathways (e.g., NF-κB). By analogy, our GBM data identify ACTN1 as a USP14 substrate, suggesting that therapeutically targeting USP14 can destabilize ACTN1 and blunt mesenchymal progression in GBM.
Our study revealed that USP14 interacts with ACTN1 in GBM, regulating its protein levels without affecting mRNA levels, suggesting a post-translational mechanism. USP14 knockdown significantly reduced ACTN1 expression, downregulated MES markers, and increased PN markers, while USP14 overexpression elevated ACTN1 and MES marker levels. Depleting USP14 also substantially reduced GBM tumorigenicity and improved survival in mice models, effects that were reversed with ectopic ACTN1 expression. This strongly supports that USP14-mediated ACTN1 stabilization is crucial for maintaining MES traits and facilitating GBM progression.
DUBs like USP14 counteract E3-ubiquitin ligases by removing ubiquitin modifications from proteins41. Our findings showed that USP14’s role as a DUB requires its catalytic activity since a catalytically inactive mutant, USP14-C114A, failed to stabilize ACTN1. To confirm this mechanism, we explored whether USP14 deubiquitinates ACTN1. USP14 knockdown increased ACTN1 ubiquitination, while USP14-WT overexpression (but not USP14-C114A) reduced it. Among the seven polyubiquitin linkages, Lys48 and Lys63 are primarily associated with proteasomal degradation42,43, and our data showed that USP14 directly cleaves Lys27-, Lys48-, and Lys63-linked chains from ACTN1. Utilizing IU1, a small-molecule inhibitor of USP14’s catalytic activity, we observed significant anti-tumor activity in GBM preclinical models, suggesting potential for USP14-targeted therapeutic development. Clinical data revealed a positive correlation between USP14 and ACTN1 levels in GBM patients, where elevated USP14 was linked to decreased patient survival, highlighting the USP14/ACTN1 axis as a potential prognostic marker in GBM. Our study identifies ACTN1 as a bona fide USP14 substrate that enforces mesenchymal programs in GBM and shows that pharmacologic DUB inhibition with IU1 provides proof of concept for the druggability of this axis. Complementing these findings, recent work in patient-derived GSCs demonstrates that USP14 also drives stemness and radioresistance by stabilizing ALKBH5, underscoring the breadth of USP14’s tumor-promoting roles in glioblastoma29. Together, these observations nominate USP14—and its substrate circuitry—as a therapeutically actionable node in GBM, supporting the development of selective, brain-penetrant USP14 inhibitors and biomarker-guided strategies to target USP14-dependent tumors.
In summary, our findings reveal USP14 as a key enzyme that stabilizes ACTN1 through deubiquitination, thereby playing an essential role in the proneural-to-mesenchymal transition of glioblastoma. By maintaining mesenchymal characteristics, the USP14/ACTN1 axis promotes tumor growth and progression. This work not only enhances our understanding of PMT at the molecular level but also highlights USP14 inhibition as a promising therapeutic approach to potentially improve outcomes for patients with GBM.
Methods
Clinical samples and database
We obtained 10 normal brain tissues (NBTs) and 64 glioblastoma samples from the Department of Neurosurgery at the Affiliated Hospital of Guizhou Medical University. NBTs were derived from patients undergoing craniotomy decompression for traumatic brain injury. Glioblastoma samples were collected from patients at the time of surgery before chemo- and radiotherapy, and two neuropathologists identified the histological specimens independently according to the WHO criteria. Of the glioblastoma samples, 25 showed high expression of MES GBM markers (YKL-40, MET, COL5A1) and low expression of PN GBM markers (Olig2, PDGFRα)31. Clinicopathological details are provided in Supplementary Table 1. All ethical regulations relevant to human research participants were followed. All procedures for tumor collection and analysis were approved by the Guizhou Medical University Institutional Research Medical Ethics Committee, with informed consent obtained from all participants.
Cell culture
U87MG, U251, LN229, and T98G cells were purchased from the American Type Culture Collection (ATCC). Human embryonic kidney HEK293T cells were obtained from Bena Culture Collection Technology (China). The primary GBM cell lines were derived from GBM surgical specimens and maintained in primary serum-free cultures grown on laminin-coated plates. All cells were cultured in FBS (10%)- and antibiotics-contained Dulbecco’s modified Eagle’s medium (DMEM) at 37 °C. Normal human astrocytes (NHAs) were originally from Lonza and maintained per the manufacturer’s instructions. All cell lines were authenticated by STR profiles and tested for mycoplasma contamination every 2 months.
RNA extraction and qRT-PCR analysis
Following the manufacturer’s protocol, total RNA was extracted with TRIzol reagent (Invitrogen), and first-strand cDNA synthesis was conducted using PrimeScript RT Master Mix (TaKaRa). Real-time quantitative PCR was then carried out with SYBR Green (Applied Biosystems), adhering to the provided guidelines. The relative expression levels of USP14 and ACTN1 mRNAs were normalized to GAPDH and calculated using the 2-ΔΔCt method. The following primers were used for qRT-PCR: USP14, 5’-TGGCTTCAGCGCAGTATATTAC-3’ (forward) and 5’-CCTTGTTCACCTTTCTCGGCA-3’ (reverse); ACTN1, 5’-TGAGGAGTGGTTGCTGAATGAG-3’ (forward) and 5’-AACTTCTCTGCCAGGTGGTCC-3’ (reverse).
Antibodies and reagents
The study utilized the following antibodies: USP14 (Proteintech, #14517-1-AP; WB 1:1000, IF 1:100), ACTN1 (Santa Cruz, #sc-17829; WB 1:500, IF 1:100), FLAG-tag (Abcam, #ab205606; WB 1:1000), His-tag (Cell Signaling Technology, #12698; WB 1:1000), HA-tag (Abcam, #ab9110; WB 1:1000), YKL-40 (Novus Biologicals, #AF2599; WB 1:1000), Olig2 (Santa Cruz, #sc-293163; WB 1:500), PDGFRα (Cell Signaling Technology, #3174; WB 1:1000), MET (Santa Cruz, #sc-8057; WB 1:200), COL5A1 (Cell Signaling Technology, #86903; WB 1:1000), and β-actin (Abcam, #ab8227; WB 1:5000). The USP14 inhibitor IU1 (#662210) and the protein synthesis inhibitor cycloheximide (CHX, #C7698) were obtained from Sigma-Aldrich, while MG132 (#S2619), a proteasome inhibitor, was sourced from Selleckchem.
Transfection
USP14, ACTN1, and control shRNAs were procured from Sigma–Aldrich, with targeting sequences detailed in Supplementary Table 2. Flag-tagged wild-type USP14 (USP14-WT), its catalytically inactive mutant (USP14-C114A), and His-tagged ACTN1 overexpression plasmids were custom-synthesized by Jikai Gene Biotechnology (Shanghai, China) and validated via DNA sequencing. Plasmids encoding various HA-tagged ubiquitin forms, including Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63 (pRK5-HA series), were obtained from Addgene. Cells were seeded in 6-well plates and incubated for 24 hours to reach 70–80% confluency before transfection. Transfections utilized Lipofectamine 3000 (Invitrogen, cat. L3000150) in accordance with the manufacturer’s protocol, and stable cell lines were subsequently selected using puromycin.
Transwell invasion and migration assays
In the invasion assay, 2 × 104 cells were seeded into the upper chambers of 24-well Transwell inserts with 8 µm pores (Corning, NY, USA) in 200 µL of serum-free medium. The lower chambers contained 500 µL of DMEM with 10% FBS to stimulate invasion. After 24 hours of incubation, cells were fixed with 4% paraformaldehyde for 15 minutes and stained with crystal violet. Six randomly selected fields were captured by microscopy for analysis. In the migration assay, filters were used without Matrigel pre-coating.
CCK-8 assay
The CCK-8 assay (Beyotime) was utilized to assess glioblastoma cell proliferation in line with the manufacturer’s protocol. Transfected cells were seeded into 96-well plates at a concentration of 5000 cells per well, and 10 µL of CCK-8 reagent was added at 24, 48, 72, and 96 hours. Cell viability was determined by measuring absorbance at 450 nm. Each experiment included three biological replicates, with measurements repeated in triplicate to ensure accuracy.
Immunoblotting (IB)
Immunoblot (IB) analysis was performed to analyze target protein expression. Cellular proteins were extracted using RIPA buffer with protease inhibitors (Roche), separated by SDS-PAGE, and transferred to PVDF membranes. The membranes were incubated overnight with primary antibodies, followed by HRP-conjugated secondary antibodies after washing. Signals were detected using a SuperSignal West Femto Maximum Sensitivity Substrate Trial Kit (Thermo Fisher Scientific).
Immunofluorescence (IF)
Cells were fixed in 4% paraformaldehyde for 20 minutes, permeabilized with 0.25% Triton X-100, and blocked with 1% BSA for 1 hour. Cells were then incubated overnight with primary antibodies, followed by washing with PBS-T and a 1-hour incubation with secondary antibodies. Nuclei were stained with DAPI, and images were captured using Zeiss LSM 5 confocal microscopy. Colocalization of USP14 and ACTN1 was quantified using the Manders overlap coefficient.
Immunohistochemistry (IHC)
Immunohistochemistry (IHC) was conducted following established protocols. Paraffin-embedded 4-µm tissue sections were baked at 60°C for 1 hour, deparaffinized in xylene, and rehydrated through graded ethanol solutions (100%, 95%, and 85%). Antigen retrieval was performed in pH 6.0 citric buffer for 20 minutes, followed by treatment with 3% hydrogen peroxide in PBS and blocking with goat serum. Sections were sequentially incubated with primary and HRP-conjugated secondary antibodies, developed with DAB chromogen (Vector Laboratories), and counterstained with hematoxylin. Staining intensity for USP14 and ACTN1 was scored from 0 to 3 under ×40 magnification. For analysis of protein expression and survival in the clinical GBM cohort, cases were dichotomized by the cohort median of the per-sample IHC intensity score: High = above the median; Low = at or below the median.
Protein half-life assay
For the ACTN1 half-life assay, GBM cells were treated with the protein synthesis inhibitor cycloheximide (CHX, Sigma-Aldrich, 100 μM) for specified time intervals before sample collection.
Co-immunoprecipitation
Cells transfected with the specified constructs were lysed in NETN buffer containing protease inhibitors. Cell lysates were pre-cleared with protein A/G agarose (Santa Cruz) at 4 °C for 1 hour, then incubated with the target antibodies overnight at 4 °C for immunoprecipitation. The resulting immunocomplexes were bound to protein A/G agarose for an additional 2 hours at 4 °C, washed four times with NETN buffer, and boiled to elute proteins. The samples were then analyzed by SDS-PAGE and IB.
GST pulldown assay
To test direct binding, GST pull-downs were performed exclusively with purified proteins. Full-length human ACTN1 was cloned into pGEX-4T-1, expressed in E. coli BL21 (DE3) at 16 °C with 0.4 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), and purified on glutathione Sepharose 4B followed by gel filtration (Superdex 200) in binding buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM DTT, 5% glycerol, 0.05% NP-40). Purification followed manufacturer protocols for GST-tagged proteins.
Human USP14 (WT or catalytic inactive mutant C114A) bearing an N-terminal Flag tag was expressed in E. coli BL21 (DE3) under the same induction conditions and purified on ANTI-FLAG® M2 affinity gel with competitive elution using 3×Flag peptide, then polished by gel filtration in the same buffer.
For pull-downs, 5 µg GST-ACTN1 (or GST control) was immobilized on 20 µL glutathione beads pre-blocked with 1% BSA (30 min, 4 °C), washed 3× with binding buffer, and incubated with 1–2 µg purified Flag-USP14 (WT or C114A) in 200 µL binding buffer for 1–2 h at 4 °C with rotation. Beads were washed 4–5 times (including two high-salt washes at 300 mM NaCl) and boiled in 2× SDS sample buffer. Bound USP14 was detected by SDS–PAGE/IB with anti-Flag; equal bait loading was verified by Coomassie staining.
In vivo and in vitro ACTN1 ubiquitylation assay
For the in vivo ACTN1 ubiquitylation assay, cells were transfected with the indicated plasmids, then treated with 20 μM MG132 for 8 hours. Cells were collected and lysed in NETN buffer plus 0.1% SDS, 20 μM MG132, and protease inhibitors. Lysates were incubated with anti-ACTN1 antibody for 3 hours and protein A/G agarose beads for a further 8 hours at 4 °C. Reactions were subjected to IB analysis.
For the in vitro ACTN1 ubiquitylation assay, HEK293T cells were transfected with His-ACTN1 and HA-ubiquitin, HA-ubiquitin-Lys27, HA-ubiquitin-Lys48, or HA-ubiquitin-Lys63, and were treated with 20 μM MG132 for 8 hours. Ubiquitylated ACTN1 was purified from the cell extracts with an anti-His affinity column and then incubated with the recombinant GST-USP14 WT or GST-USP14 C114A protein in a deubiquitylation buffer for 2 hours at 37 °C. Reactions were subjected to IB analysis.
In vivo xenografts
For intracranial xenograft studies, male BALB/c-A nude mice aged 5 to 6 weeks were randomly assigned to experimental groups (n = 10 per group). Using a stereotactic apparatus, 5 × 105 luciferase-tagged LN229 or GBM2 cells—modified as required—were injected into the right caudate putamen. Tumor initiation and growth were monitored via bioluminescence imaging (IVIS 200 Spectrum Imaging System) on days 0, 7, 14, and 28 post-implantation. For survival analysis, mice were observed up to 70 days or until severe symptoms (e.g., weight loss, impaired movement) manifested.
To assess the effects of the USP14 inhibitor IU1, ALZET micro-osmotic pumps (DURECT Corp.) were surgically implanted in tumor-bearing mice, administering IU1 (40 mg/kg) or a vehicle at a constant rate of 0.5 μL/h over seven days. Tumor progression was continuously monitored with in vivo bioluminescent imaging, with the study endpoint set at 70 days or upon the development of significant clinical signs.
Sample coding was applied to maintain blinding for the experimenters during analysis. We have complied with all relevant ethical regulations for animal use. All animal procedures adhered to national guidelines and received approval from the Guizhou Medical University Institutional Committee for Animal Research.
Statistics and reproducibility
Data are reported as mean ± s.e.m. Sample sizes and replicates were determined via G*Power based on effect size and Type I/II error levels. Normality and variance were assessed using the Shapiro–Wilk and F tests. For data with normal distribution, Student’s t-test (two groups) or one/two-way ANOVA (> two groups) was applied; otherwise, the Mann-Whitney U test was used. Outliers (>3 standard deviations from the mean) were excluded. Pearson correlation analyzed the relationship between USP14 and ACTN1 expression, and Kaplan-Meier analysis was used for survival in GBM patients and mouse models. GraphPad Prism 9 performed all analyses, with p < 0.05 as the significance threshold.
Ethics approval and consent to participate
All animal procedures adhered to national guidelines and received approval from the Guizhou Medical University Institutional Committee for Animal Research (no. 2302336). All procedures for tumor collection and analysis were approved by the Guizhou Medical University Institutional Research Medical Ethics Committee, with informed consent obtained from all participants.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Numerical source data for all figures are provided as Supplementary Data 1 (Excel); all other data are available from the corresponding author upon reasonable request.
References
Read, R. D., Tapp, Z. M., Rajappa, P. & Hambardzumyan, D. Glioblastoma microenvironment-from biology to therapy. Genes Dev. 38, 360–379 (2024).
Le Rhun, E. et al. Molecular targeted therapy of glioblastoma. Cancer Treat. Rev. 80, 101896 (2019).
Ou, A., Yung, W. K. A. & Majd, N. Molecular mechanisms of treatment resistance in Glioblastoma. Int. J. Mol. Sci. 22, https://doi.org/10.3390/ijms22010351 (2020).
Wang, Q. et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 32, 42–56.e46 (2017).
Verhaak, R. G. et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110 (2010).
Phillips, H. S. et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9, 157–173 (2006).
Guardia, G. D. A. et al. Proneural and mesenchymal glioma stem cells display major differences in splicing and lncRNA profiles. NPJ Genom. Med. 5, 2 (2020).
Kahlert, U. D., Nikkhah, G. & Maciaczyk, J. Epithelial-to-mesenchymal(-like) transition as a relevant molecular event in malignant gliomas. Cancer Lett. 331, 131–138 (2013).
Jiang, Y. et al. Prosaposin is a biomarker of mesenchymal glioblastoma and regulates mesenchymal transition through the TGF-beta1/Smad signaling pathway. J. Pathol. 249, 26–38 (2019).
Khan, A. B. et al. CXCR4 expression is associated with proneural-to-mesenchymal transition in glioblastoma. Int J. Cancer 152, 713–724 (2023).
Masita Silviana, N., Andarini, S., Lyrawati, D. & Hidayat, M. Masticatory Functional Load Increases the mRNA Expression Levels of ACTN2 and ACTN3 and the Protein Expression of alpha-Actinin-2 in Rat Masseter Muscle. Turk. J. Pharm. Sci. 18, 28–33 (2021).
Noureddine, M. et al. Structural and functional insights into alpha-actinin isoforms and their implications in cardiovascular disease. J. Gen. Physiol. 157, e202413684 (2025).
Wang, R., Gao, Y. & Zhang, H. ACTN1 interacts with ITGA5 to promote cell proliferation, invasion and epithelial-mesenchymal transformation in head and neck squamous cell carcinoma. Iran. J. Basic Med. Sci. 26, 200–207 (2023).
Xie, G. F. et al. High ACTN1 is associated with poor prognosis, and ACTN1 silencing suppresses cell proliferation and metastasis in oral squamous cell carcinoma. Drug Des. Dev. Ther. 14, 1717–1727 (2020).
Yang, X. et al. High expression levels of ACTN1 and ACTN3 indicate unfavorable prognosis in acute myeloid leukemia. J. Cancer 10, 4286–4292 (2019).
Cao, Y. et al. Oroxylin A suppresses ACTN1 expression to inactivate cancer-associated fibroblasts and restrain breast cancer metastasis. Pharm. Res. 159, 104981 (2020).
Chen, Q., Zhou, X. W., Zhang, A. J. & He, K. ACTN1 supports tumor growth by inhibiting Hippo signaling in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 40, 23 (2021).
Zhang, S. et al. alpha-Actinin1 promotes tumorigenesis and epithelial-mesenchymal transition of gastric cancer via the AKT/GSK3beta/beta-Catenin pathway. Bioengineered 12, 5688–5704 (2021).
Yang, W., Lin, L., Lu, T., Yu, H. & Zhang, S. Identification of EMT-associated prognostic features among grade II/III gliomas. Sci. Rep. 14, 2822 (2024).
Blondelle, J. et al. Cullin-3–dependent deregulation of ACTN1 represents a pathogenic mechanism in nemaline myopathy. JCI Insight 5, e125665 (2019).
Dewson, G., Eichhorn, P. J. A. & Komander, D. Deubiquitinases in cancer. Nat. Rev. Cancer 23, 842–862 (2023).
Snyder, N. A. & Silva, G. M. Deubiquitinating enzymes (DUBs): Regulation, homeostasis, and oxidative stress response. J. Biol. Chem. 297, 101077 (2021).
Park, J., Cho, J., Kim, E. E. & Song, E. J. Deubiquitinating enzymes: a critical regulator of mitosis. Int. J. Mol. Sci. 20, https://doi.org/10.3390/ijms20235997 (2019).
Zhao, C. et al. A self-amplifying USP14-TAZ loop drives the progression and liver metastasis of pancreatic ductal adenocarcinoma. Cell Death Differ. 30, 1–15 (2023).
Lee, M. Y., Kim, M. J., Jin, J. O. & Lee, P. C. USP14 inhibition regulates tumorigenesis by inducing apoptosis in gastric cancer. BMB Rep. 56, 451–456 (2023).
Shi, D. et al. USP14 promotes tryptophan metabolism and immune suppression by stabilizing IDO1 in colorectal cancer. Nat. Commun. 13, 5644 (2022).
Zhao, X., Wu, X., Wang, H., Lai, S. & Wang, J. Targeted therapy for cisplatin-resistant lung cancer via aptamer-guided nano-zinc carriers containing USP14 siRNA. MedComm. (2020) 4, e237 (2023).
Chen, Z. et al. Targeting MYH9 represses USP14-mediated NAP1L1 deubiquitination and cell proliferation in glioma. Cancer Cell Int. 23, 220 (2023).
Zhou, X. et al. USP14 modulates stem-like properties, tumorigenicity, and radiotherapy resistance in glioblastoma stem cells through stabilization of MST4-phosphorylated ALKBH5. Theranostics 15, 2293–2314 (2025).
Hou, W. et al. USP14 inhibition promotes recovery by protecting BBB integrity and attenuating neuroinflammation in MCAO mice. CNS Neurosci. Ther. 29, 3612–3623 (2023).
Qiu, W. et al. USP10 deubiquitinates RUNX1 and promotes proneural-to-mesenchymal transition in glioblastoma. Cell Death Dis. 14, https://doi.org/10.1038/s41419-023-05734-y (2023).
Liu, X. et al. Deubiquitinase OTUD6A in macrophages promotes intestinal inflammation and colitis via deubiquitination of NLRP3. Cell Death Differ. 30, 1457–1471 (2023).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Bao, Z. et al. Intratumor heterogeneity, microenvironment, and mechanisms of drug resistance in glioma recurrence and evolution. Front Med. 15, 551–561 (2021).
Gao, Z. et al. PDIA3P1 promotes Temozolomide resistance in glioblastoma by inhibiting C/EBPbeta degradation to facilitate proneural-to-mesenchymal transition. J. Exp. Clin. Cancer Res. 41, 223 (2022).
Stark, C. et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 34, D535–D539 (2006).
You, L. et al. USP14-IMP2-CXCL2 axis in tumor-associated macrophages facilitates resistance to anti-PD-1 therapy in gastric cancer by recruiting myeloid-derived suppressor cells. Oncogene 44, 2413–2426 (2025).
Lin, X. T. et al. Integrated ubiquitomics characterization of hepatocellular carcinomas. Hepatology 82, 42–58 (2025).
Zhou, L. et al. Lovastatin targets the USP14-Survivin axis to suppress triple-negative breast cancer via ubiquitin-mediated proteasomal degradation. Cells 14, https://doi.org/10.3390/cells14110816 (2025).
Morgan, E. L. et al. Inhibition of USP14 promotes TNFalpha-induced cell death in head and neck squamous cell carcinoma (HNSCC). Cell Death Differ. 30, 1382–1396 (2023).
Klaeske, K. et al. Differential regulation of myocardial E3 ligases and deubiquitinases in ischemic heart failure. Life 11, https://doi.org/10.3390/life11121430 (2021).
Tu, Y. et al. Loss of deubiquitylase USP2 triggers development of glioblastoma via TGF-beta signaling. Oncogene 41, 2597–2608 (2022).
Tu, Y. et al. Smoothened promotes glioblastoma radiation resistance via activating USP3-mediated Claspin Deubiquitination. Clin. Cancer Res. 26, 1749–1762 (2020).
Acknowledgements
This work was funded by the following grants: Project of Science and Technology Foundation of Guizhou Health Commission (Grant No. gzwkj [2022-090] and Grant No. gzwkj [2024-146]); Guizhou Provincial Science and Technology Projects (Grant No. QianKeHe Foundation-ZK [2024] General-244); Doctoral Research Launch Foundation, the Affiliated Hospital of Guizhou Medical University (Grant No. gyfybsky-2024-16). We extend our sincere appreciation to Dr. Xiaomin Cai from Xinhua Hospital Affiliated to Shanghai Jiaotong University School of Medicine for graciously supporting us.
Author information
Authors and Affiliations
Contributions
W.J.Q., P.L., and L.Z.C. conceptualized and designed the study. W.J.Q. X.P.S. and R.T.W. carried out the majority of the experiments. Y.J.H., S.B.S., and Y.M.C. gathered the clinical samples. J.Q.S., Y.G.L., and L.Z.C. conducted data analysis and interpretation. W.J.Q. and X.P.S. authored the manuscript, with H.Y., P.L., and L.Z.C. providing critical review and revisions. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Derrick Ong and Kaliya Georgieva. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Qiu, W., Shi, X., Wei, R. et al. USP14-mediated stabilization of ACTN1 maintains mesenchymal characteristics in glioblastoma. Commun Biol 8, 1680 (2025). https://doi.org/10.1038/s42003-025-09083-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-09083-8