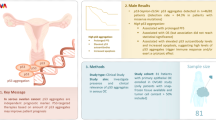
Abstract
P53 Phase separation is crucial towards amyloid aggregation and p63 and p73 have enhanced expression in tumors. This study examines the phase behaviors of p53, p63, and p73. Here we show that unlike the DNA-binding domain of p53 (p53C), the p63C and p73C undergo phase separation, but do not form amyloids under physiological temperatures. Wild-type and mutant p53C form droplets at 4°C and aggregates at 37 °C with amyloid properties. Mutant p53C promotes amyloid-like states in p63C and p73C, recruiting them into membraneless organelles. Amyloid conversion is supported by thioflavin T and Congo red binding, increased light scattering, and circular dichroism. Full-length mutant p53 and p63C (or p73C) co-transfection shows reduced fluorescence recovery after photobleaching. Heparin inhibits the prion-like aggregation of p63C and p73C induced by p53C. These findings highlight the role of p53 in initiating amyloid aggregation in p63 and p73, opening avenues for targeting prion-like conversion in cancer therapy.
Similar content being viewed by others
Introduction
The p53 gene, often dubbed the “guardian of the genome,” has been recognized as having a central role in cancer biology1,2. With mutations in this pivotal tumor suppressor occurring in over half of all malignancies, it is crucial to search for innovative treatments targeting these anomalies3. Its essential function in cancer prevention impacts cell cycle control and triggers cell death when DNA is damaged. It has been found that mutant p53 tends to undergo amyloid aggregation, which is correlated with the carcinogenic process4,5,6,7. Our group and others have contributed to this growing body of knowledge, uncovering the threatening ability of mutant p53 to confer oncogenic properties, including chemotherapy resistance, upon these aggregates4,8,9,10,11.
Protein aggregation and misfolding result in a toxic gain of function in neurodegenerative diseases; however, in cancer, mutant aggregates of p53 result in a loss of its tumor-suppressing function and increase the oncogenic potential of cells6,12,13,14,15. This discovery has identified mutant p53 as a novel pharmacological target, previously considered elusive11,12,16. Groundbreaking research conducted in different laboratories has established the foundation for understanding the pathological role of mutant p53 amyloid aggregates, which have been observed in breast cancer cells as well as other types of tumors, including skin, prostate, and ovarian carcinomas17,18,19,20.
Biophysical studies have revealed that despite the structural similarities between the DNA binding domains of p53 and its paralogous forms p63 and p73 (herein referenced as p53C, p63C, and p73C), the latter two exhibit a reduced propensity for aggregation21,22. Biophysical studies and molecular dynamics (MD) simulations have revealed specific regions of increased exposure backbone hydrogen bonds (BHBs) in p53C compared with p63C and p73C21. These regions of structural vulnerability in the p53C are new targetable sites for modulating p53 stability and aggregation, a potential approach to cancer therapy11,21. This difference pinpoints a unique aspect of p53’s structure that may be exploited therapeutically. Thus, the modulation of p53’s stability and aggregation presents a tempting target for cancer therapy, with recent studies showing how mutant forms of p53 can interact with p63 and p73 through a co-aggregation mechanism7,23. Mounting evidence confirms that p53 mutants associate with p63 and p73, impairing the tumor suppressor functions and increasing oncogenicity7,11,24. However, the interaction and oncogenic conversion mechanisms must be fully understood. P63 and p73, while less frequently mutated in cancers, are indispensable for development and play pivotal roles in tumor suppression. Despite their structural similarities, these proteins have distinct functions and are regulated through unique mechanisms25.
The phenomenon of phase separation has recently gained prominence as a crucial event in the life cycle of a cell, with implications for neurodegenerative diseases and cancer11,26,27,28,29,30,31,32,33. We have recently demonstrated that phase separation is a precursor to the amyloid aggregation of p53, presenting a significant avenue for therapeutic intervention32. The tendency to undergo phase separation and, eventually, phase transition is an intrinsic property of p5310,30,32,34,35,36. Other p53-related proteins and tumor suppressors equally undergo phase separation in the nucleus37,38. In the case of the tumor suppressor speckle-type BTB/POZ protein (SPOP), mutations have been associated with phase separation defects37. SPOP mutations disturb substrate interactions that affect SPOP phase separation and its localization to membraneless organelles. p53 has arisen as a cancer-related protein in which mutations can disturb its folding, likely leading to phase transition and, thereby, to the formation of amyloid aggregates with oncogenic properties (GoF)11,32,39.
When unraveling the intricate protein interactions within cancer, the interplay among p53, p63, and p73 emerges as a central focus of our study. Overexpression of p63/p73 and their related isoforms has been implicated in several tumors25. Further, p53 gain-of-function mutations can hijack p63/p73 into aggregates7 and, under specific conditions, form hetero-tetramers40, adding complexity to their hierarchical role in cancer. Oligomerization of all three proteins is regulated by the same S100 protein family41, suggesting reminiscent sequence conservation among the p53 family and potentially overlapping functionalities in physiology and cancer signaling. Our research aims to unravel the phase behaviors of these proteins, illuminating the complex mechanisms driving their transition from a diffuse phase to an amyloid-like solid state. Our findings contribute to understanding how the gain-of-function mutant M237I, the wild-type forms of p53, and the hotspot mutations R248Q and R249S precipitate the amyloid aggregation of p63 and p73. The M237I substitution is a gain-of-function chemoresistance mutation present in glioblastoma. It enhances the resistance of glioblastoma cells to temozolomide by up-regulating methylguanine DNA-methyltransferase42. The frequency of M237I in the UMD TP53 mutation database returned 366 occurrences, the lowest in comparison with hotspot mutations such as R248Q (2562 occurrences), R249S (895 occurrences), and R273H (2361 occurrences)11. However, glioblastoma is the most aggressive and invasive cancer of the neural system, revealing poor prognosis after treatment and low survival rates. A previous report revealed that M237I is located near the Cys3-His Zn2+-binding motif, disrupting local hydrophobicity, decreasing protein stability, and substantially increasing amyloid aggregational rates9. M237I has increased amyloid aggregation compared to other hotspot p53 mutations, such as R248Q4. The devasting clinical features of glioblastoma and the increased tendency of M237I to aggregate brought to our attention whether increased p53 aggregation could explain the worst phenotype seen in glioblastoma. Such insights offer new vantage points regarding mutant p53’s pathogenic influence in oncogenesis, setting the stage for groundbreaking therapeutic strategies. The induction of amyloid-like conformation by p53 variants in paralogous proteins holds significant cancer-causing potential. Heparin, a polyanion known for its regulatory influence on p53 aggregation, emerges as a potent inhibitor of the prion-like aggregation of p63 and p73 mediated by p53. Transfected cells with M237I mutant evidence the colocalization of p53 condensates with p73. Colocalization and a phase transition were observed in cells co-transfected with the full-length M237I mutant of p53 and p63C, manifested by reduced fluorescence recovery after photobleaching. By pinpointing the specific conditions conducive to the amyloid aggregation of p63 and p73, our work enhances the comprehension of p53’s role in cancer development. It underscores the therapeutic potential of intervening in these phase transitions as a novel anticancer strategy.
Results
The interplay between phase separation and amyloid aggregation in the p53 family of proteins
Our investigation began with a focus on the phase separation behavior of essential proteins associated with cancer, namely p63, p73, and p53. We specifically studied the DNA-binding domains of these proteins, p63C, p73C, and p53C, and their propensity to undergo phase separation at various concentrations. As shown in Fig. 1a, both p63C and p73C, as well as p53C, exhibited phase separation at different protein concentrations. This process, facilitated by polyethylene glycol (PEG) at 4 °C, was observed using differential interference contrast (DIC) microscopy (Fig. 1a–f and Supplementary Fig. 1a). Visual examination of the images revealed that p63C exhibited demixing at lower concentrations than p73C, forming larger droplets at higher concentrations (Fig. 1e). Expanding on our previous research, which detailed the phase separation of p53C leading to aggregation under diverse conditions32, we observed that the M237I mutant phase separates in the presence of PEG and transitions to a solid-like state more rapidly than the wild-type p53C (Fig. 1c, f). The increased aggregation propensity of some p53C mutations, such as the M237I, contrasts with the role of the N- and C- intrinsically disordered p53 regions (IDRs). Like the wild-type p53C32, the full-length wild-type p53 (residues 1-393) underwent phase separation rather than aggregation (Supplementary Fig. 1b), supporting that the destabilized M237I p53 mutation contributed the most to aggregation than p53 IDRs. Aggregation of the full-length wild-type p53 requires longer incubation times at 37 °C compared to p53C4. Within living cells, the full-length version of M237I p53 resulted in phase separation and phase transition within nuclear compartments, likely contributing to gain-of-function effects32. Therefore, the aggregation propensity of M237I supersedes the influence of p53 IDRs on phase separation, reinforcing the relevance of studying the p53 DNA-binding domain carrying M237I and hotspot mutations in exerting effects on p63C and p73C. The role of p63 and p73 IDRs in phase separation processes is awaiting further exploration. Figure 1c, f illustrate that in the presence of PEG, p63C and p73C also formed droplets akin to p53C; after increasing the temperature to 37 °C, wild-type and mutant p53C formed aggregates (Fig. 1c, arrows). Wild-type and M237I p53C transited from droplets to aggregates (Fig. 1f, asterisks), whereas p63C and p73C did not demonstrate intrinsic aggregation (Fig. 1c, f). Figure 1d presents the transition of p53C to a solid state at 25 °C, capturing the formation of p53C aggregates through successive bright-field DIC microscopy images and video recordings (Supplementary Fig. 2 and Supplementary Movies 1–3).
Bright-field differential interference contrast (DIC) microscopy images were collected, revealing (a) the concentration-dependent demixing of p63C and p73C, and (b) the demixing of p53C from the PEG solution. c The temperature dependence of p53C, M237I, p63C, and p73C demixing and aggregation (arrows). d The time dependence of p53C aggregation from droplets at 25 °C (see methods); Schematic representations of (e) p63C and p73C demixing concentrations, and (f) the temperature dependence of the studied proteins. Colored circles indicate the phase separation of p53C (green), M237I (gray), p63C (red), and p73C (blue). Circles filled with an asterisk depict aggregation conditions.
We analyzed Congo Red (CR) binding, a hallmark of amyloid aggregates, to p53 family proteins at 4 °C and 37 °C. The amyloid nature of p53C aggregates, whether mutant or wild-type, was readily indicated by CR binding as assessed by its fluorescence (Fig. 2a, b), a phenomenon not observed for p63C and p73C (Fig. 2c, d). Control experiments without PEG revealed no droplets and aggregation at 4 °C for p53C and M237I. At 37 °C, CR-positive aggregates were observed for p53C and M237I but not for p63C and p73C (Supplementary Fig. 3). Because p63C droplets do not bind to CR, revealing they are non-amyloidogenic, we labeled p63C using rhodamine to explore droplet behavior under different conditions. Control experiments show that the rhodamine-labeled p63C droplets disassemble upon PEG dilution, evidence of its reversible and non-amyloidogenic nature (Supplementary Fig. 4a, right panel). As for different p53 constructs30, p63C phase separation is also electrostatically driven. At higher sodium chloride concentrations (from 150 to 500 mM), PEG-induced rhodamine-labeled p63C droplets are not formed (Supplementary Fig. 4a, left panel). Further, the PEG dependency on rhodamine-labeled p63C droplet formation revealed that phase separation occurs more robustly using 15 or 20% PEG, is weakly revealed at 10% PEG, and is not observed at 5% PEG (Supplementary Fig. 4b).
Microscopy images were captured, presenting the Congo red (CR) fluorescence and bright-field channels of (a) p53C, (b) M237I, (c) p63C, and (d) p73C at 4 and 37 °C. White arrows indicate the presence of p53C and M237I aggregates. Scale bar: 20 μm; Line plots showing the area under the light scattering curve as a function of increasing temperatures (4, 25, and 37 °C) of (e) p53C (green), (f) M237I (gray), (g) p63C (light red), and (h) p73C (light blue) in the absence (colored circles) and the presence (colored squares) of PEG. Plots in (g, h) show two segments in the y-axis to highlight negligible scattering. Line plots with the filled area showing turbidity at 600 nm as a function of increasing concentrations of (i) p53C (green palette), (j) M237I (gray palette), (k) p63C (red palette), and (l) p73C (blue palette) at 4, 25, and 37 °C. Plots in (k, l) show two segments in the y-axis to highlight negligible turbidity. Red and blue palettes are not depicted in (k, l) due to similar values across studied temperatures. Scatter dot plots showing the thioflavin T fluorescence as a function of increasing temperatures (4, 25, and 37 °C) of (m) p53C, (n) M237I, (o) p63C, and (p) p73C. The color palette is the same as in (i–l). Plots show two segments in the y-axis to evidence large changes in the thioflavin T signal across studied proteins. The data are shown as the mean ± s.e.m. of n = 3 independent experiments using the same protein batch.
Light scattering (LS) assays were employed to quantitatively compare the aggregation of p53C droplets versus the non-aggregation of p63C and p73C (Fig. 2e–h). The results underscore that p63C and p73C displayed no temperature-dependent aggregation, whether in solution or the droplet state (Fig. 2g, h), a striking disparity compared to p53C and M237I. Conversely, p53C and M237I demonstrated temperature-dependent aggregation, which is evident in both the presence and absence of PEG (Fig. 2e, f). In the presence of PEG, 15% at 25 °C, p53C, and the p53C M237I revealed a 10- and 20-fold increment of the LS signal compared with p63/p73C, respectively. At 37 °C, these values raised to 15- and 40-fold, respectively. Turbidimetry assays at 600 nm corroborated the results obtained from light scattering analysis (Fig. 2i–l) and ThT binding analysis (Fig. 2m–p). LS and turbidimetry measurements indicate general aggregation processes, including amorphous and amyloid aggregation. In contrast, ThT relies specifically on amyloid aggregates. At 25 °C, the p53C and the p53C M237I revealed ca. 12- and 65-fold increase of ThT binding relative to p63/p73C. At 37 °C, these numbers reached ca. 22- and 120-fold, revealing that M237I has a greater tendency to form amyloids than wild-type p53C. P63C and p73C have shown negligible amyloid formation. The observed higher propensity of p53C for temperature-induced aggregation than p63C and p73C indicates significant differences in their aggregation behaviors, potentially bearing critical biological implications that warrant further investigation.
p53C mutants as a catalyst for prion-like aggregation in p63C and p73C protein droplets
Figure 3 explores the capability of p63C and p73C droplets to transition into aggregates in the presence of p53C aggregates, utilizing rhodamine-labeled p63C (Fig. 3a) or p73C (Fig. 3b). Droplet morphology drastically changes from homogeneously round to diffuse speckles when p53C is mixed with rhodamine-labeled p63C or p73C, as revealed by the fluorescent and bright-field channels (Fig. 3a, b). To unequivocally track the co-phase separation and aggregation of p53C with p63C or p73C, we performed dual labeling experiments using Alexa fluor 488 (AF488) to report either p53C or M237I p53C and rhodamine to report p63C or p73C (Fig. 4). Microscopic examination unveiled that at 37 °C and using stoichiometric concentrations of p53C and p63C or p73C, p53C coaggregates with p63C and p73C (Fig. 4a, c). At 4 °C, droplets of p53C/p63C and p53C/p73C persisted even using M237I p53C, indicating a co-phase separation event regardless of the presence of a high amyloidogenic mutation (Fig. 4a–d). In contrast, the coaggregation of M237I p53C/p63C and M237I p53C/p73C is significantly observed at 37 °C (Fig. 4b, d). We further tested the coaggregation ability of two AF-488-tagged hotspot p53C mutations on converting p63C and p73C droplets into aggregates. R248Q has a lower tendency to convert the p53C family members than R249S (Supplementary Fig. 5). The results show the phase and aggregation behavior of the p53 family members to associate with each other, forming coaggregates and persisted co-phases under the influence of temperature, and that wild-type and p53C mutations may induce coaggregation with typically non-amyloid p63C and p73C. These results provide insight into the gain-of-function phenomena observed in mutant p53 tumors. Figure 4e summarizes the coaggregation of p63C and p73C in the presence of p53C and M237I.
Microscopy images showing the Alexa Fluor 488 (AF488) labeled (a, c) p53C or (b, d) M237I (green), the rhodamine-labeled (Rhod) (a, b) p63C or (c, d) p73C (red) fluorescent channels together with the bright-field and merged images at 4 and 37 °C. Scale bar: 20 μm. e Schematic representation of the crosstalk within the p53 family members at 4 (dashed cyan) and 37 °C (dashed orange). Red and blue circles depict demixing with droplet formation. Red and white circles filled with an asterisk indicate a mixture of droplets and aggregates, and mainly aggregates, respectively. NA stands for not analyzed.
Previous reports show that p53 mutants can convert wild-type p53 and p53 family members p63 and p73 into amyloid species through a prion-like mechanism4,13. However, the species undergoing this conversion are unknown. To test this idea, we asked whether non-amyloid p63C droplets would be part of these conversion steps in the presence of p53C and M237I p53C seeds. Light scattering measurements evaluated the prion-like seeded aggregation of p53C (Fig. 5a–c). P53C (wild-type and M237I mutant) was initially incubated at 37 °C. Then, the aggregated protein was diluted fortyfold to be used as a seed and mixed with p63C under conditions where the p63C phase separates. P53C seeds increased p63C light scattering in the presence of PEG (Fig. 5a–c). The M237I p53C mutant exhibited a higher seeding capacity than the wild-type (Fig. 5c). The results, particularly with the M237I mutant, demonstrated that p53C seeds can trigger p63 aggregation even in significantly reduced concentrations. To evaluate the nature of the p63C aggregates seeded by M237I p53C, we performed the same seeding experiment under a microscope. We imaged the aggregates following the rhodamine-labeled p63C, the thioflavin T (ThT) fluorescence, and the intrinsic fluorescence of the aggregates upon excitation in the visible range (Fig. 5d). ThT is a well-described probe that reports amyloid structures. Additionally, the intrinsic fluorescence at the visible range has been used to report the cross-β sheet scaffold of canonical amyloids, such as amyloid 1-42, lysozyme, and the Tau protein43. During the cross-β sheet conversion, electronic states seem to become available, leading to this convenient label-free fluorescence43. The results show that seeds of M237I p53C convert rhodamine-labeled p63C droplets into aggregates that bind both ThT and have intrinsic fluorescence upon visible excitation (Fig. 5d). Further, we followed the seeded aggregation of p63C over time using the bright-field and Congo red channels (Supplementary Movies 4–8). Our data revealed a seeding-induced conversion of non-amyloid p63C droplets into aggregates presenting amyloid-like features.
Scatter dot plots showing area under the light scattering curves of (a, b) p63C (red bars), seeds of (a) p53C (green bar) or (b) p53C M237I (black bar), after mixing p63C with (a) p53C seeds (red bars colored green) or (b) p53C M237I seeds (red bars colored black), in the absence (-) or presence (+) of PEG 15%. c Changes in the light scattering (Δ area) show the p63C aggregation in the presence of PEG after subtracting the p53C (red bar colored green) or M237I (red bar colored black) seeds contribution. d Confocal images showing rhodamine-labeled p63C (red), Thioflavin T (green), and the intrinsic fluorescence (blue) of p63C aggregates.
We also investigated the impact of coaggregation with p53C and the M237I mutant on the secondary structure of p63C (Supplementary Fig. 6a, b). Far-ultraviolet circular dichroism experiments revealed a significant loss of secondary structure in p63C induced by the wild-type p53C and the M237I mutant (Supplementary Fig. 6a, b). The loss of secondary content is evident from the decrease in the absolute value of the mean residue molar ellipticity [θ]MRE. Further, a shift in the absorption valleys of p63C and p53C WT/M237I was observed after incubation at 37 °C from 215 nm to 218 nm. The changes in the secondary structure, estimated as detailed in the referenced study44, are shown in Supplementary Fig. 6c. A relative increase in β-sheet content is observed when p63C coaggregates with either wild-type p53C or the M237I mutant. It is worth mentioning that amyloid aggregates feature increased β-sheet content45. At 37 °C, p63C in solution exhibits approximately 11% α-helix and 33.2% β-sheet content. These percentages show slight changes after adding 15% PEG 4000: 12.7% α-helix and 29.5% β-sheet. However, under coaggregation conditions with the p53C M237I mutant in the presence of 15% PEG 4000, the α-helix content decreases to 2.7% while the β-sheet content increases to 41.3%.
Although the M237I mutation is located within the p53C Zn2+-binding motif, causing increased structural instability and propensity to aggregate, it is not as frequent in cancer as the hotspot mutations R248Q, R249S, and R273H. To address whether a few of these mutations recapitulate the M237I phenotype of converting p63C droplets into aggregates, we first showed that when EGFP-tagged and recombinantly enriched in solution, R249S and R273H aggregate differently in comparison to M237I. The morphology of R273H aggregates revealed less “sticky” and more diffuse than those observed for M237I and R249S (Supplementary Fig. 7a). Previous studies showed that the effects of R273H on p53C structural rigidity are weaker than that of M237I and R249S46, which may explain distinct morphologies. Next, we investigated the ability of Alexa Fluor 488-tagged p53C R248Q and R249S seeds to induce the aggregation of rhodamine-labeled p63C droplets. We observed by visual inspection that R249S p53C seeds have a greater tendency to convert p63C droplets into aggregates, as do R248Q seeds (Supplementary Fig. 7b, c). Altogether, we showed that R248Q and R249S recapitulate, to a different extent, the M237I phenotype of converting p63C droplets into aggregates, which is most likely explained by their different effects in disrupting the p53C stability.
Mutant p53C induces solid-like phase transition in full-length p63 and p73 within the nucleus
To elucidate the colocalization of the M237I p53 with endogenous p63 and p73, we employed a specialized transfection protocol on p53-null H1299 carcinoma cells. Previous studies have shown that the full-length M237I p53 tends to undergo phase separation in the nucleolus, forming gel-like droplets32. Figure 6 illustrates that transfected cells displayed p73 puncta colocalizing with EGFP-tagged p53 M237I within the nucleolar membraneless compartments (Supplementary Fig. 8). It is noted that colocalization of p73 and the p53 M237I mutant was less common in other nuclear areas. M237I p53 and p63 colocalization were sparsely visualized (data not shown).
a–e Collection of images showing endogenous p73 colocalization with exogenous full-length p53 M237I inside membraneless compartments. Five distinct regions of interest and the corresponding channels: p73 (red), EGFP-tagged p53 M237I (green), nuclei (blue), bright-field, and merged. Last panel show zoomed images from dashed rectangles to emphasize p53 and p73 colocalization spots (white and black arrowheads).
To better visualize co-phase separation of full-length M237I p53/p63C and M237I p53/p73C within cells and get an idea of their dynamics, we performed co-transfection assays to evaluate the colocalization of p53 M237I-EGFP and p63C-mCherry (Fig. 7a) and p73C-YFP (Fig. 7b) in p53-null H1299 carcinoma cells. Figure 7 shows colocalization of the two proteins in droplet-like states within the nucleus, particularly in the nucleolus. Fluorescence recovery after photobleaching (FRAP) to assess the fluidity of the p63C-mCherry and p73C-YFP signal shows the recovery is much slower and incomplete in the nucleolus (Supplementary Fig. 9), indicating that p63C and p73C are undergoing a gel- or solid-like transition typical of amyloid formation (Fig. 7c, d).
a, b Images of individually tagged p53 family members p63C and p73C co-transfected with full-length p53 M237I in H1299 p53-null cells. Arrows indicate membranelle compartments. Dot plots showing the time-dependent fluorescence recovery of (c) the p53 M237I/p63C and (d) the p53 M237I/p73C nuclear (black) and nucleolar (red) regions. The data are shown as the mean ± s.d. of n = 5 cells of the same coverslip.
Sequence analysis of the p53 family members and phase separation motif propensities
The analysis of p53, p63, and p73 reveals that these proteins share almost 50% identity and 75% conservation (yellow bar, Supplementary Fig. 10), especially within the DNA-binding domains, which is crucial for their transcriptional activity and structural integrity. Divergence is noted primarily in the N-terminal transactivation and C-terminal regulatory domains, influencing each protein’s functional roles and differential aggregation tendencies. Specific regions within the DNA-binding domains of all variants, especially around amino acids 251–257 (p53 numbering 251- ILTIITL -257), exhibit conserved hydrophobic residues contributing to aggregation47,48. The corresponding segments within the p63 and p73 are ILIIVTL and ILIIITL, respectively (dashed rectangle, Supplementary Fig. 10). The Thr-to-Ile substitution at position 253 (p53 numbering) changes a polar side chain in p53C to a highly hydrophobic one in p63C. This segment is located within the middle of a β-strand, which may increase local hydrophobicity and contribute to phase transition. Ηowever, this observation contrasts with the fact that p63C is less prone to amyloid aggregation than p53C21. The increased hydrophobicity of an already highly hydrophobic segment may have minor contributions to phase transition events. Indeed, Thr-to-Ile substitution at position 253 in p53C and Tyr-to-Phe at position 236 has shown increased p53C stability49. Therefore, other hotspot sites within the p53 are more likely to contribute to phase transitions, i.e., those related to the Zn2+-binding motif and within the DNA-binding interface. The oligomerization domains (OD) (Supplementary Fig. 10), essential for tetramer formation, show conserved and divergent features that might influence aggregation behavior differently among the proteins. The most significant difference between the proteins is the presence of the Sterile-alpha-motif (SAM) domain and the transactivation inhibitory (TI) domain, both absent in p53. The SAM domains in p63 and p73 are critical in protein-protein interactions that control their functions50. They consist of a conserved structure with five helices connected by linkers, essential for their stability and interaction abilities.
To predict sequence motifs in p53, p63, and p73 that may undergo phase separation and disorder-to-disorder transitions, we used FuzPred51,52 and FuzDrop53,54,55. Supplementary Fig. 10 shows droplet-promoting regions (DPRs) and disorder-to-disorder segments within p53, p63, and p73, respectively. DPRs in p53 are identified at residues 1–24, 28–108, 277–337, and 341–393 (blue, Supplementary Fig. 11a). Disorder-to-disorder regions (pDD) are noted at residues 1–20, 56–94, 284–319, and 346–393 (orange, Supplementary Fig. 11a). In p63, DPRs are identified at residues 36–53, 116–189, 279–296, 343-386, and 436-547 (blue, Supplementary Fig. 11b) and pDD at residues 38–64, 116–186, 339–382, and 443–539 (orange, Supplementary Fig. 11b). In p73, DPRs are identified at residues 1–16, 60–124, 157–172, 315–352, 380–499, and 596–611 (blue, Supplementary Fig. 11c) and pDD at residues 1–15, 47–120, 318–343, and 396–483 (orange, Supplementary Fig. 11c). Different from what was observed within the DNA-binding domain of p53, the p53 family members p63C and p73C revealed segments within their DNA-binding domains that may contribute to droplet formation. The transactivation domains of all p53 family members showed segments related to droplet formation. Curiously, all SAM domains of p63 and p73 are also involved in phase separation events.
Heparin blocks p53C-induced prion-like aggregation in p63C and p73C proteins
Collectively, the coaggregation phenomena depicted above in vitro and in cell highlight a potential mechanism of template conversion. The seeding ability of M237I, R248Q, and R249S on converting non-amyloid p63C droplets into amyloid aggregates underscores the capability of mutant p53C to facilitate pathological amyloid aggregation in p63C and p73C proteins, which generally do not form such aggregates. This coaggregation activity, potentially contributing to the oncogenic gain-of-function observed in tumors harboring mutant p53, was revealed through our microscopic analysis. These findings illuminate the potential for mutant p53 to modulate the phase behavior of p63 and p73, implicating it in the pathological aggregation processes tied to cancer.
The amyloidogenic propensity of p53 mutants suggests potential avenues for novel therapeutic interventions in cancer. One such approach to disrupt the amyloidogenic process involves the utilization of inhibitors targeting p53 aggregation. Our previous research found that heparin modulates p53 phase separation, inhibiting its progression to a more solid state and likely maintaining it in a dynamic, gel-like state32. Consequently, we explored whether heparin could similarly influence the prion-like seeding property of p53C (Fig. 8a–d). The data revealed that in the presence of heparin, M237I p53C seeds fail to induce the transition of p63 and p73 from droplets into aggregates (Fig. 8c, d and Supplementary Fig. 12). Light scattering measurements provided further support, demonstrating that both wild-type and mutant p53C diminished p63C aggregation in the presence of heparin while likely leaving the p63C droplets unaffected (Fig. 8a, b). These findings underscore the potential of heparin as an inhibitor of p53-mediated aggregation of p63C, highlighting its promise as a therapeutic intervention in cancer.
Bar plots show heparin’s effect on p63C aggregation when mixing p63C with (a) p53C or (b) p53C M237I in the presence of PEG. The data are shown as the mean ± s.e.m. of n = 3 independent experiments using the same protein batch. Microscopy images showing the AF488-labeled (c) p53C or (d) M237I, the Rhodamine-labeled p63C, the bright-field, and merged channels in the absence (-) or presence (+) of heparin. Scale bar: 20 μm.
Discussion
Our results feature the fundamental role of phase separation in the pathophysiology of p53-related cancers. The transition of p53 from a liquid to a solid state, observed in both mutant and wild-type forms, supports the hypothesis that protein phase behavior is intrinsically linked to cellular dysfunction in cancer11. The novel finding that p53 can induce the amyloid aggregation of its paralogs, p63, and p73, from the droplet state advances our understanding of the molecular underpinnings of oncogenesis. The results are summarized in a scheme in Fig. 9. This phenomenon highlights the dual nature of p53 as both a guardian against and a facilitator of tumorigenesis, depending on its conformational state.
The capacity of both mutant and WT p53C to induce a shift in p63C and p73C from a liquid to an amyloid-like solid state implies a potential universality in the aggregation mechanisms of these proteins. This insight is paramount as it suggests that the oncogenic potential of p53 may not be solely relegated to its mutant forms. Instead, it positions the phase behavior of p53 as a central factor in the dysregulation of tumor suppressor functions11,32,39. Safari and colleagues first explored the phase behavior of p53 in vitro, and full-length p53 phase separation was revealed as an anomalous condensate56. Kamagata et al. explored the role of p53 IDRs into phase separation30. The authors showed that full-length p53 phase separation is modulated by pH and salt30. P53 depleted by the C-terminal residues 356-393 substantially decreased PS when modulated by salt and pH. However, it was completely abrogated when the N-terminal residues 1-94 were deleted from the full-length protein30. The TET-CT itself does not phase separate as well30. Our group has demonstrated that a p53 construct comprising residues 1-320 has a much lower propensity to aggregate than the DNA-binding domain18. The TAD (residues 1-94) negatively regulates p53 aggregation, potentially because of transient p53 intramolecular interactions with the DNA-binding domain, as revealed by NMR data57. Further, Usluer et al. showed that p53 TAD2 binds to poly-PR/GR dipeptide repeats, inducing phase separation through electrostatics and hydrophobic interactions58. The role of the phosphorylated residue Ser392 at the p53 C-terminal has been addressed as an essential modulator of PS59. Other authors also revealed phase separation of the unstructured basic region (UBR), comprising residues 293-393, and that mutations within the tetramerization domain impaired phase separation60. While the IDRs of p53 are participative in the events of PS, we emphasize the importance of the structured DNA-binding domain as a pivotal trigger in dysregulating tumor suppressor functions through PS.
A droplet of p53C represents an environment in which p53 is highly concentrated, favoring amyloid aggregation. P53C is a metastable domain with a mid-physiological melting temperature61, meaning it may undergo order-to-disorder transitions that could aid in phase separation and aggregation. Destabilized p53C hotspot mutations in cancer potentially accelerate these processes and may eventually sequester p63 and p73 proteins, as shown before in vitro and in cells7. Here, we included a novel piece of evidence for this puzzle. The non-amyloidogenic p63C and p73C droplets are converted to amyloid-like aggregates upon a seeding-induced mechanism triggered by cancer-related p53C mutations. The implications of this finding open debates on the circumstances in which phase separation is indeed an on-pathway route to aggregates. In proteins linked to neurodegenerative disorders such as hnRNPA1, a low complexity domain named A1-LCD, undergoes homotypic interactions modulating metastable droplets that suppress fibril formation62. In the case of profilin, an abundant cellular protein, it binds to the N-terminal fragments of the huntingtin protein, destabilizing aggregation, and phase separation through an event known as polyphasic linkage63. In amyotrophic lateral sclerosis (ALS), mutations in TDP-43 have been shown to disrupt phase separation by inhibiting interactions and helix stability64. The mentioned works point to the scenario where phase separation is not necessarily an intermediate toward protein aggregation.
Nevertheless, contrasting results have shown, evidence of phase separation and aging to amyloid fibrils65 or at least enhanced fibrilization in protein-rich droplets66 for the same hnRNPA1 protein. For the ALS-associated FUS protein, aging to aggregates requires an on-pathway phase separation event67. In the case of p53, seeds of the cancer-related mutations M237I, R248Q, and R249S are competent to convert non-amyloidogenic p63C droplets into amyloid-like mixed aggregates.
Another important discussion is whether p53 phase separation and aggregation are triggered solely by the intrinsic feature of a destabilized aggregation-prone p53C mutated domain or whether other molecules are participative. Our co-transfection experiments using EGFP-tagged full-length p53 M237I with mCherry-tagged p63C or YFP-tagged p73C revealed co-phase separation of M237I p53/p63C and M237I p53/p73C within the nucleolus, an environment full of nucleic acids. We have reported the complex role of DNA and RNA binding to p53C and their modulatory role in p53C aggregation and phase separation68,69. Therefore, it is tempting to speculate that these molecules could bi-directionally tune p53 inside cells to move forward into aggregates or back to metastable droplets. However, we cannot underpin these questions, especially considering a complex tripartite system of DNA/p53/p63. Considering the destabilized cancer-related p53C mutations, their effects on DNA-binding capabilities and protein instability are also distinct. For instance, M237I is located within the Zn2+-binding site and directly increases the hydrophobicity near the Cys3-His motif. It drastically impacts the p53C structure integrity and DNA binding ability and is highly aggregational9,46. R248Q and R273H are DNA contact mutations that dramatically affect p53’s DNA binding abilities but do not substantially impact the structural folding of p53C46. Conversely, R249S, although not located within the DNA binding interface, nullifies DNA binding and mildly affects p53C folding46. The distinct signatures of these p53C mutations explain their distinct aggregation tendencies. However, they are all competent in seeding the conversion of p63C droplets into amyloid aggregates, indicating intrinsic features within the p53C folding prone to induce a seeding conversion mechanism.
Also, what could be the role of these p53 family condensates in regulating or enhancing transcription activity? A series of transcriptional factors (TFs), including Myc, p53, NanoG, Sox2, RARa, Gata2, and ER, have been shown to undergo phase separation with the Mediator complex through their activation domains70. It is worth mentioning that the Mediator comprises a subset of TFs required to transcribe most protein-coding genes71. For the OCT4 TF, acidic residues within its activation domain are required to phase separate with MED1-IDR droplets, a Mediator subunit, and gene activation in vivo70. Our sequence analysis returned 12, 14, and 10 for p53, p63, and p73 acidic residues (Asp + Glu) within their transactivation domains, so it is possible these residues likely participate in heterotypic phase separation. Full-length p53 is also trapped in MED1-IDR droplets, and FRAP analysis of these p53-MED1 droplets revealed rapid and dynamic reorganization70. Another study indicated that TF droplets have a neutral or inhibitory effect on transcription activation72. To our knowledge, the regulation of transcription activity by p53 droplets, either physiologically or through the enhancement of transcription in heterotypic p63/p53 droplets involving cancer-related p53 variants, remains unknown and warrants further exploration.
In non-cancerous cells, p53 is expressed at low concentration with half-time of minutes. There is an increase in p53 concentration only under stress; under some circumstances, the protein eventually undergoes phase separation into a condensate, where it exerts its function34. The observed heterotypic phase transition involving p53 and the paralogs p63 and p73 provides a new perspective on the gain of function (GoF) associated with mutant p53. It is tempting to associate the oncogenic properties of mutant p53 with its propensity to alter the phase state of other proteins rather than solely from direct genetic alterations. These findings align with emerging models of cancer that emphasize the significance of protein-protein interactions and the tumor microenvironment in disease progression. It is tempting to suggest that the aberrant condensates of mutant p53 may act as a scaffold on p63 and p73 and other transcription factors.
Recent research has revealed that heterotypic condensates composed of prion protein and α-synuclein can transition from liquid to solid. This transformation is mainly modulated by RNA, resulting in the formation of heterotypic amyloids73. The same research team has observed that tau protein and PrP droplets mature as they age, culminating in a solid-like state characterized by amorphous and amyloid-like co-aggregates74. Early fibrils formed by model peptide insulin fragments (ACC1 − 13) have been observed within individual liquid droplets, suggesting a potential pathway from LLPS to fibril formation33. In our study, this progression from liquid droplet to the amyloid state of initially non-amyloidogenic proteins p63C and p73C is facilitated by mutant p53C.
Our findings highlight the complex interactions between proteins implicated in oncogenic diseases. Our study introduces a novel perspective by demonstrating that heterotypic interactions between p53 and its paralogs, p63, and p73, can lead to amyloid aggregation. The results mark the first instance of such an effect in cancer-associated proteins, broadening our understanding of protein phase transitions in oncology. Evidence indicates that p53 amyloid aggregates can entrap p63 and p73 in both in vitro and in vivo settings7,21, but the underlying mechanisms of this sequestration remained obscure. Our findings elucidate that p53C aggregates directly engage with biomolecular condensates of p63C and p73C (Figs. 4 and 6), providing a mechanistic understanding of this interaction.
Moreover, the prion-like conversion of p63C and p73C by mutant p53C broadens the scope of potential therapeutic targets within the p53 signaling cascade11,12,75,76,77,78,79. The selectivity of these interactions provides a unique opportunity to design inhibitors that specifically disrupt the aberrant phase transitions while preserving the normal functions of these proteins. Numerous studies have identified p53 aggregation as a promising focus for anticancer therapies11,12,19,48,76,80,81,82. The recent discovery that antineoplastic drugs are preferentially localized within specific protein condensates in the nuclei of tumor cells77,78 underscores the significance of our findings regarding the conversion of non-amyloidogenic tumor suppressors p63C and p73C by mutant p53C. This observation suggests that drugs designed to target p53 aggregation might selectively accumulate in the condensates that contain p53 and its related proteins, enhancing the specificity and efficacy of therapeutic interventions.
Our experiments with heparin serve as a proof of principle, demonstrating that the seeding-induced transition of p53C to an aggregated state, affecting its paralogs p63C and p73C, can be halted. Heparin not only arrests the transition of p53C into the amyloid state but also inhibits the seeding of p53C on the liquid droplets of p63C and p73C (Fig. 8). This striking observation presents potential therapeutic avenues to be explored using similar polyanions, including specific RNA or DNA sequences and certain small molecules. While there is evidence that heparin can enter cells83,84,85, bringing direct relevance to the inhibition of p53 phase separation and aggregation in cancer, our primary intention in using this negatively charged molecule is to emphasize the electrostatic interaction that modulates PS and aggregation. DNA and RNA can avoid and modulate p53C aggregation68,69. Similarly, other compounds with polar and negatively charged groups, such as emodin86, resveratrol82, ADH-680, prima-181, and others11, have consistently avoided or reversed p53 aggregation. Further, carbohydrate-based anticancer conjugates87 and functional groups such as ~ OH, ~SO3-, COO-, and NO2- are reasonable choices for developing p53 anti-aggregational compounds.
The complexity of the nuclear environment, with its abundance of nucleic acids, appears to influence p53 phase separation and aggregation, as referenced in several studies11,68,69,88,89,90,91. Variables such as the RNA-to-protein ratio have been shown to modulate the phase transition of mutant p5332,68,69, and this modulation extends to its paralogs, as demonstrated in our heparin experiments.
Our findings demonstrate the colocalization of the M237I full-length p53 mutant with p73 in fixed cells, as well as the colocalization of M237I p53 with p63C and p73C in the nucleolus of living cells, aligning with the results from in vitro studies. In living cells, we assessed the fluidity of M237I p53/p63C condensates in both the nucleoplasm and nucleolus. While the nucleoplasmic condensates showed almost complete fluorescence recovery after photobleaching, recovery in the nucleolus was significantly slower and incomplete, indicating a stiffening characteristic of the amyloid conversion of p63C induced by mutant p53. Similar results were obtained with p73C. In summary, cells co-transfected with the M237I p53 mutant and p63C or p73C exhibit colocalization and phase transition, as evidenced by reduced fluorescence recovery after photobleaching.
While our research has advanced the understanding of p53 phase transitions, certain aspects warrant further investigation. The broader spectrum of environmental and genetic factors that may impact this process within the cancer cell milieu is ripe for detailed investigation. The role of post-translational modifications on the phase behavior of these proteins also presents a promising avenue for research. Pursuing in vivo studies will be crucial to expand upon our findings and deepen the understanding of how targeting phase transitions in the p53 protein family may hold therapeutic value.
In conclusion, the study contributes a significant piece to the puzzle of p53 in cancer biology. By delineating the conditions that precipitate the phase transition and amyloid aggregation of p63C and p73C, we have identified a potential vulnerability in cancer cells that could be exploited in developing novel anticancer therapies. Future work focusing on developing small molecules or biologics that can prevent or reverse the amyloid aggregation of p53 paralogs holds the promise of advancing cancer treatment strategies. The path ahead is challenging but filled with the potential for significant breakthroughs in the fight against cancer.
Methods
Protein expression and purification
The genes encoding p53C WT and M237I, p63C, and p73C proteins were subcloned into pET15-b (Addgene, #24866), pET28-a, and pNIC-CTHF vectors (Addgene, #39077), respectively, and were expressed in Escherichia coli BL21-CodonPlus. Cultures were grown in Luria-Bertani medium and incubated at 37 °C until the optical density (OD600) of 0.8 was reached. Protein expression was induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) overnight at 25 °C under agitation. Cells were harvested by centrifugation and resuspended in buffer (pH 7.4) containing 50 mM Tris-Cl, 500 mM NaCl, 5 mM dithiothreitol (DTT), and 1 mM phenyl-methyl-sulfoxide (PMSF) protease and disrupted by sonication. Insoluble proteins and cell debris were separated by centrifugation. Highly homogenous samples of p53C WT, p53C M237I, R248Q, R249S, and p63C were obtained with the following chromatography steps: soluble fraction of each protein was loaded onto a HisTrap FF nickel affinity column (Cytiva) or SP Sepharose (p53C R248Q and R249S) and elution was performed with a linear increasing imidazole gradient. Cleavage of the fusion tag was achieved by thrombin digestion [1:1000 w/w] at 4 °C overnight. The product was diluted and loaded onto a Heparin Sepharose FF affinity column. The p73C purification was described elsewhere21. Briefly, a soluble fraction of p73C was loaded onto the HisTrap FF nickel affinity column (Cytiva), digested with Tobacco Etch Virus (TEV) protease [1:50 w/w], reapplied in a HisTrap FF column (Cytiva) and the non-bound was collected. All final samples were dialyzed against 20 mM Tris-Cl (pH 7.4), 150 mM NaCl, 5 mM DTT, 5 µM ZnCl2, 5% glycerol and stored at −80 °C. Before each experiment, all proteins were thawed on ice, centrifuged at 14,000 g 4 °C for 15 min, and passed through a 100 kDa microcon centrifugal filter to exclude residual aggregation. Protein final concentration for each experiment was measured using absorbance at 280 nm and extinction coefficient of 18,040 M−1cm−1 for p53C WT and M237I, 15,930 M−1cm−1 for p63C, and 18,910 M−1cm−1 for p73C. The EGFP-tagged proteins p53C-M237I, -R249S, and -R273H were purified as described in ref. 92 with gentle modifications. The full-length p53 was produced as in ref. 56. SDS-PAGE gels revealed the enriched status of proteins used in this study (Supplementary Fig. 13).
Protein labeling
For some phase separation microscope experiments, p63C and p73C were labeled with NHS-Rhodamine (Thermo Scientific) and p53C WT, M237I, R248Q, and R249S with Alexa Fluor 488 (Thermo Scientific) fluorescent dyes using the manufacturer’s specifications. Each protein was dialyzed against 0.1 M sodium bicarbonate pH 8.5 and incubated on ice for 2 h with a five-fold molar excess of NHS-Rhodamine or one vial of Alexa Fluor 488. The dye excess was separated using a HiTrap Sephadex G-25 Desalting column (Cytiva), and proteins were eluted in 20 mM Tris-Cl (pH 7.4), 150 mM NaCl, 5 mM DTT, 5 µM ZnCl2, 5% glycerol. Protein concentration and labeling efficiency were calculated as described in each manufacturer’s user guide.
Differential interference contrast microscopy (DIC) and fluorescence microscopy
DIC and fluorescence images were captured using a Zeiss LSM 710 confocal laser scanning microscope equipped with a Plan-Apochromat 63x/1.4 objective lens. Supernatants were combined with a pre-chilled 50% PEG-4000 stock solution at 4 °C, resulting in a final PEG concentration of 15% for all experiments unless otherwise specified. Samples (20 μL) in 20 mM Tris-Cl (pH 7.4), 150 mM NaCl, 5 mM DTT, and 5 µM ZnCl2 were loaded onto coverslips, and imaging was conducted at room temperature. For concentration-dependent experiments involving p63C and p73C (Fig. 1a), PEG-4000 was added to protein solutions at concentrations ranging from 10 to 100 µM (at 4 °C), and images were captured using DIC microscopy. Temperature-dependent experiments were performed with 60 μM proteins at 4, 25, or 37 °C for 3 min (Fig. 1c), extending to 30, 90, or 270 min (Fig. 1d). To perform the Congo Red (CR) binding assay (Fig. 2a–d), proteins at concentrations of 60 μM were initially incubated at 4 or 37 °C for 3 min before the addition of CR (10 µM). After the CR addition, PEG-4000 was added to the sample, which was then analyzed.
The influence of wild-type p53C on p63C and p73C aggregation dynamics (Fig. 3) involved mixing NHS-Rhodamine labeled p63C or p73C (40 μM) with wild-type p53C (40 μM) and PEG-4000. Additionally, aggregation of p63C and p73C induced by wild-type p53C and M237I in the presence of PEG (Fig. 4a–d) was investigated, where NHS-Rhodamine labeled p63C or p73C (40 μM) was combined with Alexa Fluor 488-labeled wild type p53C or M237I (40 μM) and PEG-4000 for imaging at 4 and 37 °C. For seeding of p53C and heparin effects on p63C droplets conversion to aggregates (Figs. 5, 8, and supplementary videos), Alexa Fluor 488-labeled wild-type p53C or M237I seeds were prepared using a 60 μM stock solution incubated at 37 °C for 3 min. The stock was combined with NHS-Rhodamine labeled p63C (60 μM) to a 1.5 μM final seed concentration before the addition (or not) of heparin (10 µM). Then, PEG-4000 was introduced, and the sample was imaged. Image processing was performed using Fiji, a distribution package of ImageJ software.
Fluorescence recovery after photobleaching (FRAP)
FRAP experiments were performed and analyzed as described previously32.
Thioflavin T and light scattering
Light scattering (LS) and Thioflavin T experiments were performed in an ISSK2 spectrofluorometer (ISS Inc). To investigate the influence of temperature in LLPS/aggregation, wt p53C, M237I, p63C, and p73C were prepared in a buffer containing 20 mM Tris-Cl, (pH 7.4), 150 mM NaCl, 5 mM DTT, 5 µM ZnCl2, and 5% glycerol. The proteins were then subjected to pre-incubation at concentrations of 60 μM, with or without the addition of 15% (w/v) PEG-4000, for 3 min across different temperatures: 4 °C, 25 °C or 37 °C. Samples were excited at 320 nm, and emission was recorded from 300 to 340 nm. Experiments were performed with a 5 nm slit width and 90% of the iris closed. Data were expressed as the area under the LS curve. Samples in the same condition were incubated with Thioflavin T at 10 μM final concentration, excited at 450 nm, and emission was recorded from 460 to 500 nm. Thioflavin T emission was expressed as intensity at 470 nm. Light scattering and Thioflavin T co-aggregation experiments were conducted as described above with 40 μM of p53C or M237I and 40 μM p63C in the same buffer in the presence or absence of 15% (w/v) PEG-4000. Mixtures were pre-incubated at 4 °C and 37 °C for 3 min before each measurement. Heparin at a final concentration of 10 μM was used to investigate its modulatory properties during aggregation. For seeding experiments, a stock solution of p53C or M237I at 60 μM final concentration was submitted to aggregation at 37 °C for 3 min. Then, it was diluted to a final concentration of 1.5 μM and mixed with 60 μM of p63C in the presence of 15% (w/v) PEG-4000. Light scattering and Thioflavin T binding were investigated as described before.
Turbidity measurements
Turbidity assays were performed in a Varioskan Lux 3020-81205 Microplate Reader (Thermo Scientific). P53C, M237I, p63C, and p73C dissolved in a solution of 20 mM Tris-Cl (pH 7.4), 150 mM NaCl, 5 mM DTT, and 5 µM ZnCl2, with15% (w/v) PEG-4000 at concentrations ranging from 10 to 100 µM were transferred to a 96 well optical black/transparent bottom plate (Thermo Scientific) and pre-incubated for 3 min at 4 °C, 25 °C or 37 °C. Turbidity was measured as absorbance at 600 nm.
Cell culture
Human non-small lung carcinoma H1299 cells were obtained from The Rio de Janeiro Cell Bank (RJCB) and cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum. Cells were maintained at 37 °C in an atmosphere containing 5% CO2.
Cell transfection
Transfection experiments were performed following the Lipofectamine 2000 reagent (Invitrogen) protocol provided by the manufacturer. The day before transfection, 5 × 104 cells were cultured in a 24-well plate containing a glass coverslip. After reaching 70% of confluency, H1299 cells were transfected with ten µg of pEGFP-N1 vector (Clontech), containing a C-terminus EGFP-tagged sequence of the full-length M237I p53 protein (Addgene, #11770), and 4 µL of Lipofectamine 2000 reagent. After 24 h since transfection, the immunofluorescence protocol was performed. The pEGFP-N1 vector was modified for co-transfection experiments to carry mCherry-tagged p63C and YFP-tagged p73C (GenScript, Piscataway, NJ).
Immunofluorescence
The H1299 cells were washed thrice with PBS and fixed with paraformaldehyde 4% for 20 min at room temperature. Then, cells were washed with PBS and permeabilized with Triton X-100 (0.5%) for 15 min. After another wash step with PBS, cells were incubated with a 3% bovine serum albumin (BSA) (Sigma) solution for 1 h at room temperature. Cells were labeled with monoclonal anti-p73 antibodies (Sigma, #F3117) in three different dilutions: 1:50, 1:100, and 1:200 overnight at 4 °C. After that, cells were washed with PBS and incubated with a 1:1,000 dilution of Alexa Fluor 568 antibody for 1 h at room temperature in the dark. Finally, cells were washed with PBS and fixed with Vectashield Antifade Mounting Medium with DAPI. Images were acquired with an ELYRA LSM 710 confocal laser scanning microscope (Carl Zeiss, Inc.).
Circular dichroism
Far-UV circular dichroism (CD) experiments were conducted on a Jasco J-1500 Spectrophotometer using a 0.1 cm quartz cuvette. Spectra were collected between 200 and 260 nm and are represented as the average of five scans. Co-aggregation experiments were performed as described in the “Thioflavin T and light scattering measurements” section with p53C WT, M237I, and p63C dissolved in 20 mM Tris-Cl, (pH 7.4), 150 mM NaCl, 5 mM DTT, 5 µM ZnCl2. The mean residue molar ellipticity ([θ]MRW) in deg.cm2.dmol−1 was calculated as described below:
where mdeg corresponds to the raw ellipticity, l is the path length in centimeters, [c] is the molar concentration, and N is the number of peptide bonds. Equal amounts of p63C and p53C WT / M237I were used for co-aggregation experiments, and the number of peptide bonds was calculated as the average of the p63C and p53C WT/M237I peptide bonds. Changes in the absorption peaks within the far-UV range indicate changes in the secondary content of the studied species. β-sheets and α-helices are constrained by phi/psi angles as such, ca. −120°/ + 120° and −57°/−47°, respectively. Therefore, electronic absorptions of the peptide bonds are also characteristic depending on the secondary content, as follows: in β-sheets a negative band at 215 nm and positive at 198 nm refer to n–π* and π–π* transitions, respectively. Meanwhile, α-helices feature negative bands at 222 and 208 nm relying on n–π* and π–π* transitions93. Data deconvolution can track these subtle secondary changes. We performed the secondary structure deconvolution using the SELCON3 method in the DichroWeb suite44.
Sequence alignment
The sequences of p53 (UniProt ID: P04637), p63 (UniProt ID: Q9H3D4), and p73 (UniProt ID: O15350) proteins were aligned using the Multiple Alignment using Fast Fourier Transform (MAFFT) method. This alignment technique was chosen for its accuracy and efficiency in handling large datasets and ensuring precise alignments. The alignment and conservation analysis were performed using Jalview software, version 2.11.3.3. Sequences were obtained from the UniProt database, and conservation scores were calculated using the BLOSUM62 substitution matrix. The Clustal X color scheme was applied for residue coloring, enhancing the visual representation of conserved and variable regions within the aligned sequences.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data and biological materials generated in this study are available from the corresponding author upon request. The reporting summary for this article is available as a Supplementary file. The source data underlying Figs. 2e-p, 5a–c, 7c, d, and 8a, b and Supplementary Figs. 6a, b are provided in Supplementary data 1.
References
Vousden, K. H. & Prives, C. Blinded by the light: the growing complexity of p53. Cell 137, 413–431 (2009).
Kastenhuber, E. R. & Lowe, S. W. Putting p53 in context. Cell 170, 1062–1078 (2017).
Muller, P. A. & Vousden, K. H. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 25, 304–317 (2014).
Ano Bom, A. P. et al. Mutant p53 aggregates into prion-like amyloid oligomers and fibrils: implications for cancer. J. Biol. Chem. 287, 28152–28162 (2012).
Ishimaru, D. A. et al. Fibrillar aggregates of the tumor suppressor p53 core domain. Biochemistry 42, 9022–9027 (2003).
Silva, J. L., De Moura Gallo, C. V., Costa, D. C. & Rangel, L. P. Prion-like aggregation of mutant p53 in cancer. Trends Biochem. Sci. 39, 260–267 (2014).
Xu, J. et al. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat. Chem. Biol. 7, 285–295 (2011).
Ghosh, S. et al. p53 amyloid formation leading to its loss of function: implications in cancer pathogenesis. Cell Death Differ. 24, 1784–1798 (2017).
Pedrote, M. M. et al. Oncogenic gain of function in glioblastoma is linked to mutant p53 amyloid oligomers. iScience 23, 100820 (2020).
Sengupta, S. et al. p53 amyloid pathology is correlated with higher cancer grade irrespective of the mutant or wild-type form. J, Cell Sci. 136, https://doi.org/10.1242/jcs.261017 (2023).
Silva, J. L. et al. Targeting biomolecular condensation and protein aggregation against cancer. Chem. Rev. 123, 9094–9138 (2023).
Silva, J. L., Cino, E. A., Soares, I. N., Ferreira, V. F. & A P de Oliveira, G. Targeting the Prion-like aggregation of mutant p53 to combat cancer. Acc Chem. Res. 51, 181–190 (2018).
Navalkar, A. et al. Direct evidence of cellular transformation by prion-like p53 amyloid infection. J. Cell Sci. 134, https://doi.org/10.1242/jcs.258316 (2021).
Yang-Hartwich, Y. et al. p53 protein aggregation promotes platinum resistance in ovarian cancer. Oncogene 34, 3605–3616 (2015).
Zhang, Y. et al. Proteomic identification of ERP29 as a key chemoresistant factor activated by the aggregating p53 mutant Arg282Trp. Oncogene 36, 5473–5483 (2017).
Kwan, K., Castro-Sandoval, O., Gaiddon, C. & Storr, T. Inhibition of p53 protein aggregation as a cancer treatment strategy. Curr. Opin. Chem. Biol. 72, 102230 (2023).
Levy, C. B. et al. Co-localization of mutant p53 and amyloid-like protein aggregates in breast tumors. Int. J. Biochem. Cell Biol. 43, 60–64 (2011).
Melo Dos Santos, N. et al. Loss of the p53 transactivation domain results in high amyloid aggregation of the Delta40p53 isoform in endometrial carcinoma cells. J. Biol. Chem. 294, 9430–9439 (2019).
Zhang, Y. et al. Therapeutic potential of ReACp53 targeting mutant p53 protein in CRPC. Prostate Cancer Prostatic Dis. 23, 160–171 (2020).
De Smet, F. et al. Nuclear inclusion bodies of mutant and wild-type p53 in cancer: a hallmark of p53 inactivation and proteostasis remodelling by p53 aggregation. J. Pathol. 242, 24–38 (2017).
Cino, E. A., Soares, I. N., Pedrote, M. M., de Oliveira, G. A. & Silva, J. L. Aggregation tendencies in the p53 family are modulated by backbone hydrogen bonds. Sci. Rep. 6, 32535 (2016).
Wang, G. & Fersht, A. R. Propagation of aggregated p53: Cross-reaction and coaggregation vs. seeding. Proc. Natl Acad. Sci. USA 112, 2443–2448 (2015).
Kehrloesser, S. et al. Intrinsic aggregation propensity of the p63 and p73 TI domains correlates with p53R175H interaction and suggests further significance of aggregation events in the p53 family. Cell Death Differ. 23, 1952–1960 (2016).
Russo, C. et al. Protein aggregation of the p63 transcription factor underlies severe skin fragility in AEC syndrome. Proc. Natl Acad. Sci. USA 115, E906–E915 (2018).
Deyoung, M. P. & Ellisen, L. W. p63 and p73 in human cancer: defining the network. Oncogene 26, 5169–5183 (2007).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Brangwynne, C. P. et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 (2009).
Alberti, S. & Hyman, A. A. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 22, 196–213 (2021).
de Oliveira, G. A. P., Cordeiro, Y., Silva, J. L. & Vieira, T. Liquid-liquid phase transitions and amyloid aggregation in proteins related to cancer and neurodegenerative diseases. Adv. Protein Chem. Struct. Biol. 118, 289–331 (2019).
Kamagata, K. et al. Liquid-like droplet formation by tumor suppressor p53 induced by multivalent electrostatic interactions between two disordered domains. Sci. Rep. 10, 580 (2020).
Matos, C. O. et al. Liquid-liquid phase separation and fibrillation of the prion protein modulated by a high-affinity DNA aptamer. FASEB J. 34, 365–385 (2020).
Petronilho, E. C. et al. Phase separation of p53 precedes aggregation and is affected by oncogenic mutations and ligands. Chem. Sci. 12, 7334–7349 (2021).
Dec, R., Jaworek, M. W., Dzwolak, W. & Winter, R. Liquid-droplet-mediated ATP-triggered amyloidogenic pathway of insulin-derived chimeric peptides: unraveling the microscopic and molecular processes. J. Am. Chem. Soc. https://doi.org/10.1021/jacs.2c12611 (2023).
Heltberg, M. S. et al. Enhanced DNA repair through droplet formation and p53 oscillations. Cell 185, 4394–4408.e4310 (2022).
Park, S. K., Park, S., Pentek, C. & Liebman, S. W. Tumor suppressor protein p53 expressed in yeast can remain diffuse, form a prion, or form unstable liquid-like droplets. iScience 24, 102000 (2021).
Yang, D. S. et al. Mesoscopic protein-rich clusters host the nucleation of mutant p53 amyloid fibrils. Proc. Natl Acad. Sci. USA 118, https://doi.org/10.1073/pnas.2015618118 (2021).
Bouchard, J. J. et al. Cancer mutations of the tumor suppressor SPOP disrupt the formation of active, phase-separated compartments. Mol. Cell 72, 19–36.e18 (2018).
Kilic, S. et al. Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J. 38, e101379 (2019).
Marques, M. A., de Oliveira, G. A. P. & Silva, J. L. The chameleonic behavior of p53 in health and disease: the transition from a client to an aberrant condensate scaffold in cancer. Essays Biochem. 66, 1023–1033 (2022).
Joerger, A. C. et al. Structural evolution of p53, p63, and p73: implication for heterotetramer formation. Proc. Natl Acad. Sci. USA 106, 17705–17710 (2009).
van Dieck, J. et al. Molecular basis of S100 proteins interacting with the p53 homologs p63 and p73. Oncogene 29, 2024–2035 (2010).
Wang, X., Chen, J. X., Liu, Y. H., You, C. & Mao, Q. Mutant TP53 enhances the resistance of glioblastoma cells to temozolomide by up-regulating O(6)-methylguanine DNA-methyltransferase. Neurol. Sci. 34, 1421–1428 (2013).
Chan, F. T. et al. Protein amyloids develop an intrinsic fluorescence signature during aggregation. Analyst 138, 2156–2162, (2013).
Miles, A. J., Ramalli, S. G. & Wallace, B. A. DichroWeb, a website for calculating protein secondary structure from circular dichroism spectroscopic data. Protein Sci. 31, 37–46 (2022).
Tycko, R. Amyloid polymorphism: structural basis and neurobiological relevance. Neuron 86, 632–645 (2015).
Bullock, A. N., Henckel, J. & Fersht, A. R. Quantitative analysis of residual folding and DNA binding in mutant p53 core domain: definition of mutant states for rescue in cancer therapy. Oncogene 19, 1245–1256 (2000).
Ghosh, S. et al. Investigating the intrinsic aggregation potential of evolutionarily conserved segments in p53. Biochemistry 53, 5995–6010 (2014).
Soragni, A. et al. A designed inhibitor of p53 aggregation rescues p53 tumor suppression in ovarian carcinomas. Cancer Cell 29, 90–103 (2016).
Canadillas, J. M. et al. Solution structure of p53 core domain: structural basis for its instability. Proc. Natl Acad. Sci. USA 103, 2109–2114 (2006).
Thanos, C. D. & Bowie, J. U. p53 Family members p63 and p73 are SAM domain-containing proteins. Protein Sci. 8, 1708–1710 (1999).
Horvath, A., Miskei, M., Ambrus, V., Vendruscolo, M. & Fuxreiter, M. Sequence-based prediction of protein binding mode landscapes. PLoS Comput. Biol. 16, e1007864 (2020).
Miskei, M., Horvath, A., Vendruscolo, M. & Fuxreiter, M. Sequence-based prediction of fuzzy protein interactions. J. Mol. Biol. 432, 2289–2303 (2020).
Hardenberg, M., Horvath, A., Ambrus, V., Fuxreiter, M. & Vendruscolo, M. Widespread occurrence of the droplet state of proteins in the human proteome. Proc. Natl Acad. Sci. USA 117, 33254–33262 (2020).
Hatos, A., Tosatto, S. C. E., Vendruscolo, M. & Fuxreiter, M. FuzDrop on AlphaFold: visualizing the sequence-dependent propensity of liquid-liquid phase separation and aggregation of proteins. Nucleic Acids Res. 50, W337–W344 (2022).
Vendruscolo, M. & Fuxreiter, M. Sequence determinants of the aggregation of proteins within condensates generated by liquid-liquid phase separation. J. Mol. Biol. 434, 167201 (2022).
Safari, M. S. et al. Anomalous dense liquid condensates host the nucleation of tumor suppressor p53 fibrils. iScience 12, 342–355 (2019).
Krois, A. S., Dyson, H. J. & Wright, P. E. Long-range regulation of p53 DNA binding by its intrinsically disordered N-terminal transactivation domain. Proc. Natl Acad. Sci. USA 115, E11302–E11310 (2018).
Usluer, S., Spreitzer, E., Bourgeois, B. & Madl, T. p53 transactivation domain mediates binding and phase separation with Poly-PR/GR. Int. J. Mol. Sci. 22, https://doi.org/10.3390/ijms222111431 (2021).
Dai, Z., Li, G., Chen, Q. & Yang, X. Ser392 phosphorylation modulated a switch between p53 and transcriptional condensates. Biochim. Biophys. Acta Gene Regul. Mech. 1865, 194827 (2022).
Chen, C., Fu, G., Guo, Q., Xue, S. & Luo, S. Z. Phase separation of p53 induced by its unstructured basic region and prevented by oncogenic mutations in tetramerization domain. Int. J. Biol. Macromol. 222, 207–216 (2022).
Bullock, A. N. et al. Thermodynamic stability of wild-type and mutant p53 core domain. Proc. Natl Acad. Sci. USA 94, 14338–14342 (1997).
Das, T. et al. Metastable condensates suppress conversion to amyloid fibrils. bioRxiv, https://doi.org/10.1101/2024.02.28.582569 (2024).
Posey, A. E. et al. Profilin reduces aggregation and phase separation of huntingtin N-terminal fragments by preferentially binding to soluble monomers and oligomers. J. Biol. Chem. 293, 3734–3746 (2018).
Conicella, A. E., Zerze, G. H., Mittal, J. & Fawzi, N. L. ALS mutations disrupt phase separation mediated by alpha-helical structure in the TDP-43 Low-Complexity C-Terminal Domain. Structure 24, 1537–1549 (2016).
Lin, Y., Protter, D. S., Rosen, M. K. & Parker, R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 (2015).
Molliex, A. et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015).
Patel, A. et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015).
Ishimaru, D. et al. Cognate DNA stabilizes the tumor suppressor p53 and prevents misfolding and aggregation. Biochemistry 48, 6126–6135 (2009).
Kovachev, P. S. et al. Distinct modulatory role of RNA in the aggregation of the tumor suppressor protein p53 core domain. J. Biol. Chem. 292, 9345–9357 (2017).
Boija, A. et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e1816 (2018).
Richter, W. F., Nayak, S., Iwasa, J. & Taatjes, D. J. The mediator complex as a master regulator of transcription by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 23, 732–749 (2022).
Trojanowski, J. et al. Transcription activation is enhanced by multivalent interactions independent of phase separation. Mol. Cell 82, 1878–1893.e1810 (2022).
Agarwal, A., Arora, L., Rai, S. K., Avni, A. & Mukhopadhyay, S. Spatiotemporal modulations in heterotypic condensates of prion and alpha-synuclein control phase transitions and amyloid conversion. Nat. Commun. 13, 1154 (2022).
Rai, S. K., Khanna, R., Avni, A. & Mukhopadhyay, S. Heterotypic electrostatic interactions control complex phase separation of tau and prion into multiphasic condensates and co-aggregates. Proc. Natl Acad. Sci. USA 120, e2216338120 (2023).
Costa, D. C. et al. Aggregation and prion-like properties of misfolded tumor suppressors: is cancer a prion disease? Cold Spring Harb. Perspect. Biol. 8, https://doi.org/10.1101/cshperspect.a023614 (2016).
Ferretti, G. D. S., Quarti, J., Dos Santos, G., Rangel, L. P. & Silva, J. L. Anticancer therapeutic strategies targeting p53 aggregation. Int. J. Mol. Sci. 23, https://doi.org/10.3390/ijms231911023 (2022).
Klein, I. A. et al. Partitioning of cancer therapeutics in nuclear condensates. Science 368, 1386–1392 (2020).
Lemos, C. et al. Identification of small molecules that modulate mutant p53 condensation. iScience 23, 101517 (2020).
Silva, J. L., Vieira, T. C., Cordeiro, Y. & de Oliveira, G. A. P. Nucleic acid actions on abnormal protein aggregation, phase transitions and phase separation. Curr. Opin. Struct. Biol. 73, 102346 (2022).
Palanikumar, L. et al. Protein mimetic amyloid inhibitor potently abrogates cancer-associated mutant p53 aggregation and restores tumor suppressor function. Nat. Commun. 12, 3962 (2021).
Rangel, L. P. et al. p53 reactivation with induction of massive apoptosis-1 (PRIMA-1) inhibits amyloid aggregation of mutant p53 in cancer cells. J. Biol. Chem. 294, 3670–3682 (2019).
Ferraz da Costa, D. C. et al. Resveratrol prevents p53 aggregation in vitro and in breast cancer cells. Oncotarget 9, 29112–29122 (2018).
Naik, R. J., Sharma, R., Nisakar, D., Purohit, G. & Ganguli, M. Exogenous chondroitin sulfate glycosaminoglycan associate with arginine-rich peptide-DNA complexes to alter their intracellular processing and gene delivery efficiency. Biochim. Biophys. Acta. 1848, 1053–1064 (2015).
Raman, K., Mencio, C., Desai, U. R. & Kuberan, B. Sulfation patterns determine cellular internalization of heparin-like polysaccharides. Mol. Pharm. 10, 1442–1449 (2013).
Redini, F., Moczar, E., Antoine, E. & Poupon, M. F. Binding and internalization of exogenous glycosaminoglycans in weakly and highly metastatic rhabdomyosarcoma cells. Biochim. Biophys. Acta 991, 359–366 (1989).
Haque, E. et al. Blocking mutation independent p53 aggregation by emodin modulates autophagic cell death pathway in lung cancer. Int. J. Biochem. Cell Biol. 96, 90–95 (2018).
Ernst, B. & Magnani, J. L. From carbohydrate leads to glycomimetic drugs. Nat. Rev. Drug Discov. 8, 661–677 (2009).
Dai, Z. & Yang, X. The regulation of liquid-liquid phase separated condensates containing nucleic acids. FEBS J. https://doi.org/10.1111/febs.16959 (2023).
McGurk, L. et al. Poly(ADP-Ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Mol. Cell 71, 703–717.e709 (2018).
Silva, J. L. & Cordeiro, Y. The “Jekyll and Hyde” actions of nucleic acids on the prion-like aggregation of proteins. J. Biol. Chem. 291, 15482–15490 (2016).
Maharana, S. et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921 (2018).
Mayer, S., Rudiger, S., Ang, H. C., Joerger, A. C. & Fersht, A. R. Correlation of levels of folded recombinant p53 in escherichia coli with thermodynamic stability in vitro. J. Mol. Biol. 372, 268–276 (2007).
Sreerama, N. & Woody, R. W. Computation and analysis of protein circular dichroism spectra. Methods Enzymol. 383, 318–351 (2004).
Acknowledgements
Our laboratory is supported by grants from the National Council for Scientific and Technological Development (CNPq awards and the INCT program, grant no. 465395/2014-7 and 408046/ 2021-0 to J.L.S. and grant no. 313137/2021-8 to G.A.P.O.) and the Carlos Chagas Filho Foundation for research support in the state of Rio de Janeiro (FAPERJ) grant no. 210.008/2018 and 202840/2018 to J.L.S., grants E-26/201.296/2021 and E26/210.294/2022 to G.A.P.O., grants E-26/200.582/2022, E-26/210.346/2022 to M.A.M. and E-26/201.325/2021 to T.C.R.G.V.
Author information
Authors and Affiliations
Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication. E.C.P., G.C.A., G.S.S., F.P.A., M.F.M., A.V.S.G., C.H.S.P., M.C.S., H.R.S.A., and M.A.M. performed experiments; E.C.P., G.C.A., G.A.P.O., and J. L.S. writing the original draft; M.A.M., T.C.R.G.V., G.A.P.O., and J.L.S. conceptualization, data curation, funding acquisition, project administration, editing the manuscript and figures, and supervising the research.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Chemistry thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Petronilho, E.C., de Andrade, G.C., de Sousa, G.d.S. et al. Oncogenic p53 triggers amyloid aggregation of p63 and p73 liquid droplets. Commun Chem 7, 207 (2024). https://doi.org/10.1038/s42004-024-01289-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42004-024-01289-x
This article is cited by
-
Targeting FOXP1 phase separation in small cell lung cancer mechanisms of chemotherapy resistance
Communications Biology (2025)