Abstract
Fluorescence microscopy represents a highly effective tool for examining the microscopic world. Modern microscopic techniques provide advanced temporal and spatial resolutions, while placing high demands on the photostability of fluorophores. Although it is well-known that the photostability of fluorophores is sensitive to their environment, the influence of nearby residues on the photostability of organic fluorophores attached to proteins is not yet fully understood. This study focuses on the influence of residues adjacent to the maleimide-derived fluorophore, specifically attached to cysteine residues in proteins. Under our experimental conditions, methionine, tryptophan, phenylalanine, valine, and leucine were all found to reduce the photostability, whereas glutamate was found to enhance the photostability. To put these experimental observations into perspective, we performed molecular dynamics simulations that offer insights into the mechanistic effects of adjacent residues.
Similar content being viewed by others
Introduction
Fluorescence microscopy imaging is a fundamental tool in biological research, allowing the direct detection of target molecules of interest and the exploration of their dynamics in living cells and test tubes. The advancement of fluorescence microscopy has led to the development of a range of advanced imaging technologies, including super-resolution fluorescence microscopy1,2,3, fluorescence correlation spectroscopy4, single-molecule fluorescence methods5, light sheet fluorescence microscopy6,7, and so on. These technologies provide great insight into the structure, localization, dynamics, and function of biomolecules, and also place high demands on the photostability of fluorophores.
Photobleaching of fluorophores, referring to the irreversible loss of the ability to emit fluorescence, is a significant limitation in fluorescence imaging experiments, particularly in long-term and high-resolution measurements. Thus, various fluorescence proteins and organic fluorophores with improved photostability have been engineered8,9,10. The combination of oxygen scavengers11,12 and triplet-state quenchers13 in solution can significantly prolong the fluorescence of organic fluorophores. Self-healing dyes14, generated by covalently conjugating a triplet-state quencher with an organic fluorophore, exhibit enhanced photostability and reduced phototoxicity in single-molecule and live-cell imaging. Shielding fluorescence cores from photochemically induced reactive species by rotaxane15 and other similar structures16 is another feasible strategy to improve photostability.
It is well known that the photoproperties of a fluorophore are influenced by its environment. For example, the amino acids tryptophan, tyrosine, and methionine17,18,19,20,21 and the nucleobase guanine22 can quench fluorescence, leading to reduced brightness and fluorescence lifetime. Although organic fluorophore-labeled proteins have a wide range of applications in biological research, including immunostaining, super-resolution imaging, fluorescence correlation spectroscopy, and single-molecule fluorescence measurements, how nearby amino acid residues influence photostability is less well studied. Our recent work showed that the linker with thioether between fluorophore and cysteine residue causes severe photobleaching, which inspired us to use new conjugation methods to address this issue21. In this study, we examined the influence of residues adjacent to the attached fluorophore. Under all conditions we tested, methionine, tryptophan, phenylalanine, valine, and leucine reduced the photostability, whereas glutamate enhanced the photostability.
Results
Photostability influenced by adjacent amino acid residues
EF-G-A209C is used as one of our model protein molecules, which was generated by introducing a cysteine residue at the 209 site of the cysteine-free E. coli elongation factor G (EF-G)21. To investigate how neighboring residues affect the photophysical properties of the labeled fluorophore, we performed site-saturation mutagenesis on two adjacent residues—Pro-208 and Asp-210—near the labeling site. To simplify our investigation, sites 208 and 210 were mutated to the same naturally occurring amino acid, except cysteine (Tables S1 and S2). Sites located two or three residues away from the labeling site were not mutated and tested in our assays, because we have shown that these sites have little or no effect on the photoproperties21. The Alphafold2 predicted that these mutants have similar structures (Fig. S1).
All these EF-G variants were expressed, purified, and labeled by maleimide-derived fluorophores (Cy3, Cy5, and Atto488, Tables S3, S4, and S5). Labeled EF-G was attached to a passivated microscope slide via specific interactions between immobilized streptavidin and a biotinylated avi-tag at its C-terminus21. Then, a total internal reflection fluorescence (TIRF) microscope-based single-molecule fluorescence assay was used to quantify photoproperties, including the intensity, bleaching time, total emitted photon number before photobleaching, and signal-to-noise ratio (SNR), from individual single-molecule fluorescence trajectories (Figs. 1A and S2).
A A scheme of the single-molecule fluorescence assay. From single-molecule fluorescence trajectories of immobilized labeled EF-G proteins, total photon count, bleaching time, intensity, and signal-to-noise ratio (SNR) are quantified. B Total photon count, bleaching time, and intensity of Cy3 (red), Cy5 (blue), or Atto488 (light brown) labeled EF-G-A209C variants, whose abbreviations in the figures are named after the residues adjacent to the fluorophore. In particular, Pro-208 and Asp-210 are mutated to other amino acids. The standard deviation of three repeats is shown as the error. Each experimental condition comprises analysis of no fewer than 800 single-molecule traces.
All labeled EF-G variants displayed similar intensities and SNRs when the same fluorophore was attached (Figs. 1B and S3). Interestingly, adjacent Met, Trp, Phe, Val, and Leu residues significantly reduced the photostability of Cy3, Cy5, and Atto488 by increasing their photobleaching rates and decreasing total emitted photon numbers (Figs. 1B and S4–S6, S10–S12). On the other hand, adjacent Glu increased the photostability by decreasing photobleaching rates and increasing total emitted photon numbers (Fig. 1B). Arg and Tyr residues only decreased the photostability of Cy5, but not of Cy3, whereas the Ala residue only increased the photostability of Cy3. Thus, in the following study, we mainly focused on Met, Trp, Phe, Val, Leu, and Glu, which have general influences on the fluorophores we examined.
Influence of adjacent residues in the context of different secondary structures
Site 209 of EF-G is located on the boundary between an α-helix and a disorder region. To confirm the influence of adjacent residues on the photostability is a general behavior among different protein secondary structures, we picked three additional sites on EF-G. They are sites 205, 239, and 308, located on a disorder loop, an α-helix, and a β-sheet, respectively (Fig. 2A). The influences of Met, Trp, Phe, Val, Leu, and Glu on Cy3 and Cy5 attached to these sites were quantified.
A Three labeling sites of EF-G. E205, G239, and E308 were mutated to Cys, respectively, for dye labeling. Total photon count, bleaching time, and intensity of Cy3 (red) and Cy5 (blue) attached at EF-G-E205C (B), EF-G-G239C (C), and EF-G-E308 (D). Abbreviations of EF-G variants in the figures are named after the residues adjacent to the fluorophore. The standard deviation of three repeats is shown as the error. Each experimental condition comprises analysis of no fewer than 800 single-molecule traces.
Consistent with our results shown above, the influences of adjacent residues on intensities and SNRs are minor or even negligible (Figs. 2 and S3). Their influences on the photostability of Cy3 and Cy5 varied in magnitude among the three sites we tested. Residues surrounding site 209 (located at the α-helix/disordered region boundary) and site 308 (in a β-sheet) exhibited stronger effects, whereas site 205 (in a disordered region) and site 239 (in an α-helix) showed weaker effects. These results demonstrate that local secondary structures have additional functional roles. However, regardless of their local secondary structures, Met, Trp, Phe, Val, and Leu were consistently found to decrease the photostability of Cy3 and Cy5, whereas Glu was found to enhance the photostability (Figs. 2 and S7–S9, S13–S15).
Fluorescence lifetime influenced by adjacent residues
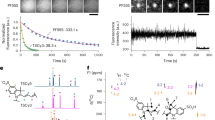
Photobleaching of fluorophores under continuous excitation is likely caused by unwanted reactions occurring in the excited states, including triplet and singlet states, leading to destruction of the chromophore. In our single-molecule assay, triplet-state quenchers, cyclooctatetraene, nitrobenzylalcohol, and trolox, are used to deplete the triplet-state to prevent photoblinking of fluorophores. Thus, we mainly examined whether fluorescence lifetime, the average time in the singlet state, is influenced by adjacent residues (Figs. 3 and S16). The fluorescence lifetime of Cy3-labeled EF-G-A209C is 0.74 ± 0.04 ns, which is barely affected by adjacent Glu (0.79 ± 0.01 ns) and Asp (0.82 ± 0.08 ns), although Glu enhances the photostability and Asp has almost no influence. Interestingly, the EF-G-A209C variants, which displayed reduced photostability, all showed prolonged fluorescence lifetime (0.99–1.25 ns).
More importantly, the fluorescence lifetimes of the Cy5 and Atto488-labeled EF-G-A209C variants also followed the same trend as the Cy3-labeled ones. Previous studies have shown that the fluorescence lifetime of cyanine dyes correlates with an increase in their activation energy for photoisomerization23. However, Atto488 is not a cyanine dye, suggesting that photoisomerization is not the only major factor here to influence fluorescence lifetime. Nevertheless, a longer fluorescence lifetime indicates that the fluorophore spends more time in its highly active singlet state, leading to a higher probability of destruction during each excitation–emission cycle and lower photostability.
Notably, previous studies have shown that free amino acids (e.g., Trp, Tyr, Met, and His) can quench the fluorescence of certain dyes through static and dynamic quenching mechanisms, leading to a reduction in fluorescence lifetime17,18,20. However, our findings reveal a different behavior: when nearby amino acid residues were mutated, the dye’s lifetime did not decrease (Fig. 3), suggesting that the observed variations in photostability are not attributable to conventional quenching effects induced by adjacent amino acids.
Local conformations of EF-G-A209C variants examined by molecular dynamics (MD) simulation
With the exception of the Met residue, which contains an active thioether atom, the side chains of other residues that affect the photostability of fluorophores, including Trp, Phe, Val, Leu, and Glu, are mostly inert. Therefore, we speculated that their influences on photostability and fluorescence lifetime are mainly caused by their different local conformations around the attached fluorophore. Thus, we carried out molecular dynamics simulations of 1–2 μs on several Cy3-labeled EF-G-A209C variants to reach steady conformations (Fig. S17). Interestingly, after superimposing these conformations (Fig. 4A), for EF-G-A209C-Met, -Trp, -Phe, -Val, and -Leu variants which showed reduced photostability and long fluorescence lifetime, the distances between Cy3 and the backbone of its nearby residues (sites 204–214) are mainly around 10 Å (Fig. 4B). In the other variants, including EF-G-A209C, EF-G-A209C-Tyr, -Glu, and -Asp, the distances between Cy3 and the backbone of its nearby residues are extended to 20–30 Å. Thus, among the variants we tested, the distance between Cy3 and the backbone of EF-G strongly correlates with their photostability and fluorescence lifetime.
A Steady conformations of Cy3-labeled EF-G-A209C variants revealed by MD simulation. The conformations of different EF-G variants were superimposed based on sites 206–240 (gray). Locations of Cy3 in different conformations are shown as spheres, labeled by the adjacent side chains and colored by total emitted photon counts. The color scale is shown. B Violin plots showing distributions of distances between the center C atom of Cy3 (C11) and the C atom of the amide groups in its nearby residues (sites 204–214).
Stacking between Cy3 and its neighboring Met, Trp, or Phe could stabilize Cy3 in conformations close to the side chains (Figs. S18, S21, and S22). For example, the benzene ring on the Phe-210 side chain can stack with the dye at a relatively close distance (within 5 Å). A comparison between the -Phe and the -Tyr variants reveals that the additional hydroxyl groups of Tyr-208 form a hydrogen bond with Gly-174, resulting in interactions between the Cy3 linker and site 172 and an extended conformation of Cy3 (Figs. S18 and S22). Comparing the -Val, -Leu, and -Ile variants, the large steric hindrance of sec-butyl on Ile-208 affects the orientation of its amide carbonyl group, leading to the formation of additional hydrogen bonds with nearby amino acids. Thus, the local conformation around the sites 208–212 of EF-G-A209C-Ile is more compact, shielding Cy3 away from the backbone (Figs. S19 and S23). Although Glu’s side chain is one carbon longer than Asp’s, only Glu enhances photostability, whereas Asp does not (Fig. 1), which is quite interesting. The MD simulations showed that linkers of Cy3 in the -Glu and -Asp variants adopt the opposite orientation, which is likely caused by the different sizes of their side chains. Linker of the EF-G-A209C-Glu is more extended (Figs. S20 and S24). Together, the MD simulations suggested that adjacent residues can modulate local conformational distributions between Cy3 and nearby residues through either direct interactions or indirect effects, such as changes in local flexibility and steric hindrance.
Conclusion
Modern fluorescence microscopy techniques are powerful tools for studying the structures and dynamics of biological systems at the cellular and molecular level, leading to new insights and discoveries in various fields. Fluorophore-labeled biomolecules with excellent photostability are always in high demand by researchers. Although the photoproperties of fluorophores are sensitive to their environment, how nearby residues and nucleobases influence the photostability of attached fluorophores is less well characterized.
The most elucidated pathways for fluorophore photobleaching predominantly involve the dissolved oxygen in solution and reactive oxygen species (ROS)24. For cyanine dyes, especially for Cy5 and Cy7, photobleaching primarily results from photolysis mediated by singlet oxygen25. The photobleaching mechanisms of rhodamine-based fluorophores involve initial formation of a free radical cation in the presence of oxygen, followed by subsequent N-dealkylation26. Therefore, for applications requiring high photon output from single fluorophores, enzymatic oxygen scavengers and triplet-state quenchers are incorporated into the solution to suppress these photobleaching pathways. This is the condition we employed in our measurements. Nevertheless, residual photobleaching persists under these optimized conditions, and the underlying pathways remain poorly understood.
The photostability of a given fluorophore can vary substantially when conjugated to different targets (e.g., proteins vs. oligonucleotides), even in identical buffer cocktails27. This observation suggests that the local microenvironment critically influences photobleaching processes. Here, we present a systematic investigation of how adjacent residues influence the photostability of three widely used fluorophores (Cy3, Cy5, and Atto488). Although cyanine dyes (Cy3, Cy5) and rhodamine derivatives (Atto488) exhibit distinct photobleaching mechanisms, we observed that Phe, Val, Met, Leu, and Trp universally reduce adjacent fluorophore photostability, while Glu enhances it. These effects were consistently observed across diverse secondary structures we tested - disordered loop, α-helix, and β-sheet - though their magnitude varied with structural context.
Previous studies have demonstrated that amino acids can quench fluorophores through either static stacking or dynamic collision mechanisms, which concomitantly reduce both fluorescence intensity and lifetime17,20,27,28. However, our measurements reveal that photostability-reducing residues exert minimal effects on fluorescence intensity while prolonging the lifetimes of cyanine dyes. These observations suggest these residues mediate photobleaching through alternative pathways rather than conventional static or dynamic quenching mechanisms. DNA conjugation of a single Trp residue to either Cy5 or Atto647N has been shown to reduce photostability while slightly decreasing brightness, consistent with our current observations27. Notably, under identical experimental conditions, Phe residues were previously reported to have no significant impact on photostability, whereas our data reveal a destabilizing effect. This discrepancy suggests Phe’s photostability effects may be context-dependent, potentially influenced by protein environment in our assays.
Intriguingly, molecular dynamics simulations reveal a strong inverse correlation between fluorophore-backbone distance and photostability. Specifically, proximal fluorophore-backbone distances (<10 Å) consistently reduce photostability, regardless of side chain properties (aromatic Phe/Trp and aliphatic hydrophobic Val/Leu/Met). Conversely, distal positioning (>10 Å) preserves fluorophore stability. The proximity of fluorophore to the backbone may hinder the photoisomerization of cyanine (Cy3, Cy5)23, thereby prolonging their fluorescence lifetime (Fig. 3). Collectively, our results demonstrate that backbone proximity influences photobleaching pathways. While the amide group - an essential backbone component - is known to mediate fluorescence quenching via photoinduced electron transfer29,30, its potential role in photobleaching remains unclear. The underlying mechanisms of backbone-modulated photobleaching require further elucidation.
In summary, this work highlights the profound influences of protein residues on the photostability of fluorophores and provides guidance for fluorophore labeling and optimization.
Materials and methods
Protein purification
Mutation sites of all Escherichia coli EF-G are listed in Table S1. All primers (Beijing Sangon) used for mutagenesis were described in Table S2. Cys at 114, 266, and 398 sites were mutated to Asp, Ala, and Ser, respectively, to generate cysteine-free EF-G to introduce an additional Cys for site-specific labeling. The EF-G variants were purified using the assays described before31. In detail, the pET-24(b)-plasmid of EF-G, which was inserted at its C terminus with the Avi-tag (GLNDIFEAQKIEWHE), was transformed into 50 μL E.coli strain BL21 (DE3) competent cells. The transformed reaction was plated onto the Kana (50 μg·mL−1) resistance LB plate and incubated overnight at 37 °C. The colony was picked up in 5 mL of LB (final concentration 50 µg·mL−1 of Kana) at 37 °C, and grew in a small-scale culture. Approximately for 20 h until the OD600 reaches 0.8, the small culture was inoculated in 1 L of large-scale culture, adding isopropyl-β-D-thiogalactoside (IPTG, 50 µM final) and grew for another 24 h at 16 °C. The cells were harvested by centrifugation at 8000 rpm for 20 min, and lysed with 40 mL lysis buffer (50 mM Tris-HCl, pH 7.5, 200 mM KCl, 1 mM TCEP, 15 μM GDP). The cells were resuspended in lysis buffer (final 1 mM of PMSF) and lysed by ultrasonication for 25–30 min (work 2 s, pause 5 s, 25% power). After centrifugation at 15,000 rpm for 40 min at 4 °C, the clear supernatant was purified on the Ni-NTA column (Qiagen).
Before purification on the Ni-NTA column, 1~2 mL of Ni-NTA Superflow resin was taken into a plastic flex column and settled. After equilibrating the column with lysis buffer twice, the protein supernatant was incubated with the Ni-NTA beads for 1 h at 4 °C. The column was washed with wash buffer (50 mM Tris-HCl, pH 7.5, 200 mM KCl, 1 mM TCEP, 15 μM GDP, 25 mM imidazole), followed by eluting the protein using elution buffer (50 mM Tris-HCl, pH 7.5, 200 mM KCl, 1 mM TCEP, 15 μM GDP, 250 mM imidazole). Then the anion exchange step was performed with a 5 mL Hitrap Q column, eluting by mixing buffer A (50 mM Tris-HCl, pH 7.5, 1 mM TCEP, 15 μM GDP) and buffer B (50 mM Tris-HCl, pH 7.5, 500 mM KCl, 1 mM TCEP, 15 μM GDP) with a linear gradient of 0~500 mM KCl. The EF-G was eluted out around 250 mM KCl. All proteins were further concentrated on the centricon MW30 in buffer with 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM TCEP, and 15 μM GDP, and were flash-frozen in liquid nitrogen and stored at −80 °C.
Protein labeling
All purified proteins and dyes, including Sulfo-Cyanine3 maleimide (Sulfo-Cy3 MAL, Lumiprobe), Sulfo-Cyanine5 maleimide (Sulfo-Cy5 MAL, Lumiprobe), Atto488 maleimide (Atto488 MAL, ATTO-TEC), were mixed at a 1:6 molar ratio in the buffer with 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM TCEP, 15 μM GDP. After incubation at room temperature for 2 h, 1 mM DTT was added. Labeled proteins were separated from excess free dyes through a desalting Sephadex G-25 column (Nap-5, GE Healthcare). The protein and fluorophore concentrations were acquired by A280, A555, A650, and A495 via a Nanodrop, based on the molar extinction coefficients of proteins in 280 nm, Sulfo-Cy3 in 555 nm, Sulfo-Cy5 in 650 nm, and Atto488 in 495 nm. The labeling efficiencies of all proteins were described in Tables S3, S4, and S5.
Preparation of PEG-passivated slides
PEG-passivated slides decorated with biotin-PEG were prepared according to the previous procedure21. Slides and coverslips were sonicated at 40 °C in the order of ethanol (10 min), 0.2 M potassium hydroxide (20 min), and ethanol (10 min). Slides and coverslips were washed with ultrapure water between every step, and then treated with amino-silane reagents (mix 150 mL methanol, 7.5 mL acetic acid, and 1.5 mL amino-silane) at room temperature overnight. After washing with ultrapure water and ethanol, slides and coverslips were incubated with PEGylation solution (every 240 μL 0.1 M sodium bicarbonate, pH 8.3 with 60 mg mPEG-Succinimidyl Valerate, MW 2000 and 7 mg biotin-PEG-SC, MW 2000) in the dark for 2.5–3 h. Wash slides and coverslips with ultrapure water, and dry them with ultrapure nitrogen gas. Put a pair of slides and coverslips in a 50 mL centrifuge tube, vacuum seal them in a plastic bag, and store at −20 °C.
Single-molecule measurements by TIRF microscopy
PEG-passivated slides decorated with biotin-PEG were incubated with 0.05 mg·mL−1 streptavidin for three minutes. After washing out unbound streptavidin, a 20-μL solution containing labeled EF-G (buffer with 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM TCEP, 15 μM GDP) was flowed into the chamber and incubated for three minutes. Unbound EF-G was washed out by the buffer containing an oxygen-scavenging system and triplet-state quenchers including 3 mg·mL−1 glucose (Sigma–Aldrich), 300 μg·mL−1 glucose oxidase (Sigma–Aldrich), 30 μg·mL−1 catalase (Roche), 1 mM cyclooctatetraene (COT, Sigma–Aldrich), 1 mM 4-nitrobenzylalcohol (4-NBA, Sigma–Aldrich) and 1.5 mM 6-hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid (Trolox, Sigma–Aldrich). All results were measured under 0.5 μW·µm−2 laser power at 25 °C, at least in three repeats.
Single-molecule fluorescent measurements were performed on a home-built objective-type TIRF microscope, based on a Nikon Eclipse Ti-E with a TIRF objective (Nikon, CFI Apochromat TIRF 100×, N.A. 1.49, Oil immersion), an EMCCD camera (Andor iXon Ultra 897), and solid-state 488 nm, 532 nm, and 640 nm excitation lasers (Coherent Inc. OBIS Smart Lasers), which can be modulated using digital signals from the EMCCD camera. Fluorescence emission from the proteins was collected by the microscope with bandpass filters ET525/50 m (for Atto488, Chroma), ET585/65 m (for Cy3, Chroma), and ET700/75 m (for Cy5, Chroma). All movies were collected by the Cell Vision software (Beijing Coolight Technology).
Data analysis for single-molecule measurements
All movies were analyzed by a custom-made software program developed as an ImageJ plugin.
Fluorescence spots were fitted by a 2-D Gaussian function. For single-molecule measurement, the spots were picked using a variant of the Hough transform. The background-subtracted total volume of the 2-D Gaussian peak was used as raw fluorescence intensity. Single-molecule trajectories containing two or more photobleaching steps or displaying gradually-decreased intensities were discarded. The time point when the signal is below 20% of the initial signal is assigned as the photobleaching time point. To calibrate the EM-gain of the EMCCD camera, values of variance and intensities were obtained by measuring pixel intensities over time with a stable light source. The actual gain of the EMCCD camera was calculated as the slope of variance vs. intensities to convert the spot intensities to the number of collected photons.
From each time-dependent fluorescence trajectory, the SNR, duration times before photobleaching, and total photon number are calculated. The distribution of duration time is fitted with an exponential decay to estimate the average dwell time before photobleaching. Photon count per frame and total emitted photons are calculated from fluorescence intensity. Three or more replicates were performed for each experiment. Standard deviation was displayed as an error bar.
Fluorescence lifetime measurements by FLIM
Labeled EF-G samples in the buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM TCEP, 15 μM GDP) with the oxygen-scavenging system and triplet-state quenchers were used. Time-correlated single-photon counting (TCSPC) system (PicoQuant) was used for fluorescence lifetime measurements. FLUOVIEW FV1200 confocal fluorescence microscope consisting of an inverted microscope (IX 83, Olympus) equipped with an Olympus UPLSAPO 60× NA 1.3 water immersion objective, and 488 nm, 561 nm, and 640 nm lasers were used to collect the fluorescence signal. All data acquisition and analysis were performed with SymPhoTime 64 software.
MD modulations
The EF-G-A209C and mutant structures were predicted by AlphaFold232, and the dye was conjugated at the C209 site using AMBER-DYES33 to obtain the initial structure. Based on the AlphaFold2-predicted mutant structures, we manually replaced the coordinates of the residues adjacent to site 209 in the EF-G-A209C structure to generate mutant structures, which were then used as different initial conformations for MD simulations. The MD setup and simulations were performed using Amber2034. The force field parameters for the dye came from Amber99SB-dyes35, and the protein force field was Amber ff14SB36. The LEaP module in the Amber20 was used to solvate the system in a truncated octahedral box with SPC/E37 water, ensuring that any atom of the protein was at least 10 Å away from the edge of the water box. Appropriate amounts of Na+ and Cl− ions were added to neutralize the system’s net charge. All systems underwent a two-stage energy minimization. In the first stage, 1000 steps of energy minimization were performed, with the first 500 steps using the steepest descent method and the remaining steps using the conjugate gradient method. During this stage, harmonic restraints with a force constant of 500 \({kcal}\cdot {{mol}}^{-1}\cdot {{{{\text{\AA }}}}}^{-2}\) were applied to the heavy atoms of the backbone to maintain rigidity. In the second stage, the entire system was energy minimized for 1000 steps without any restraints. The energy-minimized system was heated to 300 K in an NVT ensemble using Berendsen temperature control, and then was equilibrated at 300 K for 1 ns in an NPT ensemble using the Berendsen barostat. The equilibrated conformation was subjected to a 1–2 μs molecular dynamics simulation without any restraints. All MD simulations were conducted under periodic boundary conditions using an integration time step of 2 fs. The SHAKE algorithm was used to constrain all bonds involving hydrogen atoms. The cutoff distance for the non-bonded interactions was set to 9 Å, and the long-range electrostatic interactions were accounted for by the particle mesh Ewald summation method34.
Each molecular dynamics trajectory was subsampled at a uniform interval to generate 5000 frames for subsequent analysis. To remove global rotational and translational motions, all trajectory frames were aligned to the initial reference frame (Frame 0) by superimposing the backbone atoms (N, Cα, C, O) of residues 208–210 using the pytraj package (https://github.com/Amber-MD/pytraj), a Python package binding to the cpptraj program38. Spatial density distributions were then calculated for defined molecular regions using the volmap module of VMD39 software. Atomic selections included: the protein segment of residues 200–240, residue 208, residue 210, the dye linker (residue 209), and the specific dye C3W. For each group, volumetric density maps were constructed with a grid resolution of 1.0 Å. The volmap density function was executed and averaged over all aligned frames. All isosurfaces were consistently visualized at an isovalue of 0.5 to enable direct comparison between regions.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support the findings of this study are included in the manuscript and supplementary files.
References
Hell, S. W. & Wichmann, J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 19, 780–782 (1994).
Rust, M. J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–796 (2006).
Betzig, E. P. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006).
Thompson, N. L. & Axelrod, D. Immunoglobulin surface-binding kinetics studied by total internal reflection with fluorescence correlation spectroscopy. Biophys. J. 43, 103–114 (1983).
Funatsu, T., Harada, Y., Tokunaga, M., Saito, K. & Yanagida, T. Imaging of single fluorescent molecules and individual ATP turnovers by single myosin molecules in aqueous solution. Nature 374, 555–559 (1995).
Voie, A. H., Burns, D. H. & Spelman, F. A. Orthogonal-plane fluorescence optical sectioning: three-dimensional imaging of macroscopic biological specimens. J. Microsc. 170, 229–236 (1993).
Huisken, J., Swoger, J., Del Bene, F., Wittbrodt, J. & Stelzer, E. H. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007–1009 (2004).
Song, F. et al. Syntheses, spectral properties and photostabilities of novel water-soluble near-infrared cyanine dyes. J. Photochem. Photobiol. A Chem. 168, 53–57 (2004).
Toutchkine, A., Nguyen, D.-V. & Hahn, K. M. Merocyanine dyes with improved photostability. Org. Lett. 9, 2775–2777 (2007).
Chudakov, D. M. et al. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol. Rev. 90, 1103–1163 (2010).
Aitken, C. E., Marshall, R. A. & Puglisi, J. D. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 94, 1826–1835 (2008).
Swoboda, M. et al. Enzymatic oxygen scavenging for photostability without pH drop in single-molecule experiments. ACS Nano 6, 6364–6369 (2012).
Dave, R. et al. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys. J. 96, 2371–2381 (2009).
Altman, R. B. et al. Cyanine fluorophore derivatives with enhanced photostability. Nat. Methods 9, 68–71 (2012).
Buston, J. E., Young, J. R. & Anderson, H. L. Rotaxane-encapsulated cyanine dyes: enhanced fluorescence efficiency and photostability. Chem. Commun. 905–906 https://doi.org/10.1039/b001812k (2000).
Koner, A. L. & Nau, W. M. Cucurbituril encapsulation of fluorescent dyes. Supramol. Chem. 19, 55–66 (2007).
Vaiana, A. ndreaC. et al. Fluorescence quenching of dyes by tryptophan: interactions at atomic detail from combination of experiment and computer simulation. J. Am. Chem. Soc. 125, 14564–14572 (2003).
Marmé, N. et al. Inter- and intramolecular fluorescence quenching of organic dyes by tryptophan. Bioconjug. Chem. 14, 1133–1139 (2003).
Neuweiler, H. et al. Measurement of submicrosecond intramolecular contact formation in peptides at the single-molecule level. J. Am. Chem. Soc. 125, 5324–5330 (2003).
Chen, H., Ahsan, S. S., Santiago-Berrios, M. B., Abruña, H. D. & Webb, W. W. Mechanisms of quenching of Alexa fluorophores by natural amino acids. J. Am. Chem. Soc. 132, 7244–7245 (2010).
Zhang, Y. et al. General strategy to improve the photon budget of thiol-conjugated cyanine dyes. J. Am. Chem. Soc. 145, 4187–4198 (2023).
Heinlein, T. et al. Photoinduced electron transfer between fluorescent dyes and guanosine residues in DNA-hairpins. J. Phys. Chem. B 107, 7957–7964 (2003).
Harvey, B. J. & Levitus, M. Nucleobase-specific enhancement of Cy3 fluorescence. J. Fluoresc. 19, 443–448 (2009).
Zhang, Y. et al. Lumos maxima - How robust fluorophores resist photobleaching?. Curr. Opin. Chem. Biol. 79, 102439 (2024).
Nani, R. R. et al. Reactive species involved in the regioselective photooxidation of heptamethine cyanines. Chem. Sci. 6, 6556–6563 (2015).
Butkevich, A. N. et al. Triarylmethane fluorophores resistant to oxidative photobluing. J. Am. Chem. Soc. 141, 981–989 (2019).
Smit, J. H. et al. On the impact of competing intra- and intermolecular triplet-state quenching on photobleaching and photoswitching kinetics of organic fluorophores. Phys. Chem. Chem. Phys. 21, 3721–3733 (2019).
Peulen, T. O., Opanasyuk, O. & Seidel, C. A. M. Combining graphical and analytical methods with molecular simulations to analyze time-resolved FRET measurements of labeled macromolecules accurately. J. Phys. Chem. B 121, 8211–8241 (2017).
Wiesław, W. et al. Mechanism of fluorescence quenching of tyrosine derivatives by amide group. Chem. Phys. Lett. 341, 99–106 (2001).
Guzow, K. et al. Photophysical properties of tyrosine and its simple derivatives in organic solvents studied by time-resolved fluorescence spectroscopy and global analysis. Photochem. Photobiol. 81, 697–704 (2005).
Chen, C. et al. Elongation factor G initiates translocation through a power stroke. Proc. Natl. Acad. Sci. USA 113, 7515–7520 (2016).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Schepers, B. & Gohlke, H. AMBER-DYES in AMBER: Implementation of fluorophore and linker parameters into AmberTools. J. Chem. Phys. 152, 221103 (2020).
David A. et al. (2020), AMBER 2020, University of California, San Francisco.
Graen, T., Hoefling, M. & Grubmüller, H. AMBER-DYES: characterization of charge fluctuations and force field parameterization of fluorescent dyes for molecular dynamics simulations. J. Chem. Theory Comput. 10, 5505–5512 (2014).
Maier, J. A. et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015).
Berendsen, H. J. C., Grigera, J. R. & Straatsma, T. P. The missing term in effective pair potentials. J. Phys. Chem. 91, 6269–6271 (1987).
Roe, D. R. & Cheatham, T. E. III PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 9, 3084–3095 (2013).
William, H., Andrew, D. & Klaus, S. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Acknowledgements
This work was supported by the National Key R&D Program of China (Grants No. 2024YFA0916700 to C.C. and Y.X.), the National Natural Science Foundation of China (Grants No. 22425701, 22061160466, and 22277063 to C.C., and 22007054 and 32370391 to W.W.), and the Beijing Frontier Research Center for Biological Structure (to C.C. and Y.X.).
Author information
Authors and Affiliations
Contributions
C.C. and Yongzhen H. conceptualized the research. W.W. assisted Yongzhen H. with the experiments. Yongzhen H. and Yichen H. analyzed the data. Yongzhen H. carried out the MD experiments with the help of B.W. and Y.X. Yongzhen H., B.W., Y.X., and C.C. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Chemistry thanks Myung Hyun Jo and the other anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hei, Y., Wang, B., He, Y. et al. Photostability of organic fluorophore influenced by adjacent amino acid residues. Commun Chem 8, 262 (2025). https://doi.org/10.1038/s42004-025-01661-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42004-025-01661-5