Abstract
The immunoregulatory metabolite itaconate accumulates in innate immune cells upon Toll-like receptor stimulation. In response to macrophage activation by lipopolysaccharide, itaconate inhibits inflammasome activation and boosts type I interferon signalling; however, the molecular mechanism of this immunoregulation remains unclear. Here, we show that the enhancement of type I interferon secretion by itaconate depends on the inhibition of peroxiredoxin 5 and on mitochondrial reactive oxygen species. We find that itaconate non-covalently inhibits peroxiredoxin 5, leading to the modulation of mitochondrial peroxide in activating macrophages. Through genetic manipulation, we confirm that peroxiredoxin 5 modulates type I interferon secretion in macrophages. The non-electrophilic itaconate mimetic 2-methylsuccinate inhibits peroxiredoxin 5 and phenocopies immunoregulatory action of itaconate on type I interferon and inflammasome activation, providing further support for a non-covalent inhibition of peroxiredoxin 5 by itaconate. Our work provides insight into the molecular mechanism of actions and biological rationale for the predominantly immune specification of itaconate.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The bulk redox mass spectrometry proteomics data have been deposited to the MassIVE repository under dataset accession no. MSV000093232. The protein itaconation mass spectrometry proteomics data have been deposited to the ProteomeXchange repository under dataset accession no. PXD047348 or MassIVE repository under accession no. MSV000093522. The RNA sequencing data published in this paper are available from the Gene Expression Omnibus (GEO) public repository under GEO accession number GSE277689. The data that support the plots within this paper are included in source data files for each figure. Source data are provided with this paper.
Code availability
No new code has been generated for this work. Sources for the code used are cited in the Methods.
References
Cordes, T. et al. Immunoresponsive gene 1 and itaconate inhibit succinate dehydrogenase to modulate intracellular succinate levels. J. Biol. Chem. 291, 14274–14284 (2016).
Michelucci, A. et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl Acad. Sci. USA 110, 7820–7825 (2013).
Lampropoulou, V. et al. Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab. 24, 158–166 (2016).
Bambouskova, M. et al. Itaconate confers tolerance to late NLRP3 inflammasome activation. Cell Rep. 34, 108756 (2021).
Hooftman, A. et al. The immunomodulatory metabolite itaconate modifies NLRP3 and inhibits inflammasome activation. Cell Metab. 32, 468–478.e7 (2020).
Mills, E. L. et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556, 113–117 (2018).
Ryan, T. A. J. et al. Dimethyl fumarate and 4-octyl itaconate are anticoagulants that suppress tissue factor in macrophages via inhibition of type I interferon. Nat. Commun. 14, 3513 (2023).
Swain, A. et al. Comparative evaluation of itaconate and its derivatives reveals divergent inflammasome and type I interferon regulation in macrophages. Nat. Metab. 2, 594–602 (2020).
Kwai, B. X. C. et al. Itaconate is a covalent inhibitor of the Mycobacterium tuberculosis isocitrate lyase. RSC Med. Chem. 12, 57–61 (2021).
McKinney, J. D. et al. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406, 735–738 (2000).
Van Nguyen, T. et al. Itaconic acid inhibits growth of a pathogenic marine Vibrio strain: a metabolomics approach. Sci. Rep. 9, 5937 (2019).
McFadden, B. A., Williams, J. O. & Roche, T. E. Mechanism of action of isocitrate lyase from Pseudomonas indigofera. Biochemistry 10, 1384–1390 (1971).
Bambouskova, M. et al. Electrophilic properties of itaconate and derivatives regulate the IκBζ–ATF3 inflammatory axis. Nature 556, 501 (2018).
Runtsch, M. C. et al. Itaconate and itaconate derivatives target JAK1 to suppress alternative activation of macrophages. Cell Metab. 34, 487–501.e8 (2022).
Qin, W. et al. Chemoproteomic profiling of itaconation by bioorthogonal probes in inflammatory macrophages. J. Am. Chem. Soc. 142, 10894–10898 (2020).
Ryan, D. G. et al. Nrf2 activation reprograms macrophage intermediary metabolism and suppresses the type I interferon response. iScience 25, 103827 (2022).
Olagnier, D. et al. Nrf2 negatively regulates STING indicating a link between antiviral sensing and metabolic reprogramming. Nat. Commun. 9, 3506 (2018).
Gunderstofte, C. et al. Nrf2 negatively regulates type I interferon responses and increases susceptibility to herpes genital infection in mice. Front. Immunol. 10, 2101 (2019).
Wood, Z. A., Poole, L. B. & Karplus, P. A. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science 300, 650–653 (2003).
Hanley, P. J., Mickel, M., Löffler, M., Brandt, U. & Daut, J. KATP channel‐independent targets of diazoxide and 5‐hydroxydecanoate in the heart. J. Physiol. 542, 735–741 (2002).
Brault, M., Olsen, T. M., Martinez, J., Stetson, D. B. & Oberst, A. Intracellular nucleic acid sensing triggers necroptosis through synergistic type I IFN and TNF signaling. J. Immunol. 200, 2748–2756 (2018).
Su, C., Cheng, T., Huang, J., Zhang, T. & Yin, H. 4-Octyl itaconate restricts STING activation by blocking its palmitoylation. Cell Rep. 42, 113040 (2023).
Onomoto, K., Onoguchi, K. & Yoneyama, M. Regulation of RIG-I-like receptor-mediated signaling: interaction between host and viral factors. Cell Mol. Immunol. 18, 539–555 (2021).
Xu, H. et al. Structural basis for the prion-like MAVS filaments in antiviral innate immunity. Elife 3, e01489 (2014).
Widdrington, J. D. et al. Mitochondrial DNA depletion induces innate immune dysfunction rescued by IFN-γ. J. Allergy Clin. Immunol. 140, 1461–1464.e8 (2017).
Siebels, I. & Dröse, S. Q-site inhibitor induced ROS production of mitochondrial complex II is attenuated by TCA cycle dicarboxylates. Biochim. Biophys. Acta 1827, 1156–1164 (2013).
Robb, E. L. et al. Selective superoxide generation within mitochondria by the targeted redox cycler MitoParaquat. Free Radic. Biol. Med. 89, 883–894 (2015).
Murphy, M. P. et al. Guidelines for measuring reactive oxygen species and oxidative damage in cells and in vivo. Nat. Metab. 4, 651–662 (2022).
Pak, V. V. et al. Ultrasensitive genetically encoded indicator for hydrogen peroxide identifies roles for the oxidant in cell migration and mitochondrial function. Cell Metab. 31, 642–653.e6 (2020).
Dikalova, A. E. et al. Therapeutic targeting of mitochondrial superoxide in hypertension. Circ. Res. 107, 106–116 (2010).
Ramalho, T. et al. Itaconate impairs immune control of Plasmodium by enhancing mtDNA-mediated PD-L1 expression in monocyte-derived dendritic cells. Cell Metab. 36, 484–497 (2024).
Yan, S. et al. Biochemical characterization of human peroxiredoxin 2, an antioxidative protein. Acta Biochim. Biophys. Sin. (Shanghai) 44, 759–764 (2012).
Baković, J. et al. A key metabolic integrator, coenzyme A, modulates the activity of peroxiredoxin 5 via covalent modification. Mol. Cell. Biochem. 461, 91–102 (2019).
Coppo, L., Montano, S. J., Padilla, A. C. & Holmgren, A. Determination of glutaredoxin enzyme activity and protein S-glutathionylation using fluorescent eosin-glutathione. Anal. Biochem. 499, 24–33 (2016).
Holmgren, A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 254, 9627–9632 (1979).
Knoops, B., Goemaere, J., Van der Eecken, V. & Declercq, J.-P. Peroxiredoxin 5: structure, mechanism, and function of the mammalian atypical 2-Cys peroxiredoxin. Antioxid. Redox Signal 15, 817–829 (2011).
Aguirre, C., Brink, T., ten, Guichou, J.-F., Cala, O. & Krimm, I. Comparing binding modes of analogous fragments using NMR in fragment-based drug design: application to PRDX5. PLoS ONE 9, e102300 (2014).
Declercq, J.-P. et al. Crystal structure of human peroxiredoxin 5, a novel type of mammalian peroxiredoxin at 1.5 Å resolution. J. Mol. Biol. 311, 751–759 (2001).
Shekhova, E. Mitochondrial reactive oxygen species as major effectors of antimicrobial immunity. PLoS Pathog. 16, e1008470 (2020).
Dubuisson, M. et al. Human peroxiredoxin 5 is a peroxynitrite reductase. FEBS Lett. 571, 161–165 (2004).
Daniels, B. P. et al. The nucleotide sensor ZBP1 and kinase RIPK3 induce the enzyme IRG1 to promote an antiviral metabolic state in neurons. Immunity 50, 64–76.e4 (2019).
Sohail, A. et al. Itaconate and derivatives reduce interferon responses and inflammation in influenza A virus infection. PLoS Pathog. 18, e1010219 (2022).
Cho, H. et al. Differential innate immune response programs in neuronal subtypes determine susceptibility to infection in the brain by positive-stranded RNA viruses. Nat. Med. 19, 458–464 (2013).
Szeligowski, R. V. et al. Molecular evolution of IRG1 shapes itaconate production in metazoans and alleviates the “double-edged dilemma” of innate immune defense. Preprint at bioRxiv https://doi.org/10.1101/2022.06.17.496652 (2022).
Buchmann, K. Evolution of innate immunity: clues from invertebrates via fish to mammals. Front. Immunol. 5, 459 (2014).
Chen, F. et al. Citraconate inhibits ACOD1 (IRG1) catalysis, reduces interferon responses and oxidative stress, and modulates inflammation and cell metabolism. Nat. Metab. 4, 534–546 (2022).
Waqas, S. F.-H. et al. ISG15 deficiency features a complex cellular phenotype that responds to treatment with itaconate and derivatives. Clin. Transl. Med. 12, e931 (2022).
Yue, Y.-X. et al. 4-Octyl itaconate inhibits poly(I:C)-induced interferon-β secretion in mouse bone marrow-derived macrophages partially by activating Nrf2. Heliyon 9, e23001 (2023).
He, W. et al. Mesaconate is synthesized from itaconate and exerts immunomodulatory effects in macrophages. Nat. Metab. 4, 524–533 (2022).
Graham, D. B. et al. Nitric oxide engages an anti-inflammatory feedback loop mediated by peroxiredoxin 5 in phagocytes. Cell Rep. 24, 838–850 (2018).
Evavold, C. L. et al. Control of gasdermin D oligomerization and pyroptosis by the Ragulator–Rag–mTORC1 pathway. Cell 184, 4495–4511.e19 (2021).
Borin, B. N. et al. Murine norovirus protein NS1/2 aspartate to glutamate mutation, sufficient for persistence, reorients side chain of surface exposed tryptophan within a novel structured domain. Proteins 82, 1200–1209 (2014).
Slaby, I. & Holmgren, A. Thioredoxin reductase-dependent insulin disulfide reduction by phage T7 DNA polymerase reflects dissociation of the enzyme into subunits. J. Biol. Chem. 264, 16502–16506 (1989).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Wang, L., Wang, S. & Li, W. RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012).
Ewels, P., Magnusson, M., Lundin, S. & Käller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Durinck, S., Spellman, P. T., Birney, E. & Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191 (2009).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Kleverov, M. et al. Phantasus, a web application for visual and interactive gene expression analysis. eLife 13, e85722 (2024).
Korotkevich, G. et al. Fast gene set enrichment analysis. Preprint at bioRxiv https://doi.org/10.1101/060012 (2021).
Mölder, F. et al. Sustainable data analysis with Snakemake. F1000Res 10, 33 (2021).
Mertins, P. et al. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography–mass spectrometry. Nat. Protoc. 13, 1632–1661 (2018).
Meier, F. et al. Online parallel accumulation–serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol. Cell. Proteom. 17, 2534–2545 (2018).
Perkins, D. N., Pappin, D. J. C., Creasy, D. M. & Cottrell, J. S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 (1999).
Day, N. J. et al. A deep redox proteome profiling workflow and its application to skeletal muscle of a Duchenne muscular dystrophy model. Free Radic. Biol. Med 193, 373–384 (2022).
Gaffrey, M. J., Day, N. J., Li, X. & Qian, W. Resin-assisted capture coupled with isobaric tandem mass tag labeling for multiplexed quantification of protein thiol oxidation. J. Vis. Exp. 172, e62671 (2021).
Guo, J. et al. Resin-assisted enrichment of thiols as a general strategy for proteomic profiling of cysteine-based reversible modifications. Nat. Protoc. 9, 64–75 (2014).
Li, X. et al. Mass spectrometry-based direct detection of multiple types of protein thiol modifications in pancreatic beta cells under endoplasmic reticulum stress. Redox Biol. 46, 102111 (2021).
Wang, Y. et al. Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics 11, 2019–2026 (2011).
Duan, J. et al. Quantitative profiling of protein S-glutathionylation reveals redox-cependent regulation of macrophage function during nanoparticle-induced oxidative stress. ACS Nano 10, 524–538 (2016).
Kim, S. & Pevzner, P. A. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 5, 5277 (2014).
Fuhrer, T., Heer, D., Begemann, B. & Zamboni, N. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection–time-of-flight mass spectrometry. Anal. Chem. 83, 7074–7080 (2011).
Morris, G. M. et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 (2009).
Forli, S. et al. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 11, 905–919 (2016).
Laskowski, R. A. & Swindells, M. B. LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model 51, 2778–2786 (2011).
Acknowledgements
We thank C. Evavold for providing iBMDMs; E. Aladyeva, B. S. Andhey for computational support; P. Bohacova for helping with experiments; R. Xavier and E. A. Creasey for providing Prdx5−/− mice; M. S. Diamond for critical reading of the manuscript; R. Sprung, P. Erdmann Gilmore and R. Townsend from the WashU Proteomics Core for itaconate peptide MS/MS analysis. Images in some of the figures were created in BioRender.com. The study was supported in part by NIAID grant R01-A1125618 (to M.N.A.). Experiments on live-cell imaging were supported by Russian Science Foundation grant 23-75-30023 (to V.V.B.). Correspondence and requests for materials should be addressed to the corresponding author.
Author information
Authors and Affiliations
Contributions
T.P. and M.N.A. conceived and designed the study and wrote the manuscript. T.P. performed biomass preparation for RNA sequencing, metabolomics and proteomics experiments, western blot, qPCR, cytometry, cytokine assays, recombinant protein activity assays, lentiviral transduction, 3-nitrotyrosine ELISA and 3D protein visualization. B.E. performed western blot, cytometry and cytokine assays, nitrite + nitrate assay, Griess assay and recombinant protein preparation, L.P. performed cytokine and western blot assays. V.V. performed in vivo poly(I:C) injection and cytokine measurement. D.A.K. performed the docking analysis, RNA sequencing data processing and analysis. V.S. performed pathway enrichment analysis. E.P. and V.V.B. designed and performed the mitoHyPer7 oxidation experiments. J.M. performed switchSENSE fluorescence proximity sensing analysis. A.M.K. designed and performed 15N-protein nuclear magnetic resonance analysis. A.J. and S.P.D. performed cell culture experiments. D.S. performed western blot experiments. K.H. performed human monocyte isolation D.K. and S.D. prepared the recombinant proteins. M.B. performed biomass preparation for RNA sequencing and MS/MS protein itaconation experiment. C.F.L. assisted in the analysis of the MS/MS protein itaconation results. N.J.D., T.Z. and W.J.Q. performed and analysed the bulk redox proteomics experiment. C.N., R.S. and D.H.F performed recombinant protein generation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Metabolism thanks Karsten Hiller and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Christoph Schmitt, in collaboration with the Nature Metabolism team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Exogenous and endogenous itaconate regulate IFNβ related phenotypes.
a IFNβ release by activated BMDMs pretreated with itaconic acid (IA, 5 mM), or sodium itaconate (NaI, 10 mM or 20 mM) (16 h) or media (M) and stimulated with LPS (0.1 μg/ml) for 4 h (n = 8 (M, IA), n = 7 (NaI 10), n = 5 (NaI 20) biological replicates). b pH of cRPMI supplemented with 5 mM IA or malonic acid (MA) (n = 3 independent experiments). c Survival of BMDMs pretreated with IA (5 mM, 16 h), MA (5 mM, 16 h) or acidified media (HCl, 16 h) measured by LDH activity in the supernatant (left) or ATP levels in cell lysates and supernatants (right) (n = 4 (left) n = 6 (right) independent cultures) d IFNβ release from WT or Irg1-/- BMDMs stimulated with LPS (0.1 μg/ml, 24 h) (n = 6 individual cultures). e Survival of WT or IFNAR-/- mice upon lethal dose of DMXAA i.p. (n = 3 mice/group). f Serum levels of IFNβ in WT or Irg1-/- repeated experiment to Fig. 1i (n = 4 WT and n = 3 Irg1-/-). g Survival of WT or Irg1-/- female mice treated with increasing doses of DMXAA i.p. (n = 15 (WT 40 mg/kg), n = 4 (WT 50 mg/kg) n = 6 (WT 60 mg/kg), n = 5 (Irg1-/- 40 mg/kg) mice/group). h Survival of WT or Irg1-/- female mice injected with 60 mg/kg lethal dose of DMXAA i.p. (n = 3 mice/group). Data are presented as mean ±SEM. Statistical analysis: a Two sided Brown-Forsythe and Welch ANOVA test. c Two sided One sample t-test. d, f Two sided Welch’s t-test.
Extended Data Fig. 2 Itaconic acid regulates IFNβ production through mtDNA/cGAS/STING pathway.
a Representative immunoblot of STAT1 and STAT2 protein levels in BMDMs pretreated with itaconic acid (IA) (5 mM, 16 h) and co-treated with αIFNAR blocking antibody or isotype control (representative of n = 2-4). GAPDH serves as loading control and corresponding statistics. b IFNβ release by iBMDMs pretreated with media (M), IA or malonic acid (MA) or acidified media (HCl) in the presence of a gradient of RU.521 (1, 5, 10 μg/ml) or C176 (0.5, 1, 2 μM) inhibitors followed by poly(I:C) (20 μg/ml, 4 h) stimulation (n = 6 M, IA, RU.521 1 and 5; n = 4 MA, HCl, all C176; n = 2 RU.521 10 μg/ml). c Representative immunoblot of STAT1 expression in iBMDMs treated with inhibitors as in c (middle concentration) in presence of IA and corresponding statistics (n = 5). d IFNβ release from WT or MAVS-/- BMDMs pretreated as in a and stimulated with poly(I:C) as in b (n = 3 biological replicates). e mtDNA in cytosolic fraction of iBMDMs pretreated with itaconic acid as in b (n = 3 individual experiments). f IFNβ release from WT or mtDNA depleted (EtBr) iBMDMs pretreated with media or IA as in b and stimulated with DMXAA (5 μg/ml, 4 h) (n = 5 independent experiments). g-h Representative immunoblot of STAT1 and STAT2 protein levels in iBMDMs pretreated as in e and corresponding statistics (representative of n = 4 independent experiments). i-j IFNβ release from BMDMs pretreated with 3NPA (200 μM, 16 h) or media and co-treated or not with C176 (j, 1 μM, 16 h) or MitoTempo (k, 250 μM, 16 h) (n = 5). Data are represented as mean ±SEM. Statistical analysis: a, c, f, i-j Brown-Forsythe and Welsh ANOVA with Dunnet’s T3 multiple comparison test. b RM-Two-way ANOVA with Tukey’s multiple comparison test. e One sample t-test. h Ordinary One-way ANOVA with Tukey’s multiple comparison test.
Extended Data Fig. 3 Itaconic enhances hydrogen peroxide retention.
a IFNβ release from iBMDMs treated with media (M) or itaconic acid (IA) (5 mM, 16 h) in presence or absence of indicated concentrations of NAC (n = 3) or MitoTempo (n = 6). b Statistics for Fig. 3d (n = 4 independent cultures). c Relative median fluorescent intensity fold change of CellROX green in iBMDMs pretreated M, IA or Malonic acid (MA) (5 mM, 16 h) (n = 6). d Representative histograms of c and Fig. 3f, g. Dashed line represents median of media treated cells. e Oxidation status of mitoHyPer7 in Hela cells pretreated with IA or media as in a for 20 min or 4 h (dots represent individual cells, n = 3 individual experiments). f Oxidation status of mitoHyPer7 in Hela cells pretreated with IA as in a and treated with H2O2 (100 μM) followed by H2O2 removal and continued measurement for additional 30 min (representative experiment of n = 3). Data represent mean ±SEM. Statistical analysis: a RM-Two-way ANOVA with Tukey’s multiple comparison test. b, e Brown-Forsythe and Welsh ANOVA with Dunnet’s T3 multiple comparison test. c One sample t-test.
Extended Data Fig. 4 Itaconate non-covalently interacts with Prdx5.
a Itaconated peptide MS/MS spectrum of Prdx5 in BMDMs treated with itaconic acid (IA, 5 mM, 16 h). b Observed occupancy of itaconated Cys in the media (M) or IA treated samples. (n = 1) c Itaconated Prdx5 peptide with itaconate covalently bound to Cys200. d-e Activity assays of human Thioredoxin 1 (d) and human Glutaredoxin 1 (e, n = 2 technical replicates in 2 individual experiments) f t-BOOH consumption by Prdx5 in activity assay in Fig. 4a. g Activity of Prdx5 pretreated with H2O2 (100 μM, 30 min) or DTT (10 mM, 30 min) assayed in buffer or in presence of DTT (1 mM) (n = 3 independent experiments). h Standard curve of t-BOOH in buffer or presence of itaconate or malonate with linear regression equation (n = 5 independent experiments). i Activity of recombinant human Prdx5 performed as in Fig. 4c pretreated with indicated concentrations of Itaconic acid (n = 3 independent experiments). j Immunoblot of recombinant human Prdx5 redox state in presence or absence of IA (5 mM) and corresponding statistics (n = 4 independent experiments). k Representative immunoblot of Prdx5 in Ctrl or Prdx5-overexpressing iBMDMs (Prdx5-OE) (n = 3). l Representative immunoblot of Prdx5 expression in WT and Prdx5-/- BMDMs stimulated with DMXAA (5 μg/ml, 24 h) (n = 3). m-n kCal/mol (m) and Ki (n) parameters towards Prdx5 derived for the strongest binding conformation for each chemical. Data represent mean ±SEM. Statistical analysis: f Brown-Forsythe and Welsh ANOVA with Dunnet’s T3 multiple comparison test. j Two-sided One sample t-test.
Extended Data Fig. 5 Itaconate non-covalently interacts with Prdx5.
a-b Sensorgrams for the interaction between Prdx5 and metabolites from Fig. 5b,c before bulk shift correction. c Three full superimposed spectra of 15N labeled Prdx5 free (black), with 20.52 mM itaconate (red), and with 20.19 mM benzoate (blue).
Extended Data Fig. 6 Production of itaconate upon TLR stimulation leads to increased peroxide accumulation.
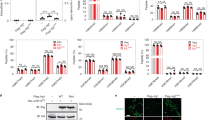
a Representative immunoblot of Irg1 expression in WT or Irg1-/- BMDMs non-stimulated or stimulated with R848 (1 μg/ml, 24 h). GAPDH serves as loading control (n = 3). b Intracellular levels of itaconate in BMDMs stimulated as in (a). To rescue itaconate levels in Irg1-/- cells were treated with itaconic acid (IA) (1 mM, at 4 h post stimulation) (n = 3 biological replicates). c-d mtPY1 levels and representative histograms in BMDMs non-stimulated or stimulated as with LPS (0.1 ug/ml) or R848 (1 μg/ml) or poly(I:C) (20 μg/ml) for 24 h (n = 6 (c), n = 5 (d) independent cultures). e Intracellular itaconate levels in WT and Irg1-/- BMDMs treated as in (c) (n = 3 biological replicates). f Nitrite+Nitrate levels in supernatant of WT or Irg1-/- BMDMs stimulated as in (c) (n = 4 biological replicates). g Nitrite levels in supernatants of BMDMs stimulated as in (c) (n = 4 independent cultures) h Randomized design assignment of TMT10 reagents used for multiplexing of enriched samples. Data represent mean ±SEM. Statistical analysis: c-d One sample t-test.
Extended Data Fig. 7 Methyl succinic acid phenocopies itaconic acid effects in activated macrophages.
a Sensogram for the interaction between Prdx5 and 2-methylsuccinate from Fig. 7h before bulk shift correction. b Survival of BMDMs pretreated with methylsuccinic acid (MetSA, 5 mM, 16 h), or media measured by LDH activity in the supernatant (left) or ATP levels (right) (n = 3-4 independent cultures) c Intracellular levels of indicated metabolites in BMDMs pretreated with itaconic acid or MetSA (5 mM, 16 h) (n = 3). d Hallmark gene set enrichment analysis in samples RNAseq data from BMDMs pretreated with metabolites as in c. e Survival of human Monocyte-derived macrophages (hMoDMs) pretreated with IA or MetSA as in c (n = 2 individual donors). f IFNβ release from hMoDMs stimulated with poly(I:C) (30 ug/ml), LPS (100 ng/ml) or STING agonis diABZI (1 μM) for 24 h (n = 4 individual donors). g IFNβ release from hMoDM pretreated with IA or MetSA as in e and stimulated with poly(I:C) as in f (n = 3 individual donors). Data represent mean ±SEM. Statistical analysis: g Brown-Forsythe and Welsh ANOVA with Dunnet’s T3 multiple comparison test.
Extended Data Fig. 8 Gating strategy to identify live singlet cells.
a First debris and dublets were removed gating on FSC-Amid FSC-Wlow. Subsequently, live cells were gated as Live/Dead NIRlow. Fluorescent intensity of select probe was determined in the Live cells population.
Supplementary information
Supplementary Table
1. Metabolomics dataset of WT and Irg1-/- BMDMs non-stimulated or stimulated with LPS, poly(I:C) or R848 for 24 h. Alternatively macrophages were pretreated with media, itaconic acid, or 2-methylsuccinic acid (5 mM, 16 h). Sample list, ion_matrix, annotation, ions. 2. List of identified peptides in WT BMDMs treated with media for 16 h. List of peptides observed in WT BMDMs treated with 5 mM itaconic acid for 16 h. List of PSMs itaconated on cysteine. 3. List of oxidized cysteines in WT and Irg1-/- BMDMs non-activated or stimulated with LPS for 24 h. 4. List of reagents and used software and algorithms.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6
Unprocessed western blots.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig.2
Statistical source data.
Source Data Extended Data Fig.2
Unprocessed western blots.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 7
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Paulenda, T., Echalar, B., Potuckova, L. et al. Itaconate modulates immune responses via inhibition of peroxiredoxin 5. Nat Metab 7, 1183–1203 (2025). https://doi.org/10.1038/s42255-025-01275-0
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s42255-025-01275-0
This article is cited by
-
Itaconate boosts type I IFN response by disrupting cytoprotection
Nature Metabolism (2025)
-
IER3 drives the transition from sepsis-associated AKI to CKD by suppressing the mitochondrial translocation of PRDX5
Cellular and Molecular Life Sciences (2025)