Abstract
Dysregulated lipid metabolism promotes persistent microglial activation and neuroinflammation in Alzheimer’s disease (AD), but the underlying pathogenic mechanisms remain to be elucidated, and druggable targets remain to be identified. Here we found that multifunctional enzyme type 2 (MFE-2), the key enzyme regulating fatty acid β-oxidation in the peroxisome, was downregulated in the microglia of humans with AD and AD model mice. Microglia-specific ablation of MFE-2 drove microglial abnormalities, neuroinflammation and Aβ deposition in AD models. Mechanistically, MFE-2 deficiency facilitated lipid accumulation, resulting in excessive arachidonic acid, mitochondrial reactive oxygen species and proinflammatory cytokine production by microglia. The compound 3-O-cyclohexane carbonyl-11-keto-β-boswellic acid (CKBA) bound to MFE-2 and restored MFE-2 levels, ameliorating AD pathology by inhibiting microglial overactivation. Collectively, our data revealed a pathogenic role of microglia with impaired lipid metabolism in AD and identified MFE-2 as a druggable target of CKBA, which restores its expression and has therapeutic potential for treating AD.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Source data is provided with this paper. The source data for all main figures have also been deposited in Zenodo and can be accessed at https://zenodo.org/records/16732189 (ref. 68). Raw scRNA-seq data have been deposited in the Genome Sequence Archive (accession code CRA017222). Bulk RNA-seq data have been deposited in OMIX (accession code OMIX011442).
Change history
24 November 2025
A Correction to this paper has been published: https://doi.org/10.1038/s43587-025-01040-8
References
2022 Alzheimer’s disease facts and figures. Alzheimers Dement. 18, 700–789 (2022).
Calsolaro, V. & Edison, P. Neuroinflammation in Alzheimer’s disease: current evidence and future directions. Alzheimers Dement. 12, 719–732 (2016).
Leng, F. & Edison, P. Neuroinflammation and microglial activation in Alzheimer disease: where do we go from here? Nat. Rev. Neurol. 17, 157–172 (2021).
Sun, N. et al. Human microglial state dynamics in Alzheimer’s disease progression. Cell 186, 4386–4403.e4329 (2023).
Bernier, L. P., York, E. M. & MacVicar, B. A. Immunometabolism in the brain: how metabolism shapes microglial function. Trends Neurosci. 43, 854–869 (2020).
Marschallinger, J. et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 23, 194–208 (2020).
Liu, L. et al. Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell 160, 177–190 (2015).
Chausse, B., Kakimoto, P. A. & Kann, O. Microglia and lipids: how metabolism controls brain innate immunity. Semin Cell Dev. Biol. 112, 137–144 (2021).
Jung, E. S. & Mook-Jung, I. New microglia on the block. Cell Metab. 31, 664–666 (2020).
Mi, Y. et al. Loss of fatty acid degradation by astrocytic mitochondria triggers neuroinflammation and neurodegeneration. Nat. Metab. 5, 445–465 (2023).
Lodhi, I. J. & Semenkovich, C. F. Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab. 19, 380–392 (2014).
Di Cara, F., Sheshachalam, A., Braverman, N. E., Rachubinski, R. A. & Simmonds, A. J. Peroxisome-mediated metabolism is required for immune response to microbial infection. Immunity 47, 93–106.e107 (2017).
Hiltunen, J. K. et al. 17B-hydroxysteroid dehydrogenases as acyl thioester metabolizing enzymes. Mol. Cell Endocrinol. 489, 107–118 (2019).
Ferdinandusse, S. et al. Mutational spectrum of D-bifunctional protein deficiency and structure-based genotype-phenotype analysis. Am. J. Hum. Genet. 78, 112–124 (2006).
Beckers, L. et al. Microglia lacking a peroxisomal β-oxidation enzyme chronically alter their inflammatory profile without evoking neuronal and behavioral deficits. J. Neuroinflammation 16, 61 (2019).
Verheijden, S., Beckers, L., De Munter, S., Van Veldhoven, P. P. & Baes, M. Central nervous system pathology in MFP2 deficiency: insights from general and conditional knockout mouse models. Biochimie 98, 119–126 (2014).
Lines, M. A. et al. Peroxisomal D-bifunctional protein deficiency: three adults diagnosed by whole-exome sequencing. Neurology 82, 963–968 (2014).
Haney, M. S. et al. APOE4/4 is linked to damaging lipid droplets in Alzheimer’s disease microglia. Nature 628, 154–161 (2024).
Victor, M. B. et al. Lipid accumulation induced by APOE4 impairs microglial surveillance of neuronal-network activity. Cell Stem Cell 29, 1197–1212.e1198 (2022).
Lin, Y. T. et al. APOE4 causes widespread molecular and cellular alterations associated with alzheimer’s disease phenotypes in human iPSC-derived brain cell types. Neuron 98, 1141–1154.e1147 (2018).
Caldeira, C. et al. Microglia change from a reactive to an age-like phenotype with the time in culture. Front. Cell Neurosci. 8, 152 (2014).
Ennerfelt, H. et al. SYK coordinates neuroprotective microglial responses in neurodegenerative disease. Cell 185, 4135–4152.e4122 (2022).
Peruzzotti-Jametti, L. et al. Mitochondrial complex I activity in microglia sustains neuroinflammation. Nature 628, 195–203 (2024).
Mukem, S., Thongbuakaew, T. & Khornchatri, K. Mito-Tempo suppresses autophagic flux via the PI3K/Akt/mTOR signaling pathway in neuroblastoma SH-SY5Y cells. Heliyon 7, e07310 (2021).
Hubbard, W. C. et al. Newborn screening for X-linked adrenoleukodystrophy (X-ALD): validation of a combined liquid chromatography-tandem mass spectrometric (LC-MS/MS) method. Mol. Genet. Metab. 97, 212–220 (2009).
Hall, N. A., Lynes, G. W. & Hjelm, N. M. Ratios for very-long-chain fatty acids in plasma of subjects with peroxisomal disorders, as determined by HPLC and validated by gas chromatography-mass spectrometry. Clin. Chem. 34, 1041–1045 (1988).
Cocco, T., Di Paola, M., Papa, S. & Lorusso, M. Arachidonic acid interaction with the mitochondrial electron transport chain promotes reactive oxygen species generation. Free Radic. Biol. Med. 27, 51–59 (1999).
Kwon, K. J., Jung, Y. S., Lee, S. H., Moon, C. H. & Baik, E. J. Arachidonic acid induces neuronal death through lipoxygenase and cytochrome P450 rather than cyclooxygenase. J. Neurosci. Res. 81, 73–84 (2005).
Hartfield, P. J. & Robinson, J. M. Arachidonic acid activates NADPH oxidase by a direct, calmodulin-regulated mechanism. Prostaglandins Other Lipid Mediat. 56, 1–6 (1998).
Hammoud, M. K. et al. Arachidonic acid impairs natural killer cell functions by disrupting signaling pathways driven by activating receptors and reactive oxygen species. Cell Commun. Signal. 22, 555 (2024).
Errede, M. et al. Microglia-derived CCL2 has a prime role in neocortex neuroinflammation. Fluids Barriers CNS 19, 68 (2022).
Bose, S. & Cho, J. Role of chemokine CCL2 and its receptor CCR2 in neurodegenerative diseases. Arch. Pharm. Res. 36, 1039–1050 (2013).
Lou, F. et al. Identification and pre-clinical investigation of 3-O-cyclohexanecarbonyl-11-keto-β-boswellic acid as a drug for external use to treat psoriasis. Br. J. Pharm. https://doi.org/10.1111/bph.16253 (2023).
Verheijden, S. et al. Peroxisomal multifunctional protein-2 deficiency causes neuroinflammation and degeneration of Purkinje cells independent of very long chain fatty acid accumulation. Neurobiol. Dis. 58, 258–269 (2013).
Fransen, M., Lismont, C. & Walton, P. The peroxisome-mitochondria connection: how and why? Int. J. Mol. Sci. https://doi.org/10.3390/ijms18061126 (2017).
Wanders, R. J., Waterham, H. R. & Ferdinandusse, S. Metabolic interplay between peroxisomes and other subcellular organelles including mitochondria and the endoplasmic reticulum. Front. Cell Dev. Biol. 3, 83 (2015).
Zhou, Y. et al. Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 26, 131–142 (2020).
Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290.e1217 (2017).
Raas, Q. et al. A microglial cell model for acyl-CoA oxidase 1 deficiency. Biochim Biophys. Acta Mol. Cell Biol. Lipids 1864, 567–576 (2019).
Nau, G. J. et al. Human macrophage activation programs induced by bacterial pathogens. Proc. Natl Acad. Sci. USA 99, 1503–1508 (2002).
Johnson, E. C. B. et al. Large-scale proteomic analysis of Alzheimer’s disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat. Med. 26, 769–780 (2020).
Rachmian, N. et al. Identification of senescent, TREM2-expressing microglia in aging and Alzheimer’s disease model mouse brain. Nat. Neurosci. 27, 1116–1124 (2024).
Zhang, F., Zhong, R. J., Cheng, C., Li, S. & Le, W. D. New therapeutics beyond amyloid-β and tau for the treatment of Alzheimer’s disease. Acta Pharm. Sin. 42, 1382–1389 (2021).
Boxer, A. L. & Sperling, R. Accelerating Alzheimer’s therapeutic development: the past and future of clinical trials. Cell 186, 4757–4772 (2023).
Jucker, M. & Walker, L. C. Alzheimer’s disease: from immunotherapy to immunoprevention. Cell 186, 4260–4270 (2023).
Bai, J. et al. Identification of a natural inhibitor of methionine adenosyltransferase 2A regulating one-carbon metabolism in keratinocytes. eBioMedicine 39, 575–590 (2019).
Moussaieff, A. & Mechoulam, R. Boswellia resin: from religious ceremonies to medical uses; a review of in-vitro, in-vivo and clinical trials. J. Pharm. Pharm. 61, 1281–1293 (2009).
Wei, C. et al. Acetyl-11-keto-β-boswellic acid ameliorates cognitive deficits and reduces amyloid-β levels in APPswe/PS1dE9 mice through antioxidant and anti-inflammatory pathways. Free Radic. Biol. Med. 150, 96–108 (2020).
Yan, X. X., Ma, C., Bao, A. M., Wang, X. M. & Gai, W. P. Brain banking as a cornerstone of neuroscience in China. Lancet Neurol. 14, 136 (2015).
Qiu, W. et al. Standardized operational protocol for human brain banking in China. Neurosci. Bull. 35, 270–276 (2019).
Mirra, S. S. et al. The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer’s disease. Neurology 41, 479–486 (1991).
Braak, H. & Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 (1991).
Oakley, H. et al. Intraneuronal β-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26, 10129–10140 (2006).
Calvo, B., Rubio, F., Fernández, M. & Tranque, P. Dissociation of neonatal and adult mice brain for simultaneous analysis of microglia, astrocytes and infiltrating lymphocytes by flow cytometry. IBRO Rep. 8, 36–47 (2020).
Lian, H., Roy, E. & Zheng, H. Protocol for primary microglial culture preparation. Bio. Protoc. https://doi.org/10.21769/BioProtoc.1989 (2016).
Long, H. et al. Interaction of RAGE with α-synuclein fibrils mediates inflammatory response of microglia. Cell Rep. 40, 111401 (2022).
Pan, X. D. et al. Microglial phagocytosis induced by fibrillar β-amyloid is attenuated by oligomeric β-amyloid: implications for Alzheimer’s disease. Mol. Neurodegener. 6, 45 (2011).
Giuliano, C. J., Lin, A., Girish, V. & Sheltzer, J. M. Generating single cell-derived knockout clones in mammalian cells with CRISPR/Cas9. Curr. Protoc. Mol. Biol. 128, e100 (2019).
Calì, T. & Brini, M. Quantification of organelle contact sites by split-GFP-based contact site sensors (SPLICS) in living cells. Nat. Protoc. 16, 5287–5308 (2021).
Li, X. et al. Transcriptional and epigenetic decoding of the microglial aging process. Nat. Aging 3, 1288–1311 (2023).
Stossi, F. & Singh, P. K. Basic image analysis and manipulation in ImageJ/Fiji. Curr. Protoc. 3, e849 (2023).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Mukhopadhyay, P. et al. Simultaneous detection of apoptosis and mitochondrial superoxide production in live cells by flow cytometry and confocal microscopy. Nat. Protoc. 2, 2295–2301 (2007).
Novikov, D. K. et al. Peroxisomal β-oxidation. Purification of four novel 3-hydroxyacyl-CoA dehydrogenases from rat liver peroxisomes. J. Biol. Chem. 269, 27125–27135 (1994).
Elmore, M. R. et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron 82, 380–397 (2014).
Kraeuter, A. K., Guest, P. C. & Sarnyai, Z. The open field test for measuring locomotor activity and anxiety-like behavior. Methods Mol. Biol. 1916, 99–103 (2019).
Leger, M. et al. Object recognition test in mice. Nat. Protoc. 8, 2531–2537 (2013).
Gao, M. Source Data for NATAGING-A06160C. Zenodo https://doi.org/10.5281/zenodo.16732188 (2025).
Acknowledgements
This work was supported by the National Natural Science Foundation of China Original Exploration Program (no. 82450903), the Experimental Animal Research Project of ‘Scientific and Technological Innovation Action Plan’ (no. 22140903100), the National Natural Science Foundation of China (no. 82073428 to J.B.), China Postdoctoral Science Foundation (2021M692114 to M.G., no. GZB20230431 to Z.W. and no. 2023M742313 to Z.W.) and the Innovative Research Team of High-Level Local Universities in Shanghai (to H.W. if not otherwise noted). We thank the Xiangya Brain Bank, School of Basic Medical Science, Central South University, for providing the brain blocks of the patients with AD and control patients. We thank X. Yan for supplying the patient materials. We also thank D. Li for providing Aβ PFF. We thank L. Shi for CKBA modification for the click chemistry experiment.
Author information
Authors and Affiliations
Contributions
Experiments were designed and conceived by H.W., M.G., F.L., Y.S., J.B. and Z.W. with the support of Y.L. and X.C. Cell and mouse experiments were performed and analyzed by H.W., M.G. and J.B., with the support of X.L., Z.Z. and F.Z. RNA-seq analysis was performed by M.G. with the support of Y.S., X.C. and Y.W. F.Z. and J.L. performed and analyzed the Airyscan microscopy data. L.F. and X.J. performed in situ staining and analysis of microglial morphology under the supervision of M.G. J.B. generated MFE-2 conditional mice. H.W. and M.G. conceived of the idea and wrote the paper with the support of F.L. All authors contributed to the editing of the paper and supported the conclusions.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Aging thanks Xu Chen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
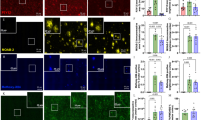
Extended Data Fig. 1 MFE-2 defect was related to microglial activation in AD mice.
a. The specificity of MFE-2 antibody used in this study was verified by immunofluorescences of MFE-2 in cultured primary microglia from Flox and ΔMFE-2 mice brain. Experiments were repeated three times. b. Representative RNAscope result of MFE-2 mRNA in microglia of 8-month-old untreated and 5xFAD mouse models (n = 3 per each group, male). Hsd17b4 in red, microglia in green and nuclei in blue. Scale bar, 25 μm. c. Schematic depicting brain microglia isolation by FACS experimental setup. And sorting strategy of brain microglia (CD45intCD11b+) by FACS. d. Western blot of MFE-2 in microglia from 8-month-old 5xFAD and untreated mice brain isolated by FACS. (Each sample contained a pool of microglia from 3 brains. Four samples per group were analyzed. Both male and female mice were used.) e. Overview of conditional MFE-2 knockout mice (ΔMFE-2) and MFE-2 knockout 5xFAD mice (5xFADΔMFE-2) production. f. Western blot of MFE-2 in isolated microglia by FACS from 8-week-old Flox and ΔMFE-2 mice brain (n = 4. Each lane counting for protein extracted from microglia of 3 brains p < 0.05. Both male and female mice were used.) g. Results of Morris water maze assays in Fig. 1g. Time in targeted quadrant and total swimming length were calculated. h. Representative immunofluorescences result of microglia distribution and morphology at early stage of AD (2 - 3-month-old) in 5xFAD and 5xFADΔMFE-2 mouse models (n = 5 per each group, male.) Mean microglial branch length was calculated by averaging the total length of branches using Leica software. Microglia in red and nuclei in blue. Scale bar, 100 μm. Statistical analyses were performed by two-tailed Student’s t-test. All data were presented as mean values ± SEM. Specific p values are indicated in the figure.
Extended Data Fig. 2 Identification of microglia clusters by scRNA-seq in Flox and ΔMFE-2 mice.
a. t-SNE plots showing the expression of selected genes that are enriched in microglia. These genes were used to identify microglia from non-microglia. Each dot represents a cell. The normalized gene expression levels of the selected genes for each cell were projected onto the t-SNE plots. b. Cell fraction distribution of pooled microglia from 8-month-old Flox, ΔMFE-2, 5xFAD and 5xFADΔMFE-2 mice in Fig. 2a. c. Heatmap to show highly expressed marker genes in 18 microglial clusters by scRNA in Fig. 2a. Z scores across microglia states were used for the plot. d. Pathways upregulated in microglia from ΔMFE-2 microglia compared to Flox mice in Fig. 2b. In blue, highlighting inflammatory pathways related to the MFE-2 knockout microglia states. The enrichment was evaluated using a two-sided hypergeometric test. e. t-SNE plots showing marker genes that were enriched in MFE-2 deficiency related clusters (cKO-related cluster III/IV). f. Pathways upregulated in MFE-2 deficiency-related microglia (cKO-related IV cluster). The enrichment was evaluated using a two-sided hypergeometric test. g. mRNA levels of Il6 in MFE-2KO and mock BV2 cell line by qPCR with or without LPS (10 μg/ml) and IFN-γ (100 ng/ml) treatment (n = 3 per each group, male). Data normalized to Gapdh and shown relative to control. Statistical analyses were performed by one-way ANOVA (g). All data were presented as mean values ± SEM. Specific p values are indicated in the figure.
Extended Data Fig. 3 Identification of microglia features in AD and ADΔMFE-2 by scRNA-seq.
a. t-SNE plots show marker genes that are enriched in DAM clusters in Fig. 3a. b. Pathways enriched in microglia DAM I cluster. The enrichment was evaluated using a two-sided hypergeometric test. c. Pathways upregulated microglia DAM II cluster. The enrichment was evaluated using a two-sided hypergeometric test. d. Flow cytometer analysis of TNF-α and IL-6 in brain microglia (CD45intCD11b+) from 8-month-old Flox, ΔMFE-2, 5xFAD and 5xFADΔMFE-2 mice. (n = 5 per group. Both male and female mice were used.) e. NS-TEM image of Aβ fibrils. Scale bar, 200 nm. Experiments were performed three independent times. Statistical analyses were performed by one-way ANOVA (d). All data were presented as mean values ± SEM. Specific p values are indicated in the figure.
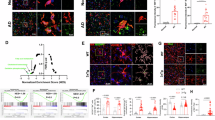
Extended Data Fig. 4 MFE-2 mediated peroxisomes and mitochondria contacts.
a. Representative immunofluorescences result of mitochondria (stained with anti-Tom20) in microglia of ΔMFE-2 and Flox mice. (n = 3 per sample, male). Mitochondria (Tom20+) in green, microglia in red and nuclei in blue. Scale bar, 25 μm. b. Flow cytometer analysis of MitoSOX of cultured microglia from P0 Flox and ΔMFE-2 mice in Fig. 4e. Rot/AA, rotenone–antimycin A. c. Primary brain microglia were isolated from P0 brains of ΔMFE-2 and Flox mice. The cultured microglia were used for the Seahorse long chain fatty acid Oxidation Stress Test combined with Seahorse XF Palmitate–BSA FAO Substrate setup. (n = 4 per each group, both male and female mice were used.) d. Overview of the construct of SPLICS Po-Mt Short P2A. And schematic showing SPLICS reporter displaying GFP over a dark background only in the regions of peroxisomes and mitochondria membrane proximity sites. e. The cultured microglia from P0 and ΔMFE-2 mice were transfected with SPLICS using lipo2000 and NATE. Another microglia group without SPLICS plasmid transfection was set as the blank control. Representative immunofluorescences result of SPLICS signal distribution in primary microglia from Flox and ΔMFE-2 mice were shown. (n = 9) Scale bar, 50 μm. Statistical analyses were performed by one-way ANOVA (e). All data were presented as mean values ± SEM. Specific p values are indicated in the figure.
Extended Data Fig. 5 MFE-2 deficiency drove dysregulated lipid β-oxidation and exacerbated microglial activation in response to Aβ.
a. Untargeted metabolism profile in FACS sorted microglia from 8-month-old ΔMFE-2 and Flox mice by LC-MS. Differential metabolites were identified using a two-sided Student’s t-test. (n = 4 in Flox group and n = 6 in MFE-2 knockout group. Microglia from three brains were pooled for each sample, male.) b. The cultured primary microglia from P0 Flox and ΔMFE-2 mice were input for FFAs quantitative evaluation by GC-MS. (n = 4 per group. Microglia from more than 10 brains were pooled for each sample. Both male and female mice were used.) Data are presented as mean values ± SEM. Differential metabolites were identified using a two-sided Student’s t-test.c. Heatmap for DEGs of cultured primary microglia from P0 Flox and ΔMFE-2 mice treated with brain extraction containing Aβ plaques from 8-month-old 5xFAD mice by bulk RNAseq. (n = 3 per sample. Microglia from more than 5 brains were pooled for each sample. Both male and female mice were used.) d. Flow cytometer analysis of inflammatory phenotype of primary microglia from 8-month-old Flox, ΔMFE-2, 5xFAD, and 5xFADΔMFE-2 mice after high-fat diet or control diet treatment. (n = 3 per each group, male) in Fig. 5f.
Extended Data Fig. 6 CKBA targeting MFE-2 without affecting its enzyme activity.
a. Representative results of CKBA-biotin pull-down of different domains and full length of MFE-2. b. Representative results of CKBA-biotin pull-down MFE-2 in BV2 cells. Experiments were repeated three times. c. Schematic diagram of synthesis of modified CKBA probe for labeling targets in living cell by click chemistry response. d. Representative immunofluorescences result of CKBA-Cy5.5 colocalization with MFE-2 (MFE-2-mCherry) in primary microglia from P0 untreated mice transfected with MFE-2-mCherry plasmid. To distinguish it from CKBA-Cy5.5 fluorescence, the fluorescence of MFE-2-mCherry was artificially set to green. Experiments were repeated three times. Scale bar, 10 μm. e. Representative immunofluorescences result of CKBA-MFE-2 labeling images stained with DBCO-Cy5.5. Scale bar, 10 μm. (n = 9) f. Measurement of the dehydrogenase activity of MFE-2 with or without CKBA by fluorometric monitoring of NAD+ reduction. And measurement of the hydratase activity of MFE-2 with or without CKBA by monitoring NADH production at 340 nm. Statistical analyses were performed by one-way ANOVA (e). All data were presented as mean values ± SEM. Specific p values are indicated in the figure.
Extended Data Fig. 7 CKBA targeting MFE-2 to resolve neuroinflammation.
a. Representative immunofluorescences result of MitoSOX stain in primary microglia treated with 100 μM AA (Arachidonic acid) combined with or without 5 μM CKBA. (n = 5 per each group). Scale bar, 100 μm. b. Timeline of the study and overview of the experimental design for vehicle or CKBA treatment in the 5xFAD mice experimental setup. c. LC-MS analysis of CKBA concentration in blood serum and brain tissue 12 hours after a single dose of CKBA administration at various dosages for wildtype mice. d. Results of Morris water maze assays of 1-year-old Vehicle-5xFAD, CKBA-5xFAD and CKBA-5xFADΔMFE-2. (n = 10 per each group. Both male and female mice were used.) Time in targeted quadrant and total swimming length were quantified. e. Representative immunofluorescences result of CD68 in sections from Vehicle-5xFAD, CKBA-5xFAD and CKBA-5xFADΔMFE-2 mice. (n = 4 per each sample, male). CD68 in green, microglia in red and nuclei in blue. Scale bar, 10 μm. Statistical analyses were performed by one-way ANOVA (a, c, d, e) and two-way ANOVA (d). All data were presented as mean values ± SEM. Specific p values are indicated in the figure.
Supplementary information
Supplementary Information
Supplementary Fig. 1.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data and unprocessed western blots.
Source Data Extended Data Fig. 1
Statistical source data and unprocessed western blots.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gao, M., Bai, J., Lou, F. et al. Loss of MFE-2 impairs microglial lipid homeostasis and drives neuroinflammation in Alzheimer’s pathogenesis. Nat Aging 5, 2279–2296 (2025). https://doi.org/10.1038/s43587-025-00976-1
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43587-025-00976-1