Abstract
Aging is characterized by amplified inflammation, including proinflammatory macrophages and increased susceptibility to endotoxemia. Here we uncover a mechanism by which macrophages maintain their inflammatory phenotype through autocrine GDF3–SMAD2/3 signaling, which ultimately exacerbates endotoxemia. We show that inflammatory adipose tissue macrophages display an age-dependent increase in GDF3, a TGFβ-family cytokine. Lifelong systemic or myeloid-specific Gdf3 deletion leads to reduced endotoxic inflammation. Using pharmacological interventions to modulate the GDF3–SMAD2/3 axis, we demonstrate its role in regulating the inflammatory adipose tissue macrophage phenotype and endotoxemia lethality in old mice. Mechanistically, single-cell RNA sequencing and assay for transposase-accessible chromatin with sequencing analyses suggest that GDF3 induces a shift toward an inflammatory state by limiting methylation-dependent chromatin compaction. Leveraging human adipose tissue samples and 11,084 participants from the atherosclerosis risk in communities study, we validate the relevance of GDF3 to aging in humans. These findings position the GDF3–SMAD2/3 axis as a critical driver of age-associated chromatin remodeling and a promising therapeutic target for mitigating macrophage-related inflammation in aging.
Similar content being viewed by others
Main
The aging of the immune system and chronic inflammation are key contributing factors to morbidity and mortality in older individuals with infection-related endotoxemia or sepsis1,2,3. Visceral adipose tissue (VAT) is an organ that exhibits early signs of immune activation during aging and acts as an immune reservoir that play a pivotal role in metabolic or infectious challenges4,5,6,7,8. Adipose tissue macrophages (ATMs) are a prevalent immune cell subset that promote chronic inflammation and tissue dysfunction via the NLRP3 inflammasome activation with age9. However, the mechanism by which aged ATMs maintain their inflammatory phenotype during endotoxemia is unknown. Previously, transcriptome analysis identified an age- and NLRP3-dependent upregulation of growth differentiation factor 3 (GDF3) in ATMs, which linked GDF3 to age-associated inflammation9. Here, we investigate the role of the GDF3–SMAD2 and SMAD3 (SMAD2/3) axis in endotoxemia-induced inflammation during aging.
GDF3, a TGFβ superfamily cytokine, has been linked to disease risk in myocardial infarction, metabolic dysfunction-associated steatohepatitis, obesity, sepsis and Alzheimer’s disease10,11,12,13,14,15. It exhibits contrasting functions—promoting metabolic dysfunction, improving muscle regeneration during injury and reducing inflammation during sepsis12,13,14,16,17,18,19,20,21,22—which probably stems from its divergent signaling capabilities. Similar to other TGFβ-members, GDF3 signals through type I and type II activin receptors to activate the transcription factors (TFs) SMAD2/3 via phosphorylation22,23. Owing to their low binding affinity to DNA, SMAD2/3 elicit divergent transcriptional outcomes, ranging from anti-inflammatory to proinflammatory effects24,25,26. Moreover, the SMAD complex can regulate chromatin remodeling through the interactions with epigenetic modifiers27. However, whether and how GDF3–SMAD2/3 axis is linked to the maintenance of inflammation and chromatin landscape during aging remains unclear.
Our data demonstrate that GDF3 induces an inflammatory phenotype in ATMs from old mice during endotoxemia. The lifelong deletion of Gdf3, at a whole-body or myeloid-specific level, reduces inflammatory macrophages, inflammatory cytokines and hypothermia. Two distinct pharmaceutical approaches targeting the GDF3–SMAD2/3 axis alleviate endotoxemia-induced inflammation in old mice. Moreover, the SMAD3 blockade protects against endotoxemia-induced lethality in old mice. In human VAT, we find that age positively correlates with GDF3 level. A large longitudinal human dataset also demonstrates a correlation between GDF3 and age-related inflammation. Mechanistically, GDF3 promotes inflammation through SMAD2/3 and alters chromatin accessibility by limiting methylation-dependent chromatin closing in aged ATMs. Together, our findings identify the GDF3–SMAD2/3 axis as a key driver shaping the inflammatory phenotype of ATMs during endotoxemia and aging and underscore its potential as a therapeutic target.
Results
Endotoxemia is associated with an increase in proinflammatory ATMs in old mice
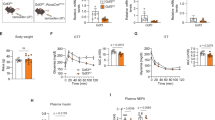
Older individuals exhibit increased lethality, cytokine storm and impaired metabolic responses to sepsis and endotoxemia, partly mediated by immune cell crosstalk within VAT5,28. To address whether the increase in inflammation is associated with a change in macrophage phenotype, we investigated the phenotype of ATMs at 4 h after administering phosphate-buffered saline (PBS) or 0.1 mg kg−1 lipopolysaccharide (LPS) via intraperitoneal (i.p.) injection to young or old wild-type (WT) mice5 (Fig. 1a, left). Consistent with previous studies5, old mice failed to maintain core body temperature (BT) with this low-dose LPS challenge (Extended Data Fig. 1a, right). CD11c was initially used as a representative marker for inflammatory phenotypes. Post LPS injection, CD11c+ ATMs (%) expanded in old mice, whereas it remained unchanged in young mice (Fig. 1b and Extended Data Fig. 1a,b). To further characterize CD11c+ ATMs, we analyzed multiple markers generally used to define macrophage phenotypes: CD11c, CD9, MHCII and CD206. The t-distributed stochastic neighbor embedding (tSNE) plot shows that ATMs are a heterogeneous population with varying combinations of markers (Fig. 1c). Notably, CD11c+ and CD206+ ATMs, commonly categorized as proinflammatory and anti-inflammatory ATMs, represent distinct populations. MHCII and CD9 were expressed on CD11c+ ATMs, further supporting CD11c as a marker for inflammatory ATMs29. These data collectively indicate that low-dose LPS exacerbates the imbalance of heterogeneous inflammatory CD11c+ ATMs in an age-dependent manner. To further investigate the phenotype of VAT immune cells, we utilized a single-cell RNA sequencing (scRNA-seq) dataset (GSE274935) containing VAT CD45+ immune cells sorted from young and old mice that were challenged with low-dose LPS. Young and old VAT immune cells were analyzed together, and a total of 23 clusters were identified, including 4 clusters of ATMs (Fig. 1d and Extended Data Fig. 2a). The proportions of each ATM cluster were altered during aging. Whereas ATM2 and ATM3 clusters expanded with age, the ATM1 cluster declined (Fig. 1e). The expanding ATM clusters exhibited higher expressions of Itgax (CD11c), Cd9 and MHCII-associated gene (H2-Ab1), while showing a lower expression of Mrc1 (CD206) and Lyve1 (Fig. 1f and Extended Data Fig. 2b). These characteristics are comparable to those of inflammatory CD11c+ ATMs identified by flow cytometry (Fig. 1c). These clusters also exhibit higher expression of Gdf3 compared with other ATM clusters, suggesting that inflammatory ATMs are the primary source of Gdf3. Notably, Gdf3 expression was particularly enriched in the Itgaxlow ATM3 cluster rather than in Itgaxhigh ATM2, supporting the notion of heterogeneity among CD11c+ ATMs.
a, Left: a schematic of the experimental design. Right: the core BT of young (3-month-old) or old (22-month-old) female WT mice after i.p. injection of 0.1 mg kg−1 LPS or PBS. n = 3 per group. b, The ratio of CD11c+ ATMs (% of CD11c+ ATMs from the LPS-injected group normalized to the PBS-injected group, all female mice). c, The tSNE plot of compiled F4/80+ ATMs in VAT from 24-month-old female WT mice (n = 5) injected with 0.1 mg kg−1 LPS; 1,000 iterations, 30 perplexities. The left axis shows tSNE2, and the bottom axes show tSNE1. d, The UMAP of VAT immune cells (CD45+ cells) from young (3-month-old, n = 2) or old (21-month-old, n = 3) female WT mice injected with 0.1 mg kg−1 LPS showing 23 clusters. e, The proportion of each ATM cluster (% of all young or old ATMs). f, The expressions of genes associated with macrophage phenotype to compare ATM clusters (ATM1, ATM2, ATM3, ATM4), normalized across all ATM clusters. g–k, Young (3–4-month-old) or old (19–22-month-old) female WT or GDF3 KO mice injected with 0.1 mg kg−1 LPS: a schematic of the experimental design (g); the gene expression of Gdf3 in VAT (h); young WT (YW), n = 5; young KO (YK), n = 5; old WT (OW), n = 8; old KO (OK), n = 8; the representative gating strategy for CD11c+ and CD206+ ATMs in VAT from OW (left) or OK (right) mouse (i); the frequency of CD11c+ ATMs (j); YW, n = 14; YK, n = 8; OW, n = 15; OK, n = 14; and mean core BT after LPS injection (k); YW, n = 14; YK, n = 8; OW, n = 8; OK, n = 8. The circle and triangle symbols represent the two independent experiments. For i, the left axis shows CD206, and the bottom axes show CD11c. l, A schematic of the experimental design. m, The gene expression of Gdf3 in positively selected B cells from spleen and peritoneal macrophages, all female mice. Control, n = 4; mKO, n = 3. n–p, Young (4-month-old) or old (21- to 24-month-old) female control or mKO mice were injected with 0.1 mg kg−1 LPS: the frequency of CD11c+ ATMs in VAT from old control (n = 11) or mKO mice (n = 6) (n); the inflammatory cytokine gene expression in VAT from old control (n = 6) or mKO mice (n = 4) (o); and the mean core BT after LPS injection (p), young control, n = 8; old control, n = 11, old mKO, n = 8. The circle and triangle symbols represent the two independent experiments. All data are presented as means ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. All in vivo experiments were repeated independently at least twice. See Extended Data Figs. 1–3 for more details. NK, natural killer; n.s., not significant; n.d., nondetected; Treg, regualtory T cell;.
Old Gdf3-deficient mice are protected from endotoxemia-induced inflammation
GDF3 is implicated in age-related adipose tissue inflammation and affects the severity or risk for multiple diseases9,10,12,14,15,21,30. We next asked whether the lifelong deficiency of Gdf3 would alter the endotoxemia-related inflammatory state of ATMs from old mice (Fig. 1g). To address this question, we generated mice with a whole-body deletion of Gdf3. We first validated the deletion of Gdf3 in VAT from young (3–4-month-old) or old (19–22-month-old) WT or Gdf3 whole-body knockout (KO) mice. A significant age-related increase in Gdf3 expression was ablated in old KO mice (Fig. 1h). To investigate the impact of lifelong Gdf3 deletion on metabolic function before endotoxic shock, we monitored various metabolic parameters, including body weights, body composition, insulin sensitivity and glucose sensitivity. However, we only observed an age effect without a genotype effect (Extended Data Fig. 1c–f). Genes related to fatty acid oxidation or synthesis also showed no differences in adipocytes isolated from VAT (Extended Data Fig. 1g). Taken together, lifelong Gdf3 deficiency has no impact on metabolic dysfunction.
Next, we assessed the impact of lifelong Gdf3 deficiency on the VAT immune cell activation and ATMs’ phenotype during endotoxemia. T cells, B cells and myeloid cells, including ATMs, exhibited the expected age-related changes in frequency (Extended Data Fig. 1h). However, no genotype effect was observed in this overall immune composition (Extended Data Fig. 1h). Next, we examined the inflammatory profile of ATMs (Fig. 1i). Notably, the deletion of Gdf3 resulted in a decreased frequency of CD11c+ ATMs, whereas CD206+ ATMs remained unaffected (Fig. 1j and Extended Data Fig. 1i). In the spleen, no differences in immune composition were observed between old WT and KO mice, indicating that the effect of Gdf3 is primarily within VAT (Extended Data Fig. 1j).
The impaired lipolysis upon aging leads to dysregulated immune response5. As GDF3 can regulate adipocyte lipolysis, we measured the free fatty acids released from VAT explants9,12,16,22. VAT explants from old KO mice exhibited improved stimulated-lipolytic capacity compared with their old WT counterparts (Extended Data Fig. 1k). Next, we assessed the severity of endotoxemia by monitoring the core BT at 0, 2 and 4 h post LPS injection. Although there were no differences in core BT in young WT and KO mice, old WT mice exhibited a greater reduction in BT compared with old KO mice (Fig. 1k). Taken together, these results suggest that GDF3 mediates endotoxemia-induced inflammation in old mice by regulating the inflammatory phenotype of ATMs, lipolysis and hypothermia.
To address how GDF3 induces the expansion of CD11c+ macrophages, we investigated whether this increase occurs via proliferation or infiltration. As GDF3 promotes the proliferation of various cell types10,20, we first hypothesized that the expansion of CD11c+ ATMs was dependent on GDF3-regulated proliferation. To perform an in vivo cell proliferation study, young (4-month-old) and old (24-month-old) WT mice were i.p. injected with 5-ethynyl-2′-deoxyuridine (EdU, 25 mg kg−1) for four consecutive days, followed by PBS or 0.1 mg kg−1 LPS injection31 (Extended Data Fig. 1l). The cell proliferation was assessed by quantifying incorporation of EdU into DNA by flow cytometry (Extended Data Fig. 1m). The frequency of EdU+CD11c+ ATMs was elevated 1.7 fold with LPS injection in old mice, whereas it remained unchanged in young mice (Extended Data Fig. 1n). Next, WT and KO mice were injected with EdU (Extended Data Fig. 1o, left). Although EdU+CD11c+ ATMs were elevated with aging, no differences between old WT and KO mice were observed (Extended Data Fig. 1o, right). Monocytes can infiltrate into VAT and differentiate into inflammatory macrophages32. To investigate whether GDF3 influences the infiltration of monocytes, we used the CCR2 as a marker for infiltrating monocytes33. Although the frequency of CCR2+ ATMs increased with age, we found no differences between old WT and KO mice (Extended Data Fig. 1p). Collectively, our data demonstrate that GDF3 induces the inflammatory phenotype of ATMs independently of proliferation or infiltration, despite both being crucial for maintaining CD11c+ ATMs during endotoxemia in old mice.
Gdf3 is not required for LPS-induced lethality in young mice
We next asked whether the protective function of Gdf3 is exclusive to old mice. Young WT and KO mice (aged 4–6 months old) were injected with a lethal dose of LPS34 (18 mg kg−1) and monitored for 6 h post LPS administration (Extended Data Fig. 3a). The core BT, immune cell composition, lymphocyte activation and inflammatory gene expression remained unaltered in young mice regardless of the presence of Gdf3, indicating that Gdf3 does not play a role in severe inflammatory responses in young mice (Extended Data Fig. 3b–e). Following a previously established GDF3 treatment regimen, the young mice were administered with recombinant GDF314 (rGDF3, 10 µg kg−1) before the lethal dose of LPS (Extended Data Fig. 3f). However, rGDF3 did not protect mice against hypothermia and VAT inflammation (Extended Data Fig. 3f). These findings suggest that the beneficial effect from Gdf3 deletion against severe inflammatory responses is specific to old mice.
To broadly address the age-related alterations in expression of Gdf3 and its family members, we first compared the gene expression of selected Tgfb-associated genes. In line with our observations (Fig. 1h), Gdf3 increased with age in VAT. Consistent with previous reports, Gdf15 was elevated but not statistically significant. There were no differences detected in other genes from the TGFβ family (Extended Data Fig. 3g). Moreover, Gdf3 expression was elevated in multiple organs of old mice, with the highest expression observed in the VAT, liver and thymus (Extended Data Fig. 3h). Lifelong B cell deficiency improves the aged VAT microenvironment by reducing inflammatory ATMs5,35. Notably, old Bnull mice exhibited reduced Gdf3 and Tgfb1 but not Gdf15 expression compared with old WT mice (Extended Data Fig. 3i). These findings suggest that Gdf3 is highly expressed in VAT and increases in a B cell-dependent manner during aging.
Myeloid-driven Gdf3 is required for endotoxemia-induced cytokine storm, inflammatory macrophages and hypothermia
As Gdf3 may be expressed in multiple cell types, we investigated its myeloid-driven function by generating and validating Gdf3 myeloid-specific KO (mKO) mice. Macrophages from the peritoneum exhibited a reduction of Gdf3 expression in mKO group. By contrast, B cells showed comparable levels of Gdf3 expression between control and mKO mice, confirming the myeloid-specific deletion of Gdf3 (Fig. 1l,m). Although old mKO mice showed no differences in body weights, they showed improved glucose sensitivity when compared with control mice (Extended Data Fig. 3j–m).
Next, we predicted that myeloid-driven Gdf3 is required for the inflammatory phenotype of macrophages during endotoxemia. At 21–24 months of age, control and mKO mice were given 0.1 mg kg−1 LPS (Fig. 1l). ATMs from the old mKO group showed a significant reduction in the frequency of CD11c+ ATMs (Fig. 1n). There was also a significant decrease in CD9+ ATMs, a slight increase in CD206+ and no change in CCR2+ ATMs (Extended Data Fig. 3n–p). The decreased frequency in CD11c+ macrophages was also seen in the subcutaneous adipose tissue, indicating the conserved function of Gdf3 across the white adipose depots (Extended Data Fig. 3q). The Gdf3 expression was significantly reduced in mKO VAT, indicating that myeloid cells are the main contributors of Gdf3 to the tissue (Fig. 1o). The inflammatory cytokine gene expression (Il1b, Il6, Tnfa) also decreased in the VAT from old mKO mice (Fig. 1o). Moreover, the stimulated-lipolytic capacity of VAT explants was improved with the myeloid-specific deletion of Gdf3 (Extended Data Fig. 3r).
In aged VAT, B cells regulate adipose tissue homeostasis. They contribute to elevated inflammation by modulating other immune cells, including ATMs, and by inhibiting stimulated lipolysis5,36. We asked if altered ATMs would affect B cells. To characterize B cells, we used IgD, IgM, programmed cell death ligand 1 (PDL1) and CD73 as phenotypic markers for dysfunctional B cells in aged VAT (Extended Data Fig. 3t). Markedly, both IgM+CD73+ and PDL1+ B cells, representing memory B cells and regulatory B cells, respectively5,37, were decreased in old mKO mice (Extended Data Figs. 3u,v). Collectively, this suggests that Gdf3 can modulate dysfunctional B cells in aging VAT.
During endotoxemia, old control mice developed hypothermia with 13% BT loss, whereas old mKO mice had significantly increased BT at 4 h post endotoxemia, showing 7% BT loss (Fig. 1p). This was also observed in male mKO mice that displayed improved glucose sensitivity and fewer CD11c+ ATMs during endotoxemia, demonstrating a conserved function of Gdf3 across sexes (Extended Data Fig. 3l,m,s,w). Taken together, our findings suggest that the lifelong myeloid-specific deletion of Gdf3 results in improved glucose sensitivity, reduced CD11c+ ATMs, improved lipolytic capacity, decreased inflammatory gene expression and protection from BT loss post LPS challenge.
BRD4-controlled Gdf3 regulates endotoxemia-induced inflammation
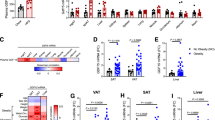
Next, we wanted to examine if the acute depletion of Gdf3 could alleviate inflammation. Bromodomain and extra terminal domain 4 (BRD4), a transcriptional and epigenetic regulator, binds to the promoter and enhancer regions of Gdf3 and facilitates the transcription of Gdf3 in obesity38. To first assess whether the expression of Gdf3 is regulated by BRD4 in older organisms, we utilized JQ1, a small molecular inhibitor of BRD439. At 23–24 months of age, the mice were intraperitoneally injected with JQ1 (12.5 mg kg−1) or vehicle for six consecutive days, followed by a 0.1 mg kg−1 LPS injection (Fig. 2a). The acute administration of JQ1 reduced VAT Gdf3 levels without affecting body weights (Fig. 2b and Extended Data Fig. 4a). We next tested whether the acute depletion of Gdf3 would have similar effects on macrophage phenotypes to those observed with a lifelong deletion of Gdf3. Although the composition and cellularity of overall immune cells in VAT were not affected by JQ1 (Extended Data Figs. 4b,c), the composition of ATMs was altered. Specifically, the frequency of CD11c+ ATMs decreased, whereas the frequency of CD206+ ATMs trended toward an increase (Fig. 2c). The frequency of CCR2+ ATMs remained unaffected (Fig. 2d). The inflammation-associated genes were reduced in VAT (Fig. 2e and Extended Data Fig. 4d). In line with the observed changes in B cell phenotypes resulting from lifelong myeloid-specific Gdf3 deficiency, PDL1+ B cells decreased following the JQ1 injection (Extended Data Fig. 4e). However, the frequency of CD73+ B cells was not affected (Extended Data Fig. 4f). At a systemic level, the JQ1-injected old mice were protected from hypothermia compared with controls (Fig. 2f).
a–f, Old (23–24-month-old) female WT mice were intraperitoneally injected with JQ1 (12.5 mg kg−1) or vehicle and LPS (0.1 mg kg−1): vehicle-injected, n = 10; JQ1-injected, n = 11; a schematic of the experimental design (a); the Gdf3 expression in VAT (b); the frequency of CD11c+, CD206+ (c) and CCR2+ (d) ATMs in VAT; the inflammatory gene expression in VAT (e); and the mean core BT after LPS injection (f). g,h, The BMDMs isolated from old (24-month) female WT mice were treated with LPS (1 µg ml−1) 4 h before vehicle or JQ1 (1 µM) treatment: a schematic of the experimental design (g) and the Gdf3 and Il1b expression in BMDMs (h). i, The pSMAD2/3, total SMAD2/3 (tSMAD2/3) and β-actin by western blot of whole VAT from young (3-month-old) or old (24-month-old) female WT and GDF3 KO mice injected with 0.1 mg kg−1 LPS. The pSMAD2/3, tSMAD2/3 and β-actin were run on separate gels. j, The fibrotic gene expression in VAT from old control (n = 5) and old mKO (n = 5) male mice injected with 0.1 mg kg−1 LPS. The dotted line represents the young control. k, The quantification of pSMAD2/3 normalized to Ponceau S staining of SVF or adipocytes from young (3-month-old) and old (22-month-old) female WT mice injected with 0.1 mg kg−1 LPS (Extended Data Fig. 5a). l, The MFI of pSMAD2/3 in T cells, B cells and ATMs in VAT from old (24-month-old) female WT mice (n = 3). m, The pSMAD2/3 MFI in CD11c+ or CD11c− ATMs from young (4-month-old) and old (23-month-old) female WT mice. Young, n = 3; old, n = 8. n–p, The positively selected ATMs from VAT of old (24-month-old, n = 6) female WT mice were treated with vehicle or rGDF3 (20 ng ml−1) in vitro: a schematic of the experimental design (n), pSMAD2/3 MFI in CD11c+ or CD206+ ATMs (o) and the frequency of CD11c+ ATMs (p). All the data are presented as means ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. See Extended Data Figs. 4 and 5 for more details. n.s., not significant.
To directly test the impact of JQ1 on macrophages, we utilized bone marrow-derived macrophages (BMDMs). BMDMs generated from marrow of old mice were pretreated with LPS (1 µg ml−1) 4 h before the vehicle or JQ1 (1 µM) treatment (Fig. 2g). Consistent with in vivo results, JQ1 reduced Gdf3 and Il1b expression in vitro (Fig. 2h). A similar phenotype was observed in BMDMs without LPS (Extended Data Figs. 4h,i). Collectively, these data indicate that the acute depletion of Gdf3 via BRD4 inhibition phenocopies the lifelong deletion of Gdf3 in old mice.
GDF3 induces phosphorylation of SMAD2/3 to promote inflammatory phenotype of ATMs during aging
Previous studies have demonstrated that GDF3 can activate SMAD2/3 signaling via phosphorylation20,38. We wanted to test if the age-related increase in phospho-SMAD2/3 (pSMAD2/3) requires GDF3. pSMAD2/3 levels were similar in in VAT from young WT and KO mice but were reduced in old KO mice compared with old WT mice (Fig. 2i). The SMAD2/3 signaling promotes fibrosis in multiple tissues including AT35. However, the expression levels of SMAD2/3 downstream fibrotic genes (Fn1, Col1a1, Mmp2, Serpine1) showed no differences between old control and mKO VAT (Fig. 2j). Considering the cellular heterogeneity of VAT, we sought to identify which cell types express pSMAD2/3. The whole VAT from young and old WT mice was separated into adipocyte fraction and stromal vascular fraction (SVF), which includes the immune cells. Interestingly, both the adipocyte fraction and SVF showed increase in pSMAD2/3 and GDF3 with age (Fig. 2k and Extended Data Fig. 5a–c). Next, phospho-flow cytometry was used to identify the immune cells with elevated pSMAD2/3 expression. The pSMAD2/3 expression, represented by the mean fluorescence intensity (MFI), was highest in ATMs compared with B cells and T cells from aged VAT (Fig. 2l). Moreover, we observed an age-dependent elevation in pSMAD2/3 levels specifically within CD11c+ ATMs, whereas CD11c− ATMs showed no significant changes (Fig. 2m).
To directly examine if GDF3 can induce phosphorylation of SMAD2/3 in macrophages, we used an in vitro system with BMDMs and ATMs (Fig. 2n and Extended Data Fig. 5d–k). First, young and old BMDMs were treated with vehicle or rGDF3 (20 ng ml−1). At baseline, pSMAD2/3 was higher in old BMDMs compared with young BMDMs (Extended Data Fig. 5e). The rGDF3 treatment increased pSMAD2/3 in old BMDMs, but not young, and decreased the frequency of CD206+ BMDMs (Extended Data Fig. 5d–f). In LPS-pretreated old BMDMs, rGDF3 increased the frequency of CD11c+ BMDMs without affecting CD206 frequencies (Extended Data Fig. 5g–i). For a more physiologically relevant in vitro model, we positively selected ATMs from young and old SVF (Fig. 2n). Consistent with BMDMs, only old ATMs showed an increased pSMAD2/3 in response to rGDF3 (Extended Data Fig. 5j–l). Similar to the increase that is induced by natural aging, CD11c+ ATMs from old mice exhibited a higher basal level of pSMAD2/3 than CD206+ ATMs in vitro (Fig. 2o). Both the CD206+ and CD11c+ ATMs showed increased pSMAD2/3 following rGDF3 treatment, but CD11c+ ATMs exhibited a more robust response with higher pSMAD2/3 MFI (Fig. 2o). In addition, rGDF3 treatment increased the frequency of CD11c+ ATMs (Fig. 2p). Taken together, our data demonstrate that GDF3 increases pSMAD2/3 and promotes the inflammatory phenotype of macrophages from old mice.
Inhibition of SMAD3 results in reduced inflammatory macrophages and hypothermia during endotoxemia
We next hypothesized that SMAD2/3 is required for inflammatory responses with age. To test this, young (4-month-old) and old (23-month-old) WT mice were injected with a specific inhibitor for SMAD3 (SIS3, three times a week, 10 mg kg−1) for 4 weeks and challenged with 0.1 mg kg−1 LPS40 (Fig. 3a). SIS3 effectively dampened pSMAD2/3 MFI and pSMAD2/3+ myeloid cells and ATMs from old mice (Fig. 3b–d). However, SIS3 had no effect on lymphocytes (gated based on size), which exhibited a lower frequency of pSMAD2/3+ cells than myeloid cells (Fig. 3b). Canonical SMAD2/3 target genes (Fn1, Serpine1) were downregulated in SIS3-injected young mice compared with vehicle controls. Interestingly, this effect was absent in old mice, suggesting that SMAD2/3 may regulate a different set of genes during aging (Fig. 3e,f).
A, A schematic of the experimental design. b,c, The frequency of pSMAD2/3+ cells (b) and pSMAD2/3+ ATMs (c) in VAT from old mice. Old vehicle (OV), n = 5; old SIS3 (OS), n = 7. d, The pSMAD2/3 MFI of ATMs from old mice. OV, n = 5; OS, n = 7. e,f, The fibrotic gene expression (Fn1 (e), Serpine1 (f)) in VAT (young vehicle (YV), n = 5; young SIS3 (YS), n = 6; OV, n = 6; OS, n = 7). g–i, The body weights (g), glucose sensitivity (h) and insulin sensitivity (i) of vehicle- and SIS3-injected old mice. j–l, The frequency of immune cells (j), CD11c+ ATMs, CD206+ ATMs (k) and CD9+ ATMs (l) in old VAT. m, The VAT gene expression involved in inflammation. YV, n = 6; YS, n = 6; OV, n = 5; OS, n = 5. n, The frequency of PDL1+ B cells in VAT. YV, n = 7; YS, n = 7; OV, n = 11; OS, n = 11. o, The mean core BT of old mice 4 h post LPS injection. OV, n = 11; OS, n = 11. In a–o, young (4-month-old) and old (23-month-old) female WT mice were intraperitoneally injected with SIS3 (10 mg kg−1, three times a week) or vehicle and LPS (0.1 mg kg−1) 4 h before the end point. YV, n = 7; YS, n = 7; OV, n = 11; OS, n = 11. p–r, Old (20-month-old) female WT mice were injected with SIS3 or vehicle as shown above and 3 mg kg−1 LPS: a schematic of the experimental design (p), the body weight loss (normalized to the 0 h time point) after the LPS injection (q) and the survival rate after LPS injection (r). s, The correlation between human VAT GDF3 level and age. t, The correlation between human VAT pSMAD2/3 level and inflammatory cytokine gene expression. All data are presented as means ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. All in vivo experiments were repeated independently twice. n.s., not significant. See Extended Data Fig. 6 for more details.
The acute blockade of SMAD3 signaling was shown to protect mice from diabetes and obesity with improved insulin or glucose sensitivity26. We asked if SIS3 could reverse age-related metabolic dysfunction. However, a month-long blockade of SMAD3 had no effect on body weight, glucose sensitivity and insulin sensitivity in old mice (Fig. 3g–i). We next investigated whether SIS3 could reduce inflammatory responses in old mice, akin to phenotypes observed in Gdf3-deleted or JQ1-injected old mice. Although the overall immune composition remained unchanged, SIS3 treatment reduced CD11c+ and CD9+ ATMs and trended toward increased CD206+ ATMs in old mice (Fig. 3j–l). SMAD3 inhibition also reduced inflammatory cytokine gene expression (Tnfa, Il1b) only in old mice (Fig. 3m). Consistent with the effects of Gdf3 genetic deletion, the PDL1+ B cells were reduced in SIS3-injected old mice (Fig. 3n). Systemically, SMAD3 inhibition protected old mice from significant core BT loss observed 4 h post LPS injection (Fig. 3o). Taken together, SMAD2/3 promotes hyperinflammatory responses in an age-specific manner during endotoxemia. Next, we tested whether SMAD3 blockade could reduce mortality in lethal LPS-induced endotoxemia34 (3 mg kg−1; Fig. 3p). Body weight loss, a noninvasive indicator of disease severity, was partially prevented in SIS3-treated old mice compared with vehicle controls (Fig. 3q). The SMAD3 blockade also improved survival, although not statistically significantly, in response to lethal endotoxemia (Fig. 3r).
A reduced SMAD3 expression has been associated with longevity in humans41. Consistent with our observations in mouse models, GDF3 expression positively correlated with age in human VAT biopsies from bariatric surgeries (body mass index (BMI) >30; Fig. 3s). Moreover, pSMAD2/3 positively correlated with IL1B but not TNF expression particularly in individuals aged over 60 years (Fig. 3t). We also utilized the data from the atherosclerosis risk in communities (ARIC) study42 to investigate the correlation between GDF3 and age-associated inflammation at a systemic level. The ARIC cohort includes 15,792 middle-aged participants (45–64 years old) who were followed longitudinally and took part in multiple clinic visits42. Notably, higher serum GDF3 levels were significantly associated with elevated level of C-reactive protein (CRP), a marker of systemic inflammation, at both visits 2 (midlife, median age 57 years, N = 11,084) and 5 (late-life, median age 76 years, N = 4,985. Extended Data Fig. 6a). After adjusting for demographics, education level and BMI, participants in the highest GDF3 quintile had 15% (β (95% confidence intervals (CI)): 1.15 (1.09–1.22)) higher CRP levels compared with those in the lowest GDF3 quintile at midlife (Extended Data Fig. 6b). This association was more pronounced at late-life, where the highest GDF3 quintile group exhibited 39% (β (95% CI): 1.39 (1.27–1.53)) higher CRP levels (Extended Data Fig. 6b). These findings underscore the link between GDF3 and age-associated inflammation in humans and support the therapeutic potential of targeting the GDF3–SMAD2/3 axis.
Gdf3 regulates chromatin accessibility and enriches SMAD4-regulated transcription
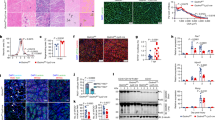
SMAD2/3 TFs have low DNA binding affinity for a specific consensus sequence, allowing them to elicit anti- or proinflammatory responses depending on the cellular context24,25,26. Genes associated with proinflammatory processes in immune cells, including myeloid cells, exhibit increased chromatin accessibility with age43. Moreover, SMAD2/3 can regulate chromatin accessibility through recruitment of chromatin/epigenetic modifiers27. We thus hypothesized that GDF3–SMAD2/3 axis could regulate chromatin accessibility and promote inflammation in ATMs with age. To assess chromatin accessibility, ATMs isolated via fluorescence-activated cell sorting (FACS) from young control, old control and old mKO mice were profiled using assay for transposase-accessible chromatin with sequencing (ATAC-seq; Fig. 4a and Extended Data Fig. 7a,b). The data quality was validated by the enrichment of ATAC-seq reads around transcription start sites (TSS) and the genomic distribution pattern of identified peaks (Extended Data Fig. 7c,d). A differential peak analysis between young and old control ATMs identified 4,680 peaks with increased accessibility and 1,011 peaks with decreased accessibility, revealing an age-specific chromatin landscape marked by widespread chromatin opening (Fig. 4b and Supplementary Table 1). Hierarchy clustering analysis further demonstrated distinct open and closed chromatin patterns between young and old ATMs (Fig. 4c). We next compared old ATMs with or without Gdf3. Interestingly, the majority of peaks were closing (n = 3,247) in old ATMs without Gdf3 (Fig. 4d). A clustering analysis also revealed distinct patterns (Fig. 4e). Out of 3,247 closing peaks with Gdf3 deletion, 50% (n = 1627) overlapped with opening peaks with aging (Extended Data Fig. 7e). A pathway enrichment analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG) and gene ontology (GO) identified several pathways associated with differentially accessible peaks across age or genotype. Age-related opening peaks were enriched in extracellular matrix (ECM)–receptor interaction, Th17 cell differentiation, AGE–RAGE signaling complication, Rap1 signaling, PI3K–Akt signaling, cytokine–cytokine receptor interaction and calcium signaling pathways (KEGG; Fig. 4f). Remarkably, several age-enriched pathways overlapped with those associated with decreased accessibility in mKO ATMs (Fig. 4g). These included the ECM–receptor interaction and Rap1 signaling pathway, previously implicated to be enriched with aging44,45.
a, A schematic of the experimental design. b,c, The differentially accessible peaks between old and young control ATMs represented by a log2 fold change (log2FC) plot (b) and hierarchy clustering heat map (c). For the log2FC plot, differentially opening and closing peaks are shown in red and blue, respectively. d,e, The differentially accessible peaks between old mKO and old control ATMs represented by a log2FC plot (d) and hierarchy clustering heat map (e). f,g, The KEGG pathway analysis of opening peaks with age (f) (old versus young control) and closing peaks with Gdf3 deletion (g) (old mKO versus old control). h,i, The prediction of TF-binding motifs enriched in the promoters of opening peaks with age (h) and closing peaks with Gdf3 deletion (i). TFs of interests are underlined. j, The IGV genome browser tracks that show chromatin accessibility of Il1r1 (left) and Il18r1 (right) in ATMs. In a–j, young (4-month-old, n = 3) or old (21–24-month-old) female control (n = 3) or mKO (n = 3) mice were injected with 0.1 mg kg−1 LPS. The TSS are highlighted with boxes. See Extended Data Fig. 7 for more details.
Next, a footprinting analysis was utilized to predict the potential TFs bound to accessible chromatin. An analysis of all identified ATAC-peaks revealed CTCF, PU.1 and JUN as the most prevalent TFs across all groups, irrespective of age or genotype (Extended Data Fig. 7h), indicating that the core TFs in ATMs remain largely unchanged. To investigate further, we predicted TFs associated with differentially accessible peaks using ShinyGO, a publicly available graphical gene set enrichment tool46. A predictive analysis highlighted SMAD4 as the TF most enriched at opening peaks upon aging and, similarly, at closing peaks upon Gdf3 deletion in old ATMs (Figs. 4h,i). Importantly, SMAD4 is required for SMAD2/3 to translocate into nucleus and initiate transcription, indicating a specific link between GDF3 and SMAD signaling in chromatin remodeling47. We observed an increased chromatin accessibility around the TSS of inflammatory cytokine receptors, Il1r1 and Il18r1, in old ATMs (Fig. 4j). Conversely, these regions became less accessible in old ATMs without Gdf3. A chromatin immunoprecipitation-enrichment analysis48 indicates that Il1r1 and Il18r1 are SMAD4-transcribed genes, suggesting a direct link between GDF3–SMAD chromatin remodeling and inflammation-associated transcriptional changes in ATMs.
The SMAD complex can regulate transcription through interactions with epigenetic cofactors27. Among the TFs enriched in both the age-dependent opening peaks and Gdf3-depedent closing peaks, SUZ12 was identified (Figs. 4h,i). SUZ12 is a key component of polycomb repressive complex 2 (PRC2), which represses gene transcription by methylating histone H3 at lysine 27 (H3K27me3). Taken together, these findings suggest that the GDF3–SMAD axis, possibly in conjunction with PRC2-mediated H3K27me3 levels, regulates chromatin remodeling and transcriptional changes in aged ATMs.
Lifelong Gdf3 deficiency reduces inflammatory ATMs by promoting chromatin remodeling and methylation signature
The loss of repressive histone mark H3K27me3 increases chromatin accessibility and promotes the transcription of inflammatory genes49. To assess age- or endotoxemia-related changes, we quantified H3K27me3 levels in VAT immune cells using flow cytometry. H3K27me3 levels in total ATMs and CD11c+ ATMs significantly decreased with age but remained unchanged following LPS treatment (Fig. 5a and Extended Data Fig. 8a), suggesting that age-dependent chromatin remodeling in ATMs occurs independently of acute inflammatory stimuli. SMAD2/3 can integrate epigenetic processes into transcription regulation by recruiting histone demethylase that counteracts the PRC2-mediated gene repression via H3K27me327,50. We therefore hypothesized that aging and GDF3 affect chromatin remodeling through the loss of H3K27me3. To determine if GDF3 influences H3K27me3 levels under basal conditions, we measured H3K27me3 in ATMs from unchallenged old (21–23-month-old) WT and GDF3 KO mice. Notably, H3K27me3 levels increased in both total ATMs and CD11c+ ATMs from KO mice (Fig. 5b), despite only a trend toward a reduction in CD11c+ ATMs (Fig. 5c). The Il1b expression in VAT was significantly reduced in KO mice (Fig. 5d), consistent with decreased chromatin accessibility and reduced inflammatory potential without Gdf3 (Fig. 4d).
a, The H3K27me3 MFI of CD11c+ ATMs from young (4-month-old) and old (22-month-old) female WT mice (n = 4, each group) injected with PBS or 0.1 mg kg−1 LPS for 4 h. b, The H3K27me3 MFI of ATMs and CD11c+ ATMs from unchallenged old (22–24-month-old) female WT (n = 6) and KO (n = 4) mice. c, The frequency of CD11c+ ATMs from unchallenged old (20–24-month-old) female WT (n = 9) and KO (n = 6) mice. d, The VAT Il1b expression from unchallenged old (20–24-month-old) female WT (n = 9) and KO (n = 6) mice. e, The UMAP of VAT immune cells (CD45+ cells) from unchallenged old (21–23-month-old) female WT (n = 3) and KO (n = 3) showing 24 clusters. f, The expressions of genes associated with GDF3–SMAD2/3 signaling, normalized across all identified clusters. g, The proportion of each ATM cluster (% of all each cluster). h, The expression of genes representing ATM phenotype, normalized across ATM clusters. i, The number of DEGs in ATM clusters (adjusted P value, Padj <0.05). j, The GSEA analysis using reactome chromatin modifying enzymes (top) and methylation-dependent heterochromatin formation (bottom) of KO versus WT ATM1 clusters. k, A volcano plot of DEGs between KO versus WT merged ATM. The inflammatory genes are highlighted in red and the chromatin remodeling-associated genes in yellow. Among chromatin remodeling-associated genes, PRC2 complex-related genes are marked with blue. All data are presented as means ± s.e.m. See Extended Data Figs. 8 and 9 for more details. NK, natural killer; Treg, regulatory T cell.
To investigate whether Gdf3-mediated chromatin remodeling alters ATM transcriptional profiles, FACS-sorted VAT immune cells from unchallenged old WT and KO mice were analyzed using scRNA-seq (n = 3 each group). WT and KO cells were analyzed together, identifying 24 clusters of cells, including three ATM clusters (Fig. 5e and Extended Data Fig. 8b–d). Consistent with Fig. 1f, ATM3, characterized with the highest Itgax (CD11c) expression, was the primary source of Gdf3 (Fig. 5f and Extended Data Fig. 8b,e–g). All three ATM clusters exhibited relatively higher expression of genes associated with the GDF3–SMAD2/3 axis compared with other VAT immune cells, including type I and type II activin receptors and signaling components (Fig. 5f). We then compared ATM frequencies within each cluster between WT and KO. Markedly, the ATM1 cluster was mainly composed of WT ATMs (Fig. 5g). ATM1 was characterized by a distinct set of inflammatory genes (S100a8/9) and inflammatory cytokine receptors (Il1r1, Il1r18), expressing low levels of both Mrc1 (CD206) or Itgax (CD11c; Fig. 5h). The flow cytometry supported this finding, showing a trend toward reduced CD206mid CD11cmid ATMs in old KO mice (Extended Data Fig. 8h). ATM2 and ATM3 represented CD206+ anti-inflammatory ATMs (Mrc1, Folr2, Lyve1, Mertk) and CD11c+ inflammatory/lipid associated ATMs (Itgax, Fabp4, Cd9, H2-Ab1, Il18), respectively (Fig. 5h). In summary, Gdf3, predominantly expressed in ATM3, is required for maintaining inflammatory ATM1 cluster during aging, indicating the local paracrine effects of Gdf3.
The number of differentially expressed genes (DEGs) revealed that ATM1 exhibited the most significant changes with lifelong Gdf3 deficiency (Fig. 5i). Gene set enrichment analysis (GSEA) on ATM1 DEGs showed enrichment in chromatin-modifying enzymes, chromatin remodeling and methylation-dependent heterochromatin formation without Gdf3 (Fig. 5j and Extended Data Fig. 9a). The adipose tissue myeloid cell aging gene signature was also reduced in KO ATM1 (Extended Data Fig. 9a). Next, the GO analysis on downregulated DEGs in KO ATM clusters identified suppressed biological processes in KO. The KO ATM1 cluster was enriched for oxidative phosphorylation pathways, whereas ATM2 and ATM3 were enriched for immune response-regulating signaling pathways (Extended Data Fig. 9b–d). To assess the overall impact of Gdf3 on all ATMs, merged ATM clusters were analyzed (Supplementary Table 2). In line with ATAC-seq results (Fig. 4 and Extended Data Fig. 7) and decreased ATM1 proportions (Fig. 5g), S100a8/9, Il18r1 and Il1r1 were significantly reduced in KO ATMs (Fig. 5k). Merged KO ATMs also showed the enrichment of gene sets related to chromatin-modifying enzymes and methylation-dependent heterochromatin formation (Extended Data Fig. 9e). Moreover, the expression of PRC2 components (Suz12, Eed, Ezh2, Jarid2) and additional repressive chromatin modifiers (Dnmt3a, Setbd2, Kmt5a) increased in KO ATMs (Fig. 5k). Out of 3,247 closing peaks in Gdf3-deficient ATMs (ATAC-seq; Fig. 4d), 1,497 peaks overlapped with downregulated genes in KO ATMs (scRNA-seq; Extended Data Fig. 9f and Supplementary Table 2). These findings demonstrate that Gdf3 regulates chromatin accessibility by counteracting histone methylation, promoting open chromatin formation that regulates transcription in ATMs during aging (Extended Data Fig. 9g).
Discussion
Older individuals have increased risk for infections and subsequent sepsis, in part owing to accumulating adiposity and a dysfunctional immune system. Gerotherapeutics that successfully improve the aged immune response are largely understudied. Our study reveals that the GDF3–SMAD2/3 axis may be a relevant pharmacologic target. GDF3 promotes the inflammatory phenotype of ATMs, contributing to the exacerbation of endotoxemia-induced inflammation in older, but not younger, organisms. GDF3 signals through SMAD2/3 and elicits proinflammatory responses in ATMs, diverging from their canonical immunoregulatory function51. Specifically, the chromatin landscape of ATMs shifts toward inflammation with age, increasing the accessibility of inflammation-associated genes. Our study demonstrates that Gdf3 deficiency can reverse the age-dependent changes in chromatin accessibility and transcription by restoring H3K27me3 levels in ATMs. Furthermore, genetic and pharmacological inhibition targeting the GDF3–SMAD2/3 axis protects against endotoxemia-induced inflammation and lethality in old mice.
The importance of VAT in aging and inflammation is corroborated by studies that highlight the immunological role of VAT during metabolic challenge or infection in older organisms4,5,6,8,9,36. Recent work indicates that B cells-derived IgG elevates macrophage expression of Tgfb, which promotes fibrosis and metabolic decline via SMAD2/3 in aged VAT35. Our work builds on this model, providing additional evidence for the importance of B cell-macrophage crosstalk in VAT5. We also provide evidence for the GDF3–SMAD2/3 axis regulating the phenotype of B cells. Although it remains unclear whether GDF3 acts synergistically with TGFβ-superfamily cytokines, our findings indicate that the mechanism governing inflammatory VAT microenvironment, driven by ATMs and B cells, may converge on SMAD2/3 signaling.
Previous work highlights the altered chromatin state with inflammation, aging or obesity29,43. Our study complements these studies by identifying increased GDF3-dependent open chromatin peaks in ATMs with age. Consistent with the GDF3 induction of pSMAD2/3 and inflammatory ATMs, predictive TF analysis points toward the SMAD complex as a critical TF that supports the inflammatory phenotype of ATMs. Although the SMAD complex is known to remodel chromatin during embryonic stem cells differentiation52, our results demonstrate it also fine-tunes the macrophage phenotype during aging. Our data (Figs. 4 and 5) suggest that Gdf3 inhibits PRC2 activity, limiting H3K27me3-mediated chromatin closing during aging. Considering that PRC2 complex can repress inflammatory genes49, and the SMAD complex can counteract PRC2 function50, the GDF3–SMAD2/3 axis may promote age-related inflammation by directly disrupting PRC2-mediated repression.
GDF3 is linked to various disease contexts. Prior studies report both protective and detrimental roles for GDF3, underscoring the complexity of its biology. scRNA-seq results suggest that Gdf3 primes ATMs to adopt inflammatory phenotypes, potentially enhancing their response during endotoxemia. Whereas GDF3 supports a proresolving phenotype of cardiac macrophages in young mice14, our findings indicates that GDF3 maintains inflammatory ATMs in old mice. We observe age-dependent macrophage responsiveness to GDF3 across multiple in vivo and in vitro systems, though the underlying mechanisms—whether due to changes in GDF3 levels, differential availability of activin receptors or downstream signaling—remain unclear. The heterogeneous expression of the type I and type II activin receptors across ATM clusters suggests that receptor-specific effects may vary by cell type, tissue or age. A deeper understanding of how GDF3 is regulated, how it interacts with diverse TGFβ family and receptor complexes and how their signaling output is dependent on cellular and disease context will be critical in the future studies.
We quantified the GDF3 in adipose tissue and serum from humans, supporting the relevance of the GDF3–SMAD2/3 axis and highlighting BRD4, GDF3 and SMAD2/3 as a potential therapeutic target in older individuals. Whether chromatin remodeling is a required component for the therapeutic effect is unknown. It is noteworthy that JQ1 and SIS3, as gerotherapeutics, probably have broader effects beyond the GDF3–SMAD2/3 axis. Prior studies have revealed that BRD4 contributes to atherosclerosis-induced macrophage senescence53, as well as bacteria-induced macrophage chromatin remodeling54. These findings suggest that BRD4 may play multiple roles during aging, promoting inflammation through both GDF3-independent and GDF3-dependent mechanisms. The blockade of SMAD2/3 signaling through activin receptor inhibitors, proposed for treating multiple diseases including cancer and kidney diseases, also shows promise55. In obese mice, the inhibition of type I activin receptor leads to a reduction in CD11c+ macrophages and the expression of Il1b, Nlrp3 and Gdf3 in VAT56. Lastly, SMAD2/3-targeting inhibitors, are currently under clinical development in phase I–III trials as cancer treatments57. Given these precedents, similar strategies could be repurposed to alleviate inflammation in the aging population.
Limitations
There are several limitations of this study. Due to the complexity of acquiring human VAT, we were unable to determine whether human ATMs express GDF3 and how this is regulated with age. Moreover, LPS will regulate chromatin accessibility in macrophages at later time points58, but whether GDF3 alters this pathway is unclear and will require additional studies. Further validations of transcriptional and protein expression of the ATM clusters that are regulated by GDF3 will be necessary. We reasoned that CD11cmid CD206mid population ATMs represents ATM1 population, which is high in S100a8/9, Il1r1 and Il18r1 and is reduced in the old GDF3 KO mice (seen in Fig. 5); however, we were unable to validate the protein levels with flow cytometry.
Methods
All experiments and animal use were conducted in compliance with the National Institute of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at the University of Minnesota. All human samples were obtained with written informed consent from participants and approved by the institutional review board at the University of Minnesota. The ARIC study was conducted under protocols approved by the Institutional Review Boards of participating institutions (University of Minnesota, Wake Forest University School of Medicine, University of Mississippi Medical Center and Johns Hopkins University).
Animal care
All mice were housed in specific pathogen-free facilities in cage racks that deliver high efficiency particulate air-filtered air to each cage with free access to sterile water at the University of Minnesota. Sentinel mice in our animal rooms tested negative for currently tested standard murine pathogens at various times while the studies were performed. Female C57Bl6/J (WT) mice were bred from our colony, purchased from The Jackson Laboratory, or received from the National Institute of Aging Rodent colony. Littermate WT, Gdf3−/−, Gdf3fl/fl and LysM-Cre mice were bred and aged in our facility. Young mice were defined as 3–6 months old, and old mice were defined as 18–26 months old in all experiments. Mice were fed with a normal chow diet (2018 Teklad Global 18% Protein Rodent Diets) and housed under a controlled temperature (21–23 °C), humidity (30–70%) and 12-h light–dark cycles. Old mice with extreme frailty, tumor, severe dermatitis or other age-related pathologies were excluded from the study.
Generation of Gdf3 fl/fl and Gdf3 −/− mice
The generation of Gdf3fl/fl mice was accomplished via CRISPR–Cas-mediated genome editing. The potential Cas9 target guide (protospacer) sequences in introns 1 and 3 were screened using the online tool CRISPOR, and candidates were selected. The templates for single guide RNA (sgRNA) synthesis were generated by PCR from a pX330 template; sgRNAs were transcribed in vitro and purified (Megashortscript, MegaClear; Thermo Fisher). The sgRNA–Cas9 ribonucleoproteins were complexed and tested for activity by zygote electroporation, followed by the incubation of embryos to blastocyst stage and the genotype scoring of indel creation at the target sites. High-performing sgRNAs were selected, and repair template oligonucleotides (single-stranded oligodeoxynucleotide) incorporating loxP sites targeted to the Cas9 cleavage sites were synthesized by Integrated DNA Technologies.
The floxed allele was created in two steps: targeting the 3′ loxP site, followed by a generation of breeding and subsequently targeting the 5′ loxP site. The sgRNA–Cas9 ribonucleoprotein and the corresponding template oligo were electroporated into C57Bl/6J zygotes (The Jackson Laboratory). Embryos were transferred to the oviducts of pseudopregnant CD-1 foster female mice using standard techniques. Homozygous Gdf3−/− mice were generated by first breeding Gdf3fl/fl (loxP/loxP) mice with β-actin-Cre transgenic mice (B6.FVB-Tmem163Tg(ACTB−cre)2Mrt/EmsJ, The Jackson Laboratory) and a second cross back to Gdf3fl/fl mice. This resulted in the germline deletion of Gdf3 that was used to generate littermate Gdf3+/+ and Gdf3−/− littermate controls.
Mouse models
Glucose tolerance test
The mice were fasted overnight for 16 h and intraperitoneally injected with 1.5 mg kg−1 glucose. Blood glucose levels were measured at 0-, 30-, 60-, 90- and 120-min time points using a glucometer.
Insulin tolerance test
The mice were fasted for 4 h and intraperitoneally injected with 0.8 U kg−1 insulin. Blood glucose levels were measured at 0-, 15-, 30-, 60-, 90- and 120-min time points using a glucometer.
Body composition measurement
EchoMRI was used.
LPS injection
The mice were intraperitoneally injected with sterile PBS (Corning) or 0.1 mg kg−1 LPS (Escherichia coli O111-B4; Sigma, L3024) diluted in PBS. The core BT was taken using a rectal thermometer at 0-, 2- and 4-h time points after the injection. Mice were killed by cervical dislocation under isoflurane anesthesia 4 h after injection.
Lethal dose LPS injection
The mice were intraperitoneally injected with 3 mg kg−1 (old) or 18 mg kg−1 (young) LPS diluted in PBS. The core BT was taken using a rectal thermometer at indicated time points.
In vivo cell proliferation study
For the in vivo cell proliferation study, 40 mg ml−1 stock solution of EdU (Santa Cruz Biotechnology, sc-284628A) was prepared by dissolving 50 mg of EdU in 1.25 ml of dimethyl sulfoxide (DMSO) and stored at −20 °C. Before the injection, 200 µl of EdU stock solution was diluted in 1.8 ml of PBS. For the vehicle, 200 µl of DMSO was diluted in 1.8 ml of PBS. The mice were intraperitoneally injected with 25 mg kg−1 of EdU daily for four consecutive days and killed on day 3.
JQ1 injection
For the JQ1 injection, 2 mg ml−1 stock solution of JQ1 was prepared by dissolving 100 mg of JQ1 (MedChemExpress, HY13030) in 50 ml of DMSO and stored at −20 °C. Before the injection, the JQ1 stock solution or DMSO (vehicle) was diluted sequentially in 40% PEG300, 5% Tween-80 and 50% PBS. The mice were intraperitoneally injected with 12.5 mg kg−1 of JQ1 daily for six consecutive days and killed on day 5. On day 5, mice were injected with 12.5 mg kg−1 JQ1 1 h before the 0.1 mg kg−1 LPS injection and killed at 4 h post LPS.
SIS3 injection
For the SIS3 injection, 100 mg ml−1 stock solution of SIS3 was prepared by dissolving 25 mg of SIS3 (SelleckChem, S7959) in 250 μl of DMSO and stored at −20 °C. Before the injection, the SIS3 stock solution or DMSO (vehicle) was diluted in 40% PEG300, 5% Tween-80 and 50% PBS. The mice were intraperitoneally injected with 10 mg kg−1 of SIS3, three times a week, for 4 weeks. On the day of the twelfth injection, the mice were injected with 10 mg kg−1 SIS3 1 h before the 0.1 mg kg−1 LPS injection and killed at 4 h post LPS.
BMDM culture
Bone marrow cells were collected from the femur and tibia. Once collected, cells were pelleted by centrifugation at 500g for 5 min and resuspended with 2 ml of ammonium–chloride–potassium lysing buffer to remove erythrocytes. After 2 min of incubation at room temperature, 5 ml of Roswell Park Memorial Institute 1640 buffer (RPMI) supplemented with 10% fetal bovine serum (FBS) was added to quench the reaction. Then, the cells were pelleted for counting purposes and seeded to a 6-well plate or 12-well plate with RPMI supplemented with 10% FBS, 1% antibiotic–antimycotic and 25 ng ml−1 M-CSF (R&D, LS004177). On day 5, the cells were supplemented with fresh 10 ng ml−1 M-CSF. On day 7, the cells were washed with PBS and serum-starved overnight with RPMI supplemented with 1% antibiotic–antimycotic. On day 8, the cells were stimulated by 20 ng ml−1 recombination GDF3 (rGDF3; R&D, 9009-GD) or indicated dose of JQ1 for 4 h. The cells were then collected for gene expression and flow cytometry analysis.
Tissue digestion
Visceral or subcutaneous adipose tissue was mechanically digested/minced with surgical scissors, then enzymatically digested in 0.1% collagenase II (Worthington, LS004174) in Hanks’ buffered salt solution (Gibco, 14185052) for 30 min at 37°, vortexing every 10 min for 30–45 min in the water bath set at 37 °C with 800 rpm agitation and vortexed every 10 min. Following digestion, cells were centrifuged at 500g for 5 min to obtain SVF (pellet) and adipocyte fraction (floating lipid layer). The adipocyte fraction was collected for gene expression. Erythrocytes were removed from SVF as described above. Then, SVF was washed and filtered through a 40-µM filter for further use. The spleen was prepared using mechanical digestion only, and the erythrocytes were removed, washed and filtered through 100- and 40-µM filters. The tissues from control and experimental groups were digested simultaneously.
Staining for flow cytometry
For live–dead staining, cells were incubated with Ghost Dye Red 780 Viability Dye (Tonbo, 13-0865-T100) for 20 min on ice, in the dark. Then, the cells were washed and incubated with Fcblock (Thermo, 14-9161-73) and surface antibodies for 45 min on ice, in the dark. For nuclear or intracellular antibodies, the FoxP3/TFr Fix/Permeabilization kit (Invitrogen, 00-5521-00) and BD Cytofix/Cytoperm Fix/Permeabilization kit (BD, 554714) were used. For phospho-flow, the cells were incubated with or Fixable Aqua Dead Cell Stain Kit (Invitrogen, L34957) for 10 min on ice, in the dark. Then, the cells were washed and incubated with warm BD Phosflow Fix Buffer (BD, 557870). After a 10-min incubation at 37 °C, the cells were washed and permeabilized with ice-cold BD Phosflow Perm buffer III (BD, 558050) for 30 min on ice, protected from light. The analysis was performed on FACSSymphony A3 cytometers using FlowJo v10; the gating strategies are shown in the extended data figures. For the FACS of ATM, live CD45+CD11b+SiglecF−F4/80+ cells from SVF were sorted on a BDFACSAriaII into RPMI with 20% FBS. For the methylation flow, surface-stained cells were fixed and permeabilized using BD Cytofix fixation buffer (BD, 554655) as per manufacturer’s instructions. After washing, cells were stained with AF488-H3K27me3 (Cell Signaling, 5499S). The antibodies used are documented in Supplementary Table 3.
scRNA-seq analysis
Library preparation
Single-cell libraries were prepared in triplicate with >85% cell viability confirmed by trypan blue staining. The cells were resuspended in MEM with 10% FBS at 1,200 cells per microliter. The library construction used Chromium Next GEM Single Cell 3ʹ dual index Kits v3.1 with Chip G (10x Genomics). About 10,000 single cells were loaded to generate gel bead-in-emulsions and lysed, and the RNA was barcoded and reverse-transcribed. The gel bead-in-emulsions were broken, and complementary DNA (cDNA) was prepared following the manufacturer’s protocol. The cDNA quality was assessed with the Agilent Bioanalyzer and Pico-green. Libraries underwent quality control by shallow-sequencing on an Illumina NextSeq and final sequencing on an Illumina Nova Seq S6 capturing ~20,000 paired-end reads per cell. Sequencing libraries were converted into feature-barcode matrices using Cell Ranger (10x Genomics, version 5.0.0) and mapped to GRCm39.
scRNA-seq quality control and clustering
An analysis of read count matrices was performed using Seurat version 4.3.1 in R. Cells with mitochondrial RNA <7%, and an R² of 0.95 for nCount_RNA versus nFeature_RNA were included. Each sample was normalized using ‘NormalizeData’, and the top 2,000 highly variable genes were selected with ‘FindVariableFeatures’. The dimensional reduction and clustering followed the standard Seurat workflow with ‘ScaleData’ and ‘RunPCA’. The top 50 principal components, identified by ‘ElbowPlot’, were used for uniform manifold approximation and projection (UMAP) visualization via ‘RunUMAP’ and finding nearest neighbors. Clusters were identified using the Louvain algorithm with a resolution of 0.2.
scRNA-seq cluster identification and DEG list generation
DEGs were identified by comparing normalized expressed RNAs for each model to RNAs expressed in control cells via the function ‘FindMarkers’ and then merging the results. When comparing clusters, the function ‘FindAllMarkers’ was used to find marker genes that characterize a specific cluster compared with all other clusters. A minimum of 100 cells from a sample group in a cluster was required for downstream analysis. For merged ATMs, ATM1, ATM2 and ATM3 clusters were combined for GSEA. DEGs shown in heat maps are shown with data normalized to the highest expression of that gene between clusters (that is, a score of 100 indicates the highest expression, not log2FC).
Western blotting
The VAT was snap-frozen in liquid nitrogen immediately after collection. The tissue was homogenized in radioimmunoprecipitation assay buffer containing phosphatase inhibitor and protease inhibitors (Sigma-Aldrich, P0044, P5726, P8340). Lysates were left on ice for 1 h and vortexed every 15 min. Lysates were then centrifuged twice at 14,000g for 10 min at 4 °C. The protein concentration was quantified using the Bradford Protein Assay Kit (Thermo, 23246), and equal amounts of protein were run on a 4–12% Bis–Tris sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel and transferred to a polyvinylidene difluoride membrane via the semidry transfer method. Blots were probed with primary antibodies and then incubated in secondary antibodies of appropriate species. Then, Femto Maximum Sensitivity Substrate (Thermo, 34096) or ECL Western Blotting Substrate (Thermo, 32209) was used for detection. Quantification of bands was done with Thermo Fisher’s iBright analysis software.
RNA extraction and gene expression analysis
Whole tissues were homogenized in Trizol (Invitrogen,15596026) using the Next Advantage Bullet Blender Storm 24. Following the homogenization, chloroform was added to samples and incubated at room temperature for 15 min. Samples were centrifuged at 14,000g for 15 min at 4 °C. Then, 70% molecular grade ethanol was added to the aqueous phase of centrifuged homogenate for RNA extraction using PureLink RNA Mini Kits (Invitrogen, 12183025) according to the manufacturer’s instructions. For suspended cells, they were lysed in lysis buffer supplemented with 1% β-mercaptoethanol, incubated and then mixed with 70% molecular grade ethanol. The reverse transcription PCR and quantitative PCR were performed as previously described. Primer sequences used for quantitative PCR are shown in Supplementary Table 4.
ATM culture
ATMs were positively selected from freshly collected SVF using EasySep Mouse F4/80 Positive Selection Kit (Stemcell, 100-0659) according to manufacturer’s instructions. After the selection, the cells were incubated in RPMI with 10% FBS and 1% antibiotic–antimycotic and stimulated with vehicle or rGDF3 (20 ng ml−1) for 30 min for flow cytometry analysis.
Ex vivo lipolysis
VATs were collected and weighed, ranging from 15 mg to 20 mg. Then, tissues were transferred to 96-well plates and cultured in 100 µl of lipolysis buffer (Krebs buffer with 3.5% fatty acid free bovine serum albumin and 0.1% glucose) with 0.1 mM norepinephrine. The buffer was collected after a 2-h incubation at 37 °C for nonesterified fatty acid assay (Fujifilm HR Series), following the manufacturer’s instructions. The non-esterified fatty acid values were normalized to tissue weights.
Human tissues
Frozen VATs were obtained from bariatric surgeries performed at the University of Minnesota. The detailed age (years) (sex (female, F), BMI (kg m−2)) information is as follows: 25 (F, 39.26), 27 (F, 39.49), 27(F, 38.74), 30 (F, 39.84), 31 (F, 49.22), 38 (F, 52.5), 39 (F, 38.18), 42 (F, 37), 42 (F, 39.67), 43 (F, 42.510), 48 (F, 33.27), 65 (F, 35.4), 66, (F, 18.8), 66, (F, 28.5), 68 (F, 41.1), 68 (F, 22), 71 (F, 37.2).
The ARIC study
The ARIC study is an ongoing population-based study of cardiovascular disease consisting of 15,792 participants from four US communities who were middle-aged (45–64 years) when they were recruited in 1987 to 1989 (visit 1). The details of the ARIC study design have been published previously59. Data from visit 2 (1990–1992, midlife) and visit 5 (2011–2013, late-life) were used to examine the association between GDF3 and CRP cross-sectionally at each visit. High-sensitivity CRP levels were measured from blood that was collected at visits 2 and 5 and stored at −70 °C. Visit 2 CRP levels were measured with the immunoturbidimetric assay using the Roche Modular P chemistry analyzer, whereas visit 5 CRP levels were measured using an immunoturbidimetric assay on the Beckman Coulter Olympus AU400e analyzer. Owing to the distribution of CRP being right-skewed, we log-transformed CRP with a log base of 2.
GDF3 was measured using a DNA aptamer-based capture array, which measured the relative concentrations of 5,284 plasma proteins from visits 2 and 5 from plasma samples that were stored long-term at −80 °C. All plasma protein measurements, including GDF3, were log-transformed with a log base of 2 to correct for skewness.
To examine the shape of the association between GDF3 and CRP, restricted cubic spline plots were used with knots at the 5th, 50th and 95th percentile. Linear regression was used to examine the association between GDF3 and CRP. Model 1 included adjustment for age at each visit, sex, race/center and educational attainment. Model 2 further adjusted for BMI at each visit. Each beta (β) and 95% CI were exponentiated to interpret CRP as a percentage change.
Statistics and reproducibility
Statistical significance was calculated using a multiple t-test, unpaired Student’s t-test, paired t-test or analysis of variance (ANOVA) with post hoc tests for multiple comparisons; *P < 0.05; **P < 0.005; ***P < 0.001; ****P < 0.0001. A one-way ANOVA was used when there was one experimental condition, but multiple groups and two-way ANOVA was used when there were two experimental conditions (that is, age and genotype). The paired t-test was used to compare the effects of a treatment on the cells derived from the same individual or sample with different treatment conditions. For a survival experiment, the log-rank (Mantel–Cox) test was used to determine statistical significance. For correlations, Pearson’s correlation analysis was used. Statistical outliers were identified and excluded using GraphPad Prism. A 95% confidence interval was used for all tests. The data were assumed to be normally distributed unless standard deviations significantly differed between groups. All tests were performed using GraphPad Prism v10. Data are expressed as mean ± s.e.m. The details of biological replicates are in the figure legends.
No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications. Biological replicates and individual experiments are described in the Methods and figure legends. No randomization method was used to allocate animals to experimental groups. Treatment groups were distributed across multiple cages. Data collection and analysis were not performed blind to the conditions of the experiments.
ATAC-seq library preparation
An ATAC-seq kit (Activemotif, 53150) was used for the ATAC-seq library preparation. In brief, ATMs (live CD45+ CD11b+ SiglecF− F4/80+ cells) were sorted using a BDFACSAriaII instrument. The cells were washed with ice-cold PBS to remove any debris, and 106 cells were resuspended in 100 μl of ice-cold ATAC lysis buffer. Then, cells were gently pipetted up and down and incubated on ice for 5 min. These cells were centrifuged at 500g for 10 min at 4 °C, and the supernatant was discarded. Next, 50 μl of tagmentation master mix containing buffer, 10× PBS, 1% Digitonin, 10% Tween 20, H2O and assembled transposome was added to each sample. Samples were incubated at 37 °C for 30 min in a thermomixer set at 800 rpm. Then, the tagmented DNA was extracted and amplified (72 °C for 5 min; 98 °C for 30 s; 12 cycles of 98 °C for 10 s, 63 °C for 30 s, 72 °C for 1 min) for library preparation. Lastly, 1.2× solid-phase reversible immobilization (SPRI) bead solution was added to samples for left side size selection. The samples were frozen and sent to Novogene, sequenced and analyzed.
ATAC bioinformatics analysis
First, FastQC was used to assess the quality of the raw reads. Then, Skewer was used to trim and filter off the sequencing adaptors and low-quality bases (mean quality >20, N ratio <15%, post-trimming length >18 nt), and trimmed reads were mapped to the mouse genome GRCm39 using BWA. After alignment, MACS2 software was used with a 100-bp shift and a 200-bp extension for peak calling analysis. Only peaks called with a peak score (or q value) of 5% or better were kept from each sample, and the number of peaks, the peak width, its distribution and the peak related genes were determined with MACS2. The Homer software was used for motif-enrichment analysis. Consensus sequences were identified by using the sequence of 250 bp (total 500 bp) upstream and downstream of the peak. To predict the accessibility around TSS, PeakAnalyzer was utilized to analyze Peak-TSS distance distribution. The distribution of peaks in functional area was analyzed by ChIPseeker. Differential peaks identified and visualized by edgeR (FC >2, false discovery rate (FDR) <0.05) were analyzed using ShinyGo 0.80 for TF predictions and GSEAs. For the visualization of mapped reads, Integrative Genomics Viewer analyzer was used.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
References
Franceschi, C., Garagnani, P., Parini, P., Giuliani, C. & Santoro, A. Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol. 14, 576–590 (2018).
Starr, M. E. & Saito, H. Sepsis in old age: review of human and animal studies. Aging Dis. 5, 126–136 (2014).
Saito, H., Sherwood, E. R., Varma, T. K. & Evers, B. M. Effects of aging on mortality, hypothermia, and cytokine induction in mice with endotoxemia or sepsis. Mech. Ageing Dev. 124, 1047–1058 (2003).
Han, S. J. et al. White adipose tissue is a reservoir for memory T cells and promotes protective memory responses to infection. Immunity 47, 1154–1168.e1156 (2017).
Carey, A. et al. Age-associated accumulation of B cells promotes macrophage inflammation and inhibits lipolysis in adipose tissue during sepsis. Cell Rep. 43, 113967 (2024).
Camell, C. D. Adipose tissue microenvironments during aging: effects on stimulated lipolysis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1867, 159118 (2022).
Dahlquist, K. J. V. & Camell, C. D. Aging leukocytes and the inflammatory microenvironment of the adipose tissue. Diabetes 71, 23–30 (2022).
Barthelemy, J., Bogard, G. & Wolowczuk, I. Beyond energy balance regulation: the underestimated role of adipose tissues in host defense against pathogens. Front. Immunol. 14, 1083191 (2023).
Camell, C. D. et al. Inflammasome-driven catecholamine catabolism in macrophages blunts lipolysis during ageing. Nature 550, 119–123 (2017).
Masurkar, N. et al. Novel cardiokine GDF3 predicts adverse fibrotic remodeling after myocardial infarction. Circulation 147, 498–511 (2023).
Xiang, L. et al. A multi-omic landscape of steatosis-to-NASH progression. Life Metab. 1, 242–257 (2022).
Andersson, O., Korach-Andre, M., Reissmann, E., Ibanez, C. F. & Bertolino, P. Growth/differentiation factor 3 signals through ALK7 and regulates accumulation of adipose tissue and diet-induced obesity. Proc. Natl Acad. Sci. USA 105, 7252–7256 (2008).
Wang, W., Yang, Y., Meng, Y. & Shi, Y. GDF-3 is an adipogenic cytokine under high fat dietary condition. Biochem Biophys. Res Commun. 321, 1024–1031 (2004).
Wang, L. et al. GDF3 protects mice against sepsis-induced cardiac dysfunction and mortality by suppression of macrophage pro-inflammatory phenotype. Cells https://doi.org/10.3390/cells9010120 (2020).
Jaeger, P. A. et al. Network-driven plasma proteomics expose molecular changes in the Alzheimer’s brain. Mol. Neurodegener. 11, 31 (2016).
Bu, Y. et al. Insulin regulates lipolysis and fat mass by upregulating growth/differentiation factor 3 in adipose tissue macrophages. Diabetes 67, 1761–1772 (2018).
Hall, J. A. et al. Obesity-linked PPARγ S273 phosphorylation promotes insulin resistance through growth differentiation factor 3. Cell Metab. 32, 665–675.e666 (2020).
Shen, J. J. et al. Deficiency of growth differentiation factor 3 protects against diet-induced obesity by selectively acting on white adipose. Mol. Endocrinol. 23, 113–123 (2009).
Patsalos, A. et al. In vivo GDF3 administration abrogates aging related muscle regeneration delay following acute sterile injury. Aging Cell 17, e12815 (2018).
Varga, T. et al. Macrophage PPARγ, a lipid activated transcription factor controls the growth factor GDF3 and skeletal muscle regeneration. Immunity 45, 1038–1051 (2016).
Wang, P. et al. Administration of GDF3 into septic mice improves survival. Front Immunol. 12, 647070 (2021).
Kotikalapudi, N. et al. Acute regulation of murine adipose tissue lipolysis and insulin resistance by the TGFβ superfamily protein GDF3. Nat. Commun. 16, 4432 (2025).
Derynck, R. & Zhang, Y. E. SMAD-dependent and SMAD-independent pathways in TGF-β family signalling. Nature 425, 577–584 (2003).
Chen, P. Y. et al. Endothelial TGF-β signalling drives vascular inflammation and atherosclerosis. Nat. Metab. 1, 912–926 (2019).
Tan, N. S. et al. Essential role of SMAD3 in the inhibition of inflammation-induced PPARβ/δ expression. EMBO J. 23, 4211–4221 (2004).
Yadav, H. et al. Protection from obesity and diabetes by blockade of TGF-β/SMAD3 signaling. Cell Metab. 14, 67–79 (2011).
Bertero, A. et al. The SMAD2/3 interactome reveals that TGFβ controls m6A mRNA methylation in pluripotency. Nature 555, 256–259 (2018).
Bruno, M. E. C. et al. Accumulation of γδ T cells in visceral fat with aging promotes chronic inflammation. Geroscience 44, 1761–1778 (2022).
Hill, D. A. et al. Distinct macrophage populations direct inflammatory versus physiological changes in adipose tissue. Proc. Natl Acad. Sci. USA 115, E5096–E5105 (2018).
Ryu, S. et al. The matricellular protein SPARC induces inflammatory interferon-response in macrophages during aging. Immunity 55, 1609–1626.e1607 (2022).
Mukherjee, S. et al. Mechanisms of γδ T cell accumulation in visceral adipose tissue with aging. Front. Aging 4, 1258836 (2023).
Yona, S. et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91 (2013).
Boring, L. et al. Impaired monocyte migration and reduced type 1 (Th1) cytokine responses in C–C chemokine receptor 2 knockout mice. J. Clin. Invest. 100, 2552–2561 (1997).
Starr, M. E. et al. Age-dependent vulnerability to endotoxemia is associated with reduction of anticoagulant factors activated protein C and thrombomodulin. Blood 115, 4886–4893 (2010).
Yu, L. et al. IgG is an aging factor that drives adipose tissue fibrosis and metabolic decline. Cell Metab. 36, 793–807.e795 (2024).
Camell, C. D. et al. Aging induces an Nlrp3 inflammasome-dependent expansion of adipose B cells that impairs metabolic homeostasis. Cell Metab. 30, 1024–1039 e1026 (2019).
Khan, A. R. et al. PD-L1hi B cells are critical regulators of humoral immunity. Nat. Commun. 6, 5997 (2015).
Hu, X. et al. Brd4 modulates diet-induced obesity via PPARγ-dependent Gdf3 expression in adipose tissue macrophages. JCI Insight https://doi.org/10.1172/jci.insight.143379 (2021).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
Jinnin, M., Ihn, H. & Tamaki, K. Characterization of SIS3, a novel specific inhibitor of Smad3, and its effect on transforming growth factor-beta1-induced extracellular matrix expression. Mol. Pharm. 69, 597–607 (2006).
Yang, J. et al. Longevity-associated SMAD3 non-coding centenarian variant impairs a cell-type specific enhancer to reduce inflammation. Preprint at bioRxiv https://doi.org/10.1101/2023.05.17.540984 (2023).
The atherosclerosis risk in communities (ARIC) study: design and objectives. The ARIC investigators. Am. J. Epidemiol. 129, 687–702 (1989).
Márquez, E. J. et al. Sexual-dimorphism in human immune system aging. Nat. Commun. 11, 751 (2020).
Cai, Y. et al. Rap1 induces cytokine production in pro-inflammatory macrophages through NFκB signaling and is highly expressed in human atherosclerotic lesions. Cell Cycle 14, 3580–3592 (2015).
Rabhi, N. et al. Obesity-induced senescent macrophages activate a fibrotic transcriptional program in adipocyte progenitors. Life Sci. Alliance https://doi.org/10.26508/lsa.202101286 (2022).
Ge, S. X., Jung, D. & Yao, R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629 (2020).
Heldin, C. H., Miyazono, K. & ten Dijke, P. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature 390, 465–471 (1997).
Lachmann, A. et al. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 26, 2438–2444 (2010).
Salminen, A., Kaarniranta, K., Hiltunen, M. & Kauppinen, A. Histone demethylase Jumonji D3 (JMJD3/KDM6B) at the nexus of epigenetic regulation of inflammation and the aging process. J. Mol. Med. 92, 1035–1043 (2014).
Dahle, Ø, Kumar, A. & Kuehn, M. R. Nodal signaling recruits the histone demethylase Jmjd3 to counteract polycomb-mediated repression at target genes. Sci. Signal. 3, ra48 (2010).
Gong, D. et al. TGFβ signaling plays a critical role in promoting alternative macrophage activation. BMC Immunol. 13, 31 (2012).
Coda, D. M. et al. Distinct modes of SMAD2 chromatin binding and remodeling shape the transcriptional response to NODAL/Activin signaling. Elife https://doi.org/10.7554/eLife.22474 (2017).
Wang, H. et al. BRD4 contributes to LPS-induced macrophage senescence and promotes progression of atherosclerosis-associated lipid uptake. Aging 12, 9240–9259 (2020).
Hoffner O’Connor, M. et al. BET protein inhibition regulates macrophage chromatin accessibility and microbiota-dependent colitis. Front. Immunol. 13, 856966 (2022).
Kargbo, R. B. ALK inhibitors for treating cancer, blood, and kidney diseases. ACS Med. Chem. Lett. 13, 1539–1541 (2022).
Zhao, M. et al. Targeting activin receptor-like kinase 7 ameliorates adiposity and associated metabolic disorders. JCI Insight https://doi.org/10.1172/jci.insight.161229 (2023).
Ciardiello, D., Elez, E., Tabernero, J. & Seoane, J. Clinical development of therapies targeting TGFβ: current knowledge and future perspectives. Ann. Oncol. 31, 1336–1349 (2020).
Gao, Y., Wang, N. & Jia, D. H3K27 tri-demethylase JMJD3 inhibits macrophage apoptosis by promoting ADORA2A in lipopolysaccharide-induced acute lung injury. Cell Death Discov. 8, 475 (2022).
Wright, J. D. et al. The ARIC (atherosclerosis risk in communities) study: JACC focus seminar 3/8. J. Am. Coll. Cardiol. 77, 2939–2959 (2021).
Acknowledgements
We acknowledge and thank the University of Minnesota Flow Cytometry Resource, specifically P. Champoux and R. Arora, for their expertise. We thank the Yale Genome Editing Center staff for their help in generating the Gdf3fl/fl mouse and the UMN Research Animal Resources Staff for their day-to-day care of the mice. We also thank the staff and participants of the ARIC study for their important contributions. This work was supported by National Institute of Health grants grant nos. F99AG095479 (I.J.), R00AG058800 (C.D.C.), R01AG069819 (D.A.B. and C.D.C.), R01AG079913 (C.D.C.), R01HL134320, the McKnight Land-Grant Professorship (C.D.C.), the Glenn Foundation for Medical Research/AFAR 2025 Discovery Award (C.D.C.), the Diana Jacobs Kalman/AFAR Scholarships for Research in the Biology of Aging (I.J.) and the Medical Discovery Team on the Biology of Aging (C.D.C.). The Atherosclerosis Risk in Communities study has been funded in whole or in part with Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health, and Human Services, under Contract nos. (75N92022D00001, 75N92022D00002, 75N92022D00003, 75N92022D00004, 75N92022D00005). SomaLogic Inc. conducted the SomaScan assays in exchange for use of ARIC data. This work was supported in part by NIH/NHLBI grant R01 HL134320. The authors thank the staff and participants of the ARIC study for their important contributions.
Author information
Authors and Affiliations
Contributions
Conceptualization: I.J. and C.D.C. Investigation: I.J., A.C., V.K., K.N., S.C., D.S., D.A.B. and C.D.C. Data curation—formal analysis: I.J. and C.D.C. scRNA-seq data analysis: A.C. ARIC data analysis: J.R.M. and P.L.L. Mouse model generation: S.B. and T.N. Project administration: I.J. and C.D.C. Oversight: C.D.C. Writing—original draft: I.J. and C.D.C. Writing—review, editing and revision: all authors. Funding acquisition: ARIC study (P.L.L.), C.D.C. and D.A.B.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Aging thanks Xiangming Fang, Duygu Ucar and Daniel Winer, who co-reviewed with Priya Makhijani, for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Lifelong Gdf3 deficiency does not affect metabolic profile of old mice, nor the proliferation or infiltration of ATMs, related to Fig. 1.
(A-B) Frequency of CD11c+ ATMs in VAT from young (n = 7) or old (n = 15) female WT mice injected with (A) PBS or (B) 0.1 mg/kg LPS, related to Fig. 1a. (C) Body weights of female WT (n = 11) and GDF3 KO (n = 8) mice measured throughout lifetime. (D) Body composition of female YW (n = 6), YK (n = 3), OW (n = 8), and OK (n = 8) mice. (E) Insulin and (F) glucose tolerance test of 2 ~ 3- and 18 ~ 19-month female WT and KO mice. YW, n = 5; YK, n = 5; OW, n = 14; OK, n = 11. (G-K) Young (3- to 4-month-old) WT, old (19- to 22-month-old) female WT or KO mice were challenged with 0.1 mg/kg LPS. (G) Adipocyte gene expression involved in fatty acid oxidation and fatty acid synthesis. OW, n = 4; OK, n = 6. (H) Frequency of immune cells (gated as % of CD45+) in VAT. Myeloid panel (YW, n = 10; OW, n = 10; OK, n = 8), T cell panel (YW, n = 4; OW, n = 5; OK, n = 5). (I) Frequency of CD206+ ATMs. YW, n = 14; YK, n = 8; OW, n = 15; OK, n = 14. (J) Frequency of immune cells (gated as % of CD45+) in spleen. Myeloid panel (YW, n = 10; OW, n = 10; OK, n = 8), T cell panel (YW, n = 4; OW, n = 5; OK, n = 5). (K) Free fatty acid released from VAT explants. OW, n = 7; OK, n = 9. (L-N) Young (4-months-old) and old (24-months-old) female WT mice were i.p. injected with 25 mg/kg EdU for 4 consecutive days and 0.1 mg/kg LPS or PBS for 4 hours on day 3. (L) Experimental schematic of in vivo proliferation assay. (M) Representative gating strategy. (N) Frequency of EdU+CD11c+ ATMs in VAT. n = 5 per group. (O-P) Young (3- to 4-month-old) and old (19- to 22-month-old) female WT or KO mice we injected with 25 mg/kg EdU for 4 consecutive days and 0.1 mg/kg LPS. (O) Schematic of experimental design (left). Frequency of EdU+CD11c+ ATMs in VAT (right). YW, n = 6; YK, n = 5; OW, n = 6; OK, n = 6. (P) Frequency of CCR2+ ATMs in VAT. YW, n = 14; YK, n = 8; OW, n = 15; OK, n = 14 All data are presented as means ± SEM.
Extended Data Fig. 2 Single cell RNA-sequencing of VAT immune cells, related to Figure.
Figure S2 (A) Separate UMAP of VAT immune cells (CD45+ cells) from young (3-month-old, n = 2) or old (21-month-old, n = 3) female WT mice injected with 0.1 mg/kg LPS showing 23 clusters. (B) Expressions of genes associated with macrophage phenotype, normalized across all 23 clusters.
Extended Data Fig. 3 Gdf3 is not required for LPS-induced lethality in young mice but is regulated with age in a B cell-dependent manner, related to Fig. 1.
Figure S3 (A-E) Young (4- to 6-months-old) female WT and KO mice were injected with lethal dose of LPS (18 mg/kg) and VAT was collected for analysis 6 hr post-LPS injection. (A) Schematic of experimental design. (B) Frequency of ATM subsets in VAT. WT, n = 8; KO, n = 5. (C) Mean core BT after LPS injection. WT, n = 13; KO, n = 8. (D) Frequency of activated T cells and activated B cells in VAT. WT + PBS, n = 3; WT + LPS, n = 5; KO + LPS, n = 4. (E) Inflammatory gene expression in VAT. WT + PBS, n = 3; WT + LPS, n = 5; KO + LPS, n = 4. (F) Young (3-months-old) male WT and KO mice were injected with rGDF3 (10 µg/kg) 24 hours prior to lethal dose of LPS (18 mg/kg). Mean core BT was measured after LPS injection (left). VAT was collected for analysis 6 hr post-LPS injection (right). (G) TGFβ superfamily gene expression in VAT and (H) Gdf3 expression in multiple tissues from unchallenged 2-months-old (n = 4) and 21-month-old (n = 4) female WT mice. (I) TGFβ superfamily gene expression in VAT from 20- to 22-month-old female WT (n = 6) and Bnull (n = 9) mice injected with 0.1 mg/kg LPS. (J-M) Metabolic profiling of old (21- to 26-month-old) control and mKO mice. Female control, n = 11; female mKO, n = 9; male control, n = 8; male mKO, n = 5. Body weights and glucose tolerance test of female (J-K) and male (L-M) mice. (N-R) Old (21- to 24-month-old) female control and mKO mice were injected with 0.1 mg/kg LPS. Frequency of (N) CD9+ ATMs, (O) CD206+ ATMs, and (P) CCR2+ ATMs in VAT. (Q) Frequency of CD11c+ and CD206+ ATMs in SAT. Control, n = 7 ~ 14; mKO, n = 4 ~ 6. (R) Free fatty acid released from VAT explants from old female control (n = 5) and mKO (n = 4) mice. (S) Frequency of CD11c+ ATMs from VAT of male and female mice injected with 0.1 mg/kg LPS. Females ♀, males ♂. Control, n = 14; mKO, n = 9. (T) Representative gating strategy for B cells. (U-V) Frequency of (U) CD73+IgM+ B cells and (V) PDL1+ B cells in VAT of old control and mKO mice injected with 0.1 mg/kg LPS. (W) Mean core BT of young (3-month-old) or old (23- to 26-month-old) male mice post-LPS (0.1 mg/kg) injection. Young control, n = 8; old control n = 8; old mKO, n = 5. All data are presented as means ± SEM.
Extended Data Fig. 4 BRD4-regulated Gdf3 promotes inflammation with age, related to Fig. 2.
Figure S4 (A-F) Old (23- to 24-month-old) female WT mice were i.p. injected with JQ1 (12.5 mg/kg) or vehicle and LPS (0.1 mg/kg). Vehicle-injected, n = 10; JQ1-injected, n = 11. (A) Body weights of vehicle- or JQ1-injected old (23- to 24-month) female mice. Vehicle-injected, n = 10; JQ1-injected, n = 11. (B) Frequency and (C) cellularity of immune cells in VAT from vehicle- or JQ1-injected old mice. (D) S100a8, S100a9, Il1r1 and Il18r1 expression in VAT. (E-F) Frequency of (E) PDL1+ and (F) CD73+ B cells in VAT. (G-I) BMDMs isolated from old (24-month) female WT mice were treated with vehicle or JQ1 (200 nM or 1 µM). (G) Schematic of experimental design. (H) Gdf3 expression in BMDMs. (I) Casp1, Il1b and Tnfa expression in BMDMs. All data are presented as means ± SEM.
Extended Data Fig. 5 GDF3 increases pSMAD2/3 in macrophages from old mice, related to Fig. 2.
Figure S5 (A) pSMAD2/3, GDF3 and β-actin by western blot, and PonceauS staining of SVF or adipocytes from young or old female WT mice injected with 0.1 mg/kg LPS. (B) Quantification of GDF3 from Figure S5A (n = 3 each group). (C) Relative expression of Gdf3 from adipocytes and SVF of old (23-month) female WT mice (n = 3). (D-F) BMDMs from young (3-month, n = 3_) or old (24-month) female WT mice were treated with vehicle or rGDF3 (20 ng/mL) in vitro. (D) Schematic of experimental design for BMDMs. (E) pSMAD2/3 MFI in BMDMs (young n = 3; old n = 10). (F) Frequency of CD206+ BMDMs from old mice (n = 7). (G-I) BMDMs from old (23-month, n = 8) female WT mice were treated with LPS (1 µg/mL) prior to vehicle or rGDF3 (20 ng/mL) in vitro. (G) Schematic of experimental design for BMDMs. (H) Frequency of CD206+ and (I) CD11c+ BMDMs. (J-L) ATMs from young (4-month) or old (24-month) female WT mice were treated with vehicle or rGDF3 (20 ng/mL) in vitro (J) Schematic of experimental design for ATMs. (K) pSMAD2/3 MFI and CD11c (%) of ATMs from young mice (n = 5). (L) pSMAD2/3 MFI of ATMs from old mice (n = 6). All data are presented as means ± SEM.
Extended Data Fig. 6 Correlation between GDF3 and C-reactive protein, related to Fig. 3.
Figure S6 (A) Restricted cubic splines displaying the cross-sectional association between CRP and GDF3 among ARIC participants at midlife and late-life. CRP is represented as log-base2. The restricted cubic spline knots are at the 5th, 50th, and 95th percentiles. The red line represents the estimate, while the dashed lines display the 95% confidence intervals. (B) Cross-sectional association of CRP with GDF3 among ARIC participants at midlife and late-life. Significant results are in bold (p < 0.05).
Extended Data Fig. 7 ATAC sequencing of ATMs, related to Fig. 4.
Figure S7 (A) Representative gating strategy for fluorescence-activated cell sorting (FACS). (B) Schematic of bioinformatics analysis flow. (C) Transcription start site (TSS) enrichment plot showing the distribution of the reads mapped to TSS. (D) Representative peak annotation pie charts for identified ATAC-seq peaks. (E) Ven diagram depicting the overlap between opening peaks with aging (old vs young control) and closing peaks with Gdf3 deletion (old mKO vs old control) from ATAC-seq analysis. (F-G) Gene ontology (GO Biological Process) analysis of genes associated with (F) opening peaks with aging and (G) closing peaks with Gdf3 deletion. (H) De novo motif search within all identified ATAC-seq peaks from young ctrl, old ctrl and old mKO ATMs. Consensus motif, transcription factor, p-value, and percentage of targets are shown in table. * ATAC-seq was performed in 3 biological replicates (young control, n = 3; old control, n = 3; old mKO, n = 3).
Extended Data Fig. 8 scRNA-seq of VAT immune cells, related to Fig. 5.
Figure S8 (A) H3K27me3 MFI of VAT immune cells from young (4-month-old) and old (22-month-old) female WT mice (n = 4, each group) injected with PBS or 0.1 mg/kg LPS for 4 hours. (B) Heatmap showing genes used to annotate clusters, normalized across all identified clusters. (C) Proportion of each clusters comparing WT and KO (% of all each clusters). (D) Separate UMAP of VAT immune cells (CD45+ cells) from old (21- to 23-month-old) female WT (n = 3) and KO (n = 3) showing 24 clusters. (E) Feature plot showing Gdf3 expression across the dataset. (F) UMAP showing 9 subclusters from ATM3. (G) Feature plot showing Gdf3 expression in ATM3 subclusters, separated by genotype. (H) Frequency of CD11cmid CD206mid ATMs in VAT from old (20- to 24-month-old) female control (n = 9) or KO mice (n = 6). All data are presented as means ± SEM.
Extended Data Fig. 9 scRNA sequencing of ATMs, related to Fig. 5.
Figure S9 (A) GSEA analysis using Reactome Chromatin Remodeling (upper) and Tabula Muris Adipose Tissue Myeloid Cell Ageing (lower) of KO vs WT, ATM1 cluster. (B-D) Over-representation Gene Ontology analysis on downregulated DEGs in KO compared to WT in each ATM clusters. (E) GSEA analysis using Reactome Chromatin Modifying Enzymes (upper) and Methylation Dependent Heterochromatin Formation (lower) of KO vs WT, merged ATM clusters. (F) Ven diagram depicting the overlap between closing peaks (ATAC-seq, Fig. 4) and downregulated genes (scRNA-seq, Fig. 5) in KO ATMs in comparison to control ATMs. (G) Working model.
Supplementary information
Supplementary Table 1
Differential ATAC-peaks.
Supplementary Table 2
scRNA-seq ATM DEGs (WT versus KO).
Supplementary Table 3
Antibodies used for flow cytometry analysis, related to staining for flow cytometry.
Supplementary Table 4
Primer sequences used for quantitative PCR analysis.
Source data
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 2
Statistical source data for Figs. 1–3 and 5 and Extended Data Figs. 1, 3–5 and 8.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jang, I.H., Carey, A., Kruglov, V. et al. GDF3 promotes adipose tissue macrophage-mediated inflammation via altered chromatin accessibility during aging. Nat Aging 6, 127–142 (2026). https://doi.org/10.1038/s43587-025-01034-6
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43587-025-01034-6