Abstract
Background
Respiratory tract infections (RTIs) drive lung function decline in children with cystic fibrosis (CF). While the respiratory microbiota is clearly associated with RTI pathogenesis in infants without CF, data on infants with CF is scarce. We compared nasal microbiota development between infants with CF and controls and assessed associations between early-life nasal microbiota, RTIs, and antibiotic treatment in infants with CF.
Methods
We included 50 infants with CF and 30 controls from two prospective birth cohorts followed throughout the first year of life. We collected 1511 biweekly nasal swabs and analyzed the microbiota after amplifying the V3–V4 region of the 16S rRNA gene. We conducted structured weekly interviews to assess respiratory symptoms and antibiotic treatment. We calculated generalized additive mixed models and permutational analysis of variance.
Results
Here, we show that the nasal microbiota is already altered before the first RTI or antibiotic treatment in infants with CF. Microbiota diversity differs between infants with CF and controls following RTIs and/or antibiotic treatment. CF infants with lower α-diversity have a higher number of subsequent RTIs.
Conclusions
Early nasal microbiota alterations may reflect predisposition or predispose to RTIs in infants with CF, and further change after RTIs and antibiotic treatment. This highlights the potential of targeting the nasal microbiota in CF-related RTI management, while also questioning current practices in the era of novel modulator therapies.
Plain language summary
Cystic fibrosis (CF) is an inherited condition which can increase the risk of developing respiratory tract infections (RTIs). We investigated the microorganisms present in the respiratory tract of infants from birth to the age of one. We found that infants with CF had differences in the microorganisms present immediately after birth compared to infants without CF. These differences increased after development of RTIs and following antibiotic treatment. Our results suggest that infants with CF could potentially benefit from treatments that modify microorganisms present in their respiratory tract prior to development of any RTI, or from different antibiotics to those used by infants without CF.
Similar content being viewed by others
Introduction
Several studies demonstrate a relationship between the dynamics of the upper respiratory microbiota in early life and susceptibility to respiratory tract infections in healthy individuals1,2,3,4,5,6,7. However, conclusive data studying the relationship between respiratory tract infections and the microbiota among infants with cystic fibrosis (CF) is scarce8.
Opportunistic bacteria begin to colonize the airways of children with CF in early life, leading to recurrent, mostly polymicrobial respiratory tract infections (RTIs)9,10. These may lead to pulmonary exacerbations that are followed by antibiotic treatment and hospitalizations11,12 and eventually drive persistent airway inflammation, structural damage, and lung function decline9,13.
The respiratory microbiota profile is altered in infants with CF, independent of the presence of pathogens detected with culture-based techniques14,15,16,17. Early microbial development is characterized by early dominance and persistence of Staphylococcus aureus in infants with CF, shifting to Corynebacterium spp. and Streptococcus spp. dominance at age 3 months, compared with dominance of Moraxella spp., higher abundance of Dolosigranulum and Prevotella spp. in infants without CF17,18. Antibiotic use in the first year of life is associated with lower bacterial density and increased colonization with gram-negative bacteria in infants with CF18.
We have previously demonstrated that distinct nasal microbiota profiles are present in the first year of life in infants with CF compared to unaffected controls17. RTIs and antibiotic therapy are frequent even in early CF disease, hampering to disentangle the interrelations between the microbiota, respiratory infections and antibiotics. This is however of uttermost importance to be able to assess a possible preventive and therapeutic potential to target the nasal microbiota in CF-related RTI management. Thus, this study had three aims: to compare nasal microbiota development between infants with CF and matched controls before and after the first RTI and first antibiotic treatment; to assess the interrelations between early-life nasal microbiota and number of RTIs, before and after the first RTI and first antibiotic treatment in infants with CF; and to analyze in detail how the first antibiotic treatment or the first RTI influence the nasal microbiota in infants with CF. We show that the nasal microbiota is already different before the first RTI or antibiotic treatment in infants with CF compared to controls. Differences increase following RTIs and/or antibiotic treatment. We observe that CF infants with lower α-diversity have a higher number of subsequent RTIs.
Methods
Study design
This study includes data of infants from two prospective birth cohort studies: the Swiss Cystic Fibrosis Infant Lung Development (SCILD), which recruits infants with CF after newborn screening result19, and the Basel Bern Infant Lung Development (BILD) cohort, which recruits unselected infants without CF20. In both studies, participants attend their first study visit between 3 and 5 weeks after birth, and are followed throughout the first year of life21. All included infants were white, due to the study’s location and the higher incidence of CF in white populations, without consideration of other socially relevant groupings. Ethics Committee of the Canton of Bern, Switzerland approved the studies with the following Project-IDs: SCILD 2017-02139; BILD 2019-01072. We obtained written informed consent from each study participant’s parent or legal guardian.
Cohort
We included 50 infants with CF (30 of those were included in our previous study17) from nine centers around Switzerland (Supplementary Table 1) and 30 matched unaffected controls, all born between 2011 and 2019. In the following, we refer to the infants from the BILD cohort as controls. We performed weekly structured telephone interviews to obtain information about respiratory symptoms, antibiotic treatment, breastfeeding, and childcare. Parents collected biweekly anterior nasal swabs and sent them by post to the coordinating study center in Bern. Infants with CF received only therapeutic but not prophylactic antibiotic therapies.
Microbiota analysis
For transport and storage of nasal swabs UTM® system from Copan was used. DNA extraction was performed as described before22. The V3-V4 region of the 16S- ribosomal RNA gene was amplified, and PCR product purified. Paired-end sequencing was performed with Illumina NovaSeq6000 platform. The DADA2 pipeline was used to obtain amplicon sequence variants (ASVs)23 using the Silva v138.2 database5. We removed ASVs not assigned to kingdom Bacteria (family Mitochondria or class Chloroplast) or identified as contaminating ASV with the decontam package in R24. We analyzed samples with at least 3000 reads, as suggested in comparable studies5 and as a clear flattening of the rarefaction curves was observed after 3000 reads (Supplementary Fig. 1). We included 1511 samples after quality control (1557 before). α-diversity calculation (Shannon-Diversity Index (SDI)), was performed without further filtering. For other analyses, only ASVs with a prevalence at least 0.1 % or abundance in at least five samples were preserved (43205 ASVs). Microbiota data was normalized using total sum scaling into relative abundance. Distance matrix to analyze β-diversity was calculated with Bray-Curtis dissimilarity.
Outcome variables
Comparison of the nasal microbiota between infants with CF and controls
We investigated 1. β–diversity: compositional microbiota differences between samples based on a dissimilarity matrix, 2. within-subject dissimilarity: β–diversity between consecutive taken swabs with less than 3 weeks difference of the same infant, 3. α-diversity: microbiota diversity within a sample, and 4. differential bacterial abundances (%). We assessed symptom scores as described previously21 (Supplementary Table 2). We included: a. all samples of all infants; b. samples taken before the first antibiotic treatment. c. samples taken before the first RTI. d. samples taken before the first RTI, but after the first antibiotic treatment. e. samples taken before the first antibiotic treatment, but after the first RTI (Fig. 1).
This figure displays two example longitudinal courses of events in infants. In the upper part (green arrow), the rare case of antibiotic treatment before the first RTI is displayed (e.g., due to Otitis media). The lower part (ochre arrow) shows an infant with two RTIs in the first year of life, one of which was treated with antibiotics. To disentangle the interrelations between nasal microbiota, respiratory tract infections (RTIs), and antibiotic treatment, we calculated statistics overall and before/after the first antibiotic treatment or before/after the first RTI in the first year of life. Created with Biorender.com.
Investigation of the interrelations between early-life nasal microbiota and number of RTIs, first RTI, and first antibiotic treatment in infants with CF
We defined “higher number” of RTIs above the 75th percentile of RTI weeks of controls. We divided infants in two groups: Infants with a “lower number” of RTIs (0–3 weeks of RTI, n = 35) and infants with a “higher number” of RTIs (>3 weeks of RTI, n = 15). We performed analyses analogous to 1.−4. and a-c of the first part of the study.
Effect of a “first hit” on the microbiota in infants with CF
We investigated the role of first antibiotic treatment and the role of number of antibiotic treatments in the first year of life analogous to 1.−4. of the first part of the study. Infants with CF were divided into three groups depending on the number of antibiotic treatments: “Never” (0 antibiotic treatments, n = 17), “<75th Percentile” (1 antibiotic treatment, n = 25), and “≥75th Percentile” (2 or more antibiotic treatments, n = 8). We investigated the role of first RTI in the first year of life analogous to 1.−4. of the first part of the study.
Statistics and reproducibility
Statistics were performed with statistical software R (version 4.1.2). For normal distributed microbiota exposures, generalized additive mixed models (gamm function, mgcv package) were performed with autoregressive structure (lag 1) to account for temporal correlation within subjects (random effect) and corrected for before named possible confounders (fixed effect)22. The age of the infant in weeks was added in a smooth term. β-diversity was tested with permutational multivariate analysis of variance (PERMANOVA). Terms were added with the option “marginal” to account for all predictor variables equally (adonis2 function, vegan package). Differential abundance analysis of bacterial families was calculated with MaAsLin2 after total sum scaling normalization5,25. We focused on the nine most abundant families for the main manuscript and summarized the rest (“others”) due to 16S rRNA sequencing resolution. Reported q-values were corrected for multiple testing with Benjamini-Hochberg procedure as recommended in MaAsLin2 default25. To control for confounding, we corrected for sweat chloride levels in infants with CF, mode of delivery, feeding type (breastfeeding vs formula fed), parental smoking, siblings, season, and antibiotic treatment. Due to only two infants with CF attending childcare in the first year of life, we matched control cases that did not attend childcare either without correcting in the further analyses. Results with p < 0.05, q < 0.05 respectively, were considered significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Results
Studied cohort and samples
The characteristics of the study cohorts are displayed in Table 1 and the study design in Fig. 1. The two cohorts were matched regarding sex, breastfeeding, delivery mode, and childcare. Mean number of reads was lower in infants with CF compared to controls, which could be due to higher exposure to antibiotics (Table 1, Supplementary Fig. 2 and Supplementary Table 3). However, infants with CF did not receive prophylactic antibiotic treatment.
Comparison of the nasal microbiota between infants with CF and controls
β-diversity, measured by Bray-Curtis dissimilarity, was higher in infants with CF compared to controls (Table 2, Supplementary Fig. 3). This difference was not present if only antibiotic naïve swabs were considered or before the first RTI was reported (Table 2), but could only be detected after the first antibiotic treatment (e.g., treatment of asymptomatic bacterial colonization in CF infants), first RTI (prior first antibiotic treatment) or both (Table 2, Supplementary Fig. 4). The R2-values from the PERMANOVA analyses were low overall despite being statistically significant, likely due to the large intra-group variation within the nasal microbiota compositions of both CF and control groups.
Infants with CF had less stable bacterial communities than controls, reflected by higher subsequent within-subject dissimilarities (Fig. 2, Supplementary Table 4). Again, this difference was not present before the first antibiotic treatment or before the first respiratory symptoms were reported (Fig. 2, Supplementary Table 4). We did not assess within-subject dissimilarity in even smaller subgroups (in contrast to the other diversity measures) because the number of consecutive taken samples of the same infant in subgroups was too low.
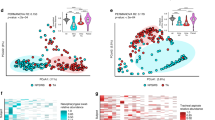
a The median within-subject dissimilarities were calculated using the Bray-Curtis dissimilarity matrix for each two consecutive swabs from all infants. These dissimilarities were analyzed with a generalized additive mixed model, adjusting for confounders. The corrected within-subject dissimilarity values are shown as points on the y-axis, while the x-axis represents the age of the infants in weeks. Trendlines (loess function) are depicted in red for infants with CF (707 consecutive swabs from 50 infants) and in green for controls (442 consecutive swabs from 30 infants). Shaded areas indicate the 95% confidence interval for the trendlines. b The same analysis was conducted for consecutive swabs taken before the first antibiotic treatment. Data for infants with CF (397 consecutive swabs from 42 infants) are shown in red, and data for controls (426 consecutive swabs from 30 infants) are shown in green.
Differences in α-diversity between infants with CF and controls could only be reported after first antibiotic treatment (higher α-diversity in CF measured by SDI) (Table 3). “Baseline” α-diversity at the beginning of life or α-diversity after first RTI (without antibiotic treatment) did not differ between the two groups (Table 3).
In microbiota differential abundance analysis, results of the most abundant bacterial families are displayed in Fig. 3 and Supplementary Table 5. Taxa annotation on genus level was not possible for all samples (Supplementary Fig. 5). In infants with CF, we detected overall more Staphylococcaceae, Propionibacteriaceae and more Micrococcaceae, less Carnobacteriaceae and less Moraxellaceae than in controls (Fig. 3a, b, Supplementary Table 5). In controls, we observed the typical shift towards a more Moraxellaceae/Carnobacteriaceae (represented by Dolosigranulum) dominated profile around age four to 5 months, while in the profile of infants with CF Staphylococcaceae dominated. When specifically examining swabs taken before the first antibiotic treatment (CF = 539, controls = 559), consistent findings were observed for Staphylococcaceae, Micrococcaceae, and Carnobacteriaceae. This emphasizes that these differences were not confounded by antibiotic treatment (Supplementary Table 5). Staphylococcaceae and Carnobacteriaceae already differed before the first RTI (CF = 435, controls = 357 swabs) (Supplementary Table 5, Fig. 3c, d). After the first antibiotic therapy alone (before the first RTI) (CF = 75, controls = 357 swabs), we observed the same result as a trend. After the first RTI but before the first antibiotic treatment, we detected more Staphylococcaceae in infants with CF (Supplementary Table 5).
a Relative abundance of the 9 most abundant families and “others” are displayed in alphabetical order and by age (months) for infants with CF (933 swabs from 50 infants) and controls (578 swabs from 30 infants). b Swabs of infants with CF (787 swabs from 42 infants) and controls (409 swabs from 21 infants) who experienced a RTI in first year of life were divided in swabs taken before and after the first RTI. c, d Results obtained from differential abundance analysis with MaAslin2 are plotted with effect size on the x-axes and FDR adjusted p-values (q-values) on the y-axes. The dashed horizontal line shows significance threshold (q = 0.05). We display differential abundance analysis (c) for all swabs (933 swabs from 50 infants with CF and 578 from 30 controls) and (d) for swabs only before the first RTI occurred (435 swabs from 48 infants with CF and 357 swabs from 30 controls).
Investigation of the interrelations between early-life nasal microbiota and number of RTIs, first RTI, and first antibiotic treatment in infants with CF
β-diversity did not differ between CF infants with a higher or lower number of RTIs in the first year of life, before the first antibiotic treatment, or before the first RTI occurred (results not shown). Microbiota communities were more stable in infants with a higher number of RTIs, reflected by lower within-subject dissimilarities between consecutive time-points (Table 4, Supplementary Fig. 6). This difference was already present in antibiotic naïve infants and before the first RTI (Table 4).
Infants with CF with a higher number of RTIs had a lower α-diversity measured by Shannon-diversity index compared to infants with a lower number of RTIs. Again, this difference was present before the first antibiotic treatment and before the first RTI occurred (Table 5, Fig. 4).
Shannon-diversity index (SDI) was compared between infants with CF with higher (300 swabs from 15 infants) and lower number (487 swabs from 27 infants) of RTIs in a generalized additive mixed model. Points display the model fitted SDI values on the y-axis, and (a) time to first RTI, or (b) time to first antibiotic treatment on the x-axis. Swabs taken before the first RTI (a), or antibiotic treatment (b), are displayed in red, and swabs taken after in green. The magenta trendline (loess function) reflects infants with a lower number of RTIs and the blue trendline displays infants with a higher number of RTIs in first year of life. The gray shadings show the 95% confidence intervals.
In infants with a higher number of RTIs, we observed reduced levels of Neisseriaceae (coefficient −1.959, q = 0.031) and Propionibacteriaceae (coefficient −1.49, q = 0.049), even prior to the occurrence of the first RTI (Neisseriaceae (coefficient −2.34, q = 0.048), Propionibacteriaceae (coefficient −2.12, q = 0.048)). Before the first antibiotic treatment, we observed this as a trend: Neisseriaceae (coefficient −2.03, q = 0.15), Propionibacteriaceae (coefficient −2.05, q = 0.068).
Effect of a “first hit” on the nasal microbiota in infants with CF
Ongoing antibiotic treatment at the time of nasal swab collection was associated with higher α-diversity (Coef 0.286; 95% CI 0.059, 0.512, p = 0.014). To investigate the effect of the first antibiotic treatment, infants with CF with at least one course of antibiotics (n = 33) were included. ß-diversity was higher after the first antibiotic treatment (PERMANOVA, R2 = 0.005, p-adj = 0.007). In addition, α-diversity increased after the first antibiotic treatment (Coef 0.396; 95% CI 0.140, 0.651; p = 0.0027) (Fig. 5). We did not analyze within-subject dissimilarity and possible further changes in the microbiota after a second course of antibiotics due to low number of samples. Low abundant bacterial families (“others”) (Coef 1.259, q = 0.000032), as well as Propionibacteriaceae (Coef 1.051, q = 0.024) and Neisseriaceae (Coef 1.259, q = 0.024) increased after the first antibiotic treatment. There was no association between the number of antibiotic treatments in first year of life and ß-diversity, within sample dissimilarity, α-diversity, or differential abundances (results not shown). However, there were only few infants with more than two courses of antibiotic therapy in our cohort.
Shannon-diversity index (SDI) was compared in a generalized additive mixed model before and after the first antibiotic treatment (635 swabs from 33 infants). Displayed are the model fitted SDI values on the y-axis and the time to first antibiotic treatment on the x-axis. Swabs taken before the first antibiotic treatment (n = 241) are displayed in red and after in green (n = 394). The trendlines (loess function) are displayed with 95% confidence intervals (gray shadings).
After the first RTI, β-diversity increased in infants with CF (PERMANOVA, p < 0.001, R2 = 0.009), reflecting a larger heterogeneity of the microbiota profiles in the CF group after the first infection. Again, R2-values were low, which points towards high variability within the CF group. Within-subject dissimilarity did not change after the occurrence of the first RTI. α- diversity also did not differ after the first RTI in infants with CF (Coef 0.001; 95% CI −0.267, 0.269; p = 0.994). In differential abundance analysis, we found an increase in abundance of Neisseriaceae (Coef 1.3, q = 0.039) after the first RTI.
Discussion
In this prospective cohort study, we investigated the nasal respiratory microbiota over the first year of life in infants with CF and controls and examined interrelations with antibiotic therapy and RTIs.
Microbiota differences between infants with CF and controls in our study are in line with previous results from studies of upper and lower respiratory samples8,17,18,26,27,28. Importantly, differences in diversity indices first occurred after initial RTI and/or antibiotic treatment. This suggests, that the microbiota community destabilizes in infants with CF following first antibiotic therapy and/or RTI compared to control individuals. This can be attributed to the event itself combined with a higher vulnerability of the CF microbiota structure, which was observed in a metagenome network analysis28. Prevaes et al. report a different β-diversity in the first 3 months of life in infants with CF compared to controls18, too. Our study adds that this difference is not only an effect of progressing disease, but due to antibiotic treatments.
Distinct bacterial families between young CF and control infants were present in antibiotic naïve samples and prior respiratory infections also indicating a “CF specific microbiota”, e.g., we demonstrated a higher abundance of Staphylococcaceae and a lower abundance of Carnobacteriaceae, in line with previous studies8,17,18. High and early abundance of Staphylococcus aureus is associated with worse respiratory outcomes even in clinically stable infants with CF, strain persistence and unbeneficial adaption29,30,31. Sequencing techniques with a higher resolution (like whole metagenome sequencing) are required to distinguish whether Staphylococcaceae dominance reflects early strain persistence of an opportunist and potential pathogen (Staphylococcus aureus)32 or whether it reflects a different composition of commensal bacteria among infants with CF in this cohort.
We report a lower α-diversity in CF infants with a higher number of RTIs, and more stable microbiota communities reflected by lower within-subject dissimilarities between consecutive time-points. The latter seems counterintuitive, but could be explained by a bacterial overgrow of certain (disadvantageous) bacterial species, which already persist in the nasal cavity in infants with CF. This might differ to individuals without CF, Bosch et al. show that bacterial community stability is lower in infants with higher number of RTIs within the first year of life6. Ahmed et al.26 report changes in β-diversity of the oropharyngeal microbiota with increasing age in CF infants, but did not report respiratory symptoms, and the increasing cumulative antibiotic dose in the first year of life, is likely to be associated with changing β-diversity.
Next to the (expected) microbial alterations through respiratory infections and/or antibiotics, in our study, a lower α-diversity (importantly: prior first RTI and antibiotic application) is associated with a higher number of subsequent RTIs in infants with CF independent from antibiotic therapy. Thus, although different additional factors contribute to disease development, certain microbiota profiles in CF infants likely predispose to respiratory disease or reflect an underlying airway biology that is followed by higher number of RTIs and antibiotic treatments. Studies in adult pwCF show an inverse relationship between microbiota diversity and disease progression, in line with our finding13,27. However, the association between higher microbiota biodiversity and beneficial health outcomes is not always consistent in humans33,34,35,36. Otitis media37,38 or chronic rhinosinusitis39 are associated with decreased α-diversity. In other studies, an increased α-diversity has been associated with disease (e.g., in elderly pneumonia patients even before antibiotic therapy)40. In CF infants, differing antibiotic treatment schemes throughout the world might further explain different outcomes for α-diversity41. A recently published longitudinal study of oropharyngeal samples of infants with CF42 observes higher numbers of distinct communities (leading to higher α-diversity) with higher cumulative antibiotic exposure in days at time of sample collection in line with our data. In contrast, this study reports a mildly decreased α-diversity in antibiotic treated infants compared to antibiotic naïve individuals at age 9 months42. An opening of microbial niche spaces with elimination of single opportunistic bacteria by antibiotics might explain our findings, e.g., a rise in gram-negative bacteria after antibiotics in CF infants has been reported18, or transient environmental species may increase after initial antibiotic treatment. Thus, an increase in α-diversity cannot not necessarily be considered as an improvement. In line, we do not observe an increase in commensal bacteria that play a key role for respiratory health (like Carnobacteriaceae or Corynebacteriaceae)3,7.
A strength of our study is the prospective study design that allowed structured collection of high-quality longitudinal data. We provide one of the largest longitudinal datasets of nasal microbiota swabs and clinical data (including respiratory symptoms and antibiotic treatment) to date. We include infants without CF from an unselected birth cohort as control subjects, who followed the same study protocol. Our data allowed us to study microbiota dynamics before and after important respiratory events (RTIs, antibiotic treatment), which is unique in infants with CF.
As limitation, we did not assess viral colonization5. Furthermore, as CF is not diagnosed at birth, information on the neonatal period was obtained retrospectively. RTI diagnoses were based on clinical assessment utilizing a previously validated respiratory symptom score (details are reported in the Supplementary Methods)43,44. Additional information (e.g., further diagnostics or treatment modalities during respiratory diseases in the infants CF study center) is not assessed systematically in our study. Fortunately, infants with CF rarely present with very severe respiratory tract symptoms (e.g., leading to hospitalizations) in Switzerland. In addition, RTIs and antibiotic treatments occur together in infants with CF and thus, it is difficult to disentangle the effects of each individually. However, the last two points are general limitations for studies among pwCF and cannot be improved by study design.
Our data raise several questions concerning the reflexive use of antibiotics in infants with CF. Each respiratory event in infants with CF contributes to airway dysbiosis, which is subsequently used to justify antibiotic treatment. Perhaps a more deliberate and selective approach to antibiotic use could help in delaying or slowing the inevitable changes in microbial community structure, leading to potentially better long-term outcomes. We propose that early alterations in nasal microbiota may contribute to increased susceptibility towards RTIs in infants with CF and may exacerbate following RTIs and antibiotic treatment. However, it is also plausible that susceptibility to respiratory events in infants is primarily driven by underlying airway biology, with consistent and aggressive antibiotic responses exacerbating dysbiosis. Could we consider dysbiosis as a marker for intervention? These questions gain growing importance in times of CFTR modulator use.
In the future, novel modulator therapies if started in infancy or early childhood are very likely to substantially decrease the frequency of exacerbations45,46,47 and thus reduce antibiotic treatment. However, CFTR modulators are not (yet) available for infants and still not applicable for 10-15% of pwCF48. In light of potential improvements through early triple modulator therapy initiation, it is even more important to understand early microbiota development to prevent early lung damage. It becomes crucial to exercise caution in treatment strategies and prioritize antimicrobial stewardship. It might be promising to treat infants with CF with probiotics to prevent destabilization of the upper airway microbiota. Some studies show promising effects of enteric probiotic therapies on clinical outcomes in individuals with CF between 2 and 44 years of age49, resulting in less frequent pulmonary exacerbations. Whether enteric probiotics influence the respiratory microbiota in CF has not been investigated so far. For a better understanding, large randomized controlled trials in infants with CF are required to study the effect of probiotic therapies on the respiratory microbiota and future RTIs and exacerbations. Metagenome analyses could provide further insights into strain adaption, persistence processes in infants with CF, and metabolomics might shed light into the mechanisms that underlie the influence of respiratory microbiota alterations on respiratory health.
In conclusion, we could show that the nasal microbiota is already altered before the first RTI or antibiotic treatment in infants with CF. It might predispose to a higher number of subsequent RTIs and is not (only) a consequence of recurrent RTIs or of antibiotic treatment in infants with CF. In the future, targeting the nasal microbiota might thus be an attractive option in CF-related RTI management.
Data availability
Sequencing data generated during this study have been stored accessible for the public in the NCBI bioproject repository (https://www.ncbi.nlm.nih.gov/bioproject/ with the accession code “PRJNA1019921”). Study participant and further metadata is available upon reasonable request to protect participant confidentiality. Requests can be sent to Insa Korten, insa.korten@insel.ch. The numerical data used to plot Fig. 3–5 (source data) can be found in Supplementary Data 1. The numerical data used to plot Fig. 2 (source data) can be found in Supplementary Data 2.
Code availability
Code used for the analysis of this data is available at https://github.com/steinbaer/microbiome-16S/. No novel code was developed for the analysis of this study.
References
Teo, S. M. et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 17, 704–715 (2015).
Neumann, R. P. et al. Nasal microbiota and symptom persistence in acute respiratory tract infections in infants. ERJ Open Res. 4, 00066–02018 (2018).
Man, W. H. et al. Bacterial and viral respiratory tract microbiota and host characteristics in children with lower respiratory tract infections: a matched case-control study. Lancet Respir. Med. 7, 417–426 (2019).
Hasegawa, K., Camargo, C. A. Jr. & Mansbach, J. M. Role of nasal microbiota and host response in infants with respiratory syncytial virus infection: Causal questions about respiratory outcomes. J. Allergy. Clin. Immunol. 149, 898–900 (2021).
de Steenhuijsen Piters, W. A. A. et al. Early-life viral infections are associated with disadvantageous immune and microbiota profiles and recurrent respiratory infections. Nat. Microbiol. 7, 224–237 (2022).
Bosch, A. et al. Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. a prospective cohort study. Am. J. Respir. Crit. Care Med. 196, 1582–1590 (2017).
Biesbroek, G. et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am. J. Respir. Crit. Care Med. 190, 1283–1292 (2014).
Frayman, K. B. et al. Differences in the lower airway microbiota of infants with and without cystic fibrosis. J. Cyst. Fibros. 18, 646–652 (2019).
Ratjen, F. et al. Cystic fibrosis. Nat. Rev. Dis. Prim. 1, 15010 (2015).
Gibson, R. L., Burns, J. L. & Ramsey, B. W. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 168, 918–951 (2003).
Harrison, F. Microbial ecology of the cystic fibrosis lung. Microbiology 153, 917–923 (2007).
Armbruster, C. R., Coenye, T., Touqui, L. & Bomberger, J. M. Interplay between host-microbe and microbe-microbe interactions in cystic fibrosis. J. Cyst. Fibros. 19(Suppl 1), S47–S53 (2020).
Cuthbertson, L. et al. Lung function and microbiota diversity in cystic fibrosis. Microbiome 8, 45 (2020).
Surette, M. G. The cystic fibrosis lung microbiome. Ann. Am. Thorac. Soc. 11(Suppl 1), S61–S65 (2014).
Thornton, C. S., Acosta, N., Surette, M. G. & Parkins, M. D. Exploring the cystic fibrosis lung microbiome: making the most of a sticky situation. J. Pediatr. Infect. Dis. Soc. 11, S13–s22 (2022).
Sibley, C. D., Rabin, H. & Surette, M. G. Cystic fibrosis: a polymicrobial infectious disease. Future Microbiol. 1, 53–61 (2006).
Mika, M. et al. The nasal microbiota in infants with cystic fibrosis in the first year of life: a prospective cohort study. Lancet Respir. Med. 4, 627–635 (2016).
Prevaes, S. M. et al. Development of the nasopharyngeal microbiota in infants with cystic fibrosis. Am. J. Respir. Crit. Care Med. 193, 504–515 (2016).
Korten, I. et al. The Swiss Cystic Fibrosis Infant Lung Development (SCILD) cohort. Swiss Med Wkly 148, w14618 (2018).
Fuchs, O., Latzin, P., Kuehni, C. E. & Frey, U. Cohort profile: the Bern infant lung development cohort. Int J. Epidemiol. 41, 366–376 (2012).
Korten, I. et al. Respiratory symptoms do not reflect functional impairment in early CF lung disease. J. Cyst. Fibros. 20, 957–964 (2021).
Gisler, A. et al. Associations of air pollution and greenness with the nasal microbiota of healthy infants: a longitudinal study. Environ. Res 202, 111633 (2021).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Davis, N. M., Proctor, D. M., Holmes, S. P., Relman, D. A. & Callahan, B. J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 226 (2018).
Mallick, H. et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput. Biol. 17, e1009442 (2021).
Ahmed, B. et al. Longitudinal development of the airway microbiota in infants with cystic fibrosis. Sci. Rep. 9, 5143 (2019).
O’Connor, J. B. et al. Divergence of bacterial communities in the lower airways of CF patients in early childhood. PLoS ONE 16, e0257838 (2021).
Pust, M. M. et al. The human respiratory tract microbial community structures in healthy and cystic fibrosis infants. NPJ Biofilms Microbiomes 6, 61 (2020).
Pillarisetti, N. et al. Infection, inflammation, and lung function decline in infants with cystic fibrosis. Am. J. Respir. Crit. Care Med. 184, 75–81 (2011).
Kahl, B. C. Impact of Staphylococcus aureus on the pathogenesis of chronic cystic fibrosis lung disease. Int. J. Med. Microbiol. 300, 514–519 (2010).
Hirschhausen, N. et al. Extended Staphylococcus aureus persistence in cystic fibrosis is associated with bacterial adaptation. Int. J. Med. Microbiol. 303, 685–692 (2013).
Long, D. R. et al. Polyclonality, shared strains, and convergent evolution in chronic cystic fibrosis Staphylococcus aureus airway infection. Am. J. Respir. Crit. Care Med. 203, 1127–1137 (2021).
Gilbert, J. A. et al. Current understanding of the human microbiome. Nat. Med. 24, 392–400 (2018).
Huttenhower, C., Kostic, A. D. & Xavier, R. J. Inflammatory bowel disease as a model for translating the microbiome. Immunity 40, 843–854 (2014).
DiGiulio, D. B., Stevenson, D. K., Shaw, G., Lyell, D. J. & Relman, D. A. Reply to Keelan and Payne: microbiota-related pathways for preterm birth. Proc. Natl. Acad. Sci. USA 112, E6415 (2015).
Fredricks, D. N., Fiedler, T. L. & Marrazzo, J. M. Molecular identification of bacteria associated with bacterial vaginosis. N. Engl. J. Med. 353, 1899–1911 (2005).
Pettigrew, M. M. et al. Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl. Environ. Microbiol. 78, 6262–6270 (2012).
Hilty, M. et al. Nasopharyngeal microbiota in infants with acute otitis media. J. Infect. Dis. 205, 1048–1055 (2012).
Man, W. H., de Steenhuijsen Piters, W. A. & Bogaert, D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat. Rev. Microbiol. 15, 259–270 (2017).
de Steenhuijsen Piters, W. A. et al. Dysbiosis of upper respiratory tract microbiota in elderly pneumonia patients. ISME J. 10, 97–108 (2016).
Pittman, J. E. et al. Association of antibiotics, airway microbiome, and inflammation in infants with cystic fibrosis. Ann. Am. Thorac. Soc. 14, 1548–1555 (2017).
Harris, J. K. et al. Upper airway microbiota development in infants with cystic fibrosis diagnosed by newborn screen. J. Cyst. Fibros. 22, 644–651 (2023).
Silverman, M., Wang, M., Hunter, G. & Taub, N. Episodic viral wheeze in preschool children: effect of topical nasal corticosteroid prophylaxis. Thorax 58, 431–434 (2003).
Latzin, P. et al. Prospectively assessed incidence, severity, and determinants of respiratory symptoms in the first year of life. Pediatr. Pulmonol. 42, 41–50 (2007).
Streibel, C. et al. Effects of elexacaftor/tezacaftor/ivacaftor therapy in children with cystic fibrosis—a comprehensive assessment using lung clearance index, spirometry, and functional and structural lung MRI. J. Cyst. Fibros. 22, 615–622 (2023).
Nichols, D. P. et al. Clinical effectiveness of elexacaftor/tezacaftor/ivacaftor in people with cystic fibrosis: a clinical trial. Am. J. Respir. Crit. Care Med. 205, 529–539 (2022).
Middleton, P. G. & Taylor-Cousar, J. L. Development of elexacaftor—tezacaftor—ivacaftor: highly effective CFTR modulation for the majority of people with Cystic Fibrosis. Expert Rev. Respir. Med. 15, 723–735 (2021).
Allen, L. et al. Future therapies for cystic fibrosis. Nat. Commun. 14, 693 (2023).
Anderson, J. L., Miles, C. & Tierney, A. C. Effect of probiotics on respiratory, gastrointestinal and nutritional outcomes in patients with cystic fibrosis: a systematic review. J. Cyst. Fibros. 16, 186–197 (2017).
Acknowledgements
We thank all parents and study participants of the SCILD and BILD study cohorts. We thank Fabienne Furrer, Sandra Lüscher, Katrin Hug, and Natalie Bürgi for their contribution to data collection. K.A.R. obtained funding from the Swiss National Science Foundation (SNF 168173). U.F. and R.S. (mobility grant) obtained funding from the Swiss National Science Foundation (SNF 182871/1). R.S. and I.K. obtained funding from Cystic Fibrosis Switzerland (CFS). M.H. was supported by grants from the Research Fund of the Swiss Lung Association, the “Stiftung Lindenhof Bern” and CFS.
Author information
Authors and Affiliations
Consortia
Contributions
R.S., I.K., P.L., and K.A.R. designed the study. R.S., I.K., E.K., B.F., C.C., J.U., A.M., D.T., I.R., S.B., D.M.S., B.K., U.F., and M.Z. involved in data acquisition. R.S. and I.K. collected clinical and metadata. M.H. and I.K. were responsible for biological samples and amplicon sequencing. N.M. and R.S. performed bioinformatics analyses. R.S. and I.K. performed and interpreted statistics. R.S., I.K., P.L., and M.H. were involved in interpretation of the results. R.S. and I.K. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Medicine thanks Donald R VanDevanter, Christopher van der Gast and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Steinberg, R., Mostacci, N., Kieninger, E. et al. Early nasal microbiota and subsequent respiratory tract infections in infants with cystic fibrosis. Commun Med 4, 246 (2024). https://doi.org/10.1038/s43856-024-00616-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s43856-024-00616-6