Abstract
Background
Age and sex significantly impact DNA methylation patterns, however, existing datasets typically include only a subset of methylation sites in the human genome, hindering our thorough understanding.
Methods
We recruited 98 generally healthy adults aged from 22 to 77 and investigated the effects of age and sex on plasma cell-free DNA (cfDNA) methylation through whole-genome bisulfite sequencing (WGBS) and association analysis.
Results
Here we show 3,047 age-associated and 1,053 sex-associated CpGs on autosomes, corresponding to 1,587 and 324 genes, respectively. To the best of our knowledge, many of these CpGs are newly discovered to be age- and sex-related at the DNA methylation level. The discovered sex-differential cfDNA methylation patterns on the X chromosome are related to XCI status. Besides, a cfDNA epigenetic clock comprising 125 CpGs is developed, demonstrating relatively high accuracy in predicting chronological age. Tissue-of-origin analysis reveals that cfDNA derived from monocytes/macrophages, granulocytes, and hepatocytes is associated with age and sex. Several individuals with abnormal cfDNA proportions of some specific cell types are found to have individual health problems.
Conclusions
Our discovered CpGs and genes help to explain age-related and sex-biased diseases such as psychiatric disorders, diabetes, and autoimmune diseases, and we demonstrate the potential of cfDNA methylation signatures as very promising biomarkers for health monitoring for the general population.
Plain language summary
Aging is a complex biological process shaping disease risks. Meanwhile, the influence of sex differences is still underexplored. There are various influencing factors, including modifications on DNA called DNA methylation which are important for gene regulations. In this study, we explore the age-related changes and sex-related differences in DNA methylation in blood samples, by recruiting 98 generally healthy adults aged from 22 to 77. Based on DNA methylation sequencing, we identify two sets of genes that help to explain some age-related and sex-biased diseases. We also develop a methylation aging clock for estimating biological age. Moreover, we demonstrate the potential of methylation signatures as very promising biomarkers for health monitoring, as they contain health risk information from immune cells and tissue injuries.
Similar content being viewed by others
Introduction
In humans and other mammals, the epigenetic modification of DNA methylation (DNAm) predominantly occurs at cytosine-phosphate-guanine (CpG) dinucleotide sites, and it is essential for regulating gene expression, growth, development, and disease1. Age-associated methylation alterations are related to diseases, such as osteoporosis, neurodegenerative disease, diabetes, and cancers2,3. One way to assess age-related disease risks is to measure biological age by epigenetic clocks4. In addition to age, sex also has a strong impact on methylation variations. The sex-associated methylation patterns contribute to sex-biases diseases, such as liver diseases5, autoimmune diseases6, and neurological disorders7. Sex-associated methylation patterns are observed in autosomes8 and the X chromosome9. In fact, DNA methylation is one of the key mechanisms in regulating X-chromosome inactivation (XCI) in females, for balancing X-lined gene dosage between the sexes10. Some genes that escape from XCI may lead to the dual expression dosage and contribute to the female bias in diseases, such as autoimmune diseases6. Since age- and sex-associated DNA methylation plays important regulatory roles in aging and diseases, it is worth in-depth studies.
Various detection methods have been developed to investigate human DNA methylome, with different coverages and resolutions. The most widely used Illumina 450 K11 and 850 K12 arrays cover approximately 450,000 and 850,000 CpGs, respectively, featuring on CpG islands and other regulatory regions13. However, these methylation arrays represent only a very small fraction (~3%) of the 28 million CpG sites in the human genome. Most studies on DNA methylation on aging and sex adopted these methylation arrays, which led to the remaining CpGs in the human genome being less known about their influence.
Moreover, methylation patterns are known to be tissue- and cell-specific14,15. The plasma cfDNA is derived from cellular processes such as apoptosis, necrosis, and active secretion16, which enables non-invasive and real-time monitoring of physiological and pathological conditions in human bodies. The deconvolution algorithms based on the DNA methylation atlas of human tissues/cell types have been used to estimate the origins of tissues/cell types of cfDNA17,18. However, previous cfDNA research has predominantly focused on disease-specific cfDNA profiles and tissue-of-origin results19,20, and less attention has been paid to the age- and sex-related cfDNA methylation profiles. Teo et al. studied on cfDNA nucleosome signals as an aging biomarker21. Shtumpf et al. constructed aging clocks based on cfDNA fragment sizes and nucleosome distances22. Li et al. utilized plasma samples from three age groups of healthy individuals to identify age-related CpGs, and they developed a cfDNA methylation age prediction model, however, they only utilized the CpG sites covered by 450 K array in their model23. Additionally, the sex-differential DNA methylation studies primarily relied on whole blood and tissue samples, and were often based on 450 K and 850 K arrays9,24,25,26.
In this study, we conduct a comprehensive profiling of CpGs methylation characteristics related to age and sex at the whole-genome level, using plasma cell-free DNA whole-genome bisulfite sequencing (WGBS) data from 98 generally healthy adults (52 females and 46 males) aged from 22 to 77. A cfDNA epigenetic clock based on 125 CpG sites is developed and validated by an independent dataset. Furthermore, we find some age and sex differences in the tissue and cell-type origins of plasma cfDNA. The detection of individuals with abnormal tissue-of-origin results show the potential of cfDNA as a biomarker for health monitoring. Our findings provide a foundation for future research on human cfDNA methylation and liquid biopsies.
Methods
Sample collection and ethics statement
This study was approved by the Institutional Review Board on Bioethics and Biosafety of BGI (BGI-IRB 21157-T2). From December 2021 to December 2022, adult participants were recruited during their physical examination, including young, middle-aged, and elderly individuals, with an almost equal number of males and females in all age groups (Fig.1a). Women who were during pregnancy or lactation; and anyone who had fever symptoms, recently had a surgical procedure, or was diagnosed with infectious diseases, cancer, or other severe diseases were all excluded from this study. A total of 98 participants (52 females and 46 males) aged from 22 to 77 were included in this study. There were about 10 males and 10 females in each 10-year age group (Fig.1a). We collected their basic information about their sex, age, and past medical history through a questionnaire. Their physical examination results were also collected. For each participant, a peripheral blood sample was collected using an EDTA blood collection tube. The study was conducted in accordance with the Declaration of Helsinki and informed consent was signed by all participants. All data were de-identified prior to analysis by removing direct identifiers such as names and medical record numbers. Each participant was assigned a unique study code for research use and to protect privacy. Written informed consent was obtained from all participants for the publication of the de-identified data.
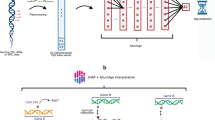
a The recruitment of participants and the workflow of whole genome bisulfite sequencing (WGBS) of plasma cfDNA. The library preparation method is based on a single-stranded library preparation technique. b Identification of age- and sex- associated CpGs on autosomes using the method of linear regression. The related genes are further analyzed based on the rank of the number of CpGs and the enriched pathways. c Sex difference analysis of methylation patterns on the X chromosome. d Tissue-of-origin analysis of cfDNA and exploration of the influences of age, sex, and individual variances. (Fig. 1 is created with Biorender.com).
Cell-free DNA extraction
Plasma isolation was performed via a two-step centrifugation procedure within 4 h after blood sampling. In the first step, the blood was centrifuged at 1600 g for 10 min at 4 °C. In the second step, the upper layer of plasma was centrifuged again at 16,000 g for 10 min at 4 °C to remove cellular debris. The resultant supernatant plasma was then stored at −80 °C before cfDNA extraction. For each sample, 0.5–1 mL plasma was used for cfDNA extraction using MagPure Circulating DNA KF Kit (Magen, China) according to the manufacturer’s instructions.
Library preparation for WGBS
The input cfDNA amount for the library preparation was 15.23 ± 6.64 ng (mean ± SD). The extracted cfDNA was bisulfite treated and purified using EZ-96 DNA Methylation Kit (Zymo Research). To evaluate bisulfite conversion efficiency, 0.05 ng of lambda DNA (New England Biolabs, #N3011S) was added to each reaction as an unmethylated control before bisulfite treatment. Subsequently, WGBS sequencing libraries were prepared utilizing a modified single-stranded library preparation method of the SPlinted Ligation Adapter Tagging (SPLAT)27,28,29. Briefly, the double-stranded DNA (dsDNA) was denatured into single-stranded DNA (ssDNA) at high temperatures; then adapters containing six random bases were annealed and ligated to both ends of ssDNA; finally, the ligation product was amplified through PCR and the barcode sequences of samples were introduced through PCR primers. Notably, the processes include the utilization of a single-stranded DNA binding protein (ET SSB) to stabilize the presence of ssDNA in solution28. A one-step adapter ligation reaction was used and the adapters were specifically tailored for the DNBSEQ platform (MGI)30. Subsequently, the libraries were subjected to 100-bp paired-end (PE) sequencing using the DNBSEQ platform with a sequencing depth of >30× for each sample.
WGBS data processing
We used Fastp (0.19.5)31 to process the raw sequencing data with default parameters, including trimming adapters, filtering out reads of low quality, and discarding reads with a high proportion of undetermined nucleotides (Ns). Subsequently, the pre-processed reads were aligned to the human reference genome (GRCh38.p14) using BitMapperBS (v1.0.2.3)32 with default settings. Following alignment, PCR duplicates were removed using sambamba (v0.8.2)33. After removing the duplicated reads, the median sequencing depth for samples was 31.19× (Supplementary Data 1). The depth calculation is based on sequencing reads and bases. The overlapping paired-end bases were calculated twice in the depth count.
Then we used MethylDackel (v0.5.1) (https://github.com/dpryan79/MethylDackel) to calculate the methylation values at each CpG site. In brief, methylation values were determined by the ratio of methylated C to the total number of reads supporting C (methylated) and T (unmethylated) at this site. Methylation values range from 0 to 1, with 0 indicating no DNA methylation and 1 indicating complete DNA methylation. CpGs with less than 5X coverage were labeled as NA, and those with more than 10% NA values in samples were removed from further analyses. The remaining NA values were imputed with the impute.knn function (using k = 10) in R language. The CpGs located on chromosome Y were not investigated in the study. Ultimately, we obtained a comprehensive DNA methylation profile consisting of 23,510,673 CpG sites on autosomes and 996,907 CpG sites on the X chromosome.
Genomic distribution analysis
Manhattan plots were generated using the R package CMplot (https://github.com/YinLiLin/R-CMplot). The annotate R package34 was utilized for annotating various features such as CpG regions and gene regions. CpG regions including open sea regions, CpG islands, CpG shelves, and CpG shores. CpG shores are defined as 2 Kb upstream/downstream from the ends of the CpG islands, less the CpG islands. CpG shelves are defined as another 2 Kb upstream/downstream of the farthest upstream/downstream limits of the CpG shores, less the CpG islands and CpG shores. The remaining genomic regions comprise the open sea annotation gene regions including 1–5 Kb upstream of the TSS (1to5kb), 3′ untranslated region (3′UTR), 5′ untranslated region (5′UTR), exons, introns, promoters (<1 Kb upstream of the TSS), and enhancers.
Age-associated CpGs analysis
We implemented a linear regression model, glm function (family=gaussian) in R language, with two-tailed t test, to identify age-associated CpG sites (Eq. 1). The p-values were adjusted using the Benjamini–Hochberg (BH) method35,36, and all CpG sites with adjusted p-value (Padj) < 0.05 were defined as age-associated.
The reported age-associated CpGs data were obtained from the EWAS Atlas database, with the traits of aging (https://ngdc.cncb.ac.cn/ewas/browse?traitList=aging).
Development of cfDNA DNAm epigenetic clock
The methylation values of the 3047 age-associated CpGs were standardized using the R language function scale() from the base package. To build the cfDNA methylation epigenetic clock we implemented an elastic net regression model, using the methodology described by Horvath37. The elastic net models were generated using the “glmnet” package in R, using the functions of cv.glmnet and predict.glmnet. The elastic net approach combines Ridge and LASSO regression with an alpha parameter of 0 for Ridge and 1 for LASSO. Here, the elastic net alpha parameter was set to 0.5. The minimal lambda was calculated using 10-fold cross-validation using the “glmnet” package. A transformed version of chronological age was regressed on DNAm levels at all included CpG sites. Given the limited sample size, we used a previously described cross-validation scheme (leave-one-out cross-validation, LOOCV) for determining unbiased estimates of the accuracy of our cfDNA methylation epigenetic clock23. The cross-validation procedure reports the unbiased estimates of age correlation r, which is defined as Pearson correlation between the actual age and the predicted value (the DNAm age), and the median absolute error (MAE).
Sex-associated CpGs analysis
We implemented a linear regression model, glm function (family=gaussian) in R language, with two-tailed t test, to identify sex-associated CpG sites (Eq. 2). Sex was coded as 0 for males and 1 for females. The p-values were adjusted using the Benjamini–Hochberg (BH) method. CpG sites on autosomes with adjusted p-value (Padj) < 0.05 were defined as sex-associated CpGs on autosomes, CpG sites on X chromosome with adjusted p-value (Padj) < 10-6 were defined as sex-associated CpGs on X chromosome.
The reported sex-associated CpGs data were obtained from the EWAS Atlas database, with the traits of gender (https://ngdc.cncb.ac.cn/ewas/browse?traitList=gender).
Enrichment analysis
We performed a negative binomial distribution model to identify genes enriched in age- or sex-associated CpGs, adjusting for gene length and total number of CpG sites per gene. The p-values were calculated and then corrected using the Benjamini-Hochberg (BH) method for multiple testing adjustments.
Functional annotation of enriched Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway was performed by Metascape (http://metascape.org), using hypergeometric test and BH correction with the default parameters.
For tissue enrichment analysis, we used Human Protein Atlas (HPA) database38 and TissueEnrich tools39 to make a tissue-specific gene enrichment of our gene sets (https://tissueenrich.gdcb.iastate.edu/), which used hypergeometric test and BH correction with the default parameters.
cfDNA tissue deconvolution analysis
We used two methods for cfDNA tissues-of-origin profiling for 98 samples based on DNA methylation atlas of human cell types17,18. The first one was developed by Moss et al., which included an extensive analysis of 7890 differentially methylated CpG sites across 25 unique human tissues and cell types17. The second one was constructed by Loyfer et al., enabling the quantification of 39 tissues/cell types18. By using wgbstools (https://github.com/nloyfer/wgbs_tools), we quantify the relative contributions of various cell types to the plasma cfDNA.
Statistics and reproducibility
There was no technical replicates for the WGBS of 98 samples. Our mathematical model for age- and sex-associated CpGs analysis, and the methods for development of cfDNA DNAm epigenetic clock, enrichment analysis, and cfDNA tissue deconvolution analysis were summarized with details in the above method details section. Two-tailed tests were used for the linear regression model. Differences among CpGs patterns in gene regions and CpG regions used chi-square test with BH correction. Wilcoxon rank-sum test (two-tailed) were used for the comparison of tissue/cell-type derived cfDNA proportions of females and males in different age groups. Spearman’s correlation was used to test the relationship between the hepatocytes-derived cfDNA proportion and ALT, AST, GGT, and HDLC.
Results
Study design and cfDNA methylation profiling
A total of 98 generally healthy volunteers, including 52 females and 46 males, are recruited in this study. Their age ranged from 22 to 77 (Fig. 1a and Supplement Data 1). We collect their basic information (age, sex, medical histories) and physical examination results. The peripheral blood samples are collected, and plasma cell-free DNA (cfDNA) is used for WGBS with a sequencing depth of ~30× for each sample (Fig. 1a). The WGBS libraries are prepared based on the SPlinted Ligation Adapter Tagging (SPLAT)27,28,29, the sequencing bias of which is relatively small compared to some conventional library preparation methods(Supplementary Fig. 1, Supplement Data 2). The fragment size distributions and the basic statistics about the overall quality of WGBS are summarized in Supplementary Fig. 1 and Supplementary Data 1 and 2.
In the cfDNA methylation analysis, CpGs with coverage below 5X are filtered out, and those CpGs with over 10% missing values in samples are excluded. After the data quality control, a comprehensive whole-genome DNA methylation profile is established, encompassing 23,510,673 CpGs on autosomes and 996,907 CpGs on the X chromosome, corresponding to about 80% CpGs in the human genome. These qualified CpGs are utilized in subsequent analyses of age- and sex-associated methylation patterns (Fig. 1b–d).
Age-associated DNA methylation patterns
We identify 3047 CpGs on autosomes and one CpG on the X chromosome with their methylation rates significantly associated with age (Padj < 0.05, linear regression) (Supplementary Data 3 and Supplementary Data 4). The visualization of the Manhattan plot is shown in Fig. 2a. For all these CpGs, their Pearson correlation coefficients of methylation rate with age |r | > 0.4 (Supplementary Data 4). The age-associated CpG on the X-chromosome chrX:97837735(GRCh38.p14) resides in the open sea region (>4 kb from CpG islands) and intergenic region (>3 kb from the nearest annotated gene). This locus is not covered by the Illumina 450 K/850 K arrays nor annotated as a regulatory element in enhancer-gene links databases (e.g., EpiMap40, ABC maps41). The specific function of this site cannot be determined. Here, we only study and discuss the autosomal age-associated CpGs.
a Manhattan plot showing the distribution of 3047 age-associated CpGs (Padj < 0.05) across all the autosomes. The linear regression model is used, and the P values for the regression coefficients are derived from two-tailed t-tests. The P values were adjusted using the Benjamini–Hochberg (BH) method. The numbers of age-associated CpGs in 10 Mb bins are shown on the chromosomes at the bottom of the Manhattan plot, with the color bar showing on the right side. The methylation rates of age-associated CpGs are negatively (94%) or positively (6%) correlated with age. Two examples of CpGs (chr10:13527445 and chr5:141040234) are shown in (b, c) with the methylation rate negatively and positively correlated with age, respectively. Methylation rate of the CpG site for each sample is shown as a dot. The red line is the regression line based on the linear regression model (with 95% confidence intervals shown as shaded areas), representing the relationship between the methylation rates and the ages. The P values are adjusted using the Benjamini–Hochberg (BH) method. d Ranking of the 1587 genes based on the number of age-associated CpGs per gene. Among the 62 genes that have more than 5 age-associated CpGs, 16 genes (such as IFT80 and RILPL1) have not previously been reported to be age-related at the DNA methylation level (EWAS Atlas database). These genes are labeled with gene names, and those genes enriched with age-associated CpGs were labeled in red. The top one gene FIGN is previously known to be age-related at the DNA methylation level, and is enriched with age-associated CpGs. e KEGG enrichment analysis of the 1587 age-associated genes using hypergeometric test (one-tailed test). The P values without adjustment are shown here, and the exact P values and adjusted P values are in Supplementary Data 7. f The cfDNA methylation epigenetic clock with 125 CpGs. The plot shows the relationship between chronological age and epigenetic age. The blue line is the regression line of epigenetic age on chronological age. Pearson’s correlation coefficients (r) and median absolute error (MAE) are denoted. Leave-one-out cross-validation (LOOCV) is used to determine the accuracy of the cfDNA methylation epigenetic clock.
Of the 3047 age-associated CpGs, 2854 CpGs exhibit negative correlations (Fig. 2b) and 193 CpGs show positive correlations with age (Fig. 2c) (Supplementary Data 4). In other words, for most of the age-associated CpGs, the methylation levels tend to decrease with age. This result is consistent with previous findings obtained through microarray analyses42,43. We further explore the genomic distributions of those CpGs in relation to their nearest genes or CpG islands. The age-associated CpGs are enriched in promoters, 5’ UTRs, enhancers, CpG islands, and CpG shores, while they are underrepresented in 3’ UTRs, introns, and open sea (Supplementary Fig. 2a, b and Supplementary Data 5).
On the one hand, we find that only 71 and 208 age-associated CpGs in this study are covered by the 450 K and 850 K arrays, respectively (Supplementary Data 4). By comparing against the EWAS Atlas database, we find that 55 of the 71 CpGs in the 450 K array and 134 of the 208 CpGs in the 850 K array are annotated as age-associated CpGs in the database (Supplementary Data 4). The consistency of age-associated CpG sites with those discovered by microarray assays indicates the reliability of our results. On the other hand, due to the limited number of CpGs coverage by the 450 K and 850 K arrays, the remaining age-associated CpGs in our study have not yet been reported as age-associated CpGs in the EWAS database. We believe our identified age-associated CpGs provide a valuable resource for future studies to elucidate epigenetic regulations in aging.
These 3047 age-associated CpGs are mapped to 1587 genes (Fig. 2d and Supplementary Data 6). The numbers of age-associated CpGs in these genes vary widely. 1168 genes (73%) contain only one age-associated CpG, 357 genes (23%) contain 2 to 4 age-associated CpGs, and 62 genes (4%) contain 5 or more age-associated CpGs. The FIGN gene has the highest number of age-associated CpGs, which is 58. Two of these 58 CpGs (cg15148145 and cg16532938) are covered in the 850 K arrays and annotated as age-related in the EWAS database (Supplementary Data 4). Among the 62 genes that had more than 5 age-associated CpGs, 46 genes are annotated as age-related in the EWAS database; while the reamaining16 genes, such as IFT80 and RILPL1, have not been previously found to be age-related at the DNA methylation level (Fig. 2d, Supplementary Data 4). IFT80 negatively regulates osteoclast differentiation44, and knock-out of IFT80 in the mouse model caused osteoporosis phenotype45,46. Considering the regulation effects of IFT80 on the osteoclast, we speculate that osteoporosis in older adults is influenced by age-associated methylation changes. Additionally, recent research has identified the relationship between RILPL1 and oculopharyngodistal myopathy (OPDM)47, a rare adult-onset hereditary muscle disease with symptoms progressively worsening with age48. The discovery that methylation levels of CpGs in RILPL1 decline with age may give some clues to explain the pathogenesis and progression of OPDM.
We also apply a negative binomial regression model to identify genes enriched in age-associated CpGs (Fig. 2d and Supplementary Data 6), adjusting for gene length and total number of CpG sites per gene. We identify 57 genes significantly associated with age, and 19 genes (e.g., FIGN, TENM2) have been previously reported in aging-related studies (Supplementary Data 4). The remaining genes that are not enriched in age-associated CpGs, or with small numbers of age-associated CpGs might still play important roles in aging. For example, the genes of NEFL, NELL1, and PDGFC contain only one age-associated CpG (Supplementary Data 4). They were already known as age-related genes with some other CpGs being reported49. Moreover, the proteins encoded by these genes are significantly dysregulated in Alzheimer’s disease (AD) patients50. Our results indicate that the DNA methylation alteration of these genes may be involved in the neurodegenerative processes. Further investigation of these age-related genes (Supplementary Data 6), especially the newly discovered genes, is important.
To understand the function and bioprocess of all the 1,587 genes, the KEGG pathway and GO enrichment analysis are performed (Fig. 2e, Supplementary Fig. 2c and Supplement Data 7). The enriched KEGG pathways included cAMP signaling51, TNF signaling52, TRP channel regulation51, cancer53, GnRH secretion54, neurodegeneration55 pathways. These pathways have previous evidence for age-associated DNA methylation alteration or are known to be related to aging.
A methylation epigenetic clock based on plasma cfDNA
Epigenetic clock based on plasma cell-free DNA (cfDNA) methylation has not extensively been explored23. Here, we develope a cfDNA methylation epigenetic clock, using an elastic net regression, based on the 3047 age-associated CpGs. Finally, a set of 125 CpG sites is included in the age-prediction model, achieving a relatively high level of accuracy. The correlation coefficient (r) is 0.91 and the median absolute error (MAE) is 3.74 years (Fig. 2f, Supplementary Data 8). The 125 CpGs and their coefficients with age are provided in Supplementary Data 9. To further validate this cfDNA methylation clock, we test the publicly available cfDNA WGBS data of 23 healthy individuals (GSE186458). The external data also show a strong correlation (r = 0.94) between the chronological age and the biological age, with an MAE of 9.44 years (Supplementary Fig. 3, Supplementary Data 10). To the best of our knowledge, the majority of sites selected for our cfDNA methylation clock are novel and not present in existing DNAm clock algorithms. Only two of the CpG sites were included in Hannum’s blood-based clock (composed of 71 DNAm sites)56, and three CpG sites included in Horvath’s Skin & Blood clock (comprising 391 DNAm sites)57 (Supplementary Data 11). The small overlap of age-associated CpGs can be attributed to several factors, including the sample materials, methylation detection methods (WGBS vs Illumina 450 K/850 K array), and population demographics (Chinese vs European/American, and different age ranges).
Sex-associated DNA methylation patterns on autosomes
We identify 1053 CpGs on autosomes with their methylation rates significantly associated with sex (Padj < 0.05, linear regression) (Supplementary Data 12). The visualization of the Manhattan plot is shown in Fig. 3a. Notably, only seven of the 1053 CpGs are also identified as age-associated CpGs (Supplementary Fig. 4, Supplementary Data 3, 12), revealing that most of the reported age- and sex-associated CpGs are independent. We find that six out of the seven CpGs are in FIGN which contains the highest number of age-associated CpGs (Fig. 2d, Supplementary Data 6). The two CpGs in FIGN (cg15148145 and cg16532938) are annotated as age- and sex- associated in the EWAS database. Here, the six CpGs not covered by the 850 K arrays are discovered to be simultaneously associated with age and sex in our study.
a Manhattan plot showing the distribution of 1053 sex-associated CpGs (Padj < 0.05) across all the autosomes. The linear regression model is used, and the P values for the regression coefficients are derived from two-tailed t-tests. The P values were adjusted using the Benjamini–Hochberg (BH) method.The numbers of sex-associated CpGs in 10 Mb bins are shown on the chromosomes at the bottom of the Manhattan plot, with the color bar showing on the right side. b–d Ranking of genes based on the number of sex-associated CpG sites per gene. The genes containing CpGs with higher methylation rates in females than those in males are shown in (b). The genes containing CpGs with higher methylation rates in males than those in females are shown in (d). The genes containing 5 or more sex-associated CpG sites are labeled with the gene names, and genes enriched with sex-associated CpGs are labeled in brown. The methylation rates of sex-associated CpGs of two example genes: c LINC01597 and e PTPRN2/LOC105375614. Box plots show median ± interquartile range (IQR) and 1.5 IQR ranges (whiskers). f KEGG and g GO enrichment analysis of 1053 autosomal sex-associated genes, using hypergeometric test (one-tailed test). The P values without adjustment are shown in (f, g). The P values and adjusted P values are in Supplementary Data 17.
The 1053 sex-associated CpGs with higher methylation rates in females are named as female-higher methylation positions (HMPs), and those with higher methylation rates in males as male-HMPs. There are 727 female-HMPs (69%) and 326 male-HMPs (31%), consistent with previous reports that most sex-associated CpG sites are more methylated in females than in males58,59. Similar to the finding about age-associated CpGs, only a limited number of these sex-associated CpGs are covered by the 450 K and 850 K arrays, 17 and 33, respectively (Supplementary Data 12). The EWAS database shows that 11 out of 17, and 14 out of 33 of CpGs have previously been found to be sex-associated by DNA methylation arrays. We evaluate the distribution of the 1053 sex-associated CpGs based on their relation to the nearest genes or CpG islands34. The sex-associated CpGs are enriched in promoters, exons, enhancers, CpG islands, and CpG shores, while they are underrepresented in the introns, open sea, and CpG shelves (Supplementary Fig. 5a, b, Supplementary Data 5).
The 1053 sex-associated CpGs are mapped to 324 genes (Supplementary Data 12). The genes containing only female HMPs are defined as female-HMGs. Likewise, the genes containing only male-HMPs are defined as male-HMGs. The remaining genes containing both female-HMPs and male-HMPs were defined as mix-HMGs. In our results, there are 236 female-HMGs (Supplementary Data 13), 82 male-HMGs (Supplementary Data 14), and 6 mix-HMGs (Supplementary Data 15). The genes enriched with female or male HMPs are labeled in brown in Fig. 3b, d, and summarized in Supplementary Data 13, Supplementary Data 14. To our knowledge, most of the CpGs are newly discovered as sex-associated in this study, however, at the gene level, a considerable number of genes are already known to be sex-associated. A previous study based on whole blood samples using the 850 K assays showed that 16 of the 236 female-HMGs have been previously reported to have higher methylation in females, and 2 of the 82 male-HMGs have been previously reported to have higher methylation in males24. Additionally, transcriptome analysis across various tissues60 has revealed that 61 of the 236 female-HMGs (19%) display sex-biased expression, and 20 of the 82 female-HMGs (24%) display sex-biased expression.
In female-HMGs, the LINC01597 gene contains the highest number of female-HMPs, which is 33 (Fig. 3b, c, Supplementary Data 13, Supplementary Data 16). Previously, the transcriptome analysis of brain tissue showed that this gene had a lower expression level in females than in males61. We speculate that the methylation pattern differences in LINC01597 between sexes may play a role in regulating gene expression. In male-HMGs, the PTPRN2/LOC105375614 gene contains the highest number of male-HMPs, which is 22 (Fig.3d, e, Supplementary Data 14, Supplementary Data 16). These CpGs are located within the coding gene PTPRN2 and the long non-coding RNA gene LOC105375614, on the minus strand and the plus strand of the genome, respectively. For the protein-coding gene, PTPRN2 is important in the secretion of hormones and neurotransmitters62. In females, but not in males, it influenced the secretion of the pituitary hormones luteinizing hormone (LH) and follicle-stimulating hormone (FSH), and thus impacted the infertility in the mouse model63. Previous research has also reported higher methylation levels in the PTPRN2 in males than those in females in the whole blood samples and the brain tissues24,64. Given that lncRNA expression is often involved the regulation of DNA methylation and gene expression65,66,67, the detailed regulation mechanism and the interaction between lncRNAs and the target genes needs further investigation.
Next, we analyze the 324 genes containing sex-associated CpGs by KEGG pathway and GO enrichment (Fig. 3f, g, Supplementary Data 17). The enriched KEGG pathways included the MAPK signaling pathway, calcium signaling pathway, salivary secretion, and morphine addiction, all of which were reported to be different between males and females at the DNA methylation level7,68,69. It is worth noting that among the enriched GO functions, four are related to neural functions (Fig. 3g). Concordantly, these genes were significantly enriched in genes predominantly expressed in the cerebral cortex (Supplementary Fig. 6a, b). Previous research has demonstrated sex differences in the brain epigenome and transcriptome of neuropsychiatric disorders61,70, and the sex-associated genes mentioned before, such as LINC01597 and PTPRN2, have displayed sex differences in DNA methylation64 and gene expression61 in neuropsychiatric disorders. Our study reveals that such methylation differences in brain-related functions exist in healthy people. In addition, PTPRN2 plays an important role in insulin secretion in response to glucose stimuli71. As we know, there are sex differences in glucose metabolism and the related disease of diabetes more frequently affects males72. This difference is also found in the enriched KEGG pathway of glycolysis/gluconeogenesis (Fig. 3f). To sum up, we find many sex-associated genes with diverse molecular and biological functions, and their functions are relevant to sex-biased diseases.
Sex-associated DNA methylation on the X chromosome
On the X chromosome, we identify 638,599 CpGs (64 %) significantly associated with sex (Padj < 0.05, linear regression). The methylation differences are related to X chromosome inactivation (XCI). When applying a more stringent threshold (Padj < 10−6), we identified 29,446 CpGs (5%) with significant sex differences. In the subsequent analysis, we focus on these more significant sex-associated CpGs.
To illustrate the differences in CpG methylation rates between sexes, we plot the average methylation rates of all CpG sites on the X chromosome in males and females on the XY axis (Fig. 4a, Supplementary Data 18). Similar to the display in a previous research9, this plot reveals five methylation patterns: red dots (pattern A, 28,464 CpGs) show significantly higher methylation in females (Padj < 10−6), likely reflecting XCI; dark blue dots (pattern B, 13,018 CpGs) indicat hypomethylation (Methylation rate < 0.25) in both sexes (Padj > 10−6), suggesting potential escape from XCI in females; orange dots (pattern C, 561,972 CpGs) represent hypermethylation (Methylation rate > 0.75) in both sexes (Padj > 10−6); purple dots (pattern D, 265,880 CpGs) show significantly higher methylation in males (Padj < 10−6); and gray dots (pattern other, 121,573 CpGs) represent methylation rate range from 0.25 to 0.75 (Padj > 10−6).
a Comparison of DNA methylation rates in females and males for the CpGs on the X chromosome, revealing CpGs under XCI significantly (Padj < 10−6) more methylated in females than in males colored red (pattern A), CpGs escaping XCI with methylation rates <0.25 in both sexes with no significant sex difference (Padj > 10−6) colors dark blue (pattern B), CpGs with methylation rates > 0.75 in both sexes with no sex difference (Padj > 10-6) colors orange (pattern C), CpGs significantly more methylated in males than in females (Padj < 10-6) colors purple (pattern D), and CpGs with methylation rates range from 0.25 to 0.75 (Padj > 10-6) colors gray (pattern other). Distribution of methylation patterns in gene regions (b) and CpG regions (c). Significant differences among patterns (chi-square test, two-tailed test, adjusted P < 0.001) exist in all tested gene regions and CpG regions (Supplementary Data 5). KEGG enrichment analysis for genes with pattern A and pattern B in 5UTRs (d) and promoters (e), using hypergeometric test (one-tailed test). The P values without adjustment are shown in (d, e). The P values and adjusted P values are in Supplementary Data 20, 21. The genes with four XCI status categories are labeled in red (XCI), yellow (escape XCI), black (variable XCI), and blue (unknown). The genes labeled in blue in pattern A and B are currently unknown for their XCI status.
We then evaluate the distribution of the methylation patterns based on their relation to the nearest genes or CpG islands (Fig. 4b, c, Supplementary Data 5)34. Patterns A and B are obviously enriched in promoters, 5’ UTRs, enhancers, CpG islands, and CpG shores, while patterns C, D, and others are obviously enriched in introns and the open sea (Fig. 4b, c, Supplementary Data 5). This distribution aligns with previous findings that loci with lower methylation in males (patterns A and B) are typically in promoters9. In fact, a considerable number of genes displayed multiple patterns including A, B, C, and D, indicating a high degree of complexity in the regulatory mechanisms of gene expression on the X chromosome (Supplementary Fig. 7, Supplementary Data 19).
Given the regulation role of XCI and the significant impact of the promoter and 5’UTR on gene expression regulation73, we utilize KEGG pathway enrichment analysis to the genes with CpGs in patterns A and B, and in the promoter and the 5’UTR region (Fig. 4d, e, Supplementary Data 20,21). These genes in pattern A are enriched in pathways such as NF-κB pathway74, Primary Immunodeficiencies75, and Polycomb repressive complexes76, which are known to be associated with XCI. Most of the genes in the enriched pathways are classified as XCI genes and some genes are defined as variable escape genes (Fig. 4d, e), according to combined survey approaches for XCI status77,78. Five genes (PABPC5, PABPC1L2A, NCBP2L, IL2RG, and GABRQ) enriched in these pathways are unknown in the current catalog of XCI status78,79. The genes in pattern B are enriched in the JAK-STAT signaling pathway and Neuroactive ligand-receptor interaction. Three genes (CSF2RA, CRLF2, IL3RA) in the JAK-STAT signaling pathway are located in the pseudoautosomal region PAR1 and one gene (IL9R) is in the PAR2 region. The detected genes in the PAR1 region are usually reported to be XCI escape genes72, however, the XCI status of CRLF2 has not been thoroughly studied and classified. Although this gene is involved in diseases, such as leukemia and autoimmune disease (MalaCards Version 5.23), there is limited research on the methylation level of this gene. Notably, The JAK-STAT pathway is implicated in various physiological and pathological processes80, including autoimmune diseases that predominantly affect women6. Three genes in another enriched pathway of Neuroactive ligand-receptor interaction, including GRPR81, P2RY882, and P2RY1083 are also involved in autoimmune diseases. Beyond these genes, numerous other genes containing sex-specific CpG sites may escape or variably escape from XCI, which need further investigations, especially for the miRNA genes that were not systematically studied for their XCI status79,84. Here, we provide a list of all genes with patterns A and B in the promoter and 5’UTR regions for future studies (Supplementary Data 22). Our results demonstrate that cfDNA methylation patterns likely reflect XCI status, and provide epigenetic evidence to support conventional understandings. The non-invasive test of cfDNA methylome may be useful for XCI status analysis for future disease studies.
Characterization of cfDNA tissue-of-origins in the generally healthy individuals
We perform cfDNA tissues-of-origin profiling for 98 samples using two deconvolution methods based on the DNA methylation atlas of human bodies developed by Moss et al.17 and Loyfer et al.18. As a result, the blood cells, such as granulocytes, erythroid progenitors, monocytes/macrophages, and NK cells, are the dominant cfDNA origins (Fig. 5a, b, Supplementary Fig. 8a, b, Supplementary Data 23). These results are also consistent with previous studies17,18. There are some differences between the deconvolution results. We focus more on the generally consistent results for the blood cells and tissues verified by both methods.
a Cell-type composition of plasma cfDNA for each individual (n = 98). b Cellular contributors to cfDNA (median value of 98 samples). c Boxplot comparing the hepatocytes-derived cfDNA proportions of females (rose red) and males (blue) in different age groups. The numbers of individual samples in each group are shown in Fig. 1a. Box plots show median ± interquartile range (IQR) and 1.5 IQR ranges (whiskers). The P values were calculated by the Wilcoxon rank-sum test (two-tailed). d–g Spearman’s correlation between the hepatocytes-derived cfDNA proportion and ALT, AST, GGT, and HDLC, respectively. h, i The rank of samples based on cfDNA derived from monocytes/macrophages, granulocytes, erythroid progenitor cells, megakaryocytes, and hepatocytes. Special triangle (Δ), square (▢), hollow circle (∘), solid circles (●), and star (✩) in (a) represent the corresponding participants in (h–l).
We find that the increase in the relative proportion of granulocyte-derived cfDNA with age exists only significantly in females but not in males (Supplementary Fig. 9a–d, Supplementary Data 23). This phenomenon may be associated with the modulatory effects of estrogen on neutrophil apoptosis85, which declines with age in women. By using the deconvolution method developed by Moss et al.17, we find that females have a higher relative proportion of monocytes/macrophages-derived cfDNA compared to males in relatively young groups (age 20–30 and 31–40), but not in older groups (age: 41–50, 51–60, and >60) (Supplementary Fig. 10a, Supplementary Data 23). The significant decrease of monocytes/macrophages-derived cfDNA with age is also discovered only in female groups (Supplementary Fig. 10b, c, Supplementary Data 23). The observed variations in monocyte/macrophage-derived cfDNA are probably associated with the sex difference in monocyte counts86 and monocyte cytotoxic activity87. However, the findings about monocyte/macrophage-derived cfDNA are not supported by the method developed by Loyfer et al.18. Therefore, it needs further investigation to get a solid conclusion.
In addition to the blood-cells-derived cfDNA, the hepatocytes-derived cfDNA is the highest among all organs. Age- and sex-related variations in cfDNA origins are found in hepatocytes (Fig. 5c, Supplementary Fig. 8c, Supplementary Data 23). Compared to the relatively young people, hepatocyte-derived cfDNA is higher in older males, but not in older females. A significant sex difference (P < 0.05) is found in the old group (age:51–60), but not in younger groups (age:20–30, 31–40, and 41–50). As we know, the prevalence of chronic liver diseases, such as metabolic dysfunction-associated steatotic liver disease (MASLD) and hepatocellular carcinoma, is increasing with age, especially for those above age 5088, and is much higher in males than in females89. Cell death and tissue injuries in old males may contribute to the higher level of hepatocytes-derived cfDNA.
Next, we compare the relative proportion of hepatocyte-derived cfDNA with the blood biochemical test results. We find that the hepatocyte-derived cfDNA shows positive correlations with the levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), and gamma-glutamyl transpeptidase (GGT) (Fig. 5d–f, Supplementary Fig. 8d–f, Supplementary Data 24), which is consistent with the positive correlations found in COVID-19 patients19. Moreover, the hepatocyte-derived cfDNA shows negative correlations with High Density Lipoprotein Cholesterol (HDLC) (Fig. 5g, Supplementary Fig. 8g), an important biomarker related to liver function. Notably, our research is based on generally healthy participants rather than patients. These results indicate that cfDNA may be a promising and sensitive biomarker in the evaluation of liver health, in both patients and generally healthy individuals.
The plasma cfDNA tissue origins exhibit some individual variances. Although the participants are generally healthy, some of them may still have some health problems or non-severe diseases, which led to abnormal cfDNA deviations. For example, some participants showed abnormally high proportions of cfDNA derived from monocytes/macrophages (EB069), granulocytes (EB080), erythroid progenitors (EB005), megakaryocytes (EB005), and hepatocytes (EB071) (Fig. 5a, h–l, Supplementary Fig. 8a, h–k). To explore the possible reasons for the outliers, we investigate their physical examination and questionnaire information.
As a result, we find that EB069 (Fig. 5h, Supplementary Fig. 8h) was the only participant who had a history of gout and was currently undergoing gout-specific medicine treatment. As monocytes are known to be involved in the inflammatory processes in gout pathology90, this participant may exhibit enhanced monocyte-mediated immune responses, which increased monocyte-derived cfDNA. For another participant EB080 (Fig. 5i, Supplementary Fig. 8i), the chest CT scan reveals chronic pulmonary inflammatory lesions. Besides, this participant had a history of hyperlipidemia and hypertension, which has also been widely recognized to trigger inflammatory pathways, resulting in heightened neutrophil production and mobilization91,92. Participant EB005 (Fig. 5j, k, Supplementary Fig. 8j) was diagnosed with thrombocytosis and is undergoing treatment. Thrombocytosis is characterized by the overproduction of platelets by megakaryocytes. The clonal expansion of hematopoietic stem cells in thrombocytosis may result in increased production of hematopoietic cell lineages93, such as erythroid progenitors. However, the connection between thrombocytosis and the overproduction of erythroid progenitors is not typically a prominent feature and more studies are needed to understand the phenomenon. Participant EB071 (Fig. 5l, Supplementary Fig. 8k) who has the highest cfDNA level of hepatocytes, has elevated liver enzyme levels, with ALT at 70.9 U/L and AST at 76.5 U/L, both exceeding the normal range (0–40 U/L).
Discussion
Our research demonstrate the great value of whole-genome study on DNA methylation. Only 7% of the age- and sex-associated CpG sites on the autosomes identified in our study are included in the 450 K/850 K arrays. To the best of our knowledge, many of the CpGs are newly discovered to be age- and sex-related at the DNA methylation level, although many related genes and pathways have been proved to be age- or sex-related through other detection methods, such as transcriptome and proteome analysis. For the very special gene of FIGN, with the largest number of age-associated CpGs and simultaneously containing 6 age- and sex-associated CpGs, previous research has reported something about its relationship with aging and sex. FIGN shows sex-specific deviations in centenarians of decelerated aging25. A methylated genomic units (DMUs) specific to long-lived-man in an intergenic regions near FIGN has also been discovered94. Its expression level is relatively high in the ovary, tibial nerve and artery (GTEx Analysis Release V10), and this gene is also associated with diseases such as polycystic ovary syndrome, Parkinson’s disease and pulmonary hypertension (MalaCards Version 5.23). Our analysis provides an epigenetic perspective to understand some sex-biased and/or aging-related diseases. Moreover, we discover that many lncRNA genes contain a lot of age- and sex-associated CpGs. The regulation of DNA methylation on lncRNA gene expression may impact the downstream regulation of lncRNA-targeted genes67,95, which displays a more complex mechanism of regulation.
Plasma cfDNA is a very special and valuable sample material for health and disease studies, for it provides a non-invasive approach to measure DNA methylation alterations in various tissues and cell types in the human body16,17. For example, early cancer screening and diagnosis are studied by detecting the tumor-derived cfDNA and localizing potential tumors16. CfDNA methylation has also been explored to evaluate tissue (e.g., neutrophils, adipocytes, heart, lung, liver, and kidney) injuries in COVID-1919,96, and many other diseases97,98,99. These results improve our understanding of cfDNA methylation signatures for identifying tissue-specific injuries and systemic pathological conditions.
In the past decade, DNA methylation patterns have been employed to measure biological age accurately100. Like other classical epigenetic clocks, we believe cfDNA methylation data can also be trained to estimate age acceleration and predict aging-associated diseases and mortality risk. In addition to the hematopoietic cell types, cfDNA carries aging signals from tissues and organs. These aging and tissue injury signatures may enable risk stratifications for the general population, facilitating personalized interventions (e.g., lifestyle modification, clinical therapies) to mitigate disease progression2.
This study has several limitations. For example, ethnic differences in human DNA methylation have been widely reported, while this study only focused on the Chinese population. The sample size in this study is relatively small, expanding the participants to a wider cohort will enhance the comprehensive understanding of aging, sex-biased, and disease-related methylation profiles. Moreover, the selection of CpGs and the predictive accuracy of the epigenetic clock can be further refined by using other modeling algorithms, and its application in disease prediction and health monitoring also needs further exploration. We anticipate that future cfDNA methylation studies will provide more thorough insight into methylation variations and promote the practical application of cfDNA signatures as biomarkers.
Conclusion
In the present study, we use cell-free DNA (cfDNA) whole-genome bisulfite sequencing (WGBS, ~30 X) to comprehensively investigate the epigenetic signatures of methylation that correlate with age and sex. Our analysis reveals 3047 CpGs and 1587 genes on autosomes that exhibit significant associations with age. We provide a list of genes with methylation alterations, including genes of IFT80 and RILPL1 that are related to aging diseases. Based on the age-associated CpGs, we developed a relatively accurate methylation epigenetic clock (R = 0.91, MAE = 3.74 years) utilizing 125 CpG sites, thereby expanding the research on epigenetic clocks by using cfDNA. Additionally, we identify 1053 sex-associated CpG sites and 324 genes on autosomes. The sex-associated genes are relevant to sex-biased pathways and diseases, including those of neural functions, psychiatric disorders101, and diabetes102. We demonstrate that cfDNA methylation patterns on the X chromosome could also indicate XCI status, and the XCI escape genes involved in modulating immune responses could be found through the analysis of methylation patterns. Furthermore, our study discover age- and sex- associated cfDNA features in the tissue-of-origin, including the relative proportion derived from granulocyte, monocytes/macrophages, and hepatocytes. Through the cfDNA profiling of the general population, we detect four samples that deviated from others. They had abnormally high relative proportions of cfDNA derived from certain cell types, which reflects their health problems. Although this analysis needs a more comprehensive health data survey with larger sample sizes for further validation, our findings highlight the importance of age and sex in influencing cfDNA characteristics and display the great potential of cfDNA methylation as a biomarker in clinical applications.
Data availability
The sequencing data and DNA methylation data have been deposited into the China National GeneBank Sequence Archive (CNSA) of China National GeneBank DataBase (CNGBdb) with the accession numbers CNP0003513 and CNP0005464 (https://db.cngb.org/cnsa/).
References
Greenberg, M. V. C. & Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 20, 590–607 (2019).
Duan, R., Fu, Q., Sun, Y. & Li, Q. Epigenetic clock: a promising biomarker and practical tool in aging. Ageing Res. Rev. 81, 101743 (2022).
Saul, D. & Kosinsky, R. L. Epigenetics of aging and aging-associated diseases. Int. J. Mol. Sci. 22, 401 (2021).
Jylhava, J., Pedersen, N. L. & Hagg, S. Biological age predictors. EBioMedicine 21, 29–36 (2017).
Ye, W., Siwko, S. & Tsai, R. Y. L. Sex and race-related DNA methylation changes in hepatocellular carcinoma. Int. J. Mol. Sci. 22, 3820 (2021).
Mousavi, M. J., Mahmoudi, M. & Ghotloo, S. Escape from X chromosome inactivation and female bias of autoimmune diseases. Mol. Med. 26, 127 (2020).
Xia, Y. et al. Sex-differential DNA methylation and associated regulation networks in human brain implicated in the sex-biased risks of psychiatric disorders. Mol. Psychiatry 26, 835–848 (2021).
Gatev, E. et al. Autosomal sex-associated co-methylated regions predict biological sex from DNA methylation. Nucleic Acids Res. 49, 9097–9116 (2021).
Li, S. et al. Exploratory analysis of age and sex dependent DNA methylation patterns on the X-chromosome in whole blood samples. Genome Med. 12, 39 (2020).
Lee, J. T. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat. Rev. Mol. Cell Biol. 12, 815–826 (2011).
Sandoval, J. et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics 6, 692–702 (2011).
Moran, S., Arribas, C. & Esteller, M. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics 8, 389–399 (2016).
Pidsley, R. et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 17, 208 (2016).
Zhu, T. et al. A pan-tissue DNA methylation atlas enables in silico decomposition of human tissue methylomes at cell-type resolution. Nat. Methods 19, 296–306 (2022).
Jaffe, A. E. & Irizarry, R. A. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol 15, R31 (2014).
Luo, H., Wei, W., Ye, Z., Zheng, J. & Xu, R. H. Liquid biopsy of methylation biomarkers in cell-free DNA. Trends Mol. Med. 27, 482–500 (2021).
Moss, J. et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 9, 5068 (2018).
Loyfer, N. et al. A DNA methylation atlas of normal human cell types. Nature 613, 355–364 (2023).
Cheng, A. P. et al. Cell-free DNA tissues of origin by methylation profiling reveals significant cell, tissue, and organ-specific injury related to COVID-19 severity. Med 2, 411–422.e415 (2021).
Stackpole, M. L. et al. Cost-effective methylome sequencing of cell-free DNA for accurately detecting and locating cancer. Nat. Commun. 13, 5566 (2022).
Teo, Y. V. et al. Cell-free DNA as a biomarker of aging. Aging Cell 18, e12890 (2019).
Shtumpf, M. et al. Aging clock based on nucleosome reorganisation derived from cell-free DNA. Aging Cell 23, e14100 (2024).
Li, S. J. et al. Cell-free DNA methylation patterns in aging and their association with inflamm-aging. Epigenomics 16, 715–731 (2024).
Grant, O. A., Wang, Y. C., Kumari, M., Zabet, N. R. & Schalkwyk, L. Characterising sex differences of autosomal DNA methylation in whole blood using the Illumina EPIC array. Clin. Epigenetics 14, 62 (2022).
McCartney, D. L. et al. An epigenome-wide association study of sex-specific chronological ageing. Genome Med. 12, 1 (2019).
Hall, E. et al. Sex differences in the genome-wide DNA methylation pattern and impact on gene expression, microRNA levels and insulin secretion in human pancreatic islets. Genome Biol. 15, 522 (2014).
Raine, A., Manlig, E., Wahlberg, P., Syvänen, A. C. & Nordlund, J. SPlinted Ligation Adapter Tagging (SPLAT), a novel library preparation method for whole genome bisulphite sequencing. Nucleic Acids Res. 45, e36 (2017).
Troll, C. J. et al. A ligation-based single-stranded library preparation method to analyze cell-free DNA and synthetic oligos. BMC Genom. 20, 1023 (2019).
Cao, B. et al. The performance of whole genome bisulfite sequencing on DNBSEQ-Tx platform examined by different library preparation strategies. Heliyon 9, e16571 (2023).
Yang, C. et al. Cell-Free DNA comprises the strand-specific characteristic associated with transcription and methylation. Clin. Chem. 71, 980–992 (2025).
Chen, S. F., Zhou, Y. Q., Chen, Y. R. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, 884–890 (2018).
Cheng, H. & Xu, Y. BitMapperBS: a fast and accurate read aligner for whole-genome bisulfite sequencing. Preprint at https://doi.org/10.1101/442798 (2018).
Tarasov, A., Vilella, A. J., Cuppen, E., Nijman, I. J. & Prins, P. Sambamba: fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034 (2015).
Cavalcante, R. G. & Sartor, M. A. annotatr: genomic regions in context. Bioinformatics 33, 2381–2383 (2017).
Zheng, Z. et al. DNA methylation clocks for estimating biological age in Chinese cohorts. Protein Cell 15, 575–593 (2024).
Bacos, K. et al. Blood-based biomarkers of age-associated epigenetic changes in human islets associate with insulin secretion and diabetes. Nat Commun 7, 11089 (2016).
Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol 14, R115 (2013).
Yu, N. Y. et al. Complementing tissue characterization by integrating transcriptome profiling from the Human Protein Atlas and from the FANTOM5 consortium. Nucleic Acids Res. 43, 6787–6798 (2015).
Jain, A. & Tuteja, G. TissueEnrich: tissue-specific gene enrichment analysis. Bioinformatics 35, 1966–1967 (2019).
Boix, C. A., James, B. T., Park, Y. P., Meuleman, W. & Kellis, M. Regulatory genomic circuitry of human disease loci by integrative epigenomics. Nature 590, 300–307 (2021).
Nasser, J. et al. Genome-wide enhancer maps link risk variants to disease genes. Nature 593, 238–243 (2021).
Jones, M. J., Goodman, S. J. & Kobor, M. S. DNA methylation and healthy human aging. Aging Cell 14, 924–932 (2015).
Gentilini, D. et al. Stochastic epigenetic mutations (DNA methylation) increase exponentially in human aging and correlate with X chromosome inactivation skewing in females. Aging 7, 568–578 (2015).
Deepak, V. et al. IFT80 negatively regulates osteoclast differentiation via association with Cbl-b to disrupt TRAF6 stabilization and activation. Proc. Natl. Acad. Sci. USA 119, e2201490119 (2022).
Yuan, X. et al. Ciliary IFT80 balances canonical versus non-canonical hedgehog signalling for osteoblast differentiation. Nat. Commun. 7, 11024 (2016).
Li, X. et al. Role of primary cilia in skeletal disorders. Stem Cells Int. 2022, 6063423 (2022).
Yang, X. et al. A large pedigree study confirmed the CGG repeat expansion of RILPL1 Is associated with oculopharyngodistal myopathy. BMC Med. Genom. 16, 253 (2023).
Maeda, M. Y., Hashimoto, T. Y., Oliveira Neto, I. C. & Neves, L. R. Oculopharyngeal muscular dystrophy or oculopharyngeal distal myopathy: case report. Braz. J. Otorhinolaryngol. 83, 235–238 (2017).
Chitrala, K. N. et al. Race-specific alterations in DNA methylation among middle-aged African Americans and Whites with metabolic syndrome. Epigenetics 15, 462–482 (2020).
Jiang, Y. et al. A blood-based multi-pathway biomarker assay for early detection and staging of Alzheimer’s disease across ethnic groups. Alzheimers Dement 20, 2000–2015 (2024).
Korkmaz, F. T. & Kerr, D. E. Genome-wide methylation analysis reveals differentially methylated loci that are associated with an age-dependent increase in bovine fibroblast response to LPS. BMC Genom. 18, 405 (2017).
Steegenga, W. T. et al. Genome-wide age-related changes in DNA methylation and gene expression in human PBMCs. Age 36, 9648 (2014).
Levine, M. E. et al. DNA methylation age of blood predicts future onset of lung cancer in the women’s health initiative. Aging 7, 690–700 (2015).
Kurian, J. R. & Terasawa, E. Epigenetic control of gonadotropin releasing hormone neurons. Front. Endocrinol. 4, 61 (2013).
Hou, Y. J. et al. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 15, 565–581 (2019).
Hannum, G. et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 49, 359–367 (2013).
Horvath, S. et al. Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria Syndrome and ex vivo studies. Aging 10, 1758–1775 (2018).
Yousefi, P. et al. Sex differences in DNA methylation assessed by 450 K BeadChip in newborns. BMC Genom. 16, 911 (2015).
Yang, Y. J. et al. Identification of an epigenetically marked locus within the sex determination region of channel catfish. Int. J. Mol. Sci. 23, 5471 (2022).
Oliva, M. et al. The impact of sex on gene expression across human tissues. Sci 369, 6509 (2020).
Hoffman, G. E. et al. Sex differences in the human brain transcriptome of cases with schizophrenia. Biol. Psychiatry 91, 92–101 (2022).
Kang, T. et al. Knockdown of Ptprn-2 delays the onset of puberty in female rats. Theriogenology 176, 137–148 (2021).
Kubosaki, A., Nakamura, S., Clark, A., Morris, J. F. & Notkins, A. L. Disruption of the transmembrane dense core vesicle proteins IA-2 and IA-2β causes female infertility. Endocrinology 147, 811–815 (2006).
Kochmanski, J., Kuhn, N. C. & Bernstein, A. I. Parkinson’s disease-associated, sex-specific changes in DNA methylation at PARK7 (DJ-1), SLC17A6 (VGLUT2), PTPRN2 (IA-2beta), and NR4A2 (NURR1) in cortical neurons. NPJ Parkinsons Dis. 8, 120 (2022).
Zhi, H. et al. Lnc2Meth: a manually curated database of regulatory relationships between long non-coding RNAs and DNA methylation associated with human disease. Nucleic Acids Res. 46, D133–D138 (2018).
Yang, Z. et al. Insights into the role of long non-coding RNAs in DNA methylation mediated transcriptional regulation. Front. Mol. Biosci. 9, 1067406 (2022).
Zhao, J. et al. The diverse mechanisms of miRNAs and lncRNAs in the maintenance of liver cancer stem cells. Biomed. Res. Int. 2018, 8686027 (2018).
Saadatmand, F., Abbas, M., Apprey, V., Tailor, K. & Kwabi-Addo, B. Sex differences in saliva-based DNA methylation changes and environmental stressor in young African American adults. PLoS ONE 17, e0273717 (2022).
Kaplan, G., Xu, H. Y., Abreu, K. & Feng, J. DNA epigenetics in addiction susceptibility. Front. Genetics 13, 806685 (2022).
Shen, L. et al. Genome-wide analysis of DNA methylation in 106 schizophrenia family trios in Han Chinese. EBioMedicine 72, 103609 (2021).
Lee, S. The association of genetically controlled CpG methylation (cg158269415) of protein tyrosine phosphatase, receptor type N2 (PTPRN2) with childhood obesity. Sci. Rep. 9, 4855 (2019).
Macotela, Y., Boucher, J., Tran, T. T. & Kahn, C. R. Sex and depot differences in adipocyte insulin sensitivity and glucose metabolism. Diabetes 58, 803–812 (2009).
Leenen, F. A., Muller, C. P. & Turner, J. D. DNA methylation: conducting the orchestra from exposure to phenotype?. Clin. Epigenet. 8, 92 (2016).
Bhattacharya, S., Sadhukhan, D. & Saraswathy, R. Role of sex in immune response and epigenetic mechanisms. Epigenet. Chromatin 17, 1 (2024).
Rispoli, F. et al. Immunity and genetics at the revolving doors of diagnostics in primary immunodeficiencies. Diagnostics 11, 532 (2021).
Blackledge, N. P. & Klose, R. J. The molecular principles of gene regulation by Polycomb repressive complexes. Nat. Rev. Mol. Cell Biol. 22, 815–833 (2021).
Cotton, A. M. et al. Landscape of DNA methylation on the X chromosome reflects CpG density, functional chromatin state and X-chromosome inactivation. Hum. Mol. Genet. 24, 1528–1539 (2015).
Tukiainen, T. et al. Landscape of X chromosome inactivation across human tissues. Nature 550, 244–248 (2017).
Balaton, B. P., Cotton, A. M. & Brown, C. J. Derivation of consensus inactivation status for X-linked genes from genome-wide studies. Biol. Sex Differ. 6, 35 (2015).
Xin, P. et al. The role of JAK/STAT signaling pathway and its inhibitors in diseases. Int. Immunopharmacol. 80, 106210 (2020).
Clarimundo, V. S. et al. Gastrin-releasing peptide and its receptor increase arthritis fibroblast-like synoviocytes invasiveness through activating the PI3K/AKT pathway. Peptides 95, 57–61 (2017).
He, Y. et al. P2RY8 variants in lupus patients uncover a role for the receptor in immunological tolerance. J. Exp. Med. 219, e20211004 (2022).
Gurusamy, M. et al. G-protein-coupled receptor P2Y10 facilitates chemokine-induced CD4 T cell migration through autocrine/paracrine mediators. Nat. Commun. 12, 6798 (2021).
Di Palo, A. et al. What microRNAs could tell us about the human X chromosome. Cell. Mol. Life Sci. 77, 4069–4080 (2020).
Molloy, E. J. et al. Sex-specific alterations in neutrophil apoptosis: the role of estradiol and progesterone. Blood 102, 2653–2659 (2003).
van Zeventer, I. A. et al. Monocytosis and its association with clonal hematopoiesis in community-dwelling individuals. Blood Adv. 6, 4174–4184 (2022).
Koopman, M. G. et al. Male-female differences in the cytotoxic activity of human monocytes in vitro. Clin. Lab. Haematol. 3, 45–50 (1981).
Shang, Y., Nasr, P., Widman, L. & Hagström, H. Risk of cardiovascular disease and loss in life expectancy in NAFLD. Hepatology 76, 1495–1505 (2022).
Yang, D. et al. Impact of sex on the survival of patients with hepatocellular carcinoma: a surveillance, epidemiology, and end results analysis. Cancer 120, 3707–3716 (2014).
Liu, L. et al. Recent insights into the role of macrophages in acute gout. Front. Immunol. 13, 955806 (2022).
Sreejit, G. et al. Neutrophils in cardiovascular disease: warmongers, peacemakers, or both?. Cardiovasc. Res. 118, 2596–2609 (2022).
Drechsler, M., Megens, R. T., van Zandvoort, M., Weber, C. & Soehnlein, O. Hyperlipidemia-triggered neutrophilia promotes early atherosclerosis. Circulation 122, 1837–1845 (2010).
Spivak, J. L. & Silver, R. T. The revised World Health Organization diagnostic criteria for polycythemia vera, essential thrombocytosis, and primary myelofibrosis: an alternative proposal. Blood 112, 231–239 (2008).
Xiao, F. H. et al. Methylome analysis in long-lived men deciphers DNA methylation modifications associated with male longevity in humans. Cell Rep. 44, 115158 (2025).
He, H. et al. lncRNA STAT4-AS1 inhibited TH17 Cell differentiation by targeting RORgammat protein. J. Immunol. Res. 2022, 8307280 (2022).
Andargie, T. E. et al. Cell-free DNA maps COVID-19 tissue injury and risk of death and can cause tissue injury. JCI Insight 6, e147610 (2021).
Magenheim, J. et al. Universal lung epithelium DNA methylation markers for detection of lung damage in liquid biopsies. Eur. Respir. J. 60, 2103056 (2022).
Peretz, A. et al. The DNA methylome of human vascular endothelium and its use in liquid biopsies. Med 4, 263–281.e264 (2023).
Diez Benavente, E., Sakkers, T. R., van der Harst, P. & den Ruijter, H. M. Atherosclerotic plaque-specific methylation biomarkers in plasma cell-free DNA of female and male patients with coronary artery disease. Eur. Heart J. 45, 2468–2470 (2024).
Rutledge, J., Oh, H. & Wyss-Coray, T. Measuring biological age using omics data. Nat. Rev. Genet. 23, 715–727 (2022).
Lin, P. et al. Consensus on potential biomarkers developed for use in clinical tests for schizophrenia. Gen. Psychiatr. 35, e100685 (2022).
Liebmann, M., Asuaje Pfeifer, M., Grupe, K. & Scherneck, S. Estradiol (E2) improves glucose-stimulated insulin secretion and stabilizes GDM progression in a prediabetic mouse model. Int. J. Mol. Sci. 23, 6693 (2022).
Acknowledgements
This work has been supported by National Key R&D Program of China (2023YFC2605400), Shenzhen Medical Research Fund (B2404004), Shenzhen Science and Technology Program (SYSPG20241211173852024), and National Natural Science Foundation of China (No. 32171441). We express our gratitude to Prof. Qiang Zeng (Health Management Institute, Chinese PLA General Hospital, Beijing 100853, China) for his valuable suggestions for health monitoring, and Dr. Yong Bai (BGI Research, Shenzhen 518083, China) for his valuable suggestions for data analysis. This study was supported by the China National GeneBank in computational resources.
Author information
Authors and Affiliations
Contributions
X.J. and Y.Z. conceived the study. Y.Z., R.O., W.C. and C.Y. designed the experiments. W.C., G.Z., C.Y., Y.W., Q.L., C.Z., X.W., W.Z. and Y.L. collected the samples and carried out the experiments. J.X., W.C., Y.Z., G.Z. and C.X. analyzed the data. H.Z., J.Y., Y.L., X.J. and W.P. gave suggestions for the analysis. W.C., Y.Z., J.X., G.Z. and C.X. wrote the manuscript. Y.Z., W.C., J.X., G.Z. and X.J. revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Medicine thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, W., Xu, J., Zeng, G. et al. Whole-genome profiling of age- and sex-associated DNA methylation signatures in human plasma cell-free DNA. Commun Med 5, 503 (2025). https://doi.org/10.1038/s43856-025-01220-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s43856-025-01220-y