Abstract
Myocardial injuries lead to cardiomyocyte loss and heart failure. Endogenous glucocorticoids, via the glucocorticoid receptor (GR), limit cardiomyocyte regeneration. Here we show that glucocorticoids suppress mammalian (murine) cardiomyocyte proliferative response to regenerative growth factors and cytokines. GR activation in neonatal cardiomyocytes upregulated MAPK–ERK inhibitors ERRFI1 and DUSP1. Using neuregulin 1 as a model, we demonstrated that glucocorticoids inhibit growth-factor-induced ERK activation, nuclear translocation and transcriptional output. Errfi1 and Dusp1 knockdown restored growth-factor-induced proliferation of glucocorticoid-exposed cardiomyocytes. Cardiac expression of DUSP1 and ERRFI1 increased postnatally, coinciding with regenerative capacity decline. In juvenile and adult cardiomyocytes, regenerative growth factors failed to induce the MAPK–ERK pathway and proliferation; however, DUSP1 inhibition restored these responses. GR antagonism enhanced growth-factor-induced cardiomyocyte protection, proliferation and cardiac function after adult myocardial injury. These findings reveal the emergence of a postnatal systemic brake on cardiomyocyte proliferative response to growth factors and support GR inhibition as a strategy to enhance growth-factor-based regenerative therapies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The datasets and uncropped scans of figures and extended data generated in this study are available as source data files. RNA-seq data generated in this study (Fig. 2a,b and Supplementary Table 3) were deposited in the Gene Expression Omnibus repository under accession nos. GSE286562 (controls in GSE202968). Published RNA-seq data analyzed in Extended Data Fig. 5d–f are available in the Gene Expression Omnibus repository under accession no. GSE144391 (ref. 68). Published RNA-seq data analyzed in Fig. 4b are available as supplementary information in the original paper69. Source data are provided with this paper.
References
Martin, S. S. et al. 2024 heart disease and stroke statistics: a report of US and global data from the American Heart Association. Circulation 149, e347–e913 (2024).
Secco, I. & Giacca, M. Regulation of endogenous cardiomyocyte proliferation: the known unknowns. J. Mol. Cell Cardiol. 179, 80–89 (2023).
Whelan, R. S., Kaplinskiy, V. & Kitsis, R. N. Cell death in the pathogenesis of heart disease: mechanisms and significance. Annu. Rev. Physiol. 72, 19–44 (2010).
Chiong, M. et al. Cardiomyocyte death: mechanisms and translational implications. Cell Death Dis. 2, e244 (2011).
Kytö, V. et al. Apoptotic cardiomyocyte death in fatal myocarditis. Am. J. Cardiol. 94, 746–750 (2004).
Zhang, Y.-W., Shi, J., Li, Y.-J. & Wei, L. Cardiomyocyte death in doxorubicin-induced cardiotoxicity. Arch. Immunol. Ther. Exp. 57, 435–445 (2009).
Octavia, Y. et al. Doxorubicin-induced cardiomyopathy: from molecular mechanisms to therapeutic strategies. J. Mol. Cell Cardiol. 52, 1213–1225 (2012).
González, A. et al. Cardiomyocyte apoptosis in hypertensive cardiomyopathy. Cardiovasc. Res. 59, 549–562 (2003).
Hein, S. et al. Progression from compensated hypertrophy to failure in the pressure-overloaded human heart: structural deterioration and compensatory mechanisms. Circulation 107, 984–991 (2003).
Yamaji, K. et al. Apoptotic myocardial cell death in the setting of arrhythmogenic right ventricular cardiomyopathy. Acta Cardiol. 60, 465–470 (2005).
Hashem, S. I. et al. Brief report: oxidative stress mediates cardiomyocyte apoptosis in a human model of Danon disease and heart failure. Stem Cells 33, 2343–2350 (2015).
Bongiovanni, C. et al. Reawakening the intrinsic cardiac regenerative potential: molecular strategies to boost dedifferentiation and proliferation of endogenous cardiomyocytes. Front. Cardiovasc. Med. 8, 750604 (2021).
Tzahor, E. & Poss, K. D. Cardiac regeneration strategies: staying young at heart. Science 356, 1035–1039 (2017).
Sadek, H. & Olson, E. N. Toward the goal of human heart regeneration. Cell Stem Cell 26, 7–16 (2020).
Eschenhagen, T. et al. Cardiomyocyte regeneration: a consensus statement. Circulation 136, 680–686 (2017).
van Berlo, J. H. & Molkentin, J. D. An emerging consensus on cardiac regeneration. Nat. Med. 20, 1386–1393 (2014).
Hashimoto, H., Olson, E. N. & Bassel-Duby, R. Therapeutic approaches for cardiac regeneration and repair. Nat. Rev. Cardiol. 15, 585–600 (2018).
Uygur, A. & Lee, R. T. Mechanisms of cardiac regeneration. Dev. Cell 36, 362–374 (2016).
Bersell, K., Arab, S., Haring, B. & Kühn, B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell 138, 257–270 (2009).
D’Uva, G. et al. ERBB2 triggers mammalian heart regeneration by promoting cardiomyocyte dedifferentiation and proliferation. Nat. Cell Biol. 17, 627–638 (2015).
Polizzotti, B. D. et al. Neuregulin stimulation of cardiomyocyte regeneration in mice and human myocardium reveals a therapeutic window. Sci. Transl. Med. 7, 281ra45 (2015).
D’Uva, G. & Tzahor, E. The key roles of ERBB2 in cardiac regeneration. Cell Cycle 14, 2383–2384 (2015).
Gao, R. et al. A phase II, randomized, double-blind, multicenter, based on standard therapy, placebo-controlled study of the efficacy and safety of recombinant human neuregulin-1 in patients with chronic heart failure. J. Am. Coll. Cardiol. 55, 1907–1914 (2010).
Jabbour, A. et al. Parenteral administration of recombinant human neuregulin-1 to patients with stable chronic heart failure produces favourable acute and chronic haemodynamic responses. Eur. J. Heart Fail. 13, 83–92 (2011).
Lenihan, D. J. et al. A phase I, single ascending dose study of cimaglermin alfa (Neuregulin 1β3) in patients with systolic dysfunction and heart failure. JACC Basic Transl. Sci. 1, 576–586 (2016).
Engel, F. B. et al. p38 MAP kinase inhibition enables proliferation of adult mammalian cardiomyocytes. Genes Dev. 19, 1175–1187 (2005).
Engel, F. B., Hsieh, P. C. H., Lee, R. T. & Keating, M. T. FGF1/p38 MAP kinase inhibitor therapy induces cardiomyocyte mitosis, reduces scarring, and rescues function after myocardial infarction. Proc. Natl Acad. Sci. USA 103, 15546–15551 (2006).
Novoyatleva, T. et al. FGF1-mediated cardiomyocyte cell cycle reentry depends on the interaction of FGFR-1 and Fn14. FASEB J. 28, 2492–2503 (2014).
Shen, H. et al. Mononuclear diploid cardiomyocytes support neonatal mouse heart regeneration in response to paracrine IGF2 signaling. eLife 9, e53071 (2020).
Schuetz, T. et al. Murine neonatal cardiac regeneration depends on Insulin-like growth factor 1 receptor signaling. Sci. Rep. 14, 22661 (2024).
Sundgren, N. C. et al. Extracellular signal-regulated kinase and phosphoinositol-3 kinase mediate IGF-1 induced proliferation of fetal sheep cardiomyocytes. Am. J. Physiol. Regul. Integr. Comp. Physiol. 285, R1481–R1489 (2003).
Koudstaal, S. et al. Sustained delivery of insulin-like growth factor-1/hepatocyte growth factor stimulates endogenous cardiac repair in the chronic infarcted pig heart. J. Cardiovasc. Transl. Res. 7, 232–241 (2014).
Bongiovanni, C. et al. BMP7 promotes cardiomyocyte regeneration in zebrafish and adult mice. Cell Rep. 43, 114162 (2024).
Vasudevarao, M. D. et al. BMP signaling promotes zebrafish heart regeneration via alleviation of replication stress. Nat. Commun. 16, 1708 (2025).
Kubin, T. et al. Oncostatin M is a major mediator of cardiomyocyte dedifferentiation and remodeling. Cell Stem Cell 9, 420–432 (2011).
Li, Y. et al. gp130 controls cardiomyocyte proliferation and heart regeneration. Circulation 142, 967–982 (2020).
Zou, Y. et al. Leukemia inhibitory factor enhances survival of cardiomyocytes and induces regeneration of myocardium after myocardial infarction. Circulation 108, 748–753 (2003).
Negoro, S. et al. Glycoprotein 130 regulates cardiac myocyte survival in doxorubicin-induced apoptosis through phosphatidylinositol 3-kinase/Akt phosphorylation and Bcl-xL/caspase-3 interaction. Circulation 103, 555–561 (2001).
Zacchigna, S. et al. Paracrine effect of regulatory T cells promotes cardiomyocyte proliferation during pregnancy and after myocardial infarction. Nat. Commun. 9, 2432 (2018).
Ock, S. et al. Receptor activator of nuclear factor-κB ligand is a novel inducer of myocardial inflammation. Cardiovasc. Res. 94, 105–114 (2012).
Tang, P. et al. Effect of interleukin-6 on myocardial regeneration in mice after cardiac injury. Biomed. Pharmacother. 106, 303–308 (2018).
Han, C. et al. Acute inflammation stimulates a regenerative response in the neonatal mouse heart. Cell Res. 25, 1137–1151 (2015).
Zogbi, C. et al. Beneficial effects of IL-4 and IL-6 on rat neonatal target cardiac cells. Sci. Rep. 10, 12350 (2020).
Fahmi, A. et al. p42/p44-MAPK and PI3K are sufficient for IL-6 family cytokines/gp130 to signal to hypertrophy and survival in cardiomyocytes in the absence of JAK/STAT activation. Cell Signal. 25, 898–909 (2013).
Wodsedalek, D. J. et al. IL-13 promotes in vivo neonatal cardiomyocyte cell cycle activity and heart regeneration. Am. J. Physiol. Heart Circ. Physiol. 316, H24–H34 (2019).
Paddock, S. J. et al. IL4Rα signaling promotes neonatal cardiac regeneration and cardiomyocyte cell cycle activity. J. Mol. Cell Cardiol. 161, 62–74 (2021).
Palmer, J. N., Hartogensis, W. E., Patten, M., Fortuin, F. D. & Long, C. S. Interleukin-1 beta induces cardiac myocyte growth but inhibits cardiac fibroblast proliferation in culture. J. Clin. Invest. 95, 2555–2564 (1995).
Przybyt, E., Krenning, G., Brinker, M. G. L. & Harmsen, M. C. Adipose stromal cells primed with hypoxia and inflammation enhance cardiomyocyte proliferation rate in vitro through STAT3 and Erk1/2. J. Transl. Med. 11, 39 (2013).
Pianca, N. et al. Glucocorticoid receptor antagonization propels endogenous cardiomyocyte proliferation and cardiac regeneration. Nat. Cardiovasc. Res. 1, 617–633 (2022).
D’Uva, G. & Lauriola, M. Towards the emerging crosstalk: ERBB family and steroid hormones. Semin. Cell Dev. Biol. 50, 143–152 (2016).
Lauriola, M. et al. Diurnal suppression of EGFR signalling by glucocorticoids and implications for tumour progression and treatment. Nat. Commun. 5, 5073 (2014).
Hattori, F. et al. Nongenetic method for purifying stem cell-derived cardiomyocytes. Nat. Methods 7, 61–66 (2010).
Porrello, E. R. et al. Transient regenerative potential of the neonatal mouse heart. Science 331, 1078–1080 (2011).
Guo, S. et al. Chronic corticosterone exposure suppresses copper transport through GR-mediated intestinal CTR1 pathway in mice. Biology 12, 197 (2023).
Guo, Y. & Pu, W. T. Cardiomyocyte maturation: new phase in development. Circ. Res. 126, 1086–1106 (2020).
Citri, A. & Yarden, Y. EGF-ERBB signalling: towards the systems level. Nat. Rev. Mol. Cell Biol. 7, 505–516 (2006).
Segatto, O., Anastasi, S. & Alemà, S. Regulation of epidermal growth factor receptor signalling by inducible feedback inhibitors. J. Cell Sci. 124, 1785–1793 (2011).
Keyse, S. M. Dual-specificity MAP kinase phosphatases (MKPs) and cancer. Cancer Metastasis Rev. 27, 253–261 (2008).
Wallace, A. D. & Cidlowski, J. A. Proteasome-mediated glucocorticoid receptor degradation restricts transcriptional signaling by glucocorticoids. J. Biol. Chem. 276, 42714–42721 (2001).
Kassel, O. et al. Glucocorticoids inhibit MAP kinase via increased expression and decreased degradation of MKP-1. EMBO J. 20, 7108–7116 (2001).
Chen, H.-F., Chuang, H.-C. & Tan, T.-H. Regulation of dual-specificity phosphatase (DUSP) ubiquitination and protein stability. Int. J. Mol. Sci. 20, 2668 (2019).
Plotnikov, A. et al. The nuclear translocation of ERK1/2 as an anticancer target. Nat. Commun. 6, 6685 (2015).
Michailovici, I. et al. Nuclear to cytoplasmic shuttling of ERK promotes differentiation of muscle stem/progenitor cells. Development 141, 2611–2620 (2014).
Avraham, R. & Yarden, Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat. Rev. Mol. Cell Biol. 12, 104–117 (2011).
Amit, I. et al. A module of negative feedback regulators defines growth factor signaling. Nat. Genet. 39, 503–512 (2007).
Zhang, L. et al. Egr1 regulates regenerative senescence and cardiac repair. Nat. Cardiovasc. Res. 3, 915–932 (2024).
Beisaw, A. et al. AP-1 contributes to chromatin accessibility to promote sarcomere disassembly and cardiomyocyte protrusion during zebrafish heart regeneration. Circ. Res. 126, 1760–1778 (2020).
Aharonov, A. et al. ERBB2 drives YAP activation and EMT-like processes during cardiac regeneration. Nat. Cell Biol. 22, 1346–1356 (2020).
Haubner, B. J. et al. Complete cardiac regeneration in a mouse model of myocardial infarction. Aging 4, 966–977 (2012).
Curigliano, G. et al. Cardiotoxicity of anticancer treatments: epidemiology, detection, and management. CA Cancer J. Clin. 66, 309–325 (2016).
Morelli, M. B. et al. Cardiotoxicity of anticancer drugs: molecular mechanisms and strategies for cardioprotection. Front. Cardiovasc. Med. 9, 847012 (2022).
Cardinale, D. et al. Early detection of anthracycline cardiotoxicity and improvement with heart failure therapy. Circulation 131, 1981–1988 (2015).
Zhang, S. et al. Identification of the molecular basis of doxorubicin-induced cardiotoxicity. Nat. Med. 18, 1639–1642 (2012).
Linders, A. N. et al. A review of the pathophysiological mechanisms of doxorubicin-induced cardiotoxicity and aging. npj Aging 10, 9 (2024).
Kim, Y. A. et al. Doxorubicin-induced heart failure in cancer patients: a cohort study based on the Korean National Health Insurance Database. Cancer Med. 7, 6084–6092 (2018).
Mitry, M. A. & Edwards, J. G. Doxorubicin induced heart failure: phenotype and molecular mechanisms. Int. J. Cardiol. Heart Vasc. 10, 17–24 (2016).
Swain, S. M., Whaley, F. S. & Ewer, M. S. Congestive heart failure in patients treated with doxorubicin: a retrospective analysis of three trials. Cancer 97, 2869–2879 (2003).
Ghigo, A., Li, M. & Hirsch, E. New signal transduction paradigms in anthracycline-induced cardiotoxicity. Biochim. Biophys. Acta 1863, 1916–1925 (2016).
Willis, M. S. et al. Doxorubicin exposure causes subacute cardiac atrophy dependent on the striated muscle-specific ubiquitin ligase MuRF1. Circ. Heart Fail. 12, e005234 (2019).
Chen, D.-S., Yan, J. & Yang, P.-Z. Cardiomyocyte atrophy, an underestimated contributor in doxorubicin-induced cardiotoxicity. Front. Cardiovasc Med. 9, 812578 (2022).
Li, M. et al. Phosphoinositide 3-kinase gamma inhibition protects from anthracycline cardiotoxicity and reduces tumor growth. Circulation 138, 696–711 (2018).
Fabiani, I., Chianca, M., Cipolla, C. M. & Cardinale, D. M. Anthracycline-induced cardiomyopathy: risk prediction, prevention and treatment. Nat. Rev. Cardiol. 22, 551–563 (2025).
Kamphuis, J. A. M. et al. Early- and late anthracycline-induced cardiac dysfunction: echocardiographic characterization and response to heart failure therapy. Cardiooncology 6, 23 (2020).
Strash, N. et al. Human Erbb2-induced Erk activity robustly stimulates cycling and functional remodeling of rat and human cardiomyocytes. eLife 10, e65512 (2021).
Anastasi, S., Lamberti, D., Alemà, S. & Segatto, O. Regulation of the ErbB network by the MIG6 feedback loop in physiology, tumor suppression and responses to oncogene-targeted therapeutics. Semin. Cell Dev. Biol. 50, 115–124 (2016).
Zhong, H. et al. Mig6 not only inhibits EGFR and HER2 but also targets HER3 and HER4 in a differential specificity: implications for targeted esophageal cancer therapy. Biochimie 190, 132–142 (2021).
Shaul, Y. D. & Seger, R. The MEK/ERK cascade: from signaling specificity to diverse functions. Biochim. Biophys. Acta 1773, 1213–1226 (2007).
von Kriegsheim, A. et al. Cell fate decisions are specified by the dynamic ERK interactome. Nat. Cell Biol. 11, 1458–1464 (2009).
Tomasso, A., Koopmans, T., Lijnzaad, P., Bartscherer, K. & Seifert, A. W. An ERK-dependent molecular switch antagonizes fibrosis and promotes regeneration in spiny mice (Acomys). Sci. Adv. 9, eadf2331 (2023).
Yun, M. H., Gates, P. B. & Brockes, J. P. Sustained ERK activation underlies reprogramming in regeneration-competent salamander cells and distinguishes them from their mammalian counterparts. Stem Cell Rep. 3, 15–23 (2014).
Missinato, M. A. et al. Dusp6 attenuates Ras/MAPK signaling to limit zebrafish heart regeneration. Development 145, dev157206 (2018).
Shakked, A. et al. Redifferentiated cardiomyocytes retain residual dedifferentiation signatures and are protected against ischemic injury. Nat. Cardiovasc. Res. 2, 383–398 (2023).
Liu, X. et al. Neuregulin-1/erbB-activation improves cardiac function and survival in models of ischemic, dilated, and viral cardiomyopathy. J. Am. Coll. Cardiol. 48, 1438–1447 (2006).
Odiete, O., Hill, M. F. & Sawyer, D. B. Neuregulin in cardiovascular development and disease. Circ. Res. 111, 1376–1385 (2012).
De Keulenaer, G. W. et al. Mechanisms of the multitasking endothelial protein NRG-1 as a compensatory factor during chronic heart failure. Circ. Heart Fail. 12, e006288 (2019).
Gu, X. et al. Cardiac functional improvement in rats with myocardial infarction by up-regulating cardiac myosin light chain kinase with neuregulin. Cardiovasc. Res. 88, 334–343 (2010).
Bian, Y. et al. Neuregulin-1 attenuated doxorubicin-induced decrease in cardiac troponins. Am. J. Physiol. Heart Circ. Physiol. 297, H1974–H1983 (2009).
Galindo, C. L., Ryzhov, S. & Sawyer, D. B. Neuregulin as a heart failure therapy and mediator of reverse remodeling. Curr. Heart Fail. Rep. 11, 40–49 (2014).
De Simone, A. et al. Control of osteoblast regeneration by a train of Erk activity waves. Nature 590, 129–133 (2021).
Wen, X., Jiao, L. & Tan, H. MAPK/ERK pathway as a central regulator in vertebrate organ regeneration. Int. J. Mol. Sci. 23, 1464 (2022).
Xiao, C. & Xiong, J.-W. ERK signaling waves via body-wall muscles guide planarian whole-body regeneration across long distances. Cell Regen. 12, 36 (2023).
Zhang, X.-S. et al. ERK-activated CK-2 triggers blastema formation during appendage regeneration. Sci. Adv. 10, eadk8331 (2024).
Han, P. et al. Hydrogen peroxide primes heart regeneration with a derepression mechanism. Cell Res. 24, 1091–1107 (2014).
Liu, P. & Zhong, T. P. MAPK/ERK signalling is required for zebrafish cardiac regeneration. Biotechnol. Lett. 39, 1069–1077 (2017).
Ohashi, A. et al. Axolotl liver regeneration is accomplished via compensatory congestion mechanisms regulated by ERK signaling after partial hepatectomy. Dev. Dyn. 250, 838–851 (2021).
Duprey-Díaz, M. V., Blagburn, J. M. & Blanco, R. E. Exogenous modulation of retinoic acid signaling affects adult rgc survival in the frog visual system after optic nerve injury. PLoS ONE 11, e0162626 (2016).
Yasumuro, H., Sakurai, K., Toyama, F., Maruo, F. & Chiba, C. Implications of a multi-step trigger of retinal regeneration in the adult newt. Biomedicines 5, 25 (2017).
Suzuki, M., Satoh, A., Ide, H. & Tamura, K. Transgenic Xenopus with prx1 limb enhancer reveals crucial contribution of MEK/ERK and PI3K/AKT pathways in blastema formation during limb regeneration. Dev. Biol. 304, 675–686 (2007).
Blassberg, R. A., Garza-Garcia, A., Janmohamed, A., Gates, P. B. & Brockes, J. P. Functional convergence of signalling by GPI-anchored and anchorless forms of a salamander protein implicated in limb regeneration. J. Cell Sci. 124, 47–56 (2011).
Fan, Y. et al. Ultrafast distant wound response is essential for whole-body regeneration. Cell 186, 3606–3618 (2023).
Hachemi, Y. et al. Molecular mechanisms of glucocorticoids on skeleton and bone regeneration after fracture. J. Mol. Endocrinol. 61, R75–R90 (2018).
Choi, S. et al. Corticosterone inhibits GAS6 to govern hair follicle stem-cell quiescence. Nature 592, 428–432 (2021).
Kyritsis, N. et al. Acute inflammation initiates the regenerative response in the adult zebrafish brain. Science 338, 1353–1356 (2012).
Bongiovanni, C. et al. Protocol for isolating and culturing neonatal murine cardiomyocytes. STAR Protoc. 5, 103461 (2024).
Omatsu-Kanbe, M., Yoshioka, K., Fukunaga, R., Sagawa, H. & Matsuura, H. A simple antegrade perfusion method for isolating viable single cardiomyocytes from neonatal to aged mice. Physiol. Rep. 6, e13688 (2018).
Ackers-Johnson, M. et al. A simplified, Langendorff-free method for concomitant isolation of viable cardiac myocytes and nonmyocytes from the adult mouse heart. Circ. Res. 119, 909–920 (2016).
Tholen, S. et al. Flattening of circadian glucocorticoid oscillations drives acute hyperinsulinemia and adipocyte hypertrophy. Cell Rep. 39, 111018 (2022).
Chung, S. et al. Cooperative roles of the suprachiasmatic nucleus central clock and the adrenal clock in controlling circadian glucocorticoid rhythm. Sci. Rep. 7, 46404 (2017).
Mi, H., Muruganujan, A., Ebert, D., Huang, X. & Thomas, P. D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426 (2019).
Acknowledgements
We acknowledge financial support under the National Recovery and Resilience Plan, Mission 4, Component 2, Investment 1.1, Call for tender no. 1409 published on 14 September 2022 by the Italian Ministry of University and Research (MUR), funded by the European Union (EU)—NextGenerationEU—Project ‘The cardiomyocyte-intrinsic role of the glucocorticoid receptor in cardiac aging’, CUP J53D23018140001 grant assignment decree no. 1369 adopted on 1 September 2023 by the MUR to G.D'U. The research was also supported by the EU’s Horizon 2020 research and innovation program under the ERA-NET on the Cardiovascular Diseases co-fund action to G.D'U. and E.T. (grant no. JCT2016-40-080), Fondazione Carisbo to G.D'U. (grant no. 2023.0210) and by the Italian Ministry of Health (grant no. RC24000858-2795994 ex 2790614 to G.D'U.). The views and opinions expressed are those of the authors only and do not necessarily reflect those of the EU or the European Commission. Neither the EU nor the European Commission can be responsible for them. S.D.P. was supported by a PON Research and Innovation 2014–2020 (FSE React-EU) PhD Scholarship (code no. DOT1303972—CUP J35F21003360006) funded by the MUR under DM 1061/2021, Action IV.4 ‘Doctorates and Research Contracts on Innovation Topics’. E.T. was supported by the European Research Council (ERC AdG, grant no. 788194). C. Batho and R.D. were funded by the British Heart Foundation (grant nos. G114642 and GBP0051 to C.W.). We thank the Centre for Applied Biomedical Research, University of Bologna for technical support.
Author information
Authors and Affiliations
Contributions
S.D.P., S.B. and G.D’U. conceived and designed the experiments. S.D.P. and S.B. carried out most of the experiments and analyzed the data. S.D.P. and S.B. performed in vivo experiments. S.B. performed echocardiographic analysis. C.M., F.S., C. Bongiovanni, I.D.B., A.A. and N.P. performed immunofluorescence and gene expression analyses. R.T. performed in vitro immunofluorescence image acquisition and time-lapse imaging. S.D.P., S.B. and G.D’U. analyzed RNA-seq data. C. Batho and R.D. designed and synthesized modRNAs. C.V., M.L., E.T. and C.H.W. supervised the experiments performed by their laboratory members. G.D’U. conceptualized the study and supervised the entire project. S.D.P., S.B. and G.D’U. wrote the paper, with editing contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
G.D’U., E.T. and A.A. are listed as co-inventors on a patent related to ERBB2-mediated cardiac regeneration (no. US20160326250A1). The other authors declare no competing interests.
Peer review
Peer review information
Nature Cardiovascular Research thanks Richard Lee and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Representative images showing cardiomyocyte proliferation after in vitro treatment with regenerative growth factors, as determined by BrdU incorporation assay.
Cardiomyocytes were identified by cardiac Troponin I (cTnI) staining and analysed by immunofluorescence for DNA synthesis (BrdU incorporation assay). White arrows point to proliferating cardiomyocytes; scale bars 50 μm.
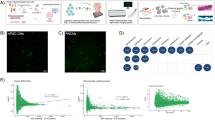
Extended Data Fig. 2 Corticosterone impairs phenotypic changes induced by caERBB2 expression.
(a) Analysis of the efficacy of lipofectamine-mediated modRNA transfection of postnatal day 1 (P1) cardiomyocytes cultured in vitro. Cardiomyocytes were identified by cardiac Troponin T (cTnT) staining and transfection was assessed through eGFP expression 24 h after modRNA delivery. A representative figure is provided; scale bar 50 μm; (b-c) Subdivision of Ki67+ postnatal day 1 cardiomyocytes analysed in Fig. 1g into mononucleated (b) and binucleated (c) cells; (d-e) Analysis of area (d) and Troponin T intensity (e) of postnatal day 1 (P1) cardiomyocytes cultured in vitro and transfected for 48 h with a modRNA encoding a constitutively active isoform of ERBB2 (caERBB2) together with corticosterone (CORT, 10−8 M). Cardiomyocytes were identified by cardiac Troponin T (cTnT) staining. Details on the number of replicates and experiments for each panel are provided in Supplementary Table 2. The values are presented as mean (error bars show standard deviation), statistical significance was determined using one-way ANOVA followed by Sidak’s test in (b), (c), (d), and (e) (comparison between pairs of treatments).
Extended Data Fig. 3 Analysis of hypertrophic changes in cardiomyocytes following in vivo administration of corticosterone in early postnatal life.
(a-i) RT-qPCR analysis of mRNA expression of (a) Myh7, (b) Myh6, (c) Tnni1, (d) Tnni3, (e) Myh7/Myh6, (f) Tnni1/Tnni3, (g) Acta1, (h) Nppa, and (i) Nppb in mouse postnatal day 2-3 (P2-3) heart lysates following in vivo delivery of water-soluble corticosterone-HBC complex (100 µg/ml) in the drinking water of the lactating mother from postnatal day 0–1 to postnatal day 2–3, for a total of 48 h; (j) Cardiomyocyte cross-sectional area evaluation by immunofluorescence analysis of WGA in heart sections of mice at postnatal day 2–3 following in vivo delivery of water-soluble corticosterone-HBC complex (100 µg/ml) in the drinking water of the lactating mother from postnatal day 0–1 to postnatal day 2–3, for a total of 48 h. Representative images are provided. Dashed outlines highlight cross-sectional areas of transversely aligned cardiomyocytes. Scale bars, 30 μm. Details on the number of replicates and experiments for each panel are provided in Supplementary Table 2. The values are presented as mean (error bars show standard deviation), statistical significance was determined using two-sided Student’s t-test in (a) (95% CI: -0.7725 to -0.3350), (b) (95% CI: -0.1838 to 0.1447), (c) (95% CI: -0.3339 to -0.005125), (d) (95% CI: -0.05248 to 0.2029), (e) (95% CI: -0.7746 to -0.3126), (f) (95% CI: -0.4603 to -0.03509), (g) (95% CI: 0.4683 to 1.389), (h) (95% CI: -0.04992 to 1.751), (i) (95% CI: -0.2244 to 0.9094), and (j) (95% CI: -4.281 to 1.119).
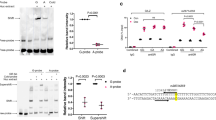
Extended Data Fig. 4 In vitro analysis of the efficacy of Errfi1 and Dusp1 gene knockdown in neonatal cardiomyocyte cultures.
(a-b) mRNA expression levels of (a) Errfi1 and (b) Dusp1 in cultured postnatal day 1 (P1) cardiomyocytes following siRNA delivery for 48 h. A pool of non-targeting scramble siRNAs (siSCR) was used as a negative transfection control; (c) Protein expression levels of ERRFI1 and DUSP1 from cultured postnatal day 1 (P1) cardiomyocytes following siRNA delivery for 72 h. A pool of non-targeting scramble siRNAs (siSCR) was used as a negative transfection control; (d) Schematic illustration of experimental set-up in Fig. 3b and Fig. 3d, e. Time-course experiments were performed in whole-cell protein and RNA lysates, with corticosterone administered 6 h before NRG1 treatment for 30, 60, and 120 min. Details on the number of replicates and experiments for each panel are provided in Supplementary Table 2. In all panels, numerical data are presented as mean (error bars show standard deviation). Statistical significance was determined using two-sided Student’s t-test in (a) (95% CI: -0.8423 to -0.2226) and (b) (95% CI: -0.7860 to -0.02741 for siDUSP1).
Extended Data Fig. 5 Activation of NRG1/ERBB2 signalling leads to the expression of IEGs in cardiomyocytes.
(a-c) RT-qPCR analysis of IEGs (Immediate Early Genes), namely (a) Fos, (b) Junb, and (c) Egr1, upon treatment with NRG1 (100 ng/ml) for 30, 60 and 120 min. (d-f) Analysis of (d) Fos, (e) Junb and (f) Egr1 expression from RNA-sequencing data of cardiac tissue of a mouse model with cardiomyocyte-restricted overexpression of a constitutively active ERBB2 (caERBB2) isoform (Aharonov et al.68). Details on the number of replicates and experiments for each panel are provided in Supplementary Table 2. In all panels, numerical data are presented as mean (error bars show standard deviation). Statistical significance was determined by one way ANOVA followed by Tukey’s test in (a-c) and by two-sided Student’s t-test in (d) (95% CI: 30.90 to 248.8), (e) (95% CI: 267.4 to 779.7) and (f) (95% CI: 1618 to 3970).
Extended Data Fig. 6 Dusp1 knockdown abolishes the ability of corticosterone to repress the proliferative potential of BMP7, FGF1 and OSM.
Analysis of cell proliferation by BrdU assay in neonatal (postnatal day 1 - P1) cardiomyocytes cultured in vitro following knock-down of Dusp1 for 48 h with/without subsequent stimulation with BMP7, FGF1 or OSM (10 ng/ml), and/or CORT (10−8 M). A pool of non-targeting scramble siRNAs (siSCR) was used as a negative transfection control. Cardiomyocytes were identified by cTnI staining and analysed by immunofluorescence for DNA synthesis (BrdU incorporation assay). Details on the number of replicates and experiments for each panel are provided in Supplementary Table 2. The values are presented as mean (error bars show standard deviation), statistical significance was determined using one-way ANOVA followed by Sidak’s test (comparison between pairs of treatments).
Extended Data Fig. 7 Glucocorticoid receptor antagonism does not increase the mitogenic activity of NRG1 in postnatal day 1 cardiomyocytes.
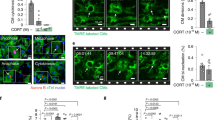
Analysis of cell proliferation by BrdU assay in neonatal (postnatal day 1 - P1) cardiomyocytes cultured in vitro following stimulation with GR antagonist RU486 (10−7 M), alone or in combination with NRG1 (100 ng/ml). Cardiomyocytes were identified by cTnI staining and analysed by immunofluorescence for DNA synthesis (BrdU incorporation assay). Representative pictures are provided; arrows point at proliferating cardiomyocyte; scale bars 50 mm. Details on the number of replicates and experiments for each panel are provided in Supplementary Table 2. The values are presented as mean (error bars show standard deviation), statistical significance was determined using one-way ANOVA followed by Sidak’s test (comparison between pairs of treatments).
Extended Data Fig. 8 Absence of overt cardiomyocyte apoptosis after three weeks of doxorubicin administration.
Analysis of caspase-3 activation (cleaved caspase-3) in heart sections from untreated control animals and from doxorubicin (DOXO)-treated animals, analyzed 21 days after the first DOXO administration. Cardiomyocytes were identified by cardiac Troponin T (cTnT) staining. Representative confocal pictures are provided; scale bars 50 μm.
Extended Data Fig. 9 Echocardiographic evaluation of additional morphological parameters of the in vivo mouse model of anthracycline-induced cardiotoxicity.
Analysis of (a) left ventricular anterior wall thickness at systole (LVAWs), (b) left ventricular end-systolic volume (LVESV), and (c) left ventricular end-diastolic volume (LVEDV) in adult mice at baseline and treated with doxorubicin (DOXO) alone or co-treated with NRG1, RU486, or NRG1 + RU486, 21 days after first DOXO injection. Details on the number of replicates and experiments for each panel are provided in Supplementary Table 2. The values are presented as mean (error bars show standard deviation), statistical significance was determined using one-way ANOVA followed by Sidak’s test in all panels (comparison between pairs of treatments).
Supplementary information
Supplementary Video 1
Time-lapse imaging of cardiomyocyte division after TMRE staining.
Supplementary Tables
Supplementary Table 1 Open reading frame (ORF), T7 promoter and poly(A) tail sequences used for modRNA synthesis. Supplementary Table 2 Details on the number of replicates and independent experiments for each panel shown in the manuscript. Supplementary Table 3 List of GR target genes in cardiomyocytes. List of significantly upregulated genes (log(fold-change) > 0, Padj < 0.05) by RNA-seq analysis of cultured neonatal cardiomyocytes treated in vitro with corticosterone (10−6 M). Supplementary Table 4 List of primers used in this manuscript.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Figs. 2e,k, 3a,b,e and 4c
Uncropped western blots.
Source Data Extended Data Fig. 4c
Uncropped western blots.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Da Pra, S., Boriati, S., Miano, C. et al. Harnessing glucocorticoid receptor antagonism to enhance the efficacy of cardiac regenerative growth factors and cytokines. Nat Cardiovasc Res (2026). https://doi.org/10.1038/s44161-026-00776-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s44161-026-00776-9