Abstract
Although invariant Vα14+ natural killer T cells (NKT cells) are thought to be generated from CD4+CD8+ double-positive (DP) thymocytes, the developmental origin of CD4–CD8– double-negative (DN) NKT cells still remains unresolved. Here we provide definitive genetic evidence obtained, through studies of mice with DP-stage-specific ablation of expression of the gene encoding the recombinase component RAG-2 (Rag2) and by a fate-mapping approach, that supports the proposal of the existence of an alternative developmental pathway through which a fraction of DN NKT cells with strong T-helper-type-1 (TH1)-biased and cytotoxic characteristics develop from late DN-stage thymocytes, bypassing the DP stage. These findings provide new insight into understanding of the development of NKT cells and propose a role for timing of expression of the invariant T cell antigen receptor in determining the functional properties of NKT cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Taniguchi, M., Harada, M., Kojo, S., Nakayama, T. & Wakao, H. The regulatory role of Vα14 NKT cells in innate and acquired immune response. Annu. Rev. Immunol. 21, 483–513 (2003).
Lantz, O. & Bendelac, A. An invariant T cell receptor α chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4–8– T cells in mice and humans. J. Exp. Med. 180, 1097–1106 (1994).
Dellabona, P., Padovan, E., Casorati, G., Brockhaus, M. & Lanzavecchia, A. An invariant Vα24-αQ/Vβ11 T cell receptor is expressed in all individuals by clonally expanded CD4-8- T cells. J. Exp. Med. 180, 1171–1176 (1994).
Borg, N.A. et al. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature 448, 44–49 (2007).
Bendelac, A., Savage, P.B. & Teyton, L. The biology of NKT cells. Annu. Rev. Immunol. 25, 297–336 (2007).
Salio, M., Silk, J.D., Jones, E.Y. & Cerundolo, V. Biology of CD1- and MR1-restricted T cells. Annu. Rev. Immunol. 32, 323–366 (2014).
Brossay, L. et al. CD1d-mediated recognition of an α-galactosylceramide by natural killer T cells is highly conserved through mammalian evolution. J. Exp. Med. 188, 1521–1528 (1998).
Lee, P.T., Benlagha, K., Teyton, L. & Bendelac, A. Distinct functional lineages of human Vα24 natural killer T cells. J. Exp. Med. 195, 637–641 (2002).
Gumperz, J.E., Miyake, S., Yamamura, T. & Brenner, M.B. Functionally distinct subsets of CD1d-restricted natural killer T cells revealed by CD1d tetramer staining. J. Exp. Med. 195, 625–636 (2002).
Crowe, N.Y. et al. Differential antitumor immunity mediated by NKT cell subsets in vivo. J. Exp. Med. 202, 1279–1288 (2005).
Gapin, L., Matsuda, J.L., Surh, C.D. & Kronenberg, M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat. Immunol. 2, 971–978 (2001).
Egawa, T. et al. Genetic evidence supporting selection of the Vα14i NKT cell lineage from double-positive thymocyte precursors. Immunity 22, 705–716 (2005).
Pellicci, D.G. et al. A natural killer T (NKT) cell developmental pathway involving a thymus-dependent NK1.1–CD4+ CD1d-dependent precursor stage. J. Exp. Med. 195, 835–844 (2002).
Benlagha, K., Wei, D.G., Veiga, J., Teyton, L. & Bendelac, A. Characterization of the early stages of thymic NKT cell development. J. Exp. Med. 202, 485–492 (2005).
Aifantis, I. et al. The E delta enhancer controls the generation of CD4–CD8– αβTCR-expressing T cells that can give rise to different lineages of αβ T cells. J. Exp. Med. 203, 1543–1550 (2006).
Baldwin, T.A., Sandau, M.M., Jameson, S.C. & Hogquist, K.A. The timing of TCR α expression critically influences T cell development and selection. J. Exp. Med. 202, 111–121 (2005).
Park, J.H. et al. Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nat. Immunol. 11, 257–264 (2010).
McCaughtry, T.M. et al. Conditional deletion of cytokine receptor chains reveals that IL-7 and IL-15 specify CD8 cytotoxic lineage fate in the thymus. J. Exp. Med. 209, 2263–2276 (2012).
Sun, Z. et al. Requirement for RORγ in thymocyte survival and lymphoid organ development. Science 288, 2369–2373 (2000).
Guo, J. et al. Regulation of the TCRα repertoire by the survival window of CD4+CD8+ thymocytes. Nat. Immunol. 3, 469–476 (2002).
Fehling, H.J., Krotkova, A., Saint-Ruf, C. & von Boehmer, H. Crucial role of the pre-T-cell receptor α gene in development of αβ but not γδ T cells. Nature 375, 795–798 (1995).
Dashtsoodol, N., Watarai, H., Sakata, S. & Taniguchi, M. Identification of CD4–CD8– double-negative natural killer T cell precursors in the thymus. PLoS One 3, e3688 (2008).
Crispe, I.N. Migration of lymphocytes into hepatic sinusoids. J. Hepatol. 57, 218–220 (2012).
Cyster, J.G. Chemokines, sphingosine-1-phosphate, and cell migration in secondary lymphoid organs. Annu. Rev. Immunol. 23, 127–159 (2005).
Jenne, C.N. et al. T-bet-dependent S1P5 expression in NK cells promotes egress from lymph nodes and bone marrow. J. Exp. Med. 206, 2469–2481 (2009).
Villey, I., de Chasseval, R. & de Villartay, J.P. RORγT, a thymus-specific isoform of the orphan nuclear receptor RORγ / TOR, is up-regulated by signaling through the pre-T cell receptor and binds to the TEA promoter. Eur. J. Immunol. 29, 4072–4080 (1999).
He, Y.W., Deftos, M.L., Ojala, E.W. & Bevan, M.J. RORγt, a novel isoform of an orphan receptor, negatively regulates Fas ligand expression and IL-2 production in T cells. Immunity 9, 797–806 (1998).
Wolfer, A. et al. Inactivation of Notch 1 in immature thymocytes does not perturb CD4 or CD8 T cell development. Nat. Immunol. 2, 235–241 (2001).
Laky, K. & Fowlkes, B.J. Presenilins regulate αβ T cell development by modulating TCR signaling. J. Exp. Med. 204, 2115–2129 (2007).
Kastner, P. et al. Bcl11b represses a mature T-cell gene expression program in immature CD4+CD8+ thymocytes. Eur. J. Immunol. 40, 2143–2154 (2010).
Bedel, R. et al. Lower TCR repertoire diversity in Traj18-deficient mice. Nat. Immunol. 13, 705–706 (2012).
Genolet, R., Stevenson, B.J., Farinelli, L., Osterås, M. & Luescher, I.F. Highly diverse TCRα chain repertoire of pre-immune CD8+ T cells reveals new insights in gene recombination. EMBO J. 31, 1666–1678 (2012).
Hendricks, D.W. & Fink, P.J. Uneven colonization of the lymphoid periphery by T cells that undergo early TCRα rearrangements. J. Immunol. 182, 4267–4274 (2009).
Djuretic, I.M. et al. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat. Immunol. 8, 145–153 (2007).
Brigl, M., Bry, L., Kent, S.C., Gumperz, J.E. & Brenner, M.B. Mechanism of CD1d-restricted natural killer T cell activation during microbial infection. Nat. Immunol. 4, 1230–1237 (2003).
Kinjo, Y. et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature 434, 520–525 (2005).
Shimizu, K. et al. Invariant NKT cells induce plasmacytoid dendritic cell (DC) cross-talk with conventional DCs for efficient memory CD8+ T cell induction. J. Immunol. 190, 5609–5619 (2013).
Kanno, R. et al. Evolution of Vα14 TCR gene family in mice. in Genetics in Wild Mice (eds. Moriwaki, K., Shiroishi, T. & Yonekawa, H.), 193–202 (Japan Scientific Societies Press, Karger, Tokyo, Basel, 1994).
Hao, Z. & Rajewsky, K. Homeostasis of peripheral B cells in the absence of B cell influx from the bone marrow. J. Exp. Med. 194, 1151–1164 (2001).
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 (2001).
Watarai, H., Nakagawa, R., Omori-Miyake, M., Dashtsoodol, N. & Taniguchi, M. Methods for detection, isolation and culture of mouse and human invariant NKT cells. Nat. Protoc. 3, 70–78 (2008).
Moro, K., Ealey, K.N., Kabata, H. & Koyasu, S. Isolation and analysis of group 2 innate lymphoid cells in mice. Nat. Protoc. 10, 792–806 (2015).
Dashtsoodol, N. et al. Generation of novel Traj18-deficient mice lacking Vα14 natural killer T cells with an undisturbed T cell receptor α-chain repertoire. PLoS One 11, e0153347 (2016).
Lefranc, M.P. IMGT, the International ImMunoGeneTics Information System for Immunoinformatics: methods for querying IMGT databases, tools, and web resources in the context of immunoinformatics. Mol. Biotechnol. 40, 101–111 (2008).
Acknowledgements
We thank A. Singer (US National Institutes of Health) for E8III-Cre mice; F. Constantini (Columbia University) for Rosa26-YFP mice; D. Littman (New York University) for Rorc–/– mice; P. Burrows for critical reading of this manuscript; T. Hasegawa for support with animal facility services; and N. Takeuchi for secretarial assistance. Supported by the Japan Society for the Promotion of Science (KAKENHI grants 24790490 to N.D. and 23229005 to M.T.).
Author information
Authors and Affiliations
Contributions
N.D. conceived of the study, performed experiments, analyzed data and wrote the paper; T.S., M.A. and R.O. assisted with experiments; S.K. and M.H. discussed experiments; T.A.E. did bioinformatics analyses; T.W. and O.O. performed next-generation sequencing experiments; M.T. directed the study and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
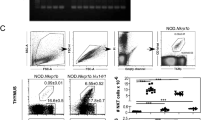
Supplementary Figure 1 Confirmation of the DN-stage origin of NKT cells with the Cd4-Cre+Rosa26-YFP mouse.
(a) YFP reporter expression within gated viable B220-TCRβ+ splenic CD4+, CD8+, and DN fractions obtained from Cd4-Cre+Rosa26-YFP mice. Numbers on flow cytometry plots show the frequency of cells within indicated gates. (b) Detection of CD1d dimer+ NKT cells within gated YFP-negative and YFP-positive TCRβ+ DN splenocytes from Cd4-Cre+Rosa26-YFP mice. Prior to the cell staining, DN splenocytes were purified by AutoMACS using anti-CD4 and anti-CD8 microbeads. Numbers on flow cytometry plots show the percentage and absolute number of CD1d dimer+ NKT cells within gated YFP-negative and YFP-positive DN TCRβ+ cells. Results shown are from two mice done in one experiment.
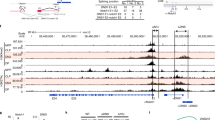
Supplementary Figure 2 TCR α-chain joining-region gene-segment usage among out-of-frame Trav11-containing sequences detected in DN thymocytes from E8III-Cre+Rag2fl/fl mice.
High-throughput sequencing of Trav11Trac transcripts was carried out using the MiSeq system, where cDNA was prepared from sorted CD4-CD8- DN thymocytes from E8III-Cre+Rag2fl/fl mice. Bars depict the frequency (mean ± SEM, n = 3 biological replicates) of Traj gene segment usage among 3,002 out-of-frame Trav11 (CDR3) sequences detected when a total of 55,056 Trav11+ sequences were analyzed as shown in Table 1. The data are from one sequencing experiment.
Supplementary Figure 3 The TCR Vβ repertoire of NKT cells of DN-stage-thymocyte origin.
Flow cytometry analysis of TCR Vβ chain usage analysis of thymic CD1d dimer+CD3ɛ+ DN NKT cells from E8III-Cre+Rag2fl/fl (black bars) and littermate control mice (gray bars). Bars represent mean percentages ± SEM of three independent experiments with at least two mice per group.
Supplementary Figure 4 Detection of Trav11Traj18 transcripts and DN NKT cells in the thymus of Cd4-Cre+Rag2fl/fl mice.
(a) Viable 7-AAD-B220-CD1d dimer-TCRβ-/lo DP and DN thymocyte fractions that were electronically gated as CD5loCD69- cells were sorted from Cd4-Cre+Rag2fl/fl mice, and Trav11Traj18 mRNA expression was determined by qPCR analysis. Relative gene expression (mean ± SEM, n = 3) with Gapdh used as internal control. Results are representative of two experiments, (n.d.) not detected. (b) Detection of CD1d dimer+ DN NKT cells in the thymus, and (c) in the liver from Cd4-Cre+Rag2fl/fl mice, where littermate mice were used as controls. MACS depletion of CD4+, CD8+ and DP cells with magnetic microbeads was done prior to the cell staining. Numbers on flow cytometry plots show the percentage and the absolute number of CD1d dimer+TCRβ+ NKT cells among electronically gated DN cells. Results shown are from one experiment with two mice per genotype analyzed.
Supplementary Figure 5 Detection of Trav11Traj18 transcripts and mature DN NKT cells in the DN thymocyte fraction of Rorc–/– mice.
(a) Trav11Traj18 mRNA expression in sorted CD4+CD8+ DP and CD4-CD8- DN thymic fractions from Rorc-/- mice, as determined by qPCR. Bars show relative expression (mean ± SEM), where Gapdh was used as an internal control. Results are representative from three independent experiments with combined total of three to six mice per group, (n.d.) not detected. (b) Detection of CD1d dimer+ mature NKT cells within DN thymocytes of Rorc-/- mice. Prior to the cell staining, DN thymocytes were purified with AutoMACS by using anti-CD4 and anti-CD8 microbeads. Numbers on plots indicate the percentage and absolute number (mean ± SEM, n=6 mice) of CD1d dimer+ NKT cells within gated DN TCRβ+ cells. Unloaded CD1d staining of DN thymocytes from B6 mice was used as a staining control. Results shown are representative from three independent experiments with two mice per group.
Supplementary Figure 6 Lack of Trav11Traj 18 transcripts as well as mature DN NKT cells in the thymus of Ptcra–/– mice.
(a) The qPCR analysis of Trav11Traj18 mRNA expression in sorted thymic CD4+CD8+ DP, and CD44+CD25- DN1, CD44+CD25+ DN2, CD44-CD25+ DN3, CD44-CD25- DN4 fractions from Ptcra-/- mice. Bars show relative expression (mean ± SEM, n = 3 biological replicates), where Gapdh was used as an internal control. Results are representative from three independent experiments, (n.d.) not detected. (b) Detection of CD1d dimer+ NKT cells within DN thymocytes from B6 and Ptcra-/- mice. Numbers on flow cytometry plots indicate the percentage of CD1d dimer+TCRβ+ NKT cells within electronically gated DN cells. Prior to the cell staining, DN thymocytes were purified with AutoMACS by using anti-CD4 and anti-CD8 microbeads. Results shown are representative from three independent experiments with two mice per group.
Supplementary Figure 7 Surface phenotype of NKT cells of DN-stage-thymus origin is similar to that of B6 wild-type stage 3 NKT cells.
Viable 7-AAD-B220-CD1d dimer+TCRβ+ DN thymic NKT cells from B6 and E8III-Cre+Rag2fl/fl mice were analyzed for the expression of CD24, CD44, and NK1.1 surface antigens by flow cytometry. Numbers on the histograms show the percentage of CD24lo DN NKT cells, and numbers on flow cytometry plots represent the percentage of DN NKT cells within the indicated quadrant gates. Results shown are representative from three independent experiments, n = 3 mice per genotype.
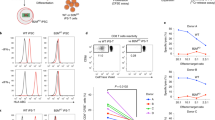
Supplementary Figure 8 Cytotoxic killing assay with YFP– and YFP+ NKT cells from E8III-Cre+Rosa26-YFP mice.
FACS-sorted YFP-negative and YFP-positive thymic DN NKT cells from E8III-Cre+Rosa26-YFP mice were activated with anti-CD3 and anti-CD28 overnight, and then were co-cultured with CellTraceTM Violet labeled YAC-1 target cells at a E/T ratio of 5:1 for 4 hrs. Samples then were FACS analyzed using annexin V and 7-aminoactinomycin D. Bars depict the mean specific cell lysis percentage ± SEM of triplicate samples done in one experiment, and were calculated by the following formula: % cytotoxicity = (target-spontaneous/maximum-spontaneous) ×100.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 (PDF 622 kb)
Rights and permissions
About this article
Cite this article
Dashtsoodol, N., Shigeura, T., Aihara, M. et al. Alternative pathway for the development of Vα14+ NKT cells directly from CD4–CD8– thymocytes that bypasses the CD4+CD8+ stage. Nat Immunol 18, 274–282 (2017). https://doi.org/10.1038/ni.3668
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ni.3668
This article is cited by
-
Loss of Zfp335 triggers cGAS/STING-dependent apoptosis of post-β selection thymocytes
Nature Communications (2022)
-
Modulation of TCR signalling components occurs prior to positive selection and lineage commitment in iNKT cells
Scientific Reports (2021)
-
Thymic iNKT cell differentiation at single-cell resolution
Cellular & Molecular Immunology (2021)
-
Liver X receptors regulate natural killer T cell population and antitumor activity in the liver of mice
Scientific Reports (2021)
-
Liver metastases
Nature Reviews Disease Primers (2021)