Abstract
Middle-aged obesity, characterized by excessive fat accumulation and systemic energy imbalance, often precedes various health complications. Recent research has unveiled a surprising link between DNA damage response and energy metabolism. Here, we explore the role of Eepd1, a DNA repair enzyme, in regulating adipose tissue function and obesity onset. Eepd1 is primarily expressed in adipose tissue, where its downregulation or deletion accelerates obesity development. We show that Eepd1 ablation hinders PKA activation, thereby inhibiting lipolysis and thermogenesis in adipose tissue. Notably, cold exposure enhances Eepd1’s myristoylation, facilitating its anchorage to adipocyte membranes and subsequent activation of PKA, while a mutation at the myristoylation site of Eepd1 disrupts this process. Moreover, individuals with obesity exhibit reduced Eepd1 expression. Pharmacological restoration of Eepd1 with Retigabine dihydrochloride effectively mitigates obesity. This study reveals Eepd1’s unexpected role in promoting adipose lipolysis and thermogenesis, underscoring its potential as a promising therapeutic target to combat obesity.
Similar content being viewed by others
Introduction
As individuals advance into their middle age, a prevalent condition known as “middle-aged spread” or “middle-aged obesity” frequently manifests. This condition is characterized by a gradual rise in body weight, accompanied by abnormal and excessive fat accumulation in adipose tissue, leading to a dysregulation of systemic energy homeostasis, particularly around the abdominal area1. This weight gain serves as a significant risk factor for various comorbidities, such as type 2 diabetes, cardiovascular diseases, and cancers2,3,4,5. Nevertheless, the biological mechanisms underlying this weight gain remain intricate and multifaceted.
Obesity, particularly middle-age obesity, has recently been linked to the induction of DNA damage in adipose tissue due to obesity-induced accumulation of reactive oxygen species and inflammation6. DNA damage triggers the cellular self-rescue processes, including homologous recombination, nonhomologous end joining, base excision repair, and nucleotide excision repair, which are referred to collectively as DNA damage response7. On the other side, cellular DNA damage can also lead to insulin resistance and other cellular metabolic disorders8,9. This vicious cycle aggravates the development of obesity as well as other metabolic diseases. Intriguingly, emerging research indicates a potential link between two seemingly unrelated aspects of life activity: the DNA damage response and systemic metabolism8. Recent research has demonstrated that the downregulation of SREBP1c, a critical transcription factor in lipid metabolism, not only exacerbates systemic insulin resistance but also aggravates adipocyte senescence by impairing poly (ADP-ribose) polymerase 1 (PARP1)-mediated DNA repair10. Moreover, mice with defects in DNA repair enzyme Neil1 exhibit metabolic disorders including obesity, fatty liver, dyslipidemia, and insulin resistance11. Similarly, studies with Ogg1-deficient mice show a connection between oxidative DNA damage repair and metabolic syndrome12. Adipocytes under obesity conditions display significant senescence phenotypes13. Furthermore, obesity or hyperinsulinemia promotes adipocyte cell cycle progression, with insulin driving mature human adipocytes to synthesize DNA and pass the S phase, rendering them more vulnerable to DNA damage stress13. These results indicate that several factors coordinate both the DNA damage response and cellular metabolism to meet life-sustaining requirements.
Adipose lipolysis and thermogenesis are essential adipose metabolic processes for maintaining energy metabolic homeostasis, and their disorder is closely related to development of middle-age obesity. However, to the best of our knowledge, the association between DNA damage response and adipose metabolism has been relatively underexplored. Within this context, it remains unclear whether DNA repair enzymes, in addition to their fundamental role in stabilizing the genome, also play a secondary function in regulating adipose metabolism.
In this work, we explored the microarray datasets of the white adipose tissues from individuals with obesity and obese mouse models to identify the genes potentially involved in both DNA damage response and metabolism. Endonuclease/exonuclease/phosphatase domain containing 1 (Eepd1) stood out among the candidate of DNA damage response genes for its distinctive genotype-phenotype associations. We revealed that Eepd1 was specifically highly expressed in adipose tissue but downregulated in middle-aged obese, diet-induced obese, and genetically obese mice compared to their respective controls. Importantly, Eepd1 ablation leads to a significant reduction in the lipolysis and thermogenesis of adipose tissue and consequently aggravated the development of obesity in mice. Eepd1 enhances lipolysis and thermogenesis primarily through PKA activation, and its myristoylation is essential for membrane localization and its ability to activate PKA. Notably, the small molecule compound Retigabine dihydrochloride markedly increases Eepd1 protein levels in adipocytes, thereby mitigating the progression of obesity.
Results
Eepd1 is highly expressed in adipose tissue but is downregulated in obesity
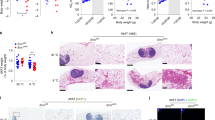
To gain a deeper understanding of the biological mechanisms responsible for middle-aged obesity, we acquired 3-month-old and 14-month-old male C57BL/6 J mice, which are roughly equivalent to 20 and 45 years old in human age, respectively14. As evident from Fig. 1, middle-aged mice exhibited significantly higher body weights compared to their younger counterparts (Fig. 1a, b), despite comparable food intake (Supplementary Fig. 1a). Moreover, adipose tissue weights, encompassing epididymal white adipose tissue (eWAT), inguinal white adipose tissue (iWAT), and brown adipose tissue (BAT), were notably elevated in middle-aged mice (Fig. 1c and Supplementary Fig. 1b). Histological analysis further revealed that adipocytes in both iWAT and eWAT were significantly enlarged in middle-aged mice (Fig. 1d–f). Strikingly, middle-aged mice displayed a substantially reduced capacity to maintain their core temperature following a 4-h exposure to cold temperatures (4 °C) (Fig. 1g). Consistent with this finding, infrared photodetection revealed a lower surface temperature in these mice (Fig. 1h). The glucose tolerance test (GTT) indicated a marked impairment in glucose disposal rate among middle-aged mice compared to young mice (Supplementary Fig. 1c, d). Similarly, the insulin tolerance test (ITT) highlighted significantly poorer whole-body insulin sensitivity in middle-aged mice (Supplementary Fig. 1e, f). Notably, the expression of genes linked to senescence-associated secretory phenotype (SASP) and aging was significantly upregulated in the eWAT of middle-aged mice (Fig. 1i). These findings indicated that middle-aged obesity was associated with disturbances in energy metabolism and glucose metabolism, accompanied by inflammation, DNA damage, and aging, compared to the young control group.
a Representative photograph of mice. b Body weight of young and middle-aged mice (n = 7). c Representative photographs of fat pad morphology. d Representative images of H&E staining from iWAT, eWAT, and BAT. Scale bar, 100 μm. e, f Quantification of adipocyte diameters from panel d: iWAT (325 cells from 3 young mice, 153 from 3 middle-aged mice) and eWAT (117 cells from 3 young mice, 111 from 3 middle-aged mice). g Core temperatures of young and middle-aged mice during cold exposure (4 °C for 4 h) (n = 8). h Representative infrared image of young and middle-aged mice exposed to 4 °C for 4 h. i Relative mRNA levels of indicated genes in eWAT of young and middle-aged mice (n = 5). j A Venn diagram depicting the DNA repair genes from the intersection of differentially expressed genes from the RNA-Seq datasets GSE70353 (human) and GSE167264 (mouse), as well as PBmice database with obesity-related genes. k Eepd1 protein expression profile in C57BL/6 J WT mouse tissues as detected by immunoblot. l Immunoblots of Eepd1 in eWAT of young and middle-aged mice. m Immunoblots of Eepd1 in iWAT and eWAT from DIO mice and WT controls of the same age. n Immunoblots of Eepd1 in iWAT and eWAT of 8-week-old male db/db mice and littermate control mice. o Immunoblots of Eepd1 in iWAT and eWAT of 8-week-old male ob/ob mice and WT mice. p Visualization of Eepd1 expression in Single cell sequencing data. q Immunoblots of Eepd1 and Fabp4 in mature adipocytes and SVF from iWAT and eWAT of C57BL/6 J mice. Data presented as mean ± SEM; Significance was assessed by two-way ANOVA (g) or unpaired two-tailed Student’s t test (b, e, f, i). OB, with obesity. iWAT, inguinal white adipose tissue. eWAT, epididymal white adipose tissue. BAT, brown adipose tissue. DIO, diet-induced obesity. SVF, stromal vascular fraction. GAPDH, β-tubulin and β-actin were used as loading controls for Immunoblots. Source data are provided as a Source Data file.
To identify DNA repair genes potentially associated with obesity and adipose thermogenesis, we retrieved microarray datasets GSE70353 (Lean participants vs. Participants with obesity) from the Gene Expression Omnibus (GEO) database and identified 137 differentially expressed DNA repair genes (Supplementary Fig. 1g). Additionally, we found 144 differentially expressed DNA repair genes in the GSE167264 dataset (Lean mice vs. Mice with obesity) (Supplementary Fig. 1h), also sourced from the GEO database. We also analyzed the hits from a screen for 37 obesity-related genes in the piggyBac mice (PBmice) database15,16. Among the intersections across these three databases, Eepd1, a DNA damage repair enzyme, emerged as a key candidate gene potentially playing a pivotal role in the development of obesity (Fig. 1j). We observed that the expression of Eepd1 is predominantly localized in the eWAT and iWAT among various tissue types (Fig. 1k and Supplementary Fig. 1i). Intriguingly, the expression levels of Eepd1 in eWAT were notably reduced in middle-aged mice compared to their younger counterparts (Fig. 1l). Consistent with these observations, Eepd1 levels were also downregulated in the iWAT and eWAT of obesity models such as diet-induced obese (DIO) mice (Fig. 1m and Supplementary Fig. 1j), leptin receptor-deficient mice (db/db) (Fig. 1n), and leptin-deficient mice (ob/ob) (Fig. 1o), when compared to their respective controls. To gain further insights into the role of Eepd1 in different cell types within adipose tissue, we retrieved adipose tissue single-nucleus RNA sequencing (snRNA-seq) data for mouse samples from the curated RDS format files available on the website https://gitlab.com/rosen-lab/white-adipose-atlas. This analysis uncovered distinct expression patterns of Eepd1, emphasizing its prominent expression in adipocytes of both wild-type (WT) and DIO mice (Fig. 1p). Remarkably, the expression levels of Eepd1 were significantly reduced in adipocytes of DIO mice compared to WT mice (Fig. 1p). To delve deeper, we isolated mature adipocytes and stromal vascular fraction (SVF) cells from the iWAT and eWAT of C57BL/6 J mice. Our results validated that Eepd1 is predominantly expressed in mature adipocytes (Fig. 1q), and its expression in these cells was significantly downregulated in DIO mice (Supplementary Fig. 1k). Collectively, these findings point to a potential metabolic role of Eepd1 in adipocytes, closely linked to the development of obesity.
Eepd1 ablation in adipose tissue facilitates middle-aged obesity development
The prominent expression of Eepd1 in mature adipocytes, coupled with its downregulation in obesity, prompted us to hypothesize that Eepd1 in adipose tissue plays a pivotal role in the development of obesity. To validate this, we generated mice with adipocyte-specific Eepd1 knockout (Eepd1AKO) by breeding Eepd1flox/flox mice with adiponectin-Cre mice (Supplementary Fig. 2a). Our results showed that Eepd1 was downregulated specifically in adipose tissues, with no changes observed in liver and skeletal muscle of Eepd1AKO mice (Supplementary Fig. 2b). Additionally, we confirmed successful knockout of Eepd1 in mature adipocytes isolated from Eepd1AKO mice (Fig. 2a). To investigate whether the downregulation of Eepd1 expression contributes to middle-age weight gain, we monitored Eepd1flox/flox and Eepd1AKO mice under a standard diet until they reached 14 months of age. Notably, despite no significant alterations in food consumption (Supplementary Fig. 2c), Eepd1AKO mice exhibited a significant increase in body weight compared to their Eepd1flox/flox counterparts (Fig. 2b, c). Furthermore, our analysis revealed a substantial increase in the volume of all three adipose tissue types in Eepd1AKO mice (Fig. 2d). Histological examinations through hematoxylin and eosin (H&E) staining demonstrated enlarged adipocytes in both the iWAT and the eWAT of Eepd1AKO mice, accompanied by an accumulation of lipids in the BAT (Fig. 2e–g). Subsequently, we exposed the mice to acute cold conditions and discovered that Eepd1AKO mice exhibited a significantly compromised ability to maintain their core body temperature under cold stress (Fig. 2h). Infrared imaging corroborated this finding, revealing lower surface temperatures in Eepd1AKO mice compared to controls, indicative of reduced heat production (Fig. 2i). Furthermore, metabolic cage measurements confirmed that Eepd1AKO mice displayed a decreased energy expenditure compared to the control group, as evidenced by analysis of covariance (ANCOVA) with body weight as a covariate (Fig. 2j, k). Importantly, this decrease could not be attributed to changes in locomotor activity (Supplementary Fig. 2d). Additionally, Eepd1AKO mice displayed a respiratory exchange rate (RER) that was comparable to that of control mice (Supplementary Fig. 2e). The plasma insulin levels were higher in Eepd1AKO mice (Fig. 2l). Moreover, the GTT and ITT revealed impairments in glucose tolerance and insulin sensitivity in middle-aged Eepd1AKO mice when compared to control mice (Fig. 2m–p). In summary, our findings demonstrate that the adipocyte-specific deletion of Eepd1 leads to increased body weight, compromised thermogenesis, and reduced energy expenditure in middle-aged Eepd1AKO mice. These observations demonstrate that the downregulation of Eepd1 expression levels may serve as a critical factor contributing to weight gain during middle-age.
Male Eepd1AKO and littermate Eepd1flox/flox control (Eepd1fl/fl) mice were maintained until 14-month-old to induce middle-aged obesity. a Immunoblots of Eepd1 in adipocytes isolated from iWAT, eWAT and BAT of Eepd1fl/fl and Eepd1AKO mice. β-actin was used as a loading control. b Representative photographs of Eepd1fl/fl and Eepd1AKO mice. c Body weights of Eepd1fl/fl and Eepd1AKO mice (n = 9 for Eepd1fl/fl, n = 8 for Eepd1AKO). d Representative photographs of fat pad morphology. e Representative image of H&E staining from iWAT, eWAT and BAT. Scale bar, 50 μm. f, g Quantification of adipocyte diameters from panel e: iWAT (281 cells from 3 Eepd1fl/fl mice, 119 from 3 Eepd1AKO mice) and eWAT (172 cells from 3 Eepd1fl/fl mice, 110 from 3 Eepd1AKO mice). h Core temperatures of Eepd1fl/fl (n = 9) and Eepd1AKO (n = 5) mice during cold exposure. i Representative infrared images of Eepd1fl/fl and Eepd1AKO mice exposed to 4 °C for 4 h. j Energy expenditure analyzed with ANCOVA using body weight as covariate for Eepd1fl/fl (n = 7) and Eepd1AKO (n = 8) mice. k ANCOVA predicted energy expenditure (EE) at a given body mass of 39 g (n = 7 for Eepd1fl/fl, n = 8 for Eepd1AKO mice). l Plasma insulin concentrations (n = 7 for Eepd1fl/fl, n = 5 for Eepd1AKO mice). m, n GTT and AUC of GTT (n = 6). o, p ITT and AUC of ITT (n = 6 for Eepd1fl/fl, n = 7 for Eepd1AKO). Data presented as mean ± SEM. Significance was assessed by two-way ANOVA (c, h, m, o), two-sided ANCOVA (j), Unpaired two-tailed Student’s t test (k, l, p), or two-sided Mann-Whitney U test (f, g, n). EE, energy expenditure. GTT, glucose tolerance test. ITT, insulin tolerance test. AUC, area under curve. Source data are provided as a Source Data file.
Adipose tissue-specific deletion of Eepd1 aggravates diet-induced obesity progression
Beyond the process of middle-aging, alterations in dietary habits are also a pivotal contributor to middle-aged obesity. Therefore, we challenged 6-week-old male Eepd1flox/flox and Eepd1AKO mice with a high-fat diet (HFD) for 3 months. Eepd1AKO mice exhibited increased body weight (Fig. 3a, b), despite maintaining a similar food intake (Fig. 3c), compared to Eepd1flox/flox mice. Nuclear magnetic resonance (NMR) analysis revealed a marked increase in fat mass in Eepd1AKO mice (Fig. 3d). A closer examination also revealed that iWAT and eWAT of Eepd1AKO mice substantially expanded after HFD feeding, compared to control mice (Fig. 3e). Histological analysis indicated a larger size of adipocytes within iWAT and eWAT of Eepd1AKO mice and more accumulated lipids in BAT of Eepd1AKO mice (Fig. 3f–h).
a Representative photograph of mice. b Body weights of Eepd1fl/fl (n = 13) and Eepd1AKO (n = 10) mice. c Food intake (n = 4). d Normalized fat mass of Eepd1fl/fl and Eepd1AKO mice via TD-NMR scans (n = 6). e Representative photographs of fat pad morphology. f Representative images of H&E staining of iWAT, eWAT and BAT. Scale bar, 100 μm. g, h Quantification of adipocyte diameters from panel f: iWAT (101 cells from 3 Eepd1fl/fl mice, 53 from 3 Eepd1AKO mice) and eWAT (62 cells from 3 Eepd1fl/fl mice, 61 from 3 Eepd1AKO mice). i Core temperatures of Eepd1fl/fl (n = 11) and Eepd1AKO (n = 7) mice during cold exposure. j Representative infrared images of cold-exposed mice (4 °C, 4 h). k Whole-body energy expenditure under basal conditions during the light and dark phases in Eepd1fl/fl and Eepd1AKO mice (n = 4). l Energy expenditure analyzed with ANCOVA using body weight as covariate for Eepd1fl/fl and Eepd1AKO mice (n = 4). m ANCOVA predicted EE at a given body mass of 36 g (n = 4). n Representative image of H&E staining and IHC of iWAT from Eepd1fl/fl and Eepd1AKO mice after 7 days of cold exposure (8°C). Scale bar, 100 μm. o Immunoblots of Eepd1 in BAT of Eepd1fl/fl and Eepd1AKO mice after 7 days of cold exposure (8°C). β-actin was used as a loading control. p, q qRT‒PCR analysis of indicated genes in iWAT and eWAT of Eepd1fl/fl (n = 13) and Eepd1AKO mice (n = 15). r, s GTT and AUC (n = 12 for Eepd1fl/fl, n = 13 for Eepd1AKO). t, u ITT and AUC (n = 13 for Eepd1fl/fl, n = 16 for Eepd1AKO). Data presented as mean ± SEM. Significance was assessed by two-way ANOVA (b, i, k, r, t), two-sided ANCOVA (l), Unpaired two-tailed Student’s t test (c, d, g, h, m, s), or two-sided Mann-Whitney U test (p, q, u). Source data are provided as a Source Data file.
We then subjected the mice to acute cold exposure (4 °C) and found that Eepd1AKO mice had an impaired capacity to maintain their core temperature during cold exposure (Fig. 3i). Consistently, the surface temperature of Eepd1AKO mice was also downregulated (Fig. 3j). To determine whether the cold intolerance of Eepd1AKO mice was attributable to decreased thermogenesis in adipose tissues, we used metabolic cages to monitor energy expenditure parameters. Strikingly, the energy expenditure of Eepd1AKO mice was significantly lower than that of control mice (Fig. 3k and Supplementary Fig. 2f). To confirm, we reanalyzed the data with ANCOVA and the differences in energy expenditure remained consistently significant even when body weight was used as a covariate (Fig. 3l, m). Intriguingly, locomotor activity and RER remained was comparable between the two groups (Supplementary Fig. 2g, h). Notably, Uncoupling protein 1 (Ucp1) abundance was downregulated in the iWAT and BAT of Eepd1AKO mice compared to controls after cold exposure (Fig. 3n, o). Furthermore, Eepd1 deletion significantly downregulated the expression of other thermogenic genes such as Peroxisome proliferator-activated receptor-γ coactivator 1α (Pgc1α) and PR domain-containing 16 (Prdm16) in adipose tissue (Fig. 3p, q), indicating an attenuation of thermogenesis activity in Eepd1AKO mice. Furthermore, The GTT demonstrated a substantial obstruction in the glucose disposal rate in Eepd1AKO mice (Fig. 3r, s). An ITT also revealed that Eepd1AKO mice had considerably worse whole-body insulin sensitivity (Fig. 3t, u). Similar findings were also observed in female Eepd1AKO mice when subjected to a HFD (Supplementary Fig. 2i-r), suggesting that there is no significant sex-dependent effect on the observed phenotype.
To validate our findings in Eepd1AKO, we also generated global Eepd1-knockout (EKO) mice (Supplementary Fig. 3a, b). When challenged with a HFD, EKO mice exhibited significantly higher body weight (Supplementary Fig. 3c, d) and total fat mass (Supplementary Fig. 3e) compared to WT littermates, despite similar food intake (Supplementary Fig. 3f). Specifically, the adipose depots were enlarged (Supplementary Fig. 3g, h), and adipocytes in the iWAT, eWAT and BAT of EKO mice were larger than controls (Supplementary Fig. 3i-k). Notably, EKO mice displayed reduced tolerance to cold conditions (Supplementary Fig. 3l), attributable to decreased thermogenesis in adipose tissues (Supplementary Fig. 3m, n). This was evidenced by a blockage in energy expenditure (Supplementary Fig. 3o–r) and downregulation of thermogenic genes in eWAT (Supplementary Fig. 3s). Furthermore, EKO mice exhibited significantly elevated blood glucose levels (Supplementary Fig. 3t) and circulating insulin concentrations (Supplementary Fig. 3u) than control mice. The GTT and ITT revealed an impairment in glucose homeostasis in EKO mice relative to control mice (Supplementary Fig. 3v–y).
Collectively, the findings indicate that the absence of Eepd1 in mice significantly impairs thermogenesis in adipose tissue, leading to obesity progression and dysregulation of glucose metabolism.
Eepd1 deletion suppressed PKA activation and thermogenesis in adipose tissue
Consistent with the in vivo observation of suppressed thermogenesis following Eepd1 deletion, our in vitro experiments revealed a decrease in the oxygen consumption rate (OCR) in Eepd1-knockdown adipocytes (Fig. 4a and Supplementary Fig. 4a, b). Additionally, Eepd1 silencing led to downregulation of the intracellular expression levels of Ucp1 (Fig. 4b). Conversely, Eepd1 overexpression significantly upregulated the OCR in adipocytes (Fig. 4c and Supplementary Fig. 4a, c and d) as well as the expression levels of Ucp1 (Fig. 4d). Lipolysis, a crucial step in non-shivering thermogenesis of adipose tissues, facilitates fuel supply and Ucp1 activation17,18. Both basal and isoproterenol (ISO)-stimulated glycerol release and non-esterified fatty acid (NEFA) release assays demonstrated that Eepd1 silencing suppressed lipolysis activity in adipocytes (Fig. 4e, f) and adipose tissues (Supplementary Fig. 4e–l). In contrast, Eepd1 overexpression enhanced lipolysis activity in vitro (Fig. 4g, h).
a C3H10T1/2-derived adipocytes were transfected with siRNAs and then OCR was measured by Seahorse assay (n = 6 biological replicates). b qRT‒PCR analysis of the indicated genes (n = 4 biological replicates). c C3H10T1/2-derived adipocytes were reverse transfected with mock or Eepd1 expressing plasmids and then OCR was measured by Seahorse assay (n = 7 biological replicates). d qRT‒PCR analysis of the indicated genes in C3H10T1/2-derived adipocytes transfected with plasmids (n = 6 biological replicates). e, f Glycerol and NEFA release assay in C3H10T1/2-derived adipocytes treated with NC or siEepd1 for 48 h (n = 6 biological replicates). g, h Glycerol and NEFA release assay in C3H10T1/2-derived adipocytes transfected with indicated plasmids for 48 h (n = 5 biological replicates). i Heatmaps of the leading gene expression from the pathway of regulation of thermogenesis and lipolysis in adipocytes. j KEGG and GO analysis of the data from RNA-seq. k, l Immunoblots of phospho-PKA substrates (RRXS*/T*) and HSL in iWAT and eWAT from WT and EKO mice fed a HFD for 12 weeks. β-actin was used as a loading control. m Immunoblots of phospho-PKA substrates (RRXS*/T*) and HSL in C3H10T1/2-derived adipocytes treated with NC or siEepd1 for 48 h. GAPDH was used as a loading control. n Immunoblots of phospho-PKA substrates (RRXS*/T*) and HSL in C3H10T1/2-derived adipocytes transfected with plasmids for 48 h. β-actin was used as a loading control. o Immunoblots of phospho-PKA substrates (RRXS*/T*) and HSL in C3H10T1/2-derived adipocytes incubated with H89 for 2 h and then transfected with indicated plasmids for 48 h. β-actin was used as a loading control. p OCR in C3H10T1/2-derived adipocytes incubated with H89 for 2 h and then infected with indicated adenovirus for 48 h (n = 7 biological replicates). Data presented as mean ± SEM. Significance was assessed by unpaired two-tailed Student’s t test (a–h), two-sided Hypergeometric test with Fisher’s exact test (j) or one-way ANOVA (p). OCR, oxygen consumption rate. ISO, isoproterenol. Source data are provided as a Source Data file.
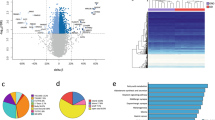
We further conducted an RNA-seq assay comparing eWAT from EKO mice and their WT littermates, uncovering that genes crucial for thermogenesis and lipolysis were among the most downregulated in adipose tissues of EKO mice (Fig. 4i). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed PKA-regulated signaling pathways, including “regulation of lipolysis in adipocytes” and “cAMP signaling pathway”, as the most enriched in EKO mice (Fig. 4j). Similarly, gene ontology (GO) pathway analysis identified PKA-related biological processes, such as “lipid metabolic process”, “brown fat cell differentiation”, and “fatty acid metabolic process”, as the most enriched in Eepd1-deleted adipose tissue (Fig. 4j). These results led us to hypothesize that Eepd1 enhances adipose tissue lipolysis and thermogenesis by activating the PKA signaling pathway. Notably, Eepd1 ablation remarkably reduced PKA substrates phosphorylation in both iWAT and eWAT of EKO mice (Fig. 4k, l). Consistently, phosphorylation of HSL, a direct downstream target of PKA, was also downregulated following Eepd1 deletion (Fig. 4k, l). In line with our in vivo observations, PKA activity was suppressed in adipocytes upon Eepd1 knockdown (Fig. 4m). Conversely, Eepd1 overexpression substantially upregulated phosphorylated PKA substrates and HSL levels (Fig. 4n, o). However, PKA inhibitors, H89 and KT5720, significantly attenuated Eepd1-mediated PKA activation (Fig. 4o and Supplementary Fig. 4m). Consistently, Seahorse analysis demonstrated that H89 treatment abrogated the Eepd1-enhanced OCR of adipocytes (Fig. 4p and Supplementary Fig. 4n). Thus, these results indicate that Eepd1 plays a pivotal role in promoting adipose tissue thermogenesis through activation of the PKA signaling pathway.
Eepd1 activates PKA independently of the activation of the homologous recombination (HR) pathway
Eepd1 is a DNA repair enzyme and functions as a gatekeeper in HR19,20. We next sought to distinguish whether Eepd1-dependent DNA repair processes were involved in Eepd1-mediated PKA activation.
First, we constructed an Eepd1 DNA-binding deficient mutant (∆35-195) by deleting its N-terminal helix-hairpin-helix (HhH) motif (Supplementary Fig. 5a), which is a RuvA-related DNA-binding domain of Eepd1 and plays a crucial role in the HR process21,22,23. Intriguingly, overexpressing Eepd1(∆35-195) also upregulated the phosphorylation levels of PKA substrates (Supplementary Fig. 5b), similar to the action of the wild type, Eepd1(WT). Hydroxyurea (HU), a ribonucleotide reductase inhibitor used to stall DNA replication, can cause DNA damage and thus upregulate phosphorylated checkpoint kinase 1 (Chk1), a crucial DNA damage checkpoint kinase downstream of Eepd121,24. We treated adipocytes with HU and observed a significant increase in the phosphorylation levels of Chk1 and γ-H2AX (Supplementary Fig. 5c). However, Eepd1 overexpression did not lead to an upregulation of p-Chk1 levels. Consistent with our previous observation, Eepd1 overexpression significantly increased the phosphorylation levels of HSL, whereas HU treatment did not enhance HSL phosphorylation (Supplementary Fig. 5c). Moreover, silencing downstream DNA damage response regulator that are involved in the HR pathway, such as ATR (ataxia-telangiectasia-mutated and Rad3-related) and Chk121,25, had little effect on the increase in HSL phosphorylation caused by Eepd1 (Supplementary Fig. 5d–f).
In addition, DAPI staining and quantitative analysis demonstrated that siRNA-mediated Eepd1 knockdown in mature adipocytes did not trigger the formation of micronuclei, a significant biomarker for DNA damage (Supplementary Fig. 5g, h). Similarly, immunostaining of γ-H2AX, a marker of DNA double-strand breaks (DSBs), showed that silencing Eepd1 did not result in an increase in DSBs (Supplementary Fig. 5i). We also included both HU-treated adipocytes and HU-treated Huh7 cells as positive controls. HU treatment did induce DSBs in some adipocytes without causing a significant increase in micronuclei (Supplementary Fig. 5g–i). In contrast, HU treatment led to a pronounced formation of both micronuclei and DSBs in Huh7 cells (Supplementary Fig. 5g–i). These findings underscore that acute deletion of Eepd1 has minimal impact on DNA stability in mature adipocytes, in stark contrast to the effects observed in rapidly dividing cells like Huh7 treated with HU.
Taken together, these findings imply that Eepd1 may perform diverse functions beyond DNA damage repair in the development of obesity.
Myristoylation of the Gly2 residue in Eepd1 is imperative for its mediated thermogenesis and lipolysis
Given that cAMP is a well-known second messenger that activates PKA, we delved into the question of whether Eepd1 exerts its influence on PKA through modulating cAMP levels in adipocytes. Intriguingly, our findings revealed that Eepd1 does not alter intracellular cAMP concentrations (Supplementary Fig. 6a). Similarly, silencing Eepd1 also yielded negligible changes in cAMP levels within adipocytes (Supplementary Fig. 6b). Further, a comparison between adipose tissues from EKO mice and their control counterparts revealed no significant disparities in cAMP levels (Supplementary Fig. 6c, d). These cumulative observations suggest that Eepd1 has a minimal impact on the processes governing cAMP synthesis and degradation.
A previous report identified Eepd1 as a protein dually fatty acylated, with myristoylation at Gly2 and palmitoylation at Cys726. Notably, these two N-terminal acylation sites of Eepd1 are evolutionarily conserved across various species (Fig. 5a). To elucidate the role of these fatty-acylated modifications in Eepd1’s effects on PKA activation and thermogenesis, we treated adipocytes overexpressing Eepd1 with the myristoylation inhibitor 2-hydroxy Myristic Acid methyl ester (2-HM) and the palmitoylation inhibitor 2-bromopalmitate (2-BP). As depicted in Figs. 5b and c, the myristoylation inhibitor 2-HM significantly impaired Eepd1-induced phosphorylation of PKA substrates, whereas the palmitoylation inhibitor 2-BP had minimal impact. Moreover, inhibition of myristoylation significantly reduced HSL phosphorylation (Fig. 5d). To further validate these findings, we created mutant versions of Eepd1: Eepd1(G2A), where Gly2 was replaced with alanine to abolish myristoylation, and Eepd1(C7S), where Cys7 was substituted with serine to prevent palmitoylation. Remarkably, Eepd1(G2A) failed to upregulate the phosphorylation levels of PKA substrates and HSL in C3H10T1/2-derived adipocytes and primary adipocytes, unlike the Eepd1(WT) (Fig. 5e and Supplementary Fig. 6e). However, Eepd1(C7S) retained its ability to promote elevations in phosphorylated PKA substrate and HSL, similar to Eepd1(WT) (Fig. 5e and Supplementary Fig. 6e). Seahorse analysis disclosed that cellular OCR was reduced in adipocytes overexpressing Eepd1(G2A) compared to the WT group (Fig. 5f and Supplementary Fig. 6f). Consistently, Eepd1(G2A) lost its capacity to stimulate lipolysis in adipocytes (Fig. 5g, h).
a Sequence alignment comparison of Eepd1’s first 10 residues across species. b-d Immunoblots of indicated proteins in C3H10T1/2-derived adipocytes reverse-transfected with mock or Eepd1 plasmids for 24 h and then treated with inhibitors for another 24 h. e Immunoblots of phospho-PKA substrates (RRXS*/T*) and HSL in C3H10T1/2-derived adipocytes reverse transfected with indicated plasmids for 48 h. f OCR in C3H10T1/2-derived adipocytes transfected with mock, Eepd1 or Eepd1 mutant plasmids for 48 h (n = 7 biological replicates). g, h Glycerol and NEFA release in C3H10T1/2-derived adipocytes transfected with plasmids for 48 h (n = 4 biological replicates). i NMT1 and NMT2 mRNA levels in human adipose tissues. BMI < 30, n = 14; BMI > 30, n = 28. j–l Immunoblots of Nmt1 and Nmt2 in eWAT from DIO, db/db and ob/ob mice comparing to respective littermate controls. m, n Nmt1 and Nmt2 mRNA levels in iWAT and eWAT from 8-week-old male C57BL/6 J mice exposure to 4 °C (n = 5) or room temperature (RT) (n = 4) for 3 days. o Nmt1 mRNA level in C3H10T1/2-derived adipocytes incubated with ISO for 6 h (n = 12 biological replicates). p Immunoblots of indicated proteins in primary adipocytes transfected with siRNAs for 24 h and then infected with Adv-Ctl or Adv-Eepd1 for another 48 h. q Quantification of p-HSL/HSL (n = 4 technical replicates/group). r Immunoblots of indicated proteins in C3H10T1/2-derived adipocytes transfected with siRNAs for 24 h and infected with adenovirus for another 24 h. s OCR in C3H10T1/2-derived adipocytes transfected with siRNAs for 24 h and then infected with adenovirus for another 24 h (n = 7). Data presented as mean ± SEM. Significance was assessed by unpaired two-tailed Student’s t test (i, n, o), one-way ANOVA (f, g, h, q, s) or two-sided Mann-Whitney U test (m). β-actin and GAPDH were used as loading controls for immunoblots. 2-HM, 2-hydroxy Myristic Acid methyl ester. 2-BP, palmitoylation inhibitor 2-bromopalmitate. Source data are provided as a Source Data file.
N-myristoyltransferases (NMTs) are crucial enzymes that facilitate the myristoylation process by attaching 14-carbon fatty acid myristates to the N-terminal glycine of target proteins27. Our findings revealed a notable decrease in the expression levels of Nmt1 and Nmt2 in adipose tissue samples collected from individuals with obesity and DIO, db/db, and ob/ob mouse models (Fig. 5i–l). Intriguingly, we found that cold exposure profoundly upregulated the expression levels of Nmt1 and Nmt2 in adipose tissues (Fig. 5l–n). Similarly, the administration of the chemical ISO, which mimics the effects of cold stimulation28, significantly enhanced Nmt1 expression in adipocytes (Fig. 5o). However, obese mice subjected to cold conditions show no corresponding increase in Nmt1 and Nmt2 expression (Fig. 5l), hinting at a potential impairment in their regulatory response to cold exposure in the context of obesity.
To further confirm the role of Eepd1 myristoylation in Eepd1-mediated PKA activation, we silenced Nmt1 and Nmt2 expression in primary adipocytes using specific siRNA. The results demonstrated that the knockdown of either Nmt1 or Nmt2 significantly reduced the Eepd1-mediated increase in phosphorylated HSL levels (Fig. 5p, q; Supplementary Fig. 6g–i). Furthermore, concurrent knockdown of both Nmt1 and Nmt2 resulted in an even more pronounced reduction in Eepd1-induced upregulation of HSL phosphorylation (Fig. 5p, q; Supplementary Fig. 6g–i). Consistent findings were also observed in adipocytes derived from C3H10T1/2 cells (Fig. 5r). Furthermore, Seahorse analysis revealed that silencing Nmt1 and Nmt2 substantially reduced the OCR in adipocytes (Fig. 5s and Supplementary Fig. 6j).
Taken together, these findings underscore the unique significance of Eepd1 myristoylation in regulating thermogenesis in mature adipocytes.
Enhanced myristoylation of Eepd1 promotes PKA activation through its association with PKA
Subsequently, we aimed to explore the interplay between the extent of Eepd1’s myristoylation modification and adipose thermogenesis, both in vivo and in vitro. In our study, sixteen 8-week-old male C57BL/6 J mice were administered adenovirus expressing Eepd1-flag into various sites of their iWAT. Following a three-day interval, each mouse was treated with myristic acid and then subjected to either a 25°C or 8°C environment for 16 h (Fig. 6a). Utilizing mass spectrometry analysis, we observed a substantial upregulation in the myristoylation level of Eepd1 in vivo under cold exposure conditions (Fig. 6b). To further assess this phenomenon, we developed a click chemistry-based assay to quantify the myristoylation status of Eepd1 in adipocytes (Fig. 6c). As expected, the G2A mutation eliminated the myristoylation of Eepd1 (Fig. 6d). Notably, the myristoylation level of Eepd1 in adipocytes was enhanced by ISO, norepinephrine (NE), and CL316,243, chemicals that mimic cold stimulation in vitro28 (Fig. 6e). This finding suggests that the myristoylation modification of Eepd1 may play a pivotal role in the activation of thermogenesis. Intriguingly, adipocytes exhibited heightened sensitivity to NE-stimulated PKA activation as indicated by a time-dependent up-regulation of phosphorylated PKA substrates, including HSL, in Eepd1-overexpressed cells compared to controls (Fig. 6f). This indicates that the enhanced myristoylation by NE augments Eepd1’s capacity to activate PKA, thus highlighting its potential role in the regulation of thermogenesis.
a Flowchart for measuring Eepd1 myristoylation levels in vivo using MS43. b MS results of Eepd1 myristoylation in iWAT of C57BL/6 J mice under 8 °C and 25 °C conditions (n = 4). c Flowchart for click chemistry-based myristoylation measurement44. d Click-chemistry-based analysis of Eepd1 myristoylation in C3H10T1/2-derived adipocytes infected with adenovirus expressing Eepd1(WT) or Eepd1(G2A) for 48 h. TCL, total cell lysate. e Click-chemistry-based analysis of the Eepd1 myristoylation in C3H10T1/2-derived adipocytes infected with adenovirus expressing Eepd1(WT) or Eepd1(G2A) for 48 h and then incubated with ISO, NE or CL316243 (CL) for 6 h. f Immunoblots of indicated proteins in mature C3H10T1/2-derived adipocytes infected with indicated adenovirus for 24 h and then incubated with NE for the indicated time. g Click-chemistry-based analysis of Eepd1 myristoylation in iWAT of WT and ob/ob mice. h Immunoblots of Eepd1 in EKO primary adipocytes infected with Adv-Eepd1(WT)-EGFP or Adv-Eepd1(G2A)-EGFP for 48 h, then treated with cycloheximide (CHX) for the indicated time. i Densitometry quantification of Eepd1/β-actin. The dotted line shows Eepd1 half-life (n = 3 technical replicates/group). j Immunoblots of Eepd1 in plasma membrane and cytoplasm of EKO primary adipocytes infected with Adv-Eepd1(WT)-EGFP or Adv-Eepd1(G2A)-EGFP for 48 h. E-cadherin and GAPDH were used as markers for cell membrane and cytoplasm. k, l Representative images of immunofluorescence for Eepd1 (green) in C3H10T1/2-derived adipocytes (k) and HeLa cells (l) infected with either Adv-Eepd1(WT)-EGFP or Adv-Eepd1(G2A)-EGFP. Scale bar, 20 μm. m–q Co-IP assay showing Eepd1 interaction with PKA subunits in 293 T cells. STE, short-time exposure. LTE, long-time exposure. r Co-IP assay showing Eepd1 interaction with HSL, p38 MAPK and CREB in adipocytes overexpressing Eepd1-Flag. s Representative immunofluorescence images of Eepd1 and PKA-RIIα in C3H10T1/2-derived adipocytes infected with the indicated adenovirus. Scale bar, 20 μm. Data presented as mean ± SEM. Significance was assessed by unpaired two-tailed Student’s t test (b) or two-way ANOVA (i). Source data are provided as a Source Data file.
Next, we performed multi-site injections of adenovirus expressing Eepd1-flag into the adipose pads of 8-week-old ob/ob mice and their littermates. Six days post-injection, the mice were administered alkyne myristic acid via gavage and subsequently exposed to an 8°C environment for 12 h (Supplementary Fig. 6k). Adipose tissue was then collected, and Eepd1-flag protein was enriched using Flag antibodies (Supplementary Fig. 6k). Through a click chemistry-based assay, we observed a decrease in the myristoylation of Eepd1 in adipocytes from obese mice (Fig. 6g).
Previous studies have demonstrated that myristoylation modifications could regulate protein stability and degradation29,30. We also observed a significant reduction in Eepd1 protein levels in obese mice (Fig. 1l-p). To this end, primary adipocytes were infected with adenoviruses expressing either WT Eepd1-EGFP or the G2A mutant Eepd1-EGFP for 48 h. Subsequently, these adipocytes were treated with Cycloheximide (CHX) for a specified duration. Our results revealed a time-dependent decrease in WT Eepd1 levels following CHX treatment (Fig. 6h, i, and Supplementary Fig. 6l, m). Notably, the G2A mutant of Eepd1 exhibited an accelerated degradation rate compared to the WT (Fig. 6h, i, and Supplementary Fig. 6l, m). Densitometric analysis of immunoblots confirmed that the half-life of the G2A Eepd1 mutant was reduced from 13.3 h to 9.4 h (p < 0.05) (Fig. 6i). Collectively, these findings suggest that the decreased expression levels of Eepd1 in obesity models are partially attributed to its impaired myristoylation.
A previous study has documented that the myristoylation of the Gly2 residue facilitates Eepd1’s anchoring to the cellular membrane26. To investigate this further, we infected mature adipocytes derived from EKO mice with Adv-Eepd1(WT)-EGFP and Adv-Eepd1(G2A)-EGFP, and then isolated the cell membrane and cytosolic components. Immunoblot analysis revealed that WT Eepd1 was predominantly localized to the cell membrane, whereas the G2A mutant exhibited a primarily cytoplasmic localization (Fig. 6j). Further immunofluorescence analysis consistently indicated that Eepd1(WT) preferentially localized to the cell membrane in various cell types, including mature adipocytes (Fig. 6k), HeLa cells (Fig. 6l), Huh7 cells (Supplementary Fig. 6n) and 293 T cells (Supplementary Fig. 6o). Conversely, the Eepd1(G2A) mutant, lacking the Gly2 myristoylation site, exhibited a diffuse distribution throughout the cytoplasm (Fig. 6k, l; Supplementary Fig. 6n, o). Taken together, these results demonstrate that Gly2 myristoylation is essential for Eepd1 to localize the cell membrane and activate PKA.
We next investigated whether Eepd1 can directly interact with PKA to regulate its activity. To validate the association between Eepd1 and subunits of PKA, we first transfected 293 T cells with Eepd1 and regulatory subunit PKA-RIIα-expressing vectors and immunoprecipitated them with Eepd1; PKA-RIIα was indeed coimmunoprecipitated with Eepd1 (Fig. 6m). Reverse coimmunoprecipitation (co-IP) by immunoprecipitating PKA-RIIα also demonstrated that Eepd1 existed in the precipitate of PKA-RIIα (Fig. 6n). Co-IP assays further revealed an association between Eepd1 and PKA-RIβ (Fig. 6o), as well as an association between Eepd1 and PKA-RIIβ (Fig. 6p). Furthermore, Eepd1 also interacted with the catalytic β subunit of PKA (PKA-Cβ) (Fig. 6q). Intriguingly, PKA substrates such as HSL, p38 MAPK and cAMP-response element binding protein (CREB) were all present in Eepd1 precipitates (Fig. 6r). Furthermore, Eepd1-EGFP and PKA-RIIα-mCherry were found to colocalize well on the cell membrane of mature adipocytes as well as 293 T cells (Fig. 6s and Supplementary Fig. 6p). Taken together, these findings confirm that Eepd1 myristoylation is essential for its localization to the cell membrane and its capacity to activate PKA.
Intervention via Eepd1 overexpression enhances thermogenesis in adipose tissue and ameliorates obesity
Given the observed deficiency of Eepd1 in mice with obesity (Fig. 1l–p), we investigated the potential of Eepd1 overexpression to mitigate obesity onset. We enhanced Eepd1 expression in adipose tissue by administering an adenovirus system via in situ injection into the iWAT of DIO mice. We injected an Eepd1-overexpressing adenovirus (Adv-Eepd1) into the right fat pad and a control adenovirus (Adv-Ctl) into the left fat pad for a week (Fig. 7a). This resulted in a significant reduction in fat mass and adipocyte size (Fig. 7b–d). Moreover, the ex vivo glycerol and NEFA release assay revealed that lipolysis was activated in adipose tissue overexpressing Eepd1 (Fig. 7e, f). Immunoblot analysis showed that Eepd1 overexpression substantially increased the abundance of Ucp1 and phosphorylated PKA substrates, including HSL (Fig. 7g).
a Workflow for administration of adenovirus in iWAT of DIO mice45. b Representative photographs of fat pad morphology. c Representative H&E staining of dissected tissues. Scale bar, 100 μm. d Quantification of iWAT adipocyte diameters from panel c: 291 cells from 3 Adv-Ctl mice, 349 from 3 Adv-Eepd1 mice. e, f Glycerol and NEFA release in iWAT under basal and ISO-stimulated conditions (n = 8 biological replicates). g Immunoblots of indicated proteins in iWAT injected with Adv-Ctl or Adv-Eepd1. h Body weight gain percentage of DIO mice subcutaneously injected with Adv-Ctl (n = 8) or Adv-Eepd1 (n = 7). i Core temperatures of Adv-Eepd1-injected mice (n = 8). j Energy expenditure under basal conditions during the light and dark phases (n = 6 for Adv-Ctl, n = 7 for Adv-Eepd1). k Energy expenditure analyzed with ANCOVA using body weight as covariate (n = 6 for Adv-Ctl, n = 7 for Adv-Eepd1). l ANCOVA predicted EE at a given body mass of 34 g (n = 6 for Adv-Ctl, n = 7 for Adv-Eepd1). m qRT‒PCR analysis of indicated genes in iWAT (n = 8 for Adv-Ctl, n = 6 for Adv-Eepd1). n Body weights of mice injected with AAV-Ctl or AAV-Eepd1 (n = 7). o, p qRT‒PCR analysis of indicated genes in iWAT (n = 7). q Immunoblots of Eepd1 in iWAT treated with Vehicle or Retigabine dihydrochloride. r Body weight of mice treated with Vehicle or Retigabine dihydrochloride for indicated time (n = 8). s Representative photographs of fat pad morphology. t Representative H&E staining of dissected tissues. Scale bar, 100 μm. u Immunoblots of indicated proteins in iWAT treated with Vehicle or Retigabine dihydrochloride. Data presented as mean ± SEM. Significance was assessed by two-way ANOVA (i, j, n, r), two-sided ANCOVA (k), unpaired two-tailed Student’s t test (e, f, h, l), or two-sided Mann-Whitney U test (d, m, o, p). Adv-Ctl, pAdEasy-EF1-MCS-3xFlag-CMV-EGFP; Adv-Eepd1, pAdEasy-EF1-MCS-Eepd1-3xFlag-CMV-EGFP; AAV-Ctl, pAAV2/9-CMV-3xFlag-P2A-mNeonGreen-tWPA; AAV-Eepd1, pAAV2/9-CMV-Eepd1-3xFlag-P2A-mNeonGreen-tWPA. Source data are provided as a Source Data file.
Next, we bilaterally administered Adv-Eepd1 into the iWAT of DIO mice, with Adv-Ctl-injected mice serving as the control group (Supplementary Fig. 7a). Notably, Adv-Eepd1-injected mice exhibited a significant reduction in body weight gain (Fig. 7h, and Supplementary Fig. 7b) and fat mass (Supplementary Fig. 7c, d). Furthermore, adipocytes within the iWAT of Adv-Eepd1-injected appeared smaller (Supplementary Fig. 7e–g). These mice also exhibited an improved tolerance to cold conditions (Fig. 7i). Additionally, they demonstrated an increased energy expenditure, which was unaffected by variations in body mass (Fig. 7j–l), and accompanied by no significant changes in locomotor activity or RER (Supplementary Fig. 7h, i). Importantly, the expression of thermogenic genes, including Ucp1, was significantly upregulated in Adv-Eepd1 mice compared to controls (Fig. 7m).
To eliminate any potential immunogenic effects of adenovirus, we additionally employed the AAV system to overexpress Eepd1 in DIO mice, administering AAV-Eepd1 bilaterally into their iWAT pads, while AAV-Ctl-injected mice served as the control group. One month following a single injection, our phenotypic analysis revealed a substantial reduction in body weight and fat mass (Fig. 7n and Supplementary Fig. 7j, k) and a significant decrease in adipocyte size (Supplementary Fig. 7l–n). Notably, AAV-Eepd1 consistently elevated HSL phosphorylation levels in the iWAT (Supplementary Fig. 7o). It is noteworthy that the overexpression of Eepd1 not only augmented the expression of thermogenic genes but also mitigated the expression of genes linked to the SASP and DNA damage (Fig. 7o, p). Collectively, these findings indicate that the gain of function in Eepd1 may serve as an effective approach to combat obesity.
In our previous research report, we performed a screen using an FDA-approved drug library and identified Retigabine dihydrochloride as small molecule capable of reinstating Eepd1 protein levels31. Here, we detected that Retigabine dihydrochloride also markedly elevated Eepd1 protein levels in adipocytes (Fig. 7q). Given that Eepd1 reduction promotes obesity development, we injected Retigabine dihydrochloride daily into the adipose tissue pads of DIO mice. Notably, within just 11 days of continuous Retigabine dihydrochloride administration, body weight of DIO mice was significantly reduced (Fig. 7r). Additionally, the fat pads of treated mice exhibited redness (Fig. 7s), and H&E staining revealed smaller adipocyte size in these mice (Fig. 7t). Immunoblot analysis further demonstrated a marked increase in phosphorylated PKA substrates, particularly HSL (Fig. 7u). Taken together, our findings indicate that Retigabine dihydrochloride mitigates obesity progression by reversing the reduction of Eepd1.
EEPD1 downregulation links to obesity susceptibility in humans
After conducting rigorous in vivo and in vitro experiments in mice, we have definitively shown that the downregulation of Eepd1 is a critical contributor to the development of obesity. To further validate this observation in humans, we analyzed a comprehensive GWAS data summary and uncovered a significant correlation between a specific EEPD1 polymorphism (rs17275193) and the adjusted waist-to-hip ratio body mass index (WHRadjBMI), thereby highlighting EEPD1 as a gene intricately associated with obesity susceptibility (Fig. 8a). Subsequently, we delved into a single cell sequencing database to examine the expression pattern of Eepd1 in the visceral adipose tissue (VAT) of human subjects. As shown in Fig. 8b, Eepd1 expression was predominantly observed in mature adipocytes, surpassing other cellular components. To confirm these observations, we isolated clinical adipose tissue samples to obtain mature adipocytes and SVF cells, followed by immunoblot analysis, which consistently revealed that EEPD1 is predominantly expressed in mature adipocytes (Fig. 8c). Intriguingly, we found that EEPD1 expression in these mature adipocytes was notably lower in individuals with obesity compared to lean individuals (Fig. 8b, d). These observations align closely with our previous findings regarding the distribution of Eepd1 expression in mice, further emphasizing the critical role of EEPD1 in adipocyte function. To strengthen our findings, we collected adipose tissue samples from patients undergoing laparoscopic surgery. Through qRT-PCR analysis, we discovered a significant negative correlation between EEPD1 mRNA levels and BMI in both subcutaneous adipose tissue (SAT) and VAT (Fig. 8e, f). Furthermore, when we classified the patients into two groups based on their BMI—the normal group with a BMI below 30 kg/m2 and the group with obesity (OB) (those with a BMI exceeding 30 kg/m2)—we observed a significant reduction in EEPD1 expression levels in both SAT and VAT of the OB group (Supplementary Fig. 8a, b). This substantial evidence underscores the critical role of EEPD1 downregulation in the progression of obesity.
a Association analysis of sequence variants with WHRadjBMI. The common SNP rs17275193 that significantly associated with WHRadjBMI in humans was indicated by a purple dot. b Single-cell RNA sequencing analysis of EEPD1 expression in VAT of human with BMI < 30 and BMI > 30. c Immunoblots of EEPD1 in SVF and adipocytes isolated from human adipose tissue. GAPDH was used as a loading control. d Violin plot showing single-cell RNA sequencing analysis of EEPD1 expression in human visceral adipose tissues. e Linear regression analysis of BMI and EEPD1 mRNA expression in human subcutaneous adipose tissues (n = 48). f Linear regression analysis of BMI and EEPD1 mRNA expression in human visceral adipose tissues (n = 46). g Immunoblots of indicated proteins in primary human adipocytes treated with NC or siEEPD1 for 48 h. β-actin was used as a loading control. h, i Glycerol and NEFA release assay in primary human adipocytes treated with NC or siEEPD1 for 48 h (n = 4 biological replicates). j qRT‒PCR analysis of indicated genes in human adipocytes transfected with siRNAs for 48 h (n = 4 biological replicates). k OCR in primary human adipocytes measured by Seahorse assay (n = 6 biological replicates). l Immunoblots of indicated proteins in primary human adipocytes transfected with mock or EEPD1-Flag vectors for 48 h. β-actin was used as a loading control. m, n Glycerol and NEFA release assay in primary human adipocytes transfected with mock or EEPD1-Flag vectors for 48 h (n = 4 biological replicates). o qRT‒PCR analysis of indicated genes in human adipocytes transfected with mock or EEPD1-Flag vectors for 48 h (n = 4 biological replicates). p OCR in primary human adipocytes measured by Seahorse assay (n = 7 biological replicates). The data are presented as the mean ± SEM. Significance was assessed by Pearson correlation analysis (e, f), unpaired two-tailed Student’s t test (d, n, p), one-way ANOVA (h, i, j, k) or two-sided Mann-Whitney U test (m, o). Source data are provided as a Source Data file.
Subsequently, we isolated precursor adipose cells from patients’ fresh adipose tissue and induced their differentiation into adipocytes in vitro. Manipulating EEPD1 expression via siRNA knockdown revealed that silencing EEPD1 reduced phosphorylation of PKA substrates and HSL in human adipocytes (Fig. 8g). This downregulation of EEPD1 not only suppressed lipolysis activity (Fig. 8h, i) but also significantly decreased the expression of UCP1 (Fig. 8j). Notably, the OCR in human adipocytes was significantly reduced following EEPD1 knockdown (Fig. 8k, and Supplementary Fig. 8c). Conversely, overexpressing EEPD1 upregulated phosphorylated PKA substrates (Fig. 8l), enhancing lipolysis (Fig. 8m, n) and increasing the expression of thermogenesis-related genes (Fig. 8o) and OCR (Fig. 8p, and Supplementary Fig. 8d). These findings emphasize the crucial role of EEPD1 in obesity pathogenesis.
Taken together, these findings suggest that EEPD1 plays a critical role in regulating adipocyte function and that increasing EEPD1 expression in the adipocytes of individuals with obesity may efficiently enhance thermogenesis and lipolysis (Fig. 9). This study highlights the potential of EEPD1 as a therapeutic target for the treatment of obesity and its related metabolic disorders.
Under physiological conditions, cold exposure upregulates Nmt1 and Nmt2, which catalyzes the myristoylation of Eepd1, enabling its anchorage to the cell membrane. This modification of Eepd1 is essential for the activation of PKA and its downstream targets, HSL and p38 MAPK. High-fat diets or aging reduce Eepd1 levels, leading to impaired PKA activation and subsequent reductions in lipolysis and thermogenesis in adipose tissue, thereby accelerating obesity development. Administration of Retigabine dihydrochloride effectively restores Eepd1 levels, enhancing energy expenditure and counteracting obesity progression46.
Discussion
The prevalence of obesity in middle-aged populations is significantly higher than in younger individuals, often attributes to a decline in basal metabolic rate, hormonal changes, and lifestyle factors. Middle-age obesity poses risks to physical appearance, mental health, and chronic diseases like type 2 diabetes and cardiovascular issues. Exploring the biological mechanisms behind this trend is crucial. Among the factors, DNA damage repair mechanisms stand out, as obesity-induced stress and inflammation damage DNA in adipose tissue, disrupting cellular metabolism6. Though a molecular link between DNA damage response and cell metabolism is emerging, the precise role of certain genes in both processes remains unclear. In our study, we discovered Eepd1 as a pivotal DNA repair enzyme that effectively triggers lipolysis and augments thermogenesis in adipose tissue, combating obesity through a myristoylated mechanism.
Protein lipidation adds an additional level of functionality to a wide range of proteins. N-myristoylation and S-palmitoylation are two primary forms of protein fatty acylation. N-myristoylation involves adding myristic acid to the N-terminal glycine of nascent polypeptides, while S-palmitoylation modifies cysteine residues with palmitic acid30. These forms of fatty acylation can occur singly or together to regulate protein function, and are implicated in a wide range of biological processes30. Although myristoylation and palmitoylation are two similar lipid modifications, we found here that G2A but not C7S mutation blunted Eepd1-mediated PKA activation and thermogenesis, suggesting a unique role of myristoylation in the regulation of adipocyte energy metabolism. Myristoylation is an irreversible attachment of myristic acid to N-terminal glycine of proteins by NMTs32. There are two N-myristoyltransferases present in human cells, known as NMT1 and NMT2. Notably, our findings indicate that NMT1 and NMT2 are significantly upregulated in adipocytes upon cold exposure or adrenergic stimulation but decreased in obese conditions when compared to WT controls. Knocking down either NMT1 or NMT2 significantly reduced the Eepd1-mediated elevation of phosphorylated HSL levels. Furthermore, the concurrent knockdown of both NMT1 and NMT2 leads to an even more substantial decrease in Eepd1-induced HSL phosphorylation. These observations hint at a functional overlap between NMT1 and NMT2 in terms of the myristoylation modification of Eepd1. Previous studies have shown that NMT1 and NMT2 share approximately 76% sequence identity in humans and exhibit overlapping biological functions and substrate selectivity33,34. This functional overlap suggests that both NMT1 and NMT2 may contribute to the myristoylation of Eepd1, thereby demonstrating redundancy in promoting Eepd1-induced HSL phosphorylation.
Myristoylation helps in protein-membrane and protein-protein interactions, and is crucial for various functions like signaling pathways30. We investigated myristoylation levels of Eepd1 using “click chemistry”-based technique and DIA-MS. Our results revealed a significant increase in the in vivo myristoylation level of Eepd1 in adipose tissue when mice were exposed to cold conditions. Consistently, the treatment of adipocytes with β3-AR agonists in vitro promotes Eepd1 myristoylation. Here, we found that myristoylation of Eepd1 plays a critical role in PKA activation. Firstly, myristoylation on the Gly2 residue of Eepd1 endows it with the capability to localize to the plasma membrane. Secondly, co-IP assays revealed a physical association between Eepd1 and PKA. Moreover, immunostaining further confirmed the colocalization of Eepd1 and PKA to cell membrane. Notably, adrenergic stimulation by NE can enhance the sensitivity of Eepd1-mediated PKA activation in adipocytes. Therefore, our findings highlight an unexpected aspect of Eepd1’s functionality in PKA activation, specifically in a myritoylation-dependent protein-membrane mechanism. Previous research has highlighted that myristoylation not only regulates protein plasma membrane anchoring but also protein stability and degradation29,30. Our results demonstrate that the myristoylation of Eepd1 was decreased in adipocytes from obese mice compared to those from WT mice. Furthermore, this decrease was accompanied by a notable reduction in Eepd1 protein levels in the adipose tissues of obesity mouse models. Additionally, mutating the myristoylation site of Eepd1 significantly compromised its protein stability. Collectively, these findings imply that the decreased protein levels of Eepd1 in obesity models are partially attributed to its impaired myristoylation.
Obesity is linked to damage to DNA and a hindered ability to repair damaged DNA. This can lead to disruptions in cellular metabolism, causing adipocyte hypertrophy and insulin resistance, worsening obesity and metabolic diseases8,10,13. Surprisingly, recent research suggests that DNA damage response and cellular metabolism are linked at a molecular level10,11,35. Aging-induced DNA breaks in skeletal muscle activate DNA-dependent protein kinase (DNA-PK), which impairs mitochondrial function and energy metabolism, thereby contributing to weight gain, fitness deterioration, and an increased risk of metabolic diseases36. In the realm of DNA damage response, Eepd1 plays a pivotal role as a gatekeeper. Cells in the S phase are prone to replication fork stalling, also known as replication stress, which is susceptible to DNA damage37. Eepd1 is responsible for the repair of stressed replication forks and initiates the HR repair pathway by cleaving the stalled replication fork21. Our research, however, uncovered an intriguing aspect of Eepd1’s functionality, revealing its role in regulating PKA activation and thermogenesis in adipocyte—a function that operates independently of its roles in DNA repair and the homologous recombination (HR) repair pathway. According to previous reports13, most of mature adipocytes are in a quiescent state or have exited the cell cycle, with minimal DNA replication or division activity, which leads to a reduced risk of replication fork stalling and DNA damage. This explains why the acute silencing of Eepd1 in mature adipocytes can inhibit PKA activation and thermogenesis without causing any apparent DNA damage, as demonstrated by micronuclei analysis and γ-H2AX staining. However, we found that in normal dietary conditions, short-term Eepd1 deficiency (within 6 months) had no effect on the weight of mice, but prolonged Eepd1 deficiency (more than 12 months) could lead to middle-age obesity, accompanied by DNA damage, inflammation, adipose senescence, and glucose metabolism disorders. To further investigate this phenomenon, we opted for Eepd1 overexpression, and remarkably, we discovered that an increase in Eepd1 levels effectively stimulates thermogenesis in adipose tissue, mitigates DNA damage, alleviates inflammation, and delays adipose senescence, thereby reducing obesity and diabetes. Consequently, we hypothesize that during the progression of middle-age obesity, the downregulation of Eepd1 exerts a dual effect by inhibiting PKA activity and accelerating DNA damage.
There are several limitations to this study. We have reported a previously unrecognized function of Eepd1 in physical association with PKA and regulation its activity; however, the details of the interaction between PKA and Eepd1 deserve further investigation by X-ray crystallography and cryogenic electron microscopy. Moreover, our results reveal that Eepd1 mitigates obesity in young mice via PKA activation and reduces adipose senescence and DNA damage in older mice, but how it coordinates these dual effects to protect against obesity and age-related metabolic disorders requires further study. Furthermore, our findings demonstrate that myristoylation of Eepd1 is indispensable for its localization to the cell membrane and its capacity to activate PKA; however, the detailed mechanisms by which Eepd1 interacts with the regulatory and catalytic subunits of PKA to activate it, as well as the subcellular localization of PKA activation, require further investigation.
Taken together, our current findings reveal that Eepd1, a DNA repair enzyme, combats obesity by activating PKA and thermogenesis in adipose tissue and that Eepd1 downregulation might be a cause of middle-age obesity. Importantly, our findings suggest that restoration of Eepd1 expression and targeting of the Eepd1-PKA pathway are promising therapeutic approaches for the treatment of obesity.
Methods
Statement of ethical regulations
All animal experiments were approved by the Institutional Animal Care and Use Committee of Shanghai Sixth People’s Hospital Affiliated with Shanghai Jiao Tong University School of Medicine (No: DWLL 2022-0599). The study protocol involving human subjects was approved by the Medical Ethics Committee of Shanghai Sixth People’s Hospital Affiliated to Shanghai Jiao Tong University School of Medicine (Approval Number: ChiCTR2300074800) and written informed consent was obtained from all participants.
Animals
The animal experiments were conducted in accordance with the Provision and General Recommendation and approved by the Animal Care Committee of Shanghai Sixth People’s Hospital affiliated to Shanghai Jiao Tong University School of Medicine (DWLL2022-0599). All mice used were bred under the C57BL/6 J background. Eepd1 global knockout (EKO) mice and Eepd1flox/flox mice were generated using wild-type C57BL/6 J mice by a CRISPR/Cas9-based protocol. Briefly, Cas9 mRNA, sgRNAs and donor plasmids were injected into fertilized zygotes. Founder EKO mice and Eepd1flox/- mice were bred with C57BL/6 J mice in two rounds. Eepd1flox/flox mice were generated by crossing Eepd1flox/- mice with each other. Mice with adipose tissue-specific knockout of Eepd1 were generated by crossing Eepd1flox/flox mice with Adiponectin-promoter-driven Cre recombinase transgenic mice. C57BL/6 J, ob/ob and db/db male mice (SPF grade) were purchased from Biocytogen Company. The animals were kept at room temperature (24-26°C) in a humidity-controlled (40%~70%) room with a 12 h light/dark cycle. A standard normal chow diet (SLACOM, P1101F-25) and sterile water were given ad libitum unless otherwise. All mice used in experiments throughout the study exhibited normal healthy conditions. Mice in different groups were allocated based on their genotype, or randomly assigned. For studies with specific diets, 6-week-old C57BL/6 J mice were given a high-fat (60% fat, Research Diet, D12492) diet for a total period of 8-14 weeks to generate DIO mice before other experiments. In Fig. 7, DIO mice were orthotopically injected with adenoviruses: pAdEasy-EF1-MCS-3xFlag-CMV-EGFP (designated as Adv-Ctl) and pAdEasy-EF1-MCS-Eepd1-3xFlag-CMV-EGFP (designated as Adv-Eepd1). Each mouse received a dose of 3 ×10^8 plaque-forming units (pfu), with injections administered twice weekly for a duration of one week. ob/ob and DIO mice were orthotopically injected with adeno-associated viruses expressing pAAV2/9-CMV-3xFlag-P2A-mNeonGreen-tWPA (designated as AAV-Ctl) and pAAV2/9-CMV-Eepd1-3xFlag-P2A-mNeonGreen-tWPA (designated as AAV-Eepd1) at a dose of 1 ×1012 viral genomes (vg) per mouse. This injection was administered once, and the phenotypic analysis was conducted four weeks later. DIO mice were administered Retigabine dihydrochloride (25 mg/kg) daily via gavage for a duration of 11 consecutive days. At the end of the experiment (for DIO mice: HFD for 8–14 weeks; for chow-diet mice: feeding for 4-12 weeks; for middle-aged mice: chow-diet for 48-56 weeks), mice were euthanized via CO2-dependent asphyxiation and tissues were harvested.
Human samples
Human samples consisting of subcutaneous and visceral adipose tissues were obtained from 32 individuals with obesity and 21 lean individuals who received laparoscopic surgery at Shanghai Sixth People’s Hospital Affiliated to Shanghai Jiao Tong University School of Medicine (ChiCTR2300074800). Sex was not considered in study design due to limitation on the number of samples. Written informed consent was obtained from all participants, and individuals’ clinical biochemical data were retrieved from medical records. The study protocol involving human subjects was approved by the Medical Ethics Committee of Shanghai Sixth People’s Hospital Affiliated to Shanghai Jiao Tong University School of Medicine (Approval Number: ChiCTR2300074800) and was conducted in accordance with the principles of the Helsinki Declaration II. Biopsies were immediately snap-frozen in liquid nitrogen and stored at -80°C until analysis. The age, sex and body mass index of each individual are provided in Supplementary Table 1.
Cell culture
C3H10T1/2 cells (ATCC, CCL-226) were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, C11995500BT) with 10% fetal calf serum (Selborne, CS-05) and 1% Penicillin/Streptomycin solution (Invitrogen) in 10% CO2 at 37°C. The cells were maintained until fully confluent and then induced to differentiate in DMEM containing 10% fetal bovine serum (FBS, Gibco, 16000-044), 0.5 mM 3-isobutyl-1-methylxanthine (IBMX, Sigma-Aldrich, I7018), 1 μM dexamethasone (Sigma-Aldrich, D4902), 10 μg/mL insulin (Roche, 11376497001) and 1 μM rosiglitazone (Sigma-Aldrich, R2408) for two days, and then the cells were cultured in DMEM (10% FBS, 10 μg/mL insulin and 1 μM rosiglitazone) for an additional 2 days. After these procedures, the cells were maintained in DMEM containing 10% FBS. To obtain mouse primary adipocytes, SVFs isolated from iWAT or eWAT of 4-week-old C57BL/6 J male mice were maintained in DMEM/F12 (Gibco, 11330033) containing 10% FBS until post-confluence. Then cells were induced to differentiate with the same procedure as C3H10T1/2. Human primary adipocytes isolated from human subcutaneous adipose tissue (sex: male; 24 < BMI < 28) (ChiCTR2300074800) were cultured in DMEM/F12 with 20% FBS until reaching post-confluence. Then cells were induced to differentiate into mature adipocytes with 1 μM rosiglitazone, 0.5 mM IBMX, 1 μM dexamethasone and 1 μg/mL insulin for 2 days before being incubated in DMEM/F12 containing 10% FBS, 1 μM rosiglitazone and 1 μg/mL insulin for another 2 days. This induction procedure was repeated several times until lipid droplets appeared. HEK293T was purchased from ATCC (CRL-1573). Huh7 (SCSP-526) and Hela (SCSP-504) were obtained from the National Collection of Authenticated Cell Cultures. These cells were cultured in a 37°C incubator with 5% CO2 and maintained in DMEM supplemented with 10% FBS (Gibco) and 1% Penicillin/Streptomycin solution (Invitrogen). Mycoplasma contamination was tested in all cells at least once a month and consistently proven to be negative.
siRNA transfection, plasmid delivery, and compound treatment
For siRNA transfection, differentiated mature adipocytes were digested with trypsin and incubated with siRNAs (GenePharma, China) using RNAiMAX (Invitrogen) following the manufacturer’s instructions. The siRNAs sequences were listed in Supplementary Table 2. For plasmids transfection, differentiated mature adipocytes were digested with trypsin and reverse transfected with plasmid expressing Eepd1 plasmids using Lipofectamine 3000 (Invitrogen) following the manufacturer’s instructions. For adenovirus infection, differentiated mature adipocytes were digested with trypsin and infected with adenovirus expressing Eepd1 or mutant Eepd1 with the help of 2 μg/mL polybrene (Sigma). For the compounds’ treatment, differentiated mature adipocytes were incubated with H89 (30 μM), KT5720 (10 μM), 2-HM (50 μM), 2-BP (20 μM), ISO (10 μM), CL316243 (1 μM), NE (10 μM) or CHX (20 μg/mL).
Adipose tissue fractionation
The iWAT and eWAT obtained from 6-week-old male C57BL/6 J mice, as well as the corresponding adipose tissues from HFD-induced obese mice and control chow-diet mice, along with SAT from humans (sex: male; 24 < BMI < 28), were minced and then digested in a collagenase buffer containing 2% bovine serum albumin (BSA) and 0.1% (w/v) collagenase I (Sigma-Aldrich, SCR103) for 45 to 60 min in a shaking incubator at 37°C. Following complete digestion, tissue digestive mixture was filtered through 70 μm mesh (BD Biosciences). The adipocytes and SVF were separated by centrifugation at 1,800 g for 5 min. After centrifugation, adipocytes in the supernatant and SVF in the pellet were collected and washed with phosphate buffer saline (PBS) twice.
Immunoblot
Adipose tissue, SVF or adipocytes were lysed with lysis buffer (50 mM Tris-HCl, 1% SDS, pH 7.4) supplemented with ethylene diamine tetraacetic acid (EDTA)-free protease inhibitor cocktail (Thermo Scientific Pierce, A32955) and phosphatase inhibitor cocktail (Thermo Scientific Pierce, A32961). The protein content was determined using bicinchoninic acid (BCA) Protein Assay (Thermo Scientific Pierce, 23225). Equal amounts of protein samples were subjected to SDS‒PAGE and incubated with the indicated primary antibodies overnight. Immunoreactivity was detected with enhanced chemiluminescent autoradiography (Millipore, WBKLS0500).
Chemiluminescence was determined using an AI600 System (GE Healthcare)
The following antibodies were used in this study: anti-PKA-Cβ (12232-1-AP, 1:1000), anti-Eepd1 (24310-1-AP, 1:1000), anti-E-cadherin (20874-1-AP, 1:1000) and anti-myc (60003-2, 1:1000) were purchased from ProteinTech; anti-p-HSL (4126, 1:1000), anti-HSL (4107, 1:1000), anti-p-PKA substrates (9624, 1:1000), anti-β-actin (8457, 1:5000), anti-GAPDH (5174, 1:2000), anti-Hsp90 (4874, 1:2000), anti-p-Chk1 (2348, 1:1000), anti-CREB (9197, 1:1000), anti-p38 MAPK (8690, 1:1000), anti-streptavidin-HRP (3999, 1:1000), anti-γH2AX (2577, 1:1000) and anti-Fabp4 (2120, 1:1000) were purchased from Cell Signaling Technology; anti-Flag (F1804, 1:1000) was purchased from Sigma-Aldrich; anti-Ucp1 (ab234430, 1:1000), anti-Nmt1 (ab186123, 1:1000), anti-Nmt2 (ab270957, 1:1000), anti-PKA-RIIβ (ab75993, 1:1000), anti-PKA-RIβ (ab167325, 1:1000) and anti-Eepd1 (ab220501, 1:1000) were purchased from Abcam; anti-PKA-RIIα (A3889, 1:1000) was purchased from Abclonal; Peroxidase-AffiniPure goat anti-rabbit IgG (FS111-035-003, 1:10000) and Peroxidase-AffiniPure goat anti-mouse IgG (FS115-035-003, 1:10000) were purchased from Jackson ImmunoResearch.
Membrane and cytoplasm protein extraction
Cell membrane and cytoplasm protein were extracted using a commercial kit (Beyotime, P0033) following the manufacturer’s instructions. Briefly, adipocytes infected with adenovirus expressing either Eepd1(WT)-EGFP or Eepd1(G2A)-EGFP were washed with PBS, harvested using a cell scraper, and centrifuged. The cell pellet was resuspended in ice-cold PBS and centrifuged again to ensure complete precipitation. The cells were then treated with PMSF-containing PBS and incubated on ice. After homogenization, the cell suspension was centrifuged to remove nuclei and unbroken cells, followed by precipitation of cell membrane fragments. The supernatant, containing cytosolic proteins, was stored at -80°C. The membrane fragments were further processed by adding DDM-containing PBS, vortexing, and incubating on ice. After centrifuging, the supernatant, now containing the membrane protein solution, was collected and stored at -80°C.
Immunoprecipitation assay
HEK293T cells were transfected with the indicated plasmids for 48 h, lysed in ice-cold IP buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 and 0.5% sodium deoxycholate) containing protease inhibitor cocktail (Thermo Fisher Scientific) for 1 h and centrifuged at 13,000 × g for 15 min. The cell lysates were then incubated with the indicated antibodies and Protein A/G-agarose (Thermo Fisher Scientific, 20421) overnight at 4 °C followed by washing in cold IP buffer. Normal mouse or rabbit IgG (Cell Signaling Technology, 3420 or 3423) was used as a negative control. The immunocomplexes were collected and subjected to immunoblotting using the indicated primary antibodies and corresponding secondary antibodies.
Detection of Eepd1 myristoylation in vitro and in vivo by click chemistry
C3H10T1/2-derived mature adipocytes expressing Flag-tagged Eepd1(WT) or Eepd1(G2A) were grown in DMEM with 2% fatty acid–free BSA containing 100 μM alkyne myristic acid (Click Chemistry Tools, 1164-25) for 16 h. Cells were washed twice with PBS and lysed on ice with IP buffer containing protease inhibitors and phosphatase inhibitors. The supernatant was incubated with Flag beads for 4 h at 4 °C with gentle rotation. After washing 3 times, the protein pellet was eluted with 0.1 M glycine. The supernatant was then neutralized with Tris-base (1.5 M, pH 9.5). The protein concentration was determined using a BCA protein assay kit (Thermo Fisher Scientific). Approximately 50 μg of protein lysates were subjected to a click chemistry reaction with 100 μL of Click-iT reaction buffer containing 60 μM Biotin Picolyl Azide (Click Chemistry Tools, 1167-5), 10 μL of copper (II) sulfate, 10 μL of reducing agent, and 20 μL of Additive 1 solution according to the manufacturer’s instructions (Click Chemistry Tools, 1262). After 30 min of incubation at room temperature, click-labeled samples were enriched using methanol protein precipitation and resolved in IP lysis buffer. The click-labeled samples were subjected to immunoblotting. Streptavidin-HRP was used for detecting azide-labeled myristoylated Eepd1. To detect the Eepd1 myristoylation in vivo, 8-week-old male WT and ob/ob mice were injected with adenovirus expressing Eepd1-Flag (3×10^8/mouse) into multiple sites in their iWAT. Six days later, each mouse was administered 50 mg/kg of alkyne myristic acid solution via oral gavage. Then the mice were exposed to 8°C for 12 h in a cold chamber. Subsequently, iWAT was collected from the mice, and proteins were lysed using IP lysis buffer. The Eepd1 protein was enriched using Flag beads, and subsequently subjected to a click chemistry assay.
Detection of Eepd1 myristoylation in vivo by data-independent acquisition mass spectrometry (DIA-MS)
Eight-week-old male C57BL/6 J mice were injected with adenovirus expressing Eepd1-Flag (3×10^8/mouse) into multiple sites in their iWAT (n = 8 mice). Three days later, each mouse was administered 50 mg/kg of myristic acid solution (prepared in 0.1% BSA) via oral gavage. Half of the mice were then exposed to 8°C for 16 h in a cold chamber, while the other half remained at room temperature (n = 4 mice per group). Subsequently, iWAT was collected from the mice, and proteins were lysed using IP lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 0.5% sodium deoxycholate). Eepd1 protein was enriched using flag beads (Smart-Lifescience). The enriched beads were processed with non-contact ultrasound, and protein concentration was determined. Proteins were reduced, alkylated, and precipitated overnight with acetone: acetic acid (100: 0.1). After centrifugation and washing, trypsin digestion was performed overnight at 37°C. The supernatant was concentrated, desalted, and dissolved in 0.1% formic acid for detection by nano-LC-IMS-MS in diaPASEF mode. MS scanning ranged from 100-1700 m/z, with 32 DIA windows of 26 Da width. Spectronaut 18 software was used for qualitative analysis, and MaxLFQ for protein group quantification. Differences between groups were analyzed by t-test with BH-corrected Q-values.
Immunofluorescence & confocal microscopy
For the immunofluorescence staining of γH2AX, C3H10T1/2-derived adipocytes and Huh7 cells were fixed with 4% paraformaldehyde at room temperature for 15 min and then washed three times with PBS. Subsequently, the cells were permeabilized with 0.1% Triton X-100 in PBS for 10 min, followed by three additional washes with PBS. Blocking was performed using 5% BSA in PBS at room temperature for 1 h. The cells were then incubated overnight at 4 °C with the primary antibody, anti-γH2AX. After incubation, the cells were washed three times with PBS. Next, the secondary antibody was added, and the cells were incubated for 1 h at room temperature in the dark, followed by three more washes with PBS. To prepare the slides for imaging, 4’,6-diamidino-2-phenylindole (DAPI, 1 μg/mL) (Sigma-Aldrich, D9542) mounting medium was added, and a coverslip was inverted onto it carefully to avoid bubbles. The edges of the coverslip were sealed with nail polish. Finally, the cells were imaged using a Zeiss LSM510 META confocal microscope with a 40x water immersion objective to visualize the location and distribution of γH2AX within the cells. In Fig. 6r, cells seeded on poly-L-lysine (Sigma-Aldrich, P8920) coated confocal dishes were infected with adenovirus expressing EGFP-tagged Eepd1 and mCherry-tagged PKA-RIIα for 48 h. Then, the cells were fixed with 3.7% formaldehyde and incubated with DAPI for 10 min. The cells were imaged on a Zeiss LSM510 META confocal microscope with a 63x water immersion objective.
Real-time PCR
Total RNA was extracted from mouse adipose tissues or adipocytes using TRIzol reagent (Thermo Fisher Scientific, 15596). cDNA was synthesized with a First Strand cDNA Synthesis Kit (Vazyme, R222-01-AB). Quantitative real-time PCR (qRT‒PCR) was performed in a LightCycler® 480 Multiwell Plate 384 PCR system (Roche) using SYBR-Green master mix (Vazyme, Q511-02). Relative gene expression was normalized to 18S RNA. qRT‒PCR primers are available in Supplementary Table 3.
RNA sequencing
High-quality RNA of all samples was obtained (OD260/280: 1.8–2, OD230/260: 0.4–0.5, RNA Integrity Number: 7–10). RNA sequencing libraries were prepared from 4 μg of total RNA using the mRNA-seq V2 Library Prep Kit for Illumina (Vazyme, NR601) following the manufacturer’s instructions. Sequencing libraries were quantified using a StepOnePlus™ Real-Time PCR System and quality controlled (QC) with an Agilent Bioanalyzer 2100 (Agilent Technologies, CA, USA). The libraries were sequenced using the HiSeq PE150 (Illumina) for 2× 50 cycles using the HiSeq PE Cluster Kit v4-cBot-HS (Illumina). One percent of the PhiX control library (Illumina) was spiked into each lane as a sequencing control.
Seahorse assay
Cellular OCR was measured using XF Cell Mito Stress Test Kit (Agilent Technologies, 103015–100) and an XF96 Analyzer (Agilent Technologies) with a modified protocol38. Briefly, primary adipocytes or C3H10T1/2 cells were differentiated for 4 days, then trypsinized and transfected with siRNAs or plasmids, or infected with adenovirus. After that, the cells were plated at 5000 cells per well in XF 96-well microplates (Agilent Technologies). Adipocytes were preincubated in prewarmed assay medium (XF base medium with 25 mM glucose, 1 mM sodium pyruvate, 2 mM L-glutamine) for 1 h in a CO2-free incubator before oxygen consumption analysis. Mitochondrial stress was measured by injecting the ATP synthase inhibitor oligomycin (4 μM), the mitochondrial uncoupler carbonyl cyanide4-(trifluoromethoxy) phenylhydrazone (FCCP, 2 μM), and the electron transport inhibitor rotenone and antimycin A (1 μM) sequentially. The OCR was adjusted by the protein amount.
Body composition
Body composition (fat and lean mass) was assessed with Minispec Fractional TD-NMR Analyzer (Bruker). The data were normalized by body weight.
Metabolic cages
The mice were acclimated to the system for 48 h before formal testing with free access to food and water throughout the process. Food intake, whole-body energy expenditure, oxygen consumption (VO2), RER and physical locomotor activity were determined using a Comprehensive Laboratory Animal Monitoring System (CLAMS; Columbus, USA). The energy expenditure was calculated by (3.815 + 1.232×RER) × VO2. The regression-based analyses of energy expenditure in mice were conducted using ANCOVA with body weight used as a covariate (https://calrapp.org)39.
Lipolysis assays
For ex vivo lipolysis, fresh iWAT and eWAT depots (~30 mg) dissected from ad libitum-fed mice were minced into small pieces and transferred into DMEM supplemented with 2% FFA-free BSA in 96-well plates for 30 min (preincubation). To analyze basal and stimulated lipolysis, adipose tissue pieces were incubated with or without 10 μM ISO (Sigma-Aldrich, I5627) for 6 h. To determine the lipolysis rate, the media was subjected to the analysis of glycerol content (Sigma-Aldrich, FG0100) and NEFA (Wako, 294-636). For each mouse, fat depot, and condition, five replicates were performed in parallel. The data were expressed as glycerol or FFA per mg tissue. For in vitro lipolysis, mature adipocytes in 6-well plates were maintained in DMEM supplemented with 2% FFA-free BSA in the presence or absence of 10 μM ISO for 6 h. Then, glycerol and free fatty acids in the medium were analyzed. The data were normalized by cells protein content.
cAMP assays and PKA activity assays
For the in vitro cAMP assay, differentiated C3H10T1/2 adipocytes in 6-cell plates were incubated in DMEM with 0.2% BSA for 6 h and subjected to 1 μM ISO stimulation for another 1 h. The ISO group was also treated with 200 μM IBMX and stimulated with ISO. For measurement of cAMP in adipose tissue extracts, mice were fasted for 16 h. cAMP levels were measured from cell or adipose tissue lysates using a cAMP Direct Immunoassay Kit (Abcam, ab65355) following the manufacturer’s instructions. PKA activity was measured from cell or adipose tissue lysates using the anti-phospho-PKA substrates (RRXS*/T*) antibody (9624, Cell Signaling).
Plasmid construction
The full-length mouse complementary DNA sequence for Eepd1 (NM_026189.3) tagged with flag or EGFP was amplified and subsequently cloned into the pLVX-IRES-Puro lentiviral vector (Clontech, 632183). Eepd1 mutant plasmids were generated using pLVX-Eepd1-flag and pLVX-Eepd1-EGFP as the template with a KOD-Plus-Mutagenesis Kit (Toyobo, SMK101) following the manufacturer’s instructions. Mouse Eepd1 adenoviral plasmids were obtained using pENTR Directional TOPO Cloning Kits (Thermo Fisher Scientific, K240020) and the gateway recombination system (Thermo Fisher Scientific, V493-20) following the manufacturer’s instructions. The primer sequences used for plasmid construction are listed in Table S3.
GTTs and ITTs
For GTTs, mice were intraperitoneally (i.p.) injected with D-glucose solution (1.0 g/kg body weight for DIO mice) after fasting for 16 h. Tail blood glucose levels were monitored following glucose injection at 0 min, 15 min, 30 min, 60 min, 90 min and 120 min. For ITTs, insulin (Novolin; 1.75 U/kg body weight for DIO mice) was i.p. administered to mice after a 6-h fast, and tail blood glucose levels were monitored at 0 min, 15 min, 30 min, 45 min, 60 min and 90 min.
Metabolic parameters
Serum insulin was measured with ELISA kit (ImmunoDiagnostics, 32270) following the manufacturer’s instructions. Serum NEFA was measured with a LabAssayTM NEFA kit (Wako, 294-636).
Cold tolerance test
Body temperatures were assessed using a rectal probe for mice (FT3400, China). Mice were exposed to 4 °C, and their rectal temperatures were measured at 0 min, 30 min, 60 min, 90 min, 120 min, 150 min and 240 min using a digital thermometer. The mice had free access to water and food.
Thermal imaging
Mice were exposed to 4 °C for 4 h and anesthetized with isoflurane. An infrared camera (T540, FLIR Systems) was used to acquire a static dorsal thermal image at a focal length of 30 cm immediately after removal from the cold room.
Histological analysis
The iWAT, eWAT and BAT were fixed in 4% (v/v) paraformaldehyde and embedded in paraffin wax. Sections (5 μm) were stained with hematoxylin and eosin. AdipoCount software40 was used for quantification of adipocyte diameters from H&E-stained sections of iWAT and eWAT collected from 3 individual mice in each group.
snRNA-seq analysis
Adipose tissue single-nucleus RNA sequencing (snRNA-seq) data for human and mouse samples were obtained from the curated RDS format files deposited on the website https://gitlab.com/rosen-lab/white-adipose-atlas. Groups were defined based on provided BMI information, and cell clustering used provided clustering information. Violin plots were generated using the VlnPlot and FeaturePlot function from the “Seurat” package in R language, which was deposited on the open research data repository Zenodo (https://doi.org/10.5281/zenodo.14271994)41.
Statistic and Reproducibility
Statistical analyses were performed with GraphPad Prism (version 9.0) software. The normal distribution assumption was assessed with the Sharpiro‒Wilk test and Kolmogorov‒Smirnov test before proceeding with the unpaired two-tailed Student’s t test, Mann-Whitney U test, one way analysis of variance (ANOVA) or two-way ANOVA followed by the Šídák’s multiple comparisons test. KEGG and GO analysis of RNA-seq data was assessed by two-sided Hypergeometric test with Fisher’s exact test. The statistical details of the experiments can be found in the figure legends. The data are presented as the mean ± standard error of the mean (SEM). p value < 0.05 was considered to indicate a statistically significant difference. The specific p values and n values are given in the figures or corresponding legends. All representative experiments (such as Immunoblot, IF, H&E, and IHC staining) are repeated at least three times independently with similar results. For mouse experiments, ‘n’ corresponds to the number of mice used. For cell and Seahorse experiments, ‘n’ represents the number of biological replicates of cultured cells. Experiments involving measurements of fat mass, lean mass, blood glucose, food intake, energy expenditure, RER and body weights of mice were performed in a blinded fashion, with randomly grouped mice based on their ear tag identification numbers. Imaging, histology and immunofluorescence were also performed and analyzed in a blinded fashion prior to the decoding of the sample identities. No statistical method was used to predetermine sample size. No data points were excluded from the study.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data supporting the findings of this study are available within the Supplementary Information and Source Data file. Source data are provided with this paper. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE42 partner repository with the dataset identifier PXD058559. The RNA-seq data generated in this study have been deposited to the Gene Expression Omnibus under accession numbers GSE283770. The RNA-Seq datasets used in Fig. 1j are public datasets available in Gene Expression Omnibus (GEO) database under accession number GSE70353 and GSE167264 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE70353 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE167264). Obesity-related genes in the PBmice database were obtained from supplementary data of published work15. Source data are provided with this paper.
Code availability
Adipose tissue snRNA-seq data for human and mouse samples were obtained from the curated RDS format files deposited on https://gitlab.com/rosen-lab/white-adipose-atlas. The computer core code used for the data analysis in this paper is available at https://github.com/Eliauk-clumsy/Eepd1.git (https://doi.org/10.5281/zenodo.14271993)41.
References
Wang, Y. C., McPherson, K., Marsh, T., Gortmaker, S. L. & Brown, M. Health and economic burden of the projected obesity trends in the USA and the UK. Lancet 378, 815–825 (2011).
Piche, M. E., Tchernof, A. & Despres, J. P. Obesity phenotypes, diabetes, and cardiovascular diseases. Circ. Res 126, 1477–1500 (2020).
Collaborators, G. B. D. O. et al. Health effects of overweight and obesity in 195 countries over 25 Years. N. Engl. J. Med 377, 13–27 (2017).
Tchernof, A. & Despres, J. P. Pathophysiology of human visceral obesity: an update. Physiol. Rev. 93, 359–404 (2013).
Speakman, J. R. & Elmquist, J. K. Obesity: an evolutionary context. Life Metab. 1, 10–24 (2022).
Wlodarczyk, M. & Nowicka, G. Obesity, DNA damage, and development of obesity-related diseases. Int. J. Mol. Sci. 20, 1146–1163 (2019).
Jiang, M. et al. Alterations of DNA damage response pathway: Biomarker and therapeutic strategy for cancer immunotherapy. Acta Pharm. Sin. B 11, 2983–2994 (2021).
Shimizu, I., Yoshida, Y., Suda, M. & Minamino, T. DNA damage response and metabolic disease. Cell Metab. 20, 967–977 (2014).
Shimizu, I. et al. p53-induced adipose tissue inflammation is critically involved in the development of insulin resistance in heart failure. Cell Metab. 15, 51–64 (2012).
Lee, G. et al. SREBP1c-PARP1 axis tunes anti-senescence activity of adipocytes and ameliorates metabolic imbalance in obesity. Cell Metab. 34, 702–718.e705 (2022).
Vartanian, V. et al. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc. Natl Acad. Sci. USA 103, 1864–1869 (2006).
Sampath, H. et al. 8-Oxoguanine DNA glycosylase (OGG1) deficiency increases susceptibility to obesity and metabolic dysfunction. PLoS One 7, e51697 (2012).
Li, Q. et al. Obesity and hyperinsulinemia drive adipocytes to activate a cell cycle program and senesce. Nat. Med 27, 1941–1953 (2021).
Dutta, S. & Sengupta, P. Men and mice: relating their ages. Life Sci. 152, 244–248 (2016).
Cui, J. et al. Disruption of Gpr45 causes reduced hypothalamic POMC expression and obesity. J. Clin. Invest 126, 3192–3206 (2016).
Sun, L. V. et al. PBmice: an integrated database system of piggyBac (PB) insertional mutations and their characterizations in mice. Nucleic Acids Res 36, D729–D734 (2008).
Schreiber, R. et al. Cold-induced thermogenesis depends on atgl-mediated lipolysis in cardiac muscle, but not brown adipose tissue. Cell Metab. 26, 753–763.e757 (2017).
Haemmerle, G. et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science 312, 734–737 (2006).
Kim, H. S. et al. Endonuclease EEPD1 Is a Gatekeeper for Repair of Stressed Replication Forks. J. Biol. Chem. 292, 2795–2804 (2017).
Liu, Y. EEPD1: breaking and rescuing the replication fork. PLoS Genet 12, e1005742 (2016).
Wu, Y. et al. EEPD1 rescues stressed replication forks and maintains genome stability by promoting end resection and homologous recombination repair. PLoS Genet 11, e1005675 (2015).
Thayer, M. M., Ahern, H., Xing, D., Cunningham, R. P. & Tainer, J. A. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 14, 4108–4120 (1995).
Belova, G. I. et al. A type IB topoisomerase with DNA repair activities. Proc. Natl Acad. Sci. USA 98, 6015–6020 (2001).
Sun, Y. et al. Radiation induces NORAD expression to promote ESCC radiotherapy resistance via EEPD1/ATR/Chk1 signalling and by inhibiting pri-miR-199a1 processing and the exosomal transfer of miR-199a-5p. J. Exp. Clin. Cancer Res 40, 306 (2021).
Nickoloff, J. A. et al. Metnase and EEPD1: DNA repair functions and potential targets in cancer therapy. Front Oncol. 12, 808757 (2022).
Moriya, K. et al. Identification of dually acylated proteins from complementary DNA resources by cell-free and cellular metabolic labeling. Anal. Biochem 511, 1–9 (2016).
Yuan, M. et al. N-myristoylation: from cell biology to translational medicine. Acta Pharm. Sin. 41, 1005–1015 (2020).
Jiang, M., Chavarria, T. E., Yuan, B., Lodish, H. F. & Huang, N. J. Phosphocholine accumulation and PHOSPHO1 depletion promote adipose tissue thermogenesis. Proc. Natl Acad. Sci. USA 117, 15055–15065 (2020).
Yuan, Y. et al. Protein lipidation in health and disease: molecular basis, physiological function and pathological implication. Signal Transduct. Target Ther. 9, 60 (2024).
Jiang, H. et al. Protein lipidation: occurrence, mechanisms, biological functions, and enabling technologies. Chem. Rev. 118, 919–988 (2018).
Xiong, J. et al. Obstructive sleep apnea syndrome exacerbates nash progression via selective autophagy-mediated eepd1 degradation. Adv Sci (Weinh), e2405955 (2024).
Udenwobele, D. I. et al. Myristoylation: An Important Protein Modification in the Immune Response. Front Immunol. 8, 751 (2017).
Giang, D. K. & Cravatt, B. F. A second mammalian N-myristoyltransferase. J. Biol. Chem. 273, 6595–6598 (1998).
Ducker, C. E., Upson, J. J., French, K. J. & Smith, C. D. Two N-myristoyltransferase isozymes play unique roles in protein myristoylation, proliferation, and apoptosis. Mol. Cancer Res 3, 463–476 (2005).
Minamino, T. et al. A crucial role for adipose tissue p53 in the regulation of insulin resistance. Nat. Med 15, 1082–1087 (2009).
Park, S. J. et al. DNA-PK promotes the mitochondrial, metabolic, and physical decline that occurs during aging. Cell Metab. 25, 1135–1146.e1137 (2017).
Liu, S. et al. Distinct roles for DNA-PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress. Nucleic Acids Res 40, 10780–10794 (2012).
Chen, S. et al. The phytochemical hyperforin triggers thermogenesis in adipose tissue via a Dlat-AMPK signaling axis to curb obesity. Cell Metab. 33, 565–580.e567 (2021).
Muller, T. D., Klingenspor, M. & Tschop, M. H. Revisiting energy expenditure: how to correct mouse metabolic rate for body mass. Nat. Metab. 3, 1134–1136 (2021).
Zhi, X. et al. AdipoCount: a new software for automatic adipocyte counting. Front Physiol. 9, 85 (2018).
Chen, S. et al. Myristoylated Eepd1 Enhances Lipolysis and Thermogenesis through PKA Activation to Combat Obesity. Zenodo, https://doi.org/10.5281/zenodo.14271993, (2024).
Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res 50, D543–D552 (2022).
Qian, Q. Created in BioRender. https://BioRender.com/v96h845 (2024).
Qian, Q. Created in BioRender. https://BioRender.com/f95e289 (2024).
Qian, Q. Created in BioRender. https://BioRender.com/k69k570 (2024).
Qian, Q. Created in BioRender. https://BioRender.com/p75t536 (2024).
Acknowledgements
We express our sincere thanks to Dr. Hui Yang and Dr. Changyang Zhou (Chinese Academy of Sciences) for the construction of Eepd1 knock-out mice and Eepd1flox/flox mice. We thank Dr. Xu-yun Zhao (College of Basic Medical Science, Shanghai Jiao Tong University) for sharing the Adiponectin-cre mice. This work was supported by the National Natural Science Foundation of China (No. 82170863 and 32471208 to S.Z.C.); Major Program of the National Natural Science Foundation of China (No. 92357303 to J.L.L.); National Key R&D Program of China (No. 2021YFA0804800 to J.L.L.); Key Program of National Natural Science Foundation of China (No. 82330026 to J.L.L.); National Natural Science Foundation of China (No. 32300997 to L.M.Y.); Innovative research team of high-level local universities in Shanghai (SHSMU-ZDCX20212501 to S.Z.C. and J.L.L.); Shanghai Rising-Star Program (No. 21QA1407000 to S.Z.C.); Shanghai Double Hundred program (No. SBR2022009 to S.Z.C.); Lingang Laboratory (No. LG-QS-202205-06 to S.Z.C.); Shanghai Sixth People’s Hospital (No. ynyq202103 to S.Z.C., No. ynjq202102 to J.L.L. and No. ynnkxyb202401 to L.M.Y.); Postdoctoral Fellowship Program of CPSF (No. 2023M742353 and No. BX20230227 to L.M.Y.).
Author information
Authors and Affiliations
Contributions
Conceptualization: S.Z.C. and J.L.L. Methodology: S.Z.C., Y.P.W., Q.Z., Q.Q.Q., Q.X.J., C.C.L., Y.L., P.H.Z., J.X., Y.Z., N.W., Y.E.L., L.M.Y. and H.Y.Y. Investigation: S.Z.C., Q.Z., Y.P.W. and Q.Q.Q. Visualization: Q.Z. and Q.Q.Q. Supervision: S.Z.C. and J.L.L. Writing—original draft: S.Z.C. and J.L.L. Writing—review & editing: S.Z.C. and J.L.L.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, S., Wang, Y., Zhou, Q. et al. Myristoylated Eepd1 Enhances Lipolysis and Thermogenesis through PKA Activation to Combat Obesity. Nat Commun 16, 651 (2025). https://doi.org/10.1038/s41467-025-56026-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-56026-2