Abstract
Cholestatic liver diseases are characterized by excessive bile acid accumulation in the liver. Endothelial cells (ECs) shape the local microenvironment in both normal conditions and liver injury, yet their role in cholestasis is unclear. Through a comparative analysis of single-cell RNA sequencing data from various murine models of liver injury, we identify distinctive Myc activation within ECs during obstructive cholestasis resulting from bile duct ligation (BDL). Myc overexpression in ECs significantly upregulates P-selectin, increasing neutrophil infiltration and worsening cholestatic liver injury. This process occurs through the FXR, activated by chenodeoxycholic acid (CDCA) and its conjugate TCDCA. Inhibiting P-selectin with PSI-697 reduces neutrophil recruitment and alleviates injury. Cholestatic patient liver samples also show elevated Myc and P-selectin in ECs, along with increased neutrophils. The findings identify ECs as key drivers of cholestatic liver injury through a Myc-driven program and suggest that targeting the CDCA/FXR/Myc/P-selectin axis may offer a therapeutic approach.
Similar content being viewed by others
Introduction
Cholestatic liver diseases encompass a range of hepatobiliary disorders arising from various etiologies, including genetic defects, bile duct obstruction mediated by intraductal or extraductal events, such as in cases of pancreatic cancer, or adverse drug reactions, all of which impairs normal bile flow1,2. Currently, treatment options for late-stage cholestasis are mainly limited to supportive care and liver transplantation. The impaired bile flow triggers an excessive accumulation of bile acids in the liver, ultimately resulting in cholestatic liver injury. The harmful effects of bile acids have traditionally been attributed to their detergent-like properties that compromise cellular membranes. However, recent studies have revealed a more complex role of bile acids as signaling molecules, acting at the crossroads of immunology3,4,5,6,7,8,9 and biochemistry10,11. These effects are mediated through diverse receptors12,13,14 and signaling pathways15,16. Nevertheless, the specific cellular entities and molecular pathways by which the immune response to accumulated bile acids exacerbates liver injury in cholestasis remain elusive.
Different liver diseases are associated with specific local microenvironments, wherein non-parenchymal cells play critical roles. Liver ECs, the most abundant non-parenchymal cell populations comprising approximately 15 to 20% of liver cells, act as gatekeepers of hepatic immunity17. Physiologically, Liver ECs regulate blood flow in the liver through vasoconstriction, and form a barrier for the transport of molecules and immune cells to the liver by modulating fenestration18. Numerous data have highlighted the crucial roles of ECs in orchestrating the immune microenvironment, such as during acute liver failure induced by acetaminophen (APAP)19, hepatic ischemia-reperfusion (HIR) injury20 and liver cancer4,21,22. However, it remains unclear whether and how ECs exert immune regulatory functions in cholestatic liver injury.
In our study, we explored the transcriptional characteristics of ECs using single-cell RNA sequencing (scRNA-seq) in murine models of liver injury induced by various etiologies23, including bile duct ligation (BDL), APAP, HIR, alcohol liver disease (ALD)-diet, methionine choline-deficient (MCD)-diet. In contrast to the BDL cholestatic model, although these liver injuries can directly or indirectly affect enterohepatic bile acid circulation and induce endothelial cell injury, the degree of bile acid accumulation varies significantly among them. Consequently, the downstream events triggered by different levels of bile acid components can differ, whether they are toxic or act as signaling molecules24,25. By comparing these models, we can gain a more comprehensive understanding of the bile acid accumulation-specific pathological biology of ECs. Our work revealed a notable and distinct activation of Myc signaling pathway within ECs. We further identified that this unique activation is regulated by chenodeoxycholic acid (CDCA)-induced activation of farnesoid X receptor (FXR) expressed in ECs. Activated ECs play a crucial role in shaping the liver’s immune microenvironment, primarily by facilitating the recruitment of neutrophils through the expression of P-selectin. These findings offer insights into a potential therapeutic strategy that targets the CDCA/FXR/Myc/P-selectin signaling axis for the treatment of obstructive cholestasis.
Results
Single-cell atlas analysis reveals a distinctive transcriptional program associated with Myc activation in ECs in murine models of cholestatic liver injury
To explore the specific transcriptional signature of ECs in cholestatic liver diseases, we conducted a comprehensive analysis of publicly available scRNA-seq datasets (GSE166178)23, including samples from control livers as well as various liver injury models such as BDL, APAP, IR, ALD and MCD diet-induced injury. After excluding low-quality cells and potential cell doublets, a total of 30,821 pure ECs were used for subsequent analyzes (Supplementary Fig. 1a–d, please refer to Methods for a detailed procedure). According to the latest classification reports26,27,28, we categorized these ECs into four subtypes, which are periportal (Endo-pp) and pericentral (Endo-pc) ECs, corresponding to ECs originating from veins and arteries, respectively, as well as two subtypes of sinusoidal ECs (LSECs) (LSEC-1 and LSEC-2) (Fig. 1a and Supplementary Fig. 1d).
a UMAP visualization showing liver EC subtypes. GSEA of hallmark gene sets (b), enrichment of transcription factor motifs in ECs (c), and (d) Myc gene expression in ECs from different liver injury models. e Immunofluorescence (IF) staining of CD31 (green) and Myc (red) in liver tissue samples from different liver injury models as indicated. Representative images illustrate the findings from three independent biological experiments with similar results.
We proceeded to conduct gene set-enrichment analysis (GSEA) to determine the most regulated gene signature in ECs from various liver injury models (Fig. 1b). ECs from healthy control livers exhibited the presence of “Mitotic spindle,” “Cell cycle,” and “G2/M checkpoint” gene sets, indicative of normal physiological turnover (Fig. 1b). Notably, ECs from BDL, APAP, and MCD showed enrichment in the “MYC” target signal, with BDL ECs displaying the most pronounced MYC signature compared to APAP and MCD models (Fig. 1b). Furthermore, an analysis of transcription factor binding site motifs within the promoters of genes upregulated in ECs again revealed a significant enrichment of MYC binding motifs in BDL, differing from other models (Fig. 1c). This was corroborated by higher transcription levels of the Myc gene in BDL ECs (Fig. 1d).
To complement the scRNA-seq data, we performed immunostaining for Myc protein. Immunofluorescence (IF) assays confirmed that Myc protein expression in ECs followed a patten consistent with the mRNA levels of Myc observed in various liver injury models from the scRNA-seq data (Fig. 1e and Supplementary Fig. 1e). Collectively, our findings indicate that cholestasis specifically induces an EC-specific transcriptional program characterized by Myc activation.
Tie2-driven endothelial Myc-overexpression drives neutrophil infiltration and exacerbates liver injury following BDL
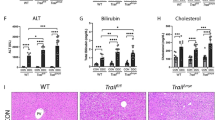
To gain a deeper understanding of the biological implications of increased Myc expression in ECs during cholestasis, we specifically overexpressed Myc in endothelium by intercrossing MycLSL/LSL mice with Tie2-Cre transgenic mice (hereafter, Tie2-MycLSL/+) (Fig. 2a). Compared to the littermate controls (MycLSL/+), liver samples from 2-month-old Tie2-MycLSL/+ mice exhibited a roughly 2-fold increase in Myc mRNA levels (Fig. 2b) and displayed nuclear staining of Myc in ECs (Fig. 2c, d). Flow cytometry analysis further revealed that Myc protein expression was specifically upregulated in ECs, but not in other cell types such as hepatocytes, CD8 T cells, CD4 T cells, or Kupffer cells, in Tie2-MycLSL/+ mice compared to WT mice (Fig. 2e and Supplementary Fig. 2a, b). Notably, the livers of Tie2-MycLSL/+ mice did not show any signs of liver injury or fibrosis (Fig. 2f). Serological analysis of aspartate aminotransferase (AST), alanine transaminase (ALT) and bilirubin levels further confirmed the absence of spontaneous liver injury in Tie2-MycLSL/+ mice (Fig. 2g and Supplementary Fig. 2e). However, alkaline phosphatase (ALP) levels were slightly elevated in Tie2-MycLSL/+ mice compared to WT mice, albeit the increase was less than 1.3-fold (Fig. 2g). Hematoxylin and eosin (H&E) staining of liver tissues from Tie2-MycLSL/+ mice did not reveal any structural abnormality in hepatic lobules (Fig. 2i). Further IHC analysis showed no significant increase in the ratio of CD45+, F4/80+, and CD8A+ cells in Tie2-MycLSL/+ livers compared to WT controls (Fig. 2i). FACS analysis revealed no changes in macrophage M1/M2 phenotypes between genotypes (Supplementary Fig. 2h, i). However, there was noticeable infiltration of Ly6G+ cells, indicating the recruitment of neutrophils in the livers of Myc overexpressing mice (Fig. 2i, j). qPCR analysis of liver samples confirmed a significant upregulation of lymphocyte antigen 6 complex locus G6D (Ly6g), neutrophilic granule protein (Ngp), and the pro-inflammatory cytokine Il6 (Fig. 2h). Additionally, the serum levels of inflammatory cytokine Il-6 and Il-1β were significantly increased in Tie2-MycLSL/+ mice (Supplementary Fig. 2f, g). Scanning electron microscopy (SEM) showed comparable structure of hepatic sinusoids and endothelial fenestration between Tie2-MycLSL/+ mice and WT mice (Supplementary Fig. 2j). IHC analysis of CD34 expression revealed no changes in the formation of the LSEC basement membrane between these two mice strains (Supplementary Fig. 2k).
a Construction strategy of EC-specific Myc-overexpression mice. b qPCR analysis for Myc gene in 2-month-old mice (n = 5). c IF staining of CD31 (green) and Myc (red) in liver tissue samples from livers of (b). d quantitative analysis of Myc-positive ECs in liver tissue sections of (c) (n = 5). e Quantitative analysis of protein expression levels of Myc in ECs from MycLSL/+ and Tie2-MycLSL/+ mice by Fluorescence-Activated Cell Sorting (FACS) (n = 8). f Macroscopic view of livers from (b). g Serum enzyme levels in mice of (b) (n = 5). h qPCR analysis of indicated genes in livers from (f). Representative images (i) and quantitative analysis (j) of H&E and IHC for immune cell markers in livers from (f), with arrows indicating positive cells in respective panels (i). k Schematic diagram of BDL surgery and serum enzyme levels in BDL-treated mice (n = 5). l Macroscopic liver phenotype of mice in (k). Representative images (m) and quantitative analysis (n and o) of H&E and IHC for immune cell markers in livers from (l), with arrows indicating CD8A-positive cells and the red dotted box indicating the necrotic area (m). Data are presented as the mean ± SEM. P values are indicated as follows: * <0.05; ** <0.01; *** <0.001; **** <0.0001, ns = no significance. P values were obtained using the unpaired two-tailed Student’s t-test. Source data are provided as a Source Data file.
To examine the impact of endothelial-specific Myc on cholestatic liver injury, we conducted BDL on Tie2-MycLSL/+ mice and WT littermate controls (Fig. 2k). Notably, we observed a more pronounced increase in serum levels of AST, ALT, and bilirubin in Tie2-MycLSL/+ mice compared to wild-type controls (Fig. 2k and Supplementary Fig. 3a). However, there were no significant changes in bile acid levels, either in serum or liver tissue, between WT and Myc transgenic mice subjected to BDL (Supplementary Fig. 3b, c). Additionally, Tie2-driven Myc overexpression resulted in further elevation of Myc expression in endothelial cells following BDL, compared to WT mice (Supplementary Fig. 2c). Upon gross inspection, Tie2-MycLSL/+ mice exhibited extensive liver necrosis (Fig. 2l). Histologically, BDL resulted in marked enlargement and an increased number of necrotic zones (bile infarcts) in Tie2-MycLSL/+ mice compared to control mice (Fig. 2m, n). IHC revealed intensified infiltration in BDL-treated Tie2-MycLSL/+ mice, characterized by significant hepatic accumulation of CD45+ and Ly6G+ cells (Fig. 2m, o), whereas no differences were observed in the intrahepatic populations of macrophages (F4/80+) and CD8A+ T cells (Fig. 2m, o). Furthermore, increased ductular reaction and liver fibrosis can be clearly observed in Tie2-MycLSL/+ compared to MycLSL/+ mice following 2 weeks of BDL (Supplementary Fig. 3g, h), although the differences were comparable when examining mice subjected to 3 days of BDL (Supplementary Fig. 3e, f).
We also evaluated EC cell expansion using the proliferation marker Ki67, IF staining showed a significantly increased proliferation level of ECs but not hepatocytes in Tie2-MycLSL/+ mice compared to WT mice (Supplementary Fig. 2d). Additionally, IHC staining for 4-hydroxynonenal (4-HNE) indicated increased oxidative damage in Tie2-MycLSL/+ mice (Supplementary Fig. 3d), which positively correlated with Myc expression (Supplementary Fig. 2c).
Overall, our findings suggest that endothelial Myc overexpression exacerbates liver injury in the context of hepatic cholestasis during both the acute phase and later stages, accompanied with an altered immune microenvironment characterized by a predominant neutrophil infiltration.
Endothelial Myc activation elicits P-selectin upregulation during cholestasis
Next, we aimed to investigate the underlying mechanisms behind the regulatory effects of endothelial Myc overexpression on intrahepatic neutrophil recruitment following BDL. The recruitment of neutrophils can be attributed to an increased level of pro-inflammatory mediators such as Il17 and the chemoattractant molecule Il8/Cxcl829. Previous studies have also demonstrated that ECs utilize a combination of specific EC receptors to facilitate leukocyte adhesion and transendothelial migration in chronic liver disease17,18,22,30. However, analysis of Il17a, Il8, Vcam1, Icam1, Icam2, and Jam1 expression in Tie2-MycLSL/+ mice showed no differences compared to controls (Fig. 3a and Supplementary Fig. 4a). We thus performed bulk RNA sequencing (RNA-seq) analysis on BDL-treated liver samples from Tie2-MycLSL/+ and WT control mice. GSEA of RNA-seq livers exhibited enrichment of signatures related to the regulation of cell adhesion, positive regulation of leukocyte migration, regulation of leukocyte adhesion to vascular ECs, and regulation of cellular extravasation in the livers of Tie2-MycLSL/+ mice (Fig. 3b). Among these gene sets associated with leukocyte adhesion to vascular EC, we observed a significant upregulation of Selp (encoding P-selectin) in mice with endothelial Myc overexpression (Fig. 3c). The selectin family (P-, E-, and L-selectin) has been implicated in the endothelial-mediated inflammation during IR, with P-selectin being involved in the initial interaction between leukocytes and ECs20,31. Unlike Sele and Sell, Selp was markedly increased in mice with endothelial Myc overexpression following BDL compared to WT controls (Fig. 3d). qPCR analysis confirmed the upregulation of Selp in both sham-treated and BDL-treated Tie2-MycLSL/+ groups compared to WT controls, while Sele was also induced in Myc transgenic mice, the increase was not statistically significant (Fig. 3e). In line, scRNA-seq data showed a noticeable expression of Selp mRNA in ECs from BDL-treated mice, while Sele levels remained similar across all animal models in the study (Fig. 3f and Supplementary Fig. 4b). IF staining confirmed the EC-specific upregulation of P-selectin expression in Myc transgenic mice (Fig. 3g, h). Overall, these findings suggest that activation of endothelial Myc leads to an increase in P-selectin expression during BDL-induced cholestasis, which may explain the recruitment of neutrophils to the liver.
a qPCR analysis of indicated genes in BDL livers (n = 5). b GSEA of the indicated gene sets. c DEGs in the indicated gene set. d Expression levels of indicated genes in RNA-seq data (n = 4). The box plot shows the data distribution, with the minimum (lower whisker), lower quartile (bottom of the box), median (center line within the box), upper quartile (top of the box), and maximum (upper whisker) indicated. e qPCR analysis of indicated genes in BDL livers (n = 5). f UMAP showing the expression of indicated genes. Representative IF images (g) and quantitative analysis (h) of IF for CD31+P-selectin+ ECs in sham- and BDL-treated livers (n = 6). Data are presented as the mean ± SEM. P values are indicated as follows: * <0.05; ** <0.01; * **** <0.0001, ns = no significance. P values were obtained using the unpaired two-tailed Student’s t-test. Source data are provided as a Source Data file.
Lyve1-driven Myc-overexpression mice phenocopy the increased susceptibility to BDL-induced liver injury in Tie2-Myc LSL/+ mice
It is important to note the potential influence of the Tie2-driven Cre system on the hematopoietic lineage32. To address this concern, we generated mice that overexpressed Myc specifically in ECs by interbreeding MycLSL/+ with transgenic mice expressing Cre recombinase driven by the lymphatic vessel endothelial hyaluronan receptor 1 (Lyve1) promoter33 (Fig. 4a). Lyve1-MycLSL/+ mice exhibited a 1.5-fold increase in Myc mRNA expression in liver tissue samples compared to WT controls (Fig. 4b). IF staining revealed prominent nuclear localization of Myc protein in ECs from Lyve1-MycLSL/+ mice (Fig. 4c, d). Flow cytometry confirmed selective Myc upregulation in ECs, showing a 1.44-fold increase in Lyve1-MycLSL/+ mice compared to WT mice, which is slightly less than the 1.75-fold increase observed in Tie2-MycLSL/+ mice (Fig. 4e and Supplementary Fig. 5a, b). There were no significant changes in Myc expression in hepatocytes, CD8 T cells, CD4 T cells, or Kupffer cells in Lyve1-MycLSL/+ mice compared to WT mice (Supplementary Fig. 5a, b). Notably, 2-month-old Lyve1-MycLSL/+ mice did not exhibit any signs of liver injury, fibrosis, or abnormal parenchymal structure (Fig. 4f, g, j and Supplementary Fig. 5e). The slight elevation of ALP levels was not observed in Lyve1-MycLSL/+ mice, likely due to the differing levels of Myc upregulation between the two Cre strains. However, similar to Tie2-MycLSL/+ mice, Lyve1-MycLSL/+ mice showed significant neutrophil infiltration in the liver, upreg0.ulation of neutrophil effectors, and increased expression of pro-inflammatory cytokine (Fig. 4h, j, k and Supplementary Fig. 5f, g). Lyve1-MycLSL/+ mice also exhibited elevated P-selectin at both the mRNA and protein levels compared to littermate controls (Fig. 4i, j, l). In addition, consistent with the observations in Tie2-MycLSL/+ mice, macrophage phenotypes and sinusoidal structure were comparable to WT mice, as evidenced by FACS analysis (Supplementary Fig. 5h, i), SEM (Supplementary Fig. 5j) and IHC staining of CD34 (Supplementary Fig. 5k), respectively. Collectively, the Lyve1-driven Myc-overexpression mice exhibit a baseline phenotype that closely mirrors the endothelial cell phenotype observed in Tie2-driven Myc-overexpression mice.
a Construction strategy for EC-specific Myc-overexpression mice. b qPCR analysis of Myc gene in the livers of 2-month-old mice (n = 5). c IF staining of CD31 (green) and Myc (red) in liver tissue samples from livers of (b). (d) quantitative analysis of Myc-positive ECs in liver tissue sections of (c) (n = 5). (e) Quantitative analysis of protein expression levels of Myc in ECs from MycLSL/+ and Lyve1-MycLSL/+ mice by FACS (n = 8). f Macroscopic view of livers from (b). g Serum enzyme levels in mice of (b) (n = 5). h, i qPCR analysis of indicated genes in livers of (f) (n = 5). j–l H&E, IHC staining of Ly6G, IF staining of CD31 (green) and P-selectin (red), and quantitative analysis (k–l) (n = 5, 6) in liver samples from the mice in (f), with arrows indicating positive cells in respective panels (j). m Schematic diagram of BDL surgery and serum enzyme levels in BDL mice (n = 5). n Macroscopic liver phenotype of the mice in (m). o H&E, IHC staining of Ly6G, IF staining of CD31 (green) and P-selectin (red), in liver samples from mice subjected to BDL. p–r Quantitative analysis of necrosis (p) (n = 5), Ly6G positive cells (q) (n = 5), and CD31+P-selectin+ cells (r) (n = 6) in liver tissue samples of (o). Data are presented as the mean ± SEM. P values are indicated as follows: * < 0.05; ** <0.01; *** <0.001; **** <0.0001, ns = no significance. P values were obtained using the unpaired two-tailed Student’s t-test. Source data are provided as a Source Data file.
When subjected to BDL-induced cholestasis, Lyve1-MycLSL/+ mice demonstrated more severe liver injury and hepatic necrosis compared to WT mice (Fig. 4m–p, and Supplementary Fig. 6a). Notably, Lyve1-driven Myc overexpression resulted in further elevation of Myc expression in endothelial cells following BDL, compared to WT mice (Supplementary Fig. 5c). Importantly, Lyve1-MycLSL/+ mice also exhibited elevated levels of P-selectin and enhanced recruitment of neutrophils within the liver after BDL (Fig. 4o, q, r). Akin to observations in Tie2-MycLSL/+ mice, (a), bile acid levels remained comparable (Supplementary Fig. 6b, c); (b) Increased ductular reaction and fibrosis were noted after 2 weeks of BDL (Supplementary Fig. 6g, h), but not after 3 days (Supplementary Fig. 6e, f); (c) increased endothelial proliferation and oxidative damage were evident (Supplementary Fig. 5d and 6d). Taken together, these results, along with the liver phenotypes observed in Tie2-MycLSL/+ mice, provide compelling evidence that Myc overexpression in ECs leading to increased P-selectin expression and neutrophil infiltration, resulting in exacerbated cholestatic liver injury induced by BDL.
P-selectin inhibition leads to reduced neutrophil infiltration and improved liver Injury in BDL-induced cholestasis
To further investigate the functional relationship between endothelial P-selectin, intrahepatic neutrophil infiltration, and liver injury induced by BDL, 2-month-old C57BL/6 J mice underwent BDL were subsequently treated with a daily intravenous injection of 35 mg/kg PSI-697, a P-selectin inhibitor, for three consecutive days (Fig. 5a). In comparison to the vehicle control group, the mice treated with PSI-697 exhibited improved liver damage with reduced hepatocyte necrosis, as evident by examination of overall liver morphology (Fig. 5b), H&E staining (Fig. 5c), and quantification of the necrotic area (Fig. 5d). In addition, the serum levels of ALT, AST, and ALP were significantly lower in the PSI-697-treated group compared to the vehicle-treated group (Fig. 5e). Notably, PSI-697 administration did not affect the expression of P-selectin at the mRNA or protein levels (Fig. 5f, g), consistent with its pharmacological mechanism of inhibiting the binding of P-selectin to its ligand34. Further analysis revealed that mice treated with PSI-697 exhibited significantly reduced mRNA levels of Ly6g and Ngp, accompanied by a notable reduction in hepatic infiltration of neutrophils as compared to the vehicle-treated group (Fig. 5h, i). Moreover, mRNA levels of proinflammatory cytokines and chemokines were significantly decreased in the livers of mice treated with PSI-697 (Fig. 5j). While PSI-697 treatment did not show significant improvement in liver fibrosis or ductular reaction after 3 days of BDL administration (Supplementary Fig. 7a–d), it did improve these parameters after 2 weeks of treatment (Supplementary Fig. 7e–h). In conclusion, our findings suggest that blocking P-selectin has a therapeutic effect on injury caused by obstructive cholestasis by inhibiting the infiltration of neutrophils in the liver.
a Inhibitor administration to BDL mice via tail vein. b Gross morphology of livers from BDL mice treated as indicated (n = 5). c H&E showing necrosis in livers from (b), with red dotted box indicating the necrotic area. (d) Quantification of necrotic areas (n = 5). e Serum enzyme levels in mice from (b) (n = 5). f qPCR analysis of Selp and Sele in livers from (b) (n = 5). g IF staining of CD31 (green) and P-selectin (red) and quantification of CD31+P-selectin+ ECs in BDL livers from (b) (n = 5). h qPCR analysis of Ly6g and Ngp in livers from (b) (n = 5). i IHC of Ly6G and quantification in liver tissue samples from (b) (n = 5). j qPCR analysis of indicated genes in livers from (b) (n = 5). Data are presented as the mean ± SEM. P values are indicated as follows: * <0.05; ** <0.01; *** <0.001; **** <0.0001, ns = no significance. P values were obtained using the unpaired two-tailed Student’s t-test. Source data are provided as a Source Data file.
CDCA is the potent trigger of the MYC/P-selectin axis in ECs through FXR
The distinct activation of the Myc/P-selectin axis in ECs in BDL-treated mice promoted us to investigate the upstream environmental cues responsible for this regulation. Cholestatic liver diseases are characterized by accumulation of bile acids in the liver, and recent studies have highlighted the diverse regulatory roles of bile acids as signaling molecules beyond their canonical functions. To investigate whether bile acids directly trigger the MYC/P-selectin signaling pathway during cholestasis, human umbilical vein endothelial cells (HUVECs) were exposed to various components of bile acids, including primary, secondary, and mixed species, as depicted in Supplementary Fig. 8a. After 48 h of treatment, we observed a notable increase in mRNA levels of MYC in HUVEC exposed to primary bile acids, while the 6-h and 24-h time points yielded comparable results to the control groups (Supplementary Fig. 8a). Interestingly, the addition of secondary bile acids with primary bile acids did not result in further enhancement of MYC expression (Supplementary Fig. 8a), implying that the primary bile acids predominantly initiated the endothelial MYC activation. Subsequently, we sought to examine the impact of specific primary bile acids, including unconjugated and conjugated CA and CDCA, on the activation of the endothelial MYC/SELP signal. Our experiments revealed that CDCA and its conjugated form, TCDCA, but not CA or TCA, strongly elicited the activation of the MYC/P-selectin signal axis (Fig. 6a–d, and Supplementary Fig. 8b–d). Spearman correlation analysis demonstrated a significant positive correlation between the protein levels of MYC and P-selectin (Fig. 6e, f).
Protein expression levels of MYC and SELP in HUVECs treated with unconjugated (a) and conjugated bile acids (c), with corresponding quantitative analyzes (b, d). Spearman correlation analysis between MYC and SELP expressions in HUVECs treated with unconjugated (e) and conjugated bile acids (f) from (a) to (c). g qPCR analysis of MYC and SELP in HUVECs treated with the FXR agonist INT747 (20 μM) or DMSO for 48 h (n = 3). Protein levels of MYC and SELP (h) and quantification (i) in HUVECs treated with INT747 (20 μM) or DMSO for 48 h (n = 3). j qPCR analysis of MYC and SELP in HUVECs treated with UDCA (200 μM) or DMSO for 48 h (n = 3). k, l Protein levels of MYC and SELP (k) and quantitative analysis (l) in HUVECs treated with UDCA (200 μM) or DMSO for 48 h (n = 3, 4). m Protein levels of MYC and SELP in siNC- or siFXR-treated HUVECs following treatment with GW4064 (5 μM) or DMSO for 48 h. A representative Western blot from n = 3 biological repeats is shown. n Protein levels of MYC and SELP in gLUC- or gMYC-treated HUVECs following GW4064 (5 μM), INT-747(20 μM) or DMSO treatment for 48 h. Western blot data are representative of at least 3 biological repeats, with 2 repeat experiments per sample. o qPCR analysis of Fxr, Myc and Selp in ECs from Fxr−/− and WT mice treated with GW4064 (5 μM) or DMSO for 48 h (n = 6). p Sorting of primary endothelial cells in Lyve1-MycLSL/+ mice by FACS. qPCR analysis of Fxr, Tgr5, Myc and Selp in ECs from Lyve1-MycLSL/+ mice treated with siFxr (q), siTgr5 (r) or NC for 48 h (n = 6). Data are presented as the mean ± SEM. P values are indicated as follows: * <0.05; ** <0.01; *** <0.001; **** <0.0001, ns = no significance. P values were obtained using the unpaired two-tailed Student’s t-test. Source data are provided as a Source Data file.
Bile acids exhibit various signaling effects by interacting with multiple receptors, particularly the nuclear receptor farnesoid X receptor (FXR) and the membrane receptor Takeda G protein-coupled receptor 5 (TGR5). Among these receptors, CDCA is the most potent agonist for FXR35. In order to determine the role of FXR in mediating the activation of the MYC/P-selectin signal by CDCA, we treated HUVECs with the FXR agonist INT-747 (also known as obeticholic acid). This resulted in a significant increase in the expression of MYC and SELP at both mRNA and protein levels compared to the control treatment (Fig. 6g–i). Conversely, when HUVECs were exposed to the FXR antagonist UDCA, the expression of MYC/SELP was markedly downregulated (Fig. 6j–l). Additionally, the specific synthetic FXR agonist GW4064 also enhanced MYC and SELP protein levels (Fig. 6m and Supplementary Fig. 8e). Silencing FXR using siRNAs abolished the upregulation of MYC/SELP induced by GW4064 (Fig.6m). In contrast, TGR5 knockdown had no effect on MYC/SELP expression in the presence of bile acids or GW4064 (Supplementary Fig. 8e), providing further support for the role of FXR, rather than TGR5, in mediating the CDCA-induced activation of the MYC/P-selectin signal. Furthermore, genetic deletion of endogenous MYC using CRISPR/Cas9 technology suggested that MYC was necessary for the induction of P-selectin expression by FXR (Fig. 6n). Additionally, the induction of Myc/P-selectin by GW4064 was also prevented in primary liver ECs from Fxr knockout mice (Fig. 6o). To explore whether the CDCA/Fxr/Myc/P-selectin signal axis play a crucial role in primary ECs, unconjugated and conjugated forms of CA and CDCA were administrated to primary ECs isolated from WT, Fxr−/−, and Tgr5−/− mice (Supplementary Fig. 9a). Consistent with the data from the HUVECs, the Myc/P-selectin signal axis was activated in the primary ECs treated with CDCA and its conjugated form, TCDCA, but not with CA or TCA (Supplementary Fig. 9a). Interestingly, TCDCA at a low concentration (10 μM), comparable to serum levels after BDL, activated the Myc/Selp signaling axis in both HUVECs and primary ECs, highlighting its potent activating role (Supplementary Fig. 9b, c). Furthermore, in primary ECs from BDL-treated mice, Fxr knockout, but not Tgr5 knockout, significantly inhibited Myc/P-selectin activation (Supplementary Fig. 9d). In addition, in Myc-overexpressing primary ECs, Fxr knockdown did not inhibit Selp expression, confirming that Fxr acts upstream of the Myc/P-selectin signaling axis (Fig. 6p–r, and Supplementary Fig. 9e). Taken together, these findings strongly support the existence of a targetable CDCA/FXR/MYC/P-selectin regulatory axis in EC biology.
Clinical relevance of the endothelial MYC/P-selectin regulatory axis in cholestasis patients
To investigate the clinical relevance of our research, we conducted scRNA-seq analysis on liver tissues from seven patients diagnosed with obstructive cholestasis, using two specimens from non-cholestatic regions of unaffected lobes as controls (Fig. 7a). Detailed clinical and pathological information is provided in Supplementary Fig. 10a. After rigorous quality control (QC), a total of 62,939 cells were selected for further analysis (Supplementary Fig. 10b). The scRNA-seq data were integrated, normalized, batch-corrected, and clustered to identify coarse cell types, which included macrophage, mast cells, plasma and B cells, T/NK cells, fibroblast cells, neutrophils, ECs, and liver parenchymal cells including hepatocytes and cholangiocytes (Fig. 7b and Supplementary Fig. 10c). Compared to the controls, the OC group exhibited a notable increase in the abundance of diverse cell types, including fibroblast population and various immune cell populations (Fig. 7c). Conversely, the abundance of parenchymal hepatocytes and cholangiocytes showed a decrease (Fig. 7b, c). Further examination of the transcriptomic alterations in ECs within OC identified 427 significantly upregulated genes when compared to the healthy group, notably including MYC and SELP (Fig. 7d). Comprehensive functional analysis of the DEGs unveiled that cell adhesion and vasculature-related signals were the most strongly enriched biological processes in OC ECs (Fig. 7e). The expression map of MYC and SELP in the overall EC populations confirmed their high expression in cholestatic livers compared to controls (Fig. 7f).
a Single-cell analysis workflow. b UMAP showing 9 major cell types. c Contour plot displaying changes in cell abundance. d Volcano plot showing significantly changed genes. e Enriched biological processes in OC ECs compared to controls. f UMAP showing the levels of MYC and SELP in ECs. g H&E and IHC for indicated proteins in liver samples, with arrows indicating Myc-positive ECs. Quantitative analysis of IHC for MYC, SELP, and MPO (n = 9, 16) (h), and Spearman correlation analysis (i). j Schematic diagram illustrating the molecular mechanism underlying the role of ECs in shaping the local immune microenvironment during obstructive cholestasis. Data are presented as the mean ± SEM. P values are indicated as follows: ** <0.01; *** <0.001; **** <0.0001. P values were obtained using the unpaired two-tailed Student’s t-test. Source data are provided as a Source Data file.
Next, we assessed the activation of the MYC/P-selectin axis and the presence of neutrophil infiltration in liver samples from 16 additionally cholestatic patients due to gallstone blockage (Supplementary Fig. 10d). We observed significant nuclear staining of MYC in liver ECs from patients, whereas very low levels of endothelial MYC staining were observed in control liver tissues from individuals who had hepatic hemangioma resection without cholestasis (Fig. 7g, h). Furthermore, we found that the expression of P-selectin in liver tissues was approximately 2-fold higher in OC samples compared to control donors (Fig. 7h). Correlation analysis revealed a significant positive correlation between the protein levels of MYC and P-selectin (Fig. 7i), supporting the potential connection between MYC and P-selectin in cholestatic disease as revealed in the above findings. Additionally, we observed profound infiltration of neutrophils in liver tissues from cholestatic patients, as assessed by myeloperoxidase (MPO) staining, whereas healthy samples showed lower and more scattered positive staining (Fig. 7g, h). Remarkably, the levels of MPO exhibited a significant positive correlation with those of MYC and P-selectin in human liver samples (Fig. 7i). These observations, along with the results from our mouse models and in vitro experiments, suggest a potential involvement of the endothelial MYC/P-selectin signal axis in the increased infiltration of neutrophils observed in human obstructive cholestasis (Fig. 7j).
Discussion
A deeper understanding of the complex interplay between the environment and the immune system holds significant promise for developing more precise and targeted treatments for a wide range of conditions characterized by abnormal immune responses and tissue destruction. In the context of cholestatic liver disease, the accumulation of BAs in the liver is known to create an inflammatory immune microenvironment1,18,36. In this study, we provide novel insights into the pathogenesis of cholestatic liver injury by focusing on liver ECs. We demonstrate that ECs acquire a distinct transcriptional profile characterized by the activation of the Myc/P-selectin signaling axis in response to the build-up of CDCA via the bile acid receptor FXR during cholestasis. This specific response results in the establishment of a pro-inflammatory microenvironment, marked by an increased infiltration of neutrophils, thus exacerbating liver damage in cholestatic conditions. Our findings shed light on the critical role of liver ECs in sensing the environmental cues and regulating the inflammatory process during cholestasis and suggest potential immuno-therapeutic targets for managing this liver disease more effectively.
Numerous advances report that cholestasis disrupts local homeostasis of liver immune microenvironment4,37,38,39,40. ECs are the most abundant non-parenchymal cell population known to play critical roles in the regulation of sinusoidal immunology through expression of various lymphocyte adhesion molecules and chemokines17,18,29. Our study reveals that cholestatic liver injury induces a unique EC transcriptional profile characterized by Myc activation, a phenomenon that appears specific to cholestasis, as evidenced in our BDL models but not observed in other liver injury models. This cholestasis-linked EC pattern notably promotes neutrophil recruitment to the liver. Patient data analysis also confirms a strong correlation between Myc/P-selectin activation in ECs and neutrophil infiltration in human cholestasis, highlighting the vital role of ECs in modulating the liver’s immune microenvironment in a disease-specific manner. Additionally, varied MYC signal activation was observed in IR, APAP, ALD, and MCD diet-induced liver injury models. In line with our results, a recent study focusing on acute liver failure by Kolodziejczyk et al.41., reported Myc activation in HSCs, ECs, and KCs in APAP model, along with an increase in the infiltration of Ly6C+ monocytes and neutrophils. Given the fact that different types of liver diseases can exhibit varying degrees of cholestasis, including drug-induced liver injury, ALD, non-alcoholic fatty liver disease (NAFLD), viral hepatitis, hepatectomy, cirrhosis, and hepatocellular carcinoma (HCC), we cautiously suggest that the activation of Myc in ECs might serve as a common mechanism in response to BAs in a wide range of liver diseases, with the degree of this process varying depending on the extent of bile acid accumulation. In addition, in our study, we observed an increase in the population of CD45+ cells in BDL-treated mice with EC-specific Myc overexpression, including cell types other than Ly6G+ cells. Based on the reports from APAP murine model, these additionally increased CD45+ cells might partially consist of Ly6C+ monocytes41 and/or eosinophils19, as these cells highly express P-selectin glycoprotein ligand-1 (PSGL-1), a special ligand for P-selectin31, which warrants further investigation to understand their potential roles in the liver’s immune response during cholestatic liver injury.
Our data highlight the CDCA/FXR pathway as a critical upstream regulator of the observed Myc/P-selectin activation in ECs. Specifically, our findings suggest that CDCA plays a predominant role in boosting MYC/P-selectin activation, rather than CA and secondary bile acids. These results align with existing studies on cholestatic liver disease, which have demonstrated a significant increase in primary bile acids, including CDCA and CA, in liver tissue42,43,44. Bile acids act as signaling molecules through their activation of the nuclear receptor FXR and the membrane receptor TGR5. Importantly, FXR shows a preference for primary bile acids for activation, with CDCA being the most potent, followed by DCA, LCA, and CA in order of potency35. On the other hand, TGR5 is preferentially activated by secondary bile acids, with LCA being the most potent, followed by DCA, CDCA, and CA in order35. Moreover, various reports have documented a significant protective phenotype in FXR-ablated mice when subjected to BDL2,45, while TGR5 knockout mice are more susceptible to BDL insult11,46. These findings, together with our observations of ECs responding to CDCA, strongly support the notion that the CDCA/FXR/Myc/P-selectin regulatory axis in ECs plays a crucial role during cholestasis. This model of action may also provide a possible explanation for the anti-inflammatory properties of UDCA or 24-norursodeoxycholic acid (norUDCA)47,48, a derivative of UDCA currently undergoing clinical trials for cholestatic liver disease treatment. Encouragingly, by inhibiting P-selectin with the pharmacological inhibitor PSI-697, we observed an improved effect on liver damage in mice with cholestasis. Therefore, we reasonably propose that the blockade of FXR downstream target P-selectin in ECs may be a promising therapeutic approach to alleviate liver injury in cholestatic conditions.
In summary, our study has identified the endothelial CDCA/FXR/Myc/P-selectin axis in the context of inflammatory liver damage associated with obstructive cholestasis. These findings not only uncover a previously unappreciated role for ECs in integrating environmental cues, inflammation, and tissue destruction, but also open up new avenues for therapeutic interventions targeting ECs and their associated signaling pathways to improve outcomes for patients suffering from cholestatic liver diseases. Continued research in this area may lead to the development of more effective and targeted treatments for patients suffering from obstructive cholestasis.
Methods
Human samples
For the scRNA-seq analysis, fresh cholestatic liver samples (n = 7) were surgically resected from patients with obstruction by biliary stones originating from the intrahepatic bile duct and/or common bile duct at Shandong Provincial Hospital. Neither ursodeoxycholic acid nor other preoperative therapies were administered. The clinical and pathological characteristics of these patients were summarized in Supplementary Fig. 10a. For comparative analysis, two control specimens were collected from regions in the unaffected lobes, distinct from the cholestatic tissues. In addition, a validation cohort was included, consisting of samples collected from 13 patients at Qilu Hospital and 3 patients at Shandong Provincial Hospital, all diagnosed with obstructive cholestasis due to gallstone blockage. Control liver tissue was obtained from 9 patients undergoing hepatic hemangioma resection without cholestasis. The clinical and pathological features of the patients in the validation cohort were summarized in Supplementary Fig. 10d. Informed consent forms were obtained from all donors or their families, and all human studies were conducted with the approval of the Ethics Committee of Shandong University (Approval No. ECSBMSSDU2021-1-19).
Animals
Eight- to ten-week-old mice were used for the experiments. The mice used in this study were fed regular chow in standard cages and maintained under a 12-hour light-dark cycle with no enrichment. EC-specific Myc-overexpressing mice, Tie2-MycLSL/+, were generated by interbreeding MycLSL/LSL mice with transgenic mice expressing Cre-recombinase driven by the tyrosine kinase 2 (Tie2, also known as Tek) promoter (Tie2-Cre). Similarly, hepatic EC-specific Myc-overexpressing mice, Lyve1-MycLSL/+, were generated by interbreeding MycLSL/LSL mice with Lyve1-Cre transgenic mice. The experiments included both male and female mice, as no significant sex-related differences were observed in terms of the phenotype or expression of the genes of interest. The Tie2-Cre mice (Cat# T003764) and Lyve1-Cre mice (Cat# T004985) were purchased from GemPharmatech Co., Ltd (Nanjing, China). The MycLSL/LSL (LSL-Myc) mice (Cat# NM-KI-00039) were purchased from Shanghai Model Organisms Center, Inc. Mice with MycLSL/+ genotype were used as controls (referred to as MycLSL/+) for the experiments using Myc transgenic mice in this study. The Fxr−/− (Nr1h4-KO) mice (Strain NO. T012640) and Tgr5−/− (Gpbar1-KO) mice (Strain NO. T012754) were constructed by GemPharmatech Co., Ltd. (Nanjing, China). The wild-type C57BL/6 J mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd All animal procedures were approved following a thorough ethical review and in compliance with guidelines set forth by the Institutional Animal Care and Use Committee at Shandong University (Approval No. ECSBMSSDU2021-2-79). Mice were euthanized using CO2 inhalation followed by cervical dislocation to facilitate tissue collection for further analysis.
Single-cell suspension preparation
Tissue samples were placed on ice in sterile Petri dishes and minced into pieces no larger than 3 mm. The minced tissue was then transferred into a gentleMACS C Tube (Cat#130-093-237, Miltenyi) containing 5 ml of digestion enzyme mix. The tube was subjected to mechanical dissociation using the gentleMACS Octo Dissociator. Following the dissociation, the samples were incubated for 30 min at 37 °C with continuous gentle shaking. The resulting suspension was filtered through a 70-μm nylon mesh (Cat#130-095-823, Miltenyi). A 10 ml portion of the cell suspension was stained with Trypan Blue to assess cell viability. The suspension was then centrifuged at 300 × g and 4 °C for 5 min, after which the supernatant was discarded. The cell pellet was resuspended in 1 ml of freezing media. To preserve cell viability and integrity, the entire dissociation process was completed within one hour, with cells kept on ice as much as possible.
cDNA library preparation and single-cell RNA sequencing
Single-cell RNA sequencing libraries were prepared using the Chromium Single Cell 3’ Reagent v2 Kits following the manufacturer’s protocol. The following kits were employed: Chromium Single Cell 3’ Library and Gel Bead Kit v2 (Cat#PN-120237, 10× Genomics), Chromium Single Cell 3’ Chip kit v2 (Cat#PN-120236, 10× Genomics), and the Chromium i7 Multiplex Kit (Cat#PN-120262, 10× Genomics). In brief, the single-cell suspensions were loaded onto the Chromium Single Cell Controller Instrument (10x Genomics), which generated single-cell gel beads in emulsions (GEMs). Once the GEMs were generated, reverse transcription reactions were performed to generate barcoded full-length cDNA. The emulsions were then disrupted using the recovery agent, and the cDNA was cleaned up using DynaBeads MyOne Silane Beads (Cat#37002D, Thermo Fisher Scientific). The cDNA was subsequently amplified by PCR with an appropriate number of cycles depending on the number of recovered cells. Following amplification, the cDNA was fragmented, end-repaired, A-tailed, and ligated with index adaptors for library preparation. The libraries were then subjected to library amplification. Finally, the libraries were sequenced on the Illumina sequencing platform (HiSeq X Ten), generating 150 bp paired-end reads.
Single-cell RNA-seq data processing
For the scRNA-seq data presented in Fig. 7a, we used the Cell Ranger software pipeline (version 2.2.0) provided by 10× Genomics. This pipeline was utilized to demultiplex cellular barcodes, align reads, and generate feature-barcode unique molecular identifier (UMI) matrices based on the human reference genome GRCh38. Regarding the scRNA-seq data processing in Figs. 1 and 3 and Supplementary Fig. 3, the raw gene expression datasets (GSE166178) were directly accessed through the GEO website (NCBI)23. The unique molecular identifier (UMI) count matrix was processed using the Scanpy (version 1.9.2) and Seurat R package (v2.4.3)49. After filtering out low-quality cells (defined as cells expressing fewer than 200 genes or having a mitochondrial genome transcript ratio > 0.2), a total of 199,093 non-parenchymal cells (NPCs) were selected for further analysis (Supplementary Fig. 1a). Among these NPCs, our focus was narrowed down to 47,761 cells that showed positive expression of Stab2 and Pecam1 (encoding CD31), which are well-established markers for ECs (Supplementary Fig. 1b). Given that the presence of other cell types could possibly be attributed to the adherence of ECs to those cells within sinusoids during single cell suspension preparation, we excluded additional cells based on marker gene expression, including neutrophils, B cells and macrophages (Supplementary Fig. 1c). The remaining cells were tested positive for Tek/Tie2 and Cdh5 (encoding VE-cadherin, a gene known to be expressed in all ECs) (Supplementary Fig. 1d), representing pure endothelial populations, were used for the subsequent analysis. Normalization of library size was performed using Scanpy on the filtered matrix to obtain the normalized count. Principal component analysis (PCA) was applied to the normalized expression matrix using highly variable genes. Cell clustering was performed using the graph-based clustering approach Leiden in Scanpy, and the results were visualized in 2-dimensions using UMAP. The likelihood ratio test which simultaneously tests for changes in mean expression and in the percentage of expressed cells was used to identify differentially expressed genes between clusters.
Bulk RNA-sequencing and data processing
Total RNA was extracted from WT and Tie2-MycLSL/+ mouse livers subjected to BDL using the Total RNA Extraction Reagent (Cat#R401-01, Vazyme) following the manufacturer’s instructions. The concentration of RNA samples was determined using a NanoDrop ND-1000 instrument. RNA libraries were prepared with the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB) according to the manufacturer’s instructions. Four independent biological replicates for each group were performed for RNA-seq. The libraries were sequenced on the Illumina sequencing platform (HiSeq X Ten), generating 150 bp paired-end reads. For sequence alignment, clean reads were aligned to the Ensembl reference genomes mm10 using HISAT2 (version 2.1.0). The FPKM (fragments per kilobase of transcript per million mapped reads) values were calculated for each detected gene using StringTie (version v1.3.3b). Differential gene expression analysis was performed using the FPKM values with the limma (version 3.56.2). Genes showing fold changes greater than 1.5 and P values less than 0.05 were considered as differentially expressed genes (DEGs).
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed to assess the variation in biological process activity using the fgsea R package (version 1.26.0). For Fig. 1b, the ranked gene list was generated by comparing the gene expression of each model to the rest of the models, and the mouse Molecular Signatures Database (MSigDB) hallmark gene sets were utilized. In Fig. 3b, the ranked gene list was generated by comparing liver tissues from Tie2-MycLSL/+ mice to control mice. The MSigDB C5 (Gene Ontology) gene sets were employed. Gene sets with a P value less than 0.05 were considered statistically significant.
Liver injury models
In this study, we utilized a range of clinically representative liver injury models for investigation and validation purposes. These included the obstructive cholestatic model induced by BDL, hepatic ischemia-reperfusion (HIR) model, drug-induced liver injury model induced by APAP, an alcohol liver disease model, as well as various diet-induced liver injury models.
BDL model
The BDL model was employed to provoke obstructive cholestasis, as previously described15,50. In brief, the bile duct was separated from the portal vein and hepatic artery with micro-serrated forceps. The 5–0 silk thread was carefully placed around the bile duct and fixed with surgical knots. A second ligation was performed in the same manner without dissecting the intervening bile ducts. In this study, mice were randomly assigned to sham or BDL group. Sham-operated and BDL-treated mice were sacrificed at 3 days, 10 days, or 2 weeks after the surgical procedure according to the experimental design. During the complete experimentation, keep the animal on a warming plate until they wake up.
HIR model
An established partial (70%) liver warm ischemia model was used in our study51. In brief, wild-type male mice (C57BL/6 J; 8 weeks of age) were first anesthetized and performed a midline laparotomy. Then, noninvasive microvascular forceps were used to block the blood supply to the left lobe and middle lobe. After ischemia for 45 min and reperfusion for 24 h, the mice were sacrificed, and liver samples and serum were collected for follow-up examination. The mice in the sham group underwent the same procedure but did not have their blood vessels operated. The warming plate was used to maintain the body temperature throughout the IR procedure.
ALD model
Wild-type male mice (C57BL/6 J; 6 weeks of age) were treated with the Gao-binge alcohol model52. Briefly, mice were acclimated to the Lieber-DeCarli liquid control diet (Cat#TP4020A, Trophic Animal Feed High-Tech Co., Ltd, China) for 5 days followed by further feeding with the liquid control or ethanol diet (5% ethanol) for 2 weeks. The volume of control diet given to mice was matched to the volume of ethanol diet consumed. On the last day of feeding, mice also were given ethanol 5 g/kg and sacrificed 8 h later. During the complete experimentation, the mice were in standard cages with normal conditions.
APAP model
Wild-type male mice (C57BL/6 J; 8 weeks of age) were used for APAP administration. In brief, mice were fasted overnight (17:00—9:00, 16 h) to reduce hepatic glutathione (GSH) levels before APAP (Cat#HY-66005, MCE) treatment. During fasting pretreatment, mice were housed in cages with wire mesh floors to prevent them from ingesting the bedding material and excreta. APAP was dissolved in warm (55 °C) phosphate-buffered saline (PBS) immediately before each experiment, cooled to 37 °C and injected intraperitoneally at a dose of 300 mg/kg dosage. PBS alone was as the vehicle control. Fasting was maintained for 4 h after APAP injection. Liver and blood samples of APAP mice were collected at 24 h time points. During the complete experimentation, the mice were in standard cages with normal conditions.
MCD model
Wild-type male mice (C57BL/6 J; 6 weeks of age) were used for the construction of the diet models. For the MCD model, mice were placed on methionine choline-deficient (MCD)-diet (Cat#TP36300, Trophic Animal Feed High-Tech Co., Ltd, China) or chow diet for 8 weeks.
P-selectin inhibition experiments
A small-molecule P-selectin antagonist, PSI-697 (Cat#HY-15526, MCE), was used for P-selectin blocking experiments. P-selectin, the largest of the selectins with a mass of 140 kDa, is a cell surface adhesion molecule that has a central role in mediating rolling and tethering of blood cells on the endothelium. When P-selectin is expressed on activated platelets and endothelial cells, interaction with its primary ligand, P-selectin glycoprotein ligand type 1 (PSGL-1), mediates the initial tethering and rolling process that precedes leukocyte transmigration through the vessel wall. PSI-697 is a commercially available P-selectin inhibitor with an IC50 of 125 μM in vitro34. PSI-697 inhibits P-selectin binding to P-selectin glycoprotein ligand-1 (PSGL-1), in a reproducible concentration-dependent manner. After the BDL surgery, the wild-type C57BL/6 J mice were administered 35 mg/kg of PSI-697 or a vehicle control via tail vein injection. This administration was performed once per day for a duration of 3 days. For the 2 weeks BDL setting, administration was performed once every other day for a duration of 2 weeks.
Liver enzyme activity
Blood was obtained from mice that were administered anesthesia via cardiac puncture. The collected blood was then subjected to centrifugation at 3000 × g for a duration of 10 min to separate the supernatant, from which the serum was collected. These serum samples were subsequently frozen and stored at a temperature of −80 °C until further analysis. The evaluation of clinically significant serum parameters was conducted using the Automated Biochemistry Analyze System (BS-240VET, Mindray).
HPLC-MS assay
The bile acid levels in serum and liver tissue samples were determined using a HPLC-MS system with a ADME column (100 mm× 2.1 mm). Frozen serum samples were thawed at 25 °C and vortex-mixed briefly. Serum in 100 µl or ~50 mg liver tissue was extracted in 600 µl acetonitrile, followed by additions of IS working solution (DCA-4d, 100 µg/ml, 10 µl) and methanol-water (5:95, v/v) (10 µl). The mixture was extracted by shocking at 4 °C for 6 min and then centrifuged at 10,000 × g for 15 min. The supernatants were transferred to another 1.5 ml Eppendorf tube and evaporated to dryness at 45 °C by a vacuum extractor. The residue was reconstituted in 100 µl of acetonitrile water containing 0.1% formic acid (20:80, v/v). Respective standards were used to generate standard calibration curves. Liver metabolite concentrations were normalized to weight. Samples were analyzed on an HPLC system hyphenated to a Qtrap 5500 (AB Sciex).
Quantitative real-time PCR
Liver samples or HUVECs were lysed using RNA isolater Total RNA Extraction Reagent (Cat#R401-01, Vazyme), following the manufacturer’s instructions. The RNA was then reverse transcribed using HiScript II Q Select RT SuperMix for qPCR (+gDNA wiper) (Cat#R233-01, Vazyme). The reverse transcribed cDNA was amplified by CFX Connect Real-time PCR detection system (Bio-Rad) using the SYBR Green PCR Master Mix (Cat#Q121-02/03, Vazyme). Data were normalized to the expression of the housekeeping gene Gapdh individually. The 2−△△Ct method was used to calculate relative expression changes. Specific primers used for qPCR assays were shown in Supplementary Table.
Histology
The mice liver tissue samples were fixed in 4% formalin overnight, processed for paraffin embedding, and sectioned into 5 μm thick sections. Subsequently, the tissue sections were stained with hematoxylin and eosin (H&E) and observed under light microscopy. Necrotic areas or areas with imminent necrosis were identified based on the presence of eosinopenia, loss of cell structure, vacuolization, cell rupture, or nucleolysis. To ensure accurate analysis, necrotic areas displaying all characteristics and areas on the verge of necrosis without nucleolysis but with most features were included in the quantitative analysis of necrosis. Histopathologic evaluation was performed by two pathologists experienced in hepatobiliary pathology.
Immunohistochemistry
Immunohistochemistry was performed on sections obtained from formalin-fixed paraffin-embedded liver tissues. The sections with a thickness of 5 μm were dewaxed, rehydrated, and then treated with sodium citrate buffer (pH 6.0) or Tris-EDTA buffer (pH 9.0) for antigenic repair. After natural cooling, they were washed with 1× PBST and incubated at room temperature for 12 min with endogenous peroxidase blockers from a commercially available kit (Cat#PV-6000, ZSGB-BIO). 1× PBST buffer solution was washed 3 times for 3 min. After incubation with 5% bovine serum albumin for 30 min at room temperature, appropriate amount of primary antibody was added and incubated at 4 °C overnight. After rinsing with PBST buffer solution, drops were added with 100 μl or appropriate amount of enzyme-labeled sheep anti-mouse/rabbit IgG polymer (Cat#PV-6000, ZSGB-BIO), and incubated at room temperature for 1 h. Appropriate amount of DAB (Cat#ZLI-9018, ZSGB-BIO) color-developing solution was added and incubated at room temperature for 1–5 min. Rinse with running water and incubate with hematoxylin solution for 7–12 s; After hydrochloric acid ethanol differentiation, rinse with tap water and return to blue. The slices were dehydrated with ethanol, transparent with xylene and finally sealed with resin. The staining results were observed and interpreted by a qualified pathologist under an optical microscope. For the quantitative analysis of 4-HNE staining, the positive staining area was defined as the percentage of the field area within a preset color range determined by the using ImageJ software. Antibodies used in this study are summarized in Supplementary Table.
Immunofluorescence (IF) staining
Primary antibodies against CD31, Myc, P-selectin, or Ki67 were used to perform IF staining with the multiplex immunohistochemistry/immunofluorescence staining kits (Cat#RK05903, Abclonal Technology Co., Ltd, China) according to the manufacturer’s instructions. The paraffin-embedded liver sections (4 µm) were blocked with 2% bovine serum albumin (Cat#A8020, Solarbio Science & Technology Co. Ltd, China) in TBST for 30 min, and overnight incubated at 4 °C sequentially with the primary antibodies. After washing with PBS, HRP-conjugated labeled anti-rabbit or anti-mouse IgG was probed at room temperature for 50 min and reacted with fluorophore-conjugated tyramine molecules (TYR-520, TYR-570, or TYR-690) for 15 min. At the end of immunofluorescence staining, the antigen was repaired with Tris/EDTA buffer (pH 9.0) after washing with PBS for 3 times before the next staining cycle. After immunofluorescence staining, the tablets were sealed with anti-fluorescence quenching tablets containing DAPI (Cat#S2110, Solarbio Science & Technology Co., Ltd, China). The fluorescent signals were photo-imaged and merged using a laser scanning confocal microscope (ZEISS, LSM880) and ZEN software (version 3.9).
Sirus red staining
Sirus Red staining was performed using a commercially available kit (Cat#BP-DL030, Sbjbio Life Science) following the manufacturer’s instructions. The positive staining area was defined as the percentage of the field area within a preset color range determined by using ImageJ software.
Flow cytometry analysis
For liver cell isolation, the liver was aseptically harvested and immediately immersed in RPMI1640 medium supplemented with 20% fetal bovine serum (FBS), DNase I (150 µg/mL), and collagenase IV (1 mg/mL). The liver tissue was finely minced and then enzymatically digested with shaking at 150 rpm at 37 °C for 30 min. The digested tissue suspension was subsequently filtered through a 100 µm cell strainer to obtain a single-cell suspension. Primary hepatocytes were isolated by centrifugation at 100 × g for 1 min at room temperature, while endothelial cells were isolated by centrifugation at 800 × g for 5 min at room temperature. Mononuclear cells were separated using density gradient centrifugation with 40%–80% Percoll.
The live and dead cells were discriminated by Zombie Aqua Fixable Viability Kit (BioLegend, San Diego, CA, USA) on PBS. CD16/32(Cat#14-0161085, eBioscience) antibody was used to block the non-specific binding to Fc receptors before surface staining. Cells were stained with surface-labeled antibodies for 25 min at 4 °C. For intracellular staining, cells were incubated with Fixation/Permeabilization at 4 °C overnight. After fixation, the cells were incubated with the indicated antibodies for 2 h at 4 °C. Flow cytometry was performed with Backman Gallios cytometers and analyzed with FlowJo software. Monoclonal antibody against CD45.2 (Cat#56-0452-82) was purchased from eBioscience (San Diego, CA, USA). Monoclonal antibodies against CD3 (Cat#102418), CD86 (Cat#105005), F4/80 (Cat#123110), CD11b (Cat#101228), and CD206 (Cat#141708) were purchased from Biolegend (San Diego, CA, USA). Monoclonal antibodies against CD8 (Cat#60-0081-U100), and CD4 (Cat#75-0041-U100) were purchased from Tonbo Biosciences (San Diego, California, USA). Monoclonal antibody against Myc (MA1-980) were purchased from Invitrogen (USA). CoraLite488-conjugated goat anti-mouse IgG(H + L) (SA00013-1) was purchased from Proteintech (China).
ELISA assay
To measure serum levels of Il-1β in mice, serum samples were collected and analyzed using ELISA kits purchased from Jiangsu Jingmei Biological Technology Co., Ltd Mouse Il-1β (Cat#JM-02323) kit was used following the manufacturer’s instructions.
Cytometric bead array
Serum cytokine (IL-2, IL-4, IL-6, IL-10, IL-17A, interferon (IFN)-γ, and tumor necrosis factor (TNF)) concentrations were measured using the Mouse Th1/Th2/Th17 Cytometric Bead Array (CBA) kit (Cat#560485, BD Biosciences, USA), following the manufacturer’s instructions. Bead analysis was performed with a FACSymphony A3 flow cytometer and FCAP Array software (BD Biosciences, USA).
Immunoblotting
Total protein was isolated from tissue samples or HUVECs using RIPA lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA) supplemented with protease inhibitor cocktail tablets (Cat#HY-K0011; MCE) and phosphatase inhibitor tablets (Cat#G2007, Servicebio). The total protein samples were loaded and separated on SDS–PAGE gels and transferred onto PVDF membranes (Cat#IPVH00010, Merck Millipore). The membranes were blocked with 5% skim milk and incubated with the specified primary antibodies overnight at 4 °C, followed by incubation with the corresponding secondary antibodies for 1 h at room temperature. The membranes were visualized using enhanced chemiluminescence (ECL) reagents (Cat#E411-03, Vazyme) and captured using a Chemiluminescence Imaging System (Tanon 5500). GAPDH was used as a loading control. Antibodies used in this study are summarized in Supplementary Table.
Scanning electron microscopy
Liver tissue samples were initially fixed in 2.5% glutaraldehyde (Cat#G1102, Servicebio) at room temperature for 2–3 h, followed by storage at 4 °C. Subsequently, the samples were fractured to expose natural surfaces. They were then washed five times with PBS (0.1 M, pH 7.4) for 15 min each. Next, the samples underwent fixation in 1% osmium tetroxide (Ted Pella) under light-avoiding conditions for 1.5 h. After two rinses with PBS and three rinses with ultrapure water (each rinses lasting 15 min), the samples were dehydrated using a gradient of ethanol concentrations (50%, 70%, 80%, 90%, and twice with 100%, each for 15 min). Following dehydration, critical point drying was performed using CO2 with Quorum K850 critical point dryer. The samples were mounted on specimen stubs with conductive adhesive, sputter-coated with gold for 60 s using an ion sputter coater (Cressington Auto108), and then imaged and photographed using Hitachi SU8010 scanning electron microscope.
Cell culture and treatment
Human umbilical vein endothelial cells (HUVECs) were obtained from Procell Life Science & Technology (Cat#CL-0122). HUVECs were cultured in high-glucose DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin under standard conditions of 37 °C and 5% CO2. For the bile acids stimulation experiments, HUVECs were seeded into 24-well plates at a density of 2.5 × 104 cells per well. They were then incubated with either a single bile acid (CA, CDCA, LCA, DCA, TCA, TCDCA) or a combination of bile acids at a respective concentration 100 μM. The duration of the incubation was 6 h, 24 h, and 48 h. In the FXR agonist stimulation assay, HUVECs were incubated with 10 μM INT-747 (Cat#S7660, Selleck) or 5 μM GW4064 (Cat#G5172, Sigma) for 48 h. UDCA was used as an FXR inhibitor, and HUVECs were incubated with 200 μM UDCA for 48 h. The primary endothelial cell culture medium was prepared as follows: RPMI-1640 supplemented with 10% (v/v) fetal bovine serum (FBS) and 1% (v/v) penicillin-streptomycin solution. A total of 3 × 104 EC cells were seeded in a 48-well plate with collagen matrices derived from acid-solubilized type I rat tail tendon collagen (Cat#C8062, Solarbio), following the manufacturer’s instructions. The ECs were treated individually with bile acids (CA, CDCA, TCA, TCDCA) at a concentration of 200 μM each for 48 h. All the bile acids used in this study are summarized in Supplementary Table.
siRNA transfection
siRNA transfections were conducted in 24-well culture dishes using a seeding density of 5.0 × 104 cells per plate. The transfection reagent utilized was Lipo8000 (Cat#C0533, Beyotime), and the transfection was carried out in antibiotic-free Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% serum. All siRNA duplexes used were purchased from Sangon Biotech. 24 hours after transfection, cells were treated with GW4064 (5 μM) or bile acid mixture (100 μM CDCA plus 100 μM TCDCA). After the initial transfection, an additional transfection cycle was carried out 48 h later (double transfection), and the samples were collected for protein analysis. The primary endothelial cells were transfected with siRNAs using Lipofectamine 2000 (Cat#11668019, Thermo Fisher Scientific). After 48 h of transfection, these cells were used in subsequent experiments. All the siRNA sequences used in this study are summarized in Supplementary Table.
Generation of knockout cell lines using the CRISPR/Cas9 technique
The sgRNAs targeting the human MYC gene were obtained from the literature (Supplementary Table) and subsequently cloned into the lentiCRISPR v2 vector (Cat#52961, Addgene). To obtain lentiviruses, lentiCRISPR v2 plasmids were cotransfected with PAX and PMD into HEK293T packaging cells using PEI. The supernatant (containing virus) was harvested at 48 and 72 h after filtering with a 0.22 μm filter membrane. For lentivirus infection, HUVECs were seeded in a 6-well plate (1 × 106 cells/well). Eight hours later, the cells were incubated with 1 ml virus solution supplemented with 100 μl FBS, 1 ml DMEM, and 2 μl polybrene (10 mg/ml). Stably expressing cells were established by puromycin (10 μg/ml) selection.
Primary endothelial cell sorting
Primary liver cells from WT, Fxr−/−, and Tgr5−/− mice were isolated from perfused livers by collagenase digestion. Briefly, mice were cannulated through the portal vein and after nicking the inferior vena cava, perfused with 20 ml of Hank’s balanced salt solution (HBSS) containing 5 mM of ethylenediaminetetraacetic acid followed by 20 ml of HBSS containing 1 mg/ml of collagenase and 1 mM of CaCl2. Livers were isolated, torn, shaken loose in DMEM with 25 mM of glucose, 20% fetal bovine serum, 100 IU/ml of penicillin G, and 100 lg/ml of streptomycin, and filtered through a 70 µm cell strainer. Cells were washed once with 1× PBS centrifuged at 800 × g for 5 min. The live and dead cells were then discriminated by Zombie Aqua Fixable Viability Kit (BioLegend) in PBS at 25 °C for 25 min. After saturating Fc receptors with CD16/32-blocking antibody, single-cell suspensions were incubated at 4 °C for 30 min with conjugated antibodies against following markers: CD45.2 (Alexa Fluor 700, eBioscience), CD31 (PE/Cyanine7, Biolegend). Liver endothelial cells were sorted using a backman Moflo Astrios EQ. Primary EC cells were cultured in a confluent 24-well dish using a solution composed of RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin solution.
Statistical analysis
The statistical significance of differences between groups was examined using unpaired two-tailed Student’s t-test. Correlations between groups were analyzed using the nonparametric Spearman’s ρ test. The exact value of n (number of biological or experimental replicates) can be found in the Figure Legend. Statistically significant differences were considered as *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001. All data are expressed as the mean ± SEM values and were analyzed using GraphPad Prism 7.0 (GraphPad Software Inc.).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The bulk RNA-seq data generated in this study have been deposited in the National Center for Biotechnology Information GEO database under accession code GSE237384. The scRNA-seq data generated in this study have been deposited in the National Center for Biotechnology Information GEO database under accession code GSE237622. The scRNA-seq data used in this study, which included samples from control livers and various liver injury models (BDL, APAP, IR, ALD and MCD), were obtained from the GEO database under accession code GSE166178. All data supporting the findings described in this manuscript are available in the article and in the Supplementary Information and from the corresponding author upon request. Source data are provided with this paper.
Code availability
All codes and scripts used for association studies are available on request.
References
Hirschfield, G. M., Heathcote, E. J. & Gershwin, M. E. Pathogenesis of cholestatic liver disease and therapeutic approaches. Gastroenterology 139, 1481–1496 (2010).
Stedman, C. et al. Benefit of farnesoid X receptor inhibition in obstructive cholestasis. Proc. Natl Acad. Sci. USA 103, 11323–11328 (2006).
Guo, C. S. et al. Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity 45, 802–816 (2016).
Ma, C. et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360, eaan5931 (2018).
Paik, D. et al. Human gut bacteria produce ΤΗ17-modulating bile acid metabolites. Nature 603, 907–912 (2022).
Arifuzzaman, M. et al. Inulin fibre promotes microbiota-derived bile acids and type 2 inflammation. Nature 611, 578–584 (2022).
Song, X. et al. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature 577, 410–415 (2020).
Cao, W. et al. The xenobiotic transporter Mdr1 enforces T cell homeostasis in the presence of intestinal bile acids. Immunity 47, 1182–1196 (2017).
Hang, S. et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature 576, 143–148 (2019).
Xiong, H. et al. Suppressed farnesoid X receptor by iron overload in mice and humans potentiates iron-induced hepatotoxicity. Hepatology 76, 387–403 (2022).
Péan, N. et al. The receptor TGR5 protects the liver from bile acid overload during liver regeneration in mice. Hepatology 58, 1451–1460 (2013).
Martinot, E. et al. Bile acids and their receptors. Mol. Asp. Med. 56, 2–9 (2017).
Chiang, J. Y. L. & Ferrell, J. M. Bile acid receptors FXR and TGR5 signaling in fatty liver diseases an therapy. Am. J. Physiol. Gastrointest. L 318, G554–G573 (2020).
Nagahashi, M. et al. Conjugated bile acid-activated S1P receptor 2 is a key regulator of sphingosine kinase 2 and hepatic gene expression. Hepatology 61, 1216–1226 (2015).
Li, S. et al. Cytoplasmic tyrosine phosphatase Shp2 coordinates hepatic regulation of bile acid and FGF15/19 signaling to repress bile acid synthesis. Cell Metab. 20, 320–332 (2014).
Perino, A., Demagny, H., Velazquez-Villegas, L. & Schoonjans, K. Molecular physiology of bile acid signaling in health, disease, and aging. Physiol. Rev. 101, 683–731 (2021).
Poisson, J. et al. Liver sinusoidal endothelial cells: physiology and role in liver diseases. J. Hepatol. 66, 212–227 (2017).
Shetty, S., Lalor, P. F. & Adams, D. H. Liver sinusoidal endothelial cells - gatekeepers of hepatic immunity. Nat. Rev. Gastroenterol. Hepatol. 15, 555–567 (2018).
Xu, L. et al. Hepatic recruitment of eosinophils and their protective function during acute liver injury. J. Hepatol. 77, 344–352 (2022).
Dar, W. A., Sullivan, E., Bynon, J. S., Eltzschig, H. & Ju, C. Ischaemia reperfusion injury in liver transplantation: cellular and molecular mechanisms. Liver Int. 39, 788–801 (2019).
Yoong, K. F. et al. Expression and function of CXC and CC chemokines in human malignant liver tumors: a role for human monokine induced by gamma-interferon in lymphocyte recruitment to hepatocellular carcinoma. Hepatology 30, 100–111 (1999).
Yoong, K. F., McNab, G., Hubscher, S. G. & Adams, D. H. Vascular adhesion protein-1 and ICAM-1 support the adhesion of tumor-infiltrating lymphocytes to tumor endothelium in human hepatocellular carcinoma. J. Immunol. 160, 3978–3988 (1998).
Wang, Z. et al. A single-cell transcriptomic atlas characterizes liver non-parenchymal cells in healthy and diseased mice. Cold Spring Harbor Laboratory https://doi.org/10.1101/2021.07.06.451396 (2021).
Blokker, B. A. et al. Fine‐tuning of sirtuin 1 expression is essential to protect the liver from cholestatic liver disease. Hepatology 69, 699–716 (2019).
Li, Y., Tang, R., Leung, P. S., Gershwin, M. E. & Ma, X. J. A. R. Bile acids and intestinal microbiota in autoimmune cholestatic liver diseases. Autoimmun. Rev. 16, 885–896 (2017).
Su, T. T. et al. Single-cell transcriptomics reveals zone-specific alterations of liver sinusoidal endothelial cells in cirrhosis. Cell Mol. Gastroenter. 11, 1139–1161 (2021).
Xiong, X. et al. Landscape of intercellular crosstalk in healthy and NASH liver revealed by single-cell secretome gene analysis. Mol. Cell 75, 644–660.e645 (2019).
Halpern, K. B. et al. Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat. Biotechnol. 36, 962–970 (2018).
Wang, Y. & Liu, Y. Neutrophil-induced liver injury and interactions between neutrophils and liver sinusoidal endothelial cells. Inflammation 44, 1246–1262 (2021).
Weber, C., Fraemohs, L. & Dejana, E. The role of junctional adhesion molecules in vascular inflammation. Nat. Rev. Immunol. 7, 467–477 (2007).
Yadav, S. S. et al. P-selectin mediates reperfusion injury through neutrophil and platelet sequestration in the warm ischemic mouse liver. Hepatology 29, 1494–1502 (1999).
Payne, S., De Val, S. & Neal, A. Endothelial-specific cre mouse models. Arterioscler Thromb. Vasc. Biol. 38, 2550–2561 (2018).
Preziosi, M., Okabe, H., Poddar, M., Singh, S. & Monga, S. P. Endothelial Wnts regulate beta-catenin signaling in murine liver zonation and regeneration: a sequel to the Wnt-Wnt situation. Hepatol. Commun. 2, 845–860 (2018).
Bedard, P. W. et al. Characterization of the novel P-selectin inhibitor PSI-697 [2-(4-chlorobenzyl)-3-hydroxy-7,8,9,10-tetrahydrobenzo[h] quinoline-4-carboxylic acid] in vitro and in rodent models of vascular inflammation and thrombosis.J Pharmacol. Exp. Ther. 324, 497–506 (2008).
Bertolini, A., Fiorotto, R. & Strazzabosco, M. Bile acids and their receptors: modulators and therapeutic targets in liver inflammation. Semin. Immunopathol. 44, 547–564 (2022).
Perino, A., Demagny, H., Velazquez-Villegas, L. & Schoonjans, K. J. P. Molecular physiology of bile acid signaling in health, disease, and aging. Physiol. Rev. 101, 683–731 (2021).
Gijbels, E., Pieters, A., De Muynck, K., Vinken, M. & Devisscher, L. Rodent models of cholestatic liver disease: a practical guide for translational research. Liver Int. 41, 656–682 (2021).
Patten, D. A., Shepherd, E. L., Weston, C. J. & Shetty, S. Novel targets in the immune microenvironment of the hepatic sinusoids for treating liver diseases. In Proc. Seminars in liver disease, 111–123 (Thieme Medical Publishers, 2019).
Sato, K. et al. Pathogenesis of Kupffer cells in cholestatic liver injury. Am. J. Pathol. 186, 2238–2247 (2016).
Gujral, J. S., Farhood, A., Bajt, M. L. & Jaeschke, H. Neutrophils aggravate acute liver injury during obstructive cholestasis in bile duct-ligated mice. Hepatology 38, 355–363 (2003).
Kolodziejczyk, A. A. et al. Acute liver failure is regulated by MYC and microbiome-dependent programs. Nat. Med. 26, 1899–1911 (2020).
Zhang, Y. et al. Effect of bile duct ligation on bile acid composition in mouse serum and liver. Liver Int. 32, 58–69 (2012).
Stiehl, A. et al. Ursodeoxycholic acid-induced changes of plasma and urinary bile acids in patients with primary biliary cirrhosis. Hepatology 12, 492–497 (1990).
Raedsch, R., Lauterburg, B. H. & Hofmann, A. F. Altered bile acid metabolism in primary biliary cirrhosis. Dig. Dis. Sci. 26, 394–401 (1981).
Wagner, M. et al. Role of farnesoid X receptor in determining hepatic ABC transporter expression and liver injury in bile duct-ligated mice. Gastroenterology 125, 825–838 (2003).
Yang, H. et al. TGR5 protects against cholestatic liver disease via suppressing the NF-kappaB pathway and activating the Nrf2/HO-1 pathway. Ann. Transl. Med. 9, 1158 (2021).
Trauner, M., Fuchs, C. D., Halilbasic, E. & Paumgartner, G. New therapeutic concepts in bile acid transport and signaling for management of cholestasis. Hepatology 65, 1393–1404 (2017).
Beuers, U., Trauner, M., Jansen, P. & Poupon, R. J. J. o.h. New paradigms in the treatment of hepatic cholestasis: from UDCA to FXR, PXR and beyond. J. Hepatol. 62, S25–S37 (2015).
Villani, A. C. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573 (2017).
Ding, B. S. et al. Divergent angiocrine signals from vascular niche balance liver regeneration and fibrosis. Nature 505, 97–102 (2014).
Xu, B. et al. Metabolic rewiring of kynurenine pathway during hepatic ischemia-reperfusion injury exacerbates liver damage by impairing NAD homeostasis. Adv. Sci. 9, e2204697 (2022).
Chao, X. et al. Impaired TFEB-mediated lysosome biogenesis and autophagy promote chronic ethanol-induced liver injury and steatosis in mice. Gastroenterology 155, 865–879.e812 (2018).
Acknowledgements
This work was supported by National Key Research and Development Program of China (2020YFA0804400); National Natural Science Foundation of China (32422024, 82270621, 82071854, 32100590, 32270820, 82373000, 82203634, 82325008); Natural Science Foundation of Shandong Province (ZR2020QH038, ZR2021QB012, ZR2022MH150); Funds for Youth Interdisciplinary and Innovation Research Groups of Shandong University (2020QNQT003 and 2020QNQT009); the Fundamental Research Funds for the Central Universities (2022JC008); and Taishan Scholars Program of Shandong Province.
Author information
Authors and Affiliations
Contributions
S.L., D.Y., J.C., P.M. and H.Q. designed experiments, interpreted data and wrote the manuscript. P.Z. and X.L. contributed to experimental design and performed the in vivo animal studies. J.L. performed the in vitro experiments. Y.Z., Y.T., R.L., X.T. and C.Z. contributed to the collection, characterization, and histological analysis of patient samples. J.S., Y.C. and P.H. contributed to the HPLC-MS assay. J.S., S.C., M.Z. and Y.Z. contributed to primary endothelial cell sorting experiment. All authors provided input and reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Edward Prochownik, Gernot Zollner and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, P., Li, X., Liang, J. et al. Chenodeoxycholic acid modulates cholestatic niche through FXR/Myc/P-selectin axis in liver endothelial cells. Nat Commun 16, 2093 (2025). https://doi.org/10.1038/s41467-025-57351-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-57351-2