Abstract
Exercise combats obesity and metabolic disorders, but the underlying mechanism is incompletely understood. KLF10, a transcription factor involved in various biological processes, has an undefined role in adipose tissue and obesity. Here, we show that exercise facilitates adipocyte-derived KLF10 expression via SIRT1/FOXO1 pathway. Adipocyte-specific knockout of KLF10 blunts exercise-promoted white adipose browning, energy expenditure, fat loss, glucose tolerance in diet-induced obese male mice. Conversely, adipocyte-specific transgenic expression of KLF10 in male mice enhanced the above metabolic profits induced by exercise. Mechanistically, KLF10 interacts with FOXO1 and facilitates the recruitment of KDM4A to form a ternary complex on the promoter regions of Pnpla2 and Lipe genes to promote these key lipolytic genes expression by demethylating H3K9me3 on their promoters, which facilitates lipolysis to defend against obesity in male mice. As a downstream effector responding to exercise, adipose KLF10 could act as a potential target in the fight against obesity.
Similar content being viewed by others
Introduction
Obesity and related chronic metabolic diseases have become a worldwide global epidemic, posing a public health crisis. It is conventionally defined as an excess of body fat causing prejudice to health. Overweight is regarded as a main fatal factor linked to diminished average life expectancy, because it is a prevalent driver of several adverse chronic diseases, such as cardiovascular disease, type 2 diabetes, fatty liver disease, and cancer1. On the basis of widespread growing population living with obesity, which may bring about detrimental consequences, precise prevention and treatment strategy is urgently required. Adipose tissue is classified based on anatomic location and cell type constituent and exerts an important role in energy homeostasis2. According to the current perception, adipose tissue could be classified into three types: white adipose tissue (WAT), brown adipose tissue (BAT) and beige adipose tissue3,4,5,6. WAT is the primary site of energy storage. BAT specializes in the dissipation of energy through the production of heat, which is via mitochondrial uncoupling of oxidative phosphorylation of free fatty acids mediated by the expression of uncoupling protein 1 (UCP1). Beige adipose tissue exhibits high UCP1 expression and respiration rates7,8,9,10. In response to cold or exercise stimulation, beige adipocytes emerge in WAT, a process known as browning. Browning of white adipose tissue is most prominent in the inguinal subcutaneous deposit and possesses the great therapeutic potential of fighting against obesity and improving glucose homeostasis11,12.
Obesity prevalence is associated with imbalanced catabolic processes of lipids13,14,15,16,17,18,19. Adipocyte lipolysis is a mobilization of excessive lipids by breakdown of triglyceride (TG) stored in fat cells and release of glycerol and free fatty acids (FFAs), which are essential biomolecules for organisms20,21. Impaired neutral TG hydrolysis contributes to surplus lipid accumulation, which is an overwhelming burden to adipose tissue micro-environment, and compromised FFAs utilization, which leads to the reduction of ATP or heat production. Two key enzymes are required in neutral TG hydrolytic processes: adipose triglyceride lipase (ATGL) and hormone-sensitive lipase (HSL). Both of them are responsible for more than 90% of TG hydrolysis. ATGL catalyzes the initial step of lipolysis, converting TG to diacylglycerol (DG), and HSL primarily catalyzes the DGs to monoacylglycerol (MG). Global and adipose-specific ATGL-knockout mice exhibited increased TG deposit and disturbed energy substrate preference22. Mice with inhibited expression of ATGL in adipose tissue are more likely sensitized to High-fat diet (HFD)-induced obesity. Also, blunted insulin sensitivity and glucose tolerance are caused by declined adipose ATGL expression23. In addition, HSL deficiency manifests in adipocyte insulin resistance due to compromised glycerol release and more severe inflammation24. Thus, ATGL and HSL, functioning as the key lipolytic enzymes, play important roles in maintaining energy homeostasis. It has been evidenced that forkhead box O-1 (FOXO1) acts as a critical transcriptional factor in facilitating lipolysis profile through its deacetylation25.
Multiple factors contribute to the epidemic of obesity, such as imbalanced diet, sedentary lifestyle, stress, endocrine disruptors and so on26,27. Regular aerobic exercise is an acknowledged strategy for resisting obesity and associated metabolic disorder28,29,30. A lot of randomized controlled trials have confirmed the effectiveness of aerobic exercise on body weight and fat mass loss, as well as BMI and waist circumference decrease31,32. Exercise, especially aerobic exercise, has been advocated as a pivotal therapeutic resource for both treatment and prevention of overweight33. It is now known that exercise training leads to less adipose tissue depots, in which enhanced lipolysis is involved. TG hydrolysis was increased after exercise. Upregulation of expression and activity of lipolysis-related genes were triggered by exercise in the adipocytes of rodents, which resulted in the prevention of access lipid accumulation34. Other than that, endurance exercise caused a strong lipolytic response in human adipose tissue35. These proofs indicate an important role of lipolysis in exercise-induced alleviation of obesity and metabolic improvement. However, few studies have demonstrated how exercise facilitates lipolysis to exert beneficial effects on metabolism and the underlying mechanism remains to be investigated further.
Krüppel-like factor 10 (KLF10), also named TGFβ-inducible-early-gene1(TIEG1), was found in human osteoblasts for the first time. KLF10 has been found to act as a downstream factor of transforming growth factor β (TGFβ)/SMAD signaling and it participates in various biological processes36. It has been reported that mice with KLF10 deficiency in CD4+ T cells exhibited more severe obesity and insulin resistance. The existence of KlF10 in preadipocytes facilitates mitotic clonal expansion and maintains an appropriate level of adipogenesis37. KLF10 can regulate gluconeogenic genes expression in liver and it is also found to inhibit lipogenesis in hepatocytes38,39. Studies also shown that KLF10 is involved in regulating cell proliferation and apoptosis, and its abnormal expression may contribute to disease processes like nonalcoholic steatohepatitis (NASH) and tumorigenesis39,40. Yet, the function of KLF10 in mature adipocytes is still unknown.
Here, it was found that mature adipocyte-specific KLF10 deficiency brought about reduced energy expenditure, higher sensitivity to diet-induced obesity (DIO), impaired WAT browning and glucose tolerance in male mice, which compromised the effects of exercise on the amelioration of DIO and metabolic disorders in male mice. Whereas, adipocyte-specific transgenic expression of KLF10 in male mice showed a synergetic effect with exercise to promote metabolic health. KLF10 in mature adipocytes was upregulated by SIRT1/FOXO1 axis in response to exercise and then interacted with FOXO1 to recruit histone lysine demethylase 4A (KDM4A) and form a ternary complex, thereby transactivating key lipolytic genes to promote lipolysis and energy expenditure. These results indicate an important role of adipose FOXO1-KLF10 reinforcing loop in promoting lipolysis, which facilitates exercise-mediated anti-obesity effects in male mice.
Results
KLF10 is upregulated in mature adipocytes in response to exercise and fasting
Among the Krüppel-like factors (KLFs), KLF3, KLF9, KLF14 and KLF15 have been reported to have relative high expression in adipose tissues and are involved in the development and metabolism of adipose tissues41. The adipose gene expression levels of the above KLFs, as well as that of KLF10, in response to exercise training in mice were investigated. Chow diet (CD)-fed wild type C57BL/6 male mice were subjected to eight-week treadmill training following an established protocol of exercise for rodents under CD feeding42. The 8 weeks of treadmill training had little effects on the adipose gene and protein expression levels of KLF3, KLF9, KLF14 and KLF15 (Supplementary Fig. 1a–i). In contrast, exercise significantly upregulated the gene and protein expression of KLF10 in iWAT and eWAT, but not in BAT. Therefore, the role of adipose KLF10 in exercise-regulated adipose tissue metabolism was investigated further.
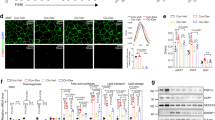
Because KLF10 is proven not only expressed in stromal vascular fraction (SVF), but also in mature adipocytes in adipose tissue (Supplementary Fig. 1j), the expression levels of KLF10, which is in response to exercise, were detected in both SVF and mature adipocytes. In both inguinal WAT (iWAT) and epididymal WAT (eWAT), exercise training led to significant upregulation of KLF10 expression in mature adipocytes, but not in SVF (Fig. 1a–f and Supplementary Fig. 1k, l). However, in BAT, exercise training had little effect on KLF10 expression level in neither mature adipocytes nor SVF (Fig. 1g, h and Supplementary Fig. 1m, n). Like exercise, fasting can also promote catabolic processes. Interestingly, Klf10 gene expression was also increased in iWAT and eWAT after overnight fasting (Supplementary Fig. 1o, p), and this was mainly because of increased Klf10 gene expression in mature adipocytes from iWAT and eWAT (Fig. 1i–l). Similarly, in BAT, neither the Klf10 gene expression in SVF nor that in mature adipocytes was obviously affected by fasting (Fig. 1m, n). These results imply that adipose KLF10 might exert a potential role under catabolic conditions, such as exercise and fasting.
From a–h Chow diet-fed 8-week-old male mice were subjected to treadmill training for 8 weeks before being sacrificed for analysis. Sed, non-exercise. Ex, exercise. a, b Messenger RNA (mRNA) levels of Klf10 in stromal vascular fraction (SVF) and mature adipocytes of inguinal white adipose tissue (iWAT), respectively. c Representative western blotting of iWAT-isolated mature adipocytes lysates. β-tubulin serves as an internal control. d, e The mRNA levels of Klf10 in SVF and mature adipocytes of epididymal white adipose tissue (eWAT), respectively. f Representative western blotting of eWAT-isolated mature adipocytes lysates. g, h The mRNA levels of Klf10 in SVF and mature adipocytes of brown adipose tissue (BAT), respectively. i–n Chow diet-fed 8-week-old male mice were fasted for 12 h before being sacrificed for analysis. i, j The mRNA levels of Klf10 in SVF and mature adipocytes of iWAT, respectively. k, l The mRNA levels of Klf10 in SVF and mature adipocytes of eWAT, respectively. m, n, The mRNA levels of Klf10 in SVF and mature adipocytes of BAT, respectively. For statistical analysis, unpaired two-tailed Student’s t tests were performed. n = 6 male mice per group in all the above experiments. All data show the means ± SD. Source data are provided as a Source Data file.
SIRT1/FOXO1 axis mediates the induction of KLF10 during exercise
Next, the mechanism of how exercise induces adipose KLF10 expression was investigated. A potential FOXO1 binding site was predicted on the gene promoter of Klf10 (Fig. 2a), suggesting that FOXO1 may regulate the transcription of Klf10 gene. Luciferase reporter analysis showed the transactivating effect of FOXO1 on Klf10 gene promoter, which was greatly weakened by mutation of FOXO1 binding site on Klf10 promoter (Fig. 2b, c). The chromatin immunoprecipitation (ChIP) result indicated that FOXO1 is capable of binding to Klf10 promoter in iWAT and eWAT, and exercise further enhanced the above binding capacity (Fig. 2d, e). FOXO1 is known to be a transcriptional factor whose nuclear localization level and transcriptional activity will be enhanced by deacetylation43. Consistently, a significant reduction of FOXO1 acetylation was observed in iWAT and eWAT after exercise (Fig. 2f and Supplementary Fig. 2a, b). Then the enzyme responsible for exercise-induced FOXO1 deacetylation was investigated. It has been reported that both SIRT1 and SIRT6 can deacetylate FOXO1 to activate its transcriptional activity25,44. In the iWAT and eWAT, the enzyme activity of SIRT1, but not that of SIRT6, was significantly enhanced by exercise training (Fig. 2g, h and Supplementary Fig. 2c, d). In 3T3-L1 adipocytes, inhibition of SIRT1 activity by Ex527 downregulated Klf10 mRNA level (Fig. 2i), while activation of SIRT1 activity by Srt1720 led to upregulation of KLF10 expression (Fig. 2j, k and Supplementary Fig. 2e). Consistently, activation of SIRT1 also inhibited FOXO1 acetylation in 3T3-L1 adipocytes (Fig. 2l and Supplementary Fig. 2f). Moreover, chromatin immunoprecipitation (ChIP) analysis in adipocytes demonstrated that the binding capacity of FOXO1 to the gene promoter of Klf10 was blunted by inhibition of SIRT1 activity, but was enhanced by activation of SIRT1 activity (Fig. 2m). Besides, knockdown of FOXO1 in adipocytes significantly impaired the role of SIRT1 activation in promoting the gene expression of Klf10 (Fig. 2n, o). Collectively, these data indicate that exercise could induce KLF10 expression in WAT via the SIRT1/FOXO1 axis.
a Schematic representation of Klf10 proximal promoter constructs used for luciferase assays. Predicted consensus of FOXO1-binding site is shown in the wild type (WT) luciferase construct. The red letters indicate mutations of the FOXO1-binding site in the FOXO1-Mut construct. b, c Luciferase activities were measured in HEK293T cells. Data were normalized to the vector group (n = 5 independent biological replicates). d–h Chow diet-fed 8-week-old male mice were treated as in Fig. 1a before being sacrificed for analysis. d, e FOXO1 enrichment on promoter of the indicated gene in iWAT and eWAT, respectively (n = 6 male mice/group). f FOXO1 acetylation was examined in iWAT and eWAT. Representative western blotting was shown. g, h SIRT1 activity in iWAT and eWAT, respectively (n = 6 mice/group). i Differentiated 3T3-L1 adipocytes were treated with or without SIRT1 antagonist Ex527 for 24 h (50 μM). Then Klf10 mRNA in the cells were determined (n = 6 independent biological replicates). j–l Differentiated 3T3-L1 adipocytes were treated with or without SIRT1 agonist Srt1720 for 24 h (2 μM). Then Klf10 mRNA levels, KLF10 protein levels and FOXO1 acetylation levels in the cells were determined, respectively (n = 6 independent biological replicates). m Differentiated 3T3-L1 adipocytes were treated with or without Ex527 or Srt1720 for 24 h. Then FOXO1 enrichment on promoter of the indicated gene in the cells were determined (n = 6 independent biological replicates). n Differentiated 3T3-L1 adipocytes were transfected with the siRNA against FOXO1, with siNC as the control. After 36 h, the Foxo1 mRNA levels in the cells were determined (n = 6 independent biological replicates). o Differentiated 3T3-L1 adipocytes with or without the knockdown of FOXO1 were cultured in the presence or absence of Srt1720 for 24 h. Then Klf10 mRNA in the cells were determined (n = 6 independent biological replicates). For statistical analysis, one-way ANOVA plus Tukey’s post hoc tests were performed in b, n; two-way ANOVA plus Tukey’s post hoc tests were performed in c–e, m, o; unpaired two-tailed Student’s t tests were performed in g–j. All data show the means ± SD. Source data are provided as a Source Data file.
Adipocyte-specific knockout of KLF10 (KLF10AKO) impairs exercise-induced browning and lipid catabolism in male mice
In order to understand the potential role of adipose KLF10 in exercise-mediated metabolic adaptation, KLF10AKO mice were constructed by crossbreeding KLF10flox/flox mice, which served as wild-type (WT) controls, with adiponectin promoter-driven Cre transgenic mice (Adiponectin-Cre+), which were fed by normal chow diet (NCD) and subjected to 8 weeks-treadmill training (Ex) or not (Sed) as shown in Fig. 3a. Adipocyte-specific knockout of KLF10 was confirmed (Supplementary Fig. 3a–c). In WT groups, exercise training significantly decreased the adipocyte size in adipose tissue (WT/Ex vs WT/Sed as shown in Fig. 3b). WT/Ex mice had stronger expression of Ucp1 protein in iWAT and BAT than WT/Sed mice (Fig. 3c and Supplementary Fig. 3d, e). Consistently, exercise promoted the expression of browning-associated genes (Cidea, Prdm16, Ucp1, Ppargc1a) and fatty acid oxidation (FAO)-related genes (Ppara, Cpt1b, Ehhadh) in iWAT (Fig. 3d). The results above indicate that exercise-induced the adipose browning process in mice. Lipolysis, which is usually activated during adipose browning, was also examined. Under cold exposure, WT/Ex mice exhibited higher levels of serum free fatty acid (FFA) and glycerol (Fig. 3e, f), which suggests enhanced lipolysis capacity by exercise. Adipose triglyceride lipase (ATGL) and hormone-sensitive lipase (HSL) are the two key enzymes for lipolysis24. In accordance with the above result, both the mRNA and protein expression levels of ATGL and HSL in both iWAT and eWAT were significantly elevated by exercise (Fig. 3g–i and Supplementary Fig. 3f, g). Nevertheless, comparing KLF10AKO/Sed mice with WT/Sed mice, KLF10AKO resulted in larger size of adipocytes and more severe lipid accumulation in adipose tissue (Fig. 3b), impaired browning processes (Fig. 3c, d and Supplementary Fig. 3d, e), and decreased lipolytic capacity (Fig. 3e-i and Supplementary Fig. 3f, g). Moreover, under KLF10AKO condition, the effect of exercise on promoting adipose browning and lipolysis capacity in mice was compromised (Fig. 3b–i and Supplementary Fig. 3–g). Therefore, adipose KLF10 could play a positive role in exercise-induced browning and lipid catabolism processes.
Chow diet (CD) fed KLF10AKO and wild type (WT) control male mice were subjected to treadmill training (Ex) or kept sedentary lifestyle (Sed) for 8 weeks before being sacrificed for analysis. a The treadmill exercise program is illustrated. The graph model was created in BioRender. Zhu, J. (2025) https://BioRender.com/z12f220. b Representative images of hematoxylin and eosin (H&E) staining of adipose tissues. Scale bars, 100 μm. Quantification of adipocyte diameter in iWAT and eWAT was shown below. c Immunohistochemistry (IHC) staining of UCP1 in iWAT and BAT. Scale bars, 50 μm. d The mRNA levels of the indicated genes in iWAT. Data were normalized to the WT/Sed group. e, f After 7 weeks of exercise training, the serum free fatty acid (FFA) and glycerol levels of CD-fed mice under 22 °C or 4 °C condition for 12 h. g The mRNA levels of the indicated genes in iWAT and eWAT. Data were normalized to the WT/Sed group. h, i Representative western blotting of iWAT and eWAT lysates. β-tubulin serves as an internal control. For statistical analysis, two-way ANOVA plus Tukey’s post hoc tests were performed in b, d, g; three-way ANOVA plus Tukey’s post hoc tests were performed in e, f. n = 6 male mice per group in all the above experiments. All data show the means ± SD. Source data are provided as a Source Data file.
The browning and lipid catabolism phenotypes of the KLF10flox/flox mice, Adiponectin-Cre+ mice and KLF10AKO mice were also examined. Under cold exposure, KLF10flox/flox mice and Adiponectin-Cre+ mice exhibited comparable levels of serum FFA and glycerol (Supplementary Fig. 3h, i), and similar levels of Pnpla2, Lipe, Ucp1 and Cpt1b mRNA expression in iWAT (Supplementary Fig. 3j), and similar Pnpla2, Lipe and Cpt1b mRNA expression in eWAT (Supplementary Fig. 3k). Compared with the KLF10flox/flox mice and Adiponectin-Cre+ mice, KLF10AKO mice had significantly lower levels of serum FFA and glycerol (Supplementary Fig. 3h, i) under cold exposure, lower mRNA expression levels Pnpla2, Lipe, Ucp1 and Cpt1b in iWAT (Supplementary Fig. 3j), and lower mRNA expression levels Pnpla2, Lipe and Cpt1b in eWAT (Supplementary Fig. 3k). These data indicate that KLF10flox/flox mice display similar phenotypes to the Adiponectin-Cre+ mice and can be used as the control in our experiments.
KLF10AKO attenuates the effects of exercise on resisting diet-induced obesity (DIO) and metabolic disorders in male mice
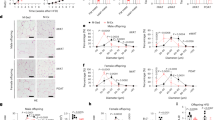
Then, we further explore whether adipocyte KLF10 takes part in exercise-mediated amelioration of DIO and related metabolic disorders. Control WT and KLF10AKO male mice were treated with high-fat-diet (HFD) for a total of 12 weeks and subjected to the established training protocol in the last 10 weeks of HFD feeding (Supplementary Fig. 4a). Incremental-load exhaustive exercise testing on a treadmill was performed to test the exercise capacity of the mice (Supplementary Fig. 4b). No difference in the treadmill exhaustion time was observed between the KLF10AKO mice and WT mice under HFD feeding (Supplementary Fig. 4c). In WT mice, 10 weeks-training considerably restrained the body weight gain, lowered fat mass, adipose tissue weights, and decreased adipocyte size (Fig. 4a–f and Supplementary Fig. 4d, e), as well as decreased liver weight and hepatic triglyceride (TG) deposit (Supplementary Fig. 4f, g). Compared with WT/Sed mice, WT/Ex mice had higher rectal temperature under cold exposure condition (Fig. 4g). Meanwhile, treadmill exercise led to lower levels of fasting blood glucose and plasma insulin, which contributed to the reduction of HOMA-IR (Fig. 4h–j). In accordance, WT/Ex mice presented higher glucose tolerance and better insulin sensitivity than those of the WT/Sed mice (Fig. 4k, l and Supplementary Fig. 4h, i). Moreover, the iWAT isolated from WT/Ex mice exhibited higher oxygen consumption rate (OCR) than that of the WT/Sedmice (Fig. 4m). By metabolic chamber measurement analysis, it was found that WT/Ex mice had stronger O2 consumption, CO2 emission and heat production than WT/Sed mice (Fig. 4n, o and Supplementary Fig. 4j), without difference in basal physical activity and food intake levels (Supplementary Fig. 4k, l). In addition, exercise training enhanced the expression of browning-related genes and FAO-related genes in WATs (Fig. 4p and Supplementary Fig. 4m), and Dio2 and Ucp1 expression in BAT (Supplementary Fig. 4n). Compared with WT/Sed mice, KLF10AKO/Sed mice displayed more severe DIO (Fig. 4a–f and Supplementary Fig. 4d, e), worse cold tolerance, glucose tolerance and insulin insensitivity (Fig. 4g–l), less energy expenditure rates (Fig. 4m–o and Supplementary Fig. 4j), and less thermogenic genes expression in WATs and BAT (Fig. 4p and Supplementary Fig. 4m, n). Importantly, unlike the case in WT mice, exercise did not effectively resist DIO and related metabolic disorders in KLF10AKO mice (Fig. 4a–p and Supplementary Fig. 4d–n). These results demonstrate an important role of adipose KLF10 in exercise-mediated metabolic benefits for the amelioration of DIO phenotypes in male mice.
The 12-week high-fat diet (HFD)-fed KLF10AKO and WT control male mice were subjected to treadmill training (Ex) or kept sedentary lifestyle (Sed) in the last 10 weeks of HFD feeding before being sacrificed for analysis. a–e Body weight, body weight gain, body composition and adipose tissue weight of the mice, respectively. f Representative images of hematoxylin and eosin (H&E) staining of adipose tissues. Scale bars, 100 μm. g Cold tolerance test was performed at the 10th week of HFD feeding. After 10 h of fasting, mice were subjected to cold exposure (4 °C) in the fasted state for 5 h, and the rectal temperatures of the mice were measured. h–j Fasting blood glucose, fasting plasma insulin and HOMA-IR of the mice, respectively. k The glucose tolerance test (GTT) were performed in mice after 11 weeks of HFD feeding. l Analysis of the GTT data in m, with subtraction of the basal glucose to generate an area of the curve (AOC). m The oxygen consumption rate of iWAT of mice were measured. n, o Regression-based analysis of absolute VO2 and VCO2 against body weight of mice, respectively. p The mRNA levels of indicated genes in iWAT of mice. Data normalized to the WT/Sed group. For statistical analysis, two-way ANOVA plus Tukey’s post hoc tests were performed in a–e, g–j, l, m, p; two-sided analysis of covariance (ANCOVA) was performed in n, o. n = 6 male mice per group in all the above experiments. All data show the means ± SD. Source data are provided as a Source Data file.
KLF10 in adipocytes facilitates lipolysis
To gain the mechanistic insight into the role KLF10 in adipose metabolism, RNA sequencing (RNA-seq) was conducted to investigate the differentially expressed genes (DEGs) in iWAT collected from HFD-fed WT and KLF10AKO mice. DEGs that were prominently upregulated and downregulated in the samples of KLF10AKO mice compared to control mice were listed (Fig. 5a). Principal component analysis (PCA) illustrated a clear global shift in KLF10AKO iWAT (Supplementary Fig. 5a). The Gene ontology (GO) analysis showed that downregulated DEGs were enriched in multiple biological processes, including lipid catabolic process, regulation of lipid metabolic process, and lipase activity (Supplementary Fig. 5b). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis indicated that the regulation of lipolysis in adipocytes pathway was enriched in downregulated DEGs (Fig. 5b, c), suggesting that adipose KLF10 may play an important role in regulating lipolysis. Then, the role of adipose KLF10 in lipolysis and exercise-mediated increase of lipolysis capacity was investigated. In WT mice, treadmill training notably raised the levels of serum glycerol and serum FFA in mice which were subjected to injection of β3-adrenergic receptor agonist (CL316,243) (Supplementary Fig. 5c, d). In the fat explant experiment, the release of FFA and glycerol were also higher in iWAT and eWAT isolated from WT/Ex mice than those from WT/Sed mice (Fig. 5d, e and Supplementary Fig. 5e, f). Moreover, mRNA and protein expression of ATGL (encoded by Pnpla2) and HSL (encoded by Lipe) in iWAT and eWAT, the two key lipolytic enzymes, were significantly upregulated after exercise (Fig. 5f, g and Supplementary Fig. 5g-i). However, when adipose KLF10 was knocked out in mice, lipolysis was impaired (KLF10AKO/Sed vs WT/Sed), and exercise could not effectively promote the lipolysis capacity of adipose tissues (KLF10AKO/Ex vs KLF10AKO/Sed) (Fig. 5d–g and Supplementary Fig. 5c–i). To confirm the role of KLF10 in lipolysis, SVFs from iWAT of KLF10flox/flox male mice were obtained and differentiated into mature adipocytes, followed by exposure to adenovirus harboring Cre (AD-Cre) to knock out KLF10 or to adenovirus harboring β-galactosidase gene z (AD-LacZ) as the control. By comparison with control adipocytes, primary adipocytes with KLF10 deficiency exhibited lower levels of FFA and glycerol release in response to isoproterenol (ISO) stimulation (Fig. 5h, i). Also, the expression of ATGL and HSL in primary adipocytes was inhibited upon KLF10 knockout (Fig. 5j, k and Supplementary Fig. 5j). On the contrary, differentiated primary adipocytes exposed to adenovirus harboring KLF10 (AD-KLF10) to overexpress KLF10 exhibited stronger lipolysis capacity compared to the AD-LacZ group (Fig. 5l–n). These results indicate the important role of adipose KLF10 in facilitating lipolysis and exercise-enhanced lipolysis capacity. Importantly, knockdown of both ATGL and HSL blunted the role of KLF10 overexpression in promoting Ucp1 and Cpt1b genes expression (Supplementary Fig. 5k). Free fatty acids (FFAs) are the products of lipolysis, which can act as PPARα ligands to activate it. Then PPARα can increase the expression of browning genes and FAO genes in adipocytes45. As indicated, increased expression of Ucp1 and Cpt1b was evidenced in KLF10 overexpressed primary adipocytes, such effect was weakened by a PPARα antagonist GW6471 (Fig. 5o). In addition, neither KLF10 deficiency nor overexpression in adipocytes affected the expression of general adipogenesis marker genes, including Cebpβ, Fabp4 and Pparγ (Supplementary Fig. 5l, m), suggesting that KLF10 is not a general regulator for adipogenesis. These results above indicate that exercise-induced adipose KLF10 promotes ATGL and HSL expression to facilitate lipolysis, which upregulates browning-associated and FAO-related genes expression at least partly through PPARα activation, thereby fueling adipose catabolism.
a Heatmaps representing 2907 upregulated and 1639 downregulated genes (KLF10AKO vs WT) in the iWAT of HFD-fed mice without exercise which was obtained from the RNA sequencing (RNA-seq) data (Padj < 0.05). b Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of downregulated genes based on RNA-seq data in a (Padj < 0.05). c The downregulated genes in lipolysis pathways based on the RNA-seq data (Padj < 0.05). d, e Mice were treated as described in Fig. 4. FFA release from iWAT and eWAT of mice under basal or CL316,243 (2 μM) stimulated conditions. f The mRNA levels of lipolysis-related genes in iWAT and eWAT of mice. g Representative western blotting of iWAT lysates. h–k Differentiated primary adipocytes containing KLF10flox/flox loci in the genome were infected with adenoviruses (AD) harboring LacZ (AD-LacZ) or Cre (AD-Cre). h, i FFA and glycerol release from the adipocytes under basal or isoproterenol (ISO, 2 μM) stimulated conditions. j The mRNA levels of indicated genes in primary adipocytes. k Representative western blotting of primary adipocytes lysates. l–n Differentiated primary adipocytes were infected with adenoviruses (AD) harboring LacZ (AD-LacZ) or KLF10 (AD-KLF10). l, m FFA and glycerol release from the adipocytes under basal or isoproterenol (ISO, 2 μM) stimulated conditions. n The mRNA levels of indicated genes in differentiated primary adipocytes. Data normalized to the AD-LacZ group. o The mRNA levels of indicated genes in differentiated primary adipocytes infected with AD-LacZ or AD-KLF10, which were then treated with GW6471 (10 μM) or vehicle for 24 h. Data normalized to the AD-LacZ+Vehicle group. n = 4 mice per group in a, c, n = 6 mice per group in d–f, n = 6 independent biological replicates in h–j, l–o. For statistical analysis, two-sided Wald tests plus Benjamini-Hochberg procedure were performed in a–c, three-way ANOVA plus Tukey’s post hoc tests were performed in d, e; two-way ANOVA plus Tukey’s post hoc tests were performed in f, h, i, l, m, o; unpaired two-tailed Student’s t tests were performed in n; two-tailed Wilcoxon test was performed in j. All data show the means ± SD. Source data are provided as a Source Data file.
KLF10AKO impairs lipolysis to exacerbate DIO and metabolic disorders in male mice
In order to confirm whether impaired lipolysis is critical for KLF10AKO-mediated phenotypes, adeno-associated viral (AAV) vectors harboring the genes encoding ATGL (AAV-ATGL), HSL (AAV-HSL) or GFP (AAV-GFP, as the control) were applied locally to overexpress the indicated genes in iWAT and eWAT of WT and KLF10AKO male mice which were fed with HFD for 12 weeks. The overexpression of ATGL and HSL was detected in both iWAT and eWAT, but not in other tissues, after the injection of AAV-ATGL and AAV-HSL (Supplementary Fig. 6a, b). In the AAV-GFP group of mice, KLF10AKO led to weaker cold tolerance (Fig. 6a), more body weight gain (Fig. 6b, c) and adiposity (Fig. 6d, e), less lipolysis capacity (Fig. 6f, g and Supplementary Fig. 6c–f), less thermogenic and FAO genes expression in fat tissues (Supplementary Fig. 6g, h), and worse glucose tolerance and insulin sensitivity (Fig. 6h–l). However, as shown in the AAV-ATGL/HSL group of mice, adipose overexpression of ATGL and HSL enhanced the lipolysis capacity as compared to the AAV-GFP group of mice (Fig. 6f, g and Supplementary Fig. 6c–f), and blunted the above phenotypes caused by KLF10AKO (Fig. 6a–l and Supplementary Fig. 6c–h). These data demonstrated that impaired lipolysis is critical for KLF10AKO-mediated exacerbation of DIO and metabolic disorders.
KLF10AKO and WT control male mice were subjected to injection of adeno-associated viral (AAV) vectors harboring the genes encoding ATGL (AAV-ATGL), HSL(AAV-HSL) or GFP (AAV-GFP, as the control) in iWATs and eWATs. One week later, mice were fed with high-fat diet (HFD) for 12 weeks before being sacrificed. a Cold tolerance test was performed at the 10th week of HFD feeding. After 10 h of fasting, mice were subjected to cold exposure (4 °C) in the fasted state for 5 h, and the rectal temperatures of the mice were measured. b–d, Body weight, body weight gain and adipose tissue weight of the mice, respectively. e Representative images of hematoxylin and eosin (H&E) staining of mice adipose tissues. Scale bars, 100 μm. Quantification of adipocyte diameter in iWAT and eWAT was shown below. f, g The serum FFA and glycerol levels of 10-week HFD-fed mice under basal or Cl316,243 stimulated condition (injection with 1 mg/kg body weight). h GTT was performed in mice after 11 weeks of HFD feeding. i Analysis of the GTT data in h, with subtraction of the basal glucose to generate an area of the curve (AOC). j ITT was performed in mice after 11 weeks of HFD feeding. k Analysis of the GTT data in j, with subtraction of the basal glucose to generate an area of the curve (AOC). l HOMA-IR of mice after 12 weeks of HFD feeding. For statistical analysis, two-way ANOVA plus Tukey’s post hoc tests were performed in a–e, i, l; three-way ANOVA plus Tukey’s post hoc tests were performed in f, g; Kruskal–Wallis test with Dunn’s correction was performed in k. n = 6 male mice per group in all the above experiments. All data show the means ± SD. Source data are provided as a Source Data file.
KLF10 forms a complex with FOXO1 and KDM4A to promote the transactivation of ATGL and HSL
Next, the underlying mechanism of how KLF10 facilitates lipolysis was investigated. FOXO1 has been shown to promote lipolysis25 and a potential FOXO1 binding site was located on the promoter of Pnpla2 gene (Fig. 7a). As expected, FOXO1 enhanced the luciferase activity driven by Pnpla2 gene promoter, and mutation of FOXO1 binding site (FOXO1 Mut) impaired the above effect mediated by FOXO1 (Fig. 7b). Interestingly, KLF10 also promoted the luciferase activity and it can cooperate with FOXO1 to further enhance the transactivation of Pnpla2 promoter (Fig. 7b). However, when FOXO1 binding site was mutated, the ability of KLF10 to promote the luciferase activity and the synergistic effect of KLF10 and FOXO1 on activating Pnpla2 gene promoter was compromised (Fig. 7b). Consistently, ChIP data indicated that KLF10 was able to bind to the gene promoter of Pnpla2 in adipocytes, but the binding was attenuated when FOXO1 was knocked down (Fig. 7c). Similar results were obtained when the above experiments were performed on Lipe gene promoter (Supplementary Fig. 7a–e). Co-immunoprecipitation (Co-IP) assay showed that KLF10 and FOXO1 interacted with each other both at the overexpression level and the endogenous level (Fig. 7d, e). In consistence with the above results, re-chromatin immunoprecipitation (Re-ChIP) assay indicated that KLF10 and FOXO1 simultaneously bound on the same region of Pnpla2 and Lipe gene promoters (Fig. 7f and Supplementary Fig. 7f). Therefore, KLF10 could act as a co-factor of FOXO1 to transactivate ATGL and HSL.
a Schematic representation of Pnpla2 proximal promoter constructs used for luciferase assays. The predicted consensus of FOXO1-binding site is shown in the WT luciferase construct. The red letters indicate mutations of the FOXO1-binding site in the FOXO1-Mut construct. b Luciferase activities were measured in HEK293T cells (n = 5 independent biological replicates). Data were normalized to the Vector group. c Differentiated C3H10T1/2 adipocytes were transfected with the indicated siRNAs. Then KLF10 enrichment on the promoter of Pnpla2 gene was determined. Data were normalized to the corresponding IgG control group. d, e Representative immunoblot of exogenous co-immunoprecipitation (Co-IP) in HEK293T cells and endogenous Co-IP in C3H10T1/2 adipocytes. WCL, whole cell lysates. f Enrichment of KLF10-interacting FOXO1 on promoter of the indicated gene by using the Re-ChIP method. g Enrichment of H3K9me3, H3K4me3, H3K27me3, H3K36me3 on promoter of the indicated gene in KLF10 knockout mice primary adipocytes. h C3H10T1/2 adipocytes were transfected with the indicated siRNAs and the mRNA level of Pnpla2 in the cells were examined. Data were normalized to the siNC group. i H3K9me3 enrichment on promoter of the indicated gene in C3H10T1/2 adipocytes. j Representative immunoblot of endogenous Co-IP in differentiated mice primary adipocytes with KLF10 knockout (AD-Cre) or overexpression (AD-KLF10). k, l Quantification of immunoblot results in j. Data were normalized to the AD-LacZ/Anti-FOXO1 group. m, n Differentiated primary adipocytes were infected with adenoviruses (AD) harboring the indicated genes. Then FOXO1 and KDM4A enrichment on promoter of the indicated gene in the cells was examined. n = 6 independent biological replicates in c–n. For statistical analysis, two-way ANOVA plus Tukey’s post hoc tests were performed in b, c, g, i, k–n; two-tailed Wilcoxon test was performed in f; one-way ANOVA plus Tukey’s post hoc tests were performed in h. All data show the means ± SD. Source data are provided as a Source Data file.
It is acknowledged that modifications of histone molecules within chromatin influences genome structure and function profoundly. Methylation of histone lysine residues is one of key modulations, which regulates the transcription of target genes46,47. Therefore, we tested some of the methylation levels on histones, including H3K9me3, H3K4me3, H3K27me3 and H3K36me3. A notable increase of H3K9me3 modification was found on Pnpla2 and Lipe gene promoters in KLF10 knockout adipocytes (Fig. 7g and Supplementary Fig. 7g). Among the histone lysine demethylases commonly recognized as regulators of H3K9me3 methylation, histone lysine demethylase 4 A (KDM4A) was found to affect the gene expression levels of Pnpla2 and Lipe, which were downregulated by the knockdown of KDM4A in adipocytes (Fig. 7h and Supplementary Fig. 7h-k). Also, higher level of H3K9me3 methylation was evidenced on Pnpla2 and Lipe gene promoters in KDM4A knockdown adipocytes (Fig. 7i and Supplementary Fig. 7l). To have a better understanding of the potential relationship among KDM4A, FOXO1, and KLF10, Co-IP was applied in primary adipocytes to show that FOXO1 could interact with both KLF10 and KDM4A whereas the interaction between FOXO1 and KDM4A was significantly inhibited when KLF10 was knocked out (Fig. 7j, k and Supplementary Fig. 7m). KLF10 overexpression promoted the above interaction in differentiated primary adipocytes (Fig. 7j, l and Supplementary Fig. 7n). Moreover, KLF10 ablation caused a considerable reduction of KDM4A binding, but not FOXO1 binding, to the promoter region of Pnpla2 and Lipe genes (Fig. 7m and Supplementary Fig. 7o), while KLF10 overexpression increased KDM4A occupation on the indicated regions in adipocytes (Fig. 7n and Supplementary Fig. 7p). Taken together, these results demonstrate that KLF10 functions as an intermediate factor to enhance FOXO1-induced transactivation of ATGL and HSL by means of recruiting KDM4A to demethylate H3K9me3 on the gene promoters of Pnpla2 and Lipe.
Adipose-specific KLF10 transgenic (KLF10ATG) promotes exercise-mediated alleviation of DIO and metabolic dysfunction in male mice
Then, the gain-of-function experiments were performed in mice by utilizing adiponectin promoter-triggered Cre expression to specifically drive the expression of KLF10 in adipocytes of fat tissues (KLF10ATG). KLF10 expression was greatly increased in the mature adipocytes of the KLF10ATG mice fat tissues (Supplementary Fig. 8a). The 8-week-old KLF10ATG and control WT mice were fed with HFD for 12 weeks and were subjected to treadmill training or sedentary lifestyle in the last 10 weeks of HFD feeding (Supplementary Fig. 8b). Similar exercise capacity was observed between KLF10ATG mice and WT mice under HFD feeding (Supplementary Fig. 8c). In comparison with WT/Sed group, treadmill training (WT/Ex) or adipocyte overexpression of KLF10 (KLF10ATG/Sed) inhibited HFD-induced body weight gain (Fig. 8a, b), adiposity (Fig. 8c–h), increased cold tolerance (Fig. 8i), improved glucose tolerance and insulin sensitivity (Fig. 8j–l and Supplementary Fig. 8d–g), promoted basal energy expenditure rates (Fig. 8m-n and Supplementary Fig. 8h) without affecting food intake and basal level of physical activity (Supplementary Fig. 8i, j), enhanced lipolytic capacity (Fig. 8o–r and Supplementary Fig. 8k–o), promoted thermogenic genes expression in fat tissues (Fig. 8s and Supplementary Fig. 8p, q). It is worth of mentioning that the combination of exercise and KLF10ATG yielded better effects on improving the above parameters (Fig. 8a–s and Supplementary Fig. 8d–q). Unlike exercise, which alleviated HFD-induced hepatosteatosis, KLF10ATG had little effect on the improvement of HFD-induced fatty liver (Supplementary Fig. 8r, s). Collectively, these data indicate that adipose KLF10ATG and exercise could synergistically protect mice from DIO and metabolic dysfunction, in which enhanced lipolysis was involved.
The 12-week high-fat diet (HFD)-fed KLF10ATG and WT control male mice were subjected to treadmill training or kept sedentary lifestyle in the last 10 weeks of HFD feeding before being sacrificed for analysis. a–e Body weight, body weight gain, body composition and adipose tissue weight of the mice. f Representative images of hematoxylin and eosin (H&E) staining of adipose tissues. Scale bars, 100 μm. g, h Quantification of adipocyte diameter in iWAT and eWAT based on the results of f. i Cold tolerance test was performed at the 10th week of HFD feeding. After 10 h of fasting, mice were subjected to cold exposure (4 °C) in the fasted state for 5 h, and the rectal temperatures of the mice were measured. j GTT were performed in mice after 11 weeks of HFD feeding. k Analysis of the GTT data in j, with subtraction of the basal glucose to generate an area of the curve (AOC). l HOMA-IR of the mice. m, n Regression-based analysis of absolute VO2 and VCO2 against body weight of mice. o FFA release in iWAT isolated from mice under basal or CL316,243 (2 μM) stimulated conditions. p The serum FFA level of HFD-fed mice under basal or Cl316,243 stimulated condition (injection with 1 mg/kg body weight). q The mRNA levels of indicate genes in iWAT and eWAT of mice. Data normalized to the WT/Sed group. r Representative western blotting of iWAT lysates. s The mRNA levels of indicate genes in iWAT of mice. Data normalized to the WT/Sed group. For statistical analysis, two-way ANOVA plus Tukey’s post hoc tests were performed in a–e, g–i, k, l, q, s; two-sided analysis of covariance (ANCOVA) was performed in m, n; three-way ANOVA plus Tukey’s post hoc tests were performed in o, p. n = 6 male mice per group in all the above experiments. All data show the means ± SD. Source data are provided as a Source Data file.
Discussion
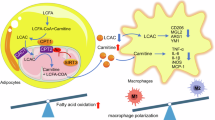
In this study, we found that exercise could induce the adipose KLF10 expression via SIRT1/FOXO1 pathway. KLF10AKO blunts exercise-mediated enhancement of adipose tissue browning and the amelioration of DIO and metabolic disorders. On the contrary, KLF10ATG and exercise reinforced the effects on facilitating adipose browning, combating DIO and metabolic dysfunction in a synergistic way. Mechanistically, KLF10 interacted with FOXO1 and recruited KDM4A to form a ternary complex, which drives exercise-activated transcription of key lipolytic genes encoding ATGL and HSL to fuel energy expenditure. Therefore, adipose KLF10 functions as an exercise effector to combat obesity and improve metabolic health (Fig.9).
KLF10 in mature adipocytes is upregulated by SIRT1/FOXO1 axis in response to exercise and then interacts with FOXO1 to recruit histone lysine demethylase 4A (KDM4A) and form a ternary complex, thereby strengthening the transcription of key lipolytic genes (Pnpla2 and Lipe) to promote lipolysis. Free fatty acids (FFAs), the products of lipolysis, could act as a ligand of PPARα to activate it. And then PPARα increases the expression of fatty acid oxidation (FAO) genes (such as Cpt1b) and browning genes (such as Ucp1) to fuel adipose catabolism. The above FOXO1-KLF10 reinforcing loop contributes to exercise-mediated anti-obesity and metabolic improvement effects in mice. The graph model was created in BioRender. Zhu, J. (2023) BioRender.com/e08n146.
Exercise is known as a preventive and therapeutic strategy for obesity and its derived metabolic syndrome. However, our understanding of the mechanisms underlying exercise-induced molecular alternation in adipose tissue remains limited. As a transcription factor, KLF10 is a member of the SP (specificity proteins)/KLF (Krüppel-like Factors) family and its role in adipose tissue is not well defined. In our study, we found that exercise triggers upregulation of KLF10 expression in white adipose tissue (Supplementary Fig. 1a-i). Although KLF10 from SVF takes a large portion of total KLF10 expression in whole adipose tissue (Supplementary Fig. 1j), exercise specifically increased the KLF10 expression level in mature adipocytes (Fig. 1), suggesting that KLF10 in mature adipocytes might respond to exercise intervention. Our results indicated that exercise-induced FOXO1 deacetylation (Fig. 2f), enhanced SIRT1 enzyme activity (Fig. 2g, h) and promoted Klf10 expression (Fig. 1b, c, e, f) in white adipose tissue. SIRT1 is known to be activated under catabolic conditions such as exercise. Then, we used SIRT1 agonist (Srt1720) to treat adipocytes in vitro. In consistence with the in vivo studies, Srt1720 led to FOXO1 deacetylation in adipocytes (Fig. 2l) and promoted KLF10 expression (Fig. 2j, k). Srt1720 enhanced, but Ex527 (a SIRT1 antagonist) inhibited, the binding capacity of FOXO1 to Klf10 gene promoter (Fig. 2m). Furthermore, knockdown of FOXO1 in adipocytes blunted the role of Srt1720 in promoting Klf10 expression (Fig. 2n, o). These data above suggest a SIRT1/FOXO1 pathway that facilitates the induction of adipose KLF10 expression by exercise.
Mounting evidence indicates that the impairment of adipose tissue lipolysis contributes to several lipid-related metabolic disorders. Mice with adipocyte-specific overexpression of G0S2, an inhibitor of ATGL, exhibited adiposity and defective cold adaptation due to compromised lipid catabolism48. Conversely, depletion of G0S2 in mice facilitated lipolysis and was conducive to the prevention of obesity, insulin resistance, and cold intolerance49. Increased phosphorylated HSL led by PH domain and leucine rich repeat protein phosphatase 2 (PHLPP2) knockout in HFD-fed mice caused higher levels of lipolysis, which greatly ameliorated insulin resistance and increased oxygen consumption50. Overexpression of diacylglycerol kinase type II isozyme (DGKδ) in mice enhances adipose lipolysis, resulting in protection against high fat diet-induced obesity and energy homeostasis51. In accordance with previous works, our experiments show that impaired lipolysis is the main driver of exacerbated DIO and metabolic disorders in mice with adipose KLF10 knockout (Fig. 6 and Supplementary Fig. 6). Furthermore, our data demonstrate an important role of adipose KLF10 in facilitating exercise-induced adipose browning, enhanced energy expenditure, metabolic improvement and anti-obesity effect in male mice (Fig. 9). Besides, increased utilization of free fatty acid (FFA) may explain the unchanged liver weight and TG accumulation in liver of KLF10ATG mice (Supplementary Fig. 8r, s). Collectively, our results support the hypothesis that KLF10 responds to exercise intervention to accelerate lipid catabolism, which subsequently fuels exercise-mediated metabolic remodeling to address the challenge of overnutrition.
KLF10AKO inhibited ATGL and HSL expression and impaired adipose lipolysis (Fig. 6f, g and Supplementary Fig. 6c–f), led to less thermogenic and fatty acid oxidation genes expression in fat tissues (Supplementary Fig. 6g-h), and exacerbated DIO and metabolic disorders (Fig. 6a–e, h–l). However, adipose overexpression of ATGL and HSL abolished KLF10AKO-mediated inhibition of adipose lipolysis (Fig. 6f, g), which is accompanied by the results that the inhibition of adipose thermogenic and fatty acid oxidation (FAO) genes expression (Supplementary Fig. 6g-h) and the exacerbation of DIO and metabolic disorders caused by KLF10AKO was blunted (Fig. 6a–e, h–l). These data suggest that impaired lipolysis plays an important role in KLF10AKO-mediated phenotypes. Furthermore, knockdown of ATGL and HSL in adipocytes blunted the role of KLF10 overexpression in promoting the expression of FAO genes (such as Cpt1b) and browning genes (such as Ucp1) in adipocytes (Supplementary Fig. 5k). FFAs are the products of lipolysis, which can act as PPARα ligands to activate it. Then PPARα increases the expression of FAO genes (such as Cpt1b) and browning genes (such as Ucp1) in adipocytes to improve metabolism. As shown in Fig. 5o, overexpression of KLF10 in adipocytes increased the expression of Cpt1b and Ucp1, and this effect was blunted by inhibiting PPARα activity using PPARα antagonist (GW6471). It should be noted that neither knockdown or overexpression of KLF10 in adipocytes had effects on the expression of general adipogenesis marker genes (Supplementary Fig. 5l, m). Therefore, our data suggest that KLF10 can transactivate ATGL and HSL expression to enhance adipose lipolysis, which would promote FAO and browning genes expression via PPARα in adipocytes. The above process would fuel the catabolism in adipocytes, thereby facilitating exercise-mediated anti-obesity effects (Fig. 9).
FOXO1, as a transcriptional factor, has been proven to play an important role in promoting lipolysis. Cytoplasmic retention of FOXO1, which resulted from its phosphorylation by protein kinase B (PKB) and CDK1, leads to suppression of lipolysis52,53. In contrast, nutritionally starved adipocytes increase FOXO1 nuclear translocation and promote its ability to transactivate the expression of lipolysis genes25,54,55. In our study, the role of KLF10 in FOXO1-induced lipolysis was demonstrated. KDM4A, which is a histone demethylase for removing H3K9me3, was found to interact with both KLF10 and FOXO1 to drive expression of lipolysis genes, and such effect relied on the KLF10-mediated recruitment of KDM4A to FOXO1, thereby forming a ternary complex on the promoters of lipolysis genes. Besides the direct binding effect, transcription factor can also act as co-regulator to regulate the transcription of target genes. SOX4 is an important transcription factor involved in maintaining stem cell characteristics and differentiation. In beige adipocytes, SOX4 facilitates transcription of the thermogenic gene Ucp1 by strengthening the binding of PR domain-containing 16 (PRDM16), zinc-finger domain-containing transcription factor and an important regulator driving adipose browning profile, to peroxisome proliferator-activated receptors (PPARγ)56. For another example, PRDM16 was revealed to suppress left ventricle (LV)-specific dilation and dysfunction via binding with transcription factor Tbx5 or Hand1 to activate the transcription of compact gene in LV57. Here, we unraveled a non-canonical function of KLF10 in transcriptional regulation by acting as a coactivator of FOXO1 and recruiting KDM4A to form a ternary complex, thereby reinforcing the transactivation of lipolysis genes. The adipose KLF10/FOXO1/KDM4A axis-regulated lipolytic program could be an important molecular mechanism underlying the effect of exercise on melioration of obesity and its related metabolic dysfunction.
Exercise has been advocated as a pivotal therapeutic resource for both treatment and prevention of overweight33,58. Exercise training is devoted to accelerating total energy expenditure thereby resulting in a negative energy balance, which is related to increased weight loss, decreased cardiovascular risk and alleviated cardiometabolic risk factors59,60. A randomized controlled trial in obese men indicated that 12-week-exercise intervention led to 7.3% of body weight loss partly owing to decreases in abdominal subcutaneous, visceral, and visceral fat-to-subcutaneous fat ratios, together with reduction of insulin resistance and improved cardiovascular fitness61. A remarkable reduction of body weight (about 10%) and lower levels of fasting and OGTT insulin, total cholesterol, LDL cholesterol and apolipoprotein B of was also evidenced in premenopausal obese women subjected to exercise training for 16 weeks62. It has been pointed out that engaging in exercise, whose level exceeds the minimum physical activity (PA) recommendations (150 min of moderate or 75 min of vigorous physical activities per week), is more likely to cause clinically significant weight loss63. However, the fact that overweight individuals tend to live a physically inactive life makes them suffer the hardship to profit from exercise64. The discrepancy of effectiveness between clinical translation of exercise and exercise model of rodents might count on low patient compliance. From this point of view, a practical strategy to enhance the benefits of exercise or drop the difficulty of exercise execution is urgently needed. In this paper, we observed that adipocyte-specific overexpression of KLF10 promoted the effect of exercise on the protection of mice from DIO and metabolic disorders (Fig.8 and Supplementary Fig. 8). This may provide strategy for obese patients with metabolic syndrome to benefit from exercise training better.
KLF10 usually functions as a transcriptional repressor. However, in some cases, it can also transactivate genes expression. For example, KLF10 can enhance the transcription of Smad2, which facilitates the activation of TGFβ/SMAD signaling pathway in AKR-2B fibroblasts65. KLF10 induces the transcription of Foxp3 gene expression in regulatory T cells, thereby protecting against airway inflammation66. Our data show that KLF10 transactivates key lipolytic genes expression to promote adipose metabolic reprogramming, which facilitates exercise-mediated amelioration of DIO in male mice. These results above indicate complicated roles of KLF10 in regulating genes expression, which is involved in multiple biological processes.
Some limitations still remain in this study. Firstly, effect of moderate-intensity continuous exercise on adipose KLF10 induction has been examined, yet the effect of different exercise schemes on the expression level of adipose KLF10 awaits further exploration. Besides, because adipose tissue functions as an important resource of various secreted factors, the potential of adipose KLF10 in mediating exercise-induced metabolic regulation in other tissues needs further investigation. Moreover, the role of adipose KLF10 in exercise-induced metabolic remodeling in female mice is also worthy of further studies.
In summary, this study demonstrates that exercise induces adipocyte KLF10 expression via SIRT1/FOXO1 pathway. KLF10 in mature adipocytes contributes to exercise-mediated alleviation of obesity and its related metabolic disorder in male mice. Moreover, exercise and adipose overexpression of KLF10 can induce metabolic benefits in DIO mice in a synergistic manner, which builds upon KLF10-induced transactivation of ATGL and HSL through a FOXO1-KLF10-KDM4A complex. These results indicate that adipose KLF10 could act as a downstream effector responding to exercise for the prevention and treatment of obesity and metabolic disorders.
Methods
Animals
All animal experiments were approved by Shanghai University of Sport Animal Care and Use Committee (Ethics No. 102772023DW004). All animal experiments involved in this study followed the National Institute of Health guidelines on the care and use of animals. Adiponectin-Cre and KLF10flox/flox mice were purchased from Shanghai Model Organisms Center to generate KLF10AKO mice (KLF10flox/flox/ Adiponectin -Cre+) by crossbreeding. Mice containing the CMV promoter-driven but stop signal-suppressed 3*Flag-tagged murine KLF10 expression cassette (CMV-Stop-KLF10 mice) were purchased from Model Animal Research Center of Nanjing University and crossbreed with Adiponectin-Cre mice to generate adipocyte specific KLF10 transgenic (KLF10ATG) mice. KLF10AKO mice and their KLF10flox/flox/Adiponectin-Cre- litter mates, together with KLF10ATG mice and their wild type (WT) litter mates were included in this study. C57BL/6 J mice were fed with high-fat diet (HFD, 60 kcal% fat, PD6001, SYSE BIO) for 12 weeks to induce obesity. The chow diet (CD, 1010086, Xietong Shengwu, China), as the control diet, was applied. C57BL/6J mice were used for all the experiments and housed at 23 ± 2 °C with a humidity of 50 ± 5% in a 12 h light/dark cycle and fed ad libitum with standard mouse feed and water throughout the experiments. Time in hours is denoted by ZT because light is a zeitgeber. ZT0 = lights on (rest onset) in the entrained cycle and ZT12 = lights off (active onset). Male mice were used in all experiments in this paper.
Treadmill training and time to exhaustion
The 8-week-old mice were acclimated to adaptive treadmill training for 3 days followed by 8-week or 10-week regular treadmill training (6 days per week). The maximum running speed was measured every 2 weeks and 60% of maximum running speed was set as training speed to maintain moderate intensity. Exercise training was performed at the end of the resting phase of mice, which is from Zeitgeber time 10 (ZT10) to ZT11. During the treadmill training, mice were supervised to keep running on the treadmill (ZhengHuabiological, ZH-PT/5S, China), ensuring the identical level of exercise training. Mice were sacrificed after the last training and their tissues were harvested for analyses.
The time to exhaustion assay is described as follows. Before an exercise bout, mice were placed in the treadmill for acclimation. Adaptive training was conducted on the first day (starting speed at 10 m/min for 10 min followed by 15 m/min for 20 min). The treadmill test was conducted on the second day. The starting speed was 5 m/min for 3 min, and the speed was increased by 5 m/min every 3 min until the speed reached 28 m/min. The maximum speed of 28 m/min was continued, and the time to exhaustion was measured.
Application of adeno-associated viral (AAV) vectors
AAV application was conducted as previously described45. AAV-ATGL and AAV-HSL were purchased from Genechem. AAV-ATGL and AAV-HSL were used to induce overexpression of ATGL and HSL in inguinal WAT (iWAT) and epididymal WAT (eWAT). AAV-GFP was used as the control. Mice iWAT pads and eWAT pads on both sides were injected with the AAVs. Mice were anesthetized with ketamine (100 mg/kg) and xylazine (10 mg/kg). For AAV delivery in iWAT, longitudinal incisions were made on the skin around the inguinal areas, followed by exposition of the fat pad using tweezers. AAV was injected into 5–8 spots in each fat pad. The total volume was 50 μl, and the total virus titer was 1 × 1011 viral genomes (vg) for each pad (1 × 1011 vg for AAV-GFP, 0.5 × 1011 vg for AAV-ATGL, and 0.5 × 1011 vg for AAV-HSL). For injection of eWAT, mice were subjected to laparotomy and AAV was injected as described above. After injection, sterile saline solution was used to rinse the abdomen, which was closed with a two-layer suture.
Analyses of biochemistry-related parameters
For GTT, mice were fasted for 6 h, which was followed by an intraperitoneal (i.p.) injection of D-glucose (1.5 mg/g body weight) at ZT 10. For ITT, mice were fasted for 6 h and then treated with human insulin (0.8 mIU/g body weight) by an intraperitoneal (i.p.) injection at ZT 10. Blood samples were collected from mice tail veins at different time points after D-glucose or insulin injection. The levels of blood glucose were measured by using a glucometer (Roche). For GTT and ITT analysis, the area of the curve (AOC) was calculated following the conventional trapezoid rule.
For cold tolerance test, mice were fasted for 10 h and placed in a temperature-controlled chamber at 4 °C. Mice were kept fasted during the test and their rectal temperature was measured by a rectal probe.
For liver TG content measurement, liver tissue was collected and follow the manufacturers’ instructions of kits from Applygen, China (#E1003).
Isolation of stromal vascular cells and adipocytes of adipose tissue
For isolation of fraction in adipose tissue, iWAT and eWAT from mice were collected following the methods described in previously50. The inguinal and epididymal adipose tissues were harvested from mice and then minced into small pieces to be digested by type I collagenase (Solarbio, 9001-12-1) for 1 h at 37 °C with gentle agitation. After filtration through the cell strainer (100 μm), the samples were subjected to centrifugation at 500 G for 5 min. The precipitation was regarded as SVF and the top layer was collected as the adipocyte fraction.
Cell culture and differentiation
For primary adipocyte culture, KLF10flox/flox male mice aged 3–4 weeks were euthanized and their SVFs from iWAT was harvested following the methods described in previously67. Isolated preadipocytes from iWAT were maintained in Dulbecco’s Modified Eagle Medium (DMEM/F12 containing 10% fetal bovine serum, FBS, Gibco). C3H10T1/2 cells and 3T3-L1 cells were cultured in DMEM (Gibco) containing 10% calf serum (Gibco). For preadipocyte differentiation, the culture medium of the 2-day post-confluent cells were replaced by DMEM (C3H10T1/2 cells and 3T3-L1 cells) or DMEM/F12 (SVFs) containing 10% FBS, 1 μM dexamethasone, 2 μg/mL insulin, 0.5 mM 3-isobutyl-1-methyl-xanthine and 1 μM rosiglitazone for 48 h. Then the cells were maintained in DMEM (C3H10T1/2 cells and 3T3-L1 cells) or DMEM/F12 (SVFs) containing 10% FBS, 2 μg/mL insulin, and 1 μM rosiglitazone for another 48 h followed by cultured in DMEM (C3H10T1/2 cells and 3T3-L1 cells) or DMEM/F12 (SVFs) containing 10% FBS for 2 days to finish differentiation process. HEK293T cells were cultured in DMEM containing 10% FBS.
Recombinant adenoviruses and RNA interference
The recombinant adenoviruses and RNA interference were performed as described previously68. Recombinant adenoviruses (AD) for LacZ and KLF10 overexpression or knockout in mouse differentiated adipocytes were generated using Vira Power Adenoviral Expression System (Invitrogen). For adenovirus infection in mouse differentiated adipocytes, cells were cultured in fresh DMEM after 24 h of viral infection. Cells were collected for further detection after 48 h of infection. The small interfering RNAs (siRNAs) were designed and synthesized by Gene Pharma. The nonspecific siRNA control (siNC) was used as a negative control. After 48 h of transfection, the cells were harvested to further detection. The transfection was conducted by using RNAiMAX (#13778-100, Invitrogen) following the manufacturer’s instruction. The nonspecific siRNA control (siNC) was used as a negative control. The target sequences for successful siRNAs (5′ to 3′) were as follows: siFoxo1-1: CCGCCAAACACCAGTCTAAAT, siFoxo1-2: TGGAAACCAGCCAGCTATAAA; siPnpla2: CATCTCCCTGACTCGTGTTTC; siLipe: CGCAGAAGACAATATGGCCTT; siKdm4a-1: CGAACATCCTACGACGATATT; siKdm4a-2: AGCACGTTGATGAGTGGAATA; siKdm4b-1: GCCCATGTGTACTTGGAAATT, siKdm4b-2: GCGCAGACAAATTAGTGTGAA; siKdm4c-1: GCAAAGTATCTTGGATCAAAT, siKdm4c-2: GCAGAGTGATAGATGTGACAT; siNC, TTCTCCGAACGTGTCACGT. The siFoxo1-1/siKdm4a-1/siKdm4b-1/siKdm4c-1 and siFoxo1-2/siKdm4a-2/ siKdm4b-2/ siKdm4c-2 target two different sites of the mouse gene of Foxo1/Kdm4a/Kdm4b/ Kdm4c, respectively.
SIRT activity measurement
The 8-week-old male mice were subjected to 8-week-treadmill exercise training. Fresh tissues were collected after the above exercise program and lysed in cold PBS. SIRT1 activity was examined using a SIRT1 Activity Assay Kit (Abcam, ab156065) according to the manufacturer’s instruction. SIRT6 activity was examined using a SIRT6 Assay Kit (BPSbioscience, 50022) according to the manufacturer’s instruction.
Measurement of oxygen consumption rate (OCR)
High-resolution respirometry (Oxygraph-2k, Oroboros, Innsbruck, Austria) was applied to detect oxygen consumption rate of adipose tissue. 40-60 mg of freshly isolated iWAT were minced into small pieces and suspended in DMEM containing 2% bovine serum albumin (BSA, Sigma) to measure the oxygen consumption rate, which was normalized to tissue weight.
Lipolysis assay
For ex vivo assays, 60–80 mg iWAT or eWAT were acquired and minced into small pieces, which were subsequently incubated in DMEM containing 2% BSA with or without β3-adrenergic agonist CL316,243 (2 μM, MCE) at 37 °C for 2 h. For cellular lipolysis assay, differentiated primary adipocytes treated by AD-LacZ, AD-Cre or AD-KLF10 and 3T3-L1 cells transfected by siNC or siKLF10 were maintained in culture medium containing 2 μM isoproterenol (ISO, MCE) or not at 37 °C for 2 h. For in vivo assays, mice were subjected to CL316,243 i.p. injection (1 mg/kg body weight) and tail vein blood was collected at 0, 20, 60 min after injection. The content of FFA and glycerol derived from culture medium or blood was examined using a liquid sample glycerol content assay kit (Applygen, E1002) and FFA detection kit (EK-bioscience, Ek-M21104), respectively. The tissue samples from iWAT were normalized to tissue weight. The cell samples were normalized to quantity of total protein levels.
Metabolic rate measurement
Indirect calorimetry (TSE-system, XYZ, 6 M/R, Germany Instruments) was utilized to monitor the concentrations of oxygen together with carbon dioxide. The oxygen consumption and carbon dioxide production were calculated by the inlet and outlet of the chambers. Food intake and physical activity were detected in the system above as well.
Chromatin immunoprecipitation (ChIP) and Re-ChIP
The implementation of ChIP followed the instruction as described previously42. Cells were cross-linked by using 1% formaldehyde for 10 min at ambient temperature before incubated with glycine for termination (125 mM). After washed twice with ice-cold PBS and centrifugation, the cells were resuspended and incubated in lysis buffer (50 mM Tris-HCl, pH 8.0, 1% SDS, 10 mM EDTA, and protease inhibitors) for 10 min. Samples were sonicated and centrifuged at 12,000 × g at 4 °C for 5 min. The supernatants were 10-fold- diluted by ChIP dilution buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, supplemented with complete protease inhibitor tablets). Next, the samples were equally divided. 10% of each sample was kept for input control. ChIP Grade Protein A/G magnetic beads (Thermo Fisher, 26162) was applied to preclear the samples for 1 h at 4 °C. The samples were immunoprecipitated with the indicated antibodies of anti-KLF10 (Santa Cruz, Cat: sc-130408, Lot: H2808), anti-FOXO1(CST, Cat: 2880 s, Lot: 3), anti-H3K9me3 (Abclonal, Cat: A22295, 3522081672), anti-H3K4me3 (Abcam, Cat: Ab8580, Lot: GR65362), H3K27me3 (Abcolonal, Cat: A22006, Lot: 3522101713), H3K36me3 (Abclonal, Cat: A20379, Lot:3561722513), KDM4A (Abclonal, Cat: A22060, Lot: 3522072734), and control IgG (Abcam, ab46540). The immune complexes were washed with washing buffer. Immunoprecipitated samples were eluted and reverse cross-linked by incubation in elution buffer at 65 °C for overnight (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS). Genomic DNA was then extracted with a PCR purification kit (Qiagen). For Re-ChIP assay, immune complexes were eluted from the first IP by incubation with 10 mM DTT at 37 °C for 30 min. Eluents were diluted 1:50 in dilution buffer and then subjected to Re-IP with the secondary antibodies. Purified DNA was applied to qPCR using primers specific to the promoters of the indicated genes. The primers for ChIP-qPCR are listed in Supplementary information. The gels for all the ChIP-PCR results are shown in Supplementary Fig. 9
Luciferase reporter analysis
Luciferase reporter assays was conducted as previously described42. The proximal promoter regions of mouse KLF10, Pnpla2, Lipe, as well as mutants of KLF10, Pnpla2, Lipe promoters were subcloned into the firefly luciferase reporter construct PGL3-basic (Promega). Firefly luciferase reporter constructs, renilla luciferase reporter plasmids, KLF10 plasmids, FOXO1 plasmids and pcDNA3.1(-) vector were transfected into HEK293T cells by utilizing Lipo6000 (#C0526, Beyotime). 48 hours after transfection, luciferase activity was measured using dual luciferase reporter kit (#RG027, Beyotime and CLARIO star by BMG LABTECH). Firefly luciferase activity was normalized to renilla activity.
Immunoprecipitation (IP) and Co-immunoprecipitation (Co-IP)
The immunoprecipitation assay was conducted as described previously69. The fat pads were homogenized in RIPA lysis buffer (Beyotime, P0013B) supplemented with 1 mM PMSF,1 mM Na3VO4, and 1 mM NaF for 1 h at 4 °C. The differentiated adipocytes were washed with cold PBS and then subjected to RIPA lysis buffer. Then the samples were centrifugated at 15000 g for 3 times, 10 min for each time. After centrifugation, the supernatant was incubated with Pan- acetylation (Proteintech, 66289-1-Ig), anti-FOXO1 (Cell Signaling, Cat: 14952 T, Lot: 3) anti-KLF10 (Santa Cruz, Cat: sc-130408, Lot: H2808) anti- DYKDDDDK (Proteintech, 66008-4-Ig), anti-HA tag (Proteintech, 66006-2-Ig) and control IgG (Abcam, ab46540) bound to the Protein A/G magnetic beads (Thermo Fisher, 26162) at 4 °C overnight. Next, the beads were washed with RIPA lysis buffer supplemeted with 1 mM PMSF, 1 mM Na3VO4, and1 mM NaF for 3 times. Proteins were separated using SDS-PAGE and transferred to PVDF membrane.
Western blot assay
Western blotting was performed as described previously for the cell and tissue samples70. The proteins were separated by SDS-PAGE gel (Absin, abs9367) to implement the western blotting by using antibodies to KLF10 (Abclonal, Cat: A2433, Lot: 4000001921), FOXO1 (Santa Cruz, Cat: sc-374427, Lot: E2623), ATGL (CST, Cat: 2138 s, Lot: 6), HSL (Proteintech, Cat: 17333-1-AP, Lot: 00103113), p-ATGL (Abcam, Cat: ab135093, Lot: GR3359112-1), p-HSL (CST, Cat: 4126 s, Lot: 9), KLF3 (Santa Cruz, Cat: sc-514500, Lot: F0524), KLF15 (Santa Cruz, Cat: sc-271675, Lot: E1124), KLF9 (Abclonal, Cat: A7196, Lot: 3560603011), KLF14 (Abclonal, Cat: A18607, Lot: 3717041101), β-tubulin (Abmart, Cat: M20005F, Lot: 334585), HSP90α/β (Santa Cruz, Cat: sc-13119, Lot: Jo722), peroxidase affiniPure goat anti-mouse IgG secondary antibody (Jackon, Cat: 111-035-003, Lot: 151083), peroxidase affiniPure goat anti-rabbit IgG secondary antibody (Jackon, Cat: 111-035-003, Lot: 153526). Tanon-5200S (BIO-RAD) and Image J was applied to develop and quantify the western blotting, and the values of target proteins were normalized to that of the internal control protein on the same membrane.
Hematoxylin-Eosin (H&E) staining and immunohistochemistry (IHC)
Mouse tissues were fixed with 4.0% paraformaldehyde overnight and then embedded in paraffin. H&E and IHC were performed as described previously4. For H&E staining and IHC staining, tissues were immediately fixed with 4.0% paraformaldehyde at 4 °C overnight, then the formaldehyde-fixed tissues were embedded in paraffin, sectioned, and stained with H&E, or with the UCP1 antibody (Abclonal, Cat: A5857, Lot: 5500009352) following standard procedures. The images of H&E staining and IHC were observed under the Olympus microscope.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Trizol reagent (Takara) was applied to extract total RNA from cells and tissues following the manufacturer’s instruction. Transcription kit (Beyotime, China) was utilized for the synthesis of complementary DNA (cDNA) from total RNA. Power SYBR green PCR master mix (Beyotime, China) was applied to detect the amplification of target genes. 18S rRNA was set as an endogenous control. The results of RT-qPCR were obtained from Quant Studio 6 Flex (Thermo Fisher). The RT-qPCR was done in triplicate and repeated at least three times. Primer information for RT-qPCR is listed in Supplementary information.
RNA-seq analysis
Total RNA was extracted from inguinal white adipose tissue of WT control mice and KLF10AKO mice fed by HFD. The transcriptome sequencing experiments and analysis were conducted by Novogene. Mus Musculus (GRCm38/mm10) is the reference genome and gene model annotation. Index of the reference genome was built using Hisat2 (v2.0.5) and paired-end clean reads were aligned to the reference genome. Pooled libraries were sequenced on an Illumina NovaSeq 6000 platform to a target depth of 20 million read pairs per sample using a paired-end 150 base pair run configuration. Differential expression analysis was performed using the edgeR R package. The resulting P values were adjusted using the Benjamini & Hochberg method. padj < =0.05 and |log2(foldchange)| >= 1 were set as the threshold for detection of expression level difference. KEGG and GO terms with corrected p-value less than 0.05 were regarded significantly enriched by differential expressed genes.
Statistical analyses
All data exhibit the means ± standard deviation (SD) of 4–6 biological replicates. The biological replicates (n) and statistical analyses were shown in the figure legends. p < 0.05 indicates a significantly statistical difference. The statistical analysis was performed by Graphpad prism (Version 9) and IBM SPSS Statistics 27. Data were subjected to normality tests (Shapiro-Wilk) and homogeneity tests (Bartlett’s test). The Student’s t test or ANOVA with Tukey post hoc tests were used for parametric test, and the Wilcoxon test or Kruskal-Wallis test with Dunn’s correction was used for nonparametric test.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw RNA-seq data generated in Fig. 5a–c and Supplementary Fig. 5a, b have been deposited in the GEO datasets under accession code GSE269020. The images in Figs. 3a, 9 and Supplementary Figs. 4a, 8b are created in BioRender. Zhu, J. (2023) BioRender.com/e08n146 & BioRender. Zhu, J. (2025) https://BioRender.com/z12f220 (agreement number: SJ27CIN962/JB280GQU4E). Source data are provided with this paper.
References
Xia, W. et al. Obesity causes mitochondrial fragmentation and dysfunction in white adipocytes due to RalA activation. Nat. Metab. 6, 273–289 (2024).
Kulaj, K. et al. Adipocyte-derived extracellular vesicles increase insulin secretion through transport of insulinotropic protein cargo. Nat. Commun. 14, 709 (2023).
Sakers, A., De Siqueira, M. K., Seale, P. & Villanueva, C. J. Adipose-tissue plasticity in health and disease. Cell 185, 419–446 (2022).
Li, B. Y., Guo, Y. Y., Xiao, G., Guo, L. & Tang, Q. Q. SERPINA3C ameliorates adipose tissue inflammation through the Cathepsin G/Integrin/AKT pathway. Mol. Metab. 61, 101500 (2022).
Lund, J. et al. The anorectic and thermogenic effects of pharmacological lactate in male mice are confounded by treatment osmolarity and co-administered counterions. Nat. Metab. 5, 677–698 (2023).
Jiang, J. et al. Thermogenic adipocyte-derived zinc promotes sympathetic innervation in male mice. Nat. Metab. 5, 481–494 (2023).
Suárez-Zamorano, N. et al. Microbiota depletion promotes browning of white adipose tissue and reduces obesity. Nat. Med. 21, 1497–1501 (2015).
Guo, Y. Y., Li, B. Y., Peng, W. Q., Guo, L. & Tang, Q. Q. Taurine-mediated browning of white adipose tissue is involved in its anti-obesity effect in mice. J. Biol. Chem. 294, 15014–15024 (2019).
Jin, L. et al. The muscle-enriched myokine Musclin impairs beige fat thermogenesis and systemic energy homeostasis via Tfr1/PKA signaling in male mice. Nat. Commun. 14, 4257 (2023).
Meng, Y. et al. Ginsenoside F1 administration promotes UCP1-dependent fat browning and ameliorates obesity-associated insulin resistance. Food Sci. Hum. Wellness 12, 2061–2072 (2023).
Hu, X. et al. Skeletal muscle-secreted DLPC orchestrates systemic energy homeostasis by enhancing adipose browning. Nat. Commun. 14, 7916 (2023).
Kim, J. S., Han, H. S., Seong, J. K., Ko, Y. G. & Koo, S. H. Involvement of a novel cAMP signaling mediator for beige adipogenesis. Metabolism 143, 155536 (2023).
Peng, W. Q. et al. l-Theanine activates the browning of white adipose tissue through the AMPK/α-ketoglutarate/Prdm16 axis and ameliorates diet-induced obesity in mice. Diabetes 70, 1458–1472 (2021).
Falk, S. et al. GLP-1 and nicotine combination therapy engages hypothalamic and mesolimbic pathways to reverse obesity. Cell Rep. 42, 112466 (2023).
Balapattabi, K. et al. Angiotensin AT(1A) receptor signal switching in Agouti-related peptide neurons mediates metabolic rate adaptation during obesity. Cell Rep. 42, 112935 (2023).
Parker, V. E. R. et al. Cotadutide promotes glycogenolysis in people with overweight or obesity diagnosed with type 2 diabetes. Nat. Metab. 5, 2086–2093 (2023).
Axelrod, C. L., Dantas, W. S. & Kirwan, J. P. Sarcopenic obesity: emerging mechanisms and therapeutic potential. Metabolism 146, 155639 (2023).
Angelidi, A. M. et al. Early metabolomic, lipid and lipoprotein changes in response to medical and surgical therapeutic approaches to obesity. Metabolism 138, 155346 (2023).
Zhu, J. Y., Chen, M., Mu, W. J., Luo, H. Y. & Guo, L. The functional role of Higd1a in mitochondrial homeostasis and in multiple disease processes. Genes Dis. 10, 1833–1845 (2023).
De Jong, K. A., Ehret, S., Heeren, J. & Nikolaev, V. O. Live-cell imaging identifies cAMP microdomains regulating β-adrenoceptor-subtype-specific lipolytic responses in human white adipocytes. Cell Rep. 42, 112433 (2023).
Sun, L. et al. Dynamic interplay between IL-1 and WNT pathways in regulating dermal adipocyte lineage cells during skin development and wound regeneration. Cell Rep. 42, 112647 (2023).
Haemmerle, G. et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science 312, 734–737 (2006).
Kuang, J. et al. Fat-Specific Sirt6 ablation sensitizes mice to high-fat diet-induced obesity and insulin resistance by inhibiting lipolysis. Diabetes 66, 1159–1171 (2017).
Grabner, G. F., Xie, H., Schweiger, M. & Zechner, R. Lipolysis: cellular mechanisms for lipid mobilization from fat stores. Nat. Metab. 3, 1445–1465 (2021).
Jung, S. M. et al. Non-canonical mTORC2 signaling regulates brown adipocyte lipid catabolism through SIRT6-FoxO1. Mol. Cell 75, 807–822.e808 (2019).
Malik, V. S. & Hu, F. B. The role of sugar-sweetened beverages in the global epidemics of obesity and chronic diseases. Nat. Rev. Endocrinol. 18, 205–218 (2022).
Furrer, R. et al. Molecular control of endurance training adaptation in male mouse skeletal muscle. Nat. Metab. 5, 2020–2035 (2023).
Cui, B. et al. Exercise alleviates neovascular age-related macular degeneration by inhibiting AIM2 inflammasome in myeloid cells. Metab.: Clin. Exp. 144, 155584 (2023).
Rosenbaum, M. & Foster, G. Differential mechanisms affecting weight loss and weight loss maintenance. Nat. Metab. 5, 1266–1274 (2023).
Stöggl, T. L. et al. Unraveling the mystery of isocaloric endurance training - Influence of exercise modality, biological sex, and physical fitness. Metabolism 144, 155582 (2023).
Berge, J. et al. Effect of aerobic exercise intensity on energy expenditure and weight loss in severe obesity-a randomized controlled trial. Obesity 29, 359–369 (2021).
van Baak, M. A. et al. Effect of different types of regular exercise on physical fitness in adults with overweight or obesity: Systematic review and meta-analyses. Obes. Rev. 22, e13239 (2021).
Zhu, J. Y., Chen, M., Mu, W. J., Luo, H. Y. & Guo, L. Higd1a facilitates exercise-mediated alleviation of fatty liver in diet-induced obese mice. Metab.: Clin. Exp. 134, 155241 (2022).
Kato, H. et al. Exercise training-enhanced lipolytic potency to catecholamine depends on the time of the day. Int. J. Mol. Sci. 21, 6920 (2020).
Petridou, A. et al. Increased triacylglycerol lipase activity in adipose tissue of lean and obese men during endurance exercise. J. Clin. Endocrinol. Metab. 102, 3945–3952 (2017).
Luo, H. Y., Zhu, J. Y., Chen, M., Mu, W. J. & Guo, L. Krüppel-like factor 10 (KLF10) as a critical signaling mediator: Versatile functions in physiological and pathophysiological processes. Genes Dis. 10, 915–930 (2023).
Liu, Y. et al. Krüppel-like factor 10 (KLF10) is transactivated by the transcription factor C/EBPβ and involved in early 3T3-L1 preadipocyte differentiation. J. Biol. Chem. 293, 14012–14021 (2018).
Guillaumond, F. et al. Kruppel-like factor KLF10 is a link between the circadian clock and metabolism in liver. Mol. Cell. Biol. 30, 3059–3070 (2010).
Luo, H. Y. et al. Hepatic Klf10-Fh1 axis promotes exercise-mediated amelioration of NASH in mice. Metab.: Clin. Exp. 155, 155916 (2024).
Yang, S. et al. KLF10 promotes nonalcoholic steatohepatitis progression through transcriptional activation of zDHHC7. EMBO Rep. 23, e54229 (2022).
Yuce, K. & Ozkan, A. I. The kruppel-like factor (KLF) family, diseases, and physiological events. Gene 895, 148027 (2024).
Chen, M. et al. Cdo1-Camkk2-AMPK axis confers the protective effects of exercise against NAFLD in mice. Nat. Commun. 14, 8391 (2023).
Xing, Y. Q. et al. The regulation of FOXO1 and its role in disease progression. Life Sci. 193, 124–131 (2018).
Cui, X. et al. Exogenous hydrogen sulfide alleviates hepatic endoplasmic reticulum stress via SIRT1/FoxO1/PCSK9 pathway in NAFLD. FASEB J. 37, e23027 (2023).
Guo, Y. Y. et al. Cdo1 promotes PPARγ-mediated adipose tissue lipolysis in male mice. Nat. Metab. 4, 1352–1368 (2022).
Klose, R. J. & Zhang, Y. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. cell Biol. 8, 307–318 (2007).
Bannister, A. J., Schneider, R. & Kouzarides, T. Histone methylation: dynamic or static? Cell 109, 801–806 (2002).
Heckmann, B. L. et al. Defective adipose lipolysis and altered global energy metabolism in mice with adipose overexpression of the lipolytic inhibitor G0/G1 switch gene 2 (G0S2). J. Biol. Chem. 289, 1905–1916 (2014).
El-Assaad, W. et al. Deletion of the gene encoding G0/G 1 switch protein 2 (G0s2) alleviates high-fat-diet-induced weight gain and insulin resistance, and promotes browning of white adipose tissue in mice. Diabetologia 58, 149–157 (2015).
Kim, K. et al. Adipocyte PHLPP2 inhibition prevents obesity-induced fatty liver. Nat. Commun. 12, 1822 (2021).
Jollet, M. et al. Diacylglycerol kinase delta overexpression improves glucose clearance and protects against the development of obesity. Metab. Clin. Exp. 158, 155939 (2024).
Homan, E. P. et al. Differential roles of FOXO transcription factors on insulin action in brown and white adipose tissue. J. Clin. Investig. 131, e143328 (2021).
Ow, J. R. et al. Remodeling of whole-body lipid metabolism and a diabetic-like phenotype caused by loss of CDK1 and hepatocyte division. eLife 9, e63835 (2020).
Lo, M. C. et al. Camptothecin activates SIRT1 to promote lipid catabolism through AMPK/FoxO1/ATGL pathway in C(2)C(12) myogenic cells. Arch. Pharm. Res. 42, 672–683 (2019).
Li, Y. et al. A global perspective on FOXO1 in lipid metabolism and lipid-related diseases. Prog. lipid Res. 66, 42–49 (2017).
Shen, H. et al. SOX4 promotes beige adipocyte-mediated adaptive thermogenesis by facilitating PRDM16-PPARγ complex. Theranostics 12, 7699–7716 (2022).
Wu, T. et al. PRDM16 is a compact myocardium-enriched transcription factor required to maintain compact myocardial cardiomyocyte identity in left ventricle. Circulation 145, 586–602 (2022).
Beals, J. W. et al. Dietary weight loss-induced improvements in metabolic function are enhanced by exercise in people with obesity and prediabetes. Nat. Metab. 5, 1221–1235 (2023).
Fiuza-Luces, C. et al. Exercise benefits in cardiovascular disease: beyond attenuation of traditional risk factors. Nat. Rev. Cardiol. 15, 731–743 (2018).
Tucker, W. J. et al. Exercise for primary and secondary prevention of cardiovascular disease: JACC focus seminar 1/4. J. Am. Coll. Cardiol. 80, 1091–1106 (2022).
Ross, R. et al. Reduction in obesity and related comorbid conditions after diet-induced weight loss or exercise-induced weight loss in men. A randomized, controlled trial. Ann. Intern. Med. 133, 92–103 (2000).
Janssen, I., Fortier, A., Hudson, R. & Ross, R. Effects of an energy-restrictive diet with or without exercise on abdominal fat, intermuscular fat, and metabolic risk factors in obese women. Diabetes Care 25, 431–438 (2002).
Swift, D. L. et al. The effects of exercise and physical activity on weight loss and maintenance. Prog. Cardiovas. Dis. 61, 206–213 (2018).
Gorostegi-Anduaga, I. et al. Effects of different aerobic exercise programmes with nutritional intervention in sedentary adults with overweight/obesity and hypertension: EXERDIET-HTA study. Eur. J. Prev. Cardiol. 25, 343–353 (2018).
Johnsen, S. A., Subramaniam, M., Katagiri, T., Janknecht, R. & Spelsberg, T. C. Transcriptional regulation of Smad2 is required for enhancement of TGFbeta/Smad signaling by TGFbeta inducible early gene. J. Cell. Biochem. 87, 233–241 (2002).
Venuprasad, K. et al. The E3 ubiquitin ligase Itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1. Nat. Immunol. 9, 245–253 (2008).
Zhang, S. Y. et al. Adipocyte-derived lysophosphatidylcholine activates adipocyte and adipose tissue macrophage nod-like receptor protein 3 inflammasomes mediating homocysteine-induced insulin resistance. EBioMedicine 31, 202–216 (2018).
Li, B. Y., Peng, W. Q., Liu, Y., Guo, L. & Tang, Q. Q. HIGD1A links SIRT1 activity to adipose browning by inhibiting the ROS/DNA damage pathway. Cell Rep. 42, 112731 (2023).
Sun, L. et al. Lactylation of METTL16 promotes cuproptosis via m(6)A-modification on FDX1 mRNA in gastric cancer. Nat. Commun. 14, 6523 (2023).
Sundaram, V. K. et al. Adipo-glial signaling mediates metabolic adaptation in peripheral nerve regeneration. Cell Metab. 35, 2136–2152.e2139 (2023).
Acknowledgements
This work was funded by the Program for Overseas High-level talents at Shanghai Institutions of Higher Learning (TP2022100 to L.G.), the National Natural Science Foundation of China (No. 32070751 and 31871435 to L.G), and was supported by Key Laboratory of Exercise and Health Sciences (Shanghai University of Sport), Ministry of Education.
Author information
Authors and Affiliations
Contributions
J.-Y.Z. was involved in study design, conducted the experiments, analyzed the data and drafted the paper. M.C., W.-J.M., H.-Y.L., Y.L., S.L., L.-J.Y., R.-Y.L., M.-T. Y., X.L., and H.-M.C. performed the experiments. L.G. conceived the idea, designed and supervised the study, obtained the funding and cowrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhu, JY., Chen, M., Mu, WJ. et al. Exercise-induced anti-obesity effects in male mice generated by a FOXO1-KLF10 reinforcing loop promoting adipose lipolysis. Nat Commun 16, 3111 (2025). https://doi.org/10.1038/s41467-025-58467-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-58467-1