Abstract
The adult kidney lacks the ability to generate new nephrons, placing individuals born with low nephron counts at greater risk for chronic kidney disease as they age. Limited nutrient availability hinders nephron formation; however, the key metabolic dependencies remain unclear. Here we show that S-adenosylmethionine (SAM) and cellular transmethylation status are crucial determinants of the kidney’s nephrogenic capacity. The RNA methyltransferase METTL3 serves as a SAM sensor and is essential for the fate determination of nephron progenitor cells (NPCs). Reducing transmethylation or inhibiting METTL3 blocks NPC differentiation and nephrogenesis, whereas enhancing transmethylation or increasing METTL3 activity facilitates an induced NPC population and increases nephron production. Additionally, we identify Lrpprc mRNA, encoding a mitochondrially enriched protein, as a key direct target of METTL3-mediated transmethylation. Accordingly, inhibiting LRPPRC negates the nephrogenic effects of SAM and METTL3. Our findings reveal a modifiable methionine-SAM-RNA transmethylation pathway that can be targeted to enhance nephron formation.
Similar content being viewed by others
Introduction
The generation of new nephrons (nephrogenesis) during kidney development stops during or shortly after gestation, meaning that adult kidneys are unable to produce new nephrons1. An average adult kidney contains approximately 900,000 nephrons; however, this figure can vary significantly, ranging from 200,000 to 2.5 million, for reasons not yet fully understood2,3. Factors contributing to lower nephron counts, particularly those below 500,000, include preterm birth and low birth weight4,5. In the United States, about 10% of infants are born prematurely6, while nearly 15% experience low birth weight globally, often due to malnutrition7. Consequently, this sizable affected group is at an increased risk of developing chronic kidney disease (CKD), hypertension, and cardiovascular issues in adulthood5,8,9,10. Conversely, some individuals possess nephron counts significantly above average, which presumably increases their lifelong resilience against CKD. This observation suggests the presence of beneficial—and potentially modifiable—interactions between environmental and genetic factors that promote nephrogenic potential in developing kidneys. Notably, it is well established that inadequate maternal nutrient availability adversely affects nephron numbers9,11. Therefore, investigating the distinct metabolic dependencies of nephron progenitor cells (NPCs) and elucidating how nutrient utilization influences gene expression to favor nephrogenesis may reveal critical pathways for enhancing nephron endowment.
RNA molecules undergo a wide array of reversible chemical modifications that significantly influence their properties, including stability, localization, translation efficiency, and splicing patterns12. This dynamic process, referred to as ‘epitranscriptomics,’ represents a novel layer of post-transcriptional gene expression regulation13. Among the various RNA modifications, N6-methyladenosine (m6A) stands out as one of the most prevalent in eukaryotic cells. It is regulated by a multi-protein complex known as the m6A writer, characterized by the METTL3–METTL14 heterodimer as its catalytic subunit. Recent studies from three independent research teams have collectively elucidated the structure and function of the METTL3–METTL14 heterodimer14,15,16. These investigations have shown that S-adenosylmethionine (SAM), the essential cofactor for methylation reactions, binds directly to METTL3. This binding is crucial, as METTL3 performs the key catalytic task of transferring a methyl group from SAM to the target mRNA. In this intricate mechanism, METTL14 plays a vital allosteric role by stabilizing the conformation of METTL3, which optimizes its binding to the mRNA substrate. Furthermore, METTL14 serves as a scaffold, effectively anchoring the mRNA during the methylation process17. Thus, inhibiting METTL3 amounts to inhibiting METTL14 and the m6A writer complex. Aberrant activation of the METTL3–m6A axis is observed in numerous diseases, including cancer and kidney disorders, where it enhances the translational efficiency of mRNAs with pathogenic potential18,19,20,21. Therapeutics targeting METTL3 have already traversed to human clinical testing22. However, the physiological relevance of this pathway remains inadequately defined, particularly regarding its normal functions during kidney development, which remains unexplored.
Reduced availability of methionine has been linked to the lower nephron counts seen with caloric restriction. The transition of NPCs from a self-renewing state to a differentiated state requires a finely tuned regulation of gene dosage and a metabolic shift from glycolysis to oxidative phosphorylation23,24,25. Our study bridges these two seemingly disparate areas by identifying METTL3 as a crucial molecular link between methionine utilization and genetic and metabolic processes that support kidney development. Using a variety of complementary approaches and nephrogenesis models, we demonstrate that the METTL3-m6A pathway plays a vital role in facilitating the differentiation of NPCs. Crucially, we establish that transmethylation represents a critical metabolic dependency for NPCs and that targeting the METTL3-dependent RNA transmethylation pathway—either nutritionally or pharmacologically—can promote nephron endowment in murine models. Furthermore, our findings suggest that the SAM-METTL3 axis influences nephrogenesis through the epitranscriptomic activation of Lrpprc, an mRNA that encodes a mitochondrial RNA-binding protein known to enhance mitochondrial function.
Results
The methionine–RNA methylation axis is activated with NPC differentiation
Our study aimed to investigate the role of the methionine-METTL3–m6A axis in the transition of NPCs from self-renewal to differentiation and its implications for nephrogenesis. We first established an in vitro model of mouse NPC fate transition by isolating self-renewing NPCs (positive for CITED1 and SIX2) from embryonic day (E) 16.5 mouse kidneys. These cells were cultured in NPEM media with either low (1.25 µM CHIR) or high (3 µM CHIR) β-catenin activity to maintain self-renewal or induce differentiation, respectively26. Using targeted metabolomics and ELISA techniques in this primary NPC culture model, we observed that the levels of methionine and SAM increased by more than 2-fold during NPC differentiation (Fig. 1A–C and Supplementary dataset 1). Correspondingly, immunoblot analysis showed that the expression of MAT2A, the enzyme responsible for converting methionine to SAM, was significantly elevated in differentiated NPCs compared to their self-renewing counterparts (Fig. 1D). We also observed upregulation of METTL3 and its interacting partner METTL14 in differentiated compared to self-renewing NPCs (Fig. 1D). Notably, the expression levels of MAT2A, METTL3, and METTL14 increased within 12 h following treatment with 3 µM CHIR. This rise in expression coincided with the downregulation of CITED1 and SIX2 but preceded the induction of PAX8 or LHX1 (Fig. 1D). Consistent with the immunoblot results, immunostaining further revealed that METTL3 expression is elevated in differentiated NPCs compared to renewing NPCs and that its expression is increased within 12-h after the treatment with 3 µM CHIR, prior to PAX8 induction (Supplementary Fig. 1). Furthermore, using orthogonal ELISA and RNA dot blot assays, we observed that total mRNA m6A levels were significantly higher in differentiated NPCs than in self-renewing cells, indicating a positive correlation between enhanced METTL3 expression, higher m6A levels, and NPC differentiation (Fig. 1E, F). Thus, the activation of the SAM–-METTL3 axis is an early event in the differentiation of cultured NPCs.
A Principal component analysis of the metabolomics data, showing segregation of renewing (rNPCs) and differentiated NPCs (dNPCs) samples (N = 4). B LC/MS-measured methionine and C ELISA-assessed SAM levels in dNPCs compared to rNPCs. D Immunoblot analysis shows the expression of indicated proteins in rNPCs compared to dNPCs. Actin serves as the loading control. Each lane represents pooled lysate from 3 to 4 biological replicate primary NPC cultures. E ELISA (N = 5; biological replicates) and F RNA dot blot analysis showing m6A enrichment in dNPCs compared to rNPCs (N = 3). Methylene blue (MB) serves as a loading control. G–I Images showing MAT2A, METTL3, and m6A immunostaining and DBA labeling in kidney sections from E16.5 embryos (N = 4). Anti-MAT2A, anti-METTL3, and anti-m6A (red); DBA (green); DAPI (blue). Scale bars 25 μm (G–I). Error bars represent the standard error of the mean (SEM). Statistical analysis: two-tailed, unpaired t-test (B, C, and E). Source data are provided as a Source Data file. N indicates biological replicates.
To corroborate our findings from primary NPC cultures to in vivo settings, we examined mouse and human in vivo NPC niches. Immunofluorescence staining, utilizing anti-MAT2A, anti-METTL3, anti-m6A, and anti-METTL14 antibodies, revealed significantly elevated levels of MAT2A, METTL3, m6A, and METTL14 signals in pre-tubular aggregates compared to self-renewing NPCs in developing E16.5 mouse kidneys (Fig. 1G–I and Supplementary Fig. 2A). Additionally, analysis of a single-cell RNA sequencing (scRNA-seq) atlas of a 17-week-old human fetal kidney27 further supported our observations, as it indicated a pronounced enrichment of METTL3, METTL14, WTAP, and MAT2A in tubular precursors and differentiation-committed NPCs (Supplementary Fig. 2B and C).
METTL3 senses the NPC transmethylation state to induce differentiation
The activation of the methionine–METTL3–m6A axis may simply represent an epiphenomenon of NPC differentiation, or it could play a crucial role in this process. To explore a potential causative relationship, we first examined the effects of SAM supplementation on primary NPCs. We found that a concentration of 100 µM SAM effectively countered the renewal effects conferred by 1.25 µM CHIR, driving renewing NPCs toward differentiation (Fig. 2A, B). In contrast, we observed that 100 µM S-adenosylhomocysteine (SAH), the immediate byproduct of methylation reactions, did not promote differentiation (Fig. 2A, B). These findings suggest that enhancing the transmethylation state via SAM supplementation is sufficient to drive the differentiation of cultured NPCs, whereas other functional aspects of the methionine cycle do not appear to play a significant role.
A Immunofluorescence and B immunoblots showing SIX2 and PAX8 expression in NPCs cultured for 48 h in 1.25 μM CHIR NPEM media supplemented with vehicle, 100 μM SAM, 100 μM SAH, or 100 μM SAM and 40 μM METTL3 inhibitor. Actin serves as the loading control. Each lane represents pooled lysate from three biological replicate NPC cultures. C Images showing GFP and PAX8 immunostaining in NPCs treated with different doses of SAM or M3A (METTL3 activator). The average percentages (mean ± SD) of NPCs that are double-positive for GFP and PAX8 are presented in relevant images (N = 3). Source data are provided as a Source Data file.
To further investigate the relationship between the transmethylation capacity and differentiation of NPCs, we isolated self-renewing NPCs from the kidneys of Six2:eGFPTGC/+ transgenic mice at E16.5. This model enabled us to monitor GFP levels as a means of tracking SIX2 expression during the transition of NPCs from self-renewal to differentiation. In alignment with our findings from wildtype NPC cultures, treatment with 1.25 µM CHIR resulted in over 90% of cells maintaining GFP expression, with a tiny fraction (~9%) also expressing PAX8. In contrast, the addition of 100 µM SAM to the 1.25 µM CHIR media induced PAX8 expression with only about 20% of the PAX-positive cells retaining GFP expression (Fig. 2C). Interestingly, when we examined lower concentrations of SAM (25 M and 50 µM), we observed an enhancement in the population of GFP-PAX8 double-positive intermediates. Specifically, 50–70% of the NPCs expressed PAX8 while still retaining GFP expression at these concentrations (Fig. 2C).
Our findings raise an intriguing question about how the SAM-induced transmethylation potential is sensed in NPCs. We hypothesized that METTL3, which exhibits increased activity during NPC differentiation, may act as a sensor for SAM and mediate its pro-differentiation effects. In support of this hypothesis, immunofluorescence analyses indicated that SAM, in a concentration-dependent manner, elevates METTL3 expression in primary NPCs (Supplementary Fig. 3A). To further explore the potential cause–effect relationship between METTL3 and NPC differentiation, we employed STM2457—a small molecule that selectively binds to the SAM-binding pocket of METTL3 with over 1000-fold specificity, thereby preventing SAM from entering and inhibiting METTL322. Treatment with STM2457 (hereafter referred to as M3i) counteracted the pro-differentiation effects of 100 µM SAM (Fig. 2A, B) and 3 µM CHIR (Supplementary Fig. 3B). In both scenarios, cells treated with M3i continued to express SIX2, suggesting that they retained their identity as renewing NPCs (Fig. 2A, B and Supplementary Fig. 3). Conversely, we sought to determine whether the activation of METTL3 could independently mimic the effects of SAM and be sufficient for promoting NPC differentiation. To test this, we utilized methyl piperazine-2-carboxylate (M3A), a small molecule that selectively activates METTL3 by enhancing its affinity for SAM28. Treatment with M3A at a concentration of 100 µM effectively induced the expression of PAX8 and negated the renewal effects conferred by 1.25 µM CHIR (Fig. 2C), resulting in only about 5% of PAX8-positive cells maintaining SIX2-GFP expression. Furthermore, at lower concentrations, M3A exhibited dose-dependent effects akin to those observed with SAM, with ~35–50% of the cells showing dual positivity for SIX2-GFP and PAX8 expression (Fig. 2C). Thus, these observations suggest that SAM and METTL3 are required for the differentiation of cultured NPCs and that differentiation commitment is titrated based on the NPC RNA transmethylation potential. Furthermore, these findings suggest that fine-tuning the SAM–METTL3 axis allows for the emergence of an induced NPC population.
Inhibiting the Methionine–METTL3 axis blocks nephrogenesis
A key next question is whether our observations of the necessity of SAM–METTL3 for NPC fate transition in an in vitro model are relevant to organ-level nephrogenesis. To rigorously investigate the methionine-SAM–METTL3 axis in embryonic kidneys, we employed a multifaceted approach that included ex vivo kidney cultures, mouse models, and both genetic and pharmacological inhibition. In our first set of experiments, we employed the ex vivo kidney culture model, which allowed us to manipulate the methionine cycle reliably. We cultured E12.5 wildtype kidneys in regular (30 mg/L methionine) or methionine-depleted organ culture media for 48 h. We observed that the methionine-depleted kidneys were markedly smaller, lacking differentiated PAX8+ nephron precursors, as well as exhibiting fewer ureteric branches (Fig. 3A). However, immunofluorescence and RNA in-situ hybridization analyses revealed that these kidneys still contained viable renewing NPCs as evidenced by SIX2-positive immunofluorescence, and Cited1 and Tafa5 positive in-situ signal in NPC niches (Supplementary Fig. 4). Thus, these observations suggest that the block in nephrogenesis was not due to a loss of self-renewing NPCs but rather due to their inability to differentiate under low methionine conditions. We next cultured E12.5 kidneys under low methionine conditions for 24 h, followed by a 24-h supplementation with either SAM, SAH, or vehicle. Remarkably, SAM completely restored nephrogenesis and ureteric branching, resulting in kidneys that appeared indistinguishable from those grown in normal media (Fig. 3B). In stark contrast, SAH supplementation failed to rescue nephrogenesis and exhibited effects similar to those seen in the low-methionine condition. These findings suggest that rather than serving solely as an amino acid for protein synthesis, methionine plays a crucial role as a methyl donor, essential for driving NPC differentiation and nephrogenesis.
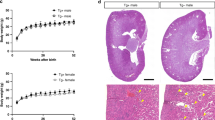
A Immunostaining for PAX8, SIX2, and E-cadherin in E12.5 kidneys cultured in regular or methionine-depleted media supplemented with vehicle, 250 μM SAM, or 100 μM SAH. The number (average ± SD) of PAX8-positive tubules and SIX2-positive NPC niches is depicted in the relevant images. B H&E-stained, SIX2, Pan-CK, and PAX8 immunostained, and DBA-labeled kidney sections from P1 mice of the indicated genotypes. White dotted circles indicate PAX8+ tubules. The graphs depict the number of glomeruli and pre-tubular structures. C PAX8, SIX2, and E-cadherin immunostaining in E12.5 Six2:eGFPTGC/+ (control) and Six2:eGFPTGC/+; Mett3F/F (Six2-Mettl3-KO) kidneys cultured ex vivo for 48 h. Six2-Mettl3-KO kidneys were grown in regular media (vehicle) or media supplemented with 250 μM SAM. Control (wildtype) kidneys were grown in regular media (vehicle) or supplemented with M3i or M3i plus SAM. The number (average ± SD) of PAX8 tubules and SIX2 NPC niches is depicted in relevant images. ** Indicates P < 0.05. Statistical analysis: one-way ANOVA with Tukey’s multiple-comparisons test (A, C); two-tailed, unpaired t-test (B). Error bars represent SEM. Source data are provided as a Source Data file.
In our second set of experiments, we determined whether the in vivo deletion of Mettl3 in NPCs reproduces the effects of methionine restriction. We generated mice lacking Mettl3 expression in SIX2-positive NPCs by intercrossing Six2:eGFPTGC/+ transgenic mice with Mett3F/F mice to produce Six2:eGFPTGC/+; Mett3F/F (Six2Cre-Mettl3-KO) mice29. Immunostaining confirmed the absence of METTL3 protein in the renewing NPCs of the Six2Cre-Mettl3-KO mice (Supplementary Fig. 5). We observed that at postnatal day (P)1, Six2Cre-Mettl3-KO mice had significantly smaller kidneys compared to age-matched littermate control mice (Six2Cre or Mett3F/F). Histological examination at P1, further demonstrated a striking reduction in nephron count, with approximately 75% fewer glomeruli observed in Six2Cre-Mettl3-KO kidneys compared to Six2Cre or Mett3F/F control kidneys (Fig. 3B). Additionally, in accordance with the temporal expression profile of the Six2:eGFPTGC/+ transgene, we found that the Six2Cre-Mettl3-KO mice initially displayed developmentally appropriate kidneys at E12.5 but experienced a decrease in nephrogenesis, evidenced by a deficit in PAX8+ tubules between E14.5 and E16.5 (Supplementary Fig. 6A and B).
The Six2:eGFPTGC/+ transgene effectively removes METTL3 not only from NPCs but also their derivatives, including the renal vesicles. Given our earlier characterization indicated the presence of METTL3 protein in renal vesicles, we aimed to investigate whether the observed nephron deficit could be attributed to the loss of METTL3 specifically in these structures. To explore this hypothesis, we employed the Wnt4Cre transgene, which selectively ablates Mettl3 from the newly differentiated PAX8+ renal vesicles while maintaining METTL3 expression in the renewing NPCs. Despite the efficient deletion of Mettl3 from the renal vesicles and later-stage kidney structures (Supplementary Fig. 5B), we found no significant effect on nephrogenesis. Wnt4-Mettl3-KO mice demonstrated normal kidney histology, with nephron counts comparable to those of age-matched littermate controls (Wnt4Cre or Mett3F/F) at both P1 and P7 (Fig. 3B and Supplementary Fig. 7). These findings imply that METTL3 influences nephrogenesis primarily through its role in the NPCs, becoming dispensable once nephron precursors have formed. This prompted a closer examination of the NPC niches in Six2Cre-Mettl3-KO mice. Using immunofluorescence and in situ RNA hybridization, we identified that the kidneys of Six2Cre-Mettl3-KO mice continued to harbor SIX2, Cited1, and Tafa5 positive self-renewing NPCs at E16.5 (Supplementary Fig. 6B) and P1 (Fig. 3B). Additionally, RNA-seq profiling demonstrated that Six2Cre-Mettl3-KO kidneys displayed a transcriptomic signature characterized by an enrichment of markers associated with renewing NPCs, alongside a reduction in markers indicative of differentiating NPCs, in comparison to control kidneys (Supplementary Fig. 6C). This indicates that, similar to the effects seen with methionine restriction, the impaired nephrogenesis resulting from Mettl3 deletion is not due to a depletion of renewing NPCs but instead appears to stem from their inability to undergo differentiation.
Finally, we reemployed the ex vivo kidney culture model to conduct epistasis analysis of SAM and METTL3. We first compared the ex vivo phenotype of E12.5 Six2Cre-Mettl3-KO kidneys and M3i-treated wildtype cultures to the in vivo phenotype observed in Six2Cre-Mettl3-KO mice. After 48 h of culture, both the Six2Cre-Mettl3-KO and M3i-treated kidney cultures exhibited a reduction in PAX8+ tubular structures and UB branches compared to wildtype controls (Fig. 3C). Notably, consistent with the in vivo observations, SIX2+ renewing NPCs remained present in both the Six2Cre-Mettl3-KO and M3i-treated kidneys. These results indicate that our ex vivo models of genetic and pharmacological inhibition of METTL3 effectively recapitulated the in vivo phenotype of the Six2Cre-Mettl3-KO mice, allowing us to use these to examine the impact of SAM in the METTL3 knockdown setting. Subsequently, we supplemented E12.5 Six2Cre-Mettl3-KO and M3i-treated wild-type kidney cultures with SAM. However, in contrast to what was observed with methionine depletion, SAM supplementation did not ameliorate the nephrogenesis and UB branching defects in either the Six2Cre-Mettl3-KO or M3i-treated kidneys (Fig. 3C). This finding suggests that both methionine and SAM exert their nephron-inducing effects, at least in part, through METTL3. Collectively, our results unveil the methionine-SAM-METTL3 axis as a critical pathway necessary for nephrogenesis.
Activating the SAM–METTL3 axis promotes nephrogenesis
Since inhibiting RNA transmethylation blocks NPC differentiation and nephrogenesis, we next asked whether activating the SAM–METTL3 axis has the opposite effect. We addressed two specific questions: (i) whether METTL3 is sufficient to drive NPC differentiation in vivo. (ii) If SAM–METTL3 axis titration allows for the expansion of induced NPCs and enhances nephrogenesis. To assess the sufficiency of METTL3 in promoting NPC differentiation, we utilized mice carrying a cre-inducible Mettl3 gain-of-function (Mettl3-OE) allele. We employed the Six2:eGFPTGC/+ driver to specifically induce Mettl3 overexpression within the renewing domain of the NPC niche. qPCR analysis confirmed that Mettl3 levels were elevated more than 2-fold in E12.5 Six2:eGFPTGC/+;Mettl3OE (Mettl3-OE) kidneys compared to age-matched littermate controls (Six2Cre or Cre-negative; Mettl3OE) (Supplementary Fig. 8A). At E12.5, both Mettl3-OE and control kidneys displayed a comparable number of SIX2-positive NPCs. However, the Mettl3-OE kidneys exhibited ectopic expression of differentiation markers C1qdc2 and Lef1, while expression of the renewal marker Cited1 was notably diminished (Fig. 4A and Supplementary Fig. 8B). This aberrant differentiation led to a depletion of SIX2+ cap mesenchyme by E16.5, resulting in impaired nephrogenesis and kidney atrophy at birth (Fig. 4B and Supplementary Fig. 8C). These findings, alongside observations from pharmacological activation of METTL3 in primary NPC cultures, validate METTL3 as a critical cell-autonomous signal that can drive NPC differentiation when genetically activated in vivo.
A Kidney sections of E12.5 control or Six2:eGFPTGC/+; Mettl3OE (Mettl3-OE) mice immunostained with SIX2 and pan-cytokeratin, or subjected to in situ hybridization using probes against Cited1 and C1qdc2. B H&E staining, SIX2 (red), and pan-cytokeratin (green) immunostaining of kidney sections from P1 control or Mettl3-OE mice. White arrowheads point to precocious PAX8 expression. C E12.5 kidneys from control, Mettl3-OE, or wildtype embryos were cultured in regular media for 48 h and co-stained for PAX8 and E-cadherin or SIX2 and E-cadherin. The media of wild-type kidneys was supplemented with a vehicle, M3A (100 or 400 μM), or SAM (250 or 1000 μM). The number (average ± SD) of PAX8 tubules and SIX2 NPC niches is depicted in relevant images. ** Indicates P < 0.05. Statistical analysis: One-way ANOVA, Tukey’s multiple comparisons (C). Source data are provided as a Source Data file.
Our transgenic approach has a key limitation: it induces ectopic and fixed-dose upregulation of Mettl3. While this method successfully established the sufficiency of METTL3, it also resulted in unintended ectopic differentiation and depletion of self-renewing NPCs, which hindered our ability to accurately assess its impact on nephrogenesis. To address these issues, we proposed a strategy involving dynamic and submaximal upregulation of the SAM–METTL3 axis through SAM supplementation and M3A treatment. Our goal was to achieve a more controlled activation that would prevent premature exhaustion of self-renewing NPCs, promote the expansion of induced NPCs—as demonstrated in our NPC culture studies—and ultimately enhance nephrogenesis. To investigate the effects of the SAM–METTL3 axis on nephrogenesis, we employed an ex vivo kidney model, enabling us to systematically assess various concentrations of SAM and M3A. Initially, we compared the impact of maximal activation of the SAM and M3A pathways with the phenotype resulting from Mettl3 transgenic overexpression. Consistent with the in vivo observations, E12.5 Mettl3-OE kidneys cultured for 48 h exhibited ectopic PAX8 expression in the self-renewing NPC domain, a depletion of SIX2+ progenitors, and overall atrophy (Fig. 4C). Likewise, wildtype E12.5 kidney cultures treated with 1000 μM SAM or 400 μM M3A, which maximally activated RNA methylation, mirrored the Mettl3-OE kidney phenotype, indicating an on-target effect. Furthermore, we also noted ectopic PAX8 expression, depletion of SIX2+ progenitors, and atrophic conditions with both SAM and M3A treatment (Fig. 4C). We next explored the effects of submaximal activation by reducing the concentrations of SAM and M3A. A 50% reduction in dosage effectively prevented atrophy, resulting in kidneys that were similar in size to those treated with the vehicle control, with only sporadic ectopic PAX8 expression (Supplementary Fig. 9). Remarkably, reducing the doses of SAM or M3A by 75% not only preserved all renewing NPC niches but also resulted in larger kidneys, with a 30% increase in both PAX8+ tubules and UB branches (Fig. 4C). Moreover, we observed a continued increase in kidney size when the culture duration of SAM-treated kidneys was extended from 48 h to 4 days (Supplementary Fig. 10). In situ hybridization analyses revealed that these longer-cultured SAM-treated samples maintained expression of the NPC renewal marker Cited1 as well as the pre-tubular aggregate marker Lhx1 even after 4 days of culture. This persistent expression indicates sustained nephrogenesis. Overall, our findings suggest that submaximal activation of RNA transmethylation not only facilitates the proliferation of SIX2 and PAX8-positive NPCs but also enhances the nephrogenesis process itself.
Neonatal activation of the SAM–METTL3 axis enhances nephron endowment
Nephrogenesis in mice persists for 4 days following birth, with nearly half of their nephrons forming during this postnatal period. However, it remains unclear whether postnatal nephrogenesis can be modulated or if interventions targeting this process ex-utero can influence nephron endowment. Building on our findings that demonstrated the pro-nephrogenic effects of the SAM–METTL3 axis in ex vivo cultures, we aimed to investigate whether these effects could be replicated in vivo by specifically targeting the postnatal nephrogenesis window in mice. We initiated our investigation by comparing the metabolic profiles of NPC niches in P3 kidneys, which are nearing the conclusion of nephrogenesis, to those in E16.5 kidneys. Using unbiased metabolomics, we observed significant differences in the metabolic landscapes of these two developmental stages (Fig. 5A, B and Supplementary Fig. 11). Notably, we found that levels of methionine and SAM were reduced by more than 70% in P3 NPC niches compared to their E16.5 counterparts, suggesting that a diminished transmethylation state may be associated with the cessation of nephrogenesis (Fig. 5C). This finding prompted us to hypothesize that enhancing the RNA transmethylation axis could rejuvenate postnatal NPC niches and potentially extend their nephrogenic capacity. To test our hypothesis, we first conducted dose-finding experiments using SAM and M3A. Newborn mice were administered daily injections from P1 to P6, receiving either two doses of SAM (25 and 50 mg/kg) or M3A (21.7 and 2.17 mg/kg). Littermates randomly assigned to receive vehicle control were included for comparison (Fig. 5D). At P10, we assessed glomerulus counts as an indicator of nephron endowment using the acid-maceration technique. In comparison to vehicle controls, administration of 21.7 mg/kg M3A led to smaller kidneys with a reduced number of glomeruli (Supplementary Fig. 12A). In contrast, a lower dose of 2.17 mg/kg M3A elicited a pro-nephrogenic response, resulting in a 13.8% increase in the kidney weight-to-body weight (KW/BW) ratio and a 26.8% increase in glomerular count compared to controls (Fig. 5E, F). Likewise, pups receiving 25 mg/kg SAM exhibited a 14.7% higher KW/BW ratio and a remarkable 53.4% increase in the number of glomeruli (Fig. 5E, F). We did not see this effect with 50 mg/kg SAM (Supplementary Fig. 12B). These findings suggest that consistent with our observations in ex vivo cultures, in vivo nephrogenesis is intricately linked to the activation levels of the SAM–METTL3 axis. Specifically, submaximal activation promotes nephrogenesis, while excessive activation appears to hinder it.
A Principal component analysis of the metabolites in E16.5 and P3 nephrogenic zone cells (NZCs). B Volcano plot showing differential metabolite abundance between P3 and E16.5, filtered by P value (horizontal dashed line). Methionine and SAM are highlighted as black circles. C LC/MS-measured methionine and SAM levels at P3 and E16.5. D Schematic of the SAM or M3A treatment regimen. Created in BioRender. Patel, V. (2025) https://BioRender.com/6239npyE Kidney-weight-to-body-weight (KW/BW) ratio and F glomerular counts of P10 mice, and G eGFR values of P40 mice treated with vehicle, SAM, or M3A. H Low- and high-magnification images of P4.5 kidneys expressing Six2:eGFP. The number (average ± SD) of GFP clusters per 1 mm² kidney section area is depicted in the relevant images. I Distribution of the number of Six2:eGFP-positive cells within NPC clusters. At least 30 clusters from three biologically independent samples were counted for each condition. J Images of P4.5 kidneys immunostained for PAX8 and GFP are shown. The percentage of PAX8- and GFP-double-positive NPC clusters is indicated. Statistical analysis: Two-tailed, unpaired t-test (C, E–G, I). Error bars indicate SEM; ** indicates P < 0.05. Source data are provided as a Source Data file.
To further demonstrate the increased nephrogenesis, we analyzed NPC niches following treatment with SAM or M3A. For enhanced visualization of these niches, we employed Six2:eGFPTGC/+ transgenic mice. At P1, P2, and P3, we administered either a vehicle, 25 mg/kg SAM, or 2.17 mg/kg M3A, and subsequently evaluated the GFP signal alongside PAX8 expression on P3, P4.5, or P5 (Fig. 5D). We honed in with higher resolution on intervals around P5 for two main reasons: (1) we sought to provide sufficient time for the drug treatments to exert their effects and (2) we recognized that timepoints closer to the conclusion of nephrogenesis at P5 were more likely to reveal significant differences. Our analyses yielded several noteworthy findings. First, we observed that NPCs were depleted by P5 in all mice. However, a notable difference emerged at P4.5, as vehicle-treated mice exhibited a decrease in NPC niches, whereas NPC niches persisted in both SAM and M3A-treated mice. Correspondingly, we observed a substantial increase in the number of SIX2-eGFP-positive NPC niches at P4.5, with a notable 50.1% rise in SAM-treated pups and a 24.4% increase in M3A-treated pups compared to their respective controls (Fig. 5H). Second, the number of niches containing between 20 and 60 Six2-eGFP-positive NPCs increased by 28% in SAM-treated pups and by 18% in M3A-treated pups by P4.5 compared to those receiving vehicle treatment (Fig. 5I). Interestingly, we observed a higher proportion of NPC niches with 20–30 cells as early as P3 in the SAM and M3A groups compared to the vehicle-treated group (Supplementary Fig. 13). Third, co-immunostaining analyses revealed a strong correlation between the increased number of NPCs and the enhanced proportion of Six2-eGFP and PAX8 double-positive induced NPCs. Nearly 70% of Six2-eGFP-positive NPCs in the SAM and M3A groups co-expressed PAX8, compared to only 35% in the vehicle-treated pups at P4.5 (Fig. 5J). Thus, the extended survival of NPCs, along with a higher proportion of induced PAX8 and SIX2 double-positive NPCs, could account for the enhanced nephrogenesis observed in the SAM and M3A treatment groups.
To further validate the enhancement of nephron endowment induced by SAM and M3A, we employed the gold-standard FITC-sinistrin transdermal assay to estimate glomerular filtration rate (eGFR), a key clinical marker for kidney function30. Newborn mice were treated with vehicle, SAM, or M3A, and GFR was measured at P40 to determine the effects of these treatments in adulthood. The vehicle-treated mice (N = 13) displayed a normal eGFR of 1602 µl/min, aligning with findings from previous studies (Fig. 5G). In stark contrast, mice receiving SAM treatment exhibited a significant 20% increase in eGFR, reaching 1930 µL/min (N = 15). M3A treatment also demonstrated beneficial effects, yielding an eGFR of 1829 µl/min at P35, which represents an 11% increase compared to the 1654 µl/min observed in vehicle-treated littermate controls (Fig. 5G). These results provide compelling evidence that both SAM and M3A treatments enhance nephron endowment, which in turn is associated with increased kidney function, as indicated by the elevated GFR measurements.
SAM–METTL3 mediates NPC differentiation and nephrogenesis via LRPPRC
METTL3 mediates its biological effects primarily through its m6A-forming catalytic activity. However, the m6A epitranscriptomic landscape of developing kidneys remains unexplored. To investigate the mechanisms downstream of the SAM–METTL3 axis, we employed an unbiased combinatorial and stepwise experimental approach that combined RNA-Seq and MeRIP-seq (m6A-modified mRNA immunoprecipitation followed by high-throughput sequencing) in control (Six2:eGFPTGC/+) and Six2Cre;Mettl3-KO kidneys (Supplementary datasets 2–4)31. Each experimental group consisted of two biological replicates, with RNA pooled from four male or female embryos. Our analysis focused on the E16.5 time point, enabling us to identify molecular targets prior to the pronounced nephrogenesis defects observed at P1. First, unbiased pathway analysis of the RNA-Seq dataset revealed that oxidative phosphorylation and the TCA cycle were among the most significantly downregulated pathways in Six2Cre; Mettl3-KO kidneys compared to control samples (Fig. 6A and Supplementary Fig. 14A). Notably, we identified a substantial number of mitochondria-related genes that were downregulated in Six2Cre;Mettl3-KO kidneys, including Ppargc1a, a nuclear-encoded co-transcription factor essential for mitochondrial biogenesis, various genes encoding components of the electron transport chain, and several mitochondrial ribosomal genes (Supplementary Fig 14A). Confirmatory qPCR analyses further validated the downregulation of the essential mitochondrial electron transport chain genes in Six2Cre;Mettl3-KO kidneys relative to control kidneys (Supplementary Fig. 15A). In contrast, we observed an increase in TOMM20 immunofluorescence in Mettl3-OE kidneys (Supplementary Fig. 15B). These findings suggest that METTL3 may promote NPC differentiation by modulating mitochondrial biogenesis and metabolism. Next, we analyzed our MeRIP-Seq dataset to define the m6A landscape of the developing kidney. Principal component analysis demonstrated that biological replicates clustered closely together, revealing consistent m6A profiles across male and female samples of the same genotype (Supplementary Fig. 14C). Importantly, control samples formed a distinct cluster, separate from the Six2Cre;Mettl3-KO samples, indicating a significant alteration in the m6A landscape following Mettl3 deletion. By employing a stringent criterion (P < 0.01 and FDR < 0.01), we identified 61 unique mRNA transcripts that exhibited a reduction of 50% or more in m6A levels in Six2Cre;Mettl3-KO kidneys compared to controls (Supplementary Fig. 14D). Among these transcripts, we focused on Lrpprc mRNA (Fig. 6B) due to its role in encoding a mitochondrially enriched protein that regulates the RNAs produced by the mitochondrial genome and influences mitochondrial metabolism32,33. While the role of LRPPRC in kidney development remains underexplored, loss-of-function mutations in this gene have been associated with Leigh syndrome, a mitochondrial disorder that can sometimes lead to developmental abnormalities in the kidneys34,35,36. Therefore, we reasoned that the downregulation of LRPPRC may, at least partially, account for the observed mitochondrial dysfunction in Six2Cre;Mettl3-KO mice.
A Pathway analysis of RNA-Seq datasets showing downregulation of mitochondrial metabolism genes in E16.5 Six2Cre-Mettl3-KO (M3KO) compared to control kidneys. B IGV tracks display the input-normalized m6A signal on Lrpprc mRNA. C Q-PCR analysis showing Lrpprc (mouse cells) and LRPPRC (human cells) transcripts are enriched in samples recovered after RNA immunoprecipitation using an anti-METTL3 antibody compared to an anti-IgG antibody. N = 3. D Immunoblots showing LRPRRC and oxidative phosphorylation complex expression in Six2Cre-Mettl3-KO (M3KO) and Mettl3-OE (M3OE) kidneys compared to their respective controls. Actin serves as a loading control. E LRPPRC, SIX2, or PAX8 immunostaining, or live-cell MitoTracker labeling of NPCs cultured for 48 h in 1.25 μM CHIR NPEM media and co-treated with 100 μM M3A or 100 μM SAM in the presence or absence of 20 μM GAA. F–H Newborn pups were injected from P1 to P4 with a vehicle, 20 mg/kg GAA, 25 mg/kg SAM, or both 20 mg/kg GAA and 25 mg/kg SAM. Kidney weight-to-body weight (KW/BW) ratio (F) and glomerular count (G) of P10 mice, and eGFR (H) of P40 mice, are shown. I–K Newborn pups were injected daily from P1 to P4 with a vehicle, 20 mg/kg GAA, 2.17 mg/kg M3A, or both 20 mg/kg GAA and 2.17 mg/kg M3A. KW/BW ratio (I) and glomerular count (J) of P10 mice, and eGFR (K) of P40 mice are shown. Statistical analysis: Fisher’s Exact Test (A), two-tailed unpaired t-test (C), and one-way ANOVA with Tukey’s multiple comparisons (F–K); error bars indicate SEM. Source data are provided as a Source Data file.
We employed a range of complementary approaches to validate Lrpprc/LRPPRC as a direct target of SAM–METTL3 that is relevant in NPC differentiation. Our initial evidence was obtained through the immunoprecipitation of mRNAs directly bound by METTL3 using an anti-METTL3 antibody in human embryonic kidney (HEK) and mouse kidney epithelial cell lines. As a positive control, we also immunoprecipitated RNA using an anti-SNRNP70 antibody, whereas RNA pull-down using an anti-IgG antibody served as a negative control. Subsequent quantitative PCR (qPCR) analysis revealed that LRPPRC mRNA was enriched up to 300-fold in samples recovered after anti-METTL3 immunoprecipitation compared to those subjected to anti-IgG immunoprecipitation, confirming that METTL3 directly binds to LRPPRC mRNA in both human and mouse cells (Fig. 6C). Additionally, immunoblot analysis indicated a substantial reduction in LRPPRC protein levels in E16.5 Six2Cre;Mettl3-KO kidneys compared to control specimens. Conversely, an increase in LRPPRC expression was observed in E12.5 Mettl3-OE kidneys (Fig. 6D). It is noteworthy that Lrpprc mRNA levels remained relatively unchanged (Supplementary Fig. 14E), suggesting that METTL3’s role is to enhance the translational efficiency of Lrpprc rather than impacting its transcriptional regulation. Further supporting our findings, immunoblot analysis using a cocktail of antibodies revealed downregulation of some components of the mitochondrial electron transport chain in E16.5 Six2Cre;Mettl3-KO kidneys, whereas we observed upregulation in E12.5 Mettl3-OE kidneys compared to their respective controls (Fig. 6D).
To further investigate the interrelationship between SAM, METTL3, and LRPPRC and to establish it as a cell-autonomous axis, we transitioned to NPC cultures. As downstream readouts of LRPPRC function, we employed MitoTracker dye to evaluate the mitochondrial membrane potential in live NPCs and immunoblots of OXPHOS protein components as a proxy of the electron transport chain. Notably, NPCs treated with a combination of 1.25 µM CHIR and differentiation-inducing doses of SAM or M3A demonstrated a significant increase in the levels of LRPPRC and OXPHOS proteins, accompanied by an enhanced MitoTracker signal, compared to those treated with 1.25 µM CHIR alone (Supplementary Fig. 16A, B). Moreover, treatment with 3 µM CHIR, known to promote METTL3 expression and NPC differentiation, was also effective in elevating LRPPRC levels, OXPHOS proteins, and MitoTracker signal (Supplementary Fig. 16C, D). Importantly, neither SAM nor 3 µM CHIR was able to enhance LRPPRC levels, OXPHOS proteins, or MitoTracker signal when NPCs were co-treated with the M3i METTL3 inhibitor (Supplementary Fig. 16A–D). These results collectively indicate that Lrpprc mRNA is a direct target of METTL3 and that the induction of LRPPRC protein during NPC differentiation occurs in a SAM-METTL3-dependent manner.
If LRPPRC is indeed a biologically relevant RNA transmethylation target in NPCs, then its inhibition should hinder SAM and METTL3-induced NPC differentiation and nephrogenesis. To address this final question, we employed gossypol acetic acid (GAA), a small molecule inhibitor of LRPPRC37,38. GAA is directly associated with the RNA binding domain of LRPPRC with high affinity (dissociation constant KD = 4.12 µM). As a result, this interaction prevents LRPPRC protein from binding to its client RNAs, leading to its degradation. We assessed various GAA doses in primary NPCs (20 µM) and in vivo studies in mice (20 and 60 mg/kg). In NPC cultures, 20 µM GAA effectively inhibited LRPPRC and OXPHOS protein levels and reduced the MitoTracker signal. Crucially, it blocked SAM, M3A, or 3 µM CHIR-induced NPC differentiation (Fig. 6E and Supplementary Fig. 17A). In the final set of experiments, we conducted daily intraperitoneal injections of either a vehicle solution, 20, or 60 mg/kg of gossypol acetic acid (GAA) in newborn mice. Analysis at P10 showed a significant reduction in glomerular counts and the kidney weight-to-body weight (KW/BW) ratio in the group receiving 60 mg/kg GAA compared to vehicle-treated controls (Supplementary Fig. 17C). However, the lower dose of 20 mg/kg did not independently impact nephrogenesis in vivo, prompting us to select it for further epistasis analysis. Recognizing that NPCs were exhausted by P5 in our previous studies, we administered either vehicle, SAM, 20 mg/kg GAA, or a combination of SAM and 20 mg/kg GAA from postnatal day 1 (P1) to P4. A second cohort received either vehicle, M3A, 20 mg/kg GAA, or M3A combined with 20 mg/kg GAA during the same period. We evaluated the mice at P10 for glomerular counts39 and at P40 for estimated glomerular filtration rate (eGFR) using the FITC-sinistrin transdermal assay30. In alignment with our previous findings, treatment with either SAM or M3A resulted in significantly larger kidneys, an elevated glomerular count, and improved eGFR when compared to vehicle-treated littermate controls (Fig. 6G–L). While the low-dose GAA treatment alone did not exert any discernible effects, its concurrent administration with SAM or M3A effectively inhibited the observed increases in kidney weight, glomerular count, and eGFR. Collectively, these results suggest that the SAM–METTL3 pathway plays a crucial role in mediating NPC differentiation and nephrogenesis, at least in part, by regulating the Lrpprc mRNA.
Discussion
We present several congruent lines of evidence suggesting that METTL3-dependent RNA transmethylation is a crucial axis governing NPC fate determination and renal nephrogenic capacity. First, we demonstrate a pronounced and synchronous increase in the levels of MAT2A, SAM, METTL3, METTL14, and m6A as NPCs transition from a state of self-renewal to differentiation. Moreover, this upregulation appears to be an early event, foreshadowing the causative role the RNA transmethylation pathway plays in initiating nephrogenesis. In contrast, we found that as the kidney approaches the conclusion of its nephrogenic potential, there is a significant decline in the levels of methionine and SAM, with a marked reduction observed in older P3 murine NPC niches compared to their younger counterparts at E16.5. This decline draws parallels to recent findings in skeletal muscle, where aged resident stem cells exhibit a diminished transmethylation capacity compared to their younger counterparts. Remarkably, restoring SAM levels in these aged muscle progenitors has been shown to rejuvenate them, enhancing their regenerative capabilities40,41.
Secondly, employing multiple orthogonal approaches and model systems, we establish both the necessity and sufficiency of the methionine–SAM–METTL3 axis in guiding the fate transition of NPCs. We noted that NPCs, akin to many cancer cells, manifest the ‘Hoffman effect’—wherein their methionine dependency stems from its pivotal role in SAM generation rather than as an amino acid component of proteins42. This methionine–SAM biological axis primarily exerts its effects through METTL3, suggesting that RNA serves as the key substrate for transmethylation in NPCs. This conclusion is further supported by recent studies indicating that mutations in the tRNA methyltransferase NSUN7 are implicated in severe cases of Congenital Anomalies of the Kidney and Urinary Tract (CAKUT)43. In addition to RNA, DNA, and proteins represent two other major substrates for methylation, suggesting that a portion of the effects of the methionine-SAM axis may also operate through these pathways. With regards to DNA, DNMT3A, and DNMT3B are the de novo methyltransferases responsible for establishing new methylation marks on previously unmethylated gene regulatory regions. DNMT1 is a ‘maintenance’ methyltransferase, as it functions in maintaining existing DNA methylation patterns after DNA replication. Interestingly, while the loss of Dnmt1 disrupts nephrogenesis in mice, recent findings indicate that de novo DNA methylation is not essential for the nephrogenic process44. This suggests a complex interplay between various methylation pathways, while also highlighting the distinct role of de novo RNA methylation in gene regulation and nephrogenesis.
Thirdly, we demonstrate that fine-tuning SAM–METTL3 activation, both ex vivo and in vivo enables a higher yield of nephrons. Surprisingly, the necessity of METTL3 is confined to the early, narrow window of NPC fate transition, becoming dispensable after the formation of pre-tubular aggregates. This exquisite essentiality presented us with a unique opportunity to target the NPC niche to enhance its nephrogenic potential without adversely impacting the later stages of nephron morphogenesis or segmentation. The cellular mechanism underlying this enhanced nephrogenesis may involve the expansion of an induced NPC population that lies between self-renewing and fully lineage-committed states. Our findings hold potential clinical implications, suggesting that targeting RNA methylation could be a promising strategy for improving nephrogenesis, particularly in the context of preterm deliveries. In such cases, nephrogenesis occurs postnatally but often fails to achieve an adequate nephron threshold, resulting in potential long-term complications45,46.
Finally, our work suggests epitranscriptomic activation of Lrpprc as a novel mechanism through which SAM and Mettl3 drive nephrogenesis. We discovered that METTL3 directly binds to and methylates Lrpprc mRNA, resulting in increased expression of the LRPPRC protein. Interestingly, LRPPRC itself functions as an RNA-binding protein that plays a critical role in the expression of mitochondrial genome-encoded RNAs32,33. Consistent with its role in promoting mitochondrial function, mutations in LRPPRC are linked to the human mitochondrial disorder Leigh syndrome34,35,36. In murine models, deletion of Lrpprc results in early embryonic lethality, while tissue-specific Lrpprc-KO mice exhibit mitochondriopathy in both hepatic and cardiac tissues47,48,49, underscoring the critical role of LRPPRC in supporting mitochondrial function. Thus, the regulation of LRPPRC by METTL3 may partially account for the well-documented metabolic transition from anaerobic glycolysis to aerobic oxidative phosphorylation, which is essential for effective nephrogenesis. However, our RNA-seq analysis reveals a significant mitochondrial biogenesis phenotype in Mettl3 knockout kidneys that cannot be attributed solely to the downregulation of LRPPRC, as it is not known to regulate nuclear-encoded mitochondrial transcription factors. This observation suggests that METTL3 may influence additional pathways related to mitochondrial function.
Our study has certain limitations and raises several unanswered questions. Firstly, although we demonstrate that the upregulation of the SAM–METTL3 axis is an early event in NPC differentiation, the precise mechanisms driving this activation remain unclear. Secondly, our findings indicate that METTL3 inhibition mimics the effects of methionine restriction, whereas METTL3 activation mirrors those of SAM supplementation. Nonetheless, we recognize the potential existence of additional METTL3-independent mechanisms that could also influence the biological effects of methionine and SAM, which our study does not address. Thirdly, while we identified Lrpprc mRNA as a novel target of METTL3, the functional interrelationship has not been definitively established in our work. Although we employed the small molecule GAA with high specificity to inhibit LRPPRC, it is important to note that GAA may elicit effects that are independent of LRPPRC. To further clarify this relationship, additional confirmatory studies using Lrpprc knockout mice are necessary. These further investigations will help us untangle the complexities of the SAM–METTL3 axis and its downstream effects on nephrogenesis. Finally, we recognize that stereological analysis offers a more technically advanced method for measuring glomerulus counts compared to the acid maceration approach we employed. However, we strengthened our evaluation of the enhanced nephrogenesis phenotype through two additional complementary strategies. Specifically, we assessed NPC niches at P3 and P4.5 using the Six2:eGFPTGC/+ transgenic mouse model, which allowed us to monitor the live SIX2-eGFP signal in postnatal NPC niches. Additionally, we measured glomerular function at P35 utilizing the gold-standard FITC-sinistrin transdermal assay to ensure a comprehensive assessment of nephrogenesis outcomes.
In summary, we demonstrate that the transmethylation status is a key metabolic dependency during the early, pivotal stages of nephrogenesis. METTL3 responds to methionine–SAM availability and facilitates a transcriptomic and metabolic output that is conducive to nephrogenesis. Notably, harnessing this axis through nutritional or pharmaceutical interventions enhances the nephrogenic potential of the developing kidney, with significant implications for cases of low nephron endowment and efforts to regenerate the kidney.
Methods
This research complies with all relevant ethical regulations of the University of Texas Southwestern Medical Center. All experiments involving animals were approved by the Institutional Animal Care and Use Committee at the University of Texas Southwestern Medical Center.
Mouse studies
This study used Six2:eGFPTGC/+29, Wnt4Cre (O’Brien, UT Southwestern Medical Center), Mettl3F/F and Mettl3Tg mouse lines19. These mice were maintained on a C57BL/6J background. Swiss Webster mice from Charles River were utilized for wild-type NPC cultures and ex vivo kidney cultures. The postnatal drug treatment studies to asses glomerular counts and eGFR measurements were performed on offspring born to Swiss Webster mice obtained from Charles River. The postnatal drug treatment studies to asses NPC niches were performed on offspring born to Six2:eGFPTGC/+ transgenic mice. All studies used equal numbers of males and females. All mice were housed in the UT Southwestern Animal Facility room, which is maintained at a temperature of 18–24 °C and a humidity level of 30–50%. All the mice were maintained on a 24-h light/dark cycle with a 6:00 AM–6:00 PM ‘light on’ schedule. Mice always had ad libitum access to food and water. The mice were fed Teklad Global 2016 16% protein diet. Mice were euthanized in accordance with approved protocols. Blood was obtained via cardiac puncture when animals were under anesthesia. The kidneys were extracted, weighed, flash-frozen, and stored at −80 °C for molecular analysis or were fixed with perfusion of 4% (wt/vol) paraformaldehyde for histological and immunofluorescence analysis.
Isolation and culture of NPCs
NPCs were isolated from kidneys at E16.5. Kidneys were decapsulated and placed in 1X PBS containing Ca2+ and Mg2+ salts. Thereafter, the kidneys were pooled and transferred to a 5 mL round-bottom tube, where they were washed twice in 1X HBSS to remove blood. HBSS was then replaced with 2 ml of room temperature nephrogenic zone cell enzymatic digestion solution (2.5 mg/ml collagenase A from Roche, catalog 11 088 793 001 and 10 mg/mL pancreatin from Sigma, catalog P1625), and the kidneys were incubated at 37 °C for 12 min while rocking on a Nutator. Following enzymatic digestion, 125 μL of fetal bovine serum was added to the cell suspension to neutralize Collagenase and Pancreatin. The cell suspension was washed twice with the AutoMACS running buffer (Miltenyi, 130-091-221) and resuspended in a fresh AutoMACS running buffer. This nephrogenic zone cell suspension was filtered through a pre-washed 40-μm filter column to remove pre-tubular aggregates and subjected to the immunoprecipitated negative depletion method to obtain pure NPCs. The NPCs were seeded at 25% density and cultured for 48 h in NPEM media containing 1.25 µM CHIR to maintain as renewing NPCs or in 3 µM CHIR to induce differentiation.
Immunofluorescent staining
(A) Cells: NPC culture cells were fixed in 4% PFA for 10 min. After three washes in 1X PBS, the cells were antigen retrieved with 0.1% TritonX containing 1X PBS for 10 min. The cells were then blocked in the blocking solution (5% goat serum in 1X PBS) and incubated with the primary antibody overnight at 4 °C. The next day, the cells were washed three times in 1X PBS and then incubated in the appropriate Alexa Fluor secondary antibodies for 45 min at room temperature. The cells were then washed three times in PBS, counterstained with DAPI, and imaged using an inverted microscope. (B) Tissue: IF staining on kidney tissues was performed on 10-μm sections of OCT- or paraffin-embedded tissue. Slides were antigen-retrieved using 0.1% Triton X-100 containing 1X PBS for 20 min for METTL3 staining, or boiled in Tris-EDTA buffer for 20 min for METTL14, MAT2a, SIX2, and PAX8. The primary antibodies were used at a 1:300 dilution, and the secondary antibodies were used at a 1:500 dilution.
Immunoblot analysis
NPCs were cultured in 24-well plates in NPEM media with or without supplementation of drugs or metabolites. At the end of the culture, the NPCs were washed once in PBS, scraped in PBS, and spun down in a tabletop centrifuge for 5 min at 2000 RPM. The protein lysis buffer (PLB) was prepared by mixing one tablet of protease-phosphatase inhibitor in 10 mL of TPer solution. 20 μL of PLB was added to the cell pellets from 500 K cells and resuspended by vortexing. The protein lysate was extracted by performing three freeze-thaw cycles in liquid nitrogen and ice. The lysate was centrifuged for 10 min at 4 °C. The supernatant was stored in a −80 °C freezer until it was ready for Western blot analysis. The protein samples were prepared using the 4X SDS Lamelli buffer. The protein was diluted using the PLB. The samples were boiled at 98 °C for 5 min, immediately cooled on ice, and loaded onto pre-made native page gels. A minimum of 6 μg was run on the gels. Semi-dry transfer for 7 min was carried out using Biorad transfer. The blotted proteins were washed in 1X TBST (1X TBS containing 0.1% Tween 20) and blocked for 1 h with 15 ml of 5% non-fat milk (Kroger) at room temperature. The blots were then incubated overnight at 4 °C with the primary antibody at a 1:1000 dilution. The next day, the blots were washed 3 times in 1X TBST, incubated with the secondary HRPs at a 1:5000 dilution for 45 min, washed thrice in 1X TBST, and imaged using the Biorad Western blot developer. For MAT2A, METTL3, METTL14, PAX8, CITED1, LHX1, and SIX2, which are between 75 and 37 kDa, the blots were treated using the stripping buffer for 20 min to remove HRP, then blocked with milk, and blotted for Actin-HRP to obtain the protein loading control. In the case of LRPPRC, a 130 kDa protein, the western blot was divided into smaller blots, and LRPPRC and Actin were blotted simultaneously.
m6A ELISA
NPCs were seeded on 24-well plates in NPEM media with or without the supplementation of drugs/metabolites. At the end of the culture, NPCs were washed once with 1X PBS, treated with QIAzol, and stored at −80 °C. RNA was extracted using the miRNeasy Mini kit from Qiagen (catalog #217004). RNA quality was confirmed through spectrophotometry and gel electrophoresis. m6A ELISA was performed using the EpiQuik m6A RNA Methylation Quantification Kit (catalog #P-9005-96; Epigentek). The readings were obtained using the BioTek Synergy H1 microplate reader.
m6A dot blot analysis
RNA from NPCs was extracted, and quality control was performed as described in the m6A ELISA section. Subsequently, RNA was diluted to 25 μg/μL using RNase-free water for each sample. The RNA samples were heated to 95 °C in a PCR machine for 3 min to disrupt secondary structures, and then immediately chilled on ice. 1 μL of each RNA sample was loaded onto an Amersham Hybond-N+membrane (GE Healthcare, catalog# RPN203B), allowed to dry, and then UV cross-linked twice at 120 mJ/cm2 (25–50 s) using the Stratalinker 2400 UV Crosslinker. The membrane was washed in a buffer containing 0.02% Tween 20 sterile 1X PBS solution for 5 min and then stained with methylene blue. Excess dye was washed off, and the membrane was imaged on the Biorad ChemiDoc imager. This image served as the loading control. The membrane was then blocked with 5% non-fat milk (Bio-Rad Laboratories, catalog number #1706404) for 45 min at room temperature while gently shaking. The membrane was then incubated overnight at 4 °C with the anti-m6A antibody diluted at 1:250 in wash buffer. The next day, the membranes were washed three times for 5 min in the wash buffer with gentle shaking. Membranes were then incubated with goat anti-rabbit IgG-HRP diluted at 1:10,000 (20 ng/ml) in the wash buffer for 45 min at room temperature with gentle shaking. After four 10-min washes at room temperature, the blots were imaged on the Biorad ChemiDoc imager. Quantification was performed using the Biorad Image Lab Software. The m6A signal was normalized to the methylene blue staining for each sample to obtain m6A values.
Metabolomics
(A) NPCs: NPCs were isolated from E16.5 embryonic kidneys and cultured in 10 cm cell culture dishes as described above. Metabolites were harvested 48 h after culture. Each plate was quickly rinsed with 5–7 mL of ice-cold normal saline solution (saline: 2F7123 Baxter 0.9% sodium chloride irrigation, USP), placed on liquid nitrogen and treated with 500 μL of 80% (vol/vol) methanol in water (cooled to −20 °C). All the plates were incubated at −20 °C for 5 min. The plates were scraped with a cell scraper on dry ice. The cell lysate/methanol mixture was transferred to a 1 mL Eppendorf tube and flash-frozen in liquid nitrogen. Then, the tubes were subjected to three freeze–thaw cycles between liquid nitrogen and 37 °C. After the third thaw, the tubes were vortexed for 1 min and then centrifuged at ~20 °C and 160×g for 15 min in a refrigerated centrifuge. The metabolite-containing supernatant was transferred to new pre-chilled tubes and left on ice to measure protein concentration. Protein was measured in the 80% methanol portion, not the pelleted protein, using the Pierce BCA kit to have the widest range curve. 80% methanol was used as the diluent for the standard curve. The volume equivalent to 10 μg of protein was transferred to a new Eppendorf tube and subjected to a speed vacuum. The samples were resuspended in 100 μL acetonitrile/water 80:20 (vol/vol) for a final concentration of [0.1 mg/mL] of protein (of the supernatant). The tubes were vortexed vigorously for 1 min and then centrifuged at ~20 °C and 160×g for 15 min in a refrigerated centrifuge. The metabolite-containing supernatant was transferred to an LC–MS vial (with insert) and cap. The samples were subjected to LC/MS. (B) Nephrogenic zone cells: The kidneys of E16.5 embryos or P3 pups were subjected to enzymatic digestion as in the NPC culture experiments. For each sample, 1 million NZCs were pelleted by centrifuging at 2000×g for 5 min, flash-frozen in liquid nitrogen, and then re-suspended in 80% (vol/vol) methanol in water (cooled to −20 °C). The remainder of the sample preparation was carried out as described for NPCs. LC/MS was performed by the Children’s Research Institute metabolomics core at UT Southwestern Medical Center. The samples were analyzed using the AB SCIEX QTRAP 6500 Triple Quadrupole liquid chromatography equipment and controlled through the AB Sciex Analyst 1.6.1 Software. Metabolite abundance was obtained from the chromatographic peak area of the precursor ion and normalized to total ion count (TIC). The plots were generated using Metaboanalyst.ca and GraphPad Prism.
SAM measurement
SAM levels in NPCs were measured using an ELISA kit (Catalog #STA671-C; Cell Biolabs) following the manufacturer’s protocol. ELISA was performed using protein lysate prepared from snap-frozen NPC pellets. The protein lysate was extracted by homogenizing the frozen cell pellets in a protein lysis buffer using the microtube hand-held homogenizer (Bel-Art F65000-0000, Fisher). The protein lysis buffer was freshly prepared with 1X PBS (#10010049, Invitrogen) and a protease inhibitor cocktail (cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail, Catalog #11836170001 from Roche) such that one tablet was added for 7 ml of 1X PBS. The samples were always maintained at 4 °C. The homogenates were centrifuged for 15 min at 10,000×g at 4 °C. The lysates were diluted to ~1 mg/ml and immediately proceeded with the ELISA. 450 nm OD readings were plotted on the standard curve obtained using the kit’s SAM-BSA standards to derive SAM levels in the test samples. Then, the SAM levels were normalized to the amount of protein loaded.
Kidney organ cultures
E12.5 kidneys were cultured on a Whatman membrane (Sigma, WHA110409) in an air-medium interface, as described previously. We used DMEM media (Thermo Fisher #12500) containing 10% Fetal Bovine Serum (FBS) and 2% PenStrep (Invitrogen #1514022). For methionine depletion experiments, one kidney from each embryo was cultured in the special DMEM growth media without methionine (Invitrogen #21013024). The other kidney was cultured in the equivalent media that contains 0.2 mM methionine (Invitrogen #11965092). l-glutamine (Sigma, #G7513) was supplemented with methionine-depleted media to match formulations with the control media. After 48 h, the media was fixed, and proceeded with IF staining. For 4-day cultures, the media was replaced after 48 h.
MeRIP-seq and RNA-seq
m6A RNA immunoprecipitation was performed using the m6A antibody precipitation approach as previously described. Briefly, RNA isolated from kidneys of Six2Cre or Six2Cre-Mettl3-KO E16.5 pups was subjected to fragmentation to obtain fragments of 100 nucleotides or smaller in length. Magnetic immunoprecipitation was performed. Two biological replicate samples per group were analyzed, each sample containing pooled fragmented RNA from 4 to 6 pairs of kidneys. The samples were subjected to Illumina’s next-generation sequencing. The raw data of the sequencing was filtered through a series of filtration methods to obtain high-quality sequencing data for subsequent analysis. The original data received from the high-throughput sequencing platforms were transformed into sequenced reads by base calling. Raw data were recorded in a FASTQ file, which contains sequenced reads and corresponding information on sequencing quality. The “e” represents the sequence error rate, and Qphred represents the base quality value, Qphred = -10log10(e). HISAT2 software was used to map the clean reads to the reference genome. Peak calling was performed using exomePeak. Peak annotation, metagene analysis, and diffpeak analysis were performed. Pathway analysis was performed using IPA software, and tracks were visualized using IGA software. A fold change heat map was generated using GraphPad Prism. featurecounts were used to calculate the counts of the Reads that mapped the genome, and FPKM was used to standardize the expression data. The DESeq2 algorithm was applied to filter the differentially expressed genes between the control and Mettl3KO samples.
RNA immunoprecipitation
RNA pulldown assay was performed on mouse kidney and HEK293 cells using the EZ Magna RIP kit (Sigma-Aldrich, Cat. #17-701) with modifications. Briefly, mouse kidney epithelial cells were grown at 33 °C in a medium containing IFN-γ until they reached 100% confluence, then switched to a medium lacking IFN-γ and cultured at 37 °C for 3 days. HEK293 cells were grown at 37 °C until confluency. Cells were washed with PBS, scraped, and pelleted at 18,000 kg. The pellets were lysed in RIP lysis buffer (supplemented with protease and RNase inhibitors) at a 1:1 buffer-to-pellet ratio, incubated on ice for 5 min, and stored at −80 °C. Next, 50 µL of Protein A/G magnetic beads were conjugated with five µg of antibody—either anti-METTL3 (Cell Signaling, Cat. #96391), anti-SNRNP70 (positive control), or IgG (negative control)—in a total volume of 350 µL RIP wash buffer. For immunoprecipitation, 900 µL of RIP buffer and 100 µL of cell lysate were added to the antibody-conjugated beads and incubated at 4 °C for 4 h. The beads were washed five times with RIP wash buffer. Ten percent of the beads were set aside for Western blot analysis to confirm METTL3 pulldown. RNA was isolated from the remaining beads by Proteinase K digestion, followed by phenol-chloroform extraction and overnight ethanol precipitation at −80 °C. The resulting RNA pellets were washed with 80% ethanol, resuspended in nuclease-free water, treated with DNase I (Invitrogen, Cat. #18068015), and reverse-transcribed into cDNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher, Cat. #K1622). Quantitative PCR (qPCR) was performed on a Bio-Rad CFX96 system using SYBR Green and primers specific for human LRPPRC (Forward: 5′-CAGTTGCATGAAGCCATCGTG-3′; Reverse: 5′-TCATAGCAGTCAATGGCGACC-3′), mouse Lrpprc (Forward: 5′-GTGACTTACCTGCTGCTCTTG-3′; Reverse: 5′-GTAATTCCCCGTCTGCAGGAA-3′), and U1 snRNA (Forward: 5′-GGGAGAATACCATGATCACGAAGGT-3′; Reverse: 5′-CCACAAATTATGCAGTCGAGTTTCC-3′). Fold enrichment was calculated using the ∆∆Cq method, comparing METTL3 or SNRNP70 pulldown to IgG controls across biological replicates.
In vivo SAM and Mettl3-activator studies
Postnatal nephrogenesis drug studies involving nephron count analysis and eGFR measurements were performed on pups born to virgin Swiss Webster dams mated with Swiss Webster males. Nephrogenic niche analysis was performed on pups aged P3 and P4.5, born to virgin Swiss Webster dams mated with Six2Cre males. On the day of birth, each pup was randomly assigned to one of three treatment groups: vehicle, 50 mg/kg SAM, or 25 mg/kg SAM. The pups were subjected to daily injections from P1 to P6. One cohort of pups was sacrificed at P10; body weights were measured, and kidneys were harvested and weighed. The kidneys were then subjected to acid maceration or fixation. A second cohort of pups was allowed to age till P35 and subjected to eGFR measurement. The M3A and GAA studies were identical to the SAM studies. 0, 2.17 and 21.7 mg/kg doses of M3A were injected.
Acid maceration for glomerular counts
Acid maceration was performed as described39. The decapsulated kidneys were chopped on a clean microscopic slide using a razor to obtain about 16 pieces. The pieces were added to a 15 ml Falcon tube containing 5 ml of 6 M HCL and incubated for 90 min on a rotator at 37 °C. At the end of the incubation, the acid-treated samples were pipetted several times until the big chunks of tissue broke down into a macerate. 1X PBS was added to the samples to make up the volume to 50 ml and incubated overnight at 4 °C. The samples were mixed well, and 100 μL of the macerate was pipetted on a flat-bottomed 96-well tissue culture plate, which was then imaged on the inverted Lumar microscope. The glomeruli were counted using the Zeiss software. The graphs were plotted using GraphPad Prism.
eGFR measurement
eGFR measurement was performed following the protocol published in ref. 30. The mice were shaved using a razor while under anesthesia. Then, the transdermal GFR monitoring device was clamped onto the animal and wrapped using a bandage. 3–5 min later, a baseline reading was obtained. Next, 60 μL of FITC-Sinistrin was administered via retro-orbital injection, and the mice were placed in an empty cage for 60–90 min. The mice were unwrapped; the device was separated from the battery and connected to the device reader on the computer to download the data. The data was analyzed using the Medibeacon software, and the half-time and eGFR were obtained. The values were plotted, and graphs were obtained using GraphPad Prism.
Nephrogenic niche analysis
The pups subjected to SAM or M3A treatment were carefully dissected in 1X PBS to obtain kidneys for imaging. The dissected kidneys were placed in 24-well tissue culture plates and imaged using the Leica microscope to obtain images of GFP + NPC niches of the kidneys. The niche counts were obtained using the Zeiss software.
Antibodies
The following antibodies were used: Actin (Sigma, catalog# a3854), Cited (Santa Cruz, catalog# sc-393585), Cytokeratin (Sigma, catalog# C2562), DBA (Vector Labs, catalog# B-1035-5), E-cadherin (Thermo Fisher, catalog# 610182), GFP (Thermo Fisher, catalog# NC9510598), Lef1 (Cell Signaling catalog# 2230S), Lhx1 (Developmental Studies Hybridoma Bank, catalog# 4F2), Lrpprc (Proteintech, catalog# 21175-1-AP), m6A (Abcam, catalog# ab151230 1:250), Mat2A (Proteintech, catalog# 55309-1-AP), Mettl3 (Thermo Fisher, catalog# MA5-27527), Mettl14 (Sigma, catalog# HPA038002), Pax8 (Abcam, catalog# ab189249), Six2 (Proteintech, catalog# 11562-1), Tomm20 (Proteintech, catalog# 11802-1-AP), goat anti-rabbit IgG-HRP (Thermofisher, catalog# G21234), goat anti-mouse-HRP (Thermofisher, catalog# G-21040) and Total Oxphos Rodent Cocktail (Abcam, catalog# ab110413) consisting of complex I- NDUFB8, complex II- SDHB, complex III-UQCRC2, complex IV-MTCO1 and complex V-ATP5A.
Primers
Mettl3 Forward 5’-GTC AGT CAG GAG ATC CTA GAG CTA TT-3’ Reverse 5’-GTC AGC GTC ACT GGC TTT C-3’ Etfb Forward 5’-CTGTCAAGAGGTCATCGACT-3’ Reverse 5’-CACAGAAGGGGTTCATGGAGT-3’ Etfdh Forward 5’-GTGCGACTAACCCTGTC-3’ Reverse 5’-GGATGAACAGTGTAGTGAGTGG-3’
Statistics and reproducibility
For in vivo experiments, N is the number of mice analyzed. For in vitro and ex vivo experiments, N refers to the number of biological replicates. A two-tailed Student’s t-test or Mann–Whitney test was used for comparing two groups, and analysis of variance (ANOVA) was followed by Tukey’s post hoc test for comparing three or more groups. A p-value of <0.05 was considered statistically significant. The sample size, type of statistical analysis, and p values are mentioned in the figure graphs, figure legends, and the results section. The mouse pups were randomly assigned to the drug groups. Investigators were not blinded to the treatment or the genotypes of animals. Immunofluorescence staining on NPCs or tissues was performed at least three times, with a minimum of three biological replicates in each round.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The processed m6A RIP-Seq datasets generated in this study are available as supplementary datasets. The raw sequencing files have been deposited in the NCBI Gene Expression Omnibus repository under accession number GSE294483. All data supporting the findings described in this manuscript are available in the article and in the Supplementary Information and from the corresponding author upon request. Source data are provided with this paper.
References
Ryan, D. et al. Development of the Human Fetal Kidney from Mid to Late Gestation in Male and Female Infants. EBioMedicine 27, 275–283 (2018).
Puelles, V. G. et al. Glomerular number and size variability and risk for kidney disease. Curr. Opin. Nephrol. Hypertens. 20, 7–15 (2011).
Hughson, M., Farris, A. B. 3rd, Douglas-Denton, R., Hoy, W. E. & Bertram, J. F. Glomerular number and size in autopsy kidneys: the relationship to birth weight. Kidney Int. 63, 2113–2122 (2003).
Luyckx, V. A. & Brenner, B. M. The clinical importance of nephron mass. J. Am. Soc. Nephrol. 21, 898–910 (2010).
Wood-Bradley, R. J., Barrand, S., Giot, A. & Armitage, J. A. Understanding the role of maternal diet on kidney development; an opportunity to improve cardiovascular and renal health for future generations. Nutrients 7, 1881–1905 (2015).
Walani, S. R. Global burden of preterm birth. Int. J. Gynaecol. Obstet. 150, 31–33 (2020).
Thapa, P. et al. Prevalence of low birth weight and its associated factors: hospital based cross sectional study in Nepal. PLoS Glob. Public Health 2, e0001220 (2022).
Bertram, J. F., Douglas-Denton, R. N., Diouf, B., Hughson, M. D. & Hoy, W. E. Human nephron number: implications for health and disease. Pediatr. Nephrol. 26, 1529–1533 (2011).
Lee, Y. Q., Collins, C. E., Gordon, A., Rae, K. M. & Pringle, K. G. The relationship between maternal nutrition during pregnancy and offspring kidney structure and function in humans: a systematic review. Nutrients 10, https://doi.org/10.3390/nu10020241 (2018).
Luyckx, V. A. & Brenner, B. M. Low birth weight, nephron number, and kidney disease. Kidney Int. Suppl. 97, S68–S77 (2005).
Zohdi, V. et al. Low birth weight due to intrauterine growth restriction and/or preterm birth: effects on nephron number and long-term renal health. Int. J. Nephrol. 2012, 136942 (2012).
Nachtergaele, S. & He, C. Chemical modifications in the life of an mRNA transcript. Annu. Rev. Genet. 52, 349–372 (2018).
Saletore, Y. et al. The birth of the epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 13, 175 (2012).
Wang, X. et al. Structural basis of N(6)-adenosine methylation by the METTL3–METTL14 complex. Nature 534, 575–578 (2016).
Sledz, P. & Jinek, M. Structural insights into the molecular mechanism of the m(6)A writer complex. Elife 5, https://doi.org/10.7554/eLife.18434 (2016).
Wang, P., Doxtader, K. A. & Nam, Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63, 306–317 (2016).
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G. & Rottman, F. M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247 (1997).
Deng, X., Qing, Y., Horne, D., Huang, H. & Chen, J. The roles and implications of RNA m(6)A modification in cancer. Nat. Rev. Clin. Oncol. 20, 507–526 (2023).
Ramalingam, H. et al. A methionine–Mettl3–N(6)-methyladenosine axis promotes polycystic kidney disease. Cell Metab. 33, 1234–1247 e1237 (2021).
Tsai, Y. C. et al. METTL3-mediated N 6-methyladenosine mRNA modification and cGAS-STING pathway activity in kidney fibrosis. J. Am. Soc. Nephrol. 35, 1312–1329 (2024).
Wang, J. N. et al. Inhibition of METTL3 attenuates renal injury and inflammation by alleviating TAB3 m6A modifications via IGF2BP2-dependent mechanisms. Sci. Transl. Med. 14, eabk2709 (2022).
Yankova, E. et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 593, 597–601 (2021).
Cargill, K. et al. Von Hippel–Lindau acts as a metabolic switch controlling nephron progenitor differentiation. J. Am. Soc. Nephrol. 30, 1192–1205 (2019).
Liu, J. et al. Regulation of nephron progenitor cell self-renewal by intermediary metabolism. J. Am. Soc. Nephrol. 28, 3323–3335 (2017).
Tortelote, G. G., Colon-Leyva, M. & Saifudeen, Z. Metabolic programming of nephron progenitor cell fate. Pediatr. Nephrol. 36, 2155–2164 (2021).
Brown, A. C., Muthukrishnan, S. D. & Oxburgh, L. A synthetic niche for nephron progenitor cells. Dev. Cell 34, 229–241 (2015).
Lindstrom, N. O. et al. Spatial transcriptional mapping of the human nephrogenic program. Dev. Cell 56, 2381–2398 e2386 (2021).
Selberg, S. et al. Discovery of small molecules that activate RNA methylation through cooperative binding to the METTL3–14-WTAP complex active site. Cell Rep. 26, 3762–3771 e3765 (2019).
Kobayashi, A. et al. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell 3, 169–181 (2008).
Scarfe, L. et al. Transdermal measurement of glomerular filtration rate in mice. J. Vis. Exp. https://doi.org/10.3791/58520 (2018).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Cui, J., Wang, L., Ren, X., Zhang, Y. & Zhang, H. LRPPRC: a multifunctional protein involved in energy metabolism and human disease. Front. Physiol. 10, 595 (2019).
Xu, F., Morin, C., Mitchell, G., Ackerley, C. & Robinson, B. H. The role of the LRPPRC (leucine-rich pentatricopeptide repeat cassette) gene in cytochrome oxidase assembly: mutation causes lowered levels of COX (cytochrome c oxidase) I and COX III mRNA. Biochem. J. 382, 331–336 (2004).
Debray, F. G. et al. LRPPRC mutations cause a phenotypically distinct form of Leigh syndrome with cytochrome c oxidase deficiency. J. Med. Genet. 48, 183–189 (2011).
Han, V. X., Tan, T. S., Wang, F. S. & Tay, S. K. Novel LRPPRC mutation in a boy with mild Leigh syndrome, French-Canadian type outside of Quebec. Child Neurol. Open 4, 2329048X17737638 (2017).
Sasarman, F. et al. Tissue-specific responses to the LRPPRC founder mutation in French Canadian Leigh Syndrome. Hum. Mol. Genet. 24, 480–491 (2015).
Jiang, R. et al. A traditional gynecological medicine inhibits ovarian cancer progression and eliminates cancer stem cells via the LRPPRC–OXPHOS axis. J. Transl. Med. 21, 504 (2023).
Zhou, W. et al. The RNA-binding protein LRPPRC promotes resistance to CDK4/6 inhibition in lung cancer. Nat. Commun. 14, 4212 (2023).
Peterson, S. M. et al. Estimation of nephron number in whole kidney using the acid maceration method. J. Vis. Exp. https://doi.org/10.3791/58599 (2019).
Kang, J. et al. Depletion of SAM leading to loss of heterochromatin drives muscle stem cell ageing. Nat. Metab. 6, 153–168 (2024).
Rajabian, N. et al. Methionine adenosyltransferase2A inhibition restores metabolism to improve regenerative capacity and strength of aged skeletal muscle. Nat. Commun. 14, 886 (2023).
Kaiser, P. Methionine dependence of cancer. Biomolecules 10, https://doi.org/10.3390/biom10040568 (2020).
Harris, M., Schuh, M. P., McKinney, D., Kaufman, K. & Erkan, E. Whole exome sequencing in a population with severe congenital anomalies of kidney and urinary tract. Front. Pediatr. 10, 898773 (2022).
Guan, Y. et al. Dnmt3a and Dnmt3b-decommissioned fetal enhancers are linked to kidney disease. J. Am. Soc. Nephrol. 31, 765–782 (2020).
Carpenter, J., Yarlagadda, S., VandenHeuvel, K. A., Ding, L. & Schuh, M. P. Human nephrogenesis can persist beyond 40 postnatal days in preterm infants. Kidney Int. Rep. 9, 436–450 (2024).
Rodriguez, M. M. et al. Histomorphometric analysis of postnatal glomerulogenesis in extremely preterm infants. Pediatr. Dev. Pathol. 7, 17–25 (2004).
Mourier, A., Ruzzenente, B., Brandt, T., Kuhlbrandt, W. & Larsson, N. G. Loss of LRPPRC causes ATP synthase deficiency. Hum. Mol. Genet 23, 2580–2592 (2014).
Ruzzenente, B. et al. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 31, 443–456 (2012).
Cuillerier, A. et al. Loss of hepatic LRPPRC alters mitochondrial bioenergetics, regulation of permeability transition and trans-membrane ROS diffusion. Hum. Mol. Genet. 26, 3186–3201 (2017).
Acknowledgements
The work is supported by the National Institutes of Health (5R01DK102572) to V.P. We thank the UT Southwestern Histology Core, the CRI Metabolomics Core, and CD Genomics for providing critical reagents and services. We thank Dr. Chris Lu and Keng-Mean Lin for their help with eGFR measurements.
Author information
Authors and Affiliations
Contributions
H.R., J.A., A.F., P.C-S., N.F., E.G., R.L, K.A., T.C., and V.P. designed the experiments, performed the experiments, and/or analyzed the data. H.R. and V.P. prepared the figures and wrote the manuscript. All the authors contributed critical edits to the paper.
Corresponding author
Ethics declarations
Competing interests
V.P. has served as a scientific consultant for Otsuka Pharmaceuticals, Maze Therapeutics, Travere Therapeutics, and Regulus Therapeutics. V.P. serves as the chair of the Scientific Advisory Panel for the PKD Foundation. V.P. lab has a sponsored research agreement with Regulus Therapeutics, which is unrelated to this work. V.P. has licensed patents (US11168325B2) involving anti-miR-17 for the treatment of ADPKD (Inventors: John R. Androsavich, B. Nelson Chau, and Vishal D. Patel. Current Assignee: Regulus Therapeutics Inc. and University of Texas System), which is unrelated to the current manuscript. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Christoph Freyer and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ramalingam, H., Alvarez, J., Flaten, A. et al. An RNA transmethylation pathway governs kidney nephrogenic potential. Nat Commun 16, 4930 (2025). https://doi.org/10.1038/s41467-025-60097-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-60097-6