Abstract
Cytokine-induced apoptosis inhibitor 1 (CIAPIN1) is a protein that regulates apoptosis and programmed cell death. This research aims to evaluate its potential role in inhibiting breast cancer cell proliferation, migration, and glycolysis and uncover its underlying molecular mechanism. We collected breast cancer tissue samples from eight patients between January 2019 and June 2023 in our Hospital to analyse CIAPIN1 expression. We transfected human breast cancer cell lines (MCF7, MDA-MB-231, MDA-MB-453, and MDA-MB-468) with siRNA of CIAPIN1. Finally, we determined protein expression using RT-qPCR and Western blotting. CIAPIN1 expression was elevated in both breast cancer tissue and serum. Overexpression of CIAPIN1 detected in the breast cancer cell lines MCF7 and MDA-MB-468. In addition, CIAPIN1 overexpression increased cell proliferation and migration rate. CIAPIN1 downregulation suppressed cell proliferation while elevated cellular apoptosis, reactive oxygen species (ROS) production and oxidative stress in breast cancer cells. Moreover, CIAPIN1 inhibition remarkably suppressed pyruvate, lactate and adenosine triphosphate (ATP) production and reduced the pyruvate kinase M2 (PKM2) protein expression and phosphorylation of signal transducer and activator of transcription 3 (STAT3) in breast cancer cells. Downregulation of CIAPIN1 suppresses cell proliferation, migration and glycolysis capacity in breast cancer cells by inhibiting the STAT3/PKM2 pathway.
Similar content being viewed by others
Introduction
Breast cancer is the leading cause of cancer-related death in women worldwide, accounting for 260,000 cases in the past two years1. In China, over three million women are diagnosed with breast cancer annually. While early non-metastatic breast cancer patients have a cure rate of approximately 70–80%, late-stage breast cancer patients with distant organ metastasis are currently considered untreatable by available therapies2. Various studies have shown significant differences in the mortality rates of breast cancer between different countries. Developed countries have lower rates of breast cancer mortality compared to underdeveloped regions. This could be primarily attributed to early detection strategies, timely diagnosis, and improved access to effective treatments1,3. Breast cancer is currently detected through a combination of blood tumor marker screening, X-ray, and ultrasound imaging, which results in an accuracy rate of over 80%4,5. However, early diagnosis of breast cancer is still a significant issue in underdeveloped regions that lack access to advanced diagnostic and therapeutic methods. Therefore, identifying potential biomarkers that indicate the presence of breast cancer is crucial in reducing its incidence and improving treatment efficacy.
STAT3 is an oncogenic transcription factor active in various human malignancies and controls many cellular processes, including cell proliferation, apoptosis, survival, immune response, angiogenesis, and chemotherapy resistance6. A previous study has shown that the pathways associated with STAT3 are a crucial downstream mechanism of the VEGFR2 receptor7. An early study has reported promising anti-cancer properties of apatinib by inhibiting the VEGFR2/STAT3 signaling pathway8. In addition, STAT3 has been found to promote aerobic glycolysis and stimulate the growth of glioblastoma, bladder carcinoma, and breast cancer9,10,11. Prior investigations have indicated that STAT3 can enhance the glycolytic metabolism of multiple cancerous growths by controlling the expression of key glycolytic enzymes and regulators of glycolysis12. However, there is a need to explore natural compounds that can inhibit STAT3 phosphorylation to target cancer progression with minimal side effects.
Malignant tumor cells behave differently than normal cells by upregulating aerobic glycolysis and producing large amounts of lactate and macromolecular precursors regardless of their oxygen status, known as the “Warburg effect”13. Studies have shown that glycolysis plays a significant role in regulating tumorigenesis and therapeutic sensitivity in melanoma14. For instance, glycerol 3-phosphate dehydrogenase facilitates glycolysis, thereby regulating melanoma cells’ tumorigenesis15. Additionally, the transcription factor 4 was found to sensitize melanoma cells to vemurafenib by inhibiting glucose transporter 3-mediated glycolysis. Glucose transporter 1 (GLUT 1), a rate limiting factor of glucose transport, is frequently upregulated in multiple solid tumors and associated with aggressive phenotypes and poor survival16,17. The cytokine-induced apoptosis inhibitor 1 (CIAPIN1), also known as anamorsin, is a protein that prevents apoptosis or programmed cell death. It is naturally present in various organs, particularly those that can regenerate or proliferate18. CIAPIN1 has shown potential as a diagnostic and therapeutic target in several human malignancies19. Its expression has been linked to oncogenes or tumor suppressors in different types of solid tumors19. However, expression, biological roles, and mechanism of CIAPIN1 in breast cancer remain unexplored. Additionally, its involvement in breast cancer is unknown. Therefore, the research aimed to investigate the potential impact of CIAPIN1 in breast cancer and its underlying molecular mechanism on breast cancer.
Materials and methods
Clinical specimens
We collected eight (8) cases of breast cancer tissues and the adjacent tissues, which were from surgically resected samples of breast cancer patients from January 2019 to June 2023 in our Hospital. We also obtained serum samples from 15 breast cancer patients and 15 healthy controls. The patients had not previously received either radiation or chemotherapy. The samples were collected and preserved in liquid nitrogen. The experimental protocols were approved by Shanghai Pudong Hospital; the Fudan University Pudong Medical Centre Institutional Review Board (IRB) granted this study a waiver from full IRB review. All methods were performed in accordance with the relevant guidelines and regulations.
Histology and IHC analysis
Breast cancer and adjacent tissues were stained with hematoxylin and eosin (HE). Primary antibodies against CIAPIN1 (1:100; ab154904, Abcam) have been applied to the sections and incubated overnight at 4 ℃. The slides were treated with a secondary goat anti-rabbit antibody for 1 h at room temperature before being stained with 3,3′-Diaminobenzidine (DAB). Expression of CIAPIN1 was evaluated as the percentage of positive cells and staining intensity. The percentage of positive cells was evaluated as follows: staining percentage ≤ 1% of total cells: 0 score; staining percentage 2 ~ 25%: 1 score, staining score 26 ~ 50%: 2 score; staining percentage 51 ~ 75%: 3 score; and staining percentage > 75%: 4 score. Intensity was graded as follows: no signal: 0 score; weak staining: 1 score; moderate staining: 2 score; and strong staining: 3 score. The total score was 0 ~ 12 which was calculated from multiplying scores of percentage of positive cells and staining intensity: negative (score 0 ~ 1), weak (score 2 ~ 4), moderate (score 5 ~ 8), or strong (score 9 ~ 12)20.
Enzyme-linked immunosorbent assay (ELISA)
Serum was collected from 15 breast cancer patients and 15 healthy controls. The concentrations of CIAPIN1 (P9446, FineTest, Wuhan, China) were measured in serum using ELISA. The concentrations were represented in µg/mL.
Cell culture
Several human breast cancer cell lines, including MCF7, MDA-MB-231, MDA-MB-453, and MDA-MB-468, were acquired from the American Type Tissue Culture Collection (ATCC) in Rockville, MD, USA. These cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 µg/mL of streptomycin, and 100 U/mL of penicillin.
Plasmid construct, siRNA, and transfection
To overexpress the CIAPIN1 gene in breast cancer cells, human CIAPIN1-cDNA sequences were cloned into lentiviral vector pCDH-CMV (LM-8070, LMAI Bio) and packaged in 293 T cells. To silence the CIAPIN1 gene, three siRNAs toward the human CIAPIN1 gene were also purchased from Genepharma: CIAPIN1-87, CIAPIN1-299 and CIAPIN1-505 (Table 1). The lentiviral vector pCDH-CMV was transfected into MCF7 and MDA-MB-468 cells, and the siRNA sequence was transfected into MDA-MB-231 and MDA-MB-453 cells by using Lipo2000 according to the manufacturer’s instructions. The effectiveness of transfection was assessed 48 h after transfection using RT-qPCR and Western blot.
MTT assay
Breast cancer cells were placed in 96-well plates at an amount of 1 × 104 cells/well. MTT solution (1 mg/mL, M1020, Solarbio, Beijing, China) was applied to the cells and incubated for 4 h at 37 degrees Celsius. The formazan crystal was dissolved with 100 μL of DMSO in each well. Finally, the amount of absorption at 570 nm wavelength was determined with a microplate reader.
5-ethynyl-20-deoxyuridine (EdU) assay
Breast cancer cells were placed in 96-well plates at an amount of 5 × 104 cells/well for the 5-ethynyl-20-deoxyuridine (EdU) assay. The cells were then incubated with EdU solution at a concentration of 50 μM and fixed with 4% paraformaldehyde. After that, the nuclei were stained with DAPI dye. Images of the cells were captured using an Olympus microscope (plate_number_1) and processed using ImageJ software from NIH Image, Bethesda, MD, USA.
Cell migration assay
The Transwell test was used to investigate the migratory capabilities of breast cancer cells. Transwell chambers (8 μm, 1 × 105 cells/mL) were filled with cells in 100-L DMEM medium (no FBS added). The bottom container held 600 μL of 10% FBS DMEM medium. Following 72 h of incubation, the cells in the upper container were cleaned using a cotton swab. 0.1% crystal violet was used to identify the moving cells. To compute the average number of moved cells, the labelled moving cells were counted from five fields under a light microscope (magnification, × 200).
Cell apoptosis
The apoptotic cells were measured using the Annexin V-FITC assay kit (CA1020, Solarbio, Beijing, China). Breast cancer cells were grown in a 24-well plate (5 × 104 cells per well) and treated for 48 h. Trypsin was then used to digest the cells, and they were treated for 15 min at room temperature with Annexin FITC and propidium iodide (PI). The rate of cell apoptosis was measured using flow cytometry.
Intracellular ROS level evaluation
The cellular ROS production was detected by dihydroethidium (DHE; Beyotime; cat. no. S0063) staining. When superoxide anions enter live cells, they dehydrogenate DHE to generate ethidium, which bonds to RNA or DNA and produces red fluorescence. The higher the degree of intracellular superoxide generation, the more intense the red glow. Breast cancer cells were seeded onto 24-well plates at a density of 1 × 105 cells per well. DHE (10 M) was then applied to the cells, which were incubated at 37 ℃ for 30 min before being treated with 100 nM Se and 200 mM HCY for 24 h. After washing with PBS, the fluorescence of the cells was evaluated using a fluorescent microscope at × 200 magnification.
Measurement of oxidative stress indicators
The breast cancer cells were homogenized (10%, w/v) for measuring oxidative stress biomarkers. The cell lysate was then frozen at −80 ℃ in the refrigerator. A commercial kit was used to assess MDA (S0131S, Beyotime, Shanghai, China), SOD (S0109, Beyotime), and CAT (S0051, Beyotime) activities, and a previously described procedure was utilized to determine the degree of oxidative stress21.
RNA isolation from breast cancer cell and tissue samples
Total RNA was extracted from 90 to 120 mg of individual breast cancer cell samples utilizing the RNeasy Lipid Mini Kit (QIAGEN, Valencia, CA) following the guidelines provided by the manufacturer. RNA extraction from each breast cancer and normal tissue sample involved 5–10 Sects. (20 μm thickness) with a combined weight of around 400 mg. Subsequently, the tissue sections were placed in five micro centrifuge tubes for RNA isolation using the RNeasy Lipid Tissue Mini Kit (QIAGEN, Valencia, CA) following the manufacturer’s specified procedures. Evaluation of RNA concentration (ng/μL) and purity (A260/280) was conducted for every RNA sample through the employment of the Nanodrop 1000 (ThermoFisher Scientific, Wilmington, DE). After assessment, all RNA samples were preserved at −80 ℃ until subjected to qRT-PCR analysis.
Real-time quantitative PCR (RT-qPCR)
To conduct the synthesis of cDNA, a quantity of 1 μg of total RNA was subjected to reverse transcription with the utilization of the High Capacity cDNA Reverse Transcriptase (RT) Kit following the guidelines provided by the manufacturer (ThermoFisher Scientific, Foster City, CA), within a final reaction volume of 20 μL. The ultimate volume of the qRT-PCR reaction amounted to 50 μL, comprising 2 μL of the cDNA sample, 1 μL of each primer mix, 25 μL of iQ SYBR Green Supermix, and 22 μL of nuclease-free water. The internal reference utilized was GAPDH. Table 2 shows the list of PCR primer sequences used in this study. The RT-qPCR settings included 7 min of denaturation at 95 ℃, 40 cycles of 15 s at 95 ℃ and 1 min at 60 ℃. The 2−ΔΔCt approach was used to assess the relative mRNA levels22.
Western blotting
The complete protein was obtained through the utilization of RIPA lysis buffer, while the nuclear protein was derived using Extraction Reagents from Pierce Biotechnology, Inc., located in Rockford, IL, USA. Protein levels were quantified utilizing a BCA protein assay kit manufactured by Beyotime Biotechnology, located in China. Protein specimens (50 μg) were separated using 10% SDS-PAGE and transferred onto PVDF membranes. The membranes were obstructed using a solution containing 5% low-fat milk and subsequently exposed to primary antibodies targeting CIAPIN1 (1:500, ab154904, rabbit polyclonal, Abcam), PKM2 (1:500, ab85555, rabbit polyclonal, Abcam), p-STAT3 (Y705) (1:500, ab267373, rabbit monoclonal, Abcam), STAT3 (1:500, ab68153, rabbit monoclonal, Abcam). The membranes underwent treatment using secondary antibodies linked with HRP. The internal control utilized in this study was GAPDH (1:2000, ab8245, mouse monoclonal, Abcam). Subsequently, the bands were detected using ECL (Thermo, Waltham, MA, USA) and processed through ImageJ software analysis.
Statistical analysis
The data were provided as mean ± SD for quantitative variables or number (percentage) for category variables. The SPSS Software 20.0 was used for all statistical analyses. Quantitative information underwent Student t-test analysis, while categorical data were subjected to χ2 test analysis. Pearson correlation was performed to analyse the correlations of CIAPIN1 with clinical indicators. In cellular studies, the one-way analysis of variance (ANOVA) was used to investigate the variation among three or more groups, followed by the Bonferroni post hoc test. P < 0.05 was considered the criteria for a statistically significant difference.
Results
CIAPIN1 expression was elevated in breast cancer tissues and serum
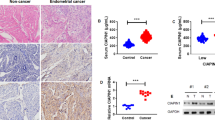
To determine the levels of CIAPIN1 in breast cancer tissues, the study carried out multiple tests, including HE staining and immunohistochemistry. The results showed that CIAPIN1 showed strong (#03) and moderate (#01, #02, #04) expression in four cancer tissues, while all the four non-cancer tissues showed weak CIAPIN1 expression. This indicates CIAPIN1 expression is higher in breast cancer than in non-cancer tissues (Fig. 1A). The mRNA levels of CIAPIN1 in breast cancer tissues and adjacent normal tissues were analysed through RT-qPCR, and increased mRNA levels of CIAPIN1 in breast cancer tissues were observed (Fig. 1B). Western blot analysis indicated higher protein levels of CIAPIN1 in breast cancer tissues than in adjacent normal breast tissues (Fig. 1C). In addition, ELISA measurement demonstrated that the serum concentrations of CIAPIN1 from breast cancer patients were elevated compared to the healthy controls (P < 0.001) (Fig. 1D).
CIAPIN1 expression was upregulated in breast cancer tissues and serum. (A) Representative image of morphological structure by HE staining and CIAPIN1 expression by immunohistochemistry in breast cancer tissues and adjacent tissue (Magnification 200 ×). (B) RT-qPCR was performed to determine the mRNA levels of CIAPIN1 in breast cancer tissues and adjacent tissue. (C) The protein levels of CIAPIN1 in breast cancer tissues (n = 4) and adjacent normal breast tissues (n = 4) were determined by Western blot. (D) ELISA was carried out to measure the serum concentrations of CIAPIN1 from breast cancer (n = 15) and healthy controls (n = 15). The impact of the low (black line) and high (red line) CIAPIN1 expression on (E) Overall survival (OS) and (F) recurrence-free survival (RFS) curves were calculated from the Kaplan‑Meier Plotter. Data are presented as mean ± SD and Dot plots and analyzed using the Student t-test. ***P < 0. 001 vs. control group. N, adjacent normal breast tissues; T, breast cancer tissues.
Moreover, CIAPIN1 accelerates the development of breast cancer and could serve as a predictive biomarker. However, the Kaplan–Meier Plotter was used to calculate the impact of low and high CIAPIN1 expression on the Overall survival (OS) and recurrence-free survival (RFS) curves (Fig. 1E,F). Therefore, our data showed that the expression of CIAPIN1 was upregulated in breast cancer tissues and serum.
Expression of CIAPIN1 in breast cancer cells
The study aimed to explore the expression of CIAPIN1 in different breast cancer cell lines, namely MCF7, MDA-MB-231, MDA-MB-453, and MDA-MB-468, using RT-qPCR and western blot analysis. The findings revealed that CIAPIN1 was more highly expressed in MDA-MB-231 and MDA-MB-453 cell lines compared to MCF7 and MDA-MB-468 cell lines, as shown in Fig. 2A,B. We then transfected MCF7 and MDA-MB-468 cells with pcDNA3.1-CIAPIN1 or vector, and after 48 h, we examined the expression levels of CIAPIN1 using RT-qPCR and western blot analysis. The results demonstrated that high levels of CIAPIN1-OE were detected in MCF7 and MDA-MB-468 cells, as shown in Fig. 2C,D.
Expression of CIAPIN1 in breast cancer cells. (A) The mRNA expression of CIAPIN1 in four breast cancer cell lines (MCF7, MDA-MB-231, MDA-MB-453, and MDA-MB-468) by RT‑qPCR. (B) The representative bands of CIAPIN1 protein in these four breast cancer cell lines. MCF7 and MDA-MB-468 cells were transfected with pcDNA3.1-CIAPIN1 or vector. (C) The levels of CIAPIN1 mRNA were detected by RT-qPCR in MCF7 and MDA-MB-468 at 48 h after transfection. (D) The levels of CIAPIN1 protein were detected by Western blot, and representative gel blots of CIAPIN1 are shown. Data are presented as mean ± SD and analyzed using the T-test. ***P < 0.01 vs. control group.
Overexpression of CIAPIN1 in breast cancer cells promotes cell proliferation and migration rate
We transfected MCF7 and MDA-MB-468 cells with pcDNA3.1-CIAPIN1 or vector and were cultured for 12, 24, 48 and 72 h. The cell viability was assessed by MTT assay in MCF7 and MDA-MB-468 cells, respectively, and an improved cell viability rate was observed with increasing transfected time (Fig. 3A,B). The EdU assay was used to investigate cell proliferation of MCF7 and MDA-MB-468 cells at 24 h after transfection. The results showed that overexpression of CIAPIN1 increased cell proliferation for the MCF7 and MDA-MB-468 cells (Fig. 3C). After quantification of EdU-positive cells relative to DAPI-positive cells, we observed a consistent cell proliferation rate of MCF7 and MDA-MB-468 cells (both, P < 0.001) (Fig. 3D).
Overexpression of CIAPIN1 in breast cancer cells promotes cell proliferation. MCF7 and MDA-MB-468 cells were transfected with pcDNA3.1-CIAPIN1 or vector, and were cultured for 12, 24 48 and 72 h. (A, B) The cell viability was determined by MTT assay in MCF7 and MDA-MB-468 cells, respectively. (C) 5-ethynyl-20-deoxyuridine (EdU) assay was used to assess cell proliferation of MCF7 and MDA-MB-468 cells at 24 h after transfection. (D) Quantification of EdU-positive cells relative to DAPI-positive cells. Data are presented as mean ± SD in triplicates and analyzed using the Student t-test. ***P < 0.01 vs. control group.
The overexpression of CIAPIN1 on cell migration ability was investigated by Transwell assay in MCF7 and MDA-MB-468 cells. The study results demonstrated that CIAPIN1 overexpression enhanced cell migration ability for the MCF7 and MDA-MB-468 cells (Fig. 4A). The same trends were found after quantification of the relative migrated cells (both, P < 0.001) (Fig. 4B).
Overexpression of CIAPIN1 promotes the migration of breast cancer cells. (A) The overexpression of CIAPIN1 on cell migration ability was investigated by Transwell assay in MCF7 and MDA-MB-468 cells. (B) The relative migrated cells were quantified. Data are presented as mean ± SD in triplicates and analyzed using the Student t-test. ***P < 0.01 vs. control group.
CIAPIN1 downregulation via siRNA suppresses cell proliferation in breast cancer cells
Three siRNAs (siRNA-87, siRNA-299 and siRNA-505) targeting CIAPIN1 mRNA were introduced into MDA-MB-231 and MDA-MB-453 cells for transient knockdown of CIAPIN1. The levels of CIAPIN1 mRNA were detected by RT-qPCR in MDA-MB-231 and MDA-MB-453 cells at 48 h after transfection. The results showed that siRNA-299 markedly suppressed the CIAPIN1 mRNA expression compared to siRNA-87 and siRNA-505 in MDA-MB-231 and MDA-MB-453 cells (P < 0.001) (Fig. 5A,B). In addition, the western blot analysis was performed to determine the levels of CIAPIN1 protein in MDA-MB-231 and MDA-MB-453 cells, and the same suppression patterns were observed in MDA-MB-231 and MDA-MB-453 cells (Fig. 5C,D). Moreover, Cell viability was suppressed by CIAPIN1 knockdown in MDA-MB-231 and MDA-MB-453 cells detected by MTT assay 72 h after transfection (Fig. 5E).
Inhibition of CIAPIN1 in breast cancer cells by siRNA suppresses cell proliferation. (A) Three siRNAs targeting CIAPIN1 mRNA (siRNA-87, siRNA-299 and siRNA-505) were introduced into MDA-MB-231 and MDA-MB-453 cells for transient knockdown of CIAPIN1. (A, B) The levels of CIAPIN1 mRNA were detected by RT-qPCR in MDA-MB-231 and MDA-MB-453 cells at 48 h after transfection. (C, D) Western blotting was performed to determine the levels of CIAPIN1 protein in MDA-MB-231 and MDA-MB-453 cells. The expression of CIAPIN1 was normalized against endogenous GAPDH levels. (E) Cell viability was suppressed by CIAPIN1 knockdown in MDA-MB-231 and MDA-MB-453 cells detected by MTT assay 72 h after transfection. Data represent the average of three independent experiments (mean ± SD), and analyzed using ANOVA, followed by the Bonferroni post hoc test for multiple-group comparison. *P < 0.05, **P < 0.01, ***P < 0.001 vs. Control group or si-NC group.
Suppression of CIAPIN1 enhances breast cancer cell apoptosis
MDA-MB-231 and MDA-MB-453 cells were transfected with siRNA (siRNA-299) targeting CIAPIN1 mRNA for a further 24 h culture. Cell apoptosis was assessed by Annexin V-FITC double staining and analysed by flow cytometry. The results demonstrated that inhibiting CIAPIN1 expression increased the apoptosis rate in MDA-MB-231 and MDA-MB-453 cells (Fig. 6A). In addition, a consistently increased apoptosis rate was observed after calculating the apoptotic rate using the sum of the early and late apoptotic cells (both, P < 0.001) (Fig. 6B).
Inhibition of CIAPIN1 enhances breast cancer cell apoptosis. MDA-MB-231 and MDA-MB-453 cells were transfected with siRNA targeting CIAPIN1 mRNA (siRNA-299) for a further 24 h culture. (A) Cell apoptosis was assessed by Annexin V-FITC double staining and analyzed by flow cytometry. The representative images of flow cytometry are shown. (B) The apoptotic rate was calculated by the sum of the early apoptotic cells (lower right quadrant ) and the late apoptotic cells (upper right quadrant). Data represent the average of three independent experiments (mean ± SD), and analyzed using Student t test.. ***P < 0.001 vs. si-NC group.
Inhibition of CIAPIN1 enhances cellular ROS production and oxidative stress in breast cancer cells
To investigate the effect of CIAPIN1 inhibition on cellular ROS production and oxidative stress in breast cancer cells, we used dihydroethidium (DHE) stained MDA-MB-231 and MDA-MB-453 cells. The study results showed that CIAPIN1 inhibition enhanced cellular ROS production in MDA-MB-231 and MDA-MB-453 cells (Fig. 7A). After quantification of DHE-positive cells relative to DAPI-positive cells, the same trends of cellular ROS production were observed (both, P < 0.001) (Fig. 7B). Three oxidative stress indicators were measured in lysate of MDA-MB-231 and MDA-MB-453 cells using the colorimetric method, including malondialdehyde (MDA), superoxide dismutase (SOD) and catalase (CAT). Our data demonstrated that CIAPIN1 inhibition increased MDA levels and decreased SOD and CAT levels in MDA-MB-231 and MDA-MB-453 cells (all, P < 0.001) (Fig. 7C–E).
Inhibition of CIAPIN1 enhances cellular ROS production and oxidative stress in breast cancer cells. (A) Representative images of DHE staining of MDA-MB-231 and MDA-MB-453 cells. (B) Quantification of DHE-positive cells relative to DAPI-positive cells. Three oxidative stress indicators were measured in the lysate of MDA-MB-231 and MDA-MB-453 cells using the colourimetric method, including (C) MDA, (D) SOD and (E) CAT. Data represent the average of three independent experiments (mean ± SD), and analyzed using Student t test.. ***P < 0.001 vs. si-NC group.
Inhibition of CIAPIN1 suppresses glycolysis in breast cancer cells through STAT3/PKM2 pathway
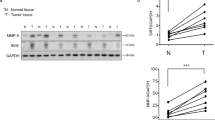
To evaluate the effect of CIAPIN1 inhibition on glycolysis in breast cancer cells through the STAT3/PKM2 pathway, we measured the energy metabolism by inhibition of CIAPIN1 in the lysate of MDA-MB-231 and MDA-MB-453 cells on pyruvate, lactate and adenosine triphosphate (ATP). The study data showed that CIAPIN1 inhibition remarkably suppressed pyruvate, lactate and ATP production in MDA-MB-231 and MDA-MB-453 cells (all, P < 0.001) (Fig. 8A–C). Next, we performed the western blot analysis to investigate the possible involvement of CIAPIN1 inhibition suppresses glycolysis through the STAT3/PKM2 pathway. Our results demonstrated that CIAPIN1 inhibition potentially suppressed the expression of PKM2 and p-STAT3 in MDA-MB-231 and MDA-MB-453 cells (Fig. 8D). Quantification analysis showed that inhibiting CIAPIN1 reduced the PKM2 protein expression and phosphoryl ation of STAT3 (all, P < 0.001) (Fig. 8E,F).
Inhibition of CIAPIN1 suppresses glycolysis in breast cancer cells through STAT3/PKM2 pathway. The energy metabolism by inhibition of CIAPIN1 was measured in the lysate of MDA-MB-231 and MDA-MB-453 cells on (A) pyruvate, (B) lactate and (C) ATP. (D) Representative gel blots depicting levels of PKM2 (normalized to GAPDH) and phosphorylated STAT3 (p-STAT3, normalized to total STAT3). Quantification analysis shows that Inhibition of CIAPIN1 reduced the PKM2 protein expression and phosphorylation of STAT3 (E, F). Data represent the average of three independent experiments (mean ± SD). ***P < 0.001 vs. si-NC group.
Overexpression of CIAPIN1 promotes glycolysis in breast cancer cells through STAT3/PKM2 pathway
We conducted an experiment using MCF7 and MDA-MB-468 cells to investigate the impact of CIAPIN1 overexpression on breast cancer cells through the STAT3/PKM2 pathway. The MCF7 and MDA-MB-468 cells were overexpressed with CIAPIN1 and then co-incubated with a STAT3 inhibitor STAT3-IN-3 (1 μM; HY-128588, MedChemExpress) for an additional 24 h. The results indicated that CIAPIN1 overexpression notably increased the expression of PKM2 and p-STAT3 in MCF7 and MDA-MB-468 cells. Conversely, the STAT3 inhibitor STAT3-IN-3 effectively decreased the levels of PKM2 and p-STAT3 (all, P < 0.001) (Fig. 9A). Further quantification analysis demonstrated that inhibiting CIAPIN1 reduced the levels of PKM2 protein and the phosphorylation of STAT3 (Fig. 9B,C).
Overexpression of CIAPIN1 promotes glycolysis in breast cancer cells through STAT3/PKM2 pathway. The MCF7 and MDA-MB-468 cells were overexpressed with CIAPIN1 and co-incubated with a STAT3 inhibitor STAT3-IN-3 (1 μM; HY-128588, MedChemExpress) for further 24 h. (A) Representative gel blots depicting levels of PKM2 (normalized to GAPDH) and phosphorylated STAT3 (p-STAT3, normalized to total STAT3). (B, C) Quantification analysis shows that inhibition of CIAPIN1 reduced the PKM2 protein expression and phosphorylation of STAT3. The energy metabolism by overexpression of CIAPIN1 was measured in the lysate of MCF7 and MDA-MB-468 cells on (D) pyruvate, (E) lactate and (F) ATP. Data represent the average of three independent experiments (mean ± SD). ***P < 0.001 vs. Vector group; ###P < 0.001 vs. CIAPIN1-OE group.
We also measured the energy metabolism in the lysate of MCF7 and MDA-MB-468 cells. Our findings revealed that overexpression of CIAPIN1 enhanced the expression of pyruvate, lactate, and ATP. At the same time, the STAT3 inhibitor STAT3-IN-3 significantly reduced the levels of pyruvate, lactate, and ATP in MCF7 and MDA-MB-468 cells (all, P < 0.001) (Fig. 9D–F).
Discussion
We conducted a study to examine the impact of CIAPIN1 on breast cancer. Our findings revealed that CIAPIN1 was elevated in breast cancer tissues, cells, and serum. Overexpression of CIAPIN1 was observed to accelerate cell proliferation and migration rate in breast cancer cells. On the other hand, when CIAPIN1 was downregulated, it suppressed cell proliferation and promoted apoptosis in breast cancer cells. In addition, CIAPIN1 inhibition increased cellular ROS production and oxidative stress in breast cancer cells. Moreover, CIAPIN1 attenuation suppressed glycolysis by inhibiting the STAT3/PKM2 pathway, whereas CIAPIN1 overexpression promotes glycolysis in breast cancer cells. Therefore, our research data demonstrated that CIAPIN1 could be a potential therapeutic target to identify breast cancer and improve treatment efficacy.
CIAPIN1 is a regulatory molecule located on the long arm of chromosome 16q21. Shibayama et al.18 discovered that it plays a role in the rat sarcoma signal transduction pathway, distinct from apoptotic B cell lymphoma 2 or the cysteine-dependent aspartate-directed protease family. CIAPIN1 is present in various healthy fetal and adult tissues and is expressed in differentiated and metabolically active tissues23. However, its expression is inhibited in specific malignant tissues, such as gastric cancer and clear cell renal cell carcinoma24,25. The current research found that CIAPIN1 was upregulated in breast cancer tissues and patient’s serum. In addition, high levels of CIAPIN1 were observed in the breast cancer cell lines, such as MCF7 and MDA-MB-468 cells.
The growth of tumors is influenced by various factors such as cell migration, invasion, cell cycle, and proliferation. In clear-cell renal cell carcinoma, DLGAP5 is up regulated, and its knockdown has been shown to suppress cell viability, proliferation, migration, and invasion26. In ovarian cancer cells, microRNA-409-5p has been found to inhibit cell proliferation and induce G2/M phase arrest and apoptosis by targeting DLGAP527. Furthermore, a study has reported that the knockdown of DLGAP5 inhibited breast cancer cell proliferation28. In the current study, we conducted an experiment where we transfected MCF7 and MDA-MB-468 cells with pcDNA3.1-CIAPIN1 or vector. We used a EdU assay to investigate the cell proliferation of MCF7 and MDA-MB-468 cells 24 h after transfection. We observed that overexpression of CIAPIN1 increased cell proliferation for both cell types, as shown in Fig. 3C. After quantifying EdU-positive cells relative to DAPI-positive cells, we observed a consistent cell proliferation rate for both cell types (Fig. 3D). Further, we investigated the effect of CIAPIN1 overexpression on cell migration ability by using Transwell assay in MCF7 and MDA-MB-468 cells. Our study showed that CIAPIN1 overexpression enhanced cell migration ability in both cell types (Fig. 4A). We found the same trends after quantifying the relative migrated cells (Fig. 4B). On the other hand, the downregulation of CIAPIN1 suppressed cell proliferation (Fig. 5) while enhancing cellular apoptosis (Fig. 6), ROS production and oxidative stress in breast cancer cells (Fig. 7).
STAT3 is an essential protein that promotes the growth and survival of cancer cells. When STAT3 is suppressed, cancer cell growth is inhibited, the cells undergo programmed cell death, and they become less invasive and more sensitive to treatment29. Our research shows that CIAPIN1, a protein inhibiting breast cancer growth, can also impede STAT3 activity by reducing phosphorylation. A recent study by Yao et al. found that STAT3 interacts with the promoter region of hnRNPA1, a protein involved in the production of PKM2. This protein promotes glycolysis and enhances its production. This finding provides new insights into how cancer cells promote their survival and growth through metabolic pathways30,32,32. However, the study results showed that CIAPIN1 inhibition reduces the PKM2 expression levels via downregulation of p-STAT3 in MDA-MB-231 and MDA-MB-453 cells. Moreover, CIAPIN1 overexpression notably increased the expression of PKM2, p-STAT3, pyruvate, lactate, and ATP. At the same time, the STAT3 inhibitor STAT3-IN-3 significantly reduced the levels of PKM2, p-STAT3, pyruvate, lactate, and ATP in MCF7 and MDA-MB-468 cells. There are some limitations in this study. Firstly, we used only four cell lines (MCF7, MDA-MB-231, MDA-MB-453, and MDA-MB-468) to evaluate the role of CIAPIN1 in breast cancer. Secondly, further in vivo validation of the function of CIAPIN and its relationship with the STAT3/PKM2 pathway is necessary.
Conclusion
The current research shows that CIAPIN1 can impede breast cancer cells' proliferation, migration and glycolysis capacity by suppressing the STAT3/PKM2 pathway. This could potentially be used as a therapeutic approach for treating breast cancer. Further research will be conducted in the future to validate these findings, using more breast cancer cell lines and in vivo settings.
Data availability
The data from this study is available upon a reasonable request from Dr. Yongping Li at liyongpingdt@sina.com.
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
Harbeck, N. et al. Breast cancer. Nat. Rev. Dis. Primers 5, 66 (2019).
Marcadis, A. R., Morris, L. G. T. & Marti, J. L. Relative survival with early-stage breast cancer in screened and unscreened populations. Mayo Clin. Proc. 97, 2316–2323 (2022).
Kratkiewicz, K., Pattyn, A., Alijabbari, N. & Mehrmohammadi, M. Ultrasound and photoacoustic imaging of breast cancer: Clinical systems, challenges, and future outlook. J. Clin. Med. 11, 1165 (2022).
Sardanelli, F. et al. Magnetic resonance imaging before breast cancer surgery: Results of an observational multicenter international prospective analysis (MIPA). Eur. Radiol. 32, 1611–1623 (2022).
Tošić, I. & Frank, D. A. STAT3 as a mediator of oncogenic cellular metabolism: Pathogenic and therapeutic implications. Neoplasia 23, 1167–1178 (2021).
Bartoli, M. et al. Vascular endothelial growth factor activates STAT proteins in aortic endothelial cells. J. Biol. Chem. 275, 33189–33192 (2000).
Zheng, B. et al. VEGFR2 promotes metastasis and PD-L2 expression of human osteosarcoma cells by activating the STAT3 and RhoA-ROCK-LIMK2 pathways. Front. Oncol. 9, 543562 (2020).
Li, H., Liang, Q. & Wang, L. Icaritin inhibits glioblastoma cell viability and glycolysis by blocking the IL-6/Stat3 pathway. J. Cell. Biochem. 120, 7257–7264 (2018).
Cao, D. et al. Retinoic acid-related orphan receptor C regulates proliferation, glycolysis, and chemoresistance via the PD-L1/ITGB6/STAT3 signaling axis in bladder cancer. Cancer Res. 79, 2604–2618 (2019).
Sola-Penna, M. et al. Serotonin activates glycolysis and mitochondria biogenesis in human breast cancer cells through activation of the Jak1/STAT3/ERK1/2 and adenylate cyclase/PKA, respectively. Br. J. Cancer 122, 194–208 (2020).
Li, M. et al. STAT3 regulates glycolysis via targeting hexokinase 2 in hepatocellular carcinoma cells. Oncotarget 8, 24777–24784 (2017).
Heiden, M. G. V., Cantley, L. C. & Thompson, C. B. Understanding the warburg effect: The metabolic requirements of cell proliferation. Science 22, 1029–1033 (2009).
Yue, S. W. et al. m6A-regulated tumor glycolysis: new advances in epigenetics and metabolism. Mol Cancer. 22(1), 137 (2023).
Ding, X. et al. Sperm-specific glycolysis enzyme glyceraldehyde-3-phosphate dehydrogenase regulated by transcription factor SOX10 to promote uveal melanoma tumorigenesis. Front. Cell Dev. Biol. 25(9), 610683 (2021).
Fu, X. et al. Mechanistic study of human glucose transport mediated by GLUT1. J. Chem. Inf. Model. 28, 517–526 (2016).
Wang, J. et al. Glucose transporter GLUT1 expression and clinical outcome in solid tumors: A systematic review and meta-analysis. Oncotarget. 8, 16875–16886 (2017).
Shibayama, H. et al. Identification of a cytokine-induced antiapoptotic molecule anamorsin essential for definitive hematopoiesis. J. Exp. Med. 199, 581–592 (2004).
Li, X., Wu, K. & Fan, D. CIAPIN1 as a therapeutic target in cancer. Expert Opin. Targets. 14, 603–610 (2010).
Shi, H. et al. Expression of CIAPIN1 in human colorectal cancer and its correlation with prognosis. BMC Cancer 10, 477 (2010).
Ma, J. Y. et al. The protective effects of echinacoside on oxidative stress injury in vascular dementia rats. Chin. Pharmacol. Bull. 30, 638–642 (2014).
Zhu, X. et al. ANGPTL4 suppresses the profibrogenic functions of atrial fibroblasts induced by angiotensin II by up-regulating PPARγ. Iran J Basic Med. Sci. 26(5), 587–593 (2023).
Hao, Z. et al. Subcellular localization of CIAPIN1. J. Histochem. Cytochem. Off. J. Histochem. Soc. 54, 1437–1444 (2006).
Li, X., Hao, Z. & Fan, R. CIAPIN1 inhibits gastric cancer cell proliferation and cell cycle progression by downregulating CyclinD1 and upregulating P27. Cancer Biol. Ther. 6, 1539–1545 (2007).
He, L., Wang, H. & Jin, H. CIAPIN1 inhibits the growth and proliferation of clear cell renal cell carcinoma. Cancer Lett. 276, 88–94 (2009).
Feng, Y. et al. Pan-cancer analysis and experiments with cell lines reveal that the slightly elevated expression of DLGAP5 is involved in clear cell renal cell carcinoma progression. Life Sci. 287, 120056 (2021).
Li, W. et al. MicroRNA-409-5p inhibits cell proliferation, and induces G(2)/M phase arrest and apoptosis by targeting DLGAP5 in ovarian cancer cells. Oncol. Lett. 24, 261 (2022).
Zhu, K., Yi, C. & Tong, C. circ_0058063 promotes breast cancer progression by upregulating DLGAP5 via sponging miR-557. Cancer Biomark. 39(1), 1–13 (2024).
Wong, A. L. et al. Do STAT3 inhibitors have potential in the future for cancer therapy?. Expert Opin. Invest. Drugs. 26(8), 883–887 (2017).
Yao, A. et al. PKM2 promotes glucose metabolism through a let-7a-5p/Stat3/hnRNP-A1 regulatory feedback loop in breast cancer cells. J. Cell. Biochem. 120(4), 6542–6554 (2019).
David, C. J., Chen, M., Assanah, M., Canoll, P. & Manley, J. L. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 63(7279), 364 (2010).
Clower, C. V. et al. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc. Natl. Acad. Sci. 107(5), 1894–1899 (2010).
Acknowledgements
We thank the patients for their specimens and consent.
Funding
The Science and Technology Development Fund of Shanghai Pudong New Area (PKJ2023-Y08) supported this study.
Author information
Authors and Affiliations
Contributions
HY: conceptualization, methodology, investigation, formal analysis, data curation. MZ, JL and ST: conceptualization, methodology, investigation, formal analysis. HZ: investigation, data curation, project administration. QW, and BP: methodology, data interpretation. YL: conceptualization, methodology, supervision. All authors contributed to the writing of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The experimental protocols were approved by Shanghai Pudong Hospital; the Fudan University Pudong Medical Centre Institutional Review Board (IRB) granted this study a waiver from full IRB review. All methods were performed in accordance with the relevant guidelines and regulations.
Consent to participate
Written informed consent was obtained from all subjects involved in the study.
Consent to publish
Patients was agreed to publish their specimen data.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yuan, H., Zhong, M., Liu, J. et al. Downregulation of CIAPIN1 regulates the proliferation, migration and glycolysis of breast cancer cells via inhibition of STAT3 pathway. Sci Rep 14, 20794 (2024). https://doi.org/10.1038/s41598-024-71405-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-71405-3