Abstract
Topologically associated domains (TADs) restrict promoter-enhancer interactions, thereby maintaining the spatiotemporal pattern of gene activity. However, rearrangements of the TADs boundaries do not always lead to significant changes in the activity pattern. Here, we investigated the consequences of the TAD boundaries deletion on the expression of developmentally important genes encoding tyrosine kinase receptors: Kit, Kdr, Pdgfra. We used genome editing in mice to delete the TADs boundaries at the Kit locus and characterized chromatin folding and gene expression in pure cultures of fibroblasts, mast cells, and melanocytes. We found that although Kit is highly active in both mast cells and melanocytes, deletion of the TAD boundary between the Kit and Kdr genes results in ectopic activation only in melanocytes. Thus, the epigenetic landscape, namely the mutual arrangement of enhancers and actively transcribing genes, is important for predicting the consequences of the TAD boundaries removal. We also found that mice without a TAD border between the Kit and Kdr genes have a phenotypic manifestation of the mutation — a lighter coloration. Thus, the data obtained shed light on the principles of interaction between the 3D chromatin organization and epigenetic marks in the regulation of gene activity.
Similar content being viewed by others
Introduction
Genome folding into topologically associated domains (TADs) and compartments is considered to be an important level of gene regulation, affecting the frequency of contacts between regulatory elements and gene promoters. TADs as functional units have been proposed to prevent unwanted promoter-enhancer contacts and narrow down the enhancer search space. The functional importance of TADs for gene regulation was emphasized by analyzing disease-associated mutations, disrupting their boundaries. Structural variants, crossing TAD boundaries, can provoke pathogenic gene misexpression through the rewiring of promoter-enhancer interactions (see review1,2). Deletions have the potential to cause TADs fusion, resulting in ectopic contacts. In T cell acute lymphoblastic leukemia disruption of CTCF-associated boundary allows the proto-oncogene activation3. Another example is limb malformation, triggered by deletion of a TAD boundary at the Ihh/Epha4/Pax3 locus4. Inversions and translocations were shown to cause TADs shuffling4,5,6,7. In some cases, this shuffling triggers an enhancer adoption followed by ectopic activation of the gene4. In other studies, the pathogenic effect of the inversion results from the loss of expression of one of the genes with no enhancer adoption8. Overall, the susceptibility of a gene to an aberrant activation by an ectopic enhancer is different and partially depends on the gene’s chromatin status6.
Although altered TADs architecture is indeed associated with some pathogenic phenotypes9,10, genome wide disappearance of TADs insulation upon CTCF or cohesin depletion does not cause global transcriptional effects11,12. Besides, in a number of studies TADs fusion had only mild effects on gene expression pattern, as enhancers inside the newly formed TAD retained their specificity, for example in Kcnj2–Sox9 locus13 and in Shh locus14. Unlike the deletion of CTCF sites, the repositioning of a TAD boundary by an inversion redirected enhancer activity and induced gene misexpression, highlighting the role of an entire TAD substructure with its epigenetic landscape in altering gene regulation13. Overall, how the regulatory environment inside the TAD affects the output of a boundary disruption remains an open question.
To test the effect of TAD boundary loss in different regulatory contexts, we investigated the Kit locus. The region comprises developmental regulators Pdgfra, Kit and Kdr, each located in the corresponding TAD, demarcated by strong domain boundaries. Mutations at this locus are often associated with malignant processes. Amplification of Pdgfra, Kit or Kdr is common among glioblastomas15,16. In a subset of gliomas, Pdgfra is aberrantly activated by an ectopic enhancer, which is associated with reduced CTCF binding10. Kit and Pdgfra mutations, resulting in the constitutive activation of these genes, are often found in gastrointestinal tumors17. Aberrant activation of Kit and Pdgfra by the oncogene-induced super-enhancers is essential for leukemic cell growth and survival18. Finally, Kdr is considered one of the most critical pro-angiogenic factors in tumor angiogenesis. Kdr expression is upregulated in cancer cells through an autocrine loop with its ligand19. Thus, maintaining the integrity of the chromatin structure within the Kit locus appears to be important, since mis-interaction of enhancers with proto-oncogenes can activate oncogenic signals.
Here, we generated a series of genome-edited mice with deletions of boundary and intra-TAD CTCF sites and used primary cell cultures to track the effect of boundary perturbations. Using cell types with contrasting activity of Pdgfra, Kit and Kdr allowed us to test the role of boundary insulation in different regulatory contexts. The level of architectural changes varied from a marginal increase of inter-TAD contacts across the Pdgfra/Kit boundary, to the disrupted insulation between Kit/Kdr TADs. Loss of insulation between Kit and Kdr TADs resulted in cell type-specific ectopic gene activation. The boundary disruption was associated with a specific phenotype in mice — a lighter coat color. Collectively, our findings support the idea that the structural and functional outcome of CTCF binding sites (CBSs) deletions depends on the epigenetic landscape of neighboring TADs.
Results
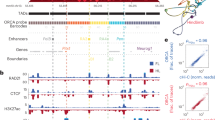
Regulation of the genes Pdgfra, Kit, Kdr is tissue-specific
The Kit gene has been attracting the attention of researchers for a long time, as its mutations induce a clearly visible dominant white-spotting phenotype20,21,22. Kit as well as its flanking genes Pdgfra and Kdr encode receptor tyrosine kinases, which participate in fundamental cellular processes in metazoans23, such as signal transduction from growth factors and cytokines, cell proliferation and differentiation, etc. Consistent with their function, each of the genes features a distinct tissue-specific expression. Pdgfra-positive cells can be found in a wide array of mesenchymal tissues24,25,26. Kit is expressed among several tissues, particularly in mast cells and melanocytes. Our transcriptome analysis reveals Kit being among the most expressed genes in mast cells (Fig. 1a). Kdr is an endothelial-specific receptor and its expression is almost exclusively restricted to the blood vessels27,28. Our data (Fig. 1a) is consistent with the expected expression patterns.
a Gene expression in mast cells, melanocytes and mouse embryonic fibroblasts (MEFs) revealed by RNA-seq (n = 3 independent primary cell cultures per group). The height of a column: an expression level in transcripts per million (TPM), the width of a column: an amount of genes with the exact expression level. Genes Pdgfra, Kit, Kdr demonstrate contrasting expression in the chosen cell types. The center mark in the box represents the median, bounded by the 25th and 75th percentiles, and the whiskers are the most extreme data point, which is no more than 1.5 times the interquartile range. Source data are provided as a Source Data file; b Hi-C maps (5 kb resolution) demonstrate that genes Pdgfra, Kit, Kdr are each located in a distinct TAD with minor features being specific to a cell type. Color bars reflect the interaction counts; c Expression and regulatory context of the locus in the cell types. Regions which we refer to as enhancers are highlighted yellow in H3K27ac track; d Research design. Using CRISPR/Cas9 we generated mutant mouse strains with a TAD boundary deletion. The mice were used to obtain primary cell cultures for a further analysis of 3D genome organization and gene expression; e Schematic view of the tissue-specific TAD structures with emphasis on enhancer localisation relative to genes and expression level (silent genes are coloured gray).
The contrast expression of the genes makes the Kit locus a good model to evaluate an enhancer hijacking. We focused on three cell types: fibroblasts (Pdgfra is expressed, Kit is inactive), mast cells (Kit is among top 0.3% of the expressed genes, Kdr is inactive), and melanocytes (Kit is among top 3% of the most expressed genes, Kdr is weakly expressed).
Three adjacent TADs define 3D structure of the Kit locus
To characterize the TADs organization we performed capture Hi-C (cHi-C) in mouse embryonic fibroblasts (MEFs), mast cells and melanocytes. Our data revealed that the region is organized into three adjacent TADs with Pdgfra, Kit and Kdr each located in a separate TAD (Fig. 1b).
Notably, the Pdgfra gene body overlaps an extended region between Pdgfra and Kit TADs, while its promoter and the first exon are located immediately upstream of the Pdgfra TAD boundary. As to Kit and Kdr, their coding regions are located within the corresponding TADs.
The Kit TAD is heterogeneous in spatial contacts with two distinct self-interacting regions in mast cells and melanocytes. We noted that in mast cells the insulation between these regions appears to coincide with the Kit terminator, while in melanocytes it corresponds to the Kit promoter. The observed insulation could be associated with transcription, as it has been shown to interfere with cohesin loop-extrusion29.
We asked whether the observed 3D genome organization of the locus — with three adjacent TADs, one for each of the genes — is conserved across vertebrates. It is worth mentioning that though a mechanism of using SMC proteins for the genome folding is conservative30, as might be averaged 3D contacts in different cell types31 or certain 3D patterns, such as the second diagonal on Hi-C maps of erythroid cells in vertebrates32, but details of the mechanism may vary between species and the exact genome coordinates of TADs vary not only between species, but in single cells of an organism. Consequently, we introduce a limitation and by conservativity we understand that Pdgfra, Kit, and Kdr are (1) in close genomic proximity, (2) each located within a separate TAD.
To estimate the evolutionary conservativity of TADs in the Kit locus, we liftovered Hi-C contacts from several vertebrate species to the mouse genome, using C-InterSecture software33, since a direct comparison of Hi-C maps from different species is inaccurate due to different genome sizes (Supplementary Fig. 1a). Resulting interaction maps demonstrate a high similarity of chromosome architecture across the Kit locus in human, mice, dog, chicken, rabbit34, and african clawed frog35 (Supplementary Fig. 1b). As we mentioned, the non-conservative nature of TADs at the precise genomic location varies among different species. However, the consistent spatial organization observed in the Kit locus implies its evolutionary importance, aligning with the critical role these genes play in early development.
The fact that Pdgfra, Kit and Kdr genes were established from a common ancestor through a series of duplications23 explains why genes with similar function are located at the same locus. But their tissue-specific activity demands an independent regulation. H3K27ac data reveals how regulatory context varies between mast cells, melanocytes and MEFs (Fig. 1c). Here we refer to enhancers based on enrichment of an enhancer-related epigenetic mark H3K27ac. In mast cells enhancers are found within the Kit TAD upstream of the Kit gene. In melanocytes enhancers within the Kit TAD surround the Kit gene. In MEFs enhancers are only found upstream of the Pdgfra gene. The Kdr TAD lacks H3K27ac enrichment in all three cell types. We assumed that TADs are crucial for insulating regulation at the locus.
We acknowledge H3K27ac enrichment is a non-exhaustive definition of an enhancer. To additionally characterize the regulatory context we addressed data from other studies on chromatin accessibility, enhancer-associated (H3K4me1) and heterochromatin-associated (H3K27me3) epigenetic marks, as well as binding of transcription factors (Supplementary Fig. 2). H3K27ac-associated enhancers were confirmed to locate in open chromatin in all the cell types, and for mast cells it demonstrated that H3K27me3 spans all the locus downstream from Kit gene. We believe that by expanding regulatory characterisation of the locus by other studies we provide more certainty to the enhancers assignment, though with a limitation that we refer to them as an enhancer upstream or downstream of a gene rather than an exact enhancer-promoter interaction.
Importantly, enhancers of the Kit gene were functionally estimated in another study through a series of deletions36. Two enhancer regions were identified upstream of Kit in mast cells and melanocytes (located at 147–154 kb and 21–28 kb, respectively). While distant regions were required for Kit expression in mast cells (upon their deletion Kit was not transcribed), a more proximal regulatory region was essential in melanocytes.
We expected the removal of the borders to result in TADs fusion, permitting enhancer hijacking. To assess whether TADs restrict an enhancer influence to its target promoter, we generated a series of mouse strains carrying different deletions of the key CBSs (Fig. 1d). We obtained primary cell cultures from the mice. Utilizing homogenous cell populations (Supplementary Fig. 3) provides an advantage of acquiring more precise functional genomic data compared to obtaining an averaged picture from animal tissues. Primary cell cultures were used to analyze spatial genome organization using cHi-C and CTCF ChIP-seq, and gene expression using H3K27ac ChIP-seq and RNA-seq.
Comparing the effects of CBSs deletions in different cell types allowed us to analyze an impact of different regulatory contexts on the same locus (Fig. 1e schematically demonstrates the tissue-specific differences of the Kit locus).
Pdgfra gene localisation at the TAD border challenges the border disruption
Pdgfra and Kit show a contrasting expression in MEFs, i.e. Pdgfra is actively transcribed, while Kit is not transcribed (Fig. 1a). According to our hypothesis, we expected TADs fusion in the absence of boundary CBSs to result in an abnormal activation of Kit by the Pdgfra enhancers.
The Pdgfra/Kit border area comprises borders of two adjacent TADs separated by a 50 kb region with the Pdgfra gene body located in that space. We characterized the architecture of the border based on our ChIP-seq data and further consensus motif identification. The Pdgfra border consists of three strong CBSs, one forward and two reverse-facing, that overlap the Pdgfra gene body (Fig. 2a, b). The Kit border consists of one forward-facing CBS.
a The studied TAD border regions illustrated on cHi-C from MEFs (gray rectangles). The CTCF binding sites are demonstrated by CTCF ChIP-seq from MEFs and mast cells. Arrows indicate forward (green) or reverse (red) orientation of a CTCF binding site. The deletions introduced to the borders are represented by purple rectangles: at the Pdgfra/Kit border (Pdgfra Δ2k, Pdgfra Δ60k) or the Kit/Kdr border (Kit Δ30k, Kit Δ30k + , Kit Δ300k). Magnification of the CTCF ChIP-seq data at (b) the Pdgfra/Kit and (c) the Kit/Kdr provides a detailed view of the borders. Colour bar reflects the interaction counts.
To disrupt the Pdgfra/Kit border we generated two mouse strains: Pdgfra Δ2k, and Pdgfra Δ60k. Using CRISPR/Cas9 we deleted the CBSs at the Pdgfra border (Pdgfra Δ2k) and the inter-TAD region up to the CBS at the Kit border (Pdgfra Δ60k). Both deletions did not affect the promoter and the first exon of the Pdgfra gene. We compared the effect of the deletions on the spatial genome organization with wild-type (Wt) cHi-C maps (Fig. 3a).
CBSs deletions Pdgfra Δ2k and Pdgfra Δ60k were insufficient to disrupt the TADs insulation between Pdgfra and Kit in MEFs, as demonstrated by comparison of the wild-type (Wt) (a), Pdgfra Δ2k mutant (b) and Pdgfra Δ60k mutant (d) Hi-C heatmaps (5 kb resolution). Colour bar reflects the interaction counts. Ectopic contacts in Pdgfra Δ60k are indicated by arrows. Spatial contact enrichment between TADs is additionally visualized by subtraction maps (10 kb resolution) for Pdgfra Δ2k (c) and Pdgfra Δ60k (e) relative to Wt. Gain of contacts in mutants is indicated in red, and loss of contacts in blue. The tracks below the subtraction maps show the estimated contact enrichment. f RNA-seq, CTCF and H3K27ac ChIP-seq signals across the Kit locus.
The Pdgfra Δ2k mouse strain carries a 2 kb deletion of the three CTCF peaks at the Pdgfra TAD border, overlapping an intron of the Pdgfra gene. The Hi-C map demonstrated that new spatial contacts between Pdgfra and Kit TADs were not established in the Pdgfra Δ2k MEFs (Fig. 3b), which is further confirmed by the subtraction map (Fig. 3c). The persistent insulation could be explained by the remaining CBSs at the Kit side of the boundary. Thus, the Pdgfra Δ2k strain represented a case of a partial disruption between two TADs.
The Pdgfra Δ60k mouse strain carries a 60 kb deletion of the entire Pdgfra/Kit TAD border, that removes the whole Pdgfra coding sequence but promoter and the first exon. Though initially we intended to simultaneously introduce two separate deletions at the Pdgfra and Kit sides of the Pdgfra/Kit border, we considered the Pdgfra Δ60k strain valid for the research purpose despite the Pdgfra knockout. Since the promoter and enhancers of the Pdgfra gene remained intact, we expected its activity to be sufficient for an enhancer hijacking.
Pdgfra is crucial for an early development, and its homozygous deletion is lethal for mouse embryos by 15 embryonic day (E15)37. Consequently, the Pdgfra Δ60k strain was maintained in a heterozygous state. Nonetheless, we were able to obtain fibroblast cultures from the E13.5 homozygous embryos.
The effect of Pdgfra Δ60k deletion was more distinct, compared to Pdgfra Δ2k. The contacts between the most distal regions became more pronounced, as a line above the TADs (Fig. 3d, arrows). Overall, the deletion caused enrichment of spatial contacts between the Pdgfra and Kit TADs (Fig. 3e). However, the changes were marginal. We believe that the TADs remained largely separated, that could be explained by a compensatory mechanism of a neighboring CTCF peak. Upon the deletion of boundary CBSs, a TAD boundary can shift towards the position of the nearest intra-TAD CTCF site38,39. CTCF ChIP-seq of Pdgfra Δ60k (Fig. 3f) revealed an increased binding of CTCF at the motif, located at a 4 kb distance from the TAD border and contained within the retained Pdgfra promoter. We have performed differential peak analysis to computationally assess the significance of our observations (Supplementary Fig. 4). We have used HOMER software which utilizes all the peaks detected across the genome to calculate the differential enrichment between Wt and mutant datasets. The Pdgfra promoter CTCF peak appeared to be differentially enriched with the p value < 0.05. This may account for the robustness of insulation between the TADs.
We further asked if the mutations changed the expression pattern. The RNA-seq analysis of the mutant and Wt MEFs revealed that within a ± 1 megabase range surrounding the deletion site, there was no significant change in the expression of any genes, with the exception of Pdgfra (Fig. 3f). This exception is attributed to the deletion removing most of the Pdgfra gene. We found that an intergenic region between Pdgfra and Kit is transcribed upon Pdgfra Δ60k deletion, starting from the intact Pdgfra promoter and continuing to an intergenic region where it stops (Supplementary Fig. 5). But for an enhancer hijacking, H3K27ac enriched regions upstream to Pdgfra remained insulated (Fig. 3f), and consequently there was no increase in Kit transcriptional activity in any of the mutant MEFs.
The obtained data shows that Pdgfra Δ60k deletion disrupted the Pdgfra/Kit TADs border with marginal enrichment of spatial contacts between the two TADs, which was insufficient for an ectopic Kit activation. We presume that an adjacent CTCF, located further from the border, played a compensatory role explaining the robustness of the border. Considering the pivotal developmental role of the Pdgfra, its positioning at the TAD boundary may offer regulatory advantages. The preservation of this boundary might require additional factors, such as compensatory effects of neighboring CBSs, to ensure its stability.
Kit/Kdr boundary CBSs deletions result in the TADs fusion
The Kit/Kdr boundary is a joint between two adjacent TADs which contains three clustered CBSs, with two sites in forward and one in reverse orientation (Fig. 2a, c). The Kit Δ30k mouse strain carries a 30 kb deletion of the three CBSs at the Kit/Kdr border. Interestingly, the Kit Δ30k deletion resulted in a phenotype change with the mice coat color changing to brown relative to black Wt.
Analyzing the first ChIP-seq data obtained from the Kit Δ30k mast cells, we noticed a strong signal of CTCF enrichment within the Kit TAD, located ~116 kb away from the boundary. To minimize any potential impact of this site in maintaining the TAD’s structure, we generated a Kit Δ30k+ strain, carrying an additional 300 bp deletion. However we found that both deletions had a similar effect on 3D genome organization in mast cells (see Supplementary Fig. 6 for Kit Δ30k+ effects in mast cells). Thus, we focused on the Kit Δ30k.
We monitored the effects of the deletion on two cell types where Kit is actively transcribed — mast cells and melanocytes. Kdr is normally inactive in mast cells and is marginally expressed in melanocytes (Fig. 1a). The resulting cell populations were highly homogeneous, as revealed by flow cytometry (Supplementary Fig. 3b), and displayed the typical morphological features. Mast cells produced heparin and histamine containing granules that can be detected by toluidine blue staining (Fig. 4a). Melanocytes demonstrated a typical morphology with granules of melanin in cytoplasm (Fig. 4b). Highly homogeneous cell cultures are crucial to gain pure sequencing data.
a Cytospin preparations of Wt (left) and Kit Δ30k (right) mast cells after 4 weeks in culture. Histamine containing granules could be detected by toluidine blue staining. Scale bar: 100 um; b Primary Wt (left) and Kit Δ30k (right) melanocytes after 2.5 weeks in culture (without staining). Scale bar: 100 um. 3D genome organization and regulation context in Wt and Kit Δ30k mast cells (c) and melanocytes (d) are presented by Hi-C maps (5 kb resolution), CTCF and H3K27ac ChIP-seq, and RNA-seq. Colour bars reflect the interaction counts. Spatial contacts newly established after the TADs disruption are indicated by arrows. Subtraction maps demonstrate changes of spatial contact frequencies in mutant mast cells (e) and melanocytes (f) relative to the Wt. The tracks below the subtraction maps show the estimated contact enrichment, indicating loss of insulation in the mutant cells. Removal of the Kit/Kdr boundary resulted in loss of inter-TAD insulation and extensive interactions across the TADs boundary.
Contact maps from the Wt mast cells denoted a nested substructure of the Kit TAD, with two self-interacting regions. Kit with its upstream enhancers are located within the centromeric half of the TAD and thus are insulated from the telomeric half (Fig. 4c). Kit Δ30k deletion disrupted the Kit/Kdr TAD boundary insulation in mast cells and led to a noticeable gain of inter-TAD contacts. Looping interactions connecting the outer boundaries of Kit and Kdr TADs were also established. However, we noted that contacts rewiring mainly involved the telomeric half of the Kit TAD, which merged with the Kdr TAD. The insulation inside the Kit TAD was not affected by the deletion, and a self-interacting region, located at the centromeric side, remained isolated. Thus, despite the TAD boundary disruption in mast cells, active Kit enhancers remained insulated from Kdr.
Our CTCF ChIP-seq data confirmed a successful deletion of CBSs. The analysis did not reveal any CTCF peaks between two self-interacting regions inside the Kit TAD (Fig. 4c), nor an emergence of a novel CTCF peak in mutant cells, as was shown for Pdgfra Δ60k. Hence, Kit enhancers are possibly restrained from the downstream genes by different mechanisms rather than CTCF restricted loop extrusion, for instance, by the Kit transcriptional activity.
We then compared the effects of the boundary CBSs deletion in mast cells and melanocytes. The inner structure of the Kit TAD was divided with Kit and its downstream enhancers being insulated from the centromeric part of the TAD (Fig. 4d). Kit Δ30k deletion resulted in extensive rewiring of chromatin contacts in melanocytes. cHi-C maps revealed loss of insulation at the Kit/Kdr boundary and concurrent merging of neighboring TADs. Notably, spatial contacts were established between Kdr TAD and the telomeric part of the Kit TAD, enabling Kit enhancers to interact with the Kdr promoter.
The establishment of inter-TAD interactions in mast cells and melanocytes was further confirmed by the subtraction maps of mutant and Wt cells with simulated deletions (Fig. 4e, f). This approach was used to estimate the contribution of distance decrease on the rewiring of chromatin contacts and is discussed in more detail in the Supplementary Notes and demonstrated in Supplementary Fig. 7.
Thus, in both cell types, removal of boundary CTCFs resulted in loss of inter-TAD insulation and extensive interactions across the TADs boundary. At the same time, the insulation between two self-interacting regions within the Kit TAD was preserved. Our results imply that this insulation is provided by the epigenetic environment, in particular, by the relative position of enhancers and actively transcribed Kit within the TAD.
The Kit/Kdr TADs fusion provides tissue-specific ectopic gene activation
We then asked whether the reorganization of contacts observed in the Kit Δ30k and Kit Δ30k+ mutant mice was functional and could trigger a transcriptional outcome. We noted that Kit Δ30k deletion was associated with a specific phenotype in adult mice. Mutant mice had brownish coat color, compared to the black Wt animals (Fig. 5a).
a Coat color difference between brown Kit Δ30k (left) and black Wt (right) mice; b A plot of normalized RNA-seq reads counts in Wt and Kit Δ30k melanocytes indicates a 6.7-fold increase of Kdr transcription. Dots represent individual replicates. p-value was estimated with DESeq2, using the two-sided Wald test; c In mast cells Kit Δ300k deletion caused an antisense Kdr transcription; d Crossing scheme used to obtain Mus musculus (black) x Mus castaneus (orange) hybrids and further RNA-seq analysis; e Expression relative to a M. castaneus (Cast) allele. To evaluate an allelic activity, reads with M. castaneus-specific SNPs were scored for Pdgfra (n = 19 SNPs), Kit (n = 12 SNPs) and Kdr (n = 21 SNPs). Data are presented as mean values ± SD. p value was estimated using two-tailed Mann–Whitney test, NS — not significant. Source data are provided as a Source Data file.
In mutant mast cells RNAseq analysis revealed no transcriptional changes in the locus or among the adjacent genes (± 1 Mb). Of note, for mast cells data from another study40 reveals that all the region from the Kit termination site to the closest active gene Srd5a3 appears to be enriched in H3K27me3, suggesting a repressive state of Kdr (Supplementary Fig. 2a).
Remarkably, the situation was different in melanocytes. In Kit Δ30k cells, we detected a 6.7-fold increase in Kdr expression level compared to the Wt data (Fig. 5b). Notably, Kdr is normally expressed in melanocytes, although at a low level, suggesting an absence of repressive epigenetic state. Ectopic transcriptional activation observed in mutant melanocytes is in agreement with the redirection of the promoter-enhancer contacts revealed by cHi-C. We highlight that Kdr is the sole gene within a ± 1 megabase range of the deletion that exhibited a significant change in expression.
Surprisingly, decrease of genomic distance between Kit and Kdr genes was also insufficient for Kdr activation in mast cells. We had performed transcriptome analysis on mast cells from a mouse strain carrying a 300 kb deletion at the Kit/Kdr TADs border, previously obtained in our laboratory41 (Kit Δ300k, Fig. 2a). We found an abnormal transcription in Kit Δ300k that could be explained by transcription started from the Kit promoter, which remained intact after the deletion, and continued to the closest termination site within Kdr (Fig. 5c). Of note, since Kdr locates on an opposing DNA strand to Kit, its transcript was antisense. The Kit Δ300k mice strain is heterozygous, therefore Kit was expressed from a normal allele. To compare effects of Kit Δ300k and Kit Δ30k deletions on 3D genome organization would be of interest, but unfortunately the Kit Δ300k is recessive lethal, so we were unable to obtain homozygous cells that are required to detect 3D genome changes.
In Kit Δ300k we did not detect a Kdr sense transcript, which potentially could be explained by the fact that an antisense transcript could inactivate a sense transcription through interference (see review42). But nor did we detect increased activity of genes downstream to Kdr which were unaffected by the antisense transcription. Thus, we believe that a high level of Kit transcription in mast cells could contribute to restriction of an enhancer hijacking rather than genomic distance.
Since Kdr expression is detected mostly in cardiac and endothelial cells, while Kit is expressed in a variety of cell types43,44, we decided to find out if Kdr activation occurred in any other tissue due to our manipulations with the 3D genome structure. To this end we performed immunostaining in E15.5 embryos and looked for double-positive cells, expressing both Kit and Kdr. We focused specifically on somites, skin, eyes, vibrissae hair bulge, liver and yolk sac, attempting to find an aberrant Kdr signal in Kit-positive cells. Numerous double-positive cells were found in the skin of Kit Δ30k embryos (Supplementary Fig. 8). These cells were located at the epidermal layer and displayed branched, dendritic morphology. Kit-positive cells with identical shape in the skin of Wt embryos were negative for Kdr signal. Kit-expressing cell types, residing in the mouse fetal skin, are melanocytes and mast cells, although the latter exclusively locate to the dermis45. Thus, the location of Kit-Kdr double-positive cells in the epidermal layer and their dendritic morphology suggests that these are melanocytes43,46.
Notably, Kit Δ30k deletion could lead to Kit activation in Kdr-expressing endothelial cells, but we did not detect such activation, at least immunohistochemically. It can be assumed that in endotheliocytes, Kit Δ30k deletion did not cause adoption of Kdr enhancers by Kit.
As we mentioned earlier, we did not find any genes within a ± 1 mb range from the deleted Kit/Kdr TADs boundary that might have significantly changed in expression. However, it can be hypothesized that in other cell types, the removal of the TAD boundary may affect more distant genes. Systematically testing this is challenging. Nevertheless, changes in the activity of genes that could produce a clearly visible phenotype are testable. We hypothesized that the key regulator of circadian rhythm, the Clock gene, located approximately 400 kb away (Supplementary Fig. 9), would be a suitable candidate for such a study. Alterations in gene activity might then influence the daily activities of animals. To test this hypothesis, we assessed changes in locomotor activity, sleep, and food and water intake in Kit Δ30k and Wt animals (Supplementary Fig. 9). We found no significant differences in these parameters, this leads us to tentatively suggest that the removal of the nearby specific TAD boundary might not significantly impact the regulation of Clock gene activity.
Mus musculus x Mus castaneus breeding further confirmed ectopic Kdr activation to result from boundary CBSs deletions
Since the melanocyte in vitro differentiation takes 4.5–7 weeks, it can be assumed that the cells obtained from Wt and Kit Δ30k mice could be exposed to the influence of confounding effects (cultivation conditions, trans-acting factors, differentiation state, etc.). Therefore, it is possible that increased Kdr expression does not reflect the difference in gene activity on the chromosome with and without deletion, but is only a consequence of different transcriptional status of cells. To test the hypothesis, we crossed Kit Δ30k and Wt M. musculus with Mus castaneus (Fig. 5d). Since almost all genes carry SNPs that distinguish M. musculus from M. castaneus (including Pdgfra, Kit, and Kdr genes), we were able to evaluate the activity of the genes using the reference M. castaneus allele as a normalization factor. Since both alleles are in the same cell, this assessment does not depend on transcriptional status of cells. We isolated primary cells from the epidermis of the hybrids, differentiated them, performed RNA-seq in melanocytes, and calculated the read counts from one of the two species based on the SNP analyses. Then, we used read counts from the M. castaneus allele as a normalization factor and compared the level of Pdgfra, Kit and Kdr expression between Wt and Kit Δ30k alleles. The level of Kdr expression was significantly lower in the control experiment with M. castaneus x M. musculus Wt hybrids, with both alleles being equally active (Fig. 5e). Thus, ectopic Kdr activation in melanocytes is truly caused by a deletion of the TAD border between the Kit and Kdr genes, and not by some confounding effects. Surprisingly, our analysis also revealed a downregulation of Kit in the KitΔ30k allele, which was modest and yet significant. This decreased Kit activity could be a result of the partial Kit enhancer retargeting47,48.
Thus, the deletions of the boundary CBSs at the Kit/Kdr TADs border induced tissue-specific effects (Fig. 6). For mast cells, despite the weakened insulation between two TADs, the Kit enhancer region did not form new contacts with downstream promoters and Kdr remained inactive. In melanocytes, TAD boundary deletion induced the rewiring of Kit enhancers, which triggered Kdr misexpression and phenotype changes. We propose that the individual epigenetic characteristics of the locus, e.g., gene activity and enhancer localisation within the TAD, determine the tissue-specific response to a boundary deletion.
Discussion
We found that TADs contribution to gene regulation depends on tissue-specific features at the Kit locus. The TAD border deletion caused fusion of the Kit and Kdr TADs in mast cells and melanocytes. Whereas an ectopic activation of Kdr occurred only in melanocytes. Several studies demonstrated that whether border CBSs deletion causes an ectopic gene activation largely depends on the locus, since the deletions provide inconsistent results (Supplementary Table 1). For the first time we provide comparison of the same locus at different regulatory landscapes in several pure primary cell cultures. Our results imply that TADs may unite different regulatory features, thus their function is not limited to an integrity of CBSs border, as was previously assumed.
Deletion of a border between two TADs causes their fusion
We examined the impact of TADs disruptions in primary cell cultures, as opposed to an averaged data from tissues largely used before4,13,49,50,51. This approach enabled us to obtain high-quality functional genomic data. The data provided a view on chromatin interactions at different regulatory landscapes of the Kit locus. Three adjacent TADs encompass the locus, that is Pdgfra, Kit and Kdr TADs.
We found that removal of border CBSs resulted in TADs fusion, though architectural changes varied from a marginal increase of inter-TAD contacts across the Pdgfra/Kit border to the full contacts enrichment across the Kit/Kdr border.
We assume the CTCF site at the Pdgfra promoter, which remained intact after the Pdgfra Δ60k deletion, to account for an incomplete loss of an insulation between Pdgfra and Kit TADs. Though an insulating strength of an individual CBS is not correlated with its CTCF occupancy52, we revealed the CTCF binding at the intact site to increase in Pdgfra Δ60k. It is in agreement with the fact that CBSs might be dispensable to maintain a TAD border, and neighboring CTCF might compensate for deletion of border CBSs14,39,53. It would be of interest to analyze how the deletion of the CBSs at the Pdgfra promoter affects 3D genome organization and gene regulation. But its deletion requires the deletion of the entire Pdgfra promoter. Loss of the target promoter was shown to retarget enhancer activity to the non-target genes located in the same TAD54. Therefore, it would be difficult to distinguish the effects of boundary deletion from the chromatin rewiring induced by the promoter loss. For this reason, we believe that deleting the CBSs at the Pdgfra promoter would trigger more extensive perturbations than just boundary deletion.
Another potential explanation for a robustness of the Pdgfra TAD to the introduced deletions is Pdgfra gene localisation at the border. Transcription from the Pdgfra promoter located near the TAD border — transcripts being the Pdgfra gene at the Pdgfra Δ2k and an intergenic region at the Pdgfra Δ60k — could strengthen the insulation. It was proposed that transcribed genic and non-genic boundaries are associated with stronger insulation due to boundary RNAs facilitating CTCF recruitment, with highly transcribed boundaries being the most CTCF-enriched55. But in case of Pdgfra Δ60k, since the level of intergenic mRNA in the Pdgfra Δ60k mutant is low, we would suspect the impact of this transcript on the CTCF binding to be rather weak. Thus, the Pdgfra TAD border was strengthened by a compensatory role of a neighboring CBSs or transcription at the border, and Pdgfra and Kit TADs remained largely separate. Since an enhancer influence on the promoter is constrained by TADs borders56, a few newly established spatial interactions were insufficient for Kit activation in MEFs.
Since the distribution of genomic contacts is determined by a distance between loci, we emphasize that studying 3D genome organization upon deletions, especially large deletions, requires elimination of decrease in genomic distance due to deletions impact on the rewiring of chromatin interactions. To tackle the problem we utilized the C-InterSecture algorithm.
A limitation of our study is that we provide an analysis of the Pdgfra/Kit border in only one cell type. Though comparing an effect in MEFs with other cell types would be of interest, as an example glia has the same expression profile of Pdgfra and Kit, we believe that we would encounter the same problem of border robustness due to mechanisms discussed above. An additional constraint we were faced with is the TAD border disruption resulting in Pdgfra knockout mice, which is lethal in homozygous state, but disrupting the borders simultaneously and locally while preserving the integrity of Pdgfra poses a challenging task.
Of note, there is another mouse strain carrying the deletion of Pdgfra gene — Patch mutation. Interestingly, it causes an ectopic Kit activation in somites and lateral mesenchyme and exhibits a distinct pigment phenotype — white abdomen57,58,59. An analysis of changes in genome and spatial organization of the Patch mutant mice is yet to be performed. We suspect that the mutation covers a larger portion of the genome, enabling TADs fusion and consequently enhancer hijacking. Considering the substantial impact of deletions at the Pdgfra/Kit TADs border on mice development, we stipulate that the mechanisms we discussed, which impart robustness to this border, might be of great importance.
Concerning the Kit/Kdr border, the removal of CBSs at the TADs border led to loss of insulation, consequently resulting in the fusion of TADs. Herewith TADs fusion was different depending on the insulation pattern within the Kit TAD in mast cells or melanocytes. We discuss the result in more detail in the next section.
Kit transcription defines tissue-specific characteristics of the TAD
Genome organization of the Kit TAD varies between the examined cell types. The TAD demonstrated homogeneous spatial contacts in MEFs, whereas in mast cells and melanocytes the Kit TAD was divided into two distinct self-interacting regions. We noted that partition of the TAD occurred at the Kit gene (which is the only gene at the TAD), and insulation point differed locating at the Kit promoter in melanocytes and the Kit terminator in mast cells. We presume Kit transcription to provide the insulation in the TAD, because Kit is inactive and an insulation is absent in MEFs. It is in agreement with the study revealing that transcription modulates chromosome folding during mouse thymocyte maturation with single activated genes forming an insulated region within TADs60. Models of cohesin loading and transcription at different states suggested RNA polymerase II (RNAP) to act as a barrier to loop extrusion29. Transcribing RNAP may disrupt chromatin interactions by displacing cohesin from CTCF sites61,62 and relocating it over long distances along DNA63. Moreover, upon depletion of RNAP many enhancer-promoter loops are lost and new loops are established at the CTCF sites, which were restricted by RNAP64.
It is of great interest that insulation within the Kit TAD at the promoter or terminator of Kit depends on the cell type. Transcription can participate in the formation of nested structures within TADs, which are established independently of CTCF-cohesin loop extrusion mechanism65. But data from Wt MEFs and mast cells revealed a CBS within the Kit promoter, which could account for the insulation within the TAD. Though Kit is expressed in both cell types, the expression level in mast cells is ten times higher than in melanocytes. High level of transcription is assumed to antagonize CTCF-cohesin loop extrusion66. Cohesin was revealed to accumulate at 3’-end of highly active genes in an absence of CBS or an efficient cohesin release by WAPL63,67. This implies that cohesin could be shifted from the CBS at the Kit promoter to its terminator resulting in 3D genome organization revealed by our Hi-C maps in mast cells, while this effect was absent in melanocytes. Thus, a level of Kit expression could influence tissue-specific features of the spatial genome organization at the TAD.
TAD disruption results in Kdr activation in melanocytes, but not in mast cells
How TADs integrity affects gene regulation is a matter of great interest, given TADs are assumed to establish enhancer-promoter interactions.
Our transcriptome analysis of Kit Δ30k cells revealed an ectopic activation of Kdr in mutant melanocytes, whereas Kdr remained silent in mutant mast cells. The plausible explanations for the cell type specific enhancers adoption by Kdr are different transcription levels of Kit and an epigenetic status of Kdr in mast cells and melanocytes.
Genomic distance is potentially another factor to influence an enhancer adoption. We were able to assess its contribution to Kdr insulation in mast cells upon a larger deletion at the Kit/Kdr TADs border (300 kb against 30 kb). Transcriptome analysis revealed the lack of Kdr transcription. Though we detected an antisense transcript including Kdr gene, which could potentially inactivate a sense transcription through interference (see review42). But nor did we detect increased activity of genes downstream to Kdr which were unaffected by an antisense transcription. Thus, we conclude that other factors rather than genomic distance restricted enhancer hijacking in mast cells.
We found Kdr to be expressed at a low level in Wt melanocytes, and silent in Wt mast cells. Taking into account Kdr enrichment of a repressive epigenetic mark H3K27me3 in mast cells, it is possible that the repressive state of Kdr makes it inaccessible to an enhancer hijacking. Our Hi-C maps revealed that spatial interactions between Kit and Kdr were not established in mast cells, and the active enhancers remained insulated from Kdr. However, whether the repressive state of Kdr could influence the spatial insulation is unclear, as well as the causation between gene activity and epigenetic state. We consider the high level of Kit transcription in mast cells (which is tenfold higher relative to melanocytes) to be a more likely explanation for the insulation, since RNAPII was revealed to act as a barrier for cohesin-mediated loop extrusion29.
We additionally confirmed Kdr activation in melanocytes to result from the Kit Δ30k deletion by breeding Kit Δ30k M. musculus with Wt M. castaneus, showing that the vast majority of the Kdr transcripts (98%) originated from a M. musculus allele. The observed difference corresponds to the 3D architecture of the locus upon Kit Δ30k deletion. Of note, we did not detect a reverse situation of Kit activation by active enhancers of Kdr in endothelial cells. Thus, cell type-specific features of the locus, such as localization of H3K27ac-marked active enhancers, underlie the TADs function, at least in case of the Kit locus.
Our M. musculus x M. castaneus breeding experiment revealed downregulation of Kit in the Kit Δ30k melanocytes. Although significant, the difference in relative Kit expression level was slight (Wt/castaneus 0.88 → Kit Δ30k/castaneus 0.75), compared to the dramatic Kdr upregulation (Wt/castaneus 0.99 → Kit Δ30k/castaneus 49.71). Downregulation of gene activity due to an enhancer retargeting may reflect the promoter competition phenomenon. There is some evidence of the dilution of the enhancer activity among multiple genes due to the TAD boundary disruption, which leads to the downregulation of target genes (see the review68). Here we might see a similar mechanism.
Cell type-specific effect of TADs disruption on gene expression was also demonstrated in comparisons of induced pluripotent stem cells (iPSCs) and iPSC-derived cardiomyocytes69, ESCs and motor neurons38. In addition, strength of TADs border may vary between cell types, as was shown for mESCs and MEFs53. Though given a generally more open chromatin state of stem cells relative to differentiated cells70, the advantage of our study is providing evidence of the effects in primary cell cultures from animals with deletions.
Studies concerning the influence of TADs disruption on expression suggest that regulation within TADs may largely depend on other factors rather than on the integrity of a TAD border only (Supplementary Table 1). We provide evidence of cell-type specific features of the Kit locus to affect a TAD’s role insulating an enhancer influence on a non-target promoter.
Phenotypic consequences of the Kit TAD disruption
In our study we observed a change in the mice phenotype as a result of the CBSs deletions at the border between Kit and Kdr TADs: the coat of the Kit Δ30k mice were lighter relative to the wild-type mice. It is well-known that mutations in Kit are implicated in the white-spotting phenotype in mice21,71,72, rabbits73,74, cats75, dogs22, horses76 and other animals. The mechanism for this phenotype is an impaired melanocyte development and migration due to the defective Kit receptor signaling. An assumption that Kit downregulation detected in melanocytes could explain a paler phenotype in Kit Δ30k mice can be made, although it faces an important contradiction. Kit mutations described in earlier studies result in a total absence of pigmentation, either locally or all over the body, while Kit Δ30k mice have lighter coloration, which is an uncharacteristic manifestation of Kit mutations. However, it is obvious that coloration is an essential object for natural selection, so even such an almost invisible change in the laboratory could be eliminated in natural conditions.
Mutations leading to TADs fusion are under negative selection77, thus, the evolutionary conservatism of TADs may be a consequence of their importance for maintaining a certain pattern of gene activity. Whereas a change in the structure of TADs can lead to the emergence of a new pattern of gene activity, which may be an evolutionary innovation. This was remarkably demonstrated by the example of moles78. One of the adaptations of moles to an underground lifestyle is to increase the concentration of testosterone in females. An aberrant pattern of Fgf9 gene activity ensures the tolerance of female oogenesis to the male hormone. To change Fgf9 activity, a chromosomal rearrangement, affecting the distal part of the Fgf9 TAD, occurred in moles. Given TADs role in insulating a regulatory pattern, they ensure an integration of a newly established gene or a change in regulation to occur without huge alterations in neighborhood79. Thus, the regulatory context provided by TADs is an important object for the evolution of organisms.
Perspective
Our findings indicate that the consequences of TAD border deletions may largely depend on the specific characteristics of an individual locus within a particular cell-type context. We revealed that an epigenetic landscape in the Kit locus, especially an active transcription and an enhancer localization, accounts for a different insulating role of the Kit TAD in mast cells and melanocytes. We propose that future research should dive into understanding the interplay and relative contributions of these features in shaping the insulation role of TADs.
Methods
Animals
Mouse Kit Δ30k line was established from a chimeric animal generated after mouse embryonic stem cells (mESCs) injection as described earlier80. Briefly, mESCs were transfected with CRISPR/Cas9 plasmids aimed at a border region between Kit and Kdr TADs (see gRNA in Supplementary Table 2). With PCR genotyping we selected a homozygous clone with Kit Δ30k deletion (see primers in Supplementary Table 2). Modified mESCs were injected into recipient blastocysts. Chimeric founders (F0) were crossed with wild-type (Wt) C57BL/6 J animals to generate heterozygous animals (F1). After 8 rounds of backcrossing to C57BL/6 J animals, a homozygous Kit Δ30k line was established.
All other mouse lines in this study were generated through cytoplasmic injection of mRNA Cas9 and gRNAs (Supplementary Table 2), as described in Korablev et al., 2019. For generation Pdgfra Δ2k and Pdgfra Δ60k lines CRISPR components were injected in C57BL/6 J zygotes. For generation Kit Δ30k+ line CRISPR components were injected in Kit Δ30k zygotes.
The daily activity of animals was assessed using a Phenomaster. For this purpose, we compared two groups of 12-week-old males from the Kit Δ30k and C57BL/6 lines. The animals were trained to use the drinking bowls and feeders over two consecutive days. Following this, they were isolated in the Phenomaster cages, where their total locomotor activity, sleep duration, number of sleep episodes, and water and food consumption were recorded over a period of 72 h.
Animals were kept in a standard environment at 24 °C temperature, 40–50% relative air humidity and 14 h light/10 h dark–light-cycle. Food and water were available ad libitum. At the end of experiments, remaining animals were euthanized by CO2. All the procedures and technical manipulations with animals were in compliance with the European Communities Council Directive of 24 November 1986 (86/609/EEC) and approved by the Bioethical Committee at the Institute of Cytology and Genetics (Permission N45 from 16 November 2018).
Cell cultures
Mouse embryonic fibroblasts (MEFs)
MEFs were obtained from E13.5 as described in81. MEFs were cultured at 37 °C under 5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM) (Thermo Fisher Scientific, 12800082), supplemented with 10% FBS (Capricorn Scientific, FBS-11A), 1x penicillin & streptomycin 10x (Capricorn Scientific, PS-B), 1x GlutaMax-I 100× (Thermo Fisher Scientific, 35050061). For subculture cells were rinsed with 1 × PBS and detached using 0.25% trypsin-EDTA at 37 °C for 3 min. Cells were typically split every 2–3 days at a 1:2 ratio. Flow cytometry showed more than 80% Pdgfra positive cells (Supplementary Fig. 3b).
Melanocyte cell culture
Melanocytes from Wt and Kit Δ30k mice were obtained according to the Murphy et al., protocol82 with modifications. In brief, 4-day-old pups were decapitated, the skin was separated, rinsed in 10x penicillin/streptomycin solution (Capricorn Scientific, PS-B), and incubated overnight in 0.25% trypsin/DMEM (Capricorn Scientific, TRY-2B). Next day, blood, adipose and muscle tissues were removed, epidermal and dermal layers were disassociated. Epidermis was chopped with scissors in 0.25% trypsin/PBS solution, and additionally incubated in 0.25% trypsin for 10–15 min at 37 °C. Then, the cell suspension was filtered through a nylon mesh to remove large tissue segments, cells were pelleted by centrifugation and pre-plated for 25–30 min. Finally, unattached cells were transferred to the plastic surface 6-well plate, covered with 0.1% Gelatin from porcine skin type B (Sigma, G9391), and cultivated in Melanocyte growth medium (PromoCell, C-39410), containing geneticin 100 ng/ml (G418) (Thermo Fisher Scientific, 11811023), for 48 h. On day 3, the medium was replaced with a fresh one, without G418. At that step individual melanocytes could be clearly visualized. In 10–14 days, the majority of the cell population represented granulated melanocytes. The cells were grown for 4–7 weeks until the required cell quantity was reached and more than 80% of cells were Kit positive (Supplementary Fig. 3b).
Mast cell culture
Mast cells were differentiated from hematopoietic cells derived from bone marrow according to the Vukman et al., protocol83. Cells were cultured for 8 to 10 passages in Iscove’s Modified Dulbecco’s Medium (IMDM) (Thermo Fisher Scientific, 12200069) supplemented with 10% FBS (Thermo Fisher Scientific, 16141002), mrSCF(10 ng/ml) (Biolegend, 579702) and mrIL-3 (10 ng/ml) (Biolegend, 575502). Mast cells were also stained by toluidine blue (Biovitrum, 07-002) — a basic thiazine metachromatic dye that stains nuclei blue and has a high affinity for acidic granules, in accordance with the manufacturer’s recommendations.
According to the flow cytometry analysis of Kit and FcεR1 expression, at the final passages mast cells accounted for ~90% of the total cell population (Supplementary Fig. 3b). For the Hi-C experiment Kit positive cells were magnetically separated (Miltenyi Biotec, 130-097-146).
Flow cytometry (FC)
FC was performed as described in84. A total of ~0.5-1 * 106 cells/ml were harvested and washed in PBS. Antibody and cell suspension were mixed in an ice-cold FC buffer (PBS, 10% FBS) in a ratio 1:300 and incubated on ice for 1 h. After incubation cells were pelleted by spinning at 300×g for 10 min. The pellet was washed in the FC buffer twice to remove unnecessary antibodies. The pellet was resuspended in a 500 μL buffer and filtered through nylon mesh to remove cell conglomerates. A negative control was prepared as a cell suspension washed in PBS without immunostaining. To exclude cells with compromised membranes cell suspension was incubated with 7-aminoactinomycin D (7-AAD) (Thermo Fisher Scientific, A1310) in a ratio 1:1000 for at least 20 min, after which the cell pellet was washed as described above. The prepared cell suspensions were analysed using BD FC Aria III (BD Biosciences). The antibodies were from BioLegend: APC/Cyanine7 anti-mouse CD117, c-Kit (#135136); APC anti-mouse CD117, c-Kit (#105812); PE anti-mouse CD140a, Pdgfra (#135905); FITC anti-mouse FcεRIα (#134306).
ChIP-seq
The anti-histone H3 (acetyl K27) ChIP-seq and input libraries were prepared for each cell type as described in Pekowska et al.85 with slight modifications. The anti-CTCF ChIP-seq was performed in MEFs and mast cells. Briefly, ~ 5 * 106 cells were harvested per experiment and crosslinked with 1% formaldehyde at RT for 10 min. Crosslinking was quenched with 125 mM glycine. Cells were lysed on ice in 0.5 ml lysis buffer (10 mM Tris pH 7.5, 1 mM EDTA, 0.4–1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, Complete Mini EDTA-free protease inhibitors (Roche, 11836170001)). Lysed chromatin was fragmented using the BANDELIN SONOPULSE until reaching a fragment size of 150–500 base pairs (5–10 cycles, 30 s/60 s on/off, 60% amplitude, 4°C). SDS concentration and sonication cycles were adjusted according to a cell type and shearing efficiency. Lysates were clarified by 16,000 g centrifugation for 10–15 min at 4 °C and diluted with 1–1.5 ml of lysis buffer without SDS to reduce SDS concentration before the immunoprecipitation step (~0.17% SDS final). Chromatin was pre-cleared with 20 μl Protein A magnetic beads (New England Biolabs, S1425S) for 1.5 h and 100 µL of the solution was saved as an input control. During this time, a 40 µl aliquot of Protein A beads was washed with PBS and rotated with the respective antibody at 4°C (1.5 − 5 µg antibody). Protein-DNA complexes were then immunoprecipitated overnight at 4 °C with rotation. The beads were washed at 4°C in the following buffers: lysis buffer (0.1% SDS, twice), lysis buffer containing 0.5 M NaCl (twice), LiCl buffer (0.25 M LiCl, 0.5% IGEPAL-630, 0.5% sodium deoxycholate, twice), TE (pH 8.0) plus 0.2% Triton X-100 (once), and TE (pH 8.0, once). Crosslinks were reversed at 65°C overnight and DNA was purified with ChIP DNA Clean & Concentrator kit (Zymo Research, D5205). Sequencing libraries were prepared using the KAPA Hyper Prep kit. The anti-histone H3 (acetyl K27) (Abcam, ab4729) and anti-CTCF (Abcam, ab70303) antibodies have been successfully validated by Western Blot, dilution 1:3000 (Supplementary Fig. 10). The experiments were performed in two biological replicates.
Capture Hi-C (cHi-C)
Hi-C from two biological replicates of each cell type were performed as previously described in86 with some modifications from87. A total of ~ 5 * 106 cells were crosslinked in 1–2% PFA/PBS buffer at RT. A cell pellet was resuspended in a lysis buffer (150 mM Tris-HCl pH 7.5, 140 mM NaCl, 0.5% NP-40, 1% Triton X-100, 1x Complete Mini EDTA-free protease inhibitors) and incubated for 15 min at 4°C followed by 15 min at RT. Extracted nuclei were spun down at 800 g then washed once in lysis buffer and once in NEBuffer3.1 (New England Biolabs, B703S). After centrifugation, nuclei pellet was resuspended in NEBuffer3.1 supplemented with 0.3% SDS, then incubated at 37 °C for 1 h followed by 10 min at 65 °C to open chromatin. 10% Triton X-100 was added to the reaction (1.8% final) to quench SDS and incubated at 37 °C for 30 min. Chromatin was digested overnight with 400U DpnII (New England Biolabs, R0543M). Following the inactivation of DpnII, the nuclei were spun down at 2500 g and resuspended in NEBuffer2.1. Biotin fill-in was performed with biotin-14-dATP (Invitrogen, 19524016) using Klenow DNA polymerase (SibEnzyme, E325) at 22 °C for 4 h. Samples were centrifuged and the pellet was resuspended in ligation mixture (1xT4 Ligation buffer (New England Biolabs, B0202S), 5% PEG, 1% Triton X-100, 0.1 mg/ml BSA and T4 ligase). Ligation was carried out in 1.2 mL volume overnight at 16 °C with mixing. The ligation products were reverse crosslinked at 65°C overnight with proteinase K (New England Biolabs, P8107S) and DNA was extracted with phenol-chloroform. Efficiency of DNA digestion and ligation was assessed with gel electrophoresis. Sonication was performed on the Covaris M220 system to obtain 200–400 bp fragments. DNA fragments were double size-selected using AMPure XP beads (Beckman Coulter, A63880). MyOne Streptavidin C1 magnetic beads (Invitrogen, 65001) were added to bind biotin-tagged fragments. Sequencing libraries were prepared using the KAPA Hyper Prep kit. The hybridization of libraries with RNA probes was performed according to the myBaits Manual v4.01 (Arbor Biosciences). Enrichment probes were designed over the region chr5:74,135,000-76,410,000.
RNA-seq
0.2*106–2*106 cells were harvested and washed twice with PBS. Total RNA was extracted using Aurum Total RNA mini kit (Bio-Rad, 7326820). Sample concentration was evaluated using Qubit RNA HS (Q32852, Invitrogen). Gel electrophoresis was used to verify the integrity of extracted RNA. Three replicates were generated for each cell type and genotype, respectively.
Immunohistochemistry (IHC)
E13.5-15.5 mice were fixed with 4% PFA solution in 1X PBS. Skin tissue sections from 4-day-old mice were harvested from head and back in 1X PBS and fixed in 4% PFA solution overnight on a roller shaker at 4 °C. Next day samples were washed in 1X PBS solution three times for 30 min. For tissue dehydration organs were sequentially incubated for at least 24 h with a 15% and 30% sucrose solution in 1X PBS at 4 °C. Next, organs were embedded in Tissue-Tek O.C.T. Compound (Sakura Finetek, 4583) and frozen. Organ sections of 50 um thickness were prepared on MICROM HM 505 N cryostat (Microm) and immediately collected on SuperFrost™ slides (Thermo Fisher Scientific). Sections were washed with 1X PBS and incubated in blocking solution: 2% BSA (Sigma Aldrich, A2153), 0.2% Triton X-100 (Amresco, X100), 5% FBS (Capricom Scientific, FBS-11A). Primary antibodies anti-Kdr (R&D, AF644) and anti-Kit (CellSignaling, 3074) were diluted in blocking solution (1:100) and incubated overnight at slow speed on an orbital shaker at room temperature. Next, slices were washed with 1X PBS three times for 20 min. Slices were stained with secondary antibodies (Jackson Immunо Research, 705-165-147 and 711-545-152) in dilution 1:500 and DAPI diluted in 1X PBS for 2 h at room temperature. Slices were washed with 1X PBS three times for 20 min and completely dried out and finally were mounted with ProLong™ Diamond Antifade Mountant (Thermo Fisher Scientific, P36965). Immunofluorescence were visualized under confocal fluorescence microscope LSM 780 NLO (Zeiss) with ZEN software (Zeiss). Microscopic analysis was carried out at the Multiple-access Center for Microscopy of Biological Subjects of the Institute of Cytology and Genetics SB RAS.
Computational methods
All computations except the RNA-seq data analysis were performed using nodes of Novosibirsk State University high-throughput computational cluster.
сHi-C data processing
The cHi-C sequencing data were processed using Juicer software Version 1.1188 and mapped to the mm10 genome. Contact maps were generated from read pairs with MAPQ ≥ 30 and normalized using the VC_SQRT method. Maps at 5 kb resolution were further visualized via the Epigenome browser89.
TAD organization analysis
The Hi-C maps for human (hg38), mice (mm10), rabbit (oryCun2), dog (canFam3), chicken (galGal5) were obtained from GSE16757934. The Hi-C map of african clawed frog (xenLae2) was obtained from35.
The conservativity of the Kit locus was estimated by the C-InterSecture software33. For the cross-species comparison, we generated maps of synteny regions from the pairwise genome alignments. At first, the genomes of interest were aligned by LastZ Version 1.04.2290. Then, the given alignments were converted into net-files by KentUtils [https://github.com/ucscGenomeBrowser/kent] and transformed into the syntenic maps by the C-InterSecture script. Additionally, with a considerable part of the genomes being not included in any synteny blocks, we mechanically filled gaps (<1 Mb) splitting codirected synteny blocks. The resulting synteny maps were used to liftover Hi-C contacts between the genomes by a “balanced” model.
Computation of the contacts enrichment
The comparison of capture Hi-C of wild type and mutant was performed by the C-InterSecture software, too. The synteny maps were manually generated according to the coordinates of deletion. Then, the Hi-C contacts of wild type were liftovered on the mutant genome by a “balanced” model, as though the deletion effects on chromosome architecture only by changing of a genomic distance. Finally, the given contacts were liftovered from mutant genome on wild type by an “easy” model accounting only synteny between locus.
The difference between the based and liftovered Hi-C map was calculated thus:
Let i, j and k - is genome coordinate in bins. Then:
\({V}_{i,\, j}\) is the contact value between bins i and j.
\({V}_{i,\,j}^{*}\) is the liftovered contact value between bins i and j.
\(D(k,f)=\overline{\log ({V}_{i,\, j}/{V}_{i,\, j}^{*})} \, {{{{{\rm{for}}}}}} \, i\in [k-f,k),\, {j}\in (k,{k}+f],{V}_{i,\, j} \, \ne \, 0,\,{V}_{i,\, j}^{*} \, \ne \, 0,\) and f is distance in bins
Then, we performed a Z-score transformation of D(k,f) for all k and chosen f.
ChIP-seq data processing
Raw paired-end reads were preprocessed with Cutadapt tool to trim adapter sequences (-a and -A flags with Illumina adapter sequences)91. Read quality was assessed using FastQC. Coverage tracks (bigWig) were generated using AQUAS ChIP-Seq pipeline1 with TF or histone parameters [https://github.com/kundajelab/chipseq_pipeline].
To determine a CTCF motif orientation we used the FIMO tool Version 5.5.0 from MEME92 with the matrices MA0139.1 and MA1929.1 from JASPAR93.
To perform a differential ChIP-seq analysis we used getDifferentialPeaksReplicates.pl tool from HOMER software Version 4.11 collection of command line programs94 with standard parameters using mouse genome of mm10 version and Tag directory. We utilized the makeTagDirectory.pl command from HOMER software Version 4.11 to generate a Tag directory. This was performed using the narrow peak files obtained from the AQUAS ChIP-Seq pipeline1 [https://github.com/kundajelab/chipseq_pipeline] for two biological replicates of both wild-type and mutant CTCF ChIP-seq experiments.
RNA-seq data processing
Sequencing data were uploaded to and analyzed on the Galaxy web platform usegalaxy.org95. Quality of sequencing data was controlled via FastQC Galaxy Version 0.7396. Reads were mapped to the mouse reference genome mm10 (GRCm38) with RNA STAR Galaxy Version 2.7.8a97 (default parameter) using the Gencode main annotation file release M1 (NCBIM37). Mapped reads were counted using featureCounts Galaxy Version 2.0.198 (default parameter). Differential gene expression and normalized counts were calculated using DESeq2 Galaxy Version 2.11.40.799 (default parameter).
To evaluate alleles activity in hybrid melanocytes we used Mus castaneus SNPs identified by the Mouse Genomes Project consortium100,101. The identified number of SNPs inside the analyzed genes was: Pdgfra — 19, Kit — 12, Kdr — 21. For each SNP, we calculated the normalized activity of the Wt or Kit Δ30k allele relative to the M. musculus allele. In order to avoid the problem of division by zero, a pseudocount (1) was added to each SNP coverage value before normalization: \({{{{{\rm{Normalized}}}}}} \; {{{{{\rm{allele}}}}}} \; {{{{{\rm{expression}}}}}}=\frac{M.{musculus} \, {{{{{\rm{SNP}}}}}} \; {{{{{\rm{coverage}}}}}}+1}{M.{castaneus}\, {{{{{\rm{SNP}}}}}} \; {{{{{\rm{coverage}}}}}}+1}\). p-value was estimated using Mann–Whitney U test.
Statistics and reproducibility
Hi-C, RNA-seq, and ChIP-seq experiments were repeated at least in two independent replicates for all cell cultures. Staining with toluidine blue of mast cells was performed at least in three independent replicates and was reproducible.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw sequencing data have been deposited in the NCBI SRA database with the following accession number PRJNA838252. Processed data, including Hi-C contact maps, RNA-seq and ChIP-seq tracks are available at https://genedev.bionet.nsc.ru/ftp/by_Project/Kit_locus_GEO. The data used for other organisms are publicly available: fibroblasts34 of human, mouse, rabbit, dog and chicken; African clawed frog fibroblasts35. Source data are provided with this paper.
References
Spielmann, M., Lupiáñez, D. G. & Mundlos, S. Structural variation in the 3D genome. Nat. Rev. Genet 19, 453–467 (2018).
Ibrahim, D. M. & Mundlos, S. Three-dimensional chromatin in disease: What holds us together and what drives us apart? Curr. Opin. Cell Biol. 64, 1–9 (2020).
Hnisz, D. et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351, 1454–1458 (2016).
Lupiáñez, D. G. et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025 (2015).
Redin, C. et al. The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat. Genet 49, 36–45 (2017).
Kraft, K. et al. Serial genomic inversions induce tissue-specific architectural stripes, gene misexpression and congenital malformations. Nat. Cell Biol. 21, 305–310 (2019).
Cova, G. et al. Combinatorial effects on gene expression at the Lbx1/Fgf8 locus resolve split-hand/foot malformation type 3. Nat. Commun. 14, 1475 (2023).
Laugsch, M. et al. Modeling the pathological long-range regulatory effects of human structural variation with patient-specific hiPSCs. Cell Stem Cell 24, 736–752.e12 (2019).
Guo, Y. A. et al. Mutation hotspots at CTCF binding sites coupled to chromosomal instability in gastrointestinal cancers. Nat. Commun. 9, 1520 (2018).
Flavahan, W. A. et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114 (2016).
Schwarzer, W. et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 https://doi.org/10.1038/nature24281 (2017)..
Nora, E. P. et al. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–944.e22 (2017)
Despang, A. et al. Functional dissection of the Sox9–Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nat. Gen. 51, 1263–1271 https://doi.org/10.1038/s41588-019-0466-z (2019).
Williamson, I. et al. Developmentally regulated Shh expression is robust to TAD perturbations. Development (Cambridge) 146, dev179523 https://doi.org/10.1242/dev.179523 (2019).
Puputti, M. et al. Amplification of KIT, PDGFRA, VEGFR2, and EGFR in gliomas. Mol. Cancer Res 4, 927–934 (2006).
Szerlip, N. J. et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc. Natl Acad. Sci. USA 109, 3041–3046 (2012).
Battochio, A. et al. Detection of c-KIT and PDGFRA gene mutations in gastrointestinal stromal tumors: comparison of DHPLC and DNA sequencing methods using a single population-based cohort. Am. J. Clin. Pathol. 133, 149–155 (2010).
Benbarche, S. et al. Screening of ETO2-GLIS2-induced Super Enhancers identifies targetable cooperative dependencies in acute megakaryoblastic leukemia. Sci. Adv. 8, eabg9455 (2022).
Perrot-Applanat, M. & Di Benedetto, M. Autocrine functions of VEGF in breast tumor cells: adhesion, survival, migration and invasion. Cell Adh Migr. 6, 547–553 (2012).
Lyon, M. F. & Glenister, P. H. A new allele sash (Wsh) at the W-locus and a spontaneous recessive lethal in mice. Genet. Res. 39, 315–322 (1982).
Geissler, E. N., Ryan, M. A. & Housman, D. E. The dominant-white spotting (W) locus of the mouse encodes the c-kit proto-oncogene. Cell 55, 185–192 (1988).
Wong, A. K. et al. A de novo mutation in KIT causes white spotting in a subpopulation of German Shepherd dogs. Anim. Genet. 44, 305–310 (2013).
Grassot, J., Gouy, M., Perrière, G. & Mouchiroud, G. Origin and Molecular Evolution of Receptor Tyrosine Kinases with Immunoglobulin-Like Domains. Mol. Biol. Evolution 23, 1232–1241 (2006).
Seifert, R. A., Alpers, C. E. & Bowen-Pope, D. F. Expression of platelet-derived growth factor and its receptors in the developing and adult mouse kidney. Kidney Int. 54, 731–746 (1998).
Gouveia, L., Betsholtz, C. & Andrae, J. Expression analysis of platelet-derived growth factor receptor alpha and its ligands in the developing mouse lung. Physiological Rep. 5, e13092 (2017).
Andrae, J., Gouveia, L., He, L. & Betsholtz, C. Characterization of platelet-derived growth factor-A expression in mouse tissues using a lacZ knock-in approach. PLOS ONE 9, e105477 (2014).
Millauer, B. et al. High affinity VEGF binding and developmental expression suggest Flk-1 as a major regulator of vasculogenesis and angiogenesis. Cell 72, 835–846 (1993).
Quinn, T. P., Peters, K. G., De Vries, C., Ferrara, N. & Williams, L. T. Fetal liver kinase 1 is a receptor for vascular endothelial growth factor and is selectively expressed in vascular endothelium. Proc. Natl Acad. Sci. 90, 7533–7537 (1993).
Banigan, E. J. et al. Transcription shapes 3D chromatin organization by interacting with loop extrusion. Proc. Natl Acad. Sci. 120, e2210480120 (2023).
Kabirova, E. et al. Function and evolution of the loop extrusion machinery in animals. Int. J. Mol. Sci. 24, 5017 (2023).
Battulin, N. et al. Comparison of the three-dimensional organization of sperm and fibroblast genomes using the Hi-C approach. Genome Biol. 16, 77 (2015).
Ryzhkova, A., Taskina, A., Khabarova, A., Fishman, V. & Battulin, N. Erythrocytes 3D genome organization in vertebrates. Sci. Rep. 11, 4414 (2021).
Nuriddinov, M. & Fishman, V. C-InterSecture—a computational tool for interspecies comparison of genome architecture. Bioinformatics 35, 4912–4921 (2019).
Li, D. et al. Comparative 3D genome architecture in vertebrates. BMC Biol. 20, 99 (2022).
Hoencamp, C. et al. 3D genomics across the tree of life reveals condensin II as a determinant of architecture type. Science 372, 984–989 (2021).
Berrozpe, G. et al. A distant upstream locus control region is critical for expression of the kit receptor gene in mast cells. Mol. Cell Biol. 26, 5850–5860 (2006).
Schatteman, G. C., Morrison-Graham, K., Koppen, A. V., Weston, J. A. & Bowen-Pope, D. F. Regulation and role of PDGF receptor a-subunit expression during embryogenesis. Development 115, 123–131 (1992).
Narendra, V. et al. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347, 1017–1021 (2015).
Paliou, C. et al. Preformed chromatin topology assists transcriptional robustness of Shh during limb development. Proc. Natl Acad. Sci. 116, 12390–12399 (2019).
Berrozpe, G. et al. Polycomb responds to low levels of transcription. Cell Rep. 20, 785–793 (2017).
Korablev, A., Lukyanchikova, V., Serova, I. & Battulin, N. On-target CRISPR/Cas9 activity can cause undesigned large deletion in mouse zygotes. Int. J. Mol. Sci. 21, 3604 (2020).
Pelechano, V. & Steinmetz, L. M. Gene regulation by antisense transcription. Nat. Rev. Genet 14, 880–893 (2013).
Bernex, F. et al. Spatial and temporal patterns of c-kit-expressing cells in WlacZ/+ and WlacZ/WlacZ mouse embryos. Development 122, 3023–3033 (1996).
Iida, M. et al. Identification of cardiac stem cells with FLK1, CD31, and VE-cadherin expression during embryonic stem cell differentiation. FASEB J. 19, 371–378 (2005).
Jacob, T. et al. Molecular and spatial landmarks of early mouse skin development. Developmental Cell 58, 2140–2162.e5 (2023).
Peters, E. M. J., Tobin, D. J., Botchkareva, N., Maurer, M. & Paus, R. Migration of melanoblasts into the developing murine hair follicle is accompanied by transient c-Kit Expression. J. Histochem Cytochem. 50, 751–766 (2002).
Cho, S. W. et al. Promoter of lncRNA Gene PVT1 is a tumor-suppressor DNA boundary element. Cell 173, 1398–1412.e22 (2018).
Cao, Y., Liu, S., Cui, K., Tang, Q. & Zhao, K. Hi-TrAC detects active sub-TADs and reveals internal organizations of super-enhancers. Nucleic Acids Res. 51, 6172–6189 (2023).
Rodríguez-Carballo, E. et al. The HoxD cluster is a dynamic and resilient TAD boundary controlling the segregation of antagonistic regulatory landscapes. Genes Dev. 31, 2264–2281 (2017).
Rodríguez-Carballo, E. et al. Chromatin topology and the timing of enhancer function at the HoxD locus. Proc. Natl Acad. Sci. 117, 31231–31241 (2020).
Rajderkar, S. et al. Topologically associating domain boundaries are required for normal genome function. Commun. Biol. 6, 1–10 (2023).
Anania, C. et al. In vivo dissection of a clustered-CTCF domain boundary reveals developmental principles of regulatory insulation. Nat. Genet 54, 1026–1036 (2022).
Barutcu, A. R., Maass, P. G., Lewandowski, J. P., Weiner, C. L. & Rinn, J. L. A TAD boundary is preserved upon deletion of the CTCF-rich Firre locus. Nat. Commun. 9, 1444 https://doi.org/10.1038/s41467-018-03614-0 (2018).
Oh, S. et al. Enhancer release and retargeting activates disease-susceptibility genes. Nature 595, 735–740 (2021).
Islam, Z. et al. Active enhancers strengthen insulation by RNA-mediated CTCF binding at chromatin domain boundaries. Genome Res. 33, 1–17 (2023).
Zuin, J. et al. Nonlinear control of transcription through enhancer-promoter interactions. Nature 604, 571–577 (2022).
Wehrle-Haller, B., Morrison-Graham, K. & Weston, J. A. Ectopic c-kit expression affects the fate of melanocyte precursors in Patch mutant embryos. Dev. Biol. 177, 463–474 (1996).
Soriano, P. The PDGF alpha receptor is required for neural crest cell development and for normal patterning of the somites. Development 124, 2691–2700 (1997).
Stephenson, D. A. et al. Platelet-derived growth factor receptor alpha-subunit gene (Pdgfra) is deleted in the mouse patch (Ph) mutation. Proc. Natl Acad. Sci. USA 88, 6–10 (1991).
Chahar, S. et al. Transcription induces context-dependent remodeling of chromatin architecture during differentiation. PLOS Biol. 21, e3002424 (2023).
Zhang, S. et al. RNA polymerase II is required for spatial chromatin reorganization following exit from mitosis. Sci. Adv. 7, eabg8205 (2021).
Heinz, S. et al. Transcription Elongation Can Affect Genome 3D Structure. Cell 174, 1522–1536.e22 (2018).
Busslinger, G. A. et al. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544, 503–507 (2017).
Zhang, S., Übelmesser, N., Barbieri, M. & Papantonis, A. Enhancer–promoter contact formation requires RNAPII and antagonizes loop extrusion. Nat. Genet 55, 832–840 (2023).
Hsieh, T.-H. S. et al. Resolving the 3D Landscape of Transcription-Linked Mammalian Chromatin Folding. Mol. Cell 78, 539–553.e8 (2020).
Reed, K. S. M. et al. Temporal analysis suggests a reciprocal relationship between 3D chromatin structure and transcription. Cell Rep. 41, 111567 (2022).
Olan, I. et al. Transcription-dependent cohesin repositioning rewires chromatin loops in cellular senescence. Nat. Commun. 11, 6049 (2020).
Uyehara, C. M. & Apostolou, E. 3D enhancer-promoter interactions and multi-connected hubs: Organizational principles and functional roles. Cell Rep. 42, 112068 (2023).
Greenwald, W. W. et al. Subtle changes in chromatin loop contact propensity are associated with differential gene regulation and expression. Nat. Commun. 10, 1054 (2019).
Gaspar-Maia, A., Alajem, A., Meshorer, E. & Ramalho-Santos, M. Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 12, 36–47 (2011).
Chabot, B., Stephenson, D. A., Chapman, V. M., Besmer, P. & Bernstein, A. The proto-oncogene c-kit encoding a transmembrane tyrosine kinase receptor maps to the mouse W locus. Nature 335, 88–89 (1988).
Nocka, K. et al. Expression of c-kit gene products in known cellular targets of W mutations in normal and W mutant mice–evidence for an impaired c-kit kinase in mutant mice. Genes Dev. 3, 816–826 (1989).
Fontanesi, L. et al. The KIT gene is associated with the english spotting coat color locus and congenital megacolon in checkered giant rabbits (Oryctolagus cuniculus). PLoS ONE 9, e93750 (2014).
Hu, S. et al. KIT is involved in melanocyte proliferation, apoptosis and melanogenesis in the Rex Rabbit. PeerJ 8, e9402 (2020).
David, V. A. et al. Endogenous retrovirus insertion in the KIT oncogene determines white and white spotting in domestic cats. G3 (Bethesda) 4, 1881–1891 (2014).
Hauswirth, R. et al. Novel variants in the KIT and PAX3 genes in horses with white-spotted coat colour phenotypes. Anim. Genet. 44, 763–765 (2013).
Huynh, L. & Hormozdiari, F. TAD fusion score: discovery and ranking the contribution of deletions to genome structure. Genome Biol. 20, 60 (2019).
Real, F. M. et al. The mole genome reveals regulatory rearrangements associated with adaptive intersexuality. Science 370, 208–214 (2020).
Ringel, A. R. et al. Repression and 3D-restructuring resolves regulatory conflicts in evolutionarily rearranged genomes. Cell 185, 3689–3704.e21 (2022).
Smirnov, A. V. et al. Evaluation of the α-casein (CSN1S1) locus as a potential target for a site-specific transgene integration. Sci. Rep. 12, 1–10 (2022).
Kruglova, A. A. et al. Embryonic stem cell/fibroblast hybrid cells with near-tetraploid karyotype provide high yield of chimeras. Cell Tissue Res. 334, 371–380 (2008).
Murphy, B. M., Weiss, T. J. & Burd, C. E. Rapid generation of primary murine melanocyte and fibroblast cultures. J. Vis. Exp. https://doi.org/10.3791/59468 (2019).
Vukman, K. V., Metz, M., Maurer, M. & O’Neill, S. M. Isolation and culture of bone marrow-derived mast cells. Bio-Protoc. 4, e1053 (2014).
Yale Flow Cytometry. Flow cytometry (FACS) staining protocol (Cell surface staining). https://medicine.yale.edu/immuno/flowcore/protocols/analysis/ (2019).
Pękowska, A. et al. Gain of CTCF-anchored chromatin loops marks the exit from naive pluripotency. Cell Syst. 7, 482–495.e10 (2018).
Belaghzal, H., Dekker, J. & Gibcus, J. H. Hi-C 2.0: An optimized Hi-C procedure for high-resolution genome-wide mapping of chromosome conformation. Methods 123, 56–65 (2017).
Ulianov, S. V. et al. Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome Res 26, 70–84 (2016).
Durand, N. C. et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 (2016).
Li, D. et al. WashU epigenome browser update 2022. Nucleic Acids Res. 50, W774–W781 (2022).
Harris, R. S. Improved pairwise alignment of genomic DNA. Ph.D. Thesis. (The Pennsylvania State University, 2007).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10 (2011).
Bailey, T. L., Johnson, J., Grant, C. E. & Noble, W. S. The MEME Suite. Nucleic Acids Res. 43, W39–W49 (2015).
Castro-Mondragon, J. A. et al. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 50, D165–D173 (2022).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Afgan, E. et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544 (2018).
Babraham Bioinformatics - FastQC A Quality Control tool for High Throughput Sequence Data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Keane, T. M. et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477, 289–294 (2011).
Doran, A. G. et al. Deep genome sequencing and variation analysis of 13 inbred mouse strains defines candidate phenotypic alleles, private variation and homozygous truncating mutations. Genome Biol. 17, 167 (2016).
Acknowledgements
This work was supported with the budget project of Institute of Cytology and Genetics SB RAS (state project FWNR-2022-0019). Experiment with M. castaneus hybrids was supported by Russian Science Foundation grant #22-14-00247. Hi-C experiments sequencing was performed using equipment of the Novosibirsk State University, supported by the Ministry of Education and Science of Russian Federation, grant #FSUS-2024-0018. Illumina data analysis was supported by the strategic academic leadership program “Priority 2030” in Novosibirsk State University. Cell culturing was performed at the Collective Center of ICG SB RAS “Collection of Pluripotent Human and Mammalian Cell Cultures for Biological and Biomedical Research”, project number FWNR-2022-0019 (https://ckp.icgen.ru/cells/; http://www.biores.cytogen.ru/brc_cells/collections/ICG_SB_RAS_CELL). The calculations were performed using computational resources of Computational Center of Novosibirsk State University.
Author information
Authors and Affiliations
Contributions
N.B. conceived and supervised the study; E.K., A.R., V.L. and A.Kh. performed Hi-C, ChIP-seq, RNA-seq experiments; M.N. performed Hi-C data analysis; P.B. performed ChIP-seq and RNA-seq data analysis; T.S. performed immunohistochemistry analysis; A.S. designed the genetic constructs; N.V.K. conducted the experiment to assess the circadian rhythms of the animals; A.Ko., G.K. and I.S. obtained genome–edited mice. All the authors contributed to the manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
: Nature Communications thanks Lila Allou, Martin Franke and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kabirova, E., Ryzhkova, A., Lukyanchikova, V. et al. TAD border deletion at the Kit locus causes tissue-specific ectopic activation of a neighboring gene. Nat Commun 15, 4521 (2024). https://doi.org/10.1038/s41467-024-48523-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-48523-7
This article is cited by
-
A 14-bp motif in the KIT active promoter region is critical for melanin accumulation in yaks, mice, and humans
BMC Biology (2025)
-
Direction and modality of transcription changes caused by TAD boundary disruption in Slc29a3/Unc5b locus depends on tissue-specific epigenetic context
Epigenetics & Chromatin (2025)
-
Structural variants in the 3D genome as drivers of disease
Nature Reviews Genetics (2025)