Abstract
T cell expansion has a crucial function in both autoimmune and chronic inflammatory diseases, with cycling T cells contributing to the pathogenesis of autoimmune diseases by causing uncontrolled immune responses and tissue damage. Yet the regulatory mechanisms governing T cell expansion remain incompletely understood. Here we show that the enzyme N-acetyltransferase 10 (NAT10) regulates T cell activation and proliferation upon antigen stimulation. T cell-specific NAT10 deficiency in mice reduces the number of mature T cells in peripheral lymphoid organs. Mechanistically, NAT10 acetylates RACK1 at K185, preventing subsequent RACK1 K48-linked ubiquitination and degradation. The increased RACK1 stability alters ribosome formation and cellular metabolism, leading to enhanced supply of energy and biosynthetic precursors and, eventually, T cell proliferation. Our findings thus highlight the essential function of NAT10 in T cell self-renewal and metabolism and elucidate NAT10 mode of action for the potential development of novel therapies for immune-related disorders.
Similar content being viewed by others
Introduction
Autoimmune and inflammatory diseases represent a spectrum of conditions characterized by disruptions in immune system homeostasis, particularly involving T-cell dysregulation1,2. Conditions such as rheumatoid arthritis (RA), multiple sclerosis (MS), and systemic lupus erythematosus (SLE) exemplify this dysregulation, where abnormal T-cell hyperactivity and unchecked proliferation lead to tissue damage and inflammatory responses3,4,5. Autoreactive T cells mistakenly recognize self-antigens as foreign, initiating an immune attack on host tissues and causing their deterioration6. Similarly, inflammatory disorders such as psoriasis and inflammatory bowel disease (IBD) also involve imbalances in T-cell regulation7. Environmental factors or microbial antigens can trigger inappropriate T-cell activation in these conditions, leading to chronic inflammation8. The resulting T-cell hyperproliferation and increased activation produce excessive pro-inflammatory cytokines and recruit additional immune cells, exacerbating inflammation9,10. Therefore, targeting autoreactive T-cell proliferation and activity is a crucial therapeutic strategy for managing autoimmune and inflammatory diseases.
Cycling T cells, or proliferating T cells, are a subset of T lymphocytes characterized by their active cell division in response to antigenic stimulation, and play a key role in the adaptive immunity11,12. However, dysregulation of this process is central to the pathogenesis of autoimmune and inflammatory disorders, where proliferating T cells attack self-antigens, leading to tissue damage and organ dysfunction13. This cellular proliferation involves repeated cycles of DNA replication and mitosis, significantly increasing cell numbers and enhancing the immune response14. These effector T cells disrupt the immune balance by releasing pro-inflammatory cytokines and promoting the development of additional inflammatory T cells15. Uncontrolled proliferation of these cells can impair regulatory T cell (Treg) function, further contributing to immune dysregulation and inflammation16. Understanding the molecular mechanisms that regulate the proliferation and differentiation of cycling T cells is critical for developing targeted immunotherapies.
Our investigation identified N-acetyltransferase 10 (NAT10) as a key regulator in cycling T cell biology. Despite this initial discovery, the role of NAT10 in T cell proliferation and its broader immunological implications remains largely unexplored. Further elucidation of NAT10 function could provide new avenues for therapeutic intervention, particularly in modulating immune responses in autoimmune and inflammatory diseases.
NAT10 plays a crucial role in various cellular physiological and pathophysiological processes17,18,19, facilitating the acetylation of a wide range of substrates by transferring an acetyl group from acetyl coenzyme A (Ac-CoA) to various targets, including proteins and smaller molecules. Alterations in NAT10 expression or genetic mutations of NAT10 have been associated with several pathological conditions, including cancers, developmental disorders, and neurodegenerative diseases. Previous studies have highlighted the acetyltransferase activity of NAT10, particularly its modification of α-tubulin, a key component of the cytoskeleton20. This post-translational modification of α-tubulin by NAT10 is essential for maintaining microtubule dynamics and structure. Furthermore, NAT10 has been recognized for its ability to enact acetylation-driven modulation of the prominent tumor suppressor complex p5321. This modulation is instrumental in orchestrating the fidelity of cell cycle governance and the DNA damage response. However, the function of NAT10 in the immune system, especially its role of T cell-mediated immune responses, remains unclear. Here in this study, we explore how NAT10 played a role in inflammatory and autoimmune diseases by controlling of T cell proliferation. We find that NAT10-deficient mice have severe defects in T cell development, which result in a significantly reduced number of mature T cells in the peripheral lymphoid organs. NAT10 contributes to the regulation of T cell proliferation in response to antigen stimulation by inhibiting the K48-linked ubiquitination and degradation of the receptor for activated C kinase 1 (RACK1). Impaired RACK1 stability impacts ribosome formation and various metabolic processes, including glycolysis and fatty acids metabolism, both of which are critical for supplying the energy and biosynthetic precursors to sustain the rapid proliferation and effector functions of activated T cells. Our research on the precise mechanisms of NAT10 function in T cells might provide insights into the regulation of immune responses and the development of new therapies for immune-related diseases.
Result
NAT10 is highly expressed in cycling and activated T cells
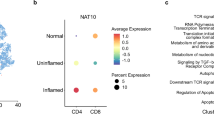
In order to comprehend the composition of infiltrated immune cells in the colons of patients with IBD and to compare the characteristics of inflamed and non-inflamed regions, we analyzed public single cell (sc)-RNA sequencing data of colon tissues obtained from the Single Cell Portal (accession code SCP1884), which encompassed 14 healthy colon tissues, 17 inflamed colon tissues, and 5 non-inflamed colon tissues from patients with Crohn’s disease (CD). To resolve the cell type alterations of CD4+ T cells, 6 subclusters were identified, including naive-like T, Activated FosB+ T, Th1, Th17, Treg and MKi67+ cycling T subpopulations, based on their marker genes (Fig. 1a, b). We further compared the proportions of cycling T cells in these three groups as above. The results revealed a higher levels of cycling T cells were observed in the inflamed regions of patients with CD compared to those in non-inflamed regions or healthy volunteers (Fig. 1c). Consistently, this population of CD4+ cycling T cells also appeared in the single cell transcriptome sequencing data of cerebrospinal fluid from multiple sclerosis patients (Supplementary Fig. 1a, b), indicating that it played a key role in the transformation of cycling T cells into other types of pathogenic T lymphocyte functional subsets. We thus proposed that this group of cycling T cells was closely related to the occurrence of autoimmune and inflammatory diseases.
a The raw single cell sequencing data containing 14 non-IBD colon tissues, 17 inflamed colon tissues, and 5 non-inflamed colon tissues from CD patients obtained from the Single Cell Portal (accession code SCP1884) and processed according to workflow as the cartoon shown. t-distributed stochastic neighbor embedding (t-SNE) plot showing the clusters of indicated immune cells. b The heatmap showing the marker genes of clusters in (a). c The percentage of cycling T cells in total CD4+ T cells of healthy (n = 14), CD non-inflamed (n = 17), CD inflamed samples (n = 5). d Retrieved upregulated expression genes between cycling and non-cycling T cells in CD samples were subjected to GO analysis, and the dot plot showing the enriched significant pathway. p value was determined by default two-tailed Stouffer test and parameters in MAST packages. e The gene signature score of G1S and G2M transition pathway were calculate by Integrate all single cell rank-based gene set enrichment analysis (irGSEA) and visualized by t-SNE plot. f Expression features of NAT10 gene in different clusters were showed as violin plots. g NAT10 expression feature of cycling T cells from healthy, CD non-inflamed, CD inflamed samples. h The different expression genes between NAT10 positive (NAT10pos) and NAT10 negative (NAT10neg) cells were subjected to GO enrichment analysis, and top enriched pathways were presented by dot plot. p value was determined by default two-tailed Stouffer test and parameters in MAST packages. i WT CD4+ (n = 4) or CD8+ (n = 3) naïve T cells were stimulated with anti-CD3/anti-CD28 for indicated time, and the abundance of Nat10 mRNA were analyzed by RT-qPCR. These qPCR data are presented as fold change relative to the Actb mRNA level and normalized by Bio-Rad CFX Manager 3.1. j WT naïve CD4+ T cells were activated for indicated time by TCR crosslinking, the cytosolic and nuclear extracts were subjected to immunoblotting with antibody against NAT10. k Naïve or activated T cells were fixed and permeabilizated, stained with anti-NAT10 antibody and fluorescein-conjugated second antibody, representative images were subsequently captured by confocal. The left scale bar indicates 10 μm, the right one indicates 1 μm. Source data are provided as a Source Data file. Data were represented as mean ± S.D. Three independent experiments at least were performed in data represented above. Statistical significance in (c) was determined by one-way ANOVA with multiple comparisons. *P < 0.05, ***P < 0.001.
Previous research suggested that this cycling CD4+ T-cell subpopulation was in a state of active cell proliferation22. Our data also demonstrated that different from other CD4+ T cell subsets, the differentially expressed genes (DEGs) in cycling CD4+ T-cell from the patients with CD were mainly enriched in cell cycle and proliferation-related pathways such as DNA replication and meiosis (Fig. 1d, e). The energy utilized by T cells during clonal expansion is primarily allocated to support genome synthesis, thereby leading to a concomitant decrease in effector function during the proliferation process. Likewise, DEGs in cycling T cells from the patients with CD also indicated an anticipated reduction in rRNA, mRNA, and miRNA metabolic processes, all of which were intricately linked to translation processes (Supplementary Fig. 1C). During T cell proliferation, there was an increased demand for energy and metabolic pathways. T cells consumed glucose through glycolysis to generate ATP needed for cell division and cytokine production23,24. Additionally, T cell activation stimulated the uptake of amino acids and lipids, which were crucial for protein synthesis and membrane expansion during proliferation25. Metabolic pathways, such as glycolysis, oxidative phosphorylation (OXPHOS), and fatty acid oxidation (FAO), were more actively engaged in cycling T cells from the patients with CD compared to other T cell subtypes (Supplementary Fig. 1d, e). This was manifested by the increased expression of metabolic enzymes associated with these pathways in cycling T cells (Supplementary Fig. 1f).
To investigate its physiological function, we first identified biomarkers of cycling CD4+ T-cell. scRNA-seq data showed that NAT10 was predominantly expressed in cycling T cells, albeit in a relatively small proportion (Fig. 1f). A similar phenotype of high NAT10 expression was also observed in cycling CD4+ T cells of MS patients (Supplementary Fig. 1g). This small proportion of NAT10+ cycling T cells was related to the heterogeneity of the cell cycle phases within the cycling T cell population. Compared with those in colons of healthy donors or the non-inflammatory regions of CD patients, the cycling CD4+ T cells in the inflammatory regions expressed higher levels of NAT10 (Fig. 1g), indicating that the NAT10-positve (NAT10pos) cycling CD4+ T cells had stronger proliferative activity associated with inflammatory responses (Fig. 1h). To clarify the relationship between NAT10 expression and T cell activation, naive CD4+ and CD8+ T cells were stimulated with anti-CD3 plus anti-CD28. The results indicated that mRNA and protein levels of NAT10 were increased rapidly in splenic naive CD4+ and CD8+ T cells stimulated with anti-CD3 plus anti-CD28 (Fig. 1i, j). Upon stimulation, a portion of NAT10 translocated from the nucleus to the cytoplasm (Supplementary Fig. 1h), mirroring the behavior observed in CD4+ T cells. Consistent with CD4+ T cells, NAT10 also predominantly resided in the nucleus of activated CD8+ T cells (Supplementary Fig. 1h). Short-term stimulation induced the relocalization of NAT10 from the nucleus to the cytoplasm during CD8+ T cells activation (Fig. 1k). These findings indicated that in activated T cells, NAT10 exerted its function not only within the nucleus but also through potential interactions with proteins or RNA in the cytoplasm, thereby regulating the functionality of these molecules.
NAT10 is necessary for T lymphocyte development and homeostasis maintenance
To elucidate the function of NAT10 in T lymphocytes and its involvement in the development of inflammatory diseases, we generated Nat10 floxed mice and crossed them with CD4-Cre mice to obtain T cell-specific knockout mice (NAT10cKO, Supplementary Fig. 2a, b). NAT10cKO mice were born in the same proportion of genotypes as Mendel’s genetic law, and their survival ratio has no difference with that of wild-type (WT) mice. We firstly analyzed the development of peripheral T lymphocytes in NAT10cKO mice in distinct immune organs using flow cytometry (FACS). As shown in Fig. 2a, we observed a significant reduction in the percentage and cell numbers of splenic T lymphocytes in NAT10cKO mice. A similar phenotype was found in the inguinal lymph node (iLN) of NAT10cKO mice (Supplementary Fig. 2c). A decrease in the number of T lymphocytes often resulted in compensatory activation of remaining T cells. In NAT10cKO mice, we also observed an upregulation in the proportion of CD44hi memory subsets of CD8+ T lymphocytes both in spleen and iLN (Fig. 2b and Supplementary Fig. 2d). By analyzing thymic maintenance, we ruled out the possibility that NAT10 deficiency led to the developmental dysfunction of T lymphocyte in the thymus, as no significant difference was observed in the proportions of double positive (DP) or CD4+/CD8+ single positive (SP) T cell subsets in the thymus between NAT10cKO and WT mice (Supplementary Fig. 2e, f). However, this phenomenon was not due to an increase in frequency of Treg cells, as their proportion in the peripheral immune organs of NAT10cKO mice was comparable to that of WT controls (Fig. 2c). Although NAT10 impacted the development of T cells, we observed a significant autoactivation of peripheral T cells, evidenced by their upregulated capacity to produce cytokines under steady-state conditions (Fig. 2d). Indeed, previous studies have demonstrated that upon arrest of T cell development, T cells exhibited an autoactivation phenotype characterized by increases in effector memory (EM) and central memory (CM) cells, as well as upregulation of activate biomarkers such as CD69 and CD4426.
a Flow cytometry (FACS) analysis of frequency of T cell and B cell subpopulation in inguinal lymph nodes (iLN) and spleen from NAT10WT and NAT10cKO mice (n = 6). b Naïve and memory subtypes of CD4+ T cell and CD8+ T cell in iLN and spleen from NAT10WT and NAT10cKO mice (n = 6). N, naïve; M, memory; CM, central memory; EM, effector memory. c FACS analysis of CD4+Foxp3+ Treg cells in iLN and spleen from NAT10WT and NAT10cKO mice (n = 6). d Frequency of IFN-γ+ or IL-17A+ CD4+ cells and IFN-γ+ CD8+ T cells in spleen and iLN of NAT10WT (n = 3) and NAT10cKO (n = 4) mice were analyzed by FACS. e Lethally radiated SJL mice (CD45.1) received mixed bone marrow from NAT10WT (CD45.1+ CD45.2+; n = 5) and NAT10cKO (CD45.2+; n = 4) and reconstructed for 8 weeks. Frequency of NAT10cKO, NAT10WT congenic CD4+ T cell and CD8+ T cell in iLN and spleen were assessed by FACS. Source data are provided as a Source Data file. Data were represented as mean ± S.D. Three independent experiments at least were performed in data represented above. Significance of the differences in all panels were identified by two-tailed unpaired t tests. ns no significance, *P < 0.05, **P < 0.01, ***P < 0.001.
To evaluate whether NAT10 regulated T lymphocyte homeostasis in an intrinsic role, we co-transplanted bone marrow (BM) cells from NAT10cKO mice (CD45.2+CD45.1-) and WT SJL mice (CD45.2+CD45.1+) into lymphocyte-deficient Rag1 KO mice. NAT10-deficient and WT T cells undergo similar development in the thymic stage (Supplementary Fig. 2g), but the proportion of mature T cell derived from BM of NAT10cKO mice exhibited a significant reduction in spleen and iLN of recipient mice (Fig. 2e). These data indicated that the regulation of peripheral T lymphocyte homeostasis by NAT10 was depending on its intrinsic function rather than being caused by changes in the cellular developmental environment. Combined with the high expression of NAT10 in cycling T cells, we proposed that cycling T cell was an indispensable state of peripheral T cell activation and expansion.
Deficiency of NAT10 in T lymphocytes alleviates the onset of inflammatory and autoimmune diseases
Based on the finding above, NAT10 played a crucial role in peripheral T lymphocyte maintenance. Therefore, NAT10 might contribute to the occurrence and progression of inflammatory diseases. Therefore, targeting NAT10 in T cells had potential implications for alleviating these diseases. To investigate the potential involvement of NAT10 in the pathogenesis of IBD, we established a T cell-mediated colitis model by using CD4+CD25-CD45RBhi naïve T cells from either WT or NAT10cKO mice and adoptively transferring them into Rag1–/– mice. The results revealed that NAT10-deficient T cells failed to induce colitis in Rag1–/– mice as their WT control. Receipt mice with NAT10-deficient T cells did not exhibit body weight loss (Fig. 3a), inflammation-driven death (Fig. 3b), intestinal barrier damage (Fig. 3c), and immune cell infiltration in the gut as those receiving WT T cells (Fig. 3d). Moreover, NAT10-deficient T cells infiltrating the intestine demonstrated reduced IFN-γ expression, suggesting their impaired activation and expansion (Fig. 3e). Dextran sulfate sodium (DSS) is a chemical compound commonly used to induce colitis in murine models. DSS disrupted the intestinal epithelial barrier, leading to increased intestinal permeability and inflammation caused by macrophages. In contrast to its critical role in T cells, specific deletion of NAT10 in myeloid cells did not show any significant difference in the severity of DSS-induced inflammation compared to WT mice (Supplementary Fig. 3a). NAT10 deficiency also had no impact on macrophage development in the bone marrow (Supplementary Fig. 3b) or on the development and homeostasis of peripheral T cells (Supplementary Fig. 3c). In vitro, NAT10 deficiency did not affect the induction of pro-inflammatory cytokines in macrophages stimulated by LPS (Supplementary Fig. 3d). These findings demonstrated that the role of NAT10 was indispensable in T lymphocyte-mediated colitis, and inhibition of NAT10 effectively alleviated the progression of colitis.
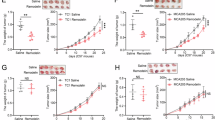
a, b Rag1–/– mice were intravenously (i.v.) injected the CD4+CD25-CD45RBhi naïve T cells isolated from NAT10WT (n = 5) or NAT10cKO mice (n = 5). The body weight (a) and survival rate (b) of receipt mice were monitored every 2 days. p value and significance were determined by two-tailed Log-rank test use default parameters. c Representative H&E staining slides of sections from proximal colon of NAT10WT (n = 3) or NAT10cKO mice (n = 5). The top scale bar indicates 200 μm, the bottom one indicates 50 μm. d FACS analysis of NAT10WT (n = 3) or NAT10cKO mice (n = 5) derived CD4+ T cells infiltration in iLN, spleen, mesenteric lymph nodes (mLN) and lamina propria (LP) of RAG1-/- recipients. e LP infiltrated immunocytes and splenocytes isolated from recipients transferred naïve T cells from NAT10WT (n = 3) or NAT10cKO mice (n = 5) were stimulated with PMA plus ionomycin, and IFN-γ production were analyzed by FACS. f NAT10WT (n = 11) and NAT10cKO (n = 7) mice were induced experimental autoimmune encephalomyelitis (EAE) model, the clinical scores were monitored from 5th day after immunization. g On the 14th day after immunization, the brains and spinal cords of the immunized mice were collected and dissociated, and immunocytes infiltration was analyzed by FACS (n = 3). The bar graphs were presented the percentage and absolute cell number of distinct immune cells subsets. h Representative H&E staining of spinal cord sections from mice immunized for 14 days. i–j Splenocytes of immunized mice were re-stimulated with 10 µg/mL MOG33-35 peptide and subjected to CCK8-based proliferation assay and ELISA-based cytokines production assay (n = 3). Source data are provided as a Source Data file. Data were represented as mean ± S.D. Three independent experiments at least were performed in data represented above. Statistical significance was determined by two-sided multiple t test (a, f), one-way ANOVA with multiple comparisons (d, h, i, j) and two-tailed t test (c, e). *P < 0.05, **P < 0.01, ***P < 0.001.
To further investigate the role of NAT10 in T cell-mediated autoimmune diseases, we utilized NAT10 cKO mice to construct a model of experimental autoimmune encephalomyelitis (EAE). We immunized both WT and cKO mice with myelin oligodendrocyte glycoprotein (MOG) peptide (MOG35-55) along with pertussis toxin (PT). Compared to WT mice, NAT10cKO mice failed to efficiently induce autoimmune EAE (Fig. 3f) and exhibited reduced cell number of infiltrating immune cell in central nervous system (CNS) (Fig. 3g, h). CD4+ T cells infiltrating the CNS derived from NAT10cKO mice displayed impaired responsiveness to MOG antigen, characterized by attenuated proliferation (Fig. 3i) and diminished production of effector cytokines (Fig. 3j). Based on these data, the absence or inhibition of NAT10 resulted in the significant alleviation of T lymphocyte-mediated autoimmune and inflammatory diseases. Therefore, we believed that NAT10 represented a potential and promising therapeutic target for drug development.
NAT10 is indispensable for the expansion of T lymphocytes
Mature T lymphocytes maintain a quiescent state in vivo for prolonged periods, and upon activation by antigens presented by antigen-presenting cells (APCs). To investigate the role of NAT10 in T lymphocyte function, we isolated T lymphocytes from both WT and NAT10cKO mice using FACS and analyzed their proliferation and activation status after stimulation with anti-CD3/anti-CD28. NAT10-deficient T lymphocytes exhibited a smaller cell size compared to WT counterparts following activation (Supplementary Fig. 4a), suggesting an inhibited growth process without NAT10 in T cells. Subsequently, we conducted an analysis of their proliferative potential using CFSE labeling. The results revealed that NAT10 deficiency led to the defect of proliferative capacity in T lymphocytes (Fig. 4a). A similar phenotype was also observed in NAT10-deficient T cells stimulated with PMA plus ionomycin (Fig. 4b). Interestingly, the inhibition of growth and proliferation in T lymphocytes caused by NAT10 deficiency was not associated with primarily activation, as the expression levels of CD69 and CD44 on the surface were similar to those of WT after stimulation (Supplementary Fig. 4b, c). Furthermore, NAT10-deficient CD4+ T cells could not efficiently differentiate into distinct functional subsets including Th1, Th17 or Treg in vitro (Fig. 4c). Compared to WT control, NAT10-deficient CD4+ T cells tended to apoptosis with absence of expansion effectively upon stimulation (Fig. 4d).
Naïve CD4+ T cell (CD44loCD62Lhi) derived from NAT10WT and NAT10cKO mice were labeled with CFSE and stimulated with anti-CD3 plus anti-CD28 (a) or PMA plus ionomycin (b) for indicated time (n = 3). The proliferation potential was assessed according CFSE dilution by FACS. c Naïve CD4+ T cells from NAT10WT and NAT10cKO mice were polarized under indicated differentiation condition (n = 3). The subset-specific cytokines or transcription factors were analyzed by FACS after 96 h. d WT and NAT10-deficient naïve CD4+ T cell was stimulated with anti-CD3 plus anti-CD28 for 24 h (n = 3). The apoptosis (Annexin V+PI+) of each group was measured by FACS. e The enrichment in significant terms of TOP20 downregulated genes in NAT10-deficient CD4+ T cells compared to those in WT control was analyzed by KEGG analysis. f WT and NAT10-deficient naïve CD4+ T cells were stimulated with anti-CD3 plus anti-CD28 for indicated time and incorporated with BrdU (n = 3). The cell cycle was analyzed by FACS. g Heatmap showing different expression level of cell cycle associated genes between NAT10WT and NAT10-deficient naïve CD4+ T cells stimulated with anti-CD3 plus anti-CD28 for 24 h. h Naïve CD4+ T cell derived from NAT10WT and NAT10cKO mice were stimulated with anti-CD3 plus anti-CD28 for 24 h (n = 3). The mRNA level of Ccne1, Cdk4, Cdkn1a, and Trp53 gene were measured by RT-qPCR. These qPCR data are presented as fold change relative to the Actb mRNA level and normalized by Bio-Rad CFX Manager 3.1. i The protein abundance of c-Myc and P53 in WT and NAT10-deficient CD4+ T cells under stimulation were analyzed by immunoblotting (IB). Source data are provided as a Source Data file. Data were represented as mean ± S.D. Three independent experiments at least were performed in data represented above. Statistical significance was determined by unpaired two-tailed t test (a–d, f, h). *P < 0.05, **P < 0.01, ***P < 0.001.
To elucidate the impact of NAT10 deficiency on biological processes, we compared the transcriptomes of activated WT and NAT10-deficient T cells. Compared to WT controls, NAT10-deficient T cells exhibited downregulation of genes primarily enriched in pathways such as DNA replication, cell cycle, DNA damage repair, and multiple metabolic pathways (Fig. 4e and Supplementary Data file 1). We next performed cell cycle analysis using BrdU incorporation to investigate the effect of NAT10 on cell cycle progression and found that NAT10 deficiency impaired the entrance of S phase in T lymphocytes during the activation (Fig. 4f). Consistently, the genes that associated with cell cycle regulation failed to be induced upon NAT10 deficiency, whereas the expression of genes those related to cell cycle arrest, including Mdm2, Cdkn1a encoding P21, and Tp53 encoding P53, showed significantly upregulation (Fig. 4g). Similar results were also confirmed by using qPCR assay and immunoblotting (IB) (Fig. 4h, i). To further investigate the stage at which NAT10 was involved in T cell proliferation and activation, we infected NAT10flox/flox T cells with retrovirus expressing Cre after activation for 24 h. The cell proliferation assay showed that NAT10 was dispensable in mid-to-late stages of T cell activation (Supplementary Fig. 4d). Furthermore, we found that silence of NAT10 after T cell activation had no effect on their differentiation into distinct functional subtypes (Supplementary Fig. 4e). Collectively, these findings indicated that NAT10 played an indispensable role in T cell normal growth, proliferation, and differentiation during the early stages of cell cycle in activated T lymphocyte.
Loss of NAT10 has no effect on the activation of various signal pathways downstream of TCR
Upon recognition of antigen by the T cell receptor (TCR), zeta-associated protein of 70 kDa (ZAP70) is recruited and activated in T cells27,28. ZAP70 then phosphorylates various downstream signaling molecules, leading to the activation of multiple signaling pathways including NF-κB, mitogen-activated protein kinase (MAPK), and nuclear factor of activated T cells (NFAT)29. These pathways drive T cell proliferation, differentiation, and effector function. Here, we employed anti-CD3 and anti-CD28 to stimulate WT and NAT10-deficient T lymphocytes and subsequently performed IB analysis of these signaling pathways during the early stages of cell activation. Although early studies have reported the recruitment of NAT10 into the T cell receptor (TCR) complex, our findings revealed that NAT10 deficiency did not alter the phosphorylation levels of ZAP70 and phospholipase C gamma (PLCγ) upon TCR stimulation (Supplementary Fig. 5a). Consistently, NAT10 deficiency had no or a mild impact on the activation of signaling pathways, including NF-κB (Fig. 5a), MAPK (Fig. 5b), and NFATs (Fig. 5c).
a–d Naïve CD4+ T cells isolated from NAT10WT and NAT10cKO mice were stimulated with anti-CD3 plus anti-CD28 for indicated time. The activation of MAPKs (a), NF-κB (b), NFAT (c) and AKT (d) signaling pathway were measured by IB. Extracellular acidification rate (ECAR) (e) and oxygen consumption rate (OCR) (f) of WT and NAT10-deficient naïve CD4+ T cells stimulated with anti-CD3 plus anti-CD28 for 18 h were measured by Agilent Seahorse XF Analyzers (n = 3). Heatmap of different expression level of glycolysis (g) and fatty acid metabolism (h) associated genes between WT and NAT10-deficient naïve T cells stimulated with anti-CD3 plus anti-CD28 for 24 h. Source data are provided as a Source Data file. Data were represented as mean ± S.D. Three independent experiments at least were performed in data represented above.
Upon antigen recognition, T cells undergo metabolic reprogramming characterized by increased glycolysis and glutaminolysis to meet the energy demand required for T cell growth and proliferation23,30. AKT, known as protein kinase B (PKB), plays a crucial role in regulating cellular metabolism. Activation of AKT signaling pathway influences multiple metabolic processes in cells. Although a similar level of AKT phosphorylation was found between WT and NAT10-deficient T cells (Fig. 5d), glycolysis assays in vitro indicated that the T cells without NAT10 were unable to effectively undergo glycolysis (Fig. 5e). Fatty acid metabolism plays a pivotal role in regulating T cell proliferation and activation31. Upon T cell activation, there is an increased demand for fatty acids as energy substrates and building blocks for lipid synthesis to support T cell growth and division25. However, NAT10-deficient T cells were also unable to produce ATP for cell energy through FAO pathway (Fig. 5f), indicating that the energy demand of NAT10-deficient T cells was significantly reduced when they did not proliferate efficiently. Furthermore, various key enzymes involved in glycolysis (Fig. 5g) and fatty acid metabolism (Fig. 5h) were significantly dysregulated in NAT10-deficient T lymphocytes compared to WT control upon activation. Due to the decline in mitochondrial oxidative respiration, the level of reactive oxygen species (ROS) in NAT10-deficient T cells also decreased (Supplementary Fig. 5b). N-acetylcysteine (NAC), often used as an antioxidant that scavenged ROS in vitro, was unable to rescue the proliferation-defective phenotype of NAT10-deficient T cells when treated with NAC or GSH (Supplementary Fig. 5c). Therefore, the decrease in proliferative capacity of these T cells was not a result of an increase in intracellular ROS production. In terms of glycolysis, acetate enters the metabolic pathway as acetyl-CoA, which can be generated via the conversion of acetate to acetyl-CoA by the enzyme acetyl-CoA synthetase (ACS). Acetyl-CoA is then utilized in the tricarboxylic acid (TCA) cycle, providing carbon units for ATP production via oxidative phosphorylation. In FAO, acetate serves as a substrate for β-oxidation, a process by which fatty acids are broken down to generate energy. Acetate is converted to acetyl-CoA, which becomes an entry point into the β-oxidation pathway. However, addition of acetate during the proliferation of NAT10-deficient T cells was unable to restore their proliferative defect (Supplementary Fig. 5d) and cell cycle arrest (Supplementary Fig. 5e), which suggested that the metabolic disruption induced by NAT10 deficiency was associated with other mechanisms rather than direct regulatory effects.
NAT10 interacts with RACK1 and promotes its acetylation
NAT10, is an acetyltransferase that regulates mRNA ac4C modification and translation processes32. Remodelin is the only known NAT10 inhibitor33. Remodelin displayed a significant reduce in T cells proliferation and cell cycle arrest (Fig. 6a, b). NAT10, possessing acetyltransferase activity, had the capacity to acetylate histones and non-histone proteins. We observed a significant elevation in pan-acetylation levels of whole protein following T cell activation (Supplementary Fig. 6a). We believed that some of the acetylation was mediated by NAT10, with only a small number of acetyltransferases was increased in transcription following T cell activation (Supplementary Fig. 6b). To explore whether NAT10 alters the homeostasis maintenance and effector function of T lymphocytes through the acetylation of specific proteins, we employed mass spectrometry to identify proteins that interact with NAT10 and undergo acetylation modifications in activated CD4+ T lymphocytes. The results indicated that 170 proteins, including RACK1, HIDG1A, ATRX, PCNA, have been identified to bind with NAT10 and displayed a significant acetylation in activated CD4+ T lymphocytes (Fig. 6c and Supplementary Data file 2–3). These NAT10-interacting acetylated proteins were enriched in various cellular processes, such as DNA replication, ribosome, cell cycle et al (Supplementary Fig. 6c). Previous literatures implied that the absence of RACK1 led to peripheral developmental disorders and proliferation defects in T lymphocytes34,35. Thus, we proposed that the diverse manifestations induced by NAT10 deficiency in T lymphocytes might be mediated through the regulation of RACK1. Furthermore, we found that the acetylation level of RACK1 was increased according to the activation of CD4+ T cell with αCD3 plus αCD28 (Fig. 6d). Within activated CD4+ T cells, a significant cytoplasmic colocalization was observed between NAT10 and RACK1 (Fig. 6e). To substantiate the interaction between these proteins, co-immunoprecipitation assays were conducted following the overexpression of FLAG-tagged NAT10 and HA-tagged RACK1 in HEK293T cells, yielding corroborative findings as depicted in Supplementary Fig. 6d. As an enzyme that possessed both RNA acetyltransferase, it was possible that NAT10 binding to RACK1 was dependent of its RNA-binding activity. We therefore pretreated these samples with RNase for co-immunoprecipitation experiments. The results indicated that, although the levels of 28S, 18S, and 5S rRNA were reduced, the interaction between NAT10 and RACK1 remained unaffected. This finding suggested that the association between NAT10 and RACK1 was independent of the association of NAT10 with RNA (Supplementary Fig. 6e). To delineate the specific domains mediating the NAT10-RACK1 interaction, a series of truncation mutants of NAT10, encompassing various functional domains, were constructed (Fig. 6f). These truncations were co-transfected with an HA-RACK1 expressing plasmid into HEK293T cells and subjected to further co-immunoprecipitation analysis. The results showed that NAT10 mutants lacking RNA helicase or tRNA binding domain failed to interact with RACK1 as well as the full-length protein (Fig. 6g). The results demonstrated that the helicase and tRNA-binding domains of NAT10 were indispensable for its interaction with RACK1.
a CFSE labeled WT naïve CD4+ T cells were activated in complete medium containing 20 µM Remodelin, and the proliferation rate was measured at 48 h and 72 h (n = 3). D, DMSO; R, Remodelin. b Cell cycle analysis of αCD3 + αCD28-activated T cells treated with 20 µM Remodelin was evaluated by BrdU incorporation (n = 3). c Venn plot showing overlapped proteins which both interacted with NAT10 and acetylated in activated T cells. d RACK1 were isolated by IP (under denaturing conditions) from whole-cell lysates of naive CD4+ T cells stimulated with αCD3 plus αCD28 and subjected to IB assays using anti-pan acetylation (top panels) antibodies. Protein lysates were also subjected to direct IB (bottom panels). e The activated T cells were fixed and permeabilizated, stained with primary antibodies and fluorescein-conjugated second antibody (Red for RACK1; Green for NAT10; Blue for DAPI), representative images were subsequently captured by confocal. The left scale bar indicates 10 μm, and the right scale bar indicates 2 μm. f Structure (top) and functional domain (bottom) of human NAT10 protein. The different colors indicated distinct domain of NAT10. g RACK1 were isolated by immunoprecipitation (IP) and subjected to IB assays using FLAG (top panels) antibodies to evaluate the interaction between RACK1 and NAT10. Protein lysates were also subjected to direct IB (bottom panels). Source data are provided as a Source Data file. Data were represented as mean ± S.D. Three independent experiments at least were performed in data represented above. Statistical significance was determined by unpaired two-tailed t test (a, b). **P < 0.01, ***P < 0.005.
NAT10 is involved in T lymphocyte proliferation by regulating the ubiquitination-dependent stability of RACK1
To ascertain the influence of NAT10 on RACK1, we conducted the protein levels of RACK1 in WT and NAT10-deficient CD4+ T cells. The data demonstrated that, relative to WT control, there was a significant diminution in the RACK1 protein concentration in NAT10-deficient CD4+ T cells (Fig. 7a and Supplementary Fig. 7a). However, the reduction in RACK1 protein levels caused by the loss of NAT10 was not due to transcriptional suppression, as the qPCR assay revealed that Rack1 mRNA levels in NAT10-deficient T cells were slightly higher than those in the WT group (Supplementary Fig. 7b). NAT10 deficiency significantly impaired the acetylation of RACK1 in activated CD4+ T cells (Supplementary Fig. 7c). RACK1 is a eukaryote-specific ribosomal protein and is required for efficient processing of the 18S rRNA 3’-end, which might be related to its role in translation initiation. Consistently, reduced RACK1 caused by NAT10 deficiency significantly led to a decrease in abundance of 18S rRNA (Supplementary Fig. 7d).
a Naïve CD4+ T cells isolated from NAT10WT and NAT10cKO mice were activated with αCD3 plus αCD28 for 24 h. The protein abundance of RACK1 was measured by IB. b WT and NAT10-deficient naïve CD4+ T cells were stimulated with anti-CD3/anti-CD28 for 22 h, and treated with 20 µM MG132 for another 2 h. The cell lysates were subjected to IB to measure protein abundance of RACK1. c, d IB analysis of ubiquitinated RACK1 (detected with anti-ubiquitin) among proteins IP with anti-HA (RACK1) from lysates of HEK293T cells transfected with FLAG-tagged wild-type ubiquitin (c) or mutant ubiquitin (d) in the presence (+) or absence (−) of expression plasmids for Myc-tagged NAT10 and HA-tagged RACK1 (top), and IB analysis of total cellular polyubiquitination (FLAG-Ub), RACK1 and TUBLIN in lysates without immunoprecipitation (below). e IB analyses of αCD3 + αCD28-triggered K48-linked ubiquitination of RACK1 in WT and NAT10-deficient CD4+ T cells. RACK1 was isolated by IP and followed by IB detection of ubiquitin. f Schematic diagram showing identified ubiquitinated lysine sites of RACK1 with or without NAT10 co-expression by mass spectrum. “-NAT10 or +NAT10” means “with or without NAT10 overexpression”. g Venn plot showing the same site of amino acid which was both detected acetylation and ubiquitination. h IB analysis of various ubiquitinated mutant RACK1 among proteins IP with HA from lysates of HEK293T cells co-transfected with Myc-tagged wild-type ubiquitin (top), and immunoblot analysis of total cellular polyubiquitination (Myc-Ub) and RACK1. i K175, K185 and K271 ubiquitination of RACK1 in activated T cells identified by mass spectrometry. j The acetylation of Strep-RACK1 mediated by recombinant FLAG-NAT10 was measured by IB. k mCherry+ WT and RACK1 K185Q mutant-transfected NAT10-deficient CD4+ T cells isolated by FACS, and treated with 20 µM MG132 for 2 h. The protein level of WT and RACK1 K185Q mutant were detected by IB. l The acetylation of Strep-RACK1 mediated by recombinant WT FLAG-NAT10 and NAT10 G641E mutant was measured by IB. m HEK293T cells were transfected with Myc-tagged ubiquitin, HA-tagged RACK1 and FLAG-NAT10 G641 mutant and its WT control, and subjected to ubiquitination assay 36 h later. n, o mCherry+ WT and RACK1 K185Q mutant-transfected naïve NAT10-deficient CD4+ T cells were labeled with CFSE, activated with anti-CD3/anti-CD28 (n = 4). The proliferation of these cells was measured with CFSE dilution by FACS and after 48 h and presented as bar graph. p–q ECAR and OCR of mCherry+ WT and RACK1 K185Q mutant-transfected naïve NAT10-deficient CD4+ T cells stimulated with anti-CD3 plus anti-CD28 for 18 h were measured by Agilent Seahorse XF Analyzers (n = 3). r, s mCherry+ WT and RACK1 K185Q mutant-transfected naïve NAT10-deficient CD4+ T cells were polarized under Th1 differentiation condition (n = 3). The expression of IFN-γ were analyzed by FACS after 96 h. Source data are provided as a Source Data file. Data were represented as mean ± S.D. Three independent experiments at least were performed in data represented above. Statistical significance was determined by one-way ANOVA with multiple comparisons (o, s). *P < 0.05, **P < 0.01, ***P < 0.005, ns no significant.
We further posited that NAT10 might be implicated in modulating the protein stability of RACK1. Upon inhibiting proteasomal activity with MG132, we discerned a substantial amelioration of RACK1 protein levels (Fig. 7b and Supplementary Fig. 7e). Given that ubiquitination is a predominant modality for regulating protein stability, we initially probed the potential influence of NAT10 on the ubiquitination levels of RACK1. We co-transfected HEK293T cells with expression vectors encoding Ubiquitin, NAT10, and RACK1, followed by an analysis of ubiquitination levels of RACK1. The results revealed a marked diminishment in ubiquitin conjugation to RACK1 concomitant with the augmentation of NAT10 (Fig. 7c and Supplementary Fig. 7f). There are several types of polyubiquitination, determined by which lysine residue of the ubiquitin molecule is used for chain formation. As the most well-known type, K48-linked polyubiquitination targets proteins for degradation by the proteasome. K63-linked polyubiquitination is involved in non-degradative processes such as DNA repair, signal transduction, and endocytosis. K6-, K27-, and K29-linked polyubiquitination are less well-characterized but are known to be involved in various cellular processes, including DNA repair, mitochondrial maintenance, and regulation of signaling pathways. K11-linked polyubiquitination is associated with cell cycle regulation and protein degradation through the endoplasmic reticulum-associated degradation (ERAD) pathway. In co-transfected HEK293T cells, NAT10 significantly suppressed RACK1 polyubiquitination, encompassing K48-, K63-, K6-, K27-, and K29-linked subtypes (Fig. 7d and Supplementary Fig. 7g). These findings suggested that the inhibition of RACK1 polyubiquitination by NAT10 was not specific to certain ubiquitin linkage types. To further elucidate the inhibitory effect of NAT10 on RACK1 K48-linked ubiquitination, naive CD4+ T cells isolated from the spleens of WT and NAT10 cKO mice were activated with anti-CD3 and anti-CD28 antibodies and treated with MG132 before IB analysis. The results indicated that, in activated CD4+ T cells, the absence of NAT10 led to a significant increase in K48-linked polyubiquitination of RACK1 (Fig. 7e and Supplementary Fig. 7h). In summary, NAT10 maintained the stability of RACK1 by suppressing its ubiquitination, especially K48-linked polyubiquitination.
Though NAT10 could augment the acetylation state of RACK1 and attenuating its ubiquitination, it itself did not possess deubiquitinase activity. Hence, we postulated that NAT10 might function through a competitive acetylation of specific lysine residues, thereby preventing subsequent ubiquitination. Utilizing high-resolution mass spectrometry, we identified nine ubiquitination sites on RACK1 that were present under steady-state conditions but disappeared in the presence of NAT10 in HEK293T cells (Fig. 7f). Among these, the lysine residues K175, K185, and K271 were also found to be acetylated in activated T cells (Fig. 7g and Supplementary Fig. 7i). To verify the effect of acetylation on potential ubiquitination sites, we used site-directed mutagenesis to replace these three lysine residues with glutamine (Q), among which the K185Q mutant manifested a pronounced decrement in ubiquitination relative to the WT control (Fig. 7h and Supplementary Fig. 7j). We further compared the acetylation status of RACK1 in WT CD4 + T cells before and after activation. The results indicated the presence of multiple acetylation sites in both resting and activated T cells, with varying trends for each site. Notably, K185 was not acetylated in resting T cells but showed significant acetylation following 15 min of anti-CD3 and anti-CD28 stimulation (Fig. 7i). Additionally, an acetylation assay in vitro confirmed that NAT10 can directly acetylate the K185 site on RACK1 (Fig. 7j and Supplementary Fig. 7k). We next investigated the protein stability of the K185Q mutant and found it to be markedly higher than that of the WT control and unaffected by proteasome inhibitors (Fig. 7k and Supplementary Fig. 7l). Furthermore, we constructed a catalytically inactive mutant of NAT10 G641E and found that it was unable to induce RACK1 acetylation or inhibit its ubiquitination (Fig. 7l, m and Supplementary Fig. 7m).
To validate the physiological significance of K185 acetylation on the functionality of RACK1, both wild-type and RACK1 K185Q mutant were ectopically expressed in NAT10-deficient CD4+ T cells via retroviral transfection (Supplementary Fig. 7n). Analytical comparisons revealed that the RACK1 K185Q mutant exhibited a higher protein abundance in comparison to its wild-type counterpart under conditions of approximate mRNA equivalence (Supplementary Fig. 7o), implying that the protein likely possessed enhanced stability. NAT10-deficient CD4+ T cells, upon overexpression of the RACK1 K185Q mutant, demonstrated an enhanced capacity for T cell proliferation (Fig. 7n, o), glycolysis and FAO metabolism (Fig. 7p, q), and cytokine production (Fig. 7r, s), when compared with cells expressing the WT allele of RACK1. In summary, NAT10 stabilized RACK1 protein in CD4+ T cells by inhibiting its K48-linked polyubiquitination and promoting lysine acetylation, particularly at K185. NAT10 deficiency impaired RACK1 protein stability and T cell proliferation, despite higher Rack1 mRNA levels in NAT10-deficient cells (Supplementary Fig. 8).
Discussion
Precise regulation of proliferating T cell kinetics and cell cycle traversal represents a quintessential therapeutic fulcrum within the context of autoimmune and inflammatory pathologies. The transcendence of controlled cellular cycling into a state of hyperproliferation disrupts immunological equilibrium, inadvertently inciting organismal injury and inflammatory pathology. Disorders rooted in autoimmunity and chronic inflammatory diseases—such as MS and IBD (Crohn’s disease and ulcerative colitis)—emanate from the disintegration of immunological forbearance, culminating in the undue activation and clonal propagation of effectors T cell subsets36. Hence, therapeutic targeting of the proliferating T cell cohort, finely tuning their cell cycle dynamics, portends an innovative approach to remedy the prevailing immune derangement, thereby mitigating the clinical manifestations of the disease. RACK1 is a multifunctional scaffold protein that plays a significant role in various cellular processes, including signal transduction, protein synthesis, and cell cycle regulation. In the context of T cell proliferation, RACK1 particularly plays a role in the regulation of the cell cycle, promoting the transition from the G1 to the S phase, which is essential for T cell proliferation. RACK1 can be ubiquitinated, which often targets proteins for degradation by the proteasome. Ubiquitination of RACK1 might regulate its stability and turnover, thereby controlling the intensity and duration of T cell signaling. However, how to precisely regulate the ubiquitination process of RACK1 is still one of the problems in the field to be solved.
Our current data revealed that NAT10, which encodes a protein known as N-acetyltransferase 10, emerged as a key player in the context of cycling T cells. It exhibited specific expression patterns and functions that were crucial for T cell proliferation and cell cycle control. In this study, we found that NAT10 acted as a key modulator of RACK1 expression levels in cycling T cells. RACK1 is a scaffolding protein involved in various cellular processes, including signal transduction, protein translation, and cell cycle control37. NAT10 positively regulated RACK1 expression, ensuring its presence at appropriate levels for proper functioning. The precise regulation of RACK1 levels is crucial for maintaining the balance between T cell activation and suppression, as dysregulation can lead to either excessive immune responses or impaired immunosurveillance. NAT10-RACK1 interaction influenced critical signaling pathways involved in T cell proliferation. Our findings suggested that NAT10 exerted a stabilizing effect on RACK1 protein by acetylating the lysine residue at position 185 (K185). This acetylation impedes subsequent K48-linked ubiquitination modifications that are typically associated with proteasomal degradation. As a result of this modulation, the protein stability of RACK1 is enhanced, thereby potentially prolonging its cellular functions. The acetylation of specific lysine residues serves as a regulatory mechanism that can interfere with the ubiquitin-proteasome pathway. Specifically, the acetylation of K185 by NAT10 created a steric hindrance or induced a conformational change in RACK1, precluding the attachment of K48-linked polyubiquitin chains, which were often recognized as a signal for proteasomal targeting. Notably, our discovery underscored the intricacies of post-translational modifications in the intricate regulation of protein stability and function. This regulation ensured proper T cell proliferation, differentiation, and effector functions, keeping immune responses under control. This regulation had implications in autoimmune diseases, where aberrant T cell migration contributed to tissue damage and chronic inflammation. Moreover, the NAT10-RACK1 axis was implicated in DNA repair mechanisms in cycling T cells. RACK1 has been shown to interact with proteins involved in DNA damage response and repair. By modulating RACK1, NAT10 indirectly affected the DNA repair capacity of cycling T cells, crucial for maintaining genomic stability and preventing the development of autoimmune disorders. Therefore, targeting this NAT10-RACK1 axis might provide new strategies to modulate T cell function while minimizing off-target effects on other cell types. For instance, small molecules or biologics that disrupted the NAT10-RACK1 interaction could be investigated as potential therapeutic agents to dampen excessive T cell responses observed in autoimmunity.
In conclusion, the role of NAT10 in regulating RACK1 expression and activity in cycling T cells is of critical importance. This regulation influences T cell proliferation, intracellular signaling, and DNA repair. Elucidating the molecular mechanisms and functional consequences of NAT10-RACK1 interaction holds great potential for developing novel therapeutic approaches to modulate T cell responses in autoimmune and inflammatory diseases. Further research is necessary to fully understand the therapeutic implications and translate these findings into clinical applications.
Methods
Mice
Nat10 floxed (Nat10fl/fl) mice (T007971) and Rag1–/– mice (T004753) were purchased from Gemphamatech, Nanjing, China. To generate NAT10 T cell specific knock out (NAT10cKO), Nat10 floxed mice were crossed with Cd4-cre mice (JAX, 022071). To obtain NAT10 myeloid cell specific knock out (NAT10MKO), Nat10 floxed mice were crossed with Lyz2-cre mice (JAX, 004781). All mice were generated on C57BL/6 genetic background. Littermates were generated by heterozygotes mice crossing. Experiments were conducted using sex- and age- matched (6-8 weeks) mice. All mice were breed and kept in specific pathogen-free (SPF) facility in a controlled environment of 50–70% humidity, with a 12/12 h light/dark cycle and 20 °C–22 °C in Laboratory Animal Center of Zhejiang University. Age and sex matched mice of indicated genotypes were randomly allocated to groups for comparison. All experiments involving animals were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee of Zhejiang University, mice were euthanized using carbon dioxide (CO2) inhalation before used for tissue collection.
Sample sizes
The sample sizes were determined based on the laboratory’s previous experience and experimental design with similar research content. No method is used to pre-determine how many samples are needed. Sample size for each experiment was indicated in figure legends and methods.
Cell culture
HEK293T was a kind gift from Dr. Yichuan Xiao, Shanghai Institute of Nutrition and Health, China. No approach was applied for HEK293T authentication. HEK293T were cultured in DMEM (11965092, Gibco) supplemented with 10% FBS (10270106, Gibco) and 1% streptomycin/penicillin (SV30010, Cytiva). After transfected with indicated plasmid, HEK293T cells were harvested at 72 h. Primary T cells were cultured in RPMI-1640 (11875093, Gibco) containing 10% FBS and 1% streptomycin/penicillin for 24 h for activation and 96 h for subset differentiation.
Constructs and antibody
For overexpression in HEK293T, full length or truncated CDS were amplified by PCR and tagged with Flag, HA or Myc and subsequently subcloned into pCDNA3.1 vector.
Antibodies used for WB and IP, including antibody against NAT10 (13365-1-AP, 1:1000) was from Proteintech. Antibodies against IκBα (C-21, SC-371; 1:500), p65 (C-20, SC-372; 1:500), ERK (K-23, SC-74; 1:500), phospho-ERK (E-4, SC-7383; 1:500), JNK (C-17, SC-827; 1:500), p38 (H-147, SC-7149; 1:500), p105/p50 (C-19, SC-1190; 1:500), AKT1 (B-1, SC-5298; 1:500), NFATc2 (7A6, SC-7294; 1:500), NFATc1 (4G6-G5, SC7296; 1:500), IKKα (H-744, SC-7218; 1:500) Lamin B (C-20, SC-6216; 1:1000) were from Santa Cruz Biotechnology. Antibodies against phospho-AKT (Ser473, D9E; 1:1000), phosphor-IκBα (Ser32, 9241; 1:1000), phospho-JNK (Thr183/Tyr185, 9251; 1:1000), phospho-p38 (Thr180/Tyr182, 9211; 1:1000), phospho-p105 (Ser933, 18E6; 1:1000), phospho-ZAP70 (Tyr319, 2701; 1:1000), and phospho-IKKα/β (Ser176/180, 16A6; 1:1000) were purchased from Cell Signaling Technology Inc. Anti-Actin (C-4, MAB1501; 1:10000), anti-HA (12AC5, 11583816001; 1 µg per IP), anti-FLAG (M2, F3165; 0.25 µg per IP), anti-HA HRP (3F10, 12013819001; 1:2000), and anti-FLAG HRP(M2, A8592; 1:10000) were from Millipore Sigma.
Flow cytometry
Single cells prepared from indicated tissue were stained with specific fluorescently labeled antibodies and subjected to flow cytometric analysis using a Beckman Cytoflex S cytometer. Antibodies used for flow cytometry, including APC anti-mouse CD3e (145-2C11, 17-0031-83; 1:100), eFluor450 anti-mouse CD4 (RM4-5, 48-0042-82; 1:100), PerCP-Cy5.5 anti-mouse CD8a (53-6.7, 45-0081-82; 1:100), APC anti-mouse CD62L (MEL-14, 17-0621-83; 1:100), APC anti-mouse IFN-γ (XMG1.2, 16-7311-85; 1:100), FITC anti-mouse Foxp3 (FJK-16s, 11-5773-82; 1:100) and PE anti-mouse CD25 (PC61.5, 12-0251-81; 1:100), PerCP-Cy5.5 anti-mouse Gr-1 (RB6-8C5, 45-5931-80; 1:100), PE anti-mouse F4/80 (BM8, 12-4801-82; 1:100), APC-eFlour 780 anti-mouse CD11b (M1/70, 47-0112-82; 1:100) were purchased from Invitrogen. FITC anti-mouse CD44 (M7, 561859; 1:100), PE anti-mouse IL17A (TC11-18H10, 561020; 1:100), PE anti-mouse B220 (RA3-6B2, 553089; 1:100), APC anti-mouse TCRβ (H57-597, 561080; 1:100), FITC anti-mouse CD69 (H1.2F3, 557392; 1:100) and PE anti-mouse CD45 (30-F11, 561087; 1:100) were from BD Biosciences. APC-Cy7 anti-mouse CD45.1 (A20, 110715; 1:100), PE anti-mouse CD45.2 (QA18A15, 111103; 1:100) and FITC anti Brdu (3D4, 364103; 1:100) were from BioLegend. For analysis of T cell development, anti-CD3e and anti-B220 were used to distinguish T cells and B cells. After stained with anti-CD4 and anti-CD8a to define CD4+ and CD8+ T cells, CD4+ T cells were gated to CD62LhiCD44lo naïve population and CD44hiCD62Llo memory population; CD8+ T cells were gated to CD62LhiCD44lo naïve population, CD62hiCD44hi central memory population and CD44hiCD62Llo effector memory population. Treg cells were characterized with CD4+Foxp3+. Specially, Thymocytes were described as following subpopulation: CD4+ single positive (CD4 SP), CD8+single positive (CD8 SP), CD4+CD8+ double positive (DP) and CD4-CD8- double negative (DN). When referred to development trajectories of DN thymocytes, group 1-5 were defined as TCRβloCD69lo, TCRβmidCD69lo, TCRβmidCD69hi, TCRβhiCD69hi and TCRβhiCD69mid.
For intracellular staining (ICS), 0.5 μg/ml PMA (P1585, Sigma) and 1 µM ionomycin (S1672, Beyotime) were used to treated T cells for 3 h accompanied with 1× monensin (00-4505-51, eBioscience). After surface staining, indicated cells were fixed and permeabilized according to the kits for instruction. Following the isolation of T cells, intracellular cytokines staining was performed by fixing and permeabilizing the cells using the Cytofix/Cytoperm Fixation/Permeabilization Kit (554714, BD). Additionally, for the detection of Foxp3, T cells were treated with Fixation/Permeabilization Concentrate (00-5223-56, eBioscience) diluted with Diluent (00-5123-43, eBioscience). Intracellular cytokines or transcription factors, such as IL-17A, IFN-γ and Foxp3 were detected through specific antibodies staining and cytometric analysis. FACS sequential gating and sorting strategies are provided in Supplementary Fig. 9.
Immunofluorescence
Naïve T cells were activated in plate-coated anti-CD3 (145-2C11, 16-0031-82, eBioscience; 1 µg/mL) and anti-CD28 (CD28.2, 16-0289-81, eBioscience; 1 µg/mL) in poly-lysine treated cell slide, washed with PBS and fixed with 2% formaldehyde. The fixed cells were permeabilized and blocked with PBS containing 0.1% Triton X-100 (T8787, Sigma) and 5% BSA (B2064, Sigma) for 1 h at 4°C, and stained with specific primary antibodies and fluorescein conjugated second antibodies over night at 4°C. After washing, the slides were mounted with ProLong Gold Antifade Mountant containing DAPI (P36931, Invitrogen). The images were captured by Zeiss LSM 880 and processed with ZEN3 software.
T cell isolation and activation
CD4+ magnetic microbeads (130-049-201, Miltenyi) coupled with LS columns (5140312025, Miltenyi) were used to isolate CD4+ T cells from spleen and LNs. After stained with fluorescence labeled CD4, CD62L and CD44 antibodies, CD4+CD44loCD62Lhi naïve T cells were sorted using BD FACS Aria II.
To activated isolated T cells, 48 well-plates were coated with anti-CD3 (1 μg/ml) and anti-CD28 (1 μg/ml) and washed 3 times with PBS before use.
Bone marrow derived macrophages (BMDMs)
Bone marrow was wash out from fibula and tibia of mice, and single bone marrow cell suspension were cultured in DMEM medium containing 20% FBS and 15% supernatant of L929 (ATCC) cell culture media for 2 days. The equal volume of culture medium was supplemented and culture for another 2 days, the mature BMDMs were then detached and used for indicated experiments.
T cell differentiation
CD4+CD62LhiCD44- naïve T cells were activated with plate coated anti-CD3 (1 μg/mL) and anti-CD28 (1 μg/mL) and differentiated for 96 h according to following conditions: 10 ng/mL IL-12 (419-ML-010, R&D) and 5 μg/mL anti–IL-4 (16-7041-81, eBioscience) for Th1, 1 ng/mL TGF-β (240-B-002, R&D), 10 ng/mL IL-6 (406-ML-005, R&D), 5 μg/mL anti-IFN-γ (16-7311-81, eBioscience), and 5 μg/mL anti-IL-4 for classical Th17, 10 ng/ml IL-1β (211-11B-10, Peprotech), 10 ng/mL IL-6, 20 ng/mL IL-23 (1887-ML-010, R&D), 5 μg/mL anti-IFN-γ, and 5 μg/mL anti-IL-4 for pathologic Th17, 1 ng/mL TGF-β and 10 ng/ml IL-2 (200-02, Peprotech) for Treg. After ICS, those cells were subjected to flow cytometry analysis.
T cell proliferation
For CFSE dilution assay, naïve T cells were labeled with 5 µM CFSE (C34554, Life) at 37 °C. After wash 3 times with PBS, CFSE labeled T cells were activated with plate coated anti-CD3 (1 μg/mL) and anti-CD28 (1 μg/mL) or with 0.5 μg/ml PMA and 1 μM ionomycin for 48 h and 72 h.
To assess cell cycle, 10 μM Brdu (B5002, Sigma) was incorporated for 18, 24 and 36 h. After fixation and permeabilization, T cells were stained with FITC-anti-Brdu antibody and subjected to cytometric analysis.
Apoptosis analysis
Naïve CD4+ T cells were activated with plate coated anti-CD3 (1 μg/mL) and anti-CD28 (1 μg/mL) for 24 h. Then the apoptosis was analyzed with cell apoptosis analysis kit (130-092-052, Miltenyi).
EAE induction
NAT10WT and NAT10cKO mice were anesthetized with pentobarbital and then immunized s.c. with MOG35-55 peptide (2568, R&D; 300 μg) mixed in CFA containing 5 mg/ml heat-killed Mycobacterium tuberculosis H37Ra (231141, Difco). 200 ng pertussis toxin (181, list labs) was i.p. injected to immunized mice subsequently and 24 h later. Mice were then monitored daily to record clinical score according to following criterion: 0, no clinical signs; 1, limp tail; 2, paraparesis (weakness, incomplete paralysis of one or 2 hind limbs); 3, paraplegia (complete paralysis of 2 hind limbs); 4, paraplegia with forelimb weakness or paralysis; 5, moribund or death. To analyzing inflammation development in CNS, brain and spinal cord of immunized mice were dissociation with trypsin. Digests were then subjected to 37% percoll (17-0891-02, Cytiva) to harvest immunocytes. Immunocytes infiltrated in CNS were distinguished as CD45+CD4+ T cells, CD45+CD8+ T cells, CD11b+CD45+ macrophages and CD11b+CD45mid microglia. Investigators who performed the experiments did not know the details of the samples until the data collection and analysis were finished.
Adoptive transfer induced colitis
To induce colitis with adoptive T cell transfer, 0.5 × 106 CD4+CD25-CD45RBhi naïve T cells sorted from NAT10WT or NAT10cKO mice were transferred to Rag1–/– mice through tail vein respectively. Recipients were monitored and weighed body weight every 2 days. Mice were scarified 4 weeks later, and colon were collected for cytometry analysis and HE staining. Investigators who performed the experiments did not know the details of the samples until the data collection and analysis were finished.
Dextran sulfate sodium (DSS) induced colitis
Initial weight of mice was monitored, then the drinking water was replaced with 3.5%(W/V) DSS (0216011010, MP Biomedicals) solution for 6 days followed by pure water for another 2 days. The weight and survival rate were monitored every day. Investigators who performed the experiments did not know the details of the samples until the data collection and analysis were finished.
Bone marrow chimera
Bone marrow cells isolated from NAT10WT mice (CD45.1.2) and NAT10cKO mice (CD45.2) were mixed in a 1:1 ratio and transferred to lethal irradiated (9.5 Gy, using X-rays as the irradiation source via an RS2000 biological irradiator) SJL mice (CD45.1, 002014, JAX) through i.v. injection. 8 weeks later, recipients were scarified and analyzed.
Seahorse extracellular flux analysis
Glycolysis stress and OXPHOS of T cells were evaluated using commercial kits from Angilent (103010100 and 103017100) according to user’s manual. Briefly, Naïve or activated T cells (8 × 104) were plated into the mini cell culture plates precoated Cell-tak (354240, Corning) for 4 h to attach. Then the cells were set in CO2 free incubator for another 1 h. Glucose, oligomycin and 2-DG were added to port A, B and C of sensor cartridge respectively for glycolysis stress analysis, and oligomycin, FCCP and rotenone/antimycin were added to the above ports for OXPHOS analysis. Prepared sensor cartridges were loaded to Agilent Seahorse XFp analyzer.
Immunoprecipitation
For immunoprecipitation in HEK293T, cells transfected with referred expression plasmids were lysed with RIPA buffer (50 mM Tris-HCl pH 7.5, 125 mM NaCl, 1% Triton X-100) containing protease inhibitors. After pre-cleaning, lysates were incubated with anti-HA or anti-FLAG antibodies overnight at 4 °C. Then the immunoprecipitants were pulled down using protein A (P2051, Beyotime) or protein G (P2053, Beyotime) agarose beads. After washing with RIPA buffer, the beads were suspended with 2× SDS loading buffer (1610737, Bio-Rad) and boiled for 5 min and subsequently subjected to SDS-PAGE and immunoblotting.
For endogenous immunoprecipitation, cells were lysed with RIPA buffer containing protease inhibitors. After pre-cleaning with protein A/G beads (P2012, Beyotime), the lysates were incubated with specific antibodies overnight at 4 °C. The recipients were washed with RIPA buffer and denatured and subjected to SDS-PAGE and immunoblotting.
Ubiquitination assay in vivo
HEK293T cells transfected with referred protein and ubiquitin expression plasmids were lysed with RIPA buffer (supplemented with protease inhibitors and 20 mM NEM (04259, Sigma)) containing 2% SDS (L3771, Sigma). Lysates were boiled at 95 °C for 10 min, diluted with 10fold volume RIPA buffer without SDS subsequently and sonicated briefly. After centrifuged at 1,2000 rpm for 15 min, the supernatants were collected and incubated with anti-HA affinity agarose beads (A2095, Sigma) overnight. Consequently, the beads were washed 4 times with RIPA buffer and denatured by boiled for 5 min in 2× loading buffer.
IP-MS
Sorted naïve T cell from NAT10WT or NAT10cKO were activated as described above for 36 h. After washed with ice cold PBS 3 times, activated T cells were lysed with RIPA buffer containing PMSF (10837091001, Sigma), aprotinin and pepstatin (10236624001, Sigma). Cell lysates were precleaned with protein G agarose beads for 1 h, and then incubated with anti-NAT10 antibody overnight. Immunoprecipitants were aggregated with protein G agarose beads for 4 h and subjected to SDS-PAGE. After staining with Coomassie blue staining and de-staining, proteins interacted with NAT10 were identified through mass spectrum.
Cytosolic and nuclear fraction
Cells were collected and washed with ice cold PBS 3 times. Cytosolic fraction was collected after lysed the cells with buffer B (10 mM HEPES pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.4% NP-40, 1 mM DTT, 0.1 mM PMSF) for 15 min and centrifugated briefly. Followed by washing 3 times with buffer B, nuclei were lysed in buffer C (20 mM HEPES pH 7.9, 0.42 M NaCl, 1 mM EDTA, 1 mM DTT, 1 mM PMSF) for 12 min and centrifuged (12,000 × g) for 5 min to collected nuclear fraction.
ELISA and RT-qPCR
For ELISA, cell culture supernatant as indicated were collect and briefly centrifuged to remove cell debris. Afterwards, the samples were analyzed with a commercial ELISA analysis system (eBioscience). For RT-qPCR experiments, TRIzol reagent (15596026, Life) was used to isolate total RNA. RNaseH- MMLV-reverse transcriptase (2641, TAKARA) was used for cDNA synthesis. Diluted cDNA was subjected to RT-qPCR in CFX96 touch machine (Bio-Rad). The expression of the indicated genes was analyzed based on relative quantification and Actb was set as reference. The sequence of primers for qPCR were shown in Supplementary Table 1.
RNA sequencing
Total RNA from 1 × 106 naïve or activated T cells were extracted with TRIzol reagent., and the RNA quality was evaluated with Agilent 2100. The mRNA was enriched by oligo(dT) magnetic beads (61002, Invitrogen) and reversed, and the library were subjected to sequencing. The differential expression genes (DEGs) were analyzed by DESeq2 package in R4.1.1. Significant genes were defined by Q-value at a cutoff of 0.05 and fold-change of 1.2.
IP-MS and acetyl-proteomics
Sorted naïve T cell from NAT10WT or NAT10cKO were activated as described above for 36 h (1 sample each). After washed with ice cold PBS 3 times, activated T cells were lysed with RIPA buffer containing PMSF, aprotinin and pepstatin. Cell lysates were precleaned with protein G agarose beads for 1 h, and then incubated with anti-NAT10 antibody overnight. Immunoprecipitants were aggregated with protein G agarose beads for 4 h and subjected to SDS-PAGE. After staining with Coomassie blue staining and de-staining, gel was cut and digest with trypsin and eluted with 0.1%TFA, followed by vacuum-drying. Peptides analysis was then performed on Q Exactive HF-X Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher) coupled with an Easy-nLC 1200 system (Thermo Fisher). Mobile phases buffer A was water with 0.1% formic acid, and phase buffer B was 80% acetonitrile with 0.1% formic acid. Peptides were loaded onto a capillary column (75 μm × 15 cm, 1.9 μm C18, 5 μm tip), and analyzed with a linear gradient (5–35% buffer B, 30 min; 35–100% buffer B, 5 min; 100% buffer B, 1 min and wash with 100% buffer B for 4 min). LC-MS data were obtained in a data dependent mode, peptides precursors were survey-scanned with range 350-1500 m/z, resolution 60000, AGC target 1 × 106 and maximum injection time 20 ms, followed by tandem MS with resolution 15000. Dynamic exclusion was set to 45 s with a 10-ppm mass tolerance around the precursor. Spectra data was searched by the MaxQuant software (version 2.0.0.0) against reviewed UniProt Mus Musculus protein database (download in October 2021) with mass tolerance values of 20 ppm, the false discovery rate (FDR) of 1% respectively and the maximum missed cleavage sites for trypsin of 2. Peptide length was set to 6–60 amino acids per peptide chain, peptide mass minimum was set to 600–6000 Da per peptide chain. Carbamidomethyl[C] was set as fixed modification and oxidation of methionine and acetyl on protein N termini were set as variable modifications.
For acetyl-proteomics, CD4+ T cells were isolated with magnetic beads, stimulated with anti-CD3 and anti-CD28 for 0, 5, 30 min, and lysed with lysis buffer (1%SDS), 1% protease inhibitors cocktail (539131, 539132, 539134, 539133, Sigma), 3 μM TSA (HY-15144, MCE), 50 mM NAM (N0636, Sigma). After briefly sonicated and 12,000 × g centrifuged for 10 min at 4 °C, the supernatant of cell lysates was collected and digested with trypsin. Peptide were then eluted with IP buffer (100 Mm NaCl, 1 mM EDTA, 0.5% NP-40, 50 mM Tris-HCl, pH 8.0) and incubated with pre-cleaned affinity gel (PTM104, PTM Bio) overnight at 4 °C. The precipitates were washed 4 times with IP buffer and twice with ddH2O and eluted with 0.1% TFA. Samples were vacuum-dried and desalted with C18 ZipTips (Millipore) and subjected to LC-MS performed by nanoElute UHPLC system (Bruker Daltonics) coupled with timsTOF Pro (Bruker Daltonics) mass spectrometry. Peptides were then dissolved in buffer A (0.1% formic acid, 2% acetonitrile in water), directly loaded onto a home-made reversed-phase analytical column (25-cm length, 75/100 μm i.d.). Peptides were separated with a gradient from 6% to 24% buffer B (0.1% formic acid in acetonitrile) over 70 min, 24 to 35% in 14 min and climbing to 80% in 3 min and holding for 3 min at a static flow rate of 450 nL/min. A 1.60 kV voltage was applied by the electrospray. Peptides precursors were analyzed with a MS/MS scan range 100-1700 m/z. The timsTOF Pro was operated in parallel accumulation serial fragmentation (PASEF) mode. Precursors with charge states 0–5 were selected for fragmentation, and 10 PASEF-MS/MS scans were acquired per cycle. The dynamic exclusion was set to 30 s.
The MS/MS data were processed using MaxQuant sofware (v.1.6.15.0). Tandem MS were searched against the mouse SwissProt database. The false discovery rate (FDR) of 1% respectively and the maximum missed cleavage sites for trypsin of 2. The mass tolerance for precursor ions was set as 20 ppm in first search and 5 ppm in main search, and the mass tolerance for fragment ions was set as 0.02 Da. Carbamidomethyl[C] was set as fixed modification and oxidation of methionine and acetyl on protein N termini were set as variable modifications.
Single cell data acquirement and analysis
Human CD atlas study of colon was acquired from Broad Single Cell Portal (SCP1884), then the data were analyzed in R4.1.1. Briefly, the samples were merged using Seurat package and filtered according to mitochondria counts. After normalization, the linear dimensional reduction and batch effect correction were performed by Harmony package. Then the data were clustered, and CD4+ T cells were picked and further clustered to activated, cycling, naïve-like, Th1, Th17 and Treg based on tSNE analysis. The sequencing data of the intestinal mucosa from patients with Crohn’s disease (CD) was acquired from the Single Cell Portal (accession number: SC1884), encompassing a limited cohort of 6 samples derived from the colonic mucosa at inflammatory sites. Furthermore, the single-cell sequencing data of cerebrospinal fluid from patients with multiple sclerosis was sourced from the GEO database (accession code: GSE138266), comprising single-cell sequencing data from 6 healthy controls and 6 individuals diagnosed with multiple sclerosis. Marker genes were calculated using the FindMarkers function of Seurat package through the Wilcox Rank-Sum test algorithm according to the following criteria: logFC > 0.25, p value < 0.05 and min. pct > 0.1.
DEGs between different cell subtypes or groups were identified by Seurat package FindMarkers function trough the Wilcox Rank-Sum test algorithm according to the following criteria: logFC > 0.25, p value < 0.05 and min. pct > 0.1. The DEGs were then subjected to KEGG enrichment for function analysis.
Statistical analysis
Graph pad prism 8 software were used for graph presentation and statistical analysis, no data were excluded. The methods of statistical significance determination were shown in the figure legends. P < 0.05 was set as criteria of significance, and represented as *P < 0.05, **P < 0.01, ***P < 0.001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw transcriptome data generated for this study have been deposited in the Genome Sequence Archive under the accession number PRJCA028331. Source data are provided with this paper.
References
Kuchroo, V. K., Ohashi, P. S., Sartor, R. B. & Vinuesa, C. G. Dysregulation of immune homeostasis in autoimmune diseases. Nat. Med. 18, 42–47 (2012).
Duan L., Rao X., Sigdel K. R. Regulation of inflammation in autoimmune disease. 2019, 2019: 7403796.
Rosetti, F., Madera-Salcedo, I. K., Rodríguez-Rodríguez, N. & Crispín, J. C. Regulation of activated T cell survival in rheumatic autoimmune diseases. Nat. Rev. Rheumatol. 18, 232–244 (2022).
Kaskow, B. J. & Baecher-Allan, C. Effector T cells in multiple sclerosis. Cold Spring Harb. Perspect. Med. 8, a029025 (2018).
Suárez-Fueyo, A., Bradley, S. J. & Tsokos, G. C. T cells in systemic lupus erythematosus. Curr. Opin. Immunol. 43, 32–38 (2016).
Santambrogio, L. & Marrack, P. The broad spectrum of pathogenic autoreactivity. Nat. Rev. Immunol. 23, 69–70 (2023).
Lee, S. H., Kwon, J. E. & Cho, M. L. Immunological pathogenesis of inflammatory bowel disease. Intestinal Res. 16, 26–42 (2018).
Davenport, M. P., Smith, N. L. & Rudd, B. D. Building a T cell compartment: how immune cell development shapes function. Nat. Rev. Immunol. 20, 499–506 (2020).
Malissen, B., Grégoire, C., Malissen, M. & Roncagalli, R. Integrative biology of T cell activation. Nat. Immunol. 15, 790–797 (2014).
Zhong, Y. et al. Hierarchical regulation of the resting and activated T cell epigenome by major transcription factor families. Nat. Immunol. 23, 122–134 (2022).
Obst, R. The timing of T cell priming and cycling. Front. Immunol. 6, 563 (2015).
Rudd, C. E. Cell cycle ‘check points’ T cell anergy. Nat. Immunol. 7, 1130–1132 (2006).
Velardi, E., Tsai, J. J. & van den Brink, M. R. M. T cell regeneration after immunological injury. Nat. Rev. Immunol. 21, 277–291 (2021).
Wells, A. D. & Morawski, P. A. New roles for cyclin-dependent kinases in T cell biology: linking cell division and differentiation. Nat. Rev. Immunol. 14, 261–270 (2014).
Ohashi, P. S. T-cell signalling and autoimmunity: molecular mechanisms of disease. Nat. Rev. Immunol. 2, 427–438 (2002).
Dominguez-Villar, M. & Hafler, D. A. Regulatory T cells in autoimmune disease. Nat. Immunol. 19, 665–673 (2018).
Jiang, X. et al. Maternal NAT10 orchestrates oocyte meiotic cell-cycle progression and maturation in mice. Nat. Commun. 14, 3729 (2023).
Yan, Q. et al. NAT10-dependent N4‐acetylcytidine modification mediates PAN RNA stability, KSHV reactivation, and IFI16-related inflammasome activation. Nat. Commun. 14, 6327 (2023).
Pan, Z. et al. Role of NAT10-mediated ac4C-modified HSP90AA1 RNA acetylation in ER stress-mediated metastasis and lenvatinib resistance in hepatocellular carcinoma. Cell Death Discov. 9, 56 (2023).
Zheng, J. et al. NAT10 regulates mitotic cell fate by acetylating Eg5 to control bipolar spindle assembly and chromosome segregation. Cell Death Differ. 29, 846–860 (2022).
Liu, X. et al. NAT10 regulates p53 activation through acetylating p53 at K120 and ubiquitinating Mdm2. EMBO Rep. 17, 349–366 (2016).
Zhao, J. et al. Single-cell RNA sequencing reveals the heterogeneity of liver-resident immune cells in human. Cell Discov. 6, 22 (2020).
Shyer, J. A., Flavell, R. A. & Bailis, W. Metabolic signaling in T cells. Cell Res. 30, 649–659 (2020).
Chapman, N. M., Boothby, M. R. & Chi, H. Metabolic coordination of T cell quiescence and activation. Nat. Rev. Immunol. 20, 55–70 (2020).
Lim, S. A., Su, W., Chapman, N. M. & Chi, H. Lipid metabolism in T cell signaling and function. Nat. Chem. Biol. 18, 470–481 (2022).
Fan, K. et al. CRL4DCAF2 is required for mature T-cell expansion via Aurora B-regulated proteasome activity. J. Autoimmun. 96, 74–85 (2019).
Swamy, M. ZAP70 holds the key to kinetic proofreading for TCR ligand discrimination. Nat. Immunol. 23, 1293–1294 (2022).
Lo, W.-L. et al. Lck promotes Zap70-dependent LAT phosphorylation by bridging Zap70 to LAT. Nat. Immunol. 19, 733–741 (2018).
Gaud, G., Lesourne, R. & Love, P. E. Regulatory mechanisms in T cell receptor signalling. Nat. Rev. Immunol. 18, 485–497 (2018).
Araujo, L., Khim, P., Mkhikian, H., Mortales, C. L. & Demetriou, M. Glycolysis and glutaminolysis cooperatively control T cell function by limiting metabolite supply to N-glycosylation. Elife 6, e21330 (2017).
Reilly, N. A., Lutgens, E., Kuiper, J., Heijmans, B. T. & Jukema, J. W. Effects of fatty acids on T cell function: role in atherosclerosis. Nat. Rev. Cardiol. 18, 824–837 (2021).
Wei, W. et al. NAT10-mediated ac4C tRNA modification promotes EGFR mRNA translation and gefitinib resistance in cancer. Cell Rep. 42, 112810 (2023).
Larrieu, D., Britton, S., Demir, M., Rodriguez, R. & Jackson, S. P. Chemical Inhibition of NAT10 Corrects Defects of Laminopathic Cells. Science 344, 527–532 (2014).
Chou, W.-C. et al. AIM2 in regulatory T cells restrains autoimmune diseases. Nature 591, 300–305 (2021).
Qiu, G. et al. Impaired autophagy and defective T cell homeostasis in mice with T cell-specific deletion of receptor for activated C kinase 1. Front. Immunol. 8, 575 (2017).
Min, B. Spontaneous T cell proliferation: A physiologic process to create and maintain homeostatic balance and diversity of the immune system. Front. Immunol. 9, 547 (2018).
Adams, D. R., Ron, D. & Kiely, P. A. RACK1, A multifaceted scaffolding protein: Structure and function. Cell Commun. Signal. 9, 22 (2011).
Acknowledgements
We thank the Life Sciences Institute core facilities, Zhejiang University, for technical assistance. This study was supported by distinguished Young Scientist Fund of NSFC (82125016), National Natural Science Foundation of China, Key Program (82230061), This research was supported by the Joint Funds of the Zhejiang Provincial Natural Science Foundation of China under Grant No. LHDMD22H100002 and LQ21H030013. This study was supported by the Natural Science Foundation of Jiangsu Province (Grants No BK20211168). This study was supported by the National Natural Science Foundation of China (82301968/82202019), by the Young Scientists Fund of the National Natural Science Foundation of China (Grant No. 82202019/82402066), the China Postdoctoral Science Foundation (Grant No. 2022M723664/ 2023M734007), and the 111 Program (D20036).
Author information
Authors and Affiliations
Contributions
Conceptualization, J.J. and Y.-y.L.; methodology, W.-p.L., X.-t.M., J.-H.X., J.-y.L., B.-q.L., L.-x. W., and B.Y.; investigation, W.-p.L., X.-t.M., J.-H.X., J.-y.L., B.-q.L., L.-x. W. and B.Y.; writing-original draft, W.-p.L., J.J., Y.-y.L.; writing-review & editing, W.-p.L., J.J. and Y.-y.L.; visualization, J.J. and Y.-y.L.; supervision, J.J. and Y.-y.L.; funding acquisition, J.J., and Y.-y.L.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, Wp., Mao, Xt., Xie, Jh. et al. N-acetyltransferase 10 is implicated in the pathogenesis of cycling T cell-mediated autoimmune and inflammatory disorders in mice. Nat Commun 15, 9388 (2024). https://doi.org/10.1038/s41467-024-53350-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-53350-x
This article is cited by
-
Emerging role of N-acetyltransferase 10 in diseases: RNA ac4C modification and beyond
Molecular Biomedicine (2025)
-
Uncovering shared genetic features between inflammatory bowel disease and systemic lupus erythematosus
Scientific Reports (2025)
-
NAT10-mediated acetylation of NIK mRNA in B cells promotes IgA production
EMBO Reports (2025)
-
RNA cytidine acetyltransferase NAT10 maintains T cell pathogenicity in inflammatory bowel disease
Cell Discovery (2025)