Abstract
Ferroptosis is a newly identified programmed cell death induced by iron-driven lipid peroxidation and implicated as a potential approach for tumor treatment. However, emerging evidence indicates that hepatocellular carcinoma (HCC) cells are generally resistant to ferroptosis and the underlying molecular mechanism is poorly understood. Here, our study confirms that S100 calcium binding protein P (S100P), which is significantly up-regulated in ferroptosis-resistant HCC cells, efficiently inhibits ferroptosis. Mechanistically, S100P facilitates lysosomal degradation of acetyl-CoA carboxylase alpha (ACC1), which is indispensable for de novo biosynthesis of lipids. Loss of S100P elevates the expression of ACC1 and promotes ferroptotic sensitivity of HCC cells. S100P-mediated ACC1 degradation relies on RAB5C, which directs ACC1 to lysosome via P62-dependent selective autophagy. Knockdown of RAB5C or P62 abrogates S100P-induced lysosomal degradation of ACC1 and restores resistance of HCC cells to ferroptosis. Our work reveals an alternative anti-ferroptosis pathway and suggests S100P as a promising druggable target for ferroptosis-related therapy of HCC.
Similar content being viewed by others
Introduction
HCC is one of the most malignant tumors with high mortality throughout the world. Though the treatment strategies including targeted therapy, immunotherapy, or combination therapy have been significantly improved in the past decade, the intrinsic and acquired drug resistance hampers the therapeutic efficacy, leading to the dismal outcome in most advanced HCC patients. Therefore, clarifying the pathogenesis of HCC and developing novel therapeutic options are urgently needed to ameliorate the morbidity and mortality of HCC patients.
Ferroptosis, characterized by accumulated peroxidized phospholipids containing long polyunsaturated fatty acid (PUFA-PLs) in an iron-dependent manner, is a newly identified programmed cell death which is distinct from apoptosis and necrosis1,2,3,4. Although the effect and mechanism of ferroptosis are not yet fully understood, recent studies demonstrate that ferroptosis can hamper the carcinogenesis in multiple tumors including HCC2,5,6,7,8,9,10,11,12,13,14,15,16,17,18. However, HCC is more resistant to ferroptosis compared with other tumors such as RCC19. The underlying molecular mechanism remains unclear. Therefore, a better understanding of the mechanism of ferroptosis resistance in HCC cells may facilitate therapeutic advancements for ferroptosis-targeted treatment of HCC.
S100P is first purified from placenta and is a member of the S100 family of small calcium-binding proteins. S100P is initially produced in the cytoplasm and can also be secreted and act through the receptor for advanced glycation end products (RAGE). Although existing literatures suggest that dysregulated expression of S100 proteins is involved in the development and progression of different cancers20,21,22, the exact role and molecular mechanism of S100P in cancers remains to be fully understood. S100P gene is specially located on human chromosome 4q16, whereas all other S100 genes are localized on human chromosome 1, indicating its unique potential role distinct from other members. Here we identified an unexpected role of S100P as a metabolic reprogramming regulator to remodel ferroptosis susceptibility in HCC cells.
In the present study, we reveal that S100P-RAB5C-P62-ACC1 axis acts as a central pathway to drive the susceptibility of ferroptosis in HCC cells independent of RAGE. S100P decreases the level of PUFA-PLs via RAB5C and P62-mediated lysosomal degradation of ACC1, whereas deficiency of S100P restores the activity of ACC1 and sensitizes HCC cells to ferroptosis. Loss of RAB5C or P62 abolishes S100P-induced ACC1 down-regulation and renders cells sensitive to ferroptosis. Moreover, knockout of ACC1 in S100P-deficient HCC cells restores ferroptosis resistance, indicating that S100P-mediated ferroptosis inhibition is dependent on inactivation of ACC1. Our findings provide an insight into the mechanism of de novo lipid synthesis-mediated ferroptosis in HCC and suggest that targeting S100P may represent a viable approach to overcome resistance of ferroptosis-targeted therapy in HCC.
Results
S100P is positively correlated with ferroptosis resistance in liver cancers
Our previous work revealed that a wide range of HCC tumor cells are highly resistant to RSL3-induced ferroptosis23. To systematically clarify the molecular mechanism underlying this general phenotype, we tested the ferroptosis sensitivity of a panel of HCC cells upon RSL3 treatment and found that none of the HCC cell lines except SNU387 cells exhibited sensitivity to ferroptotic cell death (Fig. 1a). As expected, the high level of cell death in SNU387 cells was completely abrogated by ferroptosis inhibitor ferrostatin-1 (fer-1; Fig. 1b). Since lipid peroxidation is a hallmark of ferroptosis, we next estimated the levels of lipid peroxidation in SNU387 and ferroptosis-resistant cell line HuH7 by staining of BODIPY-C11 581/591. As shown in Fig. 1c and S1a, SNU387 cells showed stronger signaling intensity of lipid peroxidation compared with HuH7 cells. To elucidate the molecular mechanism underlying the distinct ferroptosis sensitivity in these two cell lines, we analyzed the differentially expressed genes via Cancer Cell Line Encyclopedia (CCLE) database and identified top ranked genes which might be involved in ferroptosis through The Cancer Therapeutics Response Portal (CTRP) dataset (Fig. 1d).
a The ferroptotic sensitivity assessment of a panel of HCC cell lines (HuH7, HepG2, Hep3B, Li7, SNU387) using CCK-8 after the treatment of indicated concentrations of RSL3 for 12 h. b Cell death of HuH7 and SNU387 cells treated with RSL3 (200 nM) and fer-1 (2 µM) for 6 h. Dead cells were stained by Sytox Green. c Lipid peroxidation measurements of HuH7 and SNU387 cells treated with RSL3 (200 nM) and lipro-1 (2 µM) for 3 h. d Heat maps of differently expressed genes from CCLE database in HuH7 and SNU387 cells. e The relative mRNA level of PTGS2 and S100P in HuH7 cells treated with RSL3 (2 µM) and SNU387 cells treated with RSL3 (200 nM) for indicated times. f The expression of S100P of HuH7 cells treated with RSL3 (1 μM) for indicated times. g The expression of S100P of HuH7 cells treated with erastin (5 μM) for indicated times. The Western blot experiments were repeated three times independently with similar results in f and g. Data and error bars are mean ± s.d., n = 3 biological independent experiments in a–c and e. P values were calculated using two-tailed Student’s t test in b, c and e. Source data are provided as a Source Data file.
To validate this finding, we performed qRT-PCR experiments and confirmed most of the candidate genes (Fig. S1b and Fig. S1c). Further analysis revealed that S100P, APOC1 and SPOCK1 are the most potential candidates which might participate in mediating ferroptosis sensitivity, since these genes are specifically expressed in ferroptosis-resistant or sensitive HCC cells (Fig. 1d, Fig. S1d). In addition, analysis of CTRP database showed that these genes exhibited strong correlation with ferroptosis (Fig. S1e). To elucidate which gene most likely contributes to distinct characteristics of ferroptosis, we examined the change of these three genes in HuH7 and SNU387 cells upon RSL3 treatment for the indicated time. As expected, the mRNA levels of PTGS2, which is the marker of ferroptosis, was markedly increased in both cell lines (Fig. 1e). Interestingly, S100P also exhibited significant up-regulation in both cell lines (Fig. 1e), whereas APOC1 or SPOCK1 showed a mild or no increase in HuH7 and SNU387 cells upon ferroptosis induction (Fig. S1f). Moreover, the protein level of S100P was increased in a time-dependent manner when treated with erastin or RSL3 (Figs. 1f and 1g). Similar results were observed in the cells treated with sorafenib, which was used for HCC treatment and to promote ferroptosis (Fig. S1g). Further experiments demonstrated that the expression level of S100P was negatively correlated with ferroptotic cell death in a panel of tumor cell lines (Fig. S1h and i). Taken together, our data demonstrate that S100P is positively correlated with ferroptosis resistance in HCC cells, thus we focus on defining the effect and underlying mechanism of S100P-mediated ferroptosis resistance of HCC.
S100P is a ferroptosis suppressor
To examine whether S100P contributes to ferroptosis resistance of HCC cells, we established HuH7 S100P knockout (KO) cell line using CRISPR-Cas9 technology (Fig. 2a). Indeed, given their lack of S100P expression, these cells are robustly sensitive to RSL3-induced ferroptosis (Fig. 2b). To further confirm this phenotype, we generated S100P-/- HepG2 cells (Fig. 2c) and examined the level of ferroptotic cell death. In line with the finding observed in S100P-/- HuH7 cells, deficiency of S100P sensitized HepG2 cells to ferroptosis, whereas the high level of cell death could be completely abolished by fer-1 (Fig. 2d). Moreover, BODIPY-C11 staining revealed that higher levels of lipid peroxidation were detected in S100P-/- cells upon RSL3 treatment (Fig. 2e). Similar results were achieved in another HCC cell line HCCLM3 (Fig. S2a and b). Whereas WT HCCLM3 cells are robustly resistant to RSL3-induced ferroptosis, loss of S100P remarkably sensitized cells to ferroptosis. Notably, exogenous expression of S100P in S100P KO cells significantly restored ferroptosis resistance (Fig. 2f and g). Since S100P is undetectable in SNU387 and HT1080 cells (Fig. S1h-i), we suppose whether enforced expression of S100P could render these cell lines resistant to ferroptosis. As expected, overexpression of S100P decreased RSL3 or erastin induced ferroptotic cell death in SNU387 cells (Fig. 2h–j). In accordance with this finding, HT1080 S100P-overexpressing cells showed robust resistance to ferroptosis (Fig. S2c–g). We also used BODIPY 581/591 C11 to detect the level of lipid peroxidation. As shown in Fig. S2h, ectopic expression of S100P significantly decreased the level of oxidized lipids induced by RSL3. Similar phenotype was observed in HeLa cells with exogenous expression of S100P (Fig. S2i and S2j).
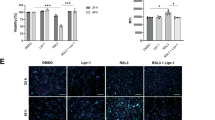
a, c Western blot analysis of HuH7 or HepG2 cells expressing sg-ctrl and sg-S100P. b, d Cell death of HuH7 or HepG2 cells derived from a or c treated with RSL3 (2 µM) and fer-1 (2 µM) for 6 h. e Lipid peroxidation of HuH7 and S100P KO cells treated with RSL3 (2 µM) and lipro-1 (2 µM) for 3 h. f Western blot analysis of S100P expression in Huh7 S100P KO cells with ectopic expression of S100P. g Cell death of Huh7 cells derived from f treated with RSL3 (2 µM) and fer-1 (2 µM) for 6 h. h Western blot analysis of SNU387 cells transfected with S100P. i Cell death of SNU387 cells derived from h treated with RSL3 (200 nM) and fer-1 (2 µM) for 6 h. j Cell death assay of SNU387 cells over-expressing S100P treated with erastin (2 µM) and fer-1 (2 µM) for 24 h. k Representative images of 3D Spheroids derived from HuH7 and S100P KO cells with the treatment of IKE (5 µM) and lipro-1 (2 µM) for 48 h. Scale bars, 10 µm. l Cell death of 3D Spheroids visualized by Sytox green positive area. m Cell death of attached and detached cells (anoikis model constructed in HuH7 cells) treated with RSL3 (2 µM) for 6 h. n Western blot analysis of S100P expression in HuH7 attached and detached cells treated with RSL3 (1 µM) for indicated times. o Anoikis model construction of HuH7 and S100P KO cells treated with RSL3 (2 µM) for 6 h. Scale bars, 10 µm. p Cell death of anoikis model constructed in HuH7 and S100P KO cells treated with RSL3 (2 µM) for 6 h. The Western blot experiments were repeated three times independently with similar results in a, c, f, h and n. Data and error bars are mean ± s.d., n = 3 biological independent experiments in d, e, g, i, j, l, m and p. n = 4 biological independent experiments in b. Statistical analysis was done with two-tailed Student’s t test. Source data are provided as a Source Data file.
As in vitro 2D cell culture often lacks many features of cancers, such as hypoxia, altered cell-cell contacts and rewired metabolism, we cultured these cells into 3D tumor spheroids to better mimic the in vivo context. Consistently, imidazole ketone erastin (IKE) triggers high level of ferroptotic cell death in S100P-/- but not WT HuH7 spheroids (Fig. 2k and 2l). Primary tumor development is accompanied by metastasis, which is often the main cause for failure of tumor treatment. Recent research indicates that ferroptosis inhibition is a major driver for cancer cell metastasis10. The beginning of tumor metastasis in vivo is represented by detachment of individual cells from the primary tumor site, we therefore mimicked this phenomenon in vitro by rendering HuH7 cells out of contact with extracellular matrix. Interestingly, we found that detached tumor cells exhibited lower level of ferroptotic cell death induced by RSL3 (Fig. 2m). Moreover, upregulation of PTGS2 and CHAC1, the hallmarks of ferroptosis, was significantly blocked in detached cells (Fig. S3a and S3b), indicating that detached tumor cells are more resistant to ferroptosis. Notably, the expression level of S100P was elevated in detached cells (Fig. 2n), suggesting that S100P might participate in the ferroptosis resistance of detached tumor cells. To validate this finding, we cultured detached WT and S100P-/- HuH7 cells and detected the ferroptotic resistance. As shown in Fig. 2o and Fig. 2p, knockout of S100P sensitized detached tumor cells to ferroptotic cell death.
Sorafenib has been identified to induce ferroptosis and suppress tumor development in vivo19,24. However, sorafenib alone is insufficient to induce ferroptosis in vitro25. In accordance with this study, our data revealed that high dose (above 1 μM) of sorafenib-induced hepatotoxicity was independent of ferroptosis (Fig. S3c), since ferroptosis inhibitor liprostatin-1 (lipro-1) was incapable of blocking the cytotoxicity. In contrast, low dose of sorafenib alone had no effect on the cell viability of HCC cells. However, it should be noted that low dose of sorafenib enhanced the ferroptosis sensitivity of HCC cells upon RSL3 treatment and this effect could be completely rescued by lipro-1 (Fig. S3d), which was consistent with previous study26. Moreover, we observed that S100P KO cells exhibited higher ferroptotic cell death than WT cells upon combination treatment of RSL3 and sorafenib (Fig. S3d). In addition, we also examined the importance of S100P in other HCC treatment and potential ferroptosis-regulating agents such as regorafenib, cabozantinib, or doxorubicin. Similar to sorafenib, although all these compounds alone were unable to induce ferroptotic cell death (Fig. S3e–g), they could efficiently sensitize HCC cells to RSL3-induced ferroptosis and S100P KO cells exhibited higher ferroptotic cell death (Fig. S3h–j). Together, these data demonstrate that S100P potently suppresses ferroptosis by various inducers in multiple types of HCC cell culture.
S100P rewires lipid metabolism to alter ferroptosis susceptibility
Previous studies showed that S100P binds with receptor for activated glycation end products (RAGE) to activate cellular signaling, thereby facilitating tumor cell proliferation and survival. To explore whether RAGE is indispensable for S100P-mediated ferroptosis suppression, RAGE inhibitor cromolyn was used to test the role of RAGE in S100P over-expressing (OE) cell lines. In line with aforementioned data, S100P overexpression rendered cells resistant to ferroptosis. However, supplementation of cromolyn exhibited no inhibitory effect on S100P-regulated ferroptosis resistance (Fig. 3a, b and Fig. S3k). To further validate the role of RAGE in ferroptosis, we knocked-out RAGE in HuH7 cells by utilizing CRISPR-Cas9 technology (Fig. S3l). As shown in Fig. S3M, N, deficiency of RAGE did not affect ferroptosis sensitivity. Moreover, loss of RAGE in S100P OE SNU387 cells did not reverse RSL3-induced cell death (Fig. 3c and d). In accordance with this finding, deficiency of RAGE in HT1080 S100P OE cells was incapable of altering ferroptosis susceptibility (Fig. 3e and f).
a Cell viability of SNU387 S100P OE cells pretreated with indicated concentrations of Cromolyn for 12 h and then treated with RSL3 (200 nM) for 6 h. b Cell viability of HT1080 S100P OE cells pretreated with indicated concentrations of Cromolyn for 12 h and then treated with RSL3 (200 nM) for 6 h. c Western blot analysis of S100P OE cells of SNU387 expressing sg-ctrl and sg-RAGE. d Cell death measurements of SNU387 S100P OE cells expressing sg-ctrl or sg-RAGE with the treatment of RSL3 (100 nM) for 6 h. e Western blot analysis of HT1080 S100P OE cells expressing sg-ctrl and sg-RAGE. f Cell death measurements of HT1080 S100P OE cells expressing sg-ctrl or sg-RAGE with the treatment of RSL3 (100 nM) for 6 h. g Western blot analysis of classical ferroptosis-related genes in HuH7 and S100P KO cells. h Western blot analysis of classical ferroptosis-related genes in HepG2 and sg-S100P cells. i Western blot analysis of classical ferroptosis-related genes in SNU387 and S100P OE cells. j Western blot analysis of classical ferroptosis-related genes in HT1080 and S100P OE cells. The Western blot experiments were repeated three times independently with similar results in c, e and g–j. Data and error bars are mean ± s.d., n = 3 biological independent experiments in a, b, d and f. Statistical analysis was done with two-tailed Student’s t test in d and f. Source data are provided as a Source Data file.
To elaborate the molecular mechanism of S100P-mediated ferroptosis resistance, we then examined whether S100P affects well-known ferroptosis-sensitive or resistant genes such as GPX4, FSP1, DHODH, GCH1, SLC7A11 and ACSL4. As shown in Fig. 3g, loss of S100P in HuH7 did not affect the status of these ferroptosis-related genes. Consistently, S100P deficiency in HepG2 had no impact on the levels of these genes (Fig. 3h). Furthermore, enforced expression of S100P in SNU387 or HT1080 cells was incapable of regulating the protein levels of ferroptosis-determinant genes (Fig. 3i and j). Taken together, these data indicated that the classical ferroptosis-regulatory genes were supposed not to be involved in S100P-mediated ferroptosis resistance, thereby arousing our great research interest to further define the possible signaling pathway in S100P-mediated ferroptosis-resistant effect.
Since lipid peroxidation is the major driver for ferroptosis induction, we therefore investigated whether S100P inhibits ferroptosis through rewiring lipid metabolism. For this purpose, we performed non-targeted lipidomics assay and found that deficiency of S100P significantly raised the abundance of PUFA-PLs (Fig. 4a), which are the major substrates of lipid peroxidation. Previous study demonstrated that phosphatidylethanolamines (PEs) carrying the fatty acylsarachidonoyl (AA, 20:4) and adrenoyl (AdA, 22:4) drive the susceptibility of lipid peroxidation. Consistent with this standpoint, S100P-/- cells displayed much higher level of PEs containing AA or AdA compared with WT cells (Fig. 4b). In addition, lack of S100P elevated the content of free AA and TAGs containing PUFA (Fig. S4a and b), suggesting that S100P might mediate free PUFA to alter the content of PUFA-PLs.
a Lipid metabolomics analysis in HuH7 WT and S100P KO cells, indicating that S100P inhibits the content of PUFAs. b Phosphatidylethanolamines (PE) contents in HuH7 WT and S100P KO cells. c Western blot analysis of genes about lipid synthesis or uptake in HuH7 WT and S100P KO cells. d Western blot analysis of genes about lipid synthesis or uptake in HepG2 cells expressing sg-ctrl or sg-S100P. e Western blot analysis of genes about lipid synthesis or uptake in SNU387 cells and S100P OE cells. f Western blot analysis of genes about lipid synthesis or uptake in HT1080 cells and S100P OE cells. g Western blot analysis in HuH7 cells treated with RSL3 (1 µM) for indicated times. h Western blot analysis in HuH7 cells treated with erastin (5 µM) for indicated times. i Western blot analysis in HEK293T cells transfected with indicated doses of S100P for detection of ACC1 expression. j Western blot analysis in HEK293T cells transfected with S100P for detection of ACC1 after the treatment with CHX. k Quantitative data of band gray density of ACC1 are shown. l Western blot analysis in SNU387 S100P OE cells treated with Chloroquine (CQ) and MG132, showing that CQ rather than MG132 can rescue the down-regulation of S100P to ACC1. m Western blot analysis in SNU387 S100P OE cells with the treatment of Bailomycin A1. The Western blot experiments were repeated three times independently with similar results in c–j and l, m. Data and error bars are mean ± s.d., n = 4 (S100P KO group) and n = 3 (WT group) biological independent experiments in b. n = 3 biological independent experiments in k. Statistical analysis was done with two-tailed Student’s t test. Source data are provided as a Source Data file.
S100P degrades ACC1 via lysosome–dependent manner
Tumor cells derive free PUFA from dietary uptake or de novo biosynthesis. CD36 is well known as a regulator of exogenous long chain fatty acid transport, whereas sterol regulatory element binding transcription factor 1 (SREBP1) transcriptionally induces the expression of genes responsible for de novo biosynthesis of long chain fatty acid. ACC1, a critical rate-limiting enzyme for lipid biosynthesis, is a classical target of SREBP1.To understand how S100P mediates the level of PUFA, we examined the protein levels of CD36, ACC1 and SREBP1 in WT and S100P KO cells. Interestingly, deficiency of S100P in HuH7, HepG2 and HCCLM3 cells resulted in a significant increase in ACC1 protein level, whereas no change in CD36 or SREBP1 (Fig. 4c and d, Fig. S4c). In contrast, ectopic expression of S100P in SNU387 and HT1080 cells efficiently down-regulated the expression of ACC1 (Fig. 4e and f). The aforementioned data (Fig. 1g and h) showed increased expression of S100P upon ferroptosis inducer treatment, we therefore speculated whether ACC1 undergoes opposite direction in this condition. As shown in Fig. 4g and h, ACC1 was greatly reduced upon RSL3 or erastin treatment. Moreover, S100P decreased the protein level of ACC1 in a dose-dependent manner (Fig. 4i). Consistent with these data, western blot analysis showed S100P and ACC1 are negatively correlated in human HCC samples (Fig. S4d and S4e). Furthermore, we found that human S100Plow HCC tumor samples had higher abundance of PUFA-PLs (Fig. S4f-S4i). To examine whether S100P mediates ACC1 expression through the transcriptional or posttranscriptional pathway, we detected the mRNA levels of ACC1 in WT and S100P-/- cells. Interestingly, S100P did not affect mRNA level of ACC1, as well as other known enzymes involved in lipid metabolism including ACLY, FASN, ACC2 and ELOVL1 (Fig. S4j and k). Cycloheximide (CHX) has been used to block de novo protein synthesis. As shown in Fig. 4j and k, S100P OE cells exhibited more rapid decrease of ACC1 upon treatment of CHX, suggesting that S100P participates in the protein degradation of ACC1.
To further understand the molecular mechanism of S100P-mediated ACC1 degradation, we treated S100P-expressing SNU387 cells with proteasome inhibitor MG132 or lysosome inhibitor chloroquine (CQ) to elucidate the exact molecular mechanism involved in S100P-induced regulation of ACC1. For proteasomal degradation and proteasome inhibitor, NRF2 is a good control which is mainly degraded in proteasome and could be restored by proteasome inhibitor. For lysosome inhibitor experiment, LC3 is a good control as inhibition of lysosome function leads to accumulation of LC3. As shown in Fig. 4l, lysosome inhibitor CQ upregulated the level of ACC1, as well as LC3B II. In contrast, proteasome inhibitor MG132 restored the expression of NRF2, but not ACC1, indicating that S100P-mediated ACC1 downregulation is dependent on lysosomal degradation. Notably, bafilomycin A1, another classical inhibitor of lysosome-mediated protein degradation, has the similar effect to restore ACC1 protein levels (Fig. 4m). Given that autophagy plays a predominant role in lysosome-mediated protein degradation, we therefore further examined the dependence of ACC1 elimination on classical autophagy pathway. We observed that knockdown of ATG7, which is crucial for autophagy-lysosomal pathway, greatly rescued S100P-induced ACC1 elimination in S100P OE cells and inhibited lipid peroxidation (Fig. S4l and m). To examine whether S100P specifically facilitates ACC1 lysosomal degradation, we examined the protein level of FTH1, which is a classical target for lysosomal degradation. The data showed that S100P is incapable of down-regulating FTH1 (Fig. S4n). Together, these findings demonstrate that S100P inhibits de novo biosynthesis of PUFA via specifically lysosomal degradation of ACC1.
P62 and RAB5C are critical for S100P-mediated ACC1 degradation
The aforementioned data prompted us to speculate whether S100P binds with ACC1 to promote its lysosomal degradation. Co-immunoprecipitation (co-IP) and confocal experiments validated that S100P co-localizes and robustly interacts with ACC1 (Fig. 5a and b). Confocal microcopy assay showed that S100P is well distributed throughout cytoplasm and colocalized with ACC1 (Fig. 5c). We also found that there is a potential interaction between S100P and ACC1 by performing molecular docking assays on the ClusPro platform (Fig. 5d). In addition, the data revealed that ACC1 protein largely colocalized with the lysosome marker LAMP1 or LAMP2 in S100P OE cells, whereas mainly localized in the cytoplasm of WT cells (Fig. S5a–c), further confirming that S100P induces lysosome localization of ACC1. Since S100P is mainly localized in cytoplasm, there might exist lysosome-localized proteins to assist S100P to induce degradation of ACC1. To further explore how S100P facilitates lysosomal degradation of ACC1, we performed mass spectrometry (MS) analysis of immunoprecipitated complexes to detect potentially S100P-interacting proteins (Fig. 5e). We found that RAB5C, which is a lysosome-related protein to enhance the lysosomal localization of ACC127, was identified in S100P immunoprecipitated complexes (Fig. 5e and f). Consistently, there was potential interaction between S100P and RAB5C by performing molecular docking assays (Fig. S5d). Furthermore, S100P increased the binding of ACC1 to RAB5C as determined by co-IP analysis (Fig. 5g and h). Knocking down RAB5C almost completely blocked S100P-mediated ACC1 degradation and rendered HCC cells sensitive to ferroptosis (Fig. 5i and j). Moreover, confocal experiments revealed that silencing RAB5C markedly reduced S100P-mediated ACC1 lysosome-localization (Fig. S5e). We also identified that RAB5C selectively facilitates lysosomal degradation of ACC1, whereas it had little effect on FTH1(Fig. S5f).
a Exogenous immunoprecipitation assay of the interaction between ACC1 and S100P. IP assay was done by anti-Flag antibody. b Exogenous immunoprecipitation assay of the interaction between ACC1 and S100P. IP assay was done by anti-GFP antibody. c Confocal microscopy assay of the colocalization of ACC1 and S100P in HT1080 cells. The nucleus was stained with DAPI (blue), and the colocalization of ACC1 (green) and S100P (red) was visualized as yellow fluorescence in the merged panel. Scale bars, 20 µm. d Views of interactions between S100P (green cartoon) and ACC1 (pink cartoon). e IP-MS strategies used to find binding proteins for S100P37. f Immunoprecipitation analysis of the binding of exogenous S100P and endogenous RAB5C. IP assay was done by anti-Flag antibody. g Immunoprecipitation analysis of the binding complex of exogenous S100P, ACC1 and endogenous RAB5C. IP assay was done by anti-Flag antibody. h Immunoprecipitation analysis of the interaction between ACC1, S100P and RAB5C. IP assay was done by anti-ACC1 antibody. i Western blot analysis in S100P OE cells to detect the protein level of ACC1 after the knockout of RAB5C. j Cell death measurements of S100P OE cells expressing sg-ctrl or sg-RAB5C with the treatment of RSL3 (200 nM) for 6 h. k Exogenous immunoprecipitation assay of the interaction between ACC1 and P62. HA-P62, Falg-ACC1 and HA-S100P plasmids were transfected into HEK293T cells. IP assay was done by anti-Flag antibody. l Western blot analysis in HEK293T cells overexpressing S100P to detect the protein level of ACC1 after the knockout of P62. The Western blot experiments were repeated three times independently with similar results in a, b, f–i and k, l. Data and error bars are mean ± s.d., n = 3 biological independent experiments in j. Statistical analysis was done with two-tailed Student’s t test. Source data are provided as a Source Data file.
However, both S100P and RAB5C are proteins with low molecular weight, which is unlikely that these two proteins could act as the cargo to transport ACC1 for lysosomal degradation. In addition, we found that nutrient starvation or mTOR inhibition, which could induce non-selective autophagy, had little effect on the expression of ACC1 and S100P (Fig. S6a–f). Moreover, S100P was unable to influence mTOR activity, indicating that S100P-RAB5C might regulate ACC1 degradation via selective autophagy. P62 participates in selective autophagy and acts as the cargo for K63-linked polyubiquitination modified proteins in autophagy-lysosomal pathway28. Indeed, we observed strong K63-linked polyubiquitination of ACC1 (Fig. S6g). We then examined whether P62 is involved in S100P-mediated ACC1 degradation. Our data revealed that S100P facilitates P62 binding to ACC1, which helped the selective autophagy of ACC1 (Fig. 5k). Moreover, deficiency of P62 largely abrogated S100P-regultaed decrease of ACC1 (Fig. 5l), indicating that S100P recruits P62 to cargo K63-linked polyubiquitination modified ACC1 for RAB5C-directed lysosomal degradation.
As we have demonstrated that S100P-mediated ACC1 degradation is a selective autophagy, we suppose that it might be hard to observe significant change of LC3B using anti-LC3B antibody compared with severe autophagy induction conditions. Indeed, we did not observe the accumulation of LC3B II by overexpression of S100P or by RSL3 treatment by using LC3B antibody (Fig. S6c). GFP-mCherry-LC3 vector is a good tool to monitor the autophagy flux and much more sensitive to detect selective autophagy than western blot analysis of LC3. The fluorescence of GFP was quenched in the lysosome due to the acidic environment, whereas the fluorescence of mCherry was not affected by acidic pH. Therefore, it is also clear to determine the process of autophagy. The LC3 puncta with the fluorescence of both GFP and mCherry is autophagosomes. In contrast, the puncta with the fluorescence of mCherry alone is autolysosomes. Almost all LC3 puncta were GFP + / mCherry+ upon inhibiting lysosomal acidic environment by CQ (Fig. S6h), further validating the efficiency of this system. When no autophagy was induced, almost all GFP uniformly localized with mCherry fluorescent protein throughout the cell. Once upon autophagy induction by nutrient starvation, the autophagosomes or autolysosomes were formed and observed (Fig. S6h). Notably, we also observed much weaker autophagy in the cells overexpressing S100P or treated with RSL3 (Fig. S6h), further demonstrating that S100P-mediated ACC1 degradation is a selective autophagy.
S100P-ACC1 axis is an alternative pathway to regulate ferroptosis sensitivity
We further investigated whether ACC1 degradation contributes to S100P-mediated ferroptosis resistance in HCC. Supplementation of PF-05175157, which is an enzymatic inhibitor of ACC, markedly decreased RSL3-induced cell death in HuH7 and HepG2 S100P KO cells (Fig. 6a and b). Consistent with this finding, high level of ferroptotic cell death in SNU387 and HT1080 cells was largely abrogated upon pharmacological inhibition of ACC1 (Fig. 6c and d). To further demonstrate the role of ACC1 in S100P-induced anti-ferroptosis activity, we utilized CRISPR-Cas9 gene editing to establish ACC1 deficient cells in HuH7 S100P KO cells (Fig. 6e). Analysis of cell death data exhibited that loss of ACC1 in S100P KO cells largely eliminated ferroptosis and exhibited similar level of ferroptotic cell death with WT cells (Fig. 6f). In addition, we also knocked out of ACC1 in S100P-null cell lines such as SNU387 and HT1080 cells. As shown in Fig. 6g–l, lack of ACC1 remarkably rescued RSL3-induced ferroptosis. To further demonstrate that ACC1 contributes to S100P-mediated ferroptosis inhibition, we used BODIPY 581/591 C11 to detect the level of oxidized lipids. As expected, lipro-1 completely blocked RSL3-induced lipid peroxidation (Fig. 6m). Whereas deficiency of S100P induced strong lipid peroxidation, knockdown of ACC1 in S100P KO cells significantly reduced lipid peroxidation (Fig. 6m). These data clearly indicate that S100P renders HCC cells resistant to ferroptosis via downregulation of ACC1, and pharmacological or genetic inhibition of ACC1 efficiently rescued ferroptotic cell death induced by S100P loss.
a Cell death measurements of HuH7 WT and S100P KO cells pretreated with PF-05175157 (50 nM) for 12 h and followed by RSL3 (2 µM) treatment for 6 h. b Cell death measurements of HepG2 WT and S100P KO cells pretreated with PF-05175157 (50 nM) for 12 h and then RSL3 (2 µM) treatment for 6 h. c Cell death measurements of SNU387 and S100P OE cells pretreated with PF-05175157 (25 nM) for 12 h and then RSL3 (200 nM) treatment for 6 h. d Cell death measurements of HT1080 and S100P OE cells pretreated with PF-05175157 (100 nM) for 12 h and then RSL3 (200 nM) treatment for 6 h. e Western blot analysis in S100P KO cells of HuH7 expressing sg-ctrl and sg-ACC1. f Cell death measurements of HuH7 KO cells expressing sg-ctrl or sg-ACC1 treated with RSL3 (2 µM) for 6 h. g Western blot analysis of SNU387 cells expressing sg-ctrl and sg-ACC1. h Cell viability assay of SNU387 cells expressing sg-ctrl or sg-ACC1 treated with RSL3 (200 nM) for 6 h. i Cell death measurements of SNU387 cells expressing sg-ctrl or sg-ACC1 treated with RSL3 (200 nM) for 6 h. j Western blot analysis of HT1080 cells expressing sg-ACC1. k Cell viability assay of HT1080 cells expressing sg-ctrl or sg-ACC1 treated with RSL3 (200 nM) for 6 h. l Cell death measurements of HT1080 cells expressing sg-ctrl or sg-ACC1 treated with RSL3 (200 nM) for 6 h. m Lipid peroxidation measurements of S100P KO cells treated with RSL3 (1 µM) and lipro-1 (2 µM) for 6 h after knockout of ACC1. The Western blot experiments were repeated three times independently with similar results in e, g and j. Data and error bars are mean ± s.d., n = 3 biological independent experiments in a–d, f, h, i and k–m. Statistical analysis was done with two-tailed Student’s t test. Source data are provided as a Source Data file.
S100P is critical for ferroptosis-targeted HCC development
To validate whether S100P promotes HCC development via ferroptosis inhibition in vivo, HCCLM3 cell line was used for construction of xenograft tumor model. Xenograft experiments revealed that knockout of S100P significantly abolished HCCLM3 tumor development, whereas administration of lipro-1 efficiently rescued the phenomenon (Fig. 7a and b). Lipro-1 has little effect on the growth of WT cells, possibly due to intrinsic resistance to ferroptotic cell death (Fig. S2b). Furthermore, S100P KO xenograft exhibited more severe lipid peroxidation stained by 4-HNE and MDA, and supplementation of lipro-1 greatly inhibited the abundance of oxidized lipids (Fig. 7c and d), indicating that S100P is essential for promoting HCC development by suppressing ferroptosis. Consistent phenotype was achieved in HuH7 xenograft HCC model (Fig. S7a–e). Zebrafish is identified as a good model to investigate tumor development, since the fish body is transparent to permit the dynamic visualization of single engrafted cells29. We therefore performed HuH7 WT and S100P KO cells xenograft experiments in zebrafish. As shown in Fig. S7f and g, RSL3 treatment significantly inhibited the development of S100P KO cells derived tumors, whereas RSL3 exhibited no effect on WT cells derived tumors. We further investigated whether inactivation of ACC1 could rescue S100P deficiency-induced ferroptosis and tumor growth arrest in vivo. As shown in Fig. 7a and b, administration of ACC1 inhibitor exhibited similar effect as lipro-1 to promote S100P KO HCCLM3 tumor progression. In addition, inactivation of ACC1 decreased lipid peroxidation in S100P KO HCCLM3 xenografts (Fig. 7c and d), indicating that S100P promotes tumor development via suppression of ACC1.
a, b Xenograft tumors of HCCLM3 or HCCLM3 S100P KO cells in nu/nu mice supplemented with ACC1 inhibitor PF-05175157(20 mg/kg, i.g. daily) and Lipro-1(10 mg/kg, i.p. once every two days). Growth kinetics of the formed tumors were monitored and analyzed. Tumor weights of the isolated tumor were analyzed and compared. n = 8 (WT, WT+Lipro-1, WT + ACC inhibitor, KO, KO+Lipro-1), n = 7 (KO + ACC inhibitor). c, d Representative image and immunochemistry intensity scoring of 4-HNE and MDA staining in above mentioned xenograft tumors. Scale bars, 20 µm. n = 8 (WT, WT+Lipro-1, WT + ACC inhibitor, KO), n = 7 (KO+Lipro-1, KO + ACC inhibitor). e The diagram HCC model induced by overexpression of oncogene Myc and deficiency of tumor suppressor gene p53 in the liver. Sorafenib (60 mg/kg) was given intragastric every three days. f Fluorescence imaging of liver to observe the tumor growth in 2 weeks and 4 weeks. n = 4 independent mice. g The image of the liver tumors was presented. h MDA was detected in above mentioned liver tumors. n = 3 mice per group. i The diagram HCC model induced by DEN and CCL4 with high fat diet. The AAV8 virus of S100P overexpression targeted liver was injected by tail vein injection. Sorafenib (60 mg/kg) was given intragastric every three days. j, k The image of the liver tumors and relative tumor area analysis were presented. n = 7 (Ctrl and Sorafenib), n = 8 (S100P OE and S100P OE+Sorafenib). l Representative image of 4-HNE, MDA, S100P, ACC1 of liver tumors from j. Scale bars, 50 µm. m The correlation of ACC1 and S100P expression of liver tumors from j. n = 5 (Ctrl), n = 8 (S100P OE). Bivariate relationship was calculated by Spearman’s rank correlation coefficient. n Immunochemistry intensity scoring of 4-HNE and MDA staining of liver tumors from j. n = 5 independent mice. Data and error bars are mean ± s.d. Statistical analysis was done with two-tailed student’s t test in a, b, d, h, k and n. Spearman’s rank analysis was used in (m). Source data are provided as a Source Data file.
The xenograft model does not well preserve the characteristic of “natural” HCC development. We therefore established autochthonous HCC model via DEN induction or genetic modifications, which could better reflect HCC development and recapitulate the critical steps followed during progressive liver deterioration. Distinct from human S100P gene, mouse S100p gene is a pseudogene, which loses the function in evolution. We therefore overexpressed human S100P in the liver of mice to investigate whether S100P-directed therapies may impact hepatotoxicity or HCC appearance with or without sorafenib treatment. The gene modifications-driven HCC model is induced by overexpression of oncogene Myc and knockdown of tumor suppressor gene p53 in the liver. Based on this model, we examined the role of S100P in HCC development and sorafenib-mediated HCC treatment (Fig. 7e). As shown in Fig. 7f–g, overexpression of S100P in the liver significantly accelerated HCC development. Moreover, whereas sorafenib efficiently inhibited HCC progression in the oncogene-driven HCC model, S100P remarkably abrogated the therapeutic efficacy of sorafenib. Furthermore, we found that sorafenib-mediated upregulation of MDA, which is the byproduct of lipid peroxidation and could reflect the occurrence of ferroptosis in vivo, was largely abolished by S100P (Fig. 7h). We further utilized DEN and high fat diet-induced HCC model to validate the role of S100P (Fig. 7i). Consistently, the mice with S100P overexpression exhibited much larger tumor size and these tumors were much more resistant to sorafenib treatment (Figs. 7j, k). We also observed that S100P OE decreased the abundance of ACC1 in the liver (Fig. 7l and m). Whereas sorafenib induced lipid peroxidation and ferroptosis in control HCC tumors, this effect was significantly weakened in S100P OE HCC tumors (Fig. 7l and n). Together, our data demonstrated that S100P is necessary for HCC development and resistant to the therapeutic efficacy of sorafenib, further implying that targeting S100P should be beneficial for clinical HCC treatment.
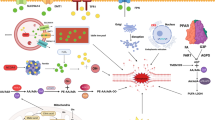
To elucidate the functional consequence of S100P clinically, we next analyzed the data from TCGA database. We found that S100P expression was significantly higher in liver cancer, as well as breast cancer, colon cancer and lung adenocarcinoma et al (Fig. S7h). We further employed the liver cancer as the model to test whether S100P is correlated with ferroptosis signature in vivo. As expected, IHC staining of S100P was highly negatively correlated with 4-HNE staining in the human liver cancer tissue microarray (Fig. 8a and b). Notably, S100P signaling intensity is higher in liver cancer than para-cancer tissues (Fig. 8c and d). Additionally, compared with non-cancerous liver tissue, the expression levels of S100P were much higher in metastatic liver cancer samples by analyzing the data from Kaplan–Meier database (Fig. 8e). Moreover, consistent with the HCC animal model data, Kaplan–Meier survival analysis demonstrated that high expression of S100P is associated with worsen overall survival in HCC patients with sorafenib treatment (Fig. 8f). Furthermore, by analyzing data from database, we found that the HCC patients with high ratio of S100P/ACC1 is correlated with worse clinical outcome. Interestingly, sorafenib treatment has much poorer therapeutic effect in the HCC patients with high ratio of S100P/ACC1 (Fig. 8g, h), suggesting that targeting S100P-ACC1 axis might promote ferroptosis-induced HCC treatment (Fig. 8i).
a Representative immunohistochemistry images of 4-HNE and S100P in the HCC tissue microarray. Scale bars, 20 µm. b The correlation of 4-HNE and S100P expression in the HCC tissue microarray from clinical HCC patients. n = 30 independent tumors. Bivariate relationship was calculated by Spearman’s rank correlation coefficient. c Representative immunohistochemistry images of S100P in HCC para-cancers and cancers. Scale bars, 20 µm. d Intensity scores analysis of S100P in HCC para-cancers and cancers. n = 30 paired liver cancer tissues and corresponding non-cancerous liver tissues. e Kaplan-Meier Plotter comparing the S100P expression in the normal, tumor and metastatic tissues of HCC patients. n = 379 (Normal), n = 806 (Tumor), n = 24 (Metastatic). The bars represent the proportions of tumor samples that show higher expression of the selected gene compared with normal samples at each of the quantile cutoff values (minimum, 1st quartile, median, 3rd quartile, maximum). f Kaplan–Meier survival curves showing the relationship between S100P expression levels and the overall survival time of liver cancer patients in the case of sorafenib treatment. g, h Kaplan–Meier survival curves showing the relationship between the ratio of S100P to ACC1 and the overall survival time of liver cancer patients with or without sorafenib treatment. i Schematic diagram of the regulatory role of S100P in ferroptosis. Data and error bars are mean ± s.d. in (d). Statistical analysis was done with two-tailed student’s t test in (d). Logrank test was used in (f–h). Kruskal-Wallis analysis was used in (e). Source data are provided as a Source Data file.
Discussion
HCC is one of the highly heterogeneous malignancies worldwide with rapidly increasing global incidence. Despite the rapid development of multidisciplinary therapeutic strategies, the overall survival of HCC patients remains poor. Increasing evidence shows that HCC cells could easily generate resistance to multiple therapy by rewiring metabolic process or altering epigenetic characteristics, thereby leading to treatment failure. It is recently reported that therapy-resistant cancer cell lines characterized by enhanced lipid metabolism are more sensitive to ferroptosis30, indicating that ferroptosis-targeted therapy might be a feasible way to cure HCC. However, emerging evidence indicates that HCC cells are generally resistant to ferroptosis and the underlying molecular mechanism is poorly understood.
Here, our work uncovers that S100P acts as an essential metabolic regulator to rewire lipid metabolism and suppress ferroptotic cell death via S100P-RAB5C- ACC1 axis. It is thought for a long time that S100P facilitates tumor development via activation of RAGE signaling pathway. However, our work elucidates a non-canonical function of S100P to mediate resistance to ferroptosis independent of RAGE. Our data clarifies that S100P is remarkably elevated in ferroptosis-resistant HCC cell lines and significantly upregulated upon ferroptosis induction, indicating the important role of S100P-mediated ferroptosis resistance in HCC. Inactivation of S100P markedly enhances ferroptosis-targeted therapeutic efficacy, revealing that S100P contributes to ferroptotic resistance of HCC. Sorafenib, a multikinase inhibitor and the first-line treatment for HCC patients, is a recognized classical ferroptosis inducer in multiple cancers including HCC31. Though there is a discrepant report about the effect of sorafenib on ferroptosis25, recent studies indicate that dysregulation of ferroptosis in HCC induced resistant response to sorafenib therapy19,32, indicating the critical regulatory effect of ferroptosis on the response to sorafenib treatment. Here our data reveals that the patients with lower S100P expression have significant survival superiority upon sorafenib treatment, which is consistent with the role of S100P-mediated ferroptosis resistance. It further provides the possibility that S100P acts a molecular subtype marker for HCC patients to define the treatment responders and the most appropriate therapeutic window for sorafenib related therapy.
Though the therapeutic strategy targeting metabolism reprograming has been identified as a potential approach for tumor manipulation, the molecular mechanism of rewiring metabolism in regulating ferroptosis of HCC remained to be clarified. By performing non-targeted lipidomics assays, our work reveals that S100P remodels lipid metabolism via down-regulating ACC1. A recent work reveals that the Trib1-COP1 complex acts as an E3 ubiquitin ligase to degrade ACC1 in proteasome-dependent manner33. Distinct from this, we clarify that S100P facilitates lysosomal degradation of ACC1 via RAB5C, which is responsible for lysosomal degradation of cargo protein. S100P facilitates RAB5C-mediated lysosomal transport of ACC1. Blocking S100P-RAB5C axis efficiently reduces the lysosomal localization and degradation of ACC1. In addition, abolishing S100P-RAB5C axis-mediated ACC1 degradation restores resistance of HCC cells to ferroptosis. Thus our work identifies the ACC1 regulatory pathway to mediate ferroptotic susceptibility of HCC cells.
Here our study defines S100P as the regulator of ferroptosis in HCC via its negative regulation of ACC1, which is crucial for de novo lipid biosynthesis. However, up to now, the role of ACC1 in the development of HCC is still in discrepancy in the literature. Certain study reports that ACC1 is overexpressed in HCC and promotes HCC development via enhancing lipid metabolism34, while liver-specific knockout of ACC1 enhances HCC development by increasing antioxidant defence35. In addition, a most recent research shows that ACC1 could efficiently suppress tumor growth via ferroptosis36. These studies elucidate that excess expression of ACC1 in tumor cells leads to severe oxidative stress and inhibits tumor development. Therefore, ferroptosis-targeted treatment of ACC1high HCC should be theoretically feasible and effective. Consistent with these researches, our work reveals that induction of ACC1 expression in HCC cells via inactivation of S100P sensitizes cells to ferroptosis and exhibits a better therapeutic efficacy upon ferroptosis treatment in vivo. In contrast, inactivation of ACC1 enhances the resistance to oxidative stress and promotes S100P KO tumor development, indicating that targeting S100P-RAB5C-ACC1 axis might be a promising ferroptosis-related therapy for HCC via enhancing lipid oxidative stress.
Methods
The study for mice is compliant with all relevant ethical regulations for animal experiments. All mouse experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee, and were approved by the Medical Ethics Committee of Shandong University. Tumor volume does not exceeding 2 (cm3) in accordance with Medical Ethics Committee of Shandong University. The study for human cancer samples complied with the ethical requirements of Shandong University.
Cell culture
HuH7, HepG2, HT1080, HELA, C33A, HCCLM3 cells were cultured in Dulbecco’s modified Eagle’s medium. SNU387, Li7, A549 were cultured in RPMI-1640 medium. Hep3B was cultured in Minimum Essential medium. All cells were cultured in a 37 °C incubator with 5% CO2 and maintained in culture medium supplemented with 10% fetal bovine serum (Biological Industries, Israel), penicillin (100 U/ml), and streptomycin (100 µg/ml). HuH7, HepG2, SNU387, HT1080, HeLa, HEK 293, HCCLM3, Li7, Hep3B, A549 and C33A cell lines were acquired from the Cell Banks of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). A2780 cells were provided by Dr. Ruihong Zhang. All the cells were free from mycoplasma and were authenticated by STR profiling.
Plasmids and lentivirus
Full-length S100P were cloned into pENTER and Lenti-FLAG-linker vectors. Full-length ACC1 were cloned into pLV3-CMV. Full-length P62 were cloned into pcDNA3.1. mCherry-GFP-LC3 plasmid was given from Dr. Rui Jia. For the sgRNAs of S100P, ACC1, RAB5C, P62 and RAGE, these sgRNAs were constructed to plenti-CRISPR-V2 vectors. For lentivirus, the sgRNA sequences were constructed into plenti-CRISPR-V2 vectors, followed by packaging lentivirus by transfection with pMD2G and pSPAX2. The medium containing lentivirus was collected to infect the tumor cell lines, and the lentivirus-infected cells were further selected by puromycin (1 μg/ml) for 3 days. The gene knockout cells and its mock control cells were cultured for further investigation.
Chemicals
Erastin (S7242), RSL3 (S8155), Ferrostatin-1 (S7243), Liproxstatin-1 (S7699), PF-05175157 (S6672), Doxorubicin (S1208), Regorafenib (S1178), Cabozantinib (S1119), Sorafenib (S7397), Torin 1 (S2827), Chloroquine (S6999), Bafilomycin A1 (S1413), MG132 (S2619) were purchased from Selleck. Cromolyn (HY-B0320A) were purchased from Med Chem Express (MCE). Imidazole ketone erastin (T5523) was purchased from TargetMol.
CRISPR-Cas9 and shRNA-mediated gene silence
The sgRNA sequences were designed from the website (https://www.addgene.org/). sgS100P: GACGGAACTAGAGACAGCCA, GACGTCTATGATCATGCCCA; sgRAGE: TCTTACGGTAGACACGGACT, CAAAACATCACAGCCCGGAT; sgACC1: GCTGGTGCTATCTATCCAT, GCTGGTGCTGTCTATCCGT. sgRAB5C: CGCGAGTGCAATGACGATGT, GGAGGCGCAGCACGACCCAA; sgP62: ACGCTACACAAGTCGTAGTC, AAGGTGAAACACGGACACTT, AGGGCTTCTCGCACAGCCGC; For shRNA-mediated gene knockdown, the following sequences were used: shATG7: GCCTGCTGAGGAGCTCTCCAT, CCCAGCTATTGGAACACTGTA.
Cell viability assay
The cells were seeded at 7000 cells per well in 96-well plates. 16 h after seeding, the cells were treated with indicated concentration of RSL3 or erastin. 6 h or 24 h later, the plates were incubated with CCK8 (selleck, Houston, USA) for 1 h and read at 450 nm. The collected values were normalized to blank well. Then the relative cell viability was normalized to the DMSO-treated wells. Graphpad Prism 9 software was used to plot the regression fit curves.
Cell death assay
For cell death analysis, the cells were seeded at 2*105 cells per well in 24-well plates. Cells were treated with chemicals, the plates were incubated with SYBR Green (S7020, Invitrogen, USA) for 30 minutes. Then the cells were observed and recorded on a fluorescence microscope and data were collected to analyze.
Lipid peroxidation assay
Cells were harvested and washed with PBS, then resuspended with PBS containing 5 μM C-11 BODIPY dye (D3861, Thermo Fisher Scientific, USA) and incubated at 37 °C for 30 min. Cells were then washed twice with PBS followed by resuspending in 200 μL PBS. Lipid ROS levels were analyzed using a Becton Dickinson FACS Calibur machine through the FL1 channel, and the data were analyzed using FlowJo. In each sample, 5000 cells were analyzed. For confocal assay, cells were embedded with coverslips, 20 h after seeding, then stained in PBS containing 5 μM C-11 BODIPY and incubated at 37 °C for 30 min. Cells were then washed twice with PBS. The stained cells were visualized and captured by confocal microscopy (Dragonfly 200, Andor, UK).
Construction of detached cell model
Poly (2-hydroxyethyl methacrylate) (P3932, Sigma-Aldrich, USA) was dissolved to 120 mg/ml. 36 mg/ml working solution was diluted with 95% ethanol, and then added 100ul working solution to the 96-plates or 1 ml to the 6-plates using UV irradiation until solidification. Cells were seeded at 7000 cells per well in 96-well plates and 2 × 105 cells per well in 6-well plates for the following assays.
Generation of 3D spheroids
S100P KO cells of HuH7 were digested and plated 1*104 cells per well into U-bottom ultra-low adherence (ULA) 96-well plates (Corning, USA), and then 3D structures were generated by centrifugation at 600 × g for 5 min followed by the addition of 2.5% (v/v) Matrigel (Corning, 354234). 3D structures were incubated for 72 h at 37 °C, 5% CO2, and 95% humidity to promote the formation of a single spheroid of cells, subsequently supplemented with IKE (5 μM) and liprostatin-1 (2 μM) for the indicated times. Sytox Green staining was used to visualize cell death.
Xenograft tumor model
To further exploring the effect of S100P on ferroptosis-mediated tumorigenesis in vivo, tumor xenograft models were established and the experimental protocols were practiced complied with the Policy on the Ethical Use and Care of Animals (School of Basic Medical Sciences, Cheeloo College of Medicine, Shandong University).
In HuH7 tumor bearing xenograft, 0.1 mL of 1x107 HuH7 or S100P KO cells that mixed with 0.1 mL matrigel was injected into the flanks of 5-week-old immunodeficient BALB/c athymic male nude mice (Beijing Vital River Laboratory Animal Technology Co., Ltd.), and then Lipro-1 (10 mg/kg) was intraperitoneal injected triple a week for 4-5 weeks. In HCCLM3 tumor bearing xenograft, 0.1 mL of 1x106 HCCLM3 or S100P KO cells that mixed with 0.1 mL matrigel was injected into the flanks of 5-week-old immunodeficient BALB/c athymic male nude mice (Beijing Vital River Laboratory Animal Technology Co., Ltd.), and then Lipro-1 (10 mg/kg) was intraperitoneal injected triple a week and PF-05175157 (20 mg/kg) was intragastrically administered every day for 4-5 weeks. Tumor growth was observed and recorded. Tumor volume was calculated: volume (cm3) = (length × width2) × 0.5. Further, mice were sacrificed and the xenograft tumors were excised for further statistical analysis. All mice were bred at the Experimental Animal Center of Shandong University under conditions without specific pathogens. All mice were raised at 26 °C, with humidity maintained at 40%-60%, and ventilated 10 times per hour. All mouse experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee, and were approved by the Medical Ethics Committee of Shandong University. The number of mice in these experiments is described in the corresponding Fig. legends (Fig. 7a, b and d).
Orthotopic liver tumor model
C57BL/6 J male mice (6-8 weeks old) were used for the construction of animal model. The gene modifications-driven HCC model is induced by overexpression of oncogene Myc and SB13 and deficiency of tumor suppressor gene p53 in the liver. pT3 vector and pT3-S100P plasmids were injected by tail vein injection together with oncogene Myc, SB13 and sg-P53. Two weeks later, Sorafenib (60 mg/kg) was given by intragastric administration every three days. Fluorescence imaging was performed weekly. Further, mice were sacrificed and liver tumors were excised for further statistical analysis. All mice were bred at the Experimental Animal Center of Shandong University under conditions without specific pathogens. All mice were raised at 26 °C, with humidity maintained at 40%-60%, and ventilated 10 times per hour. All mouse experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee, and were approved by the Medical Ethics Committee of Shandong University. The number of mice in these experiments is described in the corresponding Fig. legends (Fig. 7f, h).
Primary liver cancer model
C57BL/6 J male mice (2-3 weeks old) were used for the construction of animal model. DEN (50 mg/kg) was used by intraperitoneal injection at two weeks old. DEN (25 mg/kg) was used by intraperitoneal injection at four weeks old. Further, CCL4 (CCL4: olive oil=1:9) was used by intraperitoneal injection at eight weeks old three times every week for ten weeks. High fat diet began at six weeks old until sacrifice. AAV8 virus of overexpressing S100P targeted liver was injected by tail vein at sixteen weeks. Sorafenib (60 mg/kg) was used by intragastric administration at twenty-two weeks for one month. Further, mice were sacrificed at twenty-six weeks and liver tumors were excised for further statistical analysis. All mice were bred at the Experimental Animal Center of Shandong University under conditions without specific pathogens. All mice were raised at 26 °C, with humidity maintained at 40%-60%, and ventilated 10 times per hour. All mouse experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee, and were approved by the Medical Ethics Committee of Shandong University. The number of mice in these experiments is described in the corresponding Fig. legends (Fig. 7j–n).
Zebrafish tumor model
Zebrafish (purchased from Chinese Zebrafish Resource Center, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China) and embryos were raised at 28 °C under standard experimental conditions. 48 h after fertilization, zebrafish embryos were anesthetized with the 0.2 g/L MS-222 solution (Sigma-Aldrich, Merck, Darmstadt, Germany) and placed on a modified agarose gel for the microinjection of HCC cells. Approximately 300-500 Huh7 or S100P KO cells stained with thiazole orange were resuspended in 0.9% NaCl and 5 nL of cell solution was injected into the perivitelline space of each embryo using a Pico-liter Micro-injector (PLI-100A, Warner, MA, USA). The injected embryos were kept at 32 ◦C and were investigated by a fluorescent microscope (ZEISS SteREO Discovery.V20, Carl Zeiss, Oberkochen, Germany) 3 days after the injection. The tumor-bearing zebrafish were treated with RSL3 (2 µM) for 1-2 days. Thereafter, the tumor-bearing zebrafish were anesthetized with MS-222 and investigated under a fluorescence microscope. All zebrafish experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee and were approved by the Medical Ethics Committee of Shandong University. The number of zebrafish in these experiments is described in the corresponding Fig. legends (Fig. S7g).
Antibodies
The primary antibodies used in our study were as follows: anti-Flag antibody(1:1000, F1804, Sigma-Aldrich, USA), anti-GFP (1:1000, 66002-1-lg, Proteintech, USA), anti-HA (1:1000, TA100012, Origene, USA), anti-GAPDH (1:1000, 60004-1-Ig, Proteintech, USA), anti-β-actin (1:5000, 66009-1-Ig, Proteintech, USA), anti-Tubulin (1:1000, 10068-1-AP, Proteintech, USA), anti-CD36 (1:1000, 18836-1-AP, Proteintech, USA), anti-SREBP1 (1:1000, ab28481, Abcam, USA), anti-FTH1 (1:1000, ab75972, Abcam, USA), anti-DHODH (1:1000, 14877-1-AP, Proteintech, USA), anti-FSP1 (1:1000, 24972S, CST, USA), anti-GCH1 (1:1000, 28501-1-AP, Proteintech, USA), anti-RAGE (1:1000, 16346-1-AP, Proteintech, USA), anti-ACC1 (1:1000, 21923-1-AP, Proteintech, USA), anti-Lamp2 (1:200, 66301-1-lg, Proteintech, USA), anti-GPX4 (1:1000, ab125066, Abcam, USA), anti-ACSL4 (1:1000, ab155282, Abcam, USA), anti-NRF2 (1:1000, ab137550, Abcam, USA), anti-4-HNE (1:250, ab46545, Abcam, USA), anti-MDA (1:250, ab243066, Abcam, USA), anti-ACC1 [1:1000, C83B10, Cell Signaling Technology (CST), USA], anti-SLC7A11 (1:1000, 12691S, CST, USA), anti-ATG7 (1:1000, D12B11, CST, USA), anti-LC3B (1:1000, 3868S, CST, USA), anti-P62 (1:1000, 8025S, CST, USA), anti-p-mTOR (1:1000, 5536S, CST, USA), anti-p-S6K1 (1:1000, 9234S, CST, USA), anti-p-S6 (1:1000, 4858S, CST, USA), anti-p-4EBP1 (1:000, 2855S, CST, USA), anti-S100P (1:100, sc-374547, Santa Cruz Biotechnology, USA), anti-RAB5C (1:500, sc-365667, Santa Cruz Biotechnology, USA). Protein A/G PLUS-Agarose (sc-2003) was purchased from Santa Cruz Biotechnology.
Western blotting
Hepatocellular carcinoma tissues (n = 12) for detection S100P and ACC1 protein expression were derived from 35-75 years old male and female hepatocellular carcinoma patients collected from The Second Hospital of Shandong University, and the informed consent was obtained from involved patient. The study was approved by the Ethics Committee of Shandong University. Cellular proteins were extracted using RIPA lysis buffer supplemented with 1% protease inhibitor and phosphatase inhibitors, and protein concentration was determined by a Pierce BCA protein assay kit (Thermo, USA). Total 30 µg proteins were separated by 8%-12% SDS-polyacrylamide gel, transferred onto nitrocellulose filter membrane (Millipore Corp, Billerica, MA, USA), blocked with 5% fat-free milk for 1 h at room temperature. The primary antibodies were incubated overnight at 4 °C, sequentially the peroxidase affinipure goat anti-rabbit or mouse IgG (H + L) (Elabscience, USA) and enhanced chemiluminescence solution (Millipore Corp, Billerica, MA, USA) were used for visualizing protein expression.
Immunoprecipitation
Cells were lysed in the lysis buffer [20 mM tris-HCl (pH 8.0), 0.5% NP-40, 10 mM NaCl, 1 mM EDTA] containing complete protease inhibitor cocktail (catalog no. 04693132001, Roche, USA), followed by centrifugation at 12,000 rpm at 4 °C for 15 min. The supernatants were immunoprecipitated with protein A/G agarose (catalog no. sc-2003, Santa Cruz Biotechnology, USA), followed by incubation with anti-Flag or anti-ACC1 antibodies at 4 °C overnight. The immunoprecipitates were washed at least five times with the lysis buffer, and the protein lysate was further eluted with 2× SDS loading buffer. After heating denaturation, protein samples were analyzed by 8% and 12% SDS–polyacrylamide gel electrophoresis (SDS-PAGE) before being transferred to polyvinylidene difluoride membrane for further detection with specific antibodies.
Immunofluorescence assay
Cells were embedded with coverslips, 20 h after seeding, the cells were fixed with 4% paraformaldehyde for 15 min. After blocked with 1% bovine serum albumin, cells were incubated with specific primary antibodies overnight at 4 °C, washed with PBS, and further incubated with Alexa Fluor 594–conjugated (1:200; SA00013-3, Proteintech, USA), Alexa Fluor 488–conjugated (1:200; SA00013-1, Proteintech, USA), Alexa Fluor 594–conjugated (1:200; SA00013-4, Proteintech, USA), Alexa Fluor 488–conjugated (1:200; SA00013-2, Proteintech, USA) secondary antibodies for 1 hour at 37 °C. The nucleus was stained with DAPI (4′,6-diamidino-2-phenylindole) (catalog no. C0065, Solarbio, China) for 10 min. The stained cells were visualized and captured by confocal microscopy (Dragonfly 200, Andor, UK). The images were analyzed by Image J software.
Immunohistochemistry
The samples for IHC microarray tissues were collected from 35-75 years old male and female Hepatocellular carcinoma patients derived from Shandong Provincial Hospital. The study complied with the Ethics Committee of Shandong University, and the informed consent was obtained from involved patients. Immunohistochemistry was carried out to explore the relevance of S100P and 4-HNE in cancers and Subcutaneous tumourigenic model in nude mice. Briefly, sections were soaked in xylene for dewaxing and graded alcohol for rehydrating. Antigen retrieval was performed by placing in citrate buffer and high pressure heating to expose epitopes, and 5% BSA was added to block nonspecific epitopes. Sections were incubated with primary antibodies S100P (1:50), 4-HNE (1:250), MDA (1:250, ab243066, Abcam, USA) and ACC1 (1:200) at 4 °C overnight. Sections were cultured with biotin-labeled secondary antibody for 1 h according to the manufacturer’s instructions. Hematoxylin dye were used for antigen detection. The images were photographed by Multispectral fully electric scanning microscopy imaging system (TissueGnostics, Austria), and calculated mean optical density (MOD) values by Image Pro-plus software. This study was approved by the Ethics Committee of Shandong University. n = 30 paired liver cancer tissues and corresponding non-cancerous liver tissues.
Real-time quantitative PCR
Total RNA was extracted with Trizol Reagent (DP424, TIANGEN, China), and the reverse transcription of 1 ug RNA samples was performed with the FastKing RT Kit (catalog no. KR116, TIANGEN, China). The quantitative PCR assay was performed with SYBR Green Master Mix (catalog CW0957M, CWBIO, China). The primers were synthesized from Sango Biotech, and the sequence information was listed in Supplementary Data 1. The PCR was performed with the Bio-Rad CFX system according to the manufacturer’s instructions, and data were analyzed with Bio-Rad CFX software. Relative mRNA expression of the target genes was normalized to the internal control b-actin, and the results of these experimental groups were further normalized to the levels of the control group.
MDA measurements
Malondialdehyde concentration in liver tissue was detected with the Lipid Peroxidation (MDA) Assay Kit from Beyotime (S0131S), according to manufacturer’s instructions.
Proteome mass spectrometry
To identify the proteins that interact with S100P, S100P overexpression cells with Flag tag were lysed with the lysis buffer, and the supernatants were collected and incubated with protein A/G agarose with or without anti-Flag primary antibody at 4 °C overnight. The beads were washed with lysis buffer at least five times, boiled, and further transferred into 1× SDS loading buffer to run in 12% SDS-PAGE. After running, the gel was stained with Coomassie blue. Compared with the immunoprecipitated lane of the beads, the emerging bands in the Flag immunoprecipitated lane were selected and cut into 1 cm width pieces. Tubes were added with 1 ml ddH2O and washed for 10 minutes. The water was removed. 1 ml digestion decolorizing solution was added to each centrifuge tube. samples were washed for 10 minutes and the decolorizing solution was removed. Acetonitrile was added to dehydrate the gel until it turned completely white, and the acetonitrile was vacuum dried. Add 10 mM DTT and allow the pellet to absorb completely, the sample was incubated in a 56 °C water bath for 1 hour. After incubation, remove excess DTT liquid, add 55 mM IAM and incubate for 45 minutes away from light. After incubation, remove excess IAM liquid, add 25 mM ammonium bicarbonate, wash for 10 minutes and repeat the wash. The pellet was dehydrated with acetonitrile until the pellet turned completely white, and the acetonitrile was vacuum-dried. 1 µg/µl of enzyme reservoir solution was diluted 15-fold with 25 mM ammonium bicarbonate and added to the dehydrated pellet, and allowed to be fully absorbed by the pellet, and then added with 25 mM ammonium bicarbonate until the pellet was completely blanched, and then placed into a 37 °C water bath to digest the pellet overnight. After overnight, the digestion was terminated by adding FA at a final concentration of 0.1%. 10 µl sample was taken and detected by Thermo Fisher Scientific Q Exactive. Mass spectrometry parameters were set as followed: positive ion mode, an ion source voltage of 1800V, HCD fragmentation mode, a capillary temperature of 360 °C, and a scanning range of 350-2000 Da, which is automatically selected based on the mass-to-charge ratio of the primary ion. The mass spectrometry data were analyzed using Mascot Server. n = 1 independent experiment. The protein with peptide segments that are larger in the experimental group than in the control group is selected as the target protein.
Targerted lipidomics mass spectrometry
After the samples were thawed slowly at 4 °C, appropriate samples were taken and added with 200 μL methanol, 20 μL internal standard mixture, 800 μL methyl tert-butyl ether (MTBE), vortex mixing. Samples were shocked with ultrasound in a low temperature water bath for 20 min, then were left at room temperature for 30 min. Samples were mixed with 200 L mass spectrometry water and centrifuged at 12000 x g for 15 min. The upper organic phase was used and dried with nitrogen. 200 μL 90% isopropyl alcohol/acetonitol solution was added during mass spectrometry, vortex fully. Samples were centrifuged at 12000 x g for 15 min at 4 °C. Cell supernatant was used for analysis by UHPLC (Nexera LC-30A, SHIMADZU). Electrospray ionization (ESI) was used in positive and negative ion modes, respectively. The samples were separated by UHPLC and analyzed by mass spectrometry using an AB 6500 + QTRAP mass spectrometer (AB SCIEX). The ESI source conditions were as follows: Source temperature: 400 °C, Ion Source Gas1 (GS1): 50, Ion Source Gas2 (GS2): 55, Curtain Gas (CUR): 35, IonSpray Voltage (IS): +3000 V or -2500 V in positive or negative modes, respectively, monitored in MRM mode. The separation of the samples was performed on an LC-30AD UHPLC system using C18 and Amino columns: C18 column: column temperature 45 °C; flow rate 0.35 mL/min; mobile phase composition A: 70% acetonitrile + 30% water + 5 mM ammonium acetate, B: isopropanol solution, with the gradient elution program as follows: 0-5.0 min, B varies linearly from 20% to 60%, 5.0-13.0 min, B varies linearly from 60% to 100%, 13.1-17.0 min, B is maintained at 20%. The gradient elution program was as follows: 0-5.0 min, B varied linearly from 20% to 60%, 5.0-13.0 min, B varied linearly from 60% to 100%, 13.1-17.0 min, B was maintained at 20%. A: 2 mM ammonium acetate + 50% methanol + 50% acetonitrile, B: 2 mM ammonium acetate + 50% acetonitrile + 50% water, the gradient elution program is as follows: 0-3.0 min, B maintained at 3%, 3.0-13.0 min, B from 3% linear change to 100%, 13.0 min-17.0 min, B maintained at 100%. min, B was maintained at 100%, and 17.1 min-22.0 min, B was maintained at 3%. The samples were placed in an autosampler at 10 °C throughout the analysis. To avoid the effects caused by fluctuations in the detection signal of the instrument, a randomized sequence was used for the sequential analysis of the samples. The data were analyzed using MultiQuant. n = 7 independent samples (S100P low group) and n = 3 independent samples (S100P high group).
Non-targerted lipidomics mass spectrometry
Methanol solution mixed with 0.85% AMBIC solution (pH 7.4) was pre-cooled at -20 °C as quenched reagents. Cells were put into a test tube containing quenched reagents. The test tube was shook slightly for 10 s, then centrifuged at 1000 x g at 4 °C for 1 min. Supernatant was removed and the cells were placed in liquid nitrogen for 30 s and stored at -80 °C. The all cell samples were vortexed for 60 s with 400 μL water, and then the samples were homogenized at 45 Hz for 4 min, and sonicated for 5 min in ice-water bath. The homogenate and sonicate circle was repeated for three times. The 10 μL homogenate was mixed with 190 μL water, and then 480 μL extract solution containing internal standard was added. After 60 s vortex, the samples were sonicated for 10 min in ice-water bath. Then the samples were centrifuged at 1000 x g for 15 min at 4 °C. 250 μL of supernatant was transferred to a fresh tube. The rest of the sample was added with 250 μL of MTBE, followed with vortex, sonication and centrifugation, and another 250 μL of supernatant was taken out. This step was repeated once. And the supernatants were combined and dried in a vacuum concentrator at 37 °C. Then, the dried samples were reconstituted in 100 μL of resuspension buffer (DCM:MeOH:H2O = 60:30:4.5), the samples were vortexed for 30 s and sonicated for 10 min in ice-water bath.The constitution was then centrifuged at 14000 x g for 15 min at 4 °C, and 40 μL of supernatant was transferred to a fresh glass vial for LC/MS analysis. The quality control (QC) sample was prepared by mixing an equal aliquot of the supernatants from all of the samples. The UHPLC separation was carried out using a SCIEX ExionLC series UHPLC System. The mobile phase A consisted of 40% water, 60% acetonitrile, and 10 mmol/L ammonium formate. The mobile phase B consisted of 10% acetonitrile and 90% isopropanol, and 10 mmol/L ammonium formate. The column temperature was 40 °C. The auto-sampler temperature was 6 °C, and the injection volume was 2 μL. AB Sciex QTrap 6500+ mass spectrometer was applied for assay development. Typical ion source parameters were: IonSpray Voltage: +5500/-4500 V, Curtain Gas: 40 psi, Temperature: 350 °C, Ion Source Gas 1:50 psi, Ion Source Gas 2: 50 psi, DP: ±80 V. Skyline 20.1 Software was employed for the quantification of the target compounds. The absolute content of individuals lipids corresponding to the IS was calculated on the basis of peaks area and actual concentration of the identical lipid class internal standard (IS), and then absolute content was obtained from diverse internal standard (IS) averaged of the identical lipid class. n = 3 (WT group) and n = 4 (S100P KO group) independent samples.
Statistics analysis
Western blot, imaging results were independently repeated at least twice. For IHC assays, at least four sample size was used. All the other experiments were independently repeated at least three times. Statistical analysis was carried out using Microsoft Excel software and GraphPad Prism 8 to assess the differences between experimental groups. Logrank test was used to analyze the data of survival curve. Kruskal-Wallis analysis was used in Fig. 8e. Spearman’s rank was used to analyze the data correlation. Statistical significance was determined by using Student’s t-test with a confidence interval (CI) of 95% in all the other experiments. P ≤ 0.05 was denoted as statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The survival curve in Hepatocellular carcinoma tissue and the expression of S100P in human cancer tissues were derived from Kaplan-Meier Plotter (https://kmplot.com/analysis/). The S100P, APOC1 and SPOCK1 expression levels correlated with ferroptosis sensitivity were derived from CTRP V2 (http://portals.broadinstitute.org/ctrp/). The genes expression in SNU387 and HuH7 data were derived from CCLE. The molecular docking was operated on the ClusPro platform (https://cluspro.bu.edu/). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD056171. Metabolomics data have been deposited to the EMBL-EBI MetaboLights database (https://www.ebi.ac.uk/metabolights/) with the identifier MTBLS11347 for targeted lipidomics and MTBLS11228 for non-targeted lipidomics. Source data are provided with this paper. The remaining data are available within the Article, Supplementary Information or Source Data file. Source data are provided with this paper.
References
Dixon, S. J. et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012).
Yang, W. S. et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331 (2014).
Doll, S. et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 13, 91–98 (2017).
Kagan, V. E. et al. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol. 13, 81–90 (2017).
Chu, B. et al. ALOX12 is required for p53-mediated tumour suppression through a distinct ferroptosis pathway. Nat. Cell Biol. 21, 579–591 (2019).
Wang, W. et al. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 569, 270–274 (2019).
Ma X., et al. CD36-mediated ferroptosis dampens intratumoral CD8+ T cell effector function and impairs their antitumor ability. Cell Metab. 33, 1001–1012.e5 (2021).
Liao, P. et al. CD8(+) T cells and fatty acids orchestrate tumor ferroptosis and immunity via ACSL4. Cancer Cell 40, 365–378.e366 (2022).
Kim, R. et al. Ferroptosis of tumour neutrophils causes immune suppression in cancer. Nature 612, 338–346 (2022).
Ubellacker, J. M. et al. Lymph protects metastasizing melanoma cells from ferroptosis. Nature 585, 113–118 (2020).
Zou, Y. et al. Plasticity of ether lipids promotes ferroptosis susceptibility and evasion. Nature 585, 603–608 (2020).
Jiang, L. et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 520, 57–62 (2015).
Sun, X. et al. Activation of the P62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 63, 173–184 (2016).
Sun, J. et al. Quiescin sulfhydryl oxidase 1 promotes sorafenib-induced ferroptosis in hepatocellular carcinoma by driving EGFR endosomal trafficking and inhibiting NRF2 activation. Redox Biol. 41, 101942 (2021).
Ren, X. et al. Overcoming the compensatory elevation of NRF2 renders hepatocellular carcinoma cells more vulnerable to disulfiram/copper-induced ferroptosis. Redox Biol. 46, 102122 (2021).
Yao, F. et al. A targetable LIFR-NF-kappaB-LCN2 axis controls liver tumorigenesis and vulnerability to ferroptosis. Nat. Commun. 12, 7333 (2021).
Chen, Y. et al. CRISPR screens uncover protective effect of PSTK as a regulator of chemotherapy-induced ferroptosis in hepatocellular carcinoma. Mol. Cancer 21, 11 (2022).
Yang, M. et al. COMMD10 inhibits HIF1alpha/CP loop to enhance ferroptosis and radiosensitivity by disrupting Cu-Fe balance in hepatocellular carcinoma. J. Hepatol. 76, 1138–1150 (2022).
Gao, R. et al. YAP/TAZ and ATF4 drive resistance to Sorafenib in hepatocellular carcinoma by preventing ferroptosis. EMBO Mol. Med 13, e14351 (2021).
Arumugam, T., Simeone, D. M., Van Golen, K. & Logsdon, C. D. S100P promotes pancreatic cancer growth, survival, and invasion. Clin. Cancer Res 11, 5356–5364 (2005).
Arumugam, T. & Logsdon, C. D. S100P: a novel therapeutic target for cancer. Amino Acids 41, 893–899 (2011).
Lin, Z. et al. The lipid flippase SLC47A1 blocks metabolic vulnerability to ferroptosis. Nat. Commun. 13, 7965 (2022).
Cui, W., Liu, D., Gu, W. & Chu, B. Peroxisome-driven ether-linked phospholipids biosynthesis is essential for ferroptosis. Cell Death Differ. 28, 2536–2551 (2021).
Zhang, Z. et al. Activation of ferritinophagy is required for the RNA-binding protein ELAVL1/HuR to regulate ferroptosis in hepatic stellate cells. Autophagy 14, 2083–2103 (2018).
Zheng, J. et al. Sorafenib fails to trigger ferroptosis across a wide range of cancer cell lines. Cell Death Dis. 12, 698 (2021).
Li, Z. et al. RNA helicase DDX5 modulates sorafenib sensitivity in hepatocellular carcinoma via the Wnt/β-catenin-ferroptosis axis. Cell Death Dis. 14, 786 (2023).
Zhang, X. J. et al. Multiple omics study identifies an interspecies conserved driver for nonalcoholic steatohepatitis. Sci. Transl. Med 13, eabg8117 (2021).
He, H. et al. Selective autophagic degradation of ACLY (ATP citrate lyase) maintains citrate homeostasis and promotes oocyte maturation. Autophagy 19, 163–179 (2023).
Yan C., et al. Visualizing engrafted human cancer and therapy responses in immunodeficient zebrafish. Cell 177, 1903-1914.e14 (2019).
Viswanathan, V. S. et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 547, 453–457 (2017).
Tang, W. et al. The mechanisms of sorafenib resistance in hepatocellular carcinoma: theoretical basis and therapeutic aspects. Signal Transduct. Target Ther. 5, 87 (2020).
Sun, X. et al. Metallothionein-1G facilitates sorafenib resistance through inhibition of ferroptosis. Hepatology 64, 488–500 (2016).
Ito H., Nakamae I., Kato J. Y., Yoneda-Kato N. Stabilization of fatty acid synthesis enzyme acetyl-CoA carboxylase 1 suppresses acute myeloid leukemia development. J. Clin. Invest. 131, e141529 (2021).
Lally J. S. V., et al. Inhibition of Acetyl-CoA Carboxylase by Phosphorylation or the Inhibitor ND-654 Suppresses Lipogenesis and Hepatocellular Carcinoma. Cell Metab. 29, 174–182 (2019).
Nelson, M. E. et al. Inhibition of hepatic lipogenesis enhances liver tumorigenesis by increasing antioxidant defence and promoting cell survival. Nat. Commun. 8, 14689 (2017).
Lee, H. et al. Energy-stress-mediated AMPK activation inhibits ferroptosis. Nat. Cell Biol. 22, 225–234 (2020).
Yang, M. Created in BioRender. https://BioRender.com/a91e697 (2024).
Acknowledgements
We thank Professor Jiao Wu (Air Force Military Medical University, China) for providing suggestions about 3D tumor spheroids. We thank Professor Rui Jia for providing mCherry-GFP-LC3 plasmid. We thank Dr. Ruihong Zhang for providing A2780 cells. We thank Translational Medicine Core Facility of Shandong University for consultation and instrument availability that supported this work. The software BioRender is utilized to draw the model of Fig. 5e. This work was supported by National Key R&D Program of China (2022YFA0912600, B.C.), National Natural Science Foundation of China (32000515 and 32370800, B.C.; 82472725, 81972275 and 82171748, L.H.), Natural Science Foundation of Shandong Province (ZR2024JQ019, B.C.), Joint Fund of Shandong Provincial Natural Science Foundation (ZR2023LZL010, L.H.), Distinguished Professor of Taishan Scholars (tstp20221109, L.H.).
Author information
Authors and Affiliations
Contributions
Performing the experiments: M.Y., W.C. Methodology: M.Y., W.C., X.L., G.X., C.S., H.X., W.M., X.C., Y.P. Visualization: M.Y., W.C. Funding acquisition: B.C., L.H. Supervision: B.C., L.H. Writing – original draft: B.C. Writing – review & editing: B.C., L.H.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Stephane Gross and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, M., Cui, W., Lv, X. et al. S100P is a ferroptosis suppressor to facilitate hepatocellular carcinoma development by rewiring lipid metabolism. Nat Commun 16, 509 (2025). https://doi.org/10.1038/s41467-024-55785-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-55785-8
This article is cited by
-
A prognostic signature of ferroptosis and lipid metabolism related genes predicts survival and immunotherapy response in hepatocellular carcinoma
Discover Oncology (2026)
-
Ferroptosis: biology and role in liver disease
Journal of Gastroenterology (2025)