Abstract
Germ cell connectivity via intercellular bridges is a widely conserved feature across metazoans. However, its functional significance is poorly understood. Intercellular bridges are essential for fertility in male mice as genetic ablation of a critical bridge component, TEX14, causes spermatogenic failure, but the underlying reasons are unknown. Here we utilized a Tex14 hypomorph with reduced intercellular bridges along with Tex14-null mice that completely lack bridges to examine the roles of germ cell connectivity during spermatogenesis. We report that in males deficient for TEX14 and intercellular bridges, germ cells fail to complete meiotic DNA replication, synapsis and meiotic double-strand break repair. They also derepress retrotransposons and accumulate retrotransposon-encoded proteins during meiosis. Single-cell RNA-sequencing confirms sharing of transcripts between wild-type spermatids and demonstrates its partial attenuation in Tex14 hypomorphs, indicating that intercellular bridges enable cytoplasmic exchange between connected germ cells in testes. Our findings suggest that regulation of meiosis is non-cell-intrinsic and inform a model in which intercellular bridges influence critical meiotic events and protect germline genome integrity during spermatogenesis.
Similar content being viewed by others
Introduction
Meiosis is a pivotal event during the differentiation of germ cells. This specialized cell cycle halves the genome complement of germ cells to create haploid gametes by appending two rounds of chromosome segregation to one round of DNA replication1. During prophase I of meiosis, homologs pair, and chromosomes acquire double-strand breaks (DSBs) that are repaired by homologous recombination. Meiotic recombination is essential as it generates temporary connections between homologs that stabilize them on the metaphase I spindle. But DSBs are hazardous and failure to complete the repair properly can lead to a meiotic arrest or gamete aneuploidy2,3,4. Although it is clear that meiotic recombination is tightly regulated, our understanding of the pathways involved is incomplete, particularly in mammals.
An additional threat during meiosis is posed by transposons, that comprise a dominant portion of mammalian genomes and remain active5,6,7. This is particularly evident in the male germline, where epigenetic changes during meiosis render germline transposon repressive mechanisms less effective and make meiosis especially vulnerable to transposon activation8,9. In male mice, transposon silencing during meiosis is primarily enforced through the concerted actions of CpG DNA methylation and the piRNA pathway, loss of either of which leads to meiotic arrest and infertility10,11,12,13. How the multipronged transposon control strategies intersect with ongoing meiotic events is not fully understood. Little is known about how entry into meiosis and the subsequent progression of cells through meiotic prophase events to meiotic divisions is coordinated. Moreover, we know very little about how meiotic progression intersects with cellular structure and organization.
A conserved feature of metazoan meiosis is that it occurs in a syncytium14,15,16,17. The role of this is best understood in Drosophila females, where germ cell groups that are connected by intercellular bridges enter meiosis synchronously. During meiosis, one cell from the cyst becomes the oocyte while the remaining nurse cells transfer their cytoplasmic contents to the oocyte to promote its growth. Meiosis in male mice also occurs within a cyst of interconnected germ cells that exchange mRNA and even organelles, at least between connected post-meiotic spermatids18,19,20,21,22. Males do not employ a classical nursing mechanism, yet germ-cell connectivity is an essential feature of meiosis in male mice. Meiosis occurs after ~10 spermatogonial mitotic divisions, each of which result in the formation of a stable intercellular bridge23. During spermatogonial divisions, the germ cell factor TEX14 (testis expressed gene 14) localizes to midbodies and blocks the terminal steps of abscission, thus converting midbodies into permanent intercellular bridges that retain TEX14 as a stable component24,25,26. Knockout of Tex14 eliminates germ cell intercellular bridges and causes male infertility25. Specifically, Tex14-/- testes lacking intercellular bridges are depleted for meiotic and post-meiotic cells, demonstrating that bridges are essential for meiosis. However, the exact role of the syncytium and intercellular bridges during meiosis is not understood.
Here, we uncover the functional significance of germ cell intercellular bridges during meiosis in male mice. Intercellular bridges are important for meiotic DNA replication and their loss in Tex14-/- leads to a drastic depletion of cells entering meiotic prophase. We isolated a Tex14 mutant that is hypomorphic for intercellular bridge formation in a forward genetic screen. We named this mutant tama for Tex14 and meiosis affected; tama represents darkness in Vedic philosophy. We leverage this hypomorphic mutant to show that intercellular bridges are next required during meiotic prophase for synapsis and meiotic recombination. Strikingly, intercellular bridges are essential for the suppression of transposons and Tex14 hypomorphs derepress numerous LINE and LTR retrotransposons during late spermatocyte and spermatid stages. We propose that intercellular bridges exert influence over multiple meiotic processes and serve a role in safeguarding genome integrity during spermatogenesis.
Results
Isolation of a Tex14 mutant tama from a forward genetic screen
To discover new meiotic mutants, we performed a phenotype-driven, random mutagenesis screen in mice27,28,29,30. Briefly, mutagenesis was performed by treatment of mice with the alkylating agent N-ethyl-N-nitrosourea, which predominantly induces single nucleotide substitutions in the germline31,32. Mutagenized mice were then subjected to a three-generation breeding scheme to uncover recessive mutations (detailed in Methods), and third-generation males were screened for meiotic defects.
We screened mice by immunostaining squash preparations of testis cells for diagnostic meiotic markers that display distinctive staining patterns as cells progress through meiosis. We immunostained for SYCP3, a component of chromosome axes1,33, and for γH2AX, a phosphorylated form of the histone variant γH2AX which forms in response to meiotic DNA double-strand breaks (DSBs)34. In normal meiosis, SYCP3-positive axial elements start forming during the leptotene stage of meiotic prophase I; these elongate during the zygotene stage and homologous axes align as the first stretches of tripartite synaptonemal complex (SC) appear; SC juxtaposes homologous chromosomes along their lengths in the pachytene stage; and the SC begins to disassemble in the diplotene stage (Fig. 1a). Most DSB formation occurs in the leptotene and zygotene stages, resulting in pan-nuclear γH2AX staining; this signal diminishes as synapsis progresses and breaks are repaired by recombination (Fig. 1a). Recombination-independent γH2AX also accumulates in the sex body, a heterochromatic domain encompassing the sex chromosomes and that is particularly evident during the pachytene and diplotene stages.
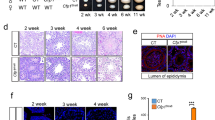
a Representative images of squashed spermatocyte preparations immunostained for SYCP3 and γH2AX. Cells were staged based on SYCP3-staining patterns: short stretches of SYCP3-positive axes develop during leptotene, elaborate into contiguous structures throughout zygotene, contiguous SYCP3-positive structures appear thicker as autosomes fully synapse during pachytene, and subsequently thinner as they desynapse during diplotene. An example of a cell with abnormal staining (nucleus-wide γH2AX along with late prophase-like tracks of SYCP3) is also shown. b Distribution of meiotic prophase stages in three tama mutants and their phenotypically wild-type littermates. c The tama and knockout alleles of Tex14. d The ratios of testes weight to body weight for 6.5- to 29.5-week-old mice. e PAS-stained sections of Bouin’s-fixed testes from 7.5- to 21-week-old Tex14m/m, Tex14-/- and Tex14m/- males along with a wild-type littermate. Zoomed-in views of the regions indicated with boxes are shown below and Sertoli cells (Sertoli), spermatogonia (Spg), spermatocytes (Spc), round spermatids (rSt) and elongated spermatids (eSt) are indicated. Two or more mice were examined for each genotype. f Representative images of TUNEL assay on adult testis sections. Red arrowheads point to TUNEL-positive cells (stained dark brown). g Quantitation of TUNEL assay. The numbers of TUNEL-positive cells counted in individual tubule sections are shown on the left. Blue horizontal lines indicate means; bars indicate standard deviations. Proportions of tubules containing varying ranges of TUNEL-positive cells are shown on the right. Mean values across mice are plotted; red bars indicate standard deviations. Source data are provided as a Source Data file.
We isolated a mutant line, which we named tama, displaying SYCP3- and γH2AX-staining patterns indicative of defects in meiotic DSB repair and/or synapsis (Fig. 1a, b). In these mutants, spermatocytes with SYCP3 staining characteristic of early prophase (leptotene and zygotene) were abundant but later stages (pachytene and diplotene) were greatly depleted, indicating a block to meiotic progression. Early prophase-like spermatocytes displayed nucleus-wide γH2AX staining consistent with that observed in wild type, indicating the formation of meiotic DSBs. However, mutants also had elevated numbers of abnormal spermatocytes with nucleus-wide γH2AX staining accompanied by long tracks of SYCP3 staining that were consistent with varying degrees of synapsis (Fig. 1a, b). These patterns are typical of mutants that cannot complete meiotic DSB repair or synapsis34, such as Dmc1-/-35,36, Msh5-/-37,38 and Sycp1-/-39. We mapped the likely causative tama mutation to an un-annotated DNA sequence variant located in the Tex14 gene (see Methods). The tama variant (Tex14m) is a A to G nucleotide transition at position Chr11:87,533,604 (GRCm38/mm10), adjacent to a splice donor site between exons 21 and 22 (Fig. 1c).
We first evaluated the impact of the tama mutation on fertility. Tex14+/m heterozygote males had normal fertility and transmitted the mutation in a near-Mendelian ratio (21.87% Tex14+/+, 54.81% Tex14+/m, and 23.32% Tex14m/m from heterozygote × heterozygote crosses; n = 343 mice; average litter size = 6; p = 0.41, Fisher’s exact test). Tex14m/m homozygote males were sterile, however. None of the three homozygous animals tested sired progeny when paired with heterozygous females for 12 weeks. Tex14m/m mutant females produced pups and had an average litter size of 5.9 (16 litters from 6 dams), comparable to Tex14+/m heterozygote females that gave an average litter size of 6 (57 litters from 14 dams). Tex14m/m homozygote females and males survived to adulthood and no obvious morphological defects were apparent. We conclude that the tama mutation causes male sterility but does not severely impact female gametogenesis or adult somatic functions.
Tex14m/m males displayed a 73.14% reduction in testes-to-body-weight ratio compared to littermates (mean ratios were 0.15% for tama mutants and 0.58% for wild-type and heterozygous littermates; p < 0.01, two-sided Student’s t-test; Fig. 1d). In histological analyses of adult testis sections, seminiferous tubules from Tex14m/m males were reduced in diameter compared to wild type and heterozygotes (Fig. 1e). Mutant tubules contained primarily Sertoli cells and early spermatogenic cells, with near-complete absence of post-meiotic spermatids. We interpret these less populated tubules as those in which apoptosis has already eliminated aberrant cells due to defects in meiotic prophase progression. Indeed, TUNEL staining showed a higher incidence of apoptosis in adult Tex14m/m males compared to wild-type and heterozygous littermates (Fig. 1f, g).
To verify that the Tex14 point mutation in the tama line is causative for the spermatogenesis defect, we compared it to a previously described Tex14 null allele (Tex14-)25. In this allele, exon 10 has been replaced with a PgkHPRT expression cassette (Fig. 1c), resulting in absence of TEX14 protein. Tex14-/- males were sterile, displayed marked hypogonadism (Fig. 1d), and showed greatly reduced numbers of spermatogenic cells (Fig. 1e) with increased levels of apoptotic cells compared to control littermates in testis histology sections (Fig. 1f, g), as previously reported25. Notably, the Tex14 null allele failed to complement the tama mutation. Tex14m/- compound heterozygotes showed a similar reduction in testes-to-body-weight ratio (73.26% reduction; 0.19% mean ratio for Tex14m/- and 0.71% for wild-type and single-heterozygous animals; p < 0.01, two-sided Student’s t-test) (Fig. 1d) and displayed a similar depletion of post-meiotic spermatids as Tex14m/m mutants (Fig. 1e). Additionally, compound heterozygotes accumulated TUNEL-positive apoptotic cells, like Tex14m/m mutants (Fig. 1f, g). We conclude that the tama mutation disrupts Tex14 function and leads to defective meiotic prophase progression, culminating in infertility.
Tex14 tama is a hypomorphic allele that causes a partial loss of intercellular bridges
TEX14 is an essential component of germ cell intercellular bridges and complete loss of TEX14 protein in Tex14-/- leads to loss of intercellular bridges25, so we evaluated the impact of the tama mutation on intercellular bridges during spermatogenesis. We first asked whether the tama mutation affects TEX14 protein abundance by immunoblotting testis extracts. Because tama mutants have a reduced number of late-stage spermatocytes (Fig. 1b), we examined juvenile males aged 10 dpp. At this age, wild-type testes predominantly contain spermatogonia and early meiotic cells, with the most advanced germ cell stage being zygotene40. TEX14 protein level was comparable between wild type and Tex14+/m heterozygotes, while Tex14m/m mutant testes showed a substantial reduction ( ~ 90%) in TEX14 protein (Fig. 2a, S1a). Since the tama mutation is located adjacent to a splice donor site (Fig. 1c), it may cause aberrant splicing, leading to a frame shift, premature translation termination, and production of a truncated version of the TEX14 protein, but this was undetectable in immunoblots (Fig. 2a, S1a). A straightforward interpretation is that the tama mutation leads to RNA and/or protein instability, and that the meiotic defect observed Tex14m/m mutants is a consequence of decreased TEX14 protein level.
a Western blot analysis of TEX14 and VINCULIN (loading control) in testis extracts from 10-dpp animals. Relative TEX14 signal intensities normalized to VINCULIN are indicated below. Two mice were examined for each genotype. b TEX14 immunofluorescence on whole-mounted seminiferous tubules rendered in 3D. Higher magnification views of the regions indicated with white boxes are shown on the right. Two Tex14+/+ and three Tex14m/m mice were examined. c Immunofluorescence of TEX14 and MKLP1 on adult testis sections. Spermatocytes (Sc), elongating spermatids (eSt), and round spermatids (rSt) are indicated. TEX14 and MKLP1 staining is highlighted with yellow, dashed circles. Higher magnification views of TEX14 and MKLP1 colocalization are shown in insets. One Tex14m/m and two Tex14-/- mice along with control littermates were examined. d Electron micrograph of intercellular bridges in testis sections. Cell boundaries are depicted with red, dashed lines, intercellular bridges are indicated with yellow, dashed boxes. A higher magnification view of intercellular bridges is shown on the right and yellow arrows point to the electron-dense lining of intercellular bridges. Two mice were examined for each genotype. Source data are provided as a Source Data file.
To assess whether the low level of TEX14 protein present in Tex14m/m mutant testes localizes to and supports intercellular bridge formation, we immunostained whole-mounted seminiferous tubules for TEX14. TEX14 forms a “ring-like” structure at the intercellular bridge connecting adjacent germ cells of a syncytium25, appearing as a “dash-like” structure when viewed from a lateral angle. In wild type, “dash-like” TEX14 staining was prominent in between DAPI-positive germ cell nuclei (Fig. 2b). “Dash-like” TEX14 staining was also detected in between DAPI-positive germ cell nuclei in Tex14m/m mutant seminiferous tubules, but these TEX14-positive structures were substantially reduced in number (Fig. 2b, S1b) and often reduced in width (Fig. S1b), consistent with the reduction in TEX14 protein level in mutants.
To evaluate whether TEX14 staining was marking intercellular bridges in mutants, we immunostained testis sections for mitotic kinesin-like protein 1 (MKLP1), a centralspindlin complex protein that localizes to the midbody and is retained as a stable intercellular bridge component when midbodies are transformed into intercellular bridges in germ cells16,26,41. MKLP1 co-localized with TEX14 forming “ring-like” structures adjacent to DAPI-positive germ cell nuclei in testis sections from Tex14+/m heterozygotes and staining was absent in Tex14-/- mice that lack intercellular bridges, as previously reported26 (Fig. 2c). We detected “ring-like” TEX14 and MKLP1 double-positive structures adjacent to DAPI-positive germ cell nuclei in Tex14m/m mutant testis sections (Fig. 2c), suggesting that Tex14m/m mutants do form few intercellular bridge-like structures. Indeed, cytoplasmic channels with electron-dense material lining the inner surface of cell membranes, characteristic of intercellular bridges18,25,41, were occasionally observed in Tex14m/m mutants by electron microscopy (Fig. 2d).
Collectively, these data lead us to conclude that Tex14m/m mutants are hypomorphic for intercellular bridge formation. While Tex14-/- males completely lack intercellular bridges25,41, Tex14m/m mutant testes contain a mixture of individualized germ cells that have completed cytokinesis and germ cells that are connected by intercellular bridge-like structures.
Tex14 mutants show a defect in entering meiotic prophase
Our initial histological analyses showed comparable defects in Tex14-/- and Tex14m/m mutants (Fig. 1d–g), so we evaluated this phenotypic similarity more closely. Because TEX14-containing intercellular bridges connect spermatogonia as they go through transit-amplifying divisions and differentiate16,25,42, we examined spermatogonia in Tex14m/m and Tex14-/-. To evaluate undifferentiated spermatogonia (i.e., As, Apr, and Aal), we immunostained adult testis sections for PLZF (a.k.a. ZBTB16)43,44 (Fig. 3a). The overall distribution of PLZF-positive undifferentiated spermatogonia counts across tubule sections appeared similar, although the number of positive cells was modestly upregulated in Tex14m/m and Tex14-/- mutants compared to wild-type or heterozygous littermates. These results suggest that loss of intercellular bridges does not severely impact undifferentiated spermatogonia numbers and are consistent with previous reports analyzing Tex14-/-25, but a small increase in the undifferentiated spermatogonial pool cannot be ruled out.
a Representative images of PLZF-stained testis sections along with quantitation of the number of positive-staining cells. b cKIT-stained testis sections along with quantitation of cKIT-positive and SYCP3-negative cells. c STRA8-stained testis sections along with quantitation of STRA8-positive cells. d SYCP3- and γH2AX-stained testis sections along with quantitation of early prophase (leptotene and zygotene) cells. Early prophase cells were identified based on SYCP3- and γH2AX-staining patterns. The numbers of positive-staining cells counted in individual tubule sections are shown in dot plots. Blue horizontal lines indicate means; bars indicate standard deviations. Proportions of tubules counted that contain varying ranges of positive-staining cells are shown in stacked bar plots. Mean values across mice are plotted; red bars indicate standard deviations. Source data are provided as a Source Data file.
We also examined spermatogonial differentiation by quantifying the KIT-positive differentiating spermatogonia population (Fig. 3b). KIT also marks preleptotene stage cells and early meiotic prophase cells45, so we did not count cells showing leptotene-like SYCP3 staining patterns to exclude meiotic prophase cells from our counts. We observed only a modest change in the distribution of KIT-positive cells in Tex14m/m and Tex14-/- compared to control littermates. The proportion of tubule sections counted that contained >15 KIT-positive cells was 24% in controls, 14% inTex14m/m and 1.33% in Tex14-/-. We attribute this variation in the distribution of KIT-positive cells in mutants to a defect during preleptotene (see below) and infer that spermatogonial populations are not drastically affected upon perturbation of TEX14-mediated intercellular bridges, in keeping with previous analyses of Tex14-/-16,25.
Because our initial results from the screen indicated a meiotic progression defect in Tex14m/m, we next asked whether loss of TEX14-mediated intercellular bridges also impacts spermatogonial cell entry into meiosis. To evaluate cells entering meiosis (preleptotene spermatocytes), we immunostained testis sections for STRA8, which is expressed during preleptotene and transiently expressed in differentiating spermatogonia in response to retinoic acid46,47,48 (Fig. 3c). In control littermates, 13.22% of seminiferous tubule sections contained high numbers of STRA8-positive cells ( > 40 cells), while most tubule sections either lacked STRA8-positive cells or had a very low number of STRA8-positive cells ( ~ 0-10 cells), reflecting synchronous initiation of meiosis. Tex14m/m and Tex14-/- mice lacked this bimodal distribution; while there were few tubule sections that had no STRA8-positive cells, the majority of tubule sections displayed a continuous range of non-zero STRA8-positive cell counts49. Importantly, the number of STRA8-positive cells in tubule sections were slightly diminished in Tex14m/m (~1-45 cells) and substantially reduced in Tex14-/- ( ~ 1-30 cells) compared to controls. This decrease in STRA8-positive preleptotene cells is probably sufficient to account for the decrease in KIT-positive cell counts in mutants.
The depletion of preleptotene spermatocytes suggests that TEX14-mediated intercellular bridges play a critical role in regulating events that occur just prior to or during meiotic entry. Consistent with this, Tex14 mutants showed a decrease in the numbers of early meiotic prophase cells (Fig. 3d). Wild-type and heterozygous littermates had a greater proportion of tubule sections containing high numbers of leptotene/zygotene cells ( > 30 cells) compared to mutants (20.33%, 2.67% and 0% in controls, Tex14m/m and Tex14-/-, respectively). Whereas Tex14m/m mostly contained tubules with fewer leptotene/zygotene cells ( ~ 1-35 cells), and a further reduction was seen in Tex14-/- (most tubule sections contained ~1-25 cells).
Collectively, these results are consistent with the hypothesis that spermatogonial divisions occur relatively normally in Tex14 mutants, but entry into meiosis is defective. Although a hitherto unknown function for TEX14 that is independent of intercellular bridges cannot be ruled out, the correlation between the reduction in TEX14-containing intercellular bridges and phenotypic severity in Tex14 mutants supports the conclusion that intercellular bridges regulate meiotic entry.
Aberrant meiotic S-phase in Tex14 mutants
Because meiosis begins with meiotic DNA replication during the preleptotene stage, and because preleptotene spermatocytes are depleted in Tex14 mutants, we hypothesized that TEX14-mediated intercellular bridges are important for proper meiotic S phase. To assess DNA replication, we subjected Tex14 mutants and their wild-type or heterozygous littermates to a 2 hr pulse label with 5-ethynyl-2’-deoxyuridine (EdU) in vivo. In whole-mounted seminiferous tubules from controls, adjacent STRA8-positive preleptotene spermatocytes homogenously incorporated EdU (Fig. 4a). This indicates that neighboring preleptotene cells within a seminiferous tubule segment, some of which are presumably connected by intercellular bridges in a syncytium, undergo meiotic DNA replication synchronously, as expected. In contrast, Tex14 mutants contained a mixture of STRA8-positive cells that incorporated EdU and those that were EdU-negative within the same seminiferous tubule segment, indicating a defect in replication (Fig. 4a). Interestingly, juxtaposed cells that appeared connected by a TEX14-positive intercellular bridge displayed similar replicative states in both wild type and Tex14m/m; i.e., connected STRA8-positive cells typically appeared to incorporate EdU uniformly where either both were EdU positive or both were EdU negative (Fig. S2a).
a STRA8 and EdU immunostaining on whole-mounted seminiferous tubules. Zoomed-in views of the regions indicated with yellow boxes are shown on the right and STRA8-positive EdU-negative cells are annotated (yellow arrows) and depicted on the far right. Two mice were examined for each genotype. b Immunofluorescence of STRA8 and EdU on testis sections. Higher magnification views of the regions indicated with white boxes are shown on the right. Three Tex14m/m and three Tex14-/- mice along with control littermates were examined. c Quantitation of the numbers of STRA8-positive EdU-negative (pink) and STRA8- EdU-double-positive (aqua) cells within tubule sections. Graphs represent individual mice and vertical bars represent individual tubules in a testis section. d Quantitation of STRA8- EdU-double-positive cells. The numbers of STRA8- EdU-double-positive cells counted in individual tubule sections that contain at least one STRA8-positive cell are shown in the dot plot above. Blue horizontal lines indicate means; bars indicate standard deviations. Proportions of tubules counted that contain varying ranges of double-positive cells are shown in the bar plot below. Mean values across mice are plotted; red bars indicate standard deviations. e Histograms of nuclei count as a function of the intensity of DAPI in STRA8-positive DMRT1-negative spermatocytes. Two mice were examined for each genotype. Source data are provided as a Source Data file.
For quantification, we examined testis sections from EdU-labeled mice (Fig. 4b). Consistent with our findings from whole mounts, most STRA8-positive tubule sections from control animals were comprised of cells that uniformly incorporated EdU (Fig. 4b, c). Whereas the majority of mutant tubule sections contained a mixture of STRA8- and EdU-double-positive cells alongside STRA8-positive and EdU-negative cells. Importantly, the numbers of double-positive cells in individual tubule sections, inferred to be cells undergoing meiotic replication, were moderately decreased in Tex14m/m (~1-40 double-positive cells) and severely depleted in Tex14-/- ( ~ 1-20 double-positive cells) compared to control littermates ( ~ 1-75 double-positive cells) (Fig. 4d). Accordingly, the proportion of STRA8-positive tubule sections containing >20 STRA8- and EdU-double-positive cells was 51.68% in controls, 11.36% in Tex14m/m and 0.58% in Tex14-/-. Our results are consistent with the hypothesis that TEX14-mediated intercellular bridges facilitate meiotic DNA replication.
This hypothesis predicts that mutant preleptotene cells would accumulate in a pre-replicative G1-like state, in contrast to wild-type preleptotene cells which transiently enter G1, replicate, and proceed to meiotic prophase. To test this prediction, we sorted STRA8-positive cells using fluorescence-activated nuclei sorting and evaluated their cell cycle profile based on DAPI fluorescence50,51. To focus our analyses on the cell population entering meiosis, we excluded STRA8-positive cells that were also positive for DMRT1, which is expressed at high levels in spermatogonia, and expression is greatly reduced upon meiotic entry52,53. In wild type, approximately one-third of STRA8-positive DMRT1-negative preleptotene cells were in G1 (31.0%; 2 C DAPI content) another one-third were in S-phase (32.3%; 2–4 C DAPI content) (Fig. 4e, S2b). A comparable profile was observed for mice lacking SPO11, where spermatocytes show no defects in replication but arrest during meiotic prophase and ultimately apoptose due to lack of meiotic DSBs50,54,55,56 (Fig. 4e). Tex14 mutants, however, showed a striking shift in cell cycle profiles (Fig. 4e). Quantification of cell cycle phases revealed that Tex14-/- contained a larger fraction of STRA8-positive DMRT1-negative preleptotene cells in G1 (46.6%) and a smaller fraction of cells in S-phase (26.9%), compared to wild type. Tex14m/m was marginally altered compared to wild type; G1-stage cells were slightly increased (36.5%) and S-phase cells were slightly diminished (30.9%). Consistent with this, Tex14m/m testes contain a larger number of meiotic prophase cells while Tex14-/- are severely depleted for prophase cells (Fig. 3d). Cell cycle profiles of DMRT1-positive spermatogonia were comparable between mutants and wild type, as expected (Fig. S2c).
Collectively, these data lead us to conclude that spermatogonia with perturbed intercellular bridges reach the preleptotene stage and switch on STRA8 expression in response to retinoic acid, however many cells are unable to undergo meiotic DNA replication. The Tex14m/m and Tex14-/- mutant phenotypes are concordant in this regard, and as expected, phenotypic severity correlates with the extent of intercellular bridge depletion. We posit that aberrant preleptotene cells are ultimately eliminated, yielding a decrease in the total number of STRA8-positive cells in mutants, although we cannot rule out the possibility that fewer mutant spermatogonia express STRA8, to begin with.
Tex14 deficiency causes synapsis and meiotic recombination defects
Our initial findings from the screen indicated that Tex14m/m mutants dysregulate the hallmark meiotic recombination and synapsis events of meiotic prophase (Fig. 1a), so we examined these events more closely. Tex14-/- also exhibit a meiotic prophase defect (Fig. S3a). However the extensive depletion of prophase cells in Tex14 null mice (Fig. 3d) makes it challenging to accurately assess and draw conclusions about prophase, therefore we focused our analyses on the hypomorphic mutant.
We assayed synapsis by immunostaining spread spermatocyte chromosomes for the chromosome axis protein SYCP3 and for the SC central region protein SYCP139. Leptotene and early zygotene appeared normal, but synapsis defects were visible in pachytene. In wild type, most autosomes are fully synapsed by pachytene, visualized by the colocalization of SYCP3 and SYCP1 along the entire lengths of chromosome axes. In Tex14m/m, 64% of pachytene cells appeared fully synapsed, however 36% showed abnormal synapsis (Fig. 5a, b). Abnormalities observed included illegitimate synapsis in the form of chromosome tangles (i.e., a combination of unsynapsed and synapsed axes with partner switches indicative of nonhomologous synapsis) and there were instances of synaptic failure where cells contained asynaptic autosomes alongside fully synapsed autosomes (Fig. 5a, b). Synapsis defects like these are associated with apoptosis57 and could explain the late-stage prophase depletion in mutants.
a Chromosome spreads of pachytene-stage cells from Tex14m/m immunostained for SYCP3 and SYCP1 depicting illegitimate synapsis or asynapsis. Higher-magnification views of the boxed regions are shown on the right and chromosomes displaying asynapsis (above) and illegitimate synapsis (below) are annotated (white arrowheads). b Quantitation of synapsis in autosomes. Cells containing chromosome tangles with a mix of synapsed and asynapsed axes with partner switches indicative of nonhomologous synapsis are categorized as illegitimate synapsis. Three mice were examined for each genotype. c Chromosome spreads of pachytene-stage cells from Tex14m/m immunostained for SYCP3 and γH2AX depicting normal staining (left), large patches of γH2AX (middle), and γH2AX flares on autosomes (right). d Quantitation of γH2AX staining patterns. Three mice were examined for each genotype. e Chromosome spreads immunostained for SYCP3 and DMC1. Time course of meiotic prophase stages is shown. Four Tex14m/m along with control littermates were examined. f Representative images of SYCP3- and DMC1-stained cells with aberrant synapsis. Aberrant cells include zygotene-like and pachytene-like cells with chromosome tangles and pachytene-like cells with autosomal asynapsis. Higher-magnification views of the boxed regions marking abnormal synapsis are depicted at the bottom and synapsis abnormalities are annotated (white arrowheads). g Quantitation of DMC1 (n = 4 mutants and control littermates) and RAD51 (n = 3 mutants and control littermates) foci during meiotic progression. Foci overlapping with SYCP3-positive axes were scored and cells were staged based on SYCP3-staining patterns: leptotene (Lept.), early zygotene (E. zyg.), mid zygotene (M. zyg.), late zygotene (L. zyg.), pachytene (Pach.), aberrant zygotene-like (Zyg.-like), aberrant pachytene-like (Pach.-like). h SYCP3- and MLH1-stained pachytene-stage cells showing chromosomes with wild-type-like foci counts (1-2 foci) and abnormal MLH1 foci counts (0 foci). Higher magnification views of single chromosomes are shown in insets. Four Tex14m/m along with control littermates were examined. i Quantification of MLH1 foci (n = 4 mutants and control littermates) on autosomes and sex chromosomes in pachytene cells. j Proportions of autosomes with 0, 1-2, or ≥3 MLH1 foci (n = 4 mutants and control littermates). Blue horizontal lines indicate means; bars indicate standard deviations. Quadruple-asterisk represents p < 0.0001, triple-asterisk represents p < 0.001, double-asterisk represents p < 0.01, and a single-asterisk represents p < 0.05 in two-sided Mann-Whitney U-tests. Exact p values: g DMC1 Lept. p = 0.132, E. zyg. p = 0.191, M. zyg. p = 0.003, L. zyg. p = 0.094, Pach. p = 4.52E-7; RAD51 Lept. p = 0.073, E. zyg. p = 0.159, M. zyg. p = 5E-4, L. zyg. p = 0.368, Pach. p = 8.47E-6. (i) MLH1 p = 0.004. Source data are provided as a Source Data file.
To begin to assess recombination, we reexamined γH2AX staining, this time on spread spermatocyte chromosomes to better resolve staining patterns and their correlations with synapsis state. We scored for the presence of γH2AX flares that appeared in a small region immediately adjacent to individual chromosomes and the presence of large patches of γH2AX staining covering regions encompassing several chromosomes. Pachytene-stage cells from Tex14m/m with apparently normal autosomal synapsis more frequently displayed high numbers of γH2AX flares and large patches of persistent γH2AX on autosomes compared to control littermates (Fig. 5c, d), in keeping with our initial analyses of γH2AX in nuclei squashes. 24% of wild-type or heterozygous pachytene cells contained patches and/or ≥4 flares, while 67% of Tex14m/m pachytene cells displayed this phenotype. These results are consistent with the idea that Tex14m/m mutants have a defect in repairing meiotic DSBs.
To test this hypothesis further and to evaluate the molecular characteristics of Tex14m/m meiotic cells, we immunostained chromosome spreads for the recombinases DMC1 and RAD5154,58,59. These proteins assemble at DSB sites and are removed as recombination progresses, so they can be used to track meiotic recombination. In normal meiosis, DMC1 and RAD51 foci appear on chromosome axes in leptotene, accumulate to maximal levels by early zygotene, then continue to decline through pachytene (Fig. 5e, g). In Tex14m/m mutants, DMC1 foci counts peaked at early zygotene, with a higher-than-normal average compared to control littermates, although this difference was not statistically significant (Fig. 5e, g). Mutant foci counts were more widely spread at this stage, with few cells showing normal counts and few with lower or higher foci counts compared to controls. DMC1 foci counts generally declined as synapsis progressed in mutants, and high levels of foci were retained in cells with aberrant synapsis (Fig. 5f, g). Importantly, during mid zygotene, mutant foci counts were significantly higher compared to controls, and this trend continued through pachytene (Fig. 5e, g). RAD51 behaved similarly in mutants, with significantly higher RAD51 foci retained in mutant pachytene cells compared to control littermates (Figs. 5g, S3b, c).
Elevated DMC1 and RAD51 foci counts in mutant pachytene cells with apparently normal synapsis suggests that the increase in recombinase foci is not simply because of synaptic errors. Based on the retention of recombinase foci in pachytene and the persistence of γH2AX on autosomes, we infer that Tex14m/m mutants are unable to complete meiotic recombination. Spermatocytes that are unable to complete recombination arrest differentiation during pachynema and apoptose60,61, so this may contribute to the depletion of late-prophase cells in Tex14 mutants.
Finally, we evaluated whether Tex14m/m mutant cells that progress to late pachytene generate crossovers by immunostaining chromosome spreads for MLH1, which cytologically marks crossover sites at pachytene62,63 (Fig. 5h). Tex14m/m pachytene cells with normal synapsis had a significant decrease in the number of MLH1 foci per cell (Fig. 5i), with 5% synapsed autosomes completely lacking crossovers (Fig. 5h, j). At least one crossover per homolog pair is crucial to ensure biorientation at the metaphase I spindle54,63. Accordingly, we observed a drastic depletion of SYCP3 and pH3 double-positive meiotic metaphase cells (Figs. S3d, e) and spermatids (Figs. S3f, g) in testis sections from Tex14m/m mutants. We conclude that synapsis and recombination are substantially dysregulated in Tex14 hypomorphs with depleted intercellular bridges.
Tex14 mutants derepress transposons during meiosis
Intercellular bridges in rat spermatids have been shown to traffic germ cell granules rich in piRNAs and transposon repression machinery20,64,65, indicating that bridges may influence transposon expression. Notably, elevated levels of LINE1 retrotransposon (L1) expression were recently reported in Tex14-/- oocytes66, although this does not impact female fertility41. Loss of proper transposon repression in male mice, for example via disruption of the piRNA pathway, manifests as a mid-prophase I arrest67,68,69. In such mutants, prophase cells accumulate transposon-encoded proteins, have defects in chromosome synapsis, arrest prior to completing meiotic recombination, and eventually apoptose, leading to male-specific infertility.
The phenotypic similarities during meiosis between Tex14tama and mutants that are defective for transposon repression led us to hypothesize that intercellular bridges play a role in enforcing proper transposon silencing. Indeed, immunostaining Tex14m/m testis sections for L1-encoded ORF1p showed transposon upregulation in SYCP3-positive meiotic cells (Fig. 6a). 58-65% of Tex14m/m seminiferous tubules contained brightly staining ORF1p-positive cells, whereas similar levels of ORF1p were not detected in wild-type littermates; 7-17% of wild-type tubules stained weakly positive for ORF1p (Fig. 6b). In mutants, bright ORF1p staining was present in some late zygotene, pachytene, and diplotene spermatocytes (Fig. 6c); we rarely observed similar levels of ORF1p staining in earlier prophase cells or in spermatogonia. Elevated ORF1p levels were also detected in immunoblots of testis extracts from adult Tex14m/m mice (Fig. 6d). ORF1p accumulation was also present in Tex14-/- meiotic cells (Fig. S4a), reinforcing the role of Tex14 in regulating transposons. But due to the severe depletion of spermatocytes in null mice, we focused our analyses on Tex14m/m.
a SYCP3 and ORF1p immunofluorescence on testis sections. Tubules with numerous bright ORF1p-positive cells are indicated with white arrows. Three mice were examined for each genotype. b Distribution of ORF1p-positive tubules. c Tubules indicated in a with higher magnification views of boxed regions at the bottom. Cells were staged based on SYCP3-staining patterns: leptotene (L), zygotene (Z), pachytene (P), diplotene (D). d. Three Tex14m/m mice were examined. d ORF1p and VINCULIN (loading control) immunoblot in testis extracts from 13-week-old mice. Two Tex14m/m and three control littermates were examined. e L1Mda_l_5end RNA FISH with ORF1p immunostaining on testis sections. Higher magnification views of regions indicated with white arrowheads are shown below. Examples of colocalization of L1Mda_l_5end RNA and ORF1p are highlighted using white arrowheads. Two Tex14m/m mice were examined. Source data are provided as a Source Data file.
To test whether the ORF1p accumulation in mutants was accompanied by an accumulation of L1 RNA transcripts, we performed sequential ORF1p immunofluorescence followed by Hybridization Chain Reaction (HCR) RNA FISH with L1 DNA probes. We probed for L1MdA_I_5end as we identified this transcript to be derepressed in Tex14m/m by single-cell RNA-sequencing (detailed below). L1 FISH signal was present in Tex14m/m ORF1p-positive cells, usually localizing to prominent ORF1p perinuclear aggregates that have previously been shown to be enriched in L1 RNA in piRNA mutants70 (Fig. 6e). We detected similar staining patterns using an additional L1 probe, recognizing an L1MdTf family transcript70 (L1spa; Fig. S4b), while probing for U6 small nuclear RNA gave strong nuclear signals in both wild type and mutants (Fig. S4c), confirming the specificity of detection. Taken together, our data are consistent with the hypothesis that TEX14-mediated intercellular bridges play a role in regulating L1 transposons in the male germline.
Elevated transposon levels in meiotic cells can lead to DNA damage independent of programmed meiotic DSBs71, so we tested this possibility by examining γH2AX staining patterns in Tex14m/m spermatocytes that also lack SPO11, the type II topoisomerase-like enzyme that generates meiotic DSBs55,56. Normally, γH2AX staining is nucleus-wide in leptotene and zygotene, then restricted to the sex chromosomes in pachytene and diplotene. Spo11-/- spermatocytes lack meiotic DSBs and lack pan-nuclear γH2AX staining but a subset shows localized staining over asynaptic regions34,60,72,73. In testis sections from Tex14m/m Spo11-/-, some spermatocytes exhibited localized γH2AX staining, likely covering asynaptic regions (Fig. S4d). We observed a few instances of bright pan-nuclear γH2AX staining that may be indicative of transposon-induced damage, however, these cells were rare in sections. In Tex14m/m Spo11-/- spread chromosomes with elongated SYCP3 axial elements, γH2AX staining was typically localized (Fig. S4e, f; patches and flares). A small subset of cells (4.92%) had bright pan-nuclear γH2AX staining. We infer that a small proportion of Tex14m/m spermatocytes may harbor SPO11-independent DSBs, while most do not accumulate substantial levels of SPO11-independent damage, and that majority of the persistent DSBs observed during pachytene likely result from deregulated meiotic recombination. Collectively, these data lead us to conclude that Tex14-defective germ cells that progress through meiotic prophase exhibit multiple abnormalities that could account for the depletion of late-prophase cells; they fail to repair meiotic DSBs and fail to silence transposons.
TEX14-mediated intercellular bridges shape the transposon expression landscape during spermatogenesis
To more broadly examine the contribution of intercellular bridges in repressing transposons, and to test whether bridges also influence germ cell transcriptomes, we performed single-cell RNA-sequencing (scRNA-seq) of testes from two Tex14m/m and two Tex14+/+ mice from a single litter using the 10x Genomics Chromium platform (see Methods). We profiled total 11,956 Tex14m/m cells and 10,132 Tex14+/+ cells after filtering. Using previously reported cell type-enriched markers53, we identified eight broadly-defined cell populations; four germ cell types (spermatogonia, spermatocytes, round spermatids, elongating spermatids) and four somatic cell types (Sertoli, endothelial, innate lymphoid, macrophages) (Fig. S5a). We next performed focused re-clustering of germ cell types and identified eight subtypes that followed the expected developmental trajectory; two spermatogonia, followed by two spermatocyte, three round spermatid, and one elongating spermatid subtype (Fig. 7a). Clusters were annotated as early/mid/late based on their position along the trajectory and the cluster containing cells with high Stra8 expression along with low Dmrt1 expression was annotated as containing preleptotene (Fig. S5b, c).
a All Tex14+/+ and Tex14m/m germ cells were positioned by batch-corrected principal component (PC) 1 and PC3. Cells are colored by their cell identity and follow the expected developmental trajectory from right to left. Spg, spermatogonia; Spg/Pre-L, spermatogonia and preleptotene cells; Early Spc, early spermatocytes; Late Spc, late spermatocytes; Early R. Sp, early round spermatids; Mid R. Sp, mid-trajectory round spermatids; Late R. Sp, late round spermatids; E. Sp, elongating spermatids. Inset panels on the right show the positions of Tex14+/+ (top) and Tex14m/m (bottom) cells. b Distribution of germ cell identities in Tex14+/+ and Tex14m/m. The height of blocks within each vertical bar corresponds to the proportion of each colored cell identity among germ cells of each genotype. c Heatmap depicting Pearson correlation of normalized pseudo-bulk expression among Tex14+/+ and Tex14m/m germ cell categories. d MA plots showing a comparison of expression of individual genes (black) and TEs (pink/green/orange) between Tex14+/+ and Tex14m/m using pseudo-bulk datasets created for each germ cell category. Pink, long interspersed nuclear element (LINE); green, long terminal repeat (LTR) retrotransposon; orange, short interspersed nuclear element (SINE). Points with high fill color intensity correspond to differentially expressed genes or TEs (FDR ≤ 0.05) and points with low color intensity correspond to non-differentially expressed genes or TEs. Black squares correspond to outlier genes whose expression ratio falls outside the bounds of the plot. e Violin plots showing the distribution of log2 transformed total LINE expression summed across all LINE families within each cell of an assigned germ cell category. Pink points correspond to the mean value for each category. Quadruple-asterisk represents p < 0.0001, triple-asterisk represents p < 0.001, double-asterisk represents p < 0.01, and a single-asterisk represents p < 0.05 in two-sided Wilcoxon tests. Exact p values: Spg p = 0.025, Spg/Pre-L p = 0.311, Early Spc p = 0.114, Late Spc p = 7.15E-26, Early R. Sp P = 8.85E-09, Mid R. Sp p = 1.23E-35, Late R. Sp p = 1.96E-12. f Scatterplots showing the number of X chromosome (Chr X) genes detected in each cell (x-axis) versus Y chromosome (Chr Y; y-axis, top rows) and versus chromosome 9 (Chr 9; y-axis, bottom rows) genes detected for late spermatocytes through late round spermatids (left to right, columns). Source data and results of statistical analyses are provided as a Source Data file.
To validate cell type assignment, we compared pseudo-bulk expression profiles for each of our germ cell subtypes from wild-type samples to pseudo-bulk expression profiles of germ cell clusters reported by a previous scRNA-seq study of adult mouse testes53 (Fig. S5d). We find broad agreement between these datasets, though the prior study identified more clusters, particularly among spermatocytes and spermatids. We propose that this is likely due to the prior study’s employment of spermatid depletion combined with spermatogonia/spermatocyte enrichment and construction of separate libraries for several stages of germ cell development.
Consistent with our histological analyses, Tex14m/m testes are depleted for late germ cell stages (Fig. 7a, b). We identified spermatogonia through round spermatids, while elongating spermatids were absent. Spermatogonia had a higher representation in Tex14m/m testes compared to wild type, likely due to a higher probability of sampling spermatogonia from mutant testes given the relative depletion of late stages. Spermatocytes undergoing meiosis and post-meiotic round spermatids were depleted in mutants compared to wild type, as expected.
We next asked whether intercellular bridges impact the overall distribution of RNAs within cells. Pseudo-bulk expression profiles derived from Tex14m/m germ-cell subtypes that are depleted for intercellular bridges are highly correlated with pseudo-bulk expression profiles from wild-type germ cells assigned to the same subclusters, suggesting that bridges do not affect the overall transcriptomic composition of germ cells (Fig. 7c). To identify specific transcriptomic differences, we performed differential expression analyses for each subcluster (see Methods). These analyses identified few differentially expressed genes in Tex14m/m early germ-cell stages (82 in spermatogonia, 5 in spermatogonia/preleptotene, and 25 in early spermatocytes; Fig. 7d, Supplementary Data 1). The number of differentially regulated transcripts drastically increased in Tex14m/m late spermatocyte and round spermatid stages (592 in late spermatocytes, and 1961, 1683 and 1398 in early, mid and late round spermatids respectively; Fig. 7d, Supplementary Data 1). We conclude that depletion of intercellular bridges modestly impacts the RNA landscape in early germ cell stages, while mutant cells that reach late meiotic prophase and later stages contain substantially altered RNA profiles.
To explore the biological pathways that are influenced by intercellular bridges during meiosis, we performed functional annotation using Gene Ontology (GO) of downregulated transcripts in Tex14m/m late spermatocytes (325 downregulated transcripts; Supplementary Data 2). Several downregulated transcripts are involved in protein translation, including transcripts encoding ribosomal proteins (e.g., Rpl18, Rpl27, Rpl34, Rps14, Rps26, Rps7), mitochondrial ribosome proteins (e.g., Mrpl16, Mrpl18, Mrpl32, Mrpl41), as well as translation initiation and elongation factors (Eif2s2, Eif2ak2, Tsfm), suggesting that loss of bridges may impact cytosolic and mitochondrial protein synthesis. Other downregulated transcripts are related to spermiogenesis, including transcripts involved in sperm motility (e.g., Akap4, Nme5, Rsph9, Cabs1, Cfap52) and histone to protamine transition (e.g., Tnp1, Tnp2, Prm1, Prm2). Thus, the depletion of bridges attenuates the levels of transcripts important for spermatid development as germ cells prepare to transition from meiosis to spermiogenesis. Our analyses identified the transcript encoding SC central element protein SYCE3 as downregulated (log2 fold change of -0.42), but no other direct regulators of meiotic recombination or synapsis were identified as downregulated, indicating that the meiotic repair and synapsis defects in Tex14m/m spermatocytes are likely not a direct result of depleted repair- and synapsis-associated transcripts.
We next evaluated transposon expression in Tex14m/m germ cells. Consistent with our immunofluorescence analyses, L1 transposons were upregulated in Tex14m/m late spermatocytes compared to wild type (Fig. 7d, Supplementary Data 1), and remained upregulated in Tex14m/m spermatids. Differentially expressed L1s in late spermatocytes spanned several L1 families, including L1MdA, L1MdF, L1MdTf, L1MdV, L1Lx, L1MdFanc, L1MdMus and L1MdN. We did not detect differential LINE expression in spermatogonia or early spermatocytes (Fig. 7d), in keeping with our immunofluorescence experiments. Long terminal repeat (LTR) retrotransposons were also upregulated in Tex14m/m germ cells compared to wild type (Fig. 7d, Supplementary Data 1). We detected a small number of differentially expressed LTRs in Tex14m/m spermatogonia, preleptotene and early spermatocytes, while LTRs were more broadly upregulated in late spermatocyte and spermatid stages.
To better understand the differential dependence of germ cell stages on intercellular bridges for repressing transposons, we examined the expression of all LINEs across spermatogenesis, irrespective of whether they were differentially expressed in mutants (Fig. 7e). In wild-type cells, mean LINE expression increased as spermatogonia transitioned to preleptotene and peaked in early spermatocytes, consistent with previously reports74. This was followed by a sharp drop in expression in late spermatocytes; this is coincident with and likely due to the robust transcriptional activation and engagement of the transposon-repressive piRNA pathway9,75. Mean LINE expression increased slightly during the late spermatocyte to spermatid transition, likely reflecting the dramatic genome remodeling and repackaging that occurs to form spermatids, and tapered to a minimum in elongating spermatids. Like wild type, mean LINE expression in Tex14m/m increased from spermatogonia, through preleptotene and early spermatocyte stages (Fig. 7e). Tex14m/m germ cells had marginally higher mean LINE expression compared to wild type during these early germ cell stages. Mean LINE expression dropped upon transition to the late spermatocyte stage, however, was substantially elevated compared to wild type, and remained elevated in subsequent spermatid stages. Nearly all detected LINE elements followed this pattern, regardless of whether they were significantly differentially expressed in any germ cell category (Fig. S5e). Therefore, perturbation of intercellular bridges renders transposon-repressive mechanisms less effective, especially during the late spermatocyte and spermatid stages.
Finally, we used single-cell expression data to validate the depletion of intercellular bridges in Tex14m/m. Intercellular bridges in the testis enable cytoplasmic sharing between connected cells, including efficient exchange of mRNAs, and this is evidenced by the presence of both X and Y chromosome transcripts in haploid spermatids19,20,21,22,53,76,77. We hypothesized that the partial depletion of intercellular bridges in Tex14m/m testes would result in a combination of individualized spermatids that exclusively contain either X or Y transcripts, and those that are interconnected by bridges and thus harbor both sex-linked transcripts. We calculated the number of X- and Y-linked genes detected in each cell in late spermatocytes through late-round spermatid categories, as well as the number of chromosome 9 genes as an autosomal control. As expected, X and Y gene expression was low in late spermatocytes due to meiotic sex-chromosome inactivation and subsequently showed a strong linear relationship in wild-type haploid spermatids (Fig. 7f), indicative of cytoplasmic sharing. Chromosome 9 and X gene expression showed a similar linear relationship in wild-type spermatids. In contrast, while some Tex14m/m spermatids harbored both X and Y transcripts, many cells showed mutually exclusive expression of X and Y (Fig. 7f). Similarly, some Tex14m/m spermatids contained both chromosome X and 9 transcripts, while others lacked X transcripts; we infer that this population is comprised of chromosome Y-containing individualized spermatids. Taken together, these data provide direct evidence of cytoplasmic sharing via intercellular bridges and its partial attenuation in Tex14m/m testes.
Discussion
Our search for new regulators of meiosis identified a Tex14 hypomorphic allele and uncovered the importance of Tex14-dependent intercellular bridges in regulating critical meiotic events. We show that intercellular bridges are important for replication, synapsis, break repair, and transposon repression during meiosis (Fig. 8). Our findings demonstrate that regulation of meiosis is non-cell-intrinsic and establish a new framework for how we think about its control in the male germline. Additionally, our identification of a Tex14 hypomorph as a meiotic recombination-defective mutant exemplifies the power of forward genetic screens in uncovering unexpected mechanisms that drive biological processes.
In wild type, meiosis occurs in a syncytium comprised of germ cells interconnected by TEX14-dependent intercellular bridges. Chromosomes are replicated during the preleptotene stage, meiotic breaks are formed and synapsis is initiated during early prophase, breaks are predominantly repaired and synapsis is complete during late meiotic prophase. In Tex14 mutants, intercellular bridges are depleted and germ cells are individualized. Many cells fail to undergo meiotic DNA replication. Cells that replicate form meiotic breaks and initiate synapsis. However, mutant cells fail to complete synapsis and fully repair breaks. Additionally, Tex14 mutants derepress transposons during late prophase, culminating in cell death and infertility.
The concomitant presence of X and Y chromosome-linked transcripts in haploid wild-type germ cells53,76,77, along with their predominantly mutually exclusive expression in Tex14 hypomorphs demonstrate that intercellular bridges enable sharing of cytoplasmic content, and lead to the obvious hypothesis that intercellular bridges influence germ cell development through sharing of regulatory factors. The molecular nature of the regulatory factors and precisely how this regulation is accomplished remains unknown, however. Non-exclusive possibilities include the sharing of regulatory small molecules, small RNAs such as piRNAs, mRNAs and/or proteins. Additionally, because intercellular bridges can be up to 3-μm wide and can channel organelles16,20, regulation may involve the sharing of organelles, such as mitochondria or specialized germ cell granules. Another non-exclusive possibility is that the intercellular bridges themselves comprise a specialized sub-cellular domain for localized cellular activities such as the translation of protein signals, akin to what has recently been described for midbodies78,79. We also cannot exclude the possibility that TEX14 may play an unknown function during meiosis that is independent of intercellular bridges, however, this is not likely given its exclusive localization at intercellular bridges in germ cells.
While direct evidence of cytoplasmic sharing via intercellular bridges in diploid mouse spermatocytes and spermatogonia is lacking, we think it unlikely that sharing, whether passive or active, is blocked in diploid cells or restricted only to the haploid stages. Instead, we favor the idea that germ cell connectivity and cytoplasmic sharing may serve functions during multiple stages of male mouse germ cell development, including non-essential functions. For instance, intercellular bridge-mediated cytoplasmic connectivity between spermatogonia may play a role that is redundant with an alternative mode of intercellular communication, which could explain why we do not detect substantial changes in the spermatogonial pool in Tex14 mutants80. Another non-exclusive possibility is that the exchange of regulatory factors, for e.g. piRNAs, may occur during pre-meiotic stages but that their perturbation manifests as a detectable phenotype during meiosis. Or connectivity may become important under specific conditions, for example, upon cellular stress14. Intercellular bridges have been postulated to synchronize development, to coordinate critical developmental transitions, to enable the sharing of essential gene products between haploids, and to create gamete equivalence thus promoting overall fitness16. Our findings along with prior studies of Tex14 knockouts25 support roles of intercellular bridges in synchronization, in mediating critical developmental transitions, and in sharing gene products between haploids, but do not exclude other models.
The meiotic replication defect in Tex14 mutants that we document here is, to our knowledge, unlike any phenotype described in known meiotic mutants. Tex14 mutant preleptotene cells express the critical meiotic entry-inducing factor STRA8, fail to replicate and presumably die, resulting in a depletion of early meiotic prophase cells. We infer from the relatively unperturbed spermatogonial numbers that mitotic replication is unaffected, and that germ cell connectivity specifically impacts meiotic replication. A comparable depletion of early meiotic prophase cells has also been reported in Stra8-knockout males, but unlike Tex14 mutants, Stra8-deficient preleptotene cells replicate their DNA81,82. Moreover, STRA8 drives the transcriptional upregulation of a large set of genes83, while we did not detect dramatic gene expression dysregulation in Tex14 mutant cells transitioning into meiotic prophase. Therefore, the meiotic depletion in Tex14 mutants cannot simply be explained by deregulation of the STRA8-dependent meiotic initiation pathway. Instead, we favor the hypothesis that intercellular bridges are required for (an unknown) aspect of DNA replication that is unique to meiosis in males. Additionally, because interconnected chains of preleptotene spermatocytes collectively transition across the blood-testis barrier through barrier remodeling84, we further hypothesize that lack of cell connectivity may impact the transit of preleptotene cells across and/or the integrity of the blood-testis barrier, which may in turn impair spermatogenesis.
The persistence of γH2AX in Tex14-deficient cells that enter meiotic prophase could reflect a mixture of problems. γH2AX most likely marks asynaptic regions in Tex14 mutants34. But γH2AX flares and recombination protein foci are retained on seemingly fully synapsed axes, indicating the presence of unrepaired DSBs at late stages when repair is predominantly complete in wild type. Florid γH2AX staining is diminished upon removal of SPO11, so the most signal is likely generated by programmed meiotic DSBs. Transposon-induced damage and replication-induced damage may also contribute. Regardless of the source of the damage, our results demonstrate that intercellular bridges serve a repair-promoting function. Moreover, the reduction of crossover sites as evidenced by the reduction of MLH1 foci in Tex14 mutants, lead us to surmise that intercellular bridges influence multiple repair-related mechanisms. While not statistically significant, numbers of DSB sites marked by recombination proteins are altered in Tex14-deficient early zygotene cells compared to wild type, so intercellular bridges may also affect programmed DSB formation, disruption of which could contribute to the stochastic defects in pairing and synapsis during meiosis57. This idea remains to be directly tested, however.
The inappropriate expression of transposons in Tex14-deficient meiosis is striking. The most straightforward explanation is that cell connectivity is essential for the proper functioning of the transposon-repressive DNA methylation and/or piRNA pathways. Perturbation of these mechanisms causes transposon derepression accompanied by synaptic defects, specifically during late meiotic prophase8,9,27,71,85,86,87,88, which is consistent with the Tex14-deficient phenotype. Inefficient methylation of newly replicated DNA during meiotic S phase has been proposed to lead to transient transposon derepression during early prophase in wild-type males74. We observe a similar trend in wild-type LINE expression during early prophase and a marginal increase in expression in Tex14-deficient cells, so it’s possible that depletion of intercellular bridges during replication exacerbates this phenomenon and further unleashes transposons during mutant meiosis. Another non-exclusive possibility that remains to be tested is that intercellular sharing of piRNAs is required to achieve the full extent of the piRNA pathway function. In support of this hypothesis, piRNA machinery-containing germ cell granules transit across intercellular bridges20,65, but whether this exchange is productive is unknown.
Disruption of small non-coding RNA pathways or the associated epigenetic landscape could also explain the broad gene expression dysregulation we observe in Tex14-deficient late-stage spermatocytes, as piRNAs and other small RNAs play diverse roles in shaping gene expression during spermatogenesis12,89. Finally, although we could not histologically evaluate spermatids due to their severe depletion in Tex14 mutants, the dramatic dysregulation of transposons and gene expression during the round spermatid stages coupled with the near-complete absence of elongating spermatids in our mutant single-cell RNA-sequencing data indicates potential roles of intercellular bridges in spermiogenesis.
The collection of meiotic defects in Tex14-deficient cells we document here leads to cell death and therefore infertility in males25. In contrast, intercellular bridge loss in Tex14-/- female mice does not cause infertility, although they have been reported to bear fewer litters41. This sexual dimorphism may reflect known sex differences in meiotic initiation, the epigenetic state during meiosis, and quality control mechanisms90. Importantly, mouse oocytes employ a nursing mechanism like flies, and oocytes undergoing meiosis develop gaps in their plasma membranes that are used as a route for cytoplasmic transfer91,92. This alternative mode of cytoplasmic sharing may alleviate the dependence on intercellular bridges for the regulation of meiotic events. Cytoplasmic transfer has been proposed to improve oocyte quality, for example, by increasing the concentration of protective factors such as piRNAs92. Another study demonstrated that intercellular bridges connecting oocytes coordinate the timing of meiotic entry through an RNA sharing mechanism66, but the meiotic functions we uncover here remain to be evaluated in females. Finally, germ cell intercellular bridges are widely conserved across metazoans, but their specific functional contributions may vary across systems14,15,16,17. Our findings establish that intercellular bridges serve a role in protecting genome integrity during mouse spermatogenesis, and we speculate that cytoplasmic sharing during germ cell development may be a widely employed mechanism to improve gamete quality, thereby promoting organismal fitness.
Methods
Mice
All experiments were performed following regulatory standards and were approved by the Rutgers University Institutional Animal Care and Use Committee. Animals were fed regular rodent chow with ad libitum access to food and water. Mice were housed using a 12-hour light/12-hour dark cycle in rooms with 70°–74 °F ambient temperature and 30–70% humidity. The Tex14- allele25 and Spo11- allele55 used here have been previously described. Tex14 mutant animals were maintained on a C57BL/6J (Jackson Laboratory) and FVB/NJ (Jackson Laboratory) mixed inbred background.
The Tex14tama allele (Tex14m) was isolated in an ENU-mutagenesis screen described previously27,28,29,30. Briefly, male mice of the C57BL/6J strain (B6) were mutagenized with ENU and crossed to wild-type females of the FVB/NJ strain (FVB) to produce founder (F1) males that were potential carriers; F1 males were crossed to wild-type FVB females to produce second-generation (G2) daughters, half of which were expected to be carriers; G2 daughters were crossed back to the F1 to generate third-generation (G3) males that were potentially homozygous. For a mutant phenotype caused by a single autosomal recessive mutation, one-eighth of the G3 males were expected to be homozygous. G3 males were screened for meiotic defects and mutant lines with interesting phenotypes were expanded for future studies.
To identify the tama mutation, we first performed rough mapping using a genetic polymorphism-based positional cloning strategy. Because mutagenesis was performed on B6 and outcrossing was done using FVB, we expected the phenotype-causing mutation to be linked to B6 SNPs. We therefore analyzed five G3 mice displaying the mutant phenotype using SNP genotyping arrays and searched for shared intervals of homozygosity for B6 SNPs. This mapped the causative mutation to a single 48-Mbp interval on chromosome 11. We next performed whole-exome sequencing and identified a single high-confidence un-annotated variant within the mapped region located at Chr11:87,533,604 (GRCm38/mm10) within the Tex14 gene.
Genotyping of Tex14m mice was done by PCR amplification of DNA extracted from toe clips using Tex14R (5′-GCTGTCCTGGAACTCACTC) and Tex14F (5′-GCCAAAGGAACTGAAAGAGAAAAATAC) primers, followed by digestion of the amplified product with AflIII restriction enzyme (NEB R0541S). The tama mutation creates an AflIII restriction site; AflIII cuts the mutant allele (184 bp and 26 bp bands) but does not cut the wild-type allele (single 210 bp band).
EdU treatment
EdU (Invitrogen, C10337) was dissolved in PBS to a final concentration of 4 μg/μl and a single dose of 20 μg/g of body weight was administered by intraperitoneal injection into 9- to 12-week-old animals. Testes were harvested two hours after the injection and processed for immunofluorescent staining according to the manufacturer’s instructions.
Histology
Testes isolated from adult mice were immersed in 4% paraformaldehyde or in Bouin’s fixative at 4 °C overnight. Bouin’s-fixed testes were washed at room temperature with gentle agitation for 1 hr in Milli-Q water, followed by two 1 hr washes in 70% ethanol. PFA-fixed testes were washed at 4 °C twice for 30 min in 1X PBS, followed by a 30 min wash in 50% EtOH. Fixed testes were stored in 70% EtOH at 4 °C for a maximum of one week. Testes were paraffin embedded, cut into 5-μm-thick sections, and mounted on slides by the Rutgers Research Pathology Services core facility. Slides were deparaffinized by three 5 min washes in xylene and rehydrated by a series of 3 min washes in 100% EtOH, 95% EtOH, 70% EtOH, and water. The immunohistochemical TUNEL assay was performed by MSKCC Molecular Cytology core facility as previously described27.
For immunofluorescence staining of testis sections, antigen retrieval was performed by treating slides for 20 min in Sodium Citrate buffer (10 mM Sodium Citrate, 0.05% Tween 20, pH 6.0) at 95°–100 °C. Slides were cooled for 20 min, blocked by one 20 min wash in blocking buffer (0.2% BSA, 0.2% gelatin from cold-water fish skin, 0.05% Tween 20 in 1X PBS), and incubated with primary antibodies. Stained slides were blocked by three 10 min washes in a blocking buffer, incubated with secondary antibodies for 1 hr at 37 °C, and blocked again by three 10 min washes in a blocking buffer. All washes were done at room temperature with gentle agitation. Slides were mounted using a mounting medium containing DAPI (Vector Laboratories). Antibodies and incubation conditions are listed within Supplementary Data 3.
Periodic acid Schiff staining was performed by the Rutgers Research Pathology Services core facility using Periodic Acid-Schiff (PAS) Kit (Richard-Allan Scientific (Thermo Fisher Scientific), #87007) according to manufacturer’s instructions. Briefly, slides were treated with 0.5% periodic acid, followed by Schiff reagent, followed by modified Mayer’s hematoxylin, and finally with bluing reagent. Slides were dehydrated using 100% EtOH, cleared in xylene, and mounted using Permount mounting medium.
EdU staining was performed using the Click-iT EdU Cell Proliferation Kit (Invitrogen, C10337) according to manufacturer’s instructions.
Cytology
Spermatocyte squashes were prepared as described27 with the following modifications. Minced testes were suspended in PFA solution (2% PFA, 1X PBS, 0.15% Triton X-100) at 75 μl of PFA solution per 25 mg testis. Fixation was allowed for 1 min and 15 μl of spermatocyte suspension was placed onto slides. To squash the spermatocytes, a coverslip was pressed down onto slides, left for 1 min, and the slides were snap-frozen in liquid nitrogen. Slides were washed in 1X PBS three times for 3 min with gentle agitation. Slides were either air dried and stored at −80 °C or stained as described93. Slides were mounted using a mounting medium containing DAPI (Vector Laboratories).
For preparation of spermatocyte spreads, isolated testes were minced on a petri dish and suspended in 200 μl of 1X DMEM/F-12 media (Gibco, #21-041-025). The spermatocyte suspension was pipetted gently, and transferred into a 15 ml tube; an additional 3.8 ml media was used to rinse the petri dish and transferred to the 15 ml tube to make the final volume 4 ml. The spermatocyte solution was allowed to settle at room temperature for 10 min, supernatant was transferred to a new 15 ml tube and centrifuged for 5 min at 250 g. The supernatant was discarded, and cells were resuspended in 1–2 ml of media. Using a Pasteur pipette, 5 drops of 4.5% sucrose were dropped onto the center of each slide, then from a height of 15–20 cm, a single drop of cell suspension was dropped onto slides, followed by one drop of 0.05% Triton X-100. Slides were incubated at room temperature for 20 min, then 8 drops of PFA solution (2% PFA, 0.02% SDS, pH 8.0) were placed onto each slide and slides were incubated at room temperature for 1 hr. Slides were dipped five times in water to wash off fixative, dipped once in 0.4% Photo-Flo 200 Solution (Kodak, #74257), then left to air dry. Immunostaining was performed as described previously93 and slides were mounted using mounting medium containing DAPI (Vector Laboratories). Unstained slides were stored at −80 °C and stained slides were stored at −20 °C.
For seminiferous tubule whole-mount preparations, isolated testes were placed in 1X PBS and the tunica albuginea was removed. Using forceps, seminiferous tubules were gently teased apart and fixed in 4% PFA at 4 °C overnight. Tubules were blocked by three 10 min washes in blocking buffer (0.2% BSA, 0.2% gelatin from cold-water fish skin, 0.05% Tween 20 in PBS) in 5 ml tubes, followed by incubation with primary antibodies in 2 ml tubes. Tubules were blocked as done previously, incubated with secondary antibodies, blocked again, and then incubated with Hoechst stain (Invitrogen, Hoechst 33342 20 mM solution) at 1:10,000 dilution at 4 °C for 30 min. All washes were done at room temperature with gentle agitation. Fluoromount-G (SouthernBiotech, #0100-01) mounting medium was placed on slides, individual tubules were placed onto slides and gently separated using forceps, and slides were coverslipped.
All slides (Superfrost (Fisherbrand, #12-544-7) or Superfrost Plus (Fisherbrand, #1255015)) for cytology were washed in 100% EtOH overnight and thoroughly rinsed with water before use. Antibodies and incubation conditions are provided as Supplementary Data 3.
Electron microscopy
Electron microscopy of testis tissue was performed by the Department of Pathology, Robert Wood Johnson Medical School Core Imaging Lab. Briefly, samples were fixed in 2.5% glutaraldehyde and 4% PFA in 0.1 M cacodylate buffer. Tissues were post-fixed in buffered 1% osmium tetroxide, dehydrated in a graded series of acetone, and embedded in Embed812 resin. Embedded testes were cut into 90-nm-thick sections on a Leica UC6 ultramicrotome and stained with a saturated solution of uranyl acetate and lead citrate. Images were digitized using a Philips CM12 transmission electron microscope with 3000× and 17000× objectives. Images were captured using an AMT (Advanced Microscopy Techniques) XR111 digital camera at 80Kv.
Immunoblot analysis
Analysis of TEX14 and ORF1p protein levels in Tex14m/m and Tex14-/- animals was adapted from a previous report94 with the following modifications. Samples were run on precast 4–15% TGX gels (Bio-Rad, #4561084) at 150 V for 40 min. Proteins were semi-dry transferred; 15 V and 30–40 min for TEX14, 15 V and 25 min for ORF1p. Membranes were rinsed with water and blocked with TBST (5% non-fat milk, 0.1% Tween in 1X PBS) at room temperature for 1 h on an orbital shaker. Blocked membranes were incubated with primary antibodies at 4 °C overnight, washed with TBST three times for 10 min at room temperature, incubated with secondary antibodies for 1 h at room temperature, washed again with TBST as done previously, and then developed with ECL solution (Kindle Biosciences, R1002) at 1:10 dilution for 1 min. The chemiluminescence signal was captured with a KwikQuant Digital Western Blot Detection System and visualized using KwikQuant Imager software. Images were produced using Photoshop. The antibodies used for western blotting analyses are listed within Supplementary Data 3.
Combined immunofluorescence and hybridization chain reaction (HCR) RNA-FISH
HCR IF and RNA-FISH were performed on paraffin-embedded testis sections using DNA probe sets and materials purchased from Molecular Instruments Inc. according to manufacturers’ instructions [v3.0]95. Testis sections were first stained for ORF1p using a polyclonal antibody at 1:1000 dilution (Abcam, ab216324), followed by HCR RNA-FISH. LINE1 RNA was detected using a custom probe against L1MdA_I_5end (Molecular Instruments, lot RTI848). We chose to probe for L1MdA_I_5end as it is derepressed in Tex14m/m. Additional probes used were L1spa RNA70 (Molecular Instruments, lot RTH655) and U6 snRNA70 (Molecular Instruments, lot RTB815). All probes were used at a final concentration of 16 nM.
Image analysis
TUNEL slides were digitized by the Rutgers Research Pathology Services core facility using Aperio CS2 slide scanner with a 40× objective. Images were produced using Aperio Image Scope software and cell counting was performed manually.
PAS-stained slides were digitized using EVOS M700 Imaging System (Invitrogen, AMF7000) with a 40× objective. For cell counting, images were visualized using QuPath software96 and manually scored.
TEX14 immunostained whole-mounted seminiferous tubules were imaged on a Leica TCS SP8 STED confocal microscope using a 63× oil-immersion or 10× objective with an equal number of optical sections along the z axis for wild type and mutants. Images were rendered in 3D and wavelet noise correction (Leica LAS X software) was applied. STRA8- and EdU-stained whole-mounted seminiferous tubules were imaged on a Leica TCS SP8 STED confocal microscope using a 63× oil-immersion objective or on a Zeiss Observer Z1 using a 20× objective.
Images of squashes and spreads were produced on a Leica TCS SP8 STED confocal microscope or on a Zeiss Observer Z1 using a 63× oil-immersion objective. Quantitation of synapsis or cell stages was done manually; foci counts were manually scored using Leica LAS X software.
Images of immunostained testis sections were captured using a Leica TCS SP8 STED confocal microscope. Alternatively, slides were digitized using either EVOS M700 Imaging System or Olympus VS-120 whole slide imaging system (40× objective), analyzed using QuPath software96 and cell counting was manually performed.
FACS analyses of spermatocytes
Nuclei were prepared for sorting as previously described51 with minor modifications. Briefly, testes from adult mice were decapsulated and fixed with 1% formaldehyde for 10 min, followed by quenching with 0.125 M Glycine for 5 min at room temperature. The fixed testes were homogenized with at least 10 strokes of a Dounce homogenizer (tight pestle) and filtered through a 70- or 100-μm cell strainer. After washing with chilled 1X PBS, nuclei were extracted using a hypotonic extraction buffer (15 mM Tris-HCl pH 7.4, 0.34 M sucrose, 15 mM NaCl, 60 mM KCl, 0.2 mM EDTA, 0.2 mM EGTA) on ice for 5 min and subsequently homogenized with 20 strokes using a loose pestle, followed by 10 strokes with a tight pestle. The nuclei were filtered through a 40-μm cell strainer and resuspended in chilled PBTB buffer (1X PBS with 0.1% Triton X-100, 5% bovine serum albumin, protease inhibitor). Nuclei were counted using a hemocytometer and the concentration adjusted to ~40 million/ml.
Nuclei were then labeled with different combinations of primary antibodies (1 μg of antibodies for 10 million nuclei) in 10% of normal serum at 4 °C overnight. Nuclei were washed, resuspended in PBTB, and then labeled with secondary antibodies (1:250 dilution) in 10% normal serum at 20 °C for 30 min. The antibodies used are listed within Supplementary Data 3.
Nuclei were filtered through a 40-μm cell strainer and stained with DAPI for 30 min or longer at room temperature before sorting. All sorting experiments were performed on a BD FACSAria Fusion flow cytometer. Debris and dead cells were excluded using both forward scatter and side scatter. We plotted the fluorescence signal of DAPI-Width and DAPI-Area to select for singlets and exclude 1 C nuclei from the analyses. To identify preleptotene spermatocytes, we gated nuclei based on the absence of DMRT1 (spermatogonial cell marker) and the presence of STRA8 (marker for meiotic cells). Then we quantified the number of nuclei based on the DNA content to derive the proportion of nuclei in each phase of the cell-cycle. Curve fitting of the different phases of the cell cycle was measured by FlowJo Software (BD Life Sciences).
Single-cell isolation and RNA-sequencing
Sequencing was performed with four 10-week-old males from a single litter; two Tex14m/m and two wild-type animals. Testes were dissected and two testes from a single animal were pooled. Testes were dissociated and cells were isolated as described previously53. Cell encapsulation, library preparation, and sequencing were performed by the Michigan Advanced Genomics core facility at the University of Michigan Medical School. Single-cell libraries were generated using the 10x Genomics Chromium Controller and following the manufacturer’s protocol for 3’ v3.1 chemistry with feature barcoding technology for cell multiplexing (10x Genomics). In brief, cells from each animal were counted and tagged with unique cell multiplexing oligo barcodes. Labeled cells from one wild type and one mutant were pooled equally and processed for cell encapsulation targeting 12,000 cells ( ~ 6000 cells per mouse), followed by library preparation. This was done in duplicate to capture ~12,000 cells per mouse. Labeled cells from the second wild type and second mutant were pooled and processed similarly. Final library quality was assessed using the LabChip GXII HT (PerkinElmer) and libraries were quantified by Qubit (ThermoFisher). Pooled libraries were subjected to 28x10x10×151 bp paired-end sequencing according to the manufacturer’s protocol (Illumina NovaSeq XPlus) to generate ~25,000 reads per cell.
Single-cell RNA-sequencing alignment and quantification
Curated Mus musculus SINE, LINE, LTR, DNA, and rolling circle transposable elements were extracted from the Dfam 3.7 database97 using famdb.py packaged with RepeatMasker v4.1.598 (command: famdb.py -i DFAM.h5 families -f fasta_name –class (TE class) --curated -ad ‘mus musculus’) installed via bioconda99. We next masked the 10X Genomics Mus musculus reference genome (refdata-gex-mm10-2020-A) with these repeats using RepeatMasker v4.1.5 (extra arguments: -e ncbi -no_is -s -lib (custom repeat fasta)). The repeat library was then appended to the masked genome, and we added additional records spanning each consensus repeat sequence to the 10X genomics Mus musculus transcriptome annotation in gtf format (refdata-gex-mm10-2020-A). We then finalized the custom 10X Genomics Cell Ranger reference using the cellranger mkref command from Cellranger v7.0.1100. Single cell expression was quantified using the cellranger multi workflow.
Single-cell RNA-sequencing analyses
Subsequent analysis was performed within R v4.2.2 using Bioconductor’s (v3.15) SingleCellExperiment v1.18.1 infrastructure101,102 in conjunction with scater v1.24.0, scuttle v1.6.3103 and scran v1.24.1104 and plotting performed via convenience functions from the same three packages or ggplot2 from the tidyverse v2.0.0105 and pheatmap v1.0.12106. Other preprocessing steps are detailed below. All steps were orchestrated via a snakemake v7.32.2 workflow107. R packages were maintained with renv v1.0.3108 on Ubuntu 24.04.4.
We imported each library’s cellranger output matrices unfiltered into R with the read10xCounts function from the DropletUtils R package v 1.16.0109,110. We removed cells that were not assigned a sample based on cellranger multi’s assignment confidence table output as well as likely empty droplets based on a kneeplot created with barcodeRanks function from DropletUtils. Doublets were removed from each library with the scDblFinder R package v1.10.0111. We identified cells with mitochondrial gene content more than three median absolute deviations (MADs) above the median or with total detected genes more than three MADs below the median using the isOutlier function from the scuttle package and removed them. All libraries were combined into a SingleCellExperiment object, and the expression matrix was normalized using the multiBatchNorm function from the batchelor 1.12.3 R package112.
We identified 10,000 highly variable features and excluded transposable elements from this list so that downstream analysis based on the variable feature subset would not be influenced by transposon expression. We computed batch-corrected principal components using the fastMNN function from the batchelor package with k = 10, d = 50 and computed Uniform Manifold Approximation and Projection coordinates using these components. We clustered cells using the clusterCells function from the bluster package v1.6.0113 using batch-corrected principal components and NNGraphParam with cluster.fun = “leiden”,k = 55. The top 100 cell identity markers for each cell identity reported in Supplementary Table 2 from Green et al.53 were used as input to garnett v0.2.20 classifier114 to identify broad somatic and germline cell identities for each cluster.
To identify fine-grained germ cell identities, wild-type cells were extracted for each broad germline cell type (spermatogonia, spermatocyte, round spermatid, elongating spermatid) to reduce the possibility that expression dysregulation in the mutant could affect clustering in unexpected ways. Each wild-type broad cell identity was reclustered using cluster cells with KmeansParam. Wild-type cells were reintegrated using fastMNN and mutant cells were assigned to each wild-type cluster using wild-type cells as a reference dataset with the SingleR R package v1.10.0115. We then reintegrated wild-type and mutant germline cells across batches with the fastMNN function.
To verify germ cell identity assignments, we compared our data to published data from Green et al.53. We created pseudo-bulk samples for each genotype within each germline subcluster using aggregateAcrossCells function from the scuttle package. Germline subclusters were then compared to pseudo-bulk expression estimates for subclusters identified in Green et al. downloaded from the Gene Expression Omnibus page for accession GSE112393 (GSE112393_MergedAdultMouseST25_12GermCellClusters_AllGeneExp.xlsx) with the correlate function from the corrr R package (method=pearson, v0.4.4)116.
Differential expression analysis was performed on pseudo-bulk samples using the pseudoBulkDGE wrapper for edgeR v3.38.4117,118,119 from the scran package. Genes and transposons identified as differentially expressed in mutants are provided in Supplementary Data 1. Gene Ontology annotations of downregulated genes (FDR ≤ 0.05) in the mutant late spermatocyte cluster were done using DAVID120,121 and results are provided in Supplementary Data 2.
Tests for differences in combined LINE expression between genotypes were performed with ggpubr v0.6.0122 (using stat_compare_means with default argument ‘wilcoxon’) and rstatix v0.7.2123 using wilcox_test() with argument alternative=’two.sided’. Inputs to this analysis and resulting statistics and p-values are available as a Source Data file.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The Tex14tama mouse strain and other reagents used are available upon request. The RNA-seq data generated in this study have been deposited in the Gene Expression Omnibus database under the accession code GSE274959. RNA-seq differential expression and Gene Ontology annotation analyses are provided as Supplementary Data files 1 and 2. Antibodies used are described within Supplementary Data 3. Source data are provided with this paper.
Code availability
All code for performing single-cell RNA-sequencing analyses are available at Github [https://github.com/orgs/JainLabTestisScRNA].
References
Zickler, D. & Kleckner, N. Meiosis: dances between homologs. Annual review genetics 57, 1–63 (2023).
Hunter, N. Meiotic Recombination: The Essence of Heredity. Cold Spring Harb Perspect Biol 7, https://doi.org/10.1101/cshperspect.a016618 (2015).
Sasaki, M., Lange, J. & Keeney, S. Genome destabilization by homologous recombination in the germ line. Nature reviews. Molecular cell biology 11, 182–195 (2010).
Hassold, T. J. & Hunt, P. A. Missed connections: recombination and human aneuploidy. Prenat Diagn 41, 584–590 (2021).
Garcia-Perez, J. L., Widmann, T. J. & Adams, I. R. The impact of transposable elements on mammalian development. Development 143, 4101–4114 (2016).
Wells, J. N. & Feschotte, C. A field guide to eukaryotic transposable elements. Annual review genetics 54, 539–561 (2020).
Kojima, K. K. Structural and sequence diversity of eukaryotic transposable elements. Genes Genet Syst 94, 233–252 (2020).
Zamudio, N. et al. DNA methylation restrains transposons from adopting a chromatin signature permissive for meiotic recombination. Genes development 29, 1256–1270 (2015).
Di Giacomo, M. et al. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Molecular cell 50, 601–608 (2013).
Yang, F. & Wang, P. J. Multiple LINEs of retrotransposon silencing mechanisms in the mammalian germline. Semin Cell Dev Biol 59, 118–125 (2016).
Deniz, O., Frost, J. M. & Branco, M. R. Regulation of transposable elements by DNA modifications. Nat Rev Genet 20, 417–431 (2019).
Ozata, D. M., Gainetdinov, I., Zoch, A., O’Carroll, D. & Zamore, P. D. PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet 20, 89–108 (2019).
Castaneda, J., Genzor, P. & Bortvin, A. piRNAs, transposon silencing, and germline genome integrity. Mutat Res 714, 95–104 (2011).
Lu, K., Jensen, L., Lei, L. & Yamashita, Y. M. Stay connected: a germ cell strategy. Trends Genet 33, 971–978 (2017).
Spradling, A. C., Niu, W., Yin, Q., Pathak, M. & Maurya, B. Conservation of oocyte development in germline cysts from Drosophila to mouse. eLife, 11, https://doi.org/10.7554/eLife.83230 (2022).
Greenbaum, M. P., Iwamori, T., Buchold, G. M. & Matzuk, M. M. Germ cell intercellular bridges. Cold Spring Harb Perspect Biol 3, a005850 (2011).
Yoshida, S. From cyst to tubule: innovations in vertebrate spermatogenesis. Wiley Interdiscip Rev Dev Biol 5, 119–131 (2016).
Dym, M. & Fawcett, D. W. Further observations on the numbers of spermatogonia, spermatocytes, and spermatids connected by intercellular bridges in the mammalian testis. Biol Reprod 4, 195–215 (1971).
Braun, R. E., Behringer, R. R., Peschon, J. J., Brinster, R. L. & Palmiter, R. D. Genetically haploid spermatids are phenotypically diploid. Nature 337, 373–376 (1989).
Ventela, S., Toppari, J. & Parvinen, M. Intercellular organelle traffic through cytoplasmic bridges in early spermatids of the rat: mechanisms of haploid gene product sharing. Mol Biol Cell 14, 2768–2780 (2003).
Morales, C. R., Wu, X. Q. & Hecht, N. B. The DNA/RNA-binding protein, TB-RBP, moves from the nucleus to the cytoplasm and through intercellular bridges in male germ cells. Dev Biol 201, 113–123 (1998).
Caldwell, K. A. & Handel, M. A. Protamine transcript sharing among postmeiotic spermatids. Proceedings National Academy Sciences USA 88, 2407–2411 (1991).
de Rooij, D. G. & Russell, L. D. All you wanted to know about spermatogonia but were afraid to ask. J Androl 21, 776–798 (2000).
Iwamori, T. et al. TEX14 interacts with CEP55 to block cell abscission. Mol Cell Biol 30, 2280–2292 (2010).
Greenbaum, M. P. et al. TEX14 is essential for intercellular bridges and fertility in male mice. Proceedings National Academy Sciences USA 103, 4982–4987 (2006).
Greenbaum, M. P., Ma, L. & Matzuk, M. M. Conversion of midbodies into germ cell intercellular bridges. Dev Biol 305, 389–396 (2007).
Jain, D. et al. rahu is a mutant allele of Dnmt3c, encoding a DNA methyltransferase homolog required for meiosis and transposon repression in the mouse male germline. PLoS genetics 13, e1006964 (2017).
Jain, D. et al. ketu mutant mice uncover an essential meiotic function for the ancient RNA helicase YTHDC2. eLife, 7, https://doi.org/10.7554/eLife.30919 (2018).
Guan, Y., Keeney, S., Jain, D. & Wang, P. J. yama, a mutant allele of Mov10l1, disrupts retrotransposon silencing and piRNA biogenesis. PLoS genetics 17, e1009265 (2021).
Petrillo, C. et al. shani mutation in mouse affects splicing of Spata22 and leads to impaired meiotic recombination. Chromosoma 129, 161–179 (2020).
Caspary, T. & Anderson, K. V. Uncovering the uncharacterized and unexpected: unbiased phenotype-driven screens in the mouse. Dev Dyn 235, 2412–2423 (2006).
Probst, F. J. & Justice, M. J. Mouse mutagenesis with the chemical supermutagen ENU. Methods Enzymol 477, 297–312 (2010).
Lammers, J. H. et al. The gene encoding a major component of the lateral elements of synaptonemal complexes of the rat is related to X-linked lymphocyte-regulated genes. Mol Cell Biol 14, 1137–1146 (1994).
Mahadevaiah, S. K. et al. Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet 27, 271–276 (2001).
Pittman, D. L. et al. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Molecular cell 1, 697–705 (1998).
Yoshida, K. et al. The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Molecular cell 1, 707–718 (1998).
de Vries, S. S. et al. Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes development 13, 523–531 (1999).
Edelmann, W. et al. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat Genet 21, 123–127 (1999).
de Vries, F. A. et al. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes development 19, 1376–1389 (2005).
Bellve, A. R. et al. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol 74, 68–85 (1977).
Greenbaum, M. P., Iwamori, N., Agno, J. E. & Matzuk, M. M. Mouse TEX14 is required for embryonic germ cell intercellular bridges but not female fertility. Biol Reprod 80, 449–457 (2009).
de Rooij, D. G. Proliferation and differentiation of spermatogonial stem cells. Reproduction 121, 347–354 (2001).
Buaas, F. W. et al. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet 36, 647–652 (2004).
Costoya, J. A. et al. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet 36, 653–659 (2004).
Schrans-Stassen, B. H., van de Kant, H. J., de Rooij, D. G. & van Pelt, A. M. Differential expression of c-kit in mouse undifferentiated and differentiating type A spermatogonia. Endocrinology 140, 5894–5900 (1999).
Koubova, J. et al. Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proceedings National Academy Sciences USA 103, 2474–2479 (2006).
Oulad-Abdelghani, M. et al. Characterization of a premeiotic germ cell-specific cytoplasmic protein encoded by Stra8, a novel retinoic acid-responsive gene. J Cell Biol 135, 469–477 (1996).
Zhou, Q. et al. Expression of stimulated by retinoic acid gene 8 (Stra8) in spermatogenic cells induced by retinoic acid. vivo study vitamin A-sufficient postnatal murine testes. Biol Reprod 79, 35–42 (2008).
Rezende-Melo, C. A. et al. Spermatogonial asynchrony in Tex14 mutant mice lacking intercellular bridges. Reproduction 160, 205–215 (2020).
Pratto, F. et al. Meiotic recombination mirrors patterns of germline replication in mice and humans. Cell 184, 4251–4267.e4220 (2021).
Lam, K. G., Brick, K., Cheng, G., Pratto, F. & Camerini-Otero, R. D. Cell-type-specific genomics reveals histone modification dynamics in mammalian meiosis. Nature communications 10, 3821 (2019).
Matson, C. K. et al. The mammalian doublesex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Dev Cell 19, 612–624 (2010).
Green, C. D. et al. A comprehensive roadmap of murine spermatogenesis defined by single-cell RNA-Seq. Dev Cell 46, 651–667.e610 (2018).
Huang, Y. & Roig, I. Genetic control of meiosis surveillance mechanisms in mammals. Front Cell Dev Biol 11, 1127440 (2023).
Baudat, F., Manova, K., Yuen, J. P., Jasin, M. & Keeney, S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Molecular cell 6, 989–998 (2000).
Romanienko, P. J. & Camerini-Otero, R. D. The mouse Spo11 gene is required for meiotic chromosome synapsis. Molecular cell 6, 975–987 (2000).
Kauppi, L. et al. Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes development 27, 873–886 (2013).
Bishop, D. K., Park, D., Xu, L. & Kleckner, N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69, 439–456 (1992).
Tarsounas, M., Morita, T., Pearlman, R. E. & Moens, P. B. RAD51 and DMC1 form mixed complexes associated with mouse meiotic chromosome cores and synaptonemal complexes. J Cell Biol 147, 207–220 (1999).
Barchi, M. et al. Surveillance of different recombination defects in mouse spermatocytes yields distinct responses despite elimination at an identical developmental stage. Mol Cell Biol 25, 7203–7215 (2005).
Pacheco, S. et al. The ATM signaling cascade promotes recombination-dependent pachytene arrest in mouse spermatocytes. PLoS genetics 11, e1005017 (2015).
Anderson, L. K., Reeves, A., Webb, L. M. & Ashley, T. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151, 1569–1579 (1999).
Gray, S. & Cohen, P. E. Control of meiotic crossovers: from double-strand break formation to designation. Annual review genetics 50, 175–210 (2016).
Lehtiniemi, T. & Kotaja, N. Germ granule-mediated RNA regulation in male germ cells. Reproduction 155, R77–R91 (2018).
Meikar, O. et al. An atlas of chromatoid body components. RNA 20, 483–495 (2014).
Soygur, B. et al. Intercellular bridges coordinate the transition from pluripotency to meiosis in mouse fetal oocytes. Sci. Adv. 7, https://doi.org/10.1126/sciadv.abc6747 (2021).
Fu, Q. & Wang, P. J. Mammalian piRNAs: Biogenesis, function, and mysteries. Spermatogenesis 4, e27889 (2014).
Ollinger, R., Reichmann, J. & Adams, I. R. Meiosis and retrotransposon silencing during germ cell development in mice. Differentiation 79, 147–158 (2010).
Zamudio, N. & Bourc’his, D. Transposable elements in the mammalian germline: a comfortable niche or a deadly trap? Heredity (Edinb) 105, 92–104 (2010).
De Luca, C., Gupta, A. & Bortvin, A. Retrotransposon LINE-1 bodies in the cytoplasm of piRNA-deficient mouse spermatocytes: Ribonucleoproteins overcoming the integrated stress response. PLoS genetics 19, e1010797 (2023).
Soper, S. F. et al. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell 15, 285–297 (2008).
Bellani, M. A., Romanienko, P. J., Cairatti, D. A. & Camerini-Otero, R. D. SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm-/- spermatocytes. J Cell Sci 118, 3233–3245 (2005).
Mahadevaiah, S. K. et al. Extensive meiotic asynapsis in mice antagonises meiotic silencing of unsynapsed chromatin and consequently disrupts meiotic sex chromosome inactivation. J Cell Biol 182, 263–276 (2008).
Gaysinskaya, V. et al. Transient reduction of DNA methylation at the onset of meiosis in male mice. Epigenetics Chromatin 11, 15 (2018).
Li, X. Z. et al. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Molecular cell 50, 67–81 (2013).
Jung, M. et al. Unified single-cell analysis of testis gene regulation and pathology in five mouse strains. eLife 8, https://doi.org/10.7554/eLife.43966 (2019).
Chen, Y. et al. Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell research 28, 879–896 (2018).
Jung, G. I. et al. An oocyte meiotic midbody cap is required for developmental competence in mice. Nature communications 14, 7419 (2023).
Park, S. et al. The mammalian midbody and midbody remnant are assembly sites for RNA and localized translation. Dev Cell 58, 1917–1932.e1916 (2023).
Niedenberger, B. A. et al. Dynamic cytoplasmic projections connect mammalian spermatogonia in vivo. Development 145, https://doi.org/10.1242/dev.161323 (2018).
Anderson, E. L. et al. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proceedings National Academy Sciences USA 105, 14976–14980 (2008).
Baltus, A. E. et al. In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat Genet 38, 1430–1434 (2006).
Kojima, M. L., de Rooij, D. G. & Page, D. C. Amplification of a broad transcriptional program by a common factor triggers the meiotic cell cycle in mice. eLife 8, https://doi.org/10.7554/eLife.43738 (2019).
Smith, B. E. & Braun, R. E. Germ cell migration across Sertoli cell tight junctions. Science 338, 798–802 (2012).
Bourc’his, D. & Bestor, T. H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96–99 (2004).
Barau, J. et al. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science 354, 909–912 (2016).
Aravin, A. A., Sachidanandam, R., Girard, A., Fejes-Toth, K. & Hannon, G. J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316, 744–747 (2007).
Carmell, M. A. et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12, 503–514 (2007).
Olotu, O., Ahmedani, A. & Kotaja, N. Small Non-Coding RNAs in Male Reproduction. Semin Reprod Med 41, 213–225 (2023).
Hunter, N. Oocyte Quality Control: Causes, Mechanisms, and Consequences. Cold Spring Harb Symp Quant Biol 82, 235–247 (2017).
Ikami, K. et al. Altered germline cyst formation and oogenesis in Tex14 mutant mice. Biol. Open 10, https://doi.org/10.1242/bio.058807 (2021).
Lei, L. & Spradling, A. C. Mouse oocytes differentiate through organelle enrichment from sister cyst germ cells. Science 352, 95–99 (2016).
Dowdle, J. A. et al. Mouse BAZ1A (ACF1) is dispensable for double-strand break repair but is essential for averting improper gene expression during spermatogenesis. PLoS genetics 9, e1003945 (2013).
Saito, Y. et al. YTHDC2 control of gametogenesis requires helicase activity but not m(6)A binding. Genes development 36, 180–194 (2022).
Schwarzkopf, M. et al. Hybridization chain reaction enables a unified approach to multiplexed, quantitative, high-resolution immunohistochemistry and in situ hybridization. Development 148, https://doi.org/10.1242/dev.199847 (2021).
Bankhead, P. et al. QuPath: Open source software for digital pathology image analysis. Sci Rep 7, 16878 (2017).
Storer, J., Hubley, R., Rosen, J., Wheeler, T. J. & Smit, A. F. The Dfam community resource of transposable element families, sequence models, and genome annotations. Mob DNA 12, 2 (2021).
Smit, A. F. A., Hubley, R. & Green, P. RepeatMasker Open-4 (2013-2015).
Gruning, B. et al. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat Methods 15, 475–476 (2018).
Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nature communications 8, 14049 (2017).
Amezquita, R. A. et al. Orchestrating single-cell analysis with Bioconductor. Nat Methods 17, 137–145 (2020).
Huber, W. et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12, 115–121 (2015).
McCarthy, D. J., Campbell, K. R., Lun, A. T. & Wills, Q. F. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186 (2017).
Lun, A. T., McCarthy, D. J. & Marioni, J. C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res 5, 2122 (2016).
Wickham, H. et al. Welcome to the Tidyverse. Journal Open Source Software 4, 1686 (2019).
Kolde, R. Pheatmap: pretty heatmaps. R package (2019).
Molder, F. et al. Sustainable data analysis with Snakemake. F1000Res 10, 33 (2021).
Ushey, K. & Wickham, H. renv: Project Environments (2024).
Griffiths, J. A., Richard, A. C., Bach, K., Lun, A. T. L. & Marioni, J. C. Detection and removal of barcode swapping in single-cell RNA-seq data. Nature communications 9, 2667 (2018).
Lun, A. T. L. et al. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol 20, 63 (2019).
Germain, P. L., Lun, A., Garcia Meixide, C., Macnair, W. & Robinson, M. D. Doublet identification in single-cell sequencing data using scDblFinder. F1000Res 10, 979 (2021).
Haghverdi, L., Lun, A. T. L., Morgan, M. D. & Marioni, J. C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol 36, 421–427 (2018).
Lun, A. bluster: Clustering Algorithms for Bioconductor. R package, (2022).
Pliner, H. A., Shendure, J. & Trapnell, C. Supervised classification enables rapid annotation of cell atlases. Nat Methods 16, 983–986 (2019).
Aran, D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 20, 163–172 (2019).
Kuhn, M., Jackson, S. & Cimentada, J. corrr: Correlations in R. R package (2022).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
McCarthy, D. J., Chen, Y. & Smyth, G. K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic acids research 40, 4288–4297 (2012).
Chen, Y., Lun, A. T. & Smyth, G. K. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res 5, 1438 (2016).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57 (2009).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research 37, 1–13 (2009).
Kassambara, A. ggpubr: ‘ggplot2’ Based Publication Ready Plots. R package (2023).
Kassambara, A. rstatix: Pipe-Friendly Framework for Basic Statistical Tests. R package (2023).
Acknowledgements
We thank Martin M. Matzuk for sharing Tex14-/- mice and the TEX14 antibody, and Scott Keeney for sharing Spo11-/- mice. We thank Diana Laird (University of California, San Francisco), Ian Adams (University of Edinburgh) and Luke Berchowitz (Columbia University) for helpful discussions. We thank Rutgers Research Pathology Services for help with tissue processing, PAS staining, and digitizing histology slides; Rajesh Patel at the Robert Wood Johnson Medical School Core Imaging Lab for assistance with electron microscopy sample preparation and imaging; Wenjin Chen at Rutgers Cancer Institute of New Jersey Biomedical Informatics Shared Resource for digitizing immunostained slides. We thank the Memorial Sloan Kettering Molecular Cytology core facility for TUNEL staining. Rutgers Cancer Institute of New Jersey Biomedical Informatics Shared Resource is supported in part by National Cancer Institute Cancer Center Support Grant P30 CA072720-5917. The forward genetic screen that yielded the Tex14m allele was conducted while D.J. was a postdoctoral fellow with Scott Keeney (Memorial Sloan Kettering) and was supported by the Howard Hughes Medical Institute (to S.K.) and the Human Frontier Science Program (to D.J.). This work was supported by National Institutes of Health grants R35 GM147130 (to D.J.), R01 GM140163 and R35 GM152168 (to C.E.E), R01 HD104680 and R01 HD113274 (to S.S.H.) and by the National Institute of Diabetes and Digestive and Kidney Diseases intramural research program (to F.P.).
Author information
Authors and Affiliations
Contributions
J.S., K.T., S.N.S., S.L., and A.A. performed mouse phenotyping. M.A.L. and M.R. performed single-cell RNA-sequencing analyses in Tex14m/m. F.P. conducted FACs analyses of spermatocytes. S.S.H. and C.E.E. supervised M.R. and M.A.L., respectively. D.J. designed the study with contributions from all authors. D.J., J.S., M.A.L., S.N.S., and F.P. wrote the paper with contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Allan Spradling and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sorkin, J., Tilton, K., Lawlor, M.A. et al. Intercellular bridges are essential for transposon repression and meiosis in the male germline. Nat Commun 16, 1488 (2025). https://doi.org/10.1038/s41467-025-56742-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-56742-9