Abstract
Obesity is regarded as a chronic inflammatory disease involving adipose tissue macrophages (ATM), but whether immunometabolic reprogramming of ATM affects obesity remains unclarified. Here we show that in ATM glutaminolysis is the fundamental metabolic flux providing energy and substrate, bridging with AMP-activated protein kinase (AMPK) activity, succinate-induced interleukin-1β (IL-1β) production, and obesity. Abrogation of AMPKα in myeloid cells promotes proinflammatory ATM, impairs thermogenesis and energy expenditure, and aggravates obesity in mice fed with high-fat diet (HFD). Conversely, IL-1β neutralization or myeloid IL-1β abrogation prevents obesity caused by AMPKα deficiency. Mechanistically, ATP generated from glutaminolysis suppresses AMPK to decrease phosphorylation of the β subunit of succinyl-CoA synthetase (SUCLA2), thereby resulting in the activation of succinyl-CoA synthetase and the overproduction of succinate and IL-1β; by contrast, siRNA-mediated SUCLA2 knockdown reduces obesity induced by HFD in mice. Lastly, phosphorylated SUCLA2 in ATM correlates negatively with obesity in humans. Our results thus implicate a glutaminolysis/AMPK/SUCLA2/IL-1β axis of inflammation and obesity regulation in ATM.
Similar content being viewed by others
Introduction
Obesity is a major risk factor for metabolic disease and is becoming a global health threat. However, effective clinical therapy is limited1,2. Obesity is regarded as a chronic inflammatory disease characterized by inflammatory infiltration in adipose tissue3. In the process of obesity, adipose tissue creates a proinflammatory microenvironment to recruit and activate macrophages for proinflammatory factor production, resulting in the exacerbation of obesity and the occurrence of related metabolic diseases4,5,6,7. Emerging evidence reveals that inflammatory factors, such as chemokines and cytokines, play indispensable effects on adipose tissue inflammation associated obesity3. However, the effectiveness of improving obesity by regulating the production of inflammatory factors in macrophages remains unclear.
The emerging field of immunometabolism highlights the importance of cellular metabolism for the maintenance and activity of macrophages, as it contributes to pathologies such as insulin resistance, diabetes, and obesity8. Glutaminolysis provides a vital energy substrate for maintaining optimal macrophage functionality in adipose tissue and is closely linked with the tricarboxylic acid cycle (TCA cycle), which includes the conversion of glutamine to α-ketoglutarate by glutaminase and glutamate dehydrogenase9,10. Succinate is converted from α-ketoglutarate by α-ketoglutarate dehydrogenase and succinyl-CoA synthetase (SCS) in the TCA cycle and is crucial for macrophage activation11,12,13. Succinate promotes the expression of interleukin-1β (IL-1β) through stabilizing hypoxia-inducible factor-1α (HIF-1α) in proinflammatory macrophages14. It has been reported that the inhibition of glutaminolysis reduces proinflammatory cytokine secretion in macrophages15. However, many questions remain unanswered regarding the associations among glutaminolysis, IL-1β derived from succinate accumulation in adipose tissue, and obesity. The regulation of glutaminolysis, the origin of succinate, and whether the inhibition of macrophage IL-1β production prevents obesity remains elusive.
AMP-activated protein kinase (AMPK) is a widely expressed classic serine/threonine protein kinase that maintains cellular energy homeostasis by regulating catabolism and anabolism16,17. It has been reported that the loss of AMPKβ1 in hematopoietic cells aggravates adipose tissue inflammation and insulin resistance following a high-fat diet (HFD)18. AMPK activity is decreased in proinflammatory macrophages17. In contrast, AMPK activated by berberine, AICAR, or metformin effectively mitigates inflammation in macrophages, providing further evidence that AMPK is essential for the proinflammatory macrophages17,19,20. However, the underlying mechanism by which AMPK activity is decreased under a proinflammatory state and how AMPK modulates inflammation in macrophages remains elusive.
In our study, we clarify the immunometabolic reprogramming effects of adipose tissue macrophages (ATM) on obesity. Glutaminolysis in ATM downregulates AMPK activity through adenosine triphosphate (ATP) generation. This reduction leads to activating SCS via the decrease in the phosphorylation of β subunit of SCS (SUCLA2) by AMPK, thereby resulting in promoting the inflammatory ATM. Furthermore, SUCLA2 knockdown via siRNA delivery into macrophages effectively mitigates ATM-related obesity. Thus, our study determines that SUCLA2, as an AMPK substrate, is essential for proinflammatory ATM via immunometabolic reprogramming and may be a promising therapeutic target for obesity treatment.
Results
Glutaminolysis couples IL-1β in macrophages and obesity development
To explore the effects of glutaminolysis on the regulation of IL-1β in macrophages, the [U-13C]-glutamine tracer was utilized. Under proinflammatory macrophage activation, we found that 95% of intracellular glutamine originated from the extracellular environment, and direct glutamine-derived contributions account for 75% of glutamate, 61% of α-ketoglutarate, 70% of succinate, 62% of fumarate, 69% of malate, 42% of citrate and 43% of cis-Aconitic acid, suggesting that glutaminolysis was an essential metabolic pathway for macrophages activation (Supplementary Fig. 1a, b). After glutaminolysis was inhibited through glutamine deprivation, the expression of HIF-1α and IL-1β was reduced in proinflammatory macrophages (Supplementary Fig. 1c). Meanwhile, we found that no secreted IL-1β was detected upon the stimulation of lipopolysaccharide (LPS), indicating that glutaminolysis affected the production rather than the secretion of IL-1β (Supplementary Fig. 1d). Consistent with these findings, glutaminolysis was inhibited by potent inhibitors, such as GPNA (glutamine transporter inhibitor), CB-839 and BPTES (glutaminase inhibitors), and R-162 (glutamate dehydrogenase inhibitor) (Supplementary Fig. 1e), and a decrease in IL-1β expression was also observed in proinflammatory macrophages, validating the crucial role of glutaminolysis in regulating IL-1β (Supplementary Fig. 1f-1i).
Glutaminolysis-derived succinate promoted IL-1β through HIF-1α in proinflammatory macrophages, however, LPS-induced inflammatory macrophage activation increased the levels of other mitochondrial metabolites such as fumarate and malate14, thus we wondered about the effects of glutaminolysis derived TCA cycle metabolites on the IL-1β expression. The [U-13C]-glutamine tracer found that glutaminolysis contributed to a significant accumulation of glutamine-linked TCA cycle metabolites, such as M5-α-ketoglutarate, M4-succinate, and M4-fumarate, in proinflammatory macrophages (Supplementary Fig. 1j–m). There was a slight increase in the glutamine-derived M4-malate, without an increase in glutamine-derived metabolites, such as M4-citrate and M4-cis-aconitic acid (Supplementary Fig. 1n–p). The inhibitory effect of α-ketoglutarate and fumarate on IL-1β, along with the promoting effect of succinate on IL-1β, indicated that different metabolites exerted diverse regulation on macrophage activation14,21. We observed that succinate and its upstream metabolite α-ketoglutarate, but not its downstream metabolite fumarate, could abolish the decreased expression of HIF-1α and IL-1β in proinflammatory macrophages when glutaminolysis was inhibited via glutamine deprivation, suggesting that the metabolic flux from glutamine to succinate was responsible for HIF-1α and IL-1β expression (Supplementary Fig. 1q–t).
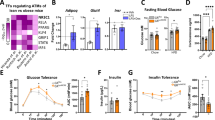
Then, the associations between glutaminolysis and obesity were examined using the collected subcutaneous white adipose tissue (ScWAT) from human subjects categorized by body mass index (BMI). We investigated the levels of α-ketoglutarate, succinate, fumarate, and glutamine in human adipose tissue (Supplementary Table 1), and observed a significant increase in the ratio of α-ketoglutarate to glutamine, succinate to glutamine, and fumarate to glutamine in adipose tissue among overweight individuals (Fig. 1a, c, e). Additionally, the succinate/glutamine ratio and fumarate/glutamine ratio in adipose tissue were positively correlated with BMI (Fig. 1b, d, f). We also performed a reanalysis of the glutamine-related proteins in the obese cohort of the Jackson Heart Study (JHS, n = 1985) based on a previously published study investigating protein-metabolite associations in human plasma22, and Gene Ontology analysis revealed significant enrichment in the inflammatory response and interleukin-1 response among glutamine-related proteins (Fig. 1g, Supplementary Table 2). Moreover, among these glutamine-related proteins, IL-1β was identified as a central protein in the protein-protein interaction analysis, suggesting that glutamine may be involved in the pathogenesis of obesity-related inflammation through IL-1β (Fig. 1h). Therefore, glutaminolysis coordinated with the expression of IL-1β in macrophages and might be involved in the development of obesity.
a–f Metabolite detection and correlation analysis of white adipose tissue of human subjects. The ratio of α-ketoglutarate to glutamine (a), succinate to glutamine (c), fumarate to glutamine (e) in subcutaneous white adipose tissue, and correlation analysis between the ratio of α-ketoglutarate to glutamine (b), succinate to glutamine (d), fumarate to glutamine (f) in subcutaneous white adipose tissue and BMI value (n (BMI < 25) = 12 research objects, n (BMI > 25) = 10 research objects). g Reanalysis of the proteins correlated with glutamine in the obese cohort of the Jackson Heart Study from a published study22, a total of 58 proteins (screen criteria: p value < 0.05 and p value adjusted for BMI > 0.05 in Table S4 of this study) were identified as potential contributors to obesity development. Gene Ontology analysis revealed significant enrichment in the inflammatory response and Interleukin-1 response among glutamine-correlated proteins. h Protein-protein Interaction analysis of the network of glutamine-related proteins in Fig. 1g through STRING, the red globules represent proteins associated with the inflammatory response. Data are presented as the mean ± SEM, and groups were compared by the unpaired two-tailed Student’s t test (a, c, e). linear regression analysis to assess the relationship between the variables, the significance of the regression models was evaluated using F-tests, these tests were conducted as two-sided tests to determine whether there were any significant differences or associations between the variables, a limited number of regression analyses was performed, and thus, no adjustments for multiple comparisons were deemed necessary (b, d, f). P < 0.05 was considered to be statistically significant.
AMPK is an adaptor that governs glutaminolysis and IL-1β in macrophages
Although the importance of glutaminolysis for the expression of IL-1β in macrophages was well established, the specific regulatory mechanisms remain elusive. Glutaminolysis in macrophages leads to the generation of two distinct byproducts involved in the regulation of IL-1β: succinate accumulation and ATP production14,23 (Supplementary Fig. 2a). To elucidate the regulatory mechanism of glutaminolysis on IL-1β, we investigated the regulatory effects of energy on IL-1β expression. We found that glutamine increased the energy demand, as evidenced by the reduction in ATP levels induced by glutamine deprivation in proinflammatory macrophages (Supplementary Fig. 2b). AMPK functions as a sensor of cellular energy status and can be activated by energy deficiency16. In response to the decrease of ATP, an increase in AMPK activity, as indicated by increased phosphorylation at Thr172 of AMPKα, was observed in proinflammatory macrophages under glutamine deprivation accompanied by the reduction in IL-1β (Supplementary Fig. 2c). These findings suggested that AMPK activity was regulated by ATP derived from glutaminolysis and might be associated with IL-1β expression in proinflammatory macrophages. We subsequently explored the regulatory effect of AMPK on IL-1β, and decreased protein expression of HIF-1α and IL-1β was observed following treatment with the AMPK activator A-769662 (Supplementary Fig. 2d). We also found that AMPK activation by A-769662 did not affect HIF-1α gene expression, which suggested that AMPK regulated HIF-1α through post-translational modification (Supplementary Fig. 2e).
Based on these findings, two processes were observed during glutaminolysis: the inhibition of AMPK by ATP and the accumulation of succinate from glutamine. Both of these processes could enhance the expression of IL-1β. The previous research indicated that succinate is a classical metabolite in stabilizing HIF-1α through post-translational modification in proinflammatory macrophages14, and the observation that AMPK regulated HIF-1α through post-translational modification (Supplementary Fig. 2d, 2e), led us to speculate that alterations in AMPK activity might affect succinate production. As anticipated, treatment of proinflammatory macrophages with A-769662 led to a reduction in cellular succinate content and the HIF-1α protein expression, with no effect on the HIF-1α gene expression (Supplementary Fig. 2f–h). Inversely, the knockdown of AMPK via siRNA resulted in elevated cellular succinate content and HIF-1α protein expression without change in gene expression of HIF-1α in proinflammatory macrophages (Supplementary Fig. 2i–k). These results suggested that AMPK linked the succinate metabolism in proinflammatory macrophages.
To investigate whether AMPK regulates succinate derived from glutamine, a [U-13C]-glutamine tracer was utilized through the manipulation of AMPK by siRNA (Supplementary Fig. 2l). An increase in [U-13C]-glutamine-derived M4-succinate, along with its downstream metabolites M4-fumarate and M4-malate was observed in proinflammatory macrophages with AMPKα knocked down (Supplementary Fig. 2m–p), suggesting that the altered AMPK activity impacted glutaminolysis-derived succinate metabolism and consequently affected HIF-1α and IL-1β expression.
To further investigate the modulatory effect of AMPK on IL-1β in macrophages, a mouse model with AMPK deficiency in myeloid cells was established (Supplementary Fig. 3a, 3b), and bone marrow-derived macrophages (BMDMs) with AMPK deficiency were obtained (Supplementary Fig. 3c). RNA-sequencing was used to analyze the regulation of gene expression by AMPK in AMPKαfl/fl and LysM-Cre, AMPKαfl/fl BMDMs in the presence or absence of LPS stimulation (Supplementary Fig. 4a). Gene expression was analyzed in AMPKαfl/fl BMDMs following stimulation with LPS to investigate the response to inflammatory stimulation. Gene Ontology analysis revealed that these upregulated genes were significantly enriched in biological processes related to the inflammatory response (Supplementary Fig. 4b, 4c), and GSEA also revealed significant upregulation of pathways associated with the inflammatory response, hypoxia and IL-1 after LPS stimulation (Supplementary Fig. 4d–f). These findings were consistent with the finding that obesity promoted proinflammatory activation of macrophage in adipose tissue24. Next, the changes in the gene expression by AMPK were analyzed in AMPKαfl/fl BMDMs and LysM-Cre, AMPKαfl/fl BMDMs following LPS treatment. A volcano plot was generated, which showed that AMPK deficiency significantly downregulated the gene expression of AMPKα1 and upregulated the gene expression of IL-1β (Supplementary Fig. 5a). Moreover, the differentially expressed genes were subjected to Gene Ontology analysis, which revealed that AMPK deficiency significantly altered the expression of genes associated with the inflammatory response in the presence of LPS (Supplementary Fig. 5b). Analysis of the interaction networks between inflammatory response-related genes via STRING revealed that AMPK deficiency significantly impacted the expression of genes associated with interleukin-1 (IL-1) and chemokine activity, and IL-1β was identified as a central player among these altered genes, highlighting the crucial role of IL-1β in the AMPK-regulated inflammatory response in macrophages (Supplementary Fig. 5c). Furthermore, GSEA also revealed significant upregulation of inflammation-related, hypoxia-related, and interleukin-1-related enrichment in LysM-Cre, AMPKαfl/fl BMDMs (Supplementary Fig. 5d–g). Notably, significantly elevated protein expression of HIF-1α was observed in LysM-Cre, AMPKαfl/fl BMDMs, however, there was no discernible difference in the gene expression of HIF-1α (Supplementary Fig. 5h, 5i), which is consistent with the upregulation of HIF-1α by succinate-derived posttranslational modification14,25. We also detected that AMPK deficiency upregulated the expression of IL-1β through glutaminolysis and HIF-1α, as evidenced by the glutaminase inhibitors (BPTES and CB-839) and the HIF-1α inhibitor (BAY 87-2243) (Supplementary Fig. 5j–l). Taken together, these results indicated that AMPK served as a pivotal adaptor in regulating glutaminolysis and IL-1β of proinflammatory macrophages.
Myeloid AMPK deficiency exacerbates HFD-induced obesity
Next, we investigated whether macrophage AMPK affects obesity development. We found that the ScWAT of mice fed an HFD exhibited a characteristic crown-like structure in which macrophages accumulate and form a distinct architecture for efficient clearance of dead adipocytes and cellular debris26 (Supplementary Fig. 6a). These changes were accompanied by decreased AMPKα phosphorylation at Thr172 and increased IL-1β expression in macrophages (Supplementary Fig. 6a), indicating that the association between macrophage AMPK and IL-1β was involved in obesity in vivo.
We utilized AMPKαfl/fl mice and LysM-Cre, AMPKαfl/fl mice, in which AMPK knockdown occurred solely in F4/80+ Stromal Vascular Fraction (SVF) cells rather than F4/80- SVF cells and mature adipocytes in adipose tissue (Supplementary Fig. 7a–j), to elucidate our hypothesis.
First, phenotypic differences in these mice fed with chow diet were investigated (Supplementary Fig. 8a). We found that AMPK deficiency in myeloid cells did not affect obesity-related metabolic characteristics, as there were no significant differences in body weight gain (Supplementary Fig. 8b, c), body fat and lean mass (Supplementary Fig. 8d), weight of metabolic organs (Supplementary Fig. 8e–h) or insulin sensitivity (Supplementary Fig. 8i) between AMPKαfl/fl and LysM-Cre, AMPKαfl/fl mice. Moreover, we also conducted a comparative analysis of gene expression between AMPKαfl/fl and LysM-Cre, AMPKαfl/fl BMDMs without LPS stimulation, indicating that AMPK deficiency did not influence the expression of genes associated with inflammation (Supplementary Fig. 8j, k), which might partially explain the mechanism that myeloid AMPK deficiency exhibited no effect on whole-body metabolism in response to a chow diet.
Next, we investigated in mice fed with HFD (Fig. 2a). We observed that the monocyte-macrophage population primarily responds to HFD rather than a standard chow diet in LysM-Cre, AMPKαfl/fl mice through flow cytometry (Supplementary Fig. 9a–h), indicating that macrophage AMPK has an essential effect on HFD-induced obesity. AMPK deficiency in myeloid cells exacerbated the development of obesity, evidenced by the increase in body weight (Fig. 2b, c), body fat mass (Fig. 2d), the weight of metabolic organs, such as the liver, brown adipose tissue (BAT) and ScWAT but not epididymal white adipose tissue (EpWAT) (Fig. 2e–h, Supplementary Fig. 10a, 10b), and increased infiltration of lipids, as indicated by increased vacuolation area in the liver and enlarged adipocyte size in BAT and ScWAT stained with Hematoxylin and Eosin (H&E) staining (Fig. 2i) in LysM-Cre, AMPKαfl/fl mice compared with AMPKαfl/fl mice. Additionally, the expression of genes related to adipocyte differentiation was reduced in the EpWAT of LysM-Cre, AMPKαfl/fl mice compared with AMPKαfl/fl mice, indicating that AMPK deficiency in myeloid cells leads to inhibited EpWAT hyperplasia (Supplementary Fig. 10c). Moreover, when subjected to the insulin tolerance test, LysM-Cre, AMPKαfl/fl mice exhibited a diminished insulin response, resulting in increased blood glucose levels compared with AMPKαfl/fl mice (Fig. 2j), suggesting that AMPK deficiency in myeloid cells exacerbates insulin resistance. We also investigated the energy metabolism of the mice at room temperature and revealed that LysM-Cre, AMPKαfl/fl mice exhibited decreased energy expenditure compared with AMPKαfl/fl mice through regression analysis of CalR27 (Fig. 2k–m). Additionally, impaired thermogenesis was observed in LysM-Cre, AMPKαfl/fl mice compared with AMPKαfl/fl mice, as evidenced by a decreased rectal temperature during cold exposure (Fig. 2n). Moreover, a reduction in UCP-1 expression was observed in BAT both at room temperature and during cold exposure (Fig. 2o, Supplementary Fig. 10d), and this change was accompanied by decreased lipid mobilization, as indicated by an increase in the adipocyte size of ScWAT following cold exposure in LysM-Cre, AMPKαfl/fl mice compared with AMPKαfl/fl mice (Fig. 2p, Supplementary Fig. 10e).
a Schematic diagram of mice feeding and this image was created in BioRender. Zang, Y. (2025) https://BioRender.com/e16k510b–r Male AMPKαfl/fl mice and LysM-Cre, AMPKαfl/fl mice with C57BL/6 background at the age of 6 weeks were fed HFD to explore the obesity and related phenotype. Body weight gain (b, n = 12 mice), representative mice image (c), relative fat and lean mass (d, n = 12 mice), the representative images of liver, BAT and ScWAT (e), the organ weight of liver (f, n = 12 mice), BAT (g, n = 12 mice) and ScWAT (h, n = 12 mice), representative H&E staining of the liver, BAT and ScWAT (i), insulin tolerance test (j, n = 11 mice in AMPKαfl/fl group and n = 12 mice in LysM-Cre, AMPKαfl/fl group), regression-based analysis of absolute metabolic rate against body mass of metabolic cage for light time (k, n = 6 mice), dark time (l, n = 6 mice) and a total day (m, n = 6 mice), the rectal temperature in cold exposure at 4 °C for different times (n, n = 9 mice), immunohistochemical staining of UCP-1 of BAT (o) and ScWAT (p) at room temperature (22 °C) and cold exposure (10 °C for 48 h), the proinflammatory genes of ScWAT (q, Ilb, Il6, Nos2 and Ccl2: n = 12 mice, Tnfa: n = 12 mice in AMPKαfl/fl group and n = 11 mice in LysM-Cre, AMPKαfl/fl group, F4/80: n = 10 mice) and immunohistochemical staining of F4/80 in BAT and ScWAT (r). Data are presented as the mean ± SEM, groups were compared by the unpaired two-tailed Student’s t test (d, f, g, h, j, n, q) or two-way ANOVA followed by Fisher’s LSD test (b). P < 0.05 was considered to be statistically significant.
ATM-derived inflammation causes the development of insulin resistance and diabetes28; therefore, we also assessed the inflammatory status in adipose tissue. Consistent with the in vitro findings that macrophages with AMPK deficiency exhibited increased expression of genes associated with inflammation and chemotaxis (Supplementary Fig. 5b), the white adipose tissue (WAT) of LysM-Cre, AMPKαfl/fl mice exhibited elevated expression of proinflammatory and chemotaxis-related genes compared with AMPKαfl/fl mice (Fig. 2q, Supplementary Fig. 10f). This was accompanied by increased infiltration of macrophages into WAT, as indicated by the increased gene expression of macrophage marker genes, such as F4/80 (Fig. 2q), and the increased protein expression of F4/80 in adipose tissue of LysM-Cre, AMPKαfl/fl mice compared with AMPKαfl/fl mice (Fig. 2r). Thus, AMPK deficiency in myeloid cells exacerbated the development of obesity and metabolic disorders.
IL-1β neutralizing prevents HFD-induced obesity caused by myeloid AMPK deficiency
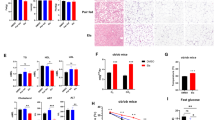
To investigate whether macrophage AMPK regulates obesity through IL-1β, we employed IL-1β neutralizing antibody in AMPKαfl/fl and LysM-Cre, AMPKαfl/fl mice to assess the progression of obesity (Fig. 3a). First, the blocking efficiency of the neutralizing antibody was assessed by immunofluorescence staining, which revealed a reduction in the IL-1β expression in adipose tissue of both AMPKαfl/fl and LysM-Cre, AMPKαfl/fl mice (Fig. 3b). Subsequently, an analysis of obesity-related phenotypes was conducted. An increase in body weight, which was promoted by myeloid AMPK deficiency from LysM-Cre, AMPKαfl/fl mice after HFD feeding, was significantly inhibited by the IL-1β neutralizing antibody (Fig. 3c). Fat mass was reduced, lean mass was increased (Fig. 3d), the weight of metabolic organs was reduced (Fig. 3e–g), the vacuolation area in the liver was reduced, the adipocyte size in BAT and ScAWAT was decreased (Fig. 3h) and the insulin sensitivity was increased (Fig. 3i) in LysM-Cre, AMPKαfl/fl mice following IL-1β neutralizing antibody treatment compared with LysM-Cre, AMPKαfl/fl mice not receiving the IL-1β neutralizing antibody. Moreover, enhanced thermogenesis was also observed, as evidenced by an increase in rectal temperature following cold exposure (Fig. 3j, k) and elevated protein expression of UCP-1 in BAT (Fig. 3l), after treatment with an IL-1β neutralizing antibody in the LysM-Cre, AMPKαfl/fl mice compared with LysM-Cre, AMPKαfl/fl mice without an IL-1β neutralizing antibody. These findings indicated that the functionality of adipose tissue improved after IL-1β blockade. Also, a reduction in inflammation was demonstrated by the downregulation of proinflammatory genes, such as Il1b, Tnfa, and Ccl2 (Fig. 3m), as well as decreased macrophage infiltration in adipose tissue due to the decreased expression of the macrophage marker F4/80 at both the gene (Fig. 3m) and protein levels in the adipose tissue (Fig. 3n) of LysM-Cre, AMPKαfl/fl mice following treatment with the IL-1β neutralizing antibody. Taken together, these findings suggested that the blockade of IL-1β through neutralizing antibodies was sufficient to mitigate the exacerbation of obesity caused by myeloid AMPK deficiency.
a Schematic diagram of mice feeding and this image was created in BioRender. Zang, Y. (2025) https://BioRender.com/e16k510. b–n Male AMPKαfl/fl mice and LysM-Cre, AMPKαfl/fl mice with C57BL/6 background at the age of 6 weeks were fed HFD with or without IL-1β neutralizing antibody (1 mg/kg, twice one week) to explore the obesity development. Immunofluorescent staining of IL-1β in BAT and ScWAT (b), body weight gain (c, n = 5 mice), relative fat and lean mass (d, n = 5 mice), the weight of Liver (e, n = 5 mice), BAT (f, n = 5 mice), and ScWAT (g, n = 5 mice), representative H&E staining of the liver, BAT and ScWAT (h), insulin tolerance test (i, n = 5 mice), the rectal temperature in cold exposure at 4 °C for different times (j, k, n = 5 mice), immunohistochemical staining of UCP-1 in BAT (l), the proinflammatory genes of ScWAT (m, Il1b, Tnfa, Nos2, Ccl2 and F4/80: n = 5 mice in each group, Il6: n = 4 mice in LysM-Cre, AMPKαfl/fl + IL-1β mAb group and n = 5 mice in other group), and the immunohistochemical staining of F4/80 in BAT and ScWAT (n). Data are presented as the mean ± SEM, groups were compared by two-way ANOVA followed by Fisher’s LSD test (c–g, i–k, m). P < 0.05 was considered to be statistically significant.
Macrophage AMPK regulates obesity in IL-1β dependent manner
IL-1β is secreted from several secretory cells, such as macrophages, dendritic cells, and adipocytes29,30, and we wondered whether the anti-obesity effects of macrophage AMPK resulted from macrophage IL-1β. We generated the mice deficient in IL-1β (LysM-Cre, IL-1βfl/fl) and the mice deficient in both IL-1β and AMPK (LysM-Cre, IL-1βfl/fl, AMPKαfl/fl) specifically in myeloid cells (Supplementary Fig. 11a). After verifying the knockout efficiency in BMDMs (Supplementary Fig. 11b), we examined the progression of obesity in response to HFD (Supplementary Fig. 11c). Our results indicated that there was no significant difference in obesity-related phenotypes between LysM-Cre, IL-1βfl/fl mice and LysM-Cre, IL-1βfl/fl, AMPKαfl/fl mice, including body weight (Fig. 4a–c), fat and lean mass (Fig. 4d), metabolic organ weight (Fig. 4e–h), hepatic vacuolation area, adipocyte size in brown BAT and ScAWAT (Fig. 4i), insulin sensitivity (Fig. 4j), rectal temperature following cold exposure as an indicator of thermogenesis (Fig. 4k, l), UCP-1 protein expression in BAT (Fig. 4m), proinflammatory gene expression of Il1b, Il6, Tnfa, Nos2, and Ccl2 (Fig. 4n–r), and macrophage content in adipose tissue (Fig. 4s, t), and these obesity-related phenotypes can be exacerbated by myeloid AMPK knockout in IL-1β normal mice (LysM-Cre, AMPKαfl/fl mice compared with AMPKαfl/fl mice and IL-1βfl/fl mice) (Fig. 4a–t). Furthermore, compared with AMPKαfl/fl and IL-1βfl/fl mice, myeloid IL-1β knockout (LysM-Cre, IL-1βfl/fl mice and LysM-Cre, IL-1βfl/fl, AMPKαfl/fl mice) exhibited an anti-obesity effect, suggesting that IL-1β plays a crucial role in obesity development and that its regulation is a key mechanism for controlling obesity. Collectively, these findings indicated that macrophage AMPK regulated obesity development in a macrophage IL-1β-dependent manner.
a–t Male LysM-Cre, IL-1βfl/fl mice, LysM-Cre, IL-1βfl/fl, AMPKαfl/fl mice, AMPKαfl/fl mice, IL-1βfl/fl mice, and LysM-Cre, AMPKαfl/fl mice with C57BL/6 background at the age of 8 weeks were fed HFD to explore the obesity and related phenotype. Body weight change (a, n = 8 mice), body weight gain (b, n = 8 mice), representative mice image (c), relative fat and lean mass (d, n = 8 mice), the representative image of liver, BAT and ScWAT (e), the metabolic organ weight of liver (f, n = 8 mice), BAT (g, n = 8 mice) and ScWAT (h, n = 8 mice), representative H&E staining of liver, BAT and ScWAT (i), insulin tolerance test (j, n = 8 mice), the rectal temperature in cold exposure at 4 °C for different times (k, l, n = 8 mice), immunohistochemical staining of UCP-1 in BAT (m), the proinflammatory genes of ScWAT (n–s, n = 8 mice) and immunohistochemical staining of F4/80 in BAT and ScWAT (t). Data are presented as the mean ± SEM, groups were compared by one-way ANOVA followed by Fisher’s LSD test (b, d, f–h, right of j, l, n–s) or two-way ANOVA followed by Fisher’s LSD test (a). P < 0.05 was considered to be statistically significant.
Macrophage IL-1β is affected by AMPK phosphorylated SUCLA2
The knockdown of AMPK in macrophages enhanced glutaminolysis in mitochondria, as evidenced by increased levels of succinate, fumarate, and malate but not α-ketoglutarate derived from glutamine through the use of [U-13C]-glutamine tracer (Supplementary Fig. 2m–p). These findings suggested that AMPK may influence the conversion of α-ketoglutarate to succinate. The conversion of α-ketoglutarate to succinate involves two steps. The initial step entails the catalysis of α-ketoglutarate dehydrogenase, which converts α-ketoglutarate to succinyl-CoA; subsequently, SCS catalyzes the transformation of succinyl-CoA to succinate23. Through a yeast two-hybrid experiment utilizing AMPKβ1 as the bait protein, potential substrates of AMPK were identified by our previous research31,32. SUCLA2, which is the β subunit of the SCS that directly mediates the formation of succinate, was found to interact with AMPK, suggesting that AMPK might regulate glutaminolysis-dependent IL-1β expression through SUCLA2 (Fig. 5a). Through mitochondrial separation, we found that AMPKα levels were comparable in the cytoplasm and mitochondria, while SUCLA2 was more highly expressed in the mitochondria of macrophages (Supplementary Fig. 12a, b). Although SUCLA2 was identified as a potential substrate of AMPK through a yeast two-hybrid experiment in which AMPKβ1 was used as the bait protein, we found that SUCLA2 also interacted with AMPKα1 and AMPKα2 in addition to AMPKβ1 and AMPKβ2 in 293 T cells by exogenously overexpressing both the AMPK subunits and SUCLA2 (Supplementary Fig. 12c–f). Moreover, the endogenous interaction between AMPKα and SUCLA2 was also investigated in macrophages, and AMPKα was shown to interact with SUCLA2 (Fig. 5b) but not with another β subunit of SCS, SUCLG2 (Fig. 5c). SUCLA2 knockdown through siRNA significantly attenuated the expression of HIF-1α and IL-1β in macrophages in the presence of LPS (Fig. 5d), and SUCLA2 knockdown inhibited the upregulation of HIF-1α and IL-1β in macrophages with AMPK deficiency, indicating that AMPK regulated IL-1β through a SUCLA2-dependent mechanism (Fig. 5e).
a Schematic representation of the potential substrate of AMPK in succinate formation, and this image was created in BioRender. Zang, Y. (2025) https://BioRender.com/e16k510. Co-immunoprecipitation analysis of the interaction of AMPKα with SUCLA2 (b) or SUCLG2 (c) in RAW 264.7 cells. d Immunoblot analysis of indicated proteins in BMDMs that are transfected with siRNA for 30 h followed by stimulation with 100 ng/mL LPS for an additional 12 h. e Immunoblot analysis of indicated proteins in AMPKαfl/fl BMDMs (Flox) and LysM-Cre, AMPKαfl/fl BMDMs (MKO) that are transfected with siRNA for 18 h followed by stimulation with 100 ng/mL LPS for an additional 12 h. f Immunoblot analysis of the expression of SUCLG1 and SUCLA2 after AMPKα was knocked down by siRNA for 48 h in RAW 264.7 cells. g The relative enzymatic activity of SUCLA2 (the direction from succinyl-CoA to succinate) was detected after AMPKα was knocked down by siRNA for 36 h followed by stimulation with 100 ng/mL LPS for an additional 12 h in RAW264.7 cells (n = 3 biological replicates). h In vitro phosphorylation analysis was performed by mixing purified His-CAMKKβ, His-AMPKα1β1γ1 and His-SUCLA2 in the presence of ATP-g-S, and immunoblot analysis of indicated proteins with indicated antibodies. i Venn diagram was used to integrate the phosphorylation site that was detected by mass spectrometry and predicted by GPS 5.0 (http://gps.biocuckoo.cn/) or HPRD. j Protein sequence alignment indicated the conservation of SUCLA2 at Ser60 across multiple species. k Immunoblot analysis of indicated proteins in the in vitro kinase assay that mixed the purified His-CAMKKβ, His-AMPKα1β1γ1 with His-SUCLA2 (WT) or His-SUCLA2 (S60A). l Immunoblot analysis of indicated proteins in BMDMs after incubation with 200 µM A-769662 for 3 h. m In vitro phosphorylation analysis was performed by mixing purified His-CAMKKβ, His-AMPKα1β1γ1 and His-SUCLA2 (WT) or His-SUCLA2 (S60A) in the presence of ATP-g-S, and immunoblot analysis of indicated proteins with indicated antibodies. The enzymatic activity (the direction from succinyl-CoA to succinate) of purified WT-SUCLA2 (n) or S60A-SUCLA2 (o) after incubation with AMPK in vitro (n = 3 biological replicates). p The enzymatic activity (the direction from succinyl-CoA to succinate) of purified WT-SUCLA2, S60A-SUCLA2, and S60D-SUCLA2 in the same protein content (n = 4 biological replicates). q–r The relative gene expression of IL-1β in RAW264.7 cells that overexpress human WT-SUCLA2, S60A-SUCLA2 and S60D-SUCLA2 through lentivirus infection for 48 h followed the treatment by 100 ng/mL LPS for 6 h in the condition of glutamine replete (q) or deprived (r) condition (n = 4 biological replicates). The relative gene expression of IL-1β in RAW264.7 cells that overexpress human WT-SUCLA2 with S60A-SUCLA2 (s) or S60D-SUCLA2 (t) through lentivirus infection for 48 h, the cells were treated with 200 µM A-769662 for 3 h in advance followed by 100 ng/mL LPS for 6 h (n = 4 biological replicates). Data are presented as the mean ± SEM, groups were compared by the unpaired two-tailed Student’s t test (g) or one-way ANOVA followed by Bonferroni’s multiple-comparisons test (q, r) or two-way ANOVA followed by Bonferroni’s multiple-comparisons test (n, o, p, s, t), representative data are shown from one of the three independent experiments (b–f, h, k–m). P < 0.05 was considered to be statistically significant.
We subsequently examined how AMPK regulates SUCLA2. There were no significant changes in the protein expression of α subunit of SCS (SUCLG1) or SUCLA2 after AMPKα knockdown (Fig. 5f). However, the increased enzymatic activity of SUCLA2 (from succinyl-CoA to succinate) was observed in proinflammatory macrophages (Fig. 5g), suggesting that AMPK might regulate SUCLA2 activity through post-translational modification directly. As expected, phosphorylated SUCLA2 was detected when the AMPKα1/β1/γ1 complex activated by CAMKKβ and the SUCLG1/SUCLA2 complex were mixed in vitro (Fig. 5h), suggesting that AMPK can phosphorylate SUCLA2 directly. Subsequent mass spectrometry analysis via in vitro kinase assay combined with bioinformatic phosphorylation site prediction via the HPRD database and GPS5.0 software revealed that AMPK is capable of phosphorylating SUCLA2 at the highly conserved residue Ser60 that exists across various species (Fig. 5i, j, Supplementary Fig. 12g–i). Moreover, this phosphorylation of SUCLA2 at Ser60 by AMPK was completely abolished when the S60A mutation was introduced into SUCLA2, providing further evidence that AMPK phosphorylates SUCLA2 at Ser60 (Fig. 5k). Except for the in vitro observation, we also observed a consistent increase in the phosphorylation levels of AMPKα at Thr172 and SUCLA2 at Ser60 following treatment with A-769662 in macrophages, suggesting AMPK mediated phosphorylation of SUCLA2 at Ser60 existed in macrophages (Fig. 5l). To further validate the specific phosphorylation of SUCLA2 by AMPK only at Ser60, an in vitro kinase assay was conducted to investigate the overall phosphorylation of SUCLA2 after incubation with AMPK. The results demonstrated that the S60A mutation abolished the overall phosphorylation of SUCLA2 by AMPK, thereby ruling out the possibility of multiple sites phosphorylated by AMPK (Fig. 5m).
To investigate the potential role of AMPK in regulating IL-1β through the phosphorylation of SUCLA2 at Ser60, the S60A and S60D mutations were introduced into SUCLA2 as phosphorylation blockade mutation and phosphomimetic mutation, respectively. We observed that active AMPK suppressed the activity of WT-SUCLA2 (Fig. 5n), but not the S60A-SUCLA2 (Fig. 5o) in the purified protein, indicating that the regulation of SUCLA2 activity by AMPK is dependent on Ser60 of SUCLA2. We also found that the S60D mutation in the purified protein, as anticipated, significantly attenuated the enzymatic activity of SUCLA2, thereby implying that phosphorylation at position Ser60 of SUCLA2 could directly regulate the levels of succinate (Fig. 5p). There was no difference in enzymatic activity between purified WT-SUCLA2 and S60A-SUCLA2 (Fig. 5p), which might result from the low phosphorylation of purified WT-SUCLA2 at Ser60 without AMPK incubation (Fig. 5k).
We further investigated the role of Ser60 of SUCLA2 in regulating IL-1β in proinflammatory macrophages. Compared to WT-SUCLA2 overexpressed macrophages, S60A-SUCLA2 overexpression exhibited increased expression of IL-1β, while S60D-SUCLA2 overexpression had the opposite effect under replete glutamine conditions (Fig. 5q). However, the regulation of Ser60 of SUCLA2 on IL-1β was abolished under conditions of glutamine deprivation, suggesting that the regulation of IL-1β by Ser60 of SUCLA2 was dependent on glutamine (Fig. 5r). Finally, we also observed that compared with WT-SUCLA2 overexpressed macrophages, the overexpression of S60A-SUCLA2 impaired the inhibitory effect of AMPK activation on IL-1β (Fig. 5s), while the overexpression of S60D-SUCLA2 impaired the stimulatory effect of LPS on IL-1β (Fig. 5t). Altogether, the phosphorylation at Ser60 of SUCLA2 exhibited a crucial role in regulating IL-1β in proinflammatory macrophages, and the regulation of IL-1β by AMPK was dependent on the phosphorylation of SUCLA2 at Ser60.
Knockdown macrophage SUCLA2 combats HFD-induced obesity
We subsequently investigated the translational significance of our findings that the AMPK-SUCLA2 axis orchestrates inflammation to drive the pathogenesis of obesity. We found that proinflammatory macrophages exhibited a decreased phosphorylation at Ser60 in SUCLA2 (Fig. 6a) and increased enzymatic activity of SUCLA2 (Fig. 6b), similar to the findings that obese WAT has reduced phosphorylation of SUCLA2 at Ser60 in macrophages (Fig. 6c), indicating a close association between macrophage SUCLA2 activity and obesity.
a Immunoblot analysis of indicated proteins in BMDMs after incubation with 100 ng/mL LPS for 3 h. b The enzymatic activity of SUCLA2 in BMDMs after incubation with 100 ng/mL LPS for 3 h (n = 3 biological replicates). c Immunofluorescent staining of F4/80 and pS60-SUCLA2 in the subcutaneous adipose tissue from lean or HFD-induced obese mice. d Schematic representation of the nanoparticle formation with p5RHH and siRNA, and this image was created in BioRender. Zang, Y. (2025) https://BioRender.com/e16k510. e The relative gene expression of SUCLA2 in BMDMs that were transfected with p5RHH-siRNA complex for 48 h (n = 4 biological replicates). f Schematic diagram of mice feeding and injection of p5RHH-siRNA complex, and this image was created in BioRender. Zang, Y. (2025) https://BioRender.com/e16k510. g–o Male C57BL/6 mice at the age of 8 weeks were fed HFD and received p5RHH-siCtrl or p5RHH-siSUCLA2 complex injection to explore the metabolic-related phenotype. Body weight gain (g, n = 10 mice), mice image (h), relative fat and lean mass (i, n = 10 mice), the organ weight of liver, EpWAT, ScWAT and BAT (j, n = 10 mice), the image of liver, EpWAT, ScWAT and BAT (k), insulin tolerance test (l, n = 10 mice in p5RHH+siCtrl group, and n = 9 mice in p5RHH+siSUCLA2 group), the rectal temperature in cold exposure at 4 °C for different times (m, n = 9 mice), and the proinflammatory genes of ScWAT (n, Il1b, Il6, Ccl2 and F4/80: n = 10 mice, Tnfa: n = 8 mice, Nos2: n = 10 mice in p5RHH+siCtrl group, and n = 9 mice in p5RHH+siSUCLA2 group) and EpWAT (o, Il1b, Il6, Nos2, Ccl2 and F4/80: n = 9 mice in p5RHH+siCtrl group, and n = 10 mice in p5RHH+siSUCLA2 group, Tnfa:n = 9 mice in each group). Data are presented as the mean ± SEM, groups were compared by the unpaired two-tailed Student’s t test (e, i, j, l, m, n, o), two-way ANOVA followed by Bonferroni’s multiple-comparisons test (b) and two-way ANOVA followed by Fisher’s LSD test (g), representative data are shown from one of the three independent experiments (a, c). P < 0.05 was considered to be statistically significant.
Then, we constructed a mouse model of macrophage SUCLA2 deficiency. A biocompatible cationic amphipathic peptide (p5RHH) that could form a 55-nm nanoparticle with siRNA and be phagocytosed by macrophages within inflammatory organs such as inflamed joints and WAT33,34,35,36 was used to deliver siRNA (Fig. 6d). p5RHH-coated SUCLA2 siRNA was utilized to achieve a significant reduction in the gene expression of SUCLA2 in macrophages, thereby confirming the efficient transfection and successful gene knockdown in macrophages using p5RHH-siRNA (Fig. 6e). Meanwhile, we detected that the knockdown of SUCLA2 mediated by p5RHH was limited in F4/80+ SVF cells compared to mature adipocytes and F4/80- SVF cells in HFD-induced mice (Supplementary Fig. 13a–d). Additionally, the p5RHH-mediated knockdown of SUCLA2 also resulted in a reduction in IL-1β expression in F4/80+ SVF cells (Supplementary Fig. 13e), indicating that targeting macrophage SUCLA2 could be a potential therapeutic approach for inflammation-related diseases.
To investigate the impact of p5RHH-coated SUCLA2 siRNA on obesity, mice fed an HFD for two weeks were administered p5RHH which coats control or SUCLA2 siRNA (Fig. 6f), following a previously published study35. Compared with those in the control group, the mice injected with the p5RHH-SUCLA2 siRNA complex exhibited a reduction in body weight gain induced by HFD (Fig. 6g, h), accompanied by a decrease in fat mass (Fig. 6i) and metabolic organ weight (Fig. 6j, k). Moreover, we also observed improvements in insulin sensitivity using the insulin resistance test (Fig. 6l), an enhanced capacity for thermogenesis opon exposed to cold (Fig. 6m), and decreased inflammation in WAT (Fig. 6n, o) in the p5RHH-SUCLA2 siRNA-treated mice compared with the control mice. Thus, our data suggested that intervention with SUCLA2 in macrophages could ameliorate obesity.
The AMPK-SUCLA2 pathway is involved in obese human ATM
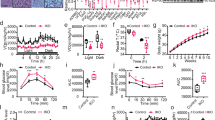
Also, we explored the AMPK-SUCLA2-IL-1β pathway in macrophages from human adipose tissue. In line with the inhibitory effect of obesity on AMPK activity in mouse macrophages (Supplementary Fig. 6a), we observed downregulated phosphorylation of AMPKα at Thr172 in macrophages from adipose tissue samples of obese individuals compared with those of lean individuals, indicating that macrophage AMPK was influenced by obesity in humans (Fig. 7a). Moreover, we observed decreased phosphorylation of SUCLA2 at Ser60 and increased expression of IL-1β in macrophages from adipose tissue samples of obese individuals compared with those from lean individuals, suggesting that SUCLA2-mediated IL-1β signaling may be involved in the pathogenesis of obesity in humans (Fig. 7a). Based on these findings, AMPK-SUCLA2-IL-1β-mediated inflammation in human adipose tissue might contribute to the pathogenesis of obesity, and targeting this axis may be a promising treatment for obesity-related disorders.
a Immunofluorescent staining of CD68, pT172-AMPK, pS60-SUCLA2 and IL-1β in the subcutaneous adipose tissue from lean (BMI = 21.5), overweight (BMI = 25.7) or obese (BMI = 32.0) subjects, representative data are shown from one of the three independent experiments. b Model of how macrophage AMPK responds to glutaminolysis and regulates glutaminolysis-coupled IL-1β expression, and this image was created in BioRender. Zang, Y. (2025) https://BioRender.com/e16k510.
Discussion
Emerging evidence shows that immune cells in adipose tissue integrate metabolic and immunological functions and constitute a pivotal pathogenic link between obesity and its metabolic sequelae37. Macrophages are a class of innate immune cells that are found in adipose tissue, and they are implicated in angiogenesis, the proliferation and differentiation of adipocyte precursors, and the secretion of inflammatory cytokines in the obesity process38,39,40. However, whether the intervention of ATM ameliorates obesity remains undetermined. Our study indicates that inhibiting the proinflammatory activation of ATM prevents obesity through decreased IL-1β production from macrophages and the promotion of energy expenditure by increasing the thermogenic capacity of both WAT and BAT. Our research provides experimental evidence that targeting ATM can alleviate obesity. IL-1β is the product of several cell types, such as macrophages, dendritic cells, and adipocytes29,30. Therefore, we use mice with myeloid IL-1β deficiency to examine the effects of these agents on obesity. Like obese mice treated with an IL-1β neutralizing antibody, mice with myeloid IL-1β deficiency can resist HFD-induced obesity. IL-1β also recruits immune cells and promotes inflammatory cytokine secretion in adipose tissue. However, the effects of other immune cells recruited by IL-1β or other inflammatory cytokines on adipose tissue inflammation need further investigation.
For decades, immunometabolism has been the focus of research on different macrophage subtypes and functions8. Importantly, immunometabolic pathways not only provide energy, such as ATP but also signaling molecules, such as succinate, to regulate the phenotypes and functions of macrophages41. In individuals with obesity, the proinflammatory profile of inflammatory macrophages in adipose tissue plays a pivotal role in the development of adipose inflammation and the aggravation of obesity4. Altered amino acid metabolism, such as the process of converting arginine into nitric oxide, is one of the first characteristics employed to define macrophage subsets42. Additionally, glutaminolysis is reported to regulate innate immunity via fumarate accumulation-derived epigenetic modifications43. Consistent with these findings, our findings indicate that glutaminolysis is an extremely important metabolic pathway for the regulation of ATM. In obese humans, we find an increase in the ratio of succinate to glutamine in adipose tissue, suggesting that glutaminolysis is induced in the adipose tissue of obese humans. Furthermore, glutaminolysis provides ATP energy to inhibit AMPK activity, promoting SCS activity and serving as a metabolic substrate for succinate accumulation. This leads to the proinflammatory activation of ATM, which further demonstrates that the dual functions of energy application and signaling molecule production in amino acid metabolism are involved in the regulation of macrophages.
AMPK is a ubiquitously expressed serine/threonine protein kinase that exerts its regulatory effect on cellular metabolism by phosphorylating multiple substrates16. However, studies on the role of AMPK in immunometabolism are rare. Previous research shows that AMPK regulates macrophage functions through the inhibition of the NF-κB pathway by upregulating the expression of SIRT1 and CREB or upregulating the expression of STAT6 and PPARγ44. Our study demonstrates that AMPK inhibits proinflammatory macrophages through decreased SCS activity via the phosphorylation of SUCLA2, thereby resulting in reduced succinate accumulation and IL-1β production. SUCLA2 is a rate-limiting enzyme in the TCA cycle, but AMPK is distributed primarily in the cytoplasm and nucleus45. It has been reported that the presence of AMPK in the mitochondrial matrix, where it phosphorylates mitochondrial proteins in 293 T cells, implies that AMPK can directly regulate mitochondrial metabolism by targeting key metabolic enzymes within the mitochondria46. We also observe abundant expression of AMPK in the mitochondria compared to the cytoplasm in macrophages, and SUCLA2, a novel substrate, served as a bridge between AMPK and immunometabolism. In our study, we observe the inhibition of AMPK activity via ATP from glutaminolysis. Glycolysis and fatty acid metabolism are also affected by AMPK activity, which regulates the proinflammatory macrophages18,47,48,49. In our research, we examine the association between macrophage function and both AMPK and glutamine-derived succinate production. Additionally, the effects of glycolysis and fatty acid metabolism regulated by AMPK on macrophages need to be further examined.
SUCLA2 functions as the β subunit of the SCS which is a heterodimer composed of two subunits: the α subunit, encoded by SUCLG1, and the β subunit, encoded by either SUCLG2 or SUCLA2. SCS catalyzes the conversion of succinyl-CoA to succinate within the TCA cycle50. The tissue-specific gene expression patterns of SUCLA2 and SUCLG2 exhibit differences. The heart and brain display a greater abundance of ATP-specific SUCLA2, whereas the kidney and liver predominantly express GTP-specific SUCLG251. Compared to patients without disease-causing mutations in SUCLG2, patients with mutations in SUCLA2 typically exhibit progressively developing encephalomyopathy characterized by sensorineural deafness, dystonia, and hypotonia52. Some SUCLA2 mutations cause a less complete disease course due to impairment of the enzymatic function of SUCLA2, indicating that alterations in the enzymatic activity of SUCLA2 might impact cellular function53. Our research reveals that the SUCLA2 phosphorylation site at Ser60 affects SCS activity. Additionally, a decrease in SUCLA2 phosphorylated at Ser60 is detected in the ATM of obese humans, suggesting that SUCLA2 may be a hallmark of adipose tissue inflammation caused by obesity. Once SUCLA2 is depleted in macrophages, inflammation and obesity are blocked, suggesting that macrophage SUCLA2 may be a potential target for obesity therapy.
In summary, our findings demonstrate that AMPK acts as a metabolic sensor of glutaminolysis to regulate succinate accumulation and IL-1β production in proinflammatory macrophages through the phosphorylation of SUCLA2 (Fig. 7b). The involvement of macrophage AMPK in the development of obesity occurs in an IL-1β-dependent manner, and suppression of macrophage SUCLA2 impedes obesity progression. Therefore, targeting macrophage SUCLA2 might be a promising strategy for treating obesity.
Methods
Human tissue sample collection and metabolite detection
The ScWAT from human subjects was investigated in this study. The studies on human adipose tissue were approved by the Medical Ethics Committee of Shanghai Yangpu District Central Hospital (Approval no. LL-2022-SCI-008). Only the subjects who provided informed written consent were included in the study, and all subjects underwent medical history inquiries before hospitalization. Human adipose tissue samples were collected from the preperitoneal fat layer of patients who underwent surgery with appendicitis, gallstone, sebaceous cyst, or lipomyoma at Shanghai Yangpu Hospital during 2022–2024, and gender was not considered in this study because of the limited sample number. All the samples were dissected and immediately frozen in liquid nitrogen for metabolite detection.
Sample preparation was performed based on the previous report with minor modification54. Briefly, adipose tissue was homogenized and derivatized with 200 mM 3-NPH in 75% methanol and 100 mM EDC in 75% methanol containing 1.5% pyridine at 30 °C for 60 min. After derivatization, 50% methanol solution was added. After centrifugation, the supernatants were injected into the LC-MS system for analysis. The Acquity UPLC I-Class system (Waters, Milford, MA, USA) was coupled with a quadrupole-time of flight mass spectrometer (Waters, Milford, MA, USA). The chromatographic separation was carried out on an Acquity UPLC HSS T3 (2.1 × 100 mm,1.8 μm, Waters) with column temperature set at 40 °C. The gradient elution was constructed with 0.1% aqueous formic acid (A) and acetonitrile: isopropanol (B, 90:10, v/v) at a flow rate of 0.4 mL/minute. Following gradient: 0–1 min, 5% B; 1–12 min, 5–95% B; 12–14 min, 95–100% B, held at 100% for 2 min, then returned to 5% within 0.1 min and held at 5% for 2 min to re-equilibrate the column pressure. The mass spectrometer was operated with the electrospray ionization source (ESI). The capillary voltages were set at 2.5 and 3 kV in negative and positive mode, respectively, and sampling cone voltage at 40 kV. The temperatures of ESI chamber and desolvation gas were at 120 °C and 450 °C, respectively. The flow rates of con gas and desolvation gas were maintained at 50 and 800 L/hour, respectively, and the nebulizing gas pressure was set at 6.5 bar. The acquisition mode was full scan with fragmentation. Q/TOF mass analyzer was operated at 21,000 mass resolution, and the scan range covered m/z 50–1,500.
Animals study
All animal experiments and protocols were approved by the Animal Ethics Committee of the Shanghai Institute of Materia Medica. All mice were housed in isolated ventilated cages with 3 to 5 mice in each cage in an animal barrier facility (specific pathogen-free/SPF) at the Shanghai Institute of Materia Medica (Shanghai, China). All mice were housed in a temperature (22 ± 2 °C) and humidity (50%) controlled room on a 12/12 h light/dark cycle. All mice with access to sterile pellet food and water ad libitum and male mice aged 6-8 weeks were used for the respective experiments. Wild-type C57BL/6 (#202) mice were purchased from Shanghai Shengchang Biotechnology Co., LTD (China). AMPKα1-floxed mice (#014141, Prkaa1fl), AMPKα2-floxed mice (#014142, Prkaa2fl), and LysM-Cre mice (#004781, LysM-cre) were purchased from Jackson Laboratory (Bar Harbor, Maine, USA). IL-1β-floxed mice (#T007839) were purchased from GemPharmatech Co., Ltd (Nanjing, China). To obtain myeloid AMPKα-knockout mice, AMPKα1/α2-floxed homozygous mice were first generated by mating homozygous AMPKα1-floxed mice with AMPKα2-floxed mice. Then, LysM-Cre, AMPKα1/α2-floxed mice were obtained by mating homozygous AMPKα1/α2-floxed mice with LysM-Cre mice. To obtain mice with IL-1β knockout in myeloid cells, LysM-Cre mice were mated with IL-1β-floxed mice. To obtain mice with AMPKα and IL-1β double knockout in myeloid cells, LysM-Cre, AMPKα1/α2-floxed mice were mated with IL-1β-floxed mice. For the mice experiment, the control mice and experimental mice were co-housed in the same experimental environment. Normal chow diet (Shanghai Shilin Biotechnology Co., LTD, China) was used to maintain the growth of mice, and high-fat diet containing 60% kcal fat (#D12492, Research Diets) was used to induce obesity. The body weight and food intake were recorded weekly. At the end of the animal experiment, the mice were euthanasia with CO2 using an SMQ-I type asphyxiation chamber for laboratory animals (Shanghai Tianhuan Science Develop Co., LTD, China).
p5RHH-siRNA nanoparticle preparation
The p5RHH peptide (VLTTGLPALISWIRRRHRRHC) was synthesized by Genscript and was dissolved at 10 mM in the distilled water without DNase, RNase, and protease, and was kept at –80 °C in 10 μL aliquots before use. The scrambled siRNA (target sequence, UUCUCCGAACGUGUCACGUTT) and SUCLA2 siRNA (target sequence, GCCUCAAAGGAGGAGUGAATT) were purchased from GenePharma, dissolved at 100 μM in DEPC water and were kept at –80 °C in 10 μL aliquots before use. The p5RHH-siRNA nanoparticles were prepared by mixing equal volumes of p5RHH peptide and siRNA at a peptide/siRNA ratio of 100:1 in 200 μL HBSS (#14025076, Gibco) and incubated on ice for 10 min. Then, the prepared p5RHH-siRNA nanoparticles were diluted with saline at 0.05 mg/mL, and the diluted p5RHH-siRNA were subjected to mice with 10 mL/kg saline solution through intravenous injection twice a week. For in vitro experiments, 40 nM siRNA was mixed with p5RHH at a peptide/siRNA ratio of 100:1 in HBSS and incubated at 37 °C for 40 min before being added into the cultural medium of macrophages.
Insulin tolerance test
The mice were given 0.75 U/kg recombinant human insulin (Novolin, Novo Nordisk) by intraperitoneal injection after 4 h of fasting with water freely, and the blood glucose level was measured from the tail before insulin administration by intraperitoneal injection and 15, 30, 60, 90, and 120 min after insulin administration using a glucometer (ACUU-CHEK, Roche).
Cold exposure
The cold exposure experiment was carried out in the incubators with constant temperature and humidity (#BD-HWS-250; Nanjing Beidi experimental instrument Co., LTD, China). The water and cages were cooled to a preset temperature overnight before the experiment. For cold exposure, mice were housed individually at 4 °C for 8 h, and the water was available ad libitum but without food. The rectal temperature was measured every two hours by the BAT-12 microprobe digital thermometer and RET-3 mouse rectal probe (Physitemp Instruments).
Metabolic analysis
The locomotor activity and oxygen consumption of mice were measured through the sixteen-chamber indirect calorimeter (TSE PhenoMaster, TSE system) according to the manufacturer’s instructions. Mice were housed individually in the chamber and were acclimated for 24 h before the measurements began, then the metabolic data of mice were collected for 48 h and analyzed. The food and water were fed ad libitum and the light kept the same 12/12 h light/dark cycle that is consistent in the home cages. Finally, the raw data was analyzed through a free-to-use online tool (CalR, https://calrapp.org).
Body mass analysis
Whole-body lean mass, fat mass, and fluid were measured using 1H-nuclear magnetic resonance spectroscopy (Minispec LF90 II, Bruker) according to the manufacturer’s instructions.
Histologic analysis and immunohistochemistry
Liver and adipose tissue were fixed in 4% neutral formalin for at least 48 h at 4 °C and the formalin was changed with 70% ethanol for at least 48 h, then the fixed tissue was desiccated and embedded in paraffin followed by being cut into 5-µm sections and stained with hematoxylin and eosin (H&E) or analyzed by immunohistochemistry (IHC). For IHC, the adipose tissue sections were heated at 95 °C in citric acid solution (#P0081, Beyotime) to extract epitopes after dewaxing. Then, endogenous peroxidases were quenched with 3% H2O2 (Hebei Jianning Pharmaceutical Co., LTD, China) followed by the blocking with 5% BSA (#B2064-100G, Sigma) in PBS for 1 h. Then the sections were incubated with anti-UCP1 antibody (#ab234430, Abcam) or anti-F4/80 antibody (#70076 s, CST) at 4 °C overnight. The next day, an HRP-conjugated secondary antibody (#GK500705, Gene Tech) was used and the sections were stained with a DAB staining kit (#P0202, Beyotime) according to the manufacturer’s instructions. Finally, the sections were visualized through teksqray SQS-600P (tissue slice in Fig.4) or PerkinElmer Vectra 3 (except for the tissue slice sections in Fig. 4).
Obtain bone marrow-derived macrophages (BMDMs)
The femurs and tibiae were collected from mice at the age of 6-8 weeks. Then, the ends of the bone were cut off using a surgical scissor and bone marrow was flushed out by PBS (9 g NaCl, 0.144 g KH2PO4, 0.421 g Na2HPO4 dissolved in 1 L of distilled water, pH 7.4) using a 1 mL syringe. The cell suspension was made using the pipette blowing the bone marrow repeatedly and the cell suspension was centrifuged at 500 × g for 5 min to remove PBS. Then, 1 mL of red blood cell lysis buffer (#B541001; Sangon Biotech (Shanghai) Co., Ltd, China) was added to the cell pellet for 5 min, and the red blood cell lysis buffer was removed by centrifuge at 500 g for 5 min to get the bone marrow cells without red blood. Finally, the bone marrow cells were resuspended in the differential medium [DMEM (#12800082, Gibco) containing 10% fetal bovine serum (#10091-148, Gibco), 100 units/mL penicillin and 100 µg/mL streptomycin (#15140-122, Gibco), and 20 ng/mL M-CSF (#416-ML-010, R&D)], and the cells were seed in cell culture plate and cultured in a humidified incubator at 37 °C with 5% CO2. The differential medium was refreshed every 2 or 3 days and mature BMDMs were obtained after culturing for 7 days.
Stromal vascular fraction (SVF) separation
The adipose tissue from C57BL/6 mice was dissected using surgical scissors and then enzymatically digested in a PBS buffer containing 2 mg/mL Collagenase D (#11088882001, Roche), 2.4 mg/mL Dispase Il (#4942078001, Roche), and 10 mM CaCl2 for 1 h at 37 °C. After the digestion process was halted by adding DMEM-F12 (#12500-039, Gibco) supplemented with 10% fetal bovine serum (#10091-148, Gibco), the resulting cell suspension was filtered through a 70-µm filter. Finally, the filtered cell suspension was centrifuged at 500 × g for 10 min to collect the floating mature adipocytes and SVF at the bottom of the centrifuge tube.
Magnetic beads separation
The F4/80 positive cells of SVF were isolated using a magnetic beads-based separation system. Briefly, after isolating SVF from adipose tissue, the cell number was determined and followed by centrifugation at 300 × g for 10 min. Subsequently, the SVF cells were resuspended in 90 µL buffer (#130-091-221-1, Miltenyi) containing 10 µL anti-F4/80 microbeads (#130-110-443, Miltenyi) every 107 total cells and incubated at 4 °C for 15 min. After washing the incubated cells with 1 mL buffer and resuspending them in 500 µL buffer, the F4/80 positive or negative cells of SVF were separated using MACS columns and MACS separators (Miltenyi).
Flow cytometry analysis
The SVF separated from adipose tissue was washed once with DPBS (#554656, BD Pharmingen). Subsequently, 1 µL of viability stain (#564406, BD Pharmingen) was added to the cells suspended in 500 µL of DPBS for a 30-min incubation at 4 °C. The cells were then centrifuged at 500 × g for 5 min to remove the viability stain, followed by a wash with DPBS. After another centrifugation at 500 × g for 5 min to remove the DPBS, 100 µL of DPBS containing 2 µL Fc Block (#553142, BD Pharmingen) was added to the cells, which were then incubated at 4 °C for an additional 15 min. Next, cells containing Fc Block were mixed with an extra 100 µL of DPBS containing 2 µL antibody and incubated at 4 °C for 30 min. Finally, the cells were washed twice and resuspended in 500 µL DPBS before flow cytometry analysis (#CytoFLEX S, Beckman Coulter).
Macrophage activation
In total, 100 ng/mL lipopolysaccharide(LPS) (#L2630, Sigma) was utilized to stimulate the proinflammatory activation of macrophages at different times for the detection of intracellular genes and proteins. To trigger the activation of the inflammasome and detect secreted IL-1β, macrophages were exposed to 2 mM ATP (#HY-B2176, MCE) for 30 min after the incubation with 100 ng/mL LPS, and the secreted IL-1β was detected by mouse IL-1β Elisa kit (#abs520001, Absin).
Western blot analysis
The cells were washed twice using PBS, and the protein samples were harvested using SDS loading [20 mL 0.5 M Tris-HCl (pH 6.8), 4 g SDS, 200 mg bromophenol blue, 20 mL glycerin, and 3.08 g DTT dissolved in 200 mL of distilled water] following by boiling at 100 °C for 10 min to denature. Then, the protein samples were separated by SDS-PAGE gels (SDS-polyacrylamide gel electrophoresis) using the running buffer (14.4 g glycine, 3.02 g Tris-base and 1 g SDS dissolved in 1 L of distilled water), and the separated protein was transferred to the NC membrane using the transfer buffer (14.4 g glycine, 3.02 g Tris-base and 200 mL methanol dissolved in 1 L of distilled water). Next, the NC membrane was blocked with 5% skim milk at room temperature for 1 h to reduce the nonspecific background. After blocking, the 5% skim milk was removed and the NC membrane was incubated with primary antibody at 4 °C overnight. The next day, the primary antibody was removed and the NC membrane was washed with TBST (2.42 g Tris-base, 8 g NaCl and 1.6 mL HCl, and 1 mL tween 20 dissolved in 1 L of distilled water, pH 7.6) three times with each time for 10 min at room temperature following by the incubated with secondary antibody for 1 h at room temperature. Finally, the NC membrane was stained with ECL detection buffer (#1705061, Bio-Rad) and was photographed by the ChemiDoc system.
RNA isolation and quantitative real-time polymerase chain reaction
Cells or tissues were lyzed on ice using 300 µL TRIzol reagent (#15596018, Invitrogen) for 15 min. 60 µL chloroform (#10006818; Sinopharm Chemical Reagent Co., Ltd, China) was added into the TRIzol reagent and vortex for 10 seconds followed by centrifuging at 12,000 × g for 15 min and collecting the supernatant containing mRNA. Then, 150 µL isopropanol (#80109218; Sinopharm Chemical Reagent Co., Ltd, China) was added to the supernatant and the mix was kept at -20 °C overnight after vortex for 10 seconds. The next day, the sample was centrifuged at 12,000 × g for 15 min to get the mRNA, and the mRNA was washed using 70% ethanol (#10009218; Sinopharm Chemical Reagent Co., Ltd, China) two times and centrifuged at 7500 × g for 10 min to obtain mRNA with less impurity. 20 µL RNAse-free water was used to dissolve mRNA and the concentration was measured using Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific). Next, 1 µg of mRNA was reverse-transcribed using ABScript III RT Master Mix for qPCR (#RK20408, ABclonal) using a 20 µL system to get cDNA. Finally, 180 µL RNAse-free water was used to dilute the cDNA.
For quantitative real-time polymerase chain reaction, a 10 µL system was used: 4 µL cDNA, 1 µL primer, and 5 µL 2X Universal SYBR Green Fast qPCR Mix (#RK21203, ABclonal). The thermal cycling conditions were 95 °C for 5 min followed by 40 cycles of 95 °C for 30 seconds and 60 °C for 30 seconds using a QuantStudio system (Thermo Fisher Scientific). Following the reactions, the raw data were analyzed using QuantStudio Software. The data were then normalized to housekeeping gene (β-actin for cells, and 36b4 for adipose tissue) expression.
siRNA-mediated knockdown assay
The target sequences (5′ to 3′) for successful mouse siRNAs were as follows: siNC: UUCUCCGAACGUGUCACGUTT; siAMPKα1: UUUGAAAGACCAAAGUCGGCU; siAMPKα2: AUCUAAACUGCGAAUCUUCUG; siSUCLA2#1: GCCUCAAAGGAGGAGUGAATT; siSUCLA2#2: GCAGCUAAUGCAGAUAUUATT. To knock down the AMPKα in RAW 264.7 cells, RAW 264.7 cells were seeded in cell culture plate overnight to 70% density before transfection. Mixing the 20 nM siRNA of AMPKα1 and 20 nM siRNA of AMPKα2, and transfect the siRNA to cells with Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. To knock down the SUCLA2 in BMDMs, mature BMDMs were harvested from bone marrow cells after culture for 7 days. 40 nM siRNA of SUCLA2 was transfected to cells with Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Stable isotope-assisted tracing
To investigate the mitochondrial anaplerosis from glutamine in inflammatory macrophage, 200 × 104 RAW 264.7 cells were seeded in a 6-cm dish overnight in DMEM (#12800082, Gibco) containing 10% fetal bovine serum (#10091-148, Gibco), 100 units/mL penicillin and 100 µg/mL streptomycin (#15140-122, Gibco). Then, the medium was changed with DMEM (#A1443001, Thermo Fisher Scientific) containing 10% fetal bovine serum (#30067334, Thermo Fisher Scientific), 100 units/mL penicillin and 100 µg/mL streptomycin (#15140-122, Gibco), 25 mM glucose (#63005518; Sinopharm Chemical Reagent Co., Ltd, China) and 10 mM [U-13C] glutamine (#CLM-1822-H-PK, Cambridge Isotope Laboratories, Inc.). Compared with the control group, the medium of the LPS group contained an extra 100 ng/mL LPS (#L2630, Sigma) to induce proinflammatory activation, and the cells were collected after the medium was changed for 12 h.
To investigate the regulation of AMPK on the mitochondrial anaplerosis from glutamine in inflammatory macrophage, 120 × 104 RAW 264.7 cells were seeded in a 6-cm dish overnight in DMEM (#12800082, Gibco) containing 10% fetal bovine serum (#10091-148, Gibco), 100 units/mL penicillin and 100 µg/mL streptomycin (#15140-122, Gibco), then, the siRNA of AMPKα1 (20 nM) and AMPKα2 (20 nM) was transfected into cells with Lipofectamine 3000 (Thermo Fisher Scientific) for 24 h according to the manufacturer’s instructions. Next, the medium was changed with DMEM (#A1443001, Thermo Fisher Scientific) containing 10% fetal bovine serum (#30067334, Thermo Fisher Scientific), 100 units/mL penicillin and 100 µg/mL streptomycin (#15140-122, Gibco), 25 mM glucose (#63005518, Sinopharm Chemical Reagent Co., Ltd), 10 mM [U-13C] glutamine (#CLM-1822-H-PK, Cambridge Isotope Laboratories, Inc.) and 100 ng/mL LPS (#L2630, Sigma). The cells were collected after the medium was changed for 12 h.
The content of TCA cycle metabolites was subjected to commercial metabolic flux analysis (Shanghai Profleader Biotech Co., Ltd, China). The cell samples were added 500 μL of 80% methanol and the mixture was processed by 5 cycles of 1 min ultra-sonication and 1 min interval in an ice-water bath. Then stood for 30 min at –40 °C and 10 min at 4 °C. After centrifugation at 15,000 × g and 4 °C for 15 min. The supernatant was evaporated to dryness under mild nitrogen and reconstituted in 20 μL of 50% acetonitrile (including 1 μg/mL phenylalanine-d5 internal standard) before performing UHPLC-HRMS/MS analysis. Quality control (QC) sample was obtained by isometrically pooling all the prepared samples. Chromatographic separation was performed on a ThermoFisher Ultimate 3000 UHPLC system with a Waters BEH Amide column (2.1 mm × 100 mm, 1.7 μm). The injection volume was 2 μL and the flow rate was 0.25 mL/minute. The column temperature was 15 °C. The mobile phases consisted of water with 0.01% formic acid and 2 mM ammonium formate (phase A) and acetonitrile (phase B). Linear gradient elution was performed with the following program: 0 min, 90% B; 4 min, 85% B; 11 min, 75% B; 14 min, 70% B, 14.5 min, 50% B and held to 17 min; 17.1 min, 90% B and held to 20.01 min. The eluents were analyzed on a ThermoFisher Q Exactive™ Hybrid Quadrupole-Orbitrap™ Mass Spectrometry (QE) in Heated Electrospray Ionization Negative (HESI-) mode, separately. Spray voltage was set to 4,000 V. Capillary and Probe Heater Temperature was separately 320 °C and 320 °C. The sheath gas flow rate was 35 (Arb, arbitrary unit), and the Aux gas flow rate was 10 (Arb). S-Lens RF Level was 50 (Arb). The full scan was operated at a high resolution of 70,000 FWHM (m/z = 200) at a range of 70–1050 m/z with AGC Target setting at 3×106. Finally, the analysis software, Xcalibur (version 4.0.27.19), of Thermo Fisher Company, was used to view the spectrum and integrate the target metabolites. Meanwhile, manual inspection was assisted to ensure the accuracy of the integral area of each target object. Data were derived to obtain the original integral area. The original data were then corrected for natural isotopes with reference to the literature method (Bioinformatics, 2019,1–4.).
Glutamine deprivation
To deprive the glutamine in the medium, the medium was changed with DMEM (#A1443001, Thermo Fisher Scientific) without glucose and glutamine. For the control group, the medium was added 25 mM glucose (#63005518; Sinopharm Chemical Reagent Co., Ltd, China) and 4 mM glutamine (#G3126, Sigma). For the glutamine deprivation group, the medium was added 25 mM glucose (#63005518; Sinopharm Chemical Reagent Co., Ltd, China) and distilled water with the same volume of glutamine in the control group.
ATP measurement
The content of ATP in cells was measured with an ATP test kit (#S0027, Beyotime) according to the manufacturer’s instructions. Briefly, the medium was removed and 200 µL lysate was added to the cell plate per well lysate cells. After cracking, the centrifuge was at 12,000 × g for 5 min, and the supernatant was taken for subsequent determination. Prepare an appropriate amount of ATP test solution at a ratio of 100 µL per sample or standard, and the ATP test solution was added to the test hole or test tube, then 40 µL sample or standard was added into the test hole or tube and the RLU was measured with a luminometer.
Intercellular succinate measurement
In total, 600 × 104 cells were combined with 100 μL of ice-cold 80% methanol/water and incubated on wet ice for 15 min. 25 μL of standard curves and cell lysis solution were mixed with 50 μL of 1 M O-Benzylhydroxylamine (#B-22984-5g, Sigma) and 50 μL of 1 M 3-(ethyliminomethy lideneamino)-N,N-dimethylpropan-1-amine (#39391-10 mL, Sigma) in pyridine buffer. After 60 min at room temperature, 300 μL of ethyl acetate was added and the plates were shaken for 20 min centrifuged at 300 × g for 5 min, and then 100 μL of the organic layer was taken into a 96-well plate. Then the aqueous layer was dried using a stream of nitrogen at 40 °C and reconstituted in 150 μL of methanol/water (50: 50, v/v). The samples were analyzed by LC-MS/MS.
RNA sequencing and bioinformatics analysis
Mature BMDMs were differentiated from bone marrow cells of AMPKαfl/fl mice (n = 3) or LysM-Cre, AMPKαfl/fl mice (n = 3) for 7 days in 10-cm dish. To induce inflammatory activation, BMDMs were incubated with LPS (#L2630, Sigma) for 12 h. 1 mL TRIzol reagent (#15596018, Life Technologies) was added into the 10-cm dish and the lysate was transferred to a 1.5 mL tube after the cell was washed by PBS twice and stored at −80 °C.
The RNA lysate was subjected to commercial RNA-seq analysis (Majorbio). Total RNA was extracted using TRIzol Reagent according to the manufacturer’s instructions. Then RNA quality was determined by 5300 Bioanalyser (Agilent) and quantified using the ND-2000 (NanoDrop Technologies). Only high-quality RNA sample (OD260/280 = 1.8–2.2, OD260/230 ≥ 2.0, RQN ≥ 6.5, 28S:18S ≥ 1.0, total RNA > 1 μg) was used to construct sequencing library. RNA purification, reverse transcription, library construction, and sequencing were performed at Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China) according to the manufacturer’s instructions. The RNA-seq transcriptome library was prepared following Illumina Stranded mRNA Prep, Ligation (San Diego, CA) using 1 μg of total RNA. Shortly, messenger RNA was isolated according to polyA selection method by oligo(dT) beads and then fragmented by fragmentation buffer firstly. Secondly double-stranded cDNA was synthesized using a SuperScript double-stranded cDNA synthesis kit (Invitrogen, CA) with random hexamer primers. Then the synthesized cDNA was subjected to end-repair, phosphorylation and adapter addition according to the library construction protocol. Libraries were size selected for cDNA target fragments of 300 bp on 2% Low Range Ultra Agarose followed by PCR amplified using Phusion DNA polymerase (NEB) for 15 PCR cycles. After quantified by Qubit 4.0, the sequencing library was performed on NovaSeq X Plus platform (PE150) using NovaSeq Reagent Kit. OR the sequencing library was performed on DNBSEQ-T7 platform(PE150) using DNBSEQ-T7RS Reagent Kit (FCL PE150) version 3.0. The raw paired end reads were trimmed and quality controlled by fastp with default parameters. Then clean reads were separately aligned to reference genome with orientation mode using HISAT2 software. The mapped reads of each sample were assembled by StringTie in a reference-based approach.
The enrichment of differential gene expression was performed on online enrichment (metascape, https://metascape.org/), gene set enrichment analysis (GSEA) was performed with the software GSEA_4.1.0 (https://www.gsea-msigdb.org/), and network analysis was performed on online analysis (STRING, https://cn.string-db.org/).
Immunofluorescence of adipose tissue
The expression of pT172-AMPKα, pS60-SUCLA2, and IL-1β in adipose tissue macrophage was detected by immunofluorescence with a multiple-color multifluorescent immunohistochemical staining kit (#D110071, WiSee Biotechnology) based on the principle of tyramine signal amplification technology according to the manufacturer’s instructions using the immunohistochemical antibodies as the primary antibody. Finally, the sections were visualized through teksqray SQS-120PRO (Fig. 6c and Fig. 7a) or PerkinElmer Vectra 3 (Fig. 3b and Supplementary Fig. 6a).
Co-immunoprecipitation
In total, 500 µL western and IP cell lysis buffer (#P0013, Beyotime) containing protease inhibitor cocktail (#HY-K0010, MCE) was added to the 10-cm dish to lyse cells after cells were washed with PBS twice. Then, the whole cell lysis was harvested by centrifugation at 9400 × g at 4 °C for 10 min and IgG or indicated antibodies were added followed by rotation at 4 °C overnight. The next day, the mixture was incubated with 50 µL protein A agarose (#P3476, Sigma) at 4 °C for 2 h. Next, the supernatant was removed, and the precipitates (bead-antibody-protein complex) were washed at 400 × g, 4 °C five times with ice-cold PBS buffer. Finally, the final precipitation was harvested by SDS-loading and detected by Western blot.
Mitochondria isolation
The mitochondria were isolated from the cell through the mitochondria Isolation kit (#C3601, Beyotime) according to the manufacturer’s instructions. Briefly, 1 mL of mitochondrial separation reagent was added into 20-50 million cells, gently suspending cells on ice for 15 min. Then, transfer the cell suspension to a glass homogenizer of appropriate size, and the cells were homogenized about 10–30 times followed by centrifuging at 600 × g at 4 °C for 10 min. Next, transfer the supernatant to another centrifuge tube and centrifuge at 11,000 × g for 10 min at 4 °C. Finally, the mitochondria were harvested after the supernatant was removed.
Measurement of succinyl-CoA synthetase activity
The succinyl-CoA synthetase activity activity was measured in the direction of the succinyl-CoA to succinate reaction. Only the enzymatic activity in RAW 264.7 cells (Fig. 5g) was determined by a commercial kit (#MAK217, Sigma) because the kit is discontinued for sale. The subsequent SUCLA2 activity of BMDMs or purified proteins was determined by the reaction in vitro. For BMDMs (Fig. 6b), 300 µL assay buffer (#MAK217A, Sigma) was added into a well in a 12-well cell plate to lyse cells after the treatment of BMDMs, and the lytic cells were transferred to the 1.5 mL tubes and kept on ice for 10 min following the centrifugation at 12,000 × g for 10 min to remove cell debris. Then, 30 µL cell lysis was added to the enzyme activity detection system in the well of the 96-well cell plate. For the purified SUCLG1-SUCLA2 complex (Fig. 5n–p), 30 µL purified protein was added to the enzyme activity detection system in the well of the 96-well cell plate. The complete assay mixture, with a volume of 100 µL, comprised of 50 mM potassium phosphate (pH 7.2), 0.2 mM succinyl-CoA, 2 mM ADP, 10 mM MgCl2, and 0.2 mM DTNB. The reactions were initiated by the rapid addition of DTNB and succinyl-CoA. After brief incubation with DTNB, the CoA-SH released from succinyl-CoA was measured spectrophotometrically at 415 nm using a microwell plate reader. The control values were subtracted to correct for nonspecific hydrolysis of the CoA derivatives in the absence of cell lysis.
In vitro kinase assay
The in vitro kinase assay contains two steps: AMPK pre-phosphorylation and substrate phosphorylation. For AMPK pre-phosphorylation, 40 nM CAMKKβ and 400 nM AMPKα1β1γ1 complex were added to kinase assay buffer (#PV3189, Thermo Fisher Scientific) containing 50 mM HEPES (pH 7.5), 10 mM MgCl2, 1 mM EGTA, and 0.01% Brij-35, 8 µM ATP (#HY-B2176, MCE) and incubated at 37 °C for 4 h. For substrate phosphorylation, 200 nM pre-phosphorylated AMPKα1β1γ1 complex and 200 nM SUCLG1-SUCLA2 complex were added to kinase assay buffer (#PV3189, Thermo Fisher Scientific) containing 50 mM HEPES (pH 7.5), 10 mM MgCl2, 1 mM EGTA, and 0.01% Brij-35, 50 µM ATP-g-S (#ab138911, Abcam) incubated at 37 °C for 4 h. Then, the samples were alkylated with 1 mM PNBM (#ab138910, Abcam) and incubated for 1 h at room temperature, and then subjected to SDS-PAGE and western blot. The phosphorylation of SUCLA2 by AMPK was detected using an anti-thiophosphate ester antibody (#ab92570, Abcam).
Purification of recombinant proteins
Escherichia coli BL21 cells containing indicated Pet28b plasmids including His-CAMKKβ, AMPKα1β1γ1 complex (α1 subunit contains His-tag) and PETDeut-1 plasmid containing SUCLG1-SUCLA2 complex (SUCLG1 subunit contains His-tag) were cultured in LB medium (10 g NaCl, 10 g tryptone and 5 g yeast extract dissolved in 1 L of distilled water) and rotated at a 37 °C, 250 rpm until the OD595 of the medium reached 0.4 to 0.6. Next, 100 μM IPTG (#HY-15921, MCE) was added to the LB medium to induce protein expression at 16 °C, 180 rpm for 16 h. Then, the bacterial medium was centrifuged at 5280 g for 15 min and the precipitation was resuspended in buffer A (500 mM NaCl, 50 mM Tris-HCl (pH 7.4), 10 mM imidazole, 1 mM β-mercaptoethanol, 10% glycerol, 0.1 mM PMSF) followed by ultrasound for indicate time and power. Next, the pellets were removed by centrifugation at 14,500 × g for 40 min and the supernatant was added to Ni Sepharose 6 Fast Flow (#100012, Yelibio. Company,) for incubation with gentle rotation at 4 °C for 2 h. Then, the globin mixture was harvested in the separation tube and washed with buffer B (500 mM NaCl, 50 mM Tris-HCl (pH 7.4), 50 mM imidazole, 1 mM β-mercaptoethanol, 10 % glycerol) to wash out the impurities. The protein was then eluted in buffer C (500 mM NaCl, 50 mM Tris-HCl (pH 7.4), 100 mM imidazole, 1 mM β-mercaptoethanol, 10 % glycerol). Finally, the protein sample was dialyzed in buffer solution D (150 mM NaCl, 50 mM Tris-HCl (pH 7.4), 1 mM β-mercaptoethanol, 10% glycerol, 1 mM EDTA) at 4 °C overnight, and the collected protein was kept at –80 °C before use.
Peptide phosphorylation detection
To identify the site of SUCLA2 phosphorylated by AMPK, 400 nM pre-phosphorylated AMPKα1β1γ1 complex and 1 µM SUCLG1-SUCLA2 complex were added to kinase assay buffer (#PV3189, Thermo Fisher Scientific) containing 50 mM HEPES (pH 7.5), 10 mM MgCl2, 1 mM EGTA, and 0.01 % Brij-35, 50 µM ATP (#HY-B2176, MCE) incubated at 37 °C overnight and the SUCLG1-SUCLA2 complex without AMPK incubation was taken as control. Then, the protein sample was subjected to commercial protein and phosphorylated site analysis (Applied Protein Technology, China). The analysis process is as follows: 1. Enzymatic hydrolysis of protein sample: After reduction and alkylation treatment, the appropriate amount of enzyme (mass ratio 1:50) was added to the sample, and incubated at 37 °C for 20 h. The enzymatic hydrolysis products were freeze-dried after desalting and redissolved in 0.1% FA solution and saved at −20 °C later use. 2. Mass spectrometry: Liquid A is an aqueous solution of 0.1% formic acid, and liquid B is an aqueous solution of 0.1% formic acid acetonitrile (84% acetonitrile). After the column is balanced with 95% liquid A, the sample is loaded into the trap column by the automatic sampler, and the samples were separated by chromatography and analyzed by mass spectrometer—the detection mode is a positive ion, parent ion scanning range is 300–1,800 m/z, primary mass spectrometry resolution is 70,000 at 200 m/z, automatic gain control target is 1e6, maximum IT is 50 ms, and dynamic exclusion time is 30.0 s. The mass charge ratio of polypeptides and polypeptide fragments is as follows methods: 20 fragment maps (MS2 scan) were collected after each full scan, MS2 activation type is HCD, isolation window is 2 m/z, secondary mass spectrometry resolution is 17500 at 200 m/z, normalized collision energy is 27 eV, underfill is 0.1%. 3. Data analysis: the identified protein result was obtained after the raw file was searched from MaxQuant 1.6.14.
Construction of plasmids and viral transfection
To overexpress SUCLA2 and AMPK in 293 T cells, the CDS sequence of human SUCLA2 (HA-tag is on the C-terminal), human AMPKα1 (myc-tag is on the C-terminal), AMPKα2 (myc-tag is on the C-terminal), human AMPKβ1 (myc-tag is on the C-terminal), and AMPKβ2 (myc-tag is on the C-terminal) were separately cloned into pcDNA3.1 plasmid. To obtain purified protein, the CDS sequence of human CAMKKβ (His-tag is on the C-terminal), human AMPKα1β1γ1 complex (only AMPKα1 subunit has His-tag on the C-terminal, and AMPKα1, β1, γ1 share the same promoter) were separately cloned into Pet28b plasmid, and human SUCLG1-SUCLA2 complex without mitochondrial localization peptide (only SUCLG1 subunit has His-tag on the C-terminal and SUCLG1 and SUCLA2 have their own promoters) were cloned into PETduet-1 double promoter plasmid.
To overexpress SUCLA2 in RAW 264.7 cells, human SUCLA2 (WT, S60A and S60D) were separately cloned into pLVX-IRES-Puro plasmid. To obtain viral particles, 4.8 μg of lentiviral construct, 3.6 μg of pSPAX2, and 2.4 μg of VSV-G were cotransfected into lenti-293T cells in a 6-cm dish using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. After transfection for 48 h and 72 h, the virus supernatants were collected and filtered through a 0.45-μm membrane (Millipore) followed by transfection on RAW 264.7 cells.
Statistical analysis
All results in this study are presented as mean ± SEM, and data were analyzed with unpaired two-tailed Student’s t test between two groups, and one-way ANOVA or two-way ANOVA among multiple groups using GraphPad Prism 8 software. P < 0.05 was considered to be statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw data of gene expression in the RNA Sequencing of AMPKfl/fl BMDMs and LysM-Cre, AMPKfl/fl BMDMs have been uploaded to Genome Sequence Archive (GSA) under accession code CRA014991. The raw data of flow cytometry analysis in this study have been uploaded to FlowRepository under accession code FR-FCM-Z8TP [https://flowrepository.org/id/RvFr7AJU9R6SxhRmCELfQSD6Tm5ahLpmOlWPQIaHENE6RMoU2ZbSansN398SkYbW]. Source data are provided with this paper. All data are included in the Supplementary Information or available from the authors, as are unique reagents used in this Article. The raw numbers for charts and graphs are available in the Source Data file whenever possible. Source data are provided with this paper.
References
Chooi, Y. C., Ding, C. & Magkos, F. The epidemiology of obesity. Metabolism 92, 6–10 (2019).
Cefalu, W. T. et al. Advances in the science, treatment, and prevention of the disease of obesity: Reflections from a diabetes care editors’ expert forum. Diab. Care 38, 1567–1582 (2015).
Chavakis, T., Alexaki, V. I. & Ferrante, A. W. Jr Macrophage function in adipose tissue homeostasis and metabolic inflammation. Nat. Immunol. 24, 757–766 (2023).
Chawla, A., Nguyen, K. D. & Goh, Y. P. S. Macrophage-mediated inflammation in metabolic disease. Nat. Rev. Immunol. 11, 738–749 (2011).
Sakamoto, T. et al. Macrophage infiltration into obese adipose tissues suppresses the induction of UCP1 level in mice. Am. J. Physiol.-Endocrinol. Metab. 310, E676–E687 (2016).
Mathis, D. Immunological goings-on in visceral adipose tissue. Cell Metab. 17, 851–859 (2013).
Mowers, J. et al. Inflammation produces catecholamine resistance in obesity via activation of PDE3B by the protein kinases IKKepsilon and TBK1. Elife 2, e01119 (2013).
Van den Bossche, J., O’Neill, L. A. & Menon, D. Macrophage Immunometabolism: Where Are We (Going)? Trends Immunol. 38, 395–406 (2017).
Stein, W. H. & Moore, S. The free amino acids of human blood plasma. J. Biol. Chem. 211, 915–926 (1954).
Merlin, J. et al. Non-canonical glutamine transamination sustains efferocytosis by coupling redox buffering to oxidative phosphorylation. Nat. Metab. 3, 1313–1326 (2021).
Ren, W. et al. Glutamine metabolism in macrophages: A novel target for obesity/type 2 diabetes. Adv. Nutr. 10, 321–330 (2019).
Feng, Y. et al. Pharmacological inhibition of glutaminase 1 attenuates alkali-induced corneal neovascularization by modulating macrophages. Oxid. Med Cell Longev. 2022, 1106313 (2022).
Yoo, H. C., Yu, Y. C., Sung, Y. & Han, J. M. Glutamine reliance in cell metabolism. Exp. Mol. Med. 52, 1496–1516 (2020).
Tannahill, G. M. et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature 496, 238–242 (2013).
Boutens, L. et al. Unique metabolic activation of adipose tissue macrophages in obesity promotes inflammatory responses. Diabetologia 61, 942–953 (2018).
Steinberg, G. R. & Hardie, D. G. New insights into activation and function of the AMPK. Nat. Rev. Mol. Cell Biol. 24, 255–272 (2023).
Yang, Z., Kahn, B. B., Shi, H. & Xue, B. Z. Macrophage alpha1 AMP-activated protein kinase (alpha1AMPK) antagonizes fatty acid-induced inflammation through SIRT1. J. Biol. Chem. 285, 19051–19059 (2010).
Galic, S. et al. Hematopoietic AMPK beta1 reduces mouse adipose tissue macrophage inflammation and insulin resistance in obesity. J. Clin. Invest 121, 4903–4915 (2011).
Jeong, H. W. et al. Berberine suppresses proinflammatory responses through AMPK activation in macrophages. Am. J. Physiol. Endocrinol. Metab. 296, E955–E964 (2009).
Kelly, B., Tannahill, G. M., Murphy, M. P. & O’Neill, L. A. Metformin Inhibits the Production of Reactive Oxygen Species from NADH:Ubiquinone Oxidoreductase to Limit Induction of Interleukin-1beta (IL-1beta) and Boosts Interleukin-10 (IL-10) in Lipopolysaccharide (LPS)-activated Macrophages. J. Biol. Chem. 290, 20348–20359 (2015).
Humphries, F. et al. Succination inactivates gasdermin D and blocks pyroptosis. Science 369, 1633–1637 (2020).
Benson, M. D. et al. Protein-metabolite association studies identify novel proteomic determinants of metabolite levels in human plasma. Cell Metab. 35, 1646–1660.e1643 (2023).
Ryan, D. G. et al. Coupling Krebs cycle metabolites to signalling in immunity and cancer. Nat. Metab. 1, 16–33 (2019).
Li, X. et al. Adipose tissue macrophages as potential targets for obesity and metabolic diseases. Front Immunol. 14, 1153915 (2023).
Selak, M. A. et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell 7, 77–85 (2005).
Cinti, S. et al. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J. Lipid Res 46, 2347–2355 (2005).
Mina, A. I. et al. CalR: A web-based analysis tool for indirect calorimetry experiments. Cell Metab. 28, 656–666 e651 (2018).
Yao, J. et al. Macrophage IRX3 promotes diet-induced obesity and metabolic inflammation. Nat. Immunol. 22, 1268–1279 (2021).
Garlanda, C., Dinarello, C. A. & Mantovani, A. The interleukin-1 family: back to the future. Immunity 39, 1003–1018 (2013).
Petrus, P. et al. Glutamine links obesity to inflammation in human white adipose tissue. Cell Metab. 31, 375–390 e311 (2020).
Li, W. et al. Screening of AMPK interacted proteins by yeast two hybrid system. China Biotechnol. 2012, 32, 1–7 (2012).
Jiang, Y. et al. AMPK-mediated phosphorylation on 53BP1 promotes c-NHEJ. Cell Rep. 34, 108713 (2021).
Zhou, H.-f et al. Peptide-siRNA nanocomplexes targeting NF-κB subunit p65 suppress nascent experimental arthritis. J. Clin. Investig. 124, 4363–4374 (2014).
Hou, K. K., Pan, H., Schlesinger, P. H. & Wickline, S. A. A role for peptides in overcoming endosomal entrapment in siRNA delivery — A focus on melittin. Biotechnol. Adv. 33, 931–940 (2015).
Zou, W. et al. Myeloid-specific Asxl2 deletion limits diet-induced obesity by regulating energy expenditure. J. Clin. Invest 130, 2644–2656 (2020).
Yan, H. et al. Suppression of NF-kappaB activity via nanoparticle-based siRNA delivery alters early cartilage responses to injury. Proc. Natl Acad. Sci. USA 113, E6199–E6208 (2016).
Schipper, H. S., Prakken, B., Kalkhoven, E. & Boes, M. Adipose tissue-resident immune cells: key players in immunometabolism. Trends Endocrinol. Metab. 23, 407–415 (2012).
Sun, K., Kusminski, C. M. & Scherer, P. E. Adipose tissue remodeling and obesity. J. Clin. Invest 121, 2094–2101 (2011).
Kamei, N. et al. Overexpression of monocyte chemoattractant protein-1 in adipose tissues causes macrophage recruitment and insulin resistance. J. Biol. Chem. 281, 26602–26614 (2006).
Huber, J. et al. CC chemokine and CC chemokine receptor profiles in visceral and subcutaneous adipose tissue are altered in human obesity. J. Clin. Endocrinol. Metab. 93, 3215–3221 (2008).
Murray, P. J. et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41, 14–20 (2014).
Van den Bossche, J. et al. Pivotal Advance: Arginase-1-independent polyamine production stimulates the expression of IL-4-induced alternatively activated macrophage markers while inhibiting LPS-induced expression of inflammatory genes. J. Leukoc. Biol. 91, 685–699 (2012).
Arts, R. J. et al. Glutaminolysis and fumarate accumulation integrate immunometabolic and epigenetic programs in trained immunity. Cell Metab. 24, 807–819 (2016).
Cui, Y. et al. The role of AMPK in macrophage metabolism, function and polarisation. J. Transl. Med 21, 892 (2023).
Garcia, D. & Shaw, R. J. AMPK: Mechanisms of cellular energy sensing and restoration of metabolic balance. Mol. Cell 66, 789–800 (2017).
Cai, Z. et al. Phosphorylation of PDHA by AMPK drives TCA cycle to promote cancer metastasis. Mol. Cell 80, 263–278 e267 (2020).
Phair, I. R. et al. AMPK integrates metabolite and kinase-based immunometabolic control in macrophages. Mol. Metab. 68, 101661 (2023).
Mounier, R. et al. AMPKalpha1 regulates macrophage skewing at the time of resolution of inflammation during skeletal muscle regeneration. Cell Metab. 18, 251–264 (2013).
Ouimet, M. et al. MicroRNA-33-dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. J. Clin. Invest 125, 4334–4348 (2015).
Huang, J. & Fraser, M. E. Structural basis for the binding of succinate to succinyl-CoA synthetase. Acta Crystallogr D. Struct. Biol. 72, 912–921 (2016).
Lambeth, D. O., Tews, K. N., Adkins, S., Frohlich, D. & Milavetz, B. I. Expression of two succinyl-CoA synthetases with different nucleotide specificities in mammalian tissues. J. Biol. Chem. 279, 36621–36624 (2004).
Gut, P. et al. SUCLA2 mutations cause global protein succinylation contributing to the pathomechanism of a hereditary mitochondrial disease. Nat. Commun. 11, 5927 (2020).
Matilainen, S. et al. Mitochondrial encephalomyopathy and retinoblastoma explained by compound heterozygosity of SUCLA2 point mutation and 13q14 deletion. Eur. J. Hum. Genet. 23, 325–330 (2014).
Xu, H. et al. Development of a simultaneous quantification method for the gut microbiota-derived core nutrient metabolome in mice and its application in studying host-microbiota interaction. Analytica Chim. Acta 1251, 341039 (2023).
Acknowledgements
We thank L.-Y. Rui (University of Michigan) and H. Zhang (Chinese Academy of Medical Sciences Fuwai Hospital) for helpful comments on this study. We thank members of the laboratory of H. Zhou (Shanghai Institute of Materia Medica) for their assistance in conducting interactive protein identification of SUCLA2 through mass spectrometry. We also thank R. Feng (Shenzhen Shengqiang Technology Co., LTD and Shanghai Lianchuan Biotechnology Co., LTD) for his assistance in the application of clinical ethics, transport of human adipose tissue samples, and section scanning of human adipose tissue. The Images in this study were created in BioRender.This work was supported by grants from the National Natural Science Foundation of China grants (82273983 to H.W.J., 32000525 to H.W.J., 82130099 to J.L., and 92253306 to J.L.), National Key R&D Program of China (2022YFA1303800 to H.W.J.), Science and Technology Commission of Shanghai Municipality (23ZR1474700 to H.W.J.), Medical Innovation Research Special Project of Shanghai (19411971500 to E.J.T.), Medical Innovation Research Special Project of Shanghai (22Y11908600 to E.J.T.), Shanghai Institute of Materia Medica, Chinese Academy of Science (No.SIMM0220231001 to J.L. and New Star project to H.W.J.), the Research Funds of Hangzhou Institute for Advanced Study, UCAS (No. 2022ZZ01013 to R.W., 2023HIAS-Y030 to R.W., 2023HIAS-V002 to J.L., and 2024HIAS-N001 to S.F.L.), Science and Technology Commission of Yangpu District (YPM202101 to E.J.T. and YPGWM202401 to E.J.T.), and China Postdoctoral Science Foundation (2024M760704 to C.P.).
Author information
Authors and Affiliations
Contributions
J.L., H.W.J., and C.P. conceived the project, conceptualized the experiments and wrote the manuscript; L.Y.J., W.H.Y., H.M., Z.L., H.Y.G., X.T.G., M.W., C.M.X., S.F.L., L.Y.Y.L., W.B.W., and J.Y.S. provided assistance in the animal experiment; H.L.W. and S.K.Y. performed in the RNA-sequencing and bioinformatics analysis; X.Z.G., Y.T.C., and C.X. were responded for the metabolite quantification of human adipose tissue; B.S., A.L.W., and E.J.T. were responded for the sample collection of human adipose tissue; R.W., Y.Z., H.W.J., and J.L. participated in the discussion and supervised the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interest.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Peng, C., Jiang, H., Jing, L. et al. Macrophage SUCLA2 coupled glutaminolysis manipulates obesity through AMPK. Nat Commun 16, 1738 (2025). https://doi.org/10.1038/s41467-025-57044-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-57044-w
This article is cited by
-
Dysregulation of the AMPK-SREBP1-FASN axis in MASLD: driving a vicious cycle of lipotoxicity and metabolic-immune crosstalk
Lipids in Health and Disease (2026)
-
Brusatol ameliorates psoriatic dyslipidemia by targeting IL-1β to restore AMPK-mediated lipid homeostasis
Chinese Medicine (2026)
-
1,1-Diethoxyethane increases insulin sensitivity and ameliorates obesity and dyslipidemia in mice fed high-fat diet
npj Metabolic Health and Disease (2026)