Abstract
The neuromuscular junction (NMJ) is the synapse formed between motor neurons and skeletal muscle fibers. Its stability relies on the continued expression of genes in a subset of myonuclei, called NMJ myonuclei. Here, we use single-nuclei RNA-sequencing (snRNA-seq) to identify numerous NMJ-specific transcripts. To elucidate how the NMJ transcriptome is regulated, we also performed snRNA-seq on sciatic nerve transected, botulinum toxin injected, and Musk knockout muscles. The data show that NMJ gene expression is not only driven by agrin-Lrp4/MuSK signaling but is also affected by electrical activity and trophic factors other than agrin. By selecting the three NMJ genes Etv4, Lrtm1, and Pdzrn4, we further characterize novel contributors to NMJ stability and function. AAV-mediated overexpression shows that Etv4 is sufficient to upregulate the expression of −50% of the NMJ genes in non-synaptic myonuclei, while AAV-CRISPR/Cas9-mediated muscle-specific knockout of Pdzrn4 induces NMJ fragmentation. Further investigation of Pdzrn4 revealed that it localizes to the Golgi apparatus and interacts with MuSK protein. Collectively, our data provide a rich resource of NMJ transcripts, highlight the importance of ETS transcription factors at the NMJ, and suggest a novel pathway for NMJ post-translational modifications.
Similar content being viewed by others
Introduction
Skeletal muscle fibers form a syncytium, with many myonuclei sharing a single cytoplasm, resulting from the fusion of mono-nucleated progenitors. In mice, muscle fibers can reach several millimeters in length, accommodating hundreds of myonuclei1. Although all myonuclei within a fiber share one cytoplasm, their gene expression repertoire is not homogenous. Based on location and gene expression signature, myonuclei can be categorized into three main groups2,3,4,5. The largest myonuclei category is distributed throughout the fiber and express a set of contractile and metabolic genes specific to their fiber type. We refer to this group as ‘body myonuclei’2 as they make up the “body” of the muscle fiber, but they have also been called bulk myonuclei4 or simply named after the fiber type they belong to3. Two substantially smaller but specialized populations, typically called myotendinous junction (MTJ) and neuromuscular junction (NMJ) myonuclei2,3,4,5, are localized at muscle-tendon and muscle-motor neuron contact sites, respectively. NMJ myonuclei, also known as subsynaptic or fundamental myonuclei6, in mice, consist on average of four nuclei clustered beneath the muscle postsynapse7,8,9. They express transcripts whose products are enriched at the postsynapse and are often of functional importance for the NMJ. Examples are the acetylcholine receptor (AChR) subunits Chrne, Chrna1, Chrnd, Chrnb110, which encode the AChRε, AChRα, AChRδ and AChRβ subunits, respectively, and Musk, which encodes the muscle-specific kinase (MuSK) that is essential for NMJ formation and maintenance11,12,13. This localized expression of NMJ genes allows postsynaptic enrichment of the requisite proteins to form and maintain the NMJ.
Spatial restriction of synaptic proteins to central regions of muscle begins during embryonic development, before motor axons reach the muscle fibers, a phenomenon known as prepatterning6,14. Innervation by motor neurons condenses and further enriches postsynaptic proteins, via neural agrin release from motor axons. Neural agrin binds to the transmembrane protein LDL receptor-related protein 4 (Lrp4) expressed by muscle fibers15,16. Agrin-Lrp4 forms a receptor complex with MuSK to promote intracellular cross-autophosphorylation and MuSK activation. Full activation of MuSK requires the adaptor protein downstream of tyrosine kinases-7 (Dok7), which additionally supports the recruitment of downstream proteins to the MuSK scaffold17. Together, these proteins locally transduce the synaptic signal from neural agrin into muscle fibers to induce NMJ gene expression in nearby myonuclei that aggregate at the nerve-muscle contact site and become NMJ myonuclei18,19.
While the precise mechanism for transmitting MuSK signal to NMJ myonuclei is not well understood, several studies identified a six-base pair DNA sequence shared among several NMJ genes20,21,22,23,24,25. This DNA sequence is known as the ‘N-box’ and works as a binding site for ETS transcription factors. ETS Variant 5 (Etv5, also known as Erm) has been shown to be a critical NMJ gene regulator as full-body null mice transcribe less Musk and Chrne, have severely fragmented NMJs, and are deficient in neuromuscular transmission26. Interestingly, Etv5 null mice also support the idea that other transcription factors must be regulating NMJ genes as NMJs still develop and certain transcripts such as Chrna1 are not significantly reduced26.
Motor neurons not only promote NMJ gene expression but also suppress certain genes in muscle fibers. Release of acetylcholine (ACh) and the resulting action potentials in muscle fibers suppress expression of NMJ genes in body myonuclei10,27. Suppressed transcripts also include the developmental Chrng isoform while agrin-Lrp4/MuSK-signaling at the NMJ initiates the expression of the adult Chrne isoform6,19. Hence, there is a close interplay between local, “trophic” signals that maintain synaptic gene expression in NMJ myonuclei and “suppression” signals that spread throughout the muscle fibers to prevent synaptic gene expression in body myonuclei19. The recent use of single-nuclei RNA-sequencing (snRNA-seq) has allowed to identify NMJ transcripts in an unbiased manner2,3,4,5. As there are still many neuromuscular disorders of unknown etiology, some of which show a “dying back” phenomenon, that can be ameliorated by overexpression of Dok728,29,30, studying the transcripts expressed by NMJ myonuclei may also provide new insights into such diseases.
In this study, we used snRNA-seq to identify NMJ transcripts in adult mouse muscle. To examine how motor neurons contribute to the enrichment of NMJ transcripts, we conducted snRNA-seq upon (i) transection of the sciatic nerve, which abrogates “trophic” support from the nerve and the “suppression” signals by electrical activity, (ii) injection of botulinum toxin A (BoTX), which prevents ACh release31 but leaves trophic support from the motor neuron intact, and (iii) knocked out Musk to abrogate agrin-Lrp4/MuSK signaling. From these snRNA-seq data, we selected three NMJ transcripts and characterized them by overexpression and muscle fiber-specific knockout studies. We show that overexpression of the transcription factor ETV4 is sufficient to drive the expression of NMJ transcripts in body myonuclei and that the Golgi-associated protein PDZRN4 affects AChR and MuSK localization at the postsynapse. Taken together, our data provide a rich resource of NMJ enriched transcripts, available for exploration on our online snRNA-seq atlas (https://ruegglab.shinyapps.io/snatlas/)32. Additionally, we provide new insights into the function of three previously uncharacterized proteins at the NMJ.
Results
Identification of transcripts expressed in NMJ myonuclei
To identify NMJ myonuclei-enriched transcripts, we developed a single nuclei isolation protocol for skeletal muscle that bypasses the need for Fluorescence-Activated Cell Sorting (FACS). Single nuclei suspensions were prepared from two tibialis anterior (TA) and two gastrocnemius (GAS) muscles of adult C57BL/6 mice. Following barcoding, library preparation, sequencing, and filtering, we generated a UMAP with ~36,000 nuclei expressing an average of ~1700 genes each (Fig. 1a, Supplementary Data 1). Using canonical cell type markers, we identified 11 mono-nucleated cell types and five myonuclei populations (Fig. 1b), similar to previous work2,3,5. Our unbiased UMAP clustering generated two clusters that have not been described in previous skeletal muscle snRNA-seq datasets. By differential expression analysis, we identified Mmrn1 and Tenm2 as transcriptional markers for these two populations (Supplementary Data 2). Using single molecule fluorescence in situ hybridization (smFISH), Mmrn1-expressing nuclei localized near larger blood vessels, visualized by immunofluorescence (IF) staining against laminin-α433,34, while Tenm2-expressing nuclei were located close to motor neuron axons, visualized by IF staining against neurofilament (Supplementary Fig. S1a). As Mmrn1+ nuclei also express Pecam1 (Fig. 1b), we concluded that they represent a subtype of endothelial cells. Tenm2+ nuclei were also identified in a single-cell sciatic nerve atlas35, suggesting that they represent a peri- and endoneurial cell type.
a UMAP of ~36,000 nuclei isolated from tibialis anterior (TA; n = 2) and gastrocnemius (GAS; n = 2) muscles from four adult wild-type male mice. b Dot-plot of marker genes used for cell/nuclear type identification. c Proportion of nuclei and myonuclei subtypes found in TA and GAS muscles. d Volcano plot visualizing differentially expressed transcripts in NMJ myonuclei compared to body myonuclei (max. p-adj. cutoff at 2.23E-308). Examples of characterized NMJ transcripts are indicated in black. NMJ transcripts characterized in this manuscript are indicated in purple. e Venn diagram showing the number of transcripts enriched/reduced in NMJ/MTJ myonuclei and their overlap (p-adj. <5E-5, log2FC ± 0.25). f Top 15 cellular component gene ontology (GO) terms associated with transcripts enriched in NMJ myonuclei (472 transcripts, p-adj. <5E-5, log2FC > 0.25). GO-term analysis was performed with DAVID. In (c) data is presented as mean ± SEM. For differential expression in (d) a Wilcoxon Rank Sum test was used. In (e), the probability of overlap was calculated using the hypergeometric probability for a representation factor >1. In (f) a modified Fisher’s exact test was used to determine significance. Source data are provided as a Source Data file.
Myonuclei were identified based on the expression of Ttn and Neb and represented ~45% and ~65% of nuclei in GAS and TA muscle, respectively (Fig. 1c, Supplementary Fig. S1b). Within the myonuclei population type IIB, IIX and IIA myonuclei were distinguishable based on the expression of Myh4, Myh1 and Myh2, respectively (Fig. 1b, c, Supplementary Fig. S1b). Interestingly, the fold change of Mylk4 was higher than Myh4 in type IIB myonuclei, suggesting that this gene is an alternative gene marker for this muscle fiber subtype (Fig. 1b, Supplementary Fig. S1b, Supplementary Data 2). MTJ and NMJ populations gave rise to additional distinct clusters and made up ~3% and ~1% of myonuclei, respectively (Fig. 1c). The distinct clustering of these two small myonuclei populations highlights their unique transcriptional identity. They are characterized by hundreds of genes that are differentially expressed in the NMJ and MTJ populations compared to body myonuclei (Fig. 1d, Supplementary Fig. S1c, Supplementary Data 3). More genes were enriched, than depleted in these specialized populations, indicating that they largely maintain their traditional myonuclear identity despite their additional specialized transcriptional responsibilities. In line with this notion, a higher transcriptional activity in NMJ myonuclei has previously been suggested based on chromatin organization36. We also explored possible similarities between NMJ and MTJ myonuclei by comparing enriched and reduced transcripts from both populations (Fig. 1e). Many commonly enriched NMJ and MTJ myonuclei transcripts coded for membrane or extracellular matrix proteins (e.g. Lama2, Col4a4 and App), indicating an increased need for locally produced structural proteins to support these crucial connection sites between the muscle fiber and motor axons and tendons, respectively. Conversely, several transcripts encoding proteins involved in muscle contraction (e.g. Tnnt3 and Atp2a1) were reduced in both populations (Fig. 1e). To estimate overall variability between previously published snRNA-seq data2,3,4,5, we determined the overlap of transcripts enriched in NMJ and MTJ nuclei (Supplementary Fig. S1d, e). Previous studies identified fewer NMJ and MTJ genes than this study. The overlap was very high, between 92% and 54% (Supplementary Fig. S1d) for the NMJ and between 84% and 71% for the MTJ genes (Supplementary Fig. S1e).
To get an overview of the NMJ-enriched transcripts, we performed Gene Ontology (GO)-term analysis. The results included terms such as basement membrane, neuromuscular junction, cell-cell junction, synapse, and axon (Fig. 1f). This indicates that NMJ myonuclei express a range of transcripts that specifically support NMJ formation and maintenance. Interestingly, Dok7 and Rapsn, encoding two essential proteins involved in forming and maintaining the NMJ37,38, were not significantly enriched in NMJ myonuclei, similar to what was observed with laser capture microdissection (LCM)39. In conclusion, the method to isolate nuclei from mouse skeletal muscles without sorting them via FACS2,3,5 or after genetic labeling4 allowed us to identify eleven mono-nucleated cell populations, some of which were not previously reported. The method also yielded 472 and 501 transcripts that were highly enriched, and 270 and 379 transcripts that were reduced in NMJ and MTJ myonuclei, respectively. Interestingly, a good number of NMJ and MTJ transcripts overlap, suggesting molecular similarities between the two sub-compartments. Our data also show that transcripts encoding most (but not all) proteins essential for the formation and maintenance of the NMJ are specifically expressed in NMJ myonuclei.
NMJ gene expression after sciatic nerve transection
The motor neuron orchestrates gene expression in myonuclei locally by secreting neural agrin, a signaling and scaffold-inducing protein, and by the release of acetylcholine, which triggers action potentials along the entire postsynaptic muscle fiber. Several lines of evidence indicate that neural agrin maintains synaptic gene expression at NMJ myonuclei through Lrp4/MuSK signaling and that electrical activity inhibits their expression in body myonuclei by calcium release into the cytoplasm19. As a consequence, many NMJ transcripts, such as Chrna1, Chrnb1, Chrnd10, Ncam140, and Musk13, are re-expressed (as during development) in body myonuclei upon denervation. This re-expression is thought to be based on the presence of ‘E-box’ domains within promoter or enhancer regions that are bound by myogenic regulatory factors, such as myogenin19. However, not all ‘E-box’ domains are functional as demonstrated for Chrne, which remains largely restricted to NMJ myonuclei upon denervation10 and where deletion of the ‘E-box’ domain does not reduce expression in primary myotubes41.
To get a more global view on the regulation of NMJ genes, we next examined gene transcription using snRNA-seq from TA and GAS muscle after sciatic nerve transection. Bulk RNA-seq data show that TA and GAS muscles exhibit denervation gene signatures already three days post nerve transection, while muscle atrophy takes seven days to become significant42,43. Hence, we chose 5 days post nerve transection as the time point of analysis. The snRNA-seq data from denervated muscles were clustered together with the “innervated” dataset from Fig. 1. Similar to previous observations44,45, five-day denervation significantly increased the proportion of macrophages and Schwann cells compared to the innervated state (Supplementary Fig. S2a-c). FAP numbers have previously been shown to increase during denervation46,47; however, at 5 days post denervation we did not observe any proportional increase using snRNA-seq. Interestingly, within the body myonuclei populations, two new clusters emerged after denervation, similar to a previous snRNA-seq study on 14-day denervated muscle focusing on atrophic transcriptional processes47. We termed the new clusters denervation 1 and 2 as both expressed NMJ genes, such as Musk and Ncam1 (Fig. 2a, b). Many fiber type-specific genes, such as Mylk4 and Myh4 were downregulated by denervation (Fig. 2b, Supplementary Fig. S2d). Denervation 1 and 2 were both devoid of Myh4, Myh1 and Myh2 transcripts (Supplementary Fig. S2d), which made it difficult to assign a specific fiber type to the clusters. Nevertheless, based on the localization of the UMAP clusters and other novel fiber type gene markers, such as Agbl1 (Fig. 2b), nuclei in the denervation 1 cluster likely derive from type IIB fibers and nuclei in denervation 2 from IIA/IIX fibers. Furthermore, both denervation 1 and 2 clusters expressed distinct genes, such as Scn5a (denervation 1) and Xirp2 (denervation 2; Fig. 2b), indicating that the denervation response differs between fiber types. Denervation significantly affected the proportions within the myonuclei population (Fig. 2c). Despite observing a shift in major myonuclei populations in the UMAP after five days of denervation, NMJ and MTJ cluster sizes and localization were not significantly altered (Fig. 2a, c), suggesting that their transcriptional identities are largely independent of innervation.
a SnRNA-seq UMAP generated from control and five days denervated muscles. Only the myonuclei (~37,000) are visualized and the UMAP is split by condition to visualize differences in clustering. n = 2 for TA and GAS for both conditions. b Feature plots showing marker genes expressed in control (left) and denervated (right) myonuclei. c Proportion of myonuclei and effect of five days denervation. d Heatmap showing example genes upregulated in denervated body myonuclei. e Heatmap showing example genes downregulated in NMJ myonuclei after sciatic nerve cut. f Pie-chart visualizing the expression changes of NMJ transcripts (472 transcripts) after sciatic nerve cut. Outer pie chart specifies the relative changes in NMJ and body nuclei of the subgroups of the inner pie chart. In (c), the data is presented as mean ± SEM and a two tailed t-test was performed (p < 0.05 = *p < 0.01 = **p < 0.001 = ***). Nerve cut and controls n = 4. In (f), transcripts were categorized as significantly changed if p-value was <0.01 and log2FC ± 0.25. Source data and p-values are provided as a Source Data file.
Denervation resulted in the presence of many NMJ transcripts, such as Chrna1, Chrnb1, Chrnd, Musk and Ncam1 in body myonuclei (Fig. 2d, Supplementary Data 4), supporting the notion that denervation resets muscle fibers into a “developmental stage” as the transcriptional inhibition via electrical activity is removed10,27,40. We observed the same pattern for other NMJ transcripts, such as Utrn and Col19a1. Utrn has previously been reported to be upregulated upon denervation48 and Col19a1 levels are reported to be elevated in muscle biopsies of ALS patients with faster disease progression49. In addition, denervation led to the re-expression of developmental genes, such as Chrng and Scn5a, that are silenced in NMJ and body myonuclei of adult innervated muscle (Fig. 2d). In contrast, denervation did not increase transcription of several prominent NMJ genes in body myonuclei and in fact lowered their expression in NMJ nuclei. Examples are Chrne, Ache, Colq, and Etv5 (Fig. 2e, Supplementary Data 4). By analyzing all NMJ-enriched transcripts (from Fig. 1, Supplementary Data 3), we found that ~22% of the NMJ transcripts were upregulated in body myonuclei and hence may harbor a functional ‘E-box’ binding domain (Fig. 2f, Supplementary Data 5). However, almost half (~44%) of the NMJ transcripts were not detected in body myonuclei upon denervation and were even downregulated in NMJ myonuclei. Another ~30% of the NMJ transcripts were not significantly changed in their expression in NMJ myonuclei while ~4% were specifically upregulated. These results suggest that either nerve-dependent “trophic” factors or the local depolarization (as a result of the release of ACh from the presynaptic nerve terminal) promote gene expression in NMJ myonuclei (Fig. 2f, Supplementary Data 5).
Trophic factors and electrical activity promote synaptic gene expression in NMJ myonuclei
While denervation by nerve transection removes the release of ACh from the nerve terminal and the suppressive function of electrical activity in body myonuclei, local agrin-Lrp4/MuSK signaling at the NMJ remains intact as agrin remains stably bound to the synaptic basal lamina50 (Supplementary Fig. S3a). To identify the upstream pathways that regulate the expression of particular NMJ genes, we next injected BoTX into TA muscles and investigated gene expression five days later, or blocked agrin-Lrp4/MuSK signaling at the NMJ by using the AAVMYO-CRISPR/Cas9 method to delete Musk for four weeks51,52. BoTX blocks ACh release from nerve terminals and thereby blocks electrical activity in postsynaptic muscle fibers, but leaves the presynaptic nerve terminal intact53,54. MuSK depletion abrogates agrin-Lrp4/MuSK signaling at the NMJ. Gene expression patterns were subsequently measured by snRNA-seq and the resulting data was clustered together with the dataset obtained from nerve transection shown in Fig. 2. The resulting UMAP similarly gave rise to denervation cluster 1; however, denervation cluster 2 remained within the type IIA myonuclei cluster (Fig. 3a, Supplementary Fig. S3b–d). Upon BoTX injection, body myonuclei showed a denervation gene signature similar to nerve transection, with Musk and Chrnb1 re-expressed in body and MTJ myonuclei (Fig. 3b). Musk knockout (KO) similarly showed re-expression of Chrnb1, however Musk transcripts were strongly depleted at the NMJ (Fig. 3b) and MuSK protein was absent at the postsynaptic side of the NMJs (Supplementary Fig. S3e), confirming successful MuSK depletion. MuSK depletion significantly downregulated a discrete set of genes, not downregulated by nerve transection or BoTX injection, including Prkar1a and Ryr3 (Fig. 3b). This shows that the reduction of MuSK protein (i.e. agrin signaling) was sufficient to affect a component of NMJ gene expression distinct from both innervation and denervation.
a SnRNA-seq was performed on BoTX-injected and Musk knockout (KO) TA muscles and respective controls, and the data were clustered together with the previous nerve cut dataset from Fig. 2. Only myonuclei (~60,000) are depicted in the UMAP. Individual conditions and their controls are visualized as split UMAPs to the right. b Violin plots visualizing the expression of selected NMJ genes in myonuclei after nerve cut, BoTX injection and Musk KO. The violin plots were generated from snRNA-seq data, UMAP is depicted in (a). Body myonuclei from all fiber types were pooled to generate an average expression value. c, d and e Pie charts showing how NMJ transcripts are regulated in NMJ myonuclei after BoTX injection and Musk KO compared to sciatic nerve transection. f Violin plots of NMJ transcripts that show different responses to nerve cut, BoTX injection or Musk KO. Nerve cut and the corresponding controls n = 4, the remaining conditions n = 1. In (b–f) DE-analysis was performed using a Wilcoxon Rank Sum test. All transcripts with a p-val. <0.01 and log2FC ± 0.25 are indicated by *. In (c–e), transcripts were categorized as significantly changed when p-val. <0.01 and log2FC ± 0.25.
After confirming successful denervation by BoTX injection and reduction of agrin-Lrp4/MuSK-signaling upon Musk KO, we examined NMJ gene expression in body and NMJ myonuclei. Out of all the NMJ genes re-expressed in body myonuclei by sciatic nerve transection, ~56% and ~35% were also re-expressed after BoTX injection and Musk KO, respectively (Fig. 3c, Supplementary Data 6). The difference in the number of NMJ genes that were re-expressed in body myonuclei between the different paradigms can have several reasons, including difference of the completeness of denervation by BoTX or Musk KO or signals from the presynaptic nerve that spread within the muscle fibers and prevent re-expression in myonuclei. As we were mainly interested in the transcriptional response in NMJ myonuclei, we next focused on the effect of BoTX injection and MuSK depletion in these nuclei. Out of the 354 transcripts that were downregulated in NMJ myonuclei after MuSK depletion, 218 (~62%) were NMJ transcripts (Supplementary Fig. S3f, Supplementary Data 7), suggesting that agrin-MuSK signaling is a major contributor to the maintenance of local transcription at the NMJ.
To distinguish how electrical activity and agrin-Lrp4/MuSK signaling contribute to the transcriptome in NMJ myonuclei, we next compared NMJ transcripts that were downregulated by nerve cut (loss of nerve and electrical activity) with BoTX injection (preservation of presynaptic nerve and loss of electrical activity) and MuSK depletion (preservation of presynaptic nerve but reduction of agrin-Lrp4/MuSK signaling). Of all the NMJ transcripts that were downregulated by nerve cut, 62% were also down in Musk KO and 27% with BoTX injection (Fig. 3d, Supplementary Data 6). This leaves 29% of NMJ transcripts that were downregulated by nerve cut only, suggesting that these transcripts require the presence of the presynaptic nerve terminal independent of ACh release and/or agrin-Lrp4/MuSK signaling.
As we observed downregulation of NMJ transcripts after both nerve cut and BoTX injection, nerve-evoked electrical activity seems to promote the expression of certain NMJ genes, such as Chrne, Rps6ka1 and Col4a4 (Fig. 3f). Ache expression has been shown to be regulated by electrical activity55 and BoTX injection similarly lead to a downregulation of its transcript (Fig. 3f). Most transcripts downregulated by BoTX were also downregulated by MuSK depletion, indicating that NMJ transcripts often require both electrical activity and agrin-Lrp4/MuSK signaling. Since we observed downregulation of certain NMJ genes by nerve cut but not by BoTX injection (e.g., Lrp5, Ctdspl, Epb41l4a and Scn4a, Fig. 3f), this supports the hypothesis that other potential trophic factors, e.g., neuregulins19, may play a role in promoting local gene expression at the NMJ. However, these potential trophic factors likely act as auxiliary proteins in addition to agrin since Musk KO in most cases also resulted in reduced gene expression. Next, we quantified the contribution of agrin-Lrp4/MuSK signaling during denervation and hence overlapped NMJ transcripts that remain at the NMJ after nerve cut to BoTX injection and Musk KO. As expected, BoTX injection showed a very similar response to nerve cut (Fig. 3e). Musk KO lead to a reduction of ~36.5% of these “denervation resistant” transcripts (Fig. 3e). This confirms that agrin persisting in the synaptic basal lamina after denervation is sufficient to promote expression of certain NMJ genes.
In conclusion, we generated a large dataset that describes the transcriptional changes in NMJ myonuclei upon experimental manipulations that affect innervation, electrical activity and/or agrin-Lrp4/MuSK signaling. These data confirm that agrin-Lrp4/MuSK signaling plays a critical role in the control of postsynaptic gene expression but they also suggest the presence of additional trophic factors that contribute to this.
Selection and confirmation of NMJ genes
Similar to other snRNA-seq datasets2,3,4, we detected numerous uncharacterized NMJ-specific and -enriched transcripts. As we were interested in better understanding the mechanisms involved in NMJ maintenance, we selected transcripts that were specific to NMJ myonuclei in innervated muscle, that were not re-expressed by body myonuclei in denervation, and whose expression was downregulated by Musk KO (Supplementary Fig. S4a, b). We ultimately selected three genes, Lrtm1, Pdzrn4, and Etv4 for a detailed functional analysis based on their expression pattern, their consistent appearance in snRNA-seq datasets2,3,4, and some additional functional criteria. Lrtm1, which was upregulated after sciatic nerve transection and BoTX injection specifically in NMJ myonuclei (Supplementary Fig. S4a), encodes a member of the leucine-rich repeat transmembrane protein family, which have been implicated in neural development56. The second candidate was Pdzrn4 (Supplementary Fig. S4a), which encodes a PDZ domain-containing protein paralogous to PDZRN3 that is expressed in body and NMJ myonuclei and has been shown to bind to MuSK and target it for degradation in cultured myotubes57. Pdzrn4 was downregulated after sciatic nerve transection and Musk KO but not after BoTX injection, indicating that it may be partially regulated by a trophic factor other than agrin. Finally, we selected Etv4, a member of the ETS family of transcription factors that have been implicated in the regulation of NMJ gene expression19. Interestingly, Etv4, similar to Etv5, showed a tendency to be downregulated in all our experimental paradigms (Supplementary Fig. S4a) indicating that they are regulated by the same factors.
First, we confirmed expression of the transcripts near NMJ myonuclei by using smFISH. Etv4, Lrtm1 and Pdzrn4 transcripts were highly enriched near NMJ myonuclei (identified by probes against Chrne) of both, fast-twitch TA and slow-twitch soleus muscles (Fig. 4a, b). All three transcripts were also enriched in NMJ regions isolated from TA muscle by laser capture microdissection58 (Fig. 4c). To assure that we would study the correct protein isoform of the three candidate genes, we used bulk RNA-seq data for TA and soleus58 to determine the predominant splice variants expressed in skeletal muscle (Fig. 4d, Supplementary Fig. S4c−e). Reads mapping to exon junctions showed the existence of different transcripts for Etv4 and Lrtm1. Interestingly, Pdzrn4 encodes two different transcripts with alternative transcription start sites, only one of which was detected in muscle (Fig. 4d, Supplementary Fig. S4d). This transcript codes for a protein that lacks the N-terminal RING domain and hence makes it unlikely that PDZRN4 acts as an E3 ubiquitin ligase as was described for PDZRN357. In conclusion, the NMJ transcripts Etv4 and Lrtm1 undergo alternative mRNA splicing with a preponderance of one particular splice variant and for Pdzrn4, only one transcript version was found.
a and b single molecule fluorescent in situ hybridization (smFISH) on TA and soleus cross-sections from adult male C57BL/6 mice. NMJ myonuclei were identified by the presence of Chrne transcript. c Box-plots showing average transcripts per million (TPM) values from laser-capture-isolated NMJ regions and non-NMJ regions (xNMJ)39. d Representative Sashimi plots generated from bulk-RNA-seq data of TA and soleus39. For Pdzrn4 and Lrtm1, all exons (e) are depicted, and for Etv4, only three exons are depicted (a region that undergoes alternative mRNA splicing). Exons are portrayed as blue boxes with the aligned reads in red. The numbers in-between splice junctions show the proportion of how frequently a particular overlapping read was detected. Exons that can be spliced alternatively at their 3’ or 5’ end are indicated as alternative exons (ae). (a) and (b) were independently repeated twice on different muscles with similar results. The box-plots in (c) show the median with the lower and upper quartiles, and the whiskers show the minimum and maximum values. A regular t-test was performed (p < 0.05 = *p < 0.01 = **p < 0.001 = ***). xNMJ and NMJ n = 8. Source data and p-values are provided as a Source Data file.
PDZRN4 overexpression mislocalizes NMJ proteins, and ETV4 induces NMJ gene expression
To characterize protein localization and function of the candidate genes in mouse skeletal muscle, we next overexpressed the most dominantly expressed muscle splice versions of ETV4, LRTM1 and PDZRN4 by intra-muscular injection of myotropic AAVMYO52 into the TA (Fig. 5a). Expression was driven by the CMV promoter and proteins were FLAG-tagged at their C-terminal end (Fig. 5a). Lower hindlimb muscles were collected one and two weeks post injection. The overexpressed proteins were localized by IF staining on whole-mount preparations (Fig. 5b) and cross-sections (Supplementary Fig. S5a). All proteins were enriched at the NMJ as visualized by AChR labeling with α-bungarotoxin (Fig. 5b). LRTM1 localized to the plasma membrane as shown by IF staining and cellular fractionation (Fig. 5b, Supplementary Fig. S5a, b). When ETV4 was overexpressed for one week, most myonuclei were positive for the FLAG-tag as shown by single fiber staining (Supplementary Fig. S5c). PDZRN4 accumulated around nuclei and formed puncta that appeared as strings within muscle fibers (Fig. 5b), These strings were organized similar to myofibrils in cross-sections (Supplementary Fig. S5a). Importantly, we detected intramuscular AChR and MuSK puncta that were associated with the overexpressed PDZRN4 (Fig. 5c, Supplementary Fig. S5d). Although PDZRN4 could be visualized throughout the entire muscle fiber, these MuSK- and AChR-positive puncta remained in close proximity of the NMJ, suggesting gene transcription and protein translation for AChR and MuSK remains specific to NMJ myonuclei.
a Experimental scheme; AAVMYO containing either Etv4, Lrtm1 or Pdzrn4 coding sequence was injected into the TA of wild-type 12 week old male mice (2.5 × 1010 vector genome copies). All three transcripts were driven by a CMV promoter and were FLAG-tagged at the C-terminal end. b One-week post injection, whole-mount preparations of the extensor digitorum longus (EDL) were stained with DAPI, α-bungarotoxin (AChR) and an anti-FLAG antibody. c Whole-mount preparations of Pdzrn4- and PBS-injected EDL muscles. The overexpression of PDZRN4 leads to extrasynaptic localization of AChRs and MuSK. d Principle component (PC) analysis of bulk RNA-seq data from TA muscles overexpressing either Etv4, Lrtm1 or Pdzrn4. e Venn diagrams comparing NMJ transcripts (from Fig. 1) with up- or downregulated transcripts induced by overexpression of the indicated gene. Numbers in red/blue indicate NMJ enriched/depleted transcripts or upregulated/downregulated genes, respectively. Numbers in black represent oppositely regulated genes. f Heatmap generated from bulk RNA-seq depicting how NMJ and denervation-associated genes are changed by overexpression. g SmFISH of NMJ marker Chrne combined with an IF staining for laminin on EDL cross-sections. The experimental scheme in (a) was created with BioRender. Both (b) and (c) were repeated twice with similar results. In (e), for statistical analysis a Wald test (two-sided) was performed. Up- and down-regulated transcripts fulfill p-adj. <0.05 and Log2FC ± 0.25. The experiment in (g) was performed once, on different muscles, with similar results.
Next, we analyzed the muscle transcriptome using bulk-RNA-seq in all samples. According to principle component (PC) analysis and the number of differentially expressed genes, both ETV4 and PDZRN4 strongly affected the muscle transcriptome, while LRTM1 had little effect (Fig. 5d, Supplementary Fig. S6a). Transcriptional changes were greatest with ETV4, increasing expression of more than 3000 transcripts and decreasing a similar number, followed by PDZRN4, which increased expression of 2000 transcripts and reduced more than 1000 (Supplementary Fig. S6a). To get an idea as to how the NMJ might be affected, we compared the list of up- and downregulated genes to that of the NMJ transcripts (from Fig. 1, Supplementary Data 3). Strikingly, overexpression of ETV4 resulted in increased expression of more than 50% of the NMJ transcripts (Fig. 5e, Supplementary Data 8). These transcripts included well-characterized transcripts, such as Chrne, Chrna1, Musk, Ache and Lrp4 (Fig. 5f). Importantly, the upregulation of NMJ genes in ETV4-overexpressing muscles was not a consequence of denervation, as denervation associated genes such as Chrng, Myog and Myod143,59 were not upregulated (Fig. 5f). In contrast, LRTM1 and PDZRN4 increased only 1.7% and 15% of the NMJ transcripts, respectively (Fig. 5e), and none of the well-known NMJ transcripts were increased in these two samples (Fig. 5f). To test, whether ETV4 increased the expression of NMJ transcripts in all myonuclei or specifically in NMJ myonuclei, we used smFISH against Chrne and Ufsp13. In PBS-injected muscle, these transcripts were expressed at only few sites, consistent with NMJ-restricted expression. However, in muscles overexpressing ETV4, widespread Chrne and Ufsp1 expression was detected (Fig. 5g, Supplementary Fig. S6b, c). Despite this increase in NMJ transcripts, no ectopic AChR clusters were observed. The absence of such clusters may be due to ETV4 not enhancing all transcripts required for cluster formation; for example, bulk-RNA-seq showed no increase in Chrnd expression (Supplementary Data 8). To test whether the ETV4 overexpression would sensitize the formation of ectopic AChR cluster formation, we denervated the muscle for one week by sciatic nerve transection followed by injection of either PBS or AAVMYO encoding ETV4. After a total of two weeks, only a few ectopic AChR clusters were observed in extensor digitorum longus (EDL) muscle under control conditions while muscles overexpressing ETV4 exhibited numerous ectopic clusters (Supplementary Fig. S6d). All in all, these experiment demonstrate that ETV4 is sufficient to stimulate an NMJ-specific transcriptional program in body myonuclei, supporting the idea that a large portion of NMJ transcripts are regulated by ETS transcription factors60.
As the top DAVID GO-term in the bulk RNA-seq samples from muscles overexpressing either ETV4 or PDZRN4, but not LRTM1, was “immune system process” (Supplementary Fig. S6e), we asked whether prolonged expression would affect muscle health. Indeed, muscle fibers overexpressing PDZRN4 or ETV4 for two weeks showed signs of muscle degeneration and regeneration, indicated by the presence of mouse IgG inside of the muscle fibers and of many centrally located myonuclei (Supplementary Fig. S6f). These results indicate that long-term ectopic expression of proteins that affect the expression (ETV4) or localization (PDZRN4) of proteins confined to the NMJ (i.e. MuSK and AChR) is not well tolerated. The fact that LRTM1 overexpression does not result in such a muscle response shows that the pathological changes are not caused by the overexpression of any protein or are a response to AAVMYO transduction.
PDZRN4 localizes to the Golgi apparatus and binds to MuSK
As PDZRN4 overexpression caused AChR and MuSK mislocalization and little is known about the protein in the context of the NMJ, we focused on elucidating the function of PDZRN4 at the NMJ. IF stainings showed PDZRN4 localizing around nuclei and forming puncta that appeared in strings resembling organelle-like structures (Fig. 5b, Supplementary Fig. S5a, d). To explore which organelle PDZRN4 associates with, PDZRN4 was overexpressed in HeLa cells and co-stained with markers for the endoplasmic reticulum, lysosomes, mitochondria, and the Golgi apparatus (Supplementary Fig. S7a). While we detected a partial overlap of the overexpressed PDZRN4 with all the organelles, only the Golgi marker GM130 co-localized perfectly with the FLAG staining. Strikingly, even when the Golgi apparatus was disassembled during mitosis, there was strong co-localization with PDZRN4 (Supplementary Fig. S7b). As the Golgi apparatus in muscle fibers not only localizes near nuclei, but extends along the microtubule network61,62,63,64, we next stained PDZRN4-overexpressing muscle fibers for the Golgi-resident protein glucose transporter type 462,63 (GLUT4) and the FLAG tag. Indeed, the anti-FLAG and anti-GLUT4 antibodies stained the same structures (Fig. 6a). We also noted that overexpression of PDZRN4 significantly increased the size of the Golgi puncta (Fig. 6b). Intriguingly, at the NMJ, the overexpressed PDZRN4 also co-localized with postsynaptic AChR clusters (Fig. 6c).
a Whole-mount staining of EDL muscle shows co-localization of PDZRN4 (FLAG) and the Golgi apparatus, stained with antibodies to glucose transporter type 4 (GLUT4). b The GLUT4-positive puncta in muscles overexpressing PDZRN4 are significantly larger compared to PBS-injected muscle. c At the NMJ, PDZRN4 (FLAG) co-localized with GLUT4 and with postsynaptic AChR clusters. For better visualization only the surrounding of GLUT4 and AChR staining is shown in the bottom right image. d Immunoprecipitation (IP) was conducted in HEK 293T cells overexpressing PDZRN4 and/or MuSK. IP against the FLAG tag of PDZRN4 resulted in co-pull down of both MuSK and GM130 whereas the Golgi-associated proteins TSPT2 and TGOLN2 were not detected. e MuSK and PDZRN4 were overexpressed in HeLa cells to study how PDZRN4 affects MuSK localization. To quantify MuSK levels at the Golgi, MuSK staining intensity was determined at the Golgi region. MuSK staining at the Golgi was compared to its total staining across the entire cell. f Quantification of the relative presence of MuSK at the Golgi apparatus in control and upon PDZRN4 overexpression. g The PDZRN4 isoform expressed in muscle contains a conventional and a truncated PDZ domain (tPDZ). Each domain was removed to investigate their respective contributions to MuSK interaction. h IP experiment using the deletion mutants of PDZRN4 shown in (g). The interaction between MuSK and PDZRN4 is independent of the conventional PDZ domain but is weakened by the deletion of the tPDZ domain. In (b) each data point represents the mean value of one mouse (n = 4). In (f) each data point represents the average of at least 15 cells from one cell culture well (n = 3 wells). The data in (b, f) is presented as mean ± SEM and a two tailed t-test was performed (p < 0.05 = *p < 0.01 = **p < 0.001 = ***). The IP in (d) was repeated once with similar results and (h) was performed once. Source data and p-values are provided as a Source Data file.
To test whether PDZRN4 interacts with postsynaptic structures and to validate the intracellular co-localization of MuSK and AChRs (Fig. 5c), we next tested whether PDZRN4 would co-immunoprecipitate (IP) with MuSK. Indeed, when MuSK and FLAG-tagged PDZRN4 were both overexpressed in HEK 293T cells, MuSK was co-enriched in the anti-FLAG IP (Fig. 6d). While the Golgi protein GM130 was also present in the IP, two other membrane proteins of the Golgi apparatus, tyrosylprotein sulfotransferase 2 (TPST2) and trans-golgi network protein 2 (TGOLN2), could not be detected (Fig. 6d). Thus, the presence of GM130 in the IP is not due to some remnants of the Golgi apparatus but rather suggests that this protein might be part of the PDZRN4-MuSK complex. To test this, we co-overexpressed and stained both MuSK and PDZRN4 in HeLa cells. IF staining revealed a significant accumulation of MuSK at the Golgi apparatus together with PDZRN4 (Fig. 6e, f).
To map the binding site of PDZRN4 to MuSK, we next generated deletion mutants. The Pdzrn4 transcript expressed in skeletal muscle encodes one conventional PDZ domain as well as a truncated PDZ domain (called herein tPDZ), close to its N-terminus (Fig. 6g, Supplementary Fig. S7c). MuSK binding in the PDZRN3 paralog has been mapped to its PDZ domain57. To test whether this would also be the case for PDZRN4, the PDZ and the tPDZ domain were removed individually as well as in combination. While removal of the conventional PDZ domain did not affect co-IP of MuSK, removal of the tPDZ domain and of both domains strongly lowered the amount of MuSK (Fig. 6h). Importantly, the deletion of the tPDZ domain strongly affected PDZRN4 association with the Golgi apparatus as observed in IF stainings in HeLa cells (Supplementary Fig. S7d). To test if removal of any other region in PDZRN4 would affect MuSK association, five additional regions of PDZRN4 were removed individually (Supplementary Fig. S7e). Co-IP depicted that all PDZRN4-FLAG versions were able to pull down MuSK, though the N-terminal region appeared to be of higher importance (Supplementary Fig. S7f). In conclusion, these results indicate that PDZRN4 associates with MuSK at the Golgi apparatus. Furthermore, our data show that PDZRN4 does not require its PDZ domain to interact with MuSK, making it distinct from PDZRN357.
Muscle-specific knockout of Pdzrn4 causes NMJ fragmentation
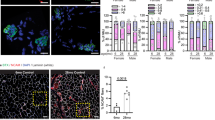
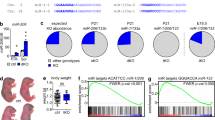
While overexpression studies test for protein sufficiency, addressing essentiality requires their removal from the organism. To see whether Etv4, Lrtm1, or Pdzrn4 were required to maintain NMJ function and structure, we used our recently established muscle fiber-specific AAVMYO-CRISPR/Cas9 system51 and designed six to seven sgRNAs for each gene (Supplementary Fig. S8a, Supplementary Data 9). Upon cloning of all sgRNAs targeting one gene into a single AAV vector, we delivered them using AAVMYO52 to mice expressing Cas9 in skeletal muscle fibers (Cas9mKI). We injected 3 × 1011 sgRNA-encoding AAVMYO (~9 × 1012 vector genomes/kg) via the tail vein into Cas9mKI mice to generate systemic muscle fiber-specific knockouts (Fig. 7a). Fifteen weeks post injection, mice were analyzed, and transcript levels for the targeted genes were measured in TA by qPCR. In all cases, targeted transcripts were significantly lower than in control TA (Fig. 7b). Interestingly, Musk mRNA was also significantly downregulated by KO of Etv4, Lrtm1, and Pdzrn4 (Fig. 7b; right), suggesting an important contribution of all genes to synaptic signaling. The extent of transcript loss differed between the targeted genes, which can be due to different extents of nonsense-mediated mRNA decay and mRNA expression in muscle-resident cells other than muscle fibers. For example, Etv4 is also expressed by tenocytes (Supplementary Fig. S4b). Functionality of the neuromuscular system was measured using all-limb and forelimb grip strength as well as balance beam performance 10-weeks post injection. None of these measures differed between the KO and controls (Fig. 7c, Supplementary Fig. S8b). Similarly, no significant decrement in action potential was observed after 10 stimulations at 40 Hz frequency using compound muscle action potential (CMAP) measurements 15-weeks post sgRNA injection (Fig. 7d). In conclusion, KO of Etv4, Lrtm1 or Pdzrn4 in adult skeletal muscle fibers does not result in a significant deficit in neuromuscular performance 15-weeks post injection. It is important to note that the NMJ is characterized by its high safety factor, which assures reliable generation of an action potential even at low AChR density65. Hence, NMJs could still be affected at the molecular level without functional consequences at this 15-week time point. To investigate whether morphological changes could be observed, we performed whole-mount IF stainings of the EDL. Imaging revealed that both Etv4 and Lrtm1 KO mice showed non-significant trends towards a higher number of AChR fragments (Fig. 7e, f). In Pdzrn4 KO mice, the number almost doubled and reached high significance (Fig. 7g). As NMJ function was seemingly not altered in any of the knockouts, it remains an open question whether NMJ fragmentation could be an early sign for its instability. It is well known that NMJ fragmentation in rodents is a hallmark of aging although this structural change also does not result in functional impairments66.
a Adult Cas9mKI mice were injected with AAVMYO encoding 6 (Lrtm1) or 7 (Etv4, Pdzrn4 and Pdzrn3) different sgRNAs (3 × 1011 vector genome copies) via the tail vein. Mice injected with Lrtm1 or Pdzrn4 sgRNAs were female and mice injected with Etv4 sgRNAs were male. b Relative mRNA expression, measured by qPCR, of the targeted genes in TA muscle 15 weeks post-injection. Note that Musk mRNA was also significantly reduced in all three groups. c All-limb and forelimb grip strength measurements were performed on mice 10 weeks post injection. d CMAP was performed on the GAS 15 weeks post-injection at different frequencies (image shows average values at 40 Hz frequency). e, f, and g representative whole-mount images of NMJs from the EDL from Etv4 (e), Lrtm1 (f), and Pdzrn4 (g) knockout (KO) mice and control littermates. AChRs are visualized by α-bungarotoxin and the presynapse by neurofilament (NF) and synaptophysin (SYP). Bar graphs next to the images show the quantification of AChR fragmentation. NMJs of Pdzrn4 KO mice are significantly fragmented. h Relative mRNA expression of the targeted genes in TA muscle 15 weeks post-injection of AAVMYO encoding sgRNA to Pdzrn3 or Pdzrn4. Double knockout for Pdzrn3 and Pdzrn4 (Pdzrn3/4 dKO) was achieved by co-injection of the two AAVMYOs. i Representative whole-mount images of NMJs from the EDL from Pdzrn3/4 dKO and control mice. j AChR clusters/NMJ were counted and quantified. Unlike the single Pdzrn4 KO, both Pdzrn3 KO and the Pdzrn3/4 dKO show no fragmentation of the postsynapse. The experimental scheme in (a) was created in BioRender. The data in (b-j) is presented as mean ± SEM. In (b–j) a two-tailed t-test was performed. p < 0.05 = *p < 0.01 = **p < 0.001 = ***. For whole-mount quantifications, a minimum of 10 NMJs per mouse were quantified (n = 4 for Etv4, Lrtm1, Pdzrn3, Pdzrn3/4 dKO and n = 5 for Pdzrn4 KO). Source data and p-values are provided as a Source Data file.
Overexpression of PDZRN3 in C2C12 myotubes reduces agrin-induced AChR clustering and disrupts NMJ development in mice, presumably by increased internalization and degradation of MuSK57. Although enrichment of Pdzrn3 transcripts at the NMJ has been reported57, our snRNA-seq data demonstrate the widespread expression in all myonuclei, which is in stark contrast to the enrichment of Pdzrn4 transcripts at NMJ myonuclei (Supplemental Fig. S8c). While PDZRN3 has been shown to act as an E3 ubiquitin ligase towards MuSK57, the PDZRN4 isoform expressed by NMJ myonuclei lacks the ubiquitination RING domain (Fig. 4d, Supplementary Fig. S7c). As PDZRN4 has a strong affinity towards the Golgi, which is a central hub for post-translational modifications and protein sorting, we hypothesized that PDZRN3 and PDZRN4 may together modify and regulate MuSK protein at the post-translational level. The double KO of Pdzrn3 and Pdzrn4 may either exacerbate the NMJ phenotype if these genes act synergistically, or it could potentially reverse the NMJ fragmentation observed in the Pdzrn4 single KO if they function in opposition. To examine this, we designed seven sgRNA targeting Pdzrn3 (Supplementary Fig. S8d, Supplementary Data 9) and used the AAVMYO-CRISPR/Cas9 system51 to generate muscle-specific Pdzrn3 single and Pdzrn3/4 double KO. Successful targeting of the two genes was confirmed by a decrease in transcript levels (Fig. 7h). No significant differences were observed in grip strength and balance beam performance 10-weeks post injection (Supplementary Fig. S8e, f). To determine whether NMJ fragmentation was exacerbated or prevented by the Pdzrn3/4 double KO, we performed whole-mount IF staining of the EDL (Fig. 7i). Quantification of AChR clusters/NMJ showed that deletion of Pdzrn3 prevented the fragmentation caused by Pdzrn4 (Fig. 7j). Our observation that depletion of PDZRN3 in adult skeletal muscle does not affect NMJ fragmentation is consistent with recent data from others utilizing conventional, inducible muscle-specific knockout mice67. These results indicate that PDZRN4 may contribute to intracellular MuSK processing in the Golgi apparatus, opposite to the postulated role of PDZRN3 in MuSK degradation by ubiquitination.
Discussion
NMJ myonuclei represent a small population of nuclei that share one cytoplasm with hundreds of other myonuclei. Research over several decades has shown that these NMJ myonuclei express genes whose protein products aggregate at the postsynaptic side of the NMJ. Among those are essential NMJ components, such as Lrp4, MuSK, and the AChR subunits. Notable exceptions are Rapsn and Dok7, which are also essential NMJ components38,68, but their transcripts do not show increased expression in NMJ myonuclei compared to body myonuclei. During development, many NMJ genes are transcribed throughout the muscle fiber. However, the formation of the NMJ and the onset of nerve-evoked electrical activity causes NMJ myonuclei to become specialized for synaptic gene transcription6,19. The continuous expression of NMJ-essential genes in NMJ myonuclei throughout life suggests the presence of nerve-derived, local signals that maintain their transcription. In addition, the nerve-evoked electrical signal, which spreads within the muscle fiber, suppresses the expression of synaptic genes in body myonuclei19. Traditional methods, such as bulk RNA-seq in conjunction with microdissection of synaptic and non-synaptic regions, fall short in distinguishing between transcripts from NMJ nuclei and those from other cell types localized at synaptic regions; e.g., myelinating and terminal Schwann cells39. To overcome this limitation, we used snRNA-seq in combination with experimental manipulations of innervation and NMJ maintenance to elucidate the signaling pathways involved in synaptic gene expression.
We identified 472 transcripts that were significantly enriched in NMJ myonuclei. These included the well-characterized NMJ-associated genes Musk, Lrp4, Ache, Colq, Ncam1, Etv5, and the different Chrn subunits, which validated our approach. Some of the transcript were enriched in both NMJ and MTJ myonuclei (Fig. 1), suggesting structural and functional similarities between the two specializations. For example, transcripts for laminin-α2 (Lama2) and collagen IV (Col4a1 and Col4a2) were enriched in both nuclei populations. Laminin-α2 is the major laminin chain and collagen IV the main collagen of the basement membrane surrounding skeletal muscle fibers69. Interestingly, mutations in LAMA2 cause a severe, early-onset congenital muscular dystrophy70. The fact that transcripts for these proteins are enriched in NMJ and MTJ nuclei, indicates that there is an increased demand of stable basement membrane in these specialized regions. In contrast, other members of the laminin and collagen protein family, such as Lamb2, Col13a1, and Colq, were selectively transcribed in NMJ nuclei. Interestingly, mutations in all three genes cause congenital myasthenic syndrome71. Similarly, Col22a1 is selectively expressed in MTJ nuclei, and the protein is well-known to be a specific marker for MTJs72.
To gain deeper insights into the regulation of synaptic gene expression in NMJ myonuclei, we also generated snRNA-seq data from denervated TA and GAS muscles five days after sciatic nerve transection (Fig. 2) and following NMJ perturbations via BoTX injection (Fig. 3). BoTX prevents ACh release from the presynaptic nerve terminal by specifically cleaving SNAP-2573, causing chemical denervation while still allowing the nerve to provide “trophic support” to the postsynaptic region. Finally, we also perturbed the agrin-Lrp4/MuSK signaling pathway by depleting MuSK, which is essential for NMJ formation and maintenance6. Denervation by nerve transection resulted in the loss of synapse-specific expression of a subset of NMJ genes, primarily due to their re-expression in body myonuclei (Fig. 2). This phenomenon is well- documented and mechanistically has been explained by the loss of nerve-evoked electrical activity, which leads to the re-expression of myogenic regulatory factors, such as myogenin and myoD59,74,75. Unlike the NMJ-specific transcription factor ETV5, which binds to the ‘N-box’, myogenic regulatory factors bind to the ‘E-box’ domain to promote NMJ gene expression19. However, we found that a large proportion of synaptic transcripts remained specifically expressed in NMJ myonuclei, with some even downregulated during denervation, suggesting that they might not be regulated via an ‘E-box’ domain. We, therefore, hypothesized that the downregulation of synaptic genes could stem from either the loss of trophic factors provided by the nerve or the loss of electrical activity.
To test this, we performed snRNA-seq on chemically denervated muscle by BoTX injection, where the motor nerve terminal remains at the NMJ53,54. Approximately 27% of NMJ genes downregulated by nerve transection were also downregulated by BoTX (Fig. 3), indicating that nerve-evoked electrical activity is important for maintaining synaptic gene expression. However, it is important to note that cleavage of SNAP-25 would likely also prevent the release of other synaptic vesicle contents besides ACh, such as ATP76. Extracellular ATP has been shown to promote gene expression of Ache and AChR subunits via P2Y purinoceptor 1 in cultured myotubes77,78. Thus, a decrease in the expression of certain NMJ genes may be a consequence of factors other than loss of innervation alone. A DNA-binding protein, FUS, which shows reduced nuclear localization during denervation, might be implicated in the reduced expression of synaptic genes. FUS has been proposed to regulate synaptic genes alongside ETV5, and mutations of Fus can cause an aggressive form of ALS79. Furthermore, we observed that certain NMJ transcripts were “resistant” to denervation as they were not expressed by body myonuclei and maintained similar expression levels in NMJ myonuclei. We suggest that continued agrin-Lrp4/MuSK signaling might be the source for maintaining their expression.
To test this hypothesis, we deleted Musk using the AAV-CRISPR/Cas9 system. Four weeks post-sgRNA injection, when Musk protein was below the detection limit, ~36% of these “denervation resistant” synaptic genes were downregulated (Fig. 3e) and ~46% of all NMJ-enriched transcripts were reduced (Supplementary Fig. S3f). These data support the notion that MuSK is a key regulator of synaptic gene expression25,80. However, ~64% of the “denervation resistant” synaptic genes remained unchanged after Musk KO. While it remains possible that four weeks were not sufficient to completely abrogate agrin-Lrp4/MuSK signaling at all NMJs, our results also imply that other neurotrophic factors, including those that have previously been implicated in the regulation of synaptic gene expression (e.g., neuregulin-ErbB), may play a role in maintaining synaptic gene expression81,82,83,84,85. Muscle-specific Erbb2/Erbb4 double KO mice show only minor effects on the NMJ86,87, but transcriptional changes cannot be excluded as only very few transcripts have been analyzed in these knockouts.
To gain further insights into the molecular mechanisms of NMJ maintenance, we addressed the function of Etv4, Lrtm1, and Pdzrn4, three NMJ-specific genes whose function has not been studied. Using AAVMYO51, we overexpressed ETV4 in mouse muscle and show that it is sufficient to trigger a synapse-specific gene expression signature in body myonuclei by upregulating more than half of the NMJ-myonuclei transcripts (Fig. 5). These results indicate that a large portion of the synaptic genes are regulated through the ‘N-box’ domain60. Overexpression of LRTM1 and PDZRN4 had less impact on NMJ-specific gene expression; however, PDZRN4 led to partial mislocalization of MuSK in proximity of the NMJ (Fig. 5). Based on these observations and the ability of PDZRN3, a paralog of PDZRN4, to bind and degrade MuSK57, we hypothesized that PDZRN4 may also be involved in MuSK regulation. We found that PDZRN4 is strongly associated to the Golgi apparatus in heterologous cells and in muscle fibers (Fig. 6 and Supplementary Fig. S7) and that PDZRN4 co-immunoprecipitated with MuSK when co-expressed in HEK 293T cells (Fig. 6). Interestingly, removal of the PDZ domain of PDZRN4 did not abolish MuSK interaction, distinguishing it from PDZRN3. Furthermore, the overexpression of PDZRN4 increased the relative amount of MuSK at the Golgi, where proteins can undergo various modifications, including acetylation, sulfation, phosphorylation, palmitoylation, O-glycosylation, and modification of their N-glycosylation88. PDZRN4 may hence be involved in the processing of MuSK in the Golgi apparatus near the NMJ, altering the post-translational modifications of its N-glycosylation89. Notably, the overexpression of PDZRN4 resulted in the appearance of a low molecular mass band for MuSK as detected by Western blot analysis. Interestingly, this lower molecular mass form of MuSK preferentially bound to PDZRN4 (Fig. 6h, Supplementary Fig. S7f). These results in conjunction with the increase of MuSK levels at the Golgi apparatus upon PDZRN4 overexpression, suggest that PDZRN4 and MuSK interact mainly in this organelle and that PDZRN4 regulates posttranslational MuSK modifications.
To test whether Etv4, Lrtm1, or Pdzrn4 contribute to NMJ maintenance, we generated muscle-specific knockouts using our recently established AAVMYO-CRISPR/Cas9 system51. Depletion of ETV4 in adult skeletal muscle did not affect NMJ function or structure. This lack of effect may be due to compensation by other ETS transcription factors, including ETV5 and GABP, both of which are expressed in skeletal muscle. Interestingly, whole-body knockout of Etv5 leads to an NMJ phenotype26. Etv5 is expressed at much higher levels than Etv4 (based on LCM data39) and as both can bind to the same ‘N-box’ domain90, Etv4 may be redundant for NMJ maintenance in the presence of Etv5. Similarly, knockout of Lrtm1 did not significantly affect muscle function or NMJ morphology, suggesting LRTM1 may also be compensated by similar proteins, as many other transmembrane proteins possess extracellular leucine-rich repeats56. Alternatively, LRTM1 may not participate in NMJ maintenance, as hypothesized, but could act as an auxiliary protein for potassium channels. A group of leucine-rich repeat-containing membrane proteins have been shown to cause negative shifts in potassium channel voltage dependence91. The most striking phenotype at the NMJ was caused by the knockout of Pdzrn4, which led to an almost doubling of fragmentation (Fig. 7g). The fact that we could not detect changes in function, tested as grip strength and fatigue resistance, is not surprising as NMJ fragmentation does not necessarily cause functional impairments66. Thus, while overexpression of the candidate genes resulted in quite striking phenotypes – in case of ETV4 and PDZRN4 - their knockout was rather mild with the exception of Pdzrn4. As a notable difference to most knockout experiments, our approach assesses gene function in adult mice and not during development. Moreover, the AAVMYO-CRISPR/Cas9 method employed here is muscle fiber-specific, whereas the proposed role of ETV5 in NMJ transcription is based on analyses of whole-body knockouts26.
We also investigated the functional relationship of PDZRN4 and PDZRN3 by generating double knockouts. Our results show that there is no compensatory interaction between the two proteins. Instead of exacerbating the NMJ fragmentation observed in Pdzrn4 KO mice, the simultaneous deletion of Pdzrn3 prevented this fragmentation (Fig. 7i, j). This suggests that the two paralogs, despite both binding to MuSK, may have opposing roles. While PDZRN3 has been implicated in promoting MuSK endocytosis and degradation57, our data indicate that PDZRN4 is predominantly localized to the Golgi apparatus, pointing to a potential role in MuSK processing. Consistent with this hypothesis, Pdzrn4 KO results in a structural NMJ deficit, whereas Pdzrn3 KO does not affect NMJ structure67. We propose that the absence of NMJ fragmentation in the Pdzrn3/4 double KOs arises from a balance where the impaired MuSK processing caused by the loss of PDZRN4 is offset by reduced MuSK degradation in the absence of PDZRN3. This interplay between the two proteins underscores their distinct yet interconnected roles in regulating MuSK stability and function.
Notably, MuSK glycosylation does not significantly influence its activation by neural agrin in cultured myotubes but does affect its self-activation89. Therefore, it is plausible that PDZRN4 is involved in the post-translational modification of additional proteins beyond MuSK, as evidenced by the presence of mislocalized MuSK vesicles containing AChRs (Fig. 5c, Supplementary Fig. S5d). Finally, the NMJ-specific expression of Pdzrn4 is confirmed by snRNA-seq data from human skeletal muscle, further supporting its critical role in NMJ maintenance and function92.
In summary, we performed snRNA-seq on skeletal muscle and identified numerous previously uncharacterized transcripts in NMJ myonuclei. We studied synaptic gene expression in NMJ and body myonuclei in denervation and found that synaptic gene expression relies not only on neurotrophic factors but also on electrical activity. Collectively, our snRNA-seq data provide a valuable resource that can be explored online on our interactive atlas (https://ruegglab.shinyapps.io/snatlas/)32. Furthermore, we characterized Etv4, Lrtm1, and Pdzrn4 in muscle through overexpression and knockout experiments, identifying PDZRN4 as a MuSK interactor at the Golgi apparatus while ETV4 was sufficient to induce a synaptic gene expression program in body myonuclei.
Methods
Mice
The mice were kept at the local animal facility (Biozentrum, Basel) under a 12-hour light to dark cycle, with ad libitum access to standard laboratory chow and water. All animal studies were performed in accordance with the law and guidelines of the Swiss authorities and were approved by the veterinary commission of canton Basel-Stadt (Schlachthofstrasse 55, 4056 Basel, Switzerland). All mice used in the experiments were 2-14 months of age. Within an experimental group, the maximum age difference between mice was ± 40%. Muscle-specific Cas9 mice (Cas9mKI) were generated by crossing Cas9 knock-in mice93 with HSA-Cre mice94.
Muscle function
Grip strength was measured using a force meter (Columbus Instruments) attached to either a triangular bar (for forelimbs) or a grid (for all limbs). Mice were held close to the bar or on top of the grid until they grasped it firmly and were then gently pulled away horizontally at a constant speed until they lost their grip. The measurement was repeated at least five times with short breaks in between, and the median of all values was used as the final performance.
For the balance beam experiments, mice were trained one day prior to the actual tests. The mice were placed on a small platform where they learned to walk up across a 12 mm wide square metal bar to another platform with a red Plexiglas house. The next day, the mice were again placed on the small platform, and the time to cross a 10 mm bar over a set 80 cm distance was measured. This was repeated three times with 30 s rest periods in between. The best value over the three trials for each mouse was used as the final performance.
CMAP measurements
CMAP was performed similarly to a previous publication39. Briefly, mice were anaesthetized, and the sciatic nerve was stimulated by trains of 10 stimulations of increasing frequencies ranging from 3 Hz to 100 Hz. Action potentials were recorded using a needle electrode inserted into the GAS muscle, with a reference electrode placed subcutaneously for grounding. Potential decrements after repetitive stimulations were quantified by measuring the change between the first stimulation and subsequent ones.
Surgical denervation and BoTX injection
Mice were anaesthetized by isoflurane inhalation, and a small incision on the skin near the sciatic nerve was made. The sciatic nerve was raised using a glass hook and a small 5 mm piece was excised. Surgical clips were used to close the wound, and the mice were returned to their cage. The mice received analgesia (Buprenorphine, 0.1 mg/kg) 1 h before, 4–6 h after and twice on the next day of the surgery.
When denervated by botulinum toxin (BoTX), mice were anaesthetized by isoflurane inhalation and received an intra-muscular injection of 25 µl Dysport® (40 U/ml in saline solution) into the TA, as previously described95. BoTX control mice were injected with 25 µl of saline solution.
AAV administration
For intra-muscular injections, one leg was shaved with an animal trimmer, and the region of the TA was disinfected. For overexpression experiments, AAVs were diluted in PBS to a concentration of 5 × 1011 vector genome copies (vg) per ml, and 50 µl (i.e. 2.5 × 1010 vg) were injected into the TA muscle. To knockout Musk in mouse muscle for snRNA-seq, 50 µl of 1.5 × 1012 vg/ml (i.e. 7.5 × 1010 vg) were injected into the TA. To generate systemic knockouts, mice were injected with 100 µl of 3 × 1012 vg/ml (i.e. 3 × 1011 vg) via the tail vein. For double knockouts, both Pdzrn3 and Pdzrn4 sgRNA encoding AAVs were pooled and injected at a concentration of 3 × 1011 vg each. All injections were made with a 30 G 0.3 ml insulin syringe, and the mice were anaesthetized by isoflurane inhalation.
Single guide RNA design and vector assembly
The sgRNAs targeting Musk were designed and tested previously51. The single guides targeting Etv4, Lrtm1, Pdzrn4, and Pdzrn3 were selected using CRISPOR96 and Geneious Prime® (version 2020.2.3) and are listed in Supplementary Data 9. Six to seven different guides were assembled into one plasmid using a multiplex CRISPR/Cas9 assembly method97. The guides were then cloned into an AAV transfer vector containing ITRs, as described previously51.
AAV production and purification
AAVs were produced as previously described51. Briefly, HEK293T (ATCC CRL-3216) cells were transfected with AAVMYO52, pAdDeltaF6 helper (a gift from J. M. Wilson, Addgene, #112867), and an AAV transfer plasmid containing an overexpression construct or sgRNA expression cassettes. Two days’ post-transfection the cell culture medium containing AAVs was collected. Three days’ post-transfection HEK293T cells were centrifuged, and the supernatant was pooled with the cell culture medium from the previous day. HEK293T cells were lysed with AAV lysis solution (50 mM Tris-HCl, 1 M NaCl, 10 mM NgCl2, pH 8.5) and 50 U of salt active nuclease (SRE0015-5KU, Sigma) for 1 h at 37 °C. The lysate was centrifuged at 3200 x g, 15 min at 4 °C, and the supernatant containing AAVs was collected. The cell culture medium containing AAVs was mixed with polyethylene glycol 8000 (89510, sigma) to a concentration of 8% (w/v) and incubated for 2 h at 4 °C followed by centrifugation at 4000 x g, 30 min at 4 °C. The AAV pellet was resuspended with 1 ml AAV lysis solution and pooled with the cell lysate. To purify AAVs the entire lysate was loaded onto a 15-25-40-60% iodixanol (Serumwerk) gradient and centrifuged at 292,000 x g (Beckman type 70 Ti rotor) for 2 h at 4 °C. Purified AAVs were collected from the 40% iodixanol phase and extracted via buffer exchange using a 100 kDa MWCO filter (Millipore). The concentration of the virus was measured by qPCR using forward (fw) primer GGAACCCCTAGTGATGGAGTT and reverse (rv) primer CGGCCTCAGTGAGCGA targeting AAV2 ITRs as previously described98.
Single nuclei isolation for snRNA-seq
Every step of single nuclei isolation was performed on ice or at 4 °C. The nuclei were always isolated from freshly dissected muscle that was kept in ice-cold PBS for a maximum 1 h after dissection. GAS snRNA-seq results were obtained from one muscle, and TA results were always a pool of two TA muscles. The tissue was individually minced with scissors (14084-08, FST) for 3 min in 2 ml microcentrifuge tubes together with 200 µl single nuclei lysis buffer (250 mM sucrose, 25 mM KCl, 4 mM MgCl2, 10 mM Tris-HCl pH 8 and 0.1% NP40 in nuclease-free water). The minced muscle was transferred to a 7 ml dounce homogenizer (D9063, Sigma-Aldrich) with an additional 3 ml single nuclei lysis buffer and carefully dounced with 8–10 strokes. The muscle slurry was transferred to a 50 ml falcon tube, and the dounce homogenizer was rinsed with 3 ml single nuclei wash buffer (1% BSA in PBS), which was pooled with the previous fraction. The muscle slurry was filtered through 70 µm (352350, Falcon) and 30 µm (130-098-458, Miltenyi Biotec) cell strainers to remove large pieces of tissue and cell debris. The filtered lysates were then centrifuged for 5 min at 500 x g in 50 ml falcon tubes, and the supernatant was removed, leaving −200–300 µl liquid in the tube. The pellet was resuspended with 1 ml of single nuclei wash buffer containing 0.2 U/µl RNase inhibitor (3335402001, Roche) and transferred to a microcentrifuge tube. After adding 1 µl Hoechst solution, the single nuclei suspension was centrifuged again for 5 min at 500 x g. Next, the entire supernatant was removed, and the nuclei pellet was resuspended in 100–400 µl of single nuclei wash buffer with RNase inhibitor, depending on the amount of starting tissue. The quality and concentration of nuclei were controlled on a fluorescent microscope using a hemocytometer (concentration would optimally range from 500–1500 nuclei/µl).
Single nuclei library construction and sequencing
To generate single nuclei libraries 10x Genomics Chromium Next GEM Single Cell 3’ kits (PN-1000121, PN-1000120, PN-1000213) and the corresponding protocol were used. We always aimed for 10,000 nuclei libraries per sample, which is the maximum recommended by the manufacturer. For the most part, the manufacturer’s instructions were followed, the only variables that had to be adjusted were the cDNA amplification and sample index PCRs (steps 2.2 and 3.5 respectively) where 12 cycles were used in both cases. Quality control was performed on an Agilent Fragment Analyzer using the HS NGS Fragment Kit (1-6000 bp). cDNA ranging from 300 to 3000 bp with a peak around 1000 bp was considered good quality. After library construction, samples were quality-controlled again and were sequenced paired-end (PE) 28/91 on a NovaSeq 6000 system (Illumina). We aimed to reach >30,000 reads per nucleus for all samples. Sequencing and quality control were performed by the genomics facility at the D-BSSE (ETH, Basel, Switzerland).
Single nuclei RNA-seq alignment, clustering, and analysis
Demultiplexing was performed with bcl2fastq (version 2.20) and was done by the genomics facility at the D-BSSE (ETH, Basel, Switzerland). FASTQ files were aligned to mouse reference mm10 (GENCODE vM23/Ensembl 98, from 2020) using cellranger count version 5.0 with the argument –include-introns. As a quality control measure, we looked at the percentage of reads mapped to intronic regions, which should range around 50–60% for nuclei.
Cellrangers output folders “filtered_feature_bc_matrix” were loaded into R (version 4.0.3) using Seurats (version 4.0.0) Read10x function. Nuclei expressing fewer than 500 features and more than 5% of mitochondrial features were removed. Next, nuclei were clustered following the fast integration (RPCA) workflow (https://satijalab.org/seurat/articles/integration_rpca). Any low-quality clusters (low feature count and high expression of mitochondrial genes) or multiplets (co-expression of several cell type-specific markers) were removed manually. After removal, the remaining nuclei were normalized and clustered again using RPCA with SCTransform normalization. For differential expression between clusters and between conditions first NormalizeData() was used on the “RNA” assay, and then the FindMarkers() function was run. For visualization Seurats DimPlot(), FeaturePlot(), VlnPlot(), and DotPlot() were used.
To generate the list of synaptic transcripts, we compared the NMJ cluster to clusters Type IIA, Type IIX, and Type IIB. To be more stringent we set a threshold for the adjusted p-value to <5E−5 and the log2FC to ±0.25. The same procedure was done to generate a list of differentially expressed genes for the MTJ cluster. To see which transcripts are changed in denervation (by nerve cut), after BoTX injection and Musk knockout, the FindMarkers() function was used and we set a threshold for the p-value to <0.01 and the log2FC to ±0.25.
Single nuclei RNA-seq atlas
To generate an interactive snRNA-seq atlas we used the ShinyCell package32 on R (version 4.4.2) and slightly modified the design of the webpage. The website https://ruegglab.shinyapps.io/snatlas/ is hosted on the shinyapps.io server, which is limited to 25 active hours per month. Alternatively the same data can be explored on https://cellxgene.cziscience.com/. The processed snRNA-seq files can also be downloaded in AnnData and Seurat format from CZ CELLxGENE [https://cellxgene.cziscience.com/collections/ef9f0fae-c12c-4f1a-bb67-63346c2ae0f1].
RNA isolation and bulk-RNA-seq analysis
TA muscles were crushed to powder on a metal block cooled with liquid nitrogen. Total RNA was extracted from the muscle powder using the RNeasy Fibrous Tissue Mini Kit (74704, Qiagen) according to the manufactures protocol. RNA concentration and integrity were measured using a Quant-iTTM RiboGreen RNA assay kit (Invitrogen) and a Bioanalyzer (Agilent), respectively. Libraries were prepared using the TruSeq stranded mRNA library kit (20020595, Illumina) starting from 200 ng of RNA. Sequencing was performed on the NovaSeq 6000 (Illumina) system (PE 2 × 51) which resulted in 26-52 mio reads per sample. FASTQ files were aligned to the indexed mouse transcriptome mm10 using Salmon (version 1.1.0) with the flags validateMappings, seqBias, and gcBias. The output quant.sf files from all samples were imported into R (version 4.1.2) using tximeta (Bioconductor) and analyzed with DESeq2 (Bioconductor). Transcript level information was summarized to gene level and genes with fewer than 5 counts across all samples were removed. The design was set to “~ Injection” and after applying DeSeq() the result for each group was exported individually together with PBS-injected controls. All genes with adjusted p-values < 0.05 and Log2FC ± 0.25 were considered as significantly changed.
Sashimi plots and bulk RNA-seq exon coverage
To generate sashimi plots bulk RNA-seq FASTQ files from TA and soleus muscles from 10-month-old male mice were downloaded from GEO (GSE139204) [9]. The FASTQ files were aligned to mouse reference GRCm39/mm39 using STAR (version 2.7.9a), and the genomic region of each gene was extracted from bam files using samtools (version 1.12). The extracted region was imported into Integrative Genomics Viewer (IGV, version 2.16.1), and Sashimi plots were generated. The Sashimi plots were extracted and adjusted for better visualization.
qPCR
Total RNA was isolated as described above. 500 ng of RNA were used to generate cDNA with the iScript cDNA Synthesis Kit (Bio-Rad). The cDNA was diluted 1:5, mixed with PowerUp SYBR Green Master Mix (Thermo Fisher), and combined with the corresponding primers at a final primer concentration of 500 nM. The qPCR reactions were run on a QuantStudioTM 5 PCR system (Thermo Fisher). For quantification all targets were normalized to housekeeper Gapdh. The primer sequences used were: Gapdh fw: ACC CAG AAG ACT GTG GAT GG, rv: GGA TGC AGG GAT GAT GTT CT; Etv4 fw: AGG AGT ACC ATG ACC CCC TG, rv: GGA CAT CTG AGT CGT AGG CG; Lrtm1 fw: CCC TTG GAT TTG TGA CTG CC, rv: GAG GAA TGG AGA GGA GAG CC; Pdzrn4 fw: CAG ATG ACT TTG AAT ATG AGG AGG; rv: TTC GGT AGC AGA CTG TCA G; Musk fw: GCT CCT GAA TCC CAC AAT GTC, rv: AGA GTC CTG GCT TTG TGA TGA; Pdzrn3 fw: GAC AGG ATT ATT GAG GTC AAC G, rv: CTT GGC TGT CTT GAA AGC T.
Immunofluorescence staining on cross-sections
Muscles were embedded in OCT, frozen in isopentane, and cut to 10 µm sections using a cryostat. Sections were fixed with 4% PFA in PBS for 5 min followed by neutralization with 100 mM glycine in PBS for 2 × 10 min. Next, sections were permeabilized and blocked with blocking solution (3% BSA, 0.4% Triton X-100 in PBS) for 30 min. Primary antibodies were diluted in 3% BSA in PBS and added to the sections for either 1 h at room temperature (RT) or overnight at 4 °C (for antibodies and dilutions, see Supplementary Data 10). This was followed by three washing steps with PBS. Secondary antibodies and DAPI were diluted in 3% BSA in PBS and added to the samples for 1 h in the dark. To finish, samples were washed with PBS and mounted with ProLongTM Gold antifade (Invitrogen). Images were taken on either a wide field IX83 microscope (Olympus) or a confocal LSM 700 (Zeiss).
HeLa cell transfection, staining, and quantification
HeLa cells (ATCC CCL-2) were cultured in DMEM (D5796, Sigma) containing 10% FBS, 1% Penicillin/Streptomycin, and 0.01 M HEPES. For IF imaging, HeLa cells were cultured on 4-well cell culture chamber slides with removable frames (Sarstedt). Transfection was performed using Lipofectamine 2000 (Invitrogen), where all wells received 260 ng/cm2 of DNA and 1.04 µl/cm2 of Lipofectamine. 24 h after transfection HeLa cells were washed with PBS and fixed for 15 min with 4% PFA in PBS. IF stainings were performed as described above. For antibodies and dilutions, see Supplementary Data 10.
To quantify MuSK accumulation at the Golgi apparatus in HeLa cells (Fig. 6e, f), MuSK staining intensity was measured at the Golgi region. In control cells, the Golgi was identified using GM130, while in PDZRN4-overexpressing cells, the Golgi was identified using PDZRN4-FLAG. The intensity of MuSK at the Golgi was then compared to the total MuSK intensity across the entire cell.
Whole-mount immunostaining and quantification
EDL muscles were carefully dissected, leaving the tendons on both ends intact. Muscles were pinned on a sylgard dish and fixed with 4% PFA in PBS for 30 min at RT, followed by three washes with PBS for 5 min each. Muscles were stored in 50% glycerol in PBS (v/v) at −20 °C until usage. Fixed muscles were cut into small bundles and permeabilized with 2% Triton X-100 in PBS for 2 h. Muscle bundles were then neutralized with 100 mM glycine in PBS and blocked in blocking solution (3% BSA, 0.2% Triton X-100 in PBS) for 30 min each. This was followed by primary antibody incubation in blocking solution overnight at 4 °C (for antibodies and dilutions, see Supplementary Data 10). On the next day, samples were washed with PBS containing 0.2% Triton X-100 four times and incubated in a secondary antibody solution and α-bungarotoxin for 2 h. Samples were washed again four times in PBS containing 0.2% Triton X-100 and then mounted with ProLongTM Gold antifade (Invitrogen). Images were taken at the Imaging Core Facility (Biozentrum, Basel, Switzerland) using a confocal LSM 700 (Zeiss) at 63x magnification.
To analyze postsynaptic fragmentation, 10-15 individual NMJs were imaged and quantified in Imaris (9.9.1) for each mouse, and the average value was used for statistics. To quantify the size of Golgi particles in mouse muscle, GLUT4 staining was rendered in 3D and quantified in Imaris (9.9.1). At least 75 GLUT4-positive puncta from a randomly selected region of each mouse were measured, and the average value was used for quantification.
Single molecule fluorescent in situ hybridization (smFISH)
For the detection of RNA on cross-sections, we used the V1 kit from RNAscope (ACDBio) and followed the manufacturers protocol. Briefly, fresh frozen 10 µm muscle cross-sections were fixed with 4% PFA for 15 min on ice, followed by an ethanol series of 50, 70 and 100% at RT to dehydrate the tissue. Next, muscle sections were digested with protease IV for 30 min, or 5 min if combined with an IF staining. After washing with PBS the cross-sections were placed in a humid chamber with hybridization probes followed by several amplification steps. The sections were stained with DAPI and mounted with ProLongTM Gold antifade (Invitrogen). Images were taken at the Imaging Core Facility (Biozentrum, Basel) using a confocal LSM 700 (Zeiss).
Single fiber isolation and staining
EDL muscles were harvested and placed in 0.2% Collagenase Type I (Sigma) in isolation media (DMEM with 1% Penicillin/Streptomycin) at 37 °C. After 1 h of digestion, the muscles were transferred to 37 °C isolation medium in a dish pre-coated with 20% horse serum (Biological Industries). The muscles were flushed using a wide-bore glass pipette to dissociate single fibers. Once a sufficient number of fibers were obtained, single fibers were transferred into a microcentrifuge tube pre-coated with 20% horse serum (Biological Industries), washed once with PBS for 5 min, fixed in 4% PFA for 10 min and washed 3 x in 0.1 M glycine in PBS (pH 7.4). Fibers were then permeabilized with 0.2% Triton X-100 in PBS for 10 min at RT before blocking with 10% goat serum (Biological Industries) in PBS for 3 hours at RT. The samples were then incubated with primary antibodies in 10% goat serum in PBS overnight at 4 °C (for antibodies and dilutions see Supplementary Data 10). The following day, the samples were washed 3 × 5 min in PBS and incubated with the corresponding secondary antibodies and α-bungarotoxin in 10% goat serum in PBS for 1 hour at RT. The fibers were then washed 3 x in PBS and mounted on glass slides with ProLong™ Gold antifade with DAPI (Invitrogen).
Differential fractionation and Western blot analysis
Frozen TA muscles were powdered on a metal block cooled with liquid nitrogen and lysed in 400 µl cytosol lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl) supplemented with protease inhibitors (11 836 170 001, Roche) for 30 min at 4 °C. Lysates were centrifuged at 100,000 x g for 30 min at 4 °C in a micro ultracentrifuge (CS 150FNX, Hitachi). The supernatant (cytosolic fraction) was transferred to a new tube, and the pellet was resuspended with 300 µl membrane lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP-40) supplemented with protease inhibitors (Roche). After 20 min of incubation, the samples were centrifuged again at 100,000 x g for 30 min at 4 °C. The supernatant (membrane fraction) was transferred to a new tube. Protein concentrations of both the cytosolic and membrane fractions were measured using the Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific) and equal amounts of protein were loaded onto a NuPAGE 4-12% Bis-Tris gel (Invitrogen). Western blots were prepared and imaged as previously described58.
Immunoprecipitation
HEK 293 T cells were cultured in 6-well plates in DMEM (D5796, Sigma) containing 10% FBS, 1% Penicillin/Streptomycin, and 0.01 M HEPES. Transfection was performed using Lipofectamine 2000 (Invitrogen), where all wells received 260 ng/cm2 of DNA and 1.04 µl/cm2 of Lipofectamine. The cells were transfected with Musk, Pdzrn4-Flag, and pac (control) plasmids, all containing a CMV promoter. 24 h after transfection cells were washed in PBS and lysed with 500 µl IP lysis buffer (20 mM Tris-HCl pH 7.4, 100 mM NaCl, 5 mM EDTA, 0.5% NP-40) supplemented with protease (Roche) and phosphatase inhibitors (Roche). The lysed cells were transferred to microcentrifuge tubes, sonicated for 3 s, and incubated on a rotating wheel for 30 min at 4 °C. The lysates were filtered through 70 µm cell strainers before measuring protein concentration using the Pierce™ BCA Protein Assay Kit. 500 µg of protein was diluted to a concentration of 1 µg/µl, and 2 µg of anti-FLAG M2 antibody (Sigma) was added and incubated overnight at 4 °C for IP. Antibodies were pulled down using magnetic DynabeadsTM Protein G (10004D, Invitrogen) and washed with IP lysis buffer. The pulled-down proteins were released from the magnetic beads by adding 50 µl 1x Laemmli buffer and heating to 95 °C for 10 min. Equal volumes were loaded onto NuPAGE 4-12% Bis-Tris gels (Invitrogen) together with whole protein lysates. Western blots were prepared and imaged as previously described58, and loading controls were run on the same blot.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data are provided with this paper. Raw single nuclei RNA-seq and bulk RNA-seq sequencing data are deposited on NCBI Gene Expression Omnibus (GEO) under the accession number GSE267913 (SubSeries GSE267910 and GSE267909). Processed single nuclei RNA-seq data is accessible for download from CZ CELLxGENE [https://cellxgene.cziscience.com/collections/ef9f0fae-c12c-4f1a-bb67-63346c2ae0f1]. Source data are provided with this paper.
References
Cramer, A. A. W. et al. Nuclear numbers in syncytial muscle fibers promote size but limit the development of larger myonuclear domains. Nat. Commun. 11, 6287 (2020).
Dos Santos, M. et al. Single-nucleus RNA-seq and FISH identify coordinated transcriptional activity in mammalian myofibers. Nat. Commun. 11, 5102 (2020).
Petrany, M. J. et al. Single-nucleus RNA-seq identifies transcriptional heterogeneity in multinucleated skeletal myofibers. Nat. Commun. 11, 6374 (2020).
Kim, M. et al. Single-nucleus transcriptomics reveals functional compartmentalization in syncytial skeletal muscle cells. Nat. Commun. 11, 6375 (2020).
Chemello, F. et al. Degenerative and regenerative pathways underlying Duchenne muscular dystrophy revealed by single-nucleus RNA sequencing. Proc. Natl Acad. Sci. USA 117, 29691–29701 (2020).
Tintignac, L. A., Brenner, H. R. & Ruegg, M. A. Mechanisms regulating neuromuscular junction development and function and causes of muscle wasting. Physiol. Rev. 95, 809–852 (2015).
Grady, R. M., Starr, D. A., Ackerman, G. L., Sanes, J. R. & Han, M. Syne proteins anchor muscle nuclei at the neuromuscular junction. Proc. Natl Acad. Sci. USA 102, 4359–4364 (2005).
Ang, S. T. J. et al. Muscle 4EBP1 activation modifies the structure and function of the neuromuscular junction in mice. Nat. Commun. 13, 7792 (2022).
Liu, W. X. et al. Loss of adult skeletal muscle stem cells drives age-related neuromuscular junction degeneration. Elife 6, e26464 (2017).
Witzemann, V., Brenner, H. R. & Sakmann, B. Neural factors regulate AChR subunit mRNAs at rat neuromuscular synapses. J. Cell Biol. 114, 125–141 (1991).
DeChiara, T. M. et al. The receptor tyrosine kinase MuSK is required for neuromuscular junction formation in vivo. Cell 85, 501–512 (1996).
Hesser, B. A., Henschel, O. & Witzemann, V. Synapse disassembly and formation of new synapses in postnatal muscle upon conditional inactivation of MuSK. Mol. Cell. Neurosci. 31, 470–480 (2006).
Valenzuela, D. M. et al. Receptor Tyrosine Kinase Specific for the skeletal-muscle lineage - expression in embryonic muscle, at the neuromuscular-junction, and after injury. Neuron 15, 573–584 (1995).
Li, L., Xiong, W. C. & Mei, L. Neuromuscular junction formation, aging, and disorders. Annu. Rev. Physiol. 80, 159–188 (2018).
Kim, N. et al. Lrp4 is a receptor for Agrin and forms a complex with MuSK. Cell 135, 334–342 (2008).
Zhang, B. et al. LRP4 serves as a coreceptor of Agrin. Neuron 60, 285–297 (2008).
Hallock, P. T. et al. Dok-7 regulates neuromuscular synapse formation by recruiting Crk and Crk-L. Gene Dev. 24, 2451–2461 (2010).
Ruegg, M. A. Organization of synaptic myonuclei by Syne proteins and their role during the formation of the nerve-muscle synapse. Proc. Natl Acad. Sci. USA 102, 5643–5644 (2005).
Belotti, E. & Schaeffer, L. Regulation of Gene expression at the neuromuscular Junction. Neurosci. Lett. 735, 135163 (2020).
Koike, S., Schaeffer, L. & Changeux, J. P. Identification of a DNA element determining synaptic expression of the mouse acetylcholine receptor delta-subunit gene. Proc. Natl Acad. Sci. USA 92, 10624–10628 (1995).
Duclert, A., Savatier, N., Schaeffer, L. & Changeux, J. P. Identification of an element crucial for the sub-synaptic expression of the acetylcholine receptor epsilon-subunit gene. J. Biol. Chem. 271, 17433–17438 (1996).
Gramolini, A. O. et al. Induction of utrophin gene expression by heregulin in skeletal muscle cells: Role of the N-box motif and GA binding protein. Proc. Natl Acad. Sci. USA 96, 3223–3227 (1999).
Khurana, T. S. et al. Activation of utrophin promoter by heregulin via the ets-related transcription factor complex GA-binding protein α/β. Mol. Biol. Cell 10, 2075–2086 (1999).
Chan, R. Y. Y., Boudreau-Larivière, C., Angus, L. M., Mankal, F. A. & Jasmin, B. J. An intronic enhancer containing an N-box motif is required for synapse- and tissue-specific expression of the acetylcholinesterase gene in skeletal muscle fibers. Proc. Natl Acad. Sci. USA 96, 4627–4632 (1999).
Lacazette, E., Le Calvez, S., Gajendran, N. & Brenner, H. R. A novel pathway for MuSK to induce key genes in neuromuscular synapse formation. J. Cell Biol. 161, 727–736 (2003).
Hippenmeyer, S., Huber, R. M., Ladle, D. R., Murphy, K. & Arber, S. ETS transcription factor Erm controls subsynaptic gene expression in skeletal muscles. Neuron 55, 726–740 (2007).
Goldman, D., Brenner, H. R. & Heinemann, S. Acetylcholine-Receptor Alpha-Subunit, Beta subunit, gamma-subunit and sigma-subunit messenger-rna levels are regulated by muscle-activity. Neuron 1, 329–333 (1988).
Miyoshi, S. et al. DOK7 gene therapy enhances motor activity and life span in ALS model mice. Embo Mol. Med. 9, 880–889 (2017).
Arimura, S. et al. DOK7 gene therapy benefits mouse models of diseases characterized by defects in the neuromuscular junction. Science 345, 1505–1508 (2014).
Ueta, R. et al. DOK7 gene therapy enhances neuromuscular junction innervation and motor function in aged mice. iScience 23, 101385 (2020).
Blasi, J. et al. Botulinum Neurotoxin-a selectively cleaves the synaptic protein Snap-25. Nature 365, 160–163 (1993).
Ouyang, J. F., Kamaraj, U. S., Cao, E. Y. & Rackham, O. J. L. ShinyCell: simple and sharable visualization of single-cell gene expression data. Bioinformatics 37, 3374–3376 (2021).
Thyboll, J. et al. Deletion of the laminin α4 chain leads to impaired microvessel maturation. Mol. Cell. Biol. 22, 1194–1202 (2002).
Frieser, M. et al. Cloning of the mouse laminin alpha 4 cDNA. Expression in a subset of endothelium. Eur. J. Biochem. 246, 727–735 (1997).
Gerber, D. et al. Transcriptional profiling of mouse peripheral nerves to the single-cell level to build a sciatic nerve ATlas (SNAT). Elife 10, e58591 (2021).
Ravel-Chapuis, A., Vandromme, M., Thomas, J. L. & Schaeffer, L. Postsynaptic chromatin is under neural control at the neuromuscular junction. EMBO J. 26, 1117–1128 (2007).
Gautam, M. et al. Failure of postsynaptic specialization to develop at neuromuscular-junctions of Rapsyn-deficient mice. Nature 377, 232–236 (1995).
Okada, K. et al. The muscle protein Dok-7 is essential for neuromuscular synaptogenesis. Science 312, 1802–1805 (2006).
Ham, D. J. et al. The neuromuscular junction is a focal point of mTORC1 signaling in sarcopenia. Nat. Commun. 11, 4510 (2020).
Covault, J. & Sanes, J. R. Neural cell-adhesion molecule (N-Cam) accumulates in denervated and paralyzed skeletal-muscles. Proc. Natl Acad. Sci. USA 82, 4544–4548 (1985).
Numberger, M., Durr, I., Kues, W., Koenen, M. & Witzemann, V. Different mechanisms regulate muscle-specific AChR gamma- and epsilon-subunit gene expression. EMBO J. 10, 2957–2964 (1991).
Ehmsen, J. T., Kawaguchi, R., Mi, R., Coppola, G. & Hoke, A. Longitudinal RNA-Seq analysis of acute and chronic neurogenic skeletal muscle atrophy. Sci. Data 6, 179 (2019).
Castets, P. et al. mTORC1 and PKB/Akt control the muscle response to denervation by regulating autophagy and HDAC4. Nat. Commun. 10, 3187 (2019).
Nicoletti, C. et al. Muscle denervation promotes functional interactions between glial and mesenchymal cells through NGFR and NGF. iScience 26, 107114 (2023).
Proietti, D. et al. Activation of skeletal muscle-resident glial cells upon nerve injury. JCI Insight 6, e143469 (2021).
Madaro, L. et al. Denervation-activated STAT3-IL-6 signalling in fibro-adipogenic progenitors promotes myofibres atrophy and fibrosis. Nat. Cell Biol. 20, 917 (2018).
Lin, H. et al. Decoding the transcriptome of denervated muscle at single-nucleus resolution. J. Cachexia Sarcopenia Muscle 13, 2102–2117 (2022).
Jasmin, B. J. et al. Expression of Utrophin and its messenger-RNA in Denervated Mdx mouse muscle. FEBS Lett. 374, 393–398 (1995).
Calvo, A. C. et al. Collagen XIX Alpha 1 improves prognosis in amyotrophic lateral sclerosis. Aging Dis. 10, 278–292 (2019).
Reist, N. E., Magill, C. & Mcmahan, U. J. Agrin-like molecules at synaptic sites in normal, denervated, and damaged skeletal-muscles. J. Cell Biol. 105, 2457–2469 (1987).
Thurkauf, M. et al. Fast, multiplexable and efficient somatic gene deletions in adult mouse skeletal muscle fibers using AAV-CRISPR/Cas9. Nat. Commun. 14, 6116 (2023).
Weinmann, J. et al. Identification of a myotropic AAV by massively parallel in vivo evaluation of barcoded capsid variants. Nat. Commun. 11, 5432 (2020).
de Paiva, A., Meunier, F. A., Molgó, J., Aoki, K. R. & Dolly, J. O. Functional repair of motor endplates after botulinum neurotoxin type A poisoning:: Biphasic switch of synaptic activity between nerve sprouts and their parent terminals. Proc. Natl Acad. Sci. USA 96, 3200–3205 (1999).
Meunier, F. A., Lisk, G., Sesardic, D. & Dolly, J. O. Dynamics of motor nerve terminal remodeling unveiled using SNARE-cleaving botulinum toxins: the extent and duration are dictated by the sites of SNAP-25 truncation. Mol. Cell. Neurosci. 22, 454–466 (2003).
Lomo, T. & Slater, C. R. Control of junctional Acetylcholinesterase by neural and muscular influences in the rat. J. Physiol.-Lond. 303, 191 (1980).
Dolan, J. et al. The extracellular leucine-rich repeat superfamily; a comparative survey and analysis of evolutionary relationships and expression patterns. BMC Genomics 8, 320 (2007).
Lu, Z. et al. Regulation of synaptic growth and maturation by a synapse-associated E3 ubiquitin ligase at the neuromuscular junction. J. Cell Biol. 177, 1077–1089 (2007).
Ham, A. S. et al. mTORC1 signalling is not essential for the maintenance of muscle mass and function in adult sedentary mice. J. Cachexia Sarcopenia Muscle 11, 259–273 (2020).
Tang, H. et al. A histone deacetylase 4/myogenin positive feedback loop coordinates denervation-dependent gene induction and suppression. Mol. Biol. Cell 20, 1120–1131 (2009).
Schaeffer, L., d’Exaerde, A. D. & Changeux, J. P. Targeting transcription to the neuromuscular synapse. Neuron 31, 15–22 (2001).
Ralston, E., Ploug, T., Kalhovde, J. & Lomo, T. Golgi complex, endoplasmic reticulum exit sites, and microtubules in skeletal muscle fibers are organized by patterned activity. J. Neurosci. 21, 875–883 (2001).
Ralston, E., Lu, Z. M. & Ploug, T. The organization of the Golgi complex and microtubules in skeletal muscle is fiber type-dependent. J. Neurosci. 19, 10694–10705 (1999).
Ploug, T., van Deurs, B., Ai, H., Cushman, S. W. & Ralston, E. Analysis of GLUT4 distribution in whole skeletal muscle fibers: Identification of distinct storage compartments that are recruited by insulin and muscle contractions. J. Cell Biol. 142, 1429–1446 (1998).
Rahkila, P., Vaananen, K., Saraste, J. & Metsikko, K. Endoplasmic reticulum to Golgi trafficking in multinucleated skeletal muscle fibers. Exp. Cell Res. 234, 452–464 (1997).
Wood, S. J. & Slater, C. R. Safety factor at the neuromuscular junction. Prog. Neurobiol. 64, 393–429 (2001).
Willadt, S., Nash, M. & Slater, C. R. Age-related fragmentation of the motor endplate is not associated with impaired neuromuscular transmission in the mouse diaphragm. Sci. Rep-Uk 6, 24849 (2016).
Kawai-Takaishi, M., Miyagawa, Y., Honda, T., Inui, M. & Hosoyama, T. Postnatal Pdzrn3 deficiency causes acute muscle atrophy without alterations in endplate morphology. Biochem Biophys. Res Commun. 696, 149542 (2024).
Bruneau, E. G. & Akaaboune, M. Dynamics of the Rapsyn scaffolding protein at the neuromuscular junction of live mice. J. Neurosci. 30, 614–619 (2010).
Sanes, J. R. The basement membrane/basal lamina of skeletal muscle. J. Biol. Chem. 278, 12601–12604 (2003).
Sarkozy A., Foley A. R., Zambon A. A., Bönnemann C. G. & Muntoni, F. LAMA2-related dystrophies: clinical phenotypes, disease biomarkers, and clinical trial readiness. Front. Mol. Neurosci. 13, https://doi.org/10.3389/fnmol.2020.00123 (2020).
McMacken, G., Abicht, A., Evangelista, T., Spendiff, S. & Lochmuller, H. The increasing genetic and phenotypical diversity of congenital myasthenic syndromes. Neuropediatrics 48, 294–308 (2017).
Koch, M. et al. A novel marker of tissue junctions, collagen XXII. J. Biol. Chem. 279, 22514–22521 (2004).
Chen, F. S. et al. Botulinum neurotoxin B inhibits insulin-stimulated glucose uptake into 3T3-L1 adipocytes and cleaves cellubrevin unlike type A toxin which failed to proteolyze the SNAP-23 present. Biochem.-Us 36, 5719–5728 (1997).
Tang, H. B. & Goldman, D. Activity-dependent gene regulation in skeletal muscle is mediated by a histone deacetylase (HDAC)-Dach2-myogenin signal transduction cascade. Proc. Natl Acad. Sci. USA 103, 16977–16982 (2006).
Eftimie, R., Brenner, H. R. & Buonanno, A. Myogenin and MyoD join a family of skeletal muscle genes regulated by electrical activity. Proc. Natl Acad. Sci. USA 88, 1349–1353 (1991).
Silinsky, E. M. & Redman, R. S. Synchronous release of ATP and neurotransmitter within milliseconds of a motor nerve impulse in the frog. J. Physiol. 492, 815–822 (1996).
Choi, R. C. et al. ATP acts via P2Y1 receptors to stimulate acetylcholinesterase and acetylcholine receptor expression: transduction and transcription control. J. Neurosci. 23, 4445–4456 (2003).
Choi, R. C. et al. Expression of the P2Y1 nucleotide receptor in chick muscle: its functional role in the regulation of acetylcholinesterase and acetylcholine receptor. J. Neurosci. 21, 9224–9234 (2001).
Picchiarelli, G. et al. FUS-mediated regulation of acetylcholine receptor transcription at neuromuscular junctions is compromised in amyotrophic lateral sclerosis. Nat. Neurosci. 22, 1793–1805 (2019).
Luo, Z. G. et al. Regulation of AChR clustering by dishevelled interacting with MuSK and PAK1. Neuron 35, 489–505 (2002).
Fu, A. K. Y. et al. Cdk5 is involved in neuregulin-induced AChR expression at the neuromuscular junction. Nat. Neurosci. 4, 374–381 (2001).
Si, J. T., Luo, Z. J. & Mei, L. Induction of acetylcholine receptor gene expression by ARIA requires activation of mitogen-activated protein kinase. J. Biol. Chem. 271, 19752–19759 (1996).
Tansey, M. G., Chu, G. C. & Merlie, J. P. ARIA/HRG regulates AChR epsilon subunit gene expression at the neuromuscular synapse via activation of phosphatidylinositol 3-kinase and Ras/MAPK pathway. J. Cell Biol. 134, 465–476 (1996).
Martinou, J. C., Falls, D. L., Fischbach, G. D. & Merlie, J. P. Acetylcholine receptor-inducing activity stimulates expression of the epsilon-subunit gene of the muscle acetylcholine-receptor. Proc. Natl Acad. Sci. USA 88, 7669–7673 (1991).
Wu, H. et al. Slit2 as a beta-catenin/Ctnnb1-dependent retrograde signal for presynaptic differentiation. Elife 4, e07266 (2015).
Escher, P. et al. Synapses form in skeletal muscles lacking neuregulin receptors. Science 308, 1920–1923 (2005).
Schmidt, N. et al. Neuregulin/ErbB regulate neuromuscular junction development by phosphorylation of alpha-dystrobrevin. J. Cell Biol. 195, 1171–1184 (2011).
Potelle, S., Klein, A. & Foulquier, F. Golgi post-translational modifications and associated diseases. J. Inherit. Metab. Dis. 38, 741–751 (2015).
Watty, A. & Burden, S. J. MuSK glycosylation restrains MuSK activation and acetylcholine receptor clustering. J. Biol. Chem. 277, 50457–50462 (2002).
Qi, T. et al. Function and regulation of the PEA3 subfamily of ETS transcription factors in cancer. Am. J. Cancer Res. 10, 3083–3105 (2020).
Yan, J. S. & Aldrich, R. W. BK potassium channel modulation by leucine-rich repeat-containing proteins. Proc. Natl Acad. Sci. USA 109, 7917–7922 (2012).
Kedlian, V. R. et al. Human skeletal muscle aging atlas. Nat. Aging 4, 727–744 (2024).
Platt, R. J. et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014).
Schwander, M. et al. beta 1 integrins regulate myoblast fusion and sarcomere assembly. Dev. Cell 4, 673–685 (2003).
Carli, L., Montecucco, C. & Rossetto, O. Assay of diffusion of different botulinum neurotoxin type a formulations injected in the mouse leg. Muscle Nerve 40, 374–380 (2009).
Concordet, J. P. & Haeussler, M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, W242–W245 (2018).
Sakuma, T., Nishikawa, A., Kume S., Chayama, K. & Yamamoto, T. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci Rep.-Uk 4, 5400 (2014).
Wang, F., Cui, X., Wang, M., Xiao, W. & Xu, R. A reliable and feasible qPCR strategy for titrating AAV vectors. Med. Sci. Monit. Basic Res. 19, 187–193 (2013).
Acknowledgements
We thank the Imaging Core Facility, sciCORE, and Animal facility of the Biozentrum for their support with imaging, computing, and mouse housing, respectively. We acknowledge Elodie Burcklen, Phillippe Demougin, and Christian Beisel of the Genomics facility of the D-BSSE (ETH) for their support in library preparation, quality control, and sequencing. We thank D. J. Ham for his advice on the manuscript and for helping acquire funding. This work was funded by the Swiss National Science Foundation (#189248), and the cantons of Basel-Stadt and Basel-Landschaft awarded to M.A.R.
Author information
Authors and Affiliations
Contributions
A.S.H. and M.A.R. conceptualized the study; A.S.H. developed the methodology; A.S.H., S.L., A.T., M.T., T.J.M., L.J., and F.O. performed the experiments, A.S.H. and S.L. quantified the data; A.S.H. analyzed the data; A.S.H. and M.A.R. interpreted the data and wrote the manuscript; M.A.R. acquired the funding.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Laurent Schaeffer and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ham, A.S., Lin, S., Tse, A. et al. Single-nuclei sequencing of skeletal muscle reveals subsynaptic-specific transcripts involved in neuromuscular junction maintenance. Nat Commun 16, 2220 (2025). https://doi.org/10.1038/s41467-025-57487-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-57487-1
This article is cited by
-
The contribution of skeletal muscle interstitial cells to myogenesis
Skeletal Muscle (2026)
-
Loss of cell-autonomously secreted laminin-α2 drives muscle stem cell dysfunction in LAMA2-related muscular dystrophy
Nature Communications (2025)
-
Loss of COL20A1 in Schwann Cells Alters Regeneration of the Neuromuscular Junction and Locomotor Activity in Mice
Journal of Molecular Neuroscience (2025)