Abstract
Autoimmune destruction of pancreatic β cells results in type 1 diabetes (T1D), with pancreatic immune infiltrate representing a key feature in this process. However, characterization of the immunological processes occurring in human pancreatic lymphatic tissues is lacking. Here, we conduct a comprehensive study of immune cells from pancreatic, mesenteric, and splenic lymphatic tissues of non-diabetic control (ND), β cell autoantibody-positive non-diabetic (AAb+), and T1D donors using flow cytometry and CITEseq. Compared to ND pancreas-draining lymph nodes (pLN), AAb+ and T1D donor pLNs display decreased CD4+ Treg and increased stem-like CD8+ T cell signatures, while only T1D donor pLNs exhibit naive T cell and NK cell differentiation. Mesenteric LNs have modulations only in CD4+ Tregs and naive cells, while splenocytes lack these perturbations. Further, T cell expression of activation markers and IL7 receptor correlate with T1D genetic risk. These results demonstrate tissue-restricted immune changes occur before and after T1D onset.
Similar content being viewed by others
Introduction
Type 1 diabetes (T1D) results in lifelong exogenous insulin dependence following autoimmune destruction of the pancreatic islet β cells1. While β cell autoimmunity may start beforehand, as suggested by the appearance of β cell-specific autoantibodies (AAb), the disease can take years to progress to overt hyperglycemia2. Evidence of β cell mass and insulin secretion can be detected even in long-standing T1D patients3,4, with clinically beneficial effects associated with residual β cell function5. T1D genetic risk is primarily associated with human leukocyte antigen (HLA) loci but also with genes involved in regulating immune responses2,6,7,8. Accordingly, T cell anergy-inducing immunotherapy shows efficacy in individuals at risk for imminent T1D onset but only delays T1D onset by ~24 months9,10. The development of therapies capable of halting autoimmune progression and preserving β cell function requires further understanding of the complex immune cell responses that occur pre- and post-T1D diagnosis.
Human immunophenotypes associated with T1D include T cell11,12,13, B cell14,15, and natural killer (NK) cell16 perturbations as well as accelerated immunological aging17 in peripheral blood mononuclear cells (PBMC). While surveying PBMCs can be informative, the immune system operates primarily within tissue microenvironments, and disease-specific signatures may not be fully recapitulated in circulating immune cells18,19,20,21,22. Indeed, pancreatic islets of T1D individuals contain β cell antigen-specific CD8+ T cells23, and the frequency of CD8+ T cells that recognize β cell antigens in the pancreas, not the blood, distinguishes T1D from healthy donors24. Surveying immunophenotypes in the pancreas itself has greatly increased our understanding of T1D pathogenesis25,26,27,28,29,30; however, the immunological processes that occur in pancreas-draining lymphoid tissues that may initiate or exacerbate T1D development and progression remain largely unclear.
Evidence suggests that pancreatic lymph nodes (pLN) harbor immune cells that participate in T1D autoimmunity31,32,33,34. Studies in the non-obese diabetic (NOD) mouse model of T1D suggest that pLN lymphadenectomy early in autoimmune development protects against diabetes31. Further, pLNs in the mouse model contain stem-like β cell-specific memory CD8+ T cells, which seed cytotoxic and terminally-differentiated effector cells that traffic to the pancreas and eliminate β cells34. In humans, pLNs from T1D donors contain insulin peptide-reactive T cells32, and are enriched for β cell antigen-reactive T cell receptor (TCR) clones33. pLNs from T1D donors also contain phenotypic alterations that suggest a more proinflammatory T cell population, specifically increased frequencies of Th17 effector CD4+ T cells with reciprocal decreases in regulatory T cell (Treg) and follicular regulatory T cell frequencies35,36. While these observations implicate the pLNs as a hub of T1D autoimmunity, due to the rarity of these sample types, there has not been a comprehensive, unbiased analysis of human pLNs that effectively captures this immune environment.
In this work, we profile human lymphoid immune perturbations across T1D autoimmune development within pLNs, mesenteric lymph nodes (mLN), and spleen from control non-diabetic autoantibody negative (ND), non-diabetic AAb+, and T1D donors. We perform deep immunophenotyping of these tissues using the complementary methods of high-parameter flow cytometry (n = 46 donors) and cellular indexing of transcriptomes and epitopes by sequencing (CITEseq; n = 18 donors). Compared to ND pLNs, AAb+ and T1D donor pLN CD4+ Tregs are reduced in frequency and have features of instability, while memory CD8+ T cells have increased stem-like signatures. Only T1D donor pLNs display increased differentiation of naive T cells and NK cells. mLNs have modulations only in CD4+ Tregs and naive T cells, while splenocytes contain few perturbations. Further, T cell expression of activation markers and the IL7 receptor correlate with T1D genetic risk. These results demonstrate that tissue-restricted immune changes, particularly in the pLNs, occur before and after T1D onset in humans.
Results
Immune profiling of pancreatic, mesenteric, and splenic lymphatic tissues
Through the Human Pancreas Analysis Program (HPAP)37, we collected ND, AAb+, and T1D donor spleens and lymph nodes that drain the pancreas, including pLNs (pancreatic head, body, and tail) and mLNs (superior mesenteric artery termed SMA mLN, and upper mesentery termed MES mLN). We were unable to profile pancreas infiltrating lymphocytes due to methodological limitations related to islet isolation. Donors were age, race, and sex matched across the disease groups (Supplementary Tables 1 and 2), with tissue samples being processed and analyzed at the same facility over the course of the study. T1D donors were diabetic for around 6 years on average, with only two donors having diabetes >8 years. We used two complementary techniques for deep immune cell profiling of available donor tissues: high-parameter flow cytometry on 256 unique samples (18 ND, 10 AAb+, and 18 T1D donors, Supplementary Table 1) and CITEseq single cell analysis on 650,704 cells post quality control (6 ND, 5 AAb+, and 7 T1D donors, Supplementary Table 2).
We first used a high-parameter flow cytometry panel to assess inter-tissue immune differences amongst all major immune cell populations across several immune lineages, including T cells, B cells, innate cells, and subsets thereof (Fig. 1a–d, Supplementary Fig. 1). The spleen showed a unique immune profile compared to LNs (Fig. 1a–d), with an increased frequency of NK cells (Supplementary Fig. 2A), monocytes, and CCR7- T cell subsets, including effector memory (Tem) and Tem CD45RA+ (Temra) T cells (Fig. 1b, d). Some subsets were evenly distributed across the spleen and LNs, including B cells (Figs. 1b, S2B). In contrast, the LNs were enriched for T cells, particularly CD4+ T cells and naive T cell subsets (Figs. 1b–d, S2C, D). The pLNs and mLNs were highly similar in immune cell composition, with few exceptions (Supplementary Fig. 2E–I). Innate lymphoid cells (ILCs) were slightly decreased in frequency within the MES mLN (Supplementary Fig. 2E), CD8+ central memory T cells (Tcm) had a slightly higher frequency occurring in the pLN-Tail (Supplementary Fig. 2F), and NK cells had a slightly decreased frequency in the pLN-Tail (Supplementary Fig. 2A).
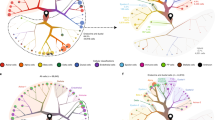
a UMAP representation of pLN-Tail lymphocytes from and splenocytes from donor HPAP-099, highlighting tissue-specific differences in immune lineage populations detected with flow cytometry. UMAPs created in OMIQ on live CD45+ cells with default settings, with individual cells colored by their immune population identity as determined by gating on lineage surface markers. b Hierarchical clustering of all samples within the flow cytometry dataset using major immune lineage populations. Coloring of the heatmap represents an individual sample’s Z-score within the respective immune population. Immune lineage population clustering patterns were labeled and partitioned at k-means clustering level 3. c PCA and d biplot of all flow cytometry samples using frequencies of major immune lineage populations. e UMAP representation of the RNA component of the CITEseq dataset colored by tissue origin, f disease status, and g cluster annotation of each cell. h Number of cells within each cluster, further subsetted by tissue type and disease state. Source data are provided as a Source Data file.
In parallel, we performed CITEseq on a subgroup of the same donors, allowing for deep resolution of immune subsets in pLNs, mLNs, and spleen samples (Fig. 1e). No specific immune population selectively resolved by disease state (Fig. 1f). Major clusters identified include B cells, T cells, and innate immune cells (Fig. 1g), with surface epitopes (Supplementary Fig. 3A) and gene transcripts (Supplementary Fig. 3B) defining subsets thereof. Importantly, lineage-defining surface markers that are in both flow cytometry and CITEseq antibody panels overlapped in their expression patterns (Supplementary Fig. 4). Some clusters in B and T cells contain over 30,000 cells, with at least a few hundred cells in less abundant populations, allowing for deep characterization of rare immune cell populations of interest (Fig. 1h). The spleen contained the majority of innate immune cells, while the pLN and mLN samples contained the majority of T cells (Fig. 1e, g, h), closely resembling the flow cytometry dataset.
AAb+ and T1D-associated shifts in immune phenotypes
To define immune cell composition and phenotypic changes in T1D development, we compared immune cell population frequencies between ND, AAb+, and T1D donors. As immune cell subsets did not differ in frequency between LN draining the pancreas head, body, and tail or the SMA or MES LNs (Supplementary Fig. 2), we binned different LNs by anatomical origin, either pLNs or mLNs, for subsequent analysis. Using hierarchical clustering, we observed three different groups of disease-associated immune cell signatures, particularly in the pLNs and mLNs (Fig. 2a). Group 1 immune populations generally decreased in frequency in AAb+ and further in T1D donors, such as CD4+ naive T cells (Tn) cells and CD25+CD4+ Tem/Temra within the pLNs and mLNs. Group 2 contained pLN and mLN immune populations, including CD25+ and CD38+ T cell subsets, that decreased in frequency in AAb+ compared to ND donors and maintained a lower frequency or rebounded to normal levels in T1D. Group 3 populations increased in frequency across disease status or were elevated only in T1D. Lymphocyte subsets following this pattern included pLN and mLN populations expressing the IL7 receptor (CD127), the putative residency marker CD69, memory CD27+ B cells, and cytotoxic CD56dimCD16+ NK cells. Overall, immune cell phenotypes are modulated in the pLNs and mLNs of AAb+ and T1D donors, with changes predominantly occurring in the pLNs.
a Heatmap of immune populations, detected by flow cytometry, from each tissue type that are significantly different in frequency in at least one comparison between disease states. Coloring of the heatmap represents the mean of Z-scores for a specific immune population within the specified tissue type. Statistical significance was calculated with robust one-way ANOVA, with post hoc testing using Hochberg’s multiple comparison adjustment. Only immune populations with a differential p-value < 0.01 in at least one disease comparison were plotted. b Modules of interest generated using WGCNA analysis on scRNAseq data from pLN lymphocytes in ND and T1D disease states. c Correlations of modules of interest between ND and T1D disease states, across all immune cell clusters. Correlation values and p values derived from the Pearson correlation with a false discovery rate threshold. For all panels, * is p < 0.05, ** is p < 0.01, *** is p < 0.001. Source data are provided as a Source Data file.
We aimed to corroborate these flow cytometry findings using an unbiased weighted gene co-expression analysis (WGCNA) approach with the transcriptome modality of the CITEseq data to uncover covarying gene modules across T1D development. We only considered ND and T1D pLNs in order to identify co-varying gene modules before and after overt autoimmunity. Seventeen gene modules covaried across the disease states, representing a diverse range of defined biological processes (Fig. 2b, c, Supplementary Fig. 5A, B). Since most modules had weak correlations with T1D status, we focused on modules containing genes that overlapped with phenotypes observed via flow cytometry. Module 14 was significantly decreased in T1D within CD4+ and CD8+ naive T cell clusters (Fig. 2b, c), similar to the decreased frequency of CD4+ Tn cells observed by flow cytometry (Fig. 2a). Module 14 was anchored by TXK, a gene critical for TCR signaling38 and a regulator of interferon signaling39, and also contained the key lymphocyte development regulator IKZF1 (Fig. 2b). Module 6 included genes associated with proinflammatory signaling and cytotoxic immune cells including CCL5, CST740, KLRD1, STAT441, NKG7, and SRGN (Fig. 2b). Module 6 was increased in the CD8+ Tcm/Tem/Temra cluster (Fig. 2c). Module 5, anchored by CD127 (IL7R), consisted of genes with known roles in T cell function and signaling such as CD247, FYB142, CAMK443, PRKCH44, ITK, and LEF1 (Fig. 2b). This module was positively correlated with T1D in CD4 Tem/activated clusters in the CITEseq dataset (Fig. 2c), similar to the observed increase in frequency of CD127+CD4+ Tem/Temra T cells in the flow cytometry dataset (Fig. 2a). Overall, unbiased analysis of the CITEseq data generated from ND and T1D donor pLN lymphocytes can recapitulate some observations within the flow cytometry dataset. Subsequent analyses focused on significant phenotypes observed by flow cytometry that could be corroborated in the smaller CITEseq cohort.
Loss and dysfunction of CD4+ Tregs in the pLNs
Immune alterations before T1D onset are of particular interest to inform the development of early immune interventions capable of retaining β cell mass. In this regard, we observed a decreased frequency of CD25+CD4+ Tem/Temra in the pLNs of AAb+ donors that was maintained in the pLNs of T1D donors (Fig. 3a). CD4+ Tregs, phenotypically defined here by high CD25 and low CD127 surface expression, have previously been shown to be reduced in the pLNs of T1D donors35, and certain single nucleotide polymorphisms (SNPs) in or nearby the CD25-encoding gene IL2RA are significantly associated with T1D risk6,45. Indeed, the frequency of CD4+CD25+CD127low Tregs was significantly reduced in both AAb+ and T1D pLNs (Fig. 3b, c), suggesting that CD4+ Tregs may be altered in the pLNs prior to and during T1D onset in a tissue-restricted manner, as previously reported in the NOD mouse46. Using CITEseq, we further explored alterations in pLNs Tregs within the CD4 Treg/Tcm cluster using a set of genes associated with various Treg signatures47,48. Core Treg genes FOXP3, IKZF4, and IL2RA were significantly reduced in the CD4 Treg/Tcm cluster in both AAb+ and T1D pLNs (Fig. 3d), showing that this signature appears prior to T1D onset. Other genes associated with Tregs, such as CTLA4, were increased in AAb+ versus ND pLNs while genes negatively associated with Tregs such as IL7R decreased in AAb+ pLNs, perhaps reflecting recent activation or attempted reconstitution of Tregs in AAb+ pLNs. To discern changes within Tregs specifically, we analyzed transcriptional differences between disease states within FOXP3+ cells in the CD4 Treg/Tcm cluster (Fig. 3e). Compared to ND Tregs, AAb+ Tregs expressed lower levels of ANTXR2, a gene critical for extracellular matrix interactions49, GPR183, a GPCR required for T cell positioning near activated T cells at the T cell zone-follicle interface50, and several MAML genes associated with Notch signaling51. Some of these genes also had decreased expression in T1D Tregs versus ND Tregs (Fig. 3e). Compared to ND pLN Tregs, Tregs in T1D pLNs expressed less FOXP1, a gene essential for Treg stability52, and RUNX1, which physically interacts with FOXP3 to upregulate Treg-associated genes53. To determine whether these signatures were unique to pLN Tregs, we examined the same signatures in mLN and splenic Tregs (Supplementary Fig. 6). We observed similar decreases in CD25+CD4+ Tem/Temra frequency in AAb+ donors, but not T1D (Supplementary Fig. 6A, B). CD25+CD127low mLN CD4+ Tregs similarly decreased in mLNs of AAb+ but not in T1D donors nor in spleen of any disease state (Supplementary Fig. 6C, D). Furthermore, we did not observe transcriptomic shifts in FOXP3 or IL2RA seen in the pLNs within mLNs or spleen (Supplementary Fig. 6E). Together, these data indicate that pLN-, and to a lesser degree mLN- or splenic-, specific effects on Tregs occur both in frequency and gene expression associated with Treg lineage stability and function before T1D onset.
a Frequency of pLN CD25+ cells or b CD25+CD127− CD4+ Treg cells within CD4+ T cell subsets, as determined by flow cytometry. Statistical significance determined by robust ANOVA with post hoc testing using Hochberg’s multiple comparison adjustment. Boxplot represents the median and interquartile range, with whiskers reaching the minima and maxima. Each data point is one pLN sample, with 52 ND, 29 AAb+, and 46 T1D samples. c Representative two-parameter density plots of CD4+ Treg cells within the pLNs, as measured by flow cytometry. d Expression of CD4+ Treg-associated genes within the CD4+ Treg/Tcm cluster from pLNs. Expression scaled within each gene. Statistical significance determined by Wilcoxon Rank Sum Test and p-value adjustment using the Bonferroni method. e Top 20 differentially expressed genes (adjusted p value < 0.05, excluding common genes (see STAR Methods)) between disease states in FOXP3+ cells within the CD4 Treg/Tcm cluster in the pLN only. Statistical significance determined by Wilcoxon Rank Sum with genes with a log fold change threshold >0.1 and p-value adjustment with the Bonferroni method across all combinatorial comparisons. For all panels in this figure, * is p < 0.05, ** is p < 0.01, *** is p < 0.001. Source data are provided as a Source Data file.
Decreased naive T cell signatures in T1D pLNs
We next characterized alterations in the naive CD4+ T cell pool in T1D donors. Naive CD4+ T cell frequency decreased in T1D versus ND and AAb+ pLNs (Fig. 4a) and mLNs (Supplementary Fig. 6G), while naive CD8+ T cell frequency in the pLNs did not change with disease status (Fig. 4b). Additionally, WGCNA Module 14, which contained genes directly involved in T cell signaling and development, was negatively correlated with T1D status in both naive CD4+ and CD8+ T cells in T1D (Figs. 2b, c, 4c). Donor age could not explain shifts in naive T cell phenotype, as age did significantly differ between disease groups, and the T1D group was slightly younger in the CITEseq cohort (Supplementary Tables 1 and 2). Within the CITEseq data we identified two distinct clusters of naive CD4+ and CD8+ T cells: the first was highly enriched for naive T cell markers, termed naive #1, while the second had comparatively lower levels of naive surface markers and appeared activated, termed naive #2 (Supplementary Figs. 2, 7). Both CD4 and CD8 naive #1 clusters in the pLNs displayed a stronger negative correlation of WGCNA Module 14 with T1D status, compared to their respective naive #2 cluster (Fig. 4c). As a percentage of total pLN CD4+ T cells, CD4 cluster naive #1 trended lower in T1D compared to ND (Fig. 4d), similar to that observed by flow cytometry (Fig. 4a). Conversely, cluster CD4 naive #2 trended toward an increasing proportion of cells in T1D (Fig. 4d). Neither of the naive CD8+ T cell clusters changed in frequency between disease states (Fig. 4b, e). Several genes within WGCNA Module 14 demonstrated significantly reduced expression in AAb+ or T1D pLNs compared to ND pLNs, particularly within the CD4 and CD8 Naive #1 clusters (Fig. 4f–h). TXK, a gene critical for TCR signaling38 and a regulator of interferon signaling39, and FKBP5, a gene that coordinates with FOXO4 to modulate cytokine production54, were significantly decreased in T1D versus ND across all naive subsets. ATM, a gene critical for DNA repair during V(D)J recombination55, was also decreased in both CD4 and CD8 naive #1 clusters but not naive #2 clusters from T1D donors. Taken together these data demonstrate a reduction in CD4+ naive T cells in T1D that is accompanied by decreased expression of genes in both CD8+ and CD4+ naive T cells involved in processes essential for T cell signaling and DNA repair.
a Frequency of pLN CD4+ Tn within total CD4+ T cells or b pLN CD8+ Tn within total CD8+ T cells, as determined by flow cytometry. Statistical significance determined by two-sided robust ANOVA with post hoc testing using Hochberg’s multiple comparison adjustment. Boxplot represents median and interquartile range, with whiskers reaching the minima and maxima. Each data point is one pLN sample, with 52 ND, 29 AAb+, and 46 T1D samples. c Mean normalized expression of the top 10 most inter-connected genes in WGCNA Module 14, plotted across the naive T cell clusters in the pLNs and disease state. d Frequency of the two naive CD4+ T cell clusters and e the two naive CD8+ T cell clusters in the pLNs, across disease states. Boxplot represents median and interquartile range. P value determined by two-sided Wilcoxon ranked sum test with Benjamini-Hochberg multiple test correction. Boxplot represents median and interquartile range, with whiskers reaching the minima and maxima. Each data point is one pLN sample, with 6 ND, 5 AAb+, and 7 T1D samples. f Normalized expression of genes of interest from WGCNA Module 14, from the pLNs only. Statistical significance determined by two-sided Wilcoxon Rank Sum Test with a log fold change threshold of 0.1 and p-value adjustment using the Bonferroni method. Boxplot represents median and interquartile range, with whiskers reaching the minima and maxima. g Differentially expressed WGCNA Module 14 genes CD4 Naive #1 and CD4 Naive #2 clusters and h CD8 Naive #1 and CD8 Naive #2 clusters from the pLNs, between ND and T1D donors. Statistical significance tested using two-sided Wilcoxon Rank Sum Test with p-value adjustment using the Bonferroni method. For all panels in this figure, * is p < 0.05, ** is p < 0.01, *** is p < 0.001. Source data are provided as a Source Data file.
Memory CD8+ T cells display a stem-like phenotype in AAb+ and T1D pLNs
As CD8+ T cells are widely thought to play a direct role in human T1D pathology by infiltrating islets and eliminating β cells1,28, we sought a deeper examination of CD8+ T cell perturbations before and after T1D onset. In human pLNs, we found multiple CD8+ T cell phenotypic changes before and after T1D onset in both flow cytometry and CITEseq datasets. We observed a marked decrease in the proportion of cells expressing the activation markers CD25 (Fig. 5a) and CD38 (Fig. 5b) in AAb+ donors compared to ND. Decreased CD25 expression was noted amongst all memory CD8+ T cell subsets in AAb+ donor pLNs, and otherwise only amongst CCR7+CD8+ memory T cell populations in mLNs (Supplementary Fig. 6I, J). The CD38+ frequency decreased in CD8+ Tn, Tn-like (Tnl), and Tcm subsets and trended lower in CD8+ Tem and Temra subsets in the pLNs of AAb+ donors. In mLNs and spleen, CD38 surface expression only decreased in splenic Tnl of AAb+ donors compared to ND. Overall, the frequency of CD25+ and CD38+CD8+ T cell subsets decreased in the pLNs, and to a lesser degree in the mLNs and spleen, of AAb+ donors in comparison to ND.
a Frequency of CD25+ or b CD38+ cells within pLN CD8+ T cell subsets, as determined by flow cytometry. Statistical significance determined by two-sided robust ANOVA with post hoc testing using Hochberg’s multiple comparison adjustment. Boxplot represents median and interquartile range, with minima and maxima whiskers. Each data point is one pLN sample, with 52 ND, 29 AAb+, and 46 T1D samples. c Mean normalized expression of WGCNA Module 6 and effector CD8+ T cell genes of interest in pLNs. d Normalized expression of CXCR3 and TOX in pLN CD8 Tcm/Tem/Temra cells. Statistical significance determined by two-sided Wilcoxon Rank Sum test with Bonferroni correction. Dashed red line indicates the exclusive threshold (>0.5) for demarcating positive gene expression. Each point represents one cell. Boxplot information follows (a). e Frequency of pLN CD8 Tcm/Tem/Temra cells expressing permutations of CXCR3 and TOX, across disease state. P value determined by two-sided Wilcoxon rank sum test with Benjamini-Hochberg multiple test correction. Boxplot information follows (a). Each data point is one pLN sample, with 6 ND, 5 AAb+, and 7 T1D samples. f Differential surface proteins enriched in CXCR3-TOX+ cells and g CXCR3+TOX- cells compared to all other permutations of CXCR3 and TOX expression within pLN CD8 Tcm/Tem/Temra cells. Other clusters in the plot are comparators. Surface markers ranked in descending order of fold change. Statistical significance determined by Wilcoxon rank sum test on ADTs with log fold change threshold >0.1 followed by Bonferroni multiple test adjustment (adjusted p value < 0.05 denotes significance). Populations of interest highlighted by black outline. h Genes differentially expressed in CXCR3-TOX+ cells and i CXCR3 + TOX- cells, compared to the CXCR3 and TOX expression permutations, within the pLN CD8 Tcm/Tem/Temra cluster. Plot parameters are the same as in (f), but with log fold change threshold >0.25 and only genes with a p < 0.0001 are plotted. For all panels, * is p < 0.05, ** is p < 0.01, *** is p < 0.001. Source data are provided as a Source Data file.
We further explored T1D-associated memory CD8+ T cell alterations in relation to disease state using CITEseq (Fig. 5c). CD25 and CD38 antibody-derived tag (ADT) signatures could not be used to corroborate phenotypes observed with flow cytometry due to low resolution of these markers in memory CD8+ T cell clusters. Therefore, we instead utilized genes within the proinflammatory and cytotoxicity associated WGCNA Module 6 that positively correlated with T1D in memory CD8+ T cells (Fig. 2b, c) and genes associated with autoimmune or tissue-targeting CD8+ effector memory T cells (Fig. 5c)34,56. Cells in the CD8 Tcm/Tem/Temra cluster included a diverse range of memory CD8+ T cell subsets, based on gene expression signals associated with stem, effector, and effector memory CD8+ T cells such as TCF7, EOMES, TOX, PRF1, GZMB, and CXCR3 (Fig. 5c). Of these genes, CXCR3 and TOX were the only genes differentially expressed in both AAb+ and T1D pLNs relative to ND pLNs, with expression of CXCR3 increasing and TOX decreasing (Fig. 5d). TOX had similar expression patterns in mLNs and spleen, while CXCR3 expression decreased in T1D versus ND mLNs (Supplementary Fig. 6M, N), suggesting that the pLNs may have a unique CXCR3 gene expression pattern compared to the other tested tissues (Supplementary Fig. 6M, N). The frequency of CXCR3-TOX+ and CXCR3+TOX- cells in the pLNs trended in the same patterns as TOX and CXCR3 gene expression (Fig. 5e), suggesting that the cells driving this gene expression pattern exclusively expressed either CXCR3 or TOX. Compared to all cells in the CD8 Tcm/Tem/Temra cluster, CXCR3-TOX+ cells had higher levels of PD1 and CCR4 surface protein (Fig. 5f). Conversely, CXCR3+TOX- cells expressed relatively higher levels of the lymph node homing receptor CD62L (L-selectin), the immune inhibitory receptor CD94 (KLRD1), and the costimulatory molecule CD26 on the cell surface (Fig. 5g). Within the CD8 Tcm/Tem/Temra cluster, CXCR3-TOX+ cells and CXCR3+TOX- cells had distinct gene expression patterns (Fig. 5h, i). Genes with higher expression in CXCR3-TOX+ cells included chemokines CCL4 and CCL4L2, inhibitory receptor TIGIT, costimulatory molecule ICOS, and transcription factors IKZF2 and BCL2 (Fig. 5h). CXCR3+TOX- cells expressed higher levels of immunosuppressive genes KLRD1 and CD55, the CD127-encoding gene IL7R, and the stemness-enforcing transcription factor BACH257,58. In total, pLN CD8+ T cells exhibited changes that manifested in AAb+ donors prior to T1D development: decreased cell surface expression of activation markers CD25 and CD38, a reduction of an effector TOX+ population, and an increase in a stem-like CXCR3+ population. The mLNs and spleen also contained some, but not all, of these phenotypic differences between disease states, specifically overlapping with pLNs in CD25 surface expression and TOX expression patterns.
Cytotoxic NK cells more prominent in T1D pLNs
We examined innate cell populations for potential changes associated with AAb+ or T1D status. While few overt differences were noted within most innate cell lineages present within the pLNs or mLNs across the different cohorts, we found that there was a proportional increase in the cytotoxic CD56dimCD16+ NK cell frequency in T1D pLNs (Figs. 2a, 6a, b). We further assessed differential gene expression within NK cell clusters between ND and T1D, focusing on genes within WGCNA Module 6, which contains genes associated with cytotoxicity, and additional NK cell cytotoxicity-associated genes from the KEGG pathway database59,60,61. Cytotoxicity-associated genes GZMB, CRTAM, and IFNG were upregulated in T1D (Fig. 6c, d). Further, the NK cell inhibitory receptor KLRB1 was the most downregulated gene by fold change in the specific gene set (Fig. 6c, e). GZMB+ NK cells in the pLNs were highly enriched in genes associated with cytotoxicity such as NKG7, KLRD1, and GNLY (Fig. 6f), while GZMB- NK cells in the pLNs expressed IL1R1 and the immunosuppressive enzyme IL4I162 (Fig. 6g). Compared to ND, GZMB expression was not increased in T1D in the mLNs or spleen, while KLRB1 decreased in expression in the mLNs (Supplementary Fig. 6O, P), indicating that the increased cytotoxic NK cell frequency and transcriptomic signature was observed in the pLNs, and not the mLNs or spleen, of T1D donors.
a Frequency of CD56dimCD16+ NK cells within the pLNs, as measured by flow cytometry. Statistical significance determined by two-sided robust ANOVA with post hoc testing using Hochberg’s multiple comparison adjustment. Boxplot represents median and interquartile range, with whiskers reaching the minima and maxima. Each data point is one pLN sample, with 52 ND, 29 AAb+, and 46 T1D samples. b Representative two-parameter density plots images of NK cell subsets within the pLNs. c List of differentially expressed genes, ranked by descending log2 fold change, of genes that are significantly more expressed in T1D pLNs. Cells are from a combination of the NK and NK/ILC clusters, from the pLNs only. Gene sets to test came from the Module 6 of the WGCNA analysis, which was significantly increased in NK cell clusters in T1D, and from the “Natural killer cell mediated cytotoxicity” gene set from Kyoto Encyclopedia of Genes and Genomes (KEGG). Statistical significance tested using Wilcoxon Rank Sum Test with p-value adjustment for multiple comparisons using the Bonferroni method. d Normalized expression of GZMB and e KLRB1 within the combined NK and NK/ILC clusters in the pLNs. Statistical significance tested using two-sided Wilcoxon Rank Sum Test with p-value adjustment using the Bonferroni method. Boxplot represents median and interquartile range, with whiskers reaching the minima and maxima. f Mean normalized expression of genes significantly increased in GZMB+ and g GZMB- cells from the combined NK and NK/ILC clusters in the pLNs. P value determined by two-sided Wilcoxon Rank Sum Test with p-value adjustment using the Bonferroni method. For all panels in this figure, * is p < 0.05, ** is p < 0.01, *** is p < 0.001. Source data are provided as a Source Data file.
Immune populations associate with HLA genetic risk and single AAb positivity
HPAP collects and publicly shares extensive clinical information on each donor, allowing for the correlation of immune phenotypes with these metadata. One such parameter is HLA type, where certain alleles of HLA-DR and HLA-DQ convey significant risk for T1D63,64,65. Higher genetic risk is associated with the rapid onset of T1D66 and can distinguish between young adults with T1D versus T2D67. Therefore, profiling the immune system in individuals with high genetic risk may uncover immune perturbations associated with T1D disease susceptibility. Using only the HLA risk score portion of Genetic Risk Score 2 (GRS2)68, we generated an HLA genetic risk score (HLA-GRS) for each donor that accounts for HLA DR-DQ haplotype interactions17,68, with a higher score indicating a higher risk for disease development. As expected, T1D donors had increased HLA-GRS compared to ND (Fig. 7a). After controlling for the effects of disease state on immune population frequency, we observed moderate correlations between HLA-GRS and the frequency of specific pLN immune populations. For example, when grouping ND and AAb+ donors, the frequency of CD8+ Tcm and Tem expressing CXCR5, HLA-DR and/or CD38 were positively correlated with HLA-GRS (Fig. 7b–d). When grouping ND and T1D donors, similar populations were associated with HLA-GRS but did not reach statistical significance, likely due to these populations being regressed out due to their increased frequency in T1D pLNs (Fig. 2a). When binning AAb+ and T1D donors, the frequency of CD69+ cells in B cells and memory T cell subsets correlated positively with HLA-GRS (Fig. 7e, f). The frequency of CD127+ memory T cell subsets also positively correlated with HLA-GRS, with a reciprocal decrease in the frequency of CD127- T cells (Fig. 7e, g). Together, these data indicate that higher-risk HLA haplotype combinations are positively associated with the frequency of certain pLN immune populations, particularly activated CD8+ memory T cells, CD127+ T cells, and CD69+ B and T cells.
a HLA-GRS score of donors in the cohort. Boxplot represents median and interquartile range. P value generated with Dunn’s test with multiple hypothesis correction adjustment using Holm’s method. * is p < 0.05. Boxplot represents median and interquartile range, with whiskers reaching the minima and maxima. Each data point is one donor, with 18 ND, 10 AAb+, and 16 T1D samples. b Immune populations that significantly correlate with HLA-GRS in ND and AAb+ donors. p values generated using the Kendall tau correlation value corrected with the Benjamini-Hochberg multiplicity adjustment. Grey line represents an adjusted p value ≥ 0.05. Dot fill represents a Kendall tau correlation value corrected for disease state effects. c, d Representative plots of HLA-GRS versus immune population frequency in ND and AAb+ donors. τc is the Kendall tau correlation value corrected for disease state effects, represented by the blue linear regression line with standard error in grey. pc is the p value adjusted for disease state effects and corrected with Benjamini-Hochberg multiplicity adjustment. The greyed area represents the 95% confidence level interval for predictions from the linear model. e Immune populations that significantly correlate with HLA-GRS in AAb+ and T1D donors. All plot parameters follow (b), (f) and (g) Representative plots of HLA-GRS versus immune population frequency in AAb+ and T1D donors. All plot parameters follow (c) and (d). Source data are provided as a Source Data file.
We further explored immune perturbations between donor categories loosely resembling stages of T1D disease progression. We binned donors into four categories consisting of AAb-negative T1D-negative (ND), single AAb+ T1D-negative (Single AAb+), multiple AAb+ T1D-negative/T1D diagnosis within the last 3 years (Onset), and T1D diagnosis more than 3 years ago (Late T1D). While limited donor numbers disallowed further subdivision, we aimed to capture low-risk AAb+ donors in the Single AAb+ group, donors with the highest likelihood of autoinflammatory activity in the Onset group, and late-stage autoimmunity in the Late T1D group. Across these categories, significantly changing immune populations in the pLNs largely resembled those that change across the original ND, AAb+, and T1D groupings but with additional insights (Supplementary Fig. 8A). For example, several immune populations are altered in the low-risk Single AAb+ group. The frequency of CD38+ T cell subsets, in both CD4+ and CD8+ T cells, decreases significantly only in Single AAb+ donors (Supplementary Fig. 8B, C), while CD25+CD4+ Tem are less frequent in Single AAb+ donors and maintain their decreased frequency in Onset and Late T1D donors (Supplementary Fig. 8D). Memory B cells are also significantly increased in Single AAb+ donors, as well as Late T1D (Supplementary Fig. 8E). Other immune changes occur in a step-wise fashion across these categories, perhaps reflecting longer-standing inflammation promoting immune differentiation, including a decrease in CD4+ Tn frequency (Supplementary Fig. 8F) and an increase in the proportion of cytotoxic CD56dimCD16 + NK cells (Supplementary Fig. 8G). Together, these data demonstrate that even low-risk Single AAb+ donors have immune perturbations in their pLNs, and that certain immune populations tend to gradually change as individuals have autoinflammation for longer periods of time.
Discussion
Our current knowledge of the landscape of immune perturbations during the ontogeny of T1D largely originates from the NOD mouse model and targeted analysis of specific immune subsets in human peripheral blood, with limited studies of human pLNs31,32,33,34,35,36. Here, we broadly profiled lymphocyte subsets from lymphatic tissues in a large cross-sectional cohort of ND, AAb+, and T1D human organ donors to define immunological perturbations occurring in pLNs prior to and after disease diagnosis. Compared to ND, AAb+ and T1D pLNs have a reduced Treg signature and increased stem-like CXCR3+CD8+ T cells. Some perturbations were T1D pLN specific, including a reduced naive T cell signature and an increased frequency of cytotoxic CD56dimCD16+ NK cells. Additionally, several immune populations in the pLNs correlated with HLA genetic risk independently of the effects of disease status, in particular CD8+ T cell activation signatures and increased CD69 and CD127 surface expression on T cells and/or B cells. Importantly, immune alterations were most readily observed in lymphoid tissues that drain the pancreas as opposed to the spleen, implying that disease-related effects may be diluted or influenced by other factors at sites more distal from autoimmune inflammation.
One of the clearest observed signatures was a marked decrease in CD25 expression on CD4+ and CD8+ T cells in the pLNs of AAb+ individuals. This was most strongly reflected by an overall decrease in CD4+ Tregs, a phenomenon observed previously in T1D35, but shown here to also manifest in AAb+ donors and in individuals with T1D. IL-2 signaling and production deficiencies have long been tied to T1D susceptibility, as PBMCs from children with recent onset T1D produce less IL-269,70, and several T1D susceptibility genomic loci are involved in IL-2 signaling6,45. IL-2 induced signaling responses are critical to maintaining T cell tolerance to self-antigen through the maintenance of CD4+ Tregs71, and IL-2 signaling strongly influences CD8+ memory T cell differentiation72. Decreases in pLN Treg frequency in AAb+ donors highlight the evolution towards an inflammatory environment that may occur in pLNs before T1D onset. Importantly, CD25+CD4+ Tem are less frequent even in single AAb+ donor pLNs, highlighting that IL-2 signaling may be altered in donors that have a low risk of T1D development. Further, while CD25+CD4+ Tem frequency in the mLNs and spleen have similar patterns across disease states compared to pLNs, a decline of a Treg signature in AAb+ or T1D disease states was not observed in mLNs or spleen, indicating that the pLNs are uniquely losing this cell population known to be critical for autoimmune control.
The loss of Tregs in AAb+ individuals was accompanied by an increase of a stem-like CD8+ T cell population closely resembling one found in multiple autoimmune contexts, including a population that resides in the pLNs and drives autoimmunity in NOD mice34 and is associated with ulcerative colitis in mice and humans73. In humans, stem-like CD8+ T cells12 and CXCR3+CD8+ T cell subsets17 in PBMCs are positively associated with T1D status. In human AAb+ and T1D pLNs, we also observed increased frequencies of less differentiated stem-like CXCR3+ memory CD8+ T cells. Importantly, CXCR3+ memory CD8+ cells did not change across disease state in the mLNs, suggesting localization of this effect to the pLNs. We further found that a TOX+ memory CD8+ T cell population defined by surface PD1 and CCR4 protein and gene expression of CCL4, IKZF2, and BCL2 reciprocally decreased in the pLNs of AAb+ individuals. While PD1+CD8+ T cells expressing TOX have been described as being functionally exhausted in some contexts, many polyfunctional non-exhausted CD8+ T cells express PD1 and TOX in human lymph nodes74. The PD1+TOX+CD8+ T cell population shares features with IKZF2+KIR+CD8+ T cells, which play a role in peripheral tolerance during inflammatory conditions75. It is unclear how these events are temporally associated with progression as T1D-associated AAb specificities develop76, but a loss of Treg activity or changes in IL2 signaling could enable the survival and expansion of T1D autoantigen-specific CD8+ T cells in the local pLN environment.
We also observed an increased proportion of cytotoxic NK cells and decreases in frequency and transcriptomic alterations in naive CD4+ and CD8+ T cells, in pLNs and mLNs of T1D donors. The greatest change in proportion of naive CD4+ T cells and cytotoxic NK cells was in longer-standing T1D donors. Although various pathways may explain the general differentiation of naive T cells and NK cells, evidence in the context of T1D implicates cytokine signaling, and in particular γ-chain cytokines, as a potential mechanism behind these observations. Long-term inflammation drives immune aging, activation, and differentiation signatures in PBMCs of T1D donors17. γ-chain cytokines IL7 and IL21 are elevated in the circulation of long-standing, and not newly diagnosed, T1D donors77,78, and IL7 and IL21 signaling are required for diabetes development in NOD mice79,80,81. IL21 and IL15 promote NK cell differentiation into cytotoxic subsets, including CD56dimCD16+ NK cells82, and γ chain cytokines promote naive T cell differentiation83, similar to phenotypic changes we observed in these immune subsets. Additionally, IL7 is known to increase the expression of its canonical receptor CD12784, and we observed that CD127 expression in the pLNs was positively associated with T1D status and HLA-associated T1D risk. Several IL2-related phenotypic changes were identified in the pLNs of AAb+ donors, implying that certain γ chain cytokines may impact earlier stages of T1D progression and others after disease establishment. Examining if γ chain cytokine signals actively drive the observed phenotypes in the T1D context, and whether they derive from the lymph node or upstream from the pancreas, will inform therapeutic strategies for immune intervention during or after T1D onset.
By calculating T1D genetic risk conveyed by HLA class II alleles, we found that the frequency of memory CD8 + T cells with HLA-DR and CD38 surface expression was positively correlated with HLA-GRS, indicating that non-T1D individuals with higher-risk HLA alleles have an increased frequency of activated memory CD8 + T cells in the pLNs. While cytotoxic CD8 + T cells infiltrate islets and eliminate β cells in T1D26,29, it is unclear how or where these autoimmune CD8 + T cells develop in humans. Evidence from the NOD mouse implicates stem-like autoimmune CD8 + T cells as a reservoir of T cells that infiltrate the pancreas34, and we observed an expansion of stem-like memory CD8 + T cells in the pLNs of AAb+ and T1D donors. It is unknown whether pLN CD8 + T cell activation observed in high-risk individuals is related to the expansion of stem-like CD8 + T cells, but a higher basal T cell activation state may potentiate the expansion of autoimmune CD8 + T cells. We found that a higher HLA-GRS in AAb+ and T1D donors positively correlates with CD127 surface expression on T cells. CD127, along with the common γ chain receptor subunit, comprises the IL7R that is critical for T cell homeostatic proliferation85, implying that pLN T cells in individuals with higher HLA-GRS may have heightened IL7 responsiveness86. AAb+ and T1D individuals with a higher HLA-GRS also had increased frequencies of CD69 + B and T cell subsets, indicating these individuals may have more tissue-resident87 or activated88 immune cells. As tissue residency is critical for local tissue immune memory responses87, further investigation is needed to explore if high-risk individuals have an expanded immune memory pool that could potentiate autoimmune responses.
Although most single AAb+ individuals do not develop T1D2,89, we found immune changes in the pLNs of this donor group. Specifically, the frequency of CD25+CD4+ Tem was decreased, as were CD38+ cells in many T cell subsets, while CD27+ memory B cells increased in frequency. While the connection between these immune changes is unclear, there is evidence of a relationship between CD38 and CD25+CD4+ Tregs. Treating human PBMCs with CD38-blocking monoclonal antibody leads to a reduction in CD25+ Treg frequencies and suppressive activity90, while another anti-CD38 monoclonal antibody reduced patient levels of immunosuppressive CD38+ Tregs and anti-inflammatory B cells91. These studies imply that CD38 is important for immunosuppressive cell activity, but the directionality of this association is uncertain. The observed increase in pLN CD27+ memory B cells further implies the pLNs of single AAb+ individuals have a pro-inflammatory environment, as evidence suggests B cells play an active role in T1D pathogenesis92. Despite the immune modulations we observed in single AAb+ donor pLNs, it is highly likely these individuals would have maintained β cell tolerance. More studies are needed to understand how low-risk single AAb+ donors can have pancreas-proximal pro-inflammatory immune changes yet maintain tolerance, and if the immune modulations we observed are associated with disease progression.
Our study was limited in a number of aspects. First, due to the complexities of donor acquisition, we were limited in our ability to acquire enough AAb+ organ donors with two or more T1D-associated AAbs, preventing a properly powered subanalysis of single versus multiple AAb+ donors. As the study design is inherently cross-sectional, we do not have longitudinal data, including glucose tolerance tests, to properly stage AAb+ individuals. This limits our ability to stage the progression of immune perturbations that we observed in our cross-sectional study. Individuals without diabetes who have multiple T1D-associated AAbs are rare, and generally progress to T1D more rapidly than those with single autoantibodies2. Furthermore, as not all single AAb+ donors develop T1D2,93, additional studies of pLNs from donors with >1AAb+ across a range of AAb specificities will be necessary to define immunological associations with pre-T1D. Second, due to blood samples being drawn well before donor organ harvest, we determined that peripheral blood lymphocytes yielded unreliable flow cytometric and transcriptomic data (data not shown). As such, we cannot directly compare our results to previous studies focusing exclusively on peripheral blood. Third, we restricted our flow cytometric analysis to freshly isolated lymphocytes, rather than frozen lymphocytes, due to evidence of cryopreservation-induced changes in certain cell surface markers94. This limits subsequent flow-based analysis to prospectively acquired cohorts. Fourth, we were unable to simultaneously assess B cell or T cell repertoires in the CITEseq analysis due to technical limitations from 3’ mRNA transcriptomic analysis. Hence, studies of T cell or B cell T1D antigen specificity are pending.
The deep immune profiling of rare and valuable tissue samples from organ donors collected across the spectrum of disease stages reported here provides an expansive resource for both the T1D community as well as individuals interested in lymph node immunology. This resource is structured as an immune cell atlas in an open data format with the goal of facilitating novel investigative approaches to understand disease mechanisms and identify novel targets to intervene in T1D development. Importantly, the conjunction of the high-dimensional analyses allowed for the observation of subtle immune changes associated with AAb positivity and T1D status. The datasets provide foundational immunobiology information for the research community that could help uncover potential therapeutic targets to delay, prevent, or ameliorate T1D.
Methods
Tissue collection
All uses of human material have been approved by the University of Pennsylvania Office of Regulatory Affairs Institutional Review Board, where the Board determined this research is exempt and does not meet criteria for human subject research. Experimental model and study participant methods are described29,95. Details on the samples obtained through the HPAP are available online at https://hpap.pmacs.upenn.edu and described37. Whole organ samples are obtained via the local organ procurement organization or via the University of Florida/nPOD. All human samples were deidentified. The consent for the procurement of research pancreata was obtained by the staff of the organ procurement organization. Donors were evaluated for autoantibody status (GAD, IA-2, IAA and ZnT8) and C-peptide levels, and underwent clinical chart review. Clinical data, including diagnosis of T1D, HbA1c, BMI, age, gender, HLA type, and more, are recorded in the HPAP database. Additional data from histology, immunophenotyping, repertoire profiling, metabolic, and transcriptomic studies are generated on all HPAP donors, providing a shared and publicly available comprehensive resource.
Tissues and cells
To isolate pancreatic and mesenteric LN lymphocytes, LNs obtained from brain-dead organ donors were submerged in 5 mL R10 media (RPMI + 10% FBS + 1% penicillin/streptomycin + 2mM L-glutamine) (RPMI—Corning #10040CM; FBS—GeminiBio #100-500-500; penicillin/streptomycin solution—Corning #30002CI; L-glutamine solution—Corning #25005CI) supplemented with 10 U/mL DNase I (Millipore Sigma #04716728001), cleaned of visceral fat, and cut into small pieces (~2 mm x ~ 2 mm). Tissue fragments were placed into a sterile 70 μm mesh (Miltenyi Biotec #130-098-462) and pushed through the mesh with the blunt end of a sterile 5 mL syringe plunger. Residual tissues and cells in the mesh were washed with 45 mL of R10 + DNAse, centrifuged at 500 xg for 7 minutes, resuspended in 10 mL of R10 + DNAse, and counted. To isolate splenocytes, approximately 30 grams of spleen tissue was placed in R10 supplemented with 10 U/mL DNase I and 3 mg/ml collagenase D (Sigma #11088866001), the capsule was removed, and remaining tissue was cut into 4 mm × 4 mm pieces. The tissue was then mechanically dissociated using a GentleMACS (Miltenyi Biotec #130-093-237) and incubated at 37 °C for 15 min with gentle inversion. After digestion, the cell suspension was filtered through a 100 μm strainer (Miltenyi Biotec #130-110-917), and erythrocytes were lysed by ACK buffer (Quality Biological #118-156-101). Post-ACK, cells were resuspended in R10 media and filtered through a sterile 70 μm mesh. Splenic mononuclear cells were isolated by density gradient centrifugation using Ficoll-Paque PLUS (Cytiva #17144002). Viable cell suspensions from LN and spleen were cryopreserved in FBS + 10% DMSO and stored at −150 °C until thawing.
Flow cytometry acquisition and analysis
All antibody staining was done on freshly isolated cells following a protocol detailed elsewhere96. After washing with phosphate-buffered saline (PBS), cells (whole blood derived leukocytes, or isolated lymphocytes) were pre-stained for the chemokine receptor CCR7 for 15 min at 37 °C 5% CO2. All following incubations were performed at room temperature. Cells were stained for viability exclusion using Live/Dead Fixable Aqua (Invitrogen #L34957) for 10 minutes, followed by a 20-minute incubation with a panel of directly conjugated antibodies and Trustain (BioLegend #422302) diluted with fluorescence-activated cell sorting (FACS) buffer (PBS containing 0.1% sodium azide and 1% bovine serum albumin) and Brilliant Stain Buffer (BD Biosciences #563794). The cells were washed in FACS buffer and fixed in PBS containing 4% paraformaldehyde (Electron Microscopy Sciences #50-980-491). Cells were stored at 4 °C in the dark until acquisition. All flow cytometry data was collected on a BD FACSymphony A5 cytometer. FlowJo software version 10.9 was used to generate population frequencies and representative flow cytometry plots. R was used for further analysis of these exported population frequencies. Antibodies used for flow cytometry analysis: Anti-CD15 FITC (BD Biosciences #555401) at 1:50 dilution, Anti-CD4 BB790 (BD Biosciences Custom Conjugation) at 1:2500 dilution, Anti-CXCR5 Alexa Fluor 647 (BD Biosciences #558113) at 1:250 dilution, Anti-CD45 Alexa Fluor 700 (BD Biosciences #560566) at 1:400 dilution, Anti-CCR7 APC-Cy7 (BioLegend #353212) at 1:67 dilution, Anti-CD11c BUV395 (BD Biosciences #563787) at 1:400 dilution, Anti-CD8 BUV496 (BD Biosciences #612942) at 1:200 dilution, Anti-CD45RA BUV563 (BD Biosciences #612927) at 1:500 dilution, Anti-CD38 BUV661 (BD Biosciences #612969) at 1:400 dilution, Anti-CD25 BUV737 (BD Biosciences #612807) at 1:200 dilution, Anti-CD3 BUV805 (BD Biosciences #612895) at 1:125 dilution, Anti-PD1 BV421 (BioLegend #565935) at 1:83 dilution, Anti-CD14 BV480 (BD Biosciences #566141) at 1:1000 dilution, Anti-CD56 BV570 (BioLegend #318330) at 1:200 dilution, Anti-HLA-DR BV605 (BD Biosciences #562845) at 1:400 dilution, Anti-CD27 BV650 (BioLegend #302828) at 1:400 dilution, Anti-CD16 BV711 (BioLegend #302044) at 1:400 dilution, Anti-CD19 BV785 (BioLegend #302240) at 1:125 dilution, Anti-CD123 PE (BD Biosciences #555644) at 1:400 dilution, Anti-CD127 PE/CF594 (BD Biosciences #562397) at 1:200 dilution, Anti-CD69 PE/Cy5 (BioLegend #310908) at 1:500 dilution, Anti-CD34 PE/Cy5.5 (Invitrogen #CD34-581-18) at 1:100 dilution, Anti-CD21 PE/Cy7 (BioLegend #354912) at 1:2000 dilution.
CITEseq sample processing and library generation
Eight samples were run simultaneously for each round of CITEseq, balancing both tissue type and disease state across runs. Cryopreserved cells were thawed by gentle shaking in a 37 °C water bath until the cell suspension was partially thawed, which was immediately decanted into cold R10 medium + DNAse. Thawed cells were centrifuged at 500 xg for 7 minutes, then resuspended in room temperature R10 + DNAse. Cells were rested in R10 + DNAse at 2 × 106 cells/mL for 2 hr at 37 °C with 5% CO2. All following cell preparation and antibody staining steps, including centrifugation, were done at 4 °C. After resting, cells were centrifuged at 500 xg for 7 minutes and resuspended in 1 mL ice cold Cell Stain Buffer (CSB, BioLegend #420201). Samples were filtered through a sterile pre-wet 70 μm mesh, followed by a 2 mL wash with CSB. 500k live cells that passed through the mesh were spun at 200 xg for 10 minutes and resuspended in CSB. Trustain was added to each sample at a 1:10 dilution and incubated on ice for 10 minutes. After, each sample received a 1:37.5 dilution of unique hashing antibody (listed below) following manufacturer’s instructions. Totalseq-A antibody cocktail (BioLegend #399907), reconstituted in CSB following the manufacturer’s protocol, was added to each sample at the manufacturer’s recommended concentration. After a 30 minute incubation on ice, cells were washed 3X with ice cold CSB, counted, and pooled in CSB at 2000 cells/μL with ~125 K cells from each sample. The pooled sample was filtered through a 40μm FlowMi strainer (Sigma BAH136800040-50EA) and counted. Pooled samples were run at 17.5k cells/well on a Chromium X using the Chromium Next GEM Single Cell 3’ HT lit v3.1 (10X Genomics #1000370) or RT kit v3.1 (10X Genomics #1000268). Single cell sequencing library preparations for the RNA modality followed the manufacturer’s protocol, with the exception of spiking in primers at step 2.2 that amplify ADT and HTO (hashtag oligonucleotide) antibody sequences. 1 μL of 0.2 μM ADT additive primer (sequence: CCTTGGCACCCGAGAATT*C*C) and 1 μL of 0.1 uΜ ΗΤΟ additive primer (sequence: GTGACTGGAGTTCAGACGTGTGCTCTTCCGAT*C*T) was spiked into each individual sample prep. Preparation of the ADT and HTO single cell sequencing libraries followed manufacturer’s protocol (BioLegend) and described elsewhere (cite-seq.com)97,98. Briefly, at step 2.3d, 60 μL of supernatant was collected, 140 μL of SPRIselect reagent (Beckman Coulter #B23318) was added to the collected supernatant, washed twice with 80% ethanol, and eluted with 90 μL EB (Qiagen #B23318). For the respective ADT and HTO modalities, antibody-derived oligomers were amplified and indexed by mixing 45 μL eluent with 50 μL of KAPA HiFi HotStart ReadyMix (Roche #07958927001) and 2.5 μL of each forward and 2.5 μL of each reverse 10 μM indexing primers (see Supplementary Data). Amplification settings for the ADT modality were as follows: 95 °C 3 min, ~8 cycles of 95 °C 20 sec, 60 °C 30 sec, and 72 °C 20 sec, followed by 72 °C for 5 min and ending with hold at 4 °C. Amplification settings for the HTO modality were the same except for a 64 °C annealing temperature. Post-amplification, ADT and HTO libraries were purified with a 1.2X SPRIselect cleanup and eluted with 30 μL EB. All final libraries were quantified using a Qubit dsDNA HS Assay kit (Invitrogen #Q32854) and a High Sensitivity D1000 DNA tape (Agilent #5067-5584) on a Tapestation D4200 (Agilent). Hashing antibodies for HTO are as follows: TotalSeq-A0251 anti-human Hashtag 1 Antibody, BioLegend #394601; TotalSeq-A0252 anti-human Hashtag 2 Antibody, BioLegend #394603; TotalSeq-A0253 anti-human Hashtag 3 Antibody, BioLegend #394605; TotalSeq-A0254 anti-human Hashtag 4 Antibody, BioLegend #394607; TotalSeq-A0256 anti-human Hashtag 6 Antibody, BioLegend #394611; TotalSeq-A0257 anti-human Hashtag 7 Antibody, BioLegend #394613; TotalSeq-A0258 anti-human Hashtag 8 Antibody, BioLegend #394615; TotalSeq-A0259 anti-human Hashtag 9 Antibody, BioLegend #394617.
CITEseq library sequencing
Sequencing runs were performed on the NovaSeq 6000 platforms (Illumina) with a target of at least 10,000 reads per cell for ADT libraries, 25,000 reads per cell for RNA libraries, and 500 reads per cell for HTO libraries.
CITEseq preprocessing
The cellranger (7.0.0; 10X Genomics) suite of tools were used for all preprocessing steps for the RNA component. Raw BCL files were demultiplexed using the cellranger mkfastq tool, resulting in fastq files for each lane used in the 10X Genomics chip. Fastq files were processed into cell by gene matrices using the cellranger count tool by aligning against the hg38 reference genome. The resultant matrices were used for downstream analyses. For ADT and HTO, raw BCL files were demultiplexed using bcl2fastq2 (Illumina) to create fastq files that correspond to each lane used in the 10X Genomics chip. Fastq files were then counted using a variation of the kite pipeline (https://github.com/pachterlab/kite) which uses kallisto and bustools99 for alignment, barcode correction, and counting to create cell by feature matrices. Code for ADT and HTO preprocessing is found here: https://github.com/betts-lab/scc-proc.
CITEseq modality processing and dehashing
The RNA filtered feature barcode matrix (as outputted from cellranger count) was loaded into R (v4.1.1) and Seurat (v4.1.1) to create a Seurat object after filtering out for wells that had low cell recovery and/or poor quality control metrics as determined by cellranger. Cells were then further filtered by the following criteria for initial quality checking: number of features >200 & number of features <6000 & percentage of mitochondrial reads <12.5%. RNA counts were then normalized on a per cell basis. The ADT barcode matrix (as outputted from kallisto bustools) was loaded into the emptyDrops function (DropletUtils v1.14.2) with a minimum threshold of 100 UMI counts to determine what barcodes are associated with empty droplets (false discovery rate <0.01). The empty droplets were filtered out of the ADT matrix. The intersection of usable barcodes was determined between the QC filtered RNA modality, the QC filtered ADT modality, and the HTO barcode matrix. The Seurat object was filtered to retain only barcodes that were detected/passing QC in all three modalities. After adding the ADT and HTO modalities as separate Assay objects, we normalized the HTO data within each hashtag. The HTODemux() function in Seurat with the argument (positive.quantile = 0.99) was used to demultiplex samples based on hashtag. Samples that were classified as singlets were kept for downstream analyses.
Modality integration and clustering
All runs were merged into a single Seurat object. ADT data were scaled and normalized using the centered-log ratio method by cell. Principal component analysis was performed for RNA and ADT separately and resultant principal components were each used as input for batch effect correction using Harmony100 with grouping variables set by donor and by each chip used for droplet emulsions. The batch effect corrected RNA-based principal components were used to generate a shared nearest neighbor (sNN) graph. Using the sNN graph, clustering was performed with the Leiden algorithm and the following settings for the FindClusters method in Seurat (method = “igraph” and resolution = 1). A uniform manifold approximation and projection (UMAP) representation of the data was constructed from the sNN graph for visualization purposes. Using both the ADT and RNA modalities, clusters were manually annotated. Clusters with multiplet signatures or phenotypes indicative of poor sample quality/viability were discarded. Given the known viability difficulties with human biopsy samples, we used a heat shock protein (HSP) gene set list (Group 582) from the HUGO Gene Nomenclature Committee (HGNC)101. These genes were fed into the AddModuleScore function in Seurat to generate a heat shock module. To remove confounding effects associated with heat shock response, cells with a score less than the 95th percentile of overall heat shock module scores were kept for downstream analysis.
WGCNA for pancreatic lymph node samples
The dataset after clustering and quality control filtering were filtered for cells from the pancreatic lymph nodes and from donors associated with ND or T1D disease status for a more targeted analysis without confounding effects from other tissue sites and the inherent diversity of donors labeled as autoantibody positive. A WGCNA was constructed using the WGCNA102 and hdWGCNA103 packages where metacells were formed based on the following grouping variables: donor, disease status, and condensed phenotype from manual annotations. The resulting topological co-expression network was used to identify modules and assess correlations with disease status and manual phenotypes. All specific function parameters and steps for WGCNA are detailed in the study GitHub repository (see Code availability).
HLA-GRS calculation and correlation to immune populations
Phasing of HLA-DQA1 and HLA-DQB1 genotypes was inferred via comparison with published haplotype frequencies from European Americans104. HLA-DQA1-DQB1 haplotypes were used to calculate the HLA component of the T1D polygenic risk score, GRS2, according to methods developed by Sharp et al.68. Scoring considered additive odds ratios for 14 individual haplotypes and interactions for 18 non-additive haplotype combinations as in the Polygenic Risk Score (PRS) Toolkit for HLA (v0.22a)68. Correlations between HLA-GRS and immune population frequency were computed using partial correlation analysis from the ppcor R package.
Graphics
All figures were made in R with the following packages: grid, ggplot2105, ComplexHeatmap, Seurat, hdWGCNA, and patchwork. All code to produce figures can be found in the study GitHub repository (see Code availability). Annotations were added to the R-generated figures using Adobe Illustrator (v.22.2.3).
Statistical analyses
For flow cytometry, all statistical analyses were run in R with the rstatix, ggpubr, and multicomp packages. Non-parametric analyses between two groups were performed with a two sample Wilcoxon test, with multiple hypothesis correction using Holm’s method when appropriate, using the rstatix package. Non-parametric analyses between 3 or more groups were performed with Dunn’s test, with Holm’s method for multiple hypothesis correction adjustment, using the rstatix package. Parametric comparisons between 3 or more groups were performed with robust one-way ANOVA in the WRS2 package, with post hoc testing using Hochberg’s multiple comparison adjustment and tr = 0.1. If cold ischemia was found to affect the frequency of an immune population, ANCOVA was used to control for the effect of cold ischemia on immune population frequency while comparing 3 or more groups, with post hoc analysis performed with Tukey’s test, using the multicomp package. Each dot in a plot depicting summarized flow cytometry data is an individual sample. Sample distributions represented by a boxplot, with the median depicted as the center value, quartile ranges forming the box edges, and whiskers depicting the minimum to maximum distribution. Correlations between HLA-GRS and immune population frequency were run using the partial Kendall rank correlation while controlling for the confounding variables of disease state and cold ischemia time. Correlation p-values were adjusted for multiple comparisons using Benjamini-Hochberg multiple test correction. For CITEseq analysis, samples were quality controlled as explained in the “CITEseq modality processing and dehashing” section. To help elucidate more specific cell type differences in differential gene analysis, a common set of differentially expressed genes was computed for each manually annotated cluster between disease states using the Wilcoxon Rank Sum implementation in Seurat FindAllMarkers with a log fold change threshold of 0.1. Any differential genes that were found in more than 12 clusters were labeled as common genes that were largely independent of cell phenotype. Combinatorial differential expression testing was performed with Seurat FindMarkers (Wilcoxon Rank Sum) followed by Bonferroni correction for multiple tests calculated across all tests performed across the combinations. Unless otherwise stated in text or figure legends, significance by statistical test was set as adjusted p value < 0.05. * is p < 0.05, ** is p < 0.01, *** is p < 0.001 for all figures. Refer to figure legends for the application and details of each test.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All de-identified human flow cytometry and clinical data have been deposited in Zenodo under the digital object identifier 10.5281/zenodo.15001886 [https://zenodo.org/records/15001886], and on PANC-DB (https://hpap.pmacs.upenn.edu/). Raw sequencing-based data are also deposited in Genbank under accession number GSE221787. All data are publicly available as of the date of publication. All data are included in the Supplementary Information or available from the authors, as are unique reagents used in this Article. The raw numbers for charts and graphs are available in the Source Data file whenever possible. Source data are provided with this paper.
Code availability
All original code has been deposited on GitHub [https://github.com/betts-lab/hpap-tissue-citeseq] and in Zenodo under the digital object identifier 10.5281/zenodo.15027610 [https://zenodo.org/records/15027610]. Exceptions are the code used to generate HLA-GRS [https://github.com/USF-HII/hla-prs-toolkit] version 0.22a68, and code for ADT and HTO preprocessing [https://github.com/betts-lab/scc-proc]. All code are publicly available as of the date of publication.
References
Atkinson, M. A., Eisenbarth, G. S. & Michels, A. W. Type 1 diabetes. Lancet 383, 69–82 (2014).
Insel, R. A. et al. Staging presymptomatic type 1 diabetes: a scientific statement of JDRF, the Endocrine Society, and the American Diabetes Association. Diab. Care 38, 1964–1974 (2015).
Oram, R. A. et al. Most people with long-duration type 1 diabetes in a large population-based study are insulin microsecretors. Diab. Care 38, 323–328 (2015).
Williams, G. M. et al. Beta cell function and ongoing autoimmunity in long-standing, childhood onset type 1 diabetes. Diabetologia 59, 2722–2726 (2016).
Gubitosi-Klug, R. A. et al. Residual β cell function in long-term type 1 diabetes associates with reduced incidence of hypoglycemia. J. Clin. Invest. 131, e143011 (2021).
Todd, J. A. et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 39, 857–864 (2007).
Nejentsev, S. et al. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature 450, 887–892 (2007).
Shapiro, M. R. et al. De-coding genetic risk variants in type 1 diabetes. Immunol. Cell Biol. 99, 496–508 (2021).
Sims, E. K. et al. Teplizumab improves and stabilizes beta cell function in antibody-positive high-risk individuals. Sci. Transl. Med. 13, eabc8980 (2021).
Long, S. A. et al. Partial exhaustion of CD8 T cells and clinical response to teplizumab in new-onset type 1 diabetes. Sci. Immunol. 1, eaai7793 (2016).
Kenefeck, R. et al. Follicular helper T cell signature in type 1 diabetes. J. Clin. Invest. 125, 292–303 (2015).
Abdelsamed, H. A. et al. Beta cell-specific CD8+ T cells maintain stem cell memory-associated epigenetic programs during type 1 diabetes. Nat. Immunol. 21, 578–587 (2020).
Pesenacker, A. M. et al. Treg gene signatures predict and measure type 1 diabetes trajectory. JCI Insight 4, e123879 (2019).
Hanley, P. et al. Circulating B cells in type 1 diabetics exhibit fewer maturation-associated phenotypes. Clin. Immunol. 183, 336–343 (2017).
Stensland, Z. C. et al. Identification of an anergic BND cell-derived activated B cell population (BND2) in young-onset type 1 diabetes patients. J. Exp. Med. 220, e20221604 (2023).
Xhonneux, L.-P. et al. Transcriptional networks in at-risk individuals identify signatures of type 1 diabetes progression. Sci. Transl. Med. 13, eabd5666 (2021).
Shapiro, M. R. et al. Human immune phenotyping reveals accelerated aging in type 1 diabetes. JCI Insight 8, e170767 (2023).
Poon, M. M. L. et al. Tissue adaptation and clonal segregation of human memory T cells in barrier sites. Nat. Immunol. 24, 309–319 (2023).
Buggert, M. et al. The identity of human tissue-emigrant CD8+ T cells. Cell 183, 1946–1961.e15 (2020).
Buggert, M., Price, D. A., Mackay, L. K. & Betts, M. R. Human circulating and tissue-resident memory CD8+ T cells. Nat. Immunol. 24, 1076–1086 (2023).
Akkaya, M., Kwak, K. & Pierce, S. K. B cell memory: building two walls of protection against pathogens. Nat. Rev. Immunol. 20, 229–238 (2020).
Hashemi, E. & Malarkannan, S. Tissue-resident NK cells: development, maturation, and clinical relevance. Cancers 12, 1553 (2020).
Babon, J. A. B. et al. Analysis of self-antigen specificity of islet-infiltrating T cells from human donors with type 1 diabetes. Nat. Med. 22, 1482–1487 (2016).
Culina, S. et al. Islet-reactive CD8 + T cell frequencies in the pancreas, but not in blood, distinguish type 1 diabetic patients from healthy donors. Sci. Immunol. 3, eaao4013 (2018).
Opie, E. L. On the relation of chronic interstitial pancreatitis to the islands of Langerhans and to diabetes melutus. J. Exp. Med. 5, 397–428 (1901).
Coppieters, K. T. et al. Demonstration of islet-autoreactive CD8 T cells in insulitic lesions from recent onset and long-term type 1 diabetes patients. J. Exp. Med. 209, 51–60 (2012).
Leete, P. et al. Differential insulitic profiles determine the extent of β-cell destruction and the age at onset of type 1 diabetes. Diabetes 65, 1362–1369 (2016).
Campbell-Thompson, M. et al. Insulitis and β-cell mass in the natural history of type 1 diabetes. Diabetes 65, 719–731 (2016).
Wang, Y. J. et al. Multiplexed in situ imaging mass cytometry analysis of the human endocrine pancreas and immune system in type 1 diabetes. Cell Metab. 29, 769–783.e4 (2019).
Damond, N. et al. A map of human type 1 diabetes progression by imaging mass cytometry. Cell Metab. 29, 755–768.e5 (2019).
Gagnerault, M.-C., Luan, J. J., Lotton, C. & Lepault, F. Pancreatic lymph nodes are required for priming of beta cell reactive T cells in NOD mice. J. Exp. Med. 196, 369–377 (2002).
Kent, S. C. et al. Expanded T cells from pancreatic lymph nodes of type 1 diabetic subjects recognize an insulin epitope. Nature 435, 224–228 (2005).
Seay, H. R. et al. Tissue distribution and clonal diversity of the T and B cell repertoire in type 1 diabetes. JCI Insight 1, e88242 (2016).
Gearty, S. V. et al. An autoimmune stem-like CD8 T cell population drives type 1 diabetes. Nature 602, 156–161 (2022).
Ferraro, A. et al. Expansion of Th17 cells and functional defects in T regulatory cells are key features of the pancreatic lymph nodes in patients with type 1 diabetes. Diabetes 60, 2903–2913 (2011).
Vecchione, A. et al. Reduced follicular regulatory T cells in spleen and pancreatic lymph nodes of patients with type 1 diabetes. Diabetes 70, 2892–2902 (2021).
Kaestner, K. H., Powers, A. C., Naji, A., HPAP Consortium & Atkinson, M. A. NIH initiative to improve understanding of the pancreas, islet, and autoimmunity in type 1 diabetes: the Human Pancreas Analysis Program (HPAP). Diabetes 68, 1394–1402 (2019).
Readinger, J. A., Mueller, K. L., Venegas, A. M., Horai, R. & Schwartzberg, P. L. Tec kinases regulate T-lymphocyte development and function: new insights into the roles of Itk and Rlk/Txk. Immunol. Rev. 228, 93–114 (2009).
Takeba, Y., Nagafuchi, H., Takeno, M., Kashiwakura, J.-I. & Suzuki, N. Txk, a member of nonreceptor tyrosine kinase of Tec family, acts as a Th1 cell-specific transcription factor and regulates IFN-gamma gene transcription. J. Immunol. 168, 2365–2370 (2002).
Kos, J. et al. Cystatin F as a regulator of immune cell cytotoxicity. Cancer Immunol. Immunother. 67, 1931–1938 (2018).
O’Shea, J. J., Lahesmaa, R., Vahedi, G., Laurence, A. & Kanno, Y. Genomic views of STAT function in CD4+ T helper cell differentiation. Nat. Rev. Immunol. 11, 239–250 (2011).
Kliche, S. et al. The ADAP/SKAP55 signaling module regulates T-cell receptor-mediated integrin activation through plasma membrane targeting of Rap1. Mol. Cell. Biol. 26, 7130–7144 (2006).
Ferretti, A. P., Bhargava, R., Dahan, S., Tsokos, M. G. & Tsokos, G. C. Calcium/calmodulin kinase IV controls the function of both T cells and kidney resident cells. Front. Immunol. 9, 2113 (2018).
Fu, G. et al. Protein kinase C η is required for T cell activation and homeostatic proliferation. Sci. Signal 4, ra84 (2011).
Vella, A. et al. Localization of a type 1 diabetes locus in the IL2RA/CD25 region by use of tag single-nucleotide polymorphisms. Am. J. Hum. Genet. 76, 773–779 (2005).
Grinberg-Bleyer, Y. et al. IL-2 reverses established type 1 diabetes in NOD mice by a local effect on pancreatic regulatory T cells. J. Exp. Med. 207, 1871–1878 (2010).
Ohkura, N. & Sakaguchi, S. Transcriptional and epigenetic basis of Treg cell development and function: its genetic anomalies or variations in autoimmune diseases. Cell Res. 30, 465–474 (2020).
Shevyrev, D. & Tereshchenko, V. Treg heterogeneity, function, and homeostasis. Front. Immunol. 10, 3100 (2019).
Dowling, O. et al. Mutations in capillary morphogenesis gene-2 result in the allelic disorders juvenile hyaline fibromatosis and infantile systemic hyalinosis. Am. J. Hum. Genet. 73, 957–966 (2003).
Li, J., Lu, E., Yi, T. & Cyster, J. G. EBI2 augments Tfh cell fate by promoting interaction with IL-2-quenching dendritic cells. Nature 533, 110–114 (2016).
Lin, S.-E., Oyama, T., Nagase, T., Harigaya, K. & Kitagawa, M. Identification of new human mastermind proteins defines a family that consists of positive regulators for notch signaling. J. Biol. Chem. 277, 50612–50620 (2002).
Ghosh, S., Roy-Chowdhuri, S., Kang, K., Im, S.-H. & Rudra, D. The transcription factor Foxp1 preserves integrity of an active Foxp3 locus in extrathymic Treg cells. Nat. Commun. 9, 4473 (2018).
Ono, M. et al. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature 446, 685–689 (2007).
Han, K. et al. Fasting-induced FOXO4 blunts human CD4+ T helper cell responsiveness. Nat. Metab. 3, 318–326 (2021).
Vacchio, M. S., Olaru, A., Livak, F. & Hodes, R. J. ATM deficiency impairs thymocyte maturation because of defective resolution of T cell receptor α locus coding end breaks. Proc. Natl Acad. Sci. USA 104, 6323–6328 (2007).
Groom, J. R. & Luster, A. D. CXCR3 in T cell function. Exp. Cell Res. 317, 620–631 (2011).
Roychoudhuri, R. et al. BACH2 regulates CD8(+) T cell differentiation by controlling access of AP-1 factors to enhancers. Nat. Immunol. 17, 851–860 (2016).
Yao, C. et al. BACH2 enforces the transcriptional and epigenetic programs of stem-like CD8+ T cells. Nat. Immunol. 22, 370–380 (2021).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–D592 (2023).
Boulland, M.-L. et al. Human IL4I1 is a secreted L-phenylalanine oxidase expressed by mature dendritic cells that inhibits T-lymphocyte proliferation. Blood 110, 220–227 (2007).
Pociot, F. et al. Genetics of type 1 diabetes: what’s next? Diabetes 59, 1561–1571 (2010).
Noble, J. A. & Erlich, H. A. Genetics of type 1 diabetes. Cold Spring Harb. Perspect. Med. 2, a007732 (2012).
Noble, J. A. & Valdes, A. M. Genetics of the HLA region in the prediction of type 1 diabetes. Curr. Diab Rep. 11, 533–542 (2011).
Johnson, M. B. et al. Type 1 diabetes can present before the age of 6 months and is characterised by autoimmunity and rapid loss of beta cells. Diabetologia 63, 2605–2615 (2020).
Oram, R. A. et al. A type 1 diabetes genetic risk score can aid discrimination between type 1 and type 2 diabetes in young adults. Diab. Care 39, 337–344 (2016).
Sharp, S. A. et al. Development and standardization of an improved type 1 diabetes genetic risk score for use in newborn screening and incident diagnosis. Diab. Care 42, 200–207 (2019).
Roncarolo, M. G. et al. Interleukin-2 production and interleukin-2 receptor expression in children with newly diagnosed diabetes. Clin. Immunol. Immunopathol. 49, 53–62 (1988).
Tomoda, T., Kurashige, T. & Taniguchi, T. Imbalance of the interleukin 2 system in children with IDDM. Diabetologia 37, 476–482 (1994).
Malek, T. R. & Bayer, A. L. Tolerance, not immunity, crucially depends on IL-2. Nat. Rev. Immunol. 4, 665–674 (2004).
Kalia, V. & Sarkar, S. Regulation of effector and memory CD8 T cell differentiation by IL-2-A balancing act. Front. Immunol. 9, 2987 (2018).
Li, Y. et al. Stem-like T cells are associated with the pathogenesis of ulcerative colitis in humans. Nat. Immunol. 25, 1231–1244 (2024).
Sekine, T. et al. TOX is expressed by exhausted and polyfunctional human effector memory CD8+ T cells. Sci. Immunol. 5, eaba7918 (2020).
Li, J. et al. KIR+CD8+ T cells suppress pathogenic T cells and are active in autoimmune diseases and COVID-19. Science 376, eabi9591 (2022).
Achenbach, P. et al. Mature high-affinity immune responses to (pro)insulin anticipate the autoimmune cascade that leads to type 1 diabetes. J. Clin. Invest. 114, 589–597 (2004).
Hoffmann, M. et al. Interleukin-7 and soluble Interleukin-7 receptor levels in type 1 diabetes—impact of IL7RA polymorphisms, HLA risk genotypes and clinical features. Clin. Immunol. 235, 108928 (2022).
Schroderus, A.-M. et al. Evaluation of plasma IL-21 as a potential biomarker for type 1 diabetes progression. Front. Immunol. 14, 1157265 (2023).
Lee, L.-F. et al. Anti-IL-7 receptor-α reverses established type 1 diabetes in nonobese diabetic mice by modulating effector T-cell function. Proc. Natl Acad. Sci. USA 109, 12674–12679 (2012).
Penaranda, C. et al. IL-7 receptor blockade reverses autoimmune diabetes by promoting inhibition of effector/memory T cells. Proc. Natl Acad. Sci. USA 109, 12668–12673 (2012).
Sutherland, A. P. R. et al. Interleukin-21 is required for the development of type 1 diabetes in NOD mice. Diabetes 58, 1144–1155 (2009).
Meazza, R., Azzarone, B., Orengo, A. M. & Ferrini, S. Role of common-gamma chain cytokines in NK cell development and function: perspectives for immunotherapy. J. Biomed. Biotechnol. 2011, 861920 (2011).
Rochman, Y., Spolski, R. & Leonard, W. J. New insights into the regulation of T cells by gamma(c) family cytokines. Nat. Rev. Immunol. 9, 480–490 (2009).
Monti, P. & Bonifacio, E. Interleukin-7 and type 1 diabetes. Curr. Diab Rep. 14, 518 (2014).
Carrette, F. & Surh, C. D. IL-7 signaling and CD127 receptor regulation in the control of T cell homeostasis. Semin. Immunol. 24, 209–217 (2012).
Vignali, D. & Monti, P. Targeting homeostatic T cell proliferation to control beta-cell autoimmunity. Curr. Diab Rep. 16, 40 (2016).
Szabo, P. A., Miron, M. & Farber, D. L. Location, location, location: tissue resident memory T cells in mice and humans. Sci. Immunol. 4, eaas9673 (2019).
Cibrián, D. & Sánchez-Madrid, F. CD69: from activation marker to metabolic gatekeeper. Eur. J. Immunol. 47, 946–953 (2017).
Jacobsen, L. M. et al. The risk of progression to type 1 diabetes is highly variable in individuals with multiple autoantibodies following screening. Diabetologia 63, 588–596 (2020).
Feng, X. et al. Targeting CD38 suppresses induction and function of T regulatory cells to mitigate immunosuppression in multiple myeloma. Clin. Cancer Res. 23, 4290–4300 (2017).
Krejcik, J. et al. Daratumumab depletes CD38+ immune regulatory cells, promotes T-cell expansion, and skews T-cell repertoire in multiple myeloma. Blood 128, 384–394 (2016).
Smith, M. J., Simmons, K. M. & Cambier, J. C. B cells in type 1 diabetes mellitus and diabetic kidney disease. Nat. Rev. Nephrol. 13, 712–720 (2017).
Orban, T. et al. Pancreatic islet autoantibodies as predictors of type 1 diabetes in the diabetes prevention trial-type 1. Diab. Care 32, 2269–2274 (2009).
Li, B. et al. Comprehensive evaluation of the effects of long-term cryopreservation on peripheral blood mononuclear cells using flow cytometry. BMC Immunol. 23, 30 (2022).
Japp, A. S. et al. HPAP Consortium, Atkinson, TCR+/BCR+ dual-expressing cells and their associated public BCR clonotype are not enriched in type 1 diabetes. Cell 184, 827–839.e14 (2021).
Kuri-Cervantes, L., Pampena, M. B. & Betts, M. R. Phenotypic characterization of SLex+ and CLA+ CD4+ T Cells. STAR Protoc. 1, 100154 (2020).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Stoeckius, M. et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 19, 224 (2018).
Melsted, P. et al. Modular, efficient and constant-memory single-cell RNA-seq preprocessing. Nat. Biotechnol. 39, 813–818 (2021).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Seal, R. L. et al. Genenames.org: the HGNC resources in 2023. Nucleic Acids Res. 51, D1003–D1009 (2023).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinforma. 9, 559 (2008).
Morabito, S., Reese, F., Rahimzadeh, N., Miyoshi, E. & Swarup, V. hdWGCNA identifies co-expression networks in high-dimensional transcriptomics data. Cell Rep. Methods 3, 100498 (2023).
Klitz, W. et al. New HLA haplotype frequency reference standards: high-resolution and large sample typing of HLA DR-DQ haplotypes in a sample of European Americans. Tissue Antigens 62, 296–307 (2003).
Hadley, W. Ggplot2: Elegrant Graphics for Data Analysis (Springer, 2016).
Acknowledgements
UC4-DK-112217 (to K.H.K., T.M.B., E.T.L.P., A.N., and M.R.B.), Penn IDOM Internal Pilot Grant 5-P30-DK-019525 (to M.R.B.), JDRF 3-SRA-2022-1237-S-B (to M.R.B.), JDRF PDF 3-PDF-2023-1323-A-N (to G.J.G.), Human Islet Research Network (HIRN). This manuscript used data acquired from the Human Pancreas Analysis Program (HPAP-RRID:SCR_016202), and the Human Islet Research Network (RRID:SCR_014393) consortium (UC4-DK-112217, U01-DK-123594, UC4-DK-112232, and U01-DK-123716). This research was performed with the support of the Network for Pancreatic Organ donors with Diabetes (nPOD; RRID: SCR_014641), a collaborative type 1 diabetes research project sponsored by the Juvenile Diabetes Research Foundation (JDRF) (nPOD: 5-SRA-2018-557-Q-R), and The Leona M. & Harry B. Helmsley Charitable Trust (grant 2018PG-T1D053). Genetic risk scoring data was analyzed with support from NIAID P01 to T.M.B. (AI042288). Flow cytometry data was generated in the Penn Cytomics and Cell Sorting Shared Resource Laboratory at the University of Pennsylvania (RRID: SCR_022376), which is partially supported by the Abramson Cancer Center NCI Grant (P30 016520). We thank K. Trihemasava, J. Shoush, and J.M.L Nordin for their assistance processing and running flow cytometry on HPAP donors, the Betts lab for their support with annotating the CITEseq dataset, and B.M.W. Golden, B.B. Golden, and B. Golden for their support.
Author information
Authors and Affiliations
Consortia
Contributions
G.J.G., A.S.J., M.B.P., L.K.C., and M.R.B. designed flow cytometry experiments. G.J.G., A.S.J., M.B.P., L.K.C., and J.T.H. performed flow cytometry experiments. G.J.G. analyzed the flow cytometry data. G.J.G., V.W., J.T.H., K.A., A.S.J., C.L., and J.S.G. processed HPAP tissue samples. G.J.G., V.W., and M.R.B. designed the CITEseq experiments. G.J.G. performed the CITEseq experiments. G.J.G., V.W., J.T.H., M.B.P., J.J.K., E.T.L.P., and M.R.B. annotated the CITEseq dataset. G.J.G., V.W., K.A., and M.R.B. analyzed and interpreted the CITEseq dataset. M.R.S. generated the HLA-GRS. J.T.H. created tables summarizing donor information. G.J.G., V.W., and K.A. performed statistical analyses. M.A.A., K.H.K., and A.N. oversaw subject selection, clinical data acquisition, clinical data analysis, research data reporting for study subjects, and editing the manuscript. G.J.G., V.W., T.M.B., E.T.L.P., A.N., and M.R.B. formulated the ideas and direction of the project. G.J.G., V.W., and M.R.B. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Golden, G.J., Wu, V.H., Hamilton, J.T. et al. Immune perturbations in human pancreas lymphatic tissues prior to and after type 1 diabetes onset. Nat Commun 16, 4621 (2025). https://doi.org/10.1038/s41467-025-59626-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-59626-0
This article is cited by
-
Imbalance of stem-like and effector T cell states in children with early type 1 diabetes across conventional and regulatory subsets
Nature Communications (2025)
-
CXCR3 expression on antigen-experienced B cells is systemically dysregulated in type 1 diabetes
Diabetologia (2025)
-
Profiling associations of interactive ligand–receptors (HLA class I and KIR gene products) with the progression to type 1 diabetes among seroconverted participants
Diabetologia (2025)
-
Transient tissue residency and lymphatic egress define human CD56bright NK cell homeostasis
Nature Immunology (2025)