Abstract
Lysosomes are essential organelles for cellular homeostasis and signaling, with dysfunction linked to neurological disorders, lysosomal storage diseases, and cancer. While proteomics has advanced our understanding of lysosomal composition, the structural characterization of lysosomal membrane proteins in their native environment remains a significant challenge. Here, we developed a cryo electron tomography workflow to visualize lysosomal membrane proteins within intact, native lysosomal membranes. We isolated endolysosomes by independently targeting two lysosomal membrane proteins, transient receptor potential mucolipin 1 and transmembrane protein 192, enriching organelles that exhibited the expected morphology and proteomic composition of the endolysosomal system. Sub-tomogram averaging enabled the structural refinement of key membrane and membrane-associated proteins, including V-ATPase, Flotillin, and Clathrin, directly within the lysosomal membrane, revealing their heterogeneous distribution across endolysosomal organelles. By integrating proteomics with structural biology, our workflow establishes a powerful platform for studying lysosomal membrane protein function in health and disease, paving the way for future discoveries in membrane-associated lysosomal mechanisms.
Similar content being viewed by others
Introduction
The endolysosomal system is a complex network of intracellular membranous organelles essential for maintaining cellular homeostasis. This dynamic and specialized system includes early endosomes, recycling endosomes, late endosomes, and lysosomes, each characterized by distinct morphological and biological features that define their function. Early endosomes serve as the initial sorting site for cargo after endocytosis, while the recycling endosomes, derived from the early endosomes, reclaim and return cargo to the plasma membrane or Golgi1,2. As early endosomes mature into late endosomes, they become a central hub for both incoming and outgoing traffic from various pathways and organelles, ultimately leading to the lysosome, which functions as the cell’s primary degradation station following fusion with autophagosomes, resulting in the formation of an autolysosome1,3,4. Overall, the endolysosomal system is a highly intricate network where endocytotic, biosynthetic, and degradation pathways converge, playing a critical role in cellular function and homeostasis.
For decades, lysosomes have been recognized as the cell’s central hub for recycling and degradation, largely due to their rich concentration of luminal acid hydrolases and their highly acidic environment5,6. However, the discovery of key lysosomal membrane proteins and their associated complexes has shifted this perspective, positioning the lysosome as a vital regulator of metabolic signaling, gene expression, lysosomal trafficking, and calcium homeostasis7,8,9,10. The critical importance of these organelles is underscored by the fact that lysosomal dysfunction not only causes rare lysosomal storage diseases but also contributes to more prevalent and debilitating neurodegenerative conditions, including Parkinson’s and Alzheimer’s disease, and various cancers11,12,13,14,15,16,17,18. By deepening our understanding of how lysosomal membrane proteins and their associated complexes function and interact in their native cellular environment, we can gain valuable insights into their role in these devastating diseases.
Since their discovery, lysosomal morphology and biochemical composition have been extensively studied using light microscopy, transmission electron microscopy (TEM), and mass spectrometry. Commonly used lysosomal isolation methods, such as Lysosome Immunoprecipitation (LysoIP), superparamagnetic iron oxide nanoparticles (SPIONS), and density gradient centrifugation, have been valuable for metabolomic and proteomic profiling of immunopurified lysosomes via mass spectrometry1,19,20,21,22,23,24,25. However, despite these technological advancements, this method has yet to demonstrate the ability to maintain the ultrastructure of intact lysosomes allowing for the direct visualization of membrane proteins and associated complexes using structural-based techniques.
Recent advancements in single-particle cryo electron microscopy (cryoEM) have enabled high-resolution structural characterization of isolated lysosomal membrane proteins and their associated complexes, providing critical insights into their regulatory mechanisms26,27,28. Additionally, cryo electron tomography (cryoET) has facilitated the direct visualization of membrane protein complexes within their native cellular environments29,30,31,32,33,34,35. The use of isolated cell-derived vesicles has led to the sub-nanometer resolution of membrane proteins34, while a breakthrough method for isolating native synaptic vesicles has enabled the complete molecular mapping of their architecture31, including the visualization of V-ATPase and a protein binding partner35. Despite these advances, no existing approach has successfully combined high-resolution structural analysis with the preservation of intact lysosomal membranes, leaving a fundamental gap in our understanding of lysosomal organization and function.
In this work, we develop an approach to isolate intact lysosomes while maintaining their native membrane architecture for single-particle cryoEM and cryoET imaging. By independently targeting two lysosomal membrane proteins, transient receptor potential mucolipin 1 (TRPML1), a key lysosomal calcium ion channel36, and transmembrane protein 192 (TMEM192), a lysosomal membrane protein commonly used in the LysoIP method21,22,23,24, and utilizing the Rho1D4 monoclonal antibody, we successfully immunopurify intact organelles. Our analysis demonstrates that these isolated organelles maintain their typical endolysosomal morphology, as observed in situ, and proteomic composition of the endolysosomal system. CryoET reveals that organelle purification using two independent lysosomal membrane protein targets results in highly similar pools of endolysosomal organelles. Sub-tomogram averaging enables structural refinement of key membrane-associated complexes, including V-ATPase, Flotillin, and Clathrin, directly within the lysosomal membrane. This analysis uncovers their heterogeneous distribution across endolysosomal compartments. Unlike conventional enrichment methods, our approach provides a detailed structural visualization of lysosomal membrane proteins in their native environment.
Results
Method for isolating intact TRPML1-mNeonGreen-1D4 and TMEM192-mCherry-1D4 immunopurified lysosomes and subsequent cryoET
The goal of our study was to establish a method to isolate intact lysosomes for structural characterization of lysosomal membrane proteins and associated membrane complexes using single-particle cryoEM and cryoET (Fig. 1a). The LysoIP method, which employs the lysosomal membrane protein TMEM192, of unknown function, tagged with an HA epitope and isolated using HA-conjugated magnetic beads, has been widely applied for lysosomal metabolomic and proteomic profiling21,22,23,24. However, this approach has not been validated by electron microscopy to confirm the presence of intact lysosomes, nor has it been optimized for elution conditions compatible with cryoEM and cryoET-based structural studies. To address these gaps, we generated a HEK 293 cell line stably expressing TMEM192-3xHA and confirmed its localization with the late endosome/lysosome marker LAMP1 (Pearson’s Correlation Coefficient (PCC) = 82.2%) (Supplementary Fig. 1a, b). TMEM192-3xHA-positive organelles were then immunopurified using an HA peptide for downstream structural analysis. However, the eluted fractions contained a low abundance of organelles with characteristic lysosomal or autolysosomal morphology and exhibited substantial contamination (Supplementary Fig. 1c). Due to the limited quantity and quality of purified organelles, structural analysis could not be performed using this approach.
a Schematic of TRPML1-mNeonGreen-1D4 and TMEM192-mCherry-1D4 lysosomes immunopurified from HEK 293 cells, eluted, and plunge frozen on cryoEM grids for cryoET and TRPML1-mNeonGreen-1D4 and TMEM192-mCherry-1D4 HEK 293 cells cultured and plunge frozen on cryoEM grids for cryo confocal microscopy, cryoFIB-SEM, and cryoET. Created in BioRender. McVeigh, B. (https://BioRender.com/ns2gzr5). b Representative fluorescence microscopy image of HEK 293 cells stably overexpressing TRPML1-mNeonGreen-1D4 (green) and a lysosomal marker, LAMP1, (magenta). Scale bar is 10 μm and is 5 μm in MERGE image (n = 3). Pearson’s correlation coefficient measured for TRPML1-mNeonGreen-1D4 and LAMP1 colocalization. Data are presented as a violin plot (n = 5, cells examined over one experiment). Source data are provided as a Source Data file. c Representative fluorescence microscopy image of HEK 293 cells stably overexpressing TMEM192-mCherry-1D4 (green and a lysosomal marker, LAMP1, (magenta). Scale bar is 10 μm and is 5 μm in MERGE image (n = 3). Pearson’s correlation coefficient measured for TMEM192-mCherry-1D4 and LAMP1 colocalization. Data are presented as a violin plot (n = 5, cells examined over one experiment). Source data are provided as a Source Data file. d Fluorescence signal was aligned with and overlaid on a TEM overview of the lamellae using mitochondria (M) as fiducials and a virtual slice through reconstructed tomogram of lysosome/ autolysosome (L/AL) in non-transfected HEK 293 cells (green, LysoTracker Red) and mitochondria (magenta, MitoTracker DR). e TRPML1-mNeonGreen-1D4 HEK 293 cells (green, TRPML1-mNeonGreen-1D4-positive lysosomes) and mitochondria (magenta, MitoTracker DR). f TMEM192-mCherry-1D4 HEK 293 cells (green, TMEM192-mCherry-1D4-positive lysosomes) and mitochondria (magenta, MitoTracker DR). Scale bar of aligned TEM image with fluorescence signal is 500 nm and of virtual slice of reconstructed tomogram is 100 nm. Experiment was performed one time.
To improve lysosome isolation, we targeted TRPML1, a well-characterized, non-selective, cation-permeable ion channel localized to late endosomes and lysosomes, whose dysfunction is associated with the rare neurodegenerative lysosomal storage disease Mucolipidosis type IV36,37,38. As an additional target, we used TMEM192. To ensure optimal yield of isolated lysosomes we generated two HEK 293 cell lines that stably overexpressed TRPML1-mNeonGreen-1D4 or TMEM192-mCherry-1D4. Instead of the HA tag, we employed the 9-amino-acid 1D4 epitope derived from the C-terminus of bovine rhodopsin39, to facilitate organelle isolation (Fig. 1a). This epitope has been widely used for efficient one-step purification of membrane proteins using Rho1D4 monoclonal antibody-conjugated Sepharose CN-Br activated beads, enabling high-resolution single-particle cryoEM39,40.
First, we evaluated the proper localization of stably overexpressed TRPML1-mNeonGreen-1D4 and TMEM192-mCherry-1D4 in HEK 293 cells using fluorescence microscopy. Co-localization analysis with the late endosome/lysosome marker LAMP1 showed a PCC of 91.5% and 81.5%, respectively (Fig. 1b, c). Next, the HEK 293 cell lines stably expressing TRPML1-mNeonGreen-1D4 or TMEM192-mCherry-1D4 were cultured on cryoEM grids. Cryo-confocal microscopy was used to identify TRPML1-mNeonGreen-1D4- and TMEM192-mCherry-1D4-positive lysosomes, followed by the preparation of thin lamellae around these regions using cryo focused ion beam scanning electron microscopy (cryoFIB-SEM) for cryoET imaging (Fig. 1a). In addition, we used non-transfected HEK 293 cells loaded with LysoTracker Red for the visualization of native lysosomes in situ to qualitatively compare endolysosomal morphology to the HEK 293 cell lines that stably overexpressed either TRPML1-mNeonGreen-1D4 or TMEM192-mCherry-1D4.
To examine the morphological features of endogenous TRPML1-mNeonGreen-1D4- and TMEM192-mCherry-1D4-positive lysosomes in situ, we based our assessments on established ultrastructural characterizations of endolysosomal organelles obtained through TEM of chemically fixed or freeze-substituted mammalian cells1,25. According to these studies, early endosomes typically lack electron-dense intraluminal content, exhibit a tubular-vesicular structures, and range from 100 to 500 nm in diameter. Late endosomes, also known as multivesicular bodies (MVBs), contain multiple intraluminal vesicles (~50 nm in diameter) and have overall diameters of 250 to 1000 nm. Lysosomes and autolysosomes exhibit heterogeneous electron-dense content, including membrane whorls and remnants of cytoplasmic organelles, with sizes ranging from 200 to 1500 nm. Autophagosomes, distinguished by their characteristic double-membrane structure, typically range from 500 to 1500 nm in diameter1,25.
Our in situ cryoET analysis of TRPML1-mNeonGreen-1D4- and TMEM192-mCherry-1D4-positive organelles revealed lysosomes/autolysosomes ~600 nm in diameter, displaying heterogeneous intraluminal content consistent with membrane whorls, typical features of lysosomes, as previously reported1,25 and observed in non-transfected HEK 293 cells (Fig. 1d–f). Importantly, the membrane architecture of these organelles was well preserved, including the consistent spacing between the membrane layers. These findings indicate that the overexpression of lysosomal membrane proteins TRPML1 and TMEM192 does not alter native lysosomal morphology.
Characterization of TRPML1-mNeonGreen-1D4 and TMEM192-mCherry-1D4 immunopurified lysosomes
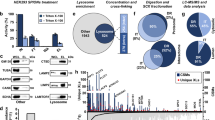
To evaluate the effectiveness of 1D4-mediated immunopurification of TRPML1-mNeonGreen-1D4- and TMEM192-mCherry-1D4-positive organelles, we performed Western blot analysis and revealed that the 1D4-immunopurified organelles were significantly enriched in lysosomal markers (LAMP1, NPC1, ATP6V0A1, LAMTOR1, FLOT1, and FLOT2) with minimal detection of markers from other cellular compartments, consistent with previous reports21,23,24 (Fig. 2a, b). Next, we characterized the proteomic profile of immunopurified TRPML1-mNeonGreen-1D4- and TMEM192-mCherry-1D4-positive organelles using quantitative mass spectrometry (Supplementary Data 1). We performed triplicate 1D4 immunopurifications for both control (TRPML1 without mNeonGreen or 1D4 epitope HEK 293 cell line) and TRPML1-mNeonGreen-1D4 HEK 293 cell line, followed by protein analysis using 6-plex TMT. Principal component analysis (PCA) revealed a clear separation between the immunopurified samples from control and TRPML1-mNeonGreen-1D4 immunopurified organelle replicates (Supplementary Fig. 2a). We conducted gene ontology (GO) enrichment analysis to categorize proteins into their respective organelle groups, with significant enrichment defined as a greater than 2-fold change (Log2FC > 1.0) and with a p-value < 0.05. Our analysis identified and quantified 1,256 proteins, including 280 proteins typically associated with endosomes and lysosomes (Fig. 2c). We observed substantial enrichment in lysosomal luminal hydrolases (CTSZ, HEXA), membrane proteins (LAMP1, LAMP2), and membrane-associated protein complexes (mTORC1, Flotillin) (Fig. 2c). Among these, key lysosomal proteins linked to lysosomal storage diseases were identified, such as CLN341, NPC142, and CLCN743. We also detected the multi-subunit domains of the V-ATPase, which is crucial for acidification of endolysosomal organelles44, proteins that are members of the mTORC1 megacomplex, which regulates cellular growth and metabolism45, and lysosomal motor proteins, kinesin, dynein, and dynactin, which facilitate in lysosomal movement along microtubules46. Additionally, we identified two members of the Flotillin family, FLOT1 and FLOT2, while these proteins are typically abundant on the plasma membrane they have also been shown to localize and function at endosomal and lysosomal membranes47,48,49,50, consistent with our Western blot analysis results (Fig. 2a, c).
a Immunoblotting for protein markers of several subcellular compartments measured in the post-nuclear supernatant (PNS) and 1D4 immunopurified (1D4 IP) lysosomes of lysates prepared from HEK 293 cells stably overexpressing TRPML1-mNeonGreen-1D4 or TRPML1, as a control and (b) TMEM192-mCherry-1D4 or TMEM192-3xHA, as a control. Experiment was performed one time. Source data are provided as a Source Data file. c Volcano plot of fold-enrichment of proteins identified between control (TRPML1) and TRPML1-mNeonGreen-1D4 triplicate 1D4 immunopurifications and (d) control (TMEM192-3xHA) and TMEM192-mCherry-1D4 triplicate 1D4 immunopurifications. Dashed lines indicate threshold of Log2FC > 1.0 and < -1.0 and a p-value < 0.05. The green and red dots indicate proteins associated with the endosome (GO:0005768) and lysosome (GO:0005764). Volcano plots were generated using a two-tailed t-test without any p-adjustment methods. Source data are provided as a Source Data file.
To further validate the efficiency of 1D4-mediated immunopurification, we conducted triplicate immunopurifications for TMEM192-mCherry-1D4 HEK 293 cell line and duplicate HA immunopurifications for TMEM192-3xHA HEK 293 cell line, as a control. Next, we performed quantitative mass spectrometry and protein analysis using 10-plex TMT. In total, we identified and quantified 6,706 proteins, with 844 proteins associated with endosomes and lysosomes in both TMEM192 1D4- or HA-based immunopurifications (Fig. 2d and Supplementary Fig. 2d). We observed clear separations between the control and immunopurified samples (Supplementary Fig. 2b, c). Comparing the enrichment of endosomal and lysosomal proteins (Log2FC > 1.0 and p-value < 0.05) among TRPML1-mNeonGreen-1D4, TMEM192-mCherry-1D4-, and TMEM192-3xHA-immunopurified organelles, we found that 1D4-immunopurified organelles exhibited 85% and 75% enrichment of endolysosomal proteins, respectively (Fig. 2c, d), while HA-immunopurified organelles showed only 5% enrichment (Supplementary Fig. 2d). These results confirm that the 1D4 epitope is a highly effective tool for immunopurification of TRPML1- and TMEM192-positive organelles, providing a robust approach for organelle isolation.
To characterize in greater detail the purity and morphology of immunopurified organelles, we prepared cryoEM grids of TRPML1-mNeonGreen-1D4 and TMEM192-mCherry-1D4 immunopurified samples and collected 227 zero-tilt cryoEM projection images of each. The images showed a pronounced enrichment of endolysosomal organelles, with no detectable contamination of mitochondria, Golgi, or endoplasmic reticulum (Fig. 3a, b) consistent with our Western blot and mass spectrometry results (Fig. 2). The TRPML1-mNeonGreen-1D4 immunopurified organelles ranged in size from 100 to 2000 nm, with 43% classified as early endosomes, 36% lysosomes and autolysosomes, 18% late endosomes, and 3% autophagosomes of the total classified endolysosomal organelles (Fig. 3c). In contrast, TMEM192-mCherry-1D4 immunopurified organelles predominantly comprised early endosomes (31%), with smaller populations of lysosomes and autolysosomes (31%), late endosomes (26%), and autophagosomes (12%), with organelle sizes ranging from 100 to 3500 nm (Fig. 3d). In addition to canonical endolysosomal organelles, we observed vesicles in TRPML1-mNeonGreen-1D4 and TMEM192-mCherry-1D4 immunopurified samples that did not meet traditional classification criteria, which may represent smaller transport carriers associated with the endolysosomal system, potentially involved in delivering key lysosomal enzymes and other cargo25 (Fig. 3).
a CryoEM image of immunopurified and eluted TRPML1-mNeonGreen-1D4 positive organelles (n = 227) and (b) TMEM192-mCherry-1D4 positive organelles (n = 237). Early endosome (EE), late endosome (LE), lysosome/ autolysosome (L/AL), autophagosome (AP), and other (O). Scale bar is 100 nm. c Violin plot of diameters measured on organelles observed in the 1D4 immunopurification and elution of TRPML1-mNeonGreen-1D4 organelles (n = 2151) and (d) TMEM192-mCherry-1D4 organelles (n = 2618). Source data are provided as a Source Data file.
Overall, these findings, together with our in situ results, confirm the success of 1D4-mediated immunopurification of native endolysosomes enriched with key lysosomal membrane proteins, providing a robust platform for structural investigations using cryoEM and cryoET.
Proteomic landscape of TRPML1-mNeonGreen-1D4- and TMEM192-mCherry-1D4-positive lysosomal membranes
Once we confirmed that our approach efficiently isolates intact TRPML1-mNeonGreen-1D4- and TMEM192-mCherry-1D4-positive lysosomes with high protein content, we utilized it for structural analysis of lysosomal membrane proteins in their native environment using cryoET. We collected 617 tilt series from isolated TRPML1-mNeonGreen-1D4-positive organelles and 56 tilt series from isolated TMEM192-mCherry-1D4-positive organelles as a control (Fig. 4). The reconstructed tomograms displayed characteristic heterogeneous membrane whorls and the presence of intraluminal vesicles within lysosomes, consistent with our in situ tomograms (Figs. 1d–f, 4) and previous reports1,25,51. Upon closer inspection, we readily identified three distinct membrane-associated protein densities corresponding to V-ATPase, Flotillin, and Clathrin in both TRPML1-mNeonGreen-1D4- and TMEM192-mCherry-1D4-positive organelles (Figs. 4, 5 and Supplementary Movies 1-6), these findings were further corroborated by our Western blot analysis and mass spectrometry results (Fig. 2).
a 2D tomographic slices of organelles immunopurified and eluted using TRPML1-mNeonGreen-1D4 as the target (top) (n = 617) and their corresponding 3D reconstructions after segmentation (bottom). Membranes are shown in green. b 2D tomographic slices of organelles immunopurified and eluted using TMEM192-mCherry-1D4 as the target (top) (n = 55) along with their segmented 3D reconstructions (bottom), where membranes are shown in slate blue. Arrowheads indicate V-ATPase (purple), Flotillin (orange), and Clathrin (gold). Scale bar is 50 nm.
a Examples of V-ATPase (n = 1325), (c) Flotillin (n = 2927), and (e) Clathrin particles (n = 2024) used for sub-tomogram averaging. Scale bar is 20 nm. (b) Density maps of V-ATPase, (d) Flotillin, and (f) Clathrin obtained from sub-tomogram averaging, overlaid with their respective coordinate models: V-ATPase (PDB: 9BRT), Flotillin (PDB: 9BQ2), and Clathrin (PDB: 1XI4).
V-ATPase, which is essential for endolysosomal acidification44, was localized with its V1 domain facing on the outer side of the lysosomal membrane, distributed across the membrane without a distinct localization pattern, though occasional clustering was noted (Fig. 4a, and Supplementary Fig. 4a, Supplementary Movies 7–10). Flotillin, an essential protein in the endocytic pathway that facilitates cargo trafficking to endosomes and lysosomes47,48,49,50 and which co-localized with LAMP1 (Supplementary Fig. 3), was randomly distributed across the lysosomal membrane surface (Fig. 4a, Supplementary Fig. 4b, Supplementary Movies 11-14). The larger radius of Flotillin was consistently positioned against the lysosomal membrane, while the smaller radius faced outward (Fig. 4, Supplementary Fig. 4), consistent with previous findings33,50. Interestingly, lysosomes with the highest Flotillin surface density exhibited an abundance of membrane whorls, whereas those with high V-ATPase density showed elevated electron density (Fig. 4and Supplementary Fig. 4). Additionally, Clathrin, a major regulator of vesicle formation and endocytic trafficking52,53,54, was observed in an open cage-like configuration on convex membranes surfaces (Figs. 4, 5e Supplementary Movies 1, 4), suggesting we captured it during vesicle formation or membrane coating.
Next, we applied template matching55 to tomograms collected on TRPML1-mNeonGreen-1D4-positive lysosomes, identifying 3459 V-ATPase, 2927 Flotillin, and 2024 Clathrin subunits on the membrane surface, corresponding to an average of 3.5, 6.9 and 15.1 particles per organelle, respectively (Supplementary Fig. 5a, b). Subsequent sub-tomogram averaging yielded density maps at resolutions of 23.4 Å, 23.5 Å, and 33.0 Å for V-ATPase, Flotillin, and Clathrin, respectively, without applied symmetry (Fig. 5, Supplementary Fig. 5). Further 3D classification of Flotillin particles produced densities consistent with the expected size of FLOT1/233, while Clathrin structures exhibited the characteristic hexagonal geometry of the Clathrin coat31,52,53,54 (Fig. 5c–f, Supplementary Fig. 5). As anticipated, resolution was limited by particle number, as well as protein mobility and flexibility. Despite these constraints, the V-ATPase reconstruction retained its characteristic membrane-embedded shape34,35,56 (Fig. 5b, PDB:9BRT). The Flotillin structure, refined with C44 symmetry, achieved a final resolution of 13.8 Å, aligning closely with the published structure33 (Fig. 5d, PDB:9BQ2). Similarly, the Clathrin structure, refined with C6 symmetry, achieved a resolution of 26.6 Å, faithfully reproducing its known architecture31,53,54 (Fig. 5f, PDB:1XI4).
While analyzing the tomograms, we identified a consistent ~30 nm separation between the outermost whorl and the lysosomal membrane of TRPML1-mNeonGreen-1D4-positive lysosomes. This region contained dense rod-like structures likely resembling the lysosomal lipid transporter, vacuolar protein sorting 13 homolog C (VPS13C) (Supplementary Fig. 6a), implicated in lipid transfer between organelles30. Similarly, filamentous structures were observed 20-30 nm from the lysosomal membrane, anchored via distinct protein densities that likely correspond to kinesin, a motor protein responsible for lysosomal anterograde transport along microtubules46 (Supplementary Fig. 6b). Additionally, we observed a protein density physically linking two organelles, potentially resembling the HOPS complex, which plays a critical role in the tethering and fusion of endosomal and lysosomal membranes57 (Supplementary Fig. 6c). However, due to the limited number of particles and the substantial heterogeneity, we were unable to perform sub-tomogram averaging.
Overall, these findings demonstrate that our 1D4-tag-based immunopurification method for TRPML1-enriched organelles enables the identification and structural analysis of lysosomal membrane and membrane-associated proteins in these isolated lysosomes.
Discussion
In this study, we present an approach for isolating intact TRPML1-mNeonGreen-1D4- and TMEM192-mCherry-1D4-positive lysosomes while preserving their native membrane architecture. By integrating cryoET with quantitative mass spectrometry, we characterized the structural landscape of lysosomes and identified key membrane proteins, including V-ATPase, Flotillin, and Clathrin, which are essential for lysosomal function and intracellular trafficking30,31,33,35,50,52,53,54,57.
Our analysis revealed striking morphological heterogeneity among the isolated organelles, ranging from organelles with minimal luminal content to multilamellar structures containing concentric membranes and protein remnants. This diversity suggests the existence of functional subpopulations within the lysosomal compartment, potentially reflecting maturation stages, cargo specificity (e.g., lipids vs. proteins), or responses to cellular stress1,2,58. Notably, lysosomes enriched in Flotillin exhibited multilamellar architectures with ~4–10 concentric membrane layers. Given the role of Flotillin in lipid raft organization and membrane fusion47, we believe that these proteins may function as structural organizers, facilitating membrane stacking through interactions with lipids (e.g., cholesterol) or curvature-inducing proteins50. Interestingly, similar observations have been reported on epithelial cells59. Conversely, late endosomes or lysosomes with a high density of V-ATPases often contained abundant protein remnants and undegraded material. Since V-ATPases are essential for lysosomal acidification44, this observation may indicate that an acidic microenvironment is established, but degradation in impaired, potentially due to sample preparation artifacts. Alternatively, the accumulation of undegraded content may reflect substrate overload, as seen during exacerbated autophagy in metabolic stress conditions60. Future studies should investigate whether the spatial correlation between V-ATPases and Flotillin suggests a coordinated mechanism that integrates pH regulation and membrane organization, thereby linking these presence to lysosomal homeostasis under physiological and pathological conditions.
Our method effectively preserves the native lysosomal membrane environment, enabling structural analysis of membrane-associated proteins in their functional context. The consistent identification and spatial distribution of these proteins across multiple tomograms underscore the robustness of our isolation strategy in capturing lysosomes and associated endolysosomal compartments. Furthermore, this workflow not only enhances existing methodologies but also contributes to broader efforts to map the structural interactome of endosomal and lysosomal membranes49,51,61,62. By bridging the gap between in situ imaging and reconstituted systems, our approach offers a versatile platform for studying lysosomal membrane architecture under near-native conditions.
Despite these advances, our approach has some limitations arising from both biological and technical constraints. Although lysosomes in our stably overexpressing TRPML1-mNeonGreen-1D4 and TMEM192-mCherry-1D4 HEK293 cell lines are morphologically similar to those in non-transfected HEK293 cells, the overexpression of membrane-tagged proteins may contribute to membrane crowding and introduce biases that limit accurate representation of the endogenous lysosomal proteome. Endogenous tagging of lysosomal proteins could provide more physiologically representative datasets for both single particle cryoEM and cryoET, though this may require extensive data collection due to the typically low abundance of individual membrane proteins.
In addition, the physical characteristics of lysosomes pose some imaging challenges. Their large size (~200–1000 nm) and electron-dense intraluminal content, comprising internal membranes and cargo, generate considerable background, which obscures membrane-associated and luminal structures. These features also necessitate thicker vitrified ice during cryoET; in our dataset, the average ice thickness was 234.6 ± 50.3 nm, unlike smaller vesicles (sub-100 nm), such as synaptic vesicles or cell-derived extracellular vesicles31,33,63. While this did not noticeably affect tilt-series alignment, prior studies suggest that such increases in ice thickness can degrade resolution through elevated electron scattering and signal attenuation64,65. While the ice thickness used in our experiments falls within the acceptable range for standard cryoET workflows, the low endogenous abundance of lysosomal membrane proteins remains a key constraint. Based on our quantitative estimates, a 400 nm diameter lysosome embedded in 200 nm ice may contain fewer than 44 V-ATPase complexes and 15 Flotillin proteins. Proteins involved in membrane tethering, trafficking, or fusion, such as HOPS components, VPS proteins, and filaments, are typically present in even lower copy numbers. This severely limits the number of particles available for sub-tomogram averaging and was reflected in our inability to reconstruct several of these complexes. Overcoming these limitations requires a combination of high-throughput data acquisition and increasingly sophisticated computational pipelines. This integration of biological sampling with scalable computational approaches is essential for resolving low-abundance assemblies and advancing structural understanding of complex organelles like lysosomes under near-native conditions.
Overall, our method offers a powerful and versatile platform for structural investigation of lysosomal membrane proteins, bridging the gap between reconstituted systems and in situ cellular imaging. By preserving native protein–lipid and protein–protein interactions within isolated yet intact lysosomal membranes, this approach enables the study of physiologically relevant assemblies in a tractable experimental context. It retains the architectural fidelity of the organelle while allowing for structural analysis of membrane-associated complexes in their functional environment. Importantly, our open-access dataset and workflow are intended to support community-driven research, facilitate the development of computational tools, and foster collaborative efforts. Importantly, the workflow we describe is well-suited for future translational applications: extending it to other pathological contexts such as neurodegenerative diseases or cancer, where lysosomal dysfunction plays a central feature11,12,13,14,15,16,17,18, may reveal mechanistic insights into disease-associated remodeling of the lysosomal proteome. By laying the foundation for visual proteomics of isolated organelles, this work expands the potential for understanding lysosome-specific biology across diverse physiological and disease-relevant conditions.
Methods
Reagents and antibodies
Antibodies were obtained from the following sources: antibodies to MCOLN1 (15291-1-AP, lot 00118225, 1:500 dilution for WB), ATP6V0A1 (13828-1-AP, lot 00103815, 1:1000 dilution for WB), golgin 97 (12640-1-AP, lot 00071456, 1:1000 dilution for WB), TOM20 (11802-1-AP, lot not specified, 1:1000 dilution for WB), RAB5A (11947-1-AP, lot 00130406, 1:1000 dilution for WB), and mCherry (26765-1-AP, lot 00133758, 1:5000 dilution for WB) from Proteintech®; antibodies to RAB11A (71-5300, lot ZC389784, 1:250 dilution for WB), Goat anti-rabbit AlexaFluor488 (A-11008, lot 2439197, 1:500 dilution), Goat anti-rabbit AlexaFluor555 (A-21428, lot 2445825, 1:500 dilution), Goat anti-Rabbit AlexaFluor680 (A-32734, lot YB366538, 1:500 dilution), Goat anti-Mouse AlexaFluor488 (A-28175, lot 2439197, 1:500 dilution), and Goat anti-mouse AlexaFluor568 (A-11004, lot 2198584, 1:500 dilution) from Thermo Fisher Scientific; antibodies to LAMTOR1 (89755, lot 5, 1:1000 dilution for WB), LAMP1 (D2D11, lot 8, 1:1000 dilution for WB and 1:100 dilution for immunofluorescence), and HA (3724, lot 8, 1:1000 dilution for WB and 1:100 dilution for immunofluorescence) from Cell Signaling Technology; antibody to NPC1 (ab134113, lot GR3290537-3, 1:5000 dilution for WB) from Abcam; antibody to RCN1 (PA5-76525, lot WJ3411001A, 1:1000 dilution for WB) from Invitrogen; antibodies to FLOT1 (610820, lot 3138357, 1:1000 dilution for WB and 1:100 dilution for immunofluorescence) and FLOT2 (610383, lot 3313817, 1:1000 dilution for WB and 1:100 dilution for immunofluorescence) from BD Biosciences; antibodies to Rabbit IgG, HRP conjugate (W4011, lot 0000645477, 1:2500 dilution) and Mouse IgG, HRP conjugate (W4021, lot 0000626657, 1:2500 dilution) from Promega. Antibody 1D4 (produced in-house) was used for Western blot analysis.
Plasmids
The sequences for human TRPML1 and human TRPML1-mNeonGreen-1D4 were cloned into the mammalian expression vector pcDNA3.1 by Genscript. The pcDNA3.1 vector containing human TMEM192-mCherry-1D4 was constructed by standard molecular cloning. pLJC5-Tmem192-3xHA was a gift from David Sabatini (Addgene plasmid # 102930; (http://n2t.net/addgene:102930); RRID:Addgene_102930)21.
Mammalian cell culture
All cells were maintained in a humified atmosphere containing 95% O2 and 5% CO2 at 37 °C. The HEK 293 cells were purchased from American Type Cell Culture (AATC, CRL-1573TM). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Fisher Scientific, MT10113CM) supplemented with 10% fetal bovine serum (Millipore Sigma, F2242). For all experiments involving organelle immunopurification, cells were changed to fresh DMEM media 1 h prior to the start of the experiment.
Transfection and stable cell line generation
Transfection of HEK 293 cells with plasmid DNA were performed using Lipofectamine 3000 (Invitrogen, L3000001) according to the manufacturer’s protocol. To generate stable cell lines of TRPML1, TRPML1-mNeonGreen-1D4, and TMEM192-mCherry-1D4 HEK 293 cells were cultured and selected in 600 μg/mL G418 (Fisher Scientific, MT61234RG) cell culture medium and then maintained in 300 μg/mL G418 cell culture medium. To generate a stable cell line of TMEM192-3xHA HEK 293 cells were cultured, selected, and maintained in 1 μg/mL puromycin (Life Technologies Inc., A11138-03) cell culture medium.
Immunofluorescence microscopy and image analysis
For immunofluorescence experiments, HEK 293 cells were grown on poly-l-lysine-coated 12 mm glass coverslips. Cells were fixed with ice-cold 100% methanol for 20 min at -20 °C, washed three times with PBS, and blocked with 3% BSA in PBS at room temperature for 1 h. Primary antibodies were diluted in 3% BSA in PBS and cells were incubated overnight at 4 °C, washed three times with 0.2% BSA in PBS, and then incubated with the corresponding fluorochrome-conjugated secondary antibodies diluted in 3% BSA in PBS for 1 h at room temperature in the dark. Cells were washed three times with PBS and mounted on the glass cover slide with Flurosave mounting reagent. Images were acquired using a Vistech VT-iSIM confocal microscope equipped with Yokogawa CSU-X1 scan head and an Andor iXon Ultra 897 EM-CCD camera and collected under a x100 oil immersion objective. Images were acquired using VisiView software Version 6.0.0.25 and annotated using Fiji software Version 2.14.0/1.54 f. Colocalization analysis and determination of Pearson’s correlation coefficient (PCC) were performed using the JACoP plugin66.
Immunopurification of lysosomes for quantitative mass spectrometry and cryo electron microscopy
For cryoEM imaging of immunopurified and eluted lysosomes five 15 cm plates were used for each cell line per experiment. Organelles from cells expressing TRPML1-mNeonGreen-1D4, TMEM192-mCherry-1D4, and TMEM192-3xHA HEK 293 cells were grown to confluency in 15 cm dishes. All subsequent steps were performed on ice or at 4 °C unless otherwise noted. Cells were washed one time with 5 mL ice-cold KPBS buffer (126 mM KCl, 10 mM KH2PO4, 50 mM sucrose, pH 7.25 adjusted with 1 N KOH), supplemented with HaltTM Protease and Phosphatase Inhibitor Cocktail (100X) (Thermo Fisher, 78446), scraped into 10 mL KPBS supplemented with HaltTM Protease and Phosphatase Inhibitor Cocktail (100X), and collected by centrifugation at 1000 × g for 2 minutes at 4 °C. The pelleted cells were resuspended in 950 μL KPBS supplemented with HaltTM Protease and Phosphatase Inhibitor Cocktail (100X) and 3.6% (w/v) OptiPrep (Sigma, D1556) (25 µL of each cell lysate was transferred to a new tube and combined with 120 µL 0.5% NP-40 for later analysis by Western blot), and gently homogenized by a 26 G needle attached to a 1 mL syringe 5 times. The lysed cells were centrifuged at 1000 x g for 10 min at 4 °C, and the post-nuclear supernatant (PNS) was transferred to a new tube on ice (25 µL of each PNS was transferred to a new tube and combined with 120 µL 0.5% NP-40 in KPBS for later analysis by Western blot). For TRPML1-mNeonGreen-1D4 and TMEM192-mCherry-1D4 HEK 293 cell lines, 1D4 antibody coupled to CnBr-activated Sepharose beads (150 µL of bead slurry per plate for each quantitative mass spectrometry replicate and 500 µL of bead slurry per cell line for cryoEM and cryoET imaging) were washed three times with 1 mL KPBS buffer supplemented with 3.6% (w/v) OptiPrep. The PNS was combined with the pre-washed 1D4 antibody Sepharose beads and incubated at 4 °C for 2 h with gentle rotation. The beads were separated from the lysate with a brief (3 sec) spin using a bench top mini centrifuge (25 µL of each flow-through was transferred to a new tube and combined with 120 µL 0.5% NP-40 in KPBS for later analysis by Western blot) and washed two times with 1 mL high-salt KPBS buffer (KPBS buffer, supplemented with 150 mM NaCl) and two times with KPBS buffer. Samples processed for Western blot analysis or quantitative mass spectrometry were lysed in 120 µL 0.5% NP-40 in KPBS for 30 minutes on ice. Samples processed for cryoEM and cryoET imaging were eluted by the addition of 150 µL of 10 mg/mL 1D4 peptide at 4 °C for 1 h with gentle rotation, two times. For TMEM192-3xHA HEK 293 cell line, anti-HA magnetic beads (Thermo Scientific™, 88836) (150 µL of bead slurry per plate for each quantitative mass spectrometry replicate and 500 µL of bead slurry per cell line for cryoEM imaging) were washed three times with 1 mL KPBS buffer supplemented with 3.6% (w/v) OptiPrep. The PNS was combined with the pre-washed anti-HA magnetic beads and incubated at 4 °C for 2 h with gentle rotation. The beads were separated from the lysate using a magnetic stand (25 µL of each flow through was transferred to a new tube and combined with 120 µL 0.5% NP-40 in KPBS for later analysis by Western blot) and washed two times with 1 mL high-salt KPBS buffer (KPBS buffer, supplemented with 150 mM NaCl) and two times with KPBS buffer. Samples processed for Western blot analysis or quantitative mass spectrometry were lysed in 120 µL 0.5% NP-40 in KPBS for 30 minutes on ice. Samples processed for cryoEM imaging were eluted by the addition of 150 µL of 10 mg/mL HA peptide at 4 °C for 1 h with gentle rotation, two times.
Western blot
Samples that were lysed in 0.5% NP-40 were spun at 20,000 × g for 30 min at 4 °C and the clarified lysate was transferred to new tube. The sample was then mixed with 4x Laemmli sample buffer (BIO-RAD, 1610747) supplemented with β 2-mercaptoethanol, resolved by sodium dodecyl sulfate– polyacrylamide gel electrophoresis (SDS-PAGE) using Bio-Rad X (4 to 12%) at 135 V for 90 min, and transferred onto nitrocellulose membranes at 60 V for 1 h. Membranes were blocked with 5.0% nonfat dry milk prepared in Tris-buffered saline with Tween 20 (TBST) (20 mM Tris-HCl, 150 mM NaCl, 0.1% Tween-20, pH 7.5) for 1 h at room temperature and probed with the appropriate primary antibody diluted in 5.0% nonfat milk prepared in TBST overnight at 4 °C. The membranes were then washed three times with TBST for 5 min each, followed by incubation with the corresponding HRP-conjugated secondary antibodies diluted in 5.0% nonfat milk prepared in TBST for 1 h at room temperature. Membranes were washed three times with TBST for 5 min each before being visualized with ECL Western blotting substrates (Thermo ScientificTM, 32106). All uncropped and unprocessed scans of Western blots are provided in Supplementary Fig. 7.
Cryo electron tomography organelle preparation and data collection
Eluted lysosomes were incubated with 10 nm colloidal gold fiducial markers (Electron Microscopy Science) (20 μL sample + 25 μL fiducials) for 1 h on ice. 4 μL of this mixture was blotted onto glow-discharged Quantifoil R3.5/1 holey carbon film, 200 mesh gold grids (Quantifoil) in a chamber at 4 °C and 85% humidity for 5 seconds, followed by rapid freezing in liquid ethane (Leica EM GP). Cryo electron tomography tilt series and zero-tilt transmission images were collected on a Titan Krios G3i 300 kV electron microscope equipped with a Gatan K3 direct electron detector (Gatan), with SerialEM 4.2.067. Tilt series were acquired over a ± 60° range with 3° angular increments using a dose-symmetric tilt scheme. The nominal magnification was 53,000x, yielding a super resolution pixel size of 0.845 Å. The dataset was collected with a total dose per tilt of 2.4-2.7 e/Å2, resulting in a cumulative dose per tomogram of 80-111 electrons/Å2. Exposures were fractionated into 4-8 frames, with defocus ranges between -2 to -5 µm. A total of 227 zero-tilt transmission images for organelle classification were acquired at a magnification of 19,500x in low-dose mode, corresponding to a pixel size of 4.5 Å. Imaging was performed with defocus of -20 µm and a total dose of 0.9 e/Å2. Some tilt series were collected at the same positions where zero-tilt images were acquired.
Cryo electron tomography data processing for isolated organelles
All tilt series were processed using Relion 5.068. Motion correction was performed with Relion implementation, with a binning factor of 2 and a patch size of 1 × 1. CTFFind469 version 4.1.14 was used to determine CTF values. Tilt series were aligned in IMOD 4.11/Etomo70,71 using gold fiducial markers, excluding any poorly aligned or veiled tilts. Tomograms were then reconstructed in Relion and denoised using cryoCARE72 version 0.3.0 with a binned pixel size of 10 Å.
V-ATPase (PDB:6WM2) and Flotillin (PDB:9BQ2) particles were picked using template matching with pytom-match-pick73,74. ~1500 of V-ATPase particles were picked per tomogram. Applying the criterion that particles should be embedded in the outermost membrane and correctly oriented, we retained 22,044 particles. Sub-tomograms were extracted with a pixel size of 10.14 Å/px, using a box size of 104 px. These particles were used to build an initial model, which served as a reference map for three rounds of 3D classification into three to four classes. In the final classification step, we applied a mask containing the membrane-embedded V-ATPase, resulting in a distinct class with 2,226 particles. Final particles were re-extracted with a pixel size of 1.69 Å/px, using a box size of 360 px, refined and subjected to 3D classification into two classes. The best class with 1325 particles were reconstructed yielding a resolution of 24.3 Å with no symmetry applied.
Similarly, ~1000 Flotillin particles were picked via template matching. After filtering based on the criterion that particles must be positioned on the membrane, we retained 5709 particles. These were extracted with a pixel size of 10.14 Å/px, using a box size of 84 px, and used to build an initial model. The initial model was subjected to two rounds of 3D classification into four classes. Two classes from the final classification step, containing 2927 particles, were selected and re-extracted with a pixel size of 1.69 Å/px and a box size of 320 px. These particles were reconstructed and subjected to 3D refinement, yielding a resolution of 23.4 Å without symmetry and 13.8 Å in C44 symmetry.
Clathrin particles were manually picked in ChimeraX 1.3 and 1.5 using the ArtiaX tool75, identifying 2946 particles. Sub-tomograms were extracted with a pixel size of 10.14 Å/px, using a box size of 144 px. These particles were used to reconstruct an initial map, which then served as a reference for 3D classification into four classes. Two well-defined classes were selected. After removing duplicate particles, we retained 2024 particles. These were extracted and reconstructed, then subjected to 3D refinement without binning, using a pixel size of 1.69 Å/pixel and a box size of 488 px. The resulting map achieved a resolution of 33.0 Å without applied symmetry and 26.6 Å with C6 symmetry.
The zero-tilt transmission images were used for classification and to determine the radius of the organelles. The images were processed in IMOD/Etomo70,71, and the reported diameter is the average of the measurements of two perpendicular diameters.
Reconstructed tilt series were denoised using cryoCARE72 prior to segmentation. Membrane segmentation was performed using MemBrain v276 and Ais77 version 1.0.25. V-ATPase (PDB:6WM2), Flotillin (PDB:9BQ2), and Clathrin (PDB:1XI4) models were converted to maps and placed onto the segmented tomogram in ChimeraX using ArtiaX tool75. Volume slices of tomograms, segmented volumes, and movies were prepared using ChimeraX78.
Denoised tomograms were segmented using MemBrain v276. Membranes containing V-ATPase or flotillin particles were manually extracted using the napari-lasso-3D plugin version 0.5.3, converted into mesh representations with MemBrain-pick, and subsequently analyzed using MemBrain-stats to calculate nearest-neighbor distances, surface protein concentrations, and Ripley’s K function76.
Cryo electron tomography data processing for cells
Gold 200-mesh grids with R1/4 holey SiO2 film (Quantifoil) were glow discharged for 60 seconds at 15 mA using a PELCO easiGlow (Ted Pella) and placed in the 4 mm circular wells of PDMS stencils (Alvéole) for passivation, micropatterning, and functionalization steps. Grids were passivated by incubating for 30 min with 0.01% Poly-L-Lysine (Sigma-Aldrich), washing 4 times with 0.1 M HEPES, pH 8.5, then incubating for 1 h with 100 mg/ml mPEG-Succinimidyl Valerate (mPEG-SVA, MW 5000, Laysan Bio) freshly prepared in 0.1 M HEPES, pH 8.5. After washing thoroughly with Milli-Q water, PLPP gel photo-initiator (Alvéole) diluted 1:4 with 70% ethanol was added to the grids and allowed to air dry in the dark. Micropatterning was performed using the Alvéole PRIMO2 operated by the Leonardo 5.4 plugin to Micromanager 2.0.0. The PEG layer was ablated using a dose of 60 mJ/mm2 in a 3-point star pattern designed based on typical HEK cell morphology and size cultured on tissue culture plastic. Patterned grids were washed with Milli-Q water and PBS, functionalized by incubating for 30 min at 37 °C with 50 µg/ml human plasma fibronectin (MilliporeSigma, F2006), washed with PBS, then placed in cell culture media for cell seeding.
HEK 293 cell lines cultured as described above, were dissociated into a single cell suspension with Versene solution (Thermo, 15040066), seeded at low density on micropattern-functionalized EM grids, and cultured overnight. Prior to vitrification, mitochondria and lysosomes were labeled with 100 nM MitoTracker DR (Invitrogen, M22426) and 50 nM LysoTracker Green DND-26 (Invitrogen, L7526) or Red DND-99 (Invitrogen, L7528) for 1 h at 37 °C and washed twice with PBS. In the Leica EM GP chamber at 37 °C and 95% humidity, 4 µL of PBS was added to the cell side of the grid, and grids were blotted from the back side for 6 seconds, followed by plunging into liquid ethane at -180 oC. Grids were clipped into Autogrid rings for the remainder of processing.
To identify cells of interest for milling, grids containing vitrified cells were imaged on a Zeiss LSM900 confocal microscope equipped with a Linkam CMS 196 cryo-stage operated with ZEN 3.6 blue edition software. High resolution z-stacks were collected using a x100 objective (N.A. 0.75) and Airyscan 2 detector. Gallium ion FIB milling was performed on a Zeiss Crossbeam 550 equipped with a Quorum PP3010 cryo-stage and cryo-transfer system operated with SmartSEM 7.05 and SmartFIB v1.19.0. Autogrids mounted onto a 26-degree pre-tilted shuttle and transferred into the Quorum prep chamber where 2–3 nm of platinum was sputter-coated onto the grid. An organometallic platinum layer was added to the grid using the gas injection system (GIS) inside the FIB-SEM chamber. Lamellae were milled using a 30 kV gallium ion beam, thinning the sample with sequentially lower ion beam currents as follows: 700 pA to a thickness of 2 µm, 300 pA to a thickness of 1 µm, 100 pA to a thickness of 0.5 µm, and 50 pA to a final thickness of 180–200 nm. After milling, lamellae were imaged on the LSM900 to identify targets for tilt series acquisition.
Lamellae were imaged using a ThermoFisher Titan Krios G3i microscope operating at 300 kV using a Gatan K3 Summit detector and GIF energy filter operated with Serial EM 4.2.0 and PACEtomo 1.8 scripts. Montaged maps were acquired of each lamella, and confocal images were used for correlative imaging of fluorescently labeled organelles. SerialEM67 utilizing PACEtomo scripts79 was used for parallel tilt series acquisition at a magnification of 33,000 x (2.67 Å/pix), GIF slit width of 20 eV, and defocus of 4 µm. Images were collected through a tilt range of -50 to +70 or -70 to +50 degrees depending on the ±10 degree tilt of the lamellae, with a tilt increment of 3 degrees. The dose per tilt was 2.5-3 e-/Å2, fractionated into 4-6 frames, to achieve a total dose of 100-120 e-/Å2. Tilt series were aligned and reconstructed by weighted back projection in IMOD68,69. Missing wedge correction was done in IsoNet72 version 2.0. Alignment of confocal images with tomograms and visualization of the overlay was done in Zeiss ZEN software using mitochondria as internal fiducials.
Mass spectrometry
Quantitative mass spectrometry analysis was described recently80 with following modifications: Protein preparations were dissolved in SDS lysis buffer (10 mM (tris(2-carboxyethyl)phosphine) (TCEP), 2% SDS, 150 mM NaCl, 50 mM Tris, pH 8.5, 50 mM ammonium biocarbonate) and reduced cysteine residues were alkylated with iodoacetamide at a final concentration of 20 mM and incubation at room temperature in the dark for 25 minutes. Alkylation was quenched with 50 mM freshly prepared DTT. SDS was removed by SP3 protein cleanup81 and proteins were digested with Lys-C protease (Wako, Catalog Number 129–02541) and trypsin (Promega Catalog Number V5113). Control samples were labelled with TMTpro channels 126, 127 N and 127 C; TRPML1-mNeonGreen-1D4 channels with 128 N, 128 C and 129 N. Mixed TMT labelled peptides were subjected to bench-top alkaline reversed phase fractionation as described (Pierce High pH Reversed-Phase Peptide Fractionation Kit, Thermo Fisher Scientific) and run as 6 combined (1 + 7, 2 + 8, 3 + 9, 4 + 10, 5 + 11 and 6 + 12) fractions. Data were recorded on an Orbitrap Ascend instrument (Thermo Fisher Scientific) with a MS2 method using 90 min FAIMS runs (CVs -40V, -60V, -80V, cycle time 1.5 s) at an MS1 resolution of 120,000 in the orbitrap. After HCD fragmentation (25% collision energy), MS2 scans were collected at 45,000 resolution in the orbitrap with a maximal injection time of 250 ms. Data were searched with COMET82 Version 2020.01 rev.4 using additional masses of +15.994914 Da for oxidized methionine modification dynamically and +304.207145 Da on peptide N-termini and lysine residues as static modification. Volcano plots of the proteomics data were generated using two-tailed t-test without any p-adjustment methods. Changes in enrichment were visualized using GraphPad Prism 10.1.1. (270). Principal component analyses graphs were generated in GraphPad Prism 10.1.1. (270).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The proteomics data generated in this study have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository under accession code PXD055923. The sub-tomogram average of V-ATPase, Flotillin, and Clathrin maps generated in this study have been deposited in the Electron Microscopy Data Bank database under the accession codes EMD-47080, EMD-47081, and EMD-49944 respectively. CryoET raw data generated in this study are available at EMPIAR-12937. Source data are provided with this paper.
References
Klumperman, J. & Raposo, G. The complex ultrastructure of the endolysosomal system. Cold Spring Harb. Perspect. Biol. 6, a016857 (2014).
Neel, E. et al. The endolysosomal system in conventional and unconventional protein secretion. J Cell Biol 223, https://doi.org/10.1083/jcb.202404152 (2024).
Huotari, J. & Helenius, A. Endosome maturation. EMBO J. 30, 3481–3500 (2011).
Grant, B. D. & Donaldson, J. G. Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell Biol. 10, 597–608 (2009).
De Duve, C., Pressman, B. C., Gianetto, R., Wattiaux, R. & Appelmans, F. Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem J. 60, 604–617 (1955).
Xu, H. & Ren, D. Lysosomal physiology. Annu Rev. Physiol. 77, 57–80 (2015).
Lim, C. Y. & Zoncu, R. The lysosome as a command-and-control center for cellular metabolism. J. Cell Biol. 214, 653–664 (2016).
Perera, R. M. & Zoncu, R. The Lysosome as a Regulatory Hub. Annu Rev. Cell Dev. Biol. 32, 223–253 (2016).
Lawrence, R. E. & Zoncu, R. The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 21, 133–142 (2019).
Lamming, D. W. & Bar-Peled, L. Lysosome: The metabolic signaling hub. Traffic 20, 27–38 (2019).
Boustany, R. M. Lysosomal storage diseases-the horizon expands. Nat. Rev. Neurol. 9, 583–598 (2013).
Platt, F. M. Emptying the stores: lysosomal diseases and therapeutic strategies. Nat. Rev. Drug Discov. 17, 133–150 (2018).
Ballabio, A. & Gieselmann, V. Lysosomal disorders: from storage to cellular damage. Biochim Biophys. Acta 1793, 684–696 (2009).
Desnick, R. J. & Schuchman, E. H. Enzyme replacement therapy for lysosomal diseases: lessons from 20 years of experience and remaining challenges. Annu Rev. Genomics Hum. Genet 13, 307–335 (2012).
Aflaki, E., Westbroek, W. & Sidransky, E. The Complicated Relationship between Gaucher Disease and Parkinsonism: Insights from a Rare Disease. Neuron 93, 737–746 (2017).
Whyte, L. S., Lau, A. A., Hemsley, K. M., Hopwood, J. J. & Sargeant, T. J. Endo-lysosomal and autophagic dysfunction: a driving factor in Alzheimer’s disease?. J. Neurochem 140, 703–717 (2017).
Oikawa, N. & Walter, J. Presenilins and gamma-Secretase in Membrane Proteostasis. Cells 8, https://doi.org/10.3390/cells8030209 (2019).
Eriksson, I. & Ollinger, K. Lysosomes in Cancer-At the Crossroad of Good and Evil. Cells 13, https://doi.org/10.3390/cells13050459 (2024).
Aguado, C., Perez-Jimenez, E., Lahuerta, M. & Knecht, E. Isolation of Lysosomes from Mammalian Tissues and Cultured Cells. Methods Mol. Biol. 1449, 299–311 (2016).
Tharkeshwar, A. K., Demedts, D. & Annaert, W. Superparamagnetic Nanoparticles for Lysosome Isolation to Identify Spatial Alterations in Lysosomal Protein and Lipid Composition. STAR Protoc. 1, 100122 (2020).
Abu-Remaileh, M. et al. Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science 358, 807–813 (2017).
Chen, C., Sidransky, E. & Chen, Y. Lyso-IP: Uncovering Pathogenic Mechanisms of Lysosomal Dysfunction. Biomolecules 12, https://doi.org/10.3390/biom12050616 (2022).
Park, H. et al. Spatial snapshots of amyloid precursor protein intramembrane processing via early endosome proteomics. Nat. Commun. 13, 6112 (2022).
Davis, O. B. et al. NPC1-mTORC1 Signaling Couples Cholesterol Sensing to Organelle Homeostasis and Is a Targetable Pathway in Niemann-Pick Type C. Dev. Cell 56, 260–276 e267 (2021).
Barral, D. C. et al. Current methods to analyze lysosome morphology, positioning, motility and function. Traffic 23, 238–269 (2022).
de Oliveira, T. M., van Beek, L., Shilliday, F., Debreczeni, J. E. & Phillips, C. Cryo-EM: The Resolution Revolution and Drug Discovery. SLAS Discov. 26, 17–31 (2021).
Kuhlbrandt, W. Forty years in cryoEM of membrane proteins. Microsc. (Oxf.) 71, i30–i50 (2022).
Piper, S. J., Johnson, R. M., Wootten, D. & Sexton, P. M. Membranes under the Magnetic Lens: A Dive into the Diverse World of Membrane Protein Structures Using Cryo-EM. Chem. Rev. 122, 13989–14017 (2022).
Davies, K. M., Blum, T. B. & Kuhlbrandt, W. Conserved in situ arrangement of complex I and III(2) in mitochondrial respiratory chain supercomplexes of mammals, yeast, and plants. Proc. Natl. Acad. Sci. USA 115, 3024–3029 (2018).
Cai, S. et al. In situ architecture of the lipid transport protein VPS13C at ER-lysosome membrane contacts. Proc. Natl. Acad. Sci. USA 119, e2203769119 (2022).
Kravcenko, U. et al. Molecular architecture of synaptic vesicles. Proc. Natl. Acad. Sci. USA 121, e2407375121 (2024).
Zheng, W., Chai, P., Zhu, J. & Zhang, K. High-resolution in situ structures of mammalian respiratory supercomplexes. Nature 631, 232–239 (2024).
Fu, Z. & MacKinnon, R. Structure of the flotillin complex in a native membrane environment. Proc. Natl. Acad. Sci. USA 121, e2409334121 (2024).
Coupland, C. E. et al. High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles. Science 385, 168–174 (2024).
Wang, C. et al. Structure and topography of the synaptic V-ATPase-synaptophysin complex. Nature 631, 899–904 (2024).
Wang, W., Zhang, X., Gao, Q. & Xu, H. TRPML1: an ion channel in the lysosome. Handb. Exp. Pharm. 222, 631–645 (2014).
Bassi, M. T. et al. Cloning of the gene encoding a novel integral membrane protein, mucolipidin-and identification of the two major founder mutations causing mucolipidosis type IV. Am. J. Hum. Genet 67, 1110–1120 (2000).
Sun, M. et al. Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum. Mol. Genet 9, 2471–2478 (2000).
Molday, L. L. & Molday, R. S. 1D4: a versatile epitope tag for the purification and characterization of expressed membrane and soluble proteins. Methods Mol. Biol. 1177, 1–15 (2014).
Rocereta, J. A. et al. Structural insights into TRPV2 modulation by probenecid. Nat Struct Mol Biol. https://doi.org/10.1038/s41594-025-01494-9 (2025).
Isolation of a novel gene underlying Batten disease CLN3. The International Batten Disease Consortium. Cell 82, 949–957 (1995).
Carstea, E. D. et al. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science 277, 228–231 (1997).
Polovitskaya, M. M. et al. Gain-of-function variants in CLCN7 cause hypopigmentation and lysosomal storage disease. J. Biol. Chem. 300, 107437 (2024).
Forgac, M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8, 917–929 (2007).
Saxton, R. A. & Sabatini, D. M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 169, 361–371 (2017).
Pu, J., Guardia, C. M., Keren-Kaplan, T. & Bonifacino, J. S. Mechanisms and functions of lysosome positioning. J. Cell Sci. 129, 4329–4339 (2016).
Kokubo, H. et al. Ultrastructural localization of flotillin-1 to cholesterol-rich membrane microdomains, rafts, in rat brain tissue. Brain Res 965, 83–90 (2003).
Kaushik, S., Massey, A. C. & Cuervo, A. M. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J. 25, 3921–3933 (2006).
Singh, J. et al. Cross-linking of the endolysosomal system reveals potential flotillin structures and cargo. Nat. Commun. 13, 6212 (2022).
Bodin, S., Elhabashy, H., Macdonald, E., Winter, D. & Gauthier-Rouviere, C. Flotillins in membrane trafficking and physiopathology. Biol. Cell 117, e2400134 (2025).
van der Beek, J., de Heus, C., Liv, N. & Klumperman, J. Quantitative correlative microscopy reveals the ultrastructural distribution of endogenous endosomal proteins. J Cell Biol 221, https://doi.org/10.1083/jcb.202106044 (2022).
Paraan, M. et al. The structures of natively assembled clathrin-coated vesicles. Sci. Adv. 6, eaba8397 (2020).
Fotin, A. et al. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature 432, 573–579 (2004).
Cheng, Y., Boll, W., Kirchhausen, T., Harrison, S. C. & Walz, T. Cryo-electron tomography of clathrin-coated vesicles: structural implications for coat assembly. J. Mol. Biol. 365, 892–899 (2007).
Chaillet, M. L., Roet, S., Veltkamp, R. C. & Förster, F. pytom-match-pick: a tophat-transform constraint for automated classification in template matching. bioRxiv, 2024.2009.2017.613497 https://doi.org/10.1101/2024.09.17.613497 (2024).
Wang, L., Wu, D., Robinson, C. V., Wu, H. & Fu, T. M. Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly. Mol. Cell 80, 501–511 e503 (2020).
van der Beek, J., Jonker, C., van der Welle, R., Liv, N. & Klumperman, J. CORVET, CHEVI and HOPS - multisubunit tethers of the endo-lysosomal system in health and disease. J Cell Sci 132, https://doi.org/10.1242/jcs.189134 (2019).
Kuo, A. & Hla, T. Regulation of cellular and systemic sphingolipid homeostasis. Nat. Rev. Mol. Cell Biol. 25, 802–821 (2024).
Klein, S. et al. Post-correlation on-lamella cryo-CLEM reveals the membrane architecture of lamellar bodies. Commun. Biol. 4, 137 (2021).
Mizushima, N. & Komatsu, M. Autophagy: renovation of cells and tissues. Cell 147, 728–741 (2011).
Bond, C., Hugelier, S., Xing, J., Sorokina, E. M. & Lakadamyali, M. Multiplexed DNA-PAINT Imaging of the Heterogeneity of Late Endosome/Lysosome Protein Composition. bioRxiv, 2024.2003.2018.585634 https://doi.org/10.1101/2024.03.18.585634 (2024).
Gonzalez-Lozano, M. A. et al. EndoMAP.v1, a Structural Protein Complex Landscape of Human Endosomes. bioRxiv, 2025.2002.2007.636106 https://doi.org/10.1101/2025.02.07.636106 (2025).
Gonzalez-Magaldi, M. et al. Structure and organization of full-length epidermal growth factor receptor in extracellular vesicles by cryo-electron tomography. Proc. Natl. Acad. Sci. USA 122, e2424678122 (2025).
Neselu, K. et al. Measuring the effects of ice thickness on resolution in single particle cryo-EM. J. Struct. Biol. X 7, 100085 (2023).
Kelley, R. et al. Towards community-driven visual proteomics with large-scale cryo-electron tomography of Chlamydomonas reinhardtii. bioRxiv, 2024.2012.2028.630444 https://doi.org/10.1101/2024.12.28.630444 (2024).
Bolte, S. & Cordelieres, F. P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc 224, 213–232 (2006).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Burt, A. et al. An image processing pipeline for electron cryo-tomography in RELION-5. FEBS Open Bio. https://doi.org/10.1002/2211-5463.13873 (2024).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Mastronarde, D. N. & Held, S. R. Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol. 197, 102–113 (2017).
Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
Buchholz, T. O. et al. Content-aware image restoration for electron microscopy. Methods Cell Biol. 152, 277–289 (2019).
Chaillet, M. L. et al. Extensive Angular Sampling Enables the Sensitive Localization of Macromolecules in Electron Tomograms. Int J Mol Sci 24 https://doi.org/10.3390/ijms241713375 (2023).
Chaillet, M. L., Roet, S., Veltkamp, R. C. & Forster, F. pytom-match-pick: A tophat-transform constraint for automated classification in template matching. J. Struct. Biol. X 11, 100125 (2025).
Ermel, U. H., Arghittu, S. M. & Frangakis, A. S. ArtiaX: An electron tomography toolbox for the interactive handling of sub-tomograms in UCSF ChimeraX. Protein Sci. 31, e4472 (2022).
Lamm, L. et al. MemBrain v2: an end-to-end tool for the analysis of membranes in cryo-electron tomography. bioRxiv, 2024.2001.2005.574336 https://doi.org/10.1101/2024.01.05.574336 (2024).
Last, M. G. F., Abendstein, L., Voortman, L. M. & Sharp, T. H. Ais: streamlining segmentation of cryo-electron tomography datasets. eLife 13, RP98552 (2024).
Meng, E. C. et al. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Eisenstein, F. et al. Parallel cryo electron tomography on in situ lamellae. Nat. Methods 20, 131–138 (2023).
Gallo, P. N. et al. The dynamic TRPV2 ion channel proximity proteome reveals functional links of calcium flux with cellular adhesion factors NCAM and L1CAM in neurite outgrowth. Cell Calcium 121, 102894 (2024).
Hughes, C. S. et al. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 14, 68–85 (2019).
Eng, J. K. et al. A deeper look into Comet-implementation and features. J. Am. Soc. Mass Spectrom. 26, 1865–1874 (2015).
Acknowledgements
This work was supported by the National Institutes of Health: Maximizing Investigators’ Research Award (MIRA) grant (R35GM144120 to V.Y.M.-B) and Predoctoral Training Grant in Pharmacology (T32GM008076) to B.M.M. M.K. is a CPRIT scholar in Cancer Research, and M.K. and D.H.S. were supported by a CPRIT grant RR220032 to M.K.. We acknowledge the use of instruments at the Institute of Structural Biology at the University of Pennsylvania Perelman School of Medicine and at the Electron Microscopy Resource Lab and at the Beckman Center for Cryo Electron Microscopy at the University of Pennsylvania Perelman School of Medicine (RRID:SCR_022375). We thank Dr. Ricardo D. Righetto for his advice throughout the cryoET data processing. We thank members of the Moiseenkova-Bell lab for their excellent guidance, assistance and support.
Author information
Authors and Affiliations
Contributions
Conceptualization: B.M.M., J.J.dJ.-P. and V.Y.M.-B.; methodology: B.M.M., J.J.dJ.-P., E.M.M., P.G., S.K.M. and D.H.S.; validation: B.M.M., J.J.dJ.-P., E.M.M., P.G., S.K.M., D.H.S. and M.K.; formal analysis: B.M.M., J.J.dJ.-P. and D.H.S.; investigation: B.M.M., J.J.dJ.-P., E.M.M., P.G., S.K.M., D.H.S. and M.K.; visualization: B.M.M. and J.J.dJ.-P.; supervision: M.K. and V.Y.M.-B.; funding acquisition: B.M.M., V.Y.M.-B. and M.K.; writing–original draft: B.M.M., J.J.dJ.-P. and V.Y.M.-B.; writing–reviewing and editing: B.M.M., J.J.dJ.-P. and V.Y.M.-B. All authors contributed to the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
McVeigh, B.M., De Jesús-Pérez, J.J., Siepe, D.H. et al. Visualization of lysosomal membrane proteins by cryo electron tomography. Nat Commun 16, 9234 (2025). https://doi.org/10.1038/s41467-025-64314-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-64314-0