Abstract
Zygotic genome activation is tightly associated with the modulation of chromatin accessibility via maternal transcription factors. Understanding how chromatin accessibility is established and identifying key maternal regulators are crucial to comprehending this process. Here, by developing CANTAC-seq, we generate a genome-wide map of accessible chromatin of early Xenopus tropicalis embryos and find that the open chromatin landscape is progressively established at cis-regulatory elements during zygotic genome activation. Based on the motif analysis and perturbation experiments, we demonstrate that E2f1 maintains a repressive chromatin environment and inhibits zygotic gene transcription before the mid-blastula transition. Moreover, we identify that Otx1, another maternal transcriptional activator, coordinates with E2f1 in regulating chromatin accessibility and zygotic genome activation. Together, E2f1 and Otx1 determine the timely expression of a subset of genes required for zygotic gene transcription and germ layer differentiation.
Similar content being viewed by others
Introduction
Following fertilization, the sperm and the egg fuse to form a zygote, which will ultimately give rise to a new organism. Initially, the zygote genome is transcriptionally quiescent, and it only gradually becomes activated through a period called “maternal to zygotic transition”1. This onset of transcription is referred to as the zygotic genome activation (ZGA), which is a universal process tightly controlled during embryogenesis, though the timing of ZGA varies largely among different species2,3. For example, slow-developing species such as mice and humans require one or more days to finish ZGA, while fast-developing species such as zebrafish and frog can complete this process in just a few hours4. In frogs, embryos undergo 12 rapid synchronous divisions within 5 h, followed by asynchronous cleavages, with the induction of gap phases during a critical time named mid-blastula transition (MBT)5,6. While the transcription of most zygotic genes coincides with the MBT during the major wave of ZGA, some zygotic genes required for frog embryo patterning begin to express during the minor wave of ZGA that occurs before the MBT7,8.
The process of ZGA is known to coincide with extensive chromatin remodeling that drives essential cellular processes, including gene expression, DNA replication, and repair3. Accessible chromatin typically marks cis-regulatory elements such as promoters and enhancers9, which modulate the binding of specific DNA-binding transcription factors (TFs), thereby regulating the transcription of key developmental regulators during ZGA. In turn, TFs also dynamically coordinate with other chromatin modifiers to modulate the local DNA access during this process10,11. Conceptually, the accumulation of activators and/or loss of repressors during ZGA regulates local chromatin targets, and often these factors together determine the time of the activation of zygotic genes2. Therefore, characterization of chromatin accessibility dynamics and identification of responsible transcription activators and repressors during ZGA are vital for advancing our knowledge of early embryogenesis.
Recent studies utilizing ATAC-seq or DNase-seq have unveiled the dynamics of chromatin accessibility during ZGA in several model organisms12,13,14,15,16,17. These studies have demonstrated that regulatory regions often display accessible chromatin prior to gene activation and have highlighted the pivotal role of specific transcription activators in driving changes in chromatin state. For instance, the pioneer factor OCT4 has been shown to establish chromatin patterns during human ZGA, but not in mice14. In zebrafish, maternal factors Pou5f3, Sox19b, and Nanog are critical in chromatin opening and primed for transcriptional activity11,17. As to the repressors, histones have been identified as common repressors of gene transcription during ZGA in both zebrafish and frogs18,19. Additionally, DNA methyltransferase Dnmt1 and the methyl-CpG repressor Kaiso, both with low DNA-binding sequence specificity, have been demonstrated with a general repressive role in ZGA of frogs20,21. So far, the involvement of sequence- or target-specific transcription repressors in chromatin changes during this process remains elusive.
Xenopus tropicalis (X. tropicalis) is a classical model organism for exploring fundamental questions in developmental biology22. This is attributed to its abundant egg supply and easily manipulated embryos. Prior studies have examined chromatin accessibility changes via ATAC-seq and DNase-seq following the MBT during X. tropicalis development23,24,25. However, compared to other model organisms, the global chromatin accessibility landscape and its molecular dynamics prior to the MBT remain poorly understood due to technical limitations23,24. In this study, we developed CANTAC-seq, concanavalin A (ConA) bead-based nucleus capture followed by Tn5-mediated accessible chromatin assay with sequencing, to profile the chromatin accessibility landscape of early X. tropicalis embryos. We generated a genome-wide map of chromatin accessible regions from stage 7 to stage 13 (blastula to neurula), revealing the dynamics of chromatin accessibility during X. tropicalis early embryogenesis. Bioinformatics analysis revealed that E2f binding motifs are highly enriched at promoter regions opened during X. tropicalis ZGA. Further perturbation experiments showed that E2f1 plays a repressive role in the establishment of an open chromatin landscape prior to the MBT. Moreover, we found that the transcription activator Otx1 coordinated with E2f1 in regulating chromatin accessibility and the timely activation of a subset of genes required for zygotic gene transcription and germ layer differentiation.
Results
Development of CANTAC-seq
High yolk content interfered with transposase activity and made it almost impossible to apply ATAC-seq on whole X. tropicalis embryos of early developmental stages23,24. Therefore, in order to investigate the chromatin regulatory landscape during early development, we have developed CANTAC-seq, which entails the use of ConA-coated magnetic beads with high glycoprotein affinity to capture nuclei26,27, followed by yolk removal and optimized on-beads tagmentation to extract open chromatin regions for sequencing (see “Methods” section; Fig. 1a).
a A schematic view of the CANTAC-seq workflow. The process begins with a cell or nucleus (blue) where ConA-coated beads (green) bind specifically to surface glycoproteins (orange). Subsequent steps depict bead-directed tagmentation and library preparation for sequencing. b Genome-wide comparison of standard ATAC-seq data with CANTAC-seq data generated from mESCs. The points indicate accessible regions identified by ATAC-seq and CANTAC-seq. The X and Y axes represented the number of sequencing reads derived from the accessible regions. c TSS enrichment of standard ATAC-seq data and CANTAC-seq data in mESCs. d Comparison of insert sizes generated from mESCs using ATAC-seq and CANTAC-seq. e Genome browser view of ATAC-seq and CANTAC-seq normalized read counts at Nanog, Klf4, Pou5f1, and Sox2 loci in mESCs. f DNA electrophoresis of libraries of X. tropicalis whole embryos (stage 13) using ATAC-seq (left) and CANTAC-seq (right) protocols.
To validate the CANTAC-seq method, we first compared it with standard ATAC-seq on mouse embryonic stem cells (mESCs) and human K562 cells. As shown in Fig. 1b and Supplementary Fig. 1a, the chromatin accessibility profiles generated by CANTAC-seq correlated well with those obtained by standard ATAC-seq on mESCs (R = 0.99) and K562 cells (R = 0.99). Quality control metrics, such as insert size distribution and enrichment around transcriptional start sites (TSS), were almost identical between CANTAC-seq and ATAC-seq from the same cell line (Fig. 1c, d and Supplementary Fig. 1b, c). Open chromatin regions detected by both methods showed similar profiles at the promoters of pluripotent marker genes, including Nanog, Klf4, Pou5f1, and Sox2 in mESCs (Fig. 1e), as well as at the well-characterized human β-globin locus control region (LCR) and its downstream hemoglobin genes in K562 cells Supplementary Fig. 1d).
Then, we performed both methods on X. tropicalis embryos. As previously reported, standard ATAC did not produce DNA fragments of distinct sizes corresponding to accessible DNA, mono-nucleosome, and di-nucleosome. In contrast, our CANTAC-seq protocol could efficiently capture nuclei, with approximately 81–87% of nuclei recovered at the tested developmental stages (Supplementary Fig. 1e, f). Importantly, we could successfully generate these distinct DNA fragments, with an insert size distribution similar to that obtained from mESCs and K562 cells. These results collectively demonstrated that while the performance of the CANTAC-seq method is on par with standard ATAC-seq approaches for cellular samples, only CANTAC-seq enables the successful mapping of accessible chromatin of early X. tropicalis embryos with high yolk content.
Accessible chromatin landscape during X. tropicalis ZGA
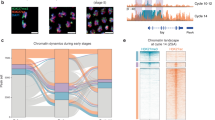
After establishing CANTAC-seq, to investigate the chromatin regulatory landscape during X. tropicalis ZGA, we mapped the global chromatin accessibility using CANTAC-seq for five developmental stages, including early, middle, and late blastula (stage 7, 8, and 9, respectively), which encompass the first major developmental transition MBT, the onset of gastrula (stage 10), and the onset of neurula (stage 13) (Fig. 2a and Supplementary Fig. 2a). These stages encompass both the minor wave (stages 7 and 8) and the major wave (stages 9–13) of ZGA processes. Each stage was analyzed in two independent biological replicates, which showed a high degree of correlation (Pearson’s correlation coefficient, 0.93–1.00, Supplementary Fig. 2b). Therefore, to increase the specificity, we combined the data and used common peaks identified from both replicates for subsequent analysis. Overall, we could identify 339, 23,731, 28,331, 44,481, and 47,726 accessible regions at stages 7, 8, 9, 10, and 13, respectively (Fig. 2b and Supplementary Data 1). This demonstrated that while the chromatin was almost inaccessible at stage 7, a drastic opening was started at stage 8, followed by a progressive increase of accessibility from stage 9 to stage 13 (Fig. 2b, c). A substantial proportion of accessible regions established during the early stage was largely maintained through later stages. Specifically, 61% (207/339) of stage 7 peaks, 44% (10,543/23,731) of stage 8 peaks, 53% (14,917/28,331) of stage 9 peaks, and 49% (21,897/44,481) of stage 10 peaks were maintained until stage 13 (Supplementary Data 1). Furthermore, we analyzed the genomic distributions of newly-emerged accessible regions at each stage and found a continuous increase of peaks at intergenic regions from stage 8 to 13 (Fig. 2d), suggesting a stronger acceleration of chromatin opening at distal regulatory regions during early development.
a A schematic view of the five developmental stages examined by CANTAC-seq. Embryos were collected at 3.5 hpf, 4 hpf, 5.5 hpf, 7 hpf, and 12 hpf to generate genome-wide chromatin accessibility profiles for stages 7, 8, and 9 (blastula), 10 (gastrula), and 13 (neurula), respectively. ZGA zygotic genome activation, MBT midblastula transition, hpf hours post-fertilization. b The bar plot illustrates the number of accessible chromatin regions detected at each of the five developmental stages during ZGA. c Heatmaps of chromatin accessibility for the five developmental stages, two biological replicates for each stage. d The bar plot illustrates the number of newly gained accessible regions at different genomic locations during ZGA. e Genome browser view of two representative genomic regions that lose (mir427 cluster, chr3: 146,260,000–146,294,000) or gain (sox17 cluster, chr6: 115,150,000–115,191,000) chromatin accessibility during ZGA. Source data are provided in the Source Data file.
Then, we compared chromatin accessibility profiles generated by CANTAC-seq with published DNase-seq data23, ATAC-seq data25, as well as ChIP-seq data of different histone marks at stages 8 and 928,29. As shown in Supplementary Fig. 2c, half of the open chromatin regions identified by CANTAC-seq overlapped with those detected by a modified DNase-seq protocol at stage 8. Moreover, our CANTAC-seq captured many more open chromatin regions compared to published ATAC-seq data at stage 9. Importantly, the open regions identified by CANTAC-seq, regardless of whether they were overlapped and non-overlapped with those detected by ATAC-seq, were marked with the active histone marks (H3K4me1/3 and H3K27ac), but not with the repressive marks (H3K27me3 and H3K9me2/3) (Supplementary Fig. 2c).
Furthermore, we checked the changes of chromatin accessibility at cis-regulatory elements for some key developmental regulators, including miR-427 cluster and sox17 gene cluster (Fig. 2e). The microRNA miR-427, which represents the ortholog of zebrafish miR-430, was involved in the deadenylation and clearance of maternal mRNAs, and was highly expressed before the MBT and then switched off at the gastrula stage30. In line with such an expression pattern, we found that the chromatin region (chr3:146,260,000–146,294,000) of the miR-427 cluster was already open before the MBT (stages 7–9), but turned closed afterwards (stages 10 and 13). In contrast, we found that chromatin accessibility at the sox17 cluster (chr6:115,150,000–115,191,000), including sox17a, sox17b.1, and sox17b.2, was gradually established, which is consistent with the expression and function of sox17 genes in germ layer differentiation31.
Finally, to reveal the potential function of increased accessible regions, we carried out gene ontology (GO) analysis on genes that gained promoter accessibility at each stage. As shown in Supplementary Fig. 2d, e, GO terms related to transcriptional regulation and GTPase activity, as well as pathways related to P53 regulation and GTPase cycle were significantly enriched at stages 8, 9 and 10, which is consistent with the fact that these biological processes and pathways are important in regulating cell migration, adhesion, as well as cell cycles during early development. Interestingly, genes with gained promoter accessibility specifically at stage 13 were enriched for those functioning in the neuronal system and encoding stimulus-sensing channels, consistent with the differentiation of nervous tissue at this stage. These findings highlighted the important functional relevance of the regulation of chromatin landscape during X. tropicalis early development.
Taken together, these analyses demonstrated that CANTAC-seq is a highly efficient method for generating accessible chromatin profiles of X. tropicalis embryos during early development.
Proximal and distal accessible chromatin in X. tropicalis early development
To understand the association between chromatin accessibility and gene expression during ZGA, we conducted transcriptome analyses at the corresponding developmental stages (Supplementary Data 2). Again, RNA-seq analyses performed on two independent biological replicates for each stage showed a high degree of correlation (Pearson’s correlation coefficient, 0.96–1.00, Supplementary Fig. 3a). We then performed principal component analyses (PCA) on the RNA-seq and CANTAC-seq datasets, which revealed a consistent pattern of developmental progression in both gene expression and chromatin accessibility profiles, as depicted in Supplementary Fig. 3b. Then, based on the promoter accessibility at each stage, we classified genes into three groups (i.e., low, medium, and high) and compared their expression levels. As illustrated in Fig. 3a, genes with more accessible promoters showed elevated expression levels at all stages examined. After the MBT, genes with high promoter accessibility showed higher expression levels than before (stages 7 and 8), and the difference in the expression level between the different groups is becoming more obvious, which is consistent with a higher impact of transcriptional regulation on RNA abundance after ZGA. To further explore the temporal relationship between chromatin opening and transcription activity, we compared promoter accessibility of genes expressed only after the MBT along the developmental stages and found that promoters are accessible prior to the onset of gene transcription (Fig. 3b). Moreover, we analyzed publicly available nascent RNA-seq data generated in X. laevis32, and identified 631 homologous zygotic transcripts that are induced at the MBT in X. tropicalis (FC > 2, FDR < 0.05). As shown in Supplementary Fig. 3c, promoters of these genes are accessible prior to the MBT. In addition, we also identified 78 homologous genes in X. tropicalis based on the list of well-known ZGA genes highly induced during MBT32. Again, promoters of these genes are accessible prior to the MBT. Taken together, these results suggested that chromatin opening precedes the transcription of these zygotic genes.
a The bar plot illustrates the expression level (Log2 (TPM + 1)) of genes with different promoter accessibility at different developmental stages. “Low”, “medium” and “high” groups were classified by separating the promoters into three groups of equal size (n = 9500 genes in each group) based on their CANTAC signal strength. Statistical significance was determined using a paired two-sided Student’s t-test. b The bar plot illustrates the expression level (Log2 (TPM + 1)) and promoter accessibility (Log2 (CPM + 1)) of the genes expressed after MBT (n = 852 genes) at different developmental stages. c, d Heatmaps of transcription factor (TF) DNA binding sequence motifs identified from proximal and distal CANTAC-seq peaks gained at each developmental stage. Stage-specific motif activity (−Log10 P value) and TF expression level (Log2 (TPM + 1)) are indicated by the size and color of the dot, respectively. Statistical significance was determined using a one-sided hypergeometric test.
As promoters and enhancers are often binding sites of TFs, we investigated whether proximal and distal CANTAC-seq peaks contained the motifs of TFs that regulate ZGA in early development. Using HOMER, we observed that TFs with enriched binding motifs for both promoters and enhancers mainly belong to the SP, KLF, and NFY protein families (Fig. 3c, d). In addition, whereas proximal regions exhibit specific enrichment of USF, ETV, and E2F motifs (Fig. 3c), distal regions are highly enriched with SOX, POU, CTCF, LHX, TFAP, and SMAD motifs (Fig. 3d). Importantly, several of the TFs identified in our motif enrichment analysis have been shown to be crucial in early development23,33,34,35,36,37,38. For instance, the maternal factor Sox3 with pioneering TF activity establishes pluripotency before the MBT by triggering chromatin remodeling23, while Ctcf is required for the de novo establishment of topologically associating domains during X. tropicalis ZGA38.
E2f1 is a repressor of minor ZGA
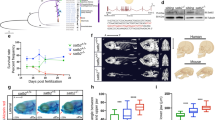
The enrichment of E2F binding motifs at promoters drew our attention, given that no function had been reported for E2F proteins during ZGA. In mammals, the E2F family consists of eight members with similar structure domains, of which E2F1/2/3 are classified as transcription activators, while E2F4/5/6/7/8 are considered to be transcription repressors39. All members of the e2f genes, except for e2f2, were identified in X. tropicalis. During early development from stages 3 to 9, e2f1, e2f3 and e2f5 exhibit high mRNA abundance, followed by a stark decrease between stages 9 and 10 (Supplementary Fig. 4a). To assess the role of the three highly expressed e2f genes in X. tropicalis early development, we designed morpholino antisense oligonucleotides (MOs) targeting e2f1 (e2f1 MO), e2f3 (e2f3 MO), and e2f5 (e2f5 MO), respectively. These MOs were separately microinjected at the one-cell stage to knock down their respective targets. We then quantified genomic DNA content in both uninjected wild-type (WT) and MO-injected embryos to assess the progression of cell division during early developmental stages. As seen in Supplementary Fig. 4b, we observed an increase in total genomic DNA content at stages 7 and 8 prior to MBT only in e2f1 MO-treated embryos, but not in e2f3 MO or e2f5 MO-treated embryos (Supplementary Fig. 4c, d). After MBT, there was no significant difference in DNA content between e2f1 MO and WT embryos at stage 9 (Supplementary Fig. 4b). Furthermore, we noted that all embryos injected with e2f1 MO exhibited delayed development and died at stage 27 (Fig. 4a and Supplementary Fig. 4e). To determine whether these outcomes resulted from e2f1 perturbation, we performed rescue experiments through co-injection of e2f1 MO and e2f1 mRNA containing mutated MO target sites. Interestingly, we found that abnormal development, as well as the increase of genomic DNA content, could be partially rescued (Fig. 4a and Supplementary Fig. 4b), with approximately 82% (76 out of 93) of embryos surviving till stage 27 (Supplementary Fig. 4e). This suggests these abnormalities are direct and specific effects of the e2f1 perturbation. In addition, we further confirmed the specificity of the e2f1 MO by co-injecting it with either HA-tagged WT e2f1 mRNA or e2f1 mRNA containing three point mutations in the MO binding site. As demonstrated in Supplementary Fig. 4f, the protein translated from WT e2f1 mRNA was significantly reduced following e2f1 MO injection. In contrast, the e2f1 mRNA with mutations in the MO binding site remained unaffected by the e2f1 MO. Furthermore, whereas WT e2f1 mRNA failed to rescue the increased DNA content phenotype induced by e2f1 MO, the mutated e2f1 mRNA successfully restored normal DNA levels at stages 7 and 8 (Supplementary Fig. 4g). Hereafter, we focused on E2f1 and elaborated on its role during X. tropicalis early development.
a Representative bright field images of embryos (stage 23) showing that e2f1 MO embryos display abnormal morphology at early development, which could be partially rescued by co-injecting e2f1 MO and e2f1 mRNA. Scale bar, 200 μm. b Heatmaps of chromatin accessibility detected by CANTAC-seq at stages 7 and 8 upon e2f1 MO, n = 2 biological replicates. c The box plot illustrates the numbers of up- and down-regulated genes upon e2f1 MO at different developmental stages. d The box plot illustrates the expression level of the genes up- or down-regulated upon e2f1 MO in uninjected WT, e2f1 MO, and e2f1 rescue embryos (e2f1 RE) at stages 7 and 8. The results are presented as the median Log2 (TPM + 1), along with the 25th and 75th quartiles, as well as the 5th and 95th percentiles. Statistical significance was determined using a paired two-sided Student’s t-test, n indicates the number of genes in each condition. e Principal component analysis (PCA) of transcriptome profiles (n = 2 biological replicates) of uninjected embryos (WT), e2f1 MO embryos (MO), as well as e2f1 RE at stages 6–8. A developmental trajectory was indicated with a dashed line for the WT embryos. f Overlap between the up-/down-regulated genes upon e2f1 MO at stage 7 and those up-/down-regulated from stage 7 to stage 8 during normal development, and GO enrichment analysis of the overlapped genes. The P values were calculated based on a one-sided hypergeometric test. Whereas no significant GO terms were enriched for down-regulated genes, a set of GO terms for up-regulated genes was enriched, and genes were listed to the right. The enriched GO terms are ranked by –Log10 (adjusted P value). The adjusted P values for enrichment of specific GO terms were calculated by using the linear step-up method of Benjamini and Hochberg. g Heatmaps of TF DNA binding sequence motifs identified from gained proximal and distal CANTAC-seq peaks upon e2f1 MO at stage 7. Motif activity (–Log10 P value) and TF expression (Log2 (TPM + 1)) are indicated by the size and color of the dot, respectively. Statistical significance was determined using a one-sided hypergeometric test. h Heatmaps depicting the expression of genes that are activated from stage 7 to stage 8 in WT embryos, as well as in embryos injected with e2f1 MO at stage 7. All the RNA samples were collected from one additional independent cross and sequenced in parallel (n = 2 biological replicates). e2f1 MO, cdkn1a, MO + cdkn1a, MO+cdkn1a, MO + e2f1 and MO + ∆DBD represent embryos injected with e2f1 MO, cdkn1a mRNA, e2f1 MO and cdkn1a mRNA, e2f1 MO and e2f1 mRNA, as well as e2f1 MO and the ∆DBD mutant mRNA, respectively. i Genome browser view of RNA-seq normalized read counts at sia1/2, mixer and mix1, and gsc loci.
Then, we conducted CANTAC-seq to analyze chromatin accessibility in e2f1 MO-treated embryos at stages 7 and 8, and compared them to the uninjected WT embryos. As shown in Fig. 4b, we observed a significant increase in chromatin accessibility across the genome at stage 7, with 11,894 regions gaining accessibility upon e2f1 MO treatment. On one hand, these accessible regions showed a high overlap (74%, 8814 out of 11,894) with regions that gained accessibility from stage 7 to stage 8 during normal development (Fig. 4b). On the other hand, we found that 66% (14,917 out of 23,731) of open chromatin regions identified in WT embryos at stage 8 were not detected in e2f1 MO embryos at stage 7. Together, these findings suggest that E2f1 exerts a repressive influence on a substantial portion (37%, 8814 out of 23,731) of chromatin accessibility during the normal ZGA process. Subsequently, we investigated how increased accessibility was accompanied by transcriptome alterations. To this end, we performed comparisons of transcriptomic profiles between e2f1 MO and WT embryos from stage 3 to stage 8 using RNA-seq. As shown in Figs. 4c and S4h, the gene expression profiles were almost identical between e2f1 MO and WT embryos before stage 7. In contrast, significant changes in gene expression were observed in e2f1 MO embryos starting from stage 7, with 743 and 610 genes up-regulated and 534 and 633 genes down-regulated at stage 7 and 8, respectively (FDR < 0.05 and |Log2 FC | > 1, Supplementary Data 3). GO terms related to gene transcription and embryo development were enriched for up-regulated genes, and GO term related to translation was enriched for down-regulated genes at stage 7 (Supplementary Fig. 4i). Importantly, co-injection of e2f1 mRNA and MO could significantly rescue the gene dysregulation observed in e2f1 MO stage 7 samples (Figs. 4d and S4j, Supplementary Data 4). Indeed, as shown in the PCA plot, e2f1 MO-treated embryos at stage 7 were closer to WT embryos of stage 8 at PC1 axis, whereas the rescued embryos moved back towards the WT stage 7 (Fig. 4e). Moreover, the e2f1 MO-induced upregulation of sia2, mix1, and gsc was almost fully rescued by co-injection of e2f1 mRNA with mutated MO binding sites, while WT e2f1 mRNA failed to restore normal expression (Supplementary Fig. 4k), further demonstrating the specificity of e2f1 MO.
In order to test whether the genes affected by e2f1 MO were also altered during normal development, we compared the genes altered upon e2f1 MO at stage 7 with differentially expressed genes during normal development between stage 7 and 8, and found a significant overlap (P < 2.2e-16, Fig. 4f). Out of the 699 ZGA genes that are normally induced from stage 7 to stage 8, 268 (38%) genes are significantly upregulated upon e2f1 MO at stage 7 (Fig. 4f). These genes were enriched for those with GO terms related to transcription, cell differentiation, morphogenesis, gastrulation, and BMP signaling (Fig. 4f). Notably, e2f1 MO caused premature expression of a set of well-known zygotic TFs, including gsc, bix1.2, sox17a/b.1/b.2, as well as pou5f3.1. In consistency with this, we observed significant enrichment of several TF motifs in the open chromatin regions from e2f1 MO-treated embryos at stage 7 (Fig. 4g), as well as from WT embryos at stage 8 (Fig. 3c, d), suggesting TFs repressed by E2f1 contribute to the chromatin opening during minor ZGA. Taken together, our analysis suggested that E2f1 functions as a maternal repressor in X. tropicalis early development.
E2f1 represses zygotic gene expression through transcriptional regulation
It is well documented that the nucleocytoplasmic ratio (N/C ratio) plays a crucial role in initiating ZGA5. More recently, Jukam et al. reported that the DNA-to-cytoplasmic ratio regulates the timing of zygotic gene expression in hybrid frog embryos40. To investigate whether the precocious initiation of zygotic gene transcription is a result of increased DNA content in e2f1 morphants, we introduced mRNA encoding the cyclin-dependent kinase inhibitor cdkn1a (Supplementary Fig. 5a), which was known to regulate DNA replication. As shown in Supplementary Fig. 5b, co-injection of cdkn1a mRNA with e2f1 MO successfully rescued the increased DNA content due to e2f1 MO alone (Supplementary Fig. 5b). Intriguingly, the precocious expression of zygotic genes at stage 7 induced by e2f1 MO was not rescued with the co-injection of cdkn1a mRNA (Fig. 4h). The observation that co-injection of cdkn1a mRNA could rescue increased DNA content, but not the precocious expression of zygotic genes induced by e2f1 MO demonstrated that the two effects of E2f1 were mechanistically independent. Given that E2f1 could directly regulate transcription by binding to target DNA via its well-characterized DNA binding domain (DBD, a.a. 119–184), to ascertain its direct repressive effect on zygotic gene activation, we performed rescue experiments using the e2f1 mutant lacking the DBD (∆DBD, Supplementary Fig. 5c). As shown in Supplementary Fig. 5d, similar to the WT e2f1, the ∆DBD mutant successfully rescued the increased DNA content. However, in contrast to WT e2f1, the mutant failed to rescue the precocious zygotic gene activation induced by e2f1 MO (Fig. 4h). As representative examples shown in Fig. 4i, zygotic genes, including sia1/2, mix1, and gsc, were activated at stage 7 by e2f1 MO. The defect could be rescued upon co-injection of WT e2f1 mRNA, but not that of the ∆DBD mutant. Similarly, co-injection of cdkn1a mRNA could not rescue the precocious expression of these zygotic genes, although it could rescue the DNA content prior to MBT.
To further establish that E2f1 represses gene expression at the transcriptional level, we injected 4′-thiouridine (4sU) to metabolically label newly synthesized zygotic transcripts and then enriched these transcripts by click-chemistry, followed by quantification of using qRT-PCR. As shown in Supplementary Fig. 5e, the abundance of 4sU-labeled zygotic transcripts derived from sia2, mix1, and gsc was significantly increased in e2f1 morphants compared to WTs at stage 7. In addition, we also performed RNA polymerase II ChIP. As shown in Fig S5f, the occupancy of RNA Polymerase II at these gene promoters was also significantly increased upon e2f1 MO at stage 7 (Supplementary Fig. 5f). Taken together, these results demonstrated that E2f1 indeed negatively regulates gene expression at the transcriptional level.
Otx1 interacts with E2f1 and functions as a transcriptional activator
To understand the molecular mechanisms underlying the E2f1-mediated repression of ZGA, we sought to identify E2f1 interaction partners. Due to the lack of specific antibodies that can recognize endogenous E2f1, we performed immunoprecipitation followed by mass spectrometry (IP-MS) analysis with the anti-HA antibody in stage 8 embryos injected with e2f1 MO and HA-tagged e2f1 mRNA. With a stringent cutoff (unique peptide > 2, coverage > 5% and Log10 FC (LFQ) > 1), we identified a total of 22 potential interaction partners with E2f1 (Table 1), including the DP family protein Tfdp1, which is known to form a heterodimeric complex with E2f1. Out of the remaining candidates, we focused on the orthodenticle homeobox transcription factor Otx1, as it was reported to be required for endoderm formation in X. tropicalis41, and its paralogue OTX2 was recently suggested to be a potential maternal regulator of human ZGA42. During X. tropicalis early development, otx1 is highly expressed while otx2 only starts to express after the MBT (Supplementary Fig. 6a). To validate the interaction between E2f1 and Otx1, we co-injected HA-tagged e2f1 and Myc-tagged otx1 mRNAs at one cell stage of X. tropicalis embryos (Fig. 5a). The interaction between E2f1 and Otx1 could be confirmed in Co-IP assays followed by western blotting (WB) with anti-HA and anti-Myc antibodies, respectively, at the blastula stage (Fig. 5a). Notably, this interaction was significantly reduced after treatment with benzonase, suggesting that the interaction between E2f1 and Otx1 is mediated largely by DNA and/or RNA.
a Co-immunoprecipitation (IP) was carried out using the antibody against HA, followed by WB to detect HA-tagged E2f1 or Myc-tagged Otx1 using anti-HA or anti-Myc antibody, respectively, in embryos expressing both HA-tagged E2f1 and Myc-tagged Otx1. Embryos expressing only HA-tagged E2f1 were used as a negative control. Protein lysates were treated with or without benzonase nuclease. Input lanes represent 1% of the total protein lysate. Three independent experiments were performed, and similar results were obtained. b Representative bright field images of uninjected WT embryos, e2f1 MO embryos, otx1 MO embryos, as well as e2f1 and otx1 double MO embryos at stages 7, 25, and 43. Scale bar, 500 μm. c The box plot illustrates the expression of the genes up- or down-regulated upon e2f1 MO at stage 7 and 8 in uninjected WT, e2f1 MO, as well as e2f1 and otx1 double MO embryos. The results are presented as the median Log2 (TPM + 1), along with the 25th and 75th quartiles, as well as the 5th and 95th percentiles. Statistical significance was determined using a paired two-sided Student’s t-test, n indicates the number of genes in each condition. d Heatmaps of chromatin accessibility detected by CANTAC-seq at stage 7 and 8 upon e2f1 MO or e2f1 and otx1 double MO, n = 2 biological replicates. e PCA of transcriptome profiles (n = 2 biological replicates) of uninjected WT embryos, e2f1 MO embryos, otx1 MO embryos, as well as e2f1 and otx1 double MO embryos at stages 7 and 8. A developmental trajectory was indicated with a dashed line for the WT embryos. f PCA of chromatin accessibility profiles (n = 2 biological replicates) of uninjected WT embryos, e2f1 MO embryos, otx1 MO embryos, as well as e2f1 and otx1 double MO embryos at stages 7 and 8. A developmental trajectory was indicated with a dashed line for the WT embryos. g The bar plot illustrates the percentage of genes (n = 699) induced between stage 7 and 8 that gain, lose, or do not change chromatin accessibility upon e2f1 MO, otx1 MO, as well as double MO at stage 7 and stage 8. Genic region: from 10 kb upstream of the Transcription Start Site to 10 kb downstream of the Transcription End Site. Source data are provided in the Source Data file.
To gain insight into the functional role of Otx1 during early development and, more importantly, to assess how Otx1 and E2f1 function together to regulate X. tropicalis ZGA, we blocked their translation by injecting MOs against e2f1 and otx1 into embryos either alone or together. As shown in Fig. 5b, otx1 MO-treated embryos were viable, whereas e2f1 & otx1 double MO-treated embryos had even more severe developmental defects than e2f1 MO-treated embryos, and all the double MO embryos died at the early tailbud stage. Next, to figure out the underlying mechanisms, we examined transcriptomic changes and chromatin accessibility alterations of otx1 MO-treated embryos, as well as e2f1 & otx1 double MO-treated embryos at stages 7 and 8 using RNA-seq and CANTAC-seq, respectively. Knockdown of otx1 alone led to 520 and 718 genes being up-regulated and 634 and 897 down-regulated at stage 7 and 8, respectively (FDR < 0.05 and |Log2 FC| > 1, Supplementary Fig. 6b and Supplementary Data 5). Similar to our approach with e2f1, the specificity of otx1 MO was validated by co-injecting it with either Myc-tagged WT otx1 mRNA or otx1 mRNA containing three point mutations in the MO binding site, followed by western blot analysis (Supplementary Fig. 6c). Additionally, the downregulation of zygotic genes sia2, mix1, and gsc upon otx1 MO injection was effectively rescued by co-injection with mutated otx1 mRNA but not WT otx1 mRNA at stage 8 (Supplementary Fig. 6d), further confirming the specificity of otx1 MO.
Then, we examined the developmental relevance of these genes and found a significant overlap (P < 2.2e-16, Supplementary Fig. 6e) between the genes down-regulated upon otx1 MO at stage 8 and the genes up-regulated between stage 7 and 8 during normal development. These overlapped genes, including several well-known zygotic TFs, were significantly enriched for the GO term related to transcription regulation (Supplementary Fig. 6e). In line with this, a slight attenuation of global chromatin accessibility upon otx1 MO was also observed at stage 8 (Supplementary Fig. 6f). Interestingly, double knockdown of e2f1 & otx1 did not affect a much larger group of genes (Supplementary Fig. 6g and Supplementary Data 6). The expression of up-regulated genes upon e2f1 MO was largely returned to WT level by double MO at both stage 7 and 8 (Fig. 5c). The prematurely opened chromatin induced by e2f1 MO at stage 7 was also partially rescued by the double MO (Fig. 5d). In contrast, the expression change induced by otx1 MO at stage 8 remained the same in double MO (Supplementary Fig. 6g). Further PCA analysis of RNA-seq and CANTAC-seq data showed that the transcriptome and the chromatin accessibility profiles of double MO embryos were indeed moving back and even lagged a bit behind the WT embryos along the developmental trajectory (Fig. 5e,f). The difference at the transcriptome level between WT embryos and double MO embryos at the PC1 axis was likely due to the effects of Otx1 independent of E2f1 (Fig. 5e). These results together suggested that Otx1 exerted a regulatory effect opposite to E2f1 during X. tropicalis minor ZGA.
Furthermore, to investigate the effect of different perturbations on chromatin accessibility of the 699 genes induced from stage 7 to stage 8, we analyzed the percent of these genes that gain, lose, or do not change accessibility upon e2f1 MO, otx1 MO or double MO (Fig. 5g). At stage 7, 57% of these genes gain accessibility at genic regions upon e2f1 MO, with 17% genes gaining accessibility at promoter regions. These together demonstrated that E2f1 exerts a dominantly repressive effect on chromatin opening at stage 7. In contrast, 95% of these genes show no change in accessibility upon otx1 MO at genic regions at stage 7. Similarly, these genes show no change of accessibility upon double MO, demonstrating that the chromatin opening induced by e2f1 MO requires Otx1 at stage 7. At stage 8, 49% and 43% genes gain and lose accessibility, respectively, at genic regions upon e2f1 MO, with 8% and 12% genes gaining and losing accessibility at promoter regions, respectively. In contrast, 63% and 64% genes lost accessibility at genic regions upon otx1 MO or double MO, with 32% and 33% losing accessibility at promoter regions. This analysis together suggested that Otx1 exerts a positive effect on chromatin opening at stage 8.
Otx1 co-regulates with E2f1 a subset of genes during minor ZGA
Both E2f1 and Otx1 are known DNA-binding TFs, we therefore asked how they bound to and localized on the genome before the MBT. Again, without a specific antibody, we performed chromatin immunoprecipitation (ChIP) followed by sequencing (ChIP-seq) of E2f1 and Otx1 with anti-HA and anti-Myc antibodies in stage 8 embryos injected with e2f1 MO and HA-tagged e2f1 mRNA or otx1 MO and Myc-tagged otx1 mRNA, respectively (Fig. 6a). In total, we identified 22,524 binding sites for E2f1, of which 30%, 42%, and 28% were located at the promoter, gene body, and intergenic region, respectively (Fig. 6b). For Otx1, we obtained 65,888 binding sites, with 6%, 52%, and 42% located at the promoter, gene body, and intergenic region, respectively (Fig. 6b). We observed a significant overlap of the DNA binding sites between E2f1 and Otx1, with almost half of E2f1 peaks overlapped with Otx1 (P < 2.2e-16, Supplementary Fig. 7a), suggesting a genome-wide co-occupancy of these two TFs. We then categorized these binding regions into three subgroups, namely E2f1 only (regions exclusively bound by E2f1, n = 12,031), E2f1&Otx1 (regions co-bound by E2f1 and Otx1, n = 10,493), and Otx1 only (regions exclusively bound by Otx1, n = 55,395). Among the common binding sites, 23% and 32% were located at promoters and intergenic regions, respectively (Supplementary Fig. 7b).
a A schematic view of the workflow of E2f1 and Otx1 ChIP-seq experiments. b Distribution of E2f1 and Otx1 binding sites at the promoter, gene body, and intergenic region. c Heatmaps of normalized read density of ChIP-seq (n = 2 biological replicates) for E2f1 and Otx1, as well as CANTAC-seq at five developmental stages. ChIP-peaks were categorized into three subgroups: E2f1-bound peaks (E2f1 only, n = 12,031), common peaks bound by E2f1 and Otx1 (E2f1&Otx1, n = 10,439), and Otx1-bound peaks (Otx1 only, n = 55,395). d Integrative heatmaps depicting the expression of E2f1 and Otx1 co-regulated genes (n = 220) in uninjected WT, e2f1 MO, otx1 MO, and double MO embryos at stages 7 and 8. Genes with E2f1 or Otx1 ChIP-peaks at promoter regions are marked in black. e Genome browser view of ChIP-seq, RNA-seq, and CANTAC-seq normalized read counts at sia1/2, mixer and mix1, and gsc loci.
To further characterize the E2f1 and Otx1 binding sites, we performed sequence motif analysis in these three categories separately. As expected, E2F and OTX motifs were exclusively enriched in E2f1-only or Otx1-only regions, respectively, while E2f1&Otx1 regions were enriched for both E2F and OTX motifs (Supplementary Fig. 7c). Moreover, FOXH, POU, SOX, and bHLH motifs were commonly enriched in all three subgroups. In addition, we observed that motifs such as KLF and ETS tend to appear in E2f1 binding regions, while motifs like GSC and T-box motifs were enriched in Otx1 binding regions. Since the chromatin accessibility was gradually established with the most dramatic increase at stage 8, we then asked whether E2f1 and Otx1 binding was associated with chromatin opening. As shown in Fig. 6c, E2f1-bound regions appeared to be more accessible than Otx1-bound regions during early development.
To further check how the co-binding of E2f1 and Otx1 regulates the gene expression during minor ZGA, we next defined their common target genes which are also developmentally relevant by the following criteria: (1) up-regulated in WT embryos from stage 7 to stage 8, (2) up-regulated upon e2f1 MO at stage 7 and (3) rescued back to the WT level by double MO at stage 7. A total of 220 genes fitting the three criteria were identified (Supplementary Data 7). Compared to genes that were up-regulated between stage 7 and 8, but not affected by e2f1 and/or otx1 perturbation, we observed a much higher enrichment of E2f1 and Otx1 binding at these 220 co-regulated target genes (Supplementary Fig. 7d) with 38.2% and 20.5% of gene promoters harboring binding sites for E2f1 and Otx1, respectively (Fig. 6d). Among these, 11.8% of gene promoters contained both E2f1 and Otx1 binding sites. Interestingly, the expression of these 220 genes was largely inhibited in otx1 MO-treated embryos at stage 8, suggesting that Otx1 is the transcription activator of these genes also at stage 8 of WT embryos.
Finally, to further explore the 220 genes co-regulated by E2f1 and Otx1, we constructed an interaction gene network based on protein–protein interactions using the STRING database43. As shown in Supplementary Fig. 7e, the resulting network comprises a higher number of transcriptional regulators and components of signaling cascades. This is also reflected by enriched GO terms related to transcription regulation, embryo development, and cell differentiation, as well as TGF-beta and WNT signaling pathways. As representative examples shown in Figs. 6e and S7f, both E2f1 and Otx1 bound at proximal and/or distal regulatory regions of germ layer differentiation-related genes such as sia1/2, mixer, mix1, and gsc, and their binding sites were highly overlapping. Consistent with their binding pattern, the up-regulation of these genes upon e2f1 MO at stage 7 was rescued by e2f1 & otx1 double MO at stage 7, and otx1 MO inhibits the expression of these genes at stage 8, indicating that E2f1 and Otx1 directly bind to and modulate the transcription of their targets in the opposite direction.
Collectively, our results propose a model wherein the coordinated actions of maternal factors E2f1 and Otx1 regulate the transcription of a subset of zygotic genes before the MBT in X. tropicalis early development. While E2f1 acts as a dominant repressor, effectively functioning like a “brake” on zygotic gene transcription, Otx1 works as an activator, functioning like an “accelerator”. Between the two factors, the negative effect of the brake E2f1 is dominant, and the accelerator Otx1 could manifest its effect only in the absence of the brake E2f1. As illustrated in Fig. 7, during the development of WT embryos, the repressive function of E2f1 holds sufficient till stage 7 and becomes too weak at stage 8 when Otx1 exerts its active effects (Fig. 7a). Knockdown of E2f1 leads to premature transcription of zygotic genes at stage 7 (Fig. 7b), whereas when the accelerator (Otx1) is removed, the transcription of these zygotic genes is not activated regardless of whether the brake (E2f1) is present or not, as observed in both the otx1 MO and double MO conditions (Fig. 7c,d). Finally, to further confirm the brake-accelerator model, we conducted E2f1 and Otx1 overexpression experiments. As shown in Supplementary Fig. 7g, while overexpression of the accelerator (Otx1) in the presence of the brake (E2f1) did not induce premature expression of these zygotic genes at stage 7, overexpression of the brake (E2f1) effectively inhibited the transcription of these genes at stage 8. These observations again demonstrated the dominant role of E2f1 in regulating zygotic gene activation.
The model illustrates how E2f1 and Otx1 co-regulate the transcription of a subset of zygotic genes before the MBT in X. tropicalis. Here, E2f1 functions as a repressor (brake), suppressing zygotic gene expression, while Otx1 acts as an activator (accelerator), promoting zygotic gene transcription. The repressive effect of E2f1 is dominant, and the effect of Otx1 on transcriptional activation becomes evident only in the absence of E2f1. In WT embryos, E2f1’s repression persists until stage 7 but diminishes at stage 8, allowing Otx1 to activate transcription (a). E2f1 knockdown causes premature zygotic gene transcription at stage 7 (b), while Otx1 knockdown prevents activation at stages 7 and 8 regardless of E2f1 status (c, d).
Discussion
Here, by developing CANTAC-seq, we systematically analyzed the dynamics of chromatin accessibility during X. tropicalis early development. We found that the chromatin accessibility was progressively established with the most dramatic increase before the MBT, and chromatin opening at cis-regulatory regions preceded zygotic gene transcription. Moreover, through analyzing these data and genetic perturbation experiments, we identified that E2f1 and Otx1 co-regulate the temporal expression pattern of their target genes during minor ZGA.
X. tropicalis has been widely used as a model organism in developmental biology, especially in studying the process of ZGA. However, high yolk content in X. tropicalis early embryos has interfered with the application of existing methods (e.g., ATAC-seq, DNase-seq) for analyzing chromatin accessibility23,24. Our CANTAC-seq method overcame this challenge and enabled robust chromatin profiling from low-input samples without the need for centrifugation. We observed a drastic opening of chromatin before the MBT, followed by the gradual increase of accessibility at later stages. Moreover, we found that the promoter accessibility of zygotic genes expressed after the MBT was already established before the MBT, which was also observed previously using a modified DNase-seq approach23. However, this finding contrasts with a previous study using ATAC-seq, which suggested that regulatory elements became accessible only after the MBT in X. tropicalis25. The inconsistency between the two studies is likely due to the superior capability of our CANTAC-seq in measuring chromatin accessibility at very early developmental stages. Furthermore, by switching ConA to antibodies recognizing specific surface markers, our CANTAC-seq approach can be easily adapted to study chromatin profiles of specific cell types without the need for prior enrichment.
Bioinformatics analysis identified maternal TFs with motifs enriched at cis-regulatory elements showing progressively accessible gains. Focusing on the previously unexplored E2F family, we demonstrated that e2f1 depletion accelerated zygotic gene transcription. Rescue experiments using cdkn1a mRNA co-injection and a DNA-binding-deficient e2f1 mutant (ΔDBD) revealed that E2f1 exerts the transcriptional repression independently of its impact on increased DNA content. Moreover, while the classical N/C ratio model provides insights for the relationship between large-scale zygotic gene transcription at the MBT5,40,44,45,46, it falls short in elucidating the initiation of minor ZGA, given that some zygotic genes transcribe before the MBT8,32,47,48. Our work reveals that E2f1-mediated repression directly controls the transcription of a subset of zygotic genes, while the transcriptional activator Otx1 works coordinately with E2f1 to regulate their timely activation. These findings establish a new paradigm where TF pairs like E2f1-Otx1 coordinate zygotic gene transcription, potentially representing a general mechanism for ZGA regulation.
It is noteworthy that E2f1 is typically recognized as a transcriptional activator that plays a key role in regulating the expression of genes crucial for the G1/S transition in mammals49,50. However, this function does not apply to the rapid, gap-phase-free pre-MBT cycles in X. tropicalis. In this study, we identified the regulatory role of E2f1 in transcription repression. A previous study using a tissue-specific E2f1 knockout mouse model has reported that E2f1-3 could form in complex with the repressor Rb to silence their targets in differentiating cells of the small intestine51. However, Rb expression in early developmental stages is extremely low52 and loss of Rb has no impact on cell cycling or differentiation of early X. laevis embryos53. Indeed, we did not identify any known transcriptional repressors in our IP-MS analysis. How E2f1 inhibits chromatin opening and zygotic gene transcription during the early development of the frog embryo awaits future elucidation.
Disruption of e2f1 and/or otx1 led to alterations in both gene expression and chromatin accessibility. As the expression of many TFs was dysregulated upon e2f1 and/or otx1 perturbation, the observed chromatin changes could be both direct and indirect effects of their perturbation. In line with a previous publication suggesting a pioneer activity of Otx1 in the establishment of functional enhancers and the specification of endoderm in the gastrula stage41, we also discovered that Otx1 played an important role in activating zygotic gene transcription and chromatin states at an earlier developmental time point in our study. More importantly, we demonstrated that Otx1 coordinated with E2f1 in regulating chromatin accessibility and the activation of a subset of genes required for zygotic gene transcription. Via ectopic expression of tagged protein, the extensive co-localization of E2f1 and Otx1 on the genome was observed using ChIP-seq, and the association between the two proteins was likely mediated by their independent DNA binding, as the effective Co-IP between the two was largely attenuated after DNA digestion. Importantly, it should be noted that the E2f1-Otx1 axis was not the only point via which E2f1-repressed genes get activated during ZGA, as not all E2f1 target genes got activated by Otx1, and half of the E2f1-bound regions were not co-localized with Otx1. Indeed, we observed several TF motifs in regions bound by E2f1 but not Otx1, indicating that E2f1 may also pair with other TFs to modulate chromatin opening and zygotic gene transcription. The same might also hold true for Otx1, i.e., it could exert its transcriptional regulation also independent of E2f1. On the other hand, as shown above for E2f1, in addition to direct transcriptional regulation, Otx1 may also play roles in other biological processes during early development.
We propose a model in which E2f1 and Otx1 function as a brake and accelerator, respectively, to determine the transcription of their target genes. Here, the negative effect of the brake is dominant over the positive effect of the accelerator, a phenomenon observed more than 60 years ago in the classical Lac operon system54. In our model, the repressive function of E2f1 holds sufficient till stage 7 and becomes too weak at stage 8 when Otx1 exerts its active effects. However, to validate this, one needs to measure the temporal protein abundance, ideally the DNA binding profile of both factors, using specific antibodies against the endogenous E2f1 and Otx1, which we unfortunately could not generate after several failed attempts. Furthermore, it would also be interesting to explore how the abundance of E2f1 and Otx1 themselves are regulated during ZGA. Deciphering both upstream and downstream factors of the E2f1/Otx1 regulatory network will largely facilitate our mechanistic understanding of ZGA during early development.
Finally, our findings could be evolutionarily conserved. In X. laevis, e2f1 and otx1 transcripts have been shown to be enriched and co-localized in pre-MBT embryos55, and both factors are translated during ZGA56,57, suggesting that E2f1 and Otx1 may regulate X. laevis ZGA in the same manner. More recently, OTX2, the paralogue of OTX1, was reported to be highly expressed upon oocyte meiotic resumption and began to decline after major ZGA in human, suggesting its functional role in human ZGA regulation42. In the same study, several E2F genes were also found to be transcribed and translated during human ZGA (Supplementary Table 1). It is therefore plausible that a similar regulatory mode might also exist in humans but may utilize different OTX and E2F family members.
Methods
X. tropicalis manipulations
This study complies with all relevant ethical regulations. All animal experiments were carried out following the animal protocols approved by the Laboratory Animal Welfare and Ethics Committee of the Southern University of Science and Technology. X. tropicalis frogs were purchased from NASCO (USA) and housed in a room with a constant temperature of 25 °C following a 12-h light-dark cycle. Sixteen pairs of one-year-old male and female adult frogs in total were used for in vitro fertilization. Embryos were obtained at different developmental stages and cultured in 0.1× MBS medium (1× MBS: 80 mM NaCl, 10 mM HEPES, 2.4 mM NaHCO3, 1 mM KCl, 0.82 mM MgSO4, 0.33 mM Ca(NO3)2, 0.41 mM CaCl2, pH 7.4) at 25 °C. The developmental stages were determined according to Nieuwkoop and Faber58. Briefly, embryos at developmental stages 3, 4, 5, 6, 7, 8, 9, 10, and 13 were collected at 1.5, 2, 2.5, 3, 3.5, 4, 5.5, 7, and 12 h post fertilization (hpf), respectively.
Plasmid construction and microinjection
MOs targeting e2f1 (5’ GTGTTCATCTTTGCTTCAAGAGTTC 3’), e2f3 (5’ CCCTTTCTCATCTTCCTGACTAG 3’), e2f5 (5’ TACGGGCCGAGCAGAGCTACAGTC 3’), and otx1 (5’ CATGCTCAAGGCTGGACAGAAACCC 3’) were obtained from Gene Tools. The open reading frames of X. tropicalis e2f1 (XM_012952820), cdkn1a (XM_002935778,) and otx1 (NM_203885) containing mutated MO target sites were cloned into the pCS2+ vector. The e2f1 ∆DBD mutant was sub-cloned via depleting the sequence coding a.a. 119–184. The capped mRNAs were in vitro transcribed using the mMESSAGE mMACHINE SP6 Transcription Kit (Invitrogen, AM1340). Afterwards, the mRNAs were purified with the RNeasy Mini Kit (QIAGEN, NC9677589) and quantified by NanoDrop (Thermo).
For microinjection, MOs were injected into 1-cell stage embryos from the animal pole with a dose of 10 ng per embryo using a pneumatic Pico Pump PV830 (WPI). The rescue experiments were carried out to confirm the specificity of MOs by co-injection of 300 pg mRNA and 10 ng corresponding MOs. Forty pg Cdkn1a mRNA was injected into 1-cell stage embryos to inhibit cell division. Injected embryos were then collected for further experiments at the desired time points. Images of the whole embryos were acquired using a stereo microscope (SMZ18, Nikon).
Cell culture
K562 cells (ATCC, CCL-243) were cultured in RPMI 1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin. MES cells were obtained from Dr. Qi Zhou and cultured in serum-free media. Briefly, 500 mL of N2B27 media was generated by including: 240 mL DMEM/F12 (Invitrogen, 11330-032), 240 mL Neurobasal (Invitrogen, 21103049), 5 mL N2 medium (Invitrogen, 17502048), 10 mL B27 medium (Invitrogen, 17504044), 1% GlutaMAX (Life Technologies, 35050-061), 1% nonessential amino acids (Life Technologies, 1140-035), 0.1 mM β-mercaptoethanol (Life Technologies, 21985-023), 1% penicillin–streptomycin (Life Technologies, 15140-122), 1 µM PD0325901 (Stemgent, 04-0006), 3 µM CHIR99021 (Stemgent, 04-0004), 5 µL mLIF (Millipore, LIF2050).
CANTAC-seq
In order to make tagmentation work on early embryos of X. tropicalis, we developed the CANTAC-seq method in which cells/nuclei were first captured and purified using ConA beads, followed by Tn5 transposition and library preparation. Briefly, X. tropicalis embryos were collected in a 1.5 mL tube at blastula stages (stages 7, 8, and 9), at the onset of gastrulation (stage 10), and at the onset of neurulation (stage 13). Embryos were pipetted up and down in 200 μL binding buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM Spermidine) with a P20 pipette tip until a homogenized lysate was formed. ConA magnetic beads (Bangs Laboratories, BP531) were pre-washed once with washing buffer (20 mM HEPES pH 7.5, 10 mM KCl, 1 mM CaCl2, 1 mM MnCl2), and 20 µL of ConA beads were added per sample and incubated at RT for 20 min. The unbound supernatant containing yolk, pigment, and lipids was removed, and the cells/nuclei-beads complex was washed once with 100 µL resuspension buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2). A small aliquot was taken for DAPI staining and nuclei counting under the microscope. Afterwards, approximately 50,000 bead-bound cells/nuclei were incubated in 100 µL lysis buffer (10 mM Tris-HCl pH7.4, 10 mM NaCl, 3 mM MgCl2, 0.1%NP40, 0.1% Tween-20, 0.01% Digitonin) for 10 min on ice, followed by washing with 600 µL lysis buffer without NP40 and Digitonin. Afterwards, an on-bead tagmentation was carried out by resuspending bead-bound cells/nucleus in 100 µL transposition mix (Vazyme, TD501) and incubated at 37 °C for 30 min with gentle shaking at 800 rpm. The tagmentation was stopped by adding 3.3 μL 0.5 M EDTA, 1 μL 10% SDS, and 1.5 μL 20 mg/mL Proteinase K to each sample and incubated at 55 °C for 1 h.
Tagmented DNA was cleaned up using the DNA Clean & Concentrator kit (Zymo, D4013), and DNA was eluted in nuclease-free water. Libraries were amplified under the following PCR conditions: 72 °C for 3 min; 98 °C for 30 s; and thermocycling at 98 °C for 15 s, 60 °C for 30 s and 72 °C for 40 s; followedby 72 °C for 5 min and hold at 4 °C. Libraries were cleaned up with VAHTS DNA Clean Beads (Vazyme, N411), followed by two times washing with 80% ethanol and eluted in nuclease-free water. The libraries were sequenced in a 2x150nt manner on the NovaSeq 6000 platform (Illumina).
ATAC-seq
For normal ATAC-seq library construction, cells were prepared as previously described with minor modifications. Briefly, 50,000 fresh cells were lysed in lysis buffer for 10 min on ice to prepare the nuclei. Immediately after lysis, nuclei were spun at 500 g for 5 min to remove the supernatant. Nuclei were then incubated with the Tn5 transposase (Vazyme, TD501) in tagmentation buffer at 37 °C for 30 min. After tagmentation, PCR was performed to amplify the library for 12 cycles under the following PCR conditions: 72 °C for 3 min; 98 °C for 30 s; and thermocycling at 98 °C for 15 s, 60 °C for 30 s, and 72 °C for 40 s; followed by 72 °C for 5 min. After the PCR reaction, libraries were purified with the DNA purification beads (Vazyme, N411). The libraries were sequenced in a 2× 150nt manner on the NovaSeq 6000 platform (Illumina).
RNA extraction, RT-qPCR, and mRNA sequencing library preparation
Total RNA was extracted from X. tropicalis embryos from different developmental stages using TRIzol™ Reagent (Invitrogen, 15596026). cDNA synthesis was conducted utilizing the Hifair III 1st Strand cDNA Synthesis Kit (YEASEN, 11141ES60). The resultant cDNAs were subsequently analyzed via qPCR using the Hieff qPCR SYBR Green Master Mix (YEASEN, 11201ES60) on the CFX Connect Thermal Cycler (BIO-RAD). The specific primers employed to assess mRNA expression levels were listed in Supplementary Table 2. For mRNA sequencing, libraries were prepared using the standard protocol provided by VAHTS mRNA-seq V2 Library Prep Kit for Illumina (Vazyme, NR601) with 1 μg total RNA. The libraries were sequenced in a 2× 150nt manner on the NovaSeq 6000 platform (Illumina).
Metabolic labeling and detection of newly synthesized transcripts
At the one-cell stage, embryos were microinjected with 2 nL of 50 mM 4′-thiouridine (4sU; MCE, HY-W113081). For RNA isolation, embryos were homogenized in TRIzol (Invitrogen, 15596026) at designated developmental stages. Newly synthesized RNA was isolated using thiol-specific biotinylation as previously described59. Briefly, 10 µg of total RNA was biotinylated with EZ-Link HPDP-Biotin (Thermo, 21341), followed by affinity purification using streptavidin-coated magnetic beads (Vazyme, N512). Purified nascent RNA was then eluted and analyzed by quantitative RT-PCR.
ChIP followed by qPCR and sequencing
The ChIP assay was performed according to the standard protocol provided by SimpleChIP Plus Sonication Chromatin IP Kit (CST, 56383) with minor modifications. Briefly, X. tropicalis embryos were fixed with 1.5% formaldehyde for 30 min. Sonication was then carried out at the Bioruptor pico (Diagenode) by applying 20 cycles of 30 s ON and 30 s OFF to obtain chromatin fragments of approximately 100–500 bp. The HA antibody (Sigma, H6908) and Myc-Tag antibody (CST, 2276) were used to pull down HA-tagged E2f1 and Myc-tagged Otx1, respectively. RNA Polymerase II was pulled down using the anti-RPB1 antibody (BioLegend, 664906). ChIP DNA was cleaned up using the ChIP DNA Clean& Concentrator kit (Zymo, D5205). For ChIP-qPCR, the enrichment was normalized to the input and represented as a relative enrichment of the material precipitated by the specified antibody on the target regions. The primers utilized in the ChIP-qPCR analysis were detailed in Supplementary Table 2. For ChIP-seq, libraries were prepared using the standard protocol provided by VAHTS Universal DNA Library Prep Kit for Illumina V3 (Vazyme, ND607). The libraries were sequenced in a 2× 150nt manner on the NovaSeq 6000 platform (Illumina).
DNA quantification
To compare the total amount of DNA in WT and MO embryos at different stages during embryogenesis, 15 embryos per stage per condition were collected. Genomic DNA was isolated together with 100,000 K562 cells as a spike-in control according to the manual of DNeasy Blood & Tissue Kit (Qiagen, 69504). For DNA quantification, real-time PCR was carried out using primer pairs that specifically target the X. tropicalis actb genomic region and the human ACTB genomic region, respectively. The relative DNA quantity was calculated by using the spiked-in human DNA as an internal control. The primers used for DNA quantification were listed in Supplementary Table 2.
Co-immunoprecipitation (Co-IP) and WB
X. tropicalis embryos at the indicated stages were collected and homogenized in lysis buffer (20 mM Tris-HCl, pH 7.4, 150 nM NaCl, 1 mM EDTA, 1% Triton, 1 mM DTT, 0.1 mM PMSF, protease inhibitor cocktail (Roche, 04693132001), 1 mM NaVO4). Protein lysates were then mixed with 2 volumes of 1,1,2-Trichlorotrifluoroethane. After centrifugation, the protein lysates in the upper layer were collected to measure concentrations using the BCA assay (Beyotime, P0011). For immunoprecipitation, the protein lysates were incubated with protein A/G magnetic beads (MCE, HY-K0202) coupled with the indicated antibodies for 4 h at 4 °C. The beads were then washed three times with TBST and eluted in loading buffer by heating at 98 °C for 5 min. The eluted protein complex was separated in 10% SDS gels and blotted on PVDF membranes by semi-dry blotting. The Spectra Multicolor Broad Range Protein Ladder (Thermo, 26634) was loaded for size estimation. The membranes were blocked in 5 % skim milk powder/TBST for 1 h at room temperature and then incubated with the primary antibody at 4 °C overnight. After washing in TBST three times, the membrane was incubated with the secondary antibody for 1 h at room temperature. After washing three times, the membranes were developed with Pierce ECL (Thermo, 32106) according to the manufacturer’s instructions. Protein bands were recorded with the ChemiDoc MP Imaging System.
Immunoprecipitation followed by mass spectrometry analysis (IP-MS)
X. tropicalis embryos injected with e2f1-HA mRNA were homogenized in lysis buffer containing protease and phosphatase inhibitors. The E2f1-associated proteins were immunoprecipitated with Protein A/G magnetic beads (MCE, HY-K0202) coupled with HA antibody (Sigma, H6908) for 4 h. Embryos injected with HA mRNA alone served as the control. Following immunoprecipitation, beads were washed with TBST, and on-bead digestion was performed overnight with sequencing-grade trypsin (Promega, V5111). The resulting peptides were analyzed using an LTQ-Orbitrap Elite mass spectrometer (Thermo Scientific). Peptides were separated on an in-house packed column (100 μm i.d. ×20 cm; ReproSil-Pur C18-AQ, 1.9 μm, 120 Å; Dr. Maisch GmbH) with a binary buffer system (buffer A: 0.1% formic acid in water; buffer B: 0.1% formic acid in acetonitrile) at a flow rate of 250 nl min−1. The effective gradient was 7–22% buffer B over 50 min, followed by an increase to 35% B over 10 min. Full MS scans were acquired in the Orbitrap (resolution: 120,000; m/z range: 350–1550). Data-dependent MS/MS scans were performed in the ion trap using an isolation window of 1.6 m/z. The method used a top-speed mode with a 3 s cycle time, a normalized collision energy of 30 for HCD fragmentation, and a dynamic exclusion duration of 30 s. The experiment was performed in two biological replicates. Raw data were processed with MaxQuant (v.1.5.5.1) for label-free quantification (LFQ) with the “match between runs” feature enabled. Database searching was performed against the UniProt X. tropicalis database, and proteins were quantified using MaxQuant’s LFQ algorithm.
CANTAC-seq and ATAC-seq analysis
For ATAC-seq analysis, fastp (v0.23.2)60 was used to trim the reads with parameters -a CTGTCTCTTATA --detect_adapter_for_pe --length_required 20 -q 30. Alignment of these reads to the X. tropicalis v10.0 reference genome was performed with Bowtie2 (version 2.4.5)61 with parameters -X 2000. Sambamba (v0.7.0)62 was used to remove the duplicated reads. Peak calling was performed by MACS2 (v2.2.7.1)63 with the parameters -g 1.435e9 --keep-dup all -q 0.05 --slocal 10000 --nomodel --nolambda -B --SPMR. The reads were counted using featrueCounts (version 2.0.1)64 and the counts were converted to counts per million (CPM) for plotting. Normalized read coverage tracks were generated with bamCoverage from the deeptools package(version 3.5.1)65 and are visualized using the Integrative Genomics Viewer (IGV, version 2.16.0)66. For motif enrichment analysis, the default motif libraries, including JSAPAR, DMMPMM, AthaMap and etc., were used by the HOMER67 tool findMotifs.pl. Peaks located within the region of −1kb ~ +0.1 kb from the transcription start site (TSS) were defined as proximal peaks, while other CANTAC peaks are classified as distal ones.
RNA-seq analysis
For RNA-sequencing data, quality control and adapter trimming were performed using fastp (v0.23.2)60 with parameters -a AGATCGGAAGAGC --detect_adapter_for_pe -w 12 --length_required 30 -q 20. Reads were mapped to the X. tropicalis v10.0 reference genome with STAR (version 2.7.0e)68. The counts for known genes were obtained using featureCounts (version 2.0.1)64 with the parameters -s 2 -p -BC. Normalized gene expression level was further calculated as transcripts per million (TPM). Differentially expressed genes were identified with the DESeq2 package69, with threshold FDR < 0.05 and Log2|FC| > 1. The normalized read coverage tracks in bigwig format were generated with bamCoverage in the deepTools package (version 3.5.1)65, with the parameters --normalizeUsing RPKM -bs 5. The tracks were further visualized using Integrative Genomics Viewer (IGV, version 2.16.0)66.
ChIP-seq analysis
To analyze ChIP-seq data, a quality check was performed using fastp with parameters --length_required 20 -q 30. The sequence reads were mapped to the X. tropicalis v10.0 reference genome by Bowtie2 (version 2.4.5)61 with parameters --very-sensitive. Duplicates were removed with Sambamba (version 0.7.0)62. Peak calling was performed by MACS2 (version 2.2.7.1)63 with parameters -g 1.435e9 --keep-dup all -q 0.01. The normalized read coverage tracks (bigwig files) were obtained by bamCoverage with parameters --normalizeUsing CPM -bs 5.
Peaks were overlapped with the Bedtools (v2.30.0)70 intersect function. Peaks within the defined promoter region were acquired using the window function with parameters -l 2000 -r 500. The calculation of peak distribution was performed by the HOMER tool67 annotatePeaks.pl. Heatmaps were plotted with normalized read coverage tracks using deepTools (version 3.5.1)65 function computeMatrix and plotHeatmap. The normalized ChIP-seq sequence tracks were visualized using the Integrative Genomics Viewer (IGV, version 2.16.0)66.
Statistics and reproducibility
All experiments were independently repeated at least three times with consistent results. All sequencing and mass spectrometry proteomics data include two replicates. The exact number of biological replicates for each experiment is detailed in the corresponding figure legend. Bar graphs are presented as mean ± standard deviation (SD). Box plots show the median, with the box representing the 25th to 75th percentiles and the whiskers extending from the 5th to the 95th percentiles. Statistical significance was determined using the appropriate tests as indicated in the figure legends, including two-sided Student’s t-test, one-sided hypergeometric test, and two-sided Fisher’s exact test. A P value of less than 0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All the sequencing data generated from this study have been submitted to the NCBI under the accession number GSE232071. The proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD051084. Source data are provided with this paper.
References
Tadros, W. & Lipshitz, H. D. The maternal-to-zygotic transition: a play in two acts. Development 136, 3033–3042 (2009).
Lee, M. T., Bonneau, A. R. & Giraldez, A. J. Zygotic genome activation during the maternal-to-zygotic transition. Annu Rev. Cell Dev. Biol. 30, 581–613 (2014).
Schulz, K. N. & Harrison, M. M. Mechanisms regulating zygotic genome activation. Nat. Rev. Genet. 20, 221–234 (2019).
Jukam, D., Shariati, S. A. M. & Skotheim, J. M. Zygotic genome activation in vertebrates. Developmental Cell 42, 316–332 (2017).
Newport, J. & Kirschner, M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell 30, 687–696 (1982).
Newport, J. & Kirschner, M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell 30, 675–686 (1982).
Tan, M. H. et al. RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. Genome Res. 23, 201–216 (2013).
Owens, N. D. L. et al. Measuring absolute RNA copy numbers at high temporal resolution reveals transcriptome kinetics in development. Cell Rep. 14, 632–647 (2016).
Klemm, S. L., Shipony, Z. & Greenleaf, W. J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 20, 207–220 (2019).
Zaret, K. S. & Carroll, J. S. Pioneer transcription factors: establishing competence for gene expression. Gene Dev. 25, 2227–2241 (2011).
Lee, M. T. et al. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503, 360–364 (2013).
Lu, F. et al. Establishing chromatin regulatory landscape during mouse preimplantation development. Cell 165, 1375–1388 (2016).
Wu, J. et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652–657 (2016).
Gao, L. et al. Chromatin accessibility landscape in human early embryos and its association with evolution. Cell 173, 248–259.e215 (2018).
Liu, G., Wang, W., Hu, S., Wang, X. & Zhang, Y. Inherited DNA methylation primes the establishment of accessible chromatin during genome activation. Genome Res. 28, 998–1007 (2018).
Wu, J. et al. Chromatin analysis in human early development reveals epigenetic transition during ZGA. Nature 557, 256–260 (2018).
Palfy, M., Schulze, G., Valen, E. & Vastenhouw, N. L. Chromatin accessibility established by Pou5f3, Sox19b and Nanog primes genes for activity during zebrafish genome activation. PLoS Genet. 16, e1008546 (2020).
Amodeo, A. A., Jukam, D., Straight, A. F. & Skotheim, J. M. Histone titration against the genome sets the DNA-to-cytoplasm threshold for the Xenopus midblastula transition. Proc. Natl. Acad. Sci. USA 112, E1086–E1095 (2015).
Joseph, S. R. et al. Competition between histone and transcription factor binding regulates the onset of transcription in zebrafish embryos. Elife 6, e2332610 (2017).
Stancheva, I. & Meehan, R. R. Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes Dev. 14, 313–327 (2000).
Ruzov, A. et al. Kaiso is a genome-wide repressor of transcription that is essential for amphibian development. Development 131, 6185–6194 (2004).
Hellsten, U. et al. The genome of the western clawed frog Xenopus tropicalis. Science 328, 633–636 (2010).
Gentsch, G. E., Spruce, T., Owens, N. D. L. & Smith, J. C. Maternal pluripotency factors initiate extensive chromatin remodelling to predefine first response to inductive signals. Nat. Commun. 10, 4269 (2019).
Esmaeili, M. et al. Chromatin accessibility and histone acetylation in the regulation of competence in early development. Dev. Biol. 462, 20–35 (2020).
Bright, A. R. et al. Combinatorial transcription factor activities on open chromatin induce embryonic heterogeneity in vertebrates. EMBO J. 40, e104913 (2021).
Skene, P. J., Henikoff, J. G. & Henikoff, S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 13, 1006–1019 (2018).
Kaya-Okur, H. S. et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930 (2019).
Hontelez, S. et al. Embryonic transcription is controlled by maternally defined chromatin state. Nat. Commun. 6, 10148 (2015).
Zhou, J. J. et al. Histone deacetylase 1 maintains lineage integrity through histone acetylome refinement during early embryogenesis. Elife. https://doi.org/10.7554/eLife.79380 (2023).
Lund, E., Liu, M. Z., Hartley, R. S., Sheets, M. D. & Dahlberg, J. E. Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. RNA 15, 2351–2363 (2009).
Mukherjee, S. et al. Sox17 and beta-catenin co-occupy Wnt-responsive enhancers to govern the endoderm gene regulatory network. Elife 9, ARTN e58029 (2020).
Chen, H. & Good, M. C. Nascent transcriptome reveals orchestration of zygotic genome activation in early embryogenesis. Curr. Biol. 32, 4314–4324 e4317 (2022).
Zhang, C., Basta, T., Jensen, E. D. & Klymkowsky, M. W. The beta-catenin/VegT-regulated early zygotic gene Xnr5 is a direct target of SOX3 regulation. Development 130, 5609–5624 (2003).
Tao, J. et al. BMP4-dependent expression of Xenopus grainyhead-like 1 is essential for epidermal differentiation. Development 132, 1021–1034 (2005).
Zhang, C., Basta, T., Fawcett, S. R. & Klymkowsky, M. W. SOX7 is an immediate-early target of VegT and regulates nodal-related gene expression in Xenopus. Dev. Biol. 278, 526–541 (2005).
Cao, Y. et al. POU-V factors antagonize maternal VegT activity and beta-Catenin signaling in Xenopus embryos. EMBO J. 26, 2942–2954 (2007).
Chiu, W. T. et al. Genome-wide view of TGFbeta/Foxh1 regulation of the early mesendoderm program. Development 141, 4537–4547 (2014).
Niu, L. et al. Three-dimensional folding dynamics of the Xenopus tropicalis genome. Nat. Genet 53, 1075–1087 (2021).
Kent, L. N. & Leone, G. The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer 19, 326–338 (2019).
Jukam, D., Kapoor, R. R., Straight, A. F. & Skotheim, J. M. The DNA-to-cytoplasm ratio broadly activates zygotic gene expression in Xenopus. Curr. Biol. 31, 4269–4281.e4268 (2021).
Paraiso, K. D. et al. Endodermal maternal transcription factors establish super-enhancers during zygotic genome activation. Cell Rep. 27, 2962–2977.e2965 (2019).
Zou, Z. et al. Translatome and transcriptome co-profiling reveals a role of TPRXs in human zygotic genome activation. Science 378, abo7923 (2022).
Szklarczyk, D. et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. https://doi.org/10.1093/nar/gkac1000 (2022).
Edgar, B. A., Kiehle, C. P. & Schubiger, G. Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development. Cell 44, 365–372 (1986).
Kane, D. A. & Kimmel, C. B. The zebrafish midblastula transition. Development 119, 447–456 (1993).
Syed, S., Wilky, H., Raimundo, J., Lim, B. & Amodeo, A. A. The nuclear to cytoplasmic ratio directly regulates zygotic transcription in Drosophila through multiple modalities. Proc. Natl. Acad. Sci. USA. https://doi.org/10.1073/pnas.2010210118 (2021).
Strong, I. J. T., Lei, X., Chen, F., Yuan, K. & O’Farrell, P. H. Interphase-arrested Drosophila embryos activate zygotic gene expression and initiate mid-blastula transition events at a low nuclear-cytoplasmic ratio. PLoS Biol. 18, e3000891 (2020).
Chan, S. H. et al. Brd4 and P300 confer transcriptional competency during zygotic genome activation. Dev. Cell 49, 867–881.e868 (2019).
Attwooll, C., Lazzerini Denchi, E. & Helin, K. The E2F family: specific functions and overlapping interests. EMBO J. 23, 4709–4716 (2004).
Chen, H. Z., Tsai, S. Y. & Leone, G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat. Rev. Cancer 9, 785–797 (2009).
Chong, J. L. et al. E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature 462, 930–934 (2009).
Destree, O. H. et al. Structure and expression of the Xenopus retinoblastoma gene. Dev. Biol. 153, 141–149 (1992).
Cosgrove, R. A. & Philpott, A. Cell cycling and differentiation do not require the retinoblastoma protein during early Xenopus development. Developmental Biol. 303, 311–324 (2007).
Jacob, F. & Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3, 318–356 (1961).
Owens, D. A. et al. High-throughput analysis reveals novel maternal germline RNAs crucial for primordial germ cell preservation and proper migration. Development 144, 292–304 (2017).
Subtelny, A. O., Eichhorn, S. W., Chen, G. R., Sive, H. & Bartel, D. P. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508, 66–71 (2014).
Peshkin, L. et al. On the relationship of protein and mRNA dynamics in vertebrate embryonic development. Dev. Cell 35, 383–394 (2015).
Nieuwkoop & Faber. Normal table of Xenopus laevis (Daudin) (Garland Publishing Inc., New York, 1994).
Sun, W. & Chen, W. Metabolic labeling of newly synthesized RNA with 4sU to in parallel assess RNA transcription and decay. Methods Mol. Biol. 1720, 25–34 (2018).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Tarasov, A., Vilella, A. J., Cuppen, E., Nijman, I. J. & Prins, P. Sambamba: fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034 (2015).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Acknowledgements
This work was supported by the National Nature Science Foundation of China (32570647 to H.C.), the National Key R&D Program of China (2022YFC3400400 to W.C.), the Shenzhen Science and Technology Program (JCYJ20240813094206009 to H.C., KQTD20180411143432337 to W.C), the Shenzhen Key Laboratory of Gene Regulation and Systems Biology (ZDSYS20200811144002008 to W.C.), and the Guangdong Basic and Applied Basic Research Foundation (2025A1515011842 to Z. Shi). We thank Dr. Zilong Wen and Dr. Xi Chen from SUSTech for the helpful discussion on the project. We also thank the Center for Computational Science and Engineering of SUSTech for the support on computational resources.
Author information
Authors and Affiliations
Contributions
W.C. and H.C. conceived the study. W.C., H.C. and W.L. designed the experiments. H.C. and W.L. developed CANTAC-seq and performed the experiments. Z. Shi and Y. C. prepared frog embryos and performed microinjections and imaging. L.Z., G.L. and C.T. performed bioinformatics analysis. R.C., D.G., X.S. and Z. Sun assisted in performing experiments. W.C., H.C., Y.C., Y.H., H.H., L.F. and Q.Z. reviewed and discussed the results. W.C. and H.C. wrote the manuscript with the input from W.L. and L.Z.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Daria Onichtchouk and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cui, H., Liang, W., Shi, Z. et al. CANTAC-seq analysis reveals E2f1 and Otx1 coordinate zygotic genome activation in Xenopus tropicalis. Nat Commun 16, 10101 (2025). https://doi.org/10.1038/s41467-025-65146-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-65146-8