Abstract
The rapid spread of SARS-CoV-2 worldwide has given rise to numerous variants. While the impact of viral mutations on antibody escape has been extensively studied, an unresolved issue concerns how emerging mutations shape HLA-restricted T-cell immune responses. Here, we analyse SARS-CoV-2 genomic variants, showing that 27% of the mutations are C > U transitions, a phenomenon common in human RNA viruses and primarily attributed to APOBEC3 enzyme-driven mutagenesis. We find that this mutation bias generally enhances viral peptide binding to human leukocyte antigen class I (HLA-I) molecules, producing immunogenic epitopes that trigger cytotoxic adaptive immune responses in most individuals across diverse populations. We also identify several HLA-I variants that are especially well-suited for presenting viral epitopes generated by these mutations. Intriguingly, individuals carrying these specific alleles are predominantly located in South and East Asia. Finally, we show that carrying HLA-I molecules that are less likely to bind C > U-induced viral peptides increases risk for severe COVID-19 disease. Our work suggests a link between C > U hypermutation and HLA-I-based presentation of viral epitopes, which may reflect the evolutionary outcome of ancient RNA virus pandemics. More broadly, our findings imply that SARS-CoV-2 diversification leads to ongoing gains of T-cell epitopes despite natural selection favouring immune escape.
Similar content being viewed by others
Introduction
Since the onset of the COVID-19 pandemic, the SARS-CoV-2 virus has accumulated mutations, which shape its ability to spread, enter cells, replicate and evade the immune system1,2,3. It is well-established that some of these viral mutations hinder the binding of antibodies to viral proteins, and thereby generate immune escape variants1,4. Emerging mutations also affect the CD8 + T cell-mediated immune responses, but their overall impact on HLA-I-associated peptide presentation have been a subject of debate. Agerer et al. found that certain mutations prevent the binding of viral peptides to HLA-A*02:01, a prevalent allele in the Caucasian population5. In another study, Stanevich et al. reported the case of a non-Hodgkin’s lymphoma patient who received rituximab, leading to a lack of neutralizing antibodies, but still had functional CD8 + T cell-mediated immunity6. During the more than three hundred day-long course of her infection, 40 different nucleotide mutations were detected in her viral samples, many of them leading to decreased HLA-I binding of viral peptides. At the same time, Hamelin et al. showed that mutations modify HLA-binding in an HLA-supertype-dependent manner7. The authors found that HLA-B*07 alleles generally bind mutated SARS-CoV-2 peptides less effectively. While the study focused on this potential immune escape in HLA-B*07-positive individuals, the opposite trend was reported for several other supertypes. Moreover, Pretti et al. showed, that some HLA-B variants bind mutant viral epitopes more effectively. For instance, individual mutations like Spike N501Y and Nucleocapsid D138Y were predicted to exhibit a stronger affinity for HLA-I than the reference sequence across diverse human populations8. The discrepancies in these findings may be attributed to the limited range of viral mutations and HLA-I variants examined.
In this study, our objective was to systematically investigate the global impact of viral mutations on HLA-I-associated T-cell immunity. To achieve this goal, we examined the dominating mutational patterns in SARS-CoV-2 evolution. In line with previous research9,10,11, we found that C > U transitions dominate the mutational landscape. We demonstrate that these mutations result in amino acid substitutions in SARS-CoV-2 proteins that generally exhibit stronger binding to common HLA-I alleles than the original sequences. As a result, the mutation-driven diversification of SARS-CoV-2 leads to ongoing gains of T-cell epitopes in most individuals across the globe. These findings bear clinical implications, as patients carrying HLA-I alleles that are less likely to bind C > U-related viral peptides exhibit a higher risk of severe COVID-19 upon infection. The results indicate a functional connection between mutagenic processes in SARS-CoV-2 and HLA-I-mediated viral epitope presentation, suggesting their synergistic effect on the adaptive immune response to coronavirus infection over evolutionary time scales. This connection may reflect selective pressure favoring HLA-I variants that efficiently present peptides generated by C > U transitions.
Results
C > U mutations enhance HLA-binding
To gain insight into the mutations acquired by SARS-CoV-2 during the pandemic, we examined the relative frequency of nucleotide substitutions using data acquired from the Nextstrain database. Importantly, this database employs a downsampling approach to mitigate the overrepresentation of samples from certain geographical regions, leading to a dataset with a seemingly modest size of 3389 strains, but with a balanced spatiotemporal distribution12,13. In addition, the dataset contains the phylogenetic relationship of these SARS-CoV-2 isolates, allowing us to track the progression of mutations along evolutionary trajectories. In accordance with previous results9,10,14,15,16, we found the dominance of C > U nucleotide substitutions (n = 2601, 27.4%) in the set of 9493 unique mutations compared to the reference Wuhan Hu-1 strain (NC_045512, Fig. 1A). We restricted our subsequent analyses to the five types of nucleotide substitutions that reached at least 10% among unique mutations (C > U, n = 2601, 27.4%; uracil to cytosine [U > C]: n = 1551, 16.3%; adenine to guanine [A > G]: n = 1359, 14.3%, guanine to uracil [G > U]: n = 1133, 11.9%; guanine to adenine [G > A]: n = 1112, 11.7%).
A Frequency of nucleotide substitutions in unique mutations. The dashed red line represents 10%, above which substitution types were further examined. B The average number of different nucleotide substitutions in isolated strains relative to the Wuhan Hu-1 reference strain is shown on a quarterly basis (number of samples are shown in Source Data Table 3). The standard deviation values are also indicated. The frequency of C > U mutations has been significantly higher compared to other nucleotide changes from 2020 till September 2024. Asterisks indicate a significantly higher prevalence of C > U mutations vs. others according to Kruskal-Wallis tests (P < 0.05) and Dunn’s post-hoc tests (specific P values are shown in Source Data Table 3). C Accumulation of specific nucleotide substitutions in the phylogenetic trajectories of SARS-CoV-2. The vertical axis represents the number of substitutions in each strain isolated at a given time point (the latter is shown on the horizontal axis). Nodes and leaves represent common ancestors and isolates, respectively, while edges represent phylogenetic relationships. Note that the transparency of edges has been increased for visualization purposes. D The effect of different nucleotide substitutions on HLA binding. The number of mutations associated with an increased or decreased number of bound peptides is indicated. Each point pair represents values belonging to a given HLA allele (n = 43). E Prevalence of different amino acid substitutions in the unique C > U mutation pool. The red dashed line represents 5% relative frequency, above which the amino acid substitution-specific HLA binding results are shown on panel (H). F Median specificities of frequent HLA-I alleles towards individual amino acids. For each amino acid–HLA allele pair, we calculated the sum of amino acid bit scores derived from sequence binding motifs, using this sum as a proxy for the allele’s specificity toward that amino acid. The median specificity across 41 frequent HLA-I alleles was then calculated for each amino acid and visualized. Amino acids on the vertical axis are ordered according to their corresponding Kyte-Doolittle hydrophobicity values. Spearman’s ρ and the two-sided correlation test P-value is shown. G The change in Kyte-Doolittle hydrophobicity is indicated for different amino acid substitutions associated with C > U mutations. Red and blue colors indicate increased and decreased hydrophobicity, respectively. H The number of mutations associated with the gain or loss of bound peptides is indicated for each amino acid substitution. Each point pair represents values belonging to a given HLA allele (n = 43). On panels (D and H), FDR-corrected P values of two-sided paired Wilcoxon’s signed-rank tests are shown. In these panels, blue color indicates that the allele is associated with binding gain for more mutations than binding loss. The opposite trend is indicated in red color. In boxplots (panels D and H), horizontal lines indicate median, boxes indicate interquartile range, and vertical lines indicate first quartile – 1.5 × IQR and third quartile + 1.5 × IQR. Source data are provided in the Source Data file.
Next, we determined the average number of each nucleotide substitution type in the isolated samples in a monthly breakdown (Fig. 1B). As expected, a significantly higher number of C > U than other mutations accumulated in SARS-CoV-2 genomes (reaching an average of 44.86 in viral strains by September 2024; standard deviation: 2.1). Importantly, this accumulation of C > U mutations was also evident when analyzing mutation events along evolutionary trajectories (Fig. 1C).
Next, we selected missense mutations from our dataset and generated all overlapping 8–11 amino acid long peptide sequences containing the mutated amino acid. Using the NetMHCpan-4.0 algorithm17, we predicted the binding of each mutated and original peptide to a set of 43 common HLA-I alleles that cover 95% of the human population18,19. For each mutation and HLA allele, we determined whether the mutation increased or decreased the total number of bound peptides to the given allele. Then, for each HLA allele, we counted the number of mutations resulting in a higher or lower number of bound peptides. We found that C > U mutations are likely to increase peptide binding for 37 of the 43 common HLA-I alleles (Fig. 1D, P-value of paired Wilcoxon’s signed-rank test: 3.93 × 10−6). Importantly, C > U mutations were associated with the largest increase of bound peptides, followed by G > U mutations with a significant, but much lower effect.
We also examined mutational patterns using a separate, extensive dataset derived from the UShER phylogenetic tree, incorporating approximately 7 million publicly available SARS-CoV-2 genomic samples20. Consistent with the findings from the Nextstrain dataset, C > U mutations were observed on the highest number of independent branches (Supplementary Fig. 1A) and were the most predominant sources of novel HLA-I-bound peptides (Supplementary Fig. 1B, see Methods for details).
Next, we focused on immunologically relevant regions of SARS-CoV-2 acquired from the Immune Epitope Database (Supplementary Fig. 2A, see Methods for details). We found the same positive effect of C > U mutations on HLA-binding. Moreover, similar trends were found for rubella, another positive single-stranded RNA virus (Supplementary Fig. 2B), which suggests that the phenomenon is not specific to SARS-CoV-2. Notably, the positive effect of C > U mutations on HLA-binding remained consistent regardless of the specific nucleotide context (Supplementary Fig. 3).
Specific amino acid substitutions are responsible for increased HLA-binding
HLA molecules bind specific amino acids at anchor positions of the mutated peptides. To identify amino acid substitutions that drive enhanced HLA binding, we summarized the number of different amino acid substitutions resulting from C > U nucleotide changes in our dataset (Fig. 1E). Threonine > isoleucine (T > I, n = 332 substitutions, 25.2%) and alanine > valine (A > V, n = 265 substitutions, 20.0%) were the most frequent substitutions, followed by proline > serine (P > S, n = 152 substitutions, 11.6%) and leucine > phenylalanine (L > F, n = 136 substitutions, 10.3%). As in the previous analysis, for each amino acid substitution, we determined the number of mutations resulting in the gain or loss of HLA-bound peptides. The trends were dominantly positive for the mentioned substitutions except for a few HLA allele–substitution pairs, like HLA-B*07:02 and P > S or P > L; and HLA-A*02 alleles and L > F (Fig. 1H and Supplementary Fig. 4).
Next, we aimed to identify biochemical properties of mutated amino acids that could explain the increased HLA-binding of peptides carrying C > U mutations. A previous study found that C > U substitutions in SARS-CoV-2 frequently cause amino acid changes resulting in elevated levels of hydrophobicity9. We found the same tendencies when focusing on changes in the Kyte-Doolittle hydrophobicity index after C > U mutations21,22. Among common substitutions, the mutated amino acids had higher hydrophobicity compared to the original ones (Fig. 1G) except for L > F, which led to a slight decrease. Notably, it was reported that many common HLA-I supertypes are specific to hydrophobic amino acids in anchor positions23. To test how general this trend is, we quantified the specificity of each HLA-I allele for different amino acids, using published immunopeptidomics data. We found that most HLA-I variants preferentially bind epitopes enriched in hydrophobic amino acids (Spearman’s ρ = 0.76, two-sided correlation test P = 1.62 × 10−4, Fig. 1F). In summary, the results suggest that the increased HLA-binding of C > U-related peptides is driven by the increased hydrophobicity of mutated amino acids and the overall higher specificity of HLA-I molecules to hydrophobic residues. This is further supported by the pattern observed for L > F substitutions, which account for 10.3% of C > U-related amino acid substitutions. Unlike other substitutions, they are not associated with increased HLA-binding as they lead to a slight decrease in hydrophobicity (Fig. 1G, H).
Alongside C > U mutations, G > U mutations also contribute to amino acid changes in epitopes that enhance their binding affinity to HLA-I molecules. However, we did not observe a consistent increase in hydrophobicity among these amino acid substitutions, suggesting that alternative mechanisms may underlie the generation of novel HLA-bound peptides in this mutation type (Supplementary Fig. 5).
C > U mutations increase the number of HLA-bound peptides in most individuals
We next sought to assess the impact of C > U mutations on HLA binding when considering the HLA genotypes of individuals in the population. Similarly to a previous report7, we found that some common HLA variants (HLA-A*30:01, HLA-A*31:01, HLA-A*33:01, HLA-B*07:02, HLA-B*27:05, HLA-B*35:01, HLA-B*53:01) are less likely to bind viral peptides produced by C > U mutations (Supplementary Fig. 6). However, the potential negative effect of these variants might be counterbalanced by others at the level of individual genotypes. To test this, we examined the HLA-I genotypes of 2599 participants (Supplementary Table 1) involved in the 1000 Genomes Project24. This dataset offers a comprehensive characterization of human genetic variation, sampling from 26 populations across five continents. For each individual, we calculated the average number of peptide-HLA complexes lost or gained when a C > U mutation is generated. Specifically, for each C > U mutation in our previous analysis, we determined the number of peptide-HLA complexes formed with the original and the mutated peptides. We then subtracted the number of original complexes from the number of mutated ones and calculated the mean of these mutation-specific values. To assess the individual contribution of each HLA locus, we performed independent analyses on the HLA-A, HLA-B, and HLA-C loci. The HLA-B locus showed the highest variability: ~ 30% of the individuals were predicted to lose peptide-HLA complexes after acquiring C > U mutations (Fig. 2A and Table 1). At the same time, HLA-A and HLA-C loci were associated with an increase in the number of predicted peptide-HLA complexes in most individuals. Moreover, when we considered all loci, C > U mutations had a positive effect on peptide binding in more than 99% of individuals worldwide. This result suggests that the negative effect of specific HLA-I alleles is compensated by others on the genotype level of individuals.
A The average change in the number of peptide-HLA complexes on the level of whole genotypes and different HLA loci after one C > U mutation. The histograms indicate the number of subjects belonging to different groups characterized by certain ranges of peptide-HLA complex gain. Dashed red lines indicate a neutral effect (zero complex gained per mutation). B, C The accumulation of HLA class I-bound peptides over time. The average number of HLA class I–bound peptides across all individuals analyzed is shown relative to the Wuhan Hu-1 reference strain over time. The analysis was carried out for all mutation types (B) and for different nucleotide substitutions (C) separately. The vertical axis represents the change between the average number of peptide-HLA complexes in different isolates relative to the root. The horizontal axes indicate the date of isolation. Nodes and leaves represent common ancestors and isolates, respectively, while edges represent phylogenetic relationships. Note that the transparency of edges has been increased for visualization purposes. D The coefficients of a linear mixed model predicting the mean genotype-level binding gains of individuals based on their geographical regions of origin (n = 686, 358, 532, 518 and 514 individuals from Africa, America, East Asia, Europe and South Asia, respectively). The models include the genomic background of individuals as random terms (see Methods). Higher values indicate that people from a particular region carry HLA alleles that are showing higher binding gains compared to HLA genotypes of individuals from Africa. Two-sided P-values calculated by t tests using Satterthwaite’s method are shown. Red color marks significant terms, the whiskers indicate the 95% confidence interval. E The magnitude of reduction in mean genotype-level binding gains in populations of South and East Asia after excluding individuals carrying specific HLA-alleles. The blue and red colors represent a decrease and an increase in mean binding gain, respectively. Alleles are ordered based on their effect on mean binding gain (alleles on the right side have the most significant positive influence on the observed trend). Source data are provided in the Source Data file.
We next investigated whether the continuous accumulation of C > U mutations in SARS-CoV-2 samples increased the number of HLA-bound peptides, considering the complete HLA-I genotypes of individuals. For this purpose, we tracked viral substitutions along evolutionary trajectories and assessed their average impact on HLA binding in the analyzed individuals. Reassuringly, we observed a temporal increase in the number of HLA-bound peptides compared to the initial viral isolate (Fig. 2B), a trend potentially explained by the accumulation of C > U mutations (Fig. 2C).
We next aimed to identify geographical regions where individuals carry HLA-I variants with particularly high gains of HLA-bound peptides (Fig. 2D). We used a linear mixed model to compare HLA binding gains among individuals from the 1000 Genomes Project while controlling for potential confounding due to genetic ancestry (see “Methods” for details). After accounting for ancestry, we found significant differences across individuals from different geographical regions. The most notable increase in HLA binding gains was observed in individuals from East Asia, followed by those from South Asia (P = 0.00893 and P = 0.0094, respectively, t tests using Satterthwaite’s method). This pattern may reflect the genomic imprint from recurrent epidemics caused by RNA viruses in this region (see Discussion). To delve deeper into the underlying factors of these binding gains, we investigated key alleles driving these trends. By sequentially removing carriers of specific HLA-I variants from the dataset, we assessed their impact on the average binding gains across the population. As illustrated in Fig. 2E, alleles HLA-A*24:02, HLA-C*14:02, and HLA-B*51:01 were found to be the strongest contributors to the pronounced binding gains observed in individuals from these regions.
Contribution of C > U mutations to SARS-CoV-2 epitopes after viral spillover to humans
We investigated whether well-known epitopes in the Wuhan Hu-1 strain of SARS-CoV-2 might have been generated by C > U mutations after its transmission to humans. The bat coronavirus RaTG13, which is considered the closest relative to SARS-CoV-2, is a likely candidate for its natural origin25. The genomes of the two viruses show 96.2% identity25 with discrepancies primarily due to C > U mutations26. We hypothesized that these mutations have generated novel HLA-bound immunogenic peptides in SARS-CoV-2. To test this, we collected SARS-CoV-2 epitopes from the Immune Epitope Database27. We focused on sequences with only one (n = 81) or two (n = 15) amino acid differences compared to the corresponding RaTG13 proteins, and analyzed the coding nucleotide sequences of both the RaTG13 and the Wuhan Hu-1 reference strains. We identified 21 instances where amino acid substitutions, likely due to C > U mutations, could account for the emergence of immunogenic epitopes in the Wuhan Hu-1 strain (Supplementary Table 2, see Methods for details).
C > U mutations generate immunogenic viral epitopes
To confirm the above results, we aimed to experimentally validate that C > U mutations are associated with the emergence of immunogenic peptides, expecting that mutated peptides are more likely to activate CD8 + T cells. We compiled two sets of peptides for analysis: one consisting of original and mutated peptide pairs based on the RaTG13 – Wuhan Hu-1 comparison (n = 15 pairs, Supplementary Table 2) and another consisting of original Wuhan Hu-1 sequences alongside their mutated counterparts that have emerged due to C > U mutations since the start of the pandemic (n = 7 pairs). We assessed the binding strength of both original and mutated peptides to common HLA-I alleles using ProImmune REVEAL assays. Notably, the predicted binding strength of these peptides agreed well with the actual binding outcomes observed in the in vitro assays (Fig. 3A). In addition, we examined whether the selected C > U mutations led to an overall gain or loss of bound peptides across the complete HLA-I genotypes of individuals in the 1000 Genome Project dataset. We selected participants (n = 79) whose allele sets were comprehensively covered by the ProImmune REVEAL assays. Similarly to our earlier analysis (Fig. 2A), we calculated the average gains in peptide binding for each subject. Our results indicate that C > U mutations generally increased the number of HLA-bound peptides in most individuals (Supplementary Fig. 8).
A The receiver-operator characteristic (ROC) curve indicates the specificity and sensitivity of binding affinity predictions (NetMHCpan 4.0 algorithm) in determining the presence or absence of in vitro binding. Empirical binding strength values were dichotomized using an established cutoff of 45, as suggested by ProImmune Ltd. The area under the curve (AUC) is also indicated. B The fraction of CD25 + CD8 + cells in PMBCs without simulation, and after treating them either with the original or the C > U-mutated peptide pools. Each point triplet represents values for the same PBMC donor (n = 14 individuals). Two-sided Friedman test P = 8.78 × 10−6, two-sided post-hoc Conover test P-values are indicated above horizontal lines. In boxplots, horizontal lines indicate median, boxes indicate interquartile range, and vertical lines indicate first quartile – 1.5 × IQR and third quartile + 1.5 × IQR. Source data are provided in the Source Data file.
To investigate the immune response to peptides generated by C > U mutations, we selected 13 pairs of original and mutated peptides that demonstrated a significant increase in HLA-binding in the ProImmune REVEAL assays (Supplementary Table 2). We evaluated their potential to activate T-cells using peripheral blood mononuclear cells (PBMCs) from HLA-matched donors. We prepared two sets of peptide pools: one with the original 13 peptides and another with their 13 mutated counterparts. We then exposed ex vivo PBMCs from 14 individuals to these peptide pools and measured CD25 expression on CD8 + T cells as an indicator of activation. Remarkably, the peptides altered by C > U mutations showed a higher propensity to activate CD8 + T cells compared to the original ones (Fig. 3B). These findings underscore the potential of C > U mutations to generate highly immunogenic viral peptides.
Enhanced capacity to present C > U mutant peptides shapes COVID-19 severity
Early detection of SARS-CoV-2 infection by the immune system is critical to prevent severe COVID-19 outcomes28,29,30,31. Numerous studies have emphasized the role of CD8 + T cell-mediated immunity in combating the virus32,33,34. Our findings suggest that C > U mutations could enhance the likelihood of recognition by the cellular adaptive immune system, potentially leading to less severe disease. Consequently, we hypothesized that COVID-19 patients carrying HLA-I molecules that are less capable of binding C > U-mutated viral peptides may experience worse disease outcomes.
To test our hypothesis, we analyzed data from the UK Biobank cohort — a large-scale, prospective study encompassing over half a million participants from the United Kingdom. This cohort offers a comprehensive dataset, including individuals’ genetic profiles, medical histories, and lifestyle factors, making it a valuable resource for examining COVID-19 disease severity risk factors. First, we calculated the genotype-level gain of HLA-bound peptides for each participant with a documented positive COVID-19 test in the UK Biobank database (baseline characteristics are provided in Supplementary Table 3). We then investigated whether participants with HLA-I molecules less likely to bind C > U-mutated viral peptides had an increased risk of developing severe COVID-19, as indicated by hospitalization. We developed a multivariate logistic regression model that incorporated variables known to affect COVID-19 outcomes, such as age (median: 65), gender, Townsend Deprivation Index, body mass index (BMI), medical history including hypertension, hyperlipidemia, diabetes, immune-related disorders, and respiratory conditions. We also considered the fraction of the UK population vaccinated at the time of the positive test as a covariate35,36. Consistent with the hypothesis, individuals with HLA-I alleles capable of binding fewer viral peptides showed a higher likelihood of severe disease (odds ratio = 1.12, P = 0.0056, P-value of two-sided Z statistics, Fig. 4). The effect remained significant when controlling for HLA alleles that are associated with disease severity and where C > U mutations exert only minor influence on HLA binding (Supplementary Fig. 9, see Methods for details). This result suggests a potential interplay between C > U mutations and HLA class I-mediated immune presentation of SARS-CoV-2 peptides in influencing disease severity.
Patients gaining a lower number of HLA-bound peptides/mutation (1st quartile, n = 4240 individuals) are more likely to have severe disease (n = 4549; total number of individuals: 16,974). The forest plot summarizes covariates of the logistic regression model, including sociodemographic and clinical factors that potentially affect COVID-19 disease outcome. The vaccination prevalence categories indicate the percentage of fully vaccinated individuals in the UK population at the time when the first positive test was reported for the patient; the odds ratios are calculated compared to COVID-19 cases of the dataset prior to the start of mass vaccination. See Methods for detailed information on the variables. BP represents blood pressure, and BMI indicates body mass index. The odds ratio with a 95% confidence interval is indicated. P-values of two-sided Z statistics are shown.
Discussion
The rapid global spread of SARS-CoV-2 has led to the emergence of numerous variants, raising critical questions about how viral mutations influence the HLA-I-associated T-cell immune response. It is well-established that some mutations in SARS-CoV-2 facilitate immune escape, potentially leading to more severe infections31 and reduced vaccine efficacy37,38,39. These mutations can impair both the antibody binding to the virus and the recognition of HLA-presented viral peptides by CD8 + T cells on the surfaces of infected cells5,40. Specifically, escape mutations often reduce CD8 + T cell recognition by interfering with the HLA presentation of viral peptides5,6. Despite the focus on escape mutations that decrease viral immune detection, less attention has been given to mutations that might enhance immune recognition of the virus.
In this study, we focused on C > U mutations, which are predominant in the genetic landscape of SARS-CoV-2 variants. The origin of these mutations remains a topic of debate. Several in silico10,11,16 and experimental41,42 studies suggest that APOBEC enzymes are important driving forces in generating C > U hypermutation. APOBEC proteins are integral to the innate immune defense against viruses and retrotransposons, and induce hypermutation in viral genomes. These enzymes have been shown to offer protective effects against several viruses, including the hepatitis B43,44, human papillomavirus45,46, and herpesviruses47,48. In HIV, the role of APOBEC3-associated mutagenesis in the adaptive immune recognition of viral peptides remains controversial. Some studies have reported reduced immune activation by APOBEC3-mutated epitopes49,50,51, while other research suggests that APOBEC3 mutagenesis can enhance viral immunogenicity in certain patient subsets52,53. In SARS-CoV-2, the lack of the characteristic nucleotide context linked to APOBEC3 around C > U mutations indicates that alternative mechanisms may also be responsible for their occurrence. For instance, Bradley et al. proposed that replication errors play a dominant role in the accumulation of these mutations54. Notably, while viral genomes isolated from Vero E6 cell lines indeed showed a lack of APOBEC3 context around C > U substitutions, mutation data from clinical isolates suggested an enrichment of nucleotide changes at APOBEC3A target sites. Importantly, we found that the effect of C > U mutations on HLA binding is independent of the surrounding sequence context (Supplementary Fig. 3), suggesting that our findings are not influenced by the source of the mutations.
We found that C > U mutations in human cells potentially counteract viral immune escape by generating novel HLA-bound viral epitopes at high frequencies (Fig. 1). In addition, numerous experimentally verified SARS-CoV-2 epitopes were most likely generated through these mutations after human transmission. Consequently, we found that individuals carrying HLA variants that can effectively present C > U-associated peptides are less likely to have severe infection (Fig. 4). Notably, a recent study indicated that the HLA-B*15:01 allele is prevalent among individuals with asymptomatic infections55. According to our analyses, this allele has the highest affinity for C > U-mutated peptides among the HLA-B variants (Supplementary Fig. 6).
Asymptomatic carriers—who typically mount strong virus-specific immune responses56—play a key role in transmission57,58,59,60, suggesting that the virus may, paradoxically, benefit from enhanced immune recognition. This raises the possibility that accumulation of C > U mutations in the viral genome could offer an evolutionary advantage by enhancing immune responses while maintaining asymptomatic infection. However, our analysis does not support this hypothesis. Using UShER-based phylogenetic analysis20, we assessed the strength and direction of selection on C > U mutations. We found no positive association between the gain of HLA-bound peptides and the fitness effect of C > U mutations (Supplementary Fig. 10). In fact, most C > U mutations predicted to increase HLA binding were found to negatively impact viral fitness. These findings are consistent with prior studies, showing that C > U mutations fix at a lower rate than other nucleotide changes61, likely due to their deleterious effects on fitness55. Thus, the accumulation of C > U mutations is likely driven by mutational pressure rather than positive selection, supporting the idea that mutational pressure can outweigh weak selection and result in suboptimal genome composition62.
A similar trend for C >U hypermutation and increased binding by HLA-I molecules was found in the rubella virus suggesting that this phenomenon may be more general (Supplementary Fig. 2B). Moreover, C > U hypermutation is widespread in other human RNA viruses, too63. These viruses have been associated with frequent host switching, providing novel emergent pathogens in the human population64,65,66, as well as exhibiting a strong selective pressure during host-pathogen co-evolution67. Given the amino acid substitution bias introduced by C > U mutations and the increased affinity of HLA-I molecules for hydrophobic residues in C > U-mutated peptides, we speculate that the HLA-I system evolved to enhance the recognition of hydrophobic amino acid residues, thereby optimizing immune responses against viral C > U mutations (Fig. 5).
Based on our results, we speculate that HLA-I molecules were selected for binding hydrophobic amino acids that are generated by C > U hypermutation in RNA viruses. Created in BioRender. Manczinger, M. (2025; https://BioRender.com/kmbfy4n).
If we consider APOBEC3 as a source of C > U mutations, our results raise the intriguing possibility that these enzymes have a dual role in antiviral immune defense. In addition to inducing lethal mutagenesis of viral genomes, APOBEC3 could give rise to immunogenic viral epitopes by increasing their hydrophobicity, an established feature of HLA-I antigen presentation and immunogenicity68. Analogously, tumors carrying APOBEC3 mutational signatures contain more hydrophobic neoantigens, are more immunogenic, and are associated with positive response to immunotherapy21,69,70,71,72,73,74. Given that APOBEC3 enzymes emerged with the appearance of placental mammals ~ 65–100 million years ago75,76, well before the evolution of HLA-I-mediated antigen presentation, it is unlikely that they were selected to enhance viral peptide recognition. Instead, we propose that the increased hydrophobicity of peptides resulting from APOBEC-induced mutations may represent an evolutionary by-product.
Interestingly, while enhanced binding of C > U-generated epitopes is observed globally, its magnitude varies across geographical regions, with the highest levels found in South and East Asia (Fig. 2D). This area has been a hotspot for viral epidemics both historically and in the present. The frequent emergence of novel pathogens in this region is driven by a combination of ecological and social factors. Historical records spanning the past 2200 years indicate that South China’s warm and humid climate, rich vegetation, and densely populated settlements created ideal conditions for pathogen emergence and spread77. Another epidemiological study linked outbreaks primarily to agrarian societies and rising population densities78. In addition, Souilmi et al. identified genomic imprints of selective sweeps in human genes, interacting with coronavirus species, suggesting an ancient coronavirus epidemic in the region approximately 20,000 years ago79. Similarly, Morris et al. found stronger signals of past selection events in individuals from the China Kadoorie Biobank compared to those in the UK Biobank80. Further research should explore the evolutionary selection pressure on C > U mutagenesis and HLA genes in these regions, potentially shedding light on a long-standing interplay between these two systems.
Methods
Statistical analysis and visualization
We used R version 4.5.181 in RStudio version 2024.09 environment for statistical analyses; the ggplot282, ggpubr83, forestplot84, pheatmap85, pROC86 and cowplot87 R libraries for visualization. Dunn’s post-hoc test was performed using the DunnTest function of the FSA R library88. Linear mixed models were created using the lmer function of the lmerTest library89 and further processed using functions from the dotwhisker90 and broom.mixed91 R libraries. Friedman and Conover tests were performed using friedman_test and frdAllPairsConoverTest functions from the rstatix92 and PMCMRplus93 libraries, respectively.
Source of viral mutation data
We acquired phylogenetic data of SARS-CoV-2 genomic isolates (including the date of isolation for each sample and the putative date of intermediate nodes) from the Nextstrain Global Analysis website on 17th October 2024. We excluded isolates from non-human origins and extracted all nucleotide single-base substitutions in each viral sample and each node of the phylogenetic tree relative to the Wuhan Hu-1 reference strain (NCBI Reference Sequence database ID: NC_045512.2, https://www.ncbi.nlm.nih.gov/nuccore/1798174254).
In addition, we utilized a further dataset published by Bloom and Neher20, which contains information on the number of independent occurrences of each mutation throughout the phylogeny. We applied the ntmut_fitness_all.csv file downloaded from their GitHub repository (https://github.com/jbloomlab/SARS2-mut-fitness) to generate all possible SARS-CoV-2 peptide variants carrying single amino acid substitutions. For this dataset, instead of focusing on unique mutations, we considered each mutation for the number of times it was found independently throughout the phylogeny.
To examine the effects of APOBEC3-generated nucleotide changes in another positive single-stranded RNA virus, rubella, we used a dataset published by Klimczak et al.94, containing 790 nucleotide changes overall, of which 226 were missense mutations.
Generation of mutated peptide fragment sequences
We used the genome of the Wuhan Hu-1 isolate (NC_045512) as a reference. We generated two types of datasets using custom R scripts. The first dataset contained information about mutations. We identified all unique mutations in the Nextstrain dataset. For each mutation, we changed the nucleotide in the reference genome and translated them to amino acid sequences. Here, our goal was to investigate the effects of individual nucleotide changes. We performed the same steps for the rubella mutation set, as well as for the SARS-CoV-2 dataset by Bloom and Neher.
The second dataset contained information about samples. For each node or isolate, we applied all nucleotide changes in its genome and translated the modified coding sequences into amino acid sequences. In the case of both datasets, we split all protein sequences into 8–11 amino acid long fragments as suggested previously95. In the sample-specific dataset, we excluded original-mutated peptide pairs that would have been located after nonsense (premature stop) mutations in the given protein.
Calculation of HLA-I binding gain and loss
We predicted the binding of each 8–11-mer peptide by common HLA-I alleles using the NetMHCpan-4.0 algorithm17. We carried out the prediction for 16 HLA-A and 13 HLA-B alleles, collected from a reference set with maximal population coverage18. In addition, since the list did not include data for HLA-C, we predicted HLA-binding by the first four-digit allele in each two-digit HLA-C allele class (n = 14)19.
We defined a given peptide as HLA-bound if the predicted “binding rank percentile” was under 0.5. We used this strict binding threshold value to minimize false positive hits. A binding gain event was defined as a change of binding rank value from ≥ 2 (not bound) to < 0.5 (strong binding), while the opposite direction was considered a binding loss. Net binding gain (NBG) was defined as the difference between the number of gained and lost peptides. Practically, this metric describes the increase/decrease in the number of HLA-bound viral peptide segments after mutations. We calculated the net binding gain value for each HLA-I allele by calculating the mean of NBG values for all unique C > U mutations from the Nextstrain dataset. As two-sided paired Wilcoxon’s signed-rank tests were performed for multiple types of nucleotide and amino acid substitutions (Fig. 1D, H and Supplementary Figs. 1, 2, 3 and 5), P-values were corrected using the method by Benjamini and Hochberg96. In case of the Bloom and Neher dataset, for each allele - instead of using absolute counts - we calculated the fraction of unique mutations associated with binding gain or loss, weighted with the number of times they appear in the phylogeny.
To investigate the effect of missense mutations on HLA-binding on the whole genotype level, we downloaded HLA-I genotype data of 2618 subjects in the 1000 Genome Project24. After excluding subjects carrying alleles that are unsupported by the prediction algorithm, we examined the HLA-binding for 2599 individuals. For each individual, we calculated the average binding gain/loss associated with C > U mutations by taking the mean of NBG values specific for the unique HLA-I variants they carry. In Fig. 2E, we investigated the effects of alleles on NBG that were present in at least 5% of individuals in all countries of the South and East Asian regions.
We performed analyses shown in Fig. 1D separately for T-cell epitope regions (Supplementary Fig. 2A). We downloaded data on epitope sequences from the Immune Epitope Database (IEDB) on 22nd November 2021. We selected HLA-I-presented linear epitopes of SARS-CoV-2 with at least one positive T-cell assay in human hosts.
To assess the specificity of HLA-alleles for different amino acids, we determined peptide binding motifs for 41 of 43 reference HLA-I alleles using on the immunopeptidomics dataset published by Sarkizova et al.97. We created information content matrix-based motifs by the universalmotif R library98. We defined the specificity of a given HLA-I allele for a given amino acid as the sum of amino acid-specific bit scores at positions 2 and 9.
Comparing HLA-I binding gain between populations
We used a linear mixed model (implemented via the lmer function in the lmerTest R package89) to examine differences in average binding gains among individuals from different geographical regions. To account for genetic similarities between individuals, we utilized the PCs_1000G dataset from the PCAmatchR R package99. This dataset contains data for the first 20 genetic principal components (PCs) of 2423 individuals from the 1000 Genome Project. We classified individuals into genetic clusters based on genetic PCs. To determine the optimal number of clusters, we applied the NbClust R function from the NbClust package (method: “ward.D2”, index: “ch”), which identified 15 optimal clusters. The resulting grouping was incorporated as a random effect in the linear mixed model, using the following formula:
Measurement of HLA-I binding and T-cell activation
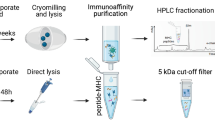
To experimentally verify in silico results, we assembled a set of original viral peptides and their mutated counterparts carrying C > U nucleotide changes (Supplementary Table 2). The final peptide set consisted of (i) T-cell epitopes of SARS-CoV-2 potentially generated by C > U mutations from RaTG13 sequences and ii) mutated Wuhan Hu-1 sequences affected by homoplasic C > U mutations in epitope-coding regions of the SARS-CoV-2 genome100. The selected peptides were synthesized, and their binding affinity levels towards a set of 19 HLA-I variants were examined with ProImmune REVEAL HLA class I binding assays, which determine binding strength based on the ability of test peptides to stabilize the peptide-HLA complex.
For the measurement of differences in T-cell activation, we selected 13 peptide pairs, where the mutated peptides showed a significant binding gain to certain HLA alleles according to experimental results (see Source Data Table 13). We generated peptide pools from the original and the mutated sequences.
We performed experimental tests following established methods101,102. Briefly, peripheral venous blood was collected in our laboratory from three HLA-matched healthy volunteers using lithium heparin-treated tubes (BD Vacutainer, Becton Dickinson, Sunnyvale, CA, USA). Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll density gradient centrifugation using Leucosep tubes (Greiner Bio-One, Kremsmünster, Austria). To increase the set of samples, commercially available HLA-characterized PBMCs (11 cases, identified with subject codes starting with “LP”) were also purchased (CTL Europe GmbH, Bonn, Germany; Source Data Table 14). The sex and age of the three healthy volunteers were self-reported, while information on the source individuals of the 11 PBMC samples was provided to us by the company.
Cells were pelleted by centrifugation at 800 g without braking for 20 minutes. The ring of PBMCs was harvested by pipetting and diluted with 15 ml PBS, then centrifuged at 350 × g for 5 min. The supernatant was removed. If necessary, red blood cells were lysed by 2 ml ACK solution (prepared in our laboratory: 0.15 M NaH4Cl, 10 mM KHCO3, 0.1 mM Na2EDTA, pH7.4, Merck, USA) at room temperature (RT) for 2 min. Cells were washed with 15 ml PBS and centrifuged at 350 × g for 5 min, and then were frozen in 90% FCS/10% DMSO (v/v%). Cells were thawed into 10 ml 37 °C RPMI-1640 cell culture media (Capricorn Scientific, Ebsdorferung, Germany) and pelleted using centrifugation at 350 × g for 5 min at RT. Cells were washed with complete RPMI-1640 (cRPMI) cell culture media containing 100 U/ml penicillin sodium salt and 100 μg/ml streptomycin sulfate salt (Merck, USA), 10 % FCS (Euroclone, Milan, Italy), 2 mM glutamine (200 mM 100x diluted Capricorn Sc.). Afterward, cell counts were determined using the Bürker-chamber and trypan blue dye (Sigma-Aldrich, Hungary).
PBMCs (5 × 105) were divided in 180 µl of cRPMI/well into 96 well plate (Greiner Bio-One, Kremsmünster, Austria) (flat-bottom TC-treated) as follows: samples (1-2) untreated, (3-4) CytoStim (Miltenyi Biotec, Bergisch Gladbach, Germany) stimulated, (5-6) peptide pool 1 (original), (7-8) peptide pool 2 (mutated). Cells were left untreated for 1 h resting period. All samples were incubated with 10 ng/ml IL-2/well. Stimulating agents were added according to the followings: (1-2) 20 µl media to the unstimulated; (3-4) 20 µl media plus 2 µl CytoStim; (5-6) the mixture of 13 “original” peptides (peptide pool 1); (7-8) the mixture of 13 “mutated” peptides (peptide pool 2).
In the case of (5-8), each peptide was dissolved in DMSO (Sigma-Aldrich) at 4 mg/ml. We prepared the mixtures of the 13 peptides pipetting 1 µl from each peptide into 87 µl cRPMI. The amount of the peptide mixture was 52 µg in one pool. Cells were treated with a 20 µl peptide pool (10.4 µg).
The stimulation period lasted 24 h. 100 µl PEB buffer (PBS-EDTA-BSA) was added to each well (0.5% BSA, 2 mM EDTA in PBS, Miltenyi). Cells were pipetted into 12 × 75 mm FACS tubes (VWR International, USA), and centrifuged at 350 G at RT for 5 min. Afterward, cells were suspended in 50 µl PBS containing 0.5 µl of the Viobility™ Fixable Dye (Ex.: 405 nm, Em.: 452 nm; 100 x diluted of the stock). After 15 min of incubation in the dark at RT, 1 ml PEB was added to each sample. Cells were centrifuged at 350 × g at RT for 5 min. Next, cells were suspended in 100 µl PEB, then 100 µl 3.7% formaldehyde was added to each sample. Subsequently, cells were incubated in the dark at RT for 20 min.
1 ml PEB was added to each sample before cells were centrifuged at 500 × g at RT for 5 min. The batch of the antibody cocktail was prepared in PEB as the followings: anti-CD3 APC 100x diluted (clone REA613, catalog number: 130-113-135), anti-CD4 VioBright B515 100x diluted (clone REA623, catalog number: 130-114-535), anti-CD8 VioGreen 50x diluted (clone REA734, catalog number: 130-110-684), anti-CD25 APCVio770 100x diluted (clone REA570, catalog number: 130-123-469). Antibodies were purchased from Miltenyi Biotec. Cells were incubated in 50 µl of the antibody cocktail at RT for 30 min. Afterward, they were washed with 1 ml PEB and centrifuged at 500 g at RT for 5 minutes. Cells were resuspended in 300 μl PEB, and 1 × 105 live single cells were acquired on Cytoflex S fluorescence-activated cell sorter (FACS; Beckman Colter, USA). Manual gating was used to determine CD8 + T cells within live CD3 + lymphocytes in CytExpert (Beckman Colter; Supplementary Fig. 7). Reactive cells were gated as CD25 + CD8 + T cells. Finally, reactive cells are shown in relation to the percentage of the parental CD8 + T cells.
Analyzing the effects of binding gain on COVID-19 outcome
We downloaded detailed sociodemographic, clinical and COVID-19 outcome data from the UK Biobank database on 19th October 2021103. Similarly to other studies104,105, we investigated subjects who had been tested positive for SARS-CoV-2 infection at least once, and whose full HLA-I genotype was known (n = 16,974). We considered patients who died of COVID-19 or tested positive in an inpatient setting as severe cases (n = 4549), while the remaining patients were classified as mild cases (n = 12,425).
We built a logistic regression model containing a set of important confounding factors associated with COVID-19 outcome105. We determined the age of the subjects by calculating the difference between the first positive COVID-19 test and the year of birth of the subject. We defined individuals with “High deprivation index” as the ones in the top quartile of the Townsend Deprivation Index variable. We considered a patient to have a certain disease based on the ICD10 codes in the dataset (UK Biobank Data Fields 41202 and 41204) according to Table 2. We considered a positive “Medication for high blood pressure and/or high cholesterol levels were used” variable as a proxy for cardiovascular disease105. We defined vaccination rates based on the percentages of fully vaccinated individuals in the United Kingdom according to Our World in Data106. We stratified patients into different classes based on the time of their first COVID-19 positivity using the following cutpoints: 10th January 2021 (vaccination program started), 7th May 2021 (25% of the population is fully vaccinated), 5th July 2021 (50% of the population is fully vaccinated).
We built additional models, also including the presence/absence of specific HLA alleles as a covariate, that are associated with disease outcome. We classified HLA-I variants according to a systematic review by Dobrijevic et al.107, especially focusing on alleles that affect hospitalization status. We only included alleles in the model that had an absolute binding gain value lower than 0.05.
Ethics
The study was carried out in compliance with the Declaration of Helsinki, and the protocol (‘Molecular phenotyping in chronic respiratory inflammation and SARS-CoV-2 infected patients’) was approved by the Ethics Committee of the National Public Health Center (Project ID: 52792-5/2021/EÜIG).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
NC_045512.2 [https://www.ncbi.nlm.nih.gov/nuccore/1798174254] was used as the reference genome for the analyses presented. HLA genotypes for individuals from the 1000 Genomes Project were downloaded from: https://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/HLA_types/20181129_HLA_types_full_1000_Genomes_Project_panel.txt. SARS-CoV-2 genomic and phylogenetic data were obtained from the Nextstrain website on 17th October, 2024 (the specific JSON file is shared on our GitHub repository). Additional mutation data were downloaded from the GitHub repository associated with the Bloom and Neher article: https://github.com/jbloomlab/SARS2-mut-fitness/blob/main/results/nt_fitness/ntmut_fitness_all.csv. The individual-level data used for the COVID-19 outcome analyses (Fig. 4 and Supplementary Fig. 9) were obtained from UK Biobank under Application ID 44917. The authors are not authorized to distribute these data. Interested researchers may apply for access via the UK Biobank Access Management System [https://www.ukbiobank.ac.uk/register-apply/]. The source data files required to reproduce the figures and tables are available in the project’s GitHub [https://github.com/lhgergo/covid-apobec] and Zenodo repositories108. Source data are provided in this paper.
Code availability
All code necessary to reproduce the results presented in this study is available in the project’s GitHub [https://github.com/lhgergo/covid-apobec] and Zenodo repositories108.
References
Thomson, E. C. et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell 184, 1171–1187.e20 (2021).
Yao, H. et al. Patient-derived SARS-CoV-2 mutations impact viral replication dynamics and infectivity in vitro and with clinical implications in vivo. Cell Discov. 6, 76 (2020).
Plante, J. A. et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 592, 116–121 (2021).
Mlcochova, P. et al. SARS-CoV-2 B.1.617.2 delta variant replication and immune evasion. Nature 599, 114–119 (2021).
Agerer, B. et al. SARS-CoV-2 mutations in MHC-I-restricted epitopes evade CD8 + T cell responses. Sci. Immunol. 6, https://doi.org/10.1126/sciimmunol.abg6461 (2021).
Stanevich, O. V. et al. SARS-CoV-2 escape from cytotoxic T cells during long-term COVID-19. Nat. Commun. 14, 149 (2023).
Hamelin, D. J. et al. The mutational landscape of SARS-CoV-2 variants diversifies T cell targets in an HLA-supertype-dependent manner. Cell Syst. 13, 143–157 (2022).
Pretti, M. A. M., Galvani, R. G., Scherer, N. M., Farias, A. S. & Boroni, M. In silico analysis of mutant epitopes in new SARS-CoV-2 lineages suggest global enhanced CD8 + T cell reactivity and also signs of immune response escape. Infect. Genet. Evol. 99, 105236 (2022).
Matyášek, R. & Kovařík, A. Mutation patterns of human SARS-CoV-2 and bat RaTG13 coronavirus genomes are strongly biased towards C>U transitions, indicating rapid evolution in theirhosts. Genes 11, 761 (2020).
Simmonds, P. Rampant C→U hypermutation in the genomes of SARS-CoV-2 and other coronaviruses: causes and consequences for their short- and long-term evolutionary trajectories. mSphere 5, e00408–e00420 (2020).
Di Giorgio, S., Martignano, F., Torcia, M. G., Mattiuz, G. & Conticello, S. G. Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2. Sci. Adv. 6, eabb5813 (2020).
Hadfield, J. et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34, 4121–4123 (2018).
Wu, A. et al. One year of SARS-CoV-2 evolution. Cell Host Microbe 29, 503–507 (2021).
Rice, A. M. et al. Evidence for strong mutation Bias toward, and selection against, U content in SARS-CoV-2: implications for vaccine design. Mol. Biol. Evol. 38, 67–83 (2021).
Sadykov, M., Mourier, T., Guan, Q. & Pain, A. Short sequence motif dynamics in the SARS-CoV-2 genome suggest a role for cytosine deamination in CpG reduction. J. Mol. Cell Biol. 13, 225–227 (2021).
Graudenzi, A., Maspero, D., Angaroni, F., Piazza, R. & Ramazzotti, D. Mutational signatures and heterogeneous host response revealed via large-scale characterization of SARS-CoV-2 genomic diversity. iScience 24, 102116 (2021).
Jurtz, V. et al. NetMHCpan-4.0: improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J. Immunol. 199, 3360–3368 (2017).
Weiskopf, D. et al. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8 + T cells. Proc. Natl. Acad. Sci. USA. 110, E2046–E2053 (2013).
Koncz, B. et al. Self-mediated positive selection of T cells sets an obstacle to the recognition of nonself. Proc. Natl. Acad. Sci. USA 118, e2100542118 (2021).
Bloom, J. D. & Neher, R. A. Fitness effects of mutations to SARS-CoV-2 proteins. Virus Evol. 9, vead055 (2024).
Boichard, A. et al. APOBEC-related mutagenesis and neo-peptide hydrophobicity: implications for response to immunotherapy. OncoImmunology 8, 1550341 (2019).
Kyte, J. & Doolittle, R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 (1982).
Sidney, J., Peters, B., Frahm, N., Brander, C. & Sette, A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 9, 1 (2008).
Abi-Rached, L. et al. Immune diversity sheds light on missing variation in worldwide genetic diversity panels. PLoS ONE 13, e0206512 (2018).
Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020).
Li, Y. et al. The divergence between SARS-CoV-2 and RaTG13 might be overestimated due to the extensive RNA modification. Future Virol. 15, 341–347 (2020).
Vita, R. et al. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 47, D339–D343 (2019).
Moss, P. The T cell immune response against SARS-CoV-2. Nat. Immunol. 23, 186–193 (2022).
Notarbartolo, S. et al. Integrated longitudinal immunophenotypic, transcriptional, and repertoire analyses delineate immune responses in patients with COVID-19. Sci. Immunol. 6, eabg5021 (2021).
Bergamaschi, L. et al. Longitudinal analysis reveals that delayed bystander CD8 + T cell activation and early immune pathology distinguish severe COVID-19 from mild disease. Immunity 54, 1257–1275 (2021).
Rydyznski Moderbacher, C. et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell 183, 996–1012.e19 (2020).
Bange, E. M. et al. CD8 + T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat. Med. 27, 1280–1289 (2021).
Westmeier, J. et al. Impaired cytotoxic CD8 + T cell response in elderly COVID-19 patients. mBio 11, e02243–20 (2020).
Szabó, E. et al. Comparison of humoral and cellular immune responses in hematologic diseases following completed vaccination protocol with BBIBP-CorV, or AZD1222, or BNT162b2 vaccines against SARS-CoV-2. Front. Med. 10, 1176168 (2023).
Mohammed, I. et al. The efficacy and effectiveness of the COVID-19 vaccines in reducing infection, severity, hospitalization, and mortality: a systematic review. Hum. Vaccin. Immunother. 18, 2027160 (2022).
Huang, C., Yang, L., Pan, J., Xu, X. & Peng, R. Correlation between vaccine coverage and the COVID-19 pandemic throughout the world: Based on real-world data. J. Med. Virol. 94, 2181–2187 (2022).
Garcia-Beltran, W. F. et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 184, 2372–2383 (2021).
Zhou, D. et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell 184, 2348–2361 (2021).
Hu, J. et al. Increased immune escape of the new SARS-CoV-2 variant of concern Omicron. Cell Mol. Immunol. 19, 293–295 (2022).
Zhang, H. et al. Profiling CD8 + T cell epitopes of COVID-19 convalescents reveals reduced cellular immune responses to SARS-CoV-2 variants. Cell Rep. 36, 109708 (2021).
Kim, K. et al. The roles of APOBEC-mediated RNA editing in SARS-CoV-2 mutations, replication and fitness. Sci. Rep. 12, 14972 (2022).
Nakata, Y. et al. Cellular APOBEC3A deaminase drives mutations in the SARS-CoV-2 genome. Nucleic Acids Res. 51, 783–795 (2023).
Chen, Y. et al. APOBEC3B edits HBV DNA and inhibits HBV replication during reverse transcription. Antivir. Res. 149, 16–25 (2018).
Turelli, P., Mangeat, B., Jost, S., Vianin, S. & Trono, D. Inhibition of hepatitis B virus replication by APOBEC3G. Science 303, 1829–1829 (2004).
Warren, C. J. et al. APOBEC3A Functions as a restriction factor of human papillomavirus. J. Virol. 89, 688–702 (2015).
Zhu, B. et al. Mutations in the HPV16 genome induced by APOBEC3 are associated with viral clearance. Nat. Commun. 11, 886 (2020).
Cheng, A. Z. et al. APOBECs and Herpesviruses. Viruses 13, 390 (2021).
Nakaya, Y., Stavrou, S., Blouch, K., Tattersall, P. & Ross, S. R. In vivo examination of mouse APOBEC3- and human APOBEC3A- and APOBEC3G-mediated restriction of parvovirus and herpesvirus infection in mouse models. J. Virol. 90, 8005–8012 (2016).
Borzooee, F., Joris, K. D., Grant, M. D. & Larijani, M. APOBEC3G Regulation of the evolutionary race between adaptive immunity and viral immune escape is deeply imprinted in the HIV genome. Front. Immunol. 9, 3032 (2019).
Monajemi, M. et al. Positioning of APOBEC3G/F mutational hotspots in the human immunodeficiency virus genome favors reduced recognition by CD8 + T Cells. PLoS ONE 9, e93428 (2014).
Kim, E.-Y. et al. Human APOBEC3 induced mutation of human immunodeficiency virus type-1 contributes to adaptation and evolution in natural Infection. PLoS Pathog. 10, e1004281 (2014).
Squires, K. D., Monajemi, M., Woodworth, C. F., Grant, M. D. & Larijani, M. Impact of APOBEC mutations on CD8 + T cell recognition of HIV epitopes varies depending on the restricting HLA. JAIDS J. Acquir. Immune Defic. Syndr. 70, 172–178 (2015).
Norman, J. M. et al. The antiviral factor APOBEC3G enhances the recognition of HIV-infected primary T cells by natural killer cells. Nat. Immunol. 12, 975–983 (2011).
Bradley, C. C. et al. Targeted accurate RNA consensus sequencing (tARC-seq) reveals mechanisms of replication error affecting SARS-CoV-2 divergence. Nat. Microbiol. 9, 1382–1392 (2024).
Augusto, D. G. et al. A common allele of HLA is associated with asymptomatic SARS-CoV-2 infection. Nature 620, 128–136 (2023).
Le Bert, N. et al. Highly functional virus-specific cellular immune response in asymptomatic SARS-CoV-2 infection. J. Exp. Med. 218, e20202617 (2021).
Oran, D. P. & Topol, E. J. Prevalence of asymptomatic SARS-CoV-2 infection: a narrative review. Ann. Intern. Med. 173, 362–367 (2020).
Johansson, M. A. et al. SARS-CoV-2 Transmission from people without COVID-19 symptoms. JAMA Netw. Open 4, e2035057 (2021).
Tan, J. et al. Transmission roles of symptomatic and asymptomatic COVID-19 cases: a modelling study. Epidemiol. Infect. 150, e171 (2022).
Emery, J. C. et al. The contribution of asymptomatic SARS-CoV-2 infections to transmission on the Diamond Princess cruise ship. ELife 9, e58699 (2020).
van Dorp, L. et al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 83, 104351 (2020).
Directional mutational pressure affects the amino acid composition and hydrophobicity of proteins in bacteria. in Contemporary Issues in Genetics and Evolution 383–391 (Springer Netherlands, Dordrecht, 1998).
Simmonds, P. & Ansari, M. A. Extensive C->U transition biases in the genomes of a wide range of mammalian RNA viruses; potential associations with transcriptional mutations, damage- or host-mediated editing of viral RNA. PLoS Pathog. 17, e1009596 (2021).
Geoghegan, J. L., Duchêne, S. & Holmes, E. C. Comparative analysis estimates the relative frequencies of co-divergence and cross-species transmission within viral families. PLoS Pathog. 13, e1006215 (2017).
Woolhouse, M. E. J. Population biology of emerging and re-emerging pathogens. Trends Microbiol. 10, s3–s7 (2002).
Kreuder Johnson, C. et al. Spillover and pandemic properties of zoonotic viruses with high host plasticity. Sci. Rep. 5, 14830 (2015).
Enard, D. & Petrov, D. A. Ancient RNA virus epidemics through the lens of recent adaptation in human genomes. Philos. Trans. R. Soc. B 375, 20190575 (2020).
Huang, L., Kuhls, M. C. & Eisenlohr, L. C. Hydrophobicity as a driver of MHC class I antigen processing: Hydrophobicity in MHC class I peptide supply. EMBO J. 30, 1634–1644 (2011).
Chen, H. et al. The immune response-related mutational signatures and driver genes in non-small-cell lung cancer. Cancer Sci. 110, 2348–2356 (2019).
Driscoll, C. B. et al. APOBEC3B-mediated corruption of the tumor cell immunopeptidome induces heteroclitic neoepitopes for cancer immunotherapy. Nat. Commun. 11, 790 (2020).
DiMarco, A. V. et al. APOBEC Mutagenesis inhibits breast cancer growth through induction of T cell–mediated antitumor immune Responses. Cancer Immunol. Res. 10, 70–86 (2022).
Mullane, S. A. et al. Correlation of Apobec Mrna expression with overall survival and pd-l1 expression in urothelial carcinoma. Sci. Rep. 6, 27702 (2016).
Smid, M. et al. Breast cancer genome and transcriptome integration implicates specific mutational signatures with immune cell infiltration. Nat. Commun. 7, 12910 (2016).
Wang, S., Jia, M., He, Z. & Liu, X.-S. APOBEC3B and APOBEC mutational signature as potential predictive markers for immunotherapy response in non-small cell lung cancer. Oncogene 37, 3924–3936 (2018).
Conticello, S. G. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 9, 229 (2008).
O’Leary, M. A. et al. The placental mammal ancestor and the post–K-Pg radiation of placentals. Science 339, 662–667 (2013).
Gong, S., Xie, H. & Chen, F. Spatiotemporal changes of epidemics and their relationship with human living environments in China over the past 2200 years. Sci. China Earth Sci. 63, 1223–1226 (2020).
Morabia, A. Epidemic and population patterns in the Chinese Empire (243 b.c.e. to 1911 c.e.): quantitative analysis of a unique but neglected epidemic catalogue. Epidemiol. Infect. 137, 1361–1368 (2009).
Souilmi, Y. et al. An ancient viral epidemic involving host coronavirus interacting genes more than 20,000 years ago in East Asia. Curr. Biol. 31, 3504–3514 (2021).
Morris, S. C. et al. Natural selection exerted by historical coronavirus epidemic(s): comparative genetic analysis in China Kadoorie Biobank and UK Biobank. Preprint at https://doi.org/10.1101/2024.02.06.579075 (2024).
Ihaka, R. & Gentleman, R. R: A language for data analysis and graphics. J. Comput. Graph. Stat. 5, 299–314 (1996).
Wickham, H. ggplot2. WIREs Comput. Stats 3, 180–185 (2011).
Kassambara, A. Ggpubr: ‘ggplot2’ Based Publication Ready Plots. (2025).
Gordon, M. & Lumley, T. Forestplot: Advanced Forest Plot Using ‘grid’ Graphics. (2025).
Kolde, R. Pheatmap: Pretty Heatmaps. (2025).
Robin, X. et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 12, 77 (2011).
Wilke, C. O. Cowplot: Streamlined Plot Theme and Plot Annotations for ‘Ggplot2’. (2025).
Ogle, D. H., Doll, J. C., Wheeler, A. P. & Dinno, A. FSA: simple fisheries stock assessment methods. https://doi.org/10.32614/CRAN.package.FSA (2015).
Kuznetsova, A., Brockhoff, P. B. & Christensen, R. H. B. lmertest package: tests in linear mixed effects models. J. Stat. Soft. 82, 1–26 (2017).
Solt, F. & Hu, Y. Dotwhisker: Dot-and-Whisker Plots of Regression Results. (2024).
Bolker, B. & Robinson, D. broom.mixed: Tidying methods for mixed models. https://doi.org/10.32614/CRAN.package.broom.mixed (2018).
Kassambara, A. rstatix: Pipe-friendly framework for basic statistical tests. https://doi.org/10.32614/CRAN.package.rstatix (2019).
Pohlert, T. PMCMRplus: Calculate pairwise multiple comparisons ofmean rank sums extended. https://doi.org/10.32614/CRAN.package.PMCMRplus (2018).
Klimczak, L. J., Randall, T. A., Saini, N., Li, J.-L. & Gordenin, D. A. Similarity between mutation spectra in hypermutated genomes of rubella virus and in SARS-CoV-2 genomes accumulated during the COVID-19 pandemic. PLoS ONE 15, e0237689 (2020).
Marty, R. et al. MHC-I Genotype restricts the oncogenic mutational landscape. Cell 171, 1272–1283 (2017).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300 (1995).
Sarkizova, S. et al. A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat. Biotechnol. 38, 199–209 (2020).
Tremblay, B. J.-M. universalmotif: An R package for biological motifanalysis. JOSS 9, 7012 (2024).
Brown, D. W., Myers, T. A. & Machiela, M. J. PCAmatchR: a flexible R package for optimal case–control matching using weighted principal components. Bioinformatics 37, 1178–1181 (2021).
van Dorp, L. et al. No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. Nat. Commun. 11, 1–8 (2020).
Honfi, D. et al. Comparison of homologous and heterologous booster SARS-CoV-2 vaccination in autoimmune rheumatic and musculoskeletal patients. IJMS 23, 11411 (2022).
Szebeni, G. J. et al. Humoral and cellular immunogenicity and safety of five different SARS-CoV-2 vaccines in patients with autoimmune rheumatic and musculoskeletal diseases in remission or with low disease activity and in healthy controls: a single center study. Front. Immunol. 13, 846248 (2022).
Sudlow, C. et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
Elliott, J. et al. COVID-19 mortality in the UK Biobank cohort: revisiting and evaluating risk factors. Eur. J. Epidemiol. 36, 299–309 (2021).
Rowlands, A. V. et al. Association between accelerometer-assessed physical activity and severity of COVID-19 in UK Biobank. Mayo Clin. Proc. Innov. Qual. Outcomes 5, 997–1007 (2021).
Mathieu, E. et al. A global database of COVID-19 vaccinations. Nat. Hum. Behav. 5, 947–953 (2021).
Dobrijević, Z. et al. The association of human leucocyte antigen (HLA) alleles with COVID-19 severity: A systematic review and meta-analysis. Rev. Med. Virol. 33, e2378 (2023).
Balogh, G. et al. C > U mutations generate immunogenic peptides in SARS-CoV-2. Zenodo https://doi.org/10.5281/ZENODO.17019282 (2025).
Acknowledgements
The work was supported by the European Union’s Horizon 2020 research and innovation program grant No. 739593 (M.M., B.K, G.M.B, and B.P); the European Union’s Horizon Europe research and innovation program under grant agreement No 101136582 Acronym ID-DarkMatter-NCD (M.M.); the European Research Council ERC-2023-ADG 101142626 FutureAntibiotics (C.P.); the National Research Development and Innovation Office K 146323 (C.P.); the National Laboratory of Biotechnology Grant 2022-2.1.1-NL-2022-00008 (C.P.); the Hungarian Scientific Research Fund grant FK-142312 (M.M.), PD-146654 (B.K.), FK22-142877 (G.J.S.) and 2023-1.1.1-PIACI_FÓKUSZ-2024-00036 (G.J.S.) from the National Research, Development, and Innovation Office (NKFI), Hungary; the National Academy of Scientist Education under the sponsorship of the Hungarian Ministry of Innovation and Technology (FEIF/646-4/2021-ITM_SZERZ), the HUN-REN (TKCS-2024/66; to B.P. and E.A.) and the National Laboratory for Health Security grant RRF-2.3.1-21-2022-00006 (B.P.). G.M.B. was supported by the ÚNKP-22-4 and ÚNKP-23-4, M.M., B.K., and G.J.S. were supported by the ÚNKP-23-5 New National Excellence Program of the Ministry for Culture and Innovation from the source of the National Research, Development and Innovation Fund. N.G. was supported by the KDP-2021 Program (C1764415) of the Ministry for Innovation and Technology from the source of the National Research, Development and Innovation Fund. M.M., B.K. and G.J.S. were supported by the Bolyai János Research Fellowship (BO/00582/22/8) of the Hungarian Academy of Sciences. COVID-19 outcome analysis was performed using data from the UK Biobank Resource under application number 44917.
Author information
Authors and Affiliations
Contributions
G.M.B., E.A., B.P., C.P., and M.M. undertook the conceptualization of the study. G.M.B., B.K., L.A., B.T.P., F.T., B.P., and M.M. were responsible for methodology and computational analysis. B.P., C.P., and M.M. supervised the project. Funding acquisition was carried out by C.P. and M.M. Project administration was carried out by G.M.B. and M.M. Experiments were conducted by N.G. and G.J.S. The manuscript was prepared by G.M.B., E.A., G.J.S., B.P., C.P. and M.M.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Anastasia Meshcheryakova and the other anonymous reviewer(s) for their contribution to the peer review of this work. [A peer review file is available].
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Balogh, G.M., Koncz, B., Asztalos, L. et al. C > U mutations generate immunogenic peptides in SARS-CoV-2. Nat Commun 16, 10156 (2025). https://doi.org/10.1038/s41467-025-65251-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-65251-8