Abstract
The broad-spectrum herbicide L-phosphinothricin (PPT) irreversibly inhibits glutamine synthetase (GS) activity. Here we present cationic amino acid transporters (CAT) as mediators of PPT accumulation and susceptibility. Comparative analysis of Arabidopsis thaliana, Marchantia polymorpha, rice, and Klebsormidium nitens reveals conserved responses to GS inhibition, with absence of efficient PPT uptake underlying resistance in M. polymorpha. Transcriptomic analysis identifies four candidate transporters in A. thaliana which, when overexpressed in M. polymorpha, confer varying levels of sensitivity corresponding with PPT accumulation. AtCAT1 and AtCAT5 confer the greatest sensitivities and are required for glutamic acid uptake and endogenous nitrogen metabolism in A. thaliana, and PPT susceptibility. Molecular dynamics simulation of AtCAT5 identifies key residues involved in PPT binding, which induce the rotational flexibility of helices H1 and H6 which form an intracellular access tunnel. These findings highlight the natural diversity underlying PPT accumulation and susceptibility, which can guide herbicide resistance management strategies.
Similar content being viewed by others
Introduction
Herbicides play a crucial role in modern agriculture, enabling effective weed control and ensuring higher crop yields. Phosphinothricin (PPT), the active compound in herbicides like glufosinate and bialaphos, has become widely utilized due to its broad-spectrum effectiveness, emerging as an alternative to the extensively employed herbicide, glyphosate. With the increasing occurrence of glyphosate resistance, PPT utilization is anticipated to further rise in the coming years, especially with the recent introduction of PPT-resistant crops1. PPT is a glutamate analogue that irreversibly inhibits glutamine synthetase (GS) in plants, disrupting nitrogen metabolism and leading to the accumulation of toxic levels of ammonia and reactive oxygen species2. While enzyme inhibition is the primary mechanism causing cellular damage and plant death, how the herbicide accesses the target enzyme remains unknown.
It is generally understood that incidence of herbicide resistant weeds positively correlates with herbicide usage. However, glufosinate remains an anomaly with comparatively few resistant weed populations despite its long history of use2. Such resistant cases were associated with target site mechanisms like altered expression or non-synonymous mutations in GS3,4. Non-target site resistance mechanisms have yet to be identified, though reported reductions in PPT uptake and PPT-induced injury by amino acid supplementation have implicated the involvement of amino acid transporters in PPT uptake5,6. For example, PPT uptake in Palmer amaranth was inhibited by glutamine but not structurally similar amino acids like glutamate, aspartate, and asparagine5, while partial inhibition by glutamate and alanine was observed in duckweed6. It was hypothesized that PPT uptake occurs through proton co-transporters capable of handling both acidic and neutral amino acids6, though how this hypothetical transporter discriminates between PPT and specific amino acids is unknown. Nonetheless, the varying efficiencies of amino acid competition with PPT uptake between species point to an inherent diversity that can be exploited for the development of resistance traits. Furthermore, loss-of-function mutations in these transporters could theoretically confer resistance, particularly in species with limited transporter redundancy.
This study leverages the evolutionary diversity across four plant clades to investigate the basis of PPT sensitivity: Arabidopsis thaliana (a dicot), rice (Oryza sativa, a monocot), liverwort (Marchantia polymorpha), and Klebsormidium nitens (a filamentous green alga). Through cross-clade comparisons integrating transcriptomic, structural, and physiological data, we first identify, then demonstrate that cationic amino acid transporters (CATs) mediate PPT accumulation and herbicide sensitivity.
Results
Species-specific susceptibility to PPT
Natural diversity hosts novel mechanisms that may otherwise remain hidden when studying model species in isolation. To identify naturally occurring herbicide resistance across clades, we compared the effects of herbicides in two phylogenetically distant plants, A. thaliana and M. polymorpha. Treatment with four herbicides with differing modes of action (Table S1) generally reduced plant size and chlorophyll content (Fig. 1A–C; Fig. S1), though differential retention of chlorophyll between species was observed such as with acetochlor. Notably, PPT demonstrated species-specific toxicity, killing A. thaliana while only slightly reducing plant size in M. polymorpha (Fig. 1A–C; Fig. S1).
A–C Response to 300 µM of acetochlor (Ace), glyphosate (Gly), metsulfuron-methyl (MetSul), or phosphinothricin (PPT) for four days (A. thaliana) and five days (M. polymorpha). Colors in the heatmap represent mean chlorophyll index (B) and plant size (C). Asterisks indicate p-value < 0.05 when compared against the no-herbicide control (Ctrl), as determined by one-way ANOVA and Dunnett’s post-hoc test. n = 3. Raw data is provided in Fig. S1 and source data. D–F Growth of A. thaliana and G–I M. polymorpha in response to three different GS inhibitors. Representative images of A. thaliana (G) grown on a concentration gradient of PPT (●), bialaphos (■), or MetSox (▲) for eight days (A. thaliana) and five days (M. polymorpha). Estimated chlorophyll content (E, H) and plant size (F, I) were quantitated from images using ImageJ. Dose response curve was estimated using a sigmoid function, and the IC50 values are listed in text of the same color. Each data point represents a single biological replicate. n = 6 (A. thaliana) and 4 (M. polymorpha). Source data are provided as a Source Data file.
To assess PPT resistance in M. polymorpha, a dose response curve was generated and compared to that of two other GS inhibitors, bialaphos and methionine sulfoximine (MetSox) (Fig. S2A). Bialaphos is metabolized by the plant to release PPT7, while MetSox is another glutamate analog that irreversibly inhibits GS. A. thaliana was highly susceptible to all three compounds (Fig. 1D–F), with reductions in plant size and chlorophyll content observed from 50 µM, 25 µM, and 5 µM for PPT, bialaphos, and MetSox, respectively. M. polymorpha exhibited similar susceptibility to bialaphos and MetSox (Fig. 1G–I); however, PPT decreased plant size only above 100 µM, and chlorophyll was retained even up to 1 mM (Fig. 1G–I). Similarly, Klebsormidium nitens, a representative streptophytic algae, showed reduced chlorophyll content in response to bialaphos and MetSox but remained unaffected up to 1 mM PPT (Fig. S2B, C). These findings suggest that PPT has reduced effectiveness in non-vascular plants.
Universal and species-specific transcriptomic responses to GS inhibition
Exogenous glutamine inhibits PPT uptake and reduces PPT-induced injury5, indicating the presence of a shared PPT-glutamine transporter and its impact on the glutamine-related metabolic pathway (Fig. S3). To identify potential overlapping feedback mechanisms between exogenous glutamine and PPT, we examined the transcriptomes of A. thaliana, M. polymorpha, rice (Oryza sativa), and K. nitens treated with glutamine, PPT, and MetSox for 48 h, when growth arrest and saturation of PPT uptake occurs5,8. While glutamine produced minimal response in all species, PPT and/or MetSox induced large transcriptomic responses corresponding to known susceptible phenotypes (Fig. 2A; Fig. S4A–D, Data S1 to 3). In angiosperm species, PPT and MetSox generally induced similar changes (Fig. S4A, C; DEG overlaps of 42% and 30% for A. thaliana and rice, respectively, Data S2 and S3).
A Total number of DEGs in each treatment and species. Numbers in black, red, and blue indicate total number, up-, and down-regulated DEGs respectively. List of DEGs provided in data S1. B–E Euler diagram showing overlap of unique DEGs from GS-inhibition response from A. thaliana (B), M. polymorpha (C), rice (D) and K. nitens (E) with public datasets associated with abiotic stress and nutrient deficiency. Data sources and DEG lists are provided in data S5. F, G Upset plots summarising the number of overlapping DEGs, and heatmap of overlap coefficients for up- (F) and down-regulated (G) one-to-one orthologues across treatments in both species. Subsets of DEGs specific to GS inhibition are indicated by the yellow (A. thaliana -specific), green (M. polymorpha -specific) and gray (universal) dots. Heatmap cells marked with asterisks indicate significant overlaps (* = p < 0.05, ** = p < 0.01, calculated using two-sided Fisher’s exact test), while gray cells indicate set size of 0 for one or more datasets. Lists of one-to-one orthologues are provided in data S2 and S3. H, I Heatmap of enriched GO parent terms present in two or more datasets (H), and enriched KEGG metabolic terms (I). Colors indicate −log10(q-value) and -log10(p-value) respectively. Biological replicates n = 3. Gray cells indicate q-value > 0.05 (H) or p-value > 0.05 (I). Lists of enriched GO and KEGG terms are provided in Data S2 and S4. Source data are provided as a Source Data file.
Across all species, the GS-inhibition response significantly overlapped with that of abiotic and nutrient stresses (p ≤ 1.90 × 10-40, Fig. 2B–E), particularly nitrate and phosphate deficiencies (Fig. S5A–D; Fig. S6A–D). Gene ontology (GO) enrichment of these intersections revealed differential responses between species that were specific to GS-inhibition and nutrient deficiency (Fig. S5E, F; Fig. S6E, F), being mainly signaling and photosynthesis-related in A. thaliana (Fig. S5E; e.g. photosynthesis, signaling), both the metabolic (e.g., organic substance biosynthetic process, protein metabolic process) and developmental (e.g., multicellular organism development, developmental process) in rice and M. polymorpha (Fig. S5F; Fig. S6E), and purely metabolic in K. nitens (e.g. biosynthetic process, amino acid metabolic process) (Fig. S6F). Collectively, this suggests a potential diversification in the responses to GS inhibition and nutrient deficiency across the different plant clades.
Closer examination of the GS inhibition response using one-to-one orthologous differentially expressed genes (DEGs) in A. thaliana and M. polymorpha revealed a universal transcriptional response in both up- and down-regulated genes (p ≤ 0.01, Fig. 2F, G, gray dots), which were enriched in GO terms associated with various metabolic, regulatory, developmental processes, and responses to stimuli (Fig. 2H; Fig. S4E, Data S4). Nonetheless, species-specific responses differences were also observed. In MetSox-treated M. polymorpha, the 1857 unique orthologous DEGs (Fig. 2F, G, green dots) were mainly related to developmental processes like system development and tissue development (Data S4). Conversely, A. thaliana-specific orthologues (Fig. 2F, G, orange dots) were predominantly enriched in metabolism-related GO terms like metabolic process and single-organism metabolic process (Data S4). Correspondingly KEGG analysis of all DEGs showed an overlap only between PPT- and MetSox-treated A. thaliana, involving processes like carbon fixation, fatty acid, and amino acid metabolisms (Fig. 2I). Only sulfur metabolism was enriched and specific to M. polymorpha (Fig. 2I), suggesting that unlike A. thaliana, the GS-inhibition response in M. polymorpha is primarily developmental rather than metabolic.
Presence of GS inhibition and absence of PPT uptake in M. polymorpha
Plants possess two GS isoforms: cytosolic GS1 and chloroplastic GS28 (Fig. S7I), the enhanced expression or mutation of which may lead to PPT resistance9. Across multiple species, PPT and MetSox treatments generally altered GS expression (Fig. S7A–H) in a feedback pattern corresponding to known susceptibilities (Fig. 1). Enzyme activity measurements confirmed that PPT inhibited M. polymorpha GS in a dose-dependent manner (Fig. 3A), corresponding with bialaphos’ toxicity to M. polymorpha (Fig. 1); these collectively indicate the absence of GS-related target site resistance mechanisms in M. polymorpha, and PPT uptake as a prerequisite for toxicity.
A Dose response curve of GS for PPT. Crude protein extracts from A. thaliana and M. polymorpha were treated with increasing concentrations of PPT. Reactions were stopped and the absorbance of y-glutamyl hydroxamate used as a measure of GS activity. Data points and error bars reflect mean and standard deviation, respectively. n = 3 biological replicates. Slopes of inhibited activity were analysed by GraphPad7 software to obtain the log10(IC50) values, which are listed in text of the same color with the upper and lower 95% confidence intervals in brackets. B, C Accumulation of PPT (B) and MetSox (C) in A. thaliana and M. polymorpha tissues. A. thaliana was treated for four days. M. polymorpha was treated with bialaphos and MetSox for five days, and PPT for eight days. Each data point represents a single biological replicate. n = 4 for all treatments except MetSox-treated M. polymorpha and PPT-treated A. thaliana, where n = 3. Bars and error bars show the mean and standard deviation. Bialaphos was not detected in any samples. Source data are provided as a Source Data file.
To confirm the contribution of uptake processes to toxicity, the accumulation of PPT, bialaphos, and MetSox in A. thaliana and M. polymorpha was measured. Bialaphos-treated samples showed no traces of bialaphos but had detectable levels of PPT (Fig. 3B), indicating bialaphos metabolization to the active compound in both species. However, while PPT-treated A. thaliana accumulated PPT, only trace levels were found in M. polymorpha (Fig. 3B), confirming that PPT-resistance in M. polymorpha is due to a lack of PPT uptake. To further support the necessity of uptake to toxicity, MetSox was detected in both MetSox-treated A. thaliana and M. polymorpha (Fig. 3C). Collectively, the presented evidence strongly suggests the lack of an efficient PPT uptake pathway in M. polymorpha.
Increased accumulation and susceptibility to PPT in AtCAT-overexpressing M. polymorpha
Competitive inhibition of PPT toxicity (Fig. S3) and uptake5 by glutamine and a few other amino acids implies the presence of a specific PPT-glutamine transporter. The AAP and CAT amino acid transporters, including eight AAP genes (Fig. S8) and nine CAT genes (Fig. S9) in A. thaliana, are involved in transporting glutamine with varying efficiencies10,11. In our transcriptome, glutamine supplementation primarily downregulated four AAP genes and one CAT gene in A. thaliana (Figs. S8A; S9A). In contrast, PPT and MetSox treatments upregulated almost all AAP genes, and both upregulated and downregulated CAT genes (Fig. 4A; Figs. S8A; S9A). Substantial inductions by PPT and MetSox were observed in AtCAT1, AtCAT5, AtAAP4, and AtAAP6 (Fig. 4A: log2(fold change) >1.4, p < 0.01), all of which localize to the plasma membrane and exhibit widespread expression patterns in A. thaliana10,11,12,13. Glutamine treatment also caused significant downregulation in AtCAT5, AtAAP4, and AtAAP6 (Fig. 4A: log2(fold change) <-0.5, p < 0.01). The response of these genes to the treatments suggests possible transcriptional feedback regulations from the treatments and, therefore, a potential role as common PPT/glutamine transporters.
A Heatmap of log2-fold change in A. thaliana and M. polymorpha AAP and CAT family genes. Asterisks indicate adjusted p-value < 0.05 (*) or 0.01 (**) as determined by Wald’s test with Benjamini-Hochberg adjustment, while gray cells indicate no gene expression. Genes highlighted in red were selected for overexpression in M. polymorpha. Gene accessions are provided in Table S2. B, C Growth of Tak-1 and transporter-overexpressing M. polymorpha lines on a concentration gradient of PPT. Plant size (B) was quantitated from images using ImageJ. Dose response curve was estimated using a sigmoid function, and the IC50 values are listed in text of the same color. Each data point represents a single biological replicate. n = 3. Only the phenotypes of selected lines (C) are shown. D Accumulation of PPT in Tak-1 and transporter-overexpressing M. polymorpha after 5 days of 100 µM PPT treatment. Data points, bars, and error bars reflect individual replicates, mean, and standard deviation, respectively. n = 3 biological replicates. The subplot shows time-dependent uptake in Tak-1 and AtCAT5ox-2 treated with 1 mM PPT up to 48 h (n = 8 biological replicates, except for Tak-1 at 6 and 12 h, and AtCAT5ox-2 at 6 and 48 h, where n = 3, 4, 7, and 4, respectively). Relevant data are presented in Fig. S7–S10. Source data are provided as a Source Data file.
To investigate PPT-transporting function of candidate transporters, AtAAP4, AtAAP6, AtCAT1, and AtCAT5 were overexpressed in M. polymorpha. Incubation with 100 µM PPT revealed no deleterious phenotypes in AtAAP6ox lines, while growth inhibition was observed in AtAAP4ox lines 2, 5, 6, and 7, where transgene expression was higher (Fig. S10). In contrast, gemma germination was arrested in all AtCAT1ox and AtCAT5ox lines even at low transgene expression levels (Fig. S10).
The PPT sensitivity of two AtCAT1 and AtCAT5 lines with different transgene expression levels, and the two most sensitive AtAAP4ox lines, AtAAP4ox-2 and AtAAP4ox-5, were then assessed. All selected lines exhibited growth inhibition at concentrations harmless to Tak-1. Plant size and estimated chlorophyll content decreased from 250 µM and 25 µM PPT for AtAAP4ox and AtCAT1ox lines, respectively (Fig. 4B, C; Fig. S11A). AtCAT5ox lines showed the highest susceptibility, with 5 µM PPT reducing plant size and estimated chlorophyll content by up to 80% and 46% respectively (Fig. 4B, C; Fig. S11A). This increased susceptibility was also observed in rockwool-grown AtCAT5ox-2 plants sprayed with 500 µM PPT (Fig. S11B). In AtCAT1ox and AtCAT5ox lines, susceptibility was positively correlated with transgene expression (Fig. S10B). Notably, AtCAT5ox-7 exhibited high PPT susceptibility despite low transgene expression (Fig. 4B, C; Fig. S10), suggesting a higher PPT affinity in AtCAT5 compared to AtCAT1 and AtAAP4. Increased PPT contents in both AtCAT1ox and AtCAT5ox lines confirmed the successful mediation of PPT accumulation by both transporters (Fig. 4D). AtCAT1 and AtCAT5 overexpression also enabled gemma growth with glutamine as the sole nitrogen source, though only AtCAT1 enabled growth with glutamate (Fig. S11C).
Interestingly, PPT was not detected in AtAAP4ox-2, indicating that AtAAP4 is likely a low-efficiency PPT transporter since minor growth inhibition was only observed above 250 µM PPT (Fig. 4C). In AtCAT5ox-2, a time-dependent accumulation in response to 1 mM PPT treatment showed an estimated accumulation rate of 0.27 ng PPT/mg of tissue/h during the first 12 h that began to plateau around 5.67 ng PPT/mg of tissue by 48 h (Fig. 4D). Low PPT accumulation was also observed in Tak-1 at approximately 0.6 ng PPT/mg of tissue (Fig. 4D), consistent with the slight susceptibility observed at 1 mM PPT (Fig. 1G–I), implying the potential presence of a low affinity transporter in M. polymorpha.
Partial PPT resistance in AtCAT1, 5, and 8 triple knockout plants
As AtCAT1 and AtCAT5, together with AtCAT8, form a distinct sub-branch in the phylogeny of plant CAT transporters (Fig. S9H) and exhibit considerable overlap in substrate range and localisations11,14, knockout of the branch or its members may hypothetically increase PPT resistance. cat5-ko and cat1,5,8-ko lines indeed exhibited higher estimated chlorophyll contents and plant sizes between 25 to 50 µM PPT (Fig. 5A–C). PPT uptake was reduced in cat1,5,8-ko, though not completely abolished. A similar, though non-significant, trend was also observed in cat1- and cat5-knockouts (Fig. 5D), suggesting that these CAT transporters contribute to PPT accumulation, alongside possible compensation by other transporters such as AAP4.
A–C Growth of cat mutants in response to a PPT concentration gradient. Representative images of plants (A) after eight days of treatment. Estimated chlorophyll content (B) and plant size (C) were quantitated from images using ImageJ. Dose response curves were estimated using a sigmoid function and the IC50 values are listed in text of the same color. Each data point represents a single biological replicate. n = 24 for Col-0 and #7-1, except for Col-0 at 0, 50, and 100 µM. n = 12 for Salk207473. n = 6 for #8-4 and #8-5. D PPT accumulation in cat mutants after 24 h of treatment. Data points, bars, and error bars reflect individual biological replicates, mean, and standard deviation, respectively. Asterisks indicate significant differences from Col-0, as determined by one-way ANOVA and Dunnett’s post-hoc test. n = 4. E–G Basal transcriptomic profile of cat mutants showing number of dysregulated genes (E) and the enriched KEGG (F) and GO terms (G). GO terms shown are those associated with the top 20 q-values from each genotype. n = 3 biological replicates. Source data are provided as a Source Data file.
To understand the physiological role of the AtCAT1,5,8 phylogenetic branch, we examined the basal transcriptional profiles of the different cat mutants. As AtCAT5 and AtCAT8 are phylogenetically close (Fig. S9) and likely share similar functions, a double mutant was used as a representative of loss of function in that sub-branch. All genotypes exhibited considerable dysregulation, with an additive effect observed when AtCAT1, 5, and 8 were knocked out (Fig. 5E, Fig. S12A–C). Similar transcriptional perturbations of various stress responses and metabolic processes were also observed (Fig. 5E–G). Examination of metabolic KEGG terms revealed common enrichment of photosynthesis – antenna proteins (Fig. 5F), suggesting dysregulation of the light harvesting mechanism across all mutants. However, various terms associated with core pathways like energy metabolism and amino acid metabolism were enriched only in cat1,5,8-ko, the dysregulation of which were also reflected in GS and AA transporter expression (Fig. 5F, Fig. S12D–F). While this alludes to the role of the collective AtCAT1,5,8 phylogenetic branch to those processes, the absent enrichment of these terms in cat1-ko and cat5,8-ko also imply functional redundancy.
Structural changes in AtCAT5 upon PPT binding
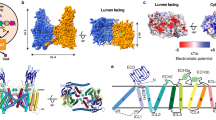
CAT transporters belong to the Solute Carrier (SLC) 7 family. Conserved in animals, plants, and bacteria, they contain 12 to 14 transmembrane helices and mediate transport of cationic amino acids across cellular membranes15,16. To determine if a similar mechanism underlies PPT and amino acid transport, a structural model of AtCAT5 was generated using AlphaFold2 and subjected to molecular dynamics simulations of 450 ns (Fig. 6A, B). Despite low sequence similarity (Identity: 27.4%, Similarity: 45.0% between AtCAT5 and GkApcT), similar manners of docking were observed between PPT in AtCAT5, and Arg and Ala in the bacterial amino acid transporter, GkApcT16 (Fig. S13). PPT-interacting residues in AtCAT5 were predominantly located in helices H1 and H6. The carboxyl group of PPT consistently formed stable polar interactions with the backbone amino groups of Gly83, Ala84, and Gly85 in H1, while interactions with the amino group of PPT were observed with Val81 from H1 and Phe269, Ala270, Gly272, and Gly273 from H6 throughout the 450 ns simulation (Fig. 6C; Fig. S13A). Like the proposed transportation mechanisms of GkApcT, an opening of the cytoplasmic tunnel was observed in AtCAT5 after 450 ns (Fig. 6B). Specifically, two adjacent helices, H1A and H6B, respectively rotated 17.8° and 17.5° from their initial positions (Fig. 6D; Fig. S14), resulting in a widening of the cytoplasmic tunnel and successful passage of PPT (Fig. S14).
A, B Surface-rendered representation of AtCAT5 at 0 (A) and 450 ns (B) showing the opening of the cytoplasmic access tunnel. PPT molecule is presented as a red ball figure. C Protein contact atlas showing weighted interaction between PPT and specific AtCAT5 residues throughout the 450 ns simulation. Edge thickness represents interaction weight. Residues G272 and G273, emphasized in dark red, are selected based on the weighted interaction metrics, pocket location, and evolutionary conservation presented in Table S6 and Fig. S13. D, E Positional changes of helices H1 (blue), H3 (gray), H6 (red), and H8 (yellow) in AtCAT5 (D) and AtCAT5-G272W/G273W mutant (E). Structures at 450 ns (solid) are superimposed over their 0 ns counterparts (translucent). The directions of major rotational changes are indicated by black arrows and the corresponding angles provided. F Distance between helices within AtCAT5 and AtCAT5-G272W/G273W mutant. Values presented are the mean distances of the first and last 20 ns of the 450 ns simulation, with distance calculated as the mean of all pairs from two sets. Relevant data are presented in Fig. S13–S14. G Response of Tak-1, AtCAT5ox lines, and AtCAT5-G272W/G273Wox lines to PPT. Additional data presented in Fig. S15. Source data are provided as a Source Data file.
The rotation and structural changes in H6B that enabled PPT passage appeared to be facilitated by Gly272 and Gly273, two consecutive glycine residues located between H6A and H6B (Fig. 6C, D). To test this hypothesis, the two glycine residues were mutated to tryptophan and molecular dynamics simulations conducted on the resulting AtCAT5-G272W/G273W protein. The mutant protein exhibited comparable affinity to PPT (AtCAT5: 6.33 ± 0.08 kcal mol-1, AtCAT5-G272W/G273W: 6.53 ± 0.08 kcal mol-1) throughout the 450 ns simulation. However, no directional movements of H1A and H6B were seen (Fig. 6E, F; Fig. S14, S16), suggesting that Gly272 and Gly273 are critical in enabling the conformational changes of H6B that could facilitate PPT passage. This was confirmed in AtCAT5-G272W/G273Wox plants which showed PPT resistance comparable to Tak-1 (Fig. 6G, Fig. S15).
Discussion
Herbicide uptake into the cellular space is major determinant of herbicide toxicity. While the molecular targets of many herbicides are well characterized, their cellular entry routes remain poorly understood. Here, we present evidence of mediation of PPT accumulation through amino acid transporters and highlight the potential mechanism through which transport occurs. In this study, the glutamate analogue PPT revealed distinct sensitivity patterns between flowering plants (A. thaliana and rice) and early-diverging lineages (M. polymorpha and K. nitens), with resistance in M. polymorpha attributable to the absence of PPT uptake (Fig. 3). This resistance was reversed by expressing A. thaliana amino acid transporters, leading to the identification of AtCAT1 and AtCAT5 as mediators of PPT accumulation.
CAT transporters, unlike broad-specificity permeases, recognize amino acids with high specificity via electrostatic interactions between conserved polar or charged residues and the substrate’s backbone or side chain16. PPT mimics this backbone and, through its phosphinic acid group, forms stable interactions with these key residues (Fig. 5). Structural modeling of AtCAT5 suggested that PPT binds via a similar mechanism to the bacterial transporter GkApcT16, inducing rotational movements in helices H1A and H6B, and opening an access tunnel to the cytoplasm (Fig. 5A). Tunnel size is regulated by the flexibility of H6B, determined by hinge residues Gly272 and Gly273 (Fig. 5A, D, E; Fig. S14), suggesting substrate selectivity may be enforced via size exclusion mechanisms at both the binding pocket and access tunnel. Mutations at these sites can reduce PPT binding and confer resistance (Fig. S15), but often compromise essential amino acid transport, imposing strong evolutionary constraints.
These functional constraints are paralleled at PPT’s target enzyme, GS. PPT resistance in angiosperms is rare and typically requires compensatory adaptations, such as transcriptional upregulation9,17,18 or a Ser59Gly substitution that enhances catalytic efficiency4. There are no known naturally occurring PPT-resistant GS, though artificially induced mutations have been generated19. The structural mimicry of glutamate by PPT, and its recognition by essential sites in both transporter and enzyme, likely explains the low incidence of PPT resistance in angiosperms despite extensive agricultural use20.
Further limiting the evolution of PPT resistance is the extensive expansion of transporter gene families in angiosperms. Both CAT and AAP transporters have undergone substantial diversification (Fig. S8G, S9H; S8A–E, S9A–G), with gene redundancy especially pronounced in angiosperms due to widespread duplication events associated with increased anatomical complexity and the need for tissue-, cell-, and subcellular-specific expression11,13. For example, while K. nitens and M. polymorpha encode only two and five CAT genes, respectively, A. thaliana and rice possess nine and seven. This redundancy reduces the likelihood of single gene mutations blocking all PPT uptake routes, decreasing the probability of resistance. Consistent with this, A. thaliana cat mutants retained partial PPT sensitivity (Fig. 5A–D), supporting the involvement of multiple transporters in mediating uptake and accumulation.
At the physiological level, GS inhibition depletes cellular glutamate and glutamine, simulating nutrient deficiency and triggering broad metabolic reprogramming2. Transcriptomic responses to GS inhibition were conserved across species and resembled those observed during nitrate starvation (Fig. S5, S6), suggesting that the regulatory network involving GS, glutamate, and glutamine – central to primary metabolism and transcriptional feedback – predates terrestrialization. Beyond core metabolic changes, M. polymorpha, A. thaliana, and rice showed broader transcriptional shifts associated with development and inter-organ communication (Fig. S5E–F, S6E–F). These land plant–specific responses likely reflect evolved mechanisms to coordinate root nutrient uptake and restrict shoot growth under stress. Tissue-specific regulation may also influence transporter expression: in A. thaliana and rice, GS inhibition led to upregulation of CAT genes, potentially promoting amino acid remobilization as an alternative nitrogen source. In contrast, M. polymorpha showed no CAT induction under GS inhibition (Fig. S9) or nitrate deficiency21, and was unable to efficiently use glutamate or glutamine as sole nitrogen sources (Fig. 11C), implying that amino acid–based nitrogen uptake played a lesser role in early-diverging lineages.
Historically, herbicide resistance mechanisms have been identified through studies of resistant weed populations, limiting insight to existing germplasm and traits. This study demonstrates how naturally occurring resistance across diverse evolutionary clades can provide mechanistic insights into herbicide uptake and action, while avoiding the confounding effects of gene redundancy. The shared uptake mechanism between PPT and endogenous amino acids (Fig. 5) likely contributes to PPT’s broad-spectrum activity and the rarity of evolved resistance. Variable PPT accumulation among resistant weeds22 further suggests that CAT transporters generally do not discriminate between PPT and amino acids. The essential nature of amino acid transport and accumulation, and the limited mutational tolerance of CAT functional residues, together constrain resistance evolution and help explain PPT’s long-term efficacy.
Methods
Plant growth conditions and determination of herbicide sensitivity in plants
Arabidopsis thaliana ecotype Col-0, Marchantia polymorpha accession Tak-1, and Klebsormidium nitens strain NIES-2285 were used as reference strains in this study. Surface-sterilized A. thaliana seeds were grown on ½ MS Basal media (0.5% sucrose, 0.05% MES, pH 5.7, 1% agar) for eight days. M. polymorpha gemma were raised for 10 days on either solid (1% agar) or liquid ½ B5 media (0.05% MES, pH 5.5). K. nitens was grown on solid Yamagami media23 under low light (approximately 3.69 µmol m-2 s-2). All three species were grown at 22 °C, 24 h light.
Herbicide sensitivity was determined by transferring plants to media supplemented with various concentrations of PPT, bialaphos, or MetSox. For A. thaliana, treatment was carried out in 24-well plates for four days. For M. polymorpha, treatment was carried out in 12-well plates for eight days (PPT) and five days (bialaphos and MetSox). For K. nitens, herbicide-containing media was inoculated with 5 µl of cell suspension. Treatment was carried out in 24-well plates for eight days. Plant size and estimated chlorophyll content was then assessed using ImageJ24. For experiments conducted on rockwool, seventeen-day old M. polymorpha plants established on rockwool imbibed with liquid ½ B5 media were sprayed with 1 ml of either water or 500 µM PPT, and the response observed after seven days.
For amino acid (AA) supplementation experiments, A. thaliana plants were grown on media containing 100 µM PPT and 5 mM AA. Prior to use, AA solutions were adjusted to pH 5.5 and filter-sterilized. Treatment was performed for seven days.
Quantification of herbicide uptake
A. thaliana seedlings and M. polymorpha gemmalings pre-cultured in liquid media were treated with 100 µM of PPT, 25 µM of bialaphos, or 5 µM of MetSox. Concentrations were based on the half-maximal inhibitory concentration (IC50) values determined in the previous section. Treatment durations are also as described in the previous section. For the comparison of M. polymorpha wildtype and overexpression lines, plants were treated for only 48 h. For comparison of A. thaliana cat mutants, plants were treated for only 24 h. Harvested plants were rinsed multiple times with sterile MilliQ water, blotted dry, then snap-frozen in liquid N and homogenized. Herbicide extraction was performed using the method described by Takano et al. 5. Briefly, 1 mL of methanol:water (75:25) was added to 40 mg of ground tissue. The mixture was vortexed vigorously for one minute, sonicated for 30 mins, then centrifuged at 4000 x g for 5 mins. The supernatant was filtered through a 0.22 µm nylon filter and analysed by liquid chromatography–tandem mass spectrometry (LC–MS/MS) using either an Agilent 6546 LC-QTOF with a Shodex Asahipak NH2P-50 4E column (4.6 ×250 mm), or Agilent 6400 QQQ with a Waters Anionic Polar Pesticides column, both with flow rates of 0.5 mL min-1. The program for the former is as follows: linear gradient of water (A) and 5% methanol and 20 mM ammonium carbonate (B): 5 min, 0% A; 10 min, 100% A; 25 min, 100% A; 26 min, 0% A. The program for the Agilent 6400 QQQ is as follows: linear gradient of 1% formic acid(A) and acetonitrile with 1% formic acid (B): 0 min, 10% A; 4 min, 85% A; 13 min, 85% A; 18.5 min, 10% A. All data was acquired in negative polarity. Quantification was performed using external standard curves and compounds identified via the respective precursor and product ions (table S3).
Preparation of crude protein extract
Plant material was snap frozen and milled to a fine powder with a mortar and pestle. Extraction buffer (100 mM Tris-HCl pH 7.4, 1 mM MgCl2, 1 mM EDTA, 10 mM β-mercaptoethanol) was added to the sample to a ratio of 10:1 and the suspension filtered with Miracloth. The filtrate was centrifuged at 20,000 x g for 30 mins. The supernatant was collected, and ammonium sulfate added to a concentration of 60% and incubated overnight with shaking at 4 °C. The sample was then centrifuged at 17,000 x g for 5 mins. The resulting pellet was resuspended in Resuspension buffer (10 mM Tris-HCl pH 7.4, 1 mM MgCl2, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride). Ammonium sulfate concentration was reduced using a Merck-Millipore Protein Concentrator ( ~ 30 kDa) and the protein was resuspended with GS Assay buffer.
Determination of GS activity
A 25 µl aliquot of crude protein extract was mixed with 25 µl of GS assay buffer (50 mM Tris-HCl pH 7.4, 50 mM sodium L-glutamate pH 7.4, 125 mM hydroxylamine-HCl pH 7.4, 20 mM ATP, 10 mM MgCl2, 1 mM β-mercaptoethanol), with PPT added to concentrations ranging from 0 to 1 mM. The reaction was incubated at room temperature for 6 h, then terminated by addition of 50 µl acidified ferric chloride (0.2 mM FeCl3, 0.4 mM HCl, 0.12 mM trichloroacetic acid). Samples were centrifuged at 15,000 x g for 1 min. Concentrations of γ-glutamyl hydroxamate levels were measured at absorbance of 535 nm and used as a measure of GS activity.
RNA sequencing and orthogrouping
A. thaliana seedlings, M. polymorpha gemmalings, and K. nitens cells were pre-cultured for eight, ten, and seven days, respectively in liquid media, then treated with 5 mM glutamine, 100 µM of PPT, or 5 µM of MetSox for 48 h. For rice (Oryza sativa ssp. Japonica cv Nipponbare), mature leaf sections were floated abaxial side down in ½ MS solutions containing the treatments for 48 h. RNA was extracted using the RNeasy Plant Mini Kit (Qiagen). Library preparation, including polyA enrichment and sequencing, was performed on the NovaSeq platform at Novogene, Singapore. Raw sequences were mapped to reference genomes (A. thaliana TAIR v10, M. polymorpha v5.1, O. sativa Phytozome v7, K. nitens v1.1) using STAR v2.5.3.
Differentially expressed genes (DEGs) were identified via the R package DESeq225, using a pairwise comparison to the expression of the untreated control with a threshold False Discovery Rate (FDR) < 0.01. Log2-fold change in gene expression was extracted using the “lfcShrink apeglm” method26.
The complete proteomes of A. thaliana and M. polymorpha were obtained from The Arabidopsis Information Resource (TAIR) and MarpolBase webservers respectively. Orthogroups were constructed from sequences of primary transcripts using OrthoFinder v2.5.4 with default settings, and one-to-one orthologues between both species identified.
Analysis of publicly available datasets
DEG lists from studies on various stresses were obtained from previous publications or the Gene Expression Omnibus (GEO)27. Microarray data from GEO were analysed using GEO2R and DEGs identified using a threshold of FDR < 0.05. Datasets were included only if materials were of the same genotype, and similar tissue type and developmental age. A total of six A. thaliana, seven M. polymorpha, four rice, and four K. nitens datasets were used. Conditions in each study and list of identified DEGs are listed in data S5.
Functional profiling of DEGs
GO and KEGG annotations were obtained from TAIR and MarpolBase. Gene ontology (GO) term enrichment and annotation was performed using PlantRegMap28, using the threshold value of p < 0.01. Redundant terms were removed via REVIGO29; where term reduction was performed, a cutoff value of 0.5 was used. KEGG enrichment of metabolic genes was performed using ClusterProfiler (v4.8.3)30. Statistical significance in the overlaps between DEGs, one-to-one orthologues, GO terms, or KEGG terms were calculated using two-sided Fisher’s exact test and visualized using the eulerr (v 7.0.0)31 and pheatmap (v1.0.12) R packages.
Phylogenetic analysis of GS and transporters
Lists of A. thaliana, M. polymorpha, and rice AAP genes were obtained from existing literature32,33, while K. nitens AAP genes were identified from Interpro using Pfam annotation PF01490. GS and CAT genes were identified using Pfam annotations PF00120, and PF13520 and PF13906, respectively. All protein sequences for A. thaliana, M. polymorpha, rice, and K. nitens were obtained from TAIR, MarpolBase, Phytozome, and Interpro respectively. Only primary transcripts were selected for further analysis.
Multiple sequence alignment was performed using MAFFT34 and duplicate sequences (100% identity) were removed. A maximum likelihood tree was constructed using the LG substitution matrix, with the optimization of tree topology, branch lengths, and rate parameters with 500 bootstrap iterations. Positions with less than 80% site coverage were removed before phylogenetic analysis.
Generation of A. thaliana AtCAT knockout mutants
The A. thaliana AtCAT knockout mutants CS924659 and SALK_207473 were obtained from ABRC. CRISPR mutants were generated through floral dip transformation of A. thaliana (Col-0) with Agrobacterium tumefaciens strain GV3101 carrying the pHSE40135 or pDU binary vectors, each containing four sgRNA sequences. The pDU vector was constructed and synthesized by VectorBuilder (www.vectorbuilder.com), with sgRNA expression driven by the A. thaliana AtU6-26 promotor and a plant codon-optimized Cas9 driven by CaMV 35S promoter. sgRNA target sequences and resulting mutations in the respective lines generated are listed in table S4. T1 plants were selected on 25 mg/L hygromycin and mutations confirmed via sequencing of target genomic regions from the T2 generation onwards. For analysis of the basal transcriptome and PPT accumulation, the lines CS924659, #12-6, and #8-5 were used as representatives for cat1, cat5,8, and cat1,5,8mutants respectively.
Generation of transporter overexpression M. polymorpha lines
The coding sequences of AtAAP4 (AT5G63850), AtAAP6 (AT5G49630), AtCAT1 (AT4G21120), and AtCAT5 (AT2G34960) were amplified from A. thaliana (Col-0) cDNA and inserted into the pMpGWB303 binary vector36. Expression of all transgenes were driven by the constitutive MpEF1α promoter. Plasmids were transformed into M. polymorpha (Tak-1) thalli using Agrobacterium tumefaciens strain GV2260, and transformants screened on ½ B5 media containing 100 µg/mL cefotaxime and 0.5 µM chlorsulfuron. Transgene expression of successful transformants was measured via qRT-PCR at the T1 generation. All physiological assays were performed using the T1 and older generations.
RNA extraction, cDNA synthesis and qRT-PCR
Total RNA extraction from plant material was performed using the QIAGEN RNeasy Plant Mini kit. cDNA was synthesized using the GoScript Reverse Transcription System (Promega). qRT-PCR was performed using the SYBR Green premix kit (Kapa Biosystems), using a reaction volume of 10 µL and two technical replicates per sample. Samples were run on a Bio-Rad CFX Connect Real Time PCR Detection System using the following program: initial denaturation at 95 °C for 10 mins, and forty cycles of 95 °C for 15 mins and 60 °C for 1 min. Gene expression was analyzed using the CFX Manager v.3.1 (Bio-Rad) and normalized to M. polymorpha ACTIN 7 (MpACT7). Primer sequences are provided in table S5.
Molecular dynamics simulation
Initial structures for AtCAT5 and the AtCAT5 G272W/G273W mutant were generated using AlphaFold237 and ColabFold38 respectively. Using the previously resolved GkApcT structure (PDB 6F34)16 as a reference, docking simulations and energy minimization were performed via the Protein+ server39 and Yasara40 respectively to obtain an inward-occluded state docked structure. Protein/membrane systems were created using CHARMM-GUI41, consisting of a lipid bilayer (DUPC:SITO:CER160, 5:5:1 ratio)42 solvated with TIP3P water supplemented with NaCl at 0.15 M using the Monte-Carlo method. The systems were subjected to 450 ns simulations with the CHARMM36 force field43 on GROMACS v2022.444. Electrostatics were treated with PME, a constant temperature of 295.15 Kelvin maintained, and a 2 fs time step used. Hydrogen bonds were constrained, and lipid bilayer restraints applied. Subsequent data analysis and representation were conducted using Visual Molecular Dynamics (VMD)45. Helices (H1A: 70-82, H3: 143-176, H6B: 273-280, H8: 340-372) were precisely defined through structural alignment with GkApcT16. Distances between specific helices at 0 ns and 450 ns were determined by computing the average distances between corresponding residues in the two structures. Three dimensional coordinates for backbone atoms, α-carbon, carbonyl-carbon, oxygen and nitrogen, were used to calculate the mean distance. To assess the distances between different helices, the average distances were calculated from all possible pairs of backbone atoms where the first atom from one helix and the second atom from another helix. Angles between helices were computed using the AngleBetweenHelices function in Pymol46. Interaction weights of residues were calculated by counting the frames where any atom of a residue came within a 5.0 Å distance of any atom of the PPT ligand; results were then visualized using Cytoscape v3.9.147. Protein 3D models were generated using ChimeraX48, Blender v3.649, and Pymol v2.4.1.
Statistics and reproducibility
Statistical analysis of data was performed as described in the specific sections. No sample size calculations were performed. All physiological and enzymatic experiments were performed at least twice with a minimum of three biological replicates to confirm reproducibility.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Gene accessions and sequences are listed in the supplementary materials. RNA-sequencing data and processed data generated in this study have been deposited at NCBI GEO under the accession code GSE245166, and at ArrayExpress under the accession codes E-MTAB-15888, E-MTAB-15889, and E-MTAB-15892. Source data are provided with this paper.
References
USDA; GAIN. China: Agricultural Biotechnology Annual. https://fas.usda.gov/data/china-agricultural-biotechnology-annual-10 (2023).
Takano, H. K. & Dayan, F. E. Glufosinate-ammonium: A review of the current state of knowledge. Pest Manag. Sci. 76, 3911–3925 (2020).
Carvalho-Moore, P. et al. Unraveling the mechanism of resistance in a glufosinate-resistant Palmer Amaranth (Amaranthus palmeri) accession. Weed Sci. 70, 370–379 (2022).
Zhang, C. et al. A naturally evolved mutation (Ser59Gly) in glutamine synthetase confers glufosinate resistance in plants. J. Exp. Bot. 73, 2251–2262 (2022).
Takano, H. K., Beffa, R., Preston, C., Westra, P. & Dayan, F. E. Physiological factors affecting uptake and translocation of glufosinate. J. Agric. Food Chem. 68, 3026–3032 (2020).
Ullrich, W. R., Ullrich-Eberius, C. I. & Köcher, H. Uptake of glufosinate and concomitant membrane potential changes in Lemna gibba G1. Pestic. Biochem. Physiol. 37, 1–11 (1990).
Ujváry, I. Chapter 3 - Pest Control Agents from Natural Products. in Hayes’ Handbook of Pesticide Toxicology (ed. Krieger, R. B. T.-H. H. of P. T. (Third E.) 119–229 (Academic Press, 2010). https://doi.org/10.1016/B978-0-12-374367-1.00003-3.
Abdeen, A. & Miki, B. The pleiotropic effects of the bar gene and glufosinate on the Arabidopsis transcriptome. Plant Biotechnol. J. 7, 266–282 (2009).
Noguera, M. M. et al. Involvement of glutamine synthetase 2 (GS2) amplification and overexpression in Amaranthus palmeri resistance to glufosinate. Planta 256, 57 (2022).
Fischer, W.-N. et al. Low and high affinity amino acid H+-cotransporters for cellular import of neutral and charged amino acids. Plant J. 29, 717–731 (2002).
Su, Y.-H., Frommer, W. B. & Ludewig, U. Molecular and functional characterization of a family of amino acid transporters from Arabidopsis. Plant Physiol. 136, 3104–3113 (2004).
Koh, E., Goh, W., Julca, I., Villanueva, E. & Mutwil, M. PEO: Plant Expression Omnibus – a comparative transcriptomic database for 103 Archaeplastida. Plant J. n/a, (2023).
Yang, H., Postel, S., Kemmerling, B. & Ludewig, U. W. E. Altered growth and improved resistance of Arabidopsis against Pseudomonas syringae by overexpression of the basic amino acid transporter. Plant. Cell Environ. 37, 1404–1414 (2014).
Yang, H., Bogner, M., Stierhof, Y.-D. & Ludewig, U. H+-independent glutamine transport in plant root tips. PLoS One 5, e8917 (2010).
Wipf, D. et al. Conservation of amino acid transporters in fungi, plants and animals. Trends Biochem. Sci. 27, 139–147 (2002).
Jungnickel, K. E. J., Parker, J. L. & Newstead, S. Structural basis for amino acid transport by the CAT family of SLC7 transporters. Nat. Commun. 9, 550 (2018).
Xia, J.-Q. et al. Knockout of OsSPL10 confers enhanced glufosinate resistance in rice. Plant Commun. 5, 100731 (2024).
Xia, J.-Q. et al. Loss of OsARF18 function confers glufosinate resistance in rice. Mol. Plant 16, 1355–1358 (2023).
Ren, B. et al. Three novel alleles of OsGS1 developed by base-editing-mediated artificial evolution confer glufosinate tolerance in rice. Crop J. 11, 661–665 (2023).
Heap, I. The International Herbicide-Resistant Weed Database. www.weedscience.org.
Krishnamoorthi, S. et al. Hyperspectral imaging of liverwort Marchantia polymorpha identifies MpWRKY10 as a key regulator defining foliar pigmentation patterns. Cell Rep. 43, 114463 (2024).
Takano, H. K., Beffa, R., Preston, C., Westra, P. & Dayan, F. E. Reactive oxygen species trigger the fast action of glufosinate. Planta 249, 1837–1849 (2019).
Shikanai, Y. et al. Arabidopsis thaliana PRL1 is involved in low-calcium tolerance. Soil Sci. Plant Nutr. 61, 951–956 (2015).
Liang, Y. et al. A nondestructive method to estimate the chlorophyll content of Arabidopsis seedlings. Plant Methods 13, 26 (2017).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Zhu, A., Ibrahim, J. G. & Love, M. I. Heavy-tailed prior distributions for sequence count data: Removing the noise and preserving large differences. Bioinformatics 35, 2084–2092 (2019).
Barrett, T. et al. NCBI GEO: Archive for functional genomics data sets — update. Nucleic Acids Res 41, D991–D995 (2013).
Tian, F., Yang, D.-C., Meng, Y.-Q., Jin, J. & Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res 48, D1104–D1113 (2020).
Supek, F., Bošnjak, M., Škunca, N. & Šmuc, T. REVIGO summarizes and visualizes long lists of Gene Ontology terms. PLoS One 6, e21800 (2011).
Wu, T. et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innov 2, 100141 (2021).
Larsson, J. eulerr: Area-Proportional Euler and Venn Diagrams with Ellipses. at https://cran.r-project.org/package=eulerr (2022).
Tegeder, M. & Ward, J. M. Molecular evolution of plant AAP and LHT amino acid transporters. Front. Plant Sci. 3, 21 (2012).
Zhao, H., Ma, H., Yu, L., Wang, X. & Zhao, J. Genome-wide survey and expression analysis of amino acid transporter gene family in rice (Oryza sativa L.). PLoS One 7, e49210 (2012).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30, 3059–3066 (2002).
Xing, H.-L. et al. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327 (2014).
Ishizaki, K. et al. Development of Gateway binary vector series with four different selection markers for the liverwort Marchantia polymorpha. PLoS One 10, e0138876 (2015).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Mirdita, M. et al. ColabFold: Making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Schöning-Stierand, K. et al. ProteinsPlus: A comprehensive collection of web-based molecular modeling tools. Nucleic Acids Res 50, W611–W615 (2022).
Land, H. & Humble, M. S. YASARA: A tool to obtain structural guidance in biocatalytic investigations BT - Protein Engineering: Methods and Protocols. in (eds. Bornscheuer, U. T. & Höhne, M.) 43–67 (Springer New York, 2018). https://doi.org/10.1007/978-1-4939-7366-8_4.
Wu, E. L. et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 35, 1997–2004 (2014).
Emami, S. et al. Molecular dynamics simulations of ternary lipid bilayers containing plant sterol and glucosylceramide. Chem. Phys. Lipids 203, 24–32 (2017).
Huang, J. et al. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017).
Kutzner, C. et al. GROMACS in the cloud: A global supercomputer to speed up alchemical drug design. J. Chem. Inf. Model. 62, 1691–1711 (2022).
Humphrey, W., Dalke, A. & Schulten, K. V. M. D. Visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Schrödinger, L. The PyMOL Molecular Graphics System. at (2015).
Shannon, P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 (2003).
Pettersen, E. F. et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Bruns, N. Blender. Unfallchirurg 123, 747–750 (2020).
Acknowledgements
We would like to thank Dr Yuan Cheng-Hui and the NUS Chemical, Molecular and Materials Analysis Centre (CMMAC) for their assistance with herbicide quantification. This study was supported by the Agency for Science, Technology and Research (A*STAR) Singapore under the industry alignment fund pre-positioning program; High Performance Precision Agriculture system (A19E4a0101), and by the National Research Foundation (NRF) Singapore under its Campus for Research Excellence and Technological Enterprise (CREATE) program. Both grants were granted to DU.
Author information
Authors and Affiliations
Contributions
Conceptualization: G.Z.H.T., D.U., Methodology: G.Z.H.T., D.U. Investigation: G.Z.H.T., H.Y.K.K., C.C.O., Z.Y.P., Y.T., K.G., S.K. Visualization: G.Z.H.T., C.C.O., D.U. Funding acquisition: D.U. Project administration: D.U. Supervision: D.U., J.R.B. Writing – original draft: G.Z.H.T., D.U. Writing – review & editing: C.C.O., S.K., Z.Y.P., K.G., and Y.T.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Joshua Mylne and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tan, G.Z.H., Koh, H.Y.K., Poh, Z.Y. et al. Cationic amino acid transporters (CAT) enhance accumulation and susceptibility to the systemic herbicide L-phosphinothricin. Nat Commun 17, 146 (2026). https://doi.org/10.1038/s41467-025-66840-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66840-3