Abstract
Vascular endothelial cells (ECs) play pivotal roles in maintaining metabolic tissue homeostasis, and EC dysfunction is associated with obesity and metabolic disorders. The mammalian ER stress sensor IRE1α kinase/RNase responds to metabolic cues, but it remains unclear whether endothelial IRE1α is implicated in controlling systemic metabolism. Here we show that genetic depletion of IRE1α in ECs leads to maladaptation of pancreatic islets under obesity-associated metabolic stress. We find that in high-fat diet-fed male mice, loss of IRE1α in ECs has no significant impact upon adiposity, but unexpectedly results in glucose intolerance with impaired insulin secretion, accompanied by blunted intra-islet angiogenesis and compensatory islet growth. Mechanistically, IRE1α RNase decays the mRNA encoding the endogenous anti-angiogenic factor thrombospondin-1 (THBS1/TSP1) in islet ECs. These findings thus uncover a critical role of the endothelial IRE1α suppression of THBS1 in governing the vascular support that enables the functional adaptation of islets to metabolic stress.
Similar content being viewed by others
Introduction
Vascular endothelial cells (ECs) form a highly plastic and dynamic monolayer that interfaces circulating blood and tissues, and they can readily shift from dormant to angiogenic states during tissue revascularization according to physiologic demands1. ECs play pivotal roles in tissue homeostasis via governing oxygen and nutrient supply, hormone transport, immune cell trafficking, and waste disposal between blood and perfused organs. To meet specific local needs and functions, ECs exhibit heterogeneous structures and phenotypes in different organs/tissues2,3. Recent single-cell transcriptomics studies have also revealed remarkable heterogeneity and molecular signatures of tissue-specific ECs, reflecting their local functional characteristics under the influence of specific tissue microenvironments4,5. In highly vascularized metabolic organs like adipose tissue, ECs are being recognized as critical gatekeepers and active regulators in the systemic control of metabolism via adjusting tissue-specific transport of nutrients such as glucose and fatty acids as well as hormones like insulin6,7,8. Constantly exposed to various circulating factors and metabolites, ECs can be viewed as energy/nutrient sensors and are susceptible to metabolic perturbations and pathogenic stimuli, which may lead to disruption of their angiocrine interactions with other cell types. Indeed, emerging evidence indicates that EC dysfunction is closely associated with metabolic dysregulation that underlies obesity and related metabolic conditions such as type 2 diabetes, cardiovascular diseases and steatohepatitis9,10. For instance, angiogenesis is a critical contributor in supporting adipose tissue expansion remodeling and its metabolic function in response to environmental changes such as nutrient excess, while impaired angiogenic capacity and decreased vascularity are thought to promote adipose inflammation and dysfunction in obesity8,11,12,13. Moreover, multiple mechanisms have been proposed to underlie the pathological EC dysfunction in various disease settings, including hypoxia, oxidative stress, and endoplasmic reticulum (ER) stress as well8,14,15.
ER stress arises from an overload of unfolded/misfolded proteins at the ER and is connected to the pathogenic progression of many metabolic and inflammatory diseases16. ER stress triggers the activation of three canonical branches of the unfolded protein response (UPR), which are mediated by three ER-localized transmembrane signal transducers, inositol-requiring enzyme 1 (IRE1), PKR-like ER kinase (PERK), and activating transcription factor 6 (ATF6)16,17. IRE1 is the most conserved ER stress sensor with a cytoplasmic segment possessing dual protein kinase/endoribonuclease (RNase) activities18,19. Upon activation of its RNase activity through autophosphorylation and oligomerization, IRE1 not only catalyzes the unconventional splicing of the mRNA encoding X-box binding protein (XBP1) to generate an active transcription factor XBP1s20,21, but also degrades select mRNAs through a process termed “regulated IRE1-dependent decay” (RIDD)22,23. Two isoforms of IRE1, IRE1α and IRE1β, are present in mammals, with IRE1α found to be ubiquitously expressed across different tissues and cells19. Many studies have shown that mammalian IRE1α serves as a multi-functional metabolic sensor that regulates many aspects of energy metabolism in different metabolic organs24, including thermogenic fat remodeling and adipose inflammation through its cell type-dependent RNase actions in adipocytes and macrophages, respectively25,26. Moreover, ER stress has also been implicated in regulating angiogenesis27, and in particular, the IRE1α-XBP1 pathway in ECs has been documented to cross-talk with the vascular endothelial growth factor (VEGF) receptor signaling and regulate endothelial proliferation and angiogenesis in the state of ischemia28. It remains largely unknown, however, whether IRE1α in ECs can respond to distinct local stress signals from tissue microenvironments and exert tissue-specific regulatory actions upon metabolic homeostasis.
In this study, we generated a mouse model in which IRE1α was specifically depleted in ECs in adults and investigated whether endothelial IRE1α could regulate systemic metabolism under nutritional stress, presumably impacting the metabolic function of adipose tissue via influencing the angiogenic capacity. Surprisingly, we found an unanticipated role of endothelial IRE1α in specifically regulating the adaptive response of pancreatic islets to overnutrition-induced metabolic stress.
Results
Endothelial IRE1α abrogation leads to impaired glucose tolerance with lower insulin production upon diet-induced obesity in male mice
To investigate the metabolic function of IRE1α in ECs under the stress of excess nutrition, we intercrossed floxed IRE1α (Ern1fl/fl) mice29 with the VE-cadherin-CreERT2 line30 to generate a mouse model with tamoxifen (Tam)-induced EC-specific IRE1α knockout (Ern1EC-KO) in adults (Fig. 1a), and challenged them to high-fat diet (HFD) feeding (Fig. 1b). Immunoblot analysis revealed apparent ablation of IRE1α protein in CD31-positive ECs (CD31 + ) but not in CD31-negative cells (CD31-) isolated from the livers, lungs and heart of Ern1EC-KO mice relative to that from Ern1fl/fl mice, indicating efficient endothelial IRE1α depletion (Fig. 1c). When maintained on a normal chow (NC) diet or following chronic HFD feeding, male Ern1EC-KO mice and their Ern1fl/fl littermates showed no significant differences in their body weight gain (Fig. 1d), daily food intake (Supplementary Fig. 1a), oxygen consumption (Supplementary Fig. 1b), body fat mass (Fig. 1e), or the tissue weight of liver, epididymal and subcutaneous white adipose tissue (epiWAT and scWAT) and brown adipose tissue (BAT) (Supplementary Fig. 1c). H&E staining revealed no overt changes in liver fat accumulation or adipocyte enlargement in scWAT/BAT of HFD-fed Ern1EC-KO mice relative to their Ern1fl/fl counterparts (Supplementary Fig. 1d). Interestingly, while exhibiting slightly more increases in serum triglyceride (TG) and total cholesterol (TC) levels, Ern1EC-KO mice had significantly higher elevations in their hepatic TG and TC content following HFD feeding (Supplementary Fig. 1e–h). This implies an impact of endothelial IRE1α deficiency upon hepatic lipid metabolism. More importantly, glucose tolerance test (GTT) showed that HFD-fed male Ern1EC-KO mice had significantly reduced ability to control blood glucose levels relative to their Ern1fl/fl littermates (Fig. 1f). Unexpectedly, insulin tolerance test (ITT) revealed no differences in insulin-induced lowering of blood glucose levels (Fig. 1g), suggesting that IRE1α deficiency in ECs did not affect the insulin sensitivity in Ern1EC-KO mice following HFD feeding. Consistently, we observed similar levels of insulin-stimulated Akt phosphorylation in the livers, scWAT or muscle from NC- or HFD-fed Ern1EC-KO and Ern1fl/fl mice (Supplementary Fig. 2a, b). This then led us to test if they had different insulin production capacities in the face of obesity. Indeed, we detected significantly blunted hyperinsulinemia with comparable serum glucose levels in fasted HFD-fed Ern1EC-KO mice relative to their Ern1fl/fl counterparts (Fig. 1h). In accordance, HFD-fed Ern1EC-KO mice also exhibited marked reductions in their serum insulin levels following glucose stimulation, despite significantly higher glucose levels in the blood (Fig. 1i). By contrast, female Ern1EC-KO and Ern1fl/fl mice showed similarly lower degree of body weight gain than their male counterparts upon HFD feeding for 12 weeks with comparable glucose intolerance and no overt changes in serum insulin levels (Supplementary Fig. 3a–d), reflecting the phenotypic sexual dimorphism in their sensitivity to HFD-induced obesity, including pancreatic islet adaptation as reported in documented studies31. In addition, without tamoxifen treatment, male Ern1fl/fl;VE-cad-CreERT2 mice and their Ern1fl/fl littermates had no phenotypic differences in their metabolic parameters following HFD feeding, including glucose tolerance, fasted serum insulin levels, and pancreatic islet β cell mass (Supplementary Fig. 4a–g). This excludes the possibility that the metabolic changes in HFD-fed Ern1EC-KO mice resulted merely from Cre expression in the absence of IRE1α depletion. Together, these results demonstrate a critical role of endothelial IRE1α in the control of glucose homeostasis under obesity-associated metabolic stress in male mice, predominately through influencing the capacity of insulin production in pancreatic islets.
a Schematic illustration of the creation of Tamoxifen (Tam)-inducible endothelial cell (EC)-specific IRE1α knock-out (Ern1EC-KO) mice in adult Ern1fl/fl;VE-cadherin-CreERT2 mice produced by intercrossing floxed IRE1α mice (Ern1fl/fl) with the VE-cadherin-CreERT2 line. b–i Phenotypic analyses of male Ern1EC-KO mice with diet-induced obesity. b Experimental procedure. Male Ern1fl/fl;VE-cadherin-CreERT2 mice and age-matched Ern1fl/fl littermates at 4 weeks of age were injected daily with 2 mg Tam for 5 consecutive days. Mice were maintained on a normal chow (NC, 10% fat) diet, or fed a high-fat diet (HFD, 60% fat) starting at 8 weeks of age. Metabolic phenotyping was conducted at the indicated time. c Immunoblot analysis of IRE1α and CD31 protein in CD31-positive (CD31 + ) ECs and CD31-negative (CD31-) control cells isolated using anti-CD31 antibody and Dynabeads from the liver, lung and heart tissues from 8-week-old animals. β-Actin or α-Tubulin was used as the loading control. Shown also is the quantification of IRE1α protein levels in CD31+ ECs and CD31- control cells (n = 4 per genotype). d Body weight monitoring (n = 20 per group). e Body fat content and lean mass relative to body weight (BW) after NC- or HFD-feeding for 12 weeks (n = 12 per group). f Glucose tolerance test (GTT) following 12 weeks of NC or HFD feeding (NC, n = 23 per group; HFD, n = 25 per group). Shown also is the incremental area under curve (iAUC). g Insulin tolerance test (ITT) following 14 weeks of NC or HFD feeding (NC, n = 21 per group; HFD, n = 25 per group). h Serum insulin (NC, n = 12 per group; HFD, n = 19 per group) and glucose levels (NC, n = 20 per group; HFD, n = 25 per group) after a 4-hour fast following 16 weeks of NC or HFD feeding. i Glucose-stimulated insulin secretion. Serum insulin levels were measured following intraperitoneal injection of glucose after a 16-hour fast, and blood glucose levels were also simultaneously monitored for HFD-fed mice (NC, n = 10 per group; HFD, n = 15 per group). Data are presented as mean ± SEM by unpaired two-tailed Student’s t-test (c, f, i) or two-way ANOVA (e, h).
IRE1α ablation in ECs dampens the compensatory growth of pancreatic islets and β cells in male mice
Because obesity-associated insulin resistance triggers physiological compensatory growth of pancreatic islets with robustly increased β cell mass32, we then examined whether endothelial IRE1α deficiency affected the adaptive changes of pancreatic islets in handling obesity-associated metabolic stress. Immunoblot analysis further revealed the efficient depletion of IRE1α protein in CD31+ ECs from pancreas, presumably including intra-islet ECs, of Ern1EC-KO mice relative to that from Ern1fl/fl mice (Fig. 2a). Histochemical and morphometric analyses showed no apparent alterations in the pancreatic architecture or the structural organization of islets in NC- or HFD-fed male Ern1fl/fl and Ern1EC-KO mice (Fig. 2b). However, marked increases in the number (by ~56.3%), averaged size of islets (by ~42.8%) and β cell mass were observed in HFD-fed Ern1fl/fl mice relative to their NC-fed counterparts, all of which were substantially blunted in Ern1EC-KO mice (Fig. 2c, d), including the number of small as well as large islets (Fig. 2e). Next, we asked if islet cell proliferation or cell death is involved. Whole-mount Ki67 immunostaining of islets33 showed that HFD-fed Ern1EC-KO mice, relative to their HFD-fed Ern1fl/fl littermates, had significant reductions in Ki67-positive (Ki67+) cells from Ins+ β cells either in total islets or in islets of varying sizes (Fig. 2f, g). Moreover, TUNEL analyses also revealed significantly higher number of TUNEL-positive (TUNEL+) nuclei from β cells (Ins+) in HFD-fed Ern1EC-KO islets (Fig. 2h, i). Thus, these data suggest that loss of endothelial IRE1α leads to defective compensatory growth of islets in male mice, which is in parallel with decreased β cell proliferation and increased β cell death under metabolic stress conditions.
a Immunoblot analysis of IRE1α and CD31 protein in CD31-positive (CD31 + ) ECs and CD31-negative (CD31-) control cells isolated using anti-CD31 antibody and Dynabeads from pancreas tissues from 8-week-old NC-fed Ern1EC-KO mice and their Ern1fl/fl control littermates. β-Actin was used as the loading control. Shown also is the quantification of IRE1α protein levels in CD31+ ECs and CD31- control cells (n = 3 per genotype). b–i Male Ern1EC-KO mice and their Ern1fl/fl littermates were fed a NC diet or HFD for 16 weeks. b Representative immunohistochemistry (IHC) staining of insulin (Ins) from pancreas sections. Scale bar, 200 µm. Shown also are magnified graphs of the indicated islets. Scale bar, 10 µm. c Quantification of islet number and average islet area from the IHC staining (3 sections per mouse, n = 6 mice per group for islet number, and n = 5 per group for islet area). d Quantification of β cell mass by normalization to pancreatic mass (n = 6 per group). e Distribution of islets of various sizes in mice of the indicated group (n = 6 per group). f Representative images of whole-mount immunofluorescent Ki67/insulin staining of isolated primary islets from HFD-fed mice. Insulin (Ins, red), Ki67 (green) and DAPI (blue). Scale bar, 50 μm. Shown also are magnifications of the demarcated regions from the merged images. Scale bar, 20 μm. g Quantification of the percentage of total Ki67 + /Ins+ cells per islet (44 islets from 3 Ern1fl/fl mice, 31 islets from 3 Ern1EC-KO mice). Shown in the lower panel are percentages of Ki67 + /Ins+ cells in islets of differing sizes. h Representative images of whole-mount immunofluorescent TUNEL/insulin staining of isolated islets from HFD-fed mice. Insulin (Ins, red), TUNEL (green) and DAPI (blue). Scale bar, 50 μm. Magnifications of the demarcated regions are also shown from the merged images. Scale bar, 20 μm. i Quantification of the number of Ins + /TUNEL+ cells per islet (26 islets from 3 Ern1fl/fl mice, 34 islets from 3 Ern1EC-KO mice). Data are presented as mean ± SEM by unpaired two-tailed Student’s t-test (a, g, i) or two-way ANOVA (c, d, e).
IRE1α deficiency in ECs causes islet dysfunction under dietary obesity in male mice
To examine if IRE1α depletion in ECs had an impact upon the cellular composition of islets, we performed immunostaining analysis of insulin and glucagon in pancreatic sections (Fig. 3a). We observed significantly elevated areas of β cells (Ins+) and slightly increased areas of α cells (Gcg+) in HFD-fed versus NC-fed Ern1fl/fl islets, which were significantly and appreciably, respectively, lowered in HFD-fed Ern1EC-KO islets (Fig. 3b, c). However, no significant changes in the area percentage of β cells (Ins+) or α cells (Gcg+) were seen in NC- or HFD-fed Ern1EC-KO versus Ern1fl/fl islets (Fig. 3d, e). We then wondered if endothelial IRE1α deficiency affected the secretory function of pancreatic islets. Whereas isolated primary islets from HFD-fed Ern1fl/fl mice showed higher capacity of glucose-stimulated insulin secretion than their NC-fed counterparts, islets from HFD-fed Ern1EC-KO mice exhibited nearly a total loss of such augmentation (Fig. 3f), while no alterations in the insulin content were detected in these islets (Fig. 3g). Notably, quantitative RT-PCR profiling showed appreciable decreases in HFD feeding-induced upregulation of insulin 1 (Ins1) and insulin 2 (Ins 2) mRNA expression in Ern1EC-KO islets (Supplementary Fig. 5a), while no significant changes were seen in the expression of critical transcription regulators34 involved in insulin biogenesis or β cell development and regeneration, including pancreas/duodenum homeobox protein 1 (Pdx1), Neurogenin 3 (Ngn3) or musculoaponeurotic fibrosarcoma oncogene family protein A (Mafa) (Supplementary Fig. 5b). Interestingly, suggesting an islet angiogenic response such as VEGF signaling35 is involved, we detected a significant upregulation of Flk1 (fetal liver kinase 1, i.e., murine VEGFR2), but not Flt1 (FMS-like tyrosine kinase 1, i.e., murine VEGFR1) or Vegfa mRNA expression in HFD-fed Ern1fl/fl islets in comparison to their NC-fed counterparts, which was abrogated in Ern1EC-KO mice (Supplementary Fig. 5c). Furthermore, we characterized insulin secretion dynamics via islet perifusion assays and observed significant reductions in both phase I and phase II secretion following glucose stimulation as well as upon KCl stimulation in HFD-fed Ern1EC-KO islets (Fig. 3h, i). In line with their defective secretory capacity, fluorescent imaging analysis also revealed decreased calcium mobilization in HFD-fed Ern1EC-KO islets following high glucose stimulation (Fig. 3j, k). Collectively, these ex vivo data demonstrate that IRE1α ablation in ECs could impair the enhancement of the secretory function of islets, and particularly β cells, in the face of obesity-induced metabolic stress.
Male Ern1EC-KO and Ern1fl/fl mice were fed a NC diet or HFD for 16 weeks. a Representative immunofluorescent staining of insulin/glucagon from pancreas sections. Insulin (Ins, red), Glucagon (Gcg, green), DAPI (blue). Scale bar, 50 μm. b, c Quantification of (b) Ins+ β-cell and (c) Gcg+ α-cell area per islet (n = 6 per NC group and n = 9 per HFD group). d, e Quantification of the area percentage of (d) Ins+ β-cells and (e) Gcg+ α-cells per islet (n = 6 per NC group and n = 9 per HFD group). f–k Primary islets were isolated after a 4-hour fast. f Glucose stimulated insulin secretion (GSIS) analysis. Islets pooled from 3 mice of each group were divided into the indicated replicates (30-50 islets per replicate) before incubation at 2.8 mM or 16.8 mM glucose for 1 hour. Insulin levels were then measured and are shown after normalization to the islet protein content. Data are representative of 2 independent experiments. g Islet insulin content was measured following GSIS, shown after normalization to islet protein content (n = 6 per group). h Perifusion analysis of insulin secretion dynamics for pooled islets from HFD-fed mice (n = 3 per group). Islets were stimulated sequentially by 16.8 mM glucose and KCl, and insulin levels are shown as fold changes relative to the unstimulated starting basal value. p.I: Phase I; p.II: Phase II. Shown are representative results from 2 independent experiments. i Quantification of glucose-stimulated biphasic insulin secretion and KCl-induced insulin release in (h) after normalization to the islet protein content (n = 6 mice per group from the 2 independent experiments). j Glucose-stimulated Ca2+ influx in whole islets from HFD-fed mice (n = 80 islets from 6 mice per genotype). Shown are representative fluorescent calcium signals in islets maintained at 2.8 mM glucose or following stimulation with 16.8 mM glucose for 150 seconds. Scale bar, 100 μm. k Dynamic monitoring of Ca2+ intensity in response to high glucose stimulation in (j). Data are presented as mean ± SEM by two-way ANOVA (b, f) or unpaired two-tailed Student’s t-test (i, k).
IRE1α depletion in ECs limits HFD feeding-induced increase of intra-islet vascular density in male mice
Mature pancreatic islets are known to be highly vascularized and possess a dense capillary network, ~5 times denser than the surrounding exocrine area, which not only supports the efficient secretion of islet hormones like insulin into the circulation, but also acts to regulate β cell mass and function36,37,38. Therefore, we went on to investigate if IRE1α deficiency in ECs disrupted the adaptive compensation of islets through affecting intra-islet vascular remodeling such as angiogenesis during metabolic stress. Immunostaining analysis of CD31 as well as CD3439 in pancreatic sections showed a significant increase (by ~23.9%) of capillary density in islets from HFD-fed male Ern1fl/fl mice relative to that from NC-fed group, which was markedly diminished in Ern1EC-KO animals (Fig. 4a, b, Supplementary Fig. 6a, b). Quantitative RT-PCR analysis also revealed blunted upregulation of the expression of Pecam1 (encoding platelet and endothelial cell adhesion molecule 1, i.e., CD31) in Ern1EC-KO islets following HFD feeding relative to that in Ern1fl/fl islets (Fig. 4c). We then performed whole-mount immunofluorescent staining33 of CD31+ ECs in isolated islets for more thorough visualization of total intra-islet vascular network (Fig. 4d). Consistently, a significant increase of intra-islet CD31+ ECs was observed in Ern1fl/fl mice following HFD feeding, which was completely diminished in Ern1EC-KO islets (Fig. 4e). This is also in parallel with significantly blunted HFD feeding-elicited increase of Ins+ β cells in islets from Ern1EC-KO mice as compared to Ern1fl/fl control mice (Fig. 4e). Moreover, significant reductions were observed in CD31+Ki67+ double-positive signals from Ern1EC-KO relative to Ern1fl/fl islets after HFD feeding (Fig. 4f, g), whereas no TUNEL signals were detected within CD31+ ECs in these islets (Fig. 4h). By contrast, we observed no significant alterations in the abundance of CD31+ or CD34+ ECs in liver sections of NC- or HFD-fed Ern1EC-KO and Ern1fl/fl mice (Supplementary Fig. 6c,d). Nor did we detect significant changes of Pecam1 mRNA abundance in Ern1EC-KO livers relative to that in Ern1fl/fl livers (Supplementary Fig. 6e). Whole-mount immunostaining of CD31 in epWAT or scWAT (Supplementary Fig. 6f,g) as well as IHC staining of CD34 in scWAT (Supplementary Fig. 6h) also revealed no significant changes in their vascular density in NC- or HFD-fed Ern1EC-KO and Ern1fl/fl mice. In addition, real-time blood flow measurements40 exhibited no apparent differences in the blood perfusion within pancreas or inguinal WAT of NC- or HFD-fed male Ern1EC-KO mice and their Ern1fl/fl littermates (Supplementary Fig. 7a–d). Although it remains unknown if the blood perfusion within the islets was affected, these results suggest that IRE1α in ECs can respond to the unique microenvironmental cues of metabolically stressed islets, promoting intra-islet endothelial proliferation/angiogenesis for adaptive vascular remodeling to enable islet’s compensatory growth and augmented secretory capacity.
Male Ern1EC-KO mice and their Ern1fl/fl littermates were fed a NC diet or HFD for 16 weeks. a Representative IHC staining of CD31 from pancreas sections of mice. Scale bar, 200 µm. b Quantification of intra-islet CD31+ signals in (a) (n = 6 per group). c Quantitative RT-PCR analysis of the abundance of Pecam1 mRNA from isolated islets (n = 5 per group). d Representative images of whole-mount immunofluorescent staining of CD31/Insulin in isolated primary islets from mice. Insulin (Ins, green), CD31 (red). Scale bar, 50 µm. e Quantification of intra-islet CD31+ signals relative to Ins+ signals ( ~ 15 islets per mouse, n = 3 mice per group). f Representative images of whole-mount immunofluorescent staining of CD31/Ki67 in isolated islets from HFD-fed mice. CD31 (red), Ki67 (green). Scale bar, 50 μm. Shown also are magnifications of the demarcated regions from the merged images. Scale bar, 20 μm. g Quantification of CD31+/Ki67+ double-positive signals per islet ( ~ 15 islets per mouse, n = 3 per group). h Representative images of whole-mount immunofluorescent staining of CD31/TUNEL in isolated islets from HFD-fed mice (40 islets out of 3 mice per group). CD31 (red), TUNEL (green). Scale bar, 50 μm. Magnifications of the demarcated regions are also shown from the merged images. Scale bar, 20 μm. Data are presented as mean ± SEM by unpaired two-tailed Student’s t-test (g) or two-way ANOVA (b, c, e).
Endothelial IRE1α depletion substantially impacts islet transcriptome with enhanced Thbs1 expression in ECs
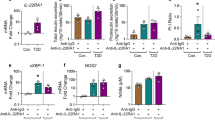
To gain further insight into how IRE1α promotes intra-islet angiogenesis under metabolic stress, we performed bulk RNA-seq analysis of primary islets isolated from Ern1fl/fl and Ern1EC-KO mice following 16-weeks of HFD feeding. Notably, rMATS analysis used for assessing alternative splicing events revealed overt exon 2 deletion in Ern1 mRNA from Ern1EC-KO islets (Supplementary Fig. 8a), confirming effective IRE1α knockout in islet ECs. Comparison of gene expression profiles between Ern1fl/fl and Ern1EC-KO islets showed 573 genes whose expression was influenced by IRE1α abrogation in ECs. Gene ontology (GO) biological process (BP) analysis revealed that IRE1α ablation in ECs affected a variety of regulatory processes in islets, including intercellular interactions, nutrient and hypoxia responses, inflammation, insulin secretion, as well as EC proliferation/migration and ER stress (Fig. 5a). Notably, among enriched genes related to insulin secretion (Supplementary Fig. 8b) are downregulated expression in Ern1EC-KO islets of Cartpt (Cocaine- and amphetamine-regulated transcript protein) and Gpr119 (G protein-coupled receptor 119), two crucial regulators of islet or β cell secretion function41,42,43 whose upregulation in response to HFD feeding was significantly dampened in islets by endothelial IRE1α deficiency (Supplementary Fig. 8c). Importantly, among top-ranked genes whose expression was most robustly upregulated or downregulated as a result of IRE1α ablation (Fig. 5b, c) and from the enriched genes implicated in regulation of EC proliferation (Fig. 5d), we identified Thbs1 coding for Thrombospondin 1 (TSP1), a well-established endogenous antiangiogenic factor that not only inhibits EC proliferation and its proangiogenic responses, but also exerts multiple regulatory effects upon glucose and lipid metabolism through affecting the function of adipose tissue as well as pancreatic islets44,45,46. Then we confirmed by quantitative RT-PCR that Thbs1 expression was indeed significantly enhanced in HFD-fed Ern1EC-KO islets (Fig. 5e), which was in parallel with higher content of TSP1 protein in islets with endothelial IRE1α deficiency (Fig. 5f). By contrast, while we observed increased serum TSP1 levels in HFD-fed mice, we did not detect significant changes resulting from endothelial IRE1α ablation (Supplementary Fig. 9a,b). This indicates that ECs are most likely not the major production source for circulating TSP1 levels.
a–d Global gene expression analysis by RNA-seq using total RNA extracts from isolated primary islets of Ern1fl/fl and Ern1EC-KO mice after 16 weeks of HFD feeding (n = 3 per group). a Gene ontology (GO) biological process (BP) analysis of 573 genes (average FPKM > 1; |log2 fold-change (FC) | > 0.5) whose expression was significantly different as a result of IRE1α deficiency in ECs. b Volcano plot depicting the log2(FC) values (Ern1EC-KO versus Ern1fl/fl) versus -log10(P-values) for genes (FPKM > 1) from the RNA-seq analysis of islets. Highlighted are up-regulated (red) or down-regulated (blue) genes ( | log2(FC) | > 0.5; P Value < 0.01) resulting from endothelial IRE1α deficiency. c List of top-ranked genes whose expression was significantly upregulated or downregulated in islets with EC IRE1α deficiency (FPKM > 1; |log2(FC) | > 0.5; P Value < 0.01). d Heat maps of 7 genes (average FPKM > 1; |log2(FC) | > 0.5; P Value < 0.01) involved in regulation of endothelial proliferation, which were differentially expressed in islets from HFD-fed Ern1EC-KO mice versus their Ern1fl/fl counterparts (n = 3 mice per group). e, f Primary islets isolated from Ern1fl/fl and Ern1EC-KO mice after 16 weeks of HFD feeding. e Quantitative RT-PCR analysis of the mRNA abundance of islet Thbs1 (n = 7 per group) and Egfl7 (Ern1fl/fl, n = 6; Ern1EC-KO, n = 7). f Immunoblot analysis of islet TSP1 protein levels (Ern1fl/fl, n = 5; Ern1EC-KO, n = 6). α-Tubulin was used as a loading control. Data are shown as mean ± SEM by unpaired two-tailed Student’s t-test (e, f).
Since TSP1 has been shown to be expressed in the endothelium of islets47,48, we further analyzed the single-cell RNA-seq data (accession No. GSE203376) from primary pancreatic islets in normal chow- versus HFD-fed mice49 and isolated the EC cluster by dimensionality reduction along with other cell types (Supplementary Fig. 10a,b). Remarkably, we also observed that the expression of Thbs1, but not Klf6 (Krüppel-like factor 6) or Sncg (Synuclein gamma), was specifically restricted to ECs along with the EC marker Cdh5 and an angiogenic transcription factor Sox18 (SRY-related HMG-box 18) (Supplementary Fig. 10c). Moreover, up-regulated Thbs1 expression was seen in HFD-fed intra-islet ECs as compared to that in normal chow-fed control group (Supplementary Fig. 10d). In addition, GO analysis of HFD feeding-regulated genes in intra-islet ECs, using the Findmarkers function with |log2FC | >0.25, revealed an enrichment of pathways related to response to unfolded protein or protein folding as well as angiogenesis (Supplementary Fig. 10e). Therefore, it is most likely that the observed upregulation of Thbs1 in HFD-fed Ern1EC-KO islets occurs specifically in islet ECs, suggesting the existence of a unique IRE1α-TSP1 axis in ECs that regulates intra-islet angiogenesis and the compensatory response of islets under metabolic stress.
IRE1α suppresses the expression of Thbs1 to promote endothelial cell proliferation
Next, we tested if IRE1α regulates the proliferation of ECs through altering TSP1 expression using the murine islet-derived MS1 endothelial cell line50, which was CD31-positive by immunofluorescent staining (Supplementary Fig. 11a), as well as primary human umbilical vein endothelial cells (HUVECs). Given that glucose and its metabolism are the primary factor driving compensatory β-cell proliferation in the face of insulin resistance32,51 and that high glucose was documented to inhibit angiogenesis in isolated human pancreatic islets with increased TSP1 expression levels52, we first examined if high glucose regulates Thbs1 expression in MS1 cells and HUVECs. Indeed, high glucose stimulation resulted in significant elevations in Thbs1/THBS1 mRNA and TSP1 protein expression levels along with markedly higher secretion of TSP1 protein from both MS1 cells and HUVECs, and lentiviral shRNA knockdown of the expression of IRE1α significantly augmented this glucose upregulation of TSP1 production (Fig. 6a–f). Moreover, CCK-8 and cell-count assays showed that IRE1α knockdown led to significantly reduced growth and viability of MS1 cells following high glucose stimulation (Fig. 6g), whereas lentiviral shRNA knockdown of TSP1 expression in the face of IREα knockdown (Fig. 6h) effectively reversed this growth defect of MS1 cells resulting from IREα deficiency (Fig. 6i). These data suggest that IREα may act to sustain endothelial cell proliferation through suppressing Thbs1 expression to block its autocrine antiangiogenic activity.
a–f MS1 endothelial cells (a, c, e) or primary HUVECs (b, d, f) were infected with lentiviruses encoding a shRNA directed against IRE1α (shErn1 or shERN1) or a scramble control (shCtrl) for 48 hours. a, b Quantitative RT-PCR analysis of Xbp1 mRNA splicing and Thbs1/THBS1 mRNA abundance in MS1 cells (a) or HUVECs (b) pre-cultured for 4 hours at 5 mM glucose or following stimulation for 24 hours with high glucose at 16 mM (n = 3 independent experiments). c, d Immunoblot analysis of IRE1α and TSP1 protein levels in MS1 cell lysates (c) or HUVEC lysates (d) when cultured at 5 mM versus 16 mM glucose for 24 hours. α-Tubulin was used as a loading control (n = 3 independent experiments). e, f Immunoblot analysis of TSP1 protein in culture medium of MS1 cells (e) or HUVECs (f). Cell lysate α-Tubulin was used as a loading control (n = 3 independent experiments). g Cell proliferation and viability analysis by cell-count assay or using the CCK-8 Kit for lentivirus-infected MS1 cells cultured at 16 mM glucose (n = 2 independent experiments). h, i MS1 cells were infected with shCtrl, shErn1, or both shErn1 and shThbs1 lentiviruses for 48 hours. h Immunoblot analysis of IRE1α and TSP1 proteins in MS1 cell lysates. i MS1 cell proliferation analysis by cell-count assay or using the CCK-8 Kit when cultured at 16 mM glucose (n = 2 independent experiments). j Proliferation analysis by the CCK-8 Kit of MS1 cells that were cultured at 16 mM glucose and treated with PBS (Vehicle, Veh.) or 1 μg/mL recombinant human TSP1 protein in the absence or presence of 1 μg/mL anti-CD47 neutralizing antibody for 48 hours (n = 6 independent treatment experiments). Data are shown as mean ± SEM by two-way ANOVA (a–d, g, i, * indicates shCtrl versus shErn1; # indicates shErn1 versus shErn1/shThbs1 in i) or unpaired two-tailed Student’s t-test (e, f, j).
Then, we tested the direct effect of TSP1 protein upon EC proliferation and interrogated the potential mechanism(s) that mediates its antiangiogenic actions. CCK-8 assay showed that treatment for 48 hours with recombinant human TSP1 protein significantly reduced the proliferation of MS1 cells (Supplementary Fig. 11b) but had no impact upon Min6 β cells (Supplementary Fig. 11c). As it has been documented that CD36, CD47 or SIRPα (signal-regulatory protein alpha) may mediate TSP1’s suppressive effects upon endothelial and vascular functions53,54,55, we analyzed the islet single-cell RNA-seq data (accession No. GSE203376) and found that Cd36 was poorly expressed in islet cells, while Cd47 but not Sirp was most abundantly expressed in islet ECs (Supplementary Fig. 11d). Quantitative RT-PCR analyses revealed that IRE1α deficiency did not affect the expression of Cd47 or Sirp in MS1 cells (Supplementary Fig. 11e). Given the reported role of the TSP1-CD47 axis in age-related EC deterioration in angiogenesis56, we asked if CD47 mediates TSP1 suppression of EC proliferation. Indeed, we observed that the CD47 neutralizing antibody reversed, at least partially, the inhibitory effect of TSP1 protein upon MS1 cell viability from the CCK-8 assay (Fig. 6j). In addition, because TSP1 has been shown to impede eNOS activation in arterial endothelium57, we wondered if IRE1α regulation of TSP1 might also have such an effect in islet ECs. Notably, we observed decreased levels of eNOS protein as well as its phosphorylation activation in HFD-fed Ern1EC-KO islets relative to their Ern1fl/fl counterparts (Supplementary Fig. 12a,b), but detected no significant changes in eNOS protein or phosphorylation levels in TSP1-treated MS1 cells with or without insulin stimulation (Supplementary Fig. 12c,d). Thus, while the TSP1-CD47 axis may largely account for the impaired intra-islet angiogenesis resulting from endothelial IRE1α ablation, defective eNOS activation is also likely to serve as a contributing factor.
IRE1α RNase promotes Thbs1 mRNA decay in endothelial cells
To determine how IRE1α negatively regulates TSP1 expression in ECs, we first asked if its RNase activity was involved. Treatment of MS1 cells with 4μ8C, a chemical inhibitor of IRE1α RNase activity58,59, potently blocked Xbp1 mRNA splicing and significantly augmented glucose-induced upregulation of Thbs1 mRNA abundance (Fig. 7a). In IRE1α-knockout HEK-293T cells, IRE1α overexpression resulted in a reduction of TSP1 protein level, which could be blocked by 4μ8C treatment, whereas XBP1s-Flag overexpression did not have a suppressive effect (Fig. 7b). Furthermore, overexpression of the wild-type (WT) human IRE1α, but not its RNase-deficient mutant IRE1α-K907A60, resulted in robust Xbp1 mRNA splicing with significant reductions in the mRNA abundance of THBS1 as well as BLOC1S1, a typical RIDD substrate of IRE1α (Fig. 7c). Consistently, overexpression of IRE1α-WT but not IRE1α-K907A led to a lower level of co-expressed TSP1 protein (Fig. 7d). To affirm this IRE1α RNase-dependent suppression of TSP1 expression, we established an experimental ER stress condition by treating MS1 cells with the chemical ER stressor tunicamycin (Tm), which induced the activation of IRE1α RNase activity as indicated by efficient Xbp1 mRNA splicing along with significant decreases in the mRNA abundance of both Bloc1s1 and Thbs1 (Supplementary Fig. 13a). Moreover, Tm-induced Xbp1 mRNA splicing and downregulation of Bloc1s1 and Thbs1 mRNA expression were significantly attenuated by lentiviral shRNA knockdown of IRE1α expression or by treatment with 4μ8C (Supplementary Fig. 13a,b). In addition, we performed mRNA stability analysis and observed sustained stability of Thbs1 mRNA along with Bloc1s1 mRNA as a result of IRE1α deficiency or 4μ8C inhibition of IRE1α RNase following actinomycin D treatment of ER stressed MS1 cells (Supplementary Fig. 13c). These results demonstrate that IRE1α RNase controls Thbs1 mRNA stability, likely through its RIDD activity. Furthermore, we identified the presence of a conserved hairpin structure containing a putative IRE1α cleavage site for typical RIDD substrates23 within both human and mouse Thbs1 mRNAs (Fig. 7e). Using in vitro synthesized RNA transcripts corresponding to nucleotide (nt) 1-1770 and 1771-3510 of the coding region of human THBS1 mRNA, we were able to detect their cleavages by recombinant human IRE1α protein, which could be effectively blocked by 4μ8C. Interestingly, more efficient cleavages were seen for the nucleotide 1771-3510 substrate of THBS1 RNA containing a predictable stem-loop hairpin than the nucleotide 1-1770 substrate without such a RIDD signature structure (Fig. 7f), suggesting that the nucleotide 1-1770 substrate may represent an in vivo example of RNAs undergoing IRE1α-dependent cleavage through the so-called “RIDD lacking endomotif” (RIDDLE)61. To further confirm that the nucleotide 1771-3510 substrate is a bona fade RIDD substrate of IRE1α, we synthesized a mutant RNA molecule (THBS1Mut) that harbors a G-to-C mutation at the putative IRE1α cleavage site within the hairpin structure (Fig. 7g). Whereas we detected a clear IRE1α-mediated cleavage of the wild-type THBS1WT RNA substrate, which again could be inhibited by 4μ8C, a much fainter cleavage was observed for the THBS1Mut RNA substrate (Fig. 7g). Together, these data suggest that IRE1α employs its RNase activity to promote Thbs1 mRNA decay via RIDD(LE) in ECs.
a MS1 cells were pre-cultured at 5 mM glucose for 4 hours before stimulation with 16 mM glucose for 12 hours in the absence or presence of 10 μM 4μ8C. Quantitative RT-PCR analysis of Xbp1 mRNA splicing and Thbs1 mRNA abundance (n = 3 independent experiments). b HEK293T cells with IRE1α depletion (HEK293T-KO) were transiently transfected for 36 hours with vector control (-) or THBS1-Myc plasmid, or co-transfected with THBS1-Myc plus IRE1α or XBP1s-Flag plasmids. Cells transfected as indicated were also treated with 10 μM 4μ8C for 12 hours. Immunoblot analysis of IRE1α, XBP1s and TSP1-Myc protein. Shown also is quantification of TSP1-Myc protein level after normalization to β-Actin control (n = 3 independent experiments). c, d HEK293T-KO cells were transiently transfected for 36 hours with vector control, IRE1α-WT or its RNase-deficient K907A mutant plasmids. c Quantitative RT-PCR analysis of human XBP1s, BLOC1S1 and THBS1 mRNA levels (n = 3 independent experiments). d Immunoblot analysis of IRE1α, TSP1-Myc and XBP1 protein with α-Tubulin as a loading control. Shown also is quantification of TSP1-Myc protein levels (n = 3 independent experiments). e Consensus sequence (underlined) of the putative RIDD region in human and mouse TSP1 mRNAs within the stem-loop structure predicted by RNAstructure Version 6.2. f Agarose gel analysis of IRE1α-mediated cleavage of human THBS1 mRNA. In vitro transcription-derived mRNA fragments of THBS1 (nt 1-1770 and nt 1771-3510) were incubated for 1 hour with recombinant human IRE1α protein (1 μg) in the presence of DMSO (-) or 10 μM 4μ8C, followed by 3% agarose gel analysis. Human XBP1 mRNA was used as a positive control. Red arrows indicate the probable major RNA cleavage products. g Synthetic RNA substrates for wild-type THBS1WT (nt 2771-2808) or THBS1Mut with the indicated G-to-C mutation were incubated for 1 hour with human IRE1α protein (1 μg) pre-mixed for 1 hour with DMSO (-) or 10 μM 4μ8C, followed by 20% TBE-Urea PAGE gel analysis. The red arrow indicates the RNA cleavage product. Data are representative of 2 independent experiments in (f, g). Results are shown as mean ± SEM by unpaired two-tailed Student’s t-test (a–d).
TSP1 abrogation in ECs corrects pancreatic islet dysfunction resulting from endothelial IRE1α deficiency in male mice
Finally, we generated tamoxifen-inducible Ern1EC-KO;Thbs1EC-KO double-knockout mice to verify the metabolic importance in vivo of IRE1α-directed suppression of TSP1 in ECs during dietary obesity. Immunoblot analysis of ECs from the livers, lungs and hearts showed efficient depletion of both IRE1α and TSP1 following tamoxifen treatment (Supplementary Fig. 14a). In comparison to HFD-fed Ern1fl/fl;Thbs1fl/fl control mice, HFD-fed Ern1 EC-KO;Thbs1EC-KO mice exhibited similar body weight gain (Fig. 8a) and daily food intake (Fig. 8b), slightly improved glucose tolerance (Fig. 8c) and insulin-responsive glycemic control (Fig. 8d). Moreover, no significant changes were detected in their serum insulin or blood glucose levels following glucose injection (Fig. 8e). These phenotypes are in sharp contrast to those of HFD-fed Ern1EC-KO mice that displayed impaired glucose tolerance and insulin secretion capacity relative to their Ern1fl/fl littermates (Fig. 1). Furthermore, unlike in HFD-fed Ern1EC-KO mice, we observed no reductions in the intra-islet vascular density (Fig. 8f), islet Pecam1 mRNA abundance (Fig. 8g), islet sizes or numbers (Fig. 8h, i), or islet’s glucose-stimulated insulin secretion capacity (Fig. 8j, k) in HFD-fed Ern1EC-KO;Thbs1EC-KO mice relative to their Ern1fl/fl;Thbs1fl/fl counterparts. Comparable vascular density was also observed in scWAT of HFD-fed Ern1EC-KO;Thbs1EC-KO mice and Ern1fl/fl;Thbs1fl/fl littermates (Supplementary Fig. 14b). These data demonstrate that endothelial ablation of TSP1 normalized the pancreatic islet dysfunction caused by IRE1α deficiency in ECs during dietary obesity. This suggests that TSP1 in islet ECs serves, in large part, as a key effector molecule mediating IRE1α’s regulatory actions in maintaining the endothelial support for the adaptive compensation of islet function under obesity-associated metabolic stress.
Male Ern1EC-KO;Thbs1EC-KO mice and their Ern1fl/fl;Thbs1fl/fl littermates were fed an HFD for 14-18 weeks. a Body weight monitoring (Ern1fl/fl;Thbs1fl/fl, n = 8; Ern1EC-KO;Thbs1EC-KO, n = 10). b Averaged daily food intake (Ern1fl/fl;Thbs1fl/fl, n = 8; Ern1EC-KO;Thbs1EC-KO, n = 10). c, d GTT along with iAUC (c) and ITT (d) analyses (Ern1fl/fl;Thbs1fl/fl, n = 7; Ern1EC-KO;Thbs1EC-KO, n = 10). e Serum insulin levels along with blood glucose levels were measured upon intraperitoneal glucose injection (Ern1fl/fl;Thbs1fl/fl, n = 6; Ern1EC-KO;Thbs1EC-KO, n = 8). f Primary islets were isolated from mice following 16 weeks of HFD feeding. Representative images of whole-mount immunofluorescent staining of CD31/insulin in islets. Scale bar, 50 µm. Shown also is quantification of intra-islet CD31+ relative to Ins+ signals (10 islets per mouse, n = 3 mice per group). g Quantitative RT-PCR analysis of the mRNA abundance of Pecam1 and Thbs1 in islets (n = 4 per group). h Representative IHC staining of insulin in pancreas sections from HFD-fed mice. Scale bar, 200 µm. Shown also are magnifications of the indicated islets. Scale bar, 20 µm. i Quantification of the distribution of islets in the indicated range of sizes, shown as the islet number per mouse for each group ( ~ 600 islets; 6 slides per mouse; n = 3 mice per group). j GSIS analysis of isolated primary islets (pooled from 3 mice, 5 replicates for each group). Insulin levels were normalized to islet protein content. k Insulin content was measured by ELISA from islets after GSIS, shown after normalization to islet protein content (pooled from 3 mice, 5 replicates for each group). Data are shown as mean ± SEM by unpaired two-tailed Student’s t-test (g) or two-way ANOVA (j).
Discussion
Anabolic insulin is essential for the maintenance of glucose homeostasis as well as whole body energy storage. Insulin resistance and obesity-associated factors stimulate islet expansion, thereby increasing insulin levels to counteract insulin resistance and glucose intolerance. Compensatory islet expansion is impaired in diabetes, driving hyperglycemia and glucose intolerance; however, regulatory mechanisms underlying islet expansion remains poorly understood. We found, unexpectedly, that pancreatic islet endothelial cells can sense metabolic stress, presumably elicited upon exposure to higher levels of glucose, insulin and fatty acids in the state of obesity-associated insulin resistance, and they critically regulate islet expansion and insulin secretion through an IRE1α/thrombosondin-1 (THBS1/TSP1) pathway. As a key UPR signaling transducer that governs ER homeostasis and cell fate decision, IRE1α regulates many metabolic pathways in tissue- or cell type-specific manners24,25,26. In this study, our findings demonstrate a crucial role of endothelial IRE1α-directed suppression of TSP1 in governing the vascular support that enables the homeostatic adaptation of islets in coping with overnutrition-associated metabolic stress, and loss of IRE1α in ECs leads to maladaptation of islets with impaired insulin secretion capacity during dietary obesity (Supplementary Fig. 15). It is particularly intriguing that out of all the ECs in various metabolic tissues, IRE1α deficiency engendered a main functional defect specifically in pancreatic islets during obesity, presumably reflecting its unique impact upon the metabolic properties of intra-islet ECs. This also indicates the peculiar microenvironmental changes in islets during obesity-associated stress that influences IRE1α’s activity in ECs, supporting the notion that vascular ECs in different organs possess distinct vulnerability to obesity as shown by a recent single-cell profiling study62.
Our findings highlight not only the physiological importance of endothelial IRE1α signaling in regulating the functional adaptation of metabolically stressed pancreatic islets, but also the critical role of intra-islet ECs in supporting this adaptive process. In pancreatic islet β cells, IRE1α has been shown to respond to glucose stimulation and regulate insulin biosynthesis as well as the compensatory islet growth in the state of obesity63,64,65, while it is also implicated in β cell failure and apoptosis in autoimmune type 1 diabetes66,67. Herein, our results from genetic mouse models revealed an important role of IRE1α in islet ECs in maintaining islet’s adaptive capacity upon metabolic stress, reflecting the multi-faceted, cell type-dependent actions of this ER stress sensor in the homeostatic control of pancreatic islets and β cells. In addition, our data also recapitulate the essential importance of ECs in supporting the adaptive remodeling of islets according to metabolic demand, which can be appreciated based on the critical role of ECs in supporting the long-term expansion of primary islet organoids from resident islet progenitors33,68.
Despite that a host of potential RIDD(LE) mRNA substrates of IRE1α RNase have been documented22,23,61, only recently were physiologically important ones being uncovered in animal models. Importantly, they were often found to display cell type-specific features, as exemplified by the mRNA encoding Myostatin in skeletal muscles69 and PGC1α in beige adipocytes25. Our mechanistic interrogation in islet ECs identified another RIDD(LE) mRNA substrate encoding TSP1, the endogenous inhibitor of angiogenesis44,45,46 that has been reported as a circulating biomarker in association with obesity and metabolic syndrome as well as pre-diabetes70,71,72. Interestingly, TSP1 has been implicated in regulating not only adipose tissue inflammation during dietary obesity73, but also pancreatic islet vascularization and β cells function47,74,75. It is worth noting that TSP1 was shown to be a key effector in mediating the anti-obesity effects of endothelial Argonaute 1 depletion in mice, whereby suppression of TSP1 was thought to underlie the enhanced vascularity in promoting the thermogenic remodeling of adipose tissue during dietary obesity76. This supports the idea that EC-derived TSP1 may exert crucial tissue-specific local autocrine and paracrine actions in response to metabolic stress, as our results also suggest that the IRE1α-TSP1 axis in islet ECs performs critical functions in regulating islet vascularity, compensatory growth and secretory capacity in the state of dietary obesity. However, we did not observe significant changes of Thbs1 expression in WAT of our Ern1EC-KO mice, presumably owing to more predominant expression of Thbs1 in other cell types like macrophages than in ECs. Alternatively, this may also imply the tissue-dependent nature of endothelial IRE1α suppression of TSP1, which is analogous to the cell type-selective RIDD action of IRE1α in regulating scWAT versus BAT thermogenesis25. TSP1 is known to exert its anti-angiogenic activity and a multitude of other functions through its two major binding receptors CD36 and CD4753,54,56, including regulating inflammation and apoptosis45,77. While it has yet to be deciphered in vivo if islet EC-derived TSP1 can directly influence β-cell survival via its paracrine action, our results suggest a possibility that the TSP1-CD47 signaling axis may exist in islet ECs to control its vascularity. It remains to be further dissected, however, whether the TSP1-CD47 pathway can disrupt the compensatory activation of islet progenitors during obesity, as in the case of aging-associated dysfunctional muscle stem cells78.
Our results from EC-specific double knockout mouse model demonstrated the metabolic importance in vivo of endothelial IRE1α suppression of TSP1 in maintaining the vascular support of islet adaptation to obesity and metabolic stress. However, our transcriptomics analysis of islets also revealed profound effects of IRE1α ablation in ECs upon islet gene expression programs implicated in diverse biological processes, including cell proliferation, nutrient sensing, as well as protein secretion. In addition to Thbs1, other regulatory factors may also be involved in contributing to the endothelial defects resulting from IRE1α deficiency, e.g., the eNOS, the angiogenic transcription factor Sox18 that is specifically expressed in islet ECs, and Flk1 in VEGF signaling, whose expression was also downregulated in HFD-fed Ern1EC-KO islets. It warrants further exploring how endothelial IRE1α deficiency may affect, through TSP1 or other elusive factors to be identified, the functional cross-talk or communication between islet ECs and β cells under metabolic stress. Since vascular endothelial defects have been connected with islet dysfunction and β cell loss in type 2 diabetes38, it remains to be investigated whether dysregulation of the IRE1α-TSP1 pathway in islet ECs is clinically associated with impairment of pancreatic islet function and β cell failure in humans. Strategies to enhance IRE1α-directed endothelial support of the compensatory adaptation of islets and β-cells to metabolic stress may offer new therapeutic avenues in the treatment of diabetes.
Methods
Animals
All animal studies were conducted in accordance with the Institutional Guidelines for the humane treatment of animals, with experimental protocols (No. WDSKY0201701−2_18120D) approved by the Committee on Ethics in the Care and Use of Laboratory Animals, College of Life Sciences, Wuhan University. Ern1fl/fl mice, in which the 121-nucleotide exon 2 of the Ern1 allele was flanked by loxP, were generated at Shanghai Research Center for Model Organisms and had been backcrossed into the genetic background of C57BL/6 J29. To create adult Ern1EC-KO mice with EC-specific IRE1α knockout, we first intercrossed Ern1fl/fl mice with VE-cadherin-Cre-ERT2 mice expressing the Cre recombinase under the control of the VE-cadherin gene promoter30 to generate Ern1fl/fl;VE-cadherin-Cre-ERT2 mice. Ern1EC-KO mice were subsequently produced after daily intraperitoneal injections of 2 mg tamoxifen (dissolved in a 10:1 sunflower seed oil/ethanol mixture at 10 mg/mL) for 5 consecutive days at the age of 4 weeks, with Ern1fl/fl mice treated likewise as the control group. Thbs1fl/fl mice (Strain No. T013681) were obtained from GemPharmatech (Nanjing, China). Ern1fl/fl;Thbs1 fl/fl;VE-cadherin-Cre-ERT2 mice were generated as the second filial generation from Ern1fl/fl;VE-cadherin-Cre-ERT2 and Thbs1fl/fl mice. Ern1EC-KO;Thbs1EC-KO double-knockout mice were subsequently produced after tamoxifen injections. Animals were housed in laboratory cages at 22 ± 3 °C with a humidity of 35 ± 5% under a 12-h dark/light cycle (lights on at 7:00 a.m.). For diet-induced obesity, age-matched male and female mice of different genotypes were randomly allocated to groups (2 mice per cage) and fed a high-fat diet (HFD, 60% kcal fat; Research Diets) for 16 weeks, starting at 8 weeks of age. For animal euthanasia, mice were sacrificed by cervical dislocation under anesthesia via i.p. injection of 2,2,2-Tribromoethanol (250 mg/kg body weight for mice, Sigma-Aldrich, T48402).
Metabolic phenotyping
Whole-body fat mass was measured by 1H-nuclear magnetic resonance (NMR) spectroscopy (Minispec LF50, BRUKER). To analyze energy expenditure, single-housed mice were acclimated for 24 h to an OxyMax Comprehensive Laboratory Animal Monitoring System (CLAMS, Columbus Instruments) before oxygen consumption and physical activity were monitored for 24 h. Oxygen consumption (VO2) was calculated after normalization to the lean mass. For glucose tolerance test (GTT), mice were fasted for 16 h before intraperitoneal injection of glucose at 1.0 g/kg body weight for NC-fed mice or 1.5 g/kg body weight for HFD-fed mice. For insulin tolerance test (ITT), mice were fasted for 4 h and then injected with insulin (Roche) at 0.75 U/kg or 1.00 U/kg body weight for NC- or HFD-fed mice, respectively. Blood glucose was determined with a glucometer (FreeStyle) before injection (0 min) or at 15, 30, 60 or 90 min after injection. For glucose stimulated insulin secretion (GSIS) analysis in vivo, mice were fasted for 16 h before intraperitoneal injection of glucose as in GTT analysis. Blood was collected from tail vein for insulin measurement, and serum levels of insulin were measured by the insulin ELISA kit (Millipore, EZRMI-13K) according to the manufacturers’ instructions. For measurement of serum TSP1 levels, tail-vein blood was collected and allowed to stand at room temperature for 45 min, followed by centrifugation twice for 10 min at 1000 × g. The supernatant was used to determine serum TSP1 levels with the ELISA kit (JONLNBIO, JL30686) according to the manufacturer’s instructions.
Isolation of mouse liver, lung, heart and pancreas endothelial cells
For isolation of lung, and heart ECs, tissues were harvested from mice, minced and then digested with 1.5 mg/mL collagenase type I (Sigma-Aldrich, C0130; added in RPMI 1640 with 5%FBS, 25 mM HEPES and 1% Pen/Strep) at 37 °C for 45 min. Tissues were then resuspended in Red-Blood-Cell lysis buffer for 5 min and terminated with double volume of DMEM, followed by filtering through a 100-µm nylon mesh. Cell pellets were resuspended in PBS with 5% FBS and incubated with the fluorochrome-conjugated antibody FITC-anti-CD31 (BD, 553372). ECs were then sorted by Dynabeads (Thermo Fisher Scientific, 11035) before being collected for Western immunoblot analysis.
For isolation of liver ECs, mice were anesthetized and livers were perfused with Kreb’s buffer (7 mg/mL NaCl, 2 mg/mL NaHCO3 and 3.6 mg/mL glucose) containing 0.5 mg/mL collagenase I (Yeasen, 40507ES) successively through the inferior vena cava. Livers were dissected and transferred into tubes through a 40-μm cell mesh, and then the volume was adjusted to 50 mL with RPMI 1640. Hepatocytes were removed by three cycles of low-speed centrifugation at 60 xg for 2 min, and the supernatants were then centrifuged at 400 xg for 10 min to separate hepatic nonparenchymal cells. Pelleted cells were re-suspended in 3.3 mL of RPMI 1640, and then gently layered on the top of 6 mL of 25% Percoll (Sigma-Aldrich, 17-0891-02) over 5 mL of 50% Percoll. After gradient centrifugation at 900 xg for 20 min at room temperature with the brake set to 0, cells at the interlayer between the 25% and 50% Percoll solution were transferred and re-suspended in 50 mL of RPMI 1640. Following incubation with anti-CD31, ECs were similarly sorted by Dynabeads and collected for subsequent Western immunoblot analysis.
For isolation of pancreas ECs, mice were anesthetized and whole-body perfused through heart with 50 mL Kreb’s buffer containing 0.5 mg/mL collagenase II (Yeasen, 40508ES60), 0.5 mg/mL collagenase IV (Yeasen, 40510ES60), 0.5 mg/mL dispase II (Yeasen, 40104ES60) and 0.2 mg/mL trypsin inhibitor (Beyotime, SG2031). Engorged pancreas was harvested and minced, and exocrine and endocrine cells were removed by two cycles of low-speed centrifugation at 60 xg for 2 min. Supernatants were then centrifuged at 400 xg for 10 min. Pellets were resuspended and incubated with anti-CD31, and ECs were sorted by Dynabeads and collected for subsequent Western immunoblot analysis.
Histology, immunohistochemistry and β cell mass analysis
Pancreas, liver, brown and white adipose tissues were isolated from mice and fixed in 10% neutral formalin for at least 48 h at 4 °C. Tissue were then desiccated and embedded in paraffin before being sliced into 4-µm thick sections for hematoxylin and eosin (H&E) (Servicebio) staining or immunohistochemistry (IHC). For IHC analysis, pancreas, liver and adipose tissue sections were mounted on adhesion slides, deparaffinized and rehydrated using a standard protocol (2 x Xylene, 2 x 100% Ethanol, 95% v/v Ethanol, 70% v/v Ethanol, 50% v/v Ethanol, and water; each for 5 min). Antigen retrieval was done by boiling the slides in the Citrate buffer (10 mM sodium citrate, 0.05% v/v Tween 20, pH 6.0) in a microwave oven for 10 min. Slides were then cooled down to room temperature for 20 min, rinsed with PBS and then placed in a humidity chamber for blocking and permeabilization in the blocking buffer (2% v/v goat serum, 1% w/v BSA, 0.1% v/v Triton X-100, 0.05% v/v Tween 20 in PBS) for 1.5 h at room temperature. Slides were then incubated overnight at 4 °C with anti-Insulin (ABclonal, A19066; diluted 1:400), anti-CD31 (RD systems, AF3628-SP; diluted 1:200) or anti-CD34 (ABclonal, A19015; diluted 1:200) primary antibody in PBS supplemented with 1% BSA and 0.05% Tween-20 with IgG staining as negative control. Slides were washed three times with PBS and then incubated with secondary antibodies (ABclonal, AS014 or AS029) at a 1:500 dilution for 1 h at room temperature. After washing three times with PBS, slides were stained using a 3,3’-diaminobenzidine (DAB) substrate kit (Abcam, ab64238) following the manufacturer’s instructions. After rinsing with PBS to stop the chromogenic reaction, slides were counterstained with hematoxylin QS (Vector Laboratories) for 2 min before analysis by microscopy (Lecia Aperio VERSA 8).
For whole-mount immunofluorescence staining, freshly isolated islets, scWAT or epiWAT tissue lumps were fixed in 4% PFA at 4 °C for 1 h, and permeabilized with PBST buffer (0.1% Triton X-100 in PBS) for 30 min. Islets were blocked in PBST buffer with 10% FBS for 2 h, and WAT tissues were blocked in MABT buffer (11.6 g/L Maleic acid, 8.7 g/L NaCl, 0.1% Tween 20) with 10% FBS for 2 h. Tissues were then incubated with anti-Insulin (Abcam, ab10983), anti-CD31 (BD Pharmingen, 553372), anti-Perilipin-1 (CST, 3470), or anti-Ki67 (BD, 612472; CST, 9127) primary antibody in the blocking buffer at 4 °C for 24 h, followed by washing 3 times with PBST buffer before incubation with the secondary antibody (Thermo Fisher Scientific, A-11012) at a 1:200 dilution at 4 °C for 24 h. Tissues were mounted with Antifade Reagent (Vector laboratories, H-1200), and imaging analysis was done using Leica TCS SP8 Confocal Microscopy.
The islet number and size in each pancreas section along with β cell mass in each mouse were quantified from a total of 15–18 pancreas sections analyzed by IHC per mouse group (3 sections/mouse, at least 90-μm apart; 5-6 mice/group). The average islet size in each section was calculated by dividing the area of insulin-positive (Ins+) region by islet number. For β cell mass analysis, total Ins+ area and total pancreas area for each section were measured, and β cell mass was then determined through multiplying the pancreas weight with the ratio of Ins+ area/pancreas area. The percentage of CD31+ or CD34+ area in each islet was calculated by dividing CD31+ or CD34+ area by islet area. All IHC quantification was done using the CaseViewer Software (Servicebio).
The glucagon-positive (Gcg+) α and insulin-positive (Ins+) β cell areas were quantified from immunofluorescent staining of pancreas sections, and the area percentage of α and β cells per islet for each mouse was calculated by dividing the Gcg+ or Ins+ area by islet area (12-20 islets per section, 1 section/mouse, 6-9 mice/group). The number of CD31 and Ki67 double-positive (CD31+/Ki67+) cells, insulin and Ki67 double-positive (Ins+/Ki67+) cells, and insulin and TUNEL double-positive (Ins+/TUNEL+) cells in each islet was also quantified, all using the ImageScope Software (Leica v12.3.3.5048) and Image J Software.
Animals blood perfusion analysis
Mice were anaesthetized with sevoflurane while their body temperature was kept at 37 °C. The blood flow in the pancreas and inguinal white adipose tissue was measured using moorFLPI-2 Laser Speckle Contrast Imager (Moor Instruments, Axminster, Devon, UK) following the manufacturer’s instructions. Colored images were acquired from 1500 frames during the 1-minute capture time.
TUNEL Assay and Cell Viability Analysis
Cell death in islets was analyzed using the DeadEndTM Fluorometric TUNEL System (Promega, G3250) according to the manufacturer’s instructions. TUNEL signals were visualized by fluorescence microscopy.
Murine islet-derived MS1 endothelial cells were from ATCC (CRL-2279) and cultured in DMEM with 5% FBS. MS1 cells were cultured in DMEM with 16 mM glucose and no FBS for cell proliferation assays. Cells were treated with recombinant human TSP1 protein (ABclonal, RP01197) in the absence or presence of anti-CD47 neutralizing antibody (Bio X Cell-InVivoMAb, BE0283; Clone MIAP410) or IgG control (Bio X Cell-InVivoMAb, BE0083; Clone MOPC-21). MIN6 cells were kindly provided by Dr. Peter Arvan from University of Michigan Medical School and were cultured in RPMI 1640 with 10% FBS and 0.05 mM β-mercaptoethanol. MIN6 cells maintained in RPMI 1640 with 2000 mg/L glucose and no FBS were treated with TSP1 protein prior to viability analysis. Cell viability was assessed by the Enhanced Cell Counting Kit-8 (CCK8) according to the manufacturer’s instructions (Beyotime Biotechnology, C0041).
Primary islet isolation and insulin secretion analysis
Mice were euthanized and injected with Hanks-balanced salt solution containing Collagenase V (Sigma-Aldrich, C9263) into their common bile duct. Distended pancreas was then isolated and digested for 20 min at 37 °C. For GSIS experiments, pancreases from 3 mice per group were pooled and filtered, followed by gentle centrifugation in Histopaque®-1119 (Sigma-Aldrich, 11191). Islets were then isolated by handpick under a dissecting microscope and collected in RPMI medium containing 10% fetal bovine serum, 50 μg/mL streptomycin, and 50 U/mL penicillin. Islets were cultured overnight at 37 °C in a humidified, 5% CO2 atmosphere before further experiments. For GSIS analysis, islets pooled from 3 mice and divided in desired replicates were incubated for 30 min in HEPES-balanced Krebs-Ringer bicarbonate (KRBH) buffer (135 mM NaCl, 3.6 mM KCl, 0.5 mM MgSO4·7H2O, 0.5 mM NaH2PO4, 2 mM NaHCO3, 10 mM HEPES, 1.5 mM CaCl2 and 0.1% BSA) without glucose. After incubation for 1 h in KRBH containing low glucose (2.8 mM), islets were then stimulated in KRBH containing high glucose (16.8 mM) for 1 h. Supernatants were collected, and islets were homogenized in lysis buffer for protein extraction at 4 °C. Secreted insulin and islet insulin content were measured by ELISA (Millipore, EZRMI-13K) and normalized to the total islet protein content.
Islet perifusion and calcium imaging analyses
Primary islets were isolated from mice and cultured overnight before perifusion experiments. For each assay, perifusion chambers were loaded with 150 IEQ islets, and islets were first perifused for 30 min with KRBH buffer containing 2.8 mM glucose, followed by perifusion at <1 mL/min flow rate with KRBH buffer containing 2.8 mM glucose, 16.8 mM glucose, 2.8 mM glucose and 30 mM KCl for 25, 30, 20 and 20 min, respectively. Fractions were collected every minute, and islets were recollected from the chamber after the assay for total protein content measurement using the BCA protein assay. Insulin secretion at each time point was determined using the HTRF insulin assay kit (Cisbio, 62INSPEC) according to the manufacturer’s instructions. For analyzing the biphasic insulin secretion, phase I and phase II of insulin secretion were based on the plateau formation of the two phases and quantified as the area under curve (AUC). Insulin levels were normalized to the average baseline insulin secretion at low glucose (2.8 mM) and presented as fold insulin secretion.
For calcium imaging analysis, isolated primary islets were cultured overnight and washed twice with KRBH buffer containing 2 mM glucose, followed by loading for 45 min with KRBH buffer containing 5.6 µM Fluo-4 AM (Thermo Fisher Scientific, F14201), 0.4‰ Pluronic F127 (Sigma-Aldrich, P2443), and 2 mM glucose. Islets were then washed twice with KRBH buffer containing 2 mM glucose, seeded on 35-mm glass-bottom dish (MatTek), and cultured for another 15 min. Islets were imaged in a humidified 37 °C cell culture chamber with 5% CO2 that was mounted on a Zeiss LSM 800 inverted confocal microscope. Continuous images were acquired with 10-s intervals, starting at 2 min after glucose supplementation to reach a final concentration of 16.8 mM until fluorescent plateaus were reached. Fluorescent intensity dynamics were analyzed using ImageJ.
Isolation of primary human umbilical vein endothelial cells (HUVECs)
Human umbilical cords were obtained from Wuhan Maternal and Child Health Hospital of Hubei province, Tongji Medical College, Huazhong University of Science and Technology with ethical approval from the Clinical Trial Ethics Committee and written consent from patients. HUVECs were isolated from umbilical cord vein with PBS containing 1 mg/mL Collagenase 1 (Yeasen, 40507ES) and cultured in VascuLife EnGS-Mv Endothelial Medium (Lifeline Cell Technology, LL-0004) in T-25 flasks which were coated with 1% gelatin for 30 min. Cells were cultured at 37 °C with 5% CO2 overnight and maintained in fresh medium before further analysis.
Gene expression analysis
Total RNA from tissue or cultured cells was extracted and purified with TRIzol reagent (Sigma-Aldrich, 93289). cDNA was synthesized by M-MLV reverse transcriptase (Thermo Fisher Scientific, M1682). Relative abundance of mRNA was determined by qPCR using SYBR Green (Abclonal, RK21203) on StepOne Real-Time PCR Systems (Applied Biosystems, 4376357), with values normalized to Actb or Rplp0 as the internal control using the ΔΔ-Ct method. Oligonucleotide primer pairs used are listed in Supplementary Table 1 and all oligonucleotides purchased from Sangon Biotech.
Virus construction and production
For knockdown of the expression of IRE1α or TSP1, lentivirus particles were produced by co-transfection of HEK-293T cells with 5 μg psPAX2 and 5 μg pMD2.G packaging plasmid together with 10 μg pLKO.1-shScramble-Puro, pLKO.1-shScramble-Hygro, pLKO.1-shErn1-Puro, pLKO.1-shERN1-Puro or pLKO.1-shThbs1-Hygro plasmid. Cell culture medium containing lentivirus particles was harvested at 48 h post transfection and used to incubate overnight with MS1 cells or HUVECs plus polybrene (5 μg/mL), followed by culturing in complete medium containing puromycin (1 μg/mL) or hygromycin (100 μg/mL) for 48 h. The target sequences for shRNAs against IRE1α and TSP1 are listed in Supplementary Table 2.
Expression constructs and transient transfection
IRE1α knockout HEK-293T cells were generated by CRISPR/Cas9 with Lenti-CRISPR-v2 plasmid, with 5’-GTTGTTGTTTGTGTCAACGC-3’ used as the guide sequence. The IRE1αWT and IRE1αK907A constructs were subcloned from plasmids previously described63 with pLVX-IRES-Neo backbone. The expression constructs for THBS1 and XBP1s were generated by cloning the respective segments of coding sequence produced by PCR into the pLVX-IRES-Myc-Neo or pCMV-tag4A backbone. For protein overexpression, transient transfection experiments were performed using Lipofectamine 2000 (Invitrogen, 11668019) according to the manufacturer’s instructions.
Western immunoblot analysis
Tissue samples or cells were lysed in RIPA buffer (1% Triton X-100, 1% NP-40, 150 mM NaCl, 0.5 % sodium deoxycholate, 0.1 % SDS, and 50 mM Tris-HCL, PH7.4) containing complete protease inhibitor cocktail and phosphatase inhibitor cocktail (Sigma-Aldrich). The supernatant of cell lysates was transferred into new microtubes and mixed with one volume of MPSB (62.5 mM Tris-HCl, 15% SDS, 8 M Urea, 10% glycerol and 100 mM DTT, pH=6.8) and 20% volume of 6 x loading buffer (300 mM Tris-HCl, 12% SDS, 0.6% bromophenol blue, 60% glycerol and 600 mM DTT, pH=7.4) after 1,2000 xg centrifugation for 10 min. The mixture was then incubated for 30 min at 37 °C. Proteins were separated by SDS-PAGE, transferred to nitrocellulose membrane, and probed with the desired antibody. Protein levels were quantified with NIH ImageJ software. Antibodies were diluted in the blocking solution and used as follows and are listed in Supplementary Table 4: anti-IRE1α, 1:1000 (CST, 3294); anti-XBP1, 1:1000 (ABcam, ab220783); anti-Flag, 1:2000 (ABclonal, AE005); anti-Myc, 1:2000 (ABclonal, AE010); anti-β-Actin, 1:50000 (ABclonal, AC026); anti-phospho-IRE1α-S724, 1:1000 (ABclonal, AP1146); anti-Akt, 1:2000 (CST, 9272); anti-phospho-Akt-S473, 1:2000 (CST, 4060); anti-TSP1, 1:1000 (CST, 37879); anti-eNOS, 1:1000 (CST, 32027); anti-phospho-eNOS-S1177, 1:1000 (CST, 9570) and anti-α-Tubulin, 1:5000 (Sigma-Aldrich, T6199).
RNA-seq analysis
Total RNA was extracted from isolated islets of HFD-fed Ern1fl/fl and Ern1EC-KO mice and then subjected to commercial RNA-seq analysis (Beijing Genomics Institute). Three independent islet samples per group were analyzed. Pai red-end, 150-nt reads were obtained from each sequencing lane. The sequencing data were filtered with SOAPnuke (v 1.5.2)79 by (1) removing reads containing sequencing adapters; (2) removing reads whose low-quality base ratio (base quality less than or equal to 5) is more than 20%; (3) removing reads whose unknown base (‘N’ base) ratio is more than 5%. Clean reads were obtained and stored in the FASTQ format, and were mapped to the mouse genome, UCSC mm10 (~94.03% alignment), using the HISAT2 (v 2.0.4)80. The Bowtie2 (v 2.2.5)81 was applied to align the clean reads to the reference coding gene set, and the expression level of target gene was calculated by the RSEM (v 1.2.12)82. Differential expression analysis was performed using the DESeq2(v 1.4.5)83. Differential expression P values are obtained from DESeq2 two-sided Wald tests of the null hypothesis that log2 fold-change=0 for each gene, with Benjamini-Hochberg FDR correction across all genes. The criteria for considering a gene being differentially regulated were based on a log2(fold-change) greater than 0.5 (either direction) and a significant P value < 0.01 versus control analyzed by two-sided hypergeometric (Fisher’s exact) tests with Benjamini-Hochberg FDR correction. GO and pathway grouping and enrichment analysis were performed by the clusterProfiler (v 3.16.1)84. For detection of genetic knockout events, Ern1 mRNA of islets from HFD-fed Ern1fl/fl and Ern1EC-KO mice (n = 3 per group) was analyzed by rMATS (v 4.1.0)85. Exon 2 inclusion levels were shown as IncLevel. Islet RNA-seq data have been deposited in the NCBI Gene Expression Omnibus with the accession number GSE263586.
For scRNA-seq analysis, cells with less than 200 detectable genes or greater than 20% mitochondrial genes (calculated with the Percentage Feature Set function in the Seurat package86) were excluded from further analysis. Data were normalized using the Normalize Data function in the Seurat package to identify genes with high variability. “Anchors” between datasets were identified and data from different samples were integrated using the Find Integration Anchors and Integrate Data in the Seurat package, respectively. Data were scaled, and principal component analysis (PCA) was performed based on the top 2000 high-variability genes. Top 7 principals were used for distributed stochastic neighbor embedding (t-SNE) construction. Feature plots and violin plots were generated by functions in Seurat package.
Actinomycin D Pulse-chase Analysis
The stability of mRNAs of interest in MS1 cells was determined by addition of 5 μg/mL actinomycin D (Act D) (MedChemExpress, HY-17559) together with 10 μg/mL tunicamycin (Tm) after cells were pre-treated with DMSO or 10 μM 4μ8C. Total RNA was extracted at the indicated time for qRT-PCR analysis. The amount of the target mRNA at each time point was determined relative to Rplp0 mRNA as the internal control, and plotted relative to its value at time 0 that was considered as 1.
RNA decay analysis
Oligonucleotide primers containing a minimal T7 RNA polymerase promoter (5’-TAATACGACTCACTATAGG-3’) were used for PCR from expression plasmid DNA templates to generate DNA fragments containing T7 promoter sequence upstream of the 1-1770 or 1771-3510 nt segment of the coding sequence of human THBS1, or the full coding region of human XBP1. In vitro transcription reactions were performed using the T7 high-yield RNA synthesis kit (TransGen Biotech, JT101) following the manufacturer’s recommendations. The transcribed RNAs were treated for 20 min with DNase and purified by EasyPure RNA Purification Kit (TransGen Biotech, ER701). For RNA decay analysis, ~2 μg transcribed RNA or ~0.1 nmole synthetic RNA substrates were incubated for 1 h with 1 μg recombinant hIRE1α protein plus 10 μM 4μ8C or DMSO in the RNA cleavage buffer (40 mM HEPES pH 7.0, 10 mM Mg(OAc)2, 50 mM KOAc, 5 mM DDT). RNA samples were then analyzed by 3% Agarose gel or 20% TBE-Urea PAGE gel electrophoresis, followed by visualization using YeaRed Nucleic Acid Gel Stain (Yeasen, 10202ES76). The oligonucleotide primers for PCR production of THBS1 and XBP1 templates for in vitro transcription are listed in Supplementary Table 3.
Statistical analysis
All data are presented as the mean ± SEM. Statistical analysis was performed with unpaired two-tailed Student’s t-test or two-way analysis of variance (ANOVA) with Fisher’s LSD test using GraphPad Prism 7.0. P < 0.05 was considered to be statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw RNA-seq data generated in this study have been deposited in the GEO database under the accession code GSE263586. In addition, we also analyzed previously published single-cell RNA-seq data GSE203376 from primary pancreatic islets in normal chow- versus HFD-fed mice. Source data are provided with this paper.
References
Ricard, N., Bailly, S., Guignabert, C. & Simons, M. The quiescent endothelium: signalling pathways regulating organ-specific endothelial normalcy. Nat. Rev. Cardiol. 18, 565–580 (2021).
Augustin, H. G. & Koh, G. Y. Organotypic vasculature: From descriptive heterogeneity to functional pathophysiology. Science (New York, N.Y.) 357 https://doi.org/10.1126/science.aal2379 (2017).
Potente, M. & Mäkinen, T. Vascular heterogeneity and specialization in development and disease. Nat. Rev. Mol. Cell Biol. 18, 477–494 (2017).
Kalucka, J. et al. Single-cell transcriptome atlas of murine endothelial cells. Cell 180, 764–779.e720 (2020).
Paik, D. T. et al. Single-cell RNA sequencing unveils unique transcriptomic signatures of organ-specific endothelial cells. Circulation 142, 1848–1862 (2020).
Pi, X., Xie, L. & Patterson, C. Emerging roles of vascular endothelium in metabolic homeostasis. Circ. Res 123, 477–494 (2018).
Hasan, S. S. & Fischer, A. The Endothelium: an active regulator of lipid and glucose homeostasis. Trends cell Biol. 31, 37–49 (2021).
AlZaim, I., de Rooij, L., Sheikh, B. N., Börgeson, E. & Kalucka, J. The evolving functions of the vasculature in regulating adipose tissue biology in health and obesity. Nat. Rev. Endocrinol. 19, 691–707 (2023).
Graupera, M. & Claret, M. Endothelial cells: new players in obesity and related metabolic disorders. Trends Endocrinol. Metab. 29, 781–794 (2018).
Cooke, J. P. Endotheliopathy of Obesity. Circulation 142, 380–383 (2020).
Cao, Y. Angiogenesis and vascular functions in modulation of obesity, adipose metabolism, and insulin sensitivity. Cell Metab. 18, 478–489 (2013).
Gealekman, O. et al. Depot-specific differences and insufficient subcutaneous adipose tissue angiogenesis in human obesity. Circulation 123, 186–194 (2011).
Crewe, C., An, Y. A. & Scherer, P. E. The ominous triad of adipose tissue dysfunction: inflammation, fibrosis, and impaired angiogenesis. J. Clin. Invest 127, 74–82 (2017).
Lenna, S., Han, R. & Trojanowska, M. Endoplasmic reticulum stress and endothelial dysfunction. IUBMB Life 66, 530–537 (2014).
Battson, M. L., Lee, D. M. & Gentile, C. L. Endoplasmic reticulum stress and the development of endothelial dysfunction. Am. J. Physiol. Heart Circulatory Physiol. 312, H355–h367 (2017).
Hetz, C., Zhang, K. & Kaufman, R. J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 21, 421–438 (2020).
Walter, P. & Ron, D. The unfolded protein response: from stress pathway to homeostatic regulation. Sci. (N. Y.) 334, 1081–1086 (2011).
Cox, J. S., Shamu, C. E. & Walter, P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73, 1197–1206 (1993).
Tirasophon, W., Welihinda, A. A. & Kaufman, R. J. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 12, 1812–1824 (1998).
Sidrauski, C. & Walter, P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90, 1031–1039 (1997).
Yoshida, H., Matsui, T., Yamamoto, A., Okada, T. & Mori, K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891 (2001).
Hollien, J. et al. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. cell Biol. 186, 323–331 (2009).
Maurel, M., Chevet, E., Tavernier, J. & Gerlo, S. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem Sci. 39, 245–254 (2014).
Huang, S., Xing, Y. & Liu, Y. Emerging roles for the ER stress sensor IRE1α in metabolic regulation and disease. J. Biol. Chem. 294, 18726–18741 (2019).
Chen, Y. et al. Adipocyte IRE1α promotes PGC1α mRNA decay and restrains adaptive thermogenesis. Nat. Metab. 4, 1166–1184 (2022).
Shan, B. et al. The metabolic ER stress sensor IRE1α suppresses alternative activation of macrophages and impairs energy expenditure in obesity. Nat. Immunol. 18, 519–529 (2017).
Binet, F. & Sapieha, P. ER stress and angiogenesis. Cell Metab. 22, 560–575 (2015).
Zeng, L. et al. Vascular endothelial cell growth-activated XBP1 splicing in endothelial cells is crucial for angiogenesis. Circulation 127, 1712–1722 (2013).
Shao, M. et al. Hepatic IRE1α regulates fasting-induced metabolic adaptive programs through the XBP1s-PPARα axis signalling. Nat. Commun. 5, 3528 (2014).
Monvoisin, A. et al. VE-cadherin-CreERT2 transgenic mouse: a model for inducible recombination in the endothelium. Dev. Dyn. 235, 3413–3422 (2006).
Casimiro, I., Stull, N. D., Tersey, S. A. & Mirmira, R. G. Phenotypic sexual dimorphism in response to dietary fat manipulation in C57BL/6J mice. J. Diabetes Complications 35, 107795 (2021).
Aguayo-Mazzucato, C. & Bonner-Weir, S. Pancreatic β Cell Regeneration as a Possible Therapy for Diabetes. Cell Metab. 27, 57–67 (2018).
Wang, D. et al. Long-Term expansion of pancreatic islet organoids from resident procr(+) progenitors. Cell 180, 1198–1211.e1119 (2020).
Conrad, E., Stein, R. & Hunter, C. S. Revealing transcription factors during human pancreatic β cell development. Trends Endocrinol. Metab. 25, 407–414 (2014).
Staels, W., Heremans, Y., Heimberg, H. & De Leu, N. VEGF-A and blood vessels: a beta cell perspective. Diabetologia 62, 1961–1968 (2019).
Richards, O. C., Raines, S. M. & Attie, A. D. The role of blood vessels, endothelial cells, and vascular pericytes in insulin secretion and peripheral insulin action. Endocr. Rev. 31, 343–363 (2010).
Eberhard, D., Kragl, M. & Lammert, E. Giving and taking’: endothelial and beta-cells in the islets of Langerhans. Trends Endocrinol. Metab. 21, 457–463 (2010).
Hogan, M. F. & Hull, R. L. The islet endothelial cell: a novel contributor to beta cell secretory dysfunction in diabetes. Diabetologia 60, 952–959 (2017).
Lee, Y. S. et al. The fractalkine/CX3CR1 system regulates β cell function and insulin secretion. Cell 153, 413–425 (2013).
Obata, A. et al. Vascular endothelial PDPK1 plays a pivotal role in the maintenance of pancreatic beta cell mass and function in adult male mice. Diabetologia 62, 1225–1236 (2019).
Abels, M. et al. CART is overexpressed in human type 2 diabetic islets and inhibits glucagon secretion and increases insulin secretion. Diabetologia 59, 1928–1937 (2016).
Ghislain, J. & Poitout, V. Targeting lipid GPCRs to treat type 2 diabetes mellitus - progress and challenges. Nat. Rev. Endocrinol. 17, 162–175 (2021).
Panaro, B. L. et al. β-cell inactivation of Gpr119 unmasks incretin dependence of GPR119-mediated glucoregulation. Diabetes 66, 1626–1635 (2017).
Lawler, P. R. & Lawler, J. Molecular basis for the regulation of angiogenesis by thrombospondin-1 and -2. Cold Spring Harb. Perspect. Med 2, a006627 (2012).
Gutierrez, L. S. & Gutierrez, J. Thrombospondin 1 in Metabolic Diseases. Front Endocrinol. (Lausanne) 12, 638536 (2021).
Isenberg, J. S. et al. Thrombospondin-1 inhibits endothelial cell responses to nitric oxide in a cGMP-dependent manner. Proc. Natl. Acad. Sci. USA 102, 13141–13146 (2005).
Olerud, J. et al. Thrombospondin-1: an islet endothelial cell signal of importance for β-cell function. Diabetes 60, 1946–1954 (2011).
Erdem, N. et al. Thrombospondin-1, CD47, and SIRPα display cell-specific molecular signatures in human islets and pancreata. Am. J. Physiol. Endocrinol. Metab. 324, E347–e357 (2023).
Fu, Q. et al. Single-cell RNA sequencing combined with single-cell proteomics identifies the metabolic adaptation of islet cell subpopulations to high-fat diet in mice. Diabetologia 66, 724–740 (2023).
Arbiser, J. L. et al. Oncogenic H-ras stimulates tumor angiogenesis by two distinct pathways. Proc. Natl. Acad. Sci. USA 94, 861–866 (1997).
Porat, S. et al. Control of pancreatic β cell regeneration by glucose metabolism. Cell Metab. 13, 440–449 (2011).
Dubois, S. et al. Glucose inhibits angiogenesis of isolated human pancreatic islets. J. Mol. Endocrinol. 45, 99–105 (2010).
Dawson, D. W. et al. CD36 mediates the in vitro inhibitory effects of thrombospondin-1 on endothelial cells. J. Cell Biol. 138, 707–717 (1997).
Isenberg, J. S. et al. CD47 is necessary for inhibition of nitric oxide-stimulated vascular cell responses by thrombospondin-1. J. Biol. Chem. 281, 26069–26080 (2006).
Yao, M. et al. Thrombospondin-1 activation of signal-regulatory protein-α stimulates reactive oxygen species production and promotes renal ischemia reperfusion injury. J. Am. Soc. Nephrology: JASN 25, 1171–1186 (2014).
Ghimire, K. et al. CD47 promotes age-associated deterioration in angiogenesis, blood flow and glucose homeostasis. Cells https://doi.org/10.3390/cells9071695 (2020).
Bauer, E. M. et al. Thrombospondin-1 supports blood pressure by limiting eNOS activation and endothelial-dependent vasorelaxation. Cardiovascular Res. 88, 471–481 (2010).
Volkmann, K. et al. Potent and selective inhibitors of the inositol-requiring enzyme 1 endoribonuclease. J. Biol. Chem. 286, 12743–12755 (2011).
Cross, B. C. et al. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc. Natl. Acad. Sci. USA 109, E869–E878 (2012).
Tirasophon, W., Lee, K., Callaghan, B., Welihinda, A. & Kaufman, R. J. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev. 14, 2725–2736 (2000).
Le Thomas, A. et al. Decoding non-canonical mRNA decay by the endoplasmic-reticulum stress sensor IRE1α. Nat. Commun. 12, 7310 (2021).
Bondareva, O. et al. Single-cell profiling of vascular endothelial cells reveals progressive organ-specific vulnerabilities during obesity. Nat. Metab. 4, 1591–1610 (2022).
Qiu, Y. et al. A crucial role for RACK1 in the regulation of glucose-stimulated IRE1alpha activation in pancreatic beta cells. Sci. Signal. 3, ra7 (2010).
Xu, T. et al. The IRE1α-XBP1 pathway regulates metabolic stress-induced compensatory proliferation of pancreatic β-cells. Cell Res 24, 1137–1140 (2014).
Hassler, J. R. et al. The IRE1α/XBP1s pathway is essential for the glucose response and protection of β cells. PLoS Biol. 13, e1002277 (2015).
Morita, S. et al. Targeting ABL-IRE1α signaling spares ER-stressed pancreatic β cells to reverse autoimmune diabetes. Cell Metab. 25, 883–897.e888 (2017).
Lee, H. et al. Beta cell dedifferentiation induced by IRE1α deletion prevents type 1 diabetes. Cell Metab. 31, 822–836.e825 (2020).
Wang, J. et al. Isolation of mouse pancreatic islet Procr(+) progenitors and long-term expansion of islet organoids in vitro. Nat. Protoc. 17, 1359–1384 (2022).
He, S. et al. IRE1α regulates skeletal muscle regeneration through Myostatin mRNA decay. J. Clin. Invest. https://doi.org/10.1172/jci143737 (2021).
von Toerne, C. et al. MASP1, THBS1, GPLD1 and ApoA-IV are novel biomarkers associated with prediabetes: the KORA F4 study. Diabetologia 59, 1882–1892 (2016).
Matsuo, Y. et al. Thrombospondin 1 as a novel biological marker of obesity and metabolic syndrome. Metabolism 64, 1490–1499 (2015).
Roberts, D. D. & Isenberg, J. S. CD47 and thrombospondin-1 regulation of mitochondria, metabolism, and diabetes. Am. J. Physiol. Cell Physiol. 321, C201–c213 (2021).
Li, Y., Tong, X., Rumala, C., Clemons, K. & Wang, S. Thrombospondin1 deficiency reduces obesity-associated inflammation and improves insulin sensitivity in a diet-induced obese mouse model. PloS one 6, e26656 (2011).
Olerud, J., Johansson, M., Lawler, J., Welsh, N. & Carlsson, P. O. Improved vascular engraftment and graft function after inhibition of the angiostatic factor thrombospondin-1 in mouse pancreatic islets. Diabetes 57, 1870–1877 (2008).
Cunha, D. A. et al. Thrombospondin 1 protects pancreatic β-cells from lipotoxicity via the PERK-NRF2 pathway. Cell Death Differ. 23, 1995–2006 (2016).
Tang, X. et al. Suppression of Endothelial AGO1 Promotes Adipose Tissue Browning and Improves Metabolic Dysfunction. Circulation 142, 365–379 (2020).
Jiménez, B. et al. Signals leading to apoptosis-dependent inhibition of neovascularization by thrombospondin-1. Nat. Med 6, 41–48 (2000).
Porpiglia, E. et al. Elevated CD47 is a hallmark of dysfunctional aged muscle stem cells that can be targeted to augment regeneration. Cell Stem Cell 29, 1653–1668.e1658 (2022).
Li, R., Li, Y., Kristiansen, K. & Wang, J. SOAP: short oligonucleotide alignment program. Bioinformatics 24, 713–714 (2008).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma. 12, 323 (2011).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16, 284–287 (2012).
Shen, S. et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 111, E5593–E5601 (2014).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Acknowledgements
We thank Qian Liu and Mingliang Tang for assistance at the animal and microscopy core facilities. This work was supported by grants from the National Natural Science Foundation of China (82230026, 32021003, 92357307, 91857204) and from the Ministry of Science and Technology of China (National Key R&D Program of China 2024YFA1802800 and 2018YFA0800700) to Y Liu, as well as from the National Natural Science Foundation of China (32501063) and the Postdoctoral Fellowship Program of CPSF (GZC20251881) to X.Z. Also supported by Shenzhen Medical Research Fund (B2402004) to Jie Liu and Y Liu.
Author information
Authors and Affiliations
Contributions
X.Z., J.L. and Y.Liu. conceived and designed the studies. X.Z., S.H. and P.C. performed most of the experiments and analyzed the data. Z.Z., J.C., Ting.Y., Z.X., Y.C., M.G., Z.W., J.H., Y.Liao. and S.Y. conducted the cell or animal experiments. Q.F. and T.Y. assisted with scRNA-seq data. Q.Y., Tailang Y., K.S., Jie L, S.D., L.R., Y.A.Z., and Z.M. provided essential technical support and assisted with HUVEC isolation and islet analysis. X.Z., S.H., J.L. and Y.Liu wrote and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, X., Huang, S., Chen, P. et al. Endothelial IRE1α promotes thrombospondin-1 mRNA decay and supports metabolic stress adaptation of pancreatic islets. Nat Commun 17, 1552 (2026). https://doi.org/10.1038/s41467-025-68276-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-68276-1