Abstract
Iron–sulfur (Fe–S) clusters are essential metallocofactors that perform a multitude of biological functions1,2,3,4,5,6,7. Their synthesis is tightly regulated and defects in this process lead to severe diseases8,9,10, such as Friedreich’s ataxia, which is caused by defective expression of frataxin (FXN)11. However, the underlying mechanisms that regulate this process remain unclear. Here we show that efficient Fe–S cluster assembly requires a fine-tuned balance in the ratio of FXN and ferredoxin-2 (FDX2). Fe–S clusters are assembled on the scaffold protein ISCU2; sulfur is provided as a persulfide by NFS1, and the persulfide is cleaved into sulfide by FDX2 (refs. 12,13). FXN stimulates the whole process by accelerating the transfer of persulfide to ISCU2 (refs. 12,14,15). Using an in-vitro-reconstituted human system, we show that any deviation from a close-to-equal amount of FXN and FDX2 downregulates Fe–S cluster synthesis. Structure–function investigation reveals that this is due to competition between FXN and FDX2 and their similar affinities for the same binding site on the NFS1–ISCU2 complex, with higher levels of FXN impairing the persulfide reductase activity of FDX2 and higher levels of FDX2 slowing the FXN-accelerated transfer of persulfide to ISCU2. We also find that FDX2 directly hinders persulfide generation and transfer to ISCU2 by interacting with the persulfide-carrying mobile loop of NFS1. We further show that knocking down the expression of FDX2 increases fly lifespan in a Drosophila model of Friedreich’s ataxia. Together, this work highlights a direct regulation of Fe–S cluster biosynthesis through antagonistic binding of FXN and FDX2, and suggests that decreasing FDX2 in the context of FXN deficiency in Friedreich’s ataxia might constitute a novel therapeutic axis.
Similar content being viewed by others
Main
Fe–S clusters are essential metallocofactors that make up the active site of many proteins involved in electron transfer, catalysis and signalling1,2,3,4,5,6,7. They are initially synthesized in mitochondria in the form of [2Fe–2S] clusters, which serve as building blocks for the assembly of [4Fe–4S] clusters16,17. Synthesis occurs through a sequential process on the scaffold protein ISCU2; the first step is iron binding, which triggers the acquisition of a persulfide provided by the cysteine desulfurase complex (NFS1–ISD11–ACP)2, followed by cleavage of the persulfide by FDX2 leading to a [1Fe–1S] precursor, and finally the formation of a [2Fe–2S] cluster after ISCU2 dimerization12,13,18,19. The whole process is enhanced by FXN, which accelerates the transfer of persulfide between NFS1 and ISCU2 (refs. 12,14,15). Any defect in the synthesis of Fe–S clusters leads to severe diseases8,9,10, the most frequent of which is Friedreich’s ataxia, a neurodegenerative and cardiac disease caused by defective FXN expression11. Therapeutic strategies for Friedreich’s ataxia have focused on increasing the levels of FXN, with gene therapy being the most promising approach20,21,22. However, the acute toxicity associated with FXN overexpression presents challenges23,24,25,26. This toxicity has been shown to rely on defective Fe–S cluster biosynthesis in vivo27, suggesting that a feedback loop exists in which FXN negatively affects the system that it stimulates. Indeed, biochemical data show that FXN and FDX2 share the same binding site in the NFS1–ISCU2 complex28,29,30, which suggests that these two proteins could interfere with each other’s activity. Here we combine biochemical and biophysical assays with structure–function studies to show that FXN and FDX2 mutually interfere with the respective reactions that they control. We also show that FDX2 has a direct inhibitory effect, disrupting the generation of persulfide on NFS1 and its subsequent transfer to ISCU2. Complementing the work on Caenorhabditis elegans by Meisel et al.31, our study shows that knocking down the orthologue of human FDX2 in a Drosophila model of Friedreich’s ataxia reduces the Fe–S cluster defect. Collectively, our data highlight an antagonistic cross-regulation of [2Fe–2S] cluster synthesis by FXN and FDX2, which fine-tunes the efficiency of Fe–S cluster synthesis as a function of protein level.
Kinetics of [2Fe–2S] cluster biosynthesis
To assess the effect of FXN and FDX2 on [2Fe–2S] cluster biosynthesis (Extended Data Fig. 1a), we used an in vitro reconstitution of the human mitochondrial iron–sulfur cluster machinery containing the scaffold protein ISCU2, the cysteine desulfurase complex (NFS1–ISD11–ACP)2 (denoted (NIA)2), the ferredoxin FDX2 and its cognate reductase FDXR, FXN, Fe2+, l-cysteine and NADPH (ref. 12). The assembly of [2Fe–2S] clusters was initiated by adding l-cysteine, and the kinetics of the reaction were monitored by following the absorbance of the [2Fe–2S] cluster in ISCU2 at 456 nm by ultraviolet (UV)-visible spectroscopy (Extended Data Fig. 1b, green line). In the absence of FXN, the rate of assembly was significantly decreased (Extended Data Fig. 1b,c, blue line), as previously reported12. Notably, a tenfold excess of FXN also slowed the reaction (Extended Data Fig. 1b,c, red line). This suggests that excess FXN directly interferes with [2Fe–2S] cluster assembly.
To assess whether this relates to competitive binding between FXN and FDX2, we monitored rates of [2Fe–2S] cluster synthesis across a titration of FXN concentrations at fixed amounts of FDX2. The titrations showed that increasing the amount of FXN proportionally increased the rate of [2Fe–2S] synthesis up to an optimal value, after which the rate proportionally decreased as the concentration of FXN continued to increase (Fig. 1a). Likewise, titrating FDX2 increased the rate of [2Fe–2S] synthesis up to an optimal value, beyond which the rate decreased (Fig. 1b). The amount of FXN required to reach the optimal rate of Fe–S cluster synthesis also increased proportionally with increasing amounts of FDX2, when FDX2 was present at concentrations higher than 0.6 molar equivalents (eq.) (Fig. 1a and Extended Data Fig. 1d). Similarly, the amount of FDX2 required for optimal cluster synthesis scaled linearly with increasing amounts of FXN (Fig. 1b and Extended Data Fig. 1e). Furthermore, when the concentration of both FXN and FDX2 was sufficient to occupy all binding sites on the (NFS1–ISD11–ACP–ISCU2)2 (denoted (NIAU)2) complex (that is, for concentrations higher than 0.6 eq.), the amounts of FXN and FDX2 required to reach optimal rates were close to a 1:1 ratio throughout the titrations (Fig. 1a,b and Extended Data Fig. 1d,e), suggesting that FXN and FDX2 operate at similar strengths on [2Fe–2S] cluster synthesis. Collectively, these data point towards a competitive effect between FXN and FDX2, leading to suboptimal rates of [2Fe–2S] cluster synthesis when the ratio of the two proteins is unbalanced. Notably, even in the complete absence of FXN, increasing the amount of FDX2 reduced the overall rate of [2Fe–2S] cluster synthesis (Fig. 1b,c), suggesting that excess levels of FDX2 also inhibit the assembly of Fe–S clusters through a non-competitive mechanism.
a, Kinetic rate of [2Fe–2S] synthesis monitored by UV-visible spectroscopy for the recombinant human ISC machinery across a titration of FXN (from 0 to 50 eq.) at discrete levels of FDX2 as indicated, measured in absorbance units (AU) (at 456 nm) per min. b, Re-ordered data from a, showing rates of [2Fe–2S] synthesis measured through a titration of FDX2 at discrete levels of FXN. c, Magnification of the rate of [2Fe–2S] synthesis across a titration of FDX2 in the absence of FXN (0 eq.) from b. The maximum rate of [2Fe–2S] synthesis per discrete level of FDX2 (a) or FXN (b,c) is highlighted as a green bar. Data in a–c are mean ± s.d., n = 3 independent experiments.
Competitive binding examined by FIDA and ITC
To characterize competitive binding between FDX2 and FXN in the human ISC system, we used flow-induced dispersion analysis (FIDA)32. FIDA is a biophysical technique based on measuring the Taylor dispersion profile of proteins and their complexes under laminar flow in a capillary, which is used to calculate the hydrodynamic radius (Rh) through the Stokes–Einstein equation (Methods and Extended Data Fig. 2a). FXN and FDX2 were labelled with an Alexa-like compound (ALC 480) by conjugation to the primary amine of solvent-accessible lysine residues, and are denoted as FXNALC and FDX2ALC, respectively. We used a metallated form of ISCU2 to stabilize its structured state12,18,33,34 and assess competitive binding under conditions that closely resemble those of the assembly reaction. As FIDA is routinely performed under aerobic conditions, oxidation and loss of the Fe2+ ion from ISCU2 would occur. We therefore used Zn2+ as a redox-inert Fe2+ surrogate, which adopts the same structure as Fe2+ in the assembly site12. However, to assess the effect of reduced FDX2, which is the active form for Fe–S cluster synthesis, we also specifically implemented FIDA under anaerobic conditions using sealed vials and dithionite to fully reduce FDX2 and maintain it in its reduced state.
First, we titrated unlabelled (NIAU)2 into a fixed concentration of FXNALC. As the concentration of (NIAU)2 increased, the apparent hydrodynamic radius (Rapp) of FXNALC increased from 2.11 nm to around 5 nm (Fig. 2a and Extended Data Fig. 2c,d), indicating the formation of a complex between FXN and the (NIAU)2 complex. After titration of unlabelled FDX2 to a fixed concentration of the preformed FXNALC–(NIAU)2 complex, the Rapp decreased until it reached a value of around 2.11 nm, corresponding to free FXNALC, with both the oxidized (Fig. 2b) and reduced (Extended Data Fig. 2f) forms of FDX2. This indicated that FDX2 competes with FXN for binding to the (NIAU)2 complex. Conversely, when FDX2ALC was titrated by (NIAU)2, the Rapp increased from 2.35 nm to higher than 5 nm (Fig. 2c and Extended Data Fig. 2c,e), indicating the formation of a complex between FDX2 and the (NIAU)2 complex. After titration of unlabelled FXN into the FDX2ALC–(NIAU)2 complex, the Rapp decreased until it reached a value of around 2.35 nm, corresponding to free FDX2ALC (Fig. 2d), indicating that FXN competes with FDX2 for the (NIAU)2 complex. These data indicate that human FXN and FDX2 compete for the same binding site.
a–d, FIDA measurements showing the change in the apparent hydrodynamic radius (Rapp) of: FXNALC after titration of the (NIAU)2 complex (a), the preformed FXNALC–(NIAU)2 complex after titration of FDX2 (b), FDX2ALC after titration of the (NIAU)2 complex (c) and the preformed FDX2ALC–(NIAU)2 complex after titration of FXN (d). e–g, ITC measurements monitoring the interaction between: FXN and the (NIAU)2 complex (e), FDX2 and the (NIA)2 complex (f) and FDX2 and the (NIAU)2 complex (g). Heat of injection error bars correspond to unbiased NITPIC-assigned integration errors of thermal spikes. Data in a–d are mean ± s.d., n = 3 independent experiments, presented in Extended Data Table 1; data in e–g are representative experiments from n = 3 independent experiments with similar results, presented in Extended Data Table 2.
The interaction of (NIAU)2 with FXNALC was best-fitted to a one-site binding model, yielding a complex size of 5.12 nm and a dissociation constant (Kd) of 102 nM (Extended Data Table 1). Fitting the binding curve of (NIAU)2 with oxidized FDX2ALC to a one-site binding model yielded a complex size of 5.14 nm and a Kd of 16 μM, whereas Kd values of 125 nM and 144 nM were obtained for oxidized and reduced FDX2, respectively, on the basis of competition experiments using unlabelled FDX2 with the FXNALC–(NIAU)2 complex (Extended Data Table 1). This suggests that the fluorescent label interfered with the binding of FDX2, and that the redox state of FDX2 does not modify its affinity for the (NIAU)2 complex, in agreement with findings reported for the yeast ISC system29.
To further characterize the interactions of FXN and FDX2 with the (NIAU)2 complex, we performed measurements by isothermal titration calorimetry (ITC). We first titrated apo-ISCU2 and zinc-loaded ISCU2 into the (NIA)2 complex (Extended Data Fig. 3a,b). The fit of the data yielded Kd values of 1,624 nM and 1,014 nM (Extended Data Table 2), indicating a slight increase in binding when in the presence of the metal. We next analysed FXN binding. The ITC data showed that FXN did not bind to the (NIA)2 complex (Extended Data Fig. 3c) or to ISCU2 alone (Extended Data Fig. 3d), but bound to the preformed (NIAU)2 complex (Fig. 2e). Similarly, FDX2 did not bind to ISCU2 alone (Extended Data Fig. 3e), but, unlike FXN, it bound to both the (NIA)2 complex (Fig. 2f) and the preformed (NIAU)2 complex (Fig. 2g). The ITC binding curve of FXN with the (NIAU)2 complex was fitted to a two-site model assuming symmetric binding sites, which yielded a refined Kd value of 146 nM (Fig. 2e and Extended Data Table 2), consistent with the 102 nM value determined by FIDA. The ITC binding curves of FDX2 with (NIA)2 and (NIAU)2 both exhibited a biphasic profile, characterized by a tight-binding exothermic phase followed by a weaker-binding endothermic phase (Fig. 2f,g and Extended Data Table 2). The presence of two distinct binding modes could be attributed to the proximal and distal conformations observed by cryo-electron microscopy (cryo-EM) in the NIAU–FDX2 complex30, with the high-affinity exothermic binding mode associated with the proximal conformation resulting from a larger interaction network between FDX2 and the (NIAU)2 complex. The fit of the ITC curve yielded a Kd value of 102 nM for the tight-binding exothermic mode (Extended Data Table 2), in good agreement with the value of 125 nM mined through FIDA competition assays using unlabelled oxidized FDX2. This confirmed that labelling by ALC 480 specifically perturbs FDX2 binding to the (NIAU)2 complex, but does not affect FXN binding.
Altogether, our data indicate that FXN and FDX2, both oxidized and reduced, bind in a competitive manner with near-equivalent affinities to the (NIAU)2 complex, consistent with an optimal rate of Fe–S cluster synthesis at close to a 1:1 ratio of FDX2 and FXN. Furthermore, the FIDA data show that FXN and FDX2 do not bind simultaneously to form a (NIAU–FXN–FDX2)2 complex (Fig. 2b,d), and ITC analysis revealed that FXN and FDX2 do not interact with each other (Extended Data Fig. 3f). This points towards a passive mode of competition between FXN and FDX2 for the (NIAU)2 complex, relying on the probability of occupancy of both proteins at the binding site, as opposed to an active mode that would occur through direct interaction and mutual displacement.
FXN and FDX2 interference analysed by ARBS
To assess the effect of competition between FXN and FDX2 on the key steps of the assembly process—persulfide formation by NFS1, persulfide transfer from NFS1 to ISCU2 and persulfide reduction—we used the alkylation-reduction band-shift (ARBS) assay12,14,35 with Fe2+-loaded ISCU2 under anaerobic conditions. In this assay, the proteins are incubated with an alkylating agent (maleimide–polyethylene glycol, herein termed mal-dPEG) of a sufficient size to conjugate to thiols, allowing them to be visualized through a molecular-mass band-shift after analysis with SDS–PAGE. ISCU2 and NFS1 contain four and seven cysteine residues, respectively, which are susceptible to alkylation by the mal-dPEG agents denoted ISCU2+4 and NFS1+7 (Supplementary Fig. 1). The cysteine–mal-dPEG conjugate forms a succinimidyl thioether that is intrinsically resistant to modification under reducing conditions. However, the alkylation of a persulfide incorporates a disulfide bond, which can be cleaved by a reducing agent, thereby liberating one mal-dPEG unit and leading to a shift to ISCU2+3 or NFS1+6 species on gel (Supplementary Fig. 1).
The ARBS assays showed that up to a tenfold excess of FXN did not affect the rate of persulfide formation on NFS1 in the (NIAU)2 complex containing the apo form of ISCU2 (Fig. 3a and Extended Data Fig. 4a), which still binds to NFS1 (Extended Data Fig. 3a) but is not able to receive a persulfide. In agreement with a previous report12, we observed that FXN accelerates the transfer of persulfide from NFS1 to iron-loaded ISCU2 (Fe–ISCU2) (Fig. 3b and Extended Data Fig. 4b). In the presence of up to 10 eq. of FXN, the transfer of persulfide was slightly faster (Fig. 3b and Extended Data Fig. 4b), indicating that conditions of excess FXN do not have an inhibitory effect on persulfide transfer. Persulfide reduction on ISCU2 was monitored by following a shift in the persulfidated ISCU2+3 form back to a non-persulfidated ISCU2+4 species after the addition of FDX2, FDXR and NADPH. We observed that the rate of persulfide reduction by FDX2 was significantly decreased when in the presence of excess FXN (Fig. 3c and Extended Data Fig. 4c). To determine whether the inhibitory effect of FXN on FDX2-mediated persulfide reduction also affects the global rate of Fe–S cluster synthesis, we analysed the effects of various FXN concentrations on the Fe–S cluster assembly reaction after persulfide transfer, monitored with UV-visible spectroscopy at 456 nm. In this assay, we first generated a persulfide on Fe–ISCU2 by incubation with (NIA)2, l-cysteine and various concentrations of FXN. After 5 min of reaction, leading to the full persulfidation of ISCU2, persulfide reduction was initiated with the addition of a preincubated mixture of FDX2, FDXR and NADPH, and [2Fe–2S] synthesis was monitored in the presence of 1, 10 or 50 eq. of FXN relative to FDX2. The kinetic assays showed that the rate of [2Fe–2S] synthesis decreased proportionally with the amount of FXN present (Fig. 3d), indicating that the interference of FXN in the FDX2-catalysed persulfide reduction step affects the global rate of [2Fe–2S] synthesis.
a–c, Kinetic profiles measured by ARBS assays monitoring persulfide generation on NFS1 in the NFS1–apo-ISCU2 complex in the absence of FXN or in the presence of 10 eq. FXN (a), persulfide transfer from NFS1 to Fe–ISCU2 in the absence of FXN or in the presence of 1, 5 or 10 eq. FXN (b) and persulfide reduction on Fe–ISCU2 by a mixture containing FDX2, FDXR and NADPH in the absence of FXN or in the presence of 1 eq. or 10 eq. FXN (c). d, Kinetics of [2Fe–2S] cluster synthesis after persulfide transfer, monitored by UV-visible spectroscopy at 456 nm after pre-incubation of the (NIAU)2 complex with l-cysteine for 5 min in the presence of 1, 10, or 50 eq. FXN, and initiation of the assembly reaction by adding FDX2, FDXR and NADPH. e–h, Kinetic profiles of ARBS assays monitoring persulfide generation on NFS1 (top legend) or on NFS1 in the NFS1-apo-ISCU2 complex (bottom legend) in the absence of FDX2 or in the presence of 10 eq. FDX2 (e), persulfide transfer from pre-persulfidated NFS1 to Fe–ISCU2 in the absence of FDX2 or in the presence of 3, 5 or 10 eq. FDX2 (f), persulfide transfer from NFS1 to Fe–ISCU2 in the absence of FDX2 or in the presence of 1, 3, 5, 7, or 10 eq. FDX2 (g) and persulfide transfer from NFS1 to Fe–ISCU2 with 1 eq. FXN present and either in the absence of FDX2 or in the presence of 1 or 10 eq. FDX2 (h). The schemes above each panel describe the reactions between non-persulfidated (-SH) and persulfidated (-SSH) proteins. Kinetic profiles monitor percentage of persulfidated NFS1 (NFS1–SSH) (a,e) or persulfidated ISCU2 (ISCU2–SSH) (b,c,f–h). Data in a–c,e–h are mean ± s.d., n = 3 independent experiments; data in d are representative kinetics selected from n = 3 independent experiments with similar results.
We next examined whether FDX2 interferes with persulfide formation and transfer within the (NIAU)2 complex. A slight inhibitory effect was observed when monitoring the rate of persulfide generation on NFS1 in the (NIA)2 complex in the presence of 10 eq. of FDX2 (Fig. 3e and Extended Data Fig. 4a). In the presence of the apo form of ISCU2, the rate of persulfide generation in the (NIAU)2 complex was further decreased in the presence of FDX2 (Fig. 3e and Extended Data Fig. 4a). In a sequential reaction, in which a persulfide was first generated on NFS1, followed by the addition of FDX2, and then the addition of Fe–ISCU2 to initiate persulfide transfer, the presence of increasingly high amounts of FDX2 led to proportionally decreased rates of persulfide transfer to ISCU2 (Fig. 3f and Extended Data Fig. 4d). Consistently, when monitoring the overall reaction combining persulfide generation and transfer by initiating the reaction with l-cysteine, the rate of persulfide transfer was also affected (Fig. 3g and Extended Data Fig. 4e). In the presence of FXN, persulfide transfer was also slowed by the addition of FDX2 (Fig. 3h and Extended Data Fig. 4f), probably because of the inhibitory effect of FDX2 on persulfide generation and transfer, and its competitive binding with FXN. These data thus indicate that in addition to competitive binding with FXN, FDX2 directly interferes with persulfide generation and subsequent transfer to ISCU2, which would be expected to affect Fe–S cluster synthesis because persulfide transfer is the rate-limiting step of the whole process12.
Structural mapping of the FDX2–NIAU interface
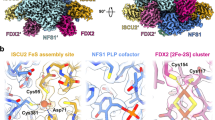
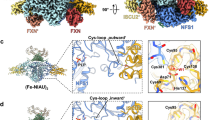
To identify the residues that contribute to both the interaction of FDX2 with the (NIAU)2 complex and the inhibitory effect of FDX2 on persulfide transfer, we generated a model using AlphaFold (Fig. 4a,b). This model shared similarities with the proximal conformation of the FDX2–(NIAU)2 cryo-EM structure30. The FDX2–(NIAU)2 interaction seemed to be the product of a concerted electrostatic salt-bridge network between aspartate and glutamate residues on helix F of FDX2, with a conserved patch of densely packed arginine residues on NFS1 (Fig. 4a). In addition, the disordered C terminus of FDX2 interacts with the mobile-loop region of NFS1, which, in this model, seems to position the catalytic cysteine C381 at the PLP cofactor and away from the ISCU2 assembly site (Fig. 4a). We reasoned that this interaction might result in the disruption of persulfide transfer from NFS1 to ISCU2. To analyse this model, we performed an alanine-scanning mutagenesis study based on the identification of interacting residues and hotspots predicted by the PLIP36 and SpotOn37 web servers, respectively. Five residues were selected to specifically target the interactions of FDX2 with the arginine patch on NFS1: four from helix F (E134, D137, D138 and D141) and one in close proximity to but not directly on helix F (E148). Three key residues from the C terminus of FDX2 were selected to perturb the interaction with the mobile loop of NFS1: Y177, D179 and H181 (Fig. 4a).
a, AlphaFold snapshot of interacting residues at the FDX2–(NIAU)2 interface. Cofactors (PLP, [2Fe–2S] and Fe) were inserted using AlphaFill42. FDX2 is shown in magenta, the first NFS1 subunit in dark green (left), the second NFS1 subunit in light green (centre top) and ISCU2 in salmon. Blue dashes represent hydrogen bonding, yellow dashes represent a salt bridge and orange dashes represents a π-cation interaction (H181FDX2–R393NFS1). FDX2 residues selected for mutation are annotated on FDX2 helix F (E134, D137, D138, D141 and E148) and the C terminus (Y177, D179 and H181). Predicted hotspot residues selected by SpotOn are marked with an asterisk (D137*, D138*, D141* and Y177*). b, Overview of the FDX2–(NIAU)2 complex predicted by AlphaFold with subunit colouring as in a. c, FIDA-performed competition assays of the FDX2 mutants titrated into a preformed complex of FXNALC–(NIAU)2. d, Plot of the Kd value of the wild-type (WT) FDX2 and mutant constructs interacting with the (NIAU)2 complex as determined by FIDA in c, and with the (NIA)2 complex as determined using ITC. e–h, Effect of FDX2 mutants on the rate of [2Fe–2S] synthesis measured under conditions of excess FXN (10 eq. FXN and 1 eq. FDX2) (e), equal FXN and FDX2 (1 eq. FDX2 and 1 eq. FDX2) (f) and excess FDX2 (10 eq. FDX2 and 1 eq. FXN) (g) and in the absence of FXN (0 eq. FXN) and in the presence of FDX2 (1 eq. FDX2) (h). In c–h, data for helix F and C terminus mutant constructs are highlighted in blue and green, respectively. In c, WT FDX2, mean ± s.d., n = 3 independent experiments, FDX2 mutant constructs n = 1. In d, FIDA data: Kd as determined from c; ITC data: WT FDX2, Kd determined using representative experiment from n = 3 independent experiments with similar results; FDX2 mutant constructs, Kd determined using n = 1 experiment per construct shown in Extended Data Fig. 5a. Data in e–h are mean ± s.d., n = 3 independent experiments.
Kd values of the mutants were determined by ITC (Extended Data Fig. 5a and Extended Data Table 3) and compared with macroscopic Kd values determined by FIDA competition assays of FDX2 with the FXNALC–(NIAU)2 complex (Fig. 4c,d). The values determined by ITC and FIDA were consistently in close agreement (Fig. 4d and Extended Data Table 3). The Kd values of all helix F mutants (E134A, D137A, D138A and D141A) were higher than the wild-type value, and FIDA competition assays indicated that these mutants had a lesser ability to compete with FXN, owing to their weaker affinities for NFS1. By contrast, the Kd values of the C-terminal mutants (Y177A, D179A and H181A), alongside that of the E148A mutant, either were not significantly changed, or were slightly lower than the wild-type value. The Y177A mutant was nearly equivalent to wild-type FDX2, and the E148A, D179A and H181A mutants had slightly stronger binding affinities. These findings indicate that the four residues on helix F (E134, D137, D138 and D141) that coordinate the network of salt-bridges with the arginine patch of NFS1 are crucial for tight binding with NFS1. By contrast, E148 and the residues in the C-terminal tail (Y177, D179 and H181) seem to be dispensable. Notably, ITC measurements revealed that all of the helix F mutants had lost the endothermic interaction associated with the distal conformation, whereas the mutants in the C-terminal tail region generally retained the biphasic binding profile and the Kd value remained close to the wild-type value (Extended Data Fig. 5a), in agreement with the cryo-EM structure showing that the distal conformation exclusively relies on loose interactions via helix F30.
The kinetics of [2Fe–2S] cluster synthesis showed that all helix F mutants had a diminished overall synthesis rate in the presence of excess FXN, when compared with wild-type FDX2 (Fig. 4e). The most pronounced effects were observed for the D138A and D141A mutants, with near-dead kinetics. The overall rates of the helix F mutants increased in conditions of equal FDX2 and FXN, but the rates of D138A and D141A were still slower than the wild-type rate (Fig. 4f), and were close to wild-type —or slightly higher—under conditions of excess FDX2 (Fig. 4g). These effects correlated with the weaker binding affinities of these mutants (Fig. 4d), leading to an increased sensitivity to the presence of competing amounts of FXN. Of note, the D137A mutant, and, more noticeably, the D141A and E148A mutants, had higher-than-wild-type rates of Fe–S cluster synthesis in the complete absence of FXN (Fig. 4h). This pointed to a higher intrinsic efficiency for persulfide reduction, or a lower inhibitory effect on persulfide generation and transfer, or both. In the complete absence of FXN, the transfer of persulfide is the slowest step of the whole process and is thus rate-limiting12. Therefore, any increase in the rate of persulfide reduction under these conditions would not increase the overall rate of Fe–S cluster synthesis. This suggests that it is rather a lesser repressive effect of these mutants on persulfide transfer that enhances [2Fe–2S] cluster synthesis, consistent with their lower affinity for the (NIAU)2 complex.
The C-terminal FDX2 mutants targeting the interaction with the mobile loop of NFS1 had either similar (Y177A and H181A) or diminished (D179A) rates of [2Fe–2S] synthesis under conditions of excess FXN (Fig. 4e), and had similar rates to wild-type FDX2 with equal amounts of FDX2 and FXN (Fig. 4f). These effects correlated with Kd values close to wild-type FDX2. Notably, all of the C-terminal mutants had higher-than-wild-type rates of Fe–S synthesis when FDX2 was present in excess (Fig. 4g), consistent with a previous study examining C-terminal truncations of human FDX2 including either H181 or all three residues30. The C-terminal mutants also had higher-than-wild-type rates of Fe–S synthesis in the complete absence of FXN (Fig. 4h), indicating that the C-terminal tail medites the direct repressive effect of FDX2. Because persulfide transfer is the rate-limiting step in the absence of FXN, the latter effect can only be attributed to an attenuation in the inhibition of persulfide generation and/or transfer, caused by a decrease in the overall affinity of FDX2 for the NFS1–ISCU2 complex and/or a looser interaction with the mobile loop of NFS1. Among the three C-terminal mutants, the D179A mutant had a higher affinity for NFS1 than did wild-type FDX2, with Kd values determined by FIDA and ITC of 24 nM and 39 nM, respectively (Fig. 4d and Extended Data Table 3). Its enhanced rate of Fe–S cluster synthesis therefore could not be the result of weaker binding, but could only be due to a looser interaction with the mobile loop of NFS1. To assess this hypothesis, we examined the effect of the D179A mutant on persulfide generation and transfer by ARBS assays in the absence and presence of FXN. The ARBS assays performed in the absence of FXN showed that the D179A mutant had no significant effect on persulfide generation (Extended Data Fig. 5b,c), but a lesser inhibitory effect on persulfide transfer (Extended Data Fig. 5d,e), thus confirming the reduced disturbance of the mobile loop movement induced by this mutation. The D179A mutant was also less inhibitory than the wild-type protein in the presence of FXN (Extended Data Fig. 5f,g), despite having a slightly higher ability to compete with FXN (Fig. 4d and Extended Data Table 3), indicating that the disruption of the interaction with the mobile loop of NFS1 predominates over its increased competitive effect with FXN. These data thus indicate that the C-terminal tail of FDX2—and, more specifically, D179—mediates a repressive effect on persulfide transfer by interfering with the movement of the mobile loop of NFS1.
Targeting FDX2 in a fly model of Friedreich’s ataxia
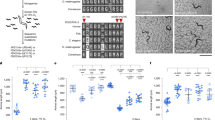
Thus far, our in vitro data suggests that decreasing the levels of FDX2 relative to FXN could reduce the competitive effect with FXN and alleviate the constraint on the mobile loop of NFS1 to enhance Fe–S cluster synthesis. To assess whether decreasing FDX2 levels could be beneficial in vivo under conditions of frataxin deficiency, we studied the effect of modulating ferredoxin levels in Drosophila melanogaster. Frataxin is a highly conserved protein, meaning that Drosophila is a suitable in vivo model for Friedreich’s ataxia38. We generated a model termed fh-GAAs, containing 42 GAA repeats inserted in the first intron of the fh gene (also known as CG8971) encoding Drosophila frataxin. This model exhibits a strong decrease in fh expression and a more than 75% decrease in mean lifespan39. Fdx1 (encoded by Fdx1, also known as CG4205), the Drosophila orthologue of FDX2, shares 62% identity with its human counterpart, with the eight amino acids studied above being conserved (Fig. 5a). We first analysed the levels of Fdx1 expression in fh-GAAs flies. At the third instar larval stage, we observed a 62% increase in Fdx1 transcript levels when fh expression was reduced by 90% (Fig. 5b, L3 larvae). In young adults, Fdx1 transcripts were also increased by 80%, with fh expression reduced to 3% of the control level (Fig. 5b, adults). This is in agreement with our previous study on a Drosophila model with a longer GAA expansion, in which Fdx1 overexpression was detected by RNA-seq40.
a, Sequence alignment of C-terminal regions of Homo sapiens (Hs) FDX2 and Drosophila melanogaster (Dm) Fdx1 (P37193). Conserved residues are highlighted in blue and the eight residues studied in vitro are in magenta. b, Relative quantification of Fdx1 and fh transcripts in control w1118 and fh-GAAs male third instar larvae and three-day-old adults. c, Relative quantification of Fdx1 transcripts in fh-GAAs; daGS>UAS-Fdx1 RNAi male third instar larvae that were untreated or treated with various concentrations of RU486. d,e, Survival curves (d) and mean lifespan (e) of fh-GAAs; daGS>UAS-Fdx1 RNAi male flies that were untreated or treated with various concentrations of RU486 during the developmental stage and a constant concentration of 10 μg ml−1 at the adult stage. Labels Dev-Adult indicate RU486 concentrations (μg ml−1) in the fly food during development and adult stages. In b,c, dots correspond to independent biological samples (n = 4), each obtained from 20 individuals. Data are mean ± s.d. In d,e, the total numbers of flies per condition of RU486 treatment and the P values obtained by log rank tests are provided in Extended Data Table 4. For each condition, flies were housed in tubes in groups of mostly 28 to 30 individuals. Each dot in e corresponds to one tube, with mean ± s.d. Number of tubes per condition: 0-0, n = 9; 2-10, n = 12; 5-10, n = 11; 10-10, n = 11; 20-10, n = 14. Increased Fdx1 expression in fh-GAAs flies was observed in three and two independent experiments at the larval and adult stages, respectively. Increased survival associated with Fdx1 inactivation in frataxin-deficient flies was observed in four independent longevity experiments. Two-sided non-parametric Mann–Whitney tests in b; one-way ANOVA followed by post-hoc Tukey analysis for multiple comparisons in c,e; significant differences are indicated: *P < 0.05, **P < 0.01, ***P < 0.001. Precise number of flies per tube (e) and P values (b,c,e) are provided in the source data.
Following findings in C. elegans31, we aimed to determine whether lowering the levels of Fdx1 could be protective in Drosophila in vivo. We previously reported that knockdown of Fdx1 by RNA interference (RNAi) under the control of the constitutive ubiquitous daGAL4 driver led to a developmental arrest at the third larval stage41. This phenotype was associated with a 66% decrease in Fdx1 transcript levels in whole-body extracts of third instar larvae (Extended Data Fig. 6a). We checked that the RNAi transgene was specific of Fdx1 and did not affect the transcription level of Fdx2 (also known as CG1319), the Drosophila ferredoxin orthologous to human FDX1 (Extended Data Fig. 6b). Then, to induce lower levels of Fdx1 knock down, we used the RU486-inducible daGS GeneSwitch driver instead of the constitutive daGAL4 driver. RU486 was incorporated into the fly food at various concentrations, leading to a dose-dependent decrease of Fdx1 mRNA levels in fh-GAAs; daGS>Fdx1 RNAi larvae (Fig. 5c). We observed a significant 31% increase in the mean lifespan of frataxin-deficient flies treated with 2 µg ml−1 and 10 µg ml−1 RU486 during the developmental and adult stages, respectively (Fig. 5d,e and Extended Data Table 4). Of note, developmental RU486 treatments at higher concentrations were less efficient, indicating that an optimal concentration of Fdx1 exists (Fig. 5d,e). As a control, we checked that RU486 treatments did not affect the lifespan of fh-GAAs flies that did not carry the daGS and Fdx1 RNAi transgenes (Extended Data Fig. 6c). We also evaluated the effect of RU486 treatments during development for concentrations lower than 2 µg ml−1 and at the adult stage for concentrations up to 100 µg ml−1. Although increasing the concentration of RU486 during development proportionally increased lifespan, with the highest effect observed at 1.6 µg ml−1, changing the concentration of RU486 at the adult stage in untreated flies or those treated with 1.6 µg ml−1 RU486 during development did not affect fly survival or have a toxic effect (Extended Data Fig. 6d,e and Extended Data Table 4). Together, this indicates that reducing Fdx1 to an optimal level during developmental stages is protective in frataxin-deficient flies. Furthermore, the Fdx1 overexpression induced by frataxin deficiency seems to worsen the survival defect of these flies, owing to the unbalanced levels of the two proteins.
Conclusion
Our findings reveal that FXN and FDX2 have direct and dose-dependent repressive effects on [2Fe–2S] cluster biosynthesis, which are driven by the mutually exclusive binding of FXN and FDX2 to the same site on the (NIAU)2 complex. These results provide a mechanism to explain the toxicity of FXN overexpression in vivo; as the concentration is raised, FXN competes with FDX2 and thereby prevents the reaction from operating at its optimal rate. These mutual repressive effects are exacerbated by the comparable affinities of FXN and FDX2, both reduced and oxidized, for their binding site in the (NIAU)2 complex, making the repression sensitive to any variation in the amount of either protein. Our data also show that FDX2 has a direct repressive effect on persulfide generation and transfer to ISCU2, which worsens the competitive effect with FXN. The structure–function correlation with FDX2 point mutants indicates that FDX2 interferes with persulfide formation and transfer by interacting with the mobile loop of NFS1, which is likely to restrain its motion within the PLP binding site for persulfide generation and toward ISCU2 for persulfide delivery.
We also report that frataxin deficiency induces the expression of Fdx1, the FDX2 orthologue, in a Drosophila model of Friedreich’s ataxia. The mechanisms involved in this transcriptional deregulation are unknown, but might represent an attempt to enhance Fe–S cluster biosynthesis, which, in the context of frataxin deficiency and in light of our findings, would constitute a maladaptive response. Notably, knocking down Fdx1 expression at the developmental stage to an optimal level markedly improves the lifespan of the flies, as was also found in a C. elegans Friedreich’s ataxia model31, suggesting that Fdx1 overexpression does not enhance but instead negatively affects Fe–S cluster biosynthesis. The positive effect of Fdx1 knockdown in Drosophila, and the existence of an optimal concentration of Fdx1 that has a positive effect on lifespan, are both highly consistent with the in vitro data, indicating that this effect relies on the alleviation of the repressive effects of Fdx1. These data suggest that in the context of frataxin deficiency in individuals with Friedreich’s ataxia, decreased levels of FDX2 could attenuate competition with FXN and reduce interference with the motion of the NFS1 mobile loop, which should overall improve Fe–S cluster assembly. Our in vitro and in vivo data thus highlight new pathways for the regulation of Fe–S cluster biosynthesis, and suggest that targeting FDX2 could be explored as a therapeutic axis for Friedreich’s ataxia.
Methods
Protein purification
Homo sapiens ISCU2 (Q9H1K1-1), (NIA)2 (Q9Y697, Q9HD34 and O14561), FXN (Q16595), FDXR (P22570) and wild-type and mutant FDX2 (Q6P4F2) proteins were prepared as previously described18. BL21 Star (DE3) cells were transformed with the corresponding plasmids: pCDFDuet-His6-Thr-NFS1 (site 1), ISD11 (site 2) with pET21(+)-ACP for (NIA)2, pET28a(+)-His6-Thr-ISCU2 for ISCU2, pET28a(+)-His6-Thr-FDX2 for wild-type and mutant FDX2 constructs, pET28a(+)-His6-Thr-FXN for FXN, and pCDFDuet-His6-FDXR with pET28a(+)-GroEL for FDXR, where His6 corresponds to a 6× histidine tag and Thr corresponds to a thrombin cleavage site with sequence SQDLVPRGS. Cells were cultured in LB with appropriate antibiotics at 37 °C at 180 rpm. Cell culture transformed with wild-type and mutant FDX2 were supplemented with FeSO4 to a final concentration of 10 µM, and FDXR growth was supplemented with riboflavin to a final concentration of 3.75 µM. Protein overexpression was induced by IPTG to a final concentration of 1 mM. (NIA)2 and FDXR were induced when the optical density at 600 nm (OD600) reached 0.6, and incubated overnight at 25 °C (FDXR) or 18 °C ((NIA)2). ISCU2, FXN and FDX2 were induced when the OD600 reached 0.7 and incubated for 3.5 h at 30 °C. Cells were collected by centrifugation at 4,500 rpm for 20 min at 4 °C with a Beckman Coulter JLA-10.500 rotor, and resuspended in His buffer A (20 mM Tris-HCl, 250 mM NaCl and 5 mM imidazole, pH 8.0) with the addition of protease inhibitor (cOmplete Protease Inhibitor Cocktail). Cells were lysed by sonication and cell debris was removed by centrifugation at 45,000 rpm for 45 min at 4 °C with a Beckman Coulter Type 70 Ti rotor. Supernatant was loaded to a Cytiva 5-ml HisTrap HP pre-equilibrated with His buffer A and eluted over a linear gradient of 0–100% His buffer B (20 mM Tris-HCl, 250 mM NaCl and 500 mM imidazole, pH 8.0). Fractions were pooled and concentrated by ultracentrifugation using Amicon Ultra-15 Centrifugal Filter Units with a molecular-mass cut-off of 10 kDa (ISCU2, FXN, FDX2 and FDXR) or 50 kDa for (NIA)2. Proteins (ISCU2, FXN, FDX2 and (NIA)2) were buffer-exchanged into SEC buffer (20 mM Tris-HCl and 250 mM NaCl, pH 8.0) using a NAP-5 desalting column, and FDXR was buffer-exchanged into ion-exchange buffer A (20 mM Tris-HCl and 25 mM NaCl, pH 8.0). ISCU2, FXN and FDX2 were treated with recombinant thrombin protease to remove the His6-tag. ISCU2 was incubated with 50 molar eq. DTT and 100 eq. DTPA for 60 min at 20 °C. (NIA)2 was incubated with 50 eq. DTT, 2 eq. PLP and 4 eq. TCEP for 60 min at 20 °C. Proteins were subjected to size-exclusion chromatography using HiLoad 16/600 columns packed with Superdex 75 pg (ISCU2, FXN and FDX2) or 200 pg for (NIA)2 pre-equilibrated with SEC buffer (FXN and FDX2) or SEC buffer + 10 mM DTT (ISCU2 and (NIA)2). Proteins were eluted and fractions were pooled. FDXR was loaded onto a Cytiva 5-ml HiTrap Q-Sepharose column and eluted with a linear gradient of 0–100% ion-exchange buffer B (20 mM Tris-HCl, 500 mM NaCl, pH 8.0). Proteins were concentrated to 0.5 ml using the corresponding Amicon Ultra-15 Centrifugal Filter Unit and buffer-exchanged under anaerobic conditions (< 2 ppm O2) using a NAP-5 column into either degassed Tris buffer (20 mM Tris-HCl and 100 mM NaCl, pH 8.0) or sodium phosphate buffer (50 mM Na2HPO4 and 150 mM NaCl, pH 8.0) sparged with argon for 60 min. Protein concentrations were determined by UV-visible spectroscopy at 280 nm in urea buffer (100 mM Tris-HCl and 8 M urea, pH 7.5) using absorption coefficients of 52,260 M−1 cm−1, 9,970 M−1 cm−1, 26,930 M−1 cm−1 and 44,920 M−1 cm−1 for (NIA)2, ISCU2, FXN and FDXR, respectively. The FDX2 protein concentration was determined by UV-visible spectroscopy at 456 nm in Tris buffer using an absorption coefficient of 10,000 M−1 cm−1 based on the concentration of the [2Fe–2S] cluster as determined using the ferrozine method. Final preparations were aliquoted, flash-frozen in liquid N2 and stored in liquid N2.
Assays of [2Fe–2S] cluster assembly
Fe–S cluster assembly reactions were performed under anaerobic conditions (< 2 ppm O2) in a glove box. Kinetic assays were routinely composed of 20 µM apo-ISCU2 incubated with 20 µM ferrous ammonium sulfate (Fe-(NH4)2(SO4)2), 2 µM of the (NFS1–ISD11–ACP) complex, 1 µM FDXR and 40 µM NADPH. Titrations were performed with 0–50 molar equivalents of FXN (corresponding to 0–100 µM, where one equivalent is relative to the concentration of the (NFS1–ISD11–ACP) complex), and 0–10 eq. of either wild-type or mutant FDX2 (0–20 µM). The reaction mix was transferred into a 384-well plate and incubated at 25 °C in a Tecan Spark microplate reader. Biosynthesis of [2Fe–2S] was initiated by the injection of 30 µM -cysteine to a final volume of 100 µl and kinetics were measured at 456 nm at 25 °C. The data were collected using SparkControl Magellan 3.0. The kinetic rates of [2Fe–2S] were calculated on the basis of the slope at the start of the curve corresponding to the maximum rate.
ARBS assays
Assays were routinely performed in sodium phosphate buffer (50 mM Na2HPO4 and 150 mM NaCl, pH 8.0) at 25 °C with a concentration of ISCU2 and equimolar ferrous ammonium sulfate and/or the (NFS1–ISD11–ACP) complex at 7.5 µM. Concentrations of FDX2 and FXN ranged from 0–10 eq. (0–75 µM) as indicated within the text. For studies following persulfide formation on NFS1, reactions were initiated with 7.5 µM l-cysteine. Assays following persulfide transfer from NFS1 to ISCU2 were initiated with 15 µM l-cysteine. Persulfide reduction assays were initiated with 7.5 µM l-cysteine followed by the addition of a preincubated mixture of FDX2, FDXR and NADPH to final concentrations of 7.5 µM, 3.75 µM and 40 µM, respectively. Reactions were sampled at respective timepoints by mixing 15 µl of the reaction mix with 5 µl of a stop mix making up a fivefold molar excess of mal-dPEG relative to the total thiol concentration (including l-cysteine and total cysteine residues), a 2.5-fold molar excess of EDTA relative to the total iron concentration and a final concentration of 1% SDS. To monitor the persulfidation state of ISCU2 in the presence of FDX2 and prevent overlap of alkylated ISCU2 and FDX2 on gel, which both contain four cysteine residues and have similar molecular weights of 14.4 kDa and 14.3 kDa, respectively, we identified an optimal concentration of SDS in which the solvent-accessible cysteine residues of ISCU2 were available for alkylation, whereas the cysteine residues of FDX2 ligating the [2Fe–2S] cofactor remained partially buried and shielded from alkylation (Supplementary Fig. 1). A final concentration of 0.08% SDS was found to be optimal for this detection. After 30 min of reaction with the stop mix, leading to full alkylation of ISCU2 and/or NFS1, 10 µl of reducing loading dye (60 mM Tris-HCl, 25% glycerol, 2% SDS, 700 mM 2-mercaptoethanol and 0.1% bromophenol blue) was added. Reaction aliquots were analysed by SDS–PAGE on 8% (NFS1) and 14% (ISCU2) acrylamide/bis-acrylamide 19:1 gels. The gels were imaged using an Odyssey Clx scanner (Li-COR) and the data were collected and analysed using Image Studio 5.2.
FIDA
Stock solutions of labelled FXN and FDX2, referred to as FXNALC and FDX2ALC, were prepared by conjugation with ALC 480 using the corresponding Fidabio Protein Labelling Kit (Fida Biosystems). For labelling reactions, FXN and FDX2 were first buffer-exchanged into sodium phosphate buffer (50 mM Na2HPO4 and 150 mM NaCl, pH 8.0) using a NAP-5 column. Then, 75 µl of protein sample (FXN or FDX2) at 100 µM was incubated with 7.5 µl 1 M sodium bicarbonate. A 4 mg ml−1 reactive dye stock solution was prepared by mixing 100 µg ALC 480 with 25 µl DMSO. Then, 7.5 µl of the dye stock solution was incubated at 21 °C for 30 min with the protein sample, protected from light, to induce labelling using a fivefold molar ratio of dye to protein. Excess dye was removed and proteins were transferred to Tris buffer (20 mM Tris-HCl and 100 mM NaCl, pH 8.0) using a NAP-5 column. The concentration of FXNALC was measured by UV-visible spectroscopy at 280 nm using an absorption coefficient of 26,930 M−1 cm−1. Because the ALC 480 dye interferes at wavelengths 400–540 nm, the concentration of FDX2ALC was measured by UV-visible spectroscopy at 320 nm using an absorption coefficient of 20,450 M−1 cm−1 as determined by the ferrozine method. Labelled proteins were aliquoted, flash-frozen in liquid N2 and stored in liquid N2. As a control to ensure that the label was not interfering with protein function, we performed [2Fe–2S] cluster assembly kinetics, and observed that the efficiencies of both FXNALC and FDX2ALC were comparable with those of the unlabelled proteins (Extended Data Fig. 2b).
Binding curves of FXNALC or FDX2ALC with the (NIAU)2 complex were generated using a premix method in which the indicator sample (the labelled protein) is preincubated with the analyte (the binding partner (NIAU)2) before analysis by FIDA. For the FXNALC binding curve, data points are gathered by incubating a fixed concentration of 20 nM FXNALC with a series of (NIAU) concentrations spanning 0–16 µM (corresponding to 0–8 µM of the dimerized (NIAU)2 complex) in a final volume of 20 µl. The data points of FDX2ALC with the (NIAU)2 complex is prepared by incubating a fixed concentration of 20 nM FDX2ALC with a titration of 0–128 µM of the (NIAU) complex (corresponding to 0–64 µM of the dimerized (NIAU)2 complex) in a final volume of 20 µl. Protein samples were prepared in Tris buffer (20 mM Tris-HCl and 100 mM NaCl, pH 8.0) and samples were incubated for a minimum of 10 min to ensure binding equilibrium is reached before assay.
For competition between FXNALC and either wild-type or mutant FDX2 constructs for the (NIAU)2 complex, a fixed concentration of FXNALC (20 nM) and (NIAU) (2 µM) was titrated by unlabelled FDX2 spanning 0–64 µM. For measurements with reduced FDX2, deoxygenated solutions of the analyte, indicator and buffer were prepared under anaerobic conditions in a glove box. Then, 10 molar eq. of dithionite were added to each FDX2 sample to reduce and keep it reduced throughout the experiment. The full reduction of FDX2 was checked by UV-visible spectroscopy at 456 nm. The analyte, indicator and buffer were loaded into capped vials sealed with tape and placed into a septum sealed tube before analysis. We checked by UV-visible spectroscopy that FDX2 remained reduced under the conditions and duration of FIDA analysis after removal of the tape on top of the cap while keeping tape around the seal. For the competition between FDX2ALC and FXN for the (NIAU)2 complex, a fixed concentration of FDX2ALC (20 nM) and (NIAU) (16 µM) was titrated by 0–256 µM unlabelled FXN.
Binding experiments were performed using a Fida 1 instrument (Fida Biosystems) using laser-induced fluorescence detection with an excitation wavelength of 480 nm. The data were collected using Fida 1 instrument software (v.2.42). A permanently coated capillary (outer diameter 375 µm, inner diameter 75 µm, length to detector 84 cm; Fida Biosystems) was routinely used with constant temperature control of 25 °C. The capillary was equilibrated with the analyte at 3,500 mbar for 30 s. After this, a small plug of the premixed indicator sample was injected at 50 mbar for 10 s. Finally, the indicator was mobilized by injecting the analyte at 400 mbar for 180 s and fluorescence was measured at the detector window. Competition binding curves were performed using a premix method with the same mobilization protocol.
The data points were analysed using Fida analysis software (v.2.32) with the single-species fit to determine the corresponding hydrodynamic radius. Binding curves were then fit using GraphPad Prism (v.8.0.2) to a single-site binding model43 (FXNALC with (NIAU)2 or FDX2ALC with (NIAU)2) or a competitive binding model (FXNALC–(NIAU)2 or FDX2ALC–(NIAU)2 titrated with unlabelled FDX2 or FXN) (Supplementary Equation 1).
ITC
ITC measurements were performed at 25 °C with a MicroCal PEAQ-ITC system (Malvern Panalytical). Zn–ISCU2 was prepared by incubating apo-ISCU2 with 5 eq. DTT and 1.5 eq. ZnSO4 for 60 min and then exchanged to remove excess DTT and ZnSO4 using a NAP-5 column. All protein samples used in ITC experiments were buffer-exchanged into the same preparation of Tris buffer (20 mM Tris-HCl and 100 mM NaCl, pH 8.0) using a NAP-5 column.
The 0.24-ml sample cell was loaded with 20 µM of the (NIA)2 or (NIAU–Zn)2 complex (corresponding to 40 µM of NFS1). Aliquots (2 µl) of the partner proteins (apo-ISCU2, Zn-loaded ISCU2, FDX2 or FXN) were placed in the 40-µl syringe at 400 µM and injected into the sample cell every 180 s under a stirring speed of 500 rpm using the MicroCal PEAQ-ITC Control Software (v.1.41). For the interaction of FDX2 or FXN with ISCU2–Zn, the sample cell was loaded with 40 µM ISCU2–Zn and the syringe was loaded with 400 µM FDX2 or FXN. For the interaction of FXN with FDX2, the sample cell was loaded with 40 µM FXN and the syringe was loaded with 400 µM FDX2. For experiments measuring the interactions of the mutant FDX2 constructs with (NIA)2, the cell was loaded with 10 µM of the (NIA)2 complex, and the syringe with 200 µM of the corresponding FDX2 mutant.
Raw thermograms were integrated using NITPIC (v.2.1.0)44,45. The integrated ITC data were analysed using SEDPHAT (v.15.2b)46. With respect to SEDPHAT nomenclature, interactions of (NIA)2–Zn–ISCU2, (NIA)2–apo-ISCU2 or (NIAU)2–FXN were fitted using the model ‘A + B + B <-> {AB} + B <-> ABB; with two symmetric sites, macroscopic K’, where A corresponds to (NIA)2 or (NIAU)2, and B corresponds to apo-ISCU2, Zn–ISCU2 or FXN. The interaction between (NIA)2 or (NIAU)2 with FDX2 was modelled using ‘A + B + B <-> AB + B <-> BA + B <-> BAB; with 2 non-symmetric sites, microscopic K’, where A corresponds to (NIA)2 or (NIAU)2, and B corresponds to FDX2. The 68.3% error intervals determined during parameter fitting within SEDPHAT were calculated as previously described47. ITC figures were produced using GUSSI (v.2.1.0)48.
AlphaFold
We used our in-house implementation of ColabFold 1.349, which incorporates AlphaFold 2.250, to generate models for the ISC complex composed of NFS1, ISD11, ACP, ISCU2 and FDX2, with the corresponding Uniprot IDs Q9Y697, Q9HD34, O14561 (69–156), Q9H1K1 (35–167) and Q6P4F2 (56-186), respectively. For the construction of multimeric models, we used the multimer version of AlphaFold 2.251 on a local workstation equipped with an RTXA6000. Amino acid sequences were used to query the UniRef30 database (June 2021) and the ColabFold environmental database (August 2021) using the MMseqs2 webserver (v.2)52. The resulting alignments were then used as input for AlphaFold, using ColabFold’s scripts, to generate five models per input. Each model generation involved six cycles and final refinements ranked according to local confidence score using predicted local distance difference test (pLDDT) and predicted aligned error (PAE).
Drosophila lines and culture methods
The fh-GAAs and daGS lines were previously described39,53. The w1118 control and UAS-Fdx1 RNAi (v104499) lines were obtained from the Vienna Drosophila Resource Center (VDRC) and the daGAL4 line (PGAL4-da.G32-2) from the Bloomington Stock Center. The composition of the fly food, methods for RU486 treatments and lifespan analysis were as previously described39. For survival experiments, male flies of the same genotype were affected randomly to various doses of RU-486 treatments. Dead flies were counted concurrently with the transfer of flies to fresh medium containing the specified RU486 concentration, blinding was not applicable.
Quantification of transcripts by quantitative PCR with reverse transcription (RT–qPCR)
Total RNA was extracted from 20 third instar male larvae or adults and treated with dsDNase (Thermo Fisher Scientific) before cDNA synthesis using SuperScript III Reverse Transcriptase (Thermo Fisher Scientific). Quantitative PCR was performed with the qPCR Mix (Promega) on a LightCycler 480 (Roche). The ribosomal gene rp49 was used as an internal reference for normalization. The primers used for amplifications were 5′-CCGCTTCAAGGGACAGTATCT-3′ and 5′-CACGTTGTGCACCAGGAACTT-3’ for rp49 (CG7939), 5′-GTCCACGGATGAAATAGTGAACA-3′ and 5′-GGGCCAAGTACAGAACATTGTC-3 or 5′-CTACACACCCCACAATGCTCT-3′ and 5′-TCACTATTTCATCCGTGGACTTG-3 for Fdx1, 5′-GGCCCTGAAAACGAAAGATGT-3′ and 5′-GCCCCGAAACCATCCAGAT-3 for Fdx2 and 5′-ACACCCTGGACGCACTGT-3′ and 5′-GTTGATCACATAGGTGCCGTG-3 for fh. Ct values were obtained using the second derivative method. Quantifications were made on four to six independent biological samples with four technical replicates for each sample. The data were collected and analysed using LightCycler 480 Software release 1.5.0. Statistical analysis were performed with Mann–Whitney tests or one-way ANOVA tests followed by post-hoc Tukey analysis for multiple comparisons.
Software
Unless otherwise stated, the data were analysed and plotted using GraphPad Prism 8.0.2 and Excel 2016. Protein structures were visualized using PyMOL 3.0. Figure layouts were created using PowerPoint 2016.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the findings of this study are included in the Article and its Supplementary Information. The structure of the NFS1–ISD11–ACP–ISCU2–FXN complex (Protein Data Bank (PDB): 6NZU) used in Fig. 2 and Extended Data Figs. 2 and 3 is available at the PDB website (https://www.rcsb.org/). Source data are provided with this paper.
References
Shi, R., Hou, W., Wang, Z.-Q. & Xu, X. Biogenesis of iron–sulfur clusters and their role in DNA metabolism. Front. Cell Dev. Biol. 9, 735678 (2021).
Honarmand Ebrahimi, K. et al. Iron–sulfur clusters as inhibitors and catalysts of viral replication. Nat. Chem. 14, 253–266 (2022).
Kimura, S. & Suzuki, T. Iron–sulfur proteins responsible for RNA modifications. Biochim. Biophys. Acta 1853, 1272–1283 (2015).
Nicolet, Y. Structure–function relationships of radical SAM enzymes. Nat. Catal. 3, 337–350 (2020).
Stiban, J., So, M. & Kaguni, L. S. Iron–sulfur clusters in mitochondrial metabolism: multifaceted roles of a simple cofactor. Biochemistry 81, 1066–1080 (2016).
Flint, D. H. & Allen, R. M. Iron−sulfur proteins with nonredox functions. Chem. Rev. 96, 2315–2334 (1996).
Crack, J. C., Green, J., Thomson, A. J. & Brun, N. E. L. Iron–sulfur clusters as biological sensors: the chemistry of reactions with molecular oxygen and nitric oxide. Acc. Chem. Res. 47, 3196–3205 (2014).
Cardenas-Rodriguez, M., Chatzi, A. & Tokatlidis, K. Iron–sulfur clusters: from metals through mitochondria biogenesis to disease. J. Biol. Inorg. Chem. 23, 509–520 (2018).
Beilschmidt, L. K. & Puccio, H. M. Mammalian Fe–S cluster biogenesis and its implication in disease. Biochimie 100, 48–60 (2014).
Camponeschi, F., Ciofi-Baffoni, S., Calderone, V. & Banci, L. Molecular basis of rare diseases associated to the maturation of mitochondrial [4Fe–4S]-containing proteins. Biomolecules 12, 1009 (2022).
Monfort, B., Want, K., Gervason, S. & D’Autreaux, B. Recent advances in the elucidation of frataxin biochemical function open novel perspectives for the treatment of Friedreich’s ataxia. Front. Neurosci. 16, 838335 (2022).
Gervason, S. et al. Physiologically relevant reconstitution of iron-sulfur cluster biosynthesis uncovers persulfide-processing functions of ferredoxin-2 and frataxin. Nat. Commun. 10, 3566 (2019).
Gervason, S. et al. The ISC machinery assembles [2Fe–2S] clusters by formation and fusion of [1Fe–1S] precursors. Nat. Chem. Biol. 21, 767–778 (2025).
Parent, A. et al. Mammalian frataxin directly enhances sulfur transfer of NFS1 persulfide to both ISCU and free thiols. Nat. Commun. 6, 5686 (2015).
Bridwell-Rabb, J., Fox, N. G., Tsai, C. L., Winn, A. M. & Barondeau, D. P. Human frataxin activates Fe–S cluster biosynthesis by facilitating sulfur transfer chemistry. Biochemistry 53, 4904–4913 (2014).
Lill, R. From the discovery to molecular understanding of cellular iron–sulfur protein biogenesis. Biol. Chem. 401, 855–876 (2020).
Want, K. & D’Autreaux, B. Mechanism of mitochondrial [2Fe–2S] cluster biosynthesis. Biochim. Biophys. Acta Mol. Cell Res. 1871, 119811 (2024).
Srour, B. et al. Iron insertion at the assembly site of the ISCU scaffold protein is a conserved process initiating Fe–S cluster biosynthesis. J. Am. Chem. Soc. 144, 17496–17515 (2022).
Freibert, S.-A. et al. N-terminal tyrosine of ISCU2 triggers [2Fe–2S] cluster synthesis by ISCU2 dimerization. Nat. Commun. 12, 6902 (2021).
Saini, A. K. et al. Recent advances in the treatment strategies of Friedreich’s ataxia: a review of potential drug candidates and their underlying mechanisms. Curr. Pharm. Des. 30, 1472–1489 (2024).
Ocana-Santero, G., Díaz-Nido, J. & Herranz-Martín, S. Future prospects of gene therapy for Friedreich’s ataxia. Int. J. Mol. Sci. 22, 1815 (2021).
Gunther, K. & Lynch, D. R. Pharmacotherapeutic strategies for Friedreich ataxia: a review of the available data. Expert Opin. Pharmacother. 25, 529–539 (2024).
Seguin, A. et al. Overexpression of the yeast frataxin homolog (Yfh1): contrasting effects on iron–sulfur cluster assembly, heme synthesis and resistance to oxidative stress. Mitochondrion 9, 130–138 (2009).
Navarro, J. A. et al. Overexpression of human and fly frataxins in Drosophila provokes deleterious effects at biochemical, physiological and developmental levels. PLoS One 6, e21017 (2011).
Vannocci, T. et al. Adding a temporal dimension to the study of Friedreich’s ataxia: the effect of frataxin overexpression in a human cell model. Dis. Model Mech. 11, dmm032706 (2018).
Belbellaa, B., Reutenauer, L., Messaddeq, N., Monassier, L. & Puccio, H. High levels of frataxin overexpression lead to mitochondrial and cardiac toxicity in mouse models. Mol. Ther. Methods Clin. Dev. 19, 120–138 (2020).
Huichalaf, C. et al. In vivo overexpression of frataxin causes toxicity mediated by iron-sulfur cluster deficiency. Mol. Ther. Methods Clin. Dev. 24, 367–378 (2022).
Yan, R. et al. Ferredoxin competes with bacterial frataxin in binding to the desulfurase IscS. J. Biol. Chem. 288, 24777–24787 (2013).
Uzarska, M. A. et al. During FeS cluster biogenesis, ferredoxin and frataxin use overlapping binding sites on yeast cysteine desulfurase Nfs1. J. Biol. Chem. 298, 101570 (2022).
Steinhilper, R. et al. Two-stage binding of mitochondrial ferredoxin-2 to the core iron-sulfur cluster assembly complex. Nat. Commun. 15, 10559 (2024).
Meisel, J. D. et al. Mutations in mitochondrial ferredoxin FDX2 suppress frataxin deficiency. Nature https://doi.org/10.1038/s41586-025-09821-2 (2025).
Jensen, H. & Østergaard, J. Flow induced dispersion analysis quantifies noncovalent interactions in nanoliter samples. J. Am. Chem. Soc. 132, 4070–4071 (2010).
Iannuzzi, C. et al. The role of zinc in the stability of the marginally stable IscU scaffold protein. Protein Sci. 23, 1208–1219 (2014).
Kim, J. H. et al. Structure and dynamics of the iron–sulfur cluster assembly scaffold protein IscU and its interaction with the cochaperone HscB. Biochemistry 48, 6062–6071 (2009).
Gervason, S., Srour, B. & D’Autréaux, B. in Fe–S Proteins: Methods in Molecular Biology Vol. 2353 (ed. Dos Santos, P. C.) 191–205 (Springer, 2021).
Adasme, M. F. et al. PLIP 2021: expanding the scope of the protein-ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 49, W530–W534 (2021).
Moreira, I. S. et al. SpotOn: high accuracy identification of protein–protein interface hot-spots. Sci. Rep. 7, 8007 (2017).
Monnier, V., Llorens, J. V. & Navarro, J. A. Impact of Drosophila models in the study and treatment of Friedreich’s ataxia. Int. J. Mol. Sci. 19, 1989 (2018).
Jullian, E. et al. Glial overexpression of Tspo extends lifespan and protects against frataxin deficiency in Drosophila. Biochimie 224, 71–79 (2024).
Russi, M. et al. A Drosophila model of Friedreich ataxia with CRISPR/Cas9 insertion of GAA repeats in the frataxin gene reveals in vivo protection by N-acetyl cysteine. Hum. Mol. Genet. 29, 2831–2844 (2020).
Palandri, A., L’Hote, D., Cohen-Tannoudji, J., Tricoire, H. & Monnier, V. Frataxin inactivation leads to steroid deficiency in flies and human ovarian cells. Hum. Mol. Genet. 24, 2615–2626 (2015).
Hekkelman, M. L., De Vries, I., Joosten, R. P. & Perrakis, A. AlphaFill: enriching AlphaFold models with ligands and cofactors. Nat. Methods 20, 205–213 (2023).
Pedersen, M. E., Gad, S. I., Ostergaard, J. & Jensen, H. Protein characterization in 3D: size, folding, and functional assessment in a unified approach. Anal. Chem. 91, 4975–4979 (2019).
Keller, S. et al. High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal. Chem. 84, 5066–5073 (2012).
Scheuermann, T. H. & Brautigam, C. A. High-precision, automated integration of multiple isothermal titration calorimetric thermograms: new features of NITPIC. Methods 76, 87–98 (2015).
Zhao, H., Piszczek, G. & Schuck, P. SEDPHAT—a platform for global ITC analysis and global multi-method analysis of molecular interactions. Methods 76, 137–148 (2015).
Brautigam, C. A. Fitting two- and three-site binding models to isothermal titration calorimetric data. Methods 76, 124–136 (2015).
Brautigam, C. A. Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzym. 562, 109–133 (2015).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Evans, R. et al. Protein complex prediction with AlphaFold-Multimer. Preprint at bioRxiv https://doi.org/10.1101/2021.10.04.463034 (2021).
Steinegger, M. & Söding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 35, 1026–1028 (2017).
Tricoire, H. et al. The steroid hormone receptor EcR finely modulates Drosophila lifespan during adulthood in a sex-specific manner. Mech. Ageing Dev. 130, 547–552 (2009).
Poulsen, N. N. et al. Flow induced dispersion analysis rapidly quantifies proteins in human plasma samples. Analyst 140, 4365–4369 (2015).
Acknowledgements
This work was supported by the French Agence Nationale de la Recherche (FeSAm ANR-22-CE11-0006-04), the Friedreich’s Ataxia Research Alliance USA (FARA research grants 202212 and 272722) and Ataxia UK. The PhD of K.W. was funded by the Commissariat à l'énergie atomique et aux énergies alternatives (CEA). We thank K. Sahni Ray and H. Jensen for technical assistance with FIDA experiments and analysis, and J. Meisel, V. Mootha and G. Ruvkun for discussions. The work benefited from the PIM platform at I2BC, supported by French Infrastructure for Integrated Structural Biology (FRISBI) ANR-10-INBS-05.
Author information
Authors and Affiliations
Contributions
K.W. designed and performed all in vitro experiments unless otherwise noted. H.G. performed [2Fe–2S] kinetics assays with FDX2 at concentrations of 5 eq., 10 eq. and 50 eq., performed the FIDA competition assay using reduced FDX2 and performed the ARBS assays of pre-persulfidated NFS1 persulfide transfer to ISCU2 in the presence of varying FDX2 equivalents, and ARBS assays with the FDX2(D179A) mutant. E.T. performed all in vivo experiments unless otherwise noted. M.N. assisted with the ITC and FIDA experiments. B.M. performed the initial [2Fe–2S] FXN titration that showed an inhibitory effect. R.M.-G. assisted with protein expression, protein purification and the set-up of anaerobic conditions for FIDA. T.T. performed the AlphaFold modelling. E.J. helped to produce genetic constructs and perform survival experiments. V.M. designed, supervised and analysed in vivo experiments. B.D. managed funds, coordinated collaborations and designed, supervised and analysed in vitro experiments. B.D., K.W. and V.M. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Cole Haynes and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Mechanistic scheme and assay of [2Fe–2S] cluster biosynthesis.
a, Scheme of the [2Fe–2S] cluster assembly process by the eukaryotic ISC machinery. The reaction is initiated by the binding of ferrous iron into the assembly site of apo-ISCU2 (step 1). ISCU2, with or without iron, binds to the NFS1–ISD11–ACP dimeric complex (ISD11–ACP are omitted for clarity, the figure is showing Fe–ISCU2 binding to NFS1) (step 2). The pyridoxal phosphate cofactor of NFS1 uses L-cysteine as a sulfur source to generate a persulfide on the catalytic cysteine carried by a mobile loop (step 3). The persulfide carried by the mobile loop of NFS1 is delivered to the receptor cysteine (Cys104) of ISCU2 (shown here as “-SH”) (step 4). This persulfide transfer reaction is accelerated by FXN (step 4). FDX2 delivers electrons to ISCU2, which reduces the persulfide of ISCU2 into sulfide (step 5). FDX2 is reduced by FDXR using electrons from NADPH. The reduction of persulfide into sulfide leads to a [1Fe–1S] precursor and the ISCU2 subunit carrying this precursor dissociates from NFS1 (step 6) and dimerizes to generate a bridging [2Fe–2S] intermediate (step 7). Finally, after dissociation of the dimer, the bridging [2Fe–2S] cluster is retained in one of the ISCU2 subunits, leading to formation of apo-ISCU2 and a monomer holding the [2Fe–2S] cluster (step 8). b, Kinetic synthesis of [2Fe–2S] by the recombinant H. sapiens ISC system monitored at 456 nm including 1 eq. (green) or 10 eq. (red) FXN, or in the absence of FXN (blue). c, Rates of [2Fe–2S] synthesis determined based on the slopes at the start of the curves from b. d,e, Plot of the optimal amount of FXN (d) or FDX2 (e) from Fig. 1a producing the maximum rate of [2Fe–2S] cluster synthesis as a function of the amount of FDX2 or FXN, expressed as equivalents of FXN and FDX2 relative to one subunit of NIAU. (b, representative kinetics selected from n = 3 independent experiments with similar results; c–e, mean, ± s.d., n = 3 independent experiments).
Extended Data Fig. 2 Analysis of ISC components by FIDA.
a, Illustration of the experimental principle of FIDA. The capillary is first equilibrated with an analyte, followed by injection of the fluorescently labelled indicator (adapted from ref. 54 under a Creative Commons licence CC BY 3.0). Indicator flow along the capillary is induced and the fluorescent profile is measured at the detector window. b, Kinetics of [2Fe–2S] cluster synthesis by the recombinant ISC system using FXN and FDX2 either unlabelled (FXN, FDX2) or labelled (FXNALC, FDX2ALC). c, FIDA Taylorgram profiles of FXNALC and FDX2ALC alone or after saturation with the (NIAU)2 complex. d,e, Corresponding hydrodynamic radius of labelled proteins and their complexes determined by FIDA mapped as a sphere alongside the structure of FXN (PDB: 6NZU) and the FXN–(NIAU)2 complex (PDB: 6NZU) (d) or FDX2 (AlphaFold v.2.2) and the FDX2–(NIAU)2 complex (AlphaFold v.2.2) (e). f, FIDA competition assays of unlabelled oxidized (blue) and reduced (red) FDX2 titrated into a preformed FXNALC–(NIAU)2 complex. (b, representative data selected from n = 3 independent experiments with similar results; f, mean, ± s.d., n = 3 independent experiments).
Extended Data Fig. 3 Measuring the interactions between components of the ISC system with ITC.
a–f, Monitoring the interaction between: (a) apo-ISCU2 and the (NIA)2 complex, (b) Zn-loaded holo-ISCU2 and the (NIA)2 complex, (c) FXN and the (NIA)2 complex, (d) FXN and Zn-loaded ISCU2, (e) FDX2 and Zn-loaded ISCU2, (f) FDX2 and FXN (heat of injection error bars correspond to unbiased NITPIC-assigned integration errors of thermal spikes; (a,b) representative experiments selected from n = 3 independent experiments with similar results; (c–f) n = 1).
Extended Data Fig. 4 ARBS assays to monitor the effect of FXN and FDX2 on persulfide generation, transfer and reduction.
a–f, ARBS assays monitoring: (a) persulfide generation on NFS1 in the absence (0 eq. FDX2) or presence of 10 eq. FDX2, and persulfide generation on NFS1 in the NFS1-apo-ISCU2 complex in the absence of FDX2 and FXN (0 eq. FDX2/FXN) or presence of 10 eq. FDX2 or 10 eq. FXN. (b) persulfide transfer from NFS1 to ISCU2 in the absence (0 eq. FXN) or presence of 1, 5 or 10 eq. FXN, (c) persulfide reduction on ISCU2 by the addition of a preincubated FDX2-FDXR-NADPH complex in the absence (0 eq. FXN) or presence of 1 or 10 eq. FXN. NP and P labelling corresponds to non-persulfidated (before the addition of L-cysteine) and persulfidated (after the addition of L-cysteine), respectively, (d) persulfide transfer from pre-persulfidated NFS1 to ISCU2 in the absence (0 eq. FDX2) or presence of 3, 5, or 10 eq. FDX2, (e) persulfide transfer from NFS1 to ISCU2 in either the absence (0 eq. FDX2) or presence of 1,3, 5, 7, or 10 eq. FDX2, (f) persulfide transfer from NFS1 to ISCU2 in the presence of 1eq FXN and either the absence (0 eq. FDX2) or presence of 1 or 10 eq. FDX2. Persulfidated protein bands are annotated as NFS1–SSH and ISCU2–SSH, and non-persulfidated protein bands are annotated as NFS1-SH and ISCU2-SH. ISCU2 is loaded with iron unless otherwise stated. For gel source data, see Supplementary Figs. 2 and 3. (a–f, representative ARBS assays from n = 3 independent experiments with similar results).
Extended Data Fig. 5 ITC and ARBS assays of FDX2 mutants.
a, ITC analysis monitoring the interaction between FDX2 mutants and the (NIA)2 complex. The binding curves are fit to a two-site symmetric binding model (E134A, D137A, D138A, D141A) or a two-site non-symmetric binding model (E148A, Y177A, D179A, H181A). b–g, ARBS assays monitoring: (b,c) persulfide generation on NFS1 in the presence of 10 eq. FDX2 WT or 10 eq. FDX2 D179A, (b) representative ARBS gel used for the quantifications of persulfidated NFS1 (NFS1-SSH) presented in (c), (d,e) persulfide transfer from NFS1 to ISCU2 in the absence of FDX2 (No FDX2) or in the presence of either 3 eq. or 5 eq. FDX2 WT or FDX2 D179A, (d) representative ARBS assay gel used for the quantifications of persulfidated ISCU2 (ISCU-SSH) presented in (e), (f,g) monitoring persulfide transfer from NFS1 to ISCU2 in the presence of 1 eq. FXN and either absence of FDX2 (No FDX2) or in the presence of either 5 eq. FDX2 WT or FDX2 D179A, (f) representative ARBS assay gel used for the quantifications of persulfidated ISCU2 (ISCU-SSH) presented in (g). For gel source data from b,d,f, see Supplementary Fig. 4. (a, heat of injection error bars corresponds to unbiased NITPIC-assigned integration errors of thermal spikes; n = 1; c,e,g, mean, ± s.d.; n = 3 independent experiments).
Extended Data Fig. 6 Quantification of Fdx1 and Fdx2 transcripts and survival of frataxin-deficient flies with or without RNAi-induced Fdx1 knockdown.
a,b, Quantification of (a) Fdx1 and (b) Fdx2 transcript levels following RNAi-induced Fdx1 knockdown with the constitutive daGAL4 driver. Relative quantifications were performed by RT–qPCR on whole-body extracts of third instar larvae. c, Mean lifespan of fh-GAAs male flies. RU treatments did not affect the fly survival. d,e, Survival curves and mean lifespan of fh-GAAs; daGS>UAS-Fdx1 RNAi flies. d, Male flies were either untreated or treated with various concentrations of RU486 during development from 0.1 to 1.6 μg ml−1, with a constant RU486 concentration of 10 μg ml−1 at the adult stage. e, Male flies were untreated or treated with 1.6 μg ml−1 RU486 during development and either untreated or treated with 10 μg ml−1 or 100 μg ml−1 at the adult stage (datasets for 0-0 and 1.6-10 conditions are similar in d and e). Labels Dev-Adult indicate RU486 concentrations (μg ml−1) in the fly food during development and adult stages. a,b, Dots (n = 6 except for daGAL4>+, n = 5) correspond to independent biological samples each obtained from 20 individuals. Mean values ± s.d. are indicated. a.u: arbitrary units. Two-sided non-parametric Mann–Whitney tests were used. (c–e) The total numbers of flies per condition and the p values obtained by log rank tests are provided in Extended Data Table 4. For each condition, the flies were housed in tubes by groups of mainly 28 to 30 individuals. For mean lifespan analysis, each dot corresponds to one tube (n = 5 in (c), n = 4 in (d,e) except for the 0–100 (n = 3) and the 1.6-0 treatments (n = 5)). Mean values ± s.d. are indicated. One-way Anova followed by Post-hoc Tukey analysis for multiple comparisons was used. Precise number of flies per tube and P values are provided in the source data. *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary information
Supplementary Information
This Supplementary Information file contains 4 Supplementary Figures and 1 Supplementary Equation. Supplementary Fig. 1 presents the gels for optimizing the concentration of SDS to prevent the overlap of alkylated ISCU2 and FDX2. Supplementary Figs. 2-4 present the uncropped versions of all the gels of ARBS assays presented in the main and Extended Data Figures. Supplementary Equation 1 presents the equations for establishing the FIDA data fitting model for competitive binding.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Want, K., Gorny, H., Turki, E. et al. Cross-regulation of [2Fe–2S] cluster synthesis by ferredoxin-2 and frataxin. Nature 649, 721–728 (2026). https://doi.org/10.1038/s41586-025-09822-1
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-025-09822-1