Abstract
Type 1 conventional dendritic cells (cDC1s) are unique in their efferocytosis1 and cross-presenting abilities2, resulting in antigen-specific T cell immunity3 or tolerance4,5,6,7,8. However, the mechanisms that underlie cDC1 tolerogenic function remain largely unknown. Here we show that the erythropoietin receptor (EPOR) acts as a critical switch that determines the tolerogenic function of cDC1s and the threshold of antigen-specific T cell responses. In total lymphoid irradiation-induced allograft tolerance9,10, cDC1s upregulate EPOR expression, and conditional knockout of EPOR in cDC1s diminishes antigen-specific induction and expansion of FOXP3+ regulatory T (Treg) cells, resulting in allograft rejection. Mechanistically, EPOR promotes efferocytosis-induced tolerogenic maturation7,11 of splenic cDC1s towards late-stage CCR7+ cDC1s characterized by increased expression of the integrin β8 gene12 (Itgb8), and conditional knockout of Itgb8 in cDC1s impairs tolerance induced by total lymphoid irradiation plus anti-thymocyte serum. Migratory cDC1s in peripheral lymph nodes preferentially express EPOR, and their FOXP3+ Treg cell-inducing capacity is enhanced by erythropoietin. Reciprocally, loss of EPOR enables immunogenic maturation of peripheral lymph node migratory and splenic CCR7+ cDC1s by upregulating genes involved in MHC class II- and class I-mediated antigen presentation, cross-presentation and costimulation. EPOR deficiency in cDC1s reduces tumour growth by enhancing anti-tumour T cell immunity, particularly increasing the generation of precursor exhausted tumour antigen-specific CD8+ T cells13 in tumour-draining lymph nodes and supporting their maintenance within tumours, while concurrently reducing intratumoural Treg cells. Targeting EPOR on cDC1s to induce or inhibit T cell immune tolerance could have potential for treating a variety of diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All transcriptional data generated in the current study were deposited at the NCBI Gene Expression Omnibus (GEO) and are publicly available through the following accession numbers: GSE253056 (bulk RNA-seq) and GSE284080 (scRNA-seq), respectively. Source data are provided with this paper.
Code availability
The scripts for replicating the RNA-seq analyses presented are accessible on GitHub (https://github.com/chansigit/Epor-cDC1-bulkRNAseq). Scripts for reproducing all scRNA-seq analyses presented are accessible on GitHub (https://github.com/chris-mcginnis-ucsf/epor_dc_tolerance) and associated processed data objects are available on Synapse (https://synapse.org/Synapse:syn64330568).
References
Roquilly, A., Mintern, J. D. & Villadangos, J. A. Spatiotemporal adaptations of macrophage and dendritic cell development and function. Annu. Rev. Immunol. 40, 525–557 (2022).
Ohara, R. A. & Murphy, K. M. The evolving biology of cross-presentation. Semin. Immunol. 66, 101711 (2023).
Anderson, D. A. 3rd & Murphy, K. M. Models of dendritic cell development correlate ontogeny with function. Adv. Immunol. 143, 99–119 (2019).
Idoyaga, J. et al. Specialized role of migratory dendritic cells in peripheral tolerance induction. J. Clin. Invest. 123, 844–854 (2013).
Ardouin, L. et al. Broad and largely concordant molecular changes characterize tolerogenic and immunogenic dendritic cell maturation in thymus and periphery. Immunity 45, 305–318 (2016).
Wohn, C. et al. Absence of MHC class II on cDC1 dendritic cells triggers fatal autoimmunity to a cross-presented self-antigen. Sci. Immunol. 5, eaba1896 (2020).
Bosteels, V. et al. LXR signaling controls homeostatic dendritic cell maturation. Sci. Immunol. 8, eadd3955 (2023).
Iberg, C. A., Jones, A. & Hawiger, D. Dendritic cells as inducers of peripheral tolerance. Trends Immunol. 38, 793–804 (2017).
Scandling, J. D., Busque, S., Shizuru, J. A., Engleman, E. G. & Strober, S. Induced immune tolerance for kidney transplantation. N. Engl. J. Med. 365, 1359–1360 (2011).
Scandling, J. D. et al. Tolerance and chimerism after renal and hematopoietic-cell transplantation. N. Engl. J. Med. 358, 362–368 (2008).
Bosteels, V. & Janssens, S. Striking a balance: new perspectives on homeostatic dendritic cell maturation. Nat. Rev. Immunol. 25, 125–140 (2024).
Travis, M. A. et al. Loss of integrin αvβ8 on dendritic cells causes autoimmunity and colitis in mice. Nature 449, 361–365 (2007).
Im, S. J. et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016).
Billingham, R. E., Brent, L. & Medawar, P. B. Actively acquired tolerance of foreign cells. Nature 172, 603–606 (1953).
Bluestone, J. A. & Anderson, M. Tolerance in the age of immunotherapy. N. Engl. J. Med. 383, 1156–1166 (2020).
Mehrotra, P. & Ravichandran, K. S. Drugging the efferocytosis process: concepts and opportunities. Nat. Rev. Drug Discov. 21, 601–620 (2022).
Zelenay, S. et al. The dendritic cell receptor DNGR-1 controls endocytic handling of necrotic cell antigens to favor cross-priming of CTLs in virus-infected mice. J. Clin. Invest. 122, 1615–1627 (2012).
Anderson, D. A. 3rd, Dutertre, C. A., Ginhoux, F. & Murphy, K. M. Genetic models of human and mouse dendritic cell development and function. Nat. Rev. Immunol. 21, 101–115 (2021).
Ferris, S. T. et al. cDC1 prime and are licensed by CD4+ T cells to induce anti-tumour immunity. Nature 584, 624–629 (2020).
Schulz, O. & Reis e Sousa, C. Cross-presentation of cell-associated antigens by CD8alpha+ dendritic cells is attributable to their ability to internalize dead cells. Immunology 107, 183–189 (2002).
Theisen, D. & Murphy, K. The role of cDC1s in vivo: CD8 T cell priming through cross-presentation. F1000Res. 6, 98 (2017).
Mellman, I., Chen, D. S., Powles, T. & Turley, S. J. The cancer-immunity cycle: Indication, genotype, and immunotype. Immunity 56, 2188–2205 (2023).
Schenkel, J. M. et al. Conventional type I dendritic cells maintain a reservoir of proliferative tumor-antigen specific TCF-1+ CD8+ T cells in tumor-draining lymph nodes. Immunity 54, 2338–2353.e6 (2021).
Spranger, S., Dai, D., Horton, B. & Gajewski, T. F. Tumor-residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell 31, 711–723.e4 (2017).
Zagorulya, M. & Spranger, S. Once upon a prime: DCs shape cancer immunity. Trends Cancer 9, 172–184 (2023).
Murphy, T. L. & Murphy, K. M. Dendritic cells in cancer immunology. Cell. Mol. Immunol. 19, 3–13 (2022).
Meiser, P. et al. A distinct stimulatory cDC1 subpopulation amplifies CD8+ T cell responses in tumors for protective anti-cancer immunity. Cancer Cell 41, 1498–1515.e10 (2023).
Bottcher, J. P. & Reis e Sousa, C. The role of type 1 conventional dendritic cells in cancer immunity. Trends Cancer 4, 784–792 (2018).
Broz, M. L. et al. Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell 26, 938 (2014).
Roberts, E. W. et al. Critical Role for CD103+/CD141+ dendritic cells bearing CCR7 for tumor antigen trafficking and priming of T cell immunity in melanoma. Cancer Cell 30, 324–336 (2016).
Balan, S., Radford, K. J. & Bhardwaj, N. Unexplored horizons of cDC1 in immunity and tolerance. Adv. Immunol. 148, 49–91 (2020).
Silva-Sanchez, A. et al. Activation of regulatory dendritic cells by Mertk coincides with a temporal wave of apoptosis in neonatal lungs. Sci. Immunol. 8, eadc9081 (2023).
Liu, K. et al. Immune tolerance after delivery of dying cells to dendritic cells in situ. J. Exp. Med. 196, 1091–1097 (2002).
Canesso, M. C. C Identification of antigen-presenting cell–T cell interactions driving immune responses to food. Science 387, eado5088 (2024).
Rudnitsky, A. et al. A coordinated cellular network regulates tolerance to food. Nature 644, 231–240 (2025).
Gargaro, M. et al. Indoleamine 2,3-dioxygenase 1 activation in mature cDC1 promotes tolerogenic education of inflammatory cDC2 via metabolic communication. Immunity 55, 1032–1050.e1014 (2022).
Blanco, T. et al. Conventional type I migratory CD103+ dendritic cells are required for corneal allograft survival. Mucosal Immunol. 16, 711–726 (2023).
Hongo, D., Tang, X., Zhang, X., Engleman, E. G. & Strober, S. Tolerogenic interactions between CD8+ dendritic cells and NKT cells prevent rejection of bone marrow and organ grafts. Blood 129, 1718–1728 (2017).
Slavin, S., Strober, S., Fuks, Z. & Kaplan, H. S. Long-term survival of skin allografts in mice treated with fractionated total lymphoid irradiation. Science 193, 1252–1254 (1976).
Crozat, K. et al. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8α+ dendritic cells. J. Exp. Med. 207, 1283–1292 (2010).
Hildner, K. et al. Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. Science 322, 1097–1100 (2008).
Grajales-Reyes, G. E. et al. Batf3 maintains autoactivation of Irf8 for commitment of a CD8α+ conventional DC clonogenic progenitor. Nat. Immunol. 16, 708–717 (2015).
Satpathy, A. T. et al. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J. Exp. Med. 209, 1135–1152 (2012).
Wu, X. et al. Mafb lineage tracing to distinguish macrophages from other immune lineages reveals dual identity of Langerhans cells. J. Exp. Med. 213, 2553–2565 (2016).
Zhang, H. et al. EpoR-tdTomato-Cre mice enable identification of EpoR expression in subsets of tissue macrophages and hematopoietic cells. Blood 138, 1986–1997 (2021).
Kuhrt, D. & Wojchowski, D. M. Emerging EPO and EPO receptor regulators and signal transducers. Blood 125, 3536–3541 (2015).
Alaluf, E. et al. Heme oxygenase-1 orchestrates the immunosuppressive program of tumor-associated macrophages. JCI insight 5, e133929 (2020).
Consonni, F. M. et al. Heme catabolism by tumor-associated macrophages controls metastasis formation. Nat. Immunol. 22, 595–606 (2021).
Doran, A. C., Yurdagul, A. Jr & Tabas, I. Efferocytosis in health and disease. Nat. Rev. Immunol. 20, 254–267 (2020).
Luo, B. et al. Erythropoeitin signaling in macrophages promotes dying cell clearance and immune tolerance. Immunity 44, 287–302 (2016).
Dikiy, S. & Rudensky, A. Y. Principles of regulatory T cell function. Immunity 56, 240–255 (2023).
Scandling, J. D. et al. Macrochimerism and clinical transplant tolerance. Hum. Immunol. 79, 266–271 (2018).
Ehst, B. D., Ingulli, E. & Jenkins, M. K. Development of a novel transgenic mouse for the study of interactions between CD4 and CD8 T cells during graft rejection. Am. J. Transplant. 3, 1355–1362 (2003).
Hashimoto, K., Joshi, S. K. & Koni, P. A. A conditional null allele of the major histocompatibility IA-beta chain gene. Genesis 32, 152–153 (2002).
Strober, S. Use of hematopoietic cell transplants to achieve tolerance in patients with solid organ transplants. Blood 127, 1539–1543 (2016).
Moon, J. J. et al. Naive CD4+ T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 27, 203–213 (2007).
Shao, T. Y. et al. Reproductive outcomes after pregnancy-induced displacement of preexisting microchimeric cells. Science 381, 1324–1330 (2023).
Liu, F. T. & Stowell, S. R. The role of galectins in immunity and infection. Nat. Rev. Immunol. 23, 479–494 (2023).
Gonzales, G. A. et al. The pore-forming apolipoprotein APOL7C drives phagosomal rupture and antigen cross-presentation by dendritic cells. Sci. Immunol. 9, eadn2168 (2024).
Wild, A. B. et al. CD83 orchestrates immunity toward self and non-self in dendritic cells. JCI Insight 4, e126246 (2019).
Sisirak, V. et al. Digestion of chromatin in apoptotic cell microparticles prevents autoimmunity. Cell 166, 88–101 (2016).
Mucida, D. et al. Retinoic acid can directly promote TGF-β-mediated Foxp3+ Treg cell conversion of naive T cells. Immunity 30, 471–472 (2009).
Larange, A. & Cheroutre, H. Retinoic acid and retinoic acid receptors as pleiotropic modulators of the immune system. Annu. Rev. Immunol. 34, 369–394 (2016).
Wu, R. et al. Mechanisms of CD40-dependent cDC1 licensing beyond costimulation. Nat. Immunol. 23, 1536–1550 (2022).
Forster, R., Davalos-Misslitz, A. C. & Rot, A. CCR7 and its ligands: balancing immunity and tolerance. Nat. Rev. Immunol. 8, 362–371 (2008).
Ohl, L. et al. CCR7 governs skin dendritic cell migration under inflammatory and steady-state conditions. Immunity 21, 279–288 (2004).
Azukizawa, H. et al. Steady state migratory RelB+ langerin+ dermal dendritic cells mediate peripheral induction of antigen-specific CD4+CD25+ Foxp3+ regulatory T cells. Eur. J. Immunol. 41, 1420–1434 (2011).
Brown, H., Komnick, M. R., Brigleb, P. H., Dermody, T. S. & Esterhazy, D. Lymph node sharing between pancreas, gut, and liver leads to immune crosstalk and regulation of pancreatic autoimmunity. Immunity 56, 2070–2085.e11 (2023).
Cruz de Casas, P., Knopper, K., Dey Sarkar, R. & Kastenmuller, W. Same yet different — how lymph node heterogeneity affects immune responses. Nat. Rev. Immunol. 24, 358–374 (2023).
Maier, B. et al. A conserved dendritic-cell regulatory program limits antitumour immunity. Nature 580, 257–262 (2020).
Dixon, K. O. et al. TIM-3 restrains anti-tumour immunity by regulating inflammasome activation. Nature 595, 101–106 (2021).
Kretzer, N. M. et al. RAB43 facilitates cross-presentation of cell-associated antigens by CD8α+ dendritic cells. J. Exp. Med. 213, 2871–2883 (2016).
Roche, P. A. & Furuta, K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 15, 203–216 (2015).
Jinushi, M. et al. MFG-E8-mediated uptake of apoptotic cells by APCs links the pro- and antiinflammatory activities of GM-CSF. J. Clin. Invest. 117, 1902–1913 (2007).
Lei, X. et al. CD4+ helper T cells endow cDC1 with cancer-impeding functions in the human tumor micro-environment. Nat. Commun. 14, 217 (2023).
Bonacina, F. et al. Myeloid apolipoprotein E controls dendritic cell antigen presentation and T cell activation. Nat. Commun. 9, 3083 (2018).
Kool, M. et al. The ubiquitin-editing protein A20 prevents dendritic cell activation, recognition of apoptotic cells, and systemic autoimmunity. Immunity 35, 82–96 (2011).
Reith, W., LeibundGut-Landmann, S. & Waldburger, J. M. Regulation of MHC class II gene expression by the class II transactivator. Nat. Rev. Immunol. 5, 793–806 (2005).
Theisen, D. J. et al. WDFY4 is required for cross-presentation in response to viral and tumor antigens. Science 362, 694–699 (2018).
Mortier, E. et al. Macrophage- and dendritic-cell-derived interleukin-15 receptor alpha supports homeostasis of distinct CD8+ T cell subsets. Immunity 31, 811–822 (2009).
Pittet, M. J., Di Pilato, M., Garris, C. & Mempel, T. R. Dendritic cells as shepherds of T cell immunity in cancer. Immunity 56, 2218–2230 (2023).
Prokhnevska, N. et al. CD8+ T cell activation in cancer comprises an initial activation phase in lymph nodes followed by effector differentiation within the tumor. Immunity 56, 107–124.e105 (2023).
Huang, Q. et al. The primordial differentiation of tumor-specific memory CD8(+ ) T cells as bona fide responders to PD-1/PD-L1 blockade in draining lymph nodes. Cell 185, 4049–4066.e4025 (2022).
Jansen, C. S. et al. An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature 576, 465–470 (2019).
Siddiqui, I. et al. Intratumoral Tcf1+PD-1+CD8+ T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 50, 195–211.e110 (2019).
Miller, B. C. et al. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20, 326–336 (2019).
Rahim, M. K. et al. Dynamic CD8+ T cell responses to cancer immunotherapy in human regional lymph nodes are disrupted in metastatic lymph nodes. Cell 186, 1127–1143.e18 (2023).
Borst, J., Ahrends, T., Babala, N., Melief, C. J. M. & Kastenmuller, W. CD4+ T cell help in cancer immunology and immunotherapy. Nat. Rev. Immunol. 18, 635–647 (2018).
Zagorulya, M. et al. Tissue-specific abundance of interferon-gamma drives regulatory T cells to restrain DC1-mediated priming of cytotoxic T cells against lung cancer. Immunity 56, 386–405.e10 (2023).
Ramirez, D. E. & Turk, M. J. Th1-like Treg cells are dressed to suppress anti-tumor immunity. Immunity 56, 1437–1439 (2023).
Moreno Ayala, M. A. et al. CXCR3 expression in regulatory T cells drives interactions with type I dendritic cells in tumors to restrict CD8+ T cell antitumor immunity. Immunity 56, 1613–1630.e5 (2023).
Wei, X. et al. Erythropoietin protects against murine cerebral malaria through actions on host cellular immunity. Infect. Immun. 82, 165–173 (2014).
Zhang, Q. et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell 179, 829–845.e20 (2019).
Magen, A. et al. Intratumoral dendritic cell-CD4+ T helper cell niches enable CD8+ T cell differentiation following PD-1 blockade in hepatocellular carcinoma. Nat. Med. 29, 1389–1399 (2023).
Mair, F. et al. Extricating human tumour immune alterations from tissue inflammation. Nature 605, 728–735 (2022).
Wu, R. & Murphy, K. M. DCs at the center of help: origins and evolution of the three-cell-type hypothesis. J. Exp. Med. 219, e20211519 (2022).
Kim, J. M., Rasmussen, J. P. & Rudensky, A. Y. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 8, 191–197 (2007).
Nakawesi, J. et al. alphavbeta8 integrin-expression by BATF3-dependent dendritic cells facilitates early IgA responses to Rotavirus. Mucosal Immunol. 14, 53–67 (2021).
Weckel, A. et al. Long-term tolerance to skin commensals is established neonatally through a specialized dendritic cell subgroup. Immunity 56, 1239–1254.e7 (2023).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Gu, Z. Complex heatmap visualization. iMeta 1, e43 (2022).
Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021).
Liberzon, A. et al. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 1, 417–425 (2015).
Schurch, C. M. et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell 182, 1341–1359.e19 (2020).
McGinnis, C. S. et al. MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nat. Methods 16, 619–626 (2019).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Zhu, Q., Conrad, D. N. & Gartner, Z. J. deMULTIplex2: robust sample demultiplexing for scRNA-seq. Genome Biol. 25, 37 (2024).
Alquicira-Hernandez, J. & Powell, J. E. Nebulosa recovers single-cell gene expression signals by kernel density estimation. Bioinformatics 37, 2485–2487 (2021).
Phipson, B. et al. propeller: testing for differences in cell type proportions in single cell data. Bioinformatics 38, 4720–4726 (2022).
Acknowledgements
We thank J. Idoyaga for providing Xcr1cre-mTFP1 mice; E. B. Rankin for providing Eporflox/flox mice; V. K. Kuchroo for providing MC38-OVA and B16F10-OVA tumour cell lines; P. Giang for preparing CD45.1+ Foxp3DTR/DTR spleens; NIH Tetramer Core Facility (NIH Contract 75N93020D00005 and RRID:SCR_026557) for providing I-Ab|mouse 2W1S|EAWGALANWAVDSA|PE-labelled tetramer; C. Zhu for processing the scRNA-seq FASTQ data and assistance with data analysis; C. Brown for assistance with resource acquisition; T. L. Roth for valuable discussion; L. L. Tolentino, K. Nguyen, C. Barclay and J. N. Delos Reyes for expert technical support in flow cytometry and fluorescence-activated cell sorting; the DIMC Core of the Stanford Diabetes Research Center; and Breakthrough T1D Center of Excellence for support. This work was supported by the following grants: U54 CA274511, CA251174, CA244114 (E.G.E.) and P01HL149626 (X.A.).
Author information
Authors and Affiliations
Contributions
X.Z. and E.G.E. conceived the study. X.Z. designed and performed the experiments, analysed data, interpreted the results and wrote the manuscript with E.G.E. C.S.M. and S.C. conducted the scRNA-seq analyses and wrote the scRNA-seq results and methods sections together with X.Z. and E.G.E. K.J.H.-G. and W.Y. prepared the scRNA-seq libraries. S.C., P.Z., N.E.R.-F. and X.Z. carried out the RNA-seq analyses. C.M.S. and J.W.H. performed the CODEX experiments. G.Y., W.G. and J.Q. assisted with flow cytometry staining and cell sorting, in vitro cell culture, T cell adoptive transfers, tumour growth studies and data recording. A.M. contributed to the flow cytometry analysis of cDC1 Epor-tdT expression in the brain and assisted with tissue preparation for in vivo studies. I.L.L. aided in the in vivo tumour studies. H.Y. and T.H. performed heart transplantation. V.M.T., W.Q. and D.B.-V. assisted with Aldh1a2 and Itgb8 animal models. B.Y. made DEC205-OVA. A.T.S. supervised the scRNA-seq analyses. K.J.H.-G., X.A., Y.X., H.P., T.C.S., M.A., D.S., H.C., A.T.S., S.S.W., B.M. and S.S. provided critical intellectual insights. E.G.E. supervised the study. All authors provided feedback on the manuscript draft.
Corresponding authors
Ethics declarations
Competing interests
X.Z. is a cofounder and shareholder of ImmunEdge Inc. E.G.E. is a founder, shareholder and board member of ImmunEdge Inc. B.Y. is a shareholder of ImmunEdge Inc. X.Z and E.G.E. are Stanford-affiliated inventors of PCT/US2023/063997, entitled ‘Epo Receptor Agonists and Antagonists’. C.S.M. holds patents related to MULTI-seq. C.M.S. is a cofounder and scientific advisor of Vicinity Bio GmbH and is on the scientific advisory board of and has received research funding from Enable Medicine Inc., all outside the current work. T.C.S. is a scientific advisory board member for Concerto Biosciences. M.A. is a consultant, board member, and shareholder in Ionpath Inc. D.S. is a founder of Pliant Therapeutics and Glial Biosciences and is on the Genentech Scientific Review Board and the Amgen Inflammation Scientific Review Board, and an advisor to Lila Biologics, Arda Therapeutics and TCGFB Inc. H.C. is a consultant for Kumquat Biosciences and TCura Bioscience. A.T.S. is a founder of Immunai, Cartography Biosciences and Prox Biosciences, an advisor to Zafrens and Wing Venture Capital, and receives research funding from Merck Research Laboratories. The other authors declare no competing interests.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 XCR1+CD8α+ cDC1s in the spleen following TLI/ATS are bona fide cDC1s.
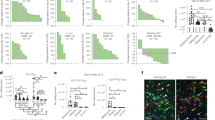
a, Total splenocyte frequency per spleen (UNT n=5; TLI/ATS n=10). b, Gating of CD11chighMHCIIhigh cDCs from the splenocytes. Summary graph of the frequency of cDCs in total splenocytes (UNT, n = 6; TLI/ATS, n = 7). c, Gating of CD8α+CD11b− cDC1s and CD8α−CD11b+ cDC2s; frequencies in cDCs (UNT, n = 6; TLI/ATS, n = 6). d, XCR1+ and e, Ki67+ cDC1 frequencies (UNT, n = 5; TLI/ATS, n = 5). f, qPCR of Batf3 and Irf8 in XCR1+CD8α+ cDC1s (n = 5/group). g, IRF8 and h, Zbtb46-GFP expression (UNT, n = 6; TLI/ATS, n = 5). i-k, MFI of MafB-mCherry expression and % of MafB-mCherry+ cells in XCR1+CD8α+ cDC1s (i), cDC2s (j) and red pulp macrophages (RPMΦs) (k). Day 0 (UNT), n = 4; TLI/ATS Day 5, n = 5; TLI/ATS Day 9, n = 5 (i,j,k). Data are shown from one experiment, representative of at least two independent experiments with similar results (a-k). Statistical analysis was performed using unpaired two-tailed Student’s t-test (a,b,c,d,e,h), or one-way ANOVA with Tukey’s multiple-comparison test (f,g,i,j,k). Data are mean ± s.e.m. (a-k).
Extended Data Fig. 2 TLI/ATS leads to widespread apoptosis and extramedullary erythropoiesis in the spleen and a marked rise in serum EPO.
a, TUNEL staining of spleen sections, UNT vs. TLI/ATS (scale bar = 200 µm). b, Spleen cell composition after ATS, TLI, or TLI/ATS; pie chart shows mean frequencies of indicated populations (n = 3). T cells (TCRβ+CD19−NK1.1−), B cells (CD19+TCRβ−NK1.1−), erythroid progenitors (CD11c−TER119+CD71+), cDCs (CD3ε−B220−SiglecH−PDCA-1−CD11chighMHCIIhigh), other myeloid cells are subdivided into CD11b+Ly6C−Ly6G+, CD11b+Ly6G −Ly6C+ and CD11bintF4/80+. c, Serum EPO levels over time after TLI/ATS (ELISA, n = 8). d, CD71+TER119+ erythroid progenitors in spleen (day 6) (upper) and in spleen/BM (lower) over time after TLI/ATS (n = 3). e, Co-detection by indexing (CODEX) imaging of WT C57BL/6 spleen (UNT vs. 1 day after TLI; scale bar = 500 µm). f, Scheme of EPO treatment in Epor-tdTomato-Cre mice (i.p. × 5 days). g,h, Flow cytometry showing splenic cDC1 frequency among cDCs (g) and Epor-tdT+ cDC1 frequency/MFI (h) (+PBS, n = 5; +EPO, n = 5). i-j, Frequencies of Epor-tdT+ and Epor-tdT− TER119+ erythroid cells (i) and Epor-tdT+CD11bintF4/80+Epor-tdT+ RPMΦs (j), (+PBS, n = 5; +EPO, n = 5). k, CCR7 vs. Epor-tdT expression in XCR1+CD8α+ cDC1s that were gated as live-dead aqua−CD3ε−CD19−B220− SiglecH−PDCA-1−CD11chighMHCIIhigh; histogram overlay for CCR7+ cDC1s (UNT, n = 5; TLI/ATS, n = 5). Data are shown from one experiment, representative of at least two independent experiments with similar results (a,b,c,d,g,h,i,j,k) or from one experiment (e). Statistical analysis was performed using unpaired two-tailed Student’s t-test (g,h,i,j,k). Data are mean ± s.e.m. (d,g-k). The diagram in f was created in BioRender. Zhang, X. (2025) https://BioRender.com/cx0n3vn.
Extended Data Fig. 3 EPO-EPOR downstream signaling is activated in cDC1s following TLI/ATS.
a-c, Gene Set Enrichment Analysis (GSEA) of transcriptional profiles using the Hallmark gene set of MSigDB. NES, normalized enrichment score; FDR, false discovery rate. Red: upregulated; Blue: downregulated. TLI/ATS vs. UNT. b, Upregulated gene sets. c, Downregulated gene sets. d-e, Intracellular phospho-flow cytometric analysis of EPO-EPOR downstream signaling in live-dead blue−Lin−SiglecH−PDCA-1−CD11chighMHCIIhigh. Spleens were harvested on the next day following the last dose of TLI or TLI/ATS. UNT (n = 4) vs. TLI (n = 4) vs. TLI/ATS (n = 4). d, XCR1+CD8α+ cDC1s and e, XCR1−CD8α− cDC2s. f,g, Histograms and MFI of the indicated EPO-EPOR downstream signaling molecules with fluorescence minus one (FMO) as controls by intracellular phospho-flow staining on the next day following the last dose of TLI/ATS treatment. Eporflox/flox (n = 4) vs. EporΔXcr1 (n = 5). cDC1s (f) and cDC2s (g). h, Ex vivo analysis of EPO-EPOR downstream signaling in splenic cDC1s. Splenic cDCs were MACS-purified with a pan-DC isolation kit and cultured at 5 × 106 cells/ml, then rested overnight. Cells were isolated from UNT or TLI/ATS-treated Eporflox/flox (n = 4; n = 4) and EporΔXcr1 (n = 4; n = 4) mice. cDCs from TLI/ATS-treated mice were stimulated ex vivo with EPO (10 IU/200 μl) or PBS (control) overnight. Phosphorylation of downstream signaling molecules was assessed by flow cytometry, after gating on XCR1+SIRPα− splenic cDC1s. Data are shown from one experiment, representative of at least two independent experiments with similar results (d-h). Statistical analysis was performed using unpaired two-tailed Student’s t-test (f,g), or one-way ANOVA Tukey’s multiple-comparison test (d, e and h left), or paired two-tailed Student’s t-test (h right). Data are mean ± s.e.m. (d-h).
Extended Data Fig. 4 FOXP3+ Tregs play an indispensable role in TLI/ATS-induced cDC1 EPOR-dependent immune tolerance.
a, Representative pseudocolor plots showing FOXP3+ Treg depletion efficiency in recipient mice on day 6 after DT treatment (DT injections on days 0, 2, and 4). b,c, Representative pseudocolor plots of C57BL/6, Batf3−/−, or EporΔXcr1 recipient conventional CD4+ T cell percentages and FOXP3+ Treg percentages in CD4+ T cells. d,e, Absolute cell number of indicated cell populations. b,d, Day 0 (UNT, n = 5; n = 5; n = 5 and TLI/ATS, n = 6; n = 5; n = 4) and c,e, Day 14 of UNT or TLI/ATS-treated groups post allo-BM infusion (UNT, n = 11; n = 6; n = 9 and TLI/ATS, n = 3; n = 11; n = 10). f, MHCII expression on cDC1s and cDC2s from MHCIIflox/flox and MHCIIΔXcr1 spleens. g,h,i, MHCIIflox/flox (n = 6) and MHCIIΔXcr1 (n = 6) recipients were given TLI/ATS and i.v. infused with BALB/c donor BM cells. 14 days post BM infusion, the percentages of donor type (H2Kd+) cells among leukocyte populations were determined in the peripheral blood of hosts. g,i, Recipient MHCI (H-2Kb)+TCRβ+CD4+ T cell frequency among total live cells and FOXP3+ frequency among CD4+ T cells were analyzed on day 14. j, CD45.2+FOXP3WTEporflox/flox (+ PBS/without DT, n = 8; +DT, n = 8) or EporΔXcr1 (+ PBS/ without DT, n = 8; +DT, n = 8) mice were injected with 30 million CD45.1+FOXP3DTR CD4+ T cells isolated by MACS. Two consecutive doses of DT or PBS were given on each of the following 2 days. Subsequently, the mice were treated with TLI/ATS, and 2W1S-BALB/c donor BM cells were infused i.v., and 14 days later, 2W1S-tetramer+CD44+H-2Kb+TCRβ+CD4+ T cells from the spleens were analyzed for FOXP3 expression by flow cytometry. FOXP3 expression in CD45.1+ or CD45.2+2W1S-tetramer+ CD4+ T cells is shown. One experiment (j) or one of two independent experiments with similar results are shown (a-i). Statistical analysis was performed using unpaired two-tailed Student’s t-test (g,i), two-way ANOVA with Tukey’s multiple-comparison test (d,e,j). Data are mean ± s.e.m. (d,e,g,i,j). The diagram in j was created in BioRender. Zhang, X. (2025) https://BioRender.com/cx0n3vn.
Extended Data Fig. 5 Differentially expressed genes (DEGs) in cDC1s in scRNA-seq analysis and ex vivo TGFβ-dependent Ag-specific FOXP3+ Treg induction by CCR7+ cDC1s.
a, UMAP of splenic cDC1 gene expression by sample identity. b, Dot plots of top condition-specific DEGs in Eporflox/flox and EporΔXcr1 mice (TLI/ATS vs. UNT). c, Absolute cDC1 numbers per spleen in UNT vs. TLI/ATS-treated Eporflox/flox (n = 5/condition) and EporΔXcr1 (n = 5/condition) mice. d, UMAPs of cDC1 subtypes in Epor-tdT+ and Epor-tdT– cells. e-g, Dot plots of top condition-specific DEGs in Epor-tdT+ and Epor-tdT– cDC1s (TLI/ATS) and in Eporflox/flox and EporΔXcr1 mice (TLI/ATS vs. UNT). Dot color = expression, size = % of indicated gene expressed cells (b,e-g). h, Bar charts showing cDC subtype (d) proportions in Epor-tdT+ and Epor-tdT– cDC1s following TLI/ATS. i, Role of TGFβ in FOXP3+ Treg induction by CCR7+ cDC1s: 12 h after apoptotic Act-mOVA injection, CCR7+ cDC1s (1×104) were cocultured with CD45.1+ CTV-labeled naïve OT-II cells ± anti-TGFβ; FOXP3 expression was analyzed by flow cytometry (n = 5/group). j,k, Representative flow cytometry analysis and l,m, Absolute cell number of indicated cell populations of Fig. 3j, Itgb8ΔXcr1 vs. littermate controls. j,l, Day 0 and k,m, Day 14 of UNT (n = 5; n = 5) or TLI/ATS-treated (n = 5; n = 5) groups post allo-BM infusion. n,o, Aldh1a2ΔCD11c: Batf3−/− (n = 6) vs. Aldh1a2flox/flox: Batf3−/− (n = 7) BM chimeric recipient mice (CD45.1+) were given TLI/ATS. 1 day after the last dose of TLI/ATS, 2W1S-BALB/c donor BM cells were infused i.v., and 14 days later, the percentages of donor type (H2Kd+) cells among leukocyte populations in the peripheral blood of hosts were determined (n) and 2W1S-tetramer+CD44+H-2Kb+TCRβ+CD4+ T cells from the spleens were analyzed for FOXP3 expression by flow cytometry and FOXP3+ Tregs were counted (o). Data are representative of at least three independent experiments with similar results (c,i) or one experiment (j-o). Statistical analysis was performed using unpaired two-tailed Student’s t-test (c,i,n,o), or two-way ANOVA followed by Tukey’s multiple-comparison test with P values adjusted (l,m), or propeller test, two-sided, no multiple-comparison correction (b), or wilcoxon rank sum test, two-sided, Bonferroni correction (h). Data are mean ± s.e.m. (c,i,l,m,n,o). The diagram in i was created in BioRender. Zhang, X. (2025) https://BioRender.com/rq2yp2e.
Extended Data Fig. 6 Absence of EPOR on cDC1s gives rise to immunogenic cDC1s that promote both CD8+ T cell cross-priming and CD4+ T cell priming to cell-associated Ags.
a, MFI of indicated molecules on gated cDC1s with fluorescence minus one (FMO) as controls. b, MFI of indicated molecules on gated cDC2s with fluorescence minus one (FMO) as controls. c, Percentages of cDC2s in splenic cDCs. d, Representative flow gating of CCR7+XCR1+SIRPα– cDC1s in splenic cDC1s (Upper), and percentages and absolute numbers of CCR7+ cDC1s (Lower). a-d, Eporflox/flox (n = 5) vs. EporΔXcr1 (n = 5) mice. e, MFI of indicated molecules on CCR7+ vs. CCR7– cDC1s. CD40 and PD-L1: Eporflox/flox (n = 5); EporΔXcr1 (n = 5). CD80 and CD86: Eporflox/flox (n = 6); EporΔXcr1 (n = 6). f, Cross-presentation assay: apoptotic Act-mOVA thymocytes injected into Eporflox/flox (n = 5) or EporΔXcr1 (n = 5) mice 1 day after transfer of CTV-labeled naïve CD45.1+ naïve OT-I cells; spleens analyzed on day 4 for OT-I expansion and proliferation. g, Same setup with OT-II cells; percentages and absolute numbers of OT-II cells and proliferating OT-II cells were assessed. Ag-specific CD4+ T cell response: Ag-specific CD4+ T cell immune response following i.v. injection of apoptotic Act-mOVA thymocytes 1 day after i.v. injection of CTV-labeled naïve CD45.1+ naïve OT-II cells. Spleens were analyzed at day 4 for OT-II expansion and proliferation. Eporflox/flox (n = 5) and EporΔXcr1 (n = 5) mice. Data are shown from one experiment, representative of at least three independent experiments with similar results (a-g). Statistical analysis was performed using unpaired two-tailed Student’s t-test (a,b,c,d,f,g) and two-way ANOVA followed by Tukey’s multiple-comparison test (e). Data are mean ± s.e.m. (a-g). The diagrams in f,g were created in BioRender. Zhang, X. (2025) https://BioRender.com/bth22u6.
Extended Data Fig. 7 Phenotypes of T cells in the spleens of Eporflox/flox vs. EporΔXcr1 mice and role of EPOR in cDC1-mediated cell-associated Ag-specific CD4+ T cell priming and proliferation and FOXP3+ Treg induction.
a-e, Percentages and absolute numbers of CD4+ T cells (a), FOXP3+CD25+ Tregs in CD4+ T cells (b), CD44highCD62Llow effector cells and CD44lowCD62Lhigh naïve cells in CD4+ T cells (c), CD8+ T cells (d), and CD44highCD62Llow effector cells and CD44lowCD62Lhigh naïve cells in CD8+ T cells (e) in the spleens of EporΔXcr1 and littermate Eporflox/flox control mice with representative flow cytometric plots. a-e, Eporflox/flox, n = 5; EporΔXcr1, n = 5. f,g, Flow cytometry-based measurement of cell-associated Ag-specific CD4+ T cell immune response in the spleen following i.v. injection of apoptotic Act-mOVA thymocytes into mice of the indicated genotypes 1 day after i.v. injection of CTV-labeled naïve CD45.1+ OT-II cells. f, WT C57BL/6 (n = 5) and Batf3−/− (n = 7). g, FOXP3+ Treg induction in Eporflox/flox and EporΔXcr1 mice. Recombinant EPO or PBS was administered daily, from Day −3 to Day 4. +PBS: Eporflox/flox (n = 5), EporΔXcr1 (n = 5); +EPO: Eporflox/flox (n = 5), EporΔXcr1 (n = 5) mice. Data are shown from one experiment, representative of at least three independent experiments with similar results (a-e), or two independent experiments with similar results (f,g). Statistical analysis was performed using unpaired two-tailed Student’s t-test (a-f), or two-way ANOVA followed by Tukey’s multiple-comparison test (g). Data are mean ± s.e.m. (a-g). The diagrams in f,g were created in BioRender. Zhang, X. (2025) https://BioRender.com/bth22u6.
Extended Data Fig. 8 Epor-tdT expression on XCR1+ cDC1s in selected organs and tolerogenic phenotype of Epor-tdT+ migratory cDC1s in PLN.
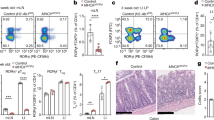
a,b,c CCR7- and Batf3-dependent Epor-tdT expression on migratory cDCs in pLNs. Migratory cDCs were gated as CD11cintMHCIIhigh from live-dead aqua−Lin−SiglecH−PDCA-1−EpCAM− cells, and resident cDCs were gated as CD11chighMHCIIint from live-dead aqua−Lin−SiglecH−PDCA-1− cells. pLNs including inguinal, axillary, brachial, and superficial cervical LNs were combined for analysis by flow cytometry (a,b). Ccr7−/−EportdT/+, Batf3−/−EportdT/+ and WT C57BL/6 mice (a). Histogram overlay of Epor-tdT expression on migratory or resident cDCs from individual mouse strains (b). Epor-tdT expression on migratory cDCs from individual pLNs of EportdT/+ mice (c). d, Epor-tdT expression on cDC1s obtained from the indicated organs in Zbtb46GFP/+EportdT/+ mice. cDCs were gated in CD45+ cells as CD11c+Zbtb46-GFP+, in which cDC1s were further gated as XCR1+CD103+. e, Flow cytometric analysis of tolerance associated cell-surface molecules on PLN Epor-tdT+ migratory cDC1s compared with Epor-tdT− cDCs and resident cDCs with FMO serving as controls (n = 8). Data are representative of at least two independent experiments with similar results (a-e). Statistical analysis was performed using one-way ANOVA Tukey’s multiple-comparison test (e). Data are mean ± s.e.m. (e). The diagrams in a,e were created in BioRender. Zhang, X. (2025) https://BioRender.com/5sr2iny.
Extended Data Fig. 9 Induction of Ag-specific CD4+FOXP3+ Tregs by PLN migratory Epor-tdT+ cDC1s.
a, FACS-sorted PLN Epor-tdT+ (n = 4) or Epor-tdT− (n = 4) XCR1+ migratory cDC1s from Epor-tdT mice were cocultured for 5 days with CTV-labeled naïve OT-II cells + DEC205-OVA (ratio 1:5); FOXP3 expression in OT-II cells was analyzed by flow cytometry. b, Same setup as (a) but with apoptotic Act-mOVA thymocytes (ratio 1:5:2) ± EPO (20 IU per well per day for 5 days); FOXP3 expression in OT-II cells was measured. Epor-tdT+ (+ PBS or w/o, n = 4; +EPO, n = 4) or Epor-tdT− (+ PBS or w/o, n = 4; +EPO, n = 3) c, FACS-sorted PLN Epor-tdT+ or Epor-tdT− migratory cDC1s were cocultured for 12 h with apoptotic CD45.1+ thymocytes ± EPO; MFIs of surface markers were analyzed. Epor-tdT+: n = 2. Epor-tdT–: n = 2. d, Migratory cDC1s from Eporflox/flox or EporΔXcr1 mice were cocultured with naïve CTV-labeled OT-II cells and Act-mOVA thymocytes (1:5:2) ± EPO (20 IU per well per day for 5 days); FOXP3 induction was assessed. n = 6/group. e, Efferocytosis of PKH67-labeled apoptotic thymocytes by migratory cDC1s and cDC2s in dLNs 12 h post-injection. f, Act-mOVACD45.1/CD45.2 mice were reconstituted with either Eporflox/flox (n = 5) or EporΔXcr1 (n = 6) BM cells after lethal irradiation. 8 weeks post-reconstitution, naïve CTV-labeled OT-II cells were i.v. infused (day 0), and EPO was administered on days −2 to 2. FOXP3 induction in OT-II cells was assessed in inguinal LNs on day 9. Data are shown from one experiment, representative of two independent experiments with similar results (a-e) or one (f) independent experiment. Statistical analysis was performed using unpaired two-tailed Student’s t-test (a,b,f), two-way ANOVA with Tukey’s multiple-comparison test (d). Data are mean ± s.e.m. (a,b,c,d,f). The diagrams in a,b,c,e,f were created in BioRender. Zhang, X. (2025) https://BioRender.com/u560oi2.
Extended Data Fig. 10 Phenotypes of T cells in the PLNs of Eporflox/flox vs. EporΔXcr1 mice.
a-e, Percentages and absolute numbers of CD4+ T cells (a), FOXP3+CD25+ Tregs in CD4+ T cells (b), CD44highCD62Llow effector cells and CD44lowCD62Lhigh naïve cells in CD4+ T cells (c), CD8+ T cells (d), and CD44highCD62Llow effector cells and CD44lowCD62Lhigh naïve cells in CD8+ T cells (e) in the PLNs of EporΔXcr1 and littermate Eporflox/flox control mice with representative flow cytometry plots. a-e, Eporflox/flox (n = 9) and EporΔXcr1 (n = 9). Data are shown from one experiment, representative of at least three independent experiments with similar results (a-e). Statistical analysis was performed using unpaired two-tailed Student’s t-test (a-e). Data are mean ± s.e.m. (a-e).
Extended Data Fig. 11 EPOR expression on tumor-infiltrating leukocytes (TILs), tumor Ag-carrying migratory cDC1s in TDLNs and tumors, and the correlation of tumor growth with systemic EPO levels.
a, Zbtb46GFP/+EportdT/+ mice were implanted s.c. with MC38 or B16F10 tumor, or EO771 tumor in the mammary fat pad. On day 12, tumors were harvested for flow cytometric analysis of Epor-tdT on cDCs (live-dead aqua−CD45+Zbtb46-GFP+CD11c+); cDC1s were gated as XCR1+ and non-cDC1s as XCR1−. b-d, Mice implanted s.c. with MC38-OVAdim or B16F10-OVA; on day 10, tumors were analyzed for Epor-tdT expression in TILs. b, Representative gating strategy of individual live-dead blue− TIL populations. c,d, Histogram overlay showing Epor-tdT expression in individual cell populations. e, Zbtb46GFP/+EportdT/+ mice with MC38-OVAdim tumors (day 12) were analyzed for Epor-tdT on tumor-infiltrating cDCs; CCR7+ (population I) and CCR7− (populations II/III by Ly6A) subsets were gated, with XCR1/CD103 staining to define cDC1s and cDC2s. f, Quantification of Epor-tdT expression on individual tumor infiltrating cDC subsets. MC38-OVAdim (n = 4) and B16F10-OVA (n = 4) tumors were harvested on day 12 post-s.c. implantation for flow cytometry. Gating strategy as in Fig. 5a and Extended Data Fig. 11e. g, Flow cytometry analysis of Epor-tdT expression on TDLN migratory cDC1s. Overlay of migratory cDC1s with Lin− live cells to show Epor-tdT expression levels. h, Serum EPO levels were measured by ELISA on the indicated days after s.c. implantation of MC38-OVAdim (n = 6) or B16F10-OVA (n = 5) tumors in WT mice. i,j,k, B16F10-OVA-ZsGreen cells were s.c. implanted into EportdT/+ mice, and tdLN and tumor were analyzed on day 9 after inoculation. j, Flow cytometry analysis of Epor-tdT expression on tdLN ZsGreen+ migratory and resident XCR1+ cDC1s. Overlay of migratory ZsGreen+ cDC1s or resident ZsGreen+ cDC1s with Lin−live cells to show Epor-tdT expression levels. k, Flow cytometry analysis of Epor-tdT expression on tumor infiltrating ZsGreen+ cDC1s. Data are shown from one experiment, representative of at least two independent experiments with similar results (a-e,f,g,h,j,k). Statistical analysis was performed using one-way ANOVA with Dunnett’s multiple-comparison test (f). Data are mean ± s.e.m. (f,h). The diagrams in a,b,e,g,i were created in BioRender. Zhang, X. (2025) https://BioRender.com/gjjtedh.
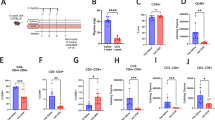
Extended Data Fig. 12 Loss of EPOR in cDC1s limits tumor growth and promotes immunogenic function of tumor Ag-carrying cDC1s in both TDLN and tumor.
a, Growth of MC38-OVAdim tumor cells implanted s.c. into Eporflox/flox (n = 8) and EporΔXcr1 mice (n = 9). b, Growth of B16F10-OVA tumor cells implanted s.c. into Eporflox/flox (n = 6) and EporΔXcr1 mice (n = 7). c, Experimental design for phenotyping tumor-Ag carrying ZsGreen+ cDC1s in tdLN and tumors in Eporflox/flox vs. EporΔXcr1 mice. B16F10-OVA-ZsGreen cells were implanted s.c. into EportdT/+ mice, and TDLNs and tumors were analyzed on day 9 after implantation. d, Flow cytometry analysis of Epor-tdT expression on ZsGreen+ migratory XCR1+SIRPα− cDC1s in tdLNs with summary graph of statistical quantification. Eporflox/flox (n = 7) and EporΔXcr1 (n = 8). e, Flow cytometry analysis of CD40, CD80 and CD86 expression on tumor infiltrating ZsGreen+ cDC1s with summary graph of statistical quantification. Eporflox/flox (n = 7) and EporΔXcr1 (n = 8) mice. (f-l) MC38-OVAdim tumors were s.c. implanted into Eporflox/flox (n = 8) and EporΔXcr1 (n = 7) and 10 days later TILs were analyzed. f. Percentages of CD45+ live immune cells and CD8+ or CD4+ T cells in CD45+ TILs. g, Frequency of OVA257-264-dextramer+CD8+ T cells among CD8+ T cells. h, Representative flow plots and quantification of CD8+ T cells expressing TIM-3 and PD-1. i, Representative flow plots and quantification of TCF1+TIM-3−CD8+ T cells. j, Representative histograms and quantification of perforin, granzyme-B, IFNγ and TNFα expression in tumor-infiltrating CD8+ T cells. k, Percentage of FOXP3+ Tregs in CD4+ T cells with representative flow plots (Left). Absolute number of Tregs (Right). l, Representative flow plots and percentages of T-bet+CXCR3+ Tregs in CD4+ FOXP3+ Tregs. (f-l). Data are shown from one experiment, representative of at least two independent experiments with similar results (a,b,d,e,f-l). Statistical analysis was performed using two-way ANOVA with Šídák’s multiple comparison test (a,b), or two-tailed unpaired Student’s t-test (d,e,f,g,i,j,k,l), or two-way ANOVA with Tukey’s multiple-comparison test (h). Data are mean ± s.e.m. (a,b,d,e-l). The diagram in c was created in BioRender. Zhang, X. (2025) https://BioRender.com/3jod9q7.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2, which contain the full gene list used to generate the signature and a list of detected differentially expressed genes (related to Fig. 4).
Source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, X., McGinnis, C.S., Yu, G. et al. Erythropoietin receptor on cDC1s dictates immune tolerance. Nature (2025). https://doi.org/10.1038/s41586-025-09824-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41586-025-09824-z