Abstract
Transcript diversity including splicing and alternative 3′ end usage is crucial for cellular identity and adaptation, yet its spatial coordination remains poorly understood. Here we present SPLISOSM (spatial isoform statistical modeling), a method for detecting isoform-resolution patterns from spatial transcriptomics data. SPLISOSM uses multivariate testing with nonparametric kernels to account for spot-level and isoform-level dependencies, achieving high statistical power on sparse data. In the mouse brain, we identify over 1,000 spatially variable transcript diversity events, primarily in synaptic signaling pathways linked to neuropsychiatric disorders, and uncover both known and previously unknown regulatory relationships with region-specific RNA binding proteins. We further show that these patterns are evolutionarily conserved between mouse and human prefrontal cortex. Analysis of human glioblastoma highlights pervasive transcript diversity in antigen presentation and adhesion genes associated with specific microenvironmental conditions. Together, we present a comprehensive spatial splicing analysis in the brain under normal and neoplastic conditions.
Similar content being viewed by others
Main
Almost all human genes undergo orchestrated pre-messenger RNA (pre-mRNA) processing with precise spatiotemporal regulation1,2. The brain exhibits particularly high transcript diversity3, where region-specific expression of RNA binding proteins (RBPs) drives specialized local transcript landscapes that control neurogenesis4,5 and synaptic plasticity6,7. Disruption of this delicate regulatory balance contributes to numerous neurological disorders8,9,10 and cancer progression11,12,13. Despite its critical importance, our understanding of how RNA isoforms are spatially organized within tissues remains limited.
Modern spatial transcriptomics (ST) platforms naturally provide some isoform-specific information, whether through 3′ fragments in short-read (SR) approaches14,15,16, full-length transcripts in long-read sequencing17,18 or exon probes in imaging-based platforms19,20 (Fig. 1a). Despite this, most ST analysis pipelines aggregate expression at the gene level, discarding rich transcript information. Few computational approaches21,22,23 detect spatial patterns in isoform distribution, creating a notable gap in our understanding of the molecular complexity and its regulation in shaping tissue architecture.
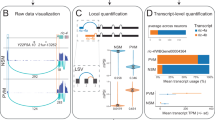
a, Alternative pre-mRNA processing generates multiple isoforms from the same gene, creating varying molecular transcript diversity detectable by major ST platforms. b, Isoform quantification from ST data is multivariate, sparse and confounded by gene expression and spatial autocorrelation. c, SPLISOSM addresses challenges in spatial isoform analysis by recasting SV and DU detections as multivariate independence tests. For SV testing, isoform usage and spatial coordinates are projected into feature spaces to assess nonlinear dependencies using the HSIC with optimized spatial and isoform kernels. For DU testing, nonparametric conditional HSIC test and parametric GLMM control for confounding spatial autocorrelation and reduce spurious associations. d, SPLISOSM enables cell-type-agnostic detection of SVP genes and their potential regulators. Figure created using BioRender.com.
Analyzing spatial isoform patterns presents several formidable challenges (Fig. 1b). First, isoforms from the same gene are interdependent, with expression constrained by total transcribed pre-mRNAs. This multivariate and compositional nature invalidates standard statistical approaches designed for gene expression analysis. Early attempts either inappropriately treated isoforms as independent18 or relied on oversimplified univariate metrics21,22, substantially compromising statistical power (Supplementary Note). Second, data sparsity—already problematic in ST—is exacerbated at the isoform level when gene counts are subdivided across multiple transcripts. Regional expression variation can lead to zero total unique molecular identifier (UMI) counts at many locations, making isoform ratios undefined. Even when detected, these ratios frequently appear binary despite representing continuous biological preferences24,25, complicating operations such as log-ratio transformations. Common workarounds such as pseudo-counts introduce additional biases that distort relationships between observations26. Finally, spatial autocorrelation violates the independent observation assumptions fundamental to most differential association tests, generating false positives from spurious associations that merely reflect mutual dependence on spatial coordinates.
To address these challenges, we present SPLISOSM (spatial isoform statistical modeling), a method for analyzing isoform usage in ST data. Across multiple datasets of adult mouse and human brain, we identify thousands of genes with spatially variable processing. To uncover regulatory mechanisms, we test for conditional association between isoform ratios and potential regulators such as RBPs, confirming known connections and discovering region-specific regulation. Finally, we extend our analysis to glioblastoma cohorts, revealing how transcript diversity is shaped by both normal brain architecture and disease-specific alterations in the tumor microenvironment.
Results
Designing isoform-level SV and association tests
Spatial gene variability arises from both cell-type distribution patterns and location-specific variation within cell types. While several methods exist for identifying spatially variably expressed (SVE) genes27,28,29,30,31, they prioritize different spatial patterns, resulting in limited consensus32. Moreover, these approaches cannot be easily adapted for multivariate isoform analysis due to their reliance on specific model assumptions (Supplementary Note). Drawing on nonparametric statistics’ effectiveness in modeling total expression from sparse ST data30, we reformulated spatial variability (SV) detection and differential usage (DU) analysis as multivariate independence testing. Specifically, SPLISOSM transforms isoform compositions, spatial coordinates and other covariates of interest into feature spaces, capturing their complex relationships through the kernel-based Hilbert–Schmidt independence criterion (HSIC)33,34 (Fig. 1c). Significant patterns are identified by comparing against the null hypothesis that a gene’s isoform preference is independent of either spatial locations or a given covariate such as RBP expression (Methods).
Our approach introduces two key technical innovations: first, we mathematically prove that all low-rank approximations of kernel tests—including those in SPARK-X30—inevitably sacrifice statistical power (Theorem 1 in Supplementary Note). This insight led us to develop enhanced spatial kernels35 using spectral graph theory that prioritize signals by their spatial frequency, limiting power losses to high-frequency fluctuations representing noise when processing large-scale data. Second, we established a framework for sparse isoform data with a new compositional kernel that handles undefined ratios (Theorem 2 in Supplementary Note), optimizing efficiency while maintaining test validity through a mean replacement strategy that uses all spatial observations.
Based on these theoretical advances, SPLISOSM implements three complementary SV tests: HSIC-GC for total gene expression, HSIC-IR for isoform usage ratios and HSIC-IC for isoform counts. For genes showing SV in isoform preference, we apply conditional DU tests to identify potential RBP regulators (Methods). Using SV, SPLISOSM enables cell-type-agnostic discovery (Fig. 1d), complementing existing single-cell isoform studies36,37,38 while offering unique advantages in contexts where cell typing is challenging, such as in tumor microenvironments.
SPLISOSM yields well-calibrated P values in simulation
We first validated SPLISOSM through simulations reflecting both gene expression and isoform preference variation (Fig. 2a and Methods). Our SV tests target distinct biological signals with well-calibrated statistics (Fig. 2b). Among them, HSIC-IR correctly identified spatial patterns in isoform usage while ignoring changes in total gene expression (scenario 4 versus 5). The modified linear kernel with mean replacement provided optimal power without introducing artifacts (Supplementary Fig. 1a,b). Complementarily, HSIC-IC detected both isoform usage (scenario 1 versus 3) and gene expression spatial variation (scenario 1 versus 4). We also observed improvements in the gene-level HSIC-GC compared to SPARK-X through the optimized spatial kernel (scenarios 4–6), validating our theoretical predictions.
a, Six simulation scenarios with SV at different layers. Regional variability was generated from two binary and two continuous spatial covariates Iso, isoform. b, Q–Q plots of SV test P values across 1,000 simulated genes per scenario. Regional gene expression (scenarios 4–6) was included to introduce artifacts in the observed ratios. c, Q–Q plots of DU test P values aggregated from 2,000 tests (1,000 genes × 2 covariates) per covariate type. Variable isoform usage independent of covariates (scenarios 2 and 5) was sampled from a Gaussian process (GP) to mimic spurious differential associations. For binary covariates, two-sided t-tests were performed on individual isoforms with P values combined at the gene level using Fisher’s method. For GLM and GLMM, null models with zero effect size were fitted and tested using the score statistic. Red text indicates components with spatial variability.
For association analysis, we focus on the challenge of correlated noise, which creates patterns that falsely align with covariates (scenarios 1 versus 2). Conventional approaches, including discrete t-tests, continuous multivariate correlation and generalized linear models (GLMs), all exhibited considerable P value inflation under spatial confounding (Fig. 2c, scenarios 1 versus 2 and 4 versus 5). SPLISOSM overcomes this limitation by introducing spatial conditioning within existing differential analysis frameworks (Methods). Simulations confirmed that our nonparametric conditional HSIC maintains proper null calibration while preserving sensitivity to subtle covariate effects (Fig. 2c and Supplementary Fig. 1c–e), and that our parametric generalized linear mixed model (GLMM) partially controlled false positives compared to standard GLMs.
Spatial splicing in adult mouse brain targets synaptic and membrane trafficking genes
Recent studies17,18 have generated spatial isoform maps of postnatal and adult mouse brains by combining 10X Visium spatial barcoding with Oxford Nanopore (ONT) sequencing. While able to resolve full-length transcripts, these long-read ST technologies are limited by throughput and data sparsity, providing a natural testbed to demonstrate SPLISOSM’s utility in biological discovery. We reanalyzed two SiT18 coronal brain section samples (CBS1, CBS2) from adult mouse (Fig. 3a). SPLISOSM detected 150 spatially variably processed (SVP) genes shared by both replicates (adjusted P < 0.05, Fig. 3b and Supplementary Figs. 2 and 3)—more than double the originally reported 61 genes—without relying on region annotations. Our SV tests demonstrated strong consistency between replicates (Spearman’s ρ of P values = 0.52, 0.39, 0.43 for HSIC-IR, HSIC-GC and HSIC-IC, respectively), while SPARK-X biased toward the sample with higher sequencing coverage (Spearman’s ρ = 0.26). These SVP genes were enriched in membrane trafficking and synaptic signaling (Clta, Cltb, Snap25, Stxbp1, Ap2a1, Atp6v1e1, Napa), with many implicated in neurodegenerative diseases (Fig. 3c). We also observed widespread isoform variability in ribosomal genes (Rps24, Rps6, Rpl13a, Rpl5, Rps9), aligning with previous studies showing cell-type and tissue-specific ribosome composition39. Transcript usage variability alone proved sufficient to define brain regions with comparable spatial resolution to expression-based clusters (Fig. 3d and Methods), and isoform preference could be grouped into region-specific programs by gene-wise clustering (Fig. 3e and Methods).
a, Workflow overview. CA, cornu ammonis; DG, dentate gyrus. b, Comparison of SV in gene expression (x axis) versus isoform usage (y axis) in the CBS2 sample. Points colored by expression perplexity (effective number of isoforms per gene). Inset: number of events significant in both CBS1 and CBS2 replicates (adjusted P < 0.05), categorized as SVP or spatially variably expressed but not variably processed (SVENP) genes. c, Pathway enrichment analysis comparing SVP versus SVENP genes, with terms arranged by precision differences between the two groups. d, Unsupervised clustering based on gene-level expression of SVE genes (left) or relative isoform ratios of SVP genes (right). Isoform counts were locally smoothed to compensate for data sparsity. e, Selected isoform usage programs identified through hierarchical clustering of SVP genes. f, Distributions of AS event types among SVP and SVE genes. g, Selected exon features of 214 skipping or mutually exclusive exons in SVP and 1,349 exons in SVENP genes, showing per-exon median relative position (left) and median exon number of transcripts containing each exon. Boxplots show median (center line), interquartile range (box) and 1.5× interquartile range (whiskers). P values from two-sided t-test. h, Unsupervised clustering based on 1,028 SVE RBP expression from SR sequencing data. i, Top RBPs ranked by number of significantly associated targets, color-coded by availability of documented motifs (CISBP-RNA) or CLIP data (POSTAR3). Right: log-normalized expression of selected RBPs. j, Differential isoform usage P values for Rbfox3 (left) and Celf5 (right) in two 50-μm spaced adjacent technical replicates. Red points indicate validated targets with RBP binding near regulated events in brain-CLIP data. k, Gnas transcript structure and spatial log-normalized expression patterns. HSPC, hematopoietic stem and progenitor cell. Panel a created using BioRender.com.
At the structural level, SVP genes showed enrichment for exon skipping particularly near the 3′ end, as well as alternative first exons (Fig. 3f,g). To identify potential modulators behind these events, we systematically searched for differential association with spatially variably expressed RBPs, which collectively reproduced major brain compartments (Fig. 3h and Supplementary Fig. 4) and may act through direct binding or as indirect cofactors to drive transcript regional specificity. Top RBPs prioritized in our analysis included well-established neural splicing regulators such as QKI, RBFOX and CELF families and less-studied ones such as ARPP21 (Fig. 3i). To evaluate these relationships, we examined available brain-CLIP datasets40, confirming four of nine significant Rbfox3 associations. Although CELF5-specific CLIP data remain unavailable, eight of ten of its associated target genes showed binding by other CELF family members, supporting CELF5’s role as a major neural splicing modulator in adult mouse brain (Fig. 3j).
Among SVP genes, Gnas showed particularly intriguing associations (Fig. 3k). Its exon 3, which is spliced out specifically in midbrain and fiber tracts, generates a hyperactive G-alpha protein driving differentiation defects and myeloid malignancies when included in SRSF2 and U2AF1 mutants41. Our analysis reveals that RBFOX and CELF families potentially regulate Gnas exon 3 skipping in the brain, raising the possibility that neural RBP dysregulation in nonneural tissues could contribute to disease pathogenesis.
Cross-platform validation of brain transcript variability
The substantial spatial transcript processing identified from long-read data prompted us to examine whether these patterns could be detected across different ST platforms, each recognizing distinct aspects of molecular variation. In addition to alternative splicing (AS), mammalian nervous systems exhibit active alternative polyadenylation (APA)42 that generates transcript diversity near the 3′ end (transcriptome 3′ end diversity, TREND43), which can be captured by 3′-based ST sequencing protocols. In a separate 10X Visium sample44 of adult mouse brain, we identified 815 SVP genes with spatially variable TREND usage (adjusted P < 0.01, Fig. 4a,b and Methods). The increased detection power reflected the higher sequencing depth of SR Visium, as confirmed through down-sampling (Supplementary Fig. 5a). Analysis of a near-single-cell Slide-seqV2 dataset of mouse hippocampus16 revealed similar albeit fewer TREND patterns due to its sparsity (Extended Data Fig. 1a). In total, 95 SVP genes overlapped between long-read and SR datasets despite their technical differences (Extended Data Fig. 2a,b). Furthermore, genes using variable isoforms across space also show significantly higher SV in their 3′ end regions detectable by SR sequencing (Extended Data Fig. 2c and Supplementary Fig. 6), suggesting coordinated transcript processing across the full transcript length45.
a, Workflow overview. b, Comparison of SV in gene expression (x axis) versus TREND usage (y axis) in the SR coronal brain section sample. Points colored by expression perplexity (effective number of isoforms per gene). Inset: significant events (adjusted P < 0.01). c, TREND event annotation with overlapping with genomic features. Alternative exons (Alt-exon) are defined as exons located within introns of other transcript variants. Bold numbers are the absolute number of events. CDS, coding sequence. De novo motif enrichment results shown at bottom. P value computed using a one-sided chi-square test. d, Pathway enrichment analysis comparing SVP and SVENP genes, arranged by precision. e, Spatial log-normalized expression of the neuronal (chr. 9:110005229) and glial (chr. 9:110083783) Map4 variants. f, Unsupervised clustering based on gene-level expression of top 200 SVE genes (left), relative TREND ratios of top 200 SVP genes (middle) and gene expression of all SVE RBPs (right). g, Gnao1 transcript structure (left) and spatial log-normalized expression patterns (middle). Right: the likely protein sequence variation, color-coded by solvent-accessible surface area. FDR, false discovery rate. h, Arpp21 transcript structure and spatial log-normalized expression patterns. aa, amino acid. i, Top RBPs associated with TREND usage in Arpp21, Celf2 and Pcbp2. Dashed line indicates P = 0.01. Panel a created using BioRender.com.
To obtain orthogonal validation, we reanalyzed a 10X Xenium Prime 5K dataset46 of adult mouse brain with subcellular spatial resolution through multiplexed in situ hybridization (Methods). Although most predesigned Xenium probes do not target sequencing-based variable regions, we observed strong gene-level agreement: 55.6% (20 of 36) of ONT-detected and 39.8% (100 of 251) of SR-detected SVP genes showed significant spatial exon usage variability, both far exceeding the baseline rate of 14.9% (Extended Data Fig. 2d–g). At single-cell resolution, we could pinpoint cellular origins of spatial transcript patterns. For example, Xenium confirmed that Dtnbp1 isoform switching detected by long-read in specific brain regions was driven by distinct neuronal subpopulations: Nptxr+/Slc17a7+ excitatory neurons in olfactory and hippocampus-CA3 and GABAergic interneurons in isocortex (Extended Data Fig. 3). Similarly, we validated the Zdhhc8 3′ end variation and its isocortex and hippocampus-dentate gyrus specificity (Supplementary Fig. 7).
This cross-platform convergence demonstrates that spatial transcript diversity represents a fundamental organizing principle of brain architecture, detectable across diverse technical approaches (Extended Data Fig. 4 and Supplementary Fig. 8). It also establishes SPLISOSM as a robust and cell-type-agnostic framework for uncovering transcript patterns invisible at gene level from ST data.
Spatial 3′ end transcript diversity in adult mouse brain shows functional convergence on signaling pathways
Although over 80% of spatially variable TREND events mapped to 3′ untranslated regions (UTRs), we found significant localization at junctions and alternative exons (Fig. 4c). Some non-UTR events represented internal poly(A) priming sites, supported by A-rich motif enrichment, allowing us to detect upstream diversity deeper into transcripts. The functional importance of TREND became evident as 25% of events overlapped coding regions. Of seven previously reported protein-altering poly(A) site shifts between neurons and glia47, we recapitulated six (Map4, Atp2a2, Cdc42, Klc1, Itsn1, Ptpn2) while providing new insights into their spatial distribution (Fig. 4e and Extended Data Fig. 1b). SVP genes converged on key neural signaling pathways, including glutamatergic and adrenergic synaptic transmission and Rap1, Ca2+, cAMP and MAPK cascades (Fig. 4d). We observed additional protein-coding changes in genes involved in diverse cellular functions (Extended Data Fig. 1c), including proteostasis (Hsp90aa1, Hsp90ab1), membrane trafficking and secretion (Septin8, Septin11, Ap1s2, Olfm1, Lgi3) and enzymatic activities (Kalrn, Rexo2, Ppp2r5c). Finally, major brain regions could be defined using TREND preference alone (Fig. 4f and Supplementary Fig. 9), in agreement with our long-read analysis.
To understand the spatial regulation of TREND, we performed association analysis between event usage and RBP expression. Contrary to conventional thinking, SV in core alternative polyadenylation machinery components was insufficient to explain the observed 3′ end diversity patterns (Extended Data Fig. 1d). Instead, neural splicing regulators from our ONT analysis such as QKI, RBFOX and CELF families also emerged as top potential regulators for APA, supporting their central role in shaping the adult mouse brain transcriptome48. A prominent RBFOX target is Gnao1, encoding a subunit of the heterotrimeric G proteins. Our analysis uncovered previously unrecognized spatial divergence in Gnao1 last exon usage between cerebral nuclei and fiber tracts versus other brain regions, generating two protein isoforms with different C termini (Fig. 4g). Using public CLIP40 and knockout data49, we confirmed that RBFOX binds and promotes Gnao1 downstream exon usage. Regional isoform specificity in Gnas and Gnao1 extends to other G proteins (Gnai2, Gnas, Gnal, Gnaq) and G-protein-coupled receptors (Gabbr1, Grm5, Adgrl1), indicating a role of alternative transcript processing in fine-tuning signal transduction for spatially diverse physiological functions.
Our catalog also brought to light self-regulatory mechanisms driving transcript diversity in RBP genes themselves. For instance, the last exon usage of Arpp21 correlated with its total expression (Fig. 4h), consistent with a regulatory model where miR-1l.,28-2, produced from the longer isoform’s second-to-last intron, inhibits Arpp21 expression50. We also verified self-regulation in the 3′ UTR of CELF family51 (Fig. 4i) and discovered additional spatially variable 3′ UTR lengths across RBPs (Elavl2, Elavl3, Qk, Rbfox1, Hnrnpa2b1, Hnrnpk). One particularly intriguing new finding involves Pcbp2, which showed enriched usage of a minor upstream last exon in isocortex and thalamus. This alternative exon encodes a protein variant with an extended disordered C terminus (Extended Data Fig. 1e), whose usage inversely correlated with overall Pcbp2 expression in both mouse brain and across human tissues (Extended Data Fig. 1f). CLIP data52 revealed stronger human PCBP2 binding to this minor last exon compared to the canonical one, suggesting selective auto-regulatory feedback contributing to spatial specialization.
RBFOX and partners cooperatively regulate RNA processing
Causal inference from observational studies faces drawbacks from unknown confounding factors. Since SPLISOSM’s differential test captures both direct and indirect associations in the general form, it is not immune to such confounding effects. By comparing spatial RBP associations with perturbation studies in specific cellular environments, we can nevertheless extract meaningful biological insights.
A revealing example is the spatial regulation of Clta isoforms (Extended Data Fig. 5a). Knockout studies demonstrated that RBFOX proteins suppress Clta exon 5 and 6 inclusion in developing motor neurons49, yet our spatial test revealed a surprising positive correlation between Rbfox3 expression and the Clta-205 isoform containing these exons. Analysis of exon inclusion in a single-cell neocortex atlas initially confirmed this positive relationship across brain cell types (Extended Data Fig. 5b). The perceived contradiction resolved, however, when examining only neuronal subtypes: within GABAergic and far-projecting glutamatergic neurons, Rbfox3 negatively correlates with exon inclusion, aligning with the knockout results. This context-dependent relationship implies the presence of other cell-type-specific factors, countering RBFOX, that drive up Clta exon 5 usage in neurons. We found several additional genes (Myl6, Cltb, Tln1, Klc1; Supplementary Fig. 10) where RBFOX knockout effects contradicted spatial associations. Supporting our hypothesis, previous experiments have shown that repressive CELF binding can override RBFOX enhancement at MYL6 exon 6 in human T cells53.
To systematically identify potential RBFOX coregulators, we assessed agreement between RBP DU results and spatial coexpression patterns (Extended Data Fig. 5c). The top candidates included RBPs with varying reported RBFOX interactions: CELF family proteins, known to act antagonistically with RBFOX in muscle and heart tissue53; ELAVL2, sharing targets with RBFOX1 in human neurons54 and QKI, recently confirmed to affect RBFOX function by minigene assay55. Expression of these collaborating RBPs remained stable after RBFOX depletion (Supplementary Fig. 10a), pointing toward direct cooperation rather than cross-regulation feedback. Sequence motif analysis provided further validation, showing significant enrichment of ELAVL, CELF and QKI binding motifs near Rbfox-associated variable exons detected in the ONT data, while KHDRBS3 (a QKI homolog) and ELAVL motifs were enriched near Rbfox-associated TREND regions in SR data (Extended Data Fig. 5d). Consistently, we observed position-dependent overrepresentation of CELF and ELAVL motifs around exons responding to RBFOX triple knockout in the perturbation data49, supporting the collaborative regulation between multiple RBPs on the same targets (Extended Data Fig. 5e).
Conserved synaptic gene transcript diversity in human prefrontal cortex
Given the critical role of splicing and APA in brain functions, we hypothesized that spatial transcript diversity patterns would be highly preserved throughout mammalian evolution. To investigate this, we reanalyzed 12 10X Visium samples56 from healthy human brain (Fig. 5a and Methods), identifying 861 SVP genes in the dorsolateral prefrontal cortex (DLPFC) (adjusted P < 0.05). Power to detect SV has not yet saturated with respect to sequencing depth (Fig. 5b and Extended Data Fig. 6a), so improved RNA capture per spot would likely reveal additional variable events. When evaluating performance across replicates, SPLISOSM’s expression-based tests (HSIC-GC and HSIC-IC) demonstrated better power and consistency compared to SPARK-X, which was hampered by suboptimal kernel design (Fig. 5c). The HSIC-IR test showed weaker concordance, primarily due to data sparsity and noise, although this improved significantly for genes with higher expression.
a, Workflow overview. b, Relationship between total read coverage in TREND regions per spot (x axis) and number of significant SVP (top, HSIC-IR adjusted P < 0.01) or variably expressed (SVE, bottom, HSIC-GC adjusted P < 0.01) genes across datasets. Line and shading indicate fitted linear model and 95% confidence interval. c, P value correlation distributions between technical replicates (left), different sections from same donor (middle) and different donors (right). Genes are stratified by average TREND read counts per spot and group means are compared using the Kruskal–Wallis test. Boxplots show median (center line), interquartile range (box) and 1.5× interquartile range (whiskers). d, SVP genes ranked by recurrence (number of significant samples, x axis) and the minimal HSIC-IR P values across samples (y axis). Highlighted genes have mouse homologs that are also spatially variably spliced (HSIC-IR adjusted P < 0.01). e, Distributions of average conservation scores in TREND regions across gene categories: nonvariable controls (n = 12,107), human-specific SVENP (n = 8,044) or SVP (n = 1,319) and human-mouse shared SVENP (n = 4,022) or SVP (n = 401). Boxplots show median (center line), interquartile range (box) and 1.5× interquartile range (whiskers). Pairwise group means are compared using two-sided t-test. f, Pathway enrichment analysis comparing SVP (orange) versus SVENP (dark blue) genes conserved between human and mouse. g, RBP-SVP associations ranked by recurrence (x axis) and minimal GLMM P values across samples (y axis). Top potential regulators of SEPTIN8 are highlighted and colored by whether the association is conserved in mouse (P < 0.01 for both GLMM and HSIC-based DU tests). h, SEPTIN8 transcript structure and spatial log-normalized expression in mouse (top) and human (bottom, sample 151673). i, SEPTIN8-long and SEPTIN8-short protein variants with potentially different cellular functions. Panels a and i created using BioRender.com.
Despite these technical limitations, we found strong evidence for cross-species conservation at both individual gene and functional levels. Of the human SVP genes, 178 had mouse homologs that also exhibited SV (Fig. 5d and Extended Data Fig. 6b,c). Moreover, conserved TREND regions displayed significantly higher sequence conservation than invariant regions (Fig. 5e). Functional enrichment analyses in both species highlighted the importance of isoform-level regulation in synaptic signaling and chaperone-mediated autophagy, processes implicated in various neurodegenerative disorders (Fig. 5f and Extended Data Fig. 6d).
The molecular regulation governing spatial isoform diversity also showed remarkable cross-species similarities. MAP4, which organizes microtubules in muscles and neurons, exhibited patterns corresponding to neuronal-versus-glial usage in both species (Fig. 4e and Extended Data Fig. 6e). While PTBP2 was previously identified as a regulator of MAP4 last exon usage in embryonic brain cortex47, its limited SV in adult brain suggested the involvement of additional factors in region-specific regulation (Extended Data Fig. 6f). Our DU analyses pointed to RBFOX1, QKI and CELF families as likely cofactors, with CLIP data revealing conserved binding sites supporting this hypothesis (Extended Data Fig. 6g).
SEPTIN8 provides another compelling example of evolutionarily conserved regulation. This gene encodes a GTP-binding protein that polymerizes into filaments essential for cellular architecture. Our spatial analysis revealed a distinct usage of SEPTIN8 upstream last exon, presumably producing a shorter isoform predominating in mouse fiber tracts and human DLPFC white matter (Fig. 5h). This truncated protein lacks the extra C-terminal region that interacts with VAMP2 to facilitate SNARE-complex assembly57, which also features a filopodia-inducing motif that enhances dendritic arborization when palmitoylated58 (Fig. 5i). The downstream Septin8 last exon is promoted by NOVA2 in mouse motor neurons58. Our analysis indicates that NOVA2 likely cooperates with QKI and CELF proteins jointly to achieve the precise spatial patterning in the brain (Fig. 5g).
Microenvironment shapes glioma spatial transcript diversity
The capability to detect patterns without cell-type or region annotations makes SPLISOSM especially valuable for complex tissues with ambiguous cellular states. Glioblastomas are characterized by their infiltrative nature and heterogeneous spatial structures59. To explore isoform distributions in this challenging context, we systematically mapped spatial transcript usage across 24 human glioma samples (Fig. 6a and Methods), identifying 306 SVP genes in 11 ONT samples60 and 2,828 SVP genes in 13 SR samples59 (adjusted P < 0.05, Fig. 6b). The most frequently recurring SR-SVP genes overlapped substantially with those found in healthy brain tissue, reflecting compositional changes of nontumor cells (Fig. 6c and Extended Data Fig. 7a,b). As in healthy brain, approximately 25% of all variable transcript events in glioma had potential implications for protein structure, with alternating exon accounting for much of this variation (Extended Data Fig. 7d). Beyond RNA processing, glioma-specific transcript diversity also emerged from genomic alterations. In one pediatric sample, we detected variable immunoglobulin transcripts resulting from variable–diversity–joining recombination, revealing clonality in B cell infiltration (Extended Data Fig. 7c).
a, Workflow overview. b, SVP genes ranked by recurrence and the minimal HSIC-IR P values across samples in ONT (left) and SR (right) cohorts. Orange indicates glioma-specific genes not variable (HSIC-IR adjusted P ≥ 0.05) in all healthy DLPFC samples. c, Distributions of recurrent and glioma-specific SVP genes. d, Pathway enrichment analysis comparing recurrent SVP (HSIC-IR adjusted P < 0.05 in ≥2 samples) genes versus recurrent SVENP (non-SVP and HSIC-GC adjusted P < 0.05 in ≥4 samples) genes. e, Spatial distribution of HLA-DRB1 isoforms in the ONT sample GBM1. f, B2M and CD74 isoform expression in GBM1, comparing tumor (n = 3,706) versus immune-infiltrated (n = 898) spots. Three novel CD74 intron-retention isoforms are pooled together for visualization. Boxplots show median (center line), interquartile range (box) and 1.5× interquartile range (whiskers). P values from two-sided t-test. g, TPM3 transcript structure, RNA-seq coverage, and spatial patterns in the SR sample ZH916bulk. Event counts were locally smoothed when computing ratios, same in l,m. h, Glioblastoma metaprogram distribution in regions with high or low TPM3-event 1 usage. P value from one-sided chi-square test. Right: log-normalized expression (logExpr) of selected markers: SNAP25 (neuron), ACTB (glial), IGKC (B cell), NDRG1 (hypoxia). AC, astrocyte-like; MES, mesenchymal-like; MES.Ast, astrocytic-like mesenchymal; MES.Hyp, MES-hypoxia; Vasc, endothelia cells and pericytes. i, FTH1 transcript structure in ONT sample DMG3 and its RNA-seq coverage in SR sample ZH8811Abulk. j, Hypoxia marker expression and association with FTH1-211 usage in DMG3. P value from one-sided chi-square test. k, Histology (left), metaprogram distribution (middle, P value from one-sided chi-square test) and hemoglobin expression (right) in spots with high (n = 438) or low (n = 1,945) FTH1-event 2 usage in ZH8811Abulk. Mac, macrophage/microglia; OPC, oligodendrocyte progenitor-like; NPC, neural progenitor-like. Boxplots show median (center line), interquartile range (box) and 1.5× interquartile range (whiskers), with P values from two-sided t-test. l, Spatial distribution of FTH1 isoforms in DMG3. m, Spatial distribution of FTH1 TREND usage in ZH8811Abulk. Panel a created using BioRender.com.
The importance of immune interactions was further underscored by consistent variability in antigen presentation components across samples, often correlated with immune infiltration patterns (Fig. 6d and Extended Data Fig. 7e–g). This included human leukocyte antigen (HLA) class I (HLA-A, B, C) and class II (HLA-DRA, DRB1, DPB1) genes, whose sequence variation likely stems from both alternative splicing and germline variability61,62 (Fig. 6e). For nonpolymorphic elements such as B2M and CD74, we found preferential usage of intron-retaining, potentially nonfunctional transcripts in tumor regions (Fig. 6f). While the SR data could not capture identical variability events due to its 3′ end bias, it demonstrated similar functional convergence toward immune-related pathways, particularly in infectious disease response and viral infection pathways (Fig. 6d and Extended Data Fig. 7g).
Furthermore, we detected SV in signal transduction, focal adhesion and cytoskeleton regulation pathways (Extended Data Fig. 7h), reflecting the intricate cellular interactions and extracellular matrix remodeling within tumors. TPM3, a tropomyosin family member with well-documented cancer-associated splicing patterns across tumor types63, exhibited complex and distinct splicing variation (Fig. 6g). We identified an upstream TPM3 event (event 4) that partially overlapped with neuronal and hypoxia signatures, while the canonical form (event 1) associated with glial programs and increased actin expression while negatively correlating with B cell infiltration (Fig. 6h). These results resonate with recent reports of differential splicing of actin cytoskeleton components between peripheral and core glioblastoma cells64. In addition, we observed spatially variable usage of the last exon of GFAP, encoding two variants of an astrocytic intermediate filament protein whose ratio is linked to glioma grade and malignancy65,66 (Extended Data Fig. 7i). The location-specific distribution of these cytoskeleton gene isoforms indicates dynamic responses to local microenvironmental cues, modulating migration potentials and driving glioblastoma invasion.
Microenvironmental influences extended to ribosomal proteins as well. We observed SV in transcripts encoding ribosomal proteins (including RPS8, RPS24, RPL5, RPLP1, RPS6, RPS12) associated with tumor infiltration, frequently involving retained introns or truncated 3′ ends (Supplementary Fig. 11). While some such as RPS8 also exhibited variability in normal brain, the altered read distributions clearly displayed glioblastoma-specific changes (Supplementary Fig. 11a), aligning with previous findings that ribosomal isoform switching occurs in response to microenvironmental cues and contributes to metabolic reprogramming phenotypic plasticity in glioblastoma67.
FTH1 offers another prominent example of microenvironment-driven transcript usage (Fig. 6i–m). This iron-storage protein, upregulated under hypoxic conditions, protects cells from ferroptosis by reducing reactive iron68. We discovered that FTH1 upregulation in hypoxic tumor regions coincided with increased expression of 3′ truncated transcripts lacking the final exon–exon junction (Fig. 6j). In SR data, we identified a minor event spanning FTH1 exons 2 and 3, predominantly in regions with erythrocytes and overlapped with the chromatin regulation metaprogram (Fig. 6k). Our findings suggest coordination between oxygen-sensing, iron metabolism and stress response pathways through transcript-level modulation of FTH1.
Discussion
Spatial omics techniques are transforming our understanding of tissue organization, yet few studies have explored molecular diversity at the isoform level. Here we present SPLISOSM, a computational method designed for robust detection of spatial transcript processing and its regulation in ST data. Through theory-grounded nonparametric kernel tests, our approach achieves high statistical power in sparse data while delivering well-calibrated, permutation-free P values. SPLISOSM demonstrates broad applicability across ST platforms and spatial resolutions. We show that standard 3′ end sequencing protocols and predesigned in situ panels can capture considerable transcript diversity, enabling researchers to extract new insights from existing datasets. Our implementation of low-rank kernel approximation allows testing hundreds of thousands of locations within minutes on standard hardware with minimal power loss (Supplementary Note).
Using SPLISOSM, we have uncovered thousands of evolutionarily conserved spatial transcript diversity events in mouse and human brain. However, the spectrum of detectable events varies across ST platforms (Fig. 1a). While SPLISOSM remains agnostic to event type, its performance depends on platform-specific limitations (Extended Data Fig. 8). Sequencing-based platforms suffer primarily from false negatives due to 3′ bias, read length and sequencing depth, particularly affecting detection of longer transcripts (Extended Data Fig. 4b). Current depths have not saturated SVP detection, suggesting that improvements in RNA capture efficiency and isoform quantification especially for novel transcripts can further expand the landscape. In contrast, imaging-based technologies face false positives from off-target probe binding, exemplified by spurious glial-specific patterns in negative control probes (Extended Data Fig. 2e). Nevertheless, our analysis revealed hundreds of variable exon usage events in mouse brain using predesigned Xenium 5K panel, though mapping these events to full-length isoforms requires future efforts.
Our integrative regulatory analyses shed light on the complex network governing spatial splicing. Neural RBPs, including RBFOX and CELF family members, may orchestrate transcript diversity through precise spatiotemporal activation69. However, important limitations temper our conclusions. First, ST data sparsity limits association test power, potentially missing lowly expressed but functionally important RBPs. Second, tissue and developmental stage mismatches between spatial data (adult mouse brain) and available functional perturbation data (often in vitro cell culture) complicate direct validation. Evidence for cooperative regulation derived from coexpression, motif enrichment and CLIP binding remains largely correlative. Future perturbation experiments in appropriate neural contexts are essential to validate and elucidate the molecular basis of the proposed cooperative mechanisms. Third, while our DU test accounts for spatial autocorrelation, they still assess correlation rather than causation and treat RBPs independently without explicitly controlling for coexpression. Future extensions could incorporate causal graphical models to test conditional independence among multiple regulators.
In glioblastoma, our analysis reveals how spatial transcript diversity is shaped by microenvironmental composition and metabolic conditions. The upregulation of truncated and intron-retaining transcripts in antigen presentation genes implies that tumors may employ alternative processing for immune evasion62. Targeting these events may enhance immune response and improve immunotherapy benefits. We also observed noteworthy diversity in ribosomal transcripts, suggesting translational regulation could play an underappreciated role in tumor heterogeneity and thus exposing new therapeutic vulnerabilities67.
In summary, SPLISOSM introduces an isoform-centric perspective to ST data analysis. The spatial organization of transcript diversity we mapped in healthy and malignant brain tissues established a foundation for understanding how alternative isoform processing contributes to specialized functions and disease pathology.
Methods
SPLISOSM overview
SPLISOSM analyzes isoform-level ST data through a two-step approach: first detecting SV, then identifying DU by associating isoform patterns with potential regulatory factors. The framework accommodates quantification results of both full-length isoforms from long-read sequencing and local transcript diversity events from SR sequencing platforms including 10X Visium and Slide-seqV2.
Unlike gene-level analysis, SPLISOSM recognizes that multiple isoforms (q > 1) from the same gene are interdependent. Their relative proportions form a multivariate statistical variable across the spatial random field of n locations
Together with the 2D spatial coordinates \(\{{Y}_{i}\in {{\mathbb{R}}}^{2},i=1,\cdots ,n\}\) and covariates such as RNA binding protein expression \(\{{Z}_{i}\in {\mathbb{R}},i=1,\cdots ,n\}\), we can formulate SV detection and differential association analysis as multivariate independence tests T(X, Y) and T(X|Y, Z|Y). The spatial conditioning in DU tests is critical for eliminating spurious associations caused by spatial autocorrelated noise and hidden confounders.
Below we outline SPLISOSM’s key components, with comprehensive methodology, theoretical analysis and implementation details available in the Supplementary Note.
Unconditional kernel independence test
A commonly used statistic for multivariate correlation analysis is the RV coefficient, defined as the normalized matrix norm of the cross-covariance matrix \({C}_{{XY}}\in\mathbb{R}^{q \times 2}, \, {C}_{{ij}}=\text{Cov}\left({X}^{i},{Y}^{j}\right)\) between X and Y
To capture complex, nonlinear spatial patterns beyond linear correlation, SPLISOSM uses kernel transformations to map variables into reproducing kernel Hilbert spaces. In these potentially infinite-dimensional feature spaces, we measure statistical dependence using the HSIC33, defined as the Hilbert–Schmidt norm of the cross-covariance operator \({C}_{{XY}}:{\mathcal{G}}\to {\mathcal{F}}\) that connects the respective feature spaces \({\mathcal{F}}\) and \({\mathcal{G}}\) of X and Y,
Similar to the univariate gene expression SV test SPARK-X30, we build calibrated, permutation-free testing procedures based on the empirical HSIC estimator,
Here, KX and \({L}_{Y}\) are the n-by-n isoform and spatial kernel matrices that specify feature maps for X and Y, and \(H=I-\frac{1}{n}1{1}^{T}\) is the centering matrix. Under the null hypothesis of statistical independence, THSIC follows an asymptotic distribution of chi-square mixture34, with weights derived from the eigenvalues {λi} and {μj} of KX and \({L}_{Y}\),
The statistical power of HSIC-based nonparametric test depends heavily on effective kernel design. To profile spatial isoform patterns in sparse data, SPLISOSM introduces two technical innovations:
First, we prove that any low-rank approximation of the spatial kernel significantly reduces test power (Theorem 1 in Supplementary Note), highlighting a key limitation of existing SV tests such as SPARK-X. Based on this insight, we developed a new full-rank spatial kernel using the intrinsic conditional autoregressive (ICAR) model35,
where W is the n-by-n binary adjacency weight matrix of the mutual k-nearest-neighbor spatial graph, \(D=\text{diag}\{{\sum }_{j}{W}_{{ij}}\}\) is the degree matrix and \(\rho \in [0,1)\) is the spatial autocorrelation coefficient ensuring invertibility.
In essence, this approach represents spatial patterns as signals on the spatial graph to be decomposed onto the eigenvectors of \({L}_{Y}\) through graph Fourier transform (GFT)70, allowing us to prioritize patterns by frequency in the graph spectral domain without manually selecting spatial bandwidths. Furthermore, the sparse structure of D and W enhances computational efficiency. For large-scale datasets (for example, hundreds of thousands of spots) where full-rank spatial kernels become computationally prohibitive, we developed GFT-based low-rank kernels that approximate the kernel inverse \({L}_{Y}^{-1}\) through sparse eigen-decomposition, preserving biologically meaningful low-frequency patterns while limiting power loss to less important high-frequency signals.
Second, to handle sparse isoform data where many spots have zero gene coverage, we designed a zero-padded centered kernel for isoform ratios Xn with n − m undefined values,
where Xm represents isoform ratios at m spots with nonzero gene coverage, and \({K}_{{X}_{m}}:= \phi \left({X}_{m}\right)\phi {\left({X}_{m}\right)}^{T}\) is the isoform kernel with transformation ɸ for compositional data (for example log-ratio transformations). Our empirical analysis showed that a simple linear kernel \(k\left(x,y\right)=\left\langle x,y\right\rangle\) (with identity transformation) delivers optimal performance.
The zero-padded centered version of a linear kernel is equivalent to a two-step procedure where the n − m undefined ratios in Xn are first replaced with global averages per isoform before constructing the n-by-n isoform kernel for independence test. The NA replacement allows us to reuse the n-by-n spatial kernel across genes with varying sparsity patterns, substantially improving computational efficiency. We mathematically prove that this approach provides a bounded approximation to testing procedures that simply omit spots with undefined ratios (Theorem 2 in Supplementary Note).
Detailed implementations of the ICAR kernel and SPLISOSM’s three SV tests—HSIC-GC (gene expression), HSIC-IR (isoform usage ratio) and HSIC-IC (isoform expression)—are available as Algorithms 1–4 in the Supplementary Note. The GFT-based spatial kernel construction is presented in Algorithm 5.
Conditional kernel independence test
Linking isoform usage X with potential regulators Z requires addressing a fundamental challenge: X and Z may appear related simply because both follow similar spatial patterns irrelevant to splicing regulation. To learn the causal relationship, we need to condition on spatial coordinates Y and all other confounders if available. However, ST data provide only snapshots: at each location Yi, we observed only one \(\left({X}_{i},{Z}_{i}\right)\) pair, making direct estimation of conditional distributions for \({X|Y}\) and \({Z|Y}\) impossible.
We address this using the conditional independence framework from ref. 34, which shares information across observations by learning regression functions \({f}_{X}:Y\to X\) and \({f}_{Z}:Y\to Z\), then testing kernel independence of the residuals.
For linear ridge regression with L2 regularization λ, we can compute the residuals as
where \(Y\in \mathbb{R}^{n\times 2}\) represents spatial coordinates and \(X\in \mathbb{R}^{n\times {\rm{d}}}\) the spatial variable to predict. By substituting the linear kernel \(Y{Y}^{T}\) with general spatial kernels \({K}_{Y}\), we extend the solution to nonlinear relationships
This allows us to construct a kernel for the conditional variable \({X|Y}\) as \({K}_{{X|Y}}={R}_{Y}^{X}{K}_{X}{R}_{Y}^{X}\) where \({R}_{Y}^{X}=\lambda {\left({K}_{Y}+\lambda I\right)}^{-1}\) is learned from the data. The parameter \(\lambda\) controls conditioning strength: at λ = 0, X is fully explained by coordinates Y, leaving no information for X|Y; as \(\lambda \to \infty\), the residuals \(X-\hat{X}\to X\), which gives the unconditional test. Since genes have diverse spatial patterns, we estimate λ and \({K}_{Y}\) for each variable X (and Z) individually using Gaussian process regression.
Using linear kernels KX and KZ, our conditional association test statistic becomes
where \(\tilde{X}\) and \(\tilde{Z}\) are column-wise centered isoform usage ratios and covariates, and \({R}_{Y}^{X}\) and \({R}_{Y}^{Z}\) are the residual operators estimated from Gaussian process regression.
Detailed implementations of the HSIC-based unconditional and conditional DU tests are provided as Algorithms 6 and 7 in the Supplementary Note. For parametric approaches, we can also introduce spatial conditioning through GLMMs with spatial random effects. Specifically, we implemented the score test for DU where null GLMM models without covariates are first fitted to account for background spatial patterns. Covariate significance is then assessed by computing the gradient (that is, score statistic) at zero effect size. Details on model fitting and hypothesis testing are available in Chapter 3 of the Supplementary Note.
General test configurations
For all isoform SV tests, we used raw UMI counts from isoform quantification results (that is, isoform counts in ONT data and TREND event counts in SR data). Input data varied by method: HSIC-GC and SPARK-X used summed isoform UMI counts (a scalar) per gene per spot, HSIC-IC used the vector of isoform counts per gene per spot and HSIC-IR used observed isoform ratios. For spots with zero gene coverage (undefined ratios), we substituted with the global mean ratio (justified in Theorem 2).
For DU testing with RBP expression, we used log1p-normalized Visium RBP gene expression (library size 1 × 104) as covariates. We first identified spatially variably expressed RBPs (adjusted P < 0.01 in both HSIC-GC and SPARK-X tests) to include as covariates, excluding ribosomal proteins and noncanonical RBPs. SVE tests were conducted on Visium-based RBP gene raw counts. For human DLPFC samples, we restricted our analysis to potential splicing modulators (RBPs with motif or CLIP binding data). Only SVP genes with average total ONT or TREND counts >0.5 per spot were investigated for DU with RBP. Specific numbers of tested RBPs and SVP genes per sample are detailed in Supplementary Table 1. For HSIC-based DU tests, we used observed isoform ratios with mean replacement for undefined values. Gaussian process regression was implemented using GaussianProcessRegressor from the scikit-learn71 package with learnable RBF and white noise kernels. For parametric models (GLM and GLMM), we used isoform count vectors as inputs, grouping genes with identical isoform numbers to enable parallel processing. We calculated P values using score tests on fitted null models under asymptotic chi-square distribution (one-sided).
Except where otherwise noted, all SPLISOSM SV results were computed with the full-rank ICAR kernel with ρ = 0.99 and the number of mutual nearest neighbors k = 4 for constructing the spatial graph. SV results in the Xenium Prime 5K adult mouse brain sample were computed with the low-rank kernel (r = 100) for computational efficiency. HSIC-based test P values were calculated using Liu’s method72 to approximate the chi-square mixture null distribution (one-sided). Multiple testing correction was applied using the Benjamini–Hochberg method73.
External datasets of RBP
We obtained mouse and human RBP lists from EuRBPDB74. For motif analysis, we obtained RBP binding motifs from CISBP-RNA75 and refined them using the R package universalmotif76 to trim low-information peripheral positions before scanning with FIMO from the MEME suite77 (P threshold 1 × 10−3, background model computed using the default fasta-get-markov). RBP CLIP data were downloaded from the POSTAR340 database and the ENCODE78 portal (for PCBP2 human eCLIP). In our downstream analysis, we prioritized RBPs with validated binding motifs or CLIP evidence as potential splicing regulators. APA-related RBPs were collected based on regulators previously characterized in a high-throughput screening study79.
Data simulation
We generated in silico spatial isoform quantification data using a hierarchical generative model following two steps: first sampling total gene expression and expected isoform usage per spot, then sampling observed isoform counts from spot-specific multinomial distributions.
For total gene expression, we sampled from Poisson distributions with spot-specific means. In scenarios 1–3 (without spatial gene expression variability), these means were drawn from independent Gaussian distributions (μ = 5, σ2 = 1). In scenarios 4–6 (with regional gene expression patterns), means were sampled from multivariate Gaussian distributions with binary covariate-controlled means and covariance matrices combining spatial autocorrelation (ICAR)35 and white noise.
For expected isoform usage ratios, we sampled in the log-ratio space with q − 1 degrees of freedom where q = 3 is the number of isoforms per gene. In scenarios 1 and 4 (without spatial isoform variability), log-ratios were sampled from independent Gaussian distributions (μ = 0, σ2 = 0.2) and transformed via softmax to obtain expected ratios. In scenarios 2 and 5 (with Gaussian process variability), log-ratios were sampled from multivariate Gaussian distributions with ICAR-based spatial covariance. In scenarios 3 and 6 (with regional isoform usage), log-ratios were determined by binary covariates with added ICAR-based spatial autocorrelated noise.
To evaluate statistical power, we created intermediate scenarios by interpolating between key configurations. For the SV test HSIC-IR, we varied the proportion of spatially autocorrelated noise between scenario 4 (regional expression, no isoform usage variability) and scenario 5 (regional expression, 50% spatially autocorrelated noise + 50% white noise in the Gaussian process). For DU tests, we adjusted covariate effect sizes between scenario 5 (regional expression, no isoform usage association) to scenario 6 (regional expression, covariate effect size 0.5).
SiT data of adult mouse brain
We downloaded processed spatial isoform transcriptomics (SiT)18 isoform quantification results as well as Visium gene expression data of mouse olfactory bulb and two coronal brain section samples (CBS1, CBS2) from Gene Expression Omnibus (GSE153859). The SiT ONT datasets contain only reference isoforms mapped to Mouse Gencode vM24. We removed isoforms expressed in <1% of spots and genes with fewer than 2 detected isoforms.
To reduce sparsity for visualization and clustering, we applied local smoothing by borrowing 20% of UMIs from six nearest neighbors per spot, then calculated isoform ratios with mean-padding for remaining undefined values. The resulting ratios may amplify noise and artifacts and should therefore be interpreted with caution. For spatial clustering, we first identified significant SVE and SVP genes using unsmoothed raw data, then performed dimensionality reduction (principal components analysis, 50 components) and Leiden clustering on either smoothed gene expression or isoform ratios. RBP-based clustering used log1P-normalized Visium expression of SVE RBPs. Clustering resolutions were individually chosen such that the resulting cluster number were similar. For SVP usage program hierarchical clustering, we computed pairwise gene similarity using RV coefficients between multivariate smoothed isoform usage ratios.
For splicing type annotation, we used SUPPA80 to split isoforms into local alternative splicing events. Skipping exons, mutually exclusive exons and their flanking introns were analyzed for motif enrichment using MEME suite77 tools (XSTREME and SEA), with additional exon sequence features extracted using Matt81.
Visium and Slide-seqV2 data of adult mouse brain
We downloaded RNA-seq .bam files of the 10X Visium44 data of a fresh frozen adult mouse coronal brain section sample from https://www.10xgenomics.com/datasets/adult-mouse-brain-coronal-section-fresh-frozen-1-standard, and the Slide-seqV2 (ref. 16) data of adult mouse hippocampus from https://singlecell.broadinstitute.org/single_cell/study/SCP815/sensitive-spatial-genome-wide-expression-profiling-at-cellular-resolution#study-summary.
TREND was quantified using Sierra82 with Mouse Gencode v.M10 reference annotation, with minimal peak cutoffs in FindPeaks set to zero to recover minor events in the quantification stage. TREND events expressed in <1% of spots and genes with fewer than 2 TREND regions were later filtered out. Events were annotated using Sierra’s AnnotatePeaksFromGTF with annotation_correction=FALSE to allow multiple annotations per region. Alternative exons (Alt-Exon) were defined as exons located within introns of other transcript variants. Motif enrichment analysis was performed on exons of TREND regions using MEME suite77 tools (XSTREME and SEA).
For SV tests on gene expression (HSIC-GC and SPARK-X), we used the sum of TREND reads per gene per spot rather than Visium’s Space Ranger quantification (except for the RBP SVE test in which we always used the latter) to maintain consistency with HSIC-IC/IR tests and ONT data analysis. Slide-seqV2 data were analyzed using HSIC-GC/IC/IR with full-rank spatial kernels. See Supplementary Note for low-rank test performance.
For spatial clustering, we use unsmoothed TREND-based gene expression and isoform usage from top 200 SVE and SVP genes for respective clusters, and Visium-based expression of all SVE RBPs for RBP clusters. We performed dimensionality reduction (principal components analysis, 50 components) and Leiden clustering following the same approach as in our SiT ONT data analysis.
Xenium Prime 5K data of adult mouse brain
We obtained the Xenium output bundle of a fresh frozen adult mouse brain hemisphere profiled using 10X Xenium Prime 5K Mouse Pan Tissue and Pathways Panel46 from https://www.10xgenomics.com/datasets/xenium-prime-fresh-frozen-mouse-brain and reran the Xenium Ranger (v.3.1.1) relabel pipeline with default parameters. Transcripts passing sequence quality filter (Q ≥ 20) from genes with multiple detected codewords were spatially binned into 20 × 20-μm regions. This yielded a sparse count matrix comprising 10,476 codewords from 4,993 genes across 92,202 spatial locations. SVP significance threshold (adjusted P < 2.5 × 10−17) was determined by the minimum P value observed in negative control and intergenic probe sets. Cell segmentation, clustering and per-cluster differential gene expression analysis were performed as part of the Xenium Ranger pipeline with default parameters. For colocalization analysis, we quantified the number of cells from different clusters within each 20 × 20-μm bin. To identify molecular heterogeneity within cluster 2 (excitatory neurons), we performed an additional differential codeword expression analysis using a two-sided t-test, comparing Dtnbp1-11516-only bins against the remaining cluster 2 bins. This analysis revealed Nptxr as a marker for the Dtnbp1-206-positive excitatory neuron subpopulation.
Single-cell splicing quantification and RBFOX triple knockout data
We analyzed processed exon and splicing quantification data from the adult mouse neocortex single-cell atlas83 using results from ref. 84. Cassette exon Percent Spliced-In was quantified at cell-type level using Quantas4 by aggregating cells of identical types. Average RBP expression per cell type was log normalized and standardized to zero means.
For RBFOX triple knockout analysis, we used processed Quantas-based splicing quantification results of mouse day 5 and 10 motor neurons in vitro differentiated from mouse embryonic stem cells from ref. 49. The alternative splicing event, Percent Spliced-in and statistical significance were all determined by the original study. All RBFOX tKO events shown in the paper are statistically significant after multi-test correction. Raw RNA sequencing (RNA-seq) data were downloaded from the Sequence Read Archive (SRP128054) to run RBP map and positional motif enrichment analysis using Matt81.
Visium data of human DLPFC
We downloaded Visium RNA-seq bam files of 12 human DLPFC samples56 through Globus (endpoint: jhpce#HumanPilot10x). TREND was quantified using Sierra82 with 10X’s GRCh38-2024-A reference annotation and default parameters. To create a shared set of TREND events across samples, we merged per-sample peaks using MergePeakCoordinates with default settings and quantified them with CountPeaks. We obtained conservation scores (phyloP100way and phastCons20way) from the UCSC Genome Browser and calculated average conservation scores specifically for exons within the TREND regions.
ONT and Visium data of human glioma samples
We analyzed two independent cohorts of human glioma samples using long-read and SR ST platforms.
For the cohort from ref. 60, we obtained processed ONT isoform quantification results, matched Visium gene expression data and region annotations for 13 human glioma samples directly from the study authors, as the raw data were restricted by human genetic resources regulations. ONT transcripts were error corrected, collapsed by shared exon–exon junctions and quantified using DEMINERS85 by the original study. For spatial analysis, we included only reference isoforms matching Gencode v.32 (hg38) and novel isoforms from known genes. We further filtered out isoforms expressed in <5% of spots and genes with fewer than 2 detected isoforms. SUPPA80 was used to categorize alternative isoform usage into local alternative splicing events.
For the cohort from ref. 59, we processed Visium data from 13 IDH wild-type glioblastoma samples downloaded from SRA (PRJNA994130). Raw RNA-seq reads were aligned to the human reference genome (hg38) using Space Ranger v.3.0.1. Metaprograms and spatial annotations were obtained from the authors’ GitHub repository (github.com/tiroshlab/Spatial_Glioma). TREND was quantified using Sierra82 with 10X’s GRCh38-2024-A reference annotation and default parameters. Due to sample heterogeneity, we processed each sample independently with sample-specific TREND regions rather than merging across samples. For spatial analysis, we filtered out TREND events expressed in <1% of spots, genes with fewer than 2 TREND regions and novel genes with symbols beginning with ‘ENSG’.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
This paper analyzes existing, publicly available data without generating new primary data. References and accession details to raw data are provided in Supplementary Table 1 and in the corresponding Methods. 10X Visium44 data of a fresh frozen adult mouse coronal brain section sample are available from 10X Genomics via https://www.10xgenomics.com/datasets/adult-mouse-brain-coronal-section-fresh-frozen-1-standard, the Slide-seqV2 (ref. 16) data of adult mouse hippocampus are available from Broad Institute via https://singlecell.broadinstitute.org/single_cell/study/SCP815/sensitive-spatial-genome-wide-expression-profiling-at-cellular-resolution#study-summary and 10X Xenium Prime 5K Mouse Pan Tissue and Pathways Panel46 are available from 10X Genomics via https://www.10xgenomics.com/datasets/xenium-prime-fresh-frozen-mouse-brain. Per-cohort isoform quantification results, figure source data and other data necessary to reproduce results described in this study can be downloaded from Zenodo at https://doi.org/10.5281/zenodo.17625643 (ref. 86). Processed and formatted per-sample data and SPLISOSM test results can be downloaded from Zenodo at https://doi.org/10.5281/zenodo.16905935 (ref. 87).
Code availability
The SPLISOSM Python package is available via GitHub from https://github.com/JiayuSuPKU/SPLISOSM under BSD-3 License. Code and scripts to reproduce this paper, as well as Google Colab notebooks for interactive data exploration and visualization, are available via GitHub from https://github.com/JiayuSuPKU/SPLISOSM_paper.
References
Chen, M. & Manley, J. L. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 10, 741–754 (2009).
Tian, B. & Manley, J. L. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 18, 18–30 (2017).
Wang, E. T. et al. Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 (2008).
Weyn-Vanhentenryck, S. M. et al. Precise temporal regulation of alternative splicing during neural development. Nat. Commun. 9, 2189 (2018).
Zhang, X. et al. Cell-type-specific alternative splicing governs cell fate in the developing cerebral cortex. Cell 166, 1147–1162 e15 (2016).
Ule, J. et al. Nova regulates brain-specific splicing to shape the synapse. Nat. Genet. 37, 844–852 (2005).
Beffert, U. et al. Modulation of synaptic plasticity and memory by Reelin involves differential splicing of the lipoprotein receptor Apoer2. Neuron 47, 567–579 (2005).
Li, Y. I. et al. RNA splicing is a primary link between genetic variation and disease. Science 352, 600–604 (2016).
Walker, R. L. et al. Genetic control of expression and splicing in developing human brain informs disease mechanisms. Cell 179, 750–771.e22 (2019).
Raj, T. et al. Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer’s disease susceptibility. Nat. Genet. 50, 1584–1592 (2018).
Liu, Z. et al. Mutations in the RNA splicing factor SF3B1 promote tumorigenesis through MYC stabilization. Cancer Discov. 10, 806–821 (2020).
Zhang, Y., Qian, J., Gu, C. & Yang, Y. Alternative splicing and cancer: a systematic review. Signal Transduct. Target Ther. 6, 78 (2021).
Jbara, A. et al. RBFOX2 modulates a metastatic signature of alternative splicing in pancreatic cancer. Nature 617, 147–153 (2023).
Stahl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Liu, Y. et al. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell 183, 1665–1681.e18 (2020).
Stickels, R. R. et al. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat. Biotechnol. 39, 313–319 (2021).
Joglekar, A. et al. A spatially resolved brain region- and cell type-specific isoform atlas of the postnatal mouse brain. Nat. Commun. 12, 463 (2021).
Lebrigand, K. et al. The spatial landscape of gene expression isoforms in tissue sections. Nucleic Acids Res. 51, e47 (2023).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Cohen, L. et al. Whole-transcriptome-scale and isoform-resolved spatial imaging of single cells in complex tissues. Preprint at bioRxiv https://doi.org/10.1101/2025.08.27.672533 (2025).
Ji, G. et al. stAPAminer: mining spatial patterns of alternative polyadenylation for spatially resolved transcriptomic studies. Genomics Proteomics Bioinformatics 21, 601–618 (2023).
Olivieri, J. & Salzman, J. Analysis of RNA processing directly from spatial transcriptomics data reveals previously unknown regulation. Preprint at bioRxiv https://doi.org/10.1101/2023.03.13.532412 (2023).
Fu, Y. et al. Single cell and spatial alternative splicing analysis with Nanopore long read sequencing. Nat. Commun. 16, 6654 (2025).
Buen Abad Najar, C. F., Yosef, N. & Lareau, L. F. Coverage-dependent bias creates the appearance of binary splicing in single cells. eLife 9, e54603 (2020).
Isaev, K. & Knowles, D. A. Investigating RNA splicing as a source of cellular diversity using a binomial mixture model. In Proc. 18th Machine Learning in Computational Biology (ed. Knowles, D. A & Mostafavi, S.) 163–175 (PMLR, 2024).
Rich, J. M. et al. The impact of package selection and versioning on single-cell RNA-seq analysis. Preprint at bioRxiv https://doi.org/10.1101/2024.04.04.588111 (2024).
Edsgard, D., Johnsson, P. & Sandberg, R. Identification of spatial expression trends in single-cell gene expression data. Nat. Methods 15, 339–342 (2018).
Svensson, V., Teichmann, S. A. & Stegle, O. SpatialDE: identification of spatially variable genes. Nat. Methods 15, 343–346 (2018).
Sun, S., Zhu, J. & Zhou, X. Statistical analysis of spatial expression patterns for spatially resolved transcriptomic studies. Nat. Methods 17, 193–200 (2020).
Zhu, J., Sun, S. & Zhou, X. SPARK-X: non-parametric modeling enables scalable and robust detection of spatial expression patterns for large spatial transcriptomic studies. Genome Biol. 22, 184 (2021).
Dries, R. et al. Giotto: a toolbox for integrative analysis and visualization of spatial expression data. Genome Biol. 22, 78 (2021).
Chen, C., Kim, H. J. & Yang, P. Evaluating spatially variable gene detection methods for spatial transcriptomics data. Genome Biol. 25, 18 (2024).
Gretton, A. et al. A kernel statistical test of independence. Adv. Neur. Inform. Proc. Syst. 20, 585–592 (2007).
Zhang, K., Peters, J., Janzing, D. & Schölkopf, B. Kernel-based conditional independence test and application in causal discovery. In Proc. 27th Conference on Uncertainty in Artificial Intelligence 804–813 (AUAI, 2011).
Su, J. et al. Smoother: a unified and modular framework for incorporating structural dependency in spatial omics data. Genome Biol. 24, 291 (2023).
Booeshaghi, A. S. et al. Isoform cell-type specificity in the mouse primary motor cortex. Nature 598, 195–199 (2021).
Joglekar, A. et al. Single-cell long-read sequencing-based mapping reveals specialized splicing patterns in developing and adult mouse and human brain. Nat. Neurosci. 27, 1051–1063 (2024).
Patowary, A. et al. Developmental isoform diversity in the human neocortex informs neuropsychiatric risk mechanisms. Science 384, eadh7688 (2024).
Olivieri, J. E. et al. RNA splicing programs define tissue compartments and cell types at single-cell resolution. eLife 10, e70692 (2021).
Zhao, W. et al. POSTAR3: an updated platform for exploring post-transcriptional regulation coordinated by RNA-binding proteins. Nucleic Acids Res. 50, D287–D294 (2022).
Wheeler, E. C. et al. Integrative RNA-omics discovers GNAS alternative splicing as a phenotypic driver of splicing factor–mutant neoplasms. Cancer Discov. 12, 836–855 (2022).
Miura, P., Shenker, S., Andreu-Agullo, C., Westholm, J. O. & Lai, E. C. Widespread and extensive lengthening of 3′ UTRs in the mammalian brain. Genome Res. 23, 812–825 (2013).
Ogorodnikov, A. et al. Transcriptome 3′end organization by PCF11 links alternative polyadenylation to formation and neuronal differentiation of neuroblastoma. Nat. Commun. 9, 5331 (2018).
Adult mouse brain coronal section (fresh frozen) obtained from BioIVT Asterand, spatial gene expression dataset analyzed using Space Ranger 2.1.0 (10X Genomics, accessed 7 November 2024); https://www.10xgenomics.com/datasets/adult-mouse-brain-coronal-section-fresh-frozen-1-standard
Alfonso-Gonzalez, C. et al. Sites of transcription initiation drive mRNA isoform selection. Cell 186, 2438–2455.e22 (2023).
Mouse brain coronal section (fresh frozen) from healthy 9-week-old C57BL/6 mouse, In situ gene expression dataset analyzed using Xenium Onboard Analysis 3.0.0 (10X Genomics, accessed 30 July 2025); https://www.10xgenomics.com/datasets/xenium-prime-fresh-frozen-mouse-brain
Hwang, H. W. et al. cTag-PAPERCLIP reveals alternative polyadenylation promotes cell-type specific protein diversity and shifts Araf isoforms with microglia activation. Neuron 95, 1334–1349 e5 (2017).
Fisher, E. & Feng, J. RNA splicing regulators play critical roles in neurogenesis. Wiley Interdiscip. Rev. RNA 13, e1728 (2022).
Jacko, M. et al. RBFox splicing factors promote neuronal maturation and axon initial segment assembly. Neuron 97, 853–868 e6 (2018).
Rehfeld, F. et al. The RNA-binding protein ARPP21 controls dendritic branching by functionally opposing the miRNA it hosts. Nat. Commun. 9, 1235 (2018).
Chatrikhi, R. et al. RNA binding protein CELF2 regulates signal-induced alternative polyadenylation by competing with enhancers of the polyadenylation machinery. Cell Rep. 28, 2795–2806 e3 (2019).
Van Nostrand, E. L. et al. A large-scale binding and functional map of human RNA-binding proteins. Nature 583, 711–719 (2020).
Gazzara, M. R. et al. Ancient antagonism between CELF and RBFOX families tunes mRNA splicing outcomes. Genome Res. 27, 1360–1370 (2017).
Berto, S., Usui, N., Konopka, G. & Fogel, B. L. ELAVL2-regulated transcriptional and splicing networks in human neurons link neurodevelopment and autism. Hum. Mol. Genet. 25, 2451–2464 (2016).
McGurk, M. P., McWatters, D. C. & Burge, C. B. KATMAP: Inferring splicing factor activity and regulatory targets from knockdown data. Nat. Biotechnol. https://doi.org/10.1038/s41587-025-02881-9 (2025).
Maynard, K. R. et al. Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. Nat. Neurosci. 24, 425–436 (2021).
Ito, H. et al. Sept8 controls the binding of vesicle-associated membrane protein 2 to synaptophysin. J. Neurochem. 108, 867–880 (2009).
Yuan, Y. et al. Cell type-specific CLIP reveals that NOVA regulates cytoskeleton interactions in motoneurons. Genome Biol. 19, 117 (2018).
Greenwald, A. C. et al. Integrative spatial analysis reveals a multi-layered organization of glioblastoma. Cell 187, 2485–2501 e26 (2024).
Ren, Y. et al. Spatial transcriptomics reveals niche-specific enrichment and vulnerabilities of radial glial stem-like cells in malignant gliomas. Nat. Commun. 14, 1028 (2023).
Vandiedonck, C. et al. Pervasive haplotypic variation in the spliceo-transcriptome of the human major histocompatibility complex. Genome Res. 21, 1042–1054 (2011).
Puttick, C. et al. MHC Hammer reveals genetic and non-genetic HLA disruption in cancer evolution. Nat. Genet. 56, 2121–2131 (2024).
Kahles, A. et al. Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell 34, 211–224.e6 (2018).
Song, X. et al. A single-cell atlas of RNA alternative splicing in the glioma-immune ecosystem. Genome Biol. 26, 426 (2025).
Stassen, O. et al. GFAPdelta/GFAPalpha ratio directs astrocytoma gene expression towards a more malignant profile. Oncotarget 8, 88104–88121 (2017).
Uceda-Castro, R. et al. GFAP splice variants fine-tune glioma cell invasion and tumour dynamics by modulating migration persistence. Sci. Rep. 12, 424 (2022).
Larionova, T. D. et al. Alternative RNA splicing modulates ribosomal composition and determines the spatial phenotype of glioblastoma cells. Nat. Cell Biol. 24, 1541–1557 (2022).
Fuhrmann, D. C., Mondorf, A., Beifuß, J., Jung, M. & Brüne, B. Hypoxia inhibits ferritinophagy, increases mitochondrial ferritin, and protects from ferroptosis. Redox Biol. 36, 101670 (2020).
Iijima, T., Hidaka, C. & Iijima, Y. Spatio-temporal regulations and functions of neuronal alternative RNA splicing in developing and adult brains. Neurosci. Res. 109, 1–8 (2016).
Chang, Y. et al. Graph Fourier transform for spatial omics representation and analyses of complex organs. Nat. Commun. 15, 7467 (2024).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Machine Learning Res. 12, 2825–2830 (2011).
Liu, H., Tang, Y. & Zhang, H. H. A new chi-square approximation to the distribution of non-negative definite quadratic forms in non-central normal variables. Comput. Stat. Data Anal. 53, 853–856 (2009).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995).
Liao, J. Y. et al. EuRBPDB: a comprehensive resource for annotation, functional and oncological investigation of eukaryotic RNA binding proteins (RBPs). Nucleic Acids Res. 48, D307–D313 (2020).
Ray, D. et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499, 172–177 (2013).
Tremblay, B. J.-M. universalmotif: an R package for biological motif analysis. J. Open Source Softw. 9, 7012 (2024).
Bailey, T. L., Johnson, J., Grant, C. E. & Noble, W. S. The MEME suite. Nucleic Acids Res. 43, W39–W49 (2015).
Luo, Y. et al. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res. 48, D882–D889 (2020).
Kowalski, M. H. et al. Multiplexed single-cell characterization of alternative polyadenylation regulators. Cell 187, 4408–4425 e23 (2024).
Trincado, J. L. et al. SUPPA2: fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol. 19, 40 (2018).
Gohr, A. & Irimia, M. Matt: Unix tools for alternative splicing analysis. Bioinformatics 35, 130–132 (2019).
Patrick, R. et al. Sierra: discovery of differential transcript usage from polyA-captured single-cell RNA-seq data. Genome Biol. 21, 167 (2020).
Tasic, B. et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78 (2018).
Moakley, D. F. et al. Reverse engineering neuron-type-specific and type-orthogonal splicing-regulatory networks using diverse cellular transcriptomes. Cell Rep. 44, 115898 (2025).
Song, J. et al. DEMINERS enables clinical metagenomics and comparative transcriptomic analysis by increasing throughput and accuracy of nanopore direct RNA sequencing. Genome Biol. 26, 76 (2025).
Su, J. Raw and processed data for the paper ‘A computational framework for mapping isoform landscape and regulatory mechanisms from spatial transcriptomics data’. Zenodo https://doi.org/10.5281/zenodo.17625643 (2025).
Su, J. Collection of processed isoform-level spatial transcriptomics datasets of mouse and human brain. Zenodo https://doi.org/10.5281/zenodo.16905935 (2025).
Acknowledgements
We thank C. Zhang for input on the splicing regulatory analysis and D. Moakley for assistance on the preprocessing of single-cell exon splicing data. We thank J. Song for access to processed 10X Visium-ONT glioblastoma data and transcript annotation. We thank J. H. Fung for feedback on the theoretical analysis. We thank S. Vickovic, D. McKellar and K. Isaev for helpful discussions. This work was funded by the National Institutes of Health, National Cancer Institute (grant nos. R35CA253126, U01CA243073, P01CA285250 to R.R.), the Edward P. Evans Center for MDS at Columbia University (to J.S. and R.R.) and the National Science Foundation (grant no. CAREER DBI2146398 to D.A.K.). Any opinions, findings and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the NSF or the NIH.
Author information
Authors and Affiliations
Contributions
J.S. conceived the project, designed and implemented SPLISOSM, conducted the theoretical analysis, benchmark experiments and applications on spatial transcript diversity in mouse and human samples. Y.Q. contributed to the parallelization and benchmark of the parametric models. J.S., M.S. and S.P. designed and conducted experimental validation. H.Y. applied SPLISOSM to the Slide-seqV2 hippocampus data. J.J. processed the SR glioblastoma samples. J.S. and T.L. developed the website. Q.L. contributed to the analysis of 3′ end variability in neurodegenerative diseases. T.M.N., X.F. and S.-w.C. contributed to the analysis and interpretation of transcript diversity in glioblastoma samples. D.A.K. and R.R. supervised the project. J.S. took lead in writing the article with input from all authors. All authors read and approved the final paper.
Corresponding authors
Ethics declarations
Competing interests
R.R. is a founder of Genotwin and a member of the Scientific Advisory Board of DiaTech and Flahy. None of these activities are related to the work described in this paper. The other authors declare no competing interests.
Peer review
Peer review information
Nature Biotechnology thanks Kevin Lebrigand and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Spatially variable TREND events and their regulation in adult mouse brain, related to Fig. 4.
(a) HSIC-IR test agreements between Visium CBS (y-axis) and Slide-seqV2 hippocampus (x-axis) datasets. Points colored by expression perplexity. (b) Differential protein-altering alternative polyadenylation events between neurons and glial cells. Related to Fig. 4e. (c) Additional protein-altering TREND events and their spatial log-normalized expression. (d) Top RBPs ranked by number of significantly associated targets, color-coded by available motifs (CISBP-RNA) or CLIP data (POSTAR3), core APA machinery involvement and motif enrichment near SVP TREND events. Spatial log-normalized expression of selected RBPs shown at right. (e) PCBP2 transcript structure in mouse and human, along with eCLIP data from ENCODE. (f) PCBP2 isoform expression in spatial mouse CBS data (top) and across GTEx human tissues (bottom). In each scatter plot, Pearson’s R correlation coefficient and the associated p-value are displayed along with the fitted linear model and its 95% confidence interval.
Extended Data Fig. 2 Comparison of spatial transcript diversity detection in adult mouse brain across ST platforms.
(a) Spatially variable exon overlaps between ONT and short-read (SR) detected variable events. Exons were decomposed from isoforms of 150 ONT-SVP genes (Fig. 3) and TREND events from 815 SR-SVP genes (Fig. 4). (b) Spatially variably processed (SVP) gene overlap between ONT and SR mouse CBS samples. Events were identified using the following HSIC-IR p-value thresholds: (ONT-SVP) p-adj \(\le\) 0.05; (SR-SVP) p-adj \(\le\) 0.01. 150 genes shared by both ONT replicates are considered as ONT-SVP genes. (c) Gene-level SVP test agreements between ONT and SR. Left: One-sided chi-square test comparing SVP calling results. Right: SR-based HSIC-IR p-value distributions. Boxplots show median (center line), interquartile range (box), and 1.5× interquartile range (whiskers), with p-values from two-sided Wilcoxon rank-sum test. Negative background sample sizes vary as not all genes detected/tested in both platforms. (d) Left: Exon overlaps across platforms. Right: Xenium-based HSIC-IR p-value distributions in adult mouse brain. Boxplots show median (center line), interquartile range (box), and 1.5× interquartile range (whiskers). P-values from two-sided Wilcoxon rank-sum test. (e) Assessment of Xenium technical artifacts. Left: HSIC-IR adjusted p-values for non-gene features. Boxplots show median (center line), interquartile range (box), and 1.5× interquartile range (whiskers). Group distributions were compared using one-sided Kruskal-Wallis test. Y-axis was truncated (p-adj \(\ge\) 1e-40) for visualization purpose. Right: Spatial transcript density of selected genes. NegControlProbe_00041 variable usage is driven by codeword 5642, which shows a similar glial-cell-specific pattern as Bmp2-1345 and Trp53bp1-17048. (f) Scatter plots showing HSIC-IR adjusted p-values for genes detected in both Xenium Prime 5K and either Visium-ONT (left, Spearman’s ρ = 0.19) or Visium-SR (right, Spearman’s ρ = 0.15). Red dots indicate genes identified as SVP in the Visium platforms as in (g). Dashed red lines mark Xenium significance thresholds (p-adj = 2.5e-17). (g) Overlap of SVP genes across platforms. Events were identified using the following HSIC-IR p-value thresholds: (ONT-SVP) p-adj \(\le\) 0.05 in both replicates; (SR-SVP) p-adj \(\le\) 0.01; (Xenium-SVP) p-adj ≤ 2.5e-17.
Extended Data Fig. 3 In situ validation of spatially variable isoform usage of Dtnbp1, related to Extended Data Fig. 2.
(a) Spatial distribution of log-normalized expression for Dtnbp1 isoforms in SiT-ONT CBS2 sample. (b) Gene structure of Dtnbp1 with annotated isoforms and Xenium Prime 5K probe sets. Among three ONT-detected Dtnbp1 isoforms (201, 204, 206), codeword 7864 specifically targets 201 and 204, while codeword 11516 recognizes all three. (c) Spatial density of Dtnbp1 codewords (left) and Cluster 2 cells (Slc17a7+ excitatory neurons, right) in Xenium Prime 5K mouse CBS sample. Cell segmentation, clustering and differential expression analysis were performed using Xenium Ranger with default parameters. (d) Dtnbp1-206 spatial variability driven by a subset of Nptxr+ excitatory neurons. Left: Volcano plot of Cluster 2 marker genes. P-values were computed from a negative binomial test and were adjusted via the Benjamini-Hochberg procedure as part of the Xenium Onboard Analysis pipeline. Middle: Comparison of Dtnbp1-11516-only bins (20 × 20 μm) with and without Cluster 2 cells. Right: Volcano plot of marker codewords of Dtnbp1-11516-only Cluster 2 bins.
Extended Data Fig. 4 Examples of Xenium-only spatially variably processed genes. Related to Extended Data Fig. 2.
a, Spatial density of Axin2 codewords in Xenium Prime 5K mouse CBS sample. b, Spatial density of Ryr1 codewords in the same sample.
Extended Data Fig. 5 RBFOX regulates neural transcript usage with other splicing factors cooperatively.
(a) Clta transcript structure and spatial log-normalized expression in the ONT-CBS2 sample. (b) Relationship between normalized Rbfox3 expression and Clta isoform ratio (left) or cell-type-aggregated exon PSI values (right). Line and shading indicate fitted linear model and 95% confidence interval. Spatial data was locally smoothed before calculating isoform ratios for visualization. (c) Identification of Rbfox3 co-regulators in ONT (left) and SR (right) samples. Heatmaps display pairwise correlations of gene expression (upper triangle) or conditional HSIC test p-values (lower triangle). RBPs with reported RBFOX interaction are highlighted. (d) Motif enrichment in Rbfox1/2/3 associated SVP targets using alternative exons and 150bp flanking introns (left) or TREND regions (right). P-values were calculated using the SEA algorithm as part of the MEME suite. (e) Motif enrichment in exons with ≥25% PSI changes following RBFOX triple knockout in Day 10 motor neurons, categorized as silenced (increased PSI after KD) or enhanced (decreased PSI). Sequences were binned by distance to exon boundaries (x-axis) to show positional enrichment.
Extended Data Fig. 6 Additional examples of evolutionarily conserved SVP events in human DLPFC, related to Fig. 5.
(a) Relationship between total read coverage in TREND regions per spot (x-axis) and proportion of significant spatially variably processed (SVP, top) or variably expressed (SVE, bottom) genes in all tested genes. Line and shading indicate fitted linear model and 95% confidence interval. (b) Number and proportion of human DLPFC SV genes that have spatially variable mouse homologs, grouped by recurrence (number of significant DLPFC samples, x-axis). (c) Sequencing depth of SVP and SVE genes, grouped by conservation and recurrence. Per-group sample size is indicated in (b). Boxplots show median (center line), interquartile range (box), and 1.5× interquartile range (whiskers). Group means are compared using two-sided T-test. (d) Pathway enrichment analysis comparing human SVP versus human SVENP genes. Related to Fig. 5f. (e) Spatial log-normalized expression of selected RBPs in sample 151673. (f) RBP-SVP associations ranked by recurrence (x-axis) and minimal GLMM p-values across samples (y-axis). Top potential regulators of MAP4 are highlighted and colored by whether the association is conserved in mouse. (g) MAP4 transcript structure and spatial log-normalized expression in human sample 151673.
Extended Data Fig. 7 Additional examples of transcript diversity in human glioma, related to Fig. 6.
(a) Distribution of recurrent SVP genes (left) and relationships between total isoform read coverage per spot (x-axis) and number of SVP and SVE genes (y-axis) across ONT datasets. Line and shading indicate fitted linear model and 95% confidence interval. (b) Distribution of recurrent SVP genes (left) and relationships between total TREND read coverage per spot (x-axis) and number of SVP and SVE genes (y-axis) across SR datasets. Line and shading indicate fitted linear model and 95% confidence interval. (c) IG gene isoform diversity in the ONT sample DMG2. (d) Distributions of per-sample alternative splicing types for ONT-SV genes (n-sample=11) and per-sample TREND annotations for SR-SV genes (n=13). Boxplots show median (center line), interquartile range (box), and 1.5× interquartile range (whiskers). Group means are compared using two-sided T-test. (e) Pathway enrichment analysis comparing recurrent SVP (adjusted HSIC-IR p-value < 0.05 in ≥2 samples) genes versus recurrent SVENP (non-SVP and adjusted HSIC-GC p-value < 0.05 in ≥4 samples) genes. Related to Fig. 6d. (f-h) SVP (HSIC-IR adjusted p-value < 0.05 in at least one sample) genes involved in selected KEGG pathways. Orange indicates disease-specific genes not variable in healthy DLPFC (undetected or HSIC-IR adjusted p-value ≥ 0.05 in all DLPFC samples). (i) GFAP transcript structure in the ONT sample DMG2, read coverage in the short-read sample ZH916bulk, and the respective isoform or TREND spatial distribution in each sample (right).
Extended Data Fig. 8 Comparison of common ST platforms for isoform variability detection.
Each platform presents unique technical advantages and limitations that impact false positives and detection power.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11 and Notes.
Supplementary Table 1
Sample metadata of datasets analyzed in the study.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Su, J., Qu, Y., Schertzer, M. et al. Mapping isoforms and regulatory mechanisms from spatial transcriptomics data with SPLISOSM. Nat Biotechnol (2026). https://doi.org/10.1038/s41587-025-02965-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41587-025-02965-6