Abstract
The human brain undergoes protracted postnatal maturation, guided by dynamic changes in gene expression. Most studies exploring these processes have used bulk tissue analyses, which mask cell-type-specific gene expression dynamics. Here, using single-nucleus RNA sequencing on temporal lobe tissue, including samples of African ancestry, we build a joint pediatric and adult atlas of 75 cell subtypes, which we verify with spatial transcriptomics. We explore the differences between pediatric and adult cell subtypes, revealing the genes and pathways that change during brain maturation. Our results highlight excitatory neuron subtypes, including the LTK and FREM subtypes, that show elevated expression of genes associated with cognition and synaptic plasticity in pediatric tissue. The resources we present here improve our understanding of the brain during its development and contribute to global efforts to build an inclusive brain cell map.
Similar content being viewed by others
Main
The adult human brain is a complex assembly of diverse cell types that has been defined with unprecedented accuracy using single-cell transcriptomics1,2,3,4. This adult transcriptomic signature is set up over a protracted period of development, which begins in the embryo and continues after birth. Although the single-cell diversity of the embryonic human brain has been explored5,6, little is known about how these cell-type-specific gene expression profiles change during childhood7. Most existing studies have used bulk transcriptomic approaches; these have revealed a dramatic period of global gene expression change during the late fetal to early infancy transition, which stabilizes during childhood (1 to <12 years of age) and adolescence (12 to <20 years of age)6,8,9,10,11. Bulk transcriptomics, however, cannot identify the more subtle, cell-type-specific changes in gene expression that drive brain maturation from childhood through adolescence to adulthood.
Childhood and adolescence are periods of important changes in brain structure, during which neuronal connections are refined and strengthened. Although synaptogenesis peaks in the early postnatal period, synaptic pruning activity begins during late childhood, peaks during adolescence and then gradually decreases12,13,14. These stages therefore represent periods of enhanced susceptibility to environmental influence, as well as increased neuropsychiatric risk15. Describing the typical cell-type-specific gene expression trajectories of the maturing brain will allow us to assess the effects of genetic perturbations and early adverse experiences on brain maturation. Furthermore, investigating the driving forces behind cell-type-specific maturation processes may help with the development of targeted therapies for neurological disease16.
To this end, the Pediatric Cell Atlas17, a branch of the Human Cell Atlas (HCA)3, aims to ensure that the benefits of single-cell transcriptomics are available to children as well as adults from diverse populations3,17. Africa has the most genetically diverse18 and youngest population19 worldwide. By 2050, 37% of the world’s children will grow up in Africa20. Consequently, it is essential to include the African pediatric population in the Pediatric Cell Atlas. A reference pediatric brain cell atlas that includes data from African donors will contribute to developing treatments for locally prevalent conditions, such as tuberculous meningitis (TBM) and human immunodeficiency virus21,22. In addition, studying the differences in gene expression dynamics between adult and pediatric brains may explain why manifestations of neurological conditions and responses to therapies differ across the lifespan17.
To contribute to these endeavors, we present a joint pediatric and adult temporal cortex cell atlas, including samples from eight southern African donors, annotated using the Allen Brain Map middle temporal gyrus (MTG) cell taxonomy1. We validate our annotation using spatial transcriptomics. We use de novo marker gene analysis with machine learning tools to compare our pediatric and adult datasets with the existing MTG cell taxonomy and compare markers that define pediatric versus adult cell states. Using differential gene expression analysis, we highlight 21 cell subtypes that show differential expression of genes involved in neurodevelopment and cognition. Finally, we use our datasets to define the cell-type-specific gene expression of putative site-of-disease TBM biomarkers23. Overall, we highlight subtle cell-type-specific differences between the pediatric and adult brain and expand the representation of diverse pediatric populations in the HCA.
Results
A joint pediatric and adult temporal cortex cell atlas
We generated single-nucleus RNA sequencing (snRNA-seq) libraries from five pediatric and three adult donor temporal cortex tissue samples. The majority of our samples were obtained from surgeries to treat epilepsy (Supplementary Data 1). These libraries were analyzed alongside similar published datasets24, resulting in a total of 23 snRNA-seq datasets (including technical replicates) from 12 individuals (six pediatric and six adult) (Fig. 1a). The samples were sequenced to a median depth of 19,853 reads per nucleus, with 176,012 nuclei remaining after removal of low-quality barcodes (Extended Data Fig. 1 and Supplementary Data 2). Although our new datasets had lower average sequencing depth than the coanalyzed published datasets, the average number of genes and transcripts detected across datasets was similar (Supplementary Data 2).
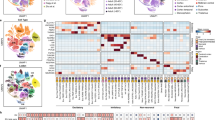
a, Data integration showing alignment of nuclei across the technical (T) and biological (B) replicates from donors ranging in age from 4 to 50 years. b, UMAP plot annotated to show the 75 cell types from the Allen Brain Map MTG atlas after filtering to retain nuclei with high-confidence annotations. Each cell type is annotated with (1) a major cell class (for example, Exc for excitatory neurons); (2) the cortical layer with which the cell is associated (for example, L2 for layer 2); (3) a subclass marker gene; and (4) a cluster-specific marker gene. The color scheme for the cell types is in accordance with the MTG cell taxonomy. c, Stacked bar plot showing the proportion of nuclei per cell type for each age category of the total number of nuclei for each group. Cell types are colored as in b. d, Validation of the high-resolution cell type annotations, showing a high degree of correspondence in the expression of known cell-type-specific marker genes (x axis) with their expected cell type (y axis) (left). The number of nuclei per cell type is shown on the right. e, Correlation plot showing cosine similarity scores assessing similarity between the annotated cell types in our dataset (y axis as in d) and the MTG reference dataset (x axis) based on the log-normalized expression counts of the top 2,000 shared highly variable features between query and reference datasets.
Using data integration and clustering, we aligned similar cell types across the 23 datasets, yielding 40 clusters (Fig. 1a and Extended Data Fig. 1h,i). Each cluster was assigned to one of the major brain cell types (level 1 annotation) based on marker gene expression (Extended Data Fig. 2a, Supplementary Data 3 and Supplementary Fig. 1). In addition, we used label transfer25 to classify each nucleus according to the Allen Brain Map MTG atlas1 (level 2 annotation) (Supplementary Data 3). Barcodes with discordant level 1 and level 2 annotations (17.94%) were removed, focusing downstream analyses on nuclei with high-confidence annotations (Supplementary Data 3). Based on marker gene analysis1 (Extended Data Fig. 2b), many of these filtered barcodes are likely to be multiplets or nuclei contaminated with ambient messenger RNA (mRNA).
All 75 reference cell types were present in the final filtered dataset of 144,438 nuclei (Fig. 1b and Supplementary Data 3) and expressed the expected marker genes1 (Fig. 1d). Both neuronal and nonneuronal cell types showed high correlation with the corresponding reference cell types1 (cosine similarity score >0.83) and lower correlation with other subtypes within their class (Fig. 1e). This pattern was maintained when considering either the pediatric or adult datasets on their own, with the majority of pediatric cell types showing only slightly lower similarity scores than the adults (Supplementary Data 3), probably owing to the reference dataset only containing adult data. The cell composition of the samples was very similar, with no significant differences in cell type proportions between pediatric and adult samples or between biological sexes (Fig. 1c, Extended Data Fig. 2c,d and Supplementary Data 3). As in the reference atlas1, oligodendrocytes were the most common nonneuronal cell type, and Exc_L2-3_LINC00507_FREM3 was the most common neuronal subtype. Neuronal clusters had a greater number of expressed genes and unique molecular identifiers (UMIs) compared with nonneuronal cells (Extended Data Fig. 3a), whereas excitatory neurons had a greater number of genes detected per nucleus than inhibitory neurons (Supplementary Data 3). When comparing the pediatric with adult cell types, there were no significant differences in the number of genes or UMIs between the age categories (Extended Data Fig. 3b,c). Overall, the quality and composition of the pediatric and adult cell atlases were very similar.
Spatial mapping of temporal cortex cytoarchitecture
Next, we used spatial transcriptomics to explore the positions of our annotated cell types within the temporal cortex. We generated Visium datasets from adult (31-year-old) and pediatric (15-year-old) temporal cortex samples (two sections each; Supplementary Data 1 and Extended Data Fig. 4). The four Visium libraries were sequenced to a median depth of 87,178 reads per spot (median of 5,878 UMIs and 2,745 genes per spot) (Supplementary Data 4).
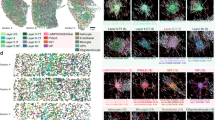
Using cell2location26, we calculated cell type abundance estimates for each Visium spot, with our annotated snRNA-seq dataset as a reference. Oligodendrocytes were the most common cell type, whereas Exc_L2_LAMP5_LTK was the most abundant neuronal cell type (Extended Data Fig. 5a). The annotated cell types mapped to their expected cortical layer locations across all tissue sections (Fig. 2a and Extended Data Fig. 5b), matching the spatial expression of known cortical layer marker genes1,27,28 (Fig. 2b). These layered expression patterns were verified for a subset of layer-specific marker genes using in situ hybridization (Extended Data Fig. 6).
a, Estimated cell type abundances (color intensity) in the 31-year-old and 15-year-old temporal cortex tissue sections for a selection of cell types including nonneuronal cells, excitatory neurons (top) and inhibitory neurons (bottom). b, Visium gene expression profiles (color intensity) for a selection of known cortical layer marker genes in the 31-year-old and 15-year-old temporal cortex tissue sections including AQP4 (layer 1), LAMP5 (layer 2), RORB (layer 4) and CLSTN2 (layers 5 and 6). c,d, Identification of colocated cell types using NMF. The dot plot (c) shows the NMF weights of the cell types (rows) across each of the NMF factors (columns), which correspond to tissue compartments. Block boxes indicate cell types that are colocated within the indicated compartments. Spatial plots (d) show NMF weights for selected NMF factors across the 31-year-old and 15-year-old temporal cortex tissue sections. Panels are displayed in the same order as the dot plot in c, with the dominant cell types for each factor indicated in parentheses. Dashed white lines and numbers indicate estimated cortical layer boundaries as indicated in the first two panels of b and d. WM, white matter. See also Extended Data Figs. 4–6.
To examine the colocation of cell types within the layered structure of the temporal cortex, nonnegative matrix factorization (NMF) was performed; this resulted in 15 cellular compartments, which were visualized across the Visium samples, revealing their spatial distribution (Fig. 2c,d and Extended Data Fig. 5c). In both the pediatric and adult datasets, there was clear colocation of the expected neuronal cell types within overlapping compartments across the cortical layers. Layer 2 was dominated by Exc_L2_LAMP5_LTK (factor 11) and Exc_L2-3_LINC00507_FREM (factor 5), layer 3 by Exc_L3-4_RORB_CARM1P1 (factor 13), layer 4 by the RORB excitatory neuron subtypes (factor 12), layer 5 by the THEMIS excitatory neuron subtypes (factor 10) and layer 6 by the FEZF2 excitatory neuron subtypes (factor 14 and factor 1), with the latter extending into the white matter (Fig. 2c,d). Inhibitory neurons were primarily associated with factors 6 and 2, which were more widely spread across the layers (Fig. 2c,d). Notably, these factors were more strongly associated with layers 5 and 6 in the adult compared with the pediatric samples. The two astrocyte subtypes were confirmed to have distinct distribution profiles, with Astro_L1-2_FGFR3_GFAP (factor_4) located primarily in layer 1 (Fig. 2c,d) and the white matter, and Astro_L1-6_FGFR3_SLC14A1 (factor_9) more widely distributed (Extended Data Fig. 5c). The remaining nonneuronal cell types were largely associated with factors located in layer 1 and the white matter (Extended Data Fig. 5c).
Overall, our spatial transcriptomic analyses provide support for our annotation approach, showing the expected spatial distribution of annotated cell types and revealing a similar tissue cytoarchitecture in adult and pediatric temporal cortex tissues.
Identification of temporal cortex cell type markers
To establish a standardized approach for defining cell types, use of the minimum combination of gene markers that can classify a cell type and distinguish it from other cell types has been proposed 29,30. Toward achieving this, Aevermann et al.29 developed a machine learning tool, NS-Forest v.2.0, which they applied to the MTG atlas. Ideally, these MTG minimal markers would be conserved in similar datasets to facilitate accurate comparisons across studies31. Indeed, we found that the majority of MTG atlas minimal markers29 (~94%) were expressed at significantly higher levels in the expected cell types than in other cell types (Extended Data Fig. 7 and Supplementary Data 5).
Application of the NS-Forest v.2.0 (ref. 29) algorithm to our downsampled snRNA-seq datasets (Methods) revealed 202 pediatric and 196 adult minimal marker genes (Fig. 3 and Supplementary Data 5). The median F-beta score per cell type (a measure of the discriminative power of a given combination of marker genes; pediatric: 0.55; adult: 0.6) and the average binary expression score (a measure of an individual gene’s classification power; pediatric: 0.9; adult: 0.89) were comparable across age groups and only slightly lower than those obtained for the MTG atlas (0.68 and 0.94, respectively)29. Forty-seven pediatric (23.3%) and 45 adult (23.0%) minimal markers overlapped with existing markers29 (Fig. 3 and Supplementary Data 5). However, there was a greater overlap in minimal markers between the pediatric and adult datasets, with 68 markers (~34%) present in both lists. MERFISH32 spatial transcriptomic analysis of a subset of minimal markers that were shared between pediatric and adult datasets confirmed their coexpression with previously described minimal markers29 in adult (31-year-old) and pediatric (15-year-old) temporal cortex samples (Extended Data Fig. 8).
a,b, Heatmap showing the scaled average normalized expression counts of the NS-Forest minimal marker genes (y axis) identified for 75 cortical cell types (x axis) across the six adult (a) and six pediatric (b) datasets. As input into NS-Forest, the nuclei of each sample were randomly downsampled to the size of the sample with the fewest nuclei. Heatmaps show gene expression values for the downsampled datasets. The minimal marker genes are annotated (color codes on the y axes) according to whether they are unique to a given cell type, whether they are coding or noncoding genes, whether they are unique to the indicated age group, whether they overlap with existing MTG minimal marker gene sets for the same cell type, and according to the cell type they define.
Our minimal marker analysis revealed improved markers for some cell types compared with the reference MTG atlas. In our datasets, long noncoding RNA LINC01331 was a minimal marker for Exc_L2-3_LINC00507_FREM3, with a beta score of 1, indicating high specificity. By contrast, one of the existing markers for this cell type, PALMD, was more highly expressed in endothelial cells in our datasets (Fig. 3 and Extended Data Fig. 9a,b). This discrepancy is probably due to the lower percentage of endothelial cells in the MTG atlas compared with our datasets (0.06% versus 0.9%)1. Similarly, one of the existing MTG atlas markers for Exc_L5-6 _THEMIS_CRABP1, OLFML2B, was more highly expressed in other layer 5 and 6 neurons in our dataset, whereas our minimal marker, POSTN, showed greater specificity (Fig. 3 and Extended Data Fig. 9c,d). In addition, Uniform Manifold Approximation and Projection (UMAP) analysis of our annotated datasets using our minimal marker gene list for each age group, in comparison with an equivalent number of random genes, resulted in better grouping of the cell subtypes into clusters, similar to the original UMAP plot (compare Fig. 1a and Fig. 4a,b). This analysis reveals that our shortlists of ~200 marker genes capture much of the underlying transcriptomic diversity in our datasets.
a,b, Annotated UMAP plots following data integration using either the minimal marker genes (left) or the equivalent number of a random set of genes (right) as anchors for the adult (a) and pediatric (b) datasets. The color scheme for the cell types is in accordance with the MTG cell taxonomy. c, Overlap of the pediatric and adult NS-Forest markers with high binary expression score (>0.7) per cell type. The bar plot shows the number of shared markers between pediatric and adult datasets (blue), the number of markers unique to the pediatric datasets (orange) and the number of markers unique to the adult datasets (gray) for each cell type.
Gene ontology (GO) analysis of our minimal marker gene lists revealed significant enrichment of GO terms related to development, cell signaling, extracellular matrix and synapse organization, when considering the pediatric and adult datasets individually or together (Supplementary Data 6). These results suggest that genes involved in neuronal development and signaling are key to neuronal identity as the brain matures and in adult life. To further assess the difference in cell type markers between our pediatric and adult datasets, we expanded our analysis to include all genes with a high NS-Forest binary expression score (>0.7)29. For most cell types, most of these top markers (>18 genes) were shared between our pediatric and adult datasets (Fig. 4c and Supplementary Data 7 and 8). Oligodendrocytes showed the highest number of shared marker genes (53) and the second highest number of pediatric-specific markers (22). Exc_L3-4_RORB_CARM1P1 had the highest number of adult-specific marker genes (30), whereas Exc_L2-4_LINC00507_GLP2R had no shared markers. GO analysis of the shared oligodendrocyte marker genes revealed driver terms related to oligodendrocyte structure and function, including ‘structural constituent of myelin sheath’, whereas the top driver terms for the pediatric-specific markers were ‘oligodendrocyte differentiation’ and ‘myelination’ (Supplementary Data 6). Overall, our expanded marker gene analysis suggests that neuronal cell types show greater dissimilarity between their pediatric and adult states than nonneuronal cells. It is likely that more diversity in the nonneuronal marker gene profiles could be revealed with subdivision into further subtypes.
Enriched developmental gene expression in pediatric samples
To identify genes that were upregulated in the pediatric cell populations and thus might be involved in brain maturation, we conducted cell-type-specific differential gene expression analysis with DESeq2 (ref. 33). We detected 165 significantly differentially expressed genes (DEGs) across 21 cell types (123 upregulated in pediatric samples and 42 downregulated), with some DEGs associated with multiple cell types (Fig. 5a and Supplementary Data 9–13). For all DEGs, the change in expression was accompanied by a corresponding change in the percentage of nuclei expressing the gene (Supplementary Data 10). BayesSpace34 analysis of a subset of DEGs in our Visium datasets confirmed that the genes were expressed at higher levels in the 15-year-old compared with the 31-year-old samples (Supplementary Fig. 2).
a, Twenty-one cell types with significant DEGs, including 12 excitatory and five inhibitory neuron subtypes, both astrocyte subtypes, oligodendrocytes and microglia. b–g, Volcano plots showing log2 fold change (x axis) and −log10 adjusted P values (y axis) for all DESeq2-tested genes in Exc_L3-5_RORB_ESR1 (b), Exc_L2-3_LINC00507_FREM3 (c), Exc_L4-5_RORB_FOLH1B (d), Exc_L2_LAMP5_LTK (e), Astro_L1-6_FGFR3_SLC14A1 (f) and Oligo_L1-6_OPALIN (g). A Wald test statistic was determined for each gene. P values were adjusted for multiple testing using the Benjamini–Hochberg method. Red dots indicate genes that were significantly upregulated in pediatric samples, whereas blue dots indicate genes that were significantly downregulated (adjusted P < 0.05 and abs(log2 fold change) > 10%). Selected genes are labeled. Red labels indicate DEGs shared between neuronal cell types. Magenta labels indicate DEGs not shared between cell types that are discussed in the text. Gray dots indicate nonsignificant genes (adjusted P > 0.05 or abs(log2 fold change) < 10%). h, Dot plot showing the scaled average normalized expression across samples for DEGs shared among Exc_L3-5_RORB_ESR1, Exc_L2-3_LINC00507_FREM3, Exc_L4-5_RORB_FOLH1B, Exc_L2_LAMP5_LTK, Exc_L3-4_RORB_CARM1P1 and Exc_L3-5_RORB_FILIP1L. i, Psupertime gene expression trajectories for selected DEGs in the indicated cell types. The x axis shows the calculated psupertime value for each cell, colored by sample of origin. The black lines are smoothened curves fit by geom_smooth in R package ggplot2. B, biological replicate.
Many of the excitatory neuron subtypes shared DEGs that are developmentally regulated in the mammalian brain (Fig. 5b–e,h). LAMC3, which encodes a subunit of the extracellular matrix protein laminin, was upregulated in three pediatric subtypes (Exc_L3-5_RORB_ESR1, Exc_L2-3_LINC00507_FREM3 and Exc_L4-5_RORB_FOLH1B) (Fig. 5b–e,h). LAMC3 has a role in cortical lamination in mouse35, and its mutations have been implicated in human brain heterotopias and gyration defects36,37. Similarly, S0X11, encoding a transcription factor that plays a part in embryonic and adult neurogenesis in the mouse brain38 and decreases in expression in the cerebral cortex during development39,40, was upregulated in pediatric Exc_L3-5_RORB_ESR1 and Exc_L2-3_LINC00507_FREM3 (Fig. 5b,c,h). FNBP1L (TOCA-1) was upregulated in Exc_L2-3_LINC00507_FREM3 and Exc_L2_LAMP5_LTK (Fig. 5c,e,h). FNBP1L promotes actin polymerization, regulating neurite outgrowth, and declines in expression over the course of brain maturation in the rat41. Two genes, STEAP2, encoding a metalloreductase, and TNF receptor gene TNFRSF25 (DR3), showed higher expression in adult Exc_L3-5_RORB_ESR1 and Exc_L4-5_RORB_FOLH1B subtypes (Fig. 5b,d,h). STEAP2 increases in expression during postnatal hippocampal maturation in mice42. TNFRSF25 is activated postnatally in the mouse brain, where it may play a part in retention of motor control during aging43. These findings indicate that previously reported expression dynamics for these genes in mammalian models are conserved in the human temporal cortex. Importantly, our analysis reveals that these patterns are specific to groups of excitatory neuron subtypes.
The majority of the DEGs were not shared across the cell types. For example, FGF13 (FHF2) and TENM1 were upregulated in pediatric Exc_L3-5_RORB_ESR1 (Fig. 5b). Fgf13 decreases in expression with age in the mouse brain, where it regulates postnatal neurogenesis44 and axonal formation45. TENM1 codes for a member of the teneurin transmembrane protein family that regulates cytoskeletal organization and neurite outgrowth, shaping synaptic connections46,47,48. KCNG1, which encodes a voltage-gated potassium channel (Kv6.1), was upregulated in pediatric Exc_L2-3_LINC00507_FREM3 neurons (Fig. 5c), whereas MYO16 (MYR8), coding for an unconventional myosin protein, was upregulated in the Exc_L2_LAMP5_LTK subtype (Fig. 5e). Both of these genes decrease in expression with age in the mammalian brain49,50.
In line with our minimal marker analyses, fewer genes were differentially expressed in nonneuronal cells (Fig. 5f,g and Supplementary Data 10). In Astro_L1-6_FGFR3_SLC14A1, PIK3R3 was upregulated in pediatric samples, whereas PFKFB2 was downregulated. PIK3R3 is involved in the PI3K–AKT growth signaling pathway, which has been implicated in brain growth disorders51. PFKFB2 encodes a bifunctional kinase/phosphatase that controls glycolysis. In contrast to our findings, PFKFB2 expression is higher in juvenile rat hippocampal astrocytes than in adults, where it may support energy demands during learning52. In oligodendrocytes, NOTCH2 and RRAS2 were both upregulated in pediatric samples. Notch2 expression decreases in the rat cortex with age53 and has been proposed to regulate glial differentiation54. These results provide additional molecular candidates to expand our understanding of the mechanisms of astrocyte and oligodendrocyte maturation.
To explore the trajectories of DEG expression, we employed psupertime pseudotime trajectory analysis55, focusing on the four excitatory neuron subtypes with the highest numbers of DEGs. In support of our DESeq2 findings, several of the identified DEGs had nonzero psupertime coefficients and therefore represent genes that are relevant to the ordering of the cells in pseudotime55 (Fig. 5i; Exc_L3-5_RORB_ESR1: 13/47 [28%], Exc_L2-3_LINC00507_FREM3: 16/38 [42%], Exc_L4-5_RORB_FOLH1B 3/27 [11%] and Exc_L2_LAMP5_LTK: 5/18 [28%]; Supplementary Data 14). When considering the pseudotime trajectories for all DEGs in these excitatory neuron subtypes, the direction of the expression matched the DESeq2 results (Supplementary Figs. 3–6). The pseudotime trajectories revealed subtle expression dynamics within the analyzed sample groups, showing that the majority of DEGs gradually increase in expression with age from childhood to adolescence, followed by a decrease in expression toward late adulthood.
Genes associated with intelligence quotient (IQ) and educational attainment (EA), as well as those associated with accelerated evolution in humans, have recently been shown to be enriched in adult temporal lobe cortical neurons, especially the Exc_L2-3_LINC00507_FREM3 subtype56. As childhood is a key period of cognitive development57, we explored whether the same genes were found among our DEGs. Of the 149 DEGs found in at least one cell type, 20 (13.42%, P = 0.02) are known to be significantly associated with EA58, six (4.02%, P = 0.7) with IQ59 and 30 (20.13%, P = 3.89 × 10−7) with accelerated evolution in humans60. These included several genes that were upregulated in pediatric samples, such as MYO16, KCNG1, FGF13 and SOX11 (Supplementary Data 10).
Overall, we highlight several genes upregulated in children and/or adolescents that have known roles in brain development and have been associated with cognitive ability. Our analysis builds on previous knowledge by implicating specific cell subtypes and provides additional candidate genes that are likely to contribute to cell-type-specific maturation processes.
Gene pathways enriched in pediatric cell types
We next used gene set enrichment analysis (GSEA) to conduct a broad analysis of the gene pathways that are differentially regulated across all brain cell types during brain maturation. We found that 2,006 GO biological process terms were enriched in the pediatric compared with the adult samples, whereas 866 were depleted (P < 0.01 and q < 0.1) (Supplementary Data 15). Of the 25 most frequently enriched terms, the majority (ten terms) were associated with cellular respiration pathways (Fig. 6 and Supplementary Data 15). Six were associated with intracellular transport, including transport of neurotransmitters, and five were linked to neurotransmitter release and synaptic plasticity. Three terms, including the top enriched term, were associated with protein translation and modification. The majority of depleted terms (ten terms) were associated with synaptic processes (Fig. 6). A further six depleted terms were connected to neuronal morphogenesis, including axon and dendrite morphogenesis. Two of the top depleted terms were associated with axon ensheathment. Neither of these terms was significantly enriched in oligodendrocytes or oligodendrocyte progenitor cells, whereas they were associated with neuronal subtypes and microglia.
GSEA heatmap showing the top 25 most frequently enriched (top 25 rows) or depleted (bottom 25 rows) terms appearing across all cell types. Only significant (P < 0.01 and q < 0.1) terms are shown. Gray indicates that a term was not significantly enriched or depleted in the indicted cell type. See also Supplementary Data 12. NES, normalized enrichment score.
Overall, our GSEA analysis points toward putative genetic pathways that may drive maturation in the pediatric brain. Cellular respiration processes needed to support the higher metabolic rates in the brain during childhood61 may be enriched. In addition, pathways related to strengthening synapses through neurotransmitter release may be enhanced. On the other hand, as synaptic pruning is underway62, pathways that promote synaptic growth may need to be suppressed.
Cell-type-specific expression of TBM biomarkers
The Pediatric Cell Atlas aims to create reference atlases that can be used to improve our understanding of cell-type-specific responses to disease in children17. Here, we used our snRNA-seq datasets to interrogate the cell-type-specific expression of putative genetic biomarkers for TBM23. These biomarkers are enriched in the ventricular cerebrospinal fluid from children with TBM in comparison with controls with meningitis caused by other brain infections23.
Sixty-six of the 76 TBM biomarkers were expressed in our dataset, with similar expression across the two age groups and genes clearly clustering according to their relative expression across the broad cell type categories (Extended Data Fig. 10). The genes with the highest relative expression in our data were expressed by nonneuronal cell types, in line with the view that immunological activity of supporting cells and their intercellular signaling interactions are important drivers of the immune response to TBM63. Several of these biomarkers (for example, FADS2, AMOT and ALDH6A1) were enriched in the two astrocyte subtypes, potentially indicating a prominent role for astrocytes in the host response to TBM.
Our analyses also clearly revealed subsets of biomarkers that are more highly expressed by neuronal than nonneuronal cell types. These included biomarkers that were associated with Exc_L2_LAMP5_LTK, Exc_L2-4_LINC00507_GLP2R and Exc_L5-6_THEMIS_C1QL3 (LYNX1, FAIM2, MAP1A, TUBB4A). This is line with the finding that neuronal excitotoxicity is elevated in TBM23 and suggests that specific excitatory neuron subtypes may contribute to this signal.
Notably, the two most enriched genes in the TBM biomarker dataset23, CXCL9 and CXCL11, were either completely absent from our datasets (CXCL11) or expressed by very few nuclei (CXCL9). The absence of these interferon-inducible chemokines in our datasets from uninfected tissue supports the proposition that they are indeed biomarkers from the site of disease64 in both adults and children with TBM and could also reflect the contribution of peripheral immune cells recruited to the brain during infection.
Discussion
The brain is the most complex organ in the human body and continuously changes as we mature. Here, we begin to unmask the molecular mechanisms guiding these processes in the temporal cortex, using single-cell and spatial transcriptomics to compare similar cell types between pediatric and adult datasets.
To facilitate accurate comparisons of cell types across age groups, we used the existing Allen Brain Map MTG atlas1 to annotate our datasets. This demonstrated that the reference atlas, generated from adult snRNA-seq datasets, is indeed generalizable31 and can be used to classify cell types from samples of different ages. This generalizability is essential for healthy human reference atlases to serve as a baseline to improve our understanding of human development and disease3. Our samples and those in the reference MTG atlas include neurosurgical tissue from donors with epilepsy, and, although the analyzed tissue is not from the site of pathology, it is important to view our findings in light of the patient diagnosis. Previous research comparing gene expression between neurosurgical and post-mortem samples used in the MTG atlas found a strong correlation of expression between cell types across conditions1. In addition, a comparison of samples from 45 adult donors with epilepsy with post-mortem samples from the MTG atlas found a similar number of genes and similar cell abundance per cell subclass across tissue sources; however, they did find more variation for these parameters in neurosurgical samples65. As more pediatric MTG samples of post-mortem and neurosurgical origin become available, it will be important to conduct similar analyses to determine whether these findings hold for the pediatric temporal cortex.
Our machine learning marker gene analysis showed that although the cell type classifications, which are based on the expression of thousands of genes, can be transferred onto new datasets, the minimal markers that define the cell types vary across datasets. Only a quarter of our NS-Forest minimal markers overlapped with the existing MTG cell atlas minimal markers29. The differences in the single-cell transcriptomics technologies used to generate our dataset and the MTG cell atlas may account for much of this discrepancy. Nonetheless, our analyses suggest that some of our markers may provide better discrimination between cell types than existing markers. These results highlight a challenge for the HCA to revise cell type markers as more datasets are made available to ensure that the cell type classification is as widely applicable as possible.
Similar to analyses of aging in the mouse66, our analyses showed little change in cell type composition within the temporal cortex during human brain maturation. However, our differential expression analysis highlighted differences in cell states between specific pediatric and adult cell subtypes. Recently, the supragranular excitatory pyramidal neurons in the MTG have been shown to have high transcriptional diversity1,67, large arborizations68 and electrophysiological properties that affect signal integration and encoding69,70,71,72 in ways that may contribute to cognition. As cognitive ability is a key feature that is established during childhood68, our analysis offers an opportunity to explore how cell-type-specific gene expression dynamics contribute to cognitive development. Notably, two of the 21 highlighted cell types were the layer 2 and 3 excitatory neurons, Exc_L2_LAMP5_LTK and Exc_L2-3_LINC00507_FREM3, that have recently been associated with human cognition56. In line with these findings, several of the DEGs associated with these cell types, including FNBP1L73 and SOX11 (ref. 74) have been implicated in cognitive ability and intelligence. Overall, our data point toward genes that may have roles in cognitive development specifically within these excitatory neurons.
The relatively low number of genes implicated in our differential expression analysis in comparison with similar studies in mouse66 suggests that the differences between the pediatric and adult brain are subtle. However, the inherent high variability in human gene expression data may mask some of the differential gene expression in our limited sample. Nonetheless, our pseudotime trajectory analyses reveal some of the expression dynamics that may occur during childhood, with the expression of many genes rising toward adolescence and dropping off in adulthood. As the HCA database for the human temporal cortex expands, it will be important to build on these analyses with more samples. Binning of samples of similar age will provide a higher-resolution analysis of cell-type-specific gene expression trajectories over the course of brain maturation.
Finally, we have provided single-nucleus gene expression datasets for the brain that include data from Black southern African donors, thereby increasing the diversity of the HCA database. We demonstrate how this resource can be used to deconvolute site-of-disease biomarker analyses for TBM, pinpointing which cell types may drive altered gene expression profiles in the brain. Importantly, these investigations have the potential to contribute to the development of effective treatments that are tailored to the specific needs of both adult and pediatric patients.
Methods
Human samples
Ethical approval was granted for the collection and use of pediatric and adult human brain tissue by the University of Cape Town Human Research Ethics Committee (UCT HREC REF 016/2018; substudies 146/2022 and 147/2022). The human brain tissue samples used to generate new datasets were obtained by informed consent for studies during temporal lobe surgical resections to treat epilepsy and/or cancer performed at the Red Cross War Memorial Children’s Hospital and Mediclinic Constantiaberg Hospital in Cape Town, South Africa. The samples used in this study were of temporal cortex origin and represent radiologically and macroscopically normal neocortex within the pathological context (details in Supplementary Data 1). Ancestry was recorded by the clinical teams based on their knowledge of the donors. The category ‘Black South African’ includes both Black and mixed race ancestries. The ‘ancestry’ descriptors in Supplementary Data 1 were used solely to ensure that the indicated population cohorts were represented in the HCA data repository. These descriptors were not used in participant accrual, study design, data analysis or data interpretation.
Following resection, samples were placed in carbogenated ice-cold artificial cerebral spinal fluid containing 110 mM choline chloride, 26 mM NaHCO3, 10 mM d-glucose, 11.6 mM sodium ascorbate, 7 mM MgCl2, 3.1 mM sodium pyruvate, 2.5 mM KCl, 1.25 mM NaH2PO4 and 0.5 mM CaCl2 (300 mOsm) and immediately transported to the laboratory (~20 min). Tissue blocks containing the full span from pia to white matter were prepared and either flash-frozen in liquid nitrogen or embedded in optimal cutting temperature compound (OCT) and stored at −80 °C. The OCT-embedded samples were flash-frozen in a 10 × 10 mm2 cryomold; this was either frozen directly in liquid nitrogen or placed in a container of isopentane (Merck), which was in turn placed in liquid nitrogen at the same level as the isopentane. The publicly available snRNA-seq datasets24 generated from samples obtained during elective surgeries performed at Universitair Ziekenhuis Leuven, Belgium, were downloaded from the Sequence Read Archive database.
Nuclei isolation for snRNA-seq
Nuclei were isolated according to a protocol adapted from ref. 75 and the 10X Genomics nuclei isolation protocol (CG000124, User Guide Rev E) (see Supplementary Notes for details).
10X Genomics snRNA-seq library preparation
The snRNA-seq library preparation was carried out using the 10X Genomics Chromium Next Gem Single Cell 3′ Reagent Kit (v.3.1) according to the manufacturer’s protocols (CG000204, User Guide Rev D), targeting 10,000 nuclei per sample. All technical replicates were derived from the same cell suspension, except for the samples generated for P0013, which were derived from two separate cell suspensions on separate days (Supplementary Data 1). At steps 2.2d and 3.5e in the protocol, the libraries were amplified using 11 cycles and 13 cycles, respectively. Library quality and concentration were assessed using either the TapeStation or Bioanalyzer (Agilent) and Qubit (Invitrogen) at the Central Analytical Facility (University of Stellenbosch). cDNA libraries were sequenced by Novogene (Singapore) on an Illumina NovaSeq system using Illumina High Output kits (150 cycles).
snRNA-seq read alignment and gene expression quantification
Fastq files were aligned to the human reference transcriptome (GRCh38) and quantified using the count function from 10X Genomics Cell Ranger (v.6.1.1) (RRID SCR_017344) (‘Code availability’, script 1 (https://zenodo.org/records/13321265)). The inclusion of introns was specified in the count function. An automatic filtering process was performed to remove barcodes corresponding to background noise that had very low UMI counts.
snRNA-seq quality control
The resulting count matrices were processed using a pipeline adapted from the Harvard Chan Bioinformatics Core (https://hbctraining.github.io/scRNA-seq_online/). The filtered gene barcode matrix for each sample was imported into R (v.4.2.0) using the Read10X function from Seurat (v.2.0)25. Nuclei that met the following criteria were retained (‘Code availability’, script 2): nUMI > 500, nGene > 250, log10GenesPerUMI > 0.8 and mitoRatio < 0.2. Gene-level filtering was performed to remove genes that had zero counts in all nuclei, genes expressed in fewer than ten nuclei and mitochondrial genes from the gene by cell counts matrix. Three doublet-removal tools, namely DoubletFinder (v.3.0)76 (‘Code availability’, script 3), DoubletDecon (v.1.1.5)77 (‘Code availability’, script 4) and Scrublet (v.0.2)77 (‘Code availability’, scripts 5 and 6), were used to identify doublets for each dataset individually. The sample-specific parameters of each of the tools were adjusted according to the specified guidelines. To achieve a balance between the false positive and false negative rates of the different doublet-detection tools, all doublets identified by DoubletFinder and the intersection of the doublets identified by DoubletDecon and Scrublet were removed77.
snRNA-seq data normalization, integration and clustering
Principal component analysis was performed to evaluate known sources of within-sample variation between nuclei, namely the mitoRatio and cell cycle phase (‘Code availability’, script 7). The UMI counts of the 3,000 most variable features were normalized and scaled on a per-sample basis by applying the SCTransform function from Seurat (v.2.0) with mitoRatio regressed out. UMAP analysis was performed on the merged object to assess whether integration was necessary. The datasets were subsequently integrated using Seurat’s SelectIntegrationFeatures, PrepSCTIntegration, FindIntegrationAnchors and IntegrateData functions (‘Code availability’, script 7). To cluster the datasets following integration, dimensionality reduction was first performed using UMAP embedding, specifying 40 dimensions (‘Code availability’, script 8). The Seurat FindClusters function was then applied at a resolution of 0.8.
snRNA-seq cluster annotation
Two levels of annotation were performed. Clusters were initially annotated as one of the major brain cell types (level 1 annotation) based on the expression of known markers genes (‘Code availability’, script 9). Label transfer was then performed using the TransferData function from Seurat (v.2.0) with the Allen Brain Map MTG atlas1 as a reference dataset (level 2 annotation) (‘Code availability’, scripts 10 and 11). This resulted in each barcode in the query dataset receiving a predicted annotation based on a similarity score to an annotated cell type in the reference. Barcodes were then filtered to remove those with discordant level 1 and level 2 annotations (for example, barcodes with ‘oligodendrocyte’ level 1 annotation and ‘Exc_L4-5_RORB_FOLH1’ level 2 annotation) (‘Code availability’, script 12). To validate the annotation, the expression of known marker genes was assessed. Cosine similarity scores were computed to compare the transcriptomic similarity of each of the annotated query cell types to the 75 reference MTG cell types using the SCP package (v.0.4.8) (https://github.com/zhanghao-njmu/SCP) (‘Code availability’, script 13). This was achieved by computing cosine similarity scores for each pair of query and reference cell types using the expression of the top 2,000 shared highly variable features between the query and reference datasets. The log-normalized expression counts were used for this purpose (RNA assay, data slot). To assess the difference between the pediatric and adult datasets relative to the reference, the above cosine similarity analysis was repeated on the pediatric and adult datasets individually (‘Code availability’, script 13).
NS-Forest machine learning marker analysis of snRNA-seq datasets
The NS-Forest tool (v.2.0)29,30 was used to identify combinations of marker genes uniquely defining each annotated cell type (‘Code availability’, scripts 14 and 15) in the pediatric and adult datasets separately. The number of nuclei per sample was randomly downsampled to that of the sample with the fewest nuclei (n = 4,865). A random-forest model was used to select a maximum of 15 marker genes per cell type based on their being both highly expressed and uniquely expressed within a cell type compared with other cell types (that is, the top Gini index-ranked features with positive expression values). The number of trees chosen for this model was 30,000, the cluster median expression threshold was set to the default value of zero, the number of genes used to rank permutations of genes by their F-beta score was 6, and the beta weight of the F score was set to 0.5. The aforementioned parameters were set according to the parameters described by Aevermann et al.29, allowing the outputs to be directly compared with their markers and with the Allen Brain Map MTG atlas minimal markers1. To assess the relevance of these markers in terms of their capacity to distinguish different cell types in a UMAP analysis, the SCTransform and integration methods were repeated using either a random set of genes or the NS-Forest markers as anchors29 (‘Code availability’, script 16).
DESeq2 age-dependent differential gene expression analysis of snRNA-seq datasets
DESeq2 (v.1.40.1)33 was used to identify genes that were differentially expressed with age (‘Code availability’, script 17) (see Supplementary Notes for details).
Pseudotime trajectory analysis with psupertime
To validate the DEGs identified with DESeq2, a pseudotime trajectory analysis was performed for a subset of excitatory neuron subtypes using the psupertime package (v.0.2.6)55 (‘Code availability’, script 18) (see Supplementary Notes for details).
Pathway enrichment analysis of snRNA-seq datasets
GO analysis of NS-Forest marker genes was performed on the gProfiler web server (2023-09-14 build)78 using default settings (adjusted P < 0.05) with ‘highlight driver terms in GO’ selected.
DEGs identified by DESeq2 (Supplementary Data 10) that were associated with EA and IQ, as well as those associated with accelerated evolution in humans (HARs), were determined by comparing the list of neuronal DEGs with the EA, IQ and HAR gene lists used by Driessens et al. (2023)56, which were subsets of lists from Lee et al. (2018)58, Savage et al. (2018)59 and Doan et al. (2016)60, respectively. A hypergeometric test was performed to test the significance of the results relative to chance (‘Code availability’, script 19).
GSEA on the DESeq2 output for all genes was performed using the Broad Institute’s GSEA software (v.2023.1) (https://www.gsea-msigdb.org/gsea/msigdb) (‘Code availability’, script 20). GSEA aggregates information from many genes to identify enriched functional pathways; this allowed us to interrogate the gene signature changes across all cell types, including those that did not show any significant DEGs66 (see Supplementary Notes for details).
Analysis of site-of-disease TBM markers
The dittoheatmap function from the dittoSeq package (v.1.13.1)79 was used to generate heatmaps for the expression of the TBM biomarkers (upregulated genes listed by Rohlwink et al.23, Supplementary Data 5) across cell types in the pediatric and adult datasets individually. In addition, Seurat’s dotplot function25 was used to visualize levels of expression and proportions of nuclei expressing the markers across cell types (‘Code availability’, script 21). Before generation of the plots, the TBM marker genes were filtered to remove those expressed in 15 nuclei or fewer across all cell types. Gene counts for each marker were aggregated across cell types and scaled. The markers were clustered according to their expression profiles using dittoheatmap’s default hierarchical clustering method (Euclidean, complete). The clustering order and dendrogram from this output for the peditaric datasets were used to generate dot plots for both peditaric and adult datasets (‘Code availability’, script 21).
snRNA-seq data plots
Plots were produced with Seurat (v.2.0)25, ggplot2 (v.3.4.2)80, ShinyCell (v.2.1.0)81 and Microsoft Excel (v.16.54).
10X Genomics Visium library preparation
Frozen OCT-embedded temporal cortex tissue samples were scored using a prechilled razor blade to fit in the Spatial Gene Expression slide capture areas. Sections (10 μm thick) were cut using a cryostat (Leica CM1860/CM1950) and collected onto the Spatial Gene Expression slide capture areas. Two replicate sections of the 15-year-old samples (10 μm apart) and two replicate sections of the 31-year-old samples (40 μm apart) were collected. The spatial Gene Expression slides with tissue sections were stored in a sealed container at −80 °C. Captured sections were stained with hematoxylin and eosin according to the 10X Genomics Demonstrated Protocol Guide (CG000160, Rev B). Brightfield images of the stained sections were captured using an EVOS M5000 microscope (Thermo Fisher Scientific) at ×20 magnification without coverslipping. Overlapping images of the sections including the fiducial frame were stitched together using Microsoft Image Composite Editor (v.2.0.3). Visium libraries were prepared from the stained tissue sections following the Visium Spatial Gene Expression Reagents Kit User Guide (CG000239, Rev D). At step 1.1, the tissue was permeabilized for 12 min according to the Visium Spatial Gene Expression Tissue Optimization User Guide (CG000238, Rev D). At step 3.2, cDNA was amplified using 20 cycles. Library quality and concentration were assessed using TapeStation (Agilent) and Qubit (Invitrogen) at the Central Analytical Facility (University of Stellenbosch). Libraries were sequenced by Novogene (Singapore) on an Illumina NovaSeq system using Illumina High Output kits (150 cycles).
Visium read alignment and gene expression quantification
The hematoxylin and eosin images were processed using the 10X Genomics Loupe Browser (v.4.0) Visium Manual Alignment Wizard. 10X Genomics Space Ranger count (10X Space Ranger v.1.3.0) was used to perform alignment of FASTQ files to the human reference transcriptome (GRCh38), tissue detection, fiducial detection and barcode/UMI counting.
cell2location analysis of Visium datasets
The average number of nuclei per Visium spot was determined using VistoSeg (v.1)82 in MATLAB R2019a (‘Code availability’, script 22). Cell2location (v.0.7a0)26 was used to spatially map the brain cell types by integrating the Visium data count matrices (Space Ranger output) with the annotated snRNA-seq datasets (‘Code availability’, script 23). To avoid mapping artifacts, mitochondrial genes were removed from the Visium datasets before spatial mapping. Reference signatures of the 75 annotated cell populations were derived using a negative binomial regression model using the default values (‘Code availability’, script 24). Unnormalized and untransformed snRNA-seq mRNA counts were used as inputs to the regression model for estimating the reference signatures (‘Code availability’, script 24. The snRNA-seq mRNA counts were filtered to 14,209 genes and 144,438 cells. The cell2location model for estimating the spatial abundance of cell populations was filtered to 14,197 genes and 14,324 cells that were shared in both the snRNA-seq and Visium data. The following cell2location parameters were used: training iterations = 30,000 cell per location, N^ = 7 (estimated using VistoSeg segmentation results), normalization (ys) alpha prior = 20 (‘Code availability’, script 25). To visualize the cell abundance in spatial coordinates, the 5% quantile of the posterior distribution was used; this represents the value of cell abundance in which the model has high confidence (‘Code availability’, script 26). Cell2location’s NMF was used to identify cellular compartments and cell types that colocated based on the cell type abundance estimates. NMF was tested using a range of factors (5 to 30) for the ‘n_fact’ parameter (‘Code availability’, script 26). n_fact = 15 was chosen as it clearly grouped the oligodendrocyte, astrocyte and excitatory neuron cell subtypes into known tissue zones, that is, the layers of the cortex (‘Code availability’, script 27).
BayesSpace analysis of Visium datasets
The raw gene expression counts from Space Ranger were normalized and log transformed, and principal component analysis was performed on the top 2,000 highly variable genes. To obtain high-resolution gene expression for selected genes, the principal component values were mapped back to their original log-transformed gene expression space (spot level) using the default BayesSpace (v.1.5.1)34 regression (‘Code availability’, script 28). To do this, the principal components from the original data were used as predictors in training the model for each gene, in which the results were the measured gene expression at the spot level. The trained model was then used to predict the gene expression at subspot level using high-resolution principal components. The high-resolution model was trained using default values except for the following parameters: seven principal components, number of clusters = 8, nrep = 100,000, burn-in = 10,000. The BayesSpace outputs for each sample were quantified for spots with expression level > 0 and displayed as box plots (‘Code availability’, script 29).
In situ hybridization chain reaction of frozen human tissue sections
Frozen sections (10 μm thick) were collected on HistoBond+ slides (Marienfeld) and stored at −20 °C. The in situ hybridization chain reaction (HCR) protocol was carried out on tissue sections as detailed by Choi et al.83 using reagents, probes and hairpins purchased from Molecular Instruments. Probes were ordered for the following genes: RELN (NM_005045.4), FABP7 (CR457057.1), AQP4 (NM_001650.5), RORB (NM_006914.4), CLSTN2 (NM_022131.3) and TSHZ2 (NM_173485.6). When necessary to quench lipofuscin autofluorescence, sections were rinsed after HCR in 1× phosphate-buffered saline and treated with 200 μl TrueBlack (Biotium) for 30 s. Slides were rinsed in phosphate-buffered saline, stained with Hoescht (Thermo Fisher) and mounted using SlowFade Gold Antifade Reagent (Invitrogen). Sections were imaged using an LSM 880 Airyscan confocal microscope (Carl Zeiss, ZEN SP 2 software) with a ×40 or ×60 objective.
MERFISH analysis of frozen temporal cortex tissue sections
Frozen sections (10 μm thick) were cut from frozen OCT-embedded temporal cortex tissue samples using a cryostat (Leica CM1950). Sections from a pediatric and an adult sample were collected onto the same MERSCOPE coverslip (VIZGEN 2040003), fixed and stored in 70% ethanol following the instructions in the VIZGEN protocol (Fresh & Fixed Frozen Tissue Sectioning & Shipping Procedure Rev A, doc. no. 91600107). The slide was processed on a VIZGEN MERSCOPE system by the MRC Weatherall Institute of Molecular Medicine Single Cell Facility (University of Oxford) within 1 month of storage. Sections were photobleached for 10 h at 4 °C and then washed in 5 ml of Sample Prep Wash Buffer (VIZGEN 20300001) in a 5-cm petri dish. Sections were incubated in 5 ml of Formamide Wash Buffer (VIZGEN 20300002) at 37 °C for 30 min and hybridized at 37 °C for 36 to 48 h using 50 μl of VIZGEN-supplied custom Gene Panel Mix according to the manufacturer’s instructions. Following hybridization, sections were washed twice in 5 ml Formamide Wash Buffer for 30 min at 47 °C. Sections were then embedded in acrylamide by polymerizing VIZGEN Embedding Premix (VIZGEN 20300004) according to the manufacturer’s instructions. Following embedding, sections were digested in Digestion Premix (VIZGEN 20300005) and RNase inhibitor (New England Biolabs M0314L) for 3 h at 37 °C and then cleared for 16 to 24 h with a mixture of VIZGEN Clearing Solution (VIZGEN 20300003) and Proteinase K (New England Biolabs P8107S) according to the manufacturer’s instructions. Following clearing, sections were washed twice for 5 min in Sample Prep Wash Buffer (PN 20300001) and then stained with VIZGEN DAPI and PolyT Stain (PN 20300021) for 15 min, followed by a 10-min wash in Formamide Wash Buffer. The Formamide Wash Buffer was removed, and sections were washed with Sample Prep Wash Buffer during MERSCOPE imaging set up. A mixture of 100 ml of RNAse Inhibitor (New England Biolabs M0314L) and 250 ml of Imaging Buffer Activator (PN 203000015) was added through the cartridge activation port to a prethawed and mixed MERSCOPE imaging cartridge (VIZGEN PN 1040004). Then, 15 ml of mineral oil (Millipore-Sigma m5904-6X500ML) was added on top of the activation port, and the MERSCOPE fluidics system was primed according to VIZGEN instructions. The flow chamber was assembled with the section coverslip according to VIZGEN specifications, and the imaging session was initiated after collection of a 10× mosaic DAPI image and selection of the 1-cm2 imaging area. MERFISH data were visualized using VIZGEN MERSCOPE Vizualizer software (v.2.3.3330.0).
Statistics and reproducibility
No statistical method was used to predetermine sample size. Low-quality nuclei were excluded as described in the ‘snRNA-seq quality control’ section. The experiments were not randomized. The investigators were not blinded to allocation during experiments or outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw sequence files and analyzed data files (integrated and annotated Seurat Object, Cell2location trained model, Visium image files) for snRNA-seq and Visium experiments have been deposited in the NCBI GEO database (snRNA-seq accession no. GSE280569; Visium accession no. GSE280570) and are also available through the HCA data repository as part of the ‘managed access’ tier at https://explore.data.humancellatlas.org/projects/ae9f439b-bd47-4d6e-bd72-32dc70b35d97. Potential users of the HCA data repository data need to be registered and signed in to the DUOS system to view the data description and can request access to the data by following the instructions on the HCA Data Portal website (https://data.humancellatlas.org/guides/requesting-access-to-controlled-access-data). A ShinyApp for exploration of the annotated snRNA-seq data is available (https://hockman-lab.shinyapps.io/shiny_app_update_final/). Source data are provided with this paper.
Code availability
All scripts used to analyze the data are indicated as numbered files in the Methods section and are available via Zenodo at https://zenodo.org/records/13321265 (ref. 84).
References
Hodge, R. D. et al. Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019).
Bakken, T. E. et al. Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature 598, 111–119 (2021).
Regev, A. et al. The Human Cell Atlas. eLife https://doi.org/10.7554/eLife.27041 (2017).
Network, B. I. C. C. A multimodal cell census and atlas of the mammalian primary motor cortex. Nature 598, 86–102 (2021).
Darmanis, S. et al. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl Acad. Sci. USA 112, 7285–7290 (2015).
Li, M. et al. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science https://doi.org/10.1126/science.aat7615 (2018).
Velmeshev, D. et al. Single-cell analysis of prenatal and postnatal human cortical development. Science 382, eadf0834 (2023).
Werling, D. M. et al. Whole-genome and RNA sequencing reveal variation and transcriptomic coordination in the developing human prefrontal cortex. Cell Rep. 31, 107489 (2020).
Kang, H. J. et al. Spatio-temporal transcriptome of the human brain. Nature 478, 483–489 (2011).
Colantuoni, C. et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature 478, 519–523 (2011).
Donertas, H. M. et al. Gene expression reversal toward pre-adult levels in the aging human brain and age-related loss of cellular identity. Sci Rep. 7, 5894 (2017).
Bourgeois, J. P. & Rakic, P. Changes of synaptic density in the primary visual cortex of the macaque monkey from fetal to adult stage. J. Neurosci. 13, 2801–2820 (1993).
Huttenlocher, P. R. & Dabholkar, A. S. Regional differences in synaptogenesis in human cerebral cortex. J. Comp. Neurol. 387, 167–178 (1997).
Petanjek, Z. et al. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc. Natl Acad. Sci. USA 108, 13281–13286 (2011).
Paus, T., Keshavan, M. & Giedd, J. N. Why do many psychiatric disorders emerge during adolescence? Nat. Rev. Neurosci. 9, 947–957 (2008).
Jaffe, A. E. et al. Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat. Neurosci. 21, 1117–1125 (2018).
Taylor, D. M. et al. The Pediatric Cell Atlas: defining the growth phase of human development at single-cell resolution. Dev. Cell 49, 10–29 (2019).
Tishkoff, S. A. et al. The genetic structure and history of Africans and African Americans. Science 324, 1035–1044 (2009).
World population datasheet 2022. PRB www.prb.org/wp-content/uploads/2022/09/2022-World-Population-Data-Sheet-Booklet.pdf (2022).
O’Malley, J., Wardlaw, T., You, D., Hug, L. & Anthony, D. Africa’s child demographics and the world’s future. Lancet 384, 730–732 (2014).
Schutte, C. M. Analysis of HIV-related mortality data in a tertiary South African neurology unit, 2006-2012. S. Afr. J. HIV Med. 14, 121–124 (2013).
Rohlwink, U. K. et al. Clinical characteristics and neurodevelopmental outcomes of children with tuberculous meningitis and hydrocephalus. Dev. Med. Child Neurol. 58, 461–468 (2016).
Rohlwink, U. K. et al. Tuberculous meningitis in children is characterized by compartmentalized immune responses and neural excitotoxicity. Nat. Commun. 10, 3767 (2019).
Thrupp, N. et al. Single-nucleus RNA-seq is not suitable for detection of microglial activation genes in humans. Cell Rep. 32, 108189 (2020).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e1821 (2019).
Kleshchevnikov, V. et al. Cell2location maps fine-grained cell types in spatial transcriptomics. Nat. Biotechnol. 40, 661–671 (2022).
Maynard, K. R. et al. Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. Nat. Neurosci. 24, 425–436 (2021).
Hintsch, G. et al. The calsyntenins—a family of postsynaptic membrane proteins with distinct neuronal expression patterns. Mol. Cell. Neurosci. 21, 393–409 (2002).
Aevermann, B. et al. A machine learning method for the discovery of minimum marker gene combinations for cell type identification from single-cell RNA sequencing. Genome Res. 31, 1767–1780 (2021).
Aevermann, B. D. et al. Cell type discovery using single-cell transcriptomics: implications for ontological representation. Hum. Mol. Genet. 27, R40–R47 (2018).
Tan, S. Z. K. et al. Brain Data Standards - a method for building data-driven cell-type ontologies. Sci. Data 10, 50 (2023).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014).
Zhao, E. et al. Spatial transcriptomics at subspot resolution with BayesSpace. Nat. Biotechnol. 39, 1375–1384 (2021).
Radner, S. et al. β2 and γ3 laminins are critical cortical basement membrane components: ablation of Lamb2 and Lamc3 genes disrupts cortical lamination and produces dysplasia. Dev. Neurobiol. 73, 209–229 (2013).
De Angelis, C. et al. Compound heterozygous variants in LAMC3 in association with posterior periventricular nodular heterotopia. BMC Med. Genomics 14, 64 (2021).
Barak, T. et al. Recessive LAMC3 mutations cause malformations of occipital cortical development. Nat. Genet. 43, 590–594 (2011).
Wang, Y., Lin, L., Lai, H., Parada, L. F. & Lei, L. Transcription factor Sox11 is essential for both embryonic and adult neurogenesis. Dev. Dyn. 242, 638–653 (2013).
Hoshiba, Y. et al. Sox11 balances dendritic morphogenesis with neuronal migration in the developing cerebral cortex. J. Neurosci. 36, 5775–5784 (2016).
Ling, K. H. et al. Molecular networks involved in mouse cerebral corticogenesis and spatio-temporal regulation of Sox4 and Sox11 novel antisense transcripts revealed by transcriptome profiling. Genome Biol. 10, R104 (2009).
Kakimoto, T., Katoh, H. & Negishi, M. Regulation of neuronal morphology by Toca-1, an F-BAR/EFC protein that induces plasma membrane invagination. J. Biol. Chem. 281, 29042–29053 (2006).
Winsky-Sommerer, R., King, H. A., Iadevaia, V., Moller-Levet, C. & Gerber, A. P. A post-transcriptional regulatory landscape of aging in the female mouse hippocampus. Front. Aging Neurosci. 15, 1119873 (2023).
Twohig, J. P. et al. Age-dependent maintenance of motor control and corticostriatal innervation by death receptor 3. J. Neurosci. 30, 3782–3792 (2010).
Yang, Q. Q. et al. Nuclear isoform of FGF13 regulates post-natal neurogenesis in the hippocampus through an epigenomic mechanism. Cell Rep. 35, 109127 (2021).
Wu, Q.-F. et al. Fibroblast growth factor 13 is a microtubule-stabilizing protein regulating neuronal polarization and migration. Cell 149, 1549–1564 (2012).
Zhang, X., Lin, P.-Y., Liakath-Ali, K. & Südhof, T. C. Teneurins assemble into presynaptic nanoclusters that promote synapse formation via postsynaptic non-teneurin ligands. Nat. Commun. 13, 2297 (2022).
Cheung, A. et al. Teneurin paralogues are able to localise synaptic sites driven by the intracellular domain and have the potential to form cis-heterodimers. Front Neurosci 16, 915149 (2022).
Beckmann, J., Schubert, R., Chiquet-Ehrismann, R. & Müller, D. J. Deciphering teneurin domains that facilitate cellular recognition, cell–cell adhesion, and neurite outgrowth using atomic force microscopy-based single-cell force spectroscopy. Nano Lett. 13, 2937–2946 (2013).
Khatir, I. et al. Decoupling of mRNA and protein expression in aging brains reveals the age-dependent adaptation of specific gene subsets. Cells https://doi.org/10.3390/cells12040615 (2023).
Patel, K. G., Liu, C., Cameron, P. L. & Cameron, R. S. Myr 8, a novel unconventional myosin expressed during brain development associates with the protein phosphatase catalytic subunits 1α and 1γ1. J. Neurosci. 21, 7954–7968 (2001).
Jansen, L. A. et al. PI3K/AKT pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia. Brain 138, 1613–1628 (2015).
Cruz, E., Bessieres, B., Magistretti, P. & Alberini, C. M. Differential role of neuronal glucose and PFKFB3 in memory formation during development. Glia 70, 2207–2231 (2022).
Irvin, D. K., Zurcher, S. D., Nguyen, T., Weinmaster, G. & Kornblum, H. I. Expression patterns of Notch1, Notch2, and Notch3 suggest multiple functional roles for the Notch-DSL signaling system during brain development. J. Comp. Neurol. 436, 167–181 (2001).
Tanaka, M., Kadokawa, Y., Hamada, Y. & Marunouchi, T. Notch2 expression negatively correlates with glial differentiation in the postnatal mouse brain. J. Neurobiol. 41, 524–539 (1999).
Macnair, W., Gupta, R. & Claassen, M. psupertime: supervised pseudotime analysis for time-series single-cell RNA-seq data. Bioinformatics 38, i290–i298 (2022).
Driessens, S. L. et al. Genes associated with cognitive ability and HAR show overlapping expression patterns in human cortical neuron types. Nat. Commun. 14, 4188 (2023).
Gauvain, M. Cognitive Development in Infancy and Childhood (Cambridge Univ. Press, 2022).
Lee, J. J. et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 50, 1112–1121 (2018).
Savage, J. E. et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat. Genet. 50, 912–919 (2018).
Doan, R. N. et al. Mutations in human accelerated regions disrupt cognition and social behavior. Cell 167, 341–354.e312 (2016).
Bentourkia, M. H. et al. Evolution of brain glucose metabolism with age in epileptic infants, children and adolescents. Brain Dev. 20, 524–529 (1998).
Huttenlocher, P. R. Synaptic density in human frontal cortex - developmental changes and effects of aging. Brain Res. 163, 195–205 (1979).
Barnacle, J. R., Davis, A. G. & Wilkinson, R. J. Recent advances in understanding the human host immune response in tuberculous meningitis. Front. Immunol. https://doi.org/10.3389/fimmu.2023.1326651 (2024).
Yang, Q. et al. IP-10 and MIG are compartmentalized at the site of disease during pleural and meningeal tuberculosis and are decreased after antituberculosis treatment. Clin. Vaccine Immunol. 21, 1635–1644 (2014).
Johansen, N. et al. Interindividual variation in human cortical cell type abundance and expression. Science 382, eadf2359 (2023).
Ximerakis, M. et al. Single-cell transcriptomic profiling of the aging mouse brain. Nat. Neurosci. 22, 1696–1708 (2019).
Berg, J. et al. Human neocortical expansion involves glutamatergic neuron diversification. Nature 598, 151–158 (2021).
Mohan, H. et al. Dendritic and axonal architecture of individual pyramidal neurons across layers of adult human neocortex. Cereb. Cortex 25, 4839–4853 (2015).
Gidon, A. et al. Dendritic action potentials and computation in human layer 2/3 cortical neurons. Science 367, 83–87 (2020).
Eyal, G. et al. Human cortical pyramidal neurons: from spines to spikes via models. Front. Cell. Neurosci. 12, 181 (2018).
Testa-Silva, G. et al. High bandwidth synaptic communication and frequency tracking in human neocortex. PLoS Biol. 12, e1002007 (2014).
Beaulieu-Laroche, L. et al. Enhanced dendritic compartmentalization in human cortical neurons. Cell 175, 643–651.e614 (2018).
Benyamin, B. et al. Childhood intelligence is heritable, highly polygenic and associated with FNBP1L. Mol. Psychiatry 19, 253–258 (2014).
Chiang, S.-Y. et al. Usp11 controls cortical neurogenesis and neuronal migration through Sox11 stabilization. Sci. Adv. 7, eabc6093 (2021).
Habib, N. et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 14, 955–958 (2017).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e324 (2019).
DePasquale, E. A. K. et al. DoubletDecon: deconvoluting doublets from single-cell RNA-sequencing data. Cell Rep. 29, 1718–1727.e1718 (2019).
Raudvere, U. et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update).Nucleic Acids Res. 47, W191–W198 (2019).
Bunis, D. G., Andrews, J., Fragiadakis, G. K., Burt, T. D. & Sirota, M. dittoSeq: universal user-friendly single-cell and bulk RNA sequencing visualization toolkit. Bioinformatics 36, 5535–5536 (2020).
Wickham, H. ggplot2: elegant graphics for data analysis (Springer, 2016).
Ouyang, J. F., Kamaraj, U. S., Cao, E. Y. & Rackham, O. J. L. ShinyCell: simple and sharable visualization of single-cell gene expression data. Bioinformatics 37, 3374–3376 (2021).
Tippani, M. et al. VistoSeg: processing utilities for high-resolution images for spatially resolved transcriptomics data. Biol. Imaging 3, e23 (2023).
Choi, H. M. et al. Mapping a multiplexed zoo of mRNA expression. Development 143, 3632–3637 (2016).
Steyn, C. Code for ‘A temporal cortex cell atlas highlights gene expression dynamics during human brain maturation’. Zenodo https://doi.org/10.5281/zenodo.13321265 (2024).
Acknowledgements
This research was supported by a Future Leaders African Independent Researcher (FLAIR) Fellowship (FLR\R1\191008, D.H.) (the FLAIR Fellowship Programme was a partnership between the African Academy of Sciences and the Royal Society funded by the UK Government’s Global Challenges Research Fund), a Royal Society/Global Challenges Research Fund FLAIR Collaboration grant (no. FCG\R1\201023, T.S.S. and D.H.), a National Research Foundation (NRF) Research Development Grant for Y-rated Researchers Award (CSRP210415595025, DH), a University of Cape Town (UCT) Building Research Active Academic Staff Grant award (D.H.) and a Fogarty International Center of the National Institutes of Health R21 award (R21TW011225, D.H., J.V.R., C.G.D. and M.J.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. C.S. was supported by a Harry Crossley Research Scholarship, an Oppenheimer Memorial Trust scholarship, an NRF scholarship and a UCT Vice Chancellor’s Research Scholarship. S.F. was supported by a DAAD-NRF joint In-country Scholarship and a UCT Vice Chancellor’s Research Scholarship. M.B.V. was supported by an EMBO long-term fellowship (ALTF 415-2018, M.B.V.) and a Claude Leon Foundation research fellowship. A.F. was supported by the NRF SARChI Chair of Clinical Neurosciences. Computations were performed using facilities provided by the University of Cape Town’s ICTS High Performance Computing team (hpc.uct.ac.za). This publication is part of the HCA (www.humancellatlas.org/publications/).
Author information
Authors and Affiliations
Contributions
C.S. and S.F. conducted the snRNA-seq experiments. C.S. conducted the majority of the snRNA-seq analyses. R.M. conducted the Visium and HCR experiments and analysis. J.M. conducted additional HCR experiments. S.Q. provided additional bioinformatics support. M.B.V. liaised with neurosurgeons and prepared all neurosurgical brain tissue samples. R.M., J.B. and J.M.N.E. conducted the neurosurgeries and provided donor metadata. C.S., T.S.S., M.G. and D.H. conceptualized, conducted and analyzed the MERFISH experiments. U.K.R., M.Z., J.V.R., C.G.D., A.F. and D.H. conceptualized the study and raised funds. D.H., C.S. and R.M. wrote the manuscript. D.H. conducted additional analyses and supervised the project. All authors read and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Nuclei quality control (QC) and clustering.
Number of doublets identified across all 23 datasets by DoubletDecon, DoubletFinder, and Scrublet. Red outline indicates the subset of barcodes called as doublets that were removed. b, Total number of nuclei per dataset before (yellow) and after (green) QC. c, Mean number of reads per nucleus (y axis) by dataset before QC split by age group (x axis). p value determined by two-tailed Welch’s t-test. d, Number of nuclei (y axis) by sample after QC split by age group (x axis). p value determined by two-sided Brunner-Munzel permutation test. e, Violin plots showing the number of unique molecular identifiers (UMIs) (top) and the number of genes detected (bottom) per nucleus per sample after QC. Black dots indicate the median value. Error bars show 95% confidence intervals. f, g, Median number of UMIs (2,263 pediatric and 2,011 adult) (f) and the median number of genes (1,372 paediatric and 1,226 adult) (g) detected per nucleus (y axes) by sample after QC split by age group (x axis). p values determined by two-tailed Brunnermunzel permutation test. h, UMAP plot for the 23 datasets prior to integration. i, UMAP plot showing the resulting clusters determined by the shared nearest neighbour algorithm. Data in all box plots represent mean ± sem for six paediatric and six adult samples. No significant differences were detected between pediatric and adult samples. B, biological replicate; NS, not significant; T, technical replicate. See also Supplementary Table 2.
Extended Data Fig. 2 Annotation and assessment of cell composition across datasets.
a, UMAP plot showing cluster annotation at the level of major brain cell types (level 1 annotation). b, Examination of known cell type-specific marker genes (x axis) after label transfer classify each nucleus according to the Allen Brain Map MTG atlas1 (level 2 annotation) (y axis) (left). Off-target gene expression is evident in several cell types (marked in red), which is likely due to multiplets or nuclei contaminated with ambient mRNA. c,d, Stacked barplots after filtering to retain nuclei with high confidence annotations showing the proportion of nuclei per cell type (y axis) for each technical replicate (c) or biological replicate (d) (x axis) out of the total number of nuclei for each group. Samples with technical replicates showed high degrees of similarity in cell composition between their replicates (c). Technical replicates from each donor were merged to allow comparisons between the 12 samples (d).
Extended Data Fig. 3 Assessment of the sequencing metrics for the annotated cell types.
a, Violin plots showing the distribution of the number of genes (left) and transcripts (right) detected per nucleus per cell type across all datasets. Black dots indicate the median value. Error bars show 95% confidence intervals. b, c, Boxplots showing the number of genes (b) and the number of UMIs (c) (y axis) detected per cell type per sample (x axis) split by age group (red: adult, grey: pediatric). Data in all box plots represent mean ± sem for six pediatric and six adult samples for each cell type. See also Supplementary Table 3 for details of statistical tests performed (Student’s two-tailed t-test, two-tailed Brunner-Munzel test or Welch two-tailed t-test). p-values were adjusted for multiple testing using the Benjamini-Hochberg method.
Extended Data Fig. 4 Visium Spatial Gene Expression samples.
a, b, 31-year-old (a) and 15-year-old (b) temporal cortex tissue blocks embedded in OCT. Black dashed boxes outline the regions collected onto the Visium Spatial Gene Expression slide. c–f, H&E stained technical replicate tissue sections used to generate Visium Spatial Gene Expression libraries for the 31-year-old (c, e) and 15-year-old (d, f) tissue samples. T, technical replicate. Scale bars are 500 µm.
Extended Data Fig. 5 Spatial mapping of cell types in the human temporal cortex.
a, Estimated cell abundance of 75 cell types across all Visium samples. Shown is a heatmap with the colour indicating the relative cell abundance of cell types (rows) across the different samples (columns). b, Estimated cell type abundances (colour intensity) in the technical replicate 31-year-old and 15-year-old temporal cortex tissue sections for a selection of cell types including non-neuronal cell types, excitatory neurons (top row) and inhibitory neurons (bottom row). c, Spatial plots show of the NMF weights for selected NMF factor/tissue compartment across the 31-year-old and 15-year-old temporal cortex tissue sections. Panels are displayed in the same order as the dotplot in Fig. 2c, with the dominant cell types for each factor indicated in brackets. T, technical replicate.
Extended Data Fig. 6 In situ HCR analysis of selected cortical layer marker genes.
Expression of a, layer 1 markers AQP4, FABP7 and RELN and b, layer 4–6 markers RORB, CLSTN2 and TSHZ2 in frozen temporal cortex tissue sections from the same 31-year-old and 15-year-old donor tissue used for Visium. High magnification views of layer 1 in a indicate AQP4/RELN-positive cells (yellow arrowheads) and FABP7 positive cells (green arrowhead). In high magnification views of layer 4 in b in the 31-year-old tissue section, RORB/CLSTN2-positive (white arrowhead) and RORB/TSHZ2-positive cells (green arrowhead) are indicated. In high magnification views of layer 4 in b in the 15-year-old tissue section RORB/CLSTN2/TSHZ2-positive cells (white arrowheads) are indicated. Dashed white lines indicate layer boundaries. Solid white line indicates tissue edge. Scale bars are 100 µm in low magnification views (tile scan at 40x) and 20 µm in high magnification views (63x). Data is representative of 2 technical replicate sections analysed per sample.
Extended Data Fig. 7 Expression of the reference MTG atlas minimal markers.
Heatmap showing the scaled average normalised expression counts of the NS-Forest minimal marker genes identified for the reference MTG cell atlas dataset (y-axis) in each of the 75 query cortical cell types identified in the combined adult and paediatric snRNA-seq datasets (x-axis). The minimal marker genes are annotated (colour codes on the y-axes) according to the cell type they define.
Extended Data Fig. 8 MERFISH spatial transcriptomics analysis of selected NS-forest markers.
a, b Low magnification views of the 31-year-old (a) and 15-year-old (b) MERFISH datasets (from the same donors used for Visium) showing the expression of known layer maker genes in the expected layers as validation of the MERFISH experiment. c–p, High magnification views of 31-year-old (c,e,g,I,k,m,o) and 15-year-old (d,f,h,j,l,n,p) MERFISH datasets showing the overlap of new NS-Forest minimal markers (green) with published NS-Forest minimal markers (magenta) in indicated cells (arrowheads). The cell type that the NS-Forest markers are associated with is indicated in the top left corner. Scale bars: 100 µm. A single section was analysed using MERFISH for each sample.
Extended Data Fig. 9 Evaluation of NS-Forest minimal marker gene expression across cell types in comparison to MTG cell taxonomy markers.
a–d, Boxplots showing the normalised expression counts for LINC01331 (a), PALMD (b), POSTN (c) and OLFML2B (d) in pediatric (top) and adult (bottom) datasets. The cell types expressing the markers at high levels are indicated in bold. Data represents mean ± sem for six paediatric and six adult samples.
Extended Data Fig. 10 Cell type-specific expression of putative TBM biomarkers.
a. Hierarchical clustering of TBM biomarker genes across the 75 cell types identified in the pediatric snRNA-seq dataset reveals clusters of genes that are expressed by specific groups of cell types. b. Analysis of the same genes across the adult snRNA-seq dataset, using the gene order in (a) reveals very similar patterns of cell-type-specific expression across the age-groups. Dashed boxes highlight gene clusters, with associated cell types indicated on the left and right of the right diagram.
Supplementary information
Supplementary Information
Supplementary Notes, Figs. 1–6 and Data 1–15.
Supplementary Data 1
Summary of snRNA-seq, Visium and MERFISH sample metadata. Samples are ordered by age. The eight ‘P00’ datasets were generated in the Hockman laboratory, whereas the four ‘Nuc’ datasets were generated by Thrupp et al. 24.
Supplementary Data 2
Summary of average quality control metrics for snRNA-seq datasets across nuclei for each sample before and after filtering. Several measures for quality control were evaluated on a per-sample basis, including the sequencing saturation, mean number of reads per nucleus, number of barcodes, median number of genes detected per nucleus, median number of UMIs detected per nucleus and number of doublets removed.
Supplementary Data 3
Label transfer annotation of snRNA-seq datasets using the Allen Brain Map MTG atlas as a reference. Sheet 1, Manual annotation of clusters into major cell types (level 1 annotation). Sheet 2, Number of nuclei per level 1 annotation category per sample after filtering discordant nuclei. Sheet 3, Number of nuclei per MTG cell type per sample after filtering discordant nuclei. The number of barcodes corresponding to each MTG cell type and sample is shown. In addition, the total, minimum and maximum numbers of nuclei per cell type and sample were computed. The number of cell types represented per sample was also determined. Sheet 4, Number of removed nuclei per level 1 annotation category per sample. Sheet 5, Number of removed nuclei per MTG cell type per sample. Sheet 6, Subtraction matrix comparing cosine similarity scores (that is, similarity score for each cell subtype compared with the MTG cell Atlas as in Fig. 1d) for the pediatric dataset to the adult dataset. Values are pediatric scores minus adult scores. Sheet 7, P values and tests performed for each cell type (Student’s two-tailed t-test, two-tailed Brunner–Munzel test or Welch two-tailed t-test) when comparing the proportion of nuclei between male and female samples. Sheets 8–10, P values and tests performed for each cell type (Student’s two-tailed t-test, two-tailed Brunner–Munzel test or Welch two-tailed t-test) when comparing the proportion of nuclei (sheet 8), number of genes (sheet 9; Extended Data Fig. 3b) and number of UMIs (sheet 10; Extended Data Fig. 3c) for each cell type between pediatric and adult samples shown. All P values were adjusted for multiple testing using the Benjamini–Hochberg method.
Supplementary Data 4
Summary of average quality control metrics for Visium datasets. Several measures for quality control were evaluated on a per-sample basis including the sequencing saturation, percentage of reads mapped to the transcriptome, number of spots under the tissue, average number of nuclei per spot as determined by VistoSeg analysis, mean reads detected per spot, median genes detected per spot, total number of genes detected, median UMI counts per spot and total number of nuclei.
Supplementary Data 5
NS-Forest minimal marker analysis. Sheet 1, Statistical tests evaluating the expression of Aevermann et al. 29 markers in our datasets. P values were determined using Seurat FindMarkers. All P values were adjusted for multiple testing using the Benjamini–Hochberg method (p_val_adj column) and the Holm method (p_adj_multiple_genes column). Sheets 2 and 3, Metadata for each feature identified by NS-Forest marker in the downsampled pediatric (sheet 2) and downsampled adult (sheet 3) datasets describing the cell type, F-beta score for each marker gene, overlap with Aevermann et al. 29 and Hodge et al. 1, uniqueness to the age group of interest, coding status and uniqueness to the associated cell type as shown in Fig. 3. As input to NS-Forest, all datasets (six pediatric and six adult) were randomly downsampled such that the total number of nuclei per sample was equal to that of the sample with the fewest nuclei.
Supplementary Data 6
gProfiler analysis of NS-forest markers. Sheets 1–3, Significantly enriched GO terms associated with the pediatric (sheet 1), adult (sheet 2) and pediatric plus adult minimal marker genes identified by NS-forest. (Sheets 3–5) Significantly enriched GO terms associated with shared (that is, associated with both adult and pediatric samples) or pediatric-specific marker genes for Oligo L1-6 OPALIN. Terms for which ‘highlighted’ is true are driver terms. gProfiler statistics were generated with g:GOSt, which uses Fisher’s one-tailed test, and the default g:SCS method for computing multiple-testing correction for P values.
Supplementary Data 7
Summary of metadata for NS-Forest markers with binary expression score >0.7 per cell type across the pediatric and adult datasets. The number of shared markers, number of markers unique to pediatric samples and number of markers unique to adult samples are shown for each cell type. The number of nuclei per cell type is shown for the combined pediatric and adult downsampled datasets, downsampled pediatric datasets and downsampled adult datasets.
Supplementary Data 8
Overlapping NS-Forest markers with a binary expression score >0.7 per cell type between the pediatric and adult datasets. NS-Forest markers with a binary expression score (>0.7) per cell type were extracted for the downsampled pediatric and downsampled adult datasets. Each sheet represents 1 of 75 cortical cell types and the NS-Forest features that were shared (in the intersection) between the pediatric and adult datasets, unique to pediatric datasets or unique to adult datasets are shown.
Supplementary Data 9
DESeq2 output of all genes tested for differential expression between pediatric and adult brains per cell type. Sheets 1–75, Differential expression analysis was performed using DESeq2’s two-tailed Wald test for each cell type separately. Genes were filtered before testing to only include those expressed in >10% of nuclei for that cell type across all pediatric and adult datasets. The associated log2 fold change values, adjusted P values (padj) and description of each feature are shown. Positive log2 fold change values indicate genes upregulated in pediatric versus adult samples. See DESeq2 documentation for an explanation of NA values (https://bioconductor.org/packages/release/bioc/vignettes/DESeq2/inst/doc/DESeq2.html#why-are-some-p-values-set-to-na).
Supplementary Data 10
DESeq2 output of significant DEGs only between pediatric and adult brains in a subset of cell types. Sheets 1–21, Significant DEGs (pPadj < 0.05) for cell types shown in Fig. 5a. Differential expression analysis was performed using DESeq2’s two-tailed Wald test for each cell type separately. The associated log2 fold change value, Padj values, description, percentage of pediatric nuclei expressing the gene, percentage of adult nuclei expressing the gene, average normalized expression across pediatric nuclei, and average normalized expression across adult nuclei are shown. The differences between pediatric and adult values for percentages of nuclei and average normalized expression are also shown. Positive log2 fold change values represent genes upregulated in pediatric versus adult datasets. See DESeq2 documentation for explanation of NA values (https://bioconductor.org/packages/release/bioc/vignettes/DESeq2/inst/doc/DESeq2.html#why-are-some-p-values-set-to-na). Sheets 22–24, DEGs associated with EA (sheet 6), IQ (sheet 7) and HARs (sheet 8) and their associated cell types.
Supplementary Data 11
Dispersion plots for DESeq2 analysis. Plots of dispersion estimates were generated for each cell type to assess the fit of the DESeq2 model to the data.
Supplementary Data 12
DEG expression compared across samples at the single-nucleus level. Violin plots for all DEGs for cell types with >6 DEGs identified by DESeq2 analysis. Black data points represent the normalized expression level for each analyzed nucleus in each sample. Blue dots indicate the mean expression level.
Supplementary Data 13
Average DEG expression compared across pediatric and adult samples. Violin plots for the all DEGs for cell types with >6 DEGs identified by DESeq2 analysis. Black data points represent the average normalized expression level.
Supplementary Data 14
Psupertime coefficients. The calculated psupertime coefficients for each gene for indicated excitatory neuron subtypes that showed the highest number of DEGs. Genes with nonzero psupertime coefficients represent genes that are relevant to the ordering of the cells in pseudotime.
Supplementary Data 15
GSEA terms associated with each cell type showing enriched or depleted pathways in pediatric versus adult samples. GSEA was performed using DESeq2’s output gene lists for each cell type ranked according to the log2 fold change × −log2 (padj) for each gene. All DESeq2-tested genes served as input into GSEA (genes were expressed in >10% of nuclei for the cell type of interest). Matrix shows the corresponding positive (sheet 1) and negative (sheet 2) NES values for each GSEA term (y axis) and cell type (x axis) based on the analysis using the ranked list of genes for each cell type. Terms were filtered to only include significantly associated terms (P < 0.01, q < 0.1). Positive NES values indicate pathways that were enriched in pediatric versus adult samples; negative NES values indicate pathways that were depleted in pediatric versus adult samples. The total number of terms per cell type and the total number of cell types associated with a given term are shown.
Source data
Source Data
Contains the following raw images for in situ hybridization data shown in Extended Data Fig. 6b: Extended_data_fig6b_15yr_40x_tilescan.czi. Extended_data_fig6b_15yr_63x.czi. Extended_data_fig6b_31yr_40x_tilescan.czi. Extended_data_fig6b_31yr_63x.czi. Extended_data_fig6a_15yr_40x_tilescan.czi.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Steyn, C., Mishi, R., Fillmore, S. et al. A temporal cortex cell atlas highlights gene expression dynamics during human brain maturation. Nat Genet 56, 2718–2730 (2024). https://doi.org/10.1038/s41588-024-01990-6
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41588-024-01990-6